Abstract
Insulin-like growth factor 2 receptor (IGF2R) is overexpressed in activated Hepatic stellate cells (HSCs) and therefore can be utilized for HSC-specific drug delivery. We recently discovered an IGF2R-specific peptide using a novel biopanning. Here, we adopted biotin-conjugated IGF2R-specific peptide, cholesterol, and vitamin A as the targeting ligands for the neutravidin-based siRNA nanocomplex to deliver PCBP2 siRNA, a potentially antifibrotic agent, to HSCs. Compared to Vitamin A and cholesterol, the IGF2R-specific peptide exhibited the highest targeting effect to human LX-2 HSC, rat HSC-T6 cell line, and activated primary rat HSCs. Accordingly, the IGF2R-specific peptide coupled nanocomplex demonstrated higher silencing activity of PCBP2 and better inhibition on the migration of activated HSCs. Compared to free siRNA and the nanocomplexes coupled with Vitamin A and cholesterol, the IGF2R-specific peptide coupled nanocomplex showed the highest uptake in the liver and lowest uptake in the lung and kidney of the rats with CCl4-induced liver fibrosis.
Keywords: Liver fibrosis, siRNA, nanocomplex, IGF2R, phage, peptide ligand, vitamin A
Graphical abstract
The IGF2R-specific peptide, cholesterol, and vitamin A are used as the targeting ligands for the neutravidin-based siRNA nanocomplex to deliver PCBP2 siRNA to fibrotic liver. Compared to free siRNA and the nanocomplexes coupled with Vitamin A (SNVP) and cholesterol (SNCP), the IGF2R-specific peptide coupled nanocomplex (SNPP) showed the highest uptake in the liver and lowest uptake in the lung and kidney of the rats with CCl4-induced liver fibrosis. IGF2R-specific peptide modified nanocomplex is therefore a promising delivery platform for antifibrotic siRNAs
BACKGROUND
Liver fibrosis is a wound healing process characterized by the accumulation of excess extracellular matrix (ECM) in the liver. It is induced by chronic liver injuries caused by nonalcoholic steatohepatitis, hepatitis, alcohol abuse, and metal poisoning.1,2 If left untreated, liver fibrosis will develop into liver cirrhosis, which is irreversible and affects nearly 633,233 adults in the United States.3
The expression of ECM increases dramatically when quiescent hepatic stellate cells (HSCs) are activated to myofibroblast-like cells.1,4–6 Although HSCs only constitute approximately 5–8% of total liver cells7, they are the major contributors for liver fibrosis and are able to cover the entire microcirculatory network of hepatic sinusoidal.8 Type I collagen is the most abundant protein in ECM, and its abnormal accumulation is primarily due to the increased half-life of its collagen α1(I) mRNA.9,10 We recently discovered an siRNA to silence the poly(rC) binding protein 2 (PCBP2) gene in HSCs to inhibit the expression of αCP2 protein, which is responsible for stabilization of the collagen α1(I) mRNA. Silencing of the PCBP2 gene reduced the expression of type I collagen in activated HSCs.11 We also developed an avidin-based siRNA nanocomplex for the PCBP2 siRNA and discovered that neutravidin is the best avidin analogue for siRNA delivery.12,13
Targeted delivery of antifibrotic agents to HSCs is a major challenge in liver fibrosis therapy. Therapeutic agents cannot easily reach HSCs because of the excessive accumulation of ECM, the closure of endothelial fenestrae, and the reduced flow exchange between sinusoid blood and liver cells.14 The delivery of antifibrotic agents to HSCs is also limited by the reduced perisinusoidal space (or space of Disse).15 One promising strategy to improve the delivery of antifibrotic agents to HSCs is to modify drug carriers with a specific ligand that binds to a moiety on activated HSCs.12,16,17 Three targeting moieties, including cellular-binding protein type I (CRBP-1), low-density lipoprotein receptor (LDLR) and insulin-like growth factor 2 receptor (IGF2R) have been studied for HSC-specific drug delivery. IGF2R, also known as mannose-6-phosphate receptor (M6PR), is a 300k Da glycoprotein that plays a critical role in the insulin-like growth signaling system. IGF2R contains three domains, including an extracellular domain, a transmembrane domain and a cytoplasmic domain18. Because its expression is upregulated during liver fibrogenesis,19,20 IGF2R has been utilized as a targeting moiety for HSC-specific drug delivery. For example, M6P was used as a ligand to deliver a triplex-forming oligonucleotides to fibrotic liver16. We recently discovered an IGF2R-specific peptide, peptide-431, using a novel combinatorial biopanning strategy. Peptide-431 and its dimeric form showed high and specific affinity to activated human and rat HSCs.20 In this study, dimeric peptide-431 was used as a targeting ligand of the siRNA nanocomplex to specifically deliver the nanocomplex to activated HSCs in fibrotic liver. Compared to free siRNA and the nanocomplexes coupled with vitamin A and cholesterol, the nanocomplex coupled with the IGF2R-specific peptide exhibited the highest silencing activity of PCBP2 and the best inhibition on the migration of activated HSCs. The IGF2R-specific peptide coupled nanocomplex also showed the highest uptake in the liver of the rats with CCl4-induced liver fibrosis.
METHODS
Materials
PCBP2 and scramble siRNAs were ordered from Invitrogen (Carlsbad, CA) and Shanghai Genepharm (Shanghai, China). Neutravidin, and BCA protein assay kit were obtained from Pierce (Rockford, IL). Protamine sulfate (salmon X grade), all-trans-retinoic acid and anti-β actin antibody were purchased from Sigma-Aldrich (St. Louis, MO). GelRed was purchased from Biotium (Hayward, CA). Anti-IGF2R antibody and anti-α-SMA antibody were purchased from R&D system (Minneapolis, MN). Biotin-PEG-amine and biotin-PEG-acid were purchased from BroadPharm (San Diego, CA). Lipofectamine® RNAiMAX, cell culture media, and all other chemical reagents were purchased from Fisher Scientific (Grand Land, NY).
Synthesis of Biotin-PEG-Ligands
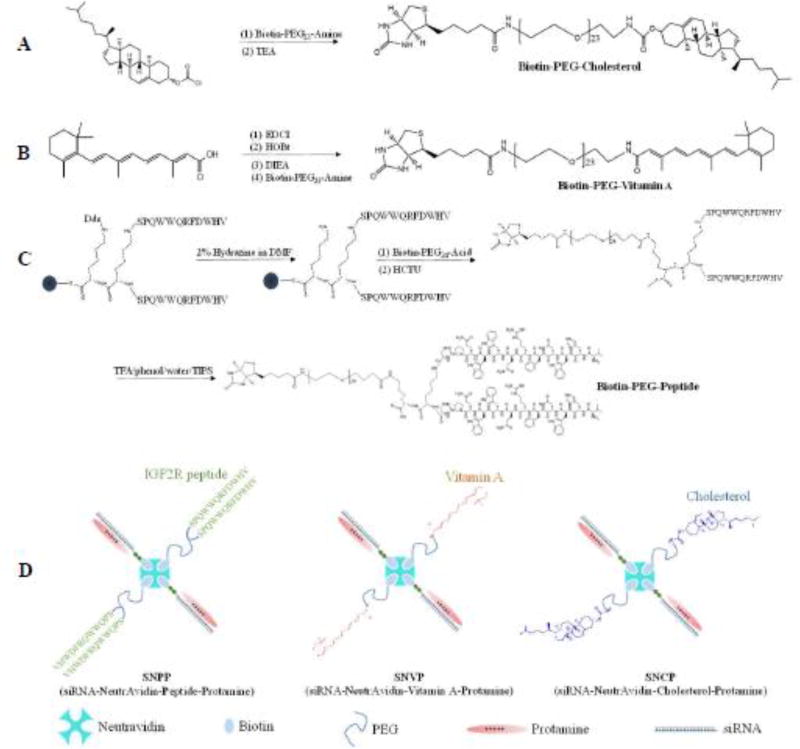
Biotin was conjugated to various ligands (IGF2R-specific peptide, vitamin A, and cholesterol) via a low molecular weight (1 kDa) polyethylene glycol (PEG) linker. The reaction schemes were presented in Figure 1 (A, B, C).
Figure 1. The synthesis and fabrication schemes of (A) biotin-conjugated cholesterol, (B) biotin-conjugated vitamin A, (C) biotin-conjugated IGF2R-speific peptide, and (D) the neutravidin-based siRNA nanocomplex.
Biotin-conjugated PCBP2 siRNA, neutravidin and biotin-conjugated ligands were mixed in a 2:1:2 ratio at room temperature to form the siRNA-neutravidin-ligand complex, followed by condensation with protamine to form the final nanocomplex
The biotin-PEG-IGF2R peptide was synthesized using solid phase peptide synthesis. Briefly, the N-(1-(4,4-dimethyl-2,6-dioxocyclohexylidene)ethyl) (Dde) protecting group in the dimeric peptide-431 (0.1 mmole) was removed by washing with 2% hydrazine in N,N-dimethylformamide (DMF) for 3 times, each for 10 minutes. Next, 2-(6-Chlor-1H-benzotriazol-1-yl)-1,1,3,3-tetramethylaminium-hexafluorophosphat (HCTU) (0.2 mmole) and biotin-PEG-acid (0.2 mmole) were added, and the solution was stirred continuously at room temperature for 24 h. After completion of the reaction, the peptide was cleaved from Wang resin with TFA/phenol/water/TIPS (88/5/5/2) at room temperature for 2 h. The biotin-PEG-peptide product was then purified by HPLC, and the molecular weight (Mw:5015.1) was confirmed by Matrix Assisted Laser Desorption/Ionization Time of Flight (MALDI-TOF).
For biotin-PEG-cholesterol, biotin-PEG-amine (0.12 mmole) and cholesterol chloroformate (0.1 mmole) were dissolved in 2 mL Dichloromethane (DCM) and stirred continuously at room temperature for 24 h. Next, 0.2 mmole Triethylamine (TEA) was added, and the reaction was continued at room temperature for another 24 h. The biotin-PEG-cholesterol product was purified by silica gel column, and its molecular weight (Mw:1729.1) was confirmed by MALDI-TOF.
For biotin-PEG-vitamin A, all-trans-retinoic acid (0.2 mmole) were dissolved in 2 mL DCM with 0.3 mmole 1-Ethyl-3-(3-dimethylaminopropyl) carbodiimide (EDC), 0.3mmole Hydroxybenzotriazole (HOBt) and 0.3 mmole N, N-Diisopropylethylamine (DIEA). The solution was stirred continuously at room temperature for 24 h. After adding 0.1 mmole biotin-PEG-amine, the reaction was continued for another 24 h, and the final product was purified by silica gel column. The molecular weight (Mw:1582.0) was confirmed by mass spectrometry (MS).
Fabrication and Characterization of the Neutravidin-based siRNA Nanocomplex
Biotin-conjugated PCBP2 siRNA (sense strand sequence: 5’-GUCAGUGUGGCUCUC UUAUtt-3’) was purchased from Shanghai Genepharm (Shanghai, China). Biotin was linked to the 3’ end of the sense strand via a disulfide linker. Biotin-siRNA, neutravidin and biotin-conjugated ligands were mixed in a 2:1:2 ratio at room temperature for 10 min to form the siRNA-neutravidin-IGF2R peptide (SNP) complex, the siRNA-neutravidin-cholesterol (SNC) complex, and the siRNA-neutravidin-vitamin A (SNV) complex. These complexes were then condensed with protamine at different N/P ratios (1:1. 2:1, 5:1 and 10:1) at room temperature for 30 min to form the multicomponent siRNA-nuetriavidin-peptide-protamine nanocomplex (SNPP), siRNA-neutravidin-cholesterol-protamine nanocomplex (SNCP), and siRNA-neutravidin-vitamin A-protamine nanocomplex (SNVP).
Formation of the nanocomplexes were confirmed using a gel retardation assay. Briefly, 10 µL of the nanocomplex was separated on 20% native polyacrylamide gel electrophoresis (PAGE) and visualized with GelRed under UV light. Particle size and zeta potential of the nanocomplexes were measured in HEPES buffer (pH 7.4) using a Malvern Zetasizer Nano-ZS (Malvern Instruments, Westborough, MA).
Serum Stability of the Nanocomplex
The nanocomplexes were incubated with 50% rat serum at 37°C for various time intervals. Half of the samples were separated by a 20 % native PAGE gel and visualized with GelRed™. To demonstrate that the siRNAs encapsulated inside the nanocomplex are still intact, another half of the samples were incubated with 40 µM heparin and 100 mM DTT for 10 min to dissociate free siRNA from the nanocomplex.21 Dissociated siRNA samples were electrophoresed in a 20% native PAGE and visualized with GelRed™.
Cytotoxicity Study of the Nanocomplex
Cytotoxicity of the sRNA nanocomplex in HSC-T6 cells was evaluated using MTT assay as described.22,23 HSC-T6 cells seeded in 96-well plate (2500 cells/well) were incubated with SNCP, SNVP and SNPP nanocomplexes at 37 °C for 24 and 48h, followed by adding MTT to measure cytotoxicity.
Cell Culture
The rat hepatic stellate cell line HSC-T6 and human hepatic stellate cell line LX-2 were kindly provided by Dr. Scott L. Friedman (Mount Sinai School of Medicine, New York University) and cultured as previously reported.20
Primary rat HSCs were isolated as we reported before.24,25 The animal protocol was approved by the Institutional Animal Care and Use Committee (IACUC), University of Missouri-Kansas City. Primary rat HSCs were cultured in DMEM with 10% FBS, 100 units/mL penicillin, and 100 µg/mL streptomycin.
Cellular Uptake of the siRNA Nanocomplex
Cellular uptake of the nanocomplexes were evaluated in HSC-T6, LX-2 and primary rat HSCs (the 5th and 16th passage generations) using flow cytometry and confocal microscopy. The 5’ end of the siRNA antisense strand was labeled with Alex Flour 647. The cells were incubated with the nanocomplexes at 37 °C for various time intervals and then washed with Opti-MEM containing 1mg/mL heparin to remove nonspecifically bound nanocomplexes.
Following heparin treatment, the cells were detached with 0.25% trypsin, washed, suspended, and subjected to fluorescence analysis using a BD FACS II flow cytometer (BD instruments, NJ). For confocal analysis, the cells were stained with 150 nM LysoTracker, fixed with 10% formalin, and examined under a confocal microscope (Leica TCS SP5, Germany).
Silencing Activity of the siRNA Nanocomplex
HSC-T6 cells, primary rat HSCs (the 5th and 16th passage generations) were transfected with the siRNA nanocomplex as described.13 SNCP, SNVP and SNPP nanocomplexes were prepared as described above with the N/P ratio of 2:1 and incubated with the cells at a final concentration of 50 nM siRNA. After incubation for 6 h, the medium was replaced with DMEM with 10% FBS, and the cells were harvested 48 h post-transfection. Silencing activity at the protein level was quantitated using western blot assay as reported.11,22
Invasion Assay
The invasion assay was performed as described with modifications.26,27 Briefly, matrigel (50 µg/mL) and type I collagen (50 µg/mL) were coated on the top and bottom, respectively, of transwell chambers. HSC-6 cells were transfected with the siRNA nanocomplexes at a concentration of 50 nM siRNA for 24 h, followed by incubation in DMEM with 0.5% FBS (for serum starvation) and 100 mM alcohol for another 24 h. The cells were detached, resuspended in DMEM with 0.5% FBS, and loaded into the transwell chambers at a density of 30,000 cells/chamber. The wells were filled with DMEM containing 10% FBS as attractant. After 4 h invasion, the migrated cells on the bottom were fixed, stained, and counted at 100× magnification under a microscope.
In-vivo Biodistribution Study
The animal protocol was approved by the Institutional Animal Care and Use Committee (IACUC) at the University of Missouri-Kansas City. Male Sprague Dawley rats were housed in a humidity and temperature controlled room with a 12 h light–dark cycle. Liver fibrosis was induced by intraperitoneal injection with the mixture of carbon tetrachloride (CCl4) and olive oil (1:1, v/v) at a dose of 1 mL/kg CCl4 twice a week for five consecutive weeks. The rats were then randomly divided into four groups and intravenously injected with Cy5-labeled siRNA or the siRNA nanocomplexes encapsulating Cy5-labeled siRNA at a dose of 0.1 mg/Kg. After 2 h, the rats were euthanized, and major organs including the liver, lungs, spleen, kidneys, heart, muscle (thigh), and blood were harvested for imaging analysis using a Bruker MS FX PRO In vivo Imaging System (Billerica, MA).
Statistical Analysis
All data were presented as the mean ± standard deviation. Statistical analysis was performed using two-way analysis of variance (ANOVA) with Tukey’s post hoc test. P<0.05 was considered statistically significant.
RESULTS
Fabrication and Characterization of the Neutravidin-based siRNA Nanocomplexes
The nanocomplexes were fabricated as illustrated in Figure 1D. Biotin-PEG-peptide-431 and biotin-labeled siRNA were mixed with neutravidin in a 2:2:1 molar ratio to form the siRNA-neutravidin-peptide complex by noncovalent neutravidin-biotin interaction, followed by condensation with protamine at different N/P ratios to form the SNPP nanocomplex. Similarly, SNCP and SNVP nanocomplexes coupled with cholesterol and vitamin A, respectively, were fabricated using the same procedure. In our previous studies, we developed an avidin-based siRNA nanocomplex at the N/P ratio 10:1, but it showed somewhat non-specific accumulation in the lung.21 This could be due to its high positive charge (+18 mV).28 Moreover, high positive charge of nanoparticles may cause significant systemic toxicity.29 Therefore, we aimed to reduce the N/P ratio of the nanocomplex to minimize its non-specific accumulation in the lung.
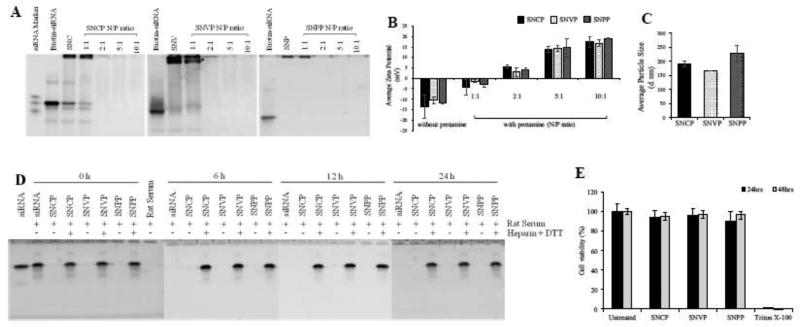
We evaluated the complexation and condensation of the siRNA nanocomplexes using gel retardation assay. As illustrated in Figure 2A, complexation of the biotin-labeled siRNA, neutravidin, and biotin-labeled ligands shifted siRNA bands, indicating a complete complexation of the siNRA, neutravidin, and biotin-labeled ligands. Condensation of the siRNA-neutravidin-ligand complex with protamine at high N/P ratios (2:1 to 10:1) completely shielded the siRNA from staining, suggesting complete encapsulation of siRNA inside the nanocomplex. Zeta potential of the siRNA-neutravidin-ligand complexes was negative, while zeta potential of the nanocomplexes was slightly positive (+4mV) at the N/P ratio 2:1 (Figure 2B). This is consistent with the gel retardation results in Figure 2A. All the three nanocomplexes, SNCP, SNVP, and SNPP, showed similar results in complexation and zeta potential, suggesting that different targeting ligands do not affect the fabrication of the s nanocomplexes. Based on these results, N/P ratio of 2:1 was selected for the neutravidin-based siRNA nanocomplex for subsequent studies.
Figure 2. Characterization and silencing activity of the nanocomplex.
(A) Gel retardation assay of the SNCP, SNVP and SNPP nanocomplexes at different N/P ratios. (B) Zeta potential of the nanocomplexes with different N/P ratios. (C) Particle size of the nanocomplexes. (SNC: siRNA-neutravidin-cholesterol complex; SNV: siRNA-neutravidin- vitamin A complex; SNP: siRNA-neutravidin-peptide-431 complex). (D) Serum Stability of the nanocomplex in 50% rat serum for 0, 6, 12, and 24h. (E) Cytotoxicity study of the nanocomplexes.
Particle size of the SNCP, SNVP and SNPP nanocomplexes was 191, 167 and 228 nm, respectively, and the polydispersity index (PDI) values were 0.132, 0.114 and 0.249, respectively (Figure 2C). The relatively large particle size of the SNPP nanocomplex is possibly because of the higher molecular weight of the peptide ligand compared to cholesterol and vitamin A.
Serum Stability and Cytotoxicity of the Neutravidin-Based siRNA Nanocomplex
Serum stability of the nanocomplexes was examined in 50% rat serum. As Figure 2D showed, free siRNA was rapidly degraded in the serum, and no siRNA was detected after 6 h. Consistent with the results in Figure 2A, the nanocomplexes shielded siRNA from GelRed staining at all time intervals, indicating a good stability of the nanocomplexes in the serum up to 24 h. Treatment of the nanocomplexes with heparin and DTT released intact siRNA even after 24 h incubation in the serum. This data revealed that the neutravidin-based nanocomplexes can efficiently protect siRNA from degradation in the serum. Moreover, MTT assay demonstrated that the SNCP, SNVP and SNPP nanocomplexes do not induce cytotoxicity in HSC-T6 cells (Figure 2E).
Cellular Uptake
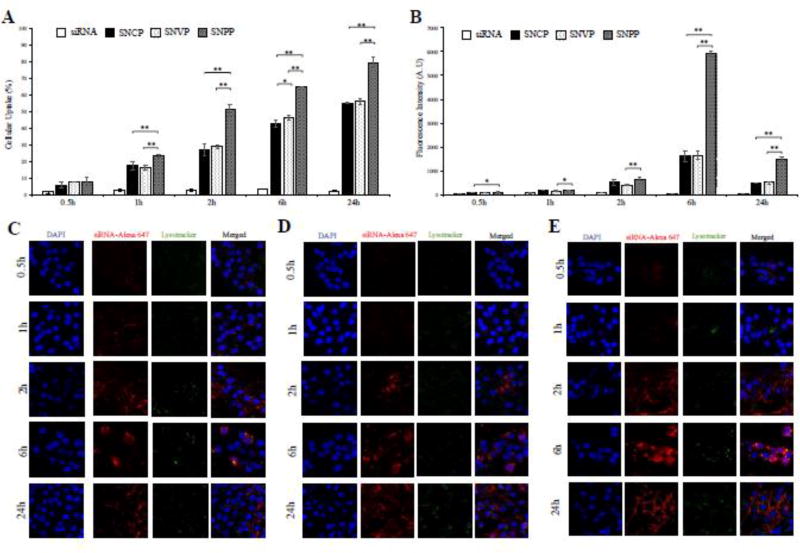
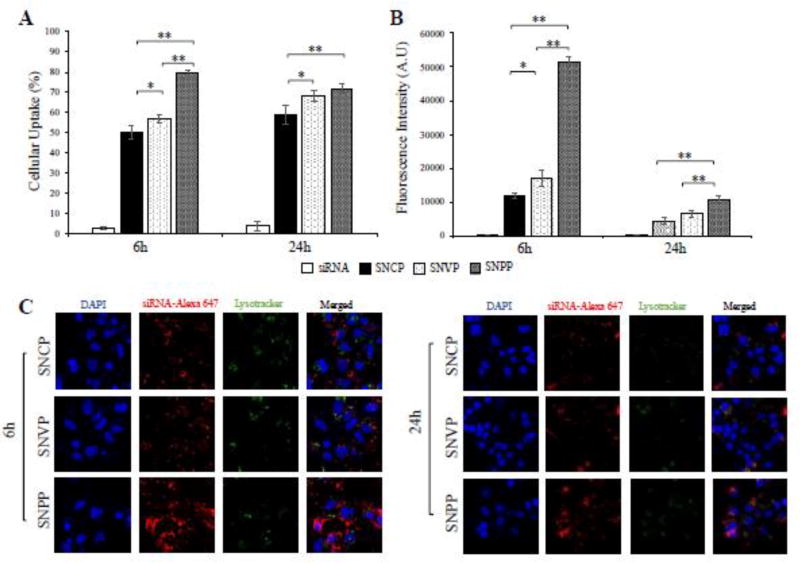
We next examined cellular uptake of the nanocomplexes in HSC-T6 cells. Flow cytometry was used to determine the percentage of cells that take up the nanocomplex (Figure 3A) and intensity of the siRNA inside the cells (Figure 3B). Compared to free siRNA, all the three nanocomplexes demonstrated substantially higher uptake in HSC-T6 cells. This is in accordance with our previous findings that neutravidin-based nanocomplexes can efficiently deliver siRNA to HSCs.21 In addition, the results indicated that targeting ligand plays an important role in the cellular uptake of the nanocomplex. Compared to nanocomplexes modified with cholesterol (SNCP) and vitamin A (SNVP), the nanocomplex modified with the IGF2R-specific peptide (SNPP) showed the highest cellular uptake, suggesting a high affinity of the IGF2R-specific peptide to HSCs. Particularly, the fluorescence intensity of the Alexa Fluor 647-labeled siRNA in SNPP transfected cells is approximately 3.6 folds higher than that in SNCP and SNVP transfected cells.
Figure 3. Quantitative cellular uptake of SNCP, SNVP and SNPP nanocomplexes in HSC-T6 cells.
siRNA was labeled with Alexa Flour 647 for fluorescence analysis using flow cytometry (A, B) and confocal microscopy (C, D, E). (A) Percent of the cells that take up the nanocomplexes. (B) Fluorescence intensity of the cells that take up the nanocomplexes. Confocal images of the cells treated with SNCP (C), SNVP (D) and SNPP (E) nanocomplexes at various time intervals. All results are presented as the mean ± SD (n=3). (*P<0.05; **P<0.01).
Subsequently, confocal microscopy was used to compare intracellular distribution of these nanocomplexes in HSC-T6 cells (Figure 3C, D, E). The results are consistent with the flow cytometry results, in which SNPP showed the highest cellular uptake. Moreover, the confocal images showed that all the three nanocomplexes efficiently deliver the siRNA into the cytoplasm with minimal entrapment inside lysosomes, indicating their distinctive capabilities to escape endosomes.
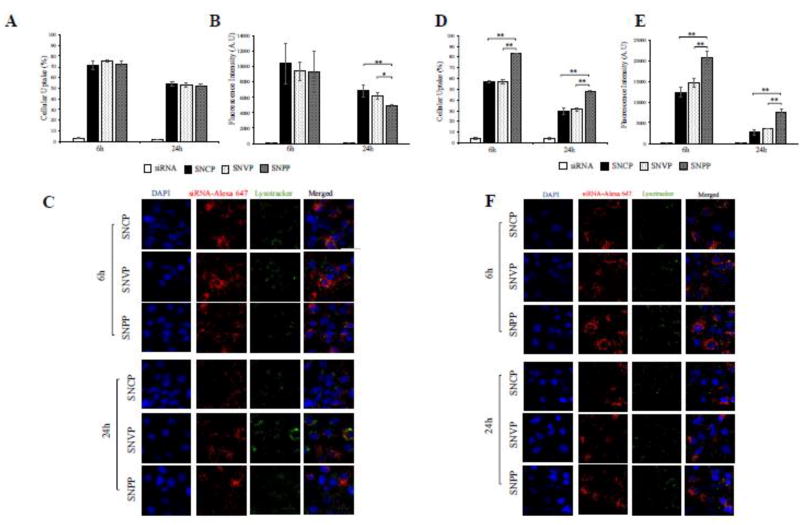
We subsequently examined cellular uptake of these nanocomplexes in quiescent primary rat HSCs (5th passage generation) and activated primary rat HSC cells (16th passage generation). As illustrated in quiescent primary rat HSCs (Figure 4 A, B, and C), SNPP nanocomplex exhibited similar uptake as SNCP and SNVP nanocomplexes. By contrast, SNPP nanocomplex showed the highest uptake in activated primary rat HSCs (Figure 4 D, E, F). These results indicated that the high uptake of SNPP nanocomplex is mediated by the overexpressed IGF2R in activated HSCs.
Figure 4.
Cellular uptake of the nanocomplexes in quiescent and activated primary rat HSCs. Cellular uptake of the SNCP, SNVP and SNPP nanocomplexes containing Alexa Flour 647 labeled siRNA were evaluated in quiescent primary rat HSCs at the 5th passage generation (A, B, and C) and activated primary rat HSCs at the 16th passage generation (D, E, and F). (A, D) Percent of the cells that take up the nanocomplexes; (B, E) Fluorescence intensity of the cells that take up the nanocomplexes; (C, F) Confocal images of the cells. All results are presented as the mean ± SD (n=3). (*P<0.05; **P<0.01).
To explore the potential applications of SNPP nanocomplex in the future for human patients, we also evaluated the uptake of these nanocomplexes in a spontaneously immortalized human hepatic human HSC cell line LX-2. As shown in Figure 6 A, after incubation for 6 and 24 h, SNPP nanocomplex demonstrated the highest cellular uptake compared to SNCP and SNVP nanocomplexes. More importantly, SNPP nanocomplex exhibited approximately 3.04-fold and 4.3-fold increase of the siRNA fluorescence intensity in the cells compared to SNCP and SNVP, respectively, at 6 h (Figure 6B). Confocal microscopy (Figure 6C) revealed the same results as flow cytometry in Figure 6A and 6B.
Figure 6. Cellular uptake of the nanocomplexes in human HSC LX-2 cells.
The SNCP, SNVP and SNPP nanocomplexes were incubated with LX-2 cells for 6 and 24h. (A) Percent of the cells that take up the nanocomplexes. (B) Fluorescence intensity of the cells that take up the nanocomplexes; (C) Confocal images of the cells. All results are presented as the mean ± SD (n=3). (*P<0.05; **P<0.01).
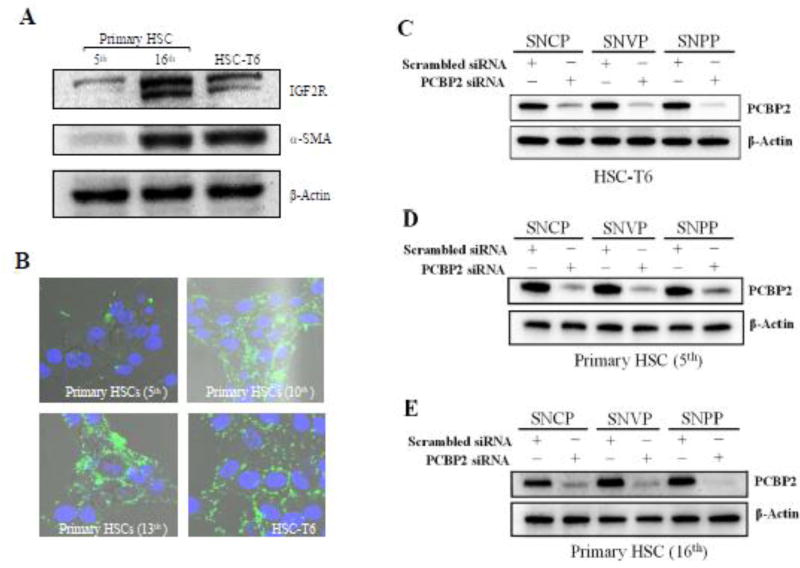
HSC-T6 is an activated HSC cell line with fibroblast-like phenotype and proliferates rapidly in cell culture.30 We hypothesized that the high uptake of SNPP nanocomplex in HSC-T6 is mainly mediated by IGF2R. We tested this hypothesis by conducting a similar cellular uptake study in quiescent and activated primary rat HSCs. Quiescent primary rat HSCs were activated by continuous passaging in cell culture. We first quantified the expression of IGF2R in primary rat HSCs at different passages. As Figure 5A showed, the expression of IGF2R and α-SMA, a marker for activated fibroblasts, in primary rat HSCs increased with the number of passaging. The primary rat HSCs at the 16th passage generation exhibited similar expression of α-SMA and higher expression of IGF2R as compared to HSC-T6 cells. Accordingly, FAM-labeled IGF2R-specific peptide-431 demonstrated higher uptake in primary rat HSCs at high passaging numbers (Figure 5B). Also, cellular uptake of the FAM-labeled peptide-431 in activated primary rat HSCs is comparable to that in HSC-T6 cells.
Figure 5. Cellular uptake and silencing activity of the nanocomplexes in activated primary rat HSCs.
(A) Quiescent primary rat HSCs were activated by continuous passaging in cell culture. The protein expressions of IGR2R and α-SMA in primary rat HSCs (the 5th and 16th passage generations) were detected using western blot. (B) Cellular uptake of Fam-labeled peptide-431 (1 µM) in primary rat HSCs after different number of passages. HSC-T6 was used as a control. (C, D, E) Silencing activity of the nanocomplexes in HSC-T6, quiescent primary rat HSCs, and activated primary rat HSCs at the protein level.
Silencing Activity
In accordance with the cellular uptake results in Figures 3 and 4, the nanocomplexes exhibited significant silencing activity at the protein level in HSC-T6 and primary rat HSCs (Figure 5 C, D, E). Compared to SNCP and SNAP, SNPP exhibited higher silencing activity in activated HSCs, such as HSC-T6 and primary rat HSCs (16th passage generations), suggesting the important role of IGF2R in the uptake of SNPP.
Invasion Assay
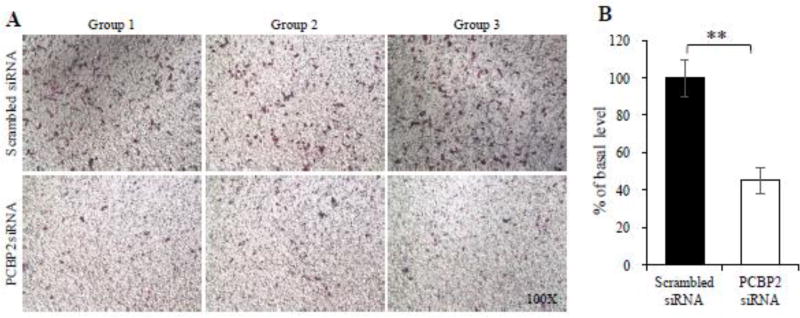
Having shown the high cellular uptake of the SNPP nanocomplex in activated HSCs, we next wanted to demonstrate its antifibrotic activity in HSCs. During liver fibrogenesis, various fibrogenic cytokines, such as TGF-β1, PDGF, and VEGF, activate HSCs and increase their migratory behavior.31,32 Alcohol abuse is another stimuli that can cause chronic liver injuries and subsequently result in liver fibrosis.4 We recently demonstrated that alcohol stimulation increased the migration capability of HSCs, and transfection of PCBP2 siRNA can efficiently reverse the migration of HSCs.22 To determine whether the SNPP nanocomplex loaded with PCBP2 siRNA inhibits the migration of HSCs, we transfected HSC-T6 cells with SNPP for 24 h and then incubated the cells with 100mM alcohol for another 24 h. As shown in Figure 7, SNPP nanocomplex inhibited approximately 55% of the invasion of the HSC-T6 cells. This result proved that SNPP nanocomplex can efficiently deliver siRNA to HSCs and subsequently exert its biological activity in the cells.
Figure 7. SNPP nanocomplex containing PCBP2 siRNA inhibits the migration effect of alcohol on HSC-T6.
HSC-T6 cells were transfected with the SNPP nanocomplex at a concentration of 50 nM siRNA for 24 h, followed by stimulation with 100 mM alcohol for another 24 h. The cells were then harvested for migration assay using transwell chambers. All results are presented as the mean ± SD (n=3). (**P<0.01).
In-vivo biodistribution
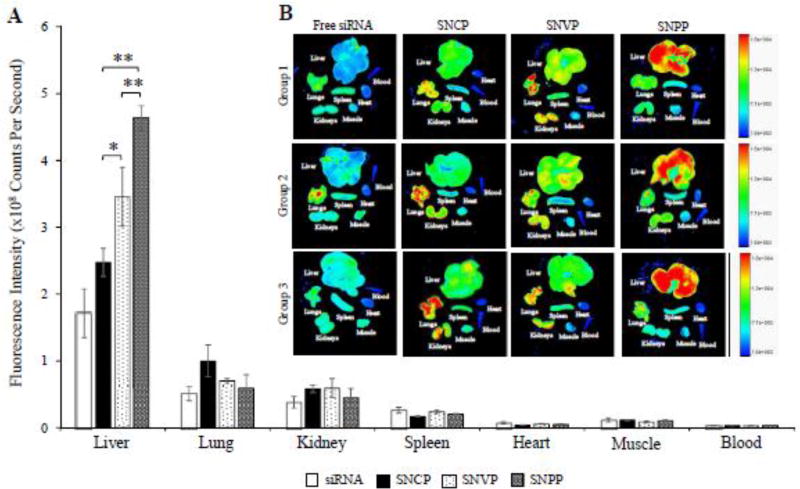
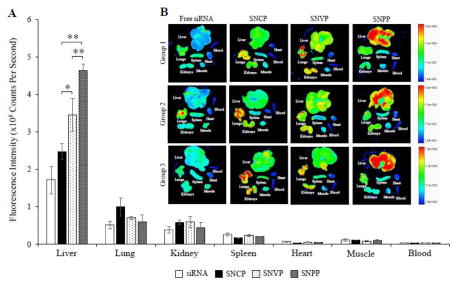
Hepatic uptake of antifibrotic agents in vivo is always a challenge because of the extensive accumulation of collagen and dramatic changes of sinusoids during liver fibrogenesis.24,25 As a result, it is of utmost importance to evaluate biodistribution of the nanocomplexes in rats with CCl4-induced liver fibrosis, which is the most widely used animal model for liver fibrosis study.4,33 The nanocomplexes were prepared with Cy5-labeled PCBP2 siRNA for fluorescence analysis. Two hours after systemic administration of the nanocomplexes, the rats were euthanized, and major organs, including the liver, lungs, kidneys, spleen, muscle, and blood were harvested for fluorescence analysis using a small animal imaging system. As illustrated in Figure 8, free siRNA was eliminated rapidly and showed low accumulation in the fibrotic liver. Compared to free siRNA, all the three nanocomplexes showed higher uptake in the liver. This is because of two reasons: the protection of siRNA by the nanocomplex, and the targeting effect of cholesterol, vitamin A, and peptide-431 for their corresponding receptors in HSCs. Consistent with the in vitro cellular uptake results in Figures 3, 5, and 6, SNPP nanocomplex showed the highest liver uptake compared to SNCP and SNVP nanocomplexes. More importantly, SNPP nanocomplex showed negligible accumulation in the lung and kidney, in which SNCP and SNVP nanocomplexes showed non-specific accumulations. Taken together, these data provided compelling evidence that the SNPP nanocomplex could be a promising carrier to specifically deliver antifibrotic siRNAs to HSCs in vitro and in vivo.
Figure 8. Biodistribution of the SNCP, SNVP and SNPP nanocomplexes in rats with CCl4-induced liver fibrotic.
Fluorescence images of the major organs from three rats were presented. Region of interest (ROI) for each organ was determined by the Bruker molecular imaging software, and fluorescence intensities with respect to the area under the ROI were plotted. All results are presented as the mean ± SD (n=3). (*P<0.05; **P<0.01).
DISCUSSIONS
In our previous studies, we prepared an avidin-based siRNA nanocomplex with an N/P ratio of 10:1. The siRNA nanocomplex showed somewhat non-specific accumulation in the lung,21 which could be due to its high positive charge (+18 mV).28 In addition, high positive charge of a carrier may cause significant systemic toxicity.29 We therefore optimized the N/P ratio and selected 2:1 as the best ratio to form the siRNA nanocomplex with a slightly positive charge (Figure 2B). By reducing the N/P ratio from 10:1 to 2:1, the amount of protamine in the nanocomplex was also reduced from 66% to 28%, leading to a higher loading of the siRNA in the nanocomplex.
Another improvement in the current siRNA nanocomplex is the incorporation of PEG (Figure 1). PEG has long been used to prolong the blood circulation time and minimize systemic toxicity of a drug carrier.34 Unmodified nanoparticles tend to accumulate in the reticuloendothelial system (RES), particularly in the Kupffer cells of the liver.35,36 For example, PEGylation of an oligonucleotide enhanced its antifibrotic activity by avoiding capture by Kupffer cells and subsequently accumulating in HSCs.37 In our study, a 1 kDa PEG was used as a linker between biotin and ligands to increase binding affinity to HSCs, prolong circulation time, and avoid nonspecific uptake by the RES.
The major challenge in liver fibrosis therapy is how to specifically deliver antifibrotic agents to HSCs in vivo, which are the major contributors for liver fibrosis but only constitute approximately 5–8% of total liver cells.7 Various receptors including LDLR, CRBP-1, and IGF2R have been exploited to enhance drug delivery to HSCs. We recently discovered an IGF2R-specific peptide with a high specificity and affinity to HSCs. The objective of this study is to compare the cellular uptake and activity of the siRNA nanocomplexes coupled with three different ligands, including cholesterol, vitamin A and the IGF2R-specific peptide (Figure 1). Despite having different ligands, the nanocomplexes exhibited similar complexation, zeta potential, particle size and serum stability (Figures 2). This clearly indicated the robustness and flexibility of the neutravidin-based siRNA nanocomplex for various ligands and applications.
The in vitro cellular uptake and activity studies revealed that the neutravidin-based nanocomplex is an efficient platform to deliver siRNA to HSCs and exert its biological activity (Figures 3–7). This is consistent with our previous studies using streptavidin-based siRNA nanocomplex.12,13 Targeting ligand also plays an important role in the cellular uptake of the nanocomplex. The nanocomplexes coupled with cholesterol and vitamin A exhibited similar uptake in activated human and rat HSCs, while the nanocomplex coupled with the IGF2R-specific peptide showed the highest cellular uptake (Figure 3). However, the nanocomplexes with different ligands exhibited similar cellular uptake in quiescent rat primary HSCs (Figure 4A). This could be explained by the low expression of IGF2R in quiescent HSCs but upregulated expression of IGF2R in activated HSCs (Figure 5A, 5B). The expression of IGF2R could be increased by ~20 folds in activated HSCs compared to quiescent HSCs.38 Moreover, approximately 16% of the IGF2R are located on the cell membrane surface, and IGF2R-mediated endocytosis is 3 times faster in activated HSCs compared to quiescent HSCs.38 All these characteristics of IGF2R contribute to its important role in HSC-specific drug delivery.
Similarly, in vivo biodistribution study revealed that IGF2R-specific peptide is the best ligand to deliver the siRNA nanocomplex to the fibrotic liver of rats with CCl4-induced liver fibrosis (Figure 8). SNPP nanocomplex showed significantly high uptake in the liver and very low uptake in other organs, indicating a high specificity of the nanocomplex to the liver. This result also provides compelling evidence that the IGF2R-specific peptide is highly specific to activated HSCs in vivo. By contrast, SNCP and SNVP nanocomplexes exhibited low uptake in the liver but high uptake in the lung and kidney.
Cholesterol has long been used as a ligand to enhance liver uptake because of the high expression of its receptors, LDLR and SR-B1, in liver cells including HSCs, hepatocytes, and Kupffer cells.12,39,40 However, the nanocomplex coupled with cholesterol showed the highest uptake in the lung of the rats with liver fibrosis. This could be due to the fact that the lung can act as a uptake organ for cholesterol.41 In addition, the physiologic changes during liver fibrogenesis also limited hepatic uptake of the nanocomplex coupled with cholesterol. On the other hand, cholesterol’s receptors, LDLR and SR-BI, are not HSC-specific and also expressed in hepatocytes and Kupffer cells, thus limiting its specificity to HSCs.24,39
Approximately 50–80% of vitamin A in the body is stored as retinyl palmitate in lipid droplets in HSCs, and they are taken up through CRBP-1.30,42 Vitamin A has therefore been utilized as a targeting ligand to deliver therapeutic agents to HSCs. For example, Sato et al. developed vitamin A coupled liposome to deliver gp46 siRNA to fibrotic liver.17 However, the expression of CRBP-1 in HSCs is downregulated during liver fibrogenesis.30,43,44 As a result, HSCs lose retinyl lipid droplets during the process of fibrosis by exocytosis.44,45 This is possibly the major reason for the moderate liver uptake of the vitamin A modified nanocomplex in the rats with liver fibrosis (Figure 8). On the other hand, other retinol-binding proteins also exist in hepatocytes and serum,46–48 which may limit the specificity of vitamin A to HSCs. Our biodistribution study revealed moderate uptake of the vitamin A coupled nanocomplex in the liver but also nonspecific uptake in the lung and kidney (Figure 8). This could be due to the expression of retinol-binding proteins in these organs. For example, lipid droplet-containing stellate cells were identified in the lung and kidney of rats.49
In this study, we demonstrated that IGF2R-specific peptide can efficiently deliver the siRNA nanocomplex to activated HSCs in vitro. Compared to nanocomplexes coupled with cholesterol and vitamin A, the IGF2R-specific peptide coupled nanocomplex showed the highest uptake in the liver and lowest uptake in the lung and kidney of the rats with CCl4-induced liver fibrosis. The IGF2R-specific peptide modified nanocomplex is therefore a promising delivery platform for antifibrotic siRNAs.
Acknowledgments
This work was supported by an award (1R01AA021510) from the National Institutes of Health.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Friedman SL. Liver fibrosis -- from bench to bedside. J Hepatol. 2003;38(Suppl 1):S38–53. doi: 10.1016/s0168-8278(02)00429-4. [DOI] [PubMed] [Google Scholar]
- 2.Lee YA, Wallace MC, Friedman SL. Pathobiology of liver fibrosis: a translational success story. Gut. 2015;64:830–841. doi: 10.1136/gutjnl-2014-306842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Scaglione S, et al. The Epidemiology of Cirrhosis in the United States: A Population-based Study. J Clin Gastroenterol. 2015;49:690–696. doi: 10.1097/MCG.0000000000000208. [DOI] [PubMed] [Google Scholar]
- 4.Cheng K, Mahato RI. Gene modulation for treating liver fibrosis. Crit Rev Ther Drug Carrier Syst. 2007;24:93–146. doi: 10.1615/critrevtherdrugcarriersyst.v24.i2.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Raghow R. The role of extracellular matrix in postinflammatory wound healing and fibrosis. FASEB J. 1994;8:823–831. doi: 10.1096/fasebj.8.11.8070631. [DOI] [PubMed] [Google Scholar]
- 6.Senoo H, Imai K, Matano Y, Sato M. Molecular mechanisms in the reversible regulation of morphology, proliferation and collagen metabolism in hepatic stellate cells by the three-dimensional structure of the extracellular matrix. J Gastroenterol Hepatol. 1998;13(Suppl):S19–32. doi: 10.1111/jgh.1998.13.s1.19. [DOI] [PubMed] [Google Scholar]
- 7.Geerts A. History, heterogeneity, developmental biology, and functions of quiescent hepatic stellate cells. Semin Liver Dis. 2001;21:311–335. doi: 10.1055/s-2001-17550. [DOI] [PubMed] [Google Scholar]
- 8.Marra F, Pinzani M. Role of hepatic stellate cells in the pathogenesis of portal hypertension. Nefrologia. 2002;22(Suppl 5):34–40. [PubMed] [Google Scholar]
- 9.Lindquist JN, Stefanovic B, Brenner DA. Regulation of collagen alpha1(I) expression in hepatic stellate cells. J Gastroenterol. 2000;35(Suppl 12):80–83. [PubMed] [Google Scholar]
- 10.Sato M, Suzuki S, Senoo H. Hepatic stellate cells: unique characteristics in cell biology and phenotype. Cell Struct Funct. 2003;28:105–112. doi: 10.1247/csf.28.105. [DOI] [PubMed] [Google Scholar]
- 11.Shukla RS, Qin B, Wan YJ, Cheng K. PCBP2 siRNA reverses the alcohol-induced pro-fibrogenic effects in hepatic stellate cells. Pharm Res. 2011;28:3058–3068. doi: 10.1007/s11095-011-0475-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shukla RS, Tai W, Mahato R, Jin W, Cheng K. Development of streptavidin-based nanocomplex for siRNA delivery. Mol Pharm. 2013;10:4534–4545. doi: 10.1021/mp400355q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shukla RS, Jain A, Zhao Z, Cheng K. Intracellular trafficking and exocytosis of a multi-component siRNA nanocomplex. Nanomedicine : nanotechnology, biology, and medicine. 2016;12:1323–1334. doi: 10.1016/j.nano.2016.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Garcia-Banuelos J, et al. Cirrhotic rat livers with extensive fibrosis can be safely transduced with clinical-grade adenoviral vectors. Evidence of cirrhosis reversion. Gene Ther. 2002;9:127–134. doi: 10.1038/sj.gt.3301647. [DOI] [PubMed] [Google Scholar]
- 15.Varin F, Huet PM. Hepatic microcirculation in the perfused cirrhotic rat liver. J Clin Invest. 1985;76:1904–1912. doi: 10.1172/JCI112186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ye Z, Cheng K, Guntaka RV, Mahato RI. Receptor-mediated hepatic uptake of M6P-BSA-conjugated triplex-forming oligonucleotides in rats. Bioconjug Chem. 2006;17:823–830. doi: 10.1021/bc060006z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sato Y, et al. Resolution of liver cirrhosis using vitamin A-coupled liposomes to deliver siRNA against a collagen-specific chaperone. Nat Biotechnol. 2008;26:431–442. doi: 10.1038/nbt1396. [DOI] [PubMed] [Google Scholar]
- 18.Morgan DO, et al. Insulin-like growth factor II receptor as a multifunctional binding protein. Nature. 1987;329:301–307. doi: 10.1038/329301a0. [DOI] [PubMed] [Google Scholar]
- 19.de Bleser PJ, et al. Insulinlike growth factor-II/mannose 6-phosphate receptor is expressed on CCl4-exposed rat fat-storing cells and facilitates activation of latent transforming growth factor-beta in cocultures with sinusoidal endothelial cells. Hepatology. 1995;21:1429–1437. doi: 10.1002/hep.1840210529. [DOI] [PubMed] [Google Scholar]
- 20.Chen Z, Jin W, Liu H, Zhao Z, Cheng K. Discovery of Peptide ligands for hepatic stellate cells using phage display. Mol Pharm. 2015;12:2180–2188. doi: 10.1021/acs.molpharmaceut.5b00177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jain A, Barve A, Zhao Z, Jin W, Cheng K. Comparison of Avidin, Neutravidin, and Streptavidin as Nanocarriers for Efficient siRNA Delivery. Mol Pharm. 2017 doi: 10.1021/acs.molpharmaceut.6b00933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.liu H, et al. Silencing of alphaCP2 reverses the alcohol- and cytokine-induced fibrogenesis in hepatic stellate cells. Liver Research. 2017 doi: 10.1016/j.livres.2017.05.003. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Barve A, Jain A, Liu H, Jin W, Cheng K. An enzyme-responsive conjugate improves the delivery of a PI3K inhibitor to prostate cancer. Nanomedicine : nanotechnology, biology, and medicine. 2016;12:2373–2381. doi: 10.1016/j.nano.2016.07.007. [DOI] [PubMed] [Google Scholar]
- 24.Cheng K, Ye Z, Guntaka RV, Mahato RI. Biodistribution and hepatic uptake of triplex-forming oligonucleotides against type alpha1(I) collagen gene promoter in normal and fibrotic rats. Mol Pharm. 2005;2:206–217. doi: 10.1021/mp050012x. [DOI] [PubMed] [Google Scholar]
- 25.Cheng K, Ye Z, Guntaka RV, Mahato RI. Enhanced hepatic uptake and bioactivity of type alpha1(I) collagen gene promoter-specific triplex-forming oligonucleotides after conjugation with cholesterol. J Pharmacol Exp Ther. 2006;317:797–805. doi: 10.1124/jpet.105.100347. [DOI] [PubMed] [Google Scholar]
- 26.Mahato R, Qin B, Cheng K. Blocking IKKalpha expression inhibits prostate cancer invasiveness. Pharm Res. 2011;28:1357–1369. doi: 10.1007/s11095-010-0351-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Qin B, Cheng K. Silencing of the IKKepsilon gene by siRNA inhibits invasiveness and growth of breast cancer cells. Breast Cancer Res. 2010;12:R74. doi: 10.1186/bcr2644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Blanco E, Shen H, Ferrari M. Principles of nanoparticle design for overcoming biological barriers to drug delivery. Nature biotechnology. 2015;33:941–951. doi: 10.1038/nbt.3330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Naha PC, Davoren M, Lyng FM, Byrne HJ. Reactive oxygen species (ROS) induced cytokine production and cytotoxicity of PAMAM dendrimers in J774A.1 cells. Toxicol Appl Pharmacol. 2010;246:91–99. doi: 10.1016/j.taap.2010.04.014. [DOI] [PubMed] [Google Scholar]
- 30.Vogel S, et al. An immortalized rat liver stellate cell line (HSC-T6): a new cell model for the study of retinoid metabolism in vitro. J Lipid Res. 2000;41:882–893. [PubMed] [Google Scholar]
- 31.Yang C, et al. Liver fibrosis: insights into migration of hepatic stellate cells in response to extracellular matrix and growth factors. Gastroenterology. 2003;124:147–159. doi: 10.1053/gast.2003.50012. [DOI] [PubMed] [Google Scholar]
- 32.Benyon RC, Arthur MJ. Mechanisms of hepatic fibrosis. J Pediatr Gastroenterol Nutr. 1998;27:75–85. doi: 10.1097/00005176-199807000-00013. [DOI] [PubMed] [Google Scholar]
- 33.Crespo Yanguas S, et al. Experimental models of liver fibrosis. Arch Toxicol. 2016;90:1025–1048. doi: 10.1007/s00204-015-1543-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Harris JM, Chess RB. Effect of pegylation on pharmaceuticals. Nat Rev Drug Discov. 2003;2:214–221. doi: 10.1038/nrd1033. [DOI] [PubMed] [Google Scholar]
- 35.Moghimi SM, Szebeni J. Stealth liposomes and long circulating nanoparticles: critical issues in pharmacokinetics, opsonization and protein-binding properties. Prog Lipid Res. 2003;42:463–478. doi: 10.1016/s0163-7827(03)00033-x. [DOI] [PubMed] [Google Scholar]
- 36.Tseng YC, Mozumdar S, Huang L. Lipid-based systemic delivery of siRNA. Adv Drug Deliv Rev. 2009;61:721–731. doi: 10.1016/j.addr.2009.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bonora GM, Ivanova E, Zarytova V, Burcovich B, Veronese FM. Synthesis and characterization of high-molecular mass polyethylene glycol-conjugated oligonucleotides. Bioconjug Chem. 1997;8:793–797. doi: 10.1021/bc970082p. [DOI] [PubMed] [Google Scholar]
- 38.Mousavi SA, Fonhus MS, Kindberg GM, Tolleshaug H, Berg T. Enhanced activity of lysosomal proteases in activated rat hepatic stellate cells is associated with a concomitant increase in the number of the mannose-6-phosphate/insulin-like growth factor II receptor. Cell Biol Int. 2013;37:703–712. doi: 10.1002/cbin.10085. [DOI] [PubMed] [Google Scholar]
- 39.Wolfrum C, et al. Mechanisms and optimization of in vivo delivery of lipophilic siRNAs. Nat Biotechnol. 2007;25:1149–1157. doi: 10.1038/nbt1339. [DOI] [PubMed] [Google Scholar]
- 40.Bijsterbosch MK, et al. bis-Cholesteryl-conjugated phosphorothioate oligodeoxynucleotides are highly selectively taken up by the liver. J Pharmacol Exp Ther. 2002;302:619–626. doi: 10.1124/jpet.302.2.619. [DOI] [PubMed] [Google Scholar]
- 41.Townsend RW, Zutshi A, Bekersky I. Biodistribution of 4-[(14)C]cholesterol-AmBisome following a single intravenous administration to rats. Drug Metab Dispos. 2001;29:681–685. [PubMed] [Google Scholar]
- 42.Higashi N, et al. Vitamin A storage in hepatic stellate cells in the regenerating rat liver: with special reference to zonal heterogeneity. Anat Rec A Discov Mol Cell Evol Biol. 2005;286:899–907. doi: 10.1002/ar.a.20230. [DOI] [PubMed] [Google Scholar]
- 43.Motoyama H, et al. Cytoglobin is expressed in hepatic stellate cells, but not in myofibroblasts, in normal and fibrotic human liver. Lab Invest. 2014;94:192–207. doi: 10.1038/labinvest.2013.135. [DOI] [PubMed] [Google Scholar]
- 44.Senoo H, et al. Hepatic stellate cell (vitamin A-storing cell) and its relative--past, present and future. Cell Biol Int. 2010;34:1247–1272. doi: 10.1042/CBI20100321. [DOI] [PubMed] [Google Scholar]
- 45.Senoo H, Mezaki Y, Fujiwara M. The stellate cell system (vitamin A-storing cell system) Anat Sci Int. 2017 doi: 10.1007/s12565-017-0395-9. [DOI] [PubMed] [Google Scholar]
- 46.Tabesh M, et al. Association of retinol-binding protein 4 with metabolic syndrome in first-degree relatives of type 2 diabetic patients. J Res Med Sci. 2017;22:28. doi: 10.4103/1735-1995.200270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Domingos MAM, Queiroz M, Lotufo PA, Bensenor IJ, Titan SMO. Serum RBP4 and CKD: Association with insulin resistance and lipids. J Diabetes Complications. 2017 doi: 10.1016/j.jdiacomp.2017.04.013. [DOI] [PubMed] [Google Scholar]
- 48.Yamaaki N, et al. Impact of serum retinol-binding protein 4 levels on regulation of remnant-like particles triglyceride in type 2 diabetes mellitus. J Diabetes Res. 2013;2013:143515. doi: 10.1155/2013/143515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nagy NE, et al. Storage of vitamin A in extrahepatic stellate cells in normal rats. J Lipid Res. 1997;38:645–658. [PubMed] [Google Scholar]