Abstract
In glioblastoma (GBM), tumor-associated macrophages (TAM) represent up to one half of the cells of the tumor mass, including both infiltrating macrophages and resident brain microglia. In an effort to delineate the temporal and spatial dynamics of TAM composition during gliomagenesis, we employed two genetically engineered mouse models where oncogenic drivers and fluorescent reporters were expressed coordinately under the control of the monocyte/microglia-selective Cx3cr1 or Ccr2 promoters, respectively. Using this approach, we demonstrated that CX3CR1LoCCR2Hi monocytes were recruited to the glioblastoma, where they transitioned to CX3CR1HiCCR2Lo macrophages and CX3CR1HiCCR2− microglia-like cells. Infiltrating macrophages/monocytes constituted ~85% of the total TAM population, with resident microglia accounting for the ~15% remaining. Bone marrow-derived infiltrating macrophages/monocytes were recruited to the tumor early during GBM initiation, where they localized preferentially to perivascular areas. In contrast, resident microglia were localized mainly to peritumoral regions. RNA-sequencing analyses revealed differential gene expression patterns unique to infiltrating and resident cells, suggesting unique functions for each TAM population. Notably, limiting monocyte infiltration via Ccl2 genetic ablation prolonged the survival of tumor-bearing mice. Our findings illuminate the unique composition and functions of infiltrating and resident myeloid cells in GBM, establishing a rationale to target infiltrating cells in this neoplasm.
Keywords: malignant glioma, microglia, inflammatory monocytes, CCR2, CX3CR1
Introduction
The most common histological type of brain tumor in adults is the glioma, with the more malignant grades dominating between the 5th and 8th decades of life. Among these clinically aggressive and deadly cancers, glioblastoma (World Health Organization grade IV) is the most frequently encountered. These tumors most often arise within the subcortical white matter of the cerebral hemispheres, where they appear as poorly demarcated masses associated with increased vascularity and tissue necrosis. Histological analysis of glioblastoma reveals highly anaplastic and mitotic tumor cells embedded with a tumor microenvironment composed of brain-resident microglia, infiltrating monocytes/macrophages, reactive astrocytes, endothelial cells, pericytes, neural stem/progenitor cells and other immune cell infiltrates (1,2). The tissue macrophage compartment contains both resident microglia and bone marrow (BM)-derived macrophages, although the exact composition and tissue origins have been incompletely characterized.
The importance of these tumor associated macrophage (TAM) populations to glioma growth is highlighted by their large numbers within these tumors, comprising as many as 30–50% of all cells in human glioblastoma (2). In addition, numerous studies focused on experimental murine low-grade glioma and glioblastoma models have revealed a critical role for these TAMs in tumor formation and maintenance. Using a neurofibromatosis type 1 (NF1) model of low-grade glioma, pharmacological or genetic silencing of microglia delays glioma formation and attenuates established tumor growth, suggesting the presence of growth factors elaborated by these TAMs (3). RNA-sequencing of TAMs in this model revealed that the CCL5 chemokine is a major microglia-derived driver of neurofibromatosis 1 glioma growth (4). In addition, a large number of elegant studies on murine high-grade glioma have demonstrated that microglia are critical for tumor growth and migration (5). These microglia are recruited by the glioma cells to establish a feed-forward cellular circuit that drives further tumor growth (5). In this regard, changing TAM polarization by inhibiting colony-stimulating factor 1 receptor signaling reduced PDGFB-driven murine glioblastoma growth (6); however, this strategy failed to show efficacy in clinical trials for recurrent glioblastoma (7).
One reason that targeting TAMs in glioblastoma has not been successful can be partially attributable to the fact that TAMs are a mixed population. Macrophages derived from infiltrating bone marrow-derived monocytes or resident microglia can be morphologically indistinguishable in tissue sections and are not reliably separated using available lineage-specific antibodies. Traditionally, investigators have relied on CD45 antibodies to distinguish resident microglia (CD45Lo) from macrophages of hematopoietic origin (CD45Hi) using FACS analysis (8). The use of CD45 to separate these populations was recently challenged by studies using head-protected irradiation chimeras, showing that microglia can increase CD45 expression to constitute a proportion of the CD45Hi population in gliomas (9). In addition, there is also a significant myeloid cell infiltration in high-grade glioma, postulated to result from total body irradiation damage to brain-blood barrier (BBB) integrity (9,10). As such, it is currently unclear to what extent glioblastoma macrophages are derived from infiltrating cells from the blood circulation or what proportion of the glioblastoma macrophages are microglia versus monocyte/macrophages. Moreover, it is not known where each population is physically localized within glioblastoma, which contains specialized cellular niches (11).
To gain insights into the populations of TAMs that comprise murine experimental glioblastoma, we leveraged a mouse model in which the Cx3cr1 and Ccr2 genes contain distinct fluorescent proteins as knock-in alleles (Cx3cr1GFP/WT; Ccr2RFP/WT mice). The use of these mice is predicated on the finding that mouse monocytes can be subdivided into Ly6C+CX3CR1IntCCR2+ inflammatory monocytes and Ly6CLo/−CX3CR1HiCCR2− circulating monocytes (12,13). Combining Cx3cr1GFP/WT;Ccr2RFP/WT knock-in mice with a genetically-engineered mouse model (GEMM) of PDGFB-driven glioblastoma, we show that the majority of inflammatory monocytes/macrophages express both CCR2 and CX3CR1, while microglia express only CX3CR1. Multi-parameter flow cytometry analyses showed that CD45Hi population consists of CCR2+CX3CR1+ cells, while the CD45Lo population only contains CX3CR1+ cells. In addition, over 85% of the TAMs within the tumors are bone marrow-derived macrophages, while microglia predominate in the peritumoral areas. Additionally, RNA-sequencing analyses revealed differential gene expression patterns unique to infiltrating and resident cells, suggesting unique functions for each TAM population. Loss of one copy of Ccl2, the ligand for CCR2, which recruits monocytes to sites of inflammation, significantly prolonged the survival of glioblastoma-bearing mice. Our results demonstrate the unique spatial and temporal differential composition, and distinct biological functions for infiltrating and resident myeloid cells in glioblastoma.
Materials and Methods
Mice
Animals were housed in the Cleveland Clinic Biological Resource Unit or Emory University Division of Animal Resources. Mice undergoing GL261 glioma cell injections were housed at the Max-Delbrück-Center of Molecular Medicine in the Helmholtz Association. All experimental procedures were approved by the Institutional Animal Care and Use Committee of the Cleveland Clinic (Animal Protocol 2013-1029) and Emory University (Protocol #2003253) and by the LaGeSo of the Helmholtz Association (G0438/12). Mice (6–10 weeks old, except animals that went through bone marrow transplant which are usually 8 weeks older) of both genders were used in all experiments. We used age and sex as criteria to equally distribute mice from different genotypes for all the experiments. Gli-Luciferase; Nestin-tv-a;Ink4a-Arf−/−;Ptenfl/fl mice were generated as previously described (1,14). B6 (Cx3cr1WT/WT;Ccr2WT/WT) and Cx3cr1GFP/WT;Ccr2RFP/WT mice were a gift from Richard Ransohoff (15,16).
Cell cultures and transfection
DF-1 cells were purchased from ATCC in 2010. Cells were grown at 39oC according to instructions from ATCC, expanded to passage 4 and stored in aliquots in liquid nitrogen. Transfection with RCAS-PDGFB-HA and RCAS-shRNA p53 were performed using a Fugene 6 transfection kit (# 11814443001 Roche, Mannheim, Germany) according to manufacturer’s instructions. Transfected cells are used for injections before they reach passage 25. Cells were tested for mycoplasma contamination by using a “PlasmoTest” kit (Invivogen). Murine GL261 glioma cells were obtained from the National Cancer Institute in 2016. They were grown in Dulbecco’s modified Eagle’s medium (DMEM) with 10% fetal calf serum, 200 mM glutamine, 100 U/mL penicillin, and 100 mg/ml streptomycin (all from Invitrogen, Carlsbad, CA, USA). Cells cultured less than 5 passages were used to inject mice to induce glioma. Because of the short history of growth and healthy appearance, no mycoplasma test was done on GL261 cells.
Generation of tumors using RCAS/Ntva system
Donors: Transgenic mice (Gli-Luciferase;Nestin-tv-a,ink4a-Arf−/−Ptenfl/fl referred as NiG) were anesthetized with intraperitoneal injections of a mixture of ketamine and xylazine (ketamine, 0.1 mg/g and xylazine, 0.02 mg/g). One microliter of 4×104 cell suspension containing RCAS-PDGF-B-HA transfected DF1 cells was delivered using a 30-gauge needle attached to a Hamilton syringe and stereotactic fixation device (Stoelting, Wood Dale, IL). Locations were determined according to the brain atlas (17). Cells were injected into the right frontal striatum with the following coordinates: bregma AP (anterior/posterior) 1.0 mm, ML (medial/lateral) 1.0 mm, and DV (dorsal/ventral) 2.0 mm. The same location was used to generate tumor in Ntva;Ccl2+/+ and Ntva;Ccl2+/−. Ccl2−/− mice were obtained from Jackson laboratory (#004434) (18) and were further crossed to B6/Ntva mice. Mice were monitored carefully and were sacrificed if they displayed lethargy due to tumor burden.
Orthotopic glioma generation
The same procedure was used as described above, except that 2.5×104 of freshly-dissociated tumor cells from RCAS/Ntva donors were injected into the frontal striatum of recipient animals.
Generation of murine gliomas using GL261 cell line
One microliter cell suspension of 2×104 GL261 cells was delivered using a 30-gauge needle attached to a Hamilton syringe (Hamilton, Reno, NV, USA). Coordinates for injections were 1 mm anterior, 2mm lateral and 3 mm deep relative to the bregma. Mice were monitored daily for the first two weeks and twice a day starting from day 15 post-injection for symptoms of tumor development (lethargy, hydrocephalus, head tilting). Mice were euthanized 21 days post-injection.
Tissue processing
Animals were anesthetized with a ketamine/xylazine mix, perfused with ice-cold Ringer’s solution, and sacrificed. Brains were removed and processed according to the different applications. For H&E tumor validation and immunohistochemistry staining, brains were fixed in 10% neutral buffered formalin for 72 hours at RT, processed in a tissue processer (Leica TP1050), embedded in paraffin, sectioned (5 μm), and slide mounted. For immunofluorescent staining, brains were fixed in 4% PFA overnight at 4°C, immersed in 30% sucrose (dissolved in PBS) for 48 hours at 4°C, embedded in Optimal Cutting Temperature (OCT, Tissue-Tek) compound, sectioned (8 μm), slide mounted, and stored at −80°C.
Immunofluorescent staining
8μm coronal sections were used for frozen sections in all histological studies. The following antibodies were used at the stated dilutions: rabbit polyclonal anti-Iba1, 1:100 (Wako Pure Chemicals, Osaka Japan); Rabbit polyclonal anti-RFP, 1:50 (Rockland), rat monoclonal anti-CD31, 1:100 (Histonova), rabbit anti-Ki67, 1:100 (Abcam). Secondary antibodies conjugated to different Alexa-Fluor dyes (488nm, 555nm, 647nm from Invitrogen) at a dilution of 1:500 in PBS/2%BSA were applied. For nuclear counterstaining, DAPI was used (Sigma). For quantification of Iba1+, GFP+ or RFP+ cells, five to ten images (20x) of tumor and peritumoral regions were taken per mouse brain using CD31 or DAPI staining as a reference on an Olympus FV1000 confocal microscope. TUNEL assay (Sigma) was performed as previously described (19). Full brain coronal sections were created by multiple area tiled scanning using the same microscope. Cell numbers were counted with FIJI and normalized to a 1 mm2 area.
Flow cytometry
Brains were digested in 0.25% Trypsin/EDTA without phenol red at 37°C for 10 minutes (glioblastoma) or 30 minutes (naïve brains, without cerebellum). Digestion was terminated by adding 2 volumes of RPMI medium containing 10% FBS. Cells were passed through a 40μm cell strainer, centrifuged and resuspended in 30% Percoll (GE Healthcare) solution and layered above a 70% Percoll layer (diluted in RPM medium with 1% FBS). Cells were separated by centrifuging at 800xg for 30 minutes at 4°C. Cells from the 30%/70% Percoll interphase were collected and washed with FACS buffer (DPBS with 0.5% BSA) and blocked with 100μl of 2× blocking solution (2% FBS, 5% normal rat serum, 5% normal mouse serum, 5% normal rabbit serum, 10 μg/ml 2.4g2 anti-FcR and 0.2% NaN3 in DPBS) on ice for 30 minutes. Cells were then stained on ice for 30 minutes and washed with FACS buffer. For enumeration of blood cell types, blood samples were taken via tail vein bleeding, lysed with RBC lysis buffer (BioLegend, cat#420301), washed with FACS buffer, counted with a hemocytometer, and stained using the same protocol described above. Antibodies used in the study include: CD45-APC, CD11b-PerCP-Cy5.5, Ly6C-PE-Cy7, F4/80-APC-Cy7 (BD Pharmingen), and Ly6G-V450 (BioLegend). All data were collected on a BD LSR flow cytometer and analyzed using FlowJo 10 software (Tree Star Inc.).
Generation of BM chimeras
Two irradiation regiments were used to ablate host hematopoietic system for BM chimera generations, the first with total body irradiation (TBI) and the second with head-protected irradiation (HPI) where the heads of the mice were protected from the ionizing X-ray. For TBI, recipient mice 4 to 5 weeks old were irradiated in a Shepherd Mark 137Cs irradiator by two doses of irradiation at 600 rads each, with a 4h interval in between. For HPI, anesthetized mice were first placed in a custom-made apparatus where their bodies were accessible by X-ray but their heads were protected by a 5-mm thick lead sheet. The apparatus was then placed in a low-energy Pantak Cabinet X-ray irradiator with totaling 950 rads delivered. Bone marrow cells were collected from femurs and tibias of donor mice. Single bone marrow cells were suspended in sterile HBSS at 2×108 cells/ml. 100 μl cell suspension (2×107 cells) was injected in each recipient mouse through the retro-orbital sinus. Recipient mice received 1.2 mg/ml gentamicin (Thermo Fisher) through drinking water for 10 days. Eight to 10 weeks after the bone marrow transplant, 50 μl of blood were collected through an incision in the tail vein and placed in 10U of heparin (Sigma). The erythrocytes were lysed and the leukocytes were washed in PBS and analyzed by flow cytometry as described above to determine the efficiency of chimeric reconstitution. To quantify the reconstitution efficiency, we took advantage of the fact that all Ly6CHi inflammatory monocytes express CCR2. A formula thus can be generated: % reconstitution efficiency = CCR2RFP cells/Ly6CHi cells X 100%.
RNA-sequencing and data analyses
Detailed description can be found in the supplemental materials online. Briefly, Naïve microglia, naïve monocytes, tumor-associated microglia, and tumor-associated monocytes were isolated by FACS sorting based on CD11b, CD45 and CX3CR1 and CCR2 combination, details are described above. Their mRNA was extracted with RNeasy Plus Mini kit (Qiagen) per manufacturer’s instructions. The quality of the mRNA was assessed by Bioanalyzer (Agilent Technology) and the RIN numbers were above 9.50. Complementary DNA (cDNA) library was established by using Ovation RNA-Seq v2 method (NuGen, San Carlos, CA). Samples were sequenced on the HiSeq 2000 at 2X 101 bp in the paired ends. Illumina HiSeq Control Software and Real-Time Analysis were used for sequencing and raw instrument data processing. Gene and isoform expression levels were calculated using Cufflinks version 2.1.1. Transcripts from mitochondrial and ribosomal RNA genes were masked and not included. The final abundances of genes in cells were computed in FPKM (Fragments Per Kilobase of exon model per Million mapped fragments). The GTF (General Transfer Format) from Ensembl release 67 was used for genome-wide transcriptome quantification of protein-coding genes, annotated in mm9, including alternative transcript isoform expression estimation. In order to compare gene and transcript expression under two conditions, the annotated GTF was fed to the Cuffdiff algorithm in Cufflinks to measure the fold change of the coding genes. Final differentially expressed genes (DEGs) were listed with their expected fragment numbers. To identify overlapping or unique DEGs, a minimum Log 2 fold change of 2 and P-value ≤ 0.01 were used as cut-off value in the pairwise comparisons. A cross comparison was then applied to identify overlapping and unique DEGs between tumor-associated microglia and macrophages. The biological processes enriched among the overlapping DEGs were searched against a variety of databases using the Reactome FIViz plugin (20) in Cytoscape (21). The significance of GO (gene ontology) terms was determined based on P-value (≤ 0.001) and FDR (≤ 0.025). The number of unique DEGs specific to tumor-associated microglia, tumor-associated macrophages, and common to both in a defined functional category are presented. The raw RNA-Seq data, along with experimental information is deposited in the NCBI Sequence Read Archive (SRA) database under accession number PRJNA349180.
Quantitative real-time PCR
RNA concentrations were measured with a NanoDrop spectrophotometer and samples were stored at -80°C. cDNA was synthesized from total RNA using the SuperScript III First-Strand Synthesis System (Life Technologies, Grand Island, NY). Quantitative real-time PCR was performed using a SsoAdvanced Universal SYBR Green Supermix (Bio-Rad, CA) according to manufacturer’s instructions. Amplification was performed on a Bio-Rad CFX96 Real-time System. Primers are purchased as off-the-shelf products from BioRad. HPRT was used as an internal control and the ΔΔCT method was used to calculate changes in fold expression.
Statistical analysis
Graphs were created using GraphPad Prism 6 (GraphPad Software Inc.) and were analyzed using an unpaired or paired parametric two-tailed t-test as appropriate, assuming equal standard deviations. One-way ANOVA was used in experiments having more than one group to compare to controls. The details are included in Figure legends (*) P < 0.05; (**) P < 0.01; (***) P < 0.001; (no asterisk) not significant.
Results
The majority of TAMs are infiltrating CD45Hi leukocytes from the blood circulation
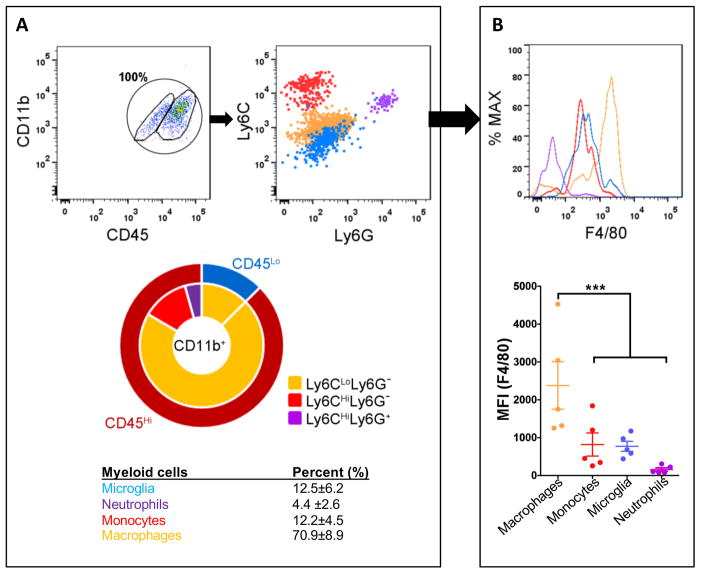
To characterize TAMs in murine PDGFB-driven glioblastoma, we initially employed standard antibody combinations for flow cytometric analysis. For these experiments, tumors were dissected from B6 mice when the mice were terminally ill from tumor burden (survival endpoint), and the total percentages of CD45+CD11b+ myeloid cells examined (Fig. 1A,B). Relative to naïve mice, where the CD45Hi population accounts for about 1.5±0.3% of the total brain myeloid cell population (Fig. 2A), the percentage of CD45Hi cells in the brain tumors was increased (87.5±6.2%, P<0.01, Fig. 1A). Next, the Ly6C and Ly6G surface markers were used to subdivide CD45Hi cells into three distinct populations (Ly6CHiLy6G− monocytes, Ly6CLoLy6G− tissue macrophages, and Ly6C+Ly6G+ neutrophils; Fig. 1A). Among all of the CD45Hi cells, the majority expressed low levels of Ly6C (Ly6CLoLy6G− macrophages). In contrast, 12.2±4.5% of these cells expressed high levels of Ly6C (Ly6CHiLy6G− inflammatory monocytes), while only 4.4±2.6% expressed both markers (Ly6C+Ly6G+ neutrophils) (Fig. 1A). CD45Lo resident microglia expressed no or very low levels of Ly6C, consistent with previous findings (22). Interestingly, when the expression of F4/80, a mature phagocytic cell marker, was examined, we found that the CD45HiLy6CLo population exhibited the highest levels of F4/80 (mean fluorescent intensity or MFI at 2.3±1.4×103, P<0.001), indicating that these cells had differentiated into tissue macrophages (Fig. 1B). In contrast, Ly6CHi inflammatory monocytes maintained low levels of F4/80 (MFI=0.83±0.68×103), confirming that they recently infiltrated from the blood circulation into the tumor. Microglial expression of F4/80 (MFI=0.77±0.29×103) was comparable to that of the Ly6CHi cells, but was much lower than that observed in the Ly6CLo cells. Neutrophil expression of F4/80 was undetectable (Fig. 1B).
Figure 1. The majority of TAMs are infiltrating CD45Hi leukocytes from the blood circulation.
(A) Representative dot plots gated on the CD11b+CD45+ cells from tumors generated in B6 mice. The total population of CD11b+CD45+ cells is considered to be 100%, with CD11b+CD45Hi (blood-derived monocytes and macrophages) and CD11+CD45Lo (resident brain microglia) populations gated separately. Their expression of Ly6C and Ly6G were analyzed and the percentage of each subpopulation quantified. (B) Quantification of F4/80 intensity for all the subpopulations. N=5. ***P<0.001 by ANOVA.
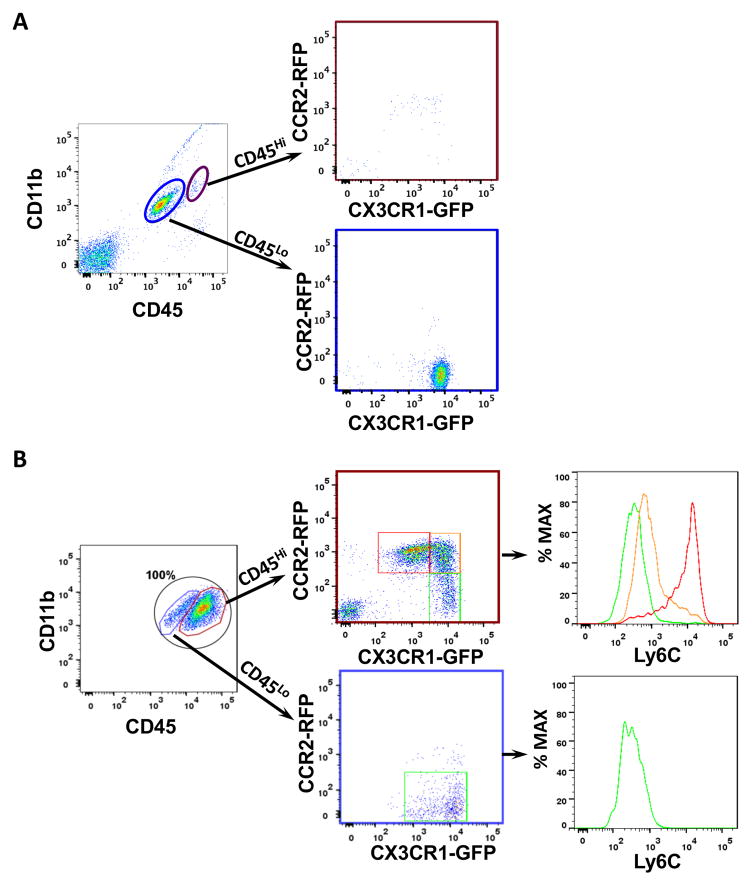
Figure 2. Cx3cr1GFP/WT;Ccr2RFP/WT knock-in mice distinguish resident microglia from BM-derived macrophages.
Representative dot plots gated on CD11b+CD45+ cells from (A) naïve and (B) tumors generated in Cx3cr1GFP/WT;Ccr2RFP/WT mice. (A) Magenta and blue circles delineate the CD11b+CD45Hi (blood-derived monocytes and macrophages) and CD11+CD45Lo (resident brain microglia) populations. Their CX3CR1-GFP and CCR2-RFP profiles are shown. (B) TAMs were identified by their CD11b and CD45 expression, and the CD45Hi and CD45Lo cells are further gated on RFP (CCR2) and GFP (CX3CR1) positivity. The CD45Hi population can be stratified into three related but distinct populations, whose Ly6C expression is examined. The CD45low population (microglia) expressed high level of CX3CR1-GFP, but little to no CCR2-RFP. Quantification of the subpopulations of CD45Hi or CD45Lo myeloid cells are described in the Result. N=3 for naïve mice and N=5 tumor bearing mice.
Cx3cr1GFP/WT;Ccr2RFP/WT knock-in mice distinguish resident microglia from BM-derived macrophages
While the above FACS marker combinations are useful for distinguishing resident microglia from BM-derived monocyte/macrophages, they are not suitable for identifying these cells in histological sections (23). To define the monocyte populations in murine glioblastoma tissues using genetic strategies, we took advantage of Cx3cr1GFP/WT;Ccr2RFP/WT mice, which express green fluorescent protein (GFP) under the control of Cx3cr1 promoter and red fluorescent protein (RFP) under the control of Ccr2 promoter. Both strains have these fluorescent protein transgenes inserted into the endogenous locus to mimic normal CX3CR1 and CCR2 expression. In these experiments, we leveraged the complementary expression intensities of CX3CR1 and CCR2 to distinguish inflammatory monocytes (CX3CR1LoCCR2Hi cells with high Ly6C levels), patrolling monocytes (CX3CR1HiCCR2Lo cells with low Ly6C levels) (24), and resident brain microglia (CX3CR1HiCCR2− cells) in murine glioblastoma.
We first demonstrated that a single copy replacement of Cx3cr1 or Ccr2 gene by GFP or RFP did not affect tumorigenesis, as determined by mouse survival and glioma penetrance (Supplementary Table S1). Consistent with previous published reports (25), naïve brains from Cx3cr1GFP/WT;Ccr2RFP/WT mice contained only CX3CR1HiCCR2− resident brain microglia (Fig. 2A). In addition, there were no differences observed in the percentages of the CD45Hi infiltrating cells when tumors from Cx3cr1GFP/WT;Ccr2RFP/WT mice were compared to those generated in wild-type B6 mice (Fig. 2B). Second, three distinct, but related monocyte populations, were observed in the murine brain tumors as stratified by their CX3CR1-GFP or CCR2-RFP expression intensities. The CX3CR1LoCCR2Hi (31.93±13.16% of total CD11b+ cells) and CX3CR1HiCCR2Lo (13.81±8.43%) populations demonstrated polarizing Ly6C expression (Fig. 2B). Surprisingly, a dominant double-positive population (36.05±8.13%) was found (gated in yellow, Fig. 2B). These double-positive cells expressed intermediate/low levels of Ly6C, but high levels of F4/80 (not shown), and correspond to the CD45HiLy6CLoF4/80Hi population in tumors generated in wild-type B6 mice (Fig. 1). However, the presence of CCR2 demonstrates that these cells are BM-derived, rather than resident microglia that upregulated CD45 expression. Lastly, the small numbers (9.83±1.42%) of CD45Lo microglia expressed abundant CX3CR1, as indicated by strong GFP signal with low to negative Ly6C expression (Fig. 2B). Collectively, these data support the use of Cx3cr1GFP/WT and Ccr2RFP/WT knock-in mice to study the temporal and spatial dynamics of TAM infiltration and function in murine glioblastoma in vivo.
Inflammatory monocytes infiltrate to glioblastoma from the blood circulation
In terminally-ill mice with glioblastoma, the number of BM-derived cells from the circulation represents the greatest portion of the TAMs. To determine whether the number of circulating BM-monocytes correlates with disease progression, we analyzed whole blood of B6 and Cx3cr1GFP/WT;Ccr2RFP/WT mice 20 days post-tumor cell transplantation (no tumors at this time point) and at death. Quantification of the numbers of monocytes and neutrophils show that there is a significant decrease in Ly6CHi blood monocytes from tumor-bearing mice at end stage compared to 20 days post-tumor cell transplantation (Supplementary Fig. S1). Identical results were observed in wild-type B6 and Cx3cr1GFP/WT;Ccr2RFP/WT mice. The total CD11b+ myeloid population (data not shown) and neutrophil numbers did not differ as a function of tumor development (Supplementary Fig. S1). Together, these data suggest that decreased number of blood monocytes is associated with their increased infiltration into glioblastoma.
BM-derived monocyte/macrophages predominate within the glioblastoma parenchyma, while microglia reside at the tumor periphery
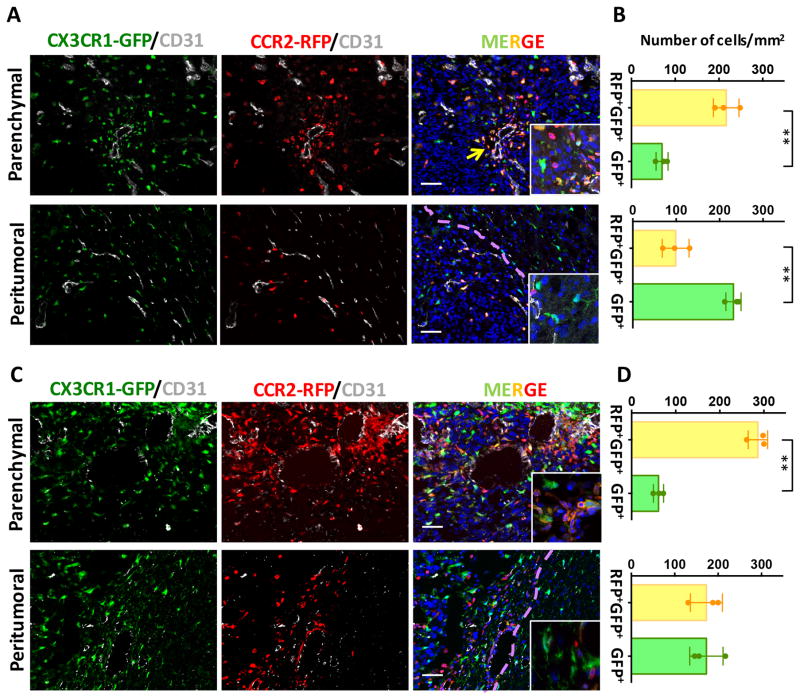
The glioblastoma microenvironment is compartmentalized into anatomically-distinct regions, referred to as “tumor niches”, which contain various populations of stromal cells as well as glioma stem cells, the most resistant population to radio- and chemotherapy (11). In this regard, it is important to distinguish differences in the spatial localization of BM-derived macrophages relative to resident microglia. To define the distribution of these TAM populations, tumor-bearing Cx3cr1GFP/WT;Ccr2RFP/WT mouse brains were immunostained with CD31 (endothelial cells) and RFP antibodies (due to the weakened signal of the RFP reporter following fixation) coupled with the endogenous GFP reporter (less affected by fixation). Most of the GFP+ cells inside of the tumor parenchyma were also positive for RFP expression. Interestingly, these GFP+RFP+ double-positive BM-derived cells formed clusters around blood vessels (arrow, Fig. 3A). In agreement with our flow cytometry data, single GFP+ (RFP−) cells were observed in much lower numbers than that of the double-positive cells (69.9±7.9 vs. 216.6±16.9, P<0.01) inside the tumor proper (Fig. 3B).
Figure 3. BM-derived monocyte/macrophages were predominant within the glioblastoma.
Brain tumor sections from Cx3cr1GFP/WT;Ccr2RFP/WT mice were visualized with GFP reporter (green), staining with anti-RFP (red) and anti-CD31 (gray) antibodies, and counterstained with DAPI (blue). Representative images demonstrate that the majority of GFP+RFP+ cells are localized in perivascular areas (yellow arrow) within the tumor, whereas in the peritumoral areas the majority myeloid cells are single GFP+ cells (A). (B) Quantification of GFP+RFP+ double positive or GFP+ single positive cells. (C) Immunofluorescence of GFP, RFP and CD31 in and around GL261-induced tumors. (D) Quantification of GFP+RFP+ double positive or GFP+ single positive cells in GL261-induced tumors. Scale bar represents 50 μm. **P<0.01 by t-test. N=3 for each group. Dotted lines mark tumor margin.
In contrast, the myeloid cells in the peritumoral regions were frequently only GFP+, in particular outside of the tumor proper (dotted line demarcates the tumor). GFP+RFP+ double-positive cells were mostly observed within the tumor parenchyma (Fig. 3A).
We also investigated the spatial distribution of these myeloid cells in another murine GBM model by implanting GL261 cells in Cx3cr1GFP/WT;Ccr2RFP/WT mice (26). The results were similar between the two models with respect to the TAM cellular distribution within the tumors (Fig. 3C, D), where the majority of TAMs within the tumor were derived from the bone marrow. In the peritumoral regions, the number of single GFP+ microglia in the GL261 model is similar to that observed in the PDGFB-driven glioma (Fig. 3D). In addition, the number of RFP+GFP+ macrophages was higher in GL261 model although this difference did not reach statistical significance (173±36.8 vs 98.7±30.9, P=0.056 by t-test).
In order to demonstrate that these double-positive cells infiltrated from the blood circulation, we generated tumors in Cx3cr1GFP/GFP;Ccr2RFP/WT mice. We have previously shown that tumors generated in this strain have significant infiltration of Ly6CHi inflammatory monocytes from the blood circulation (27). Histological examination of the tumors generated in Cx3cr1GFP/GFP;Ccr2RFP/WT mice revealed the presence of a GFP+RFP+ population in the perivascular areas of the tumors, while single GFP+ cells were found in the peritumoral locations (Supplementary Fig. S2).
Bone marrow-derived cells infiltrate early during GBM formation
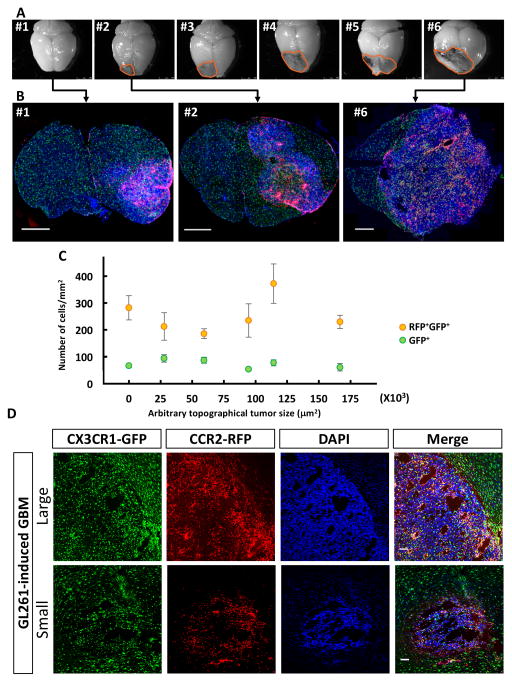
It is not known when BM-derived cells begin to infiltrate the tumor during GBM development. To gain insights into the temporal course of events, we transplanted primary tumor cells isolated from RCAS/Ntva donors into Cx3cr1GFP/WT;Ccr2RFP/WT mice, and euthanized the animals at 25 days post-injection when no tumor-induced neurological symptoms were apparent (median survival, 56 days; Supplementary Table S1). We were able to obtain tumors of various sizes in approximately half of the mice, reflecting the spectrum of stages of tumor progression (Fig. 4A). Even in the smallest tumors generated (which are invisible on the surface of the brain by gross examination; #1 in Fig. 4A), the CX3CR1-GFP/CCR2-RFP double positive cells inside the tumor mass are evident and numerous (#1 in Fig. 4B). This distribution was also observed in tumors of increasing size (Fig. 4B). When we quantified the number of these cells as a function of tumor size (Fig. 4C), we found no difference between small and large tumors, suggesting that BM-derived cells infiltrate the tumor mass early during GBM development, and maintain their presence with continued tumor growth. In naïve mice, RFP+ cells were not observed in the brain parenchyma, but only occasionally in the meninges (Supplementary Fig. S3).
Figure 4. Bone marrow-derived cells infiltrate GBM early during initial tumor formation stage.
(A) Gross examination of brains dissected from Cx3cr1GFP/WT;Ccr2RFP/WT mice euthanized 25 days after transplantation of tumors isolated form RCAS/Ntva donors. (B) Immunofluorescence of coronal brain sections containing tumors at different stages as manifested by small to large sizes. Scale bar = 1 mm. (C) Quantification of GFP+RFP+ double positive or GFP+ single positive cells inside the tumors. One-way ANOVA test does not detect statistical significance between tumors of various sizes. (D) Representative immunofluorescent sections of large (top) and small (bottom) tumors dissected from GL261 model. Scale bar = 100 μm.
Unlike PDGFB-driven glioblastoma, multiple tumors of various sizes/stages form in a single brain following the injection of GL261 glioma cells. Similarly, we found that irrespective of the tumor sizes, CX3CR1-GFP/CCR2-RFP double positive cells dominate the tumor, particularly in the perivascular spaces, whereas single GPF+ cells largely reside in the periphery of the tumor mass (Fig 4D).
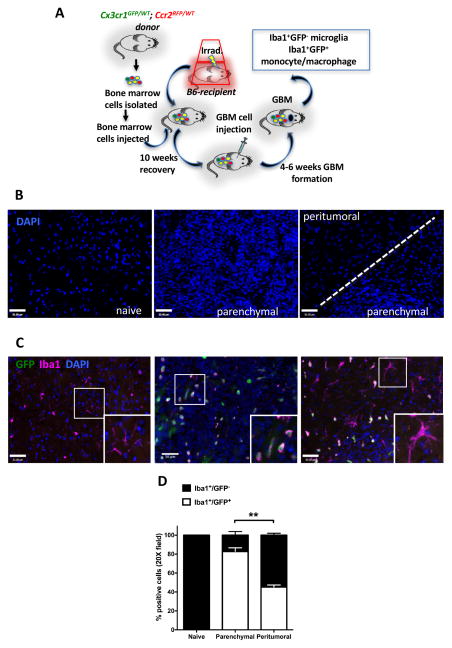
Bone marrow chimerism confirms that the majority TAMs are infiltrating monocyte/macrophages
To further confirm the relative ratios of BM-derived cells to resident microglia within the glioblastoma tumor mass, we generated BM chimeras where BM cells derived from Cx3cr1GFP/WT;Ccr2RFP/WT mice were transplanted into naïve wild-type B6 mice. In conjunction with Iba1 immunohistochemistry, these chimeric mice allow us to unambiguously distinguish donor-derived TAM as Iba1+GFP+, while host microglia as Iba1+GFP−. The procedure used to generate these BM-chimeric tumor-bearing mice is illustrated in Figure 5A. We used two irradiation regimens to ablate the host immune system, total body irradiation (TBI) and head protected irradiation (HPI), because both regiments have inherent strengths and weaknesses. Surprisingly, when we examined the reconstitution rate of the chimeras treated by HPI, we found that only ~50% of the leukocytes derived from the donor marrows (Supplementary Fig. S4). Because this low reconstitution rate yields a mixed population of GFP+ and GFP− monocytes, precluding the chimeras from being used for meaningful evaluation, cellular quantification of tissue sections obtained from HPI-treated chimeric mice was not performed.
Figure 5. Bone marrow chimerism confirms that the majority TAMs are infiltrating monocyte/macrophages.
(A) Schematic illustration of the generation of Cx3cr1GFP/WT;Ccr2RFP/WT bone marrow chimeras in Cx3cr1WT/WT;Ccr2 WT/WT (B6) mice followed by glioblastoma implantation. (B) Representative images of the DAPI staining for normal brain, tumor, and peritumoral regions of glioma-bearing chimeric mice. (C) Immunofluorescent staining for Iba1 (magenta) and endogenous GFP (green) in normal brain tissue or tumors generated in chimeric mice. (D) Quantification of Iba1+ and GFP+ cells within the tumor or in the peritumoral regions. Scale bar represents 50μm. ** P<0.01. N=5 each group.
We first examined the reconstitution efficiency of the BM following TBI treatment, and found that over 98% of the leukocytes are BM-derived cells at the time of tumor implantation (Supplementary Fig. S4). At death, tumors were dissected and analyzed by immunohistochemistry. Tumor parenchyma and peritumoral areas were identified by nuclei densities, as visualized by DAPI staining (Fig. 5B), and were used as regions-of-interest (ROIs) to quantify the numbers of Iba1+GFP+ (BM-derived) and Iba1+GFP− (resident microglia) (Fig. 5C). In non-tumor-bearing naïve chimeric mice, all of the Iba1+ cells were GFP negative, indicating their host origin. However, within the tumor parenchyma, the majority (82.8±3.8%) of the Iba1+ cells also expressed GFP, indicative of a BM origin (Fig. 5D), in agreement with flow cytometry analysis (Figs. 1, 2). In striking contrast, only 45.3±2.0% of the Iba1+ cells in the peritumoral region were BM-derived, while the majority were resident microglia (Iba1+GFP−) with ramified morphologies (Fig. 5C, D). Occasionally, Iba1−GFP+ cells were observed in the tumor (Fig. 5C), which likely represent rare CX3CR1-expressing T cells (28).
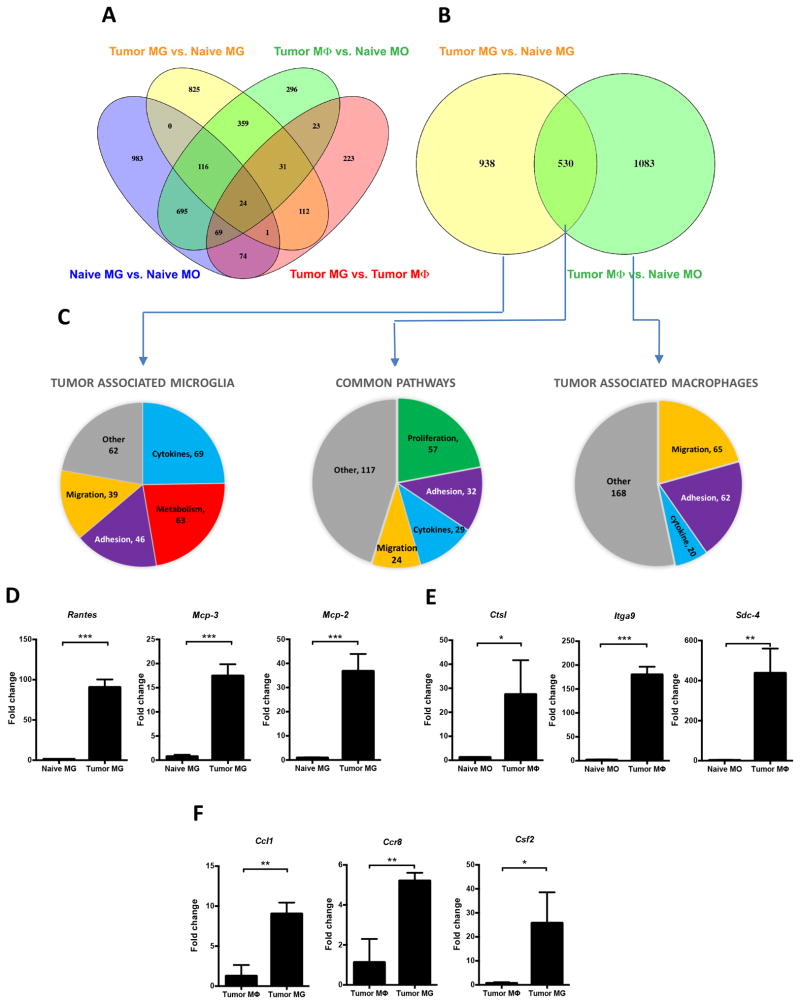
RNA sequencing analysis identifies unique gene expression profiles in infiltrating BM-derived cells versus microglia
Given their differential localizations, we sought to identify the gene expression profiles of these distinct populations. For these studies, BM-derived or resident myeloid cells were sorted from both naïve and tumor-bearing Cx3cr1GFP/WT;Ccr2RFP/WT mice according to red (Ccr2) and green (Cx3cr1) fluorescence in combination with CD45 and CD11b expression, and their transcriptome profiles analyzed by RNA-Seq. We found unique gene expression profiles from each of these four different populations (naïve microglia, naïve monocytes, tumor-associated microglia, and tumor-associated monocytes) by pair-wise comparisons (Fig. 6A). Not surprisingly, substantial differences were observed between naïve microglia and naïve monocytes (1964 up-regulated and 1144 down-regulated genes, Supplementary Fig. S5), consistent with their distinctive origins (13,29,30). When we specifically explored the differentially expressed genes (DEGs) between tumor-associated microglia and BM-derived tumor-associated macrophages, we found that 530 genes were up-regulated in both populations, but 938 and 1083 distinct genes were up-regulated in tumor-associated microglia or tumor-associated macrophages, respectively (Fig. 6B). Functional queries of these gene sets using Reactome FIViz Network analysis identified “cellular migration” as most enriched in tumor-associated macrophages; whereas genes associated with “pro-inflammatory cytokines” and “metabolism” were enriched in tumor-associated microglia (Fig. 6C). Pathways related to “cell proliferation” were significantly enriched in both populations (Fig. 6C). A complete list of these genes and pathways are included in Supplementary Tables S2–S4. We selected several deferentially regulated genes in tumor-associated microglia compared to naïve microglia (Fig. 6D), tumor-associated macrophages compared to naïve monocytes (Fig. 6E), and tumor-associated macrophages compared to tumor-associated microglia (Fig. 6F), and confirmed their RNA expression levels by quantitative RT-PCR.
Figure 6. RNA sequencing analysis identifies unique gene profiles between infiltrating macrophages and resident microglia.
(A) Venn diagram showing the number of differentially expressed genes (DEGs) revealed by pairwise comparison (Log2 fold change≥2, P<0.01) between naïve microglia (MG), tumor-associated microglia, naïve monocytes (MO), or tumor-associated macrophages (Mφ). (B) Venn diagram showing the number of DEGs (P≤0.01) between tumor-associated MG or Mφ. (C) Reactome FI Network analysis identified deferential biological functions specifically enriched in tumor-associated microglia, tumor-associated macrophage, or in both populations. Numbers indicate significantly upregulated genes contributing to that specific biological function. Quantitative RT-PCR confirms upregulation of selected molecules identified by RNAseq in either tumor-associated microglia compared to naïve microglia (D), tumor-associated macrophages compared to naïve monocytes (E), and tumor-associated macrophages compared to tumor-associated microglia (F). * P<0.05, ** P<0.01, *** P<0.001 by t-test. N=3 independent samples each group.
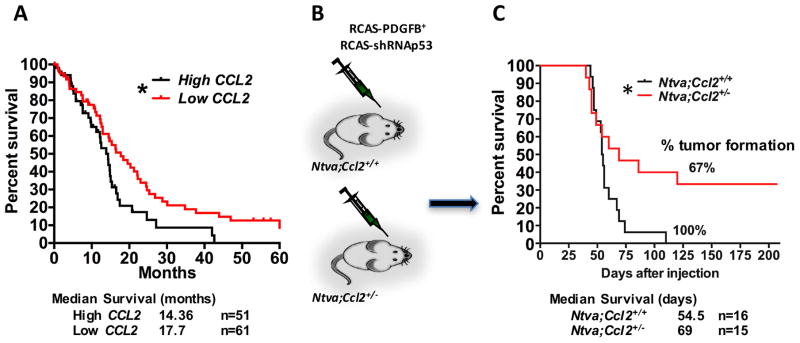
Heterozygous loss of Ccl2 prolongs the survival of glioblastoma-bearing mice
Previous studies had established that the CCL2/CCR2 axis is essential for monocyte migration into the inflamed CNS (31). In the setting of murine glioblastoma, we have shown that neoplastic cells in glioblastoma express high levels of CCL2 (a.k.a. MCP-1), which contributes to the directional infiltration of CCR2Hi inflammatory monocytes into the tumor (27). When we queried the human TCGA database for CCL2 expression and divided the patients into high and low CCL2 cohorts, we found that the patients with low tumoral CCL2 expression survived significantly longer than those with high tumoral CCL2 expression (Fig. 7A). Since CCR2Hi BM-derived cells account for over 85% of the TAM population in mouse glioblastoma (Fig. 2B), and supported by our observations in human GBM patients, we hypothesized that heterozygous loss of the Ccl2 gene would reduce CCR2Hi inflammatory monocyte infiltration and prolong the survival of glioblastoma-bearing mice. To test this hypothesis, Ntva mice were intercrossed with Ccl2−/− mice to generate Ntva;Ccl2+/− double transgenic mice for RCAS-mediated PDGFB-driven tumor induction (Fig. 7B). In agreement with a previous study (32), genetic Ccl2 reduction extended the survival time of tumor-bearing Ntva;Ccl2+/− mice relative to Ntva;Ccl2+/+ littermate controls (median survival days are 69 and 55 respectively, P<0.05, Fig. 7C). To investigate the mechanism underlying this prolonged survival conferred by Ccl2 reduction, we examined TAM accumulation, vasculature morphology and cell proliferation by histology (Supplementary Fig. S6A). There was a trend towards reduced numbers of Iba1+ macrophages in the tumors of Ntva;Ccl2+/− mice compared to Ntva;Ccl2+/+ mice (P=0.136; Supplementary Fig. S6B), in line with the previous study showing that myeloid derived suppressor cells were significantly reduced in Ccl2−/− mice in SB transposon-mediated de novo murine glioma model (32). However, there was no difference observed in total vessel area or average vessel size between these two genotypes (Supplementary Fig. S6B). We also examined the number of proliferating cells by phosphor-histone 3 and Ki67 staining, but did not observe any difference in these terminal stage tumors (Supplementary Figs. S7 & S8). Additionally, we determined whether there is difference in cell death between Ccl2+/− versus WT mice by TUNEL staining. We found no difference in cell death in the tumor proper (supplementary Fig. S9). To differentiate the source of CCL2 of being produced either by cancer cells or the stromal cells, we transplanted WT PDGFB-driven primary murine tumors into Ccl2+/+ (WT) or Ccl2+/− mice. Kaplan-Meier analyses indicate no survival advantage when Ccl2 gene expression is diminished in stromal cells (Supplementary Fig. S10), suggesting that the cancer cells are the major producers of Ccl2. We have previously demonstrated that increased levels of CCL2 expression is associated with increased infiltration of monocytes and correlates with shorter survival of tumor bearing-mice (27). Together, these data indicate that decreased level of CCL2 in neoplastic cells prolongs survival time of tumor-bearing mice.
Figure 7. Heterogeneous loss of CCL2, the CCR2 ligand, extends survival of glioblastoma bearing mice.
(A) Survival curve of GBM patients stratified according to their CCL2 expression queried from the TCGA databank. (B) Diagram illustrating the generation of tumors in Ccl2+/− mice using RCAS/Ntva system. (C) Survival curve of tumor-bearing Ntva+ mice. N is denoted in the figures. * P<0.05 by Mantel-Cox test.
Discussion
Macrophages account for 30–50% of the tumor mass in glioblastoma (2,8). However, whether these TAMs arise from peripheral monocytes or CNS resident microglia remains unclear. Here, by taking advantage of Cx3cr1GFP/WT;Ccr2RFP/WT double transgenic reporter mice, we demonstrated that infiltrating macrophages constitute ~85% of the total TAM population, with resident microglia accounting for the remaining ~15% of TAMs. Bone marrow-derived infiltrating cells preferentially localize to perivascular areas within the tumor proper, whereas the smaller population of CCR2− resident microglia are mainly localized to the peritumoral regions. By using RNA-sequencing analyses, we discovered differential gene expression patterns unique to infiltrating and resident cells, suggesting unique functions for each TAM population related to their pathobiological attributes in tumor development. Given the abundance of CCR2Hi monocytes within the tumor, we demonstrate that loss of single copy of Ccl2 from both tumor and stromal cells prolonged survival of tumor-bearing mice.
The importance of developing methods to distinguish microglia from hematopoietic myeloid cells becomes increasingly clear, with an ever-increasing number of studies revealing differential roles of these monocytic cells in various CNS diseases (33,34). Differential functions of microglial cells can be partially attributed to their unique origin from yolk sac progenitors (29) and the fact that they can maintain themselves by virtue of longevity and self-renewal (29,35,36). In this regard, resident microglia represent a distinct population of myeloid cells. Monocytes, on the other hand, originate from hematopoietic stem cells, with series of progenitors differentiating within the bone marrow and ultimately released to the blood circulation to colonize certain peripheral organs under both normal and inflammatory conditions (37).
In the context of glioblastoma, approaches to distinguishing microglia from invading monocytes have traditionally relied on the use of CD45 antibodies to separate resident microglia (CD45Lo) from macrophages of hematopoietic origin (CD45Hi) by FACS analysis (8). Analysis of human glioma samples has revealed that the CD45Hi population is greater than the CD45Lo population, suggesting that gliomas contain more recruited monocytes than microglia (10). This concept was recently challenged by a study using irradiation mouse chimeras, which demonstrated that the majority of TAMs are intrinsic microglia, and that these microglial cells upregulate their CD45 expression to constitute a significant proportion of the CD45Hi population in gliomas (9). The discrepancy between our results and those of Muller et al. may be explained by the differences in genetic compositions of the animal models used, as was initially observed and reported by Badie and Schartner (8). It is also likely that the presence of microglia in TAMs was overestimated by Muller et al., as they analyzed tumor-bearing hemispheres rather than the microdissected tumor proper. In this respect, tumor-bearing hemispheres contain large numbers of microglia that reside outside of the tumor mass (Figs. 3 to 5). Although both microglia and infiltrating monocytes can upregulate their CD45 and CD11b expressions at the height of tumor development, there remains a clear demarcation in CD45 and CD11b intensity between these two populations, as visualized by flow cytometry (Fig. 2B).
Similarly, another study employed single staining with antibodies against either CX3CR1 or CCR2 to conclude that the majority of TAMs were monocyte-derived (CX3CR1−CCR2+) macrophages (38). However, the prominent CX3CR1 and CCR2 double-positive population was not considered. It should be noted that CX3CR1 is expressed by both blood monocytes and microglia, and can be increased during monocyte differentiation into macrophages. These findings argue against the use of CX3CR1 alone as a microglia-specific marker either in the naïve mouse (12,13) or in the context of glioma (27). In addition, no widely available and well-validated antibodies to CX3CR1 or CCR2 exist to discriminate microglia, monocytes and monocyte-derived macrophages. Approaches, like those taken to identify Tmem119 (transmembrane protein 119), may reveal potential antibodies that selectively distinguish microglia from BM-derived monocytes (39). Together, the discrepant results obtained from the use of bone-marrow chimeras and standardly-employed cell surface antibodies highlight the urgent need to re-evaluate these published conclusions in myeloid cell distribution in glioblastoma and to perform lineage-tracing experiments using reporter mice that accurately distinguish microglia from monocytes/macrophages in glioblastoma.
Using Cx3cr1GFP/WT;Ccr2RFP/WT double knock-in mice, we demonstrated that during tumor development, BM-derived infiltrating cells dominate the TAM landscape (Figs. 2, 4). The fact that they preferentially locate to perivascular regions suggests that they extravasate through blood vessels within the tumor. In contrast, resident microglia do not migrate easily through brain tissue (40) to the interior of the tumor and therefore are largely observed in the peritumoral spaces. It is also interesting to note that the intensity of both CX3CR1-GFP and CCR2-RFP varies considerably among the TAMs (Fig. 2). It is well documented that CCR2Hi cells are monocytes newly arrived at the site of inflammation (41). Once homed to inflamed tissues, these cells gradually downregulate their CCR2 expression as they differentiate into macrophages (41). Here, we demonstrated that the TAMs exhibit a broad range of CCR2-RFP intensities, indicating a continuous transformation of these cells from infiltrating monocytes to mature macrophages. Remarkably, the intensity of CX3CR1-GFP in these cells also varied, and inversely correlated to CCR2-RFP expression (Fig. 2). This dynamic transition of the surface markers (Supplementary Video S1) indicates that BM-derived macrophages are highly plastic and that these cells evolve to maturation in situ following extravasation (24), although our data cannot entirely exclude the possibility that the small CCR2−CX3CR1Hi population infiltrated directly from the blood circulation. Additional linage tracing studies will be necessary to establish the ontology of this CCR2−CX3CR1Hi cells in glioblastoma.
Mature macrophages derived in situ from infiltrating monocytes play important roles in various inflammatory diseases and therefore stand as suitable targets for therapeutic interventions (42). Our gene profiling experiments revealed that pathways involved in cell migration were significantly and specifically enriched in BM-derived TAMs (Fig. 6). CCL2, a member of the MCP (monocyte chemoattractant protein) chemokine family, plays an important role in mediating monocyte migration through its receptor CCR2. It is interesting to note that low CCL2 expression in human GBM is associated with significantly prolonged patient survival (Fig. 7). Together, these findings raise the question as to whether reducing monocyte infiltration by targeting CCL2-CCR2 axis is a viable option for treating murine glioblastoma. To address this question, we showed that genetically interrupting the CCL2-CCR2 axis prolonged the survival of glioblastoma-bearing mice, in agreement with previous pharmacological studies (43,44). However, in contrast to the promising preclinical studies, neutralizing monoclonal antibodies against CCL2 administered to patients with metastatic, solid tumors did not produce favorable outcomes. Meta-analysis of the data from these clinical trials indicated that initial CCL2 inhibition may have unexpectedly caused subsequent increases in circulating CCL2 levels, possibly due to a compensatory feedback loop (45). Lack of therapeutic benefits from inhibiting this axis indicates that CCL2-CCR2 interaction represents a complex signaling network that is not well understood. Since the reduction of CCL2 did not result in statistically significant decrease of TAM infiltration, it also implies that other MCP family chemokines likely function in synergy with CCL2 to recruit monocytes into glioblastoma. In this regard, MCP-3 (CCL-7) had been shown to play a critical role in recruiting BM-derived monocytes to sites of inflammation (46).
While previous studies have revealed that BM-derived cells penetrate experimental murine GBM tumors, to our knowledge, this is the first report to provide conclusive evidence demonstrating that tumor-infiltrating BM cells are recruited early during GBM development in at least two independent murine models (Fig. 4). CCR2-RFP+ cells are found in clusters around blood vessels and spread across the entire tumor mass at all stages of tumorigenesis. This novel observation implies that BM-derived TAMs interact with neoplastic cells in the perivascular niche to promote tumor growth from the initial stage of tumor development. In light of this notion, it is plausible that the prolonged survival of the Ntva;Ccl2+/− mice reflects delayed tumor initiation as a result of diminished initial TAM infiltration due to Ccl2 reduction in tumor cells (Fig. 7).
In summary, we establish that the majority of glioblastoma-associated macrophages are BM-derived infiltrating myeloid cells. These cells are recruited early during tumor formation, preferentially localize to the perivascular niche, and contribute to tumor development. Reducing their infiltration by genetic Ccl2 (MCP-1) modulation significantly prolongs the survival of glioblastoma-bearing mice. Future studies investigating members of MCP family chemokines have potential to elucidate the contributions of these stromal cells to tumor maintenance and may one day yield effective stroma-directed therapies.
Supplementary Material
Acknowledgments
Financial Support: This work was supported by NCI grants U01CA160882 (to D. Hambardzumyan and D.H. Gutmann) and R01CA195692 (to D.H. Gutmann).
The authors are grateful to Dr. Christopher Nelson for editorial assistance. We thank the Flow Cytometry Cores at the CCF and the CHOA for technical services. This research project was supported in part by the Emory University Integrated Cellular Imaging Microscopy Core of the Emory+Children’s Pediatric Research Center. We acknowledge the excellent imaging assistance from Dr. Neil Anthony. We thank Ms. Jennifer Powers for cell sorting. We are grateful to the Biostatistics and Bioinformatics services at Washington University and Emory University. The authors thank Dr. Richard Ransohoff for providing the Cx3cr1GFP/WT;Ccr2RFP/WT mice and Dr. Helmut Kettenmann for providing GL261 tumor samples.
Footnotes
The authors declare no Conflict of Interests.
References
- 1.Becher OJ, Hambardzumyan D, Fomchenko EI, Momota H, Mainwaring L, Bleau AM, et al. Gli activity correlates with tumor grade in platelet-derived growth factor-induced gliomas. Cancer Res. 2008;68:2241–9. doi: 10.1158/0008-5472.CAN-07-6350. [DOI] [PubMed] [Google Scholar]
- 2.Charles NA, Holland EC, Gilbertson R, Glass R, Kettenmann H. The brain tumor microenvironment. Glia. 2012;60:502–14. doi: 10.1002/glia.21264. [DOI] [PubMed] [Google Scholar]
- 3.Simmons GW, Pong WW, Emnett RJ, White CR, Gianino SM, Rodriguez FJ, et al. Neurofibromatosis-1 heterozygosity increases microglia in a spatially and temporally restricted pattern relevant to mouse optic glioma formation and growth. J Neuropathol Exp Neurol. 2011;70:51–62. doi: 10.1097/NEN.0b013e3182032d37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Solga AC, Pong WW, Kim KY, Cimino PJ, Toonen JA, Walker J, et al. RNA Sequencing of Tumor-Associated Microglia Reveals Ccl5 as a Stromal Chemokine Critical for Neurofibromatosis-1 Glioma Growth. Neoplasia. 2015;17:776–88. doi: 10.1016/j.neo.2015.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hambardzumyan D, Gutmann DH, Kettenmann H. The role of microglia and macrophages in glioma maintenance and progression. Nat Neurosci. 2016;19:20–7. doi: 10.1038/nn.4185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pyonteck SM, Akkari L, Schuhmacher AJ, Bowman RL, Sevenich L, Quail DF, et al. CSF-1R inhibition alters macrophage polarization and blocks glioma progression. Nat Med. 2013;19:1264–72. doi: 10.1038/nm.3337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Butowski N, Colman H, De Groot JF, Omuro AM, Nayak L, Wen PY, et al. Orally administered colony stimulating factor 1 receptor inhibitor PLX3397 in recurrent glioblastoma: an Ivy Foundation Early Phase Clinical Trials Consortium phase II study. Neuro Oncol. 2016;18:557–64. doi: 10.1093/neuonc/nov245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Badie B, Schartner JM. Flow cytometric characterization of tumor-associated macrophages in experimental gliomas. Neurosurgery. 2000;46:957–61. doi: 10.1097/00006123-200004000-00035. [DOI] [PubMed] [Google Scholar]
- 9.Muller A, Brandenburg S, Turkowski K, Muller S, Vajkoczy P. Resident microglia, and not peripheral macrophages, are the main source of brain tumor mononuclear cells. Int J Cancer. 2015;137:278–88. doi: 10.1002/ijc.29379. [DOI] [PubMed] [Google Scholar]
- 10.Parney IF, Waldron JS, Parsa AT. Flow cytometry and in vitro analysis of human glioma-associated macrophages. Laboratory investigation. J Neurosurg. 2009;110:572–82. doi: 10.3171/2008.7.JNS08475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hambardzumyan D, Bergers G. Glioblastoma: Defining Tumor Niches. Trends in Cancer. 2015;1:252–65. doi: 10.1016/j.trecan.2015.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Geissmann F, Jung S, Littman DR. Blood monocytes consist of two principal subsets with distinct migratory properties. Immunity. 2003;19:71–82. doi: 10.1016/s1074-7613(03)00174-2. [DOI] [PubMed] [Google Scholar]
- 13.Yona S, Kim KW, Wolf Y, Mildner A, Varol D, Breker M, et al. Fate mapping reveals origins and dynamics of monocytes and tissue macrophages under homeostasis. Immunity. 2013;38:79–91. doi: 10.1016/j.immuni.2012.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hambardzumyan D, Amankulor NM, Helmy KY, Becher OJ, Holland EC. Modeling Adult Gliomas Using RCAS/t-va Technology. Transl Oncol. 2009;2:89–95. doi: 10.1593/tlo.09100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jung S, Aliberti J, Graemmel P, Sunshine MJ, Kreutzberg GW, Sher A, et al. Analysis of fractalkine receptor CX(3)CR1 function by targeted deletion and green fluorescent protein reporter gene insertion. Mol Cell Biol. 2000;20:4106–14. doi: 10.1128/mcb.20.11.4106-4114.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cook DN, Chen SC, Sullivan LM, Manfra DJ, Wiekowski MT, Prosser DM, et al. Generation and analysis of mice lacking the chemokine fractalkine. Mol Cell Biol. 2001;21:3159–65. doi: 10.1128/MCB.21.9.3159-3165.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Keith BJ, Franklin GP. The Mouse Brain in Steriotaxic Coordinates. 1997. [Google Scholar]
- 18.Lu B, Rutledge BJ, Gu L, Fiorillo J, Lukacs NW, Kunkel SL, et al. Abnormalities in monocyte recruitment and cytokine expression in monocyte chemoattractant protein 1-deficient mice. J Exp Med. 1998;187:601–8. doi: 10.1084/jem.187.4.601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen Z, Jalabi W, Hu W, Park HJ, Gale JT, Kidd GJ, et al. Microglial displacement of inhibitory synapses provides neuroprotection in the adult brain. Nat Commun. 2014;5:4486. doi: 10.1038/ncomms5486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wu G, Feng X, Stein L. A human functional protein interaction network and its application to cancer data analysis. Genome Biol. 2010;11:R53. doi: 10.1186/gb-2010-11-5-r53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Smoot ME, Ono K, Ruscheinski J, Wang PL, Ideker T. Cytoscape 2.8: new features for data integration and network visualization. Bioinformatics. 2011;27:431–2. doi: 10.1093/bioinformatics/btq675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mildner A, Schmidt H, Nitsche M, Merkler D, Hanisch UK, Mack M, et al. Microglia in the adult brain arise from Ly-6ChiCCR2+ monocytes only under defined host conditions. Nat Neurosci. 2007;10:1544–53. doi: 10.1038/nn2015. [DOI] [PubMed] [Google Scholar]
- 23.Carson MJ, Bilousova TV, Puntambekar SS, Melchior B, Doose JM, Ethell IM. A rose by any other name? The potential consequences of microglial heterogeneity during CNS health and disease. Neurotherapeutics. 2007;4:571–9. doi: 10.1016/j.nurt.2007.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dal-Secco D, Wang J, Zeng Z, Kolaczkowska E, Wong CH, Petri B, et al. A dynamic spectrum of monocytes arising from the in situ reprogramming of CCR2+ monocytes at a site of sterile injury. J Exp Med. 2015;212:447–56. doi: 10.1084/jem.20141539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mizutani M, Pino PA, Saederup N, Charo IF, Ransohoff RM, Cardona AE. The fractalkine receptor but not CCR2 is present on microglia from embryonic development throughout adulthood. J Immunol. 2012;188:29–36. doi: 10.4049/jimmunol.1100421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ausman JI, Shapiro WR, Rall DP. Studies on the chemotherapy of experimental brain tumors: development of an experimental model. Cancer Res. 1970;30:2394–400. [PubMed] [Google Scholar]
- 27.Feng X, Szulzewsky F, Yerevanian A, Chen Z, Heinzmann D, Rasmussen RD, et al. Loss of CX3CR1 increases accumulation of inflammatory monocytes and promotes gliomagenesis. Oncotarget. 2015;6:15077–94. doi: 10.18632/oncotarget.3730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cardona AE, Pioro EP, Sasse ME, Kostenko V, Cardona SM, Dijkstra IM, et al. Control of microglial neurotoxicity by the fractalkine receptor. Nat Neurosci. 2006;9:917–24. doi: 10.1038/nn1715. [DOI] [PubMed] [Google Scholar]
- 29.Ginhoux F, Greter M, Leboeuf M, Nandi S, See P, Gokhan S, et al. Fate mapping analysis reveals that adult microglia derive from primitive macrophages. Science. 2010;330:841–5. doi: 10.1126/science.1194637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Butovsky O, Jedrychowski MP, Moore CS, Cialic R, Lanser AJ, Gabriely G, et al. Identification of a unique TGF-beta-dependent molecular and functional signature in microglia. Nat Neurosci. 2014;17:131–43. doi: 10.1038/nn.3599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Boring L, Gosling J, Chensue SW, Kunkel SL, Farese RV, Jr, Broxmeyer HE, et al. Impaired monocyte migration and reduced type 1 (Th1) cytokine responses in C-C chemokine receptor 2 knockout mice. J Clin Invest. 1997;100:2552–61. doi: 10.1172/JCI119798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fujita M, Kohanbash G, Fellows-Mayle W, Hamilton RL, Komohara Y, Decker SA, et al. COX-2 blockade suppresses gliomagenesis by inhibiting myeloid-derived suppressor cells. Cancer Res. 2011;71:2664–74. doi: 10.1158/0008-5472.CAN-10-3055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ajami B, Bennett JL, Krieger C, McNagny KM, Rossi FM. Infiltrating monocytes trigger EAE progression, but do not contribute to the resident microglia pool. Nat Neurosci. 2011;14:1142–9. doi: 10.1038/nn.2887. [DOI] [PubMed] [Google Scholar]
- 34.Shemer A, Jung S. Differential roles of resident microglia and infiltrating monocytes in murine CNS autoimmunity. Semin Immunopathol. 2015;37:613–23. doi: 10.1007/s00281-015-0519-z. [DOI] [PubMed] [Google Scholar]
- 35.Ajami B, Bennett JL, Krieger C, Tetzlaff W, Rossi FM. Local self-renewal can sustain CNS microglia maintenance and function throughout adult life. Nat Neurosci. 2007;10:1538–43. doi: 10.1038/nn2014. [DOI] [PubMed] [Google Scholar]
- 36.Elmore MR, Najafi AR, Koike MA, Dagher NN, Spangenberg EE, Rice RA, et al. Colony-stimulating factor 1 receptor signaling is necessary for microglia viability, unmasking a microglia progenitor cell in the adult brain. Neuron. 2014;82:380–97. doi: 10.1016/j.neuron.2014.02.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shi C, Pamer EG. Monocyte recruitment during infection and inflammation. Nat Rev Immunol. 2011;11:762–74. doi: 10.1038/nri3070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhou W, Ke SQ, Huang Z, Flavahan W, Fang X, Paul J, et al. Periostin secreted by glioblastoma stem cells recruits M2 tumour-associated macrophages and promotes malignant growth. Nat Cell Biol. 2015 doi: 10.1038/ncb3090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bennett ML, Bennett FC, Liddelow SA, Ajami B, Zamanian JL, Fernhoff NB, et al. New tools for studying microglia in the mouse and human CNS. Proc Natl Acad Sci U S A. 2016;113:E1738–46. doi: 10.1073/pnas.1525528113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Davalos D, Grutzendler J, Yang G, Kim JV, Zuo Y, Jung S, et al. ATP mediates rapid microglial response to local brain injury in vivo. Nat Neurosci. 2005;8:752–8. doi: 10.1038/nn1472. [DOI] [PubMed] [Google Scholar]
- 41.Gordon S, Taylor PR. Monocyte and macrophage heterogeneity. Nat Rev Immunol. 2005;5:953–64. doi: 10.1038/nri1733. [DOI] [PubMed] [Google Scholar]
- 42.Getts DR, Terry RL, Getts MT, Deffrasnes C, Muller M, van Vreden C, et al. Therapeutic inflammatory monocyte modulation using immune-modifying microparticles. Sci Transl Med. 2014;6:219ra7. doi: 10.1126/scitranslmed.3007563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhu X, Fujita M, Snyder LA, Okada H. Systemic delivery of neutralizing antibody targeting CCL2 for glioma therapy. J Neurooncol. 2011;104:83–92. doi: 10.1007/s11060-010-0473-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chang AL, Miska J, Wainwright DA, Dey M, Rivetta CV, Yu D, et al. CCL2 produced by the glioma microenvironment is essential for the recruitment of regulatory T cells and myeloid-derived suppressor cells. Cancer Res. 2016;76:5671–82. doi: 10.1158/0008-5472.CAN-16-0144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lim SY, Yuzhalin AE, Gordon-Weeks AN, Muschel RJ. Targeting the CCL2-CCR2 signaling axis in cancer metastasis. Oncotarget. 2016;7:28697–710. doi: 10.18632/oncotarget.7376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tsou CL, Peters W, Si Y, Slaymaker S, Aslanian AM, Weisberg SP, et al. Critical roles for CCR2 and MCP-3 in monocyte mobilization from bone marrow and recruitment to inflammatory sites. J Clin Invest. 2007;117:902–9. doi: 10.1172/JCI29919. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.