Abstract
Environmental temperature is one of the important abiotic factors that influence the normal physiological function and productive performance of dairy cattle. Temperature stress evokes complex responses that are essential for safeguarding of cellular integrity and animal health. Post-transcriptional regulation of gene expression by miRNA plays a key role cellular stress responses. The present study investigated the differential expression of miRNA in Frieswal (Holstein Friesian × Sahiwal) crossbred dairy cattle that are distinctly adapted to environmental temperature stress as they were evolved by using the temperate dairy breed Holstein Friesian. The results indicated that there was a significant variation in the physiological and biochemical indicators estimated under summer stress. The differential expression of miRNA was observed under heat stress when compared to the normal winter season. Out of the total 420 miRNAs, 65 were differentially expressed during peak summer temperatures. Most of these miRNAs were found to target heat shock responsive genes especially members of heat shock protein (HSP) family, and network analysis revealed most of them having stress-mediated effects on signaling mechanisms. Being greater in their expression profile during peak summer, bta-miR-2898 was chosen for reporter assay to identify its effect on the target HSPB8 (heat shock protein 22) gene in stressed bovine PBMC cell cultured model. Comprehensive understanding of the biological regulation of stress responsive mechanism is critical for developing approaches to reduce the production losses due to environmental heat stress in dairy cattle.
Keywords: Heat stress, miRNAs, Frieswal, bta-miR-2898, HSPB8
Introduction
Environmental temperature is one of the important abiotic factors that influence the normal physiology of animals. The health and productivity of dairy cattle depend on the optimum conditions including the temperature range in comfort/thermo-neutral zone. Heat stress is caused by temperature above thermo-neutral range that severely impacts dairy cattle performance in terms of growth, milk production, and reproductive efficiency. The impending threats of climate change will further increase the frequency and intensity of heat waves that potentially leads to heat stress in dairy cattle. Apart from short-term managemental methods to alleviate the heat stress impacts, understanding the genetic differences and molecular mechanisms of the stress response for innate resilience and thermotolerance will be a significant long-term strategy for selection of thermotolerant animals for increasing productivity.
Heat stresses evoke complex responses that are considered essential for safeguarding the cellular integrity and survival of organisms (Belhadj Slimen et al. 2015). Multiple protein and signaling mechanisms are involved in stress response pathways. Heat shock proteins and molecular chaperones have been described as primary elements in cellular stress responses (Lindquist and Craig 1988; Hansen 2004; Lacetera et al. 2006; Paula-Lopes et al. 2003; Deb et al. 2014; Bhanuprakash et al. 2016; Sengar et al. 2017). Heat stress-induced genome wide expression studies indicated the involvement of multiple genes in stress response pathways (Mehla et al. 2014), and the transcriptional activation of genes by major factors such as hypoxia-inducing factors (HIFs) during oxidative stress and heat shock factors (HSF) during heat shock response was noted. However, the present study focuses on the post-transcriptional regulatory points that are significant in cellular stress response mechanisms.
MicroRNAs (miRNAs) are short non-coding RNA involved in post-transcriptional repression of genes by base-pairing with their target gene transcripts (Bartel 2004). During cellular stress responses, miRNA plays a key role by regulating the specificity, timing, and abundance of gene expression. Various studies (Baskerville and Bartel 2005; Harfe 2005; McKenna et al. 2010) have shown the involvement of miRNA in wide array of biological processes such as cell-fate specification, embryonic development, and metabolic pathways. In dairy cattle, miRNA-mediated regulation controls energy metabolism, lactation activity of mammary epithelial cells, reproductive functions, susceptibility to mastitis, and other infections (Fatima et al. 2014; Wang et al. 2015; Li et al. 2015; Kropp et al. 2014; Wang et al. 2016). Specific miRNA deletion mutant studies have identified mir-80, mir-229, and mir-64-66 cluster as important regulators of the heat stress response in C. elegans (Nehammer et al. 2015).
Few reports are available regarding the comprehensive list of differentially expressed miRNAs in dairy cattle (Zheng et al. 2014). The rationale behind the current study was to identify the differentially expressed miRNAs during thermal stress in Frieswal (Holstein Frisian × Sahiwal) crossbred dairy cattle. Initially, we characterized the stress response in Frieswal population during heat stress followed by identification of differentially expressed miRNAs during peak thermal stress through deep sequencing. Our study also aimed to characterize the effect of over-expressed bta-miR-2898 on its bovine target HSPB8 gene using a reporter assay.
Materials and methods
Experimental animals
All the experimental procedures involving animals were approved by the Institutional Animal Ethics Committee (IAEC). Ten healthy Frieswal bulls (Holstein Friesian × Sahiwal) of similar age group maintained under similar managemental regimen and in semen collection were randomly selected from the bull rearing unit of our institution. The samples were collected at two different environmental seasons viz., winter and summer with the temperature ranges between 15–18 °C (January–February; designated as non-heat stress/NHS) and 42–45 °C (May–June; designated as heat stress/HS), respectively. The temperature humidity index was used as heat stress indicator, irrespective of whether the animal was in heat stress or not. The animals were exposed to the natural environmental temperature for 1 h prior to collection, and when the environmental temperature was recorded within the range of 42–45 or 15–18 °C, the samples were collected at 2:00 PM. From each animal, samples were collected on four different days in each season, and the collected samples of an individual animal in each season were pooled together for further analysis. Stress response of the animals was characterized by recording physiological parameters, biochemical assays, and characterization of stressor genes (HSP70 and HSP90).
Physiological parameter recording
The animal’s rectal temperature, breathing rate, and pulse rate were recorded for both NHS and HS groups. Heat tolerance coefficient (HTC) was recorded using the formula postulated by Rhoad, 1944, HTC = 100–10 (RT–38.3) where HTC is the heat tolerance coefficient, RT is the rectal temperature, 38.3 is the physiological bovine body temperature, 10 is the correction factor to convert deviations in body temperature to a unit basis, and 100 is the perfect efficiency in maintaining temperature at 38.3 °C.
Sample preparation
Blood samples were subjected for peripheral blood mononuclear cell (PBMC) isolation and plasma separation as per standard protocol. Blood was diluted (1:2) in Alsever’s solution and centrifuged at 400×g for 30 min at 20 °C. PBMCs were then separated by density gradient centrifugation method as described earlier (Deb et al. 2014). PBMC samples were stored in liquid nitrogen till total RNA isolation for deep sequencing. Plasma samples were subjected for biochemical assays.
Biochemical parameter estimation
Immediately after separation, the plasma samples were subjected for three biochemical assays viz. thio-barbituric acid reactive substances (TBARS), catalase (CAT), and glutathinoe peroxidase (GPx) assays. TBARS assay was conducted using TBARS Assay Kit (Cayman chemical, USA) for the colorimetric measurement of lipid peroxidation by-product malondialdehyde (MDA) (μM) as per manufacturer protocol. Cayman’s catalase assay kit (Cayman chemical, USA) was used for colorimetric measurement of plasma catalase (CAT) activity (nmol/min/ml) as per the manufacturer’s instruction. Glutathione peroxidise (GPx) level in the plasma samples was estimated using Cayman’s glutathione peroxidise assay kit (Cayman chemical, USA) as per manufacturer’s standard protocol.
Real-time PCR-based relative expression of bovine HSP70 and HSP90
PBMC samples isolated from NHS and HS groups were subjected for total RNA isolation using cold trizol (Sigma Aldrich, USA) method as per manufacturer’s instruction. Extracted RNA was spectrophotometrically quantified, and the integrity was checked by visualization of 18 and 28 s ribosomal bands on an agarose gel. cDNA was synthesized from the isolated total RNA using ProtoScript first strand cDNA synthesis kit (New England Biolabs, Beverly, MA, USA) utilizing the M-MuLV reverse transcriptase and random primers. A diluted 1:10 solution of the cDNA was used for quantification using real-time qPCR (Step One, Applied Biosystems, Foster City, CA, USA). The primer sequences for HSP70, HSP90, and beta-actin gene (endogenous control) reported in our earlier studies (Deb et al. 2014; Bhanuprakash et al. 2016) were used for the present study also. The reaction was conducted in a final reaction volume of 10 μl using the SYBR Green® PCR Master Mix kit (Applied Biosystems, Foster City, CA, USA), according to manufacturer instructions. ΔΔCt method was used for quantification of the samples (Livak and Schmittgen 2001). Obtained expression values were normalized against the endogenous “housekeeping” gene, allowing the comparison of samples independently of the amount of total input cDNA. All the determinations were performed in triplicate.
RNA isolation, small RNA enrichments, and detection of RNA integrity
Total RNA was isolated from pooled PBMC samples of NHS and HS groups with PAX gene blood miRNA Kit (Qiagen, USA) according to the manufacturer’s instruction. Isolated total RNA was subjected for nucleic acid binding bead-based enrichment of small RNA with the total RNA-Seq kit v2 for small RNA libraries kit (Thermo Fisher scientific, USA). Quality and quantity of purified enriched small RNA were estimated using NanoDrop™ Spectrophotometer and Qubit quotation system (Life technologies, USA). Integrity of the RNA was checked using Agilent™ 2100 Bioanalyzer™ instrument with the Agilent™ RNA 6000 Nano Kit Bioanalyzer 2100 and Agilent™ Small RNA Kit (Agilent, USA) in RNA 6000 pico chip as per manufacturer’s instructions. The purified and characterized RNA samples were stored at − 80 °C till further use. RNA integrity number (RIN) was checked using 2100 expert software. The highest-quality library mapping statistics were obtained from input RNA with higher RIN values.
Small RNA library construction
Small RNA libraries were prepared with the total RNA-Seq kit v2 for small RNA libraries kit (Thermo Fisher scientific, USA) according to the library preparation instruction. Briefly, 3 μl of small RNA sample (1–100 ng of miRNA in ≤ 1 μg of enriched small RNA) was ligated to a 3′ adapter, and a 5′ adapter was ligated to the products, followed by reverse transcription into complementary DNA (cDNA) using SuperScript™ III reverse transcriptase enzyme (Thermo Fisher scientific, USA). Each cDNA samples were amplified via polymerase chain reaction (PCR) according to the preparation of a barcoded library PCR mix (Thermo Fisher scientific, USA). One microliter of the purified DNA was run on an Agilent™ 2100 Bioanalyzer™ instrument with the Agilent™ DNA 1000 Kit as described earlier. Using the 2100 expert software, smear analysis was performed to determine the molar concentration of cDNA libraries with size range of 50–300 bp which was subjected to “Pool barcoded small RNA library” preparation.
Pool barcoded small RNA library preparation
Molar concentration (nM) of each barcoded cDNA library (50–300 bp size range) was prepared with the Agilent™ DNA 1000 Kit. Each barcoded cDNA library was diluted to the same molar concentration (nM). An equal volume of each diluted library was mixed to prepare a pool of the barcoded libraries. The final molar concentration of the pooled library was the same for each diluted library.
Emulsion PCR and deep sequencing
The emulsion PCR was carried out applying the Ion XPress Template kit V2.0 (Life Technologies, USA) as described in the user guide (Part No. 4469004 Rev. B 07/2011) provided by the manufacturer. Quality and quantity of the enriched spheres were checked on the Guava easyCyte5 system (Millipore GmbH, Schwalbach am Taunus, Germany) as described in the appendix of Ion Xpress Template Kit User Guide. Sequencing of the amplicon libraries was carried out using the Ion Torrent Personal Genome Machine (PGM) system by using the Ion Sequencing 200 kit (Life Technologies, USA). Quality check-passed libraries were subjected to emulsion PCR using the Ion PGM 200 Xpress Template Kit (Life Technologies, USA). After bead enrichment, beads were loaded onto Ion 316 chip and deep sequenced using an Ion Torrent Personal Genome Machine (Life Technologies, Pleasanton, CA, USA).
Deep sequence data analysis
Obtained raw sequences were processed through CLC–Genomics-work bench 8.0.2 Windows, Mac OS X and Linux software (Qiagen, Denmark) to filter out the adapter sequences, low-quality, and low copy sequences, and then, the extracted small RNA sequences with 15–26 nucleotides in length were subjected to mRNA, RFam, and Repbase filter, and the remaining sequences were compared with miRBase 21 by BLASTn search to identify the conserved miRNAs in bovine (http://www.mirbase.org/). To identify the potential precursor miRNA sequences, maximum three mismatches were permitted between the identified short miRNAs and known animal miRNAs. Further, identified all mature miRNA sequences were processed for BLAST against the downloaded bovine draft genome sequences (http://www.ncbi.nlm.nih.gov/genome/82), and the hairpin RNA structures for their flanking sequences were predicated by RNA fold software (http://mfold.rit.albany.edu/?q=mfold/RNA-Folding-Form).
Quantitative real-time PCR-based validation of miRNAs
Differentially expressed eight candidate miRNAs (bta-miR-103-2, bta-miR-2898, bta-miR-150, bta-miR-2478, bta-miR-181b-2, bta-miR-2311, bta-miR-142, and bta-miR-6536-2) were selected randomly on the basis of their reads per kilo millions (RPKM) values (extreme, higher, lower, and lowest) obtained through deep sequencing (based on Log2 [HS/NHS] values). Real-time quantitative PCR (RT-qPCR) was performed using Step One, Applied Biosystems Real Time PCR machine. Isolated total RNA (2 μg) using PAX gene blood miRNA Kit (Qiagen, USA) was subjected for reverse transcription using a ProtoScript first strand cDNA synthesis kit (New England Biolabs, Beverly, MA, USA). A 1:10 diluted cDNA samples were used to perform the RT-qPCR as per the procedure described above in section “Real-time PCR-based relative expression of bovine HSP70 and HSP90.” The primers synthesized for the candidate miRNAs had the similar sequences as the Bos taurus miRNA with suitable adjustments at their 5′ ends.
Target gene prediction and miRNA-GO-network analysis
It is assumed that miRNA binding sites in the 3′UTR of their targeted genes are highly conserved among closely related species than the most distant ones (Chen and Rajewsky 2006; Rajewsky 2006). To understand the molecular function of the differentially expressed miRNAs during heat-stressed and normal animals, Target Scan (Lewis et al. 2003) software was used to predict their target mRNA related to heat stress. By combining the target gene function database with miRNA and mRNA sequence using miRNA-GO-network analysis, it was identified that multiple genes were simultaneously regulated by the same miRNA. The core of miRNAs and their centralized gene function control mechanisms were also found.
Designing target site of bta-mir-2898 and its amplification from genomic DNA
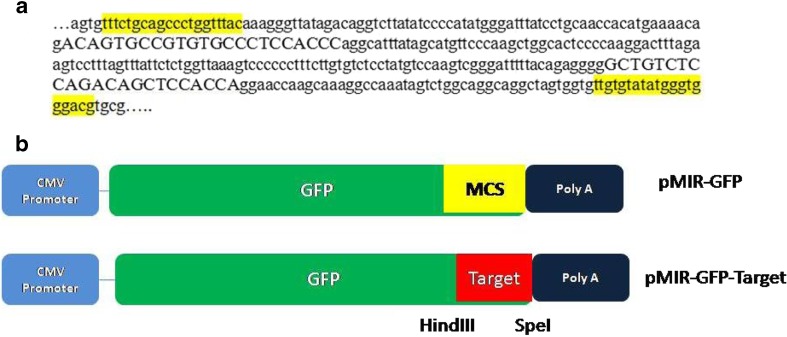
Based on the highest expression level and fold change during heat stress, bta-miR-2898 was selected as model microRNA to elucidate its in vitro effect on stressor gene in bovine PBMC. We identified the most conserved target sites of bta-miR-2898 through Target Scan software and observed that 3′ un translated region (UTR) of heat shock 22 kDa protein 8 (HSPB8) is highly conserved target for bta-miR-2898 across different species. miRNA-target interaction revealed bta-miR-2898 having two interaction sites at 3′ UTR region of HSPB8 (Fig. 8a). A set of primers were designed (HSPB 8F: TAAGCAGGATCCTTTCTGCAGCCCTGGTTTAC and HSPB8 R: TGCTTACTCGAGCGTCCCACCCATATACACAA) to flank both the target sites at the 3′ UTR region of HSPB8 CDS. We deliberately incorporated linkers viz. Hind III and Spe I at the 5′ end of HSPB 8F and HSPB8 R primers, respectively, for placing the amplified product at multiple cloning sites of the expression vector (Fig. 8b).
Fig. 8.
Schematic representation for cloning of the bta-miR-2898 target region in pMIR-GFP expression vector. a Identified target sites for bta-miR-2898 at 3′ UTR region of bovine HSPB8 (Accession Number: NM001014955); bases with capital letters are the target sites where miR2898 can interact, and highlighted bases are the sites chosen for designing primers to amplify the target regions. b Strategy for construction of pMIR-GFP target, where the bta-mir 2898 target sites at 3′ UTR region of bovine HSPB8 was cloned in pMIR-GFP expression vector. Target region was fused at 3′ UTR region of GFP. HindIII and SpeI are the linkers added at the 5′ region of forward and reverse primers, respectively, to clone at multiple cloning sites (MCS) region of the expression vector
The genomic DNA was extracted from the collected blood sample from Frieswal cattle using a QIAamp DNA Blood Mini Kit (Qiagen, USA). The polymerase chain reaction (PCR) was carried out to amplify 321 base pair fragment from a starting template of approximately 50 ng of genomic DNA in a final reaction volume of 25 μl containing 1× Taq DNA polymerase buffer (Sigma, USA), 1.5 mM MgCl2 (Sigma, USA), 200 mM dNTPs (Sigma,USA), 0.5 mM of each primer, and 1 U Taq polymerase (Sigma,USA). PCR was conducted with the initial denaturation at 94 °C for 5 min, followed by 5 cycles of 94 °C for 30 s, 55 °C for 30 s, and 72 °C for 30 s; thereafter, 30 cycles of 94 °C for 30 s, 64 °C for 30 s, and 72 °C for 30 s and a final extension 72 °C for 5 min and visualized in 1.0% agarose gel. The amplicon was confirmed by automated DNA sequencer in Sanger’s dideoxy chain termination method.
Cloning of the bta-miR-2898 target site in pMIR-GFP expression vector
The fold change values estimated by deep sequencing revealed that the differential expression of bta-mir-2898 was the highest among the highly expressed miRNAs during heat stress. Further, the results of qRT-PCR-based validation also revealed relatively highest expression of bta-mir-2898. Because of the above two reasons, we have chosen bta-mir-2898 as a model for reporter assay. Gel-purified and restriction enzyme-digested PCR product was cloned in the digested (with same sets of restriction enzymes) pMIR-GFP-purified expression vector (Cell Biolabs. Inc., USA) for in vitro reporter assay. The recombinant plasmid pMIR-GFP-target was characterized by restriction enzyme digestion and sequencing. Recombinant plasmids were purified through endo free plasmid purification kit (Qiagen, USA) and subjected for reporter assay in heat-stressed bovine PBMCs.
Reporter assay
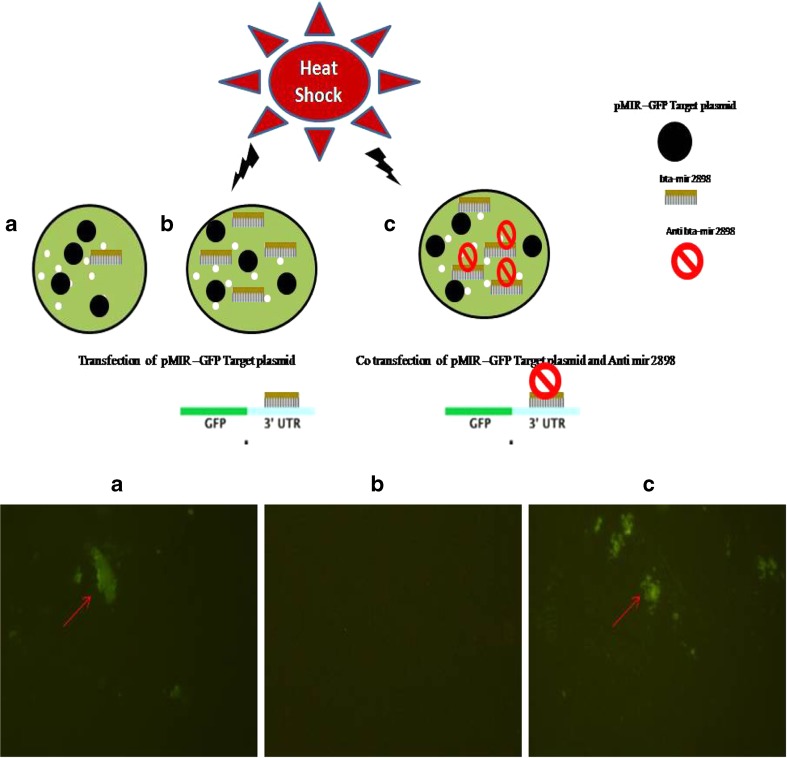
bta-miR-2898 inhibitor (anti bta-miR-2898) was custom synthesized (5 nmole with > 60% HPLC purified) from ambion life technologies, California, and 105 pmol of the inhibitor was used for the reporter assay to analyze the bta-miR-2898 function as per manufacture’s instruction. The isolated PBMC cells were given heat shock for 1 h at 42 °C in a water bath and then plated at a concentration of approximately 1 × 106 viable cells/ml in six-well plates and incubated at 37 °C in a 5% CO2 incubator. Before starting the experiment, miRNA was isolated from the stressed and normal PBMCs (as described in earlier section), and cDNA was synthesized to confirm the over-expression of bta-miR-2898 in heat-stressed PBMCs. cDNA was subjected for qRTPCR with bta-miR-2898 specific primers to quantify the miRNA expression level (data not shown here). The details of reporter assay are schematically represented in Fig. 9 (upper panel). A monolayer of 70–80% confluent PBMC cells in six-well plates was transfected with 3 μg of pMIR-GFP and pMIR-GFP-target alone or with anti-bta-miR-2898 (105 pmol) using Lipofectamine plus (Invitrogen, USA). At 48 h post-transfection, expression of GFP was visualized with a fluorescence illuminator system (Nikon ECLIPSE 80i), and images were captured at 20× magnification. Independent experiments were repeated thrice and number of fluorescent unit (%) counted in ten random fields.
Fig. 9.
pMIR–GFP reporter assay in bovine PBMC (heat shocked) for analysis of the expression of GFP after cloning of bta-mir 2898 target region of bovine HSP gene at 3′ UTR region. Upper panel depicted the experimental design and lower panel showed the level of GFP expression in different groups. a Non-heat shock group and transfected with pMIR–GFP target construct. b Heat shock group and transfected with pMIR–GFP target construct. c Heat shock group and co-transfected with pMIR–GFP target construct and anti-bta–mir 2898
Statistical analysis
Data are presented as mean ± SEM and analyzed by using an SPSS statistical program (SPSS 10.0 for Windows; SPSS, Inc., Chicago, IL, USA). Significant differences were determined by one-way ANOVA using the SPSS program. The univariate least squares analysis of variance was carried out to find the significance of differences between the expressions of miRNAs by qRTPCR. The Duncan’s multiple range test was carried out to find the significance differences between the mean values.
Results
Characterization of stress response
Physiological parameters
Rectal temperature, breathing rate, pulse rate, and heat tolerance coefficient at two different environmental temperature ranges are presented in Table 1. A rectal temperature of above 39 °C, a respiratory, and/or pulse rate of 90–120 times per minute were regarded as the indication of heat stress. The physiological parameters of HS were significantly different (p < 0.05) from the NHS group of cattle. The average rectal temperature of heat-stressed cattle was above 39 °C, and as high as 43 °C, the respiratory rate was 120 breaths per minute, pulse rate per minute was 100, and a significant decrease of HTC as compared with the normal. Rectal temperature, breathing rate, pulse rate, and HTC data showed that the cattle were under heat stress.
Table 1.
Physiological parameters recorded during different environmental temperature
| Environmental temperature ranges (°C) | Rectal temperature (°C) | Breathing rate (times/min) | Pulse rate (rate/min) | HTC |
|---|---|---|---|---|
| 15–18 °C | 38.02 ± 0.64a | 33.25 ± 0.22a | 66.63 ± 0.57a | 101.45 ± 0.17a |
| 42–45 °C | 39.66 ± 0.64b | 121.56 ± 0.22b | 105.60 ± 0.57b | 84.23 ± 0.17b |
Note: Different subscript letters indicate significant differences at P < 0.05
HTC heat tolerance coefficient
Biochemical estimations
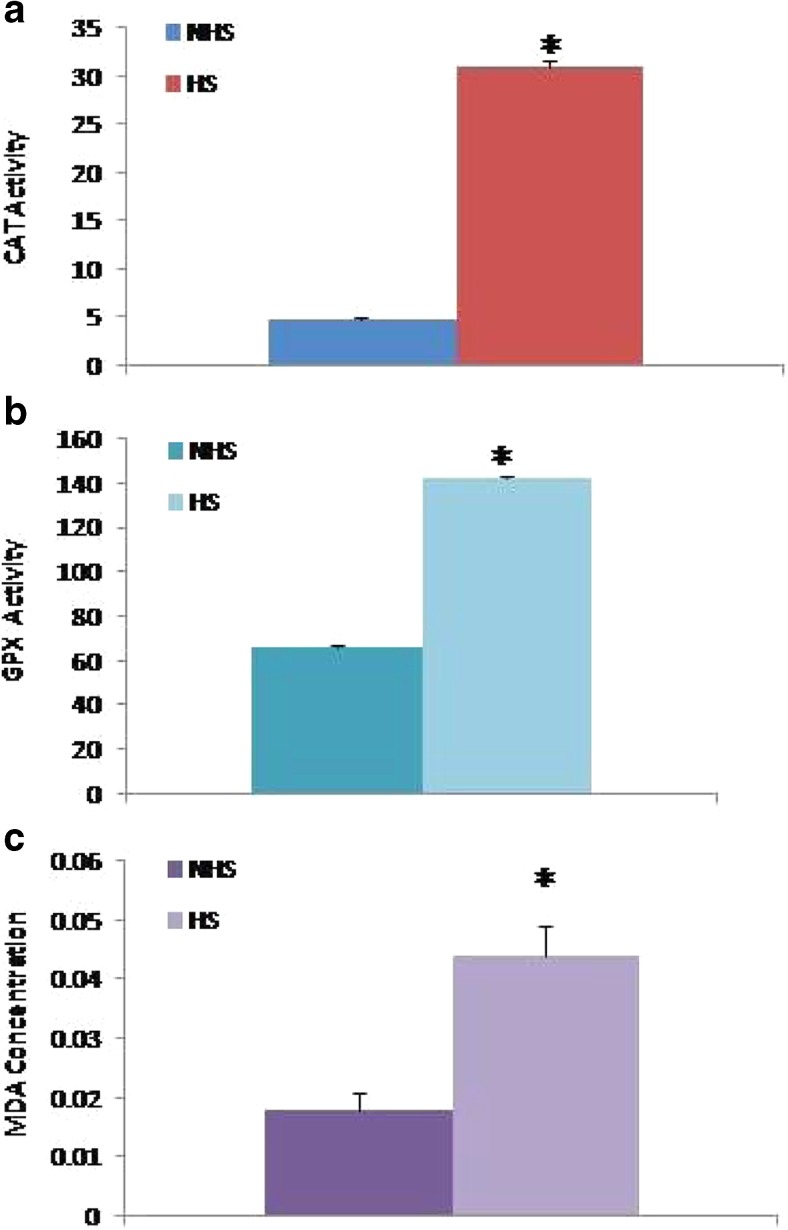
Estimated plasma CAT activity (nmol/min/ml) in heat-stressed group was 31.017 ± 0.469 that was significantly (P < 0.01) higher (p < 0.01) than non heat-stressed group (4.876 ± 0.469) and shown in Fig. 1a. Glutathione peroxidase (GPx) level (nmol/min/ml) in the plasma samples collected at different environmental temperatures revealed that with the increase of temperature GPx level changes significantly (p < 0.01). Average GPx level estimated for heat stress and non-heat-stressed groups was 142.622 ± 1.579 and 66.213 ± 1.579, respectively (Fig. 1b). Similarly, concentration of MDA (μM) significantly (p < 0.01) increased at high environmental temperatures (Fig. 1c). Estimate MDA concentration was 0.018 ± 0.004 and 0.044 ± 0.004 in non-heat-stressed and heat-stressed groups, respectively.
Fig. 1.
a Catalase (CAT) activity (nmol/min/ml). b Glutathione peroxidase (GPx) level (nmol/min/ml). c Malondialdehyed (MDA) concentration (μM) detected in plasma samples of NHS and HS groups. * indicates significant variation at P < 0.01
Expression profile of HSP70 and HSP90
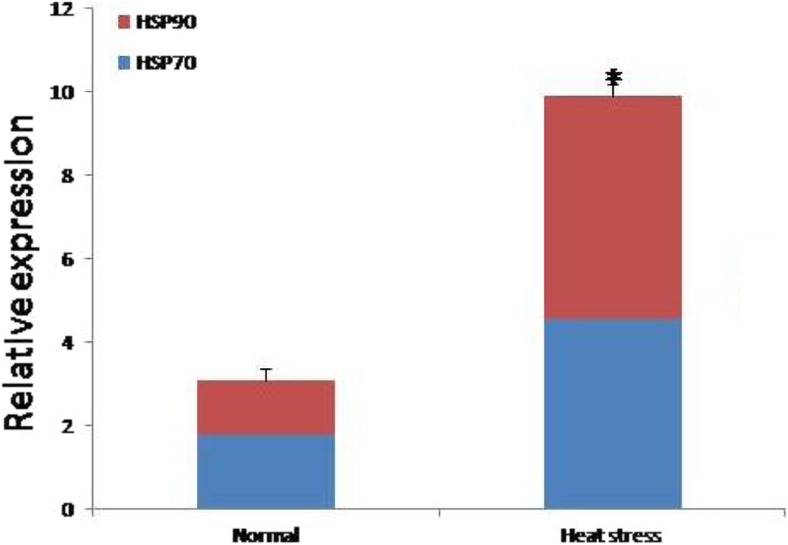
The relative expression (mean ± SE) of HSP70 and HSP90 mRNAs was significantly (p < 0.01) higher during elevated environmental temperature (HS) than the normal conditions (NHS). Obtained fold changes of HSP70 and HSP90 during 42 °C were 4.55 ± 0.28 and 5.34 ± 0.28, respectively (Fig. 2).
Fig. 2.
Fold changes of HSP70 and HSP90 in the Frieswal PBMC collected 18 °C (normal) and 45 °C (heat stress) environmental temperatures. *P < 0.01
miRNA deep sequencing
Small RNA enrichment
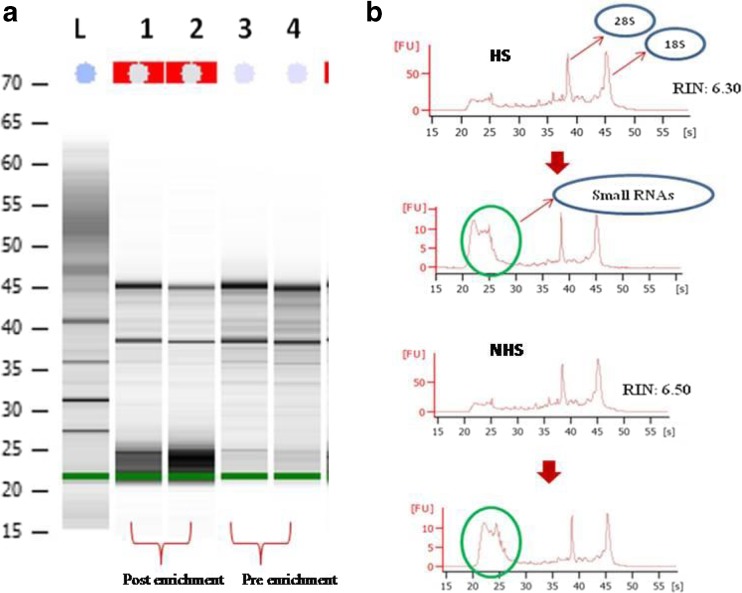
The bioanalyzer assay to determine the miRNA abundance in bovine PBMCs of HS and NHS groups showed the presence of small RNAs and their repeats after enrichments (Fig. 3a). Small RNAs which include microRNAs were visible at 20–25 nucleotide ranges on the gel showing an abundance of small RNAs based on the size of the band and lesser intensity of ribosomal RNA bands. RNA integrity numbers (RIN) ranged from 6.3 to 6.5 (Fig. 3b).
Fig. 3.
a Bioanalyzer assessment of small RNAs after enrichment. L RNA Ladder, 1 NHS post-enrichment, 2 HS post-enrichment, 3 NHS pre-enrichment, and 4 NHS pre-enrichment. b RNA Integrity Numbers (RIN) obtained through bioanalyzer reading after small RNA enrichments (enriched small RNA are shown in green round)
Small RNA library preparation
To identify miRNAs in Frieswal cattle, two independent small RNA libraries were constructed from the PBMC of normal and heat-stressed animals through Ion Torrent deep sequencing. There were 61,380 and 71,357 raw reads in the heat-stressed and normal cattle, respectively, with a length of 15–30 nucleotides after filtering out of the adapter sequences as well as low-quality sequences. In both the libraries, majority of the small RNAs were 20–25 nucleotides in length.
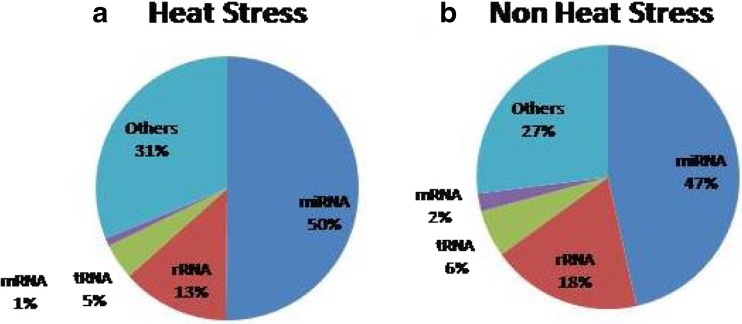
The mappable small RNA sequences were first compared to the currently known Bos taurus miRBase v20 database. Out of identified 44,873 and 30,227 small RNAs in the libraries of heat-stressed and normal groups, annotated miRNAs were 169 and 251, respectively (Table 2). Fragment analysis of small RNA libraries revealed the presence of 47 and 50% of miRNAs in the libraries of normal and heat-stressed groups, respectively (Figs. 4 and 5).
Table 2.
Statistics of small RNA sequences
| Samples | Total reads (post-filtration) | Total small RNAs | Total miRNA (annotated) |
|---|---|---|---|
| NHS | 71,357 | 30,227 | 251 |
| HS | 61,380 | 44,873 | 169 |
Fig. 4.
The distribution of fragments of mRNA, rRNA, tRNA, miRNA, and others in the small RNA libraries. a The PBMC of heat-stressed cattle. b The PBMC of non-heat-stressed cattle
Fig. 5.
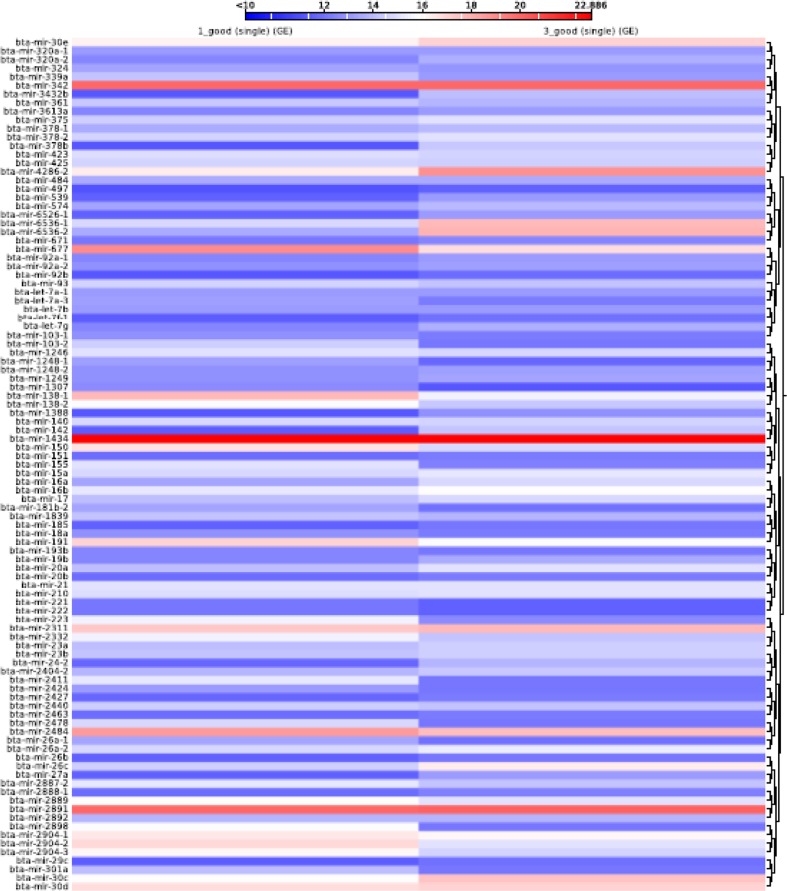
Heat map and the result of clustering to all MicroRNA detected. There are two specific groups. Column 1 represents normal groups and column 3 represents heat-stressed groups
Expression of miRNAs through deep sequencing
The increase in expression level of miRNAs is represented by varying color intensity ranging from dark blue for the lower and dark red for higher expression level in terms of RPKM values. Based on change in color intensity, the differentially expressed miRNAs were selected for further studies. The expression profiles of miRNAs were analyzed and compared with two libraries. We identified 65 miRNAs which differentially expressed among the heat-stressed and normal Frieswal cattle (Log2 HS/NHS) (Table 3).
Table 3.
The differential expression of miRNAs in PBMC of crossbred cattle by deep sequencing
| miRNA | HS (a) | NHS (b) | Fold change [Log2 (a/b)] |
|---|---|---|---|
| Highly expressed miRNA at HS | |||
| bta-mir-2898 | 52,893.92 | 4212.46 | − 3.65037 |
| bta-mir-2411 | 37,336.88 | 4212.46 | − 3.14787 |
| bta-mir-223 | 46,584.68 | 6006.655 | − 2.95522 |
| bta-mir-155 | 34,225.48 | 5148.562 | − 2.73283 |
| bta-mir-2478 | 28,002.66 | 4212.46 | − 2.73283 |
| bta-mir-103-2 | 22,066.43 | 4267.887 | − 2.37026 |
| bta-mir-138-1 | 212,409.7 | 43,470.85 | − 2.28873 |
| bta-mir-677 | 497,306.6 | 103,205.3 | − 2.26862 |
| bta-mir-150 | 100,622.9 | 25,948.75 | − 1.95522 |
| bta-mir-2904-2 | 116,366.6 | 32,435.94 | − 1.84301 |
| bta-mir-26a-1 | 10,647.93 | 3603.993 | − 1.5629 |
| bta-mir-1248-1 | 9126.794 | 3089.137 | − 1.5629 |
| bta-mir-181b-2 | 10,767.57 | 3644.488 | − 1.5629 |
| bta-mir-2904-3 | 65,028.41 | 23,168.53 | − 1.4889 |
| bta-mir-138-2 | 51,956.75 | 19,539.72 | − 1.4109 |
| bta-mir-191 | 123,738.3 | 49,901.45 | − 1.31014 |
| bta-mir-2332 | 45,137.95 | 18,803.44 | − 1.26334 |
| bta-mir-2424 | 8984.188 | 4054.492 | − 1.14787 |
| bta-let-7a-3 | 9712.635 | 4383.235 | − 1.14787 |
| bta-mir-2484 | 390,891 | 179,251.2 | − 1.12478 |
| bta-mir-2889 | 58,912.71 | 31,904.2 | − 0.88483 |
| bta-mir-193b | 5772.972 | 3907.944 | − 0.5629 |
| bta-mir-361 | 20,246.06 | 13,705.33 | − 0.5629 |
| bta-mir-93 | 24,891.26 | 16,849.84 | − 0.5629 |
| bta-mir-2904-1 | 88,986.24 | 60,238.17 | − 0.5629 |
| bta-mir-1839 | 19,964.86 | 13,514.98 | − 0.5629 |
| bta-mir-103-1 | 6654.954 | 4504.992 | − 0.5629 |
| bta-mir-2440 | 22,066.43 | 17,071.55 | − 0.37026 |
| bta-mir-423 | 30,584.47 | 24,154.42 | − 0.34051 |
| bta-mir-1246 | 32,818.95 | 26,659.68 | − 0.29987 |
| bta-mir-140 | 28,035.76 | 24,154.42 | − 0.21498 |
| bta-mir-484 | 11,408.49 | 10,297.12 | − 0.14787 |
| bta-mir-425 | 24,783.97 | 22,369.61 | − 0.14787 |
| bta-let-7b | 8873.272 | 8008.874 | − 0.14787 |
| bta-mir-2892 | 13,821.83 | 12,475.36 | − 0.14787 |
| bta-mir-320a-1 | 8765.061 | 7911.205 | − 0.14787 |
| bta-mir-21 | 33,274.77 | 31,534.94 | − 0.07748 |
| Lowly expressed miRNA at HS | |||
| bta-mir-342 | 945,569.8 | 979,979.4 | 0.051567 |
| bta-mir-1434 | 7,019,974 | 7,513,585 | 0.098036 |
| bta-mir-23b | 19,964.86 | 21,623.96 | 0.115168 |
| bta-mir-2891 | 977,370.7 | 1,061,540 | 0.119181 |
| bta-mir-30d | 106,099 | 115,842.6 | 0.126756 |
| bta-mir-210 | 29,336.12 | 33,097.9 | 0.174061 |
| bta-mir-378-2 | 25,218.77 | 29,875.21 | 0.244451 |
| bta-mir-2311 | 156,814.9 | 206,410.5 | 0.396454 |
| bta-mir-2404-2 | 14,303.19 | 19,364.74 | 0.437096 |
| bta-mir-26a-2 | 28,521.23 | 38,614.21 | 0.437096 |
| bta-mir-15a | 25,978.37 | 35,171.5 | 0.437096 |
| bta-mir-92a-1 | 6143.034 | 8316.907 | 0.437096 |
| bta-mir-23a | 16,409.48 | 22,216.4 | 0.437096 |
| bta-mir-574 | 10,727.39 | 14,523.56 | 0.437096 |
| bta-mir-17 | 17,112.74 | 27,029.95 | 0.659488 |
| bta-mir-30e | 72,915.15 | 126,923.2 | 0.799666 |
| bta-mir-20a | 16,871.71 | 31,979.1 | 0.922523 |
| bta-let-7 g | 5772.972 | 11,723.83 | 1.022058 |
| bta-mir-320a-2 | 5843.374 | 11,866.81 | 1.022058 |
| bta-mir-16a | 10,767.57 | 25,511.41 | 1.244451 |
| bta-mir-539 | 3071.517 | 8316.907 | 1.437096 |
| bta-mir-30c | 59,324.16 | 169,902.5 | 1.518016 |
| bta-mir-26c | 23,957.83 | 75,683.86 | 1.659488 |
| bta-mir-4286-2 | 83,662.28 | 427,330.6 | 2.352704 |
| bta-mir-3432b | 2469.88 | 16,719.56 | 2.759024 |
| bta-mir-142 | 2753.774 | 18,641.34 | 2.759024 |
| bta-mir-6536-1 | 26,619.82 | 212,235.2 | 2.995091 |
| bta-mir-6536-2 | 11,978.92 | 214,888.1 | 4.165016 |
Validation of the selected miRNA by real-time PCR
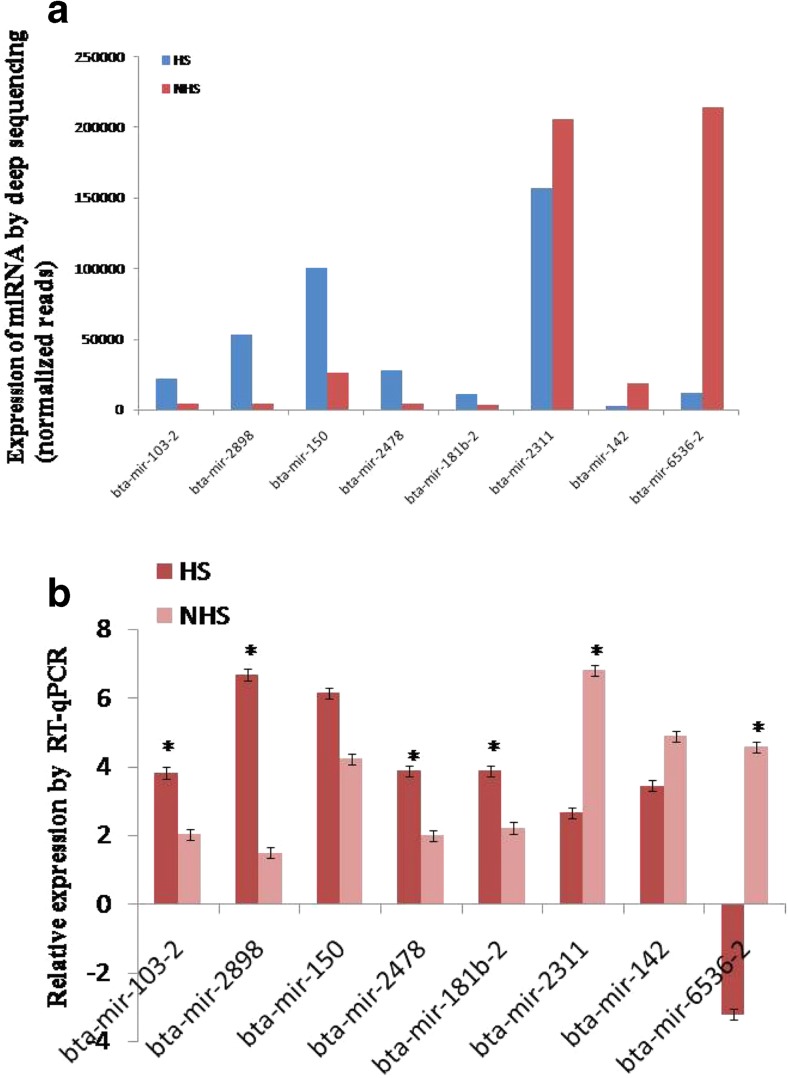
To validate the deep sequencing results, real-time PCR was conducted to measure the expression levels of five selected up-regulated miRNAs (bta-miR-103-2, bta-miR-2898, bta-miR-150, bta-miR-2478, and bta-miR-181b-2) and three down-regulated miRNAs (bta-miR-2311, bta-miR-142, and bta-miR-6536-2) in heat-stressed groups. The results showed that the expression level of these selected miRNAs measured by real-time PCR was in concordance with the normalized deep sequencing results (Fig. 6). Almost, the expression level of all the miRNAs was significantly (p < 0.01) different between the HS and NHS groups, and maximum significant variation was observed for bta-mir 2898.
Fig. 6.
Analysis of selected eight differentially expressed miRNAs in PBMC of heat-stressed and non-heat-stressed crossbred cattle by sequencing and RT-qPCR methods. a The expression of miRNAs by Ion torrent deep sequencing (normalized reads). b The relative expression of miRNAs tested RT-qPCR. The results revealed that expression of these selected miRNAs by RTqPCR were in agreement with the normalized sequencing data. *P < 0.01
Prediction of target genes and network analysis
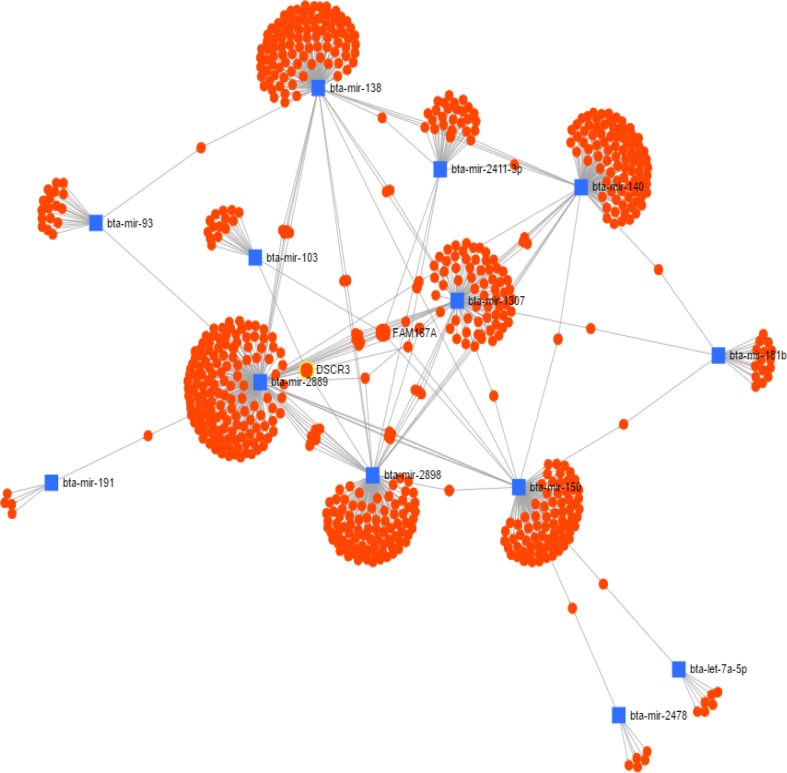
Out of 65 differentially expressed miRNAs, we identified that 30 were associated with heat stress. Target stressor genes of the selected miRNAs are shown in Table 4, among them maximum are heat shock proteins. By combining the target gene function database with miRNA and mRNA sequence analysis, it was noticed that multiple genes were simultaneously regulated by the same miRNA. The core of miRNAs and their centralized control gene functions were also found. The network analysis of differentially expressed miRNA and their target genes indicated stress-mediated effects on signaling mechanisms (Fig. 7). The metabolic pathways, PI3K-Akt signaling, MAPK signaling, and immune-regulatory (NF-Kappa B, TLR, and RIG), are majorly influenced by miRNA-mediated regulations (Table 5).
Table 4.
Predicted miRNA target genes with function related to response of heat stress
| miRNA | Count | Target genes |
|---|---|---|
| bta-miR-103-2 | 3 | heat shock 70 kDa protein 12A (HSPA12A) AHA1, activator of heat shock 90 kDa protein ATPase homolog 2 (yeast) (AHSA2) serpin peptidase inhibitor, clade H (heat shock protein 47), member 1, (collagen binding protein 1) (SERPINH) |
| bta-miR-2898 | 5 | heat shock protein, alpha crystallin-related, B6 (HSPB6) cold shock domain containing C2, RNA binding (CSDC2) heat shock 70 kDa protein 4-like (HSPA4L) heat shock 70 kDa protein 12A (HSPA12A) heat shock 22 kDa protein 8 (HSPB8) |
| bta-miR-150 | 4 | heat shock protein 90 kDa beta (Grp94), member (HSP90B1) heat shock 70 kDa protein 9 (mortalin) (HSPA9) AHA1, activator of heat shock 90 kDa protein ATPase homolog 2 (yeast) (AHSA2) |
| bta-miR-2478 | 6 | heat shock transcription factor family member 5 (HSF5) serpin peptidase inhibitor, clade H (heat shock protein 47), member 1, (collagen binding protein 1) (SERPINH1) heat shock 70 kDa protein 1B (HSPA1B) heat shock protein 90 kDa alpha (cytosolic), class A member 1 (HSP90AA1) heat shock 70 kDa protein 6 (HSPA6) heat shock transcription factor family member 5 (HSF5) |
| bta-miR-181b-2 | 4 | heat shock protein 90 kDa beta (Grp94), member 1 (HSP90B1) heat shock 70 kDa protein 5 (glucose-regulated protein, 78 kDa) (HSPA5) AHA1, activator of heat shock 90 kDa protein ATPase homolog 2 (yeast) (HSA2) heat shock protein 90 kDa beta (Grp94), member 1 (HSP90B1) |
| bta-miR-2311 | 15 | HSPB (heat shock 27 kDa) associated protein 1(HSPBAP1) heat shock protein, alpha-crystallin-related, B6 (HSPB6) heat shock 60 kDa protein 1 (chaperonin) (HSPD1) heat shock 70 kDa protein 4 (HSPA4) heat shock 70 kDa protein 9 (mortalin) (HSPA9) heat shock 70 kDa protein 4-like (HSPA4L) heat shock 70 kDa protein 6 (HSPA6) heat shock protein 90 kDa alpha (cytosolic), class A member 1 (HSP90AA1) heat shock factor binding protein 1 (HSBP1) heat shock 70 kDa protein 2 (HSPA2) AHA1, activator of heat shock 90 kDa protein ATPase homolog 2 (yeast) (AHSA2) heat shock 27 kDa protein 3 (HSPB3) heat shock 105 kDa/110 kDa protein 1 (HSPH1) heat shock 70 kDa protein 14 (HSPA14) heat shock transcription factor family member 5(HSF5) |
| bta-miR-142 | 2 | heat shock protein 70 kDa family, member 13 (HSPA13) heat shock 70 kDa protein 4-like (HSPA4L) |
| bta-miR-6536-2 | 9 | heat shock protein 70 kDa family, member 13 (HSPA13) HSPB (heat shock 27 kDa) associated protein 1(HSPBAP1) heat shock 70 kDa protein 4-like (HSPA4L) heat shock transcription factor family member 5 (HSF5) AHA1, activator of heat shock 90 kDa protein ATPase homolog 2 (yeast) (AHSA2) heat shock 27 kDa protein 3 (HSPB3) heat shock 70 kDa protein 14 (HSPA14) heat shock 70 kDa protein 2 (HSPA2) heat shock 70 kDa protein 1B (HSPA1B) |
Fig. 7.
Interaction among different selected miRNAs with their predicted targets. Blue rectangular box miRNAs and orange circles target genes
Table 5.
Pathways affected by differentially expressed miRNA in crossbred cattle during heat stress response
| Pathways affected | Number of hits | P value |
|---|---|---|
| Butanoate metabolism | 5 | 0.342 |
| Signaling pathways regulating pluripotency of stem cells | 11 | 0.342 |
| NF-kappa B signaling pathway | 8 | 0.354 |
| RIG-I-like receptor signaling pathway | 7 | 0.354 |
| Pertussis | 7 | 0.354 |
| Proteoglycans in cancer | 13 | 0.354 |
| One carbon pool by folate | 3 | 0.411 |
| Hippo signaling pathway | 10 | 0.411 |
| Prion diseases | 4 | 0.411 |
| Salmonella infection | 7 | 0.411 |
| MAPK signaling pathway | 14 | 0.44 |
| Chagas disease (American trypanosomiasis) | 8 | 0.44 |
Reporter assay
To explore the effect of differentially over-expressed bta-miR-2898 during heat stress, reporter assay was performed using pMIR-GFP expression vector. Most conserved target regions of bta-miR-2898 were identified at 3′ UTR region of bovine HSPB8 (Accession Number: NM001014955), where it can interact with two target sites of HSPB8 (Fig. 8a). A 321 base pair flanking fragment of target sites at bovine HSPB8 gene was cloned at 3′ UTR region of GFP gene in pMIR-GFP expression vector, and the clones were confirmed by restriction enzyme digestion and sequencing. GFP expression in heat-stressed PBMC transfected with pMIR-GFP alone, pMIR-GFP-target alone, and pMIR-GFP-target with anti bta–mir 2898 was studied, and the results revealed that there was negligible GFP expression in the cells treated with pMIR-GFP-target alone (Fig. 9). Number of fluorescent unit (%) counted in ten random fields of heat-stressed PBMCs during reporter assay showed that there was negligible amount of fluorescent (1–3%) within heat-stressed cells transfected with pMIR–GFP Target construct; however, more than 50% cells showed GFP expression in the non-stressed and anti bta–mir 2898-treated heat-stressed cells.
Discussion
miRNAs are an emerging layer of novel regulators in the mammalian thermal stress response (Islam et al. 2013; Place and Noonan 2014). Till today, little or no information is available regarding the role of miRNAs in heat-stressed bovine species (Zhang et al. 2016). The present study focused on differentially expressed miRNAs during elevated environmental temperatures among Frieswal crossbred cattle developed in India. The Frieswal breed of cattle is a milch breed developed by crossing the Bos indicus Sahiwal breed with the Bos taurus Holstein Friesian cattle with an exotic inheritance of 62.50%.
First line of stress response was characterized by measuring various physiological parameters like respiratory rate, pulse rate, and rectal temperature. Physiological parameters like respiration rate, heart rate, body temperature, and skin temperature give an instant response to the climatic stress (Bianca 1965). These parameters can be used as an indicator for comfort and adaptability of dairy cattle towards an adverse environment or as a receptive physiological gauge of environmental modification (Roman-Ponce et al. 1977). Physiological responses like rectal temperature, pulse rate, and respiration rate echo the degree of stress forced on animal by climatic parameters. The ability of an animal to endure the rigors of climatic stress under temperate circumstances has been assessed physiologically by measuring the changes in body temperature, respiration rate, as well as pulse rate (Charoensook et al. 2012; Liu et al. 2012). Present study revealed that, with the upshift of environmental temperature, there was a significant (p < 0.05) elevation of rectal temperature, respiratory, and pulse rate. Heat tolerance experiments by Rhoad (1944) led to the development of a formula to calculate an individual’s heat tolerance coefficient (HTC). Results showed that HTC reduced significantly (p < 0.05) during rising of environmental temperatures.
Increased production of free radicals and reactive oxygen species and dwindle in antioxidant defense mechanisms leads to oxidative stress (Trevisan et al. 2001). Thermal stress is one of the prime reasons for oxidative stress in animals during summer in tropics. Thermal stress occurs when the core body temperature of a given species exceeds its optimal range resulting from a total heat load (internal heat production and heat gained from environment) exceeding the capacity for heat dissipation (Ganaie et al. 2013). The differences in the antioxidant enzyme activity of different breeds may result in their ability to cope up with the oxidative stress during the extreme environmental temperatures. To characterize the stress response in Frieswal cattle, we evaluated the level of two enzymatic components of the antioxidative defense system viz. catalase (CAT) and gutathione peroxidase (GPX) among heat-stressed and normal populations. Our study showed that during elevated environmental temperatures, CAT and GPX levels are significantly (p < 0.01) higher than the suboptimal temperature. The antioxidant status of growing calves, heifers, and lactating Murrah buffaloes was significantly higher for catalase as well as glutathione peroxidase concentrations during summer as compared to winter in three experimental groups (Lallawmkimi 2009). During heat stress, hydrogen peroxide production was found to increase due to augmented SOD activity (Bernabucci et al. 2005), and this in turn resulted in a synchronized increase in plasma catalase and glutathione peroxidase concentrations. In addition to the measuring of antioxidant level, we also measured the lipid peroxidation by product thiobarbituric acid (TBARS)/malondialdehyde (MDA) concentration during thermal stress in Frieswal cattle. Results revealed that MDA concentration was significantly (p < 0.01) higher during thermal stress than normal temperatures. Thiobarbituric acid test is known to be a good general indicator of oxidative stress rather than a marker of lipid peroxidation (Halliwell and Chirico 1993). TBARS concentration increased in heat-exposed cattle and buffalo than the normal counterparts (Ashok et al. 2007; Bernabucci et al. 2005).
Transcriptional activation and accumulation of a set of proteins called “heat shock proteins” (HSPs) are one of the prominent cellular responses during up shift of environmental temperatures (Kregel 2002). The two major chaperones namely HSP70 and HSP90 are the most important proteins involved during cellular stress in eukaryotic cells (Lindquist 1986; Craven et al. 1996). Our earlier reports suggested that HSP70 and HSP90 are differentially expressed in native and crossbred cattle during in vitro or in vivo thermal stress (Deb et al. 2014; Bhanuprakash et al. 2016). Induction of these two specific heat shock proteins at mRNA level in non-stressed and heat-stressed cells was investigated, and the results indicated that both the HSP transcripts were differentially expressed during elevated temperature.
The deep sequencing analysis identified the differential role of miRNAs during extreme temperatures in Frieswal cattle. While examining the animals exposed at two different environmental conditions, we found that there was an abundance of miRNAs in bovine PBMCs. In total, 420 miRNAs were identified and 65 miRNAs were differentially expressed during elevated temperatures, suggesting that miRNAs may play an important role during heat stress.
In our studies, we observed that number of differentially expressed miRNAs detected in peak summer (37 miRNAs) was higher than the normal temperatures (28 miRNAs). Validation of the selected miRNA expression revealed that there was a significant (p < 0.01) upregulation of bta-miR-103-2, bta-miR-2898, bta-miR-2478, and bta-miR-181b-2 while bta-miR-2311 and bta-miR-6536-2 are significantly (P < 0.01) down-regulated during heat stress. Besides, the presence of target sites at some of the heat shock proteins, miR-103, is known to regulate CDK5R1 expression (Moncini et al. 2011) and its over-expression may decrease the rate of cell proliferation (Leung and Sharp 2010). Recently, local, systemic, or vascular-targeted delivery of miR-103 in tumor-bearing mice decreased angiogenesis and tumor growth and thus improving the efficacy of chemotherapy and radiation (Wilson et al. 2016). Consequently, bta-miR-103-2, which is over-expressed during peak summer in Frieswal cattle, may play defensive role in terms of preventing cell proliferation and maintenance of cellular homeostasis. Bovine-specific miR-2478 plays a major role in regulation of bovine energy metabolism/adipogenesis (Romao et al. 2014). Upregulation of miR-2478 in bovine muscle is associated with the expression of the target lipogenic genes SCD1 (Muroya et al. 2016). Also, miR-181b is linked with radiation and hypoxia stress responses in humans (Jacobs et al. 2013). Similarly, miR-181b is associated with stress and immune response in Holstein cattle (Zheng et al. 2014).
Numerous studies pointed out that there is an important relationship between the family of heat shock proteins and the heat stress response (McConnell et al. 2014; Deb et al. 2014; Bhanuprakash et al. 2016; Zhao et al. 2016). Functional analysis of gene targets revealed that several target genes of selected miRNAs are involved in the stress response and oxidative stress. The network analysis of differentially expressed miRNA and their target genes indicated stress-mediated effects on signaling mechanisms. The metabolic pathways, PI3K-Akt signaling, MAPK signaling, and immune-regulatory (NF-Kappa B and RIG) are greatly influenced by miRNA-mediated regulation. The findings provide insight of miRNA-mediated post-transcriptional regulation of cellular heat stress response in Frieswal cattle. The significance of these regulations and explanations for activation of distinct mechanism needs to be further explored. Earlier studies reported that miR-19a, miR-19b, miR-27b, miR-30a-5p, miR-181a, miR-181b, miR-345-3p, and miR-1246 were highly expressed in the serum of heat-stressed Holstein cows, which were involved in many pathways that may differentially regulate the expression of stress response and immune response genes (Zheng et al. 2014).
Determining the function of the significantly differentially expressed miRNAs will provide a better understanding of miRNA-mRNA interaction mechanism. Being the highly expressed miRNA from this study, bta-miR-2898 was selected for the reporter assay. Due to lack of available literatures regarding exact cell lines that over-expressed bta-miR-2898, we have chosen bovine PBMCs. We have experimentally seen that PBMCS treated at 42 °C heat stress over-expressed bta-miR-2898 as compared to the control non-stressed cells. Interactions between miRNAs and their target genes are multifaceted, and frequently, there are abundant putative miRNA recognition sites at mRNAs; however, many miRNA targets have been computationally predicted, but only a limited number of these were experimentally validated (Witkos et al. 2011). Target scan analysis identified the most conserved target sites of bta-miR-2898 at bovine HSPB8 (heat shock protein 22). HSPB8 is a small heat shock family protein that functions as a chaperone in association with Bag3. HSPB8 appears to be involved in cell proliferation and apoptosis (Suzuki et al. 2017; Marsh et al. 2015; Ghaoui et al. 2016). We identified two binding sites of bta-miR-2898 at 3′ UTR region of HSPB8. Experimental study revealed that over-expressed bta-miR-2898 in heat-stressed PBMCs can readily bind to its target region and thus the GFP expression was suppressed, while treating with anti-miR-2898 can restore GFP expression. Gamerdinger et al. (2011) hypothesized that the molecular chaperones HSPB8 and HSP70 might recognize and bind to the misfolded proteins prior to its ubiquitination with the help of E3 ubiquitin ligase. Recently, HSPB8-BAG3 complex was reported to be the preferred interaction with HSP70 in mammalian cells (Morelli et al. 2017). In consequence, HSPB8 has been shown to cooperate with the major stressor protein HSP70. Thus, it may be possible that targeting bovine HSPB8, bta-mir-2898, may indirectly alter the function of bovine HSP70. However, further experiments are needed to be conducted for drawing a pertinent conclusion.
Identifying specific miRNAs expressed in the PBMCs of Frieswal cattle at different environmental temperatures will facilitate a better understanding of the role of miRNAs on thermoregulatory mechanisms. From this study, we will be able to develop a model which will aid in understanding the role of miRNAs during thermal stress in cattle or other species. This study provides the ground work for uncovering the role of miRNA in thermal stress that may be helpful for development of miRNA-based molecular biomarker for cellular thermotolerance in dairy cattle.
Acknowledgements
The authors are thankful to the Director, ICAR-CIRC, Meerut for providing necessary facilities to carry out the present research. Authors acknowledge the Science and Engineering Research Board, Government of India, for providing financial support under the project YSS/2014/000279 to RD. We are also thankful to Military Farm, Meerut, India, for providing experimental animals; Ome Research Laboratory, Anand Agricultural University, Gujrat, India for deep sequencing.
Compliance with ethical standards
ᅟ
All the experimental procedures involving animals were approved by the Institutional Animal Ethics Committee (IAEC)
Conflicts of interests
The authors declare that they have no conflict of interest.
Footnotes
Gyanendra Singh Sengar and Rajib Deb contributed equally
Contributor Information
Gyanendra Singh Sengar, Email: gsengar71@gmail.com.
Rajib Deb, Email: drrajibdeb@gmail.com.
References
- Ashok K, Praveen K, Singh SV. Oxidative stress markers profile in erythrocytes of natural and heat exposed cattle and buffalos. Indian J Dairy Sci. 2007;60:114–118. [Google Scholar]
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism and function. Cell. 2004;116:281–297. doi: 10.1016/S0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- Baskerville S, Bartel DP. Microarray profiling of microRNAs reveals frequent coexpression with neighboring miRNAs and host genes. RNA. 2005;11(3):241–247. doi: 10.1261/rna.7240905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belhadj Slimen I, Najar T, Ghram A, Abdrrabba M. Heat stress effects on livestock: molecular, cellular and metabolic aspects, a review. J Anim Physiol Anim Nutr. 2015;100(3):401–412. doi: 10.1111/jpn.12379. [DOI] [PubMed] [Google Scholar]
- Bernabucci U, Ronchi B, Lacetera N, Nardone A. Influence of body condition score on relationships between metabolic status and oxidative stress in periparturient dairy cows. J Dairy Sci. 2005;88(6):2017–2026. doi: 10.3168/jds.S0022-0302(05)72878-2. [DOI] [PubMed] [Google Scholar]
- Bhanuprakash V, Singh U, Sengar G, Sajjanar B, Bhusan B, Raja TV, Alex R, Kumar S, Singh R, Kumar A, Alyethodi RR. Differential effect of thermal stress on Hsp70 expression, nitric oxide production and cell proliferation among native and crossbred dairy cattle. J Therm Biol. 2016;59:18–25. doi: 10.1016/j.jtherbio.2016.04.012. [DOI] [PubMed] [Google Scholar]
- Bianca W. Reviews of the progress of dairy science. Section A Physiology: Cattle in a hot environment. J Dairy Res. 1965;32:291–345. doi: 10.1017/S0022029900018665. [DOI] [Google Scholar]
- Charoensook R, Gatphayak K, Sharifi AR, Chaisongkram C, Brenig B, Knorr C. Polymorphisms in the bovine HSP90AB1 gene are associated with heat tolerance in Thai indigenous cattle. Trop Anim Health Prod. 2012;44:921–928. doi: 10.1007/s11250-011-9989-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen K, Rajewsky N. Deep conservation of microRNA-target relationships and 3′UTR motifs in vertebrates, flies, and nematodes. Cold Spring Harb Symp Quant Biol. 2006;71:149–156. doi: 10.1101/sqb.2006.71.039. [DOI] [PubMed] [Google Scholar]
- Craven RA, Egerton M, Stirling CJ. A novel Hsp70 of the yeast ER lumen is required for the efficient translocation of a number of protein precursors. EMBO J. 1996;15:2640–2650. [PMC free article] [PubMed] [Google Scholar]
- Deb R, Sajjanar B, Singh U, Kumar S, Singh R, Sengar G, Sharma A. Effect of heat stress on the expression profile of Hsp90 among Sahiwal (Bos Indicus) and Frieswal (Bos Indicus × Bos Taurus) breed of cattle: a comparative study. Gene. 2014;536(2):435–440. doi: 10.1016/j.gene.2013.11.086. [DOI] [PubMed] [Google Scholar]
- Fatima A, Waters S, O’Boyle P, Seoighe C, Morris DG. Alterations in hepatic miRNA expression during negative energy balance in postpartum dairy cattle. BMC Genomics. 2014;15(1):28. doi: 10.1186/1471-2164-15-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamerdinger M, Carra S, Behl C. Emerging roles of molecular chaperones and co-chaperones in selective autophagy: focus on BAG proteins. J Molec Med. 2011;89(12):1175–1182. doi: 10.1007/s00109-011-0795-6. [DOI] [PubMed] [Google Scholar]
- Ganaie AH, Shanker G, Bumla NA, Ghasura RS, Mir NA. Biochemical and physiological changes during thermal stress in bovines. J Vet Sci Technol. 2013;4:126. [Google Scholar]
- Ghaoui R, Palmio J, Brewer J, Lek M, Needham M, Evilä A, Hackman P, Jonson PH, Penttilä S, Vihola A, Huovinen S. Mutations in HSPB8 causing a new phenotype of distal myopathy and motor neuropathy. Neurology. 2016;86(4):391–398. doi: 10.1212/WNL.0000000000002324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halliwell B, Chirico S. Lipid peroxidation: its mechanism, measurement, and significance. Am J Clin Nutr. 1993;57(5):715–724. doi: 10.1093/ajcn/57.5.715S. [DOI] [PubMed] [Google Scholar]
- Hansen PJ (2004) Physiological and cellular adaptations of zebu cattle to thermal stress. Anim Reprod Sci. (82-83):349-360 [DOI] [PubMed]
- Harfe BD. MicroRNAs in vertebrate development. Curr Opin Genet Dev. 2005;15(4):410–415. doi: 10.1016/j.gde.2005.06.012. [DOI] [PubMed] [Google Scholar]
- Islam A, Deuster PA, Devaney JM, Ghimbovschi S, Chen Y. An exploration of heat tolerance in mice utilizing mRNA and microRNA expression analysis. PLoS One. 2013;8(8):72258. doi: 10.1371/journal.pone.0072258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs AAA, Dijkstra J, Liesman JS, VandeHaar MJ, Lock AL, van Vuuren AM, Hendriks WH, van Baal J. Effects of short- and long-chain fatty acids on the expression of stearoyl-CoA desaturase and other lipogenic genes in bovine mammary epithelial cells. Animal. 2013;7:1508–1516. doi: 10.1017/S175173111300061X. [DOI] [PubMed] [Google Scholar]
- Kregel KC. Heat shock proteins: modifying factors in physiological stress responses and acquired thermotolerance. J Appl Physiol. 2002;92:2177–2186. doi: 10.1152/japplphysiol.01267.2001. [DOI] [PubMed] [Google Scholar]
- Kropp J, Salih SM, Khatib H. Expression of microRNAs in bovine and human pre-implantation embryo culture media. Front Genet. 2014;5:91. doi: 10.3389/fgene.2014.00091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacetera N, Bernabucci U, Scalia D, Basiricò L, Morera P, Nardone A. Heat stress elicits different responses in peripheral blood mononuclear cells from Brown Swiss and Holstein cows. J Dairy Sci. 2006;89:4606–4612. doi: 10.3168/jds.S0022-0302(06)72510-3. [DOI] [PubMed] [Google Scholar]
- Lallawmkimi CM (2009) Impact of thermal stress and vitamin-E supplementation on Heat shock protein 72 and antioxidant enzymes in Murrah buffaloes. Ph. D. Thesis National Dairy Research Institute (deemed University), Karnal, India
- Leung AK, Sharp PA. MicroRNA functions in stress responses. Mol Cell. 2010;40:205–215. doi: 10.1016/j.molcel.2010.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis BP, Shih IH, Jones-Rhoades MW, Bartel DP, Burge CB. Prediction of mammalian microRNA targets. Cell. 2003;115:787–798. doi: 10.1016/S0092-8674(03)01018-3. [DOI] [PubMed] [Google Scholar]
- Li R, Beaudoin F, Ammah AA, Bissonnette N, Benchaar C, Zhao X, Lei C, Ibeagha-Awemu EM. Deep sequencing shows microRNA involvement in bovine mammary gland adaptation to diets supplemented with linseed oil or safflower oil. BMC Genomics. 2015;16(1):884. doi: 10.1186/s12864-015-1965-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindquist S. The heat-shock response. Annu Rev Biochem. 1986;55:1151–1191. doi: 10.1146/annurev.bi.55.070186.005443. [DOI] [PubMed] [Google Scholar]
- Lindquist S, Craig EA. The heat shock proteins. Ann Rev Genet. 1988;22:631–637. doi: 10.1146/annurev.ge.22.120188.003215. [DOI] [PubMed] [Google Scholar]
- Liu E, Lewis K, Al-Saffar H, Krall CM, Singh A, Kulchitsky VA, Corrigan JJ, Simons CT, Petersen SR, Musteata FM, Bakshi CS. Naturally occurring hypothermia is more advantageous than fever in severe forms of lipopolysaccharide-and Escherichia Coli-induced systemic inflammation. Am J Phys Regul Integr Comp Phys. 2012;302(12):1372–1383. doi: 10.1152/ajpregu.00023.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2− ΔΔCT method. Methods. 2001;25(4):402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Marsh NM, Wareham A, White BG, Miskiewicz EI, Landry J, MacPhee DJ. HSPB8 and the Cochaperone BAG3 are highly expressed during the synthetic phase of rat myometrium programming during pregnancy. Biol Reprod. 2015;92(5):131. doi: 10.1095/biolreprod.114.125401. [DOI] [PubMed] [Google Scholar]
- McConnell JR, Alexander LA, McAlpine SR. A heat shock protein 90 inhibitor that modulates the immunophilins and regulates hormone receptors without inducing the heat shock response. Bioorg Med Chem Lett. 2014;24:661–666. doi: 10.1016/j.bmcl.2013.11.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenna LB, Schug J, Vourekas A, McKenna JB, Bramswig NC, Friedman JR, Kaestner KH. MicroRNAs control intestinal epithelial differentiation, architecture, and barrier function. Gastroenterology. 2010;139:1654–1664. doi: 10.1053/j.gastro.2010.07.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehla K, Magotra A, Choudhary J, Singh AK, Mohanty AK, Upadhyay RC, Srinivasan S, Gupta P, Choudhary N, Antony B, Khan F. Genome wide analysis of the heat stress response in zebu (Sahiwal) cattle. Gene. 2014;533:500–550. doi: 10.1016/j.gene.2013.09.051. [DOI] [PubMed] [Google Scholar]
- Moncini S, Salvi A, Zuccotti P, Viero G, Quattrone A, Barlati S, De Petro G, Venturin M, Riva P. The role of miR-103 and miR-107 in regulation of CDK5R1 expression and in cellular migration. PLoS One. 2011;6(5):20038. doi: 10.1371/journal.pone.0020038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morelli FF, Mediani L, Heldens L, Bertacchini J, Bigi I, Carrà AD, Vinet J, Carra S. An interaction study in mammalian cells demonstrates weak binding of HSPB2 to BAG3, which is regulated by HSPB3 and abrogated by HSPB8. Cell Stress Chaperones. 2017;22(4):531–540. doi: 10.1007/s12192-017-0769-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muroya S, Hagi T, Kimura A, Aso H, Matsuzaki M, Nomura M. Lactogenic hormones alter cellular and extracellular microRNA expression in bovine mammary epithelial cell culture. J Anim Sci Biotechnol. 2016;7(1):8. doi: 10.1186/s40104-016-0068-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nehammer C, Podolska A, Mackowiak SD, Kagias K, Pocock R. Specific microRNAs regulate heat stress responses in Caenorhabditis Elegans. Sci Rep. 2015;5:8866. doi: 10.1038/srep08866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paula-Lopes FF, Chase CC, Al-Katanani YM, Krininger CE, Rivera RM, Tekin S, Majewski AC, Ocon OM, Olson TA, Hansen PJ. Genetic divergence in cellular resistance to heat shock in cattle: differences between breeds developed in temperate versus hot climates in responses of preimplantation embryos, reproductive tract tissues and lymphocytes to increased culture temperatures. Reproduction. 2003;125(2):285–294. doi: 10.1530/rep.0.1250285. [DOI] [PubMed] [Google Scholar]
- Place RF, Noonan EJ. Non-coding RNAs turn up the heat: an emerging layer of novel regulators in the mammalian heat shock response. Cell Stress Chaperones. 2014;19(2):159–172. doi: 10.1007/s12192-013-0456-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajewsky N. microRNA target predictions in animals. Nat Genet. 2006;38:8–13. doi: 10.1038/ng1798. [DOI] [PubMed] [Google Scholar]
- Roman-Ponce H, Thatcher WW, Buffington DE, Wilcox CJ, Van Horn HH. Physiological and production responses of dairy cattle to a shade structure in a subtropical Environment1. J Dairy Sci. 1977;60(3):424–430. doi: 10.3168/jds.S0022-0302(77)83882-4. [DOI] [Google Scholar]
- Romao JM, Jin W, He M, McAllister T. MicroRNAs in bovine adipogenesis: genomic context, expression and function. BMC Genomics. 2014;15(1):137. doi: 10.1186/1471-2164-15-137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sengar GS, Deb R, Raja TV, Singh U, Kant R, Bhanuprakash V, Alyethodi RR, Kumar S, Verma P, Chakraborty S, Alex R, Singh R (2017) RT-LAMP assay: an alternative approach for profiling of bovine heat shock protein 70 gene in PBMC cultured model. Molecular Biology Reports (in press) [DOI] [PubMed]
- Suzuki H, Matsushita S, Suzuki K, Yamada G. 5α-dihydrotestosterone negatively regulates cell proliferation of the periurethral ventral mesenchyme during urethral tube formation in the murine male genital tubercle. Andrology. 2017;5(1):146–152. doi: 10.1111/andr.12241. [DOI] [PubMed] [Google Scholar]
- Trevisan M, Browne R, Ram M, Muti P, Freudenheim J, Carosella AM, Armstrong D. Correlates of markers of oxidative status in the general population. Am J Epidemiol. 2001;154(4):348–356. doi: 10.1093/aje/154.4.348. [DOI] [PubMed] [Google Scholar]
- Wang XP, Luoreng ZM, Zan LS, Raza SH, Li F, Li N, Liu S. Expression patterns of miR-146a and miR-146b in mastitis infected dairy cattle. Mol Cell Probes. 2016;30(5):342–344. doi: 10.1016/j.mcp.2016.08.004. [DOI] [PubMed] [Google Scholar]
- Wang ZH, Zhang JL, Duan YL, Zhang QS, Li GF, Zheng DL. MicroRNA-214 participates in the neuroprotective effect of resveratrol via inhibiting α-synuclein expression in MPTP-induced Parkinson’s disease mouse. Biomed Pharmacother. 2015;74:252–256. doi: 10.1016/j.biopha.2015.08.025. [DOI] [PubMed] [Google Scholar]
- Witkos TM, Koscianska E, Krzyzosiak WJ. Practical aspects of microRNA target prediction. Curr Mol Med. 2011;11(2):93–109. doi: 10.2174/156652411794859250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Yuan L, Zhang X, Hamblin MH, Zhu T, Meng F, Li Y, Chen YE, Yin KJ. Altered long non-coding RNA transcriptomic profiles in brain microvascular endothelium after cerebral ischemia. Exp Neurol. 2016;277:162–170. doi: 10.1016/j.expneurol.2015.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J, He Q, Chen G, Wang L, Jin B. Regulation of non-coding RNAs in heat stress responses of plants. Front Plant Sci. 2016;7:1213. doi: 10.3389/fpls.2016.01213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Y, Chen KL, Zheng XM, Li HX, Wang GL. Identification and bioinformatics analysis of microRNAs associated with stress and immune response in serum of heat-stressed and normal Holstein cows. Cell Stress Chaperones. 2014;19(6):973–981. doi: 10.1007/s12192-014-0521-8. [DOI] [PMC free article] [PubMed] [Google Scholar]