Abstract
Background
The gene encoding a thermostable cellulase of family 12 was previously isolated from a Rhodothermus marinus through functional screening. CelA is a protein of 260 aminoacyl residues with a 28-residue amino-terminal signal peptide. Mature CelA was poorly synthesized in some Escherichia coli strains and not at all in others. Here we present an alternative approach for its heterologous production as a secreted polypeptide in Streptomyces.
Results
CelA was successfully over-expressed as a secreted polypeptide in Streptomyces lividans TK24. To this end, CelA was fused C-terminally to the secretory signal peptide of the subtilisin inhibitor protein (Sianidis et al. in J Biotechnol. 121: 498–507, 2006) from Streptomyces venezuelae and a new cloning strategy developed. Optimal growth media and conditions that stall biomass production promote excessive CelA secretion. Under optimal growth conditions in nutrient broth medium, significant amounts of mature CelA (50–90 mg/L or 100–120 mg/g of dry cell weight) are secreted in the spent growth media after 7 days. A protocol to rapidly purify CelA to homogeneity from culture supernatants was developed and specific anti-sera raised against it. Biophysical, biochemical and immmuno-detection analyses indicate that the enzyme is intact, stable and fully functional. CelA is the most thermostable heterologous polypeptide shown to be secreted from S. lividans.
Conclusion
This study further validates and extends the use of the S. lividans platform for production of heterologous enzymes of industrial importance and extends it to active thermostable enzymes. This study contributes to developing a platform for poly-omics analysis of protein secretion in S. lividans.
Electronic supplementary material
The online version of this article (10.1186/s12934-017-0847-x) contains supplementary material, which is available to authorized users.
Keywords: Cellulase, Protein translocase, Signal peptide, Secretion, Streptomyces lividans, Protein secretion biotechnology
Background
Streptomyces lividans has been used for the heterologous secretion of several polypeptides of bacterial and eukaryotic origin (for examples see [1–5]). Commonly, heterologous genes are fused to signal peptide sequences from highly expressed/secreted endogenous Streptomyces proteins [6–8]. The resulting proteins are thus targeted to the S. lividans Sec pathway and very efficiently secreted directly into the growth medium. The absence of lipopolysaccharides, the advanced genetic manipulation tools [9], the established bioprocessing regimes, the low protease activity and the avoidance of inclusion body formation, render S. lividans secretion an attractive biotechnology platform. In many instances, it can provide alternative solutions when established workhorses, like Escherichia coli, fail or are more costly in bio-processing terms [8]. Moreover, S. lividans is being developed as a powerful experimental system on which poly-omics tools can be applied to gain understanding on the molecular underpinnings of protein secretion regulation.
Previously, we demonstrated that S. lividans can efficiently secrete active trimeric murine tumor necrosis factor alpha (mTNFα) [5, 10], a Jonesia sp. xyloglucanase of 100 kDa [1] and other polypeptides such as phospholipase D [3], transglutaminase (TGase), β-1,4-endoglucanase and β-glucosidase [11] into the growth medium. One successful approach has been the use of the transcription elements and the signal peptide of the Streptomyces venezuelae CBS762.70 subtilisin inhibitor gene [12] (hereafter: vsi).
We have now extended the use of the S. lividans secretion system for the production of thermostable enzymes of industrial interest, using as an example a cellulase of glycosyl hydrolase family 12 (CelA) from Rhodothermus marinus [13]. The enzyme was previously shown to have activity on carboxymethyl cellulose and lichenan, but not on birch xylan or laminarin with a pH optimum of 6–7 and its highest measured initial activity was at 100 °C. Its structure has been solved by X-ray crystallography and revealed a beta-jelly roll fold, with an identical topology to those of mesophilic members of the family like that of the endogenous CelB2 of S. lividans [14], with an elongated groove that binds the substrates [15]. The mature region of CelA (residues 29–233) fused behind the SPvsi is efficiently secreted from S. lividans as a discrete polypeptide of ~ 26 kDa (expected size 26.05 kDa) in amounts exceeding 40 mg/L when grown in nutrient broth or double strength NB media for 48 h. Yields of 50–90 mg/L or 100–120 mg/g of dry cell weight can be produced after 7 days. Various media tested show significant differences in amounts of secreted protein. One characteristic observation is that the best performing media for secretion give the lowest amounts of biomass and unabated secretion at late growth phases. Isolation of the enzyme revealed it to be catalytically functional and stable. These data extend the use of the S. lividans secretion biotechnology platform to the production of thermostable enzymes of industrial importance and set the stage for a rational understanding of the molecular basis of protein secretion regulation using poly-omics approaches.
Methods
Bacterial strains and recombinant DNA experiments
Growth and manipulation of E. coli and Streptomyces strains were as described [9, 16].
Cloning of RmcelA in S. lividans TK24
The native Rm celA gene was amplified from R. marinus and ligated as a blunt end—PstI fragment into pBSDK0.6Sma [10] following DraII digest, Klenow modification, and a subsequent PstI digest. pBSDK0.6Sma is used to construct vsi-celA. CelA (also called Cel12A, a superfamily 12 of hydrolases) devoid of its predicted signal peptide (aa residues 29–263) was fused C-terminally to the signal peptide of Vsi (SPVsi), the subtilisin inhibitor of S. venezuelae CBS762.70 [12]. Two additional amino acids of the mature Vsi domain were maintained in the fusion protein SPVsi-EA-CelA29–263 (Additional file 1: Figure S1). Its expression was placed under the control of the strong constitutive vsi gene promoter. To this end, the region of celA that encodes residues 29–263 was amplified by PCR using the primers celA-dsp-f (5′-AAGGAACCGGAGCCTGAG) and celA-pst-mfe-r (5′-AAAACTGCAGACAATTGCTACTGCACCGTTACGGAAAAATC) and genomic DNA isolated from R. marinus DSM4253 as a template. No restriction site was included in the forward primer, while a PstI and a MfeI site were introduced in the 5′ end of the reverse primer.
The resulting plasmid containing celA downstream of the vsi promoter and SPvsi was sequenced to verify the in-frame fusion of the gene with the vsi signal peptide-encoding sequence and to verify the correct coding sequence.
To facilitate the E. coli steps of the cloning procedure into pIJ486 [17], the vector was modified and converted into the shuttle-vector pIJ486_Trueblue. This was done by cutting pIJ486 with EcoRI and BamHI and ligating the linear plasmid with pTrueBlue (genomics one) that was previously digested with EcoRI and BamHI (Additional file 1: Figure S2). Subsequently, high amounts of DNase-free pIJ486_Trueblue DNA were produced in E. coli. This DNA was used for the cloning of the target gene. The E. coli replication unit was however removed by cleavage concomitant with the insertion of the target genes prior to transformation of Streptomyces.
The vsi expression/secretion cassette from pBSDK0.6Sma [12] was isolated as a BamHI/MfeI restriction fragment and ligated into pIJ486 following BamHI and EcoRI digestion of pIJ486_Trueblue to generate plasmid pIJ486_vsi-celA. Ligation mixtures were introduced in S. lividans by PEG-mediated protoplast transformation [18] and selection for thiostrepton antibiotic resistance. Clones containing celA were verified by colony PCR using the cloning primers celA-dsp-f and celA-pst-mfe-r.
Bacterial growth and fermentation
Recombinant Streptomyces growth was in the presence of thiostrepton (10 µg/mL) to select for maintenance of plasmid pIJ486_vsi-celA. Media used in this study as described in [9] were: Phage medium [19] (per liter: 10 g glucose, 5 g tryptone, 5 g yeast extract, 5 g Lab Lemco powder, 0.74 g CaCl2·2H2O, 0.5 g MgSO4·7H2O, pH: 7.2), minimal medium (MM) [per liter: 10 g glucose, 3 g (NH4)2SO4, 2.6 g K2HPO4, 1.8 g NaH2PO4, 0.6 g MgSO4·7H2O, 25 mL] minor elements solution (per liter: 40 mg ZnSO4·7H2O, 40 mg FeSO4·7H2O, 40 mg CaCl2, 40 mg MnCl2·4H2O), minimal medium with either 5 g/L (MMC5) or 15 g/L (MMC15) bacto casamino acids, tryptic soy broth (TSB) (per liter: 30 g) [containing 17 g casein peptone (pancreatic)], [5 g NaCl, 3 g soya peptone (papain digest), 2.5 g K2HPO4, 2.5 g glucose], nutrient broth (NB) without NaCl [per liter: 8 g nutrient broth pH 6.9 (containing 5 g/L peptic digest of animal tissue, 3 g/L beef extract)], double strength nutrient broth (NB2X) without NaCl [per liter: 16 g nutrient broth pH 6.9 (containing 10 g/L peptic digest of animal tissue, 6 g/L beef extract)] and Bennet medium (Ben) (per liter: 10 g glucose, 2 g tryptone, 1 g yeast extract, 1 g beef extract). Shake-flask studies were conducted in 2 L Erlenmeyer flasks containing 1 L liquid medium (at 180 rpm; 28 °C) in a New Brunswick 44R temperature-controlled incubator. Fermentation was carried out in an Eppendorf DASGIP Parallel Bioreactor System using 2.3 L vessels containing 1 L medium (at 30 °C; fixed stirring at 500 rpm; pH 6.8 maintained with 4 M KOH and 2 M H2SO4; air supply of 1 sL/min).
Protein secretion assays
Western blotting was done using Trans-Blot® SD Semi-Dry Transfer Cell (Bio-Rad). Purified CelA protein served as reference for calibration purposes. Blotted membranes were washed and incubated overnight with the primary antibody. After washing the membrane, the secondary antibody (Jackson Immuno Research) was applied for 1 h. Detection was carried out using the GE Healthcare Amersham ECL reagents and ImageQuant LAS 4000 imager. High resolution images were processed using ImageJ.
Dot-blots were performed using the Bio-Rad Bio-Dot Microfiltration Apparatus according to the manufacturer’s instructions. A nitrocellulose Amersham Protran 0.2 membrane was sandwiched in the manifold and tightened under vacuum followed by washing with TBS buffer and gravitational filtration of diluted supernatants from reactor samples. Purified CelA protein served as reference for calibration purposes. Blotted membranes were treated and detected as previously described in western blotting.
Protein purification, chromatography and characterization
All chromatography resins and molecular weight markers were from Amersham. Purified proteins were stored at − 20 °C.
For CelA purification from S. lividans culture supernatants, cells were grown for 2 days in 500 mL Phage medium and then transferred in 4 L NB for 48 h.
Step 1: protein concentration
The S. lividans culture supernatants of the 4 L nutrient broth (NB) were concentrated into 200 mL using a rotary evaporator (HeidolphHei-VAP) at 25 °C.
Step 2: ammonium sulfate fractionation
Polypeptides in 200 mL of concentrated spent growth medium supernatant were precipitated by gradual slow addition of finely ground (NH4)2SO4 (25% then 55% saturation; 4 °C) and collected by centrifugation (Sorvall RC, F14-6 × 250y rotor, 4 °C, 20 min, 13,000 rpm). Pellets harvested by centrifugation containing CelA as a main polypeptide species and were resuspended in 4 mL of buffer A (50 mM sodium phosphate buffer pH 7.0).
Step 3: ion exchange chromatography
The 55% saturation ammonium sulfate fraction from Step 2 containing CelA was washed with 200 mL of buffer A using a Sartorius Vivaspin® 20 centrifugal concentrator (molecular weight cut-off 3000 Da) to remove the salt. The protein pellet was resuspended in 4 mL of buffer A and loaded on a Q-Sepharose column (2 mL) (equilibrated with buffer A). The column was washed with 5 column volumes of buffer A and proteins were eluted with a 0.05–1 M NaCl gradient. The fraction with the highest CelA activity was further treated by heating it at 80 °C for 4 h to remove host contaminants.
Gel permeation chromatography, circular dichroism and thermal stability
Gel permeation chromatography was as described [20]. CD spectra were recorded on a Jasco J-1500 spectrometer that was equipped with a Peltier-temperature controlled cuvette holder, as described [21, 22]. The thermal denaturation curve of CelA was obtained my monitoring helicity at 222 nm, as a function of temperature (4–82 °C; 0.8 °C/min; slits: 2.5/20). The apparent melting temperature (Tmapp) was derived by the first derivative of the melting curve, using Prism v4.0 (GraphPad).
Cellulase functional assay
CelA activity was measured by following the hydrolysis of CMC (carboxy methyl cellulose) [23, 24]. 200 μL substrate (1% w/v CMC in 0.2 M sodium phosphate buffer pH 7.0) was incubated with 0.5 μg of enzyme at 70 °C for 30 min. Subsequently, 300 μL of a DNS solution [per 100 mL: 1 g of 3,5-dinitrosalicylic acid (DNS), 1.6 g NaOH and 30 g K-Na-Tartrate] was added to the reaction mix and the solution was boiled for 5 min. The absorbance of the reaction mix was measured at 546 nm in a microtiter plate reader (Tecan Austria GmbH, Infinite 200) and the results were compared to a standard curve of glucose amounts (0.2–1.4 µmol) and absorbance linearly correlated. One unit of cellulase activity is defined as the amount of enzyme that produces 1 µmol of glucose in 1 min [13]. The catalytic activity was compared to that of a commercial crude preparation of cellulase-containing secretomes from Aspergillus niger (0.8 U/mg total crude secretome; measured at 37 °C) (22178 Sigma®). Cellulase units of the crude secretome from A. niger were calculated by assuming the amount of the unpurified main cellulase (of ~ 27 kDa) to be 1 µg in the total secretome via loading the total secretome on SDS-PAGE (15 µg/lane) and staining by Coomassie blue.
Miscellaneous
Chemicals were from Sigma. DNA enzymes were from New England Biolabs and oligonucleotides from Eurogentec. Statistical analysis was performed in R language. Differential secretion of CelA was tested using an unpaired t test without assumption of equal variance and p values were adjusted for multiple hypothesis testing error using the Benjamin Hochberg method [25]. Significant difference in secretion was considered when the adjusted p value was < 0.05. Protein aggregation was determined after breaking the cells by sonication, followed by low spin centrifugation to remove unbroken cells (10,000×g) and ultracentrifugation (at 100,000×g) and detection of proteins by western blotting.
Results
Production of recombinant RmCelA in S. lividans
High-level production of CelA was originally achieved by cloning the celA gene in E. coli in a pET23bAH vector with a C-terminal hexahistidinyl tag [13]. Over-expression of celA His6 in BL21 strains harbouring the T7 polymerase gene was poor in many of the clones tested (e.g. Fig. 1a, lanes 3 and 5). When expressed (lane 7), a percentage of the protein showed a tendency to aggregate (not shown). To explore an alternative production approach and to take advantage of extracellular production of CelA, we used S. lividans TK24 with CelA produced as a secreted polypeptide. We successfully used this system previously for the production of tumor necrosis factor α [5] and a 100 kDa xyloglucanase [1]. This approach simplifies downstream protein purification steps and can be optimized at the level of bio-processing. CelA was fused C-terminally to the Vsi signal peptide and placed behind the vsi promoter to generate pIJ486-vsi-celA (Additional file 1: Figures S1, S2).
Fig. 1.
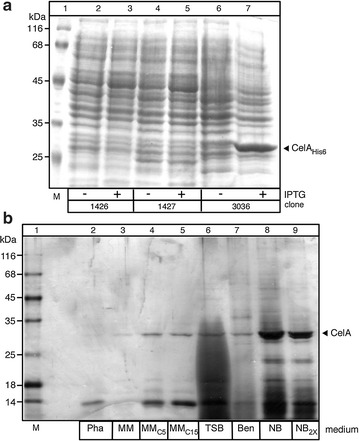
Heterologous synthesis of CelA in E. coli and synthesis/secretion in S. lividans. a CelAHis6 was synthesized in BL21 (DE3) cells. Expression of CelAHis6 carried on a pET vector was induced by 0.2 mM IPTG for 3 h at 30 °C. b Synthesis in and secretion of CelA from S. lividans. Polypeptides (1–8 µg/lane) from culture supernatants (~ 0.1 mL) that is equivalent to 0.2 mg of dry cell mass. The samples from cells grown for 2 days in the indicated media (see “Methods”) were harvested by precipitation with TCA (25%), analyzed by SDS-PAGE and stained with Coomassie blue. Lane 1, molecular weight markers: β-galactosidase (116 kDa), bovine serum albumin (66.2 kDa), ovalbumin (45 kDa), lactate dehydrogenase (35 kDa), restriction endonuclease Bsp98I (25 kDa), β-lactoglobulin (18.4 kDa), lysozyme (14.4 kDa). Pha, phage medium; NB, nutrient broth; NB2X, nutrient broth double strength; MM, minimal medium; C5 and C15, casamino acids (5 or 15 g/L); TSB, tryptic soy broth; Ben, Bennet medium
Streptomyces lividans TK24 cells harbouring pIJ486-vsi-celA were grown in 3 defined and 4 rich growth media using shake-flask cultures or lab-scale 1 L fermenters (Fig. 1b). In all cases, during growth the pH of the culture was controlled and maintained stably at 6.8 during fermentation. Supernatants were harvested at the end of their exponential phase and secreted peptides analyzed by SDS-PAGE and silver staining.
As noted previously [5], significant variation was observed in the overall protein profile of the spent growth media. Cells grown in Phage (Fig. 1b, lane 2) and Bennet (lane 7) media showed little or no secretion of CelA, moderate secretion was seen with the minimal media and TSB (lanes 3–6), while significant levels of secreted CelA were detectable in the spent NB and NB2X media (lanes 8 and 9, respectively). Recombinant CelA migrated with an apparent mass of ~ 26 kDa in SDS-PAGE gels in close agreement with the predicted size of its processed, signal-less mature domain (26 kDa) and represented ~ 50% of extracellular protein in the best producing medium (lane 8). The observed mass is suggestive of correct processing of the polypeptide and removal of the SPvsi as seen commonly with other proteins secreted in this system [1, 5]. The protein was stable in the spent growth medium at 4 °C for several days. In some cases, a minor cleavage was evident that had no effect on enzymatic activity (see below).
Kinetics and optimal conditions for CelA secretion in S. lividans
To determine optimal culture conditions and the phase that is productive for secretion, time-course experiments were performed under stably controlled pH (Fig. 2a). In NB, the amount of secreted CelA increased with time and was the highest observed yield of secreted CelA compared to other media (~ 40 mg/L) in 42 h (lanes 25 and 26). Optimal time and yields varied in the other media (Fig. 2a; Table 1), with two consistent observations: (a) biomass accumulation displayed completely different kinetics even when the different rich media were compared (Fig. 2b, d; Table 1) and yielded varying amounts of CelA at the end of the exponential phase (Fig. 2c, e and f). (b) Total endogenous protein secretion and specifically secretion of heterologous CelA, measured using dot-blots or western blots with an α-CelA antiserum, was anti-correlated with increased biomass production (panel d) and this is characteristically seen when CelA secretion from a unit of cell mass is compared (panel e). Contrary to previous observations [5, 26, 27], final CelA secretion yield was not significantly increased by addition of additional components of the NB medium, although the total yields per liter were enhanced (Fig. 2a, lanes 27–30; b, e and f). That is due to increase the amount of biomass produced in NB2X which means the amount of CelA secreted per unit of biomass decreases than NB which produce the less biomass.
Fig. 2.
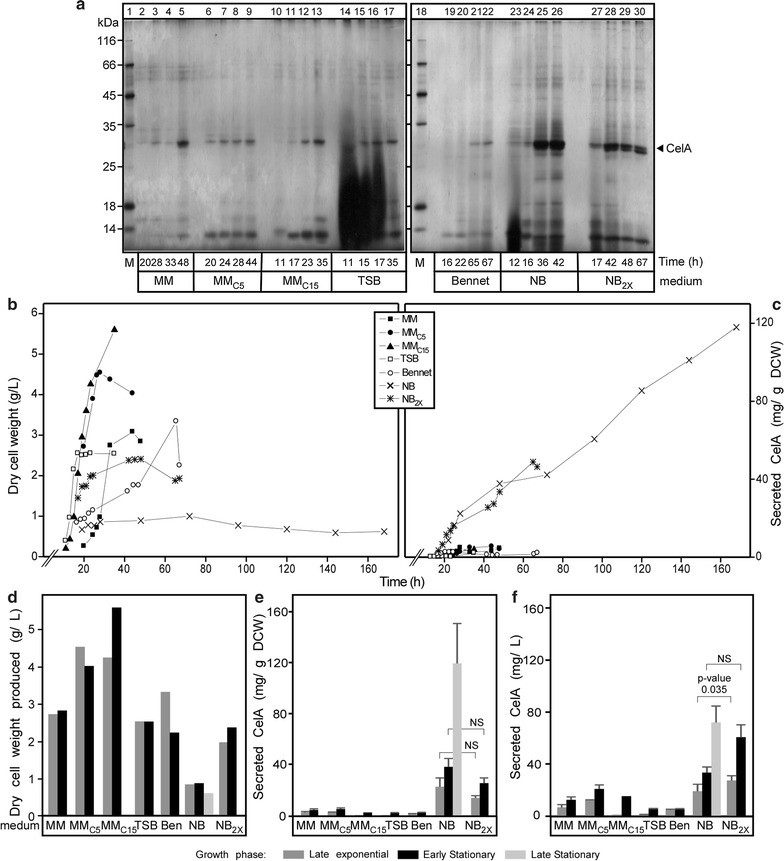
Time course of CelA secretion and the effect of additional carbon source. a Polypeptides (0.1–5 µg/lane) from culture supernatants (0.9–16 µL/lane) that is equivalent to (0.08 mg of dry cell mass) from cells grown for the indicated times in the indicated media were analyzed by SDS-PAGE and silver-stained. Lane 1, MW markers as in Fig. 1b. Secreted CelA (filled arrow) is indicated. b The growth curves of S. lividans TK24 carrying pIJ486 expressing SPVsi-CelA in the indicated media expressed as values of dry cell weight. c The dry cell weight in gram per liter produced by S. lividans TK24 with pIJ486 expressing SPVsi-CelA in the different media (as in Fig. 1b) at the indicated growth phases. Ben, Bennet medium. d The amount of CelA secreted (mg) correlated to a gram of dry cell mass by S. lividans TK24 with pIJ486 expressing SPVsi-CelA in the different media (as in Fig. 1b) for the indicated time related to its growth curves in the same media. e, f The amount of CelA secreted (mg) correlated to a gram of dry cell mass (e) or 1L of culture (f) produced by S. lividans TK24 with pIJ486 expressing SPVsi-CelA in the different media at late exponential, early stationary and late stationary phases from d difference in CelA secretion was compared using an unpaired t test without assumption of equal variance (see “Methods”; NS, non-significant difference). Error bars represent standard error of the mean (SEM). n = 3
Table 1.
Cellulase A production and yield in different fermentation media over different phases (exponential growth phase versus early stationary phase and late stationary phase)
| Medium | End of Exponential phase | Early stationary phase | Late stationary phase | ||||||
|---|---|---|---|---|---|---|---|---|---|
| CelA concentration (mg/L) |
Yield of CelA on biomass (mg/gDCW) |
Fermentation time (h) | CelA concentration (mg/L) |
Yield of CelA per unit of biomass (mg/gDCW) |
Fermentation time (h) |
CelA concentration (mg/L) |
Yield of CelA per unit of biomass (mg/gDCW) |
Fermentation time (h) |
|
| MM | 6.8 | 2.5 | 33 | 12.9 | 4.5 | 48 | – | – | – |
| MMC5 | 12.3 | 2.7 | 28 | 22.0 | 5.5 | 44 | – | – | – |
| MMC15 | 1.5 | 0.4 | 23.3 | 15.7 | 2.8 | 35 | – | – | – |
| TSB | 1.1 | 0.4 | 17 | 5.4 | 2.1 | 35 | – | – | – |
| Bennet | 4.9 | 1.5 | 65 | 4.9 | 2.2 | 67.2 | – | – | – |
| NB | 19.2* | 22.4** | 28 | 33.5* | 37.7** | 48 | 72.1 | 119.6 | 168 |
| NB2X | 27.5* | 13.9** | 23 | 60.8* | 25.2** | 42 | – | – | – |
The italics numbers indicate the difference in CelA amounts produced in NB and NB2x media
MM, minimal medium; C5 and C15, casamino acids (5 or 15 g/L); TSB, tryptic soy broth; NB, nutrient broth; NB2x, nutrient broth double strength; DCW, dry cell weight. n = 3
* The changes in CelA amount in (mg/L). The amount significantly increases with doubling the amount of media
** The changes in CelA amount in (mg/ g DCW). The amount of CelA significantly decreases with doubling the amount of the media
Effects of CelA secretion on the secretome
Time course kinetics experiments in different media (Fig. 2) suggested that the regulation of CelA production and secretion in S. lividans TK24 is complex. To gain further insight in the individual roles of the medium and of the heterologously expressed protein we carried out a detailed kinetic comparison between TK24 carrying either an empty vector or one expressing celA (Fig. 3). Growth of the two strains was practically identical, thus demonstrating that the NB medium has a fundamental influence on the cell directly, and this leads to acquisition of a stationary phase in ~ 30 h irrespective of whether SPVsi-CelA is produced or not. Moreover, the secretome profile was similar with some notable exceptions (Fig. 3b, marked with symbols). Even in the absence of SPVsi-CelA synthesis, secretome proteins followed obvious growth phase patterns with some coming up early, others late and others not changing. In contrast, SPVsi-CelA was synthesized and secreted very early on in very large amounts making it the most prominent protein of the secretome. Some endogenous secreted proteins showed reduced levels in SPVsi-CelA-producing cells (Fig. 3b, marked with “*”), while others disappeared from the secretome (Fig. 3b, open arrowhead). It is not known whether these proteins compete with SPVsi-CelA for export sites or factors or whether some more complex regulation is in effect.
Fig. 3.
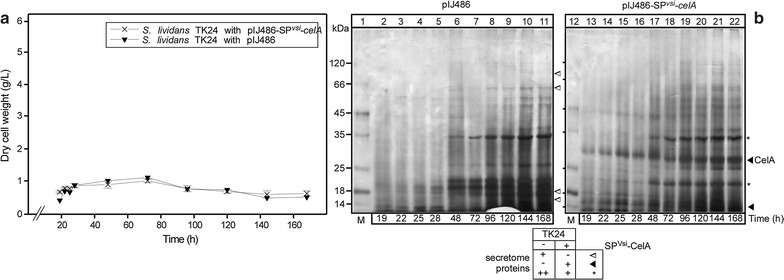
Time course of S. lividans TK24 with pIJ486-vsi-celA and S. lividans TK24 with pIJ486 (as a wild type) growth in NB medium. a The growth curves of S. lividans TK24 carrying empty pIJ486 or pIJ486 expressing SPVsi-CelA in the nutrient broth expressed as values of dry cell weight. b Polypeptides (1.2–6 µg/lane) from culture supernatants (~ 7 µL/lane; equivalent to 0.08 mg of dry cell weight) from cells grown for the indicated times in NB were analyzed by SDS-PAGE and silver-stained. Lanes 1 and 12, MW markers as in Fig. 1b. Secreted CelA (filled arrow) is indicated
Purification of CelA secreted from S. lividans
We next developed a simple 3-step purification scheme (Fig. 4; see “Methods”) for CelA secreted by S. lividans (lane 2). This involved concentration of spent growth media by rotary evaporation (lane 3) and a 25–55% ammonium sulfate fractionation (lane 4), followed by a Q-Sepharose column (lane 5), then heat denaturation at 80 °C (lane 6). More than 80% CelA could be recovered in the supernatant after ultra-centrifugation (lane 5). CelA was purified at more than 98% purity as judged by Coomassie-stained SDS-PAGE gels (lane 6).
Fig. 4.
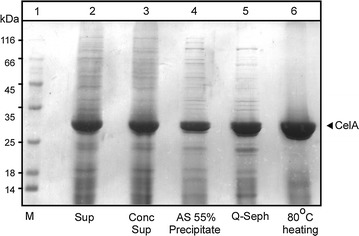
Large-scale purification of R. marinus CelA expressed and secreted in S. lividans culture supernatants. Protein samples (15 µg/lane) from the various purification steps (see “Methods”) were analyzed as in Fig. 2a and were stained with Coomassie blue. Lane 1: mass markers as in Fig. 1. Lane 2: culture supernatants from S. lividans TK24 with pIJ486 expressing SPVsi-CelA grown in NB for 48 h; lane 3: the culture supernatant after concentration with rotary evaporator; lane 4: The 35–55% ammonium sulfate fraction; lane 5: the elution fraction with the highest CelA activity from Q-Sepharose; lane 6: the fraction from lane 5 after heating at 80 °C for 4 h and centrifugal removal of precipitates
Physical and biochemical characterization of secreted recombinant CelA
To evaluate the quality of secreted CelA produced by S. lividans we determined whether it was structurally and functionally intact. Analytical size exclusion chromatography revealed that purified secreted CelA represents a monodisperse population without any visible signs of aggregation, with a native apparent molecular weight of ~ 27–30 kDa (Fig. 5a). Recombinant secreted CelA shows a main minimum in circular dichroism spectra at 213 nm, consistent with acquisition of extensive β-stranded secondary structure (Fig. 5b). It is a very stable enzyme with a non-measurable apparent T m during a temperature gradient of 15–95 °C, carried out by coincident monitoring of ellipticity by circular dichroism (Fig. 5c). This is in agreement with the thermal stability of the native protein’s enzymatic activity [23].
Fig. 5.
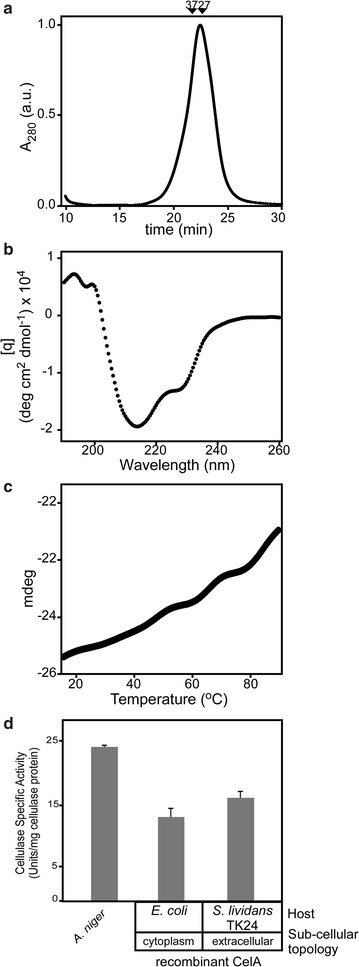
Physical and functional characterization of S. lividans-secreted CelA. a Size exclusion chromatography of recombinant purified CelA. Arrows indicate migration positions for 35 kDa (CesAB/EspA) and 27 kDa (CesAB) [39]. b Circular dichroism spectrometry. 5 µM or protein in buffer (5 mM MOPS pH 7.5, 5 mM NaCl, 1 mM DTT) 20 °C was analyzed using a 190–260 nm wavelength scan. c Thermal denaturation curves monitored by circular dichroism. Purified CelA (5 µM) in buffer (5 mM MOPS pH 7.5, 5 mM NaCl, 1 mM DTT) was exposed to gradual temperature rise and changes in ellipticity were monitored at 213 nm, as described [22]. d Biochemical activity of S. lividans-secreted CelA. Cellulase activity by CelA (20 µg/mL) was determined by hydrolysis of carboxymethyl cellulose (CMC) (see “Methods”). The activity of CelA secreted and purified from S. lividans was compared to CelAHis6 produced in E. coli and to a commercial preparation from A. niger of 24 U/mg total protein estimated to contain ~ 15 µg of unknown cellulases. n = 3; values represent mean ± SD
We next examined the catalytic activity of recombinant secreted CelA. The enzyme is highly active in hydrolyzing carboxymethyl cellulose at 70 °C. Despite the presence of secreted wild-type cellulases in TK24 (Additional file 1: Table S1), they displayed no detectable activities at this temperature. The catalytic properties of CelA are in agreement with what was previously determined for the native protein and for CelAHis6 expressed in E. coli [13]. The catalytic activity was compared to that of a crude commercial cellulase-containing A. niger secretome, measured at 25 °C as a qualitative control.
We conclude that CelA produced as a secreted polypeptide from S. lividans cells is structurally intact and enzymatically fully functional.
Discussion
We present a pilot scheme for lab-scale production and secretion of functional thermostable cellulase CelA by S. lividans. Use of this expression-secretion system was prompted on one hand by failure to establish satisfactory high-level expression of the enzyme using the traditional bacterial host E. coli and on the other by a desire to exploit the advantages of protein secretion from S. lividans in downstream processing. Use of S. lividans is an excellent alternative host strategy for protein production and production of CelA strengthens its specific application in protein secretion biotechnology.
CelA can be produced at high levels from S. lividans under specific fermentation regimes where the secreted form represents > 50% of the total protein present in the top performing spent growth media (Figs. 1b, 2). Rhodothermus marinus belongs to the Cytophaga–Flavobacterium–Bacteroides phylogenetic group and despite the evolutionary distance between the two organisms (S. lividans belongs to the Actinomycetales), high level expression of the R. marinus celA gene in S. lividans was not impeded. S. lividans also encodes and synthesizes multiple cellulase enzymes (Additional file 1: Table S1), like the highly homologous CelB2 [14], but these do not seem to be generally active under our growth and assay conditions and are not detected in spent growth media by mass spectrometry (Tsolis and Hamed, unpublished). Our previous attempts to secrete from S. lividans heterologous proteins that were carrying their own native signal peptides were unsuccessful [1] and so we systematically resort to using a native streptomycetal signal peptide for secretion. This reconfirms the importance of appropriate signal peptide mature domain combinations for optimal heterologous secretion as they are essential in regulating the cooperative binding of the preprotein to SecA as a bivalent ligand [7, 8, 22, 28].
A secretion yield of ~ 70 mg/L seen with small-scale cultures or lab-scale fermentors is among the best obtained to date from this host organism and may be further improved with bioprocess optimization. The best yields obtained to date include murine tumor necrosis factor alpha (80–120 mg/L) [5], xyloglucanase (100–150 mg/L) [1] and APA protein (alanine and proline-rich secreted protein) (80 mg/L) [29]. Significant amounts of CelA are observed after 7 days of culture and accumulated protein was observed after prolonged incubations suggesting that secreted CelA is generally proteolytically stable and remains unaffected by S. lividans secreted proteases. This was commonly observed for many heterologous proteins secreted by Streptomyces [2, 5, 30, 31] but not for others [32]. Addition of optimal carbohydrate amounts can lead to maximal product yields at reduced culture times [5]. The fermentation pattern we saw with CelA secretion was more complex (Fig. 2). Various carbon sources in the media caused reduction of CelA synthesis and/or secretion (Fig. 2). The expression/secretion of some secreted polypeptides was also affected, while that of others was not (Fig. 2a). Notably, optimal secretion was correlated with reduced media-specific growth (e.g. NB medium) and not seen with all other rich media. This suggests that certain components in one rich medium drive biomass production, while in other media, although rich, a medium component is in low abundance and presumably becomes quickly depleted. This prevents further biomass production and switches the cells to a stationary-phase state in which protein secretion is maximized. The medium components responsible, the genetic regulatory mechanisms that are responsible for switching and how protein secretion is linked to such stationary phase regulation remain unknown. We characteristically observe that certain polypeptides will begin production/secretion late into stationary phase (Fig. 3b), indicating that different levels of stationary phase control elements may exist. These findings may provide a first means to identifying the underlying metabolic connection to the secretome by using poly-omics approaches, currently under development. This and other studies [27, 33, 34] indicate that growth conditions may exert a degree of regulation of the secretory protein genes and/or the secretion pathway genes of Streptomyces. Such regulation is not known to exist in the E. coli system, where many Sec component genes are found in ribosomal operons.
It is evident that to further develop Streptomyces secretion biotechnology it is important to understand how different fermentation regimes affect protein secretion of native and heterologous proteins and this asks for a combination of transcriptomics, proteomics and fluxomics approaches. For this, the determination of the S. lividans TK24 genome [35], its comprehensive annotation and transcriptome determination (Busche et al. in preparation), the determination of the secreted proteome and its dynamics (Tsolis et al. in preparation), the determination of metabolomics tools [36] and the optimization of micro-scale culture arrays for robust biological reproducibility regimes [37] are important first steps. TK24 expressing CelA provides a suitable model to test these parameters.
The varying profiles of endogenous Streptomyces-secreted proteins under different growth conditions (Fig. 2a), makes the establishment of strict culture conditions very important for reproducibility of subsequent purification schemes. The production of highly-expressed endogenous S. lividans proteins such as the subtilisin inhibitor of ~ 14 kDa (Fig. 2a) could compete with secretion of the heterologous protein of interest and should be controlled either at the level of their synthesis or through genetic means (e.g. gene deletion).
CelA secreted by S. lividans is a stable polypeptide and is highly active in biochemical assays (Fig. 5d). Cellulose is one of the most abundant polysaccharides in nature and cellulases are important industrial enzymes with potential application in the feed industry, in the paper industry and for improving conversions in biofuel production [38]. The successful expression of this thermostable cellulase as a secretory protein from S. lividans described here is compatible with many of the established processes of industrial enzyme biotechnology and opens new areas for industrial application of cellulases.
The good secretion yield of CelA suggests that the S. lividans cell factory may be well suited for large-scale production of other thermostable polypeptides of industrial importance.
Authors’ contributions
HMB, KS, ÓS, BJSM, SK and TCK performed experiments and analyzed data, VML provided clones and maintained cells; GEE, HGO and AJ analyzed data; BK supervised fermentation work; FHO supervised cloning work; EA and KS conceived, managed and supervised the project and wrote the paper. All authors read and approved the final manuscript.
Acknowledgements
We thank colleagues of the StrepSynth consortium for useful discussions and T. Busche for sequencing pIJ486-TrueBlue. This study was supported by the European Union project (Grant QLK3-CT-2002-02056 to J.A., K.B., A.E. and O.F.) and by grants to A.E. (KU Leuven; KUL-Spa; Onderzoekstoelagen 2013; Bijzonder Onderzoeksfonds); FWO; RiMembR; Vlaanderen Onderzoeksprojecten; # G0C6814N). HMB is an Egyptian government doctoral scholar.
Competing interests
We confirm that we have read BioMed Central’s guidance on competing interests. The authors declare that they have no competing interests.
Availability of data and materials
All relevant data will be deposited in public repositories and all materials will be made available upon request.
Consent for publication
All authors of the manuscript have read and agreed to its content and are accountable for all aspects of the accuracy and integrity of the manuscript in accordance with ICMJE criteria.
The manuscript is original, has not already been published, and is not currently under consideration by another journal.
Ethics approval and consent to participate
Not applicable.
Funding
This study was supported by the European Union project (Grant QLK3-CT-2002-02056 to J.A., K.B., O.F. and A.E.) and by grants to A.E. (KU Leuven; KUL-Spa; Onderzoekstoelagen 2013; Bijzonder Onderzoeksfonds); FWO; RiMembR; Vlaanderen Onderzoeksprojecten; # G0C6814N). HMB is an Egyptian government doctoral scholar.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Additional file
Additional file 1: Figure S1. Amino acid sequence of cellulase A (CelA). Figure S2. pIJ486_Vsi-celA plasmid. Table S1. Annotated secreted cellulase-related hydrolases in the S. lividans TK24 proteome.
Footnotes
Electronic supplementary material
The online version of this article (10.1186/s12934-017-0847-x) contains supplementary material, which is available to authorized users.
Contributor Information
Mohamed Belal Hamed, Email: mohamedbelalhamed.soliman@student.kuleuven.be.
Spyridoula Karamanou, Email: lily.karamanou@KULEUVEN.BE.
Solveig Ólafsdottir, Email: solveig@matis.is.
Joana Sofia Martins Basílio, Email: joanabasilio22@gmail.com.
Kenneth Simoens, Email: kenneth.simoens@kuleuven.be.
Kostantinos C. Tsolis, Email: konstantinos.tsolis@kuleuven.be
Lieve Van Mellaert, Email: Lieve.Vanmellaert@kuleuven.be.
Eik Elísabet Guðmundsdóttir, Email: elisabet@matis.is.
Gudmundur Oli Hreggvidsson, Email: Gudmundur@matis.is.
Jozef Anné, Email: jozef.anne@kuleuven.be.
Kristel Bernaerts, Email: kristel.bernaerts@kuleuven.be.
Olafur H. Fridjonsson, Email: olafur@matis.is
Anastassios Economou, Email: tassos.economou@kuleuven.be.
References
- 1.Sianidis G, Pozidis C, Becker F, Vrancken K, Sjoeholm C, Karamanou S, Takamiya-Wik M, van Mellaert L, Schaefer T, Anne J, Economou A. Functional large-scale production of a novel Jonesia sp. xyloglucanase by heterologous secretion from Streptomyces lividans. J Biotechnol. 2006;121:498–507. doi: 10.1016/j.jbiotec.2005.08.002. [DOI] [PubMed] [Google Scholar]
- 2.Lara M, Servin-Gonzalez L, Singh M, Moreno C, Cohen I, Nimtz M, Espitia C. Expression, secretion, and glycosylation of the 45- and 47-kDa glycoprotein of Mycobacterium tuberculosis in Streptomyces lividans. Appl Environ Microbiol. 2004;70:679–685. doi: 10.1128/AEM.70.2.679-685.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ogino C, Kanemasu M, Hayashi Y, Kondo A, Shimizu N, Tokuyama S, Tahara Y, Kuroda S, Tanizawa K, Fukuda H. Over-expression system for secretory phospholipase D by Streptomyces lividans. Appl Microbiol Biotechnol. 2004;64:823–828. doi: 10.1007/s00253-003-1552-8. [DOI] [PubMed] [Google Scholar]
- 4.Hong B, Wu B, Li Y. Production of C-terminal amidated recombinant salmon calcitonin in Streptomyces lividans. Appl Biochem Biotechnol. 2003;110:113–123. doi: 10.1385/ABAB:110:2:113. [DOI] [PubMed] [Google Scholar]
- 5.Pozidis C, Lammertyn E, Politou AS, Anné J, Tsiftsoglou AS, Sianidis G, Economou A. Protein secretion biotechnology using Streptomyces lividans: large-scale production of functional trimeric tumor necrosis factor alpha. Biotechnol Bioeng. 2001;72:611–619. doi: 10.1002/1097-0290(20010320)72:6<611::AID-BIT1026>3.0.CO;2-0. [DOI] [PubMed] [Google Scholar]
- 6.Lammertyn E, Desmyter S, Schacht S, Van Mellaert L, Anne J. Influence of charge variation in the Streptomyces venezuelae alpha-amylase signal peptide on heterologous protein production by Streptomyces lividans. Appl Microbiol Biotechnol. 1998;49:424–430. doi: 10.1007/s002530051193. [DOI] [PubMed] [Google Scholar]
- 7.Lammertyn E, Anne J. Modifications of Streptomyces signal peptides and their effects on protein production and secretion. FEMS Microbiol Lett. 1998;160:1–10. doi: 10.1111/j.1574-6968.1998.tb12882.x. [DOI] [PubMed] [Google Scholar]
- 8.Anne J, Economou A, Bernaerts K. Protein secretion in Gram-positive bacteria: from multiple pathways to biotechnology. Curr Top Microbiol Immunol. 2017;404:267–308. doi: 10.1007/82_2016_49. [DOI] [PubMed] [Google Scholar]
- 9.Kieser T, Bibb MJ, Buttner MJ, Chater KF, Hopwood DA. Media, buffers and suppliers. In: John Innes Foundation, editor. Practical Streptomyces genetics. John Innes Foundation: Norwich; 2000. pp. 405–422. [Google Scholar]
- 10.Lammertyn E, Van Mellaert L, Schacht S, Dillen C, Sablon E, Van Broekhoven A, Anne J. Evaluation of a novel subtilisin inhibitor gene and mutant derivatives for the expression and secretion of mouse tumor necrosis factor alpha by Streptomyces lividans. Appl Environ Microbiol. 1997;63:1808–1813. doi: 10.1128/aem.63.5.1808-1813.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Noda S, Ito Y, Shimizu N, Tanaka T, Ogino C, Kondo A. Over-production of various secretory-form proteins in Streptomyces lividans. Protein Expr Purif. 2010;73:198–202. doi: 10.1016/j.pep.2010.05.011. [DOI] [PubMed] [Google Scholar]
- 12.Van Mellaert L, Lammertyn E, Schacht S, Proost P, Van Damme J, Wroblowski B, Anné J, Scarcez T, Sablon E, Raeymaeckers J, Van Broekhoven A. Molecular characterization of a novel subtilisin inhibitor protein produced by Streptomyces venezuelae CBS762.70. DNA Seq. 1998;9:19–30. doi: 10.3109/10425179809050021. [DOI] [PubMed] [Google Scholar]
- 13.Halldorsdottir S, Thorolfsdottir ET, Spilliaert R, Johansson M, Thorbjarnardottir SH, Palsdottir A, Hreggvidsson GO, Kristjansson JK, Holst O, Eggertsson G. Cloning, sequencing and overexpression of a Rhodothermus marinus gene encoding a thermostable cellulase of glycosyl hydrolase family 12. Appl Microbiol Biotechnol. 1998;49:277–284. doi: 10.1007/s002530051169. [DOI] [PubMed] [Google Scholar]
- 14.Crennell SJ, Hreggvidsson GO, Nordberg Karlsson E. The structure of Rhodothermus marinus Cel12A, a highly thermostable family 12 endoglucanase, at 1.8 A resolution. J Mol Biol. 2002;320:883–897. doi: 10.1016/S0022-2836(02)00446-1. [DOI] [PubMed] [Google Scholar]
- 15.Crennell SJ, Cook D, Minns A, Svergun D, Andersen RL, Nordberg Karlsson E. Dimerisation and an increase in active site aromatic groups as adaptations to high temperatures: X-ray solution scattering and substrate-bound crystal structures of Rhodothermus marinus endoglucanase Cel12A. J Mol Biol. 2006;356:57–71. doi: 10.1016/j.jmb.2005.11.004. [DOI] [PubMed] [Google Scholar]
- 16.Ausubel FM, Brent R, Kingston RE, Moore DD, Smith JA, Seidman JG, Struhl K. Current protocols in molecular biology. New York: Wiley; 1994. [Google Scholar]
- 17.Ward JM, Janssen GR, Kieser T, Bibb MJ, Buttner MJ. Construction and characterisation of a series of multi-copy promoter-probe plasmid vectors for Streptomyces using the aminoglycoside phosphotransferase gene from Tn5 as indicator. Mol Gen Genet. 1986;203:468–478. doi: 10.1007/BF00422072. [DOI] [PubMed] [Google Scholar]
- 18.Liu YC, Vidali L. Efficient polyethylene glycol (PEG) mediated transformation of the moss Physcomitrella patens. J Vis Exp. 2011;50:2560–2564. doi: 10.3791/2560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Korn F, Weingärtner B, Kutzner HJ. A study of twenty actinophages: morphology, serological relationship and host range. In: Freerksen E, Tarnak I, Thumin JH, editors. Genetics of Actinomycetales. Stuttgart: Fisher G; 1978. pp. 251–270. [Google Scholar]
- 20.Gouridis G, Karamanou S, Sardis MF, Scharer MA, Capitani G, Economou A. Quaternary dynamics of the SecA motor drive translocase catalysis. Mol Cell. 2013;52:655–666. doi: 10.1016/j.molcel.2013.10.036. [DOI] [PubMed] [Google Scholar]
- 21.Vrontou E, Karamanou S, Baud C, Sianidis G, Economou A. Global co-ordination of protein translocation by the SecA IRA1 switch. J Biol Chem. 2004;279:22490–22497. doi: 10.1074/jbc.M401008200. [DOI] [PubMed] [Google Scholar]
- 22.Chatzi KE, Sardis MF, Tsirigotaki A, Koukaki M, Šoštarić N, Konijnenberg A, Sobott F, Kalodimos CG, Karamanou S, Economou A. Preprotein mature domains contain translocase targeting signals that are essential for secretion. J Cell Biol. 2017;216:1357–1369. doi: 10.1083/jcb.201609022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hreggvidsson GO, Kaiste E, Holst O, Kristjansson JK. An extremely thermostable cellulase from the thermophilic eubacterium Rhodothermus marinus. Appl Environ Microbiol. 1996;62:3047–3049. [Google Scholar]
- 24.Ghose TK. Measurement of cellulase activities. In: Pure and applied chemistry, vol. 59. p. 2571987: 257.
- 25.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Ser B (Methodol) 1995;57:289–300. [Google Scholar]
- 26.Parro V, Mellado RP. Effect of glucose on agarase overproduction by Streptomyces. Gene. 1994;145:49–55. doi: 10.1016/0378-1119(94)90321-2. [DOI] [PubMed] [Google Scholar]
- 27.Kim E, Shin DH, Irwin DC, Wilson DB. Fed-batch production of Thermomonospora fusca endoglucanase by recombinant Streptomyces lividans. Biotechnol Bioeng. 1998;60:70–76. doi: 10.1002/(SICI)1097-0290(19981005)60:1<70::AID-BIT8>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 28.Sardis MF, Tsirigotaki A, Chatzi KE, Portaliou GA, Gouridis G, Karamanou S, Economou A. Preprotein conformational dynamics drive bivalent translocase docking and secretion. Structure. 2017;25:1056–67.e6. doi: 10.1016/j.str.2017.05.012. [DOI] [PubMed] [Google Scholar]
- 29.Vallin C, Ramos A, Pimienta E, Rodriguez C, Hernandez T, Hernandez I, Del Sol R, Rosabal G, Van Mellaert L, Anne J. Streptomyces as host for recombinant production of Mycobacterium tuberculosis proteins. Tuberculosis (Edinb) 2006;86:198–202. doi: 10.1016/j.tube.2006.02.003. [DOI] [PubMed] [Google Scholar]
- 30.Pimienta E, Fando R, Sanchez JC, Vallin C. Secretion of human interferon alpha 2b by Streptomyces lividans. Appl Microbiol Biotechnol. 2002;58:189–194. doi: 10.1007/s00253-001-0873-8. [DOI] [PubMed] [Google Scholar]
- 31.Karamanou S, Gouridis G, Papanikou E, Sianidis G, Gelis I, Keramisanou D, Vrontou E, Kalodimos CG, Economou A. Preprotein-controlled catalysis in the helicase motor of SecA. EMBO J. 2007;26:2904–2914. doi: 10.1038/sj.emboj.7601721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.DelaCruz N, Payne GF, Smith JM, Coppella SJ. Bioprocess development to improve foreign protein production from recombinant Streptomyces. Biotechnol Prog. 1992;8:307–315. doi: 10.1021/bp00016a007. [DOI] [PubMed] [Google Scholar]
- 33.Fernandez-Abalos JM, Ruiz-Arribas A, Garda AL, Santamaria RI. Effect of carbon source on the expression of celA1, a cellulase-encoding gene from Streptomyces halstedii JM8. FEMS Microbiol Lett. 1997;153:97–103. doi: 10.1016/S0378-1097(97)00241-3. [DOI] [PubMed] [Google Scholar]
- 34.Harchand RK, Singh S. Catabolite repression of cellulase biosynthesis in Streptomyces albaduncus. J Basic Microbiol. 1994;34:371–378. doi: 10.1002/jobm.3620340603. [DOI] [PubMed] [Google Scholar]
- 35.Ruckert C, Albersmeier A, Busche T, Jaenicke S, Winkler A, Friethjonsson OH, Hreggviethsson GO, Lambert C, Badcock D, Bernaerts K, et al. Complete genome sequence of Streptomyces lividans TK24. J Biotechnol. 2015;199:21–22. doi: 10.1016/j.jbiotec.2015.02.004. [DOI] [PubMed] [Google Scholar]
- 36.Muhamadali H, Xu Y, Ellis DI, Trivedi DK, Rattray NJ, Bernaerts K, Goodacre R. Metabolomics investigation of recombinant mTNFalpha production in Streptomyces lividans. Microb Cell Fact. 2015;14:157. doi: 10.1186/s12934-015-0350-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Koepff J, Keller M, Tsolis CK, Busche T, Rückert C, Hamed MB, Anne J, Kalinowski J, Wiechert W, Economou A, Oldiges M. Fast and reliable strain characterization of Streptomyces lividans through micro-scale cultivation. Biotechnol Bioeng. 2016;114(9):2011–2022. doi: 10.1002/bit.26321. [DOI] [PubMed] [Google Scholar]
- 38.Kuhad RC, Gupta R, Singh A. Microbial cellulases and their industrial applications. Enzyme Res. 2011;2011:1–10. doi: 10.4061/2011/280696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen L, Balabanidou V, Remeta DP, Minetti CA, Portaliou AG, Economou A, Kalodimos CG. Structural instability tuning as a regulatory mechanism in protein–protein interactions. Mol Cell. 2011;44:734–744. doi: 10.1016/j.molcel.2011.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All relevant data will be deposited in public repositories and all materials will be made available upon request.


