Abstract
The ring-forming Hsp104 ATPase cooperates with Hsp70 and Hsp40 molecular chaperones to rescue stress-damaged proteins from both amorphous and amyloid-forming aggregates. The ability to do so relies upon pore loops present in the first ATP-binding domain (AAA-1; loop-1 and loop-2 ) and in the second ATP-binding domain (AAA-2; loop-3) of Hsp104, which face the protein translocating channel and couple ATP-driven changes in pore loop conformation to substrate translocation. A hallmark of loop-1 and loop-3 is an invariable and mutational sensitive aromatic amino acid (Tyr257 and Tyr662) involved in substrate binding. However, the role of conserved aliphatic residues (Lys256, Lys258, and Val663) flanking the pore loop tyrosines, and the function of loop-2 in protein disaggregation has not been investigated. Here we present the crystal structure of an N-terminal fragment of Saccharomyces cerevisiae Hsp104 exhibiting molecular interactions involving both AAA-1 pore loops, which resemble contacts with bound substrate. Corroborated by biochemical experiments and functional studies in yeast, we show that aliphatic residues flanking Tyr257 and Tyr662 are equally important for substrate interaction, and abolish Hsp104 function when mutated to glycine. Unexpectedly, we find that loop-2 is sensitive to aspartate substitutions that impair Hsp104 function and abolish protein disaggregation when loop-2 is replaced by four aspartate residues. Our observations suggest that Hsp104 pore loops have non-overlapping functions in protein disaggregation and together coordinate substrate binding, unfolding, and translocation through the Hsp104 hexamer.
Keywords: AAA proteins, crystallography, heat shock proteins, molecular chaperones, site-directed mutagenesis, Saccharomyces cerevisiae
Introduction
Members of the ring-forming Hsp104/ClpB family of ATP-driven molecular chaperones are the principle protein disaggregases in fungi (Hsp104), plants (Hsp101), and eubacteria (ClpB) [1–3]. Interestingly, Hsp104 homologs are not found in animal cells [4], making members of this family a potential antimicrobial drug target. To rescue stress-damaged proteins from an aggregated state, Hsp104/ClpB disaggregases must cooperate with the cognate Hsp70/DnaK system, consisting of Hsp70 and Hsp40 in yeast and DnaK–DnaJ–GrpE in eubacteria, to form a potent bi-chaperone system. However, unlike the bacterial bi-chaperone system, a nucleotide-exchange factor such as yeast Sse1 [5] is not required for Hsp104-dependent protein disaggregation in vitro [6], but was shown to enhance its potency in yeast [7].
At the molecular level, Saccharomyces cerevisiae Hsp104 forms a homo-hexamer that is stabilized by adenine nucleotides [8–10]. Each Hsp104 monomer features two ATP-binding domains, termed AAA-1 and AAA-2, in addition to an N-terminal (N) domain and a coiled-coil motif that mediates the physical interaction with Hsp70 [11,12]. It is now widely accepted that Hsp104 facilitates the unfolding of aggregated proteins and the threading of substrate through the protein translocation channel analogous to ATP-dependent Clp proteases [13,14]. However, it remains unknown whether substrate unfolding and threading represent concerted or mechanistically distinct events. Amongst the Hsp104 domains, the functional role of the N domain is perhaps most perplexing. Although dispensable for protein disaggregation in vitro and in vivo [15–17], the N domain is essential for yeast prion dissolution [18] and curing by Hsp104 overexpression [15]. Consequently, how Hsp104 recognizes substrates and recovers stress-damaged proteins from protein aggregates has been a matter of considerable debate. Southworth and colleagues recently reported the high-resolution cryoEM structure of yeast Hsp104 bound to casein [19], an unstructured phosphoprotein that, unlike native substrates, is processed in a nucleotide-independent manner [20]. The structure confirmed a role for the conserved loop-1 and loop-3 tyrosines contacting the unfolded polypeptide, which is corroborated by an analogous cryoEM study of the bacterial homolog, ClpB [21]. However, the role of conserved aliphatic pore loop residues and the importance of loop-2 in protein disaggregation has not been investigated.
Here we present the X-ray structure of a S. cerevisiae Hsp104 fragment (Hsp1041-360) determined from a new crystal form featuring three independent copies of Hsp1041-360 in the crystallographic asymmetric unit. As each monomer has a different crystal-packing environment, consistent stereochemical features are inherent to the structure and independent of the crystal lattice. We find that both loop-1 and loop-2 form molecular interactions that resemble contacts with bound substrate. Although the structure of the physiological ring assembly was not determined, we show that the aliphatic side chains of Lys256 and Lys258 (loop-1) and Val663 (loop-3) flanking the conserved pore loop tyrosines are also involved in substrate interaction, and abolish Hsp104 function when mutated to glycine. Furthermore, our structure also suggests a previously unknown role for loop-2 in Hsp104 function. Although loop-2 shows only a small defect when all four residues are mutated to glycine or alanine, we find that loop-2 is sensitive to substitutions with aspartate. Notably, substituting loop-2 with four aspartates abolishes protein disaggregation in vitro and severely impairs thermotolerance development in vivo. Taken together, our observations suggest that loop-1 and loop-2 have distinct mechanical functions, and cooperate with loop-3 to facilitate the recovery of stress-damaged protein from aggregates.
Experimental
Protein expression and purification
S. cerevisiae Hsp104Y257A and Hsp104Y662A were generated by QuikChange site-directed mutagenesis (Agilent). All other Hsp104 pore-loop mutants were generated by overlap extension PCR followed by cassette mutagenesis. Hsp104 and its mutants were cloned into the pProEX-HTb vector (Invitrogen), which adds a tobacco etch virus protease cleavable N-terminal His6-tag, and were overexpressed in Escherichia coli BL21-CodonPlus (DE3)-RIL cells (Agilent) by isopropyl β-d-thiogalactopyranoside induction. Proteins were purified from cleared lysates by affinity chromatography on nickel-nitrilotriacetic acid (Ni-NTA) agarose column (Qiagen) and eluted in 25 mM Tris/HCl pH 7.5, 300 mM NaCl, 5% glycerol, and 5 mM β-mercaptoethanol containing 300 mM imidazole, or in TBS using a 20–800 mM imidazole gradient (Hsp1041-360). The N-terminal His6-tag was cleaved off and removed by reapplying the protein to a Ni-NTA agarose column. Hsp1041-360 was further purified by negative binding to an anion-exchange column followed by binding to a Mono-S column (GE Healthcare). His6-Ydj1 and His6-Hsp70 were purified as described [22].
Size-exclusion chromatography
Full-length Hsp104 and Hsp104 mutant proteins were further purified by size-exclusion chromatography on a Superdex 200 10/300 GL column (GE Healthcare) pre-equilibrated in 25 mM Tris/HCl pH 7.5, 150 mM NaCl, 5% glycerol, and 1 mM DTT. Size-exclusion chromatography was also used to determine the oligomeric state of Hsp104 and Hsp104 mutants. Hexamers were isolated and used for subsequent ATPase activity measurements and coupled chaperone assays.
Crystal structure determination
Crystals of Hsp1041-360 were grown by the hanging drop vapor diffusion method at 4°C by mixing 2 µl of protein solution (20 mg/ml) with 2 µl of reservoir solution containing 25% PEG 4000 (w/v), 50 mM Tris/HCl pH 8.5, and 20 mM ammonium citrate. Data were collected and processed using the HKL2000 software package [23] (Supplementary Table S1). The crystal structure of Hsp1041-360 was determined by molecular replacement using MOLREP [24] with Protein Data Bank (PDB) accession 6AMN as search model [25]. Two molecules of Hsp1041-360 were found. The calculated map revealed the location of a third molecule in the asymmetric unit, and the two domains of the third molecule were manually placed. After rigid body refinement, the N and AAA-1large domains were connected in each molecule. Cycles of rebuilding and refinement were carried out using COOT [26] and PHENIX [27], respectively. Atomic coordinates and structure factors have been deposited in the PDB with the accession number 5WBW. Protein domain motions were analyzed using DynDom [28].
ATPase activity assay
Hsp104 and variants (0.5 µM monomer) were incubated with 2 mM ATP at 22°C for 15 min. The amount of released inorganic phosphate was measured using the Malachite Green assay [29].
Coupled chaperone assay
Firefly luciferase (FFL; 10 µM) was denatured in 7 M urea in refolding buffer (25 mM HEPES-KOH pH 7.5, 150 mM potassium acetate, 10 mM magnesium acetate, and 10 mM DTT) for 30 min at 22°C, then diluted 125-fold in refolding buffer containing the bi-chaperone system (1 μM Hsp104, 1 μM hHsp70, 1 μM Ydj1), 5 mM ATP, and an ATP-regenerating system consisting of 25 mM phosphoenolpyruvate and 2 μM pyruvate kinase. β-galactosidase (β-gal; 0.4 µM) was heat aggregated in refolding buffer for 40 min at 59°C and mixed (0.2 µM final concentration) with the bi-chaperone system (1 µM each) together with 4 mM ATP, 20 mM phosphoenolpyruvate, and 2 μM pyruvate kinase. Recovered enzymatic activities were measured after 120 min (FFL) and 360 min (β-gal) as described [30].
Thermotolerance assay
Hsp104 loop-1 and loop-2 mutants were generated by excising an EcoRI-BglII fragment featuring the desired mutation and swapping it into pYS104 containing S. cerevisiae Hsp104 wild-type under control of the Hsp104 promoter. Hsp104 loop-3 mutants were generated by cassette mutagenesis. Plasmids expressing wild-type and mutant Hsp104 were transformed into S. cerevisiae OT46 (Δhsp104) and screened on synthetic defined growth medium without uracil (SD-Ura) plates [31]. Yeast cells were diluted to 0.1 D600 from overnight cultures, grown for 2.5 h at 25°C in yeast extract peptone dextrose (YPD) medium and divided into two sets. One set was treated by heat-shock at 50°C for 15 min (basal thermotolerance), while the other set was incubated at 37°C for 30 min to induce heat-shock protein synthesis prior to heat-shock (induced thermotolerance). Cells were heat-shocked and immediately chilled on ice. Five microliters of ten-fold serial dilutions were dropped on to YPD plates. Viability was scored after 2 days of incubation at 30°C.
Subunit mixing experiments
Hsp104 and mutant proteins were mixed at different ratios to achieve the indicated subunit composition in the hexamer, while keeping the total protein concentration at 10 µM. Mixtures were incubated at 22°C for 20 min to allow for subunit exchange. For urea-denatured FFL, Hsp104 hetero-hexamers were diluted ten-fold with refolding buffer containing 1 µM Hsp70 and Hsp40. For heat-aggregated β-gal, 0.3 µM of the bi-chaperone system with Hsp104 hetero-hexamers was used. As control, Hsp104, Hsp70, and Hsp40 chaperones were diluted with refolding buffer keeping their stoichiometric ratio constant. Coupled chaperone assays were performed in the presence of ATP and an ATP regenerating system as described above.
Results
Crystal structure of Hsp1041-360
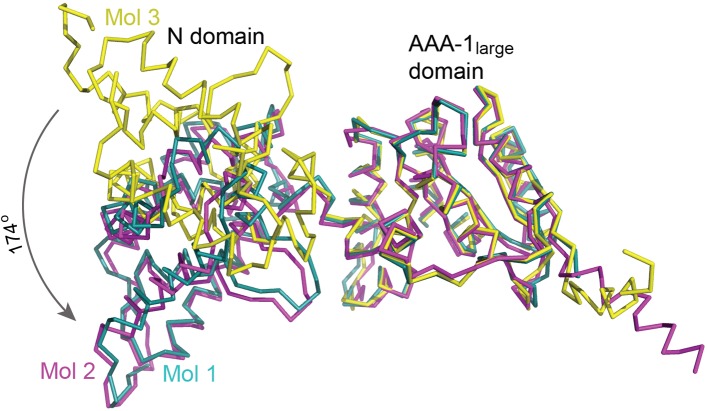
Yeast Hsp104 is a protein disaggregase that is functionally conserved with bacterial ClpB [32]. The crystal structure of Chaetomium thermophilum Hsp104 confirmed that Hsp104 and ClpB are also structurally conserved [33]. However, the atomic structures of the N- and C-terminal domains of C. thermophilum Hsp104 could not be modeled despite being part of the crystallized construct. Here we present the orthorhombic crystal structure of an N-terminal fragment of S. cerevisiae Hsp104 (Hsp1041-360) comprising the N domain (residues 4–164), the AAA-1large domain (residues 165–341), and the first α-helix of the AAA-1small domain (residues 345–356). We did not observe any unaccounted electron density that could be attributed to a bound nucleotide, even when 5 mM nucleotide (ADPNP or ADP) was added for crystallization, indicating that Hsp1041-360 was crystallized in the nucleotide-free state. The structure of Hsp1041-360 was determined by molecular replacement and was refined to a resolution of 2.6 Å (Supplementary Table S1). The crystal structure consists of three Hsp1041-360 monomers (mol 1, mol 2, and mol 3) that are structurally independent and in different physicochemical environments, which allows the identification of common structural features that may be of functional importance.
Crystal structure of Hsp1041-360 confirms the high en bloc mobility of the N domain
The atomic structures of the N and AAA-1large domains alone are nearly identical amongst the three Hsp1041-360 molecules and superimpose pairwise with an RMSD of only 0.41 ± 0.05 Å (N domain) and 0.72 ± 0.22 Å (AAA-1large). In addition, the three Hsp1041-360 molecules superimpose pairwise with the hexagonal crystal structure of one Hsp1041-360 monomer (PDB: 6AMN) [25] with an RMSD of 0.47 ± 0.01 Å (N domain) and 0.75 ± 0.01 Å (AAA-1large), and with the crystal structure of the isolated S. cerevisiae Hsp104 N domain (PDB: 5U2U) [34] with an RMSD of 0.50 ± 0.05Å calculated over all atoms. Superimposing the crystal structures of the complete Hsp1041-360 fragment through their AAA-1large domain shows that the orientation of the N domain seen in mol 1 and mol 2 is rotated by 172–174° relative to that in mol 3 (Figure 1) with residues 161–165 making up the hinge region. Interestingly, different N domain conformations are also observed in the fitted cryoEM structures of Hsp104 hexamers, with mol 1 and 2 matching the N domain conformation of the C subunit of the open conformation (PDB: 5KNE-C) [10] and the A, C, and E subunits of the closed conformation of Hsp104 with casein bound (PDB: 5VY9-A/C/E) [19]. Mol 3 matches the F subunit of the casein-bound, closed structure (PDB: 5VY9-F) [19]. It is noteworthy that the N domain conformation of the Hsp1041-360 monomer in the hexagonal crystal form (mol 4) [25] differs from the other three conformations presented here and matches the N domain conformation of the D subunit of the casein-bound, closed structure (PDB: 5VY9-D) [19]. Together, these findings indicate that the high en bloc mobility of the N domain observed in our crystal structure is likely to be of functional importance, and is also observed in physiologically relevant structures of Hsp104 hexamers.
Figure 1. Crystal structure of Hsp1041-360.
Superposition of the three Hsp1041-360 monomers shows the en bloc mobility of the N domains. Atomic structures were superimposed through their AAA-1large domain. Molecule 1 (mol 1) is shown in teal, molecule 2 (mol 2) in magenta, and molecule 3 (mol 3) in yellow.
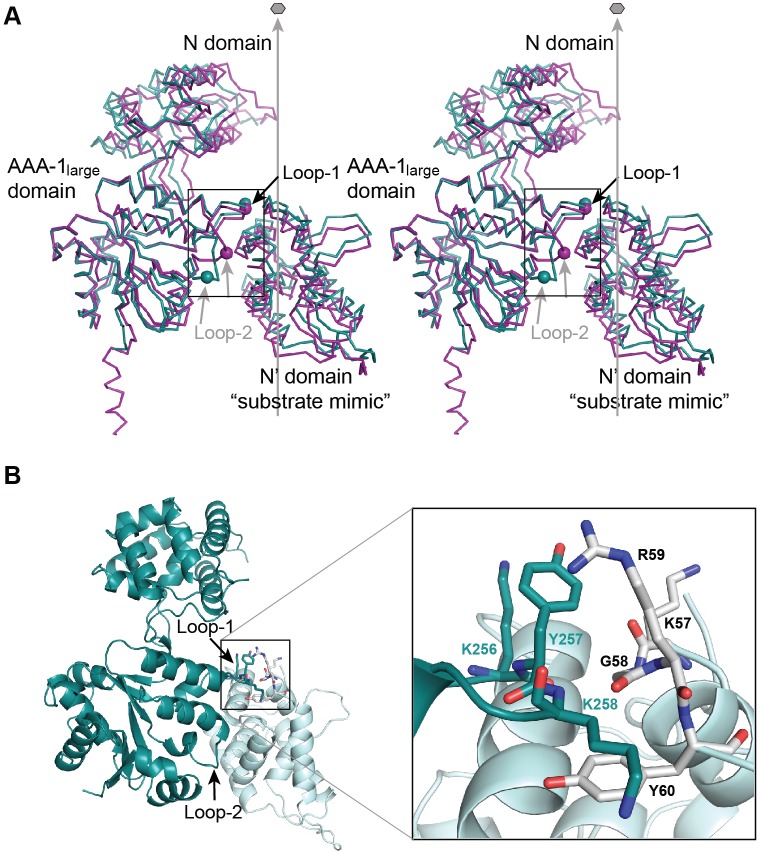
The AAA-1large domain shares the canonical α/β-fold of related AAA+ ATPases determined in their hexamer assembly [35–38], and superposes with the crystal structure of the isolated AAA-1large domain of E. coli ClpB (PDB: 1JBK) [39] with an RMSD of 1.20 ± 0.11 Å calculated over their Cα atoms. However, unlike previously determined crystal structures, we observed both pore loops in our structure. The first AAA-1 pore loop (loop-1), comprising residues 253–259, is seen in all three monomers, and loop-2, comprising residues 291–294, is ordered in two molecules. Loop-1 features the conserved Tyr257 that is sensitive to alanine mutation in Hsp104 [40], and can be site specifically cross-linked to substrate-mimicking peptides in ClpB [41]. A pore-facing tyrosine or phenylalanine is also found in many other AAA+ machines involved in protein quality control [42], and support a key role for Tyr257 in substrate binding, translocation, or both. Although no corresponding aromatic residue is found in loop-2 that features only two non-glycine residues, loop-2 was shown to be sensitive to mutation that impairs the protein unfolding activity of bacterial ClpA [43].
Hsp1041-360 monomer contacts resemble interaction with substrate
In our structure, Tyr257 makes contact with the N domain of a neighboring, non-crystallographic symmetry (NCS) related molecule (Figure 2A,B). Tyr257 is flanked by Lys256 and Lys258 that contributes to the protein–protein interface made up of hydrophobic contacts between aliphatic and aromatic side chains of loop-1 and N domain residues, Arg59 and Tyr60, with additional contributions from the main chain of residues Lys57, Gly58, and Arg59 (Figure 2B). The aforementioned hydrophobic contacts are reminiscent of a chaperone–substrate interaction, and are observed in two out of three molecules (Figure 2A). However, neither the hydroxyl group of the Tyr257 side chain nor the ε-amino group of Lys258 contributes binding energy (Figure 2B). In addition, the aliphatic side chain of Lys256 forms a stacking interaction with the Tyr257 side chain, which may orient Tyr257, while the ε-amino group of Lys256 makes an ionic interaction with the Glu146 side chain of a neighboring molecule. Our observations support a functional role for Lys256 and Lys258 in substrate interaction, and provide an explanation why aromatic residues, such as tryptophan and phenylalanine, can substitute for conserved pore loop tyrosines without marked loss of Hsp104 function [40].
Figure 2. Stereochemistry of molecular interactions between AAA-1 pore loops and a substrate mimic.
(A) Stereoview of the molecular contacts between loop-1/-2 of mol 1 (teal) and mol 2 (magenta) and the N′ domain of an NCS-related neighboring molecule that mimics a bound substrate. The Cα positions of Tyr257 (loop-1) and of Asn292 (loop-2) are shown as spheres. The protein-translocating channel that traverses the Hsp104 hexamer is indicated by the six-fold axis. The figure shows that loop-2 adopts an ‘up’ and ‘down’ configuration in the crystal, which may resemble ATP-driven conformations associated with substrate translocation. (B) Molecular interface between loop-1 and the bound substrate mimic. Loop-1 residues are shown in teal and N′ domain residues in gray. The inset shows a close-up view of the same interface.
Unlike loop-1, loop-2 is less well ordered, which prevented us from modeling side chains. In our structure, loop-2 adopts two distinct conformations in an ‘up’ and ‘down’ configuration when viewed along the six-fold axis of the Hsp104 hexamer (Figures 2A and 4A). Residue 292 (asparagine) that is non-conserved in ClpA/B proteins, is in van der Waals contact with the N domain of an NCS-related, neighboring molecule (Lys131), resembling an interaction with substrate. In support of a functional role for Asn292 in substrate binding, it was shown that mutating the analogous residue in bacterial ClpA (Ala293) from alanine to aspartate abolished binding and translocation of an unfolded model substrate [43]. It is tempting to speculate that the ‘up’ and ‘down’ configurations of loop-2 may represent conformations associated with protein unfolding or substrate translocation through the central channel of the Hsp104 hexamer, and is subject to future investigations.
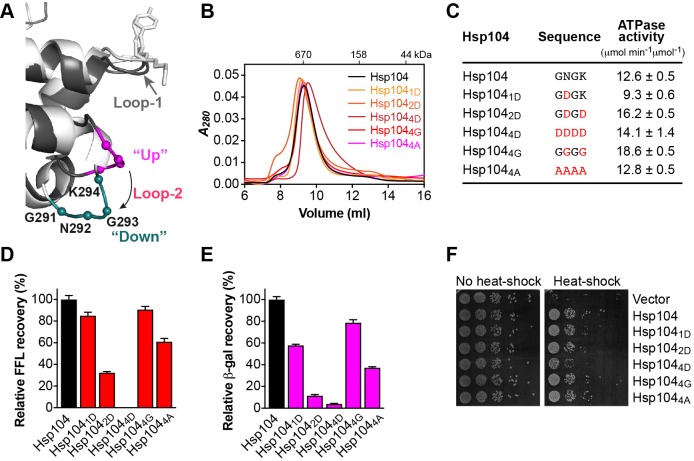
Figure 4. Loop-2 is sensitive to mutation and potentially promotes substrate unfolding.
(A) Ribbon diagram generated by superimposing the AAA-1large domain of mol 1 (light gray) and mol 2 (dark gray), showing the location of loop-2 (magenta/teal) relative to loop-1 (gray). Loop-2 adopts an ‘up’ (magenta) and ‘down’ configuration (teal) that may mimic conformations associated with substrate unfolding. Cα positions of loop-2 residues are shown as spheres. (B) Size-exclusion chromatograms of Hsp104 and loop-2 mutants. (C) ATPase activities of loop-2 mutants. Mutated residues are shown in red. (D,E) Coupled chaperone assay showing the relative recovery of enzymatic activity by loop-2 mutants in the presence of the Hsp70 chaperone system with (D) chemically denatured FFL and (E) heat-aggregated β-gal as substrate. Means of three independent measurements ± S.D. are shown. (F) Induced thermotolerance of Δhsp104 yeast expressing the empty vector, Hsp104, or loop-2 mutants.
The hydrophobicity but not aromaticity of loop-1 is crucial for protein interaction
The functional importance of conserved pore loop tyrosines in Clp/Hsp100 proteins is well established [41,43–45]. It was shown more recently that Tyr257 of Hsp104 mediates binding of an unstructured polypeptide [19]. Consistent with a role in substrate interaction, Tyr257 is sensitive to mutation that severely impaired but, interestingly, did not abolish Hsp104 function [40]. The latter suggests that other pore loop residues must also contribute toward substrate binding. The crystal structure of Hsp1041-360 revealed a previously unobserved specific interface between loop-1 and the N domain of a neighboring, NCS-related molecule involving the side chains of Lys256 and Lys258 in addition to Tyr257 (Figures 2B and 3A), contrasting the proposed role of Lys256 and Lys258 in stabilizing the hexamer assembly [19].
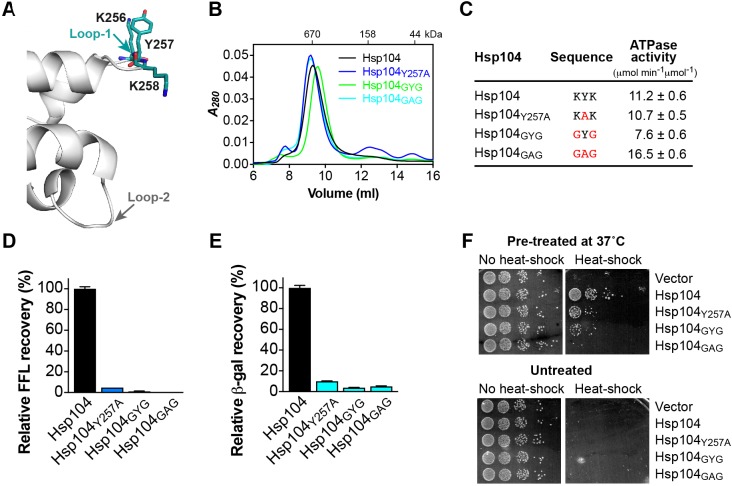
Figure 3. Loop-1 mediates protein–protein interactions essential to Hsp104 function.
(A) Ribbon diagram showing the location of loop-1 relative to loop-2 (gray). Residues of the 256Lys-Tyr-Lys258 tripeptide motif that mediate substrate interaction are colored and shown as stick model. (B) Size-exclusion chromatograms of Hsp104 and loop-1 mutants. (C) ATPase activities of loop-1 mutants. Mutated residues are shown in red. (D,E) Coupled chaperone assay showing the relative recovery of enzymatic activity by loop-1 mutants in the presence of the Hsp70 chaperone system with (D) chemically denatured FFL and (E) heat-aggregated β-gal as substrate. Means of three independent measurements ± S.D. are shown. (F) Induced (top) and basal (bottom) thermotolerance of Δhsp104 yeast expressing the empty vector, Hsp104, or loop-1 mutants.
To our knowledge, the importance of conserved aliphatic residues flanking Tyr257 has not been investigated previously. We therefore mutated Lys256 and Lys258 to glycine (Hsp104GYG) and compared the activity of Hsp104GYG with Hsp104Y257A that is functionally impaired. As expected, all of our loop-1 mutants assemble into hexamers (Figure 3B) and are functional ATPases (Figure 3C). Strikingly, we find that replacing Lys256 and Lys258 with glycine severely impaired Hsp104 function in vitro (Figure 3D,E) and in vivo (Figure 3F), even more so than Hsp104Y257A (Figure 3D–F). Because a hexamer ring assembly is a prerequisite for ATP hydrolysis [30], the ability of Hsp104GYG to hydrolyze ATP argues against a role of Lys256 and Lys258 in the formation of hexamers or stabilizing the oligomer assembly. Combining the Lys256, Tyr257, and Lys258 mutations (Hsp104GAG) completely abolished Hsp104’s ability to disaggregate chemically denatured FFL in vitro (Figure 3D) and its ability to acquire thermotolerance in vivo (Figure 3F, compared Hsp104GAG with vector control). Taken together, our observations support a role for Lys256 and Lys258 in substrate interaction that is abolished when the 256Lys-Tyr-Lys258 tripeptide is mutated to glycine and alanine, respectively.
Loop-2 is sensitive to aspartate substitutions
The crystal structure of Hsp1041-360 showed that loop-2 adopts an ‘up’ (mol 2) and ‘down’ (mol 1) configuration with the tip of loop-2 making van der Waals contact with a neighboring, NCS-related molecule (Figures 2A and 4A). The latter is suggestive of a substrate interaction and, when taken together, reminiscent of an interaction with substrate that is being translocated down the axial channel. Because a functional role for loop-2 in substrate binding or translocation has not been demonstrated previously for Hsp104/ClpB, we asked whether loop-2 is sensitive to mutation that would impact Hsp104 function.
Loop-2 is considerably shorter than other pore loops and consists of only four amino acid residues of sequence 291Gly-Asn-Gly-Lys294 (Figure 4A). It was previously reported that mutating the equivalent residue of Asn292 of E. coli ClpA (Ala293) to aspartate abolished ClpA function [43]. We therefore asked whether introducing one or more aspartates into loop-2 would have a similar impact on Hsp104 function. As anticipated, loop-2 mutants assemble into hexamers (Figure 4B and Supplementary Figure S1) and are functional ATPases (Figure 4C). We note that the Hsp1044D hexamer is right shifted in the absence of nucleotide (Figure 4B), but elutes at the expected position in the presence of ATPγS (Supplementary Figure S1). Furthermore, we found that the ATPase activity of Hsp1044D is similar to Hsp104 wild-type (Figure 4C), indicating no structural perturbations. Yet, replacing Asn292 with aspartate (Hsp1041D) reduced the recovery of chemically denatured FFL by the bi-chaperone system by 15% (Figure 4D). Introducing a second aspartate (Hsp1042D) further reduced protein disaggregation by 68%, and substituting all four residues (Hsp1044D) completely abolished Hsp104-dependent protein disaggregation in vitro (Figure 4D). The observed loss-of-function is specific to Hsp1044D because loop-2 variants featuring either four alanine (Hsp1044A) or four glycine residues (Hsp1044G) cooperate with the Hsp70 system in protein disaggregation (Figure 4D). Similar results were also obtained with heat-aggregated β-gal as the model substrate arguing against a substrate-specific defect (Figure 4E). Consistent with our in vitro observations, loop-2 mutants are also impaired in vivo, with Hsp1044D showing the largest defect in thermotolerance development (Figure 4F).
Taken together, our observations suggest that loop-2 is sensitive to aspartate substitutions that severely impair Hsp104 function when loop-2 is replaced by four aspartates. The inability of Hsp1044D to recover functional protein from aggregates could not be overcome by loop-1 (Figure 4D,E), nor could loop-2 rescue loop-1 loss-of-function mutants (Figure 3D,E), suggesting that loop-1 and loop-2 have distinct, non-overlapping roles in protein disaggregation. Although the exact nature of the functional defect of loop-2 mutants remains unclear, we speculate that the aspartate substitutions may have interfered with substrate interaction. Furthermore, the apparent lack of specificity observed with Hsp1044G that remains fully functional (Figure 4D,E) contrasts the proposed role of loop-1 as a substrate anchor that facilitates a tight interaction with substrate and is sensitive to glycine/alanine substitutions. Thus, our observations could be indicative of a more mechanical function of loop-2 in protein unfolding or translocation, which does not require a tight grip on substrate.
Loop-3 is essential for protein disaggregation
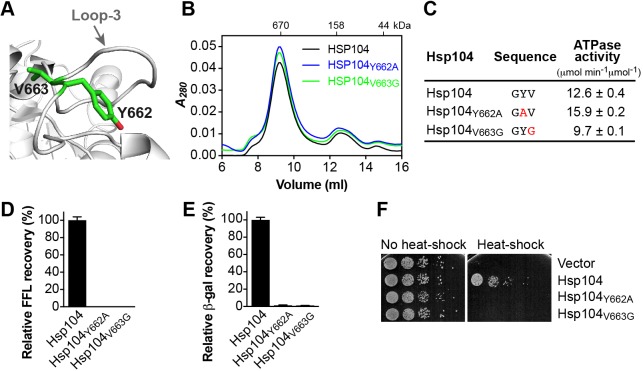
Loop-3 features a conserved aromatic amino acid (Tyr662) that is essential for substrate interaction in vitro and in vivo [40] (Figure 5A–F). Tyr662 is preceded by glycine or a small aliphatic residue and is followed by a hydrophobic amino acid (Ψ) and glycine, giving rise to a (Gly)-Tyr-Ψ-Gly motif. The latter is reminiscent to the Ψ-Tyr-Ψ motif of loop-1, which impaired Hsp104 chaperone function when Ψ is replaced with glycine (Figure 3D–F). In Hsp104, Tyr662 is followed by Val663 that shares an aliphatic side chain with Lys258. It was previously shown that the equivalent valine in heat-shock locus U (HslU) (Val92) is insensitive to isoleucine, alanine, and serine substitutions, but abolishes protein unfolding and translocation when mutated to phenylalanine or cysteine [46]. Similar observations were also made in bacterial ClpX with observed levels of impairment dependent on the substrate [44]. It is interesting to note that all of the aforementioned Clp/Hsp100 variants featuring aliphatic side chain substitutions, besides cysteine that is sensitive to oxidation, appear to be functional. We therefore asked whether replacing Val663 with glycine that lacks an aliphatic side chain would impact protein disaggregation. Strikingly, we found that protein disaggregation by Hsp104V663G was completely abolished in vitro (Figure 5D,E) and in vivo (Figure 5F), despite featuring a functional Tyr662. The observed defect was not due to an inability to self-assemble or lack of ATPase activity, which was similar to Hsp104 wild-type (Figure 5B,C). The sequence specificity and location of loop-3 near the distal end of the protein translocating channel support an essential role of loop-3 in polypeptide binding and translocation. However, it is not the conservation of Tyr662 per se but the hydrophobicity of loop-3 that is essential to Hsp104 function.
Figure 5. Loop-3 is critical for substrate binding and translocation through the distal ring.
(A) Ribbon diagram of C. thermophilum Hsp104 [33] showing the location of loop-3 with the side chains of Tyr662 and Val663 represented as green stick model. (B) Size-exclusion chromatograms of Hsp104 and loop-3 mutants. (C) ATPase activities of loop-3 mutants. Mutated residues are shown in red. (D,E) Coupled chaperone assay showing the relative recovery of enzymatic activity by loop-3 mutants in the presence of the Hsp70 chaperone system with (D) chemically denatured FFL and (E) heat-aggregated β-gal as substrate. Means of three independent measurements ± S.D. are shown. (F) Induced thermotolerance with Δhsp104 yeast cells expressing the empty vector, Hsp104, or loop-3 mutants.
Loop-1 and loop-2 cooperate in initial protein binding and unfolding
Our in vitro and in vivo experiments show that loss-of-function mutations of either loop-1 or loop-2 can abolish protein disaggregation, suggesting distinct, non-overlapping functions of AAA-1 pore loops. While Tyr257 is critically important for substrate binding [19,40,41], our structure further extends the substrate-binding interaction to the 256Lys-Tyr-Lys258 tripeptide (Figure 2B). Interestingly, both Hsp104KAK (i.e. Hsp104Y257A) and Hsp104GYG retain some chaperone activity and mutation of all three residues is required to abolish Hsp104 function (Figure 3D,F). It is worth noting that the proposed hydrophobic interaction between loop-1 and substrate is consistent with the prevailing notion of molecular chaperones in recognizing exposed hydrophobic residues to discriminate between folded and unfolded protein conformers.
The recently reported cryoEM structure of a casein-bound Hsp104 hexamer supports a threading mechanism down the central channel, necessitating cooperative interactions between adjacent subunits [19]. To determine whether pore loops of neighboring Hsp104 subunits cooperate in protein disaggregation, we used a subunit mixing assay [22] to monitor protein disaggregation by Hsp104 hexamers composed of active and inactive mutant subunits. We note that all three pore loops have distinct locations within one subunit, and are not in direct contact (Figure 6A). However, we do not rule out contacts with loops in neighboring subunits as previously proposed [19]. Figure 6B shows that protein disaggregation by Hsp104:Hsp104GAG hetero-hexamers together with the Hsp70 system was substantially impaired in the presence of only one inactive Hsp104GAG subunit. The latter suggests strong cooperativity between subunits and lend support for substrate handover between loop-1 of neighboring AAA-1 domains. A substrate handover mechanism is supported by the recent cryoEM structure of a casein-bound Hsp104 hexamer revealing direct contacts of loop-1 residues from neighboring subunits with the unfolded polypeptide [19]. A more complex pattern emerges when performing the subunit-mixing experiment with Hsp104 hexamers composed of wild-type and inactive loop-2 mutant subunits (Hsp1044D) (Figure 6C). Both cooperative and probabilistic interactions are observed depending on the nature of the substrate (Figure 6C). While a cooperative interaction between subunits was observed with heat-aggregated β-gal, a near linear decline was seen with chemically denatured FFL, indicating a probabilistic mechanism. Although it may seem that Hsp104 uses distinct modi operandi, we reasoned that only heat-aggregated and amyloid-forming substrates that are characterized by a stable secondary and/or tertiary structure [47,48] may require an additional protein unfolding step prior to substrate translocation. Because loop-2 mutants featuring either four glycines (Hsp1044G) or four alanines (Hsp1044A) are functional, and cooperate with Hsp70 and Hsp40 chaperones in protein disaggregation (Figure 4D–F), the observed defect of Hsp1044D may be indicative of a mechanical function in protein unfolding or translocation, and is reflected in the nature of the substrate used.
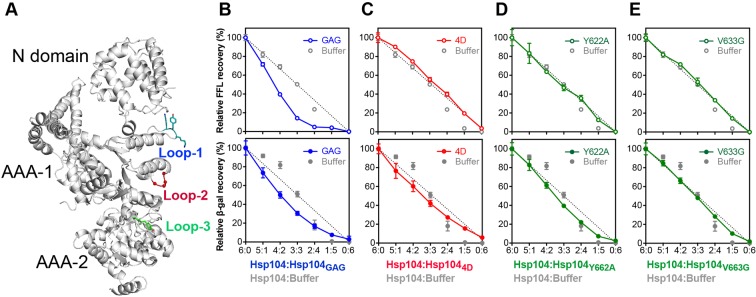
Figure 6. Protein disaggregation by Hsp104 hetero-oligomers composed of active and inactive pore loop mutant subunits.
(A) Ribbon diagram of a composite model of an Hsp104 monomer generated by superposing the AAA-1large domain of yeast Hsp1041-360 on to the crystal structure of C. thermophilum Hsp104 [33]. Channel facing loops are colored in teal (loop-1), red (loop-2), and green (loop-3). (B–E) Relative recoveries of FFL and β-gal activities by Hsp104 hetero-oligomers composed of wild-type and pore loop mutant subunits in the presence of the Hsp70 chaperone system. Means of three independent measurements ± S.D. are shown. The dashed line represents the linear decrease expected if the activity of the Hsp104 hexamer is proportional to the number of wild-type subunits present. Buffer only dilutions are also shown. (B) Hsp104:Hsp104GAG (loop-1), (C) Hsp104:Hsp1044D (loop-2), (D) Hsp104:Hsp104Y662A (loop-3), and (E) Hsp104:Hsp104V663G (loop-3).
Taken together, we propose that unstructured model substrates, such as casein and chemically unfolded FFL that are tethered to loop-1, do not require mechanical unfolding prior to substrate translocation. On the other hand, heat-aggregated substrates rely on an additional mechanical unfolding step that is dependent on cooperative interactions between loop-2 from neighboring subunits to exert a stronger pulling force.
Loop-3 is essential for substrate translocation through the distal ring
The distinct cooperative and probabilistic mechanisms observed with hetero-hexamers composed of active and inactive loop-2 subunits were intriguing. We therefore performed subunit-mixing experiments with active and inactive loop-3 variants (Hsp104Y662A and Hsp104V633G). In agreement with the literature [49], we observed a near linear decrease in FFL reactivation as the number of Hsp104Y662A subunits increased (Figure 6D). We note that the pattern differs somewhat for heat-aggregated β-gal as seen with loop-2 mutant hetero-hexamers (Figure 6C). Mixing Hsp104 with Hsp104V633G yields hetero-hexamers that do not function cooperatively in protein disaggregation (Figure 6E). How can these differences be reconciled? Loop-3 is at the distal end of the protein-translocating channel where the unfolded polypeptide emerges from the Hsp104 hexamer. Because Tyr662 has previously been shown to bind polypeptides [40,45], Tyr662 may provide a substrate anchor with Val663 adding binding energy by contacting the unfolded polypeptide. We speculate that the observed cooperativity of Hsp104:Hsp104Y662A hetero-hexamers with heat-aggregated β-gal compared with chemically denatured FFL might be the result of an additional protein unfolding step needed for native Hsp104 substrates, such as those encountered during heat-stress, and is dispensable for non-native substrates, such as chemically unfolded FFL and casein that is inherently unstructured.
Discussion
Hsp104 chaperones are protein disaggregases that recover functional protein from both amorphous and amyloid-forming aggregates. Seminal discoveries from different laboratories have provided key insights into the protein disaggregation mechanism by the Hsp104 bi-chaperone system [1,2]. It is now widely accepted that the Hsp70 chaperone system targets Hsp104 to protein aggregates in vivo [17,50] from which Hsp104 extracts one or more polypeptides. The substrate is threaded through the Hsp104 hexamer [45,51], resulting in protein unfolding. We propose that pore loop-1 facilitates the interaction with substrate at the proximal end, providing an anchoring point for initial substrate binding.
Our crystal structure of Hsp1041-360 revealed an interaction between loop-1 and an NCS-related, neighboring molecule which mimics an interaction with substrate and supports a functional role of loop-1 as an anchor for initial substrate binding. Consistent with such a role, replacing the 256Lys-Tyr-Lys258 tripeptide motif of loop-1 in full-length Hsp104 with glycine and alanine, respectively, abolishes Hsp104 function without disrupting hexamer assembly. Unlike loop-1, the role of loop-2 is more complex. Loop-2 mutants are mostly functional in protein disaggregation, and Hsp1044G that features four glycines instead of loop-2 is nearly as active as Hsp104 wild-type (Figure 4D–F). Yet, replacing loop-2 with four aspartates (Hsp1044D) abolishes protein disaggregation in vitro (Figure 4D,E) and impaired thermotolerance development in vivo (Figure 4F). Lack of sequence preference observed with Hsp1044G and Hsp1044A may be indicative of a more mechanical function of loop-2, such as what would be required to facilitate protein unfolding of heat-aggregated substrates. We speculate that loop-2, driven by ATP hydrolysis in the AAA-1 domain, moves from an ‘up’ to a ‘down’ position inside the central channel, exerting a mechanical pulling force on substrate bound to loop-1 in order to promote local unfolding. This unidirectional motion propels the substrate down the axial channel and, combined with substrate binding and pulling by loop-3, results in protein unfolding and translocation with the unfolded polypeptide emerging from the distal end.
Although our model inferred from the crystal structure of Hsp1041-360 is supported by both biochemical and functional studies in vitro and in vivo, Hsp1041-360 did not crystallize as a hexameric ring assembly. We therefore cannot exclude contacts between neighboring subunits, which may have impacted substrate binding. While our manuscript was under review, the cryoEM structures of a casein-bound Hsp104 hexamer was reported [19]. Although these structures provided a stereochemical framework to interpret our observations, any structural insight must be taken with caution because of the limited resolution and accuracy of these cryoEM reconstructions that remain to be confirmed biochemically. In the hexamer structure, both loop-1 and loop-3 make contact with the equivalent loops of neighboring subunits on both sides supporting a clockwise handover of the unfolded polypeptide when viewed top down. Furthermore, Lys258 of loop-1 interacts with residues of the neighboring subunit immediately following loop-2 (Asp295 and Asp296), which would be consistent with the complex pattern of substrate recovery observed with hexamers composed of active and inactive loop-2 subunits (Figure 6C). Taken together, we propose a coordinated interaction between pore loops with loop-1 facilitating protein binding, loop-2 potentially promoting protein unfolding by pulling down the substrate, and loop-3 mediating substrate translocation through the Hsp104 hexamer.
Supporting information
Supplementary Figure S1.
Analytical size-exclusion chromatograms of Hsp104 and Hsp1044D in the presence of nucleotide. Both Hsp104 (black) and Hsp1044D (red) form hexamer assembles with ATPγS. Molecular weight standards are shown.
Supplementary Table S1. Data collection and refinement statistics Values in parenthesis are for the highest resolution shell.
Acknowledgments
We thank the late S. Lindquist for the gift of the Hsp1041-360 bacterial expression construct, and Y. Chernoff for yeast Hsp104 (pYS104). Use of the APS-SBC 19-ID beamline was supported by the U.S. Department of Energy, Office of Biological and Environmental Research under contract DE-AC02-06CH11357.
Abbreviations
- β-gal
β-galactosidase
- FFL
firefly luciferase
- NCS
non-crystallographic symmetry
- Ni-NTA
nickel-nitrilotriacetic acid
- PDB
Protein Data Bank
- SD-Ura
synthetic defined growth medium without uracil
- YPD
yeast extract peptone dextrose
Competing interests
The authors declare that there are no competing interests associated with the manuscript.
Funding
This work was supported by the National Institutes of Health [grant numbers GM104980, GM111084]; and the Welch Foundation [grant number Q-1530].
Author contribution
J.L. and S.L. designed and performed experiments, analyzed the data, and wrote the manuscript. N.S. and L.Y. performed experiments, analyzed the data, and wrote the manuscript. C.C. performed experiments and analyzed the data. F.T.F.T. designed experiments, analyzed the data, and wrote the manuscript. All authors reviewed the manuscript.
References
- 1.Doyle S.M., Genest O. and Wickner S. (2013) Protein rescue from aggregates by powerful molecular chaperone machines. Nat. Rev. Mol. Cell Biol. 14, 617–629 [DOI] [PubMed] [Google Scholar]
- 2.Mogk A., Kummer E. and Bukau B. (2015) Cooperation of Hsp70 and Hsp100 chaperone machines in protein disaggregation. Front. Mol. Biosci. 2, 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sweeny E.A. and Shorter J. (2016) Mechanistic and structural insights into the prion-disaggregase activity of Hsp104. J. Mol. Biol. 428, 1870–1885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mosser D.D., Ho S. and Glover J.R. (2004) Saccharomyces cerevisiae Hsp104 enhances the chaperone capacity of human cells and inhibits heat stress-induced proapoptotic signaling. Biochemistry 43, 8107–8115 [DOI] [PubMed] [Google Scholar]
- 5.Liu Q. and Hendrickson W.A. (2007) Insights into Hsp70 chaperone activity from a crystal structure of the yeast Hsp110 Sse1. Cell 131, 106–120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Glover J.R. and Lindquist S. (1998) Hsp104, Hsp70, and Hsp40: a novel chaperone system that rescues previously aggregated proteins. Cell 94, 73–82 [DOI] [PubMed] [Google Scholar]
- 7.Kaimal J.M., Kandasamy G., Gasser F. and Andréasson C. (2017) Coordinated Hsp110 and Hsp104 activities power protein disaggregation in Saccharomyces cerevisiae. Mol. Cell. Biol. 37, pii: e00027–17, [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Parsell D.A., Kowal A.S. and Lindquist S. (1994) Saccharomyces cerevisiae Hsp104 protein. J. Biol. Chem. 269, 4480–4487 [PubMed] [Google Scholar]
- 9.Lee S., Sielaff B., Lee J. and Tsai F.T.F. (2010) CryoEM structure of Hsp104 and its mechanistic implication for protein disaggregation. Proc. Natl. Acad. Sci. U.S.A. 107, 8135–8140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yokom A.L., Gates S.N., Jackrel M.E., Mack K.L., Su M., Shorter J. et al. (2016) Spiral architecture of the Hsp104 disaggregase reveals the basis for polypeptide translocation. Nat. Struct. Mol. Biol. 23, 830–837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sielaff B. and Tsai F.T.F. (2010) The M-domain controls Hsp104 protein remodeling activity in an Hsp70/Hsp40-dependent manner. J. Mol. Biol. 402, 30–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Miot M., Reidy M., Doyle S.M., Hoskins J.R., Johnston D.M., Genest O. et al. (2011) Species-specific collaboration of heat shock proteins (Hsp) 70 and 100 in thermotolerance and protein disaggregation. Proc. Natl. Acad. Sci. U.S.A. 108, 6915–6920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Striebel F., Kress W. and Weber-Ban E. (2009) Controlled destruction: AAA+ ATPases in protein degradation from bacteria to eukaryotes. Curr. Opin. Struct. Biol. 19, 209–217 [DOI] [PubMed] [Google Scholar]
- 14.Sauer R.T. and Baker T.A. (2011) AAA+ proteases: ATP-fueled machines of protein destruction. Annu. Rev. Biochem. 80, 587–612 [DOI] [PubMed] [Google Scholar]
- 15.Hung G.C. and Masison D.C. (2006) N-terminal domain of yeast Hsp104 chaperone is dispensable for thermotolerance and prion propagation but necessary for curing prions by Hsp104 overexpression. Genetics 173, 611–620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lum R., Niggemann M. and Glover J.R. (2008) Peptide and protein binding in the axial channel of Hsp104: insights into the mechanism of protein unfolding. J. Biol. Chem. 283, 30139–30150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tipton K.A., Verges K.J. and Weissman J.S. (2008) In vivo monitoring of the prion replication cycle reveals a critical role for Sis1 in delivering substrates to Hsp104. Mol. Cell 32, 584–591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sweeny E.A., Jackrel M.E., Go M.S., Sochor M.A., Razzo B.M., DeSantis M.E. et al. (2015) The Hsp104 N-terminal domain enables disaggregase plasticity and potentiation. Mol. Cell 57, 836–849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gates S.N., Yokom A.L., Lin J., Jackrel M.E., Rizo A.N., Kendsersky N.M. et al. (2017) Ratchet-like polypeptide translocation mechanism of the AAA+ disaggregase Hsp104. Science 357, 273–279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li T., Weaver C.L., Lin J., Duran E.C., Miller J.M. and Lucius A.L. (2015) Escherichia coli ClpB is a non-processive polypeptide translocase. Biochem. J. 470, 39–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Deville C., Carroni M., Franke K.B., Topf M., Bukau B., Mogk A. et al. (2017) Structural pathway of regulated substrate transfer and threading through an Hsp100 disaggregase. Sci. Adv. 3, e1701726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee J., Kim J.-H., Biter A.B., Sielaff B., Lee S. and Tsai F.T.F. (2013) Heat shock protein (Hsp) 70 is an activator of the Hsp104 motor. Proc. Natl. Acad. Sci. U.S.A. 110, 8513–8518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Otwinowski Z. and Minor W. (1997) Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 [DOI] [PubMed] [Google Scholar]
- 24.Vagin A. and Teplyakov A. (1997) MOLREP: an automated program for molecular replacement. J. Appl. Cryst. 30, 1022–1025 [Google Scholar]
- 25.Lee J., Sung N., Mercado J.M., Hryc C.F., Chang C., Lee S. et al. (2017) Overlapping and specific functions of the Hsp104 N domain define its role in protein disaggregation. Sci. Rep. 7, 11184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Emsley P. and Kevin C. (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 27.Adams P.D., Afonine P.V., Bunkóczi G., Chen V.B., Davis I.W., Echols N. et al. (2010) PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Taylor D., Cawley G. and Hayward S. (2014) Quantitative method for the assignment of hinge and shear mechanism in protein domain movements. Bioinformatics 30, 3189–3196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Carter S.G. and Karl D.W. (1982) Inorganic phosphate assay with malachite green: an improvement and evaluation. J. Biochem. Biophys. Methods 7, 7–13 [DOI] [PubMed] [Google Scholar]
- 30.Biter A.B., Lee J., Sung N., Tsai F.T.F. and Lee S. (2012) Functional analysis of conserved cis- and trans-elements in the Hsp104 protein disaggregating machine. J. Struct. Biol. 179, 172–180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Derkatch I.L., Bradley M.E., Zhou P., Chernoff Y.O. and Liebman S.W. (1997) Genetic and environmental factors affecting the de novo appearance of the [PSI +] prion in Saccharomyces cerevisiae. Genetics 147, 507–519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Haslberger T., Bukau B. and Mogk A. (2010) Towards a unifying mechanism for ClpB/Hsp104-mediated protein disaggregation and prion propagation. Biochem. Cell Biol. 88, 63–75 [DOI] [PubMed] [Google Scholar]
- 33.Heuck A., Schitter-Sollner S., Suskiewicz M.J., Kurzbauer R., Kley J., Schleiffer A. et al. (2016) Structural basis for the disaggregase activity and regulation of Hsp104. Elife 5, e21516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang P., Li J., Weaver C., Lucius A. and Sha B. (2017) Crystal structures of Hsp104 N-terminal domains from Saccharomyces cerevisiae and Candida albicans suggest the mechanism for the function of Hsp104 in dissolving prions. Acta Crystallogr. D Struct. Biol. 73, 365–372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Song H.K., Hartmann C., Ramachandran R., Bochtler M., Behrendt R., Moroder L. et al. (2000) Mutational studies on HslU and its docking mode with HslV. Proc. Natl. Acad. Sci. U.S.A. 97, 14103–14108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sousa M.C., Trame C.B., Tsuruta H., Wilbanks S.M., Reddy V.S. and McKay D.B. (2000) Crystal and solution structure of an HslUV protease-chaperone complex. Cell 103, 633–643 [DOI] [PubMed] [Google Scholar]
- 37.Glynn S.E., Martin A., Nager A.R., Baker T.A. and Sauer R.T. (2009) Structures of asymmetric ClpX hexamers reveal nucleotide-dependent motions in a AAA+ protein-unfolding machine. Cell 139, 744–756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang F., Mei Z., Qi Y., Yan C., Hu Q., Wang J. et al. (2011) Structure and mechanism of the hexameric MecA-ClpC molecular machine. Nature 471, 331–335 [DOI] [PubMed] [Google Scholar]
- 39.Li J. and Sha B. (2002) Crystal structure of E. coli Hsp100 ClpB nucleotide-binding domain 1 (NBD1) and mechanistic studies on ClpB ATPase activity. J. Mol. Biol. 318, 1127–1137 [DOI] [PubMed] [Google Scholar]
- 40.Lum R., Tkach J.M., Vierling E. and Glover J.R. (2004) Evidence for an unfolding/threading mechanism for protein disaggregation by Saccharomyces cerevisiae Hsp104. J. Biol. Chem. 279, 29139–29146 [DOI] [PubMed] [Google Scholar]
- 41.Schlieker C., Weibezahn J., Patzelt H., Tessarz P., Strub C., Zeth K. et al. (2004) Substrate recognition by the AAA+ chaperone ClpB. Nat. Struct. Mol. Biol. 11, 607–615 [DOI] [PubMed] [Google Scholar]
- 42.Wang J., Song J.J., Franklin M.C., Kamtekar S., Im Y.J., Rho S.H. et al. (2001) Crystal structure of the HslVU peptidase-ATPase complex reveal an ATP-dependent proteolysis mechanism. Structure 9, 177–184 [DOI] [PubMed] [Google Scholar]
- 43.Hinnerwisch J., Fenton W.A., Furtak K.J., Farr G.W. and Horwich A.L. (2005) Loops in the central channel of ClpA chaperone mediate protein binding, unfolding, and translocation. Cell 121, 1029–1041 [DOI] [PubMed] [Google Scholar]
- 44.Siddiqui S.M., Sauer R.T. and Baker T.A. (2004) Role of the processing pore of the ClpX AAA+ ATPase in the recognition and engagement of specific protein substrates. Genes Dev. 18, 369–374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Weibezahn J., Tessarz P., Schlieker C., Zahn R., Maglica Z., Lee S. et al. (2004) Thermotolerance requires refolding of aggregated proteins by substrate translocation through the central pore of ClpB. Cell 119, 653–665 [DOI] [PubMed] [Google Scholar]
- 46.Park E., Rho Y.M., Koh O.J., Ahn S.W., Seong I.S., Song J.J. et al. (2005) Role of the GYVG pore motif of HslU ATPase in protein unfolding and translocation for degradation by HslV peptidase. J. Biol. Chem. 280, 22892–22898 [DOI] [PubMed] [Google Scholar]
- 47.Nelson R., Sawaya M.R., Balbirnie M., Madsen A.O., Riekel C., Grothe R. et al. (2005) Structure of the cross-beta spine of amyloid-like fibrils. Nature 435, 773–778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lewandowska A., Matuszewska M. and Liberek K. (2007) Conformational properties of aggregated polypeptides determine ClpB-dependence in the disaggregation process. J. Mol. Biol. 371, 800–811 [DOI] [PubMed] [Google Scholar]
- 49.Doyle S.M., Hoskins J.R. and Wickner S. (2012) DnaK-dependent disaggregation by Caseinolytic Peptidase B (ClpB) mutants reveals functional overlap in the N-terminal domain and nucleotide-binding domain-1 pore tyrosine. J. Biol. Chem. 287, 28470–28479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Winkler J., Tyedmers J., Bukau B. and Mogk A. (2012) Hsp70 targets Hsp100 chaperones to substrates for protein disaggregation and prion fragmentation. J. Cell Biol. 198, 387–404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tessarz P., Mogk A. and Bukau B. (2008) Substrate threading through the central pore of the Hsp104 chaperone as a common mechanism for protein disaggregation and prion propagation. Mol. Microbiol. 68, 87–97 [DOI] [PubMed] [Google Scholar]