Abstract
Activity-dependent transcriptional responses shape cortical function. However, we lack a comprehensive understanding of the diversity of these responses across the full range of cortical cell types, and how these changes contribute to neuronal plasticity and disease. Here we applied high-throughput single-cell RNA-sequencing to investigate the breadth of transcriptional changes that occur across cell types in mouse visual cortex following exposure to light. We identified significant and divergent transcriptional responses to stimulation in each of the 30 cell types characterized, revealing 611 stimulus-responsive genes. Excitatory pyramidal neurons exhibit inter- and intra-laminar heterogeneity in the induction of stimulus responsive genes. Non-neuronal cells demonstrated clear transcriptional responses that may regulate experience-dependent changes in neurovascular coupling and myelination. Together, these results reveal the dynamic landscape of stimulus-dependent transcriptional changes that occur across cell types in visual cortex, which are likely critical for cortical function and may be sites of de-regulation in developmental brain disorders.
Introduction
Neuronal activity shapes brain development and function by multiple mechanisms1,2. Post-translational modifications affect excitability and synapse function in the short term3. These effects are followed by activity-dependent transcriptional programs which lead to long-lasting cellular adaptations necessary for learning and circuit-level homeostasis4,5. Such transcriptional responses are evoked in the brain by a wide range of stimuli, including sensory experience, metabolic changes, circadian rhythm, stress, injury and pharmacological intervention, and are implicated in many biological responses and diseases6–10.
Early work characterizing activity-dependent transcription in cultured neurons revealed that a depolarizing stimulus rapidly induces calcium- and MAPK-dependent early-response genes (ERGs)11. These ERGs include factors like Arc, which alters synaptic transmission12, but consist mostly of early-response transcription factors (ERTFs). The ERTFs induce a second wave of late-response genes (LRGs) including neuronal modulators and secreted factors (e.g. Bdnf) that effect changes in circuitry13,14. Until recently, the study of such responses had largely been limited to neurons in vitro or bulk tissue15,16. Newer technologies, however, extended this analysis to multiple genetically defined but investigator-selected neuronal cell types in vivo13,17. From these studies, a model emerged in which sensory experience induces a common ERTF program across neuronal types, followed by distinct LRG programs that regulate synaptic plasticity in a cell-type-specific manner.
Despite this recent progress in understanding sensory experience-dependent gene expression, significant gaps in knowledge still remain. Previous approaches analyzed a limited number of inhibitory cell types and masked the full diversity of excitatory populations that form functionally and molecularly distinct layers with specialized roles within the local microcircuitry. Furthermore, neuronal activity can induce calcium waves in astrocytes, proliferation and myelination by oligodendrocytes, and structural changes in the neurovasculature18–20. Despite the considerable diversity of stimulus-responsive cell types in the cortex, we lack a comprehensive understanding of how the full complement of cells within a cortical microdomain respond to a sensory stimulus and how their response contributes to neuronal plasticity.
To address this issue, we performed unbiased high-throughput droplet-based single-cell RNA sequencing (scRNA-seq) of 114,601 individual cells in mouse visual cortex to map their transcriptional response to visual stimulation21. Remarkably, cells of all 30 transcriptionally identified types (hereafter referred to as “cell types”) responded to the visual stimulus with diverse transcriptional changes. These transcriptional cascades diverged immediately across cell populations, with the induction of different early-response transcription factors. Surprisingly, astrocytes, oligodendrocytes, and endothelial cell types were highly responsive to the visual stimulus initiating transcriptional changes indicative of structural remodeling of these non-neuronal cells. Among excitatory neurons, late-response genes were enriched for secreted factors that may modulate neural circuit connectivity and serve to demarcate inter- and intra-layer cell type organization. Together, these results indicate that in response to sensory stimuli the cortical transcriptome is much more dynamic than previously appreciated, and that the widespread changes in sensory-dependent gene expression among diverse cell types identified in this resource are likely essential for proper brain function.
Results
inDrop sequencing to study neuronal activity
We used an in vivo visual stimulus paradigm to study sensory-dependent gene expression in primary visual cortex13,16,17. 6–7 week old mice were housed in complete darkness for one week followed by either zero (control), one, or four hours of light exposure (Fig. 1a, Methods). We used fluorescence in situ hybridization (FISH) to confirm the efficacy of the light stimulus to induce expression of ERGs (Fig. 1b). Quantitative reverse transcription PCR (qRT-PCR) revealed that light induction of ERGs was specific to the visual cortex and did not occur in somatosensory, motor, or auditory cortex (Supplementary Fig. 1).
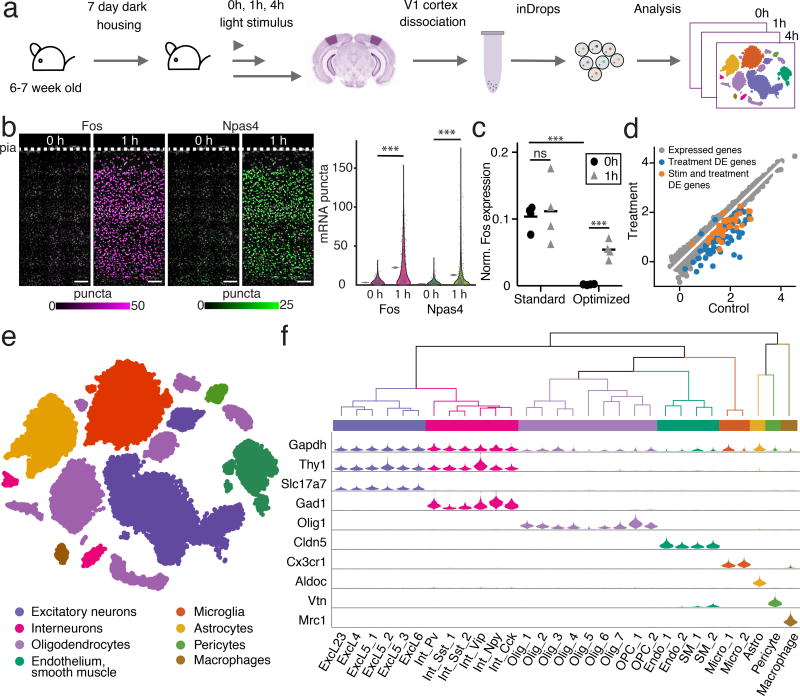
Figure 1. Workflow and identification of cell types.
(a) 6–7-week-old animals were housed in dark for 7 days and exposed to light for 0 (control), 1, or 4 h. V1 was dissociated into single cells and processed by inDrop sequencing.
(b) FISH of the immediate-early genes (IEGs) Fos and Npas4 from animals exposed to light for 0 and 1 h (left). Nuclei are pseudo-colored by expression level of Fos (magenta) or Npas4 (green) FISH probes (Online Methods). Scale bar=100 um. Quantification of FISH across time points, with mean and 95% confidence intervals denoted by gray lines (right). A random subset (10%) of the raw data was selected for visualization. For both Fos and Npas4, cells=2,667 for 0 h, cells=2,683 for 1 h. ***p<10−200, Mann-Whitney U-test, two-sided. Experiments repeated on 2 cortical slices per timepoint.
(c) qRT-PCR for Fos relative to Gapdh comparing a standard and an optimized cell dissociation protocol designed to limit IEG induction during dissociation. Mean denoted by horizontal line. ***p=2.7×10−4 and 3×10−5, ns p=0.39, (standard vs optimized 0 h, optimized 0 vs 1 h, standard 0 vs 1h respectively), n=4 animals, unpaired t-test, two-sided.
(d) RNA-Seq analysis of cocktail-treated and control cells collected from n=4 animals per condition. 114 genes that were significantly differentially expressed are denoted in blue (FDR <0.01, |fold change|>2, limma). 45 of these genes were also differentially expressed between cocktail-treated light-stimulated and light-unstimulated samples (Supplementary Fig. 2), denoted in orange. Axes units are log10 [(TMM-normalized CPM)+0.1].
(e) t-SNE plot of 47,209 cells from V1 of 23 animals. Colors denote main cell types.
(f) Dendrogram and violin plots showing the distribution of expression of selected marker genes across all 30 analyzed cell types hierarchically clustered by variable gene expression.
Immediately following light stimulation, the visual cortices were dissected and dissociated into single-cell suspensions (Fig. 1a, Online Methods). In contrast to the stimulus-dependent induction of Fos measured by FISH and qRT-PCR, RNA extracted in bulk from single-cell suspensions initially displayed high ERG expression in control samples (Fig. 1c). It was recently reported that protocols relying on enzymatic digestion induce activity-dependent gene transcription in brain cells22, making it difficult to use previously generated single cell RNA-seq data sets to identify genes whose activity is induced in vivo in response to sensory stimuli. We therefore added a cocktail of inhibitors to the dissociation solution that block neuronal activity, calcium entry, transcription, and translation (Online Methods). This reduced the expression of Fos in the control but not stimulated sample (51-fold, p=3.2×10−5, unpaired t-test), thus preserving the pattern of in vivo activity-dependent gene induction that was detected by FISH (Fig. 1b). Genome-wide analysis showed differential expression of 114 genes (FDR-corrected q<0.01, ≥ 2-fold change in expression), 110 of which were down regulated with inhibitor treatment (Supplementary Table 1). 45 of these 110 genes were also regulated by the light stimulus (FDR-corrected q<0.01) (Fig. 1d, Supplementary Fig. 2), further supporting the use of these blockers in preventing aberrant transcription during the enzymatic dissociation.
Using this optimized dissociation protocol to preserve the dynamic transcriptional state of ERGs in vivo, we conducted stimulus-dependent scRNA-seq profiling of visual cortex after light exposure of various durations. Following dissociation, single cells were captured and their mRNA barcoded using the inDrops platform21 (Online Methods). In total 28 preparations from 23 animals were sequenced (Supplementary Fig. 3), yielding 65,539 cells that passed initial quality control tests (Online Methods).
Identification of cell types in the adult mouse visual cortex
The inDrops dataset was analyzed using two independent clustering approaches to classify individual cells into cell types based on their patterns of gene expression, generating a final dataset containing 47,209 cells (Supplementary Fig. 4, Online Methods). On average, we recovered 3,234 transcripts per cell and 1,453 unique genes expressed per cell (Supplementary Fig. 5). The cells robustly classified into 8 main cell types (excitatory neurons, inhibitory neurons, oligodendrocytes and oligodendrocyte precursor cells (OPCs), astrocytes, endothelial and smooth muscle cells, pericytes, microglia, and macrophages) and 30 subtypes (Fig. 1e–f, Supplementary Fig. 6–10, Supplementary Table 2). The cluster identities are consistent with recent scRNA-seq data from the murine visual cortex (Supplementary Fig. 11)23.
Identification of sensory stimulus-regulated transcriptional programs
To identify visual experience-regulated genes within each cell type, we carried out differential gene expression analysis (Online Methods) across time points, requiring FDR <0.05 and ≥ 2-fold change in expression (example results for Exc_L23 Fig. 2a). Of 25,187 genes, 8,313 were significantly stimulus-dependent in at least one cell type and 611 passed the fold change threshold (419 upregulated, 192 downregulated, Supplementary Table 3, Supplementary Fig. 12).
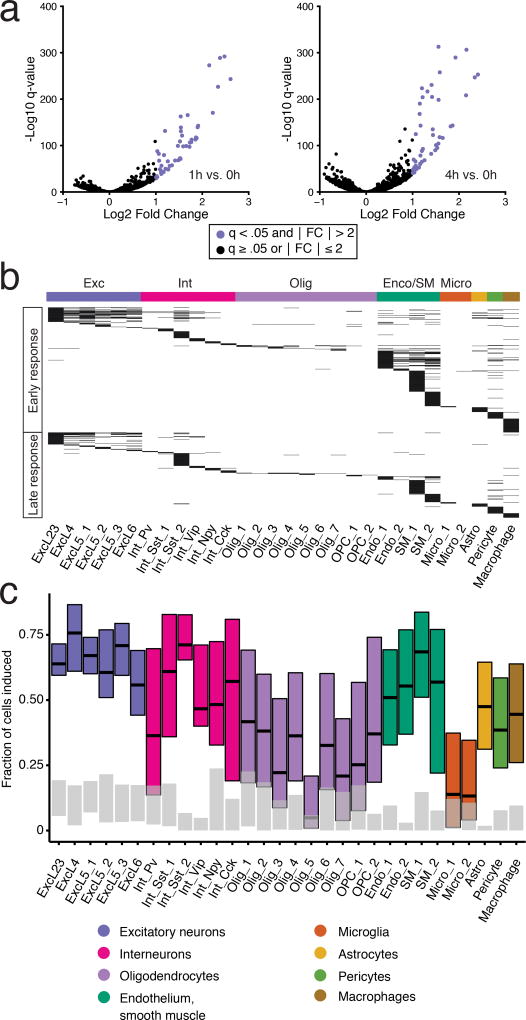
Figure 2. Identification of sensory stimulus-regulated genes.
(a) Sample volcano plots for ExcL23 cells indicating genes identified as sensory stimulus-regulated (|log2 fold change| >1 and FDR<0.05) for 1 vs 0 h (left) and 4 vs 0 h (right) comparisons. Colored dots represent sensory stimulus-regulated genes. The analysis was performed independently across thirty cell types (Supplementary Fig. 12).
(b) Heatmap of all 611 stimulus-regulated genes grouped into ERGs and LRGs by cell type. Each horizontal black line represents a stimulus-regulated gene.
(c) Estimation of the percentage of cells with stimulus-regulated transcriptional changes (Online Methods) at 1 h (colored bars) compared to 0 h (gray bars). We define induction as requiring either two, three, or four induced genes within each cell from a cell-type-specific set to consider the cell induced. These are plotted as lower, central, and upper lines of the box (Online Methods).
We identified light-induced gene expression changes in all neuronal and also, surprisingly, non-neuronal cell types in the visual cortex. Across all cell populations, the sensory experience-regulated genes were grouped into early (362) and late (249) response genes based on their temporal pattern of gene induction (Online Methods, Fig. 2b, Supplementary Table 3). Gene ontology (GO) analysis across all 611 genes showed significant enrichment for positive regulation of transcription and MAPK-signaling (additional terms in Supplementary Table 4).
The single cell resolution of the inDrops data also allowed estimation of the fraction of cells in each population exhibiting acute transcriptional responses to light stimulation (Online Methods, Fig. 2c). This analysis revealed that 49–69% of excitatory neurons exhibited light-induced transcriptional changes, irrespective of type or laminar position. By contrast, inhibitory neuronal subtypes showed variable fractions of transcriptionally responsive cells, ranging from 71% in the case of a somatostatin (Sst)-expressing subpopulation (Int_Sst_2) to only 29% of parvalbumin-expressing interneurons (Int_Pv).
Remarkably, a large fraction of non-neuronal cells also exhibited acute light-induced transcriptional responses, with endothelial and smooth muscle cell types showing the largest fraction of transcriptionally active cells (49–68%), followed by astrocytes (47%), pericytes (38%), and macrophages (38–47%). By contrast, a small fraction of oligodendrocytes and microglia showed light-induced transcriptional responses (4–37% and 11–12%, respectively). Light-exposure following dark housing thus triggers acute transcriptional responses in a large fraction of cells across the full range of V1 cell types, including a surprisingly high proportion of non-neuronal cell classes.
Diversity of experience-regulated early-response genes
One prevalent model for stimulus-dependent gene expression in the brain suggests that neuronal activity triggers several calcium-dependent signaling pathways, including the Ras/MAPK pathway, which drive the expression of a common cohort of ERGs ubiquitously across many cell types25. Many of these ERGs encode transcription factors (TFs) that have been hypothesized to trigger unique patterns of late-response gene (LRG) expression through differential binding to cell-type-specific cis-regulatory elements13,17. This hypothesis, however, is based on findings from a limited number of cell types. An alternative model is that the induction of ERTFs themselves is at least somewhat cell-type-specific and leads to the activation of distinct sets of LRGs in different cell types.
We tested these two models by examining the ERTF and LRG programs across cell types. Consistent with previous reports, LRGs were shared across fewer cell types than ERTFs (Mann-Whitney U-test, p=2×10−5) (Fig. 3a). However, of the 38 ERTFs identified, only 19 were induced in three or more cell types, suggesting that there is considerable divergence within the early stimulus-responsive gene expression program. To gain further insight into the 19 shared ERTFs, we classified them based on their expression patterns across the 30 cell types and identified four distinct gene sets (Fig. 3b). Two of these gene sets showed increased expression across most neuronal and non-neuronal cell types (between 3–14 cell types). These sets contained canonical immediate-early genes (IEGs), known to regulate the late phases of gene expression (Nr4a1, Nr4a2, Nr4a3, Fos, Fosl2, Egr1, etc.). We confirmed the induction of a subset of these IEGs across multiple cell types via FISH (visual presentation and quantification in Fig. 3c, Supplementary Fig. 13). By contrast, the final two gene sets were specific to either neuronal (Egr2, Egr4, Fosb, Junb, and Npas4) or non-neuronal cell populations (Atf3, Klf2, Klf4, Klf10, and Maff), respectively.
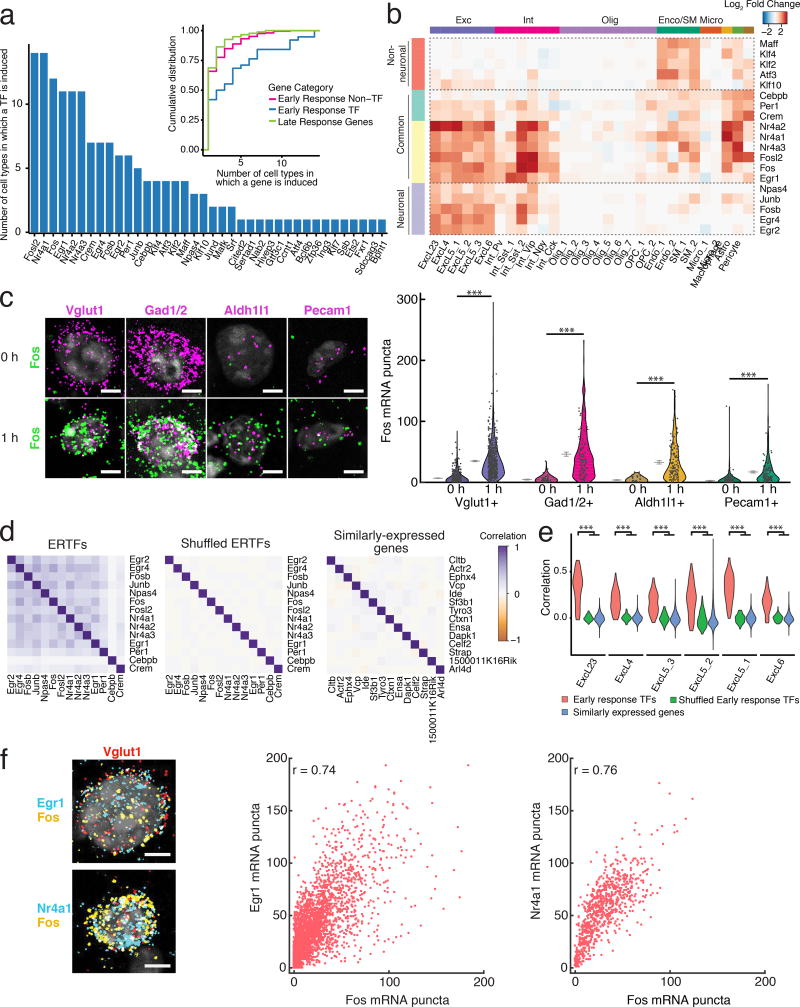
Figure 3. Diversity of experience-regulated ERGs.
(a) Number of cell types in which each ERTF is sensory experience-regulated. Inset: Cumulative distribution of the number of cell types in which each gene is experience-regulated. Stimulus-regulated LRGs were shared across fewer cell types than ERTFs (p=2×10−5, Mann–Whitney U-test, two-sided, nERTFs=38, nLRGs=176, nERnon-TF=205 genes).
(b) Heatmap of log2 fold changes between 1 and 0 h of the 19 ERTFs shared across at least 3 cell types. ERTFs are hierarchically clustered into 4 groups based on gene expression across all variable genes as in Fig. 1f.
(c) Left: Representative FISH images of Fos indicates induction across several cell types. Right: Quantification of FISH for Fos expression over cell types. A random subset (25%) of the raw data is plotted to aid visualization. For Vglut1, n0h=1,616, n1h=2,324; for Gad1/2, n0h=156, n1h=497; for Aldh1l1, n0h=121, n1h=477; for Pecam1, n0h=542, n1h=510 cells. ***p<10−39, Mann-Whitney U-test, two-sided. Mean and 95% confidence intervals denoted by gray lines. Experiments repeated on 2–8 cortical slices per timepoint.
(d) Mean pairwise Pearson correlations across individual excitatory neurons at 1 h calculated using ERTF expression (left, r=0.23±0.13, mean±s.d.), shuffled ERTF expression (middle, r=0.002±0.017), and expression-matched non-induced genes (right, r=0.002±0.029), n =91 pairwise comparisons. Correlations between ERTFs are significantly higher than between shuffled ERTFs or expression-matched non-induced genes (p=0, p=0, Mann-Whitney U-test, two-sided, respectively).
(e) Distributions of pairwise Pearson correlations of ERTFs (n=91 pairwise comparisons), ERTFs with shuffled expression, and of expression-matched non-induced genes across excitatory neuronal subtypes at 1 h. ERTFs are more highly correlated than either shuffled ERTFs or similarly expressed non-induced genes. ***p<1×10−24, Mann-Whitney U-test, two-sided.
(f) Left: 3-color FISH images of Fos, Egr1, and Nr4a1 expression at 1 h post-light stimulation in excitatory neurons (Vglut1+). Middle, Right: Quantification of FISH. Scatter plots between Egr1 and Fos, and Nr4a1 and Fos co-expression. Expression was highly correlated (Pearson r=0.74, 0.76 respectively; n=3162, 778 cells respectively). Scale bars=5 um. Data collected from 4 cortical slices for Egr1 and Fos co-expression, and 1 cortical slice for Nr4a1 and Fos co-expression.
We investigated the extent to which these ERTFs are co-expressed within individual cells of a given type to determine whether their co-expression is stochastic or reflects a deeper structure in ERTF regulation. To this end, we correlated the expression of ERTFs across individual excitatory neurons, focusing on this cell population as it represented the largest cohort of transcriptionally responsive cells. We found a significantly higher pairwise correlation between ERTFs within individual cells (Pearson r=0.23±0.13) than was observed for ERTFs shuffled across cells, or for expression-matched non-induced genes (Pearson r=0.002±0.017, r=0.002±0.029, p=0, p=0, Mann-Whitney U-test, respectively) (Online Methods, Fig. 3d–e). 3-color FISH confirmed these conclusions for Fos and Egr1 or Fos and Nr4a1 in excitatory neurons (Vglut1+), which demonstrated a high degree of pairwise correlation (Pearson’s r=0.74, r=0.76, respectively) within individual excitatory neurons (Fig. 3f).
Taken together, our findings provide a more nuanced view of diversity of experience-regulated gene expression across cell-types. Although our data confirm the existence of a core set of early-response transcription factors induced in both neuronal and non-neuronal cell types, we also find substantial divergence in ERTF expression that may contribute to late-response gene diversity between cell types. Moreover, our results suggest that, within individual excitatory neurons, the expression of ERTFs is tightly co-regulated to enable the concerted action of these factors in the regulation of late-response gene programs.
Excitatory neuronal LRGs
Excitatory neurons throughout cortical laminae are molecularly, physiologically, and hodologically diverse26; however, the diversity of late-response gene programs between excitatory neuronal subtypes has yet to be systematically explored. We identified 19 cell-type-enriched LRGs (of 55 LRGs induced in excitatory neurons), including several secreted modulators of synaptic plasticity15. For example, Cerebellin 4 (Cbln4), which encodes a secreted factor implicated in inhibitory synapse recruitment27, was enriched in layer 4 excitatory neurons, which we confirmed by FISH (Fig. 4a). Gene ontology analysis of excitatory late response genes revealed an over-representation of secreted factors and genes regulating synapse formation (Supplementary Table 4), consistent with experience-dependent transcriptional regulation of neuronal connectivity. These findings suggest that long-term adaptations to visual stimuli differ across excitatory neuronal subtypes in visual cortex and could enable distinct functions in visual processing.
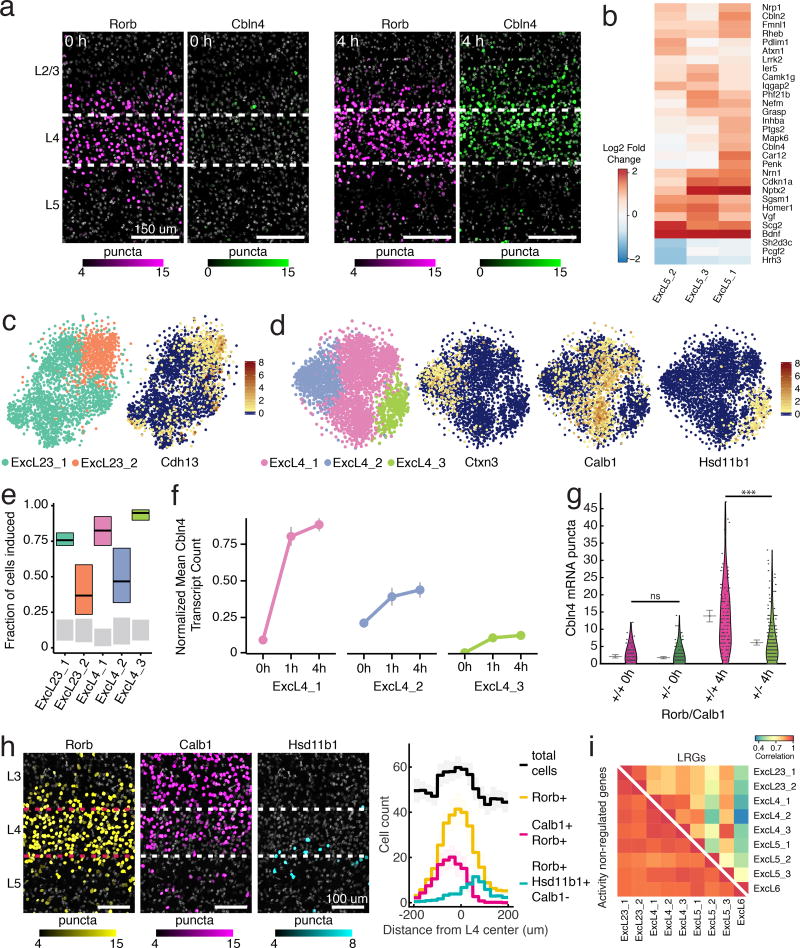
Figure 4. Sensory experience-induced transcriptional responses in V1 excitatory neurons.
(a) FISH of Cbln4 with Rorb, a layer 4-specific marker, at 0 (control) and 4 h post light stimulus. Nuclei are pseudo-colored by expression level of Cbln4 or Rorb (Online methods). Scale bars=150 um. Experiments repeated on 2 cortical slices per condition.
(b) Heatmap of log2 fold change in expression of LRGs across layer 5 subtypes between 4 and 0 h.
(c) Left: t-SNE plot showing Layer 2/3 excitatory neuronal subtypes. Right: Overlay of Cdh13 expression. Scale bar indicates normalized expression per cell. n=2941 cells.
(d) Left: t-SNE plot of Layer 4 excitatory neuronal subtypes. Right: Overlay of selected marker expression. Scale bar indicates normalized expression per cell. n=3198 cells.
(e) Estimation of the percentage of cells with stimulus-regulated transcriptional changes (Online Methods) at 1 h (colored bars), compared to 0 h (gray bars). We define induction as requiring either two, three, or four induced genes within each cell from a cell-type-specific set to consider the cell induced. These are plotted as lower, central, and upper lines of the box (Online Methods).
(f) Mean expression of Cbln4 across layer 4 subtypes (nExc4_1=732, 468, 783 cells; n Exc4_2=343, 214, 210 cells; n Exc4_3=136, 137, 175 cells; for 0, 1, 4 h timepoints respectively), S.E.M. denoted by bars.
(g) FISH quantification of Cbln4 across layer 4 subtypes (Calb1+/Rorb+ and Calb1−/Rorb+). 0 h expression not significantly different between cell types, n=97, 209 cells respectively, p=0.062, Mann-Whitney U-test, two-sided. 4 h expression significantly higher in Calb1+/Rorb+ than Calb1−/Rorb+ (n=127, 286 cells respectively), ***p<10−17, Mann-Whitney U-test, two-sided. Mean and 95% confidence intervals denoted by gray lines.
(h) Left: Rorb (yellow), Calb1 (magenta), and Hsd11b1 (cyan) expression in layer 4 by FISH, scale bars=100 um. Right: Quantification of the anatomical distribution of FISH-defined cell types across layer 4 from n=13 cortical slices. Negative/positive values on x-axis correspond to distance from the center of layer 4 towards the slice surface (negative) or towards deeper cortical layers (positive). Shaded area around lines indicates 95% confidence intervals around mean. Calb1+/Rorb+ (n=1626 cells) population is enriched superficially within layer 4 as defined by Rorb+ (n=4066 cells) and an increase in cell density (black). Hsd11b1+/Rorb+/Calb− (n=851 cells) population is enriched in ventral layer 4 and into superficial layer 5. For further analysis, see Supplementary Fig. 21.
(i) Top right: pairwise Pearson correlation between excitatory subtypes calculated using LRG expression (n=55 genes). Bottom left: Same analysis with expression-matched non-stimulus-regulated genes (n=55 genes). Non-stimulus regulated genes are more correlated across excitatory subtypes than stimulus-regulated LRGs.
We assessed whether we could detect intra-layer differences in sensory experience-dependent transcriptional responses. We focused first on layer 5 excitatory neurons, which have been classified into subtypes defined by axon projection patterns and electrophysiological properties23,26,28. We mapped three putative layer 5 excitatory neuron populations onto previously defined transcriptional cell types from mouse visual cortex23 (Supplementary Fig. 11, 14). The three populations of excitatory neurons respond to sensory stimulus by activating a similar number of genes (34–55 genes per cell type); however, despite their anatomical proximity, several of these genes are subset enriched (Fig. 4b). For example, stimulus-dependent induction of Pdlim1, a gene that encodes a protein involved in AMPA receptor trafficking29, is restricted to predicted cortico-fugal projecting ExcL5_2 neurons. Thus, even within a single cortical layer, functionally heterogeneous cell types exhibit distinct sensory-dependent transcriptional responses.
We next asked whether other cortical laminae had subpopulations that transcriptionally diverged upon sensory stimulus. Further subclustering of excitatory populations identified two layer 2/3 and three layer 4 excitatory neuron subtypes, consistent with previous transcriptionally defined cell types (Online Methods, Fig. 4c, Fig. 4d, Supplementary 15–18). We identified both sensory-experience regulated and unregulated genes as distinct across these cell types. Non-induced gene markers include Cdh13 for layer 2/3 and Ctxn3, Calb1, and Hsd11b1 for layer 4 (Fig. 4c, Fig. 4d, Supplementary Fig. 19, 20). These layer 2/3 and layer 4 excitatory neuronal cell types displayed marked differences in experience-regulated transcriptional responses (Fig. 4e). For example, Cbln4 was differentially expressed across layer 4 subtypes, with the largest light-stimulus induction occurring within the Calb1+ subtype (Fig. 4f), which was corroborated via FISH (Fig. 4g).
Finally, we asked whether these cell-types exhibiting distinct sensory-induced transcriptional responses were further organized into specific spatial arrangements within their respective layers. FISH revealed that the transcriptionally less responsive Cdh13+ excitatory cells are enriched superficially in layer 2/3 (Supplementary Fig. 21), consistent with previous observations of sparser firing in layer 2 in response to sensory stimuli compared to layer 326,30,31. In layer 4, we first looked at the two most transcriptionally responsive layer 4 subpopulations, Calb1+ and Hsd11b1+ neurons (Fig. 4e), which differed in 44 of the 90 stimulus-regulated genes identified within these subtypes. Whereas Calb1+ neurons are enriched superficially, the Hsd11b1+ neuronal population is found deep within layer 4 and spans the boundary between layers 4 and 5 (Fig. 4h, Supplementary Fig. 22). Ctxn3+ neurons, the least transcriptionally responsive population, are also enriched within superficial layer 4 and interspersed with the Calb1+ population (Supplementary Fig. 22). The anatomical organization of these cell types into sublayers, coupled with divergent transcriptional responses to a sensory stimulus, suggests unappreciated functional subdivisions within the laminae of mouse visual cortex resembling the cytoarchitecture in higher mammals32.
In summary, we observe that distinct transcriptional responses exist both between cortical layers and among neuronal subtypes within individual layers in response to light stimulation. Moreover, we find that the late response gene programs activated in response to visual stimulation are highly cell type specific and likely contribute to the cellular features that define the function of each subtype within the circuit (Fig 4i).
Inhibitory neuronal LRGs
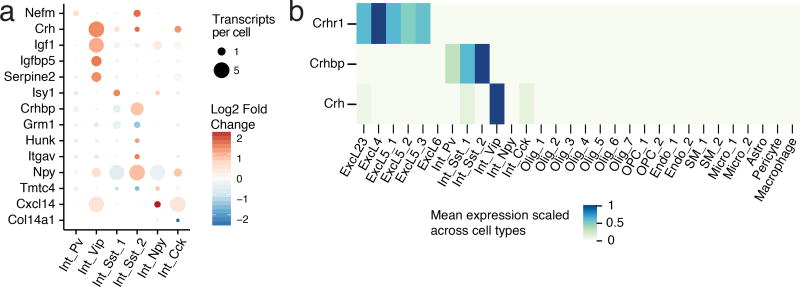
V1 interneurons were classified into six distinct subtypes (Int_Pv, Int_Vip, Int_Cck, Int_Npy, Int_Sst_1, Int_Sst_2), consistent with other scRNA-seq-based taxonomies23,33 (Fig. 1f, Supplementary Fig. 11, 23). Analysis of sensory stimulus-regulated gene expression in these cell types showed broad agreement with our prior findings based on cell-type-specific isolation of ribosome-associated mRNA13,17. Specifically, we identified 75 ERGs and 70 LRGs whose expression is light-dependent in at least one of the six inhibitory subtypes with 14 LRGs enriched in a single inhibitory subtype (Fig. 5a).
Figure 5. Inhibitory neuronal LRGs.
(a) Cell type enriched LRGs across inhibitory cell types. Transcripts per cell represent the mean depth-normalized expression across all cells. Fold change is calculated between 4 and 0 h.
(b) Scaled mean expression of Crh, Crhbp, and Crhr1 across all cell types.
Of particular interest, we observed selective induction of Corticotropin-releasing hormone (Crh) mRNA, encoding a stress hormone, in the Int_Vip population. In the hippocampus, Crh signals through the G-protein coupled receptor Crhr1 to increase the excitability of pyramidal cells34. Crhr1 expression in V1 was enriched in excitatory cell types, suggesting that Vip interneuron-derived Crh might directly modulate the excitability of pyramidal neurons in a stimulus-dependent manner (Fig. 5b). Moreover, building upon a recent study in prefrontal cortex35, we observed stimulus-dependent induction of Crh-binding protein (Crhbp), a gene that encodes a secreted factor known to negatively regulate Crh signaling, in Sst-expressing interneurons. Together, these findings suggest a possible mechanism for the control of cortical microcircuit excitability involving the opposing action of two activity-regulated signaling peptides derived from distinct inhibitory subtypes.
Experience-dependent transcriptional changes in vasculature-associated cells
Since the discovery over a century ago that neuronal activity rapidly triggers changes in local blood flow36, researchers have recognized that neurons, glia, and associated vasculature (endothelial cells, smooth muscle, and pericytes) coordinate their signaling and activity37. In addition, sensory-related neural activity restructures cortical vascular networks, although the mechanisms that regulate this process remain unknown18.
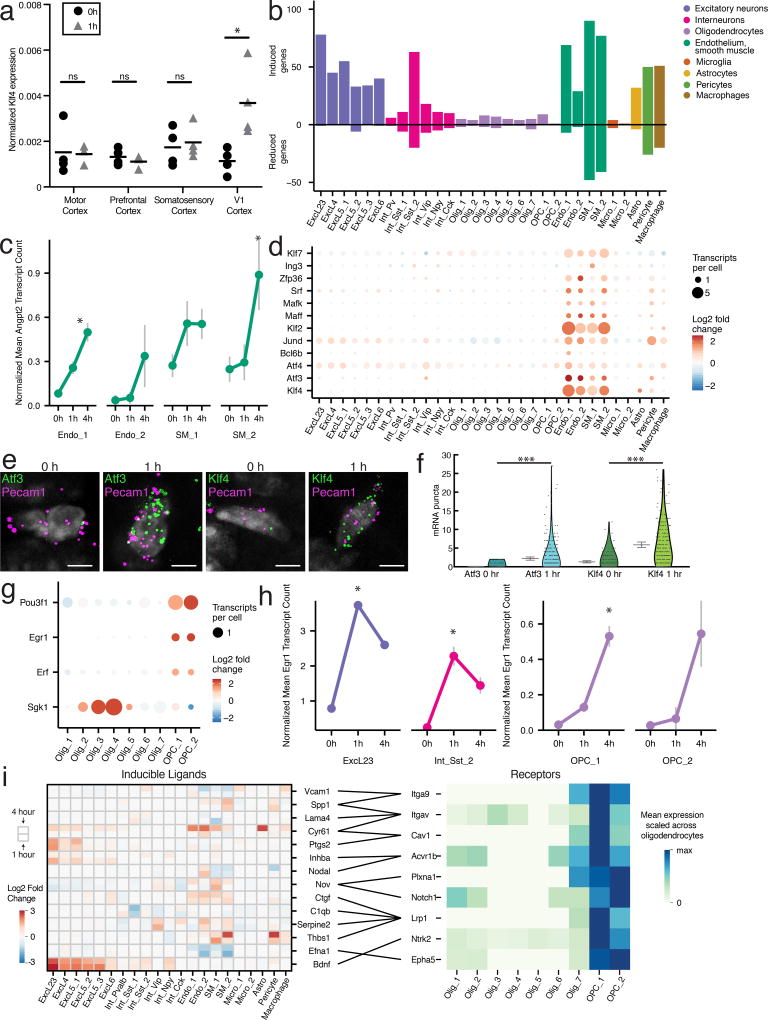
We asked whether the vascular transcriptional response was induced by local neuronal activity or by systemic, brain-wide changes in blood flow or oxygen levels. To address this question, we carried out qRT-PCR for the endothelial and smooth muscle-specific early response transcription factor Klf4 across multiple cortical regions. We found that Klf4 is induced in V1, but not other regions of the cortex indicating that the sensory stimulus-dependent changes in vascular architecture occur specifically in the region of the brain that is activated by the sensory stimulus (Fig. 6a).
Figure 6. Light-induced transcriptional changes in non-neuronal cells.
(a) qRT-PCR of Klf4 across cortical regions in 0 and 1 h light-exposed animals, mean denoted by horizontal bar. Klf4 induction is only significant in V1, p=0.02, unpaired t-test, two-sided. n=4 animals for all 0 h samples, n=3 animals for 1 h motor and prefrontal cortex, n=4 for 1 h somatosensory and visual cortex.
(b) Total number of sensory-stimulus-regulated genes across cell types.
(c) Mean expression of the LRG Angpt2 across endothelial and smooth muscle cells across all three stimulus conditions (nENDO_1=1111, 1107, 1109; nENDO_2=33, 65, 25; nSM_1=115, 92, 116; nSM_2=104, 109, 85 cells; for 0, 1, 4 h respectively). *FDR<.05, fold change>2, Monocle2. S.E.M. denoted by bars.
(d) Induction at 1 h of ERTFs enriched in vasculature-associated cells.
(e) FISH of single Pecam1+ cells at 0 and 1 h co-labeled with Atf3 and Klf4, scale bars=5 um. Experiments repeated on 2 cortical slices per timepoint.
(f) Quantification of FISH for Atf3 and Klf4 in Pecam1+ cells across 0 and 1 h (for Atf3, n0h=275, n1h=326; for Klf4, n0h=182, n1h=224 cells). p<10−22, Mann-Whitney U-test, two-sided. Mean and 95% confidence intervals denoted by gray lines. Data from 2 cortical slices per timepoint.
(g) Fold-change in expression between 0 and 4 h samples of light-induced genes in oligodendrocytes and OPCs.
(h) Mean expression of Egr1 in ExcL23, Int_Sst_2, and OPCs across all stimulus conditions (nExcL23=150, 706, 1107; nInt_Sst_2=59, 52, 70; nOPC_1=645, 472, 608; nOPC_2=31, 27, 43 cells; 0, 1, 4 h respectively). *FDR <.05, fold change>2, Monocle2. S.E.M. denoted by bars.
(i) Left: Heatmap of induction of ligands in non-oligodendrocyte cell types with receptors enriched in OPCs at both 1 and 4 h time points. Right: Scaled mean expression of OPC-enriched receptors. Lines between heatmaps denote ligand-receptor pairs.
We were surprised to identify a larger number of activity-regulated genes (257) in endothelial and smooth muscle cells than in excitatory and inhibitory neurons combined (231, Fig. 6b). The large number of induced genes in endothelial and smooth muscle cells suggests that sensory-dependent transcriptional programs play an important role in the vascular response. For example, angiopoietin 2 (Angpt2), which encodes a canonical vascular growth factor involved in blood brain barrier permeability as well as embryonic and adult angiogenesis38,39, is significantly induced more than 2-fold in Endo_1 and SM_2 cell types within 4 hours of light stimulation (Fig. 6c).
Beyond several broadly induced genes, we identified 12 sensory-regulated transcription factors (Klf2, Klf4, Klf7, Atf3, Atf4, Bcl6b, Jund, Maff, Mafk, Srf, Zfp36, and Ing3) that are selectively induced in endothelial and smooth muscle cells but not in neuronal cell types (Fig. 6d–f). A subset of these TFs have previously been implicated in structural changes to brain vasculature40. For example, aberrant over-expression of Klf2/4 in endothelial cells causes cerebral cavernous malformations41. Thus, the induction of these ERTFs is likely to be vital for vascular structural remodeling in response to sensory stimuli.
Experience-dependent transcriptional changes in oligodendrocytes
Although neuronal activity is known to regulate oligodendrogenesis and myelination, the transcriptional pathways underlying these processes remain unclear19,42. Oligodendrocytes displayed a decreased transcriptional response to light stimulus, with only 33 differentially expressed genes identified, despite an abundance of these cells in our dataset (10,158 cells). Nevertheless, we did observe in multiple oligodendrocyte populations several stimulus-induced transcriptional changes of interest, including the induction of serum/glucocorticoid-regulated kinase (Sgk1) (Fig. 6g). Sgk1 overexpression enhances the arborization of oligodendrocytes43, suggesting that sensory experience-dependent induction of Sgk1 may lead to oligodendrocyte remodeling.
We observed the induction of several transcription factors (Egr1, Pou3f1, and Erf) specifically in OPCs and not in immature, pre-myelinating, or mature oligodendrocytes suggesting that these TFs may play a role in the process of oligodendrogenesis (Fig. 6g). Interestingly, in contrast to their dynamics in other cell types, we observed delayed induction of Erf and Egr1 in OPCs consistent with the possibility that the induction of these genes in OPCs is dependent upon the prior activation of activity-regulated secreted proteins in neurons or other non-neuronal cells (Fig. 6h). Indeed, we found nine receptors44 that are both enriched in OPCs relative to other oligodendrocytes and are predicted to bind stimulus-responsive ligands induced in neurons and vasculature-associated cells including those encoded by Bdnf, Ptgs2, and Inhba (Fig. 6i, Online Methods). Together, these findings identify sensory responsive genes that might regulate OPC proliferation, differentiation, and recruitment to neuronal axons.
Discussion
The cerebral cortex is composed of a vast array of cell types whose coordinated activity is necessary for normal brain development and function. Several laboratories have recently applied scRNA-seq to characterize these cell types across brain areas8,22,23,33,45–47, but this work has largely focused on producing a static cellular taxonomy at a defined developmental time point, remaining agnostic to the transcriptional changes induced by acute sensory experience and neuronal activity. Moreover, the enzymatic brain dissociation methods applied in these previous studies likely activate stimulus-regulated transcriptional programs, which may obscure the identification of genes that are induced in the brain in response to sensory stimuli22. The resource presented here, by contrast, represents an additional dimension of cellular state that must be considered when examining existing cell type atlases and serves as a critical step towards a holistic understanding of the mechanisms by which experience modulates cortical circuits through the concerted action of diverse cell types.
The degree to which transcriptional responses vary across the panoply of experiences encountered throughout life remains unknown. We restricted our analysis to a single stimulus and cortical region. Intriguingly, despite differences in brain region, stimulus paradigm, and methodology, our 611 experience-regulated genes are highly enriched among those reported by a recent study of activity-dependent chromatin accessibility and bulk gene expression in the dentate gyrus48 (Supplementary Fig. 24). Nevertheless, similar analyses of dynamic gene expression in other brain regions in response to a multitude of stimuli will be essential in constructing a complete cell atlas of the central nervous system.
Although droplet-based single cell RNA-Seq has significant advantages enabling high-throughput and cost-effective analysis of the transcriptomes of thousands of cells, several key limitations remain. First, the anatomical location of individual cells is lost, requiring FISH or other spatially precise methods to physically map transcriptionally defined cell types. Second, the low transcript-capture efficiency21 and shallow depth of sequencing (Supplementary Fig. 3, 5, 9, 10) resolve only a fraction of transcripts within each cell, requiring the analysis of large numbers of cells to differentiate cell types. For example, targeted deep sequencing of inhibitory neurons identified 23 subtypes which are fine-grained subsets of the types described here (Supplementary Fig. 17, 23). It follows that sequencing even more cells may uncover additional rare cell types and reveal further heterogeneity of stimulus-dependent transcriptional programs within mouse visual cortex.
Through the analysis of stimulus-regulated genes, we identified excitatory neuron cell types that dramatically differ in their response to the light stimulus both between and within cortical laminae. Closer examination revealed that cell types residing in the same cortical layer had differentially evoked transcriptional responses and were found to be organized in discrete laminar positions within their respective layers. Thus, despite their proximity, these cell types may have different synaptic connectivity and could serve distinct functions within cortical circuitry. Future studies of these cell types could test whether cells displaying unique sensory-dependent transcriptional responses have distinct physiological properties and, within thalamorecipient layer 4, whether they receive different thalamocortical inputs, as has been reported for primates32.
Non-neuronal cells have functions vital to the maintenance of cortical circuitry including regulating synapse development and maturation, neurotransmitter reuptake, and metabolite and oxygen supply. We reveal that, in parallel to the experience dependent transcriptional response in neurons, a distinct and variable gene network is induced in glial, immune, and vascular cell types. The functions of many of these regulated genes are unknown, but likely contribute to the regulation of previously identified activity-dependent processes in non-neuronal cells including the remodeling of vascular networks and structural remodeling of oligodendrocytes. Future studies are necessary to determine how these transcriptional programs act in concert to remodel cortical function and circuitry in response to changes in sensory experience.
Online Methods
Mice
Animal experiments were approved by the National Institute of Health and Harvard Medical School Institutional Animal Care and Use Committee, following ethical guidelines described in the US National Institutes of Health Guide for the Care and Use of Laboratory Animals. The experiments used adult (6–8 weeks old) C57BL/6J virgin male mice (The Jackson Laboratory).
Visual stimulation and brain dissection
Mice were housed in a standard light cycle (6AM–6PM) before placement into constant darkness for 7 days. Mice were either sacrificed in the dark (0h-control condition) or light-exposed for 1 h or 4 h before sacrifice. After isofluorane anesthetization the eyes were enucleated, the animal sacrificed, the brain isolated, and desired cortical regions microdissected according to the protocols listed below.
Fluorescence in situ hybridization (FISH)
Sample preparation
Mice were light-exposed and sacrificed as above. Brains were immediately frozen on dry ice in tissue freezing medium. Brains were sliced on a cryostat (Leica CM 1950) into 20 µm sections, adhered to SuperFrost Plus slides (VWR), and immediately stored at −80C until use. Samples were processed according to the ACD RNAscope Fluorescent Multiplex Assay manual.
Sample imaging
Sections containing V1 were imaged on a Leica SP8 X confocal microscope using a 63× 1.4 NA oil immersion objective (Harvard NeuroDiscovery Center). Tiled V1 cortical areas of ~1.1 mm by ~0.5 mm, containing all cortical layers, were imaged with optical sectioning of 0.5 µm. Channels were imaged sequentially to avoid any optical crosstalk.
Image analysis pipeline
We developed automated software to segment DAPI-stained nuclei in FISH images and count the fluorescent puncta from hybridized probes within each nucleus. A labeled set of 23 images containing a total of ~2000 nuclei was used to train a stacked Random Forest (RF) classifier, using contextual "offset" features adapted from Refs.49 and50, as well as newly developed circularity features. The RF was trained on three labels: background, nuclei contour, and nuclei, therefore producing three respective probability maps. We used a watershed algorithm on the output probability maps to split neighboring nuclei and create masks. Finally, we eroded the nuclei masks to 80% of the original radius for conservative analysis of fluorescent puncta. Puncta detection was performed as described in Ref.51. Code for the image analysis pipeline is available upon request, and detailed in Ref.52.
Typically, one or two channels of a FISH experiment were devoted to cell type markers. We used a model-based minimum error thresholding method53 over all cells in an image to determine the fewest number of marker puncta necessary to classify the cell as positive for that marker. To mitigate the risk of false positives, a lower limit of 4 puncta was used if the model yielded a smaller number of puncta. A lower threshold (2 puncta) was used for the layer 4 excitatory neuron cell type marker Ctxn3, which was detected at very low abundance (Supplementary Fig. 22). The following cell-type markers were used: Vglut1 for excitatory neurons, Pecam1 for endothelial and smooth muscle cells, Aldh1l1 for astrocytes, and a combination of Gad1 and Gad2 were used to label inhibitory neurons. Layer 4 was marked by Rorb expression and a peak in the density of DAPI-stained nuclei, and layer 6 was marked by Foxp2 expression. Subtypes of excitatory neurons were marked by Cdh13, Calb1, Ctxn3, Hsd11b1 (Fig 4h, Supplementary Fig. 20–22), Gpr88, Bcl6, and Nnat (Supplementary Fig. 14).
Image presentation
For large area images (Fig. 1b, 4a and 4h, Supplementary Fig. 14, 21 22), nuclei masks created in the analysis pipeline are displayed and pseudo-colored according to the number of puncta contained within the mask. For single cell images (Fig. 3c, Fig. 3f, Fig. 6e), a Gaussian filter was used to reduce uncorrelated noise for visualization purposes only.
Generation of single-cell suspensions
Standard protocol
V1 was dissected bilaterally in ice-cold Choline solution containing: 2.1 g/l NaHCO3, 2.16 g/l glucose, 0.172 g/l, NaH2PO4 * H2O, 7.5mM MgCl2•6H2O, 2.5mM KCl, 10mM HEPES, 15.36 g/l choline chloride, 2.3 g/l ascorbic acid, and 0.34 g/l pyruvic acid. The tissue was cut into 300um slices and dissociated using the Papain dissociation system (Worthington) according to the manufacturer’s instructions with the following modifications: The EBSS solution was replaced by our Dissociation Solution: HBSS (Life Technologies), 10mM HEPES (Sigma), 172mg/l kynurenic acid (Sigma), 0.86 g/l MgCl2•6H2O (Sigma), 6.3 g/l D-glucose (Sigma), was saturated with 95% O2, 5% CO2, and was pH adjusted to 7.35. Dissociation was carried out at 37C for 1h with 20U/ml of Papain.
Protocol optimized for preserving transcriptional state
Once anesthetized the animals were transcardially perfused with ice-cold Choline solution (above) containing the following small molecule cocktail for 5 minutes: 1 uM TTX (Sigma), 100 uM AP-V (Thermo Fisher Scientific), 5 ug/ml Actinomycin D (Sigma) and 10 uM Triptolide (Sigma). V1 was then microdissected, cut into 300um slices, and incubated on ice for 30 minutes in Dissociation Solution containing 1uM TTX, 100 uM AP-V (Thermo Fisher Scientific), 5 ug/ml Actinomycin D (Sigma), 10 uM Triptolide (Sigma) and 10 ug/ml Anicomycin (Sigma). Papain was added to a final concentration of 20 U/ml and the tissue dissociated for 1h at 37°C with gentle agitation in a total volume of 3.2ml. The remaining procedures were performed per manufacturer’s instructions without the small molecule cocktail. Following gradient centrifugation the cells were washed in Dissociation Solution containing 0.04% BSA and resuspended in Dissociation Solution containing 0.04% BSA and 15% Optiprep (Sigma) for single cell RNA-Seq.
RNA isolation, reverse transcription, and qRT-PCR analysis
For quantitative reverse transcription PCR (qRT-PCR) experiments across brain regions (Fig. 6a, Supplementary Fig. 1), the dissected regions from 3–4 mice were immediately frozen for RNA isolation. For experiments comparing the expression of activity-regulated genes across dissociation procedures, the cells were dissociated using the standard and optimized dissociation methods (below) from 4 mice per condition and immediately frozen. The RNeasy Mini (Qiagen) isolation procedure was carried out according to the manufacturer’s instructions including a DNase I digestion. For qRT-PCR analysis, the RNA was reverse-transcribed using the High Capacity cDNA Reverse Transcription kit (Life Technologies). qRT-PCR was performed using the LightCycler 480 SYBR Green I Master (Roche) on the LightCycler 480 system (Roche). Reactions were run in technical duplicates which were averaged before subsequent analysis. Glyceraldehyde 3-phosphate dehydrogenase (Gapdh) was used as a normalization control for the ΔΔCt-based quantification. The sequences of real-time PCR primers for Gapdh were TGTGTCCGTCGTGGATCTGA (forward) and TTGCTGTTGAAGTCGCAGGAG (reverse). For Fos we used CTGGATTTGACTGGAGGTCTGC (forward) and TTGCTGATGCTCTTGACTGGC (reverse). For Klf4 we used GCAGTCACAAGTCCCCTCTC (forward) and TAGTCACAAGTGTGGGTGGC (reverse).
RNA-seq analysis of bulk visual cortex
RNA from three experimental conditions with four replicates each was isolated as described in the RNA isolation, reverse transcription, and qRT-PCR analysis section. The three conditions were: drug cocktail-treated and visually unstimulated, drug cocktail-untreated and visually-unstimulated, and drug cocktail-treated and visually stimulated (1 h). Sequencing libraries were prepared using the SMART-Seq® v4 Ultra® Low Input RNA Kit for Sequencing and the Nextera XT DNA sample prep kit according to manufacturer’s instructions.
Samples were sequenced on the NextSeq 500 (Illumina) and paired end reads of lengths 38,38 were obtained. Reads were trimmed with Trimmmomatic −0.33 using the following parameters: ILLUMINACLIP:NexteraPE-PE.fa:2:30:10 LEADING:3 TRAILING:3 SLIDINGWINDOW:4:20 MINLEN:3054. A reference transcriptome was built with Tophat 2.1.1 and Bowtie 1.1.1 against the GRCm38.dna_sm.primary_assembly genome with the GTF file constructed above for inDrops mapping55,56. Reads were mapped against this transcriptome with Tophat using the following options: --mate-inner-dist 500 --no-mixed --transcriptome-index [CUSTOM_TRANSCRIPTOME] --bowtie1 --no-novel-juncs. All samples showed ≥90% concordant pair alignment rate. The featureCounts57 package was used to obtain gene counts with the following command options: -p -B -C -g gene_name -a [GTF] -s 1.
Counts tables were TMM-normalized and converted to CPM using the edgeR software analysis package58,59. Any genes that were not expressed in at least 3 samples with TMM-normalized CPM>1 were dropped from further analysis. Differential expression (DE) analyses were conducted using the voom/limma analysis software packages (requiring FDR-corrected q<.01) to identify drug-dependent (DE between visually-unstimulated cocktail-treated and visually-unstimulated not-cocktail-treated, Fig. 1d) and bulk visual-stimulus-dependent (DE between visually-unstimulated cocktail-treated and visually-stimulated cocktail-treated, Supplementary Fig. 2a) genes60,61.
Single-cell RNA sequencing (inDrops)
One to two libraries of approximately 3,000 cells were collected from each animal. inDrops was performed as previously described21,62, generating indexed libraries that were then pooled and sequenced across 15 runs on a NextSeq 500 (Illumina). Four libraries were down-sampled to match sequencing depths across samples.
inDrops sequencing data processing
Transcripts were processed according to a previously published pipeline21. Briefly, this pipeline was used to build a custom transcriptome from the Ensembl GRCm38 genome and GRCm38.84 annotation using Bowtie 1.1.1,56 after filtering the annotation gtf file (gencode.v17.annotation.gtf filtered for feature_type=”gene”, gene_type="protein_coding" and gene_status="KNOWN"). Read quality control and mapping against this transcriptome then followed. Finally, unique molecular identifiers (UMIs) were used to reference sequence reads back to individual captured molecules, hereafter referred to as UMIFM counts.63 All steps of the pipeline were run using default parameters unless explicitly specified.
Quality control for the inclusion of cells
Our initial dataset contained 114,601 cells with more than 1,000 reads assigned to each cell. All mitochondrially encoded genes were removed from the dataset. Cells with fewer than 700 or more than 15,000 UMI counts were next excluded, yielding 65,539 high quality cells isolated across 23 animals. The average transcript count per cell was 3,045.
Dimensionality reduction and clustering
All 65,539 cells were combined into a single dataset. Two independent approaches were used to cluster cells (described in Supplementary Fig. 4):
1. t-SNE based approach (Approach 1)
Raw counts were first linearly normalized such that each cell in the dataset contained the same number of transcripts (3,045). Next, 4,000 most variable genes in cells derived from zero hour samples were identified as previously described in Klein et al.21. Briefly, the v-statistic for each gene (a corrected Fano Factor accounting for noise in method efficiency, and variation in cell size) was computed and the genes with the largest v-statistics were chosen as the most variable genes. Principle component analysis (PCA) based on the 4,000 most variable genes in cells derived from zero hour samples was applied to reduce the dimensionality of the dataset as described in Klein et al.21. The MATLAB implementation of the t-SNE dimensionality reduction algorithm used in this publication was next applied to position cells on a 2D coordinate system using the principal components generated above. Perplexity was set to 30, other t-SNE arguments were left as default64. A machine learning algorithm with minimal user input was used to define clusters of cells based on their proximity to one another in this 2D space as described in Rodriguez et al.65. The parameters used were: percNeigh=0.02; kernel='Gauss'; minRho=10, minDelta=3. This method resulted in 91 initial clusters.
2. Seurat-based approach (Approach 2)
The second approach applied the Seurat R package66,67. The data were log normalized and scaled to 10,000 transcripts per cell. Variable genes were identified using the MeanVarPlot() function, which calculates the average expression and dispersion for each gene, then bins genes and calculates a z-score for dispersion within each bin. The following parameters were used to set the minimum and maximum average expression and the minimum dispersion: x.low.cutoff=0.0125, x.high.cutoff=3, y.cutoff=0.5. Next, PCA was carried out and the top 30 principal components (PCs) were kept. Clustering resolution was set to 0.6. This method resulted in 32 initial clusters.
Doublet removal and additional clustering
The following analysis was carried out for each approach independently. All clusters containing fewer than 100 cells were discarded. The expression of known marker genes (Slc17a7, Gad1, Olig1, Aldoc, Cldn5, Vtn, Cx3cr1 and Mrc1) was used to assign each cluster to one of the main cell types: excitatory neurons, inhibitory neurons, oligodendrocytes, astrocytes, endothelial and smooth muscle cells, pericytes, microglia, and macrophages. Clusters with substantial expression of two or more markers were removed as they most likely represented doublet artifacts arising from the co-capture of multiple cells in one droplet.
To resolve additional diversity across these main cell classes identified with either method, all clusters assigned to a single class were then pooled and re-analyzed by reapplying the same method (t-SNE-based approach or Seurat-based approach). On occasion clusters were identified that contained a disproportional fraction of cells either from 0, 1, or 4 h samples regardless of animal of origin, suggesting that stimulus-dependent expression was a major contributor to variance in these cell types. In all such cases the clusters expressing identical cell-type markers were combined into a single cluster. This analysis resulted in 33 final clusters across 55,986 cells for the t-SNE-based approach and 48 final clusters across 56,372 cells for the Seurat-based approach.
Intersecting results obtained from the two approaches
The results from both approaches were intersected to exclude cells that were not consistently assigned. Since there were more clusters generated by Approach 2 (48), we took each of those clusters and assessed their degree of overlap with Approach 1-generated clusters (33). First, a 2D matrix was generated with Approach 1 clusters along one dimension and Approach 2 clusters along the other. The values in the matrix corresponded to the number of cells that were shared between any Approach 1 and Approach 2 cluster. 3 of the 48 Approach 2 clusters had cells distributed across several Approach 1 clusters with no Approach 1 cluster encompassing the majority (>50%) of the cells. All the cells from these three clusters were removed from the analysis as the overlap was not deemed sufficient. For each of the remaining 45 Approach 2 clusters we identified a single corresponding Approach 1 cluster that encompassed the majority (>50%) of the cells. Only the cells that were shared between each Approach 2 cluster and a single majority-corresponding Approach 1 cluster were kept for subsequent analysis. This conservative approach filtered out 12% of the cells and generated a dataset containing 48,266 cells robustly classified into 9 main cell types (excitatory neurons, inhibitory neurons, oligodendrocytes, astrocytes, endothelial and smooth muscle cells, pericytes, microglia and macrophages) and 33 subtypes.
Classification of the fine-grained cell types
For each of the 33 predicted cell types, we identified a list of enriched genes whose expression was 3-fold greater in that cell type compared to the average expression across all other cell types (Supplementary Table 2). These genes were used to assign names to the 33 identified cell types as follows:
Excitatory neurons, marked by the expression of Vesicular Glutamate Transporter 1 (Slc17a7) and Calcium/Calmodulin Dependent Protein Kinase II Beta (Camk2b) separated into layer-specific subtypes: Layer 2/3 (ExcL23), Layer 4(ExcL4), three types of Layer 5 (ExcL5_1, ExcL5_2, ExcL5_3) and Layer 6 (ExcL6). Layer 6b cells co-clustered with Layer 6 cells and were thus analyzed together. We also observed small populations of cells derived from surrounding brain regions including subiculum, hippocampus, and retrosplenial cortex33, which were removed from subsequent analyses, yielding a total of 30 final subtypes and a dataset of 47,209 cells (with an average of 3,234 transcripts per cell).
Inhibitory neurons, identified by the expression of Glutamate Decarboxylase 1, Gad1, separated into 6 subtypes based on the expression of previously described neuropeptides. Parvalbumin+ (Pvalb) interneurons (Int_Pv) have been previously described as fast spiking basket cells. Two types of Somatostatin (Sst)-expressing interneurons were identified, Int_Sst1 and Int_Sst2. Based on Allen Brain Institute in situ hybridization data, we observed that Int_Sst_2 cells are distributed throughout the cortex while Int_Sst_1 were restricted to layer 5 and layer 6. The remaining three interneuronal cell types were Neuropeptide Y (Npy)-expressing cells (Int_Npy), layer 2/3 bitufted and bipolar Vip-expressing cells (Int_Vip), and upper layer-enriched cholecystokinin-expressing interneurons (Int_Cck). Although smaller GABA-ergic subpopulations have previously been reported, we chose to focus our analysis on these 6 most abundant subtypes.
We identified one major class of astrocytes (Astro) that expressed aldolase dehydrogenase (Aldoc). Although the marker Gfap is commonly used as a marker for astrocytes, we observed, in agreement with previously published single cell data33 and in situ hybridization, that Gfap expression was restricted to a small subset of astrocytes.
Nine subtypes of Olig1-expressing cells were identified46. Two subsets, marked by Pdgfra, represent oligodendrocyte precursor cells (OPCs). A large, likely quiescent, population of OPCs expresses C1ql1 (OPC_1), while a very small Pdgfra+ population (102 cells) expresses cell-cycle associated genes and is likely actively cycling (OPC_2)33. Seven additional populations of Olig1-expressing cells could be arranged in a continuous progression corresponding to different stages of differentiation: Bmp4+ immature cells (Olig_7), premyelinating Tmem2+ cells (Olig_6), and ending with 4 separate myelinated (Mag+, Mog+, Mbp+) populations (Olig_1, Olig_2, Olig_3, Olig_4). We also observed a separate population of mature oligodendrocytes marked by Kif5a that expresses lower levels of Mog and Mag but 5-fold higher levels of Mbp (Olig_5).
Consistent with previous reports we identified microglia and macrophages as the primary immune cell types in the brain33. Two subtypes of microglia were identified, a more abundant P2yr12 high (Micro_2) population and a less abundant P2yr12 low, Ccl3/4+ population (Micro_1). Since Ccl3 and Ccl4 are markers of macrophage activation, it is possible that this second population represents activated microglia. Macrophages were identified as Cd36+, Mrc1+ cells.
Finally, we identified 2 types of endothelial cells (Endo_1, Endo_2) and 2 types of smooth muscle cells (SM_1, SM_2) expressing the tight function protein claudin 5 (Cldn5), as well as a population of pericytes that were Cldn5− and vitronectin+ (Vtn). The less abundant of the two endothelial cell types (Endo_2) expressed high levels of hemoglobin alpha (Hb–a1, Hb-a2) and hemoglobin beta (Hb–bs, Hb-bt). Although these cells were not described in previous single cell RNA-Seq datasets, prior work has shown that arterial endothelial cells express hemoglobin alpha, which is enriched at the myoendothelial junction and regulates NO-mediated vascular reactivity68. It is therefore likely that our hemoglobin-positive cells represent arterial endothelial cells.
One population of smooth muscle cells (SM_2) expresses the ATP-binding cassette Abcc9 and the inwardly rectifying voltage-gated potassium channel Kcnj8. These proteins form an ATP-sensitive potassium channel that directly links cellular ATP metabolism with membrane depolarization. The second population of smooth muscle cells (SM_1) expresses alpha smooth muscle actin (Acta2) and myosin heavy chain (Myh11) suggesting that different smooth muscle cell types may be subspecialized in their function.
Hierarchical clustering
Hierarchical clustering across all cell types was performed on the depth-normalized dataset (each cell containing 3,234 transcripts) using the top 4,000 most variable genes (from Approach 1). Mean expression for each gene was calculated across each cell type, the distance between cell types calculated using Euclidean distance and hierarchical clustering performed in R using the Ward2 algorithm.
Identification and classification of experience-regulated transcripts
To identify experience-regulated genes for each cell type, we carried out differential gene expression analysis using Monocle269 between cells isolated from the visual cortex of mice exposed to light for 0, 1, and 4 h. The data were modeled and normalized using a negative binomial distribution consistent with scRNA-seq experiments. The analysis was performed independently for each of the 30 cell types identified, comparing 1 h to 0 h and 4 h to 0 h separately. In each analysis, a gene needed to be detected in a minimum of 5% of cells in order to be included in the differential gene expression test. Genes whose false discovery rate (FDR) was less than 0.05, and whose log2 fold change in expression was either greater than 1 or smaller than −1 were considered activity-regulated. Log2 fold change was calculated from the depth-normalized data (each cell normalized to contain 3,234 transcripts) after adding 0.12 to the expression of each gene: FC=Log2(Mean1+0.12)−Log2(Mean2+0.12).
For each cell type, activity regulated genes were classified as either early or late response genes based on their pattern of expression. If the maximum expression of an induced gene was at 1h, it was classified as early, whereas if the maximum expression was at 4h, it was classified as a late response gene. Genes whose expression decreased in response to stimulus were similarly classified based on the time point of minimum expression. If a gene was classified as early response in some cell types and late response in others, its final classification was based on the most frequent pattern of induction. If the number of cell types in which a gene was an early response and a late response was the same, that gene was classified as a late response gene. All induced genes and their classifications are listed in Supplementary Table 3. These classifications are denoted by a and b (early increase and decrease respectively), and c and d (late increase and late decrease).
Determination of induced cells within a population
The top 10 genes ranked by fold change between 0 and 1 h of light stimulation were collected for each cell type. For each gene, the 0 h sample was used to define a 90th percentile expression threshold, i.e. the 10% of cells expressing the gene would be considered positive for that gene. This threshold is meant to represent the expression level present in quiescent cells; we chose 90% to account for the likely possibility that some cells may not be silent even in the unstimulated condition. To classify a single cell as induced, we evaluated the expression of all ten of the most induced genes within that cell type and required that at least three of them have an expression level greater than the threshold set by the 90th percentile of cells from the 0h condition. To provide a range for our estimate of induction, we also plot the result of the same analysis using either two or four genes within each cell meeting this requirement as the lower and upper bounds of the box.
ERTF hierarchical clustering by expression patterns across cell types
19 ERTFs that were induced in at least three cell types were hierarchically clustered based on their log2 fold change between the 0 and 1 h conditions. The distance metric between genes was Euclidean and hierarchical clustering was performed using Ward2 algorithms.
Co-expression analysis
Depth normalized gene expression across single cells at the 1 h timepoint was correlated (Pearson correlation) between 14 neuronal-induced ERTFs. Expression-matched genes were used as a control and chosen from the list of all genes whose mean expression in excitatory neurons was between the lowest and highest of the 14 ERTFs. The 14 most closely expressed genes are shown in Fig. 3d. For Fig. 3e, the distribution of similarly expressed genes was generated by randomly sampling (100 times) 14 genes that are expression matched to the 14 ERTFs. As an additional control, the expression of each ERTF was shuffled across all cells such that the expression value for that ERTF was randomly assigned to a different cell. Shuffling across cells retained the average expression and induction of each ERTF but significantly decreased the correlation of expression across ERTFs (see Fig. 3e). Statistical differences between the distributions were computed using the Mann-Whitney U-test.
Identification of excitatory layer 2/3 and 4 subtypes
Layer 2/3 and Layer 4 excitatory cells were processed independently using the Seurat algorithm. For each set the data was log normalized and scaled to 10,000 transcripts. Variable genes were identified using the following parameters: x.low.cutoff=0.0125, x.high.cutoff=3, y.cutoff=0.5. The top 30 PCs were chosen and the clustering resolution was set to 0.6. This analysis initially identified 6 clusters from ExcL23 cells. One cluster containing 22 cells was removed as it was below the 100 cell cutoff. 4 of the remaining 5 clusters contained unbalanced numbers of cells from different time-points suggesting that they were separated by stimulus-regulated genes. Consequently, these clusters were merged into a single ExcL23_1 population. ExcL4 contained 4 clusters, two of which contained unbalanced numbers of cells from different time-points and were merged into the ExcL4_1 population.
Correlation between excitatory L2/3, L4 and L5 cell types
Fig. 4i left: A union of all late response genes initially identified in ExcL23, ExcL4, ExcL5_1, ExcL5_2 and ExcL5_3 cell types was created (51 genes). Correlation (Pearson) between cell types was determined based on the mean expression of each of the LRGs in each cell type. Fig. 4i right: A random set of expression-matched non-LRGs was generated and a correlation was computed as was done for the LRGs. This analysis was repeated 100 times and the mean pairwise correlation between cell types plotted.
Correlation between excitatory L2/3 and L4 marker genes and sequencing depth
We confirmed that the cell-type-specific expression of the marker genes shown in Fig. 4c and 4d, and in Supplementary Fig. 19 was not a function of sequencing depth across L2/3 and L4 subtypes with the analysis described below (Supplementary Fig. 20).
The depth-normalized expression level (TPT) of each marker gene was calculated for each cell assigned to a given cell type. 200 cells with 5,000 ≤ total UMIFM counts/cell ≤ 10,000 were sampled from each L2/3 or L4 subtype at random. The Pearson correlation between marker gene expression and cell sequencing depth (total UMIFM counts) was then calculated for each layer’s marker gene for all subtypes of that layer using these 200 depth-matched cells.
Cross-study comparison of transcriptionally defined cell types
Expression levels from each of the two datasets were independently scaled (mean centered, unit variance) by gene within each transcriptionally defined cell type to mitigate batch effects due to different sequencing depths, cell capture approaches, normalization approaches, and library preparation across this work and Tasic et al.23. The Pearson correlation was calculated across all pairwise combinations of cell types between the two studies using only genes expressed in both datasets (normalized expression>0) in at least one cell type. The cell types were hierarchically clustered (Ward’s method) and arranged such that the distance (1 − Pearson correlation) between proximal leaves was minimized. Cell types defined in either study with shared transcriptional identity cluster closely regardless of which study they were taken from (Supplementary Fig. 11), suggesting that our classification scheme is consistent with that used by Tasic et al.23.
Cross-study comparison between similar excitatory neuron subtypes
Using all genes expressed across both studies in at least cell type is sufficient to establish gross cross-study cell type correlations, but the subtle differences in gene expression across the particularly similar excitatory subtypes are washed out by this approach. We overcame this by further calculating cross-study correlations for the excitatory cell types alone, using excitatory marker genes identified in either this study (Fig. 3b) or in Ref.23. Hierarchical clustering was conducted as described in the previous section and the results are shown in Supplementary Fig. 16.
OPC-specific receptor and induced ligand identification
We used a human ligand-receptor database44 comprising 2,422 ligand-receptor pairs as the basis for our investigation into protein signaling between OPCs and other cell types.
To identify genes enriched in OPCs relative to other oligodendrocyte populations, we required that 1) the gene’s mean expression be higher in OPCs than in any one subtype of oligodendrocytes, 2) the average expression of the gene in OPCs be at least 3-fold greater than the mean in oligodendrocytes, and 3) the gene be in the top 50% most highly expressed genes in OPCs. Intersecting this list with all receptors in the ligand-receptor database yielded 52 OPC-enriched receptors, of which nine had corresponding ligands that were induced in at least one non-oligodendrocyte cell type.
Gene Ontology (GO) analysis
GO analysis was carried out using DAVID 6.870,71. All expressed genes for the cell type being analyzed were used background. Expressed genes were defined as genes that were detected in a minimum of 5% of cells. For the GO analysis conducted to generate Supplemental Table 1, all genes expressed with TMM-normalized CPM>1 in at least three samples were used as background.
Gene set enrichment analyses using Su et al. dataset
Two gene sets were derived directly from the differential expression analyses conducted in Ref.48 on RNA-seq data (rnaseq_1 and rnaseq_4 in Supplementary Fig. 24), with the additional requirement of a minimal 2-fold change in expression between the unstimulated and electroconvulsively stimulated conditions. The remaining two gene sets were obtained by identifying the transcriptional start sites most proximal to the stimulus-dependent, differentially accessible regions identified in Ref.48 (termed atac_1 and atac_4 in Supplementary Fig. 24) using the GRCh37.p11 annotation.
A one-sided Fisher’s exact test (FET) was used to test the following null hypothesis: The fraction of induced genes identified in visual cortex in this study (611 total) that was also identified by Su et al. as being stimulus-dependent in dentate gyrus < the fraction of stimulus-independent genes identified in this study (12,458 total genes) that was also identified by Su et al. as being stimulus-dependent in dentate gyrus. The Benjamini-Hochberg procedure was applied to correct for multiple hypothesis testing.
Statistical analysis
Fig. 1c: qRT-PCR for Fos expression divided by Gapdh expression, n=4 samples from 4 different animals per condition was analyzed using a two tailed student t-test. Data distribution was assumed to be normal although this was not tested.
Supplementary Fig. 1 and Fig. 6a: qRT-PCR for Fos or Klf4 expression divided by Gapdh expression. n=3 samples (for Motor and Prefrontal cortex) and n=4 samples (for Somatosensory and Visual cortex). Each sample is taken from a different animal. Analysis was done using a two-tailed student t-test. Data distribution was assumed to be normal although this was not tested.
Fig. 2: To identify experience-regulated genes for each cell type, we carried out differential gene expression analysis using the software package Monocle2. Remaining details are reported in the Identification and classification of experience-regulated transcripts methods section.
Fig. 3a and 3d: Nonparametric Mann-Whitney U tests were used to test the difference in the distribution of values. Fig. 3a: n(ERTFs)=38, n(LRGs)=176 where n is the number of genes in each category and the values tested are the number of cell types that each gene was identified as activity-regulated. Fig. 3d: Genes were selected as indicated in: Co-expression analysis. n=176 where n is the number of pairwise correlations between two populations based on a defined set of genes.
Fig. 1b, 3c, 4g, 6f, and Supplementary Fig. 13: For quantification of FISH images, all distributions were first tested for normality via the Kolmogorov–Smirnov test. All tested distributions rejected the null hypothesis and were treated non-parametrically. Nonparametric Mann-Whitney U tests were then used to test the difference in the distribution of values.
No statistical methods were used to pre-determine sample sizes but our sample sizes are similar to those reported in previous publications8,72,73. Mice were randomly assigned to 0h, 1h and 4h time-points. No other sample randomization was performed. Experiments were not performed in a blinded fashion. More information on experimental design and reagents can be found in the Life Sciences Reporting Summary.
Data availability
The data that support the findings of this study are available from the corresponding author. Raw and processed RNA-seq data for both single-cell and bulk experiments are available at GEO accession GSE102827.
To broadly share our data, we have also created an interactive website on which the gene expression of each of the genes in our dataset can be viewed. http://greenberg.hms.harvard.edu/project/gene-database/
Code availability
Code is available upon request.
Supplementary Material
T1. GEO terms for genes induced by drug cocktail treatment
T2. Cell-type enriched genes
T3. The 611 induced genes across cell types and time points
T4. GEO terms for sensory-experience regulated genes
S1. Fos induction by visual stimulus is restricted to V1
Fos expression measured by RT-qPCR at 0 and 1 h conditions following inDrops sample preparation in 4 brain tissues. *p=0.021, unpaired t-test, two-sided. n=4 animals for all 0 h samples, n=3 animals for 1 h motor and prefrontal cortex, n=4 animals for 1 h somatosensory and visual cortex. Bar indicates mean.
S2. Drug cocktail does not grossly alter transcription in visual cortex as measured by bulk RNA-seq
(a) Scatter plot of gene expression (log10 [TMM-normalized CPM + .01]) for 12,477 genes in visual cortex from drug-cocktail-perfused animals that were dark- (0 h) or light- (1 h) exposed (n=4 animals for each condition). 168 significantly differentially expressed genes (DEG) corresponding to the bulk visual experience-dependent program are denoted in purple (FDR<.01, fold change >=2, limma, Online Methods).
(b) Venn diagram demonstrating the overlap of the following three gene sets (in clockwise order): DEG from panel A, DEG from Fig. 1d, and the 611 cell-type-specific DEG from Fig. 2b. Approximately one half of genes (56/114) that are repressed by the cocktail were identified as visual-stimulus induced in either the bulk or single-cell RNA-seq experiments. GO analysis results for all 114 genes are listed in Supplementary Table 1.
S3. inDrops sample sequencing metrics
The samples described in the manuscript comprise 28 libraries collected across 23 animals denoted by the sample names along the x-axis. All panels in this figure represent sequencing metrics across all cells with ≥1,000 total reads (cells=114,601). For each metric, whisker plots demarcate quartiles. Data outside 1.5-times the interquartile range (Q3-Q1) were labeled as dots. The median of each distribution is marked in white (left). Each distribution is also aggregated across all samples and this histogram is plotted.
(a) Number of cells collected by sample.
(b) Total reads per cell by sample.
(c) Total mapped reads per cell by sample.
(d) Reads mapping to a unique genomic location per cell by sample
(e) UMI filtered, uniquely mapping counts per cell by sample.
S4. Flowchart of bioinformatics clustering and cell-type assignment
Workflow diagram showing bioinformatic clustering and cell-type assignment pipeline (Online Methods). The number of cells passing each stage of the pipeline are indicated in light yellow.
S5. inDrops sample gene detection metrics
(a) The number of unique genes detected per cell for all cells acquired with ≥1,000 reads per cell (cells=114,601, black), and for the subset of cells that were successfully assigned a cell type designation (cells=47,209, purple) as a histogram.
(b) Same data plotted on a scatter plot showing unique genes per cell as a function of UMIFM counts per cell.
S6. Samples and stimulus conditions are evenly distributed across cell types
(a) Distribution of 23 samples across 30 cell types shown on top of total t-SNE (top) and denoted as fraction of cells for each cell type (bottom).
(b) Same plots showing distribution of stimulus paradigm across 23 samples and 30 cell types.
(c) Quantification of the distribution shown in (a) Box plot with median (Tukey). Data outside 1.5-times the interquartile range (Q3-Q1) were labeled as dots.
S7. Expression of marker genes across all cell types
Dendrogram of cell types and violin plots showing the distribution of expression of selected marker genes across all 30 analyzed cell types. Cell types are hierarchically clustered based on gene expression across all variable genes.
S8. t-SNE plots separating cell-types within each main cell class
t-SNE plots are shown for each of the eight main cell types: excitatory neurons (14287 cells), inhibitory neurons (936 cells), astrocytes (7039 cells), oligodendrocytes and oligodendrocyte precursor cells (OPCs; 10,456 cells), microglia (10158 cells), endothelial and smooth muscle cells (4071 cells), macrophages (537 cells), and pericytes (782 cells). Colors within each plot indicate different subtypes.
S9. UMIFM counts and genes detected per cell by cell type
Distributions of (a) UMIFM counts per cell and (b) unique genes detected per cell plotted by cell type as whisker plots showing quartiles (median in white) for all cells that were successfully assigned to a cell type (cells=47209, left). Summary histograms (right) show the relevant metric aggregated across all clustered cells with means denoted by the red line. Across the 47209 clustered cells, an average of 3233.6 UMIFM counts and 1452.7 genes were obtained per cell. Data outside 1.5-times the interquartile range (Q3-Q1) were labeled as dots.
S10. Dependence of clustering on sequencing depth
All excitatory neurons (L23, L4, L5_1, L5_2, L5_3 and L6, total cell number=13,230, average UMI per cell=6,020) were pooled and three additional datasets created by randomly subsampling each cell to contain ½, ¼ or ⅛ its starting number of transcripts. All four datasets were analyzed and clustered independently using Seurat. Marker genes were used to assign each new cluster back to one of the original excitatory cell types (L23, L4, L5_1, L5_2, L5_3 and L6). Graph shows the fraction of cells in each cluster derived from the subsetted data that were correctly assigned to the original cluster in the non-subsetted dataset.
S11. Correlations between inDrops- and Tasic et al.-identified cell types
Heatmap of pairwise Pearson R2 correlations between all cell types identified in Ref.23 and this study. Magnitude of correlation denoted by color intensity as shown on scale bar. For clarified representation, values are scaled such that the highest obtained R2 across each TASIC-labeled cell type is 1. Broadly defined cell types within each of the two studies are demarcated by dotted lines.
S12. Differential expression analysis results by stimulus for all cell types
Volcano plots of –log10 Monocle2 derived q-values against log2 fold change between 0 and 1 h (a) and 0 and 4 h (b) stimulus comparisons. Cells that passed FDR<.05 and |log2FC|>1 thresholds are shown in their representative colors. Number of cells for the 0, 1, and 4 h conditions respectively: ExcL23 = 1150, 706, 1107; ExcL4 = 1211, 819, 1168; ExcL5_1 = 795, 516, 665; ExcL5_2 = 162, 104, 144; ExcL5_3 = 493, 412, 502; ExcL6 = 1328, 916, 1032; Int_Pv = 87, 66, 74; Int_Sst_1 = 61, 64, 49; Int_Sst_2 = 59, 52, 70; Int_Vip = 41, 45, 40; Int_Npy = 42, 58, 37; Int_Cck = 41, 21, 29; Olig_1 = 128, 175, 307; Olig_2 = 318, 257, 389; Olig_3 = 275, 275, 296; Olig_4 = 1220, 1204, 1623; Olig_5 = 237, 225, 285; Olig_6 = 248, 236, 302; Olig_7 = 209, 182, 239; OPC_1 = 645, 472, 608; OPC_2 = 31, 27, 43; Endo_1 = 1111, 1107, 1109; Endo_2 = 33, 65, 25; SM_1 = 115, 92, 116; SM_2 = 104, 109, 85; Micro_1 = 148, 166, 235; Micro_2 = 2873, 2381, 4355; Astro = 2234, 2156, 2649; Pericyte = 295, 200, 287; Macrophage = 187, 119, 231.
S13. Nr4a1 is induced across multiple cell types as measured by FISH
Nr4a1 expression as measured by FISH in 0 and 1 h stimulus conditions in excitatory (Vglut1+, cells0h=987, cells1h=778) and inhibitory (Gad1/2+, cells0h=238, cells1h=243) neurons, astrocytes (Aldh1l1+, cells0h=220, cells1h=89), and endothelial and smooth muscle cells (Pecam1+, cells0h=82, cells1h=67). ***p<10−12, Mann-Whitney U-test, two-sided. Mean and 95% confidence intervals denoted by gray lines.
S14. Anatomical location of putative layer 5 cell types
(a) 3-color FISH for cell type enriched markers of ExcL5 subtypes (from left to right): Gpr88 (ExcL5_3), Bcl6 (ExcL5_2), and Nnat (ExcL5_1). The combined image (right) shows largely non-overlapping markers defining the ExcL5 subtypes. The Nnat marker labeled cells in the ventral section of layer 5 and layer 6. Representative images from experiments on 2 cortical slices.
(b) To confirm expression of Nnat across both layer 5 and 6, we carried out 3-color FISH (from left to right): Vglut1, Foxp2 (a canonical layer 6 marker), and Nnat. The presence of Vglut1/Nnat positive cells dorsal of and within Foxp2 defined layer 6 supports the layer 5/6 anatomical location of the ExcL5_1 population. Scale bars=100 um. Representative images from experiments on 2 cortical slices.
S15. Newly identified ExcL23 and ExcL4 subpopulations show differential gene expression
(a, c) Volcano plots of –log10 Monocle2 derived FDR against log2 fold change for all genes between the two ExcL23 subpopulations (ExcL23_1/2) described in Fig. 4c and for all three pairwise comparisons of ExcL4 subtypes (ExcL4_1,2,3) described in Fig. 4d respectively. Genes that passed FDR<.05 and |log2 FC|>1 thresholds are shown in their representative colors. nExcL23_1=774, 539, 907 cells; nExcL23_2=362, 166, 193 cells; nExcL4_3=136, 137, 175 cells; nExcL4_1=732, 468, 783 cells; nExcL4_2=343, 214, 210 cells for 0, 1, and 4 h timepoints respectively.
(b, d) Sets comprising 100 random cells were generated from each of ExcL23_1/2 or ExcL4_1/2/3 populations. The pairwise Pearson correlations between ExcL23 subpopulations (b) and ExcL4 subpopulations (d) was calculated using mean gene expression within each set. The analysis was repeated 100 times and the mean correlations between different subpopulations of the relevant layer are plotted.
S16. Excitatory subpopulation comparison between inDrops dataset and Tasic et al.
Pairwise Pearson R2 values across all combinations of Tasic et al.- and inDrops-identified excitatory subpopulations plotted as a heatmap (Online Methods)23.
S17. Summary of inDrops- and Tasic et al.-derived cell types
A summary mapping of the Tasic et al. transcriptionally-defined cell types23 and those described in this study constructed from analyses shown in Supplemental Fig. 7, 11, 16, 19, 21, 22, 23.
S18. Excitatory subpopulations are not defined by sequencing depth
Left: Distributions of UMIFM counts (a) and unique genes (b) detected per cell represented as whisker plots demarcating quartiles and the median (white) for each excitatory subtype across all successfully clustered excitatory cells (cells=13,230). Right: Aggregated distributions represented as histograms. Means of each metric are denoted by the red lines. Data outside 1.5-times the interquartile range (Q3-Q1) were labeled as dots.
S19. L23 and L4 excitatory subtype marker genes are not stimulus-regulated
Mean normalized expression for several non-stimulus regulated marker genes between (a) L23 excitatory subtypes (nExcL23_1= nExcL23_2=) and (b) layer 4 excitatory subtypes. Error bars indicate s.e.m. nExcL23_1=774, 539, 907 cells; nExcL23_2=362, 166, 193 cells; nExcL4_3=136, 137, 175 cells; nExcL4_1=732, 468, 783 cells; nExcL4_2=343, 214, 210 cells for 0, 1, and 4 h timepoints respectively.
S20. L23 and L4 subtype marker gene expression is not correlated with sequencing depth
L23 (a) and L4 (b) subtype marker gene expression (transcripts per thousand [TPT] calculated across each cell type) plotted as functions of sequencing depth (mean UMIFM counts) across 200 randomly sampled cells (with 5000≤UMIFM count/cell≤10000) within L23 and L4 respectively (markers described in Fig. 4d, 4e, Supplemental Fig. S19, Online Methods). Within each plot, the lines of best fit are shown together with the calculated Pearson correlation and the associated p-value. The color of each data point or trend-line denotes the cell type represented by that object. We find that marker gene expression is not significantly correlated with sequencing depth. We also see that for each marker gene, the relative expression of the marker is appropriately greater in its marked sublayer despite restricting the sequencing depth ranges for the cells examined. This analysis confirms that the cell-type-specific expression of these marker genes is not due to differences in sequencing depth.
S21. Cdh13+ excitatory neurons are superficially enriched in layer 2/3
(a) FISH images showing superficial enrichment of Cdh13+ cells in Layer 2/3. Nuclei are pseudo-colored by expression level of Vglut1 (green) or Cdh13 (magenta) FISH probes (Online methods). Scale bar=100 um.
(b) Quantification of FISH data showing fraction of Cdh13+ excitatory neurons (Vglut1+) as a function of distance from the pia (cells=436 Cdh13+/Vglut1+, of cells=1,624 Vglut1+ cells). Data from 8 cortical slices.
S22. FISH confirmation of L4 subtypes
(a) We carried out 3-color FISH for (from left) Rorb (yellow), Calb1 (magenta), and Ctxn3 (cyan). We found that Ctxn3+/Rorb+/Calb1− cells were intermingled with Calb1+/Rorb+ cells superficially in layer 4. Right: summary of the anatomical distribution of the Ctxn3 and Calb1 transcriptionally defined cell types. Data collected from 6 slices of mouse visual cortex. Negative/positive values on x-axis correspond to distance from the center of layer 4 towards the slice surface (negative) or towards deeper cortical layers (positive).
(b) Summary data for all layer 4 excitatory subtypes as a percentage of total cells (left), or of Rorb+ cells (right). Calb1+/Rorb+ and Ctxn3+/Rorb+/Calb1− cell types (data collected from 19 and 6 slices respectively that contained visual cortex) were enriched superficially within layer 4 by both measures, and Hsd11b1/Rorb+/Calb1− defined cells (data collected from 13 slices containing visual cortex) were enriched deep within layer 4 and peaked at the border between layer 4 and 5 (see Fig. 4h). Shaded area around lines indicate 95% confidence intervals.
(c) Quantification of cell population enrichment to superficial (−75 to 0 um from L4 center) or deep (0 to 75 um from L4 center) layer 4. The transcriptionally defined cell types Calb1+/Rorb+ and Ctxn3/Rorb+/Calb1− were significantly enriched superficially (p=0.0002, p=0.015, Mann-Whitney U-test, two-sided; data collected from 19 and 6 slices containing visual cortex, respectively), and Hsd11b1+/Rorb+/Calb1− cells were enriched deep within layer 4 (p=0.0034, Mann-Whitney U-test, two-sided; data collected from 13 slices containing visual cortex). On the other hand, the total number of cells in superficial and deep layer 4 was not significantly different (p=0.19, Mann-Whitney U-test, two-sided; data collected from 19 slices containing visual cortex). Scale bars=150 um. Shaded area around lines indicate 95% confidence intervals.
S23. Interneuron comparison between inDrops and Tasic et al. datasets
Scaled expression of interneuronal markers used to assign cell types across all interneuronal populations identified in this work and by Ref.23. Expression levels were scaled for each gene from each cell type independently (centered to mean with unit variance, Online Methods).
S24. V1-identified induced genes are highly enriched for genes associated with inducibly accessible loci in dentate gyrus
Induced gene sets from dentate gyrus comprising the genes with the most proximal TSS to inducibly accessible loci identified by Su et al. at 1 and 4 hours of stimulation (atac_1, atac_4) or the genes directly identified by Su et al. via bulk RNA-seq (rnaseq_1, rnaseq_4) were tested for enrichment among the induced genes identified in this study (Online Methods)48. The fractions of the visual cortex induced gene set identified in this study (611 genes) and its complement that were also induced in Su et al.’s paradigm are plotted. All gene sets show significant enrichment. Relative risk=1.84, 1.75, 3.67, 2.50; FDR-corrected q-value=3.2e-13, 3.4e-8, 3.4e-35, 2.7e-13 (in order shown in Figure). ***FDR-corrected q<1e-5.
Acknowledgments
We thank E. Griffith and J. Gray for critical reading of the manuscript, the HMS Single Cell Core for sample processing, and D. Harmin, A. Veres, D. Kotliar, H. Finucane, Y. Reshef, B. Hall, T. Otis, and D. Malhotra for discussions. This work was supported by grants from the National Institute of Health R01 NS028829 and the ROADS Program funded by F. Hoffmann-La Roche Ltd. to M.E.G., R01 NS046579 to B.L.S., the William F. Milton Fund to D.R.H., NIH grant T32GM007753 to M.A.N., NIH Training in the Molecular Biology of Neurodegeneration 5T32AG000222-23 to S.H., and the Burroughs Wellcome Fund Career award at the scientific interface, an Edward J. Mallinckrodt Scholarship, and NCI grant R33CA212697 to A.M.K.
Footnotes
Accession Codes
All sequencing data is deposited all GEO under accession number GSE102827.
Author Contributions
S.H., D.R.H., M.A.N. conceived the study and designed all experiments. S.H., D.R.H developed the optimized dissociation protocol. S.H., D.R.H. performed single cell experiments with assistance from R.Z., A.M.K. and A.R. M.A.N. processed all sequencing data. S.H, M.A.N., D.R.H., analyzed data with R.B.M. and A.M.K.’s assistance. D.R.H., K.R, and L.C carried out FISH experiments and imaging, D.R.H analyzed the FISH data. M.C., D.R.H. developed automated software for FISH analysis. S.H., D.R.H., M.A.N, and M.E.G. wrote the manuscript. M.E.G. and B.L.S. advised on all aspects of the study.
Competing Financial Interests
The authors declare no competing financial interests.
References
- 1.Hensch TK. Critical period plasticity in local cortical circuits. Nat. Rev. Neurosci. 2005;6:877–888. doi: 10.1038/nrn1787. [DOI] [PubMed] [Google Scholar]
- 2.Wiesel TN, Hubel DH. SINGLE-CELL RESPONSES IN STRIATE CORTEX OF KITTENS DEPRIVED OF VISION IN ONE EYE. J. Neurophysiol. 1963;26:1003–1017. doi: 10.1152/jn.1963.26.6.1003. [DOI] [PubMed] [Google Scholar]
- 3.Zucker RS, Regehr WG. Short-term synaptic plasticity. Annu. Rev. Physiol. 2002;64:355–405. doi: 10.1146/annurev.physiol.64.092501.114547. [DOI] [PubMed] [Google Scholar]
- 4.West AE, Greenberg ME. Neuronal Activity–Regulated Gene Transcription in Synapse Development and Cognitive Function. Cold Spring Harb. Perspect. Biol. 2011;3 doi: 10.1101/cshperspect.a005744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bading H. Nuclear calcium signalling in the regulation of brain function. Nat. Rev. Neurosci. 2013;14:593–608. doi: 10.1038/nrn3531. [DOI] [PubMed] [Google Scholar]
- 6.MacManus JP, et al. Translation-state analysis of gene expression in mouse brain after focal ischemia. J. Cereb. Blood Flow Metab. Off. J. Int. Soc. Cereb. Blood Flow Metab. 2004;24:657–667. doi: 10.1097/01.WCB.0000123141.67811.91. [DOI] [PubMed] [Google Scholar]
- 7.Ginty DD, et al. Regulation of CREB phosphorylation in the suprachiasmatic nucleus by light and a circadian clock. Science. 1993;260:238–241. doi: 10.1126/science.8097062. [DOI] [PubMed] [Google Scholar]
- 8.Campbell JN, et al. A molecular census of arcuate hypothalamus and median eminence cell types. Nat. Neurosci. 2017;20:484–496. doi: 10.1038/nn.4495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Renier N, et al. Mapping of Brain Activity by Automated Volume Analysis of Immediate Early Genes. Cell. 2016;165:1789–1802. doi: 10.1016/j.cell.2016.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ben-David E, Shifman S. Networks of Neuronal Genes Affected by Common and Rare Variants in Autism Spectrum Disorders. PLOS Genet. 2012;8:e1002556. doi: 10.1371/journal.pgen.1002556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Thomas GM, Huganir RL. MAPK cascade signalling and synaptic plasticity. Nat. Rev. Neurosci. 2004;5:173–183. doi: 10.1038/nrn1346. [DOI] [PubMed] [Google Scholar]
- 12.Shepherd JD, Bear MF. New views of Arc, a master regulator of synaptic plasticity. Nat. Neurosci. 2011;14:279–284. doi: 10.1038/nn.2708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mardinly AR, et al. Sensory experience regulates cortical inhibition by inducing IGF1 in VIP neurons. Nature. 2016;531:371–375. doi: 10.1038/nature17187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Harward SC, et al. Autocrine BDNF–TrkB signalling within a single dendritic spine. Nature. 2016;538:99–103. doi: 10.1038/nature19766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lin Y, et al. Activity-dependent regulation of inhibitory synapse development by Npas4. Nature. 2008;455:1198–1204. doi: 10.1038/nature07319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Majdan M, Shatz CJ. Effects of visual experience on activity-dependent gene regulation in cortex. Nat. Neurosci. 2006;9:650–659. doi: 10.1038/nn1674. [DOI] [PubMed] [Google Scholar]
- 17.Spiegel I, et al. Npas4 Regulates Excitatory-Inhibitory Balance within Neural Circuits through Cell-Type-Specific Gene Programs. Cell. 2014;157:1216–1229. doi: 10.1016/j.cell.2014.03.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lacoste B, et al. Sensory-related neural activity regulates the structure of vascular networks in the cerebral cortex. Neuron. 2014;83:1117–1130. doi: 10.1016/j.neuron.2014.07.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gibson EM, et al. Neuronal Activity Promotes Oligodendrogenesis and Adaptive Myelination in the Mammalian Brain. Science. 2014;344:1252304. doi: 10.1126/science.1252304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang X, et al. Astrocytic Ca2+ signaling evoked by sensory stimulation in vivo. Nat. Neurosci. 2006;9:816–823. doi: 10.1038/nn1703. [DOI] [PubMed] [Google Scholar]
- 21.Klein AM, et al. Droplet Barcoding for Single-Cell Transcriptomics Applied to Embryonic Stem Cells. Cell. 2015;161:1187–1201. doi: 10.1016/j.cell.2015.04.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lacar B, et al. Nuclear RNA-seq of single neurons reveals molecular signatures of activation. Nat. Commun. 2016;7:11022. doi: 10.1038/ncomms11022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tasic B, et al. Adult mouse cortical cell taxonomy revealed by single cell transcriptomics. Nat. Neurosci. 2016;19:335–346. doi: 10.1038/nn.4216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Caillard O, et al. Role of the calcium-binding protein parvalbumin in short-term synaptic plasticity. Proc. Natl. Acad. Sci. U. S. A. 2000;97:13372–13377. doi: 10.1073/pnas.230362997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wu G-Y, Deisseroth K, Tsien RW. Activity-dependent CREB phosphorylation: Convergence of a fast, sensitive calmodulin kinase pathway and a slow, less sensitive mitogen-activated protein kinase pathway. Proc. Natl. Acad. Sci. 2001;98:2808–2813. doi: 10.1073/pnas.051634198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Harris KD, Shepherd GMG. The neocortical circuit: themes and variations. Nat. Neurosci. 2015;18:170–181. doi: 10.1038/nn.3917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chacón PJ, et al. Cerebellin 4, a synaptic protein, enhances inhibitory activity and resistance of neurons to amyloid-β toxicity. Neurobiol. Aging. 2015;36:1057–1071. doi: 10.1016/j.neurobiolaging.2014.11.006. [DOI] [PubMed] [Google Scholar]
- 28.Hattox AM, Nelson SB. Layer V Neurons in Mouse Cortex Projecting to Different Targets Have Distinct Physiological Properties. J. Neurophysiol. 2007;98:3330–3340. doi: 10.1152/jn.00397.2007. [DOI] [PubMed] [Google Scholar]
- 29.Schulz TW, et al. Actin/alpha-actinin-dependent transport of AMPA receptors in dendritic spines: role of the PDZ-LIM protein RIL. J. Neurosci. Off. J. Soc. Neurosci. 2004;24:8584–8594. doi: 10.1523/JNEUROSCI.2100-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Niell CM, Stryker MP. Highly selective receptive fields in mouse visual cortex. J. Neurosci. Off. J. Soc. Neurosci. 2008;28:7520–7536. doi: 10.1523/JNEUROSCI.0623-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.de Kock CPJ, Bruno RM, Spors H, Sakmann B. Layer- and cell-type-specific suprathreshold stimulus representation in rat primary somatosensory cortex. J. Physiol. 2007;581:139–154. doi: 10.1113/jphysiol.2006.124321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nassi JJ, Callaway EM. Parallel Processing Strategies of the Primate Visual System. Nat. Rev. Neurosci. 2009;10:360–372. doi: 10.1038/nrn2619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zeisel A, et al. Cell types in the mouse cortex and hippocampus revealed by single-cell RNA-seq. Science. 2015;347:1138–1142. doi: 10.1126/science.aaa1934. [DOI] [PubMed] [Google Scholar]
- 34.Kratzer SMD, et al. Activation of CRH receptor type 1 expressed on glutamatergic neurons increases excitability of CA1 pyramidal neurons by the modulation of voltage-gated ion channels. Front. Cell. Neurosci. 2013;7 doi: 10.3389/fncel.2013.00091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li K, Nakajima M, Ibañez-Tallon I, Heintz N. A Cortical Circuit for Sexually Dimorphic Oxytocin-Dependent Anxiety Behaviors. Cell. 2016;167:60–72.e11. doi: 10.1016/j.cell.2016.08.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Roy CS, Sherrington CS. On the Regulation of the Blood-supply of the Brain. J. Physiol. 1890;11:85–158.17. doi: 10.1113/jphysiol.1890.sp000321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Attwell D, et al. Glial and neuronal control of brain blood flow. Nature. 2010;468:232–243. doi: 10.1038/nature09613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gurnik S, et al. Angiopoietin-2-induced blood–brain barrier compromise and increased stroke size are rescued by VE-PTP-dependent restoration of Tie2 signaling. Acta Neuropathol. (Berl.) 2016;131:753–773. doi: 10.1007/s00401-016-1551-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Augustin HG, Young Koh G, Thurston G, Alitalo K. Control of vascular morphogenesis and homeostasis through the angiopoietin–Tie system. Nat. Rev. Mol. Cell Biol. 2009;10:165–177. doi: 10.1038/nrm2639. [DOI] [PubMed] [Google Scholar]
- 40.Weinl C, et al. Endothelial SRF/MRTF ablation causes vascular disease phenotypes in murine retinae. J. Clin. Invest. 2013;123:2193–2206. doi: 10.1172/JCI64201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhou Z, et al. Cerebral cavernous malformations arise from endothelial gain of MEKK3-KLF2/4 signalling. Nature. 2016;532:122–126. doi: 10.1038/nature17178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hines JH, Ravanelli AM, Schwindt R, Scott EK, Appel B. Neuronal activity biases axon selection for myelination in vivo. Nat. Neurosci. 2015;18:683–689. doi: 10.1038/nn.3992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Miyata S, Hattori T, Shimizu S, Ito A, Tohyama M. Disturbance of Oligodendrocyte Function Plays a Key Role in the Pathogenesis of Schizophrenia and Major Depressive Disorder. BioMed Res. Int. 2015;2015:e492367. doi: 10.1155/2015/492367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ramilowski JA, et al. A draft network of ligand–receptor-mediated multicellular signalling in human. Nat. Commun. 2015;6:7866. doi: 10.1038/ncomms8866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Habib N, et al. Div-Seq: Single-nucleus RNA-Seq reveals dynamics of rare adult newborn neurons. Science. 2016;353:925–928. doi: 10.1126/science.aad7038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Marques S, et al. Oligodendrocyte heterogeneity in the mouse juvenile and adult central nervous system. Science. 2016;352:1326–1329. doi: 10.1126/science.aaf6463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gokce O, et al. Cellular Taxonomy of the Mouse Striatum as Revealed by Single-Cell RNA-Seq. Cell Rep. 2016;16:1126–1137. doi: 10.1016/j.celrep.2016.06.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Su Y, et al. Neuronal activity modifies the chromatin accessibility landscape in the adult brain. Nat. Neurosci. 2017;20:476–483. doi: 10.1038/nn.4494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tu Z. Computer Vision and Pattern Recognition, 2008. CVPR 2008. IEEE Conference on. IEEE; 2008. Auto-context and its application to high-level vision tasks; pp. 1–8. [Google Scholar]
- 50.Richmond D, Kainmueller D, Glocker B, Rother C, Myers G. International Conference on Medical Image Computing and Computer-Assisted Intervention. Springer; 2015. Uncertainty-driven forest predictors for vertebra localization and segmentation; pp. 653–660. [Google Scholar]
- 51.Aguet F, Antonescu CN, Mettlen M, Schmid SL, Danuser G. Advances in analysis of low signal-to-noise images link dynamin and AP2 to the functions of an endocytic checkpoint. Dev. Cell. 2013;26:279–291. doi: 10.1016/j.devcel.2013.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cicconet M, Hochbaum DR, Richmond D, Sabatini BL. Bots for Software-Assisted Analysis of Image-Based Transcriptomics. bioRxiv. 2017:172296. [Google Scholar]
- 53.Fan J. Notes on Poisson distribution-based minimum error thresholding. Pattern Recognit. Lett. 1998;19:425–431. [Google Scholar]
- 54.Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinforma. Oxf. Engl. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinforma. Oxf. Engl. 2009;25:1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Liao Y, Smyth GK, Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinforma. Oxf. Engl. 2014;30:923–930. doi: 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- 58.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinforma. Oxf. Engl. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.McCarthy DJ, Chen Y, Smyth GK. Differential expression analysis of multifactor RNA-Seq experiments with respect to biological variation. Nucleic Acids Res. 2012;40:4288–4297. doi: 10.1093/nar/gks042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Law CW, Chen Y, Shi W, Smyth GK. voom: Precision weights unlock linear model analysis tools for RNA-seq read counts. Genome Biol. 2014;15:R29. doi: 10.1186/gb-2014-15-2-r29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ritchie ME, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zilionis R, et al. Single-cell barcoding and sequencing using droplet microfluidics. Nat. Protoc. 2017;12:44–73. doi: 10.1038/nprot.2016.154. [DOI] [PubMed] [Google Scholar]
- 63.Islam S, et al. Quantitative single-cell RNA-seq with unique molecular identifiers. Nat. Methods. 2014;11:163–166. doi: 10.1038/nmeth.2772. [DOI] [PubMed] [Google Scholar]
- 64.Maaten L van der, Hinton G. Visualizing Data using t-SNE. J. Mach. Learn. Res. 2008;9:2579–2605. [Google Scholar]
- 65.Rodriguez A, Laio A. Clustering by fast search and find of density peaks. Science. 2014;344:1492–1496. doi: 10.1126/science.1242072. [DOI] [PubMed] [Google Scholar]
- 66.Satija R, Farrell JA, Gennert D, Schier AF, Regev A. Spatial reconstruction of single-cell gene expression data. Nat. Biotechnol. 2015;33:495–502. doi: 10.1038/nbt.3192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Macosko EZ, et al. Highly Parallel Genome-wide Expression Profiling of Individual Cells Using Nanoliter Droplets. Cell. 2015;161:1202–1214. doi: 10.1016/j.cell.2015.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Straub AC, et al. Endothelial cell expression of haemoglobin α regulates nitric oxide signalling. Nature. 2012;491:473–477. doi: 10.1038/nature11626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Trapnell C, et al. The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nat. Biotechnol. 2014;32:381–386. doi: 10.1038/nbt.2859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 71.Huang DW, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Shekhar K, et al. Comprehensive Classification of Retinal Bipolar Neurons by Single-Cell Transcriptomics. Cell. 2016;166:1308–1323.e30. doi: 10.1016/j.cell.2016.07.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Baron M, et al. A Single-Cell Transcriptomic Map of the Human and Mouse Pancreas Reveals Inter- and Intra-cell Population Structure. Cell Syst. 2016;3:346–360.e4. doi: 10.1016/j.cels.2016.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
T1. GEO terms for genes induced by drug cocktail treatment
T2. Cell-type enriched genes
T3. The 611 induced genes across cell types and time points
T4. GEO terms for sensory-experience regulated genes
S1. Fos induction by visual stimulus is restricted to V1
Fos expression measured by RT-qPCR at 0 and 1 h conditions following inDrops sample preparation in 4 brain tissues. *p=0.021, unpaired t-test, two-sided. n=4 animals for all 0 h samples, n=3 animals for 1 h motor and prefrontal cortex, n=4 animals for 1 h somatosensory and visual cortex. Bar indicates mean.
S2. Drug cocktail does not grossly alter transcription in visual cortex as measured by bulk RNA-seq
(a) Scatter plot of gene expression (log10 [TMM-normalized CPM + .01]) for 12,477 genes in visual cortex from drug-cocktail-perfused animals that were dark- (0 h) or light- (1 h) exposed (n=4 animals for each condition). 168 significantly differentially expressed genes (DEG) corresponding to the bulk visual experience-dependent program are denoted in purple (FDR<.01, fold change >=2, limma, Online Methods).
(b) Venn diagram demonstrating the overlap of the following three gene sets (in clockwise order): DEG from panel A, DEG from Fig. 1d, and the 611 cell-type-specific DEG from Fig. 2b. Approximately one half of genes (56/114) that are repressed by the cocktail were identified as visual-stimulus induced in either the bulk or single-cell RNA-seq experiments. GO analysis results for all 114 genes are listed in Supplementary Table 1.
S3. inDrops sample sequencing metrics
The samples described in the manuscript comprise 28 libraries collected across 23 animals denoted by the sample names along the x-axis. All panels in this figure represent sequencing metrics across all cells with ≥1,000 total reads (cells=114,601). For each metric, whisker plots demarcate quartiles. Data outside 1.5-times the interquartile range (Q3-Q1) were labeled as dots. The median of each distribution is marked in white (left). Each distribution is also aggregated across all samples and this histogram is plotted.
(a) Number of cells collected by sample.
(b) Total reads per cell by sample.
(c) Total mapped reads per cell by sample.
(d) Reads mapping to a unique genomic location per cell by sample
(e) UMI filtered, uniquely mapping counts per cell by sample.
S4. Flowchart of bioinformatics clustering and cell-type assignment
Workflow diagram showing bioinformatic clustering and cell-type assignment pipeline (Online Methods). The number of cells passing each stage of the pipeline are indicated in light yellow.
S5. inDrops sample gene detection metrics
(a) The number of unique genes detected per cell for all cells acquired with ≥1,000 reads per cell (cells=114,601, black), and for the subset of cells that were successfully assigned a cell type designation (cells=47,209, purple) as a histogram.
(b) Same data plotted on a scatter plot showing unique genes per cell as a function of UMIFM counts per cell.
S6. Samples and stimulus conditions are evenly distributed across cell types
(a) Distribution of 23 samples across 30 cell types shown on top of total t-SNE (top) and denoted as fraction of cells for each cell type (bottom).
(b) Same plots showing distribution of stimulus paradigm across 23 samples and 30 cell types.
(c) Quantification of the distribution shown in (a) Box plot with median (Tukey). Data outside 1.5-times the interquartile range (Q3-Q1) were labeled as dots.
S7. Expression of marker genes across all cell types
Dendrogram of cell types and violin plots showing the distribution of expression of selected marker genes across all 30 analyzed cell types. Cell types are hierarchically clustered based on gene expression across all variable genes.
S8. t-SNE plots separating cell-types within each main cell class
t-SNE plots are shown for each of the eight main cell types: excitatory neurons (14287 cells), inhibitory neurons (936 cells), astrocytes (7039 cells), oligodendrocytes and oligodendrocyte precursor cells (OPCs; 10,456 cells), microglia (10158 cells), endothelial and smooth muscle cells (4071 cells), macrophages (537 cells), and pericytes (782 cells). Colors within each plot indicate different subtypes.
S9. UMIFM counts and genes detected per cell by cell type
Distributions of (a) UMIFM counts per cell and (b) unique genes detected per cell plotted by cell type as whisker plots showing quartiles (median in white) for all cells that were successfully assigned to a cell type (cells=47209, left). Summary histograms (right) show the relevant metric aggregated across all clustered cells with means denoted by the red line. Across the 47209 clustered cells, an average of 3233.6 UMIFM counts and 1452.7 genes were obtained per cell. Data outside 1.5-times the interquartile range (Q3-Q1) were labeled as dots.
S10. Dependence of clustering on sequencing depth
All excitatory neurons (L23, L4, L5_1, L5_2, L5_3 and L6, total cell number=13,230, average UMI per cell=6,020) were pooled and three additional datasets created by randomly subsampling each cell to contain ½, ¼ or ⅛ its starting number of transcripts. All four datasets were analyzed and clustered independently using Seurat. Marker genes were used to assign each new cluster back to one of the original excitatory cell types (L23, L4, L5_1, L5_2, L5_3 and L6). Graph shows the fraction of cells in each cluster derived from the subsetted data that were correctly assigned to the original cluster in the non-subsetted dataset.
S11. Correlations between inDrops- and Tasic et al.-identified cell types
Heatmap of pairwise Pearson R2 correlations between all cell types identified in Ref.23 and this study. Magnitude of correlation denoted by color intensity as shown on scale bar. For clarified representation, values are scaled such that the highest obtained R2 across each TASIC-labeled cell type is 1. Broadly defined cell types within each of the two studies are demarcated by dotted lines.
S12. Differential expression analysis results by stimulus for all cell types
Volcano plots of –log10 Monocle2 derived q-values against log2 fold change between 0 and 1 h (a) and 0 and 4 h (b) stimulus comparisons. Cells that passed FDR<.05 and |log2FC|>1 thresholds are shown in their representative colors. Number of cells for the 0, 1, and 4 h conditions respectively: ExcL23 = 1150, 706, 1107; ExcL4 = 1211, 819, 1168; ExcL5_1 = 795, 516, 665; ExcL5_2 = 162, 104, 144; ExcL5_3 = 493, 412, 502; ExcL6 = 1328, 916, 1032; Int_Pv = 87, 66, 74; Int_Sst_1 = 61, 64, 49; Int_Sst_2 = 59, 52, 70; Int_Vip = 41, 45, 40; Int_Npy = 42, 58, 37; Int_Cck = 41, 21, 29; Olig_1 = 128, 175, 307; Olig_2 = 318, 257, 389; Olig_3 = 275, 275, 296; Olig_4 = 1220, 1204, 1623; Olig_5 = 237, 225, 285; Olig_6 = 248, 236, 302; Olig_7 = 209, 182, 239; OPC_1 = 645, 472, 608; OPC_2 = 31, 27, 43; Endo_1 = 1111, 1107, 1109; Endo_2 = 33, 65, 25; SM_1 = 115, 92, 116; SM_2 = 104, 109, 85; Micro_1 = 148, 166, 235; Micro_2 = 2873, 2381, 4355; Astro = 2234, 2156, 2649; Pericyte = 295, 200, 287; Macrophage = 187, 119, 231.
S13. Nr4a1 is induced across multiple cell types as measured by FISH
Nr4a1 expression as measured by FISH in 0 and 1 h stimulus conditions in excitatory (Vglut1+, cells0h=987, cells1h=778) and inhibitory (Gad1/2+, cells0h=238, cells1h=243) neurons, astrocytes (Aldh1l1+, cells0h=220, cells1h=89), and endothelial and smooth muscle cells (Pecam1+, cells0h=82, cells1h=67). ***p<10−12, Mann-Whitney U-test, two-sided. Mean and 95% confidence intervals denoted by gray lines.
S14. Anatomical location of putative layer 5 cell types
(a) 3-color FISH for cell type enriched markers of ExcL5 subtypes (from left to right): Gpr88 (ExcL5_3), Bcl6 (ExcL5_2), and Nnat (ExcL5_1). The combined image (right) shows largely non-overlapping markers defining the ExcL5 subtypes. The Nnat marker labeled cells in the ventral section of layer 5 and layer 6. Representative images from experiments on 2 cortical slices.
(b) To confirm expression of Nnat across both layer 5 and 6, we carried out 3-color FISH (from left to right): Vglut1, Foxp2 (a canonical layer 6 marker), and Nnat. The presence of Vglut1/Nnat positive cells dorsal of and within Foxp2 defined layer 6 supports the layer 5/6 anatomical location of the ExcL5_1 population. Scale bars=100 um. Representative images from experiments on 2 cortical slices.
S15. Newly identified ExcL23 and ExcL4 subpopulations show differential gene expression
(a, c) Volcano plots of –log10 Monocle2 derived FDR against log2 fold change for all genes between the two ExcL23 subpopulations (ExcL23_1/2) described in Fig. 4c and for all three pairwise comparisons of ExcL4 subtypes (ExcL4_1,2,3) described in Fig. 4d respectively. Genes that passed FDR<.05 and |log2 FC|>1 thresholds are shown in their representative colors. nExcL23_1=774, 539, 907 cells; nExcL23_2=362, 166, 193 cells; nExcL4_3=136, 137, 175 cells; nExcL4_1=732, 468, 783 cells; nExcL4_2=343, 214, 210 cells for 0, 1, and 4 h timepoints respectively.
(b, d) Sets comprising 100 random cells were generated from each of ExcL23_1/2 or ExcL4_1/2/3 populations. The pairwise Pearson correlations between ExcL23 subpopulations (b) and ExcL4 subpopulations (d) was calculated using mean gene expression within each set. The analysis was repeated 100 times and the mean correlations between different subpopulations of the relevant layer are plotted.
S16. Excitatory subpopulation comparison between inDrops dataset and Tasic et al.
Pairwise Pearson R2 values across all combinations of Tasic et al.- and inDrops-identified excitatory subpopulations plotted as a heatmap (Online Methods)23.
S17. Summary of inDrops- and Tasic et al.-derived cell types
A summary mapping of the Tasic et al. transcriptionally-defined cell types23 and those described in this study constructed from analyses shown in Supplemental Fig. 7, 11, 16, 19, 21, 22, 23.
S18. Excitatory subpopulations are not defined by sequencing depth
Left: Distributions of UMIFM counts (a) and unique genes (b) detected per cell represented as whisker plots demarcating quartiles and the median (white) for each excitatory subtype across all successfully clustered excitatory cells (cells=13,230). Right: Aggregated distributions represented as histograms. Means of each metric are denoted by the red lines. Data outside 1.5-times the interquartile range (Q3-Q1) were labeled as dots.
S19. L23 and L4 excitatory subtype marker genes are not stimulus-regulated
Mean normalized expression for several non-stimulus regulated marker genes between (a) L23 excitatory subtypes (nExcL23_1= nExcL23_2=) and (b) layer 4 excitatory subtypes. Error bars indicate s.e.m. nExcL23_1=774, 539, 907 cells; nExcL23_2=362, 166, 193 cells; nExcL4_3=136, 137, 175 cells; nExcL4_1=732, 468, 783 cells; nExcL4_2=343, 214, 210 cells for 0, 1, and 4 h timepoints respectively.
S20. L23 and L4 subtype marker gene expression is not correlated with sequencing depth
L23 (a) and L4 (b) subtype marker gene expression (transcripts per thousand [TPT] calculated across each cell type) plotted as functions of sequencing depth (mean UMIFM counts) across 200 randomly sampled cells (with 5000≤UMIFM count/cell≤10000) within L23 and L4 respectively (markers described in Fig. 4d, 4e, Supplemental Fig. S19, Online Methods). Within each plot, the lines of best fit are shown together with the calculated Pearson correlation and the associated p-value. The color of each data point or trend-line denotes the cell type represented by that object. We find that marker gene expression is not significantly correlated with sequencing depth. We also see that for each marker gene, the relative expression of the marker is appropriately greater in its marked sublayer despite restricting the sequencing depth ranges for the cells examined. This analysis confirms that the cell-type-specific expression of these marker genes is not due to differences in sequencing depth.
S21. Cdh13+ excitatory neurons are superficially enriched in layer 2/3
(a) FISH images showing superficial enrichment of Cdh13+ cells in Layer 2/3. Nuclei are pseudo-colored by expression level of Vglut1 (green) or Cdh13 (magenta) FISH probes (Online methods). Scale bar=100 um.
(b) Quantification of FISH data showing fraction of Cdh13+ excitatory neurons (Vglut1+) as a function of distance from the pia (cells=436 Cdh13+/Vglut1+, of cells=1,624 Vglut1+ cells). Data from 8 cortical slices.
S22. FISH confirmation of L4 subtypes
(a) We carried out 3-color FISH for (from left) Rorb (yellow), Calb1 (magenta), and Ctxn3 (cyan). We found that Ctxn3+/Rorb+/Calb1− cells were intermingled with Calb1+/Rorb+ cells superficially in layer 4. Right: summary of the anatomical distribution of the Ctxn3 and Calb1 transcriptionally defined cell types. Data collected from 6 slices of mouse visual cortex. Negative/positive values on x-axis correspond to distance from the center of layer 4 towards the slice surface (negative) or towards deeper cortical layers (positive).
(b) Summary data for all layer 4 excitatory subtypes as a percentage of total cells (left), or of Rorb+ cells (right). Calb1+/Rorb+ and Ctxn3+/Rorb+/Calb1− cell types (data collected from 19 and 6 slices respectively that contained visual cortex) were enriched superficially within layer 4 by both measures, and Hsd11b1/Rorb+/Calb1− defined cells (data collected from 13 slices containing visual cortex) were enriched deep within layer 4 and peaked at the border between layer 4 and 5 (see Fig. 4h). Shaded area around lines indicate 95% confidence intervals.
(c) Quantification of cell population enrichment to superficial (−75 to 0 um from L4 center) or deep (0 to 75 um from L4 center) layer 4. The transcriptionally defined cell types Calb1+/Rorb+ and Ctxn3/Rorb+/Calb1− were significantly enriched superficially (p=0.0002, p=0.015, Mann-Whitney U-test, two-sided; data collected from 19 and 6 slices containing visual cortex, respectively), and Hsd11b1+/Rorb+/Calb1− cells were enriched deep within layer 4 (p=0.0034, Mann-Whitney U-test, two-sided; data collected from 13 slices containing visual cortex). On the other hand, the total number of cells in superficial and deep layer 4 was not significantly different (p=0.19, Mann-Whitney U-test, two-sided; data collected from 19 slices containing visual cortex). Scale bars=150 um. Shaded area around lines indicate 95% confidence intervals.
S23. Interneuron comparison between inDrops and Tasic et al. datasets
Scaled expression of interneuronal markers used to assign cell types across all interneuronal populations identified in this work and by Ref.23. Expression levels were scaled for each gene from each cell type independently (centered to mean with unit variance, Online Methods).
S24. V1-identified induced genes are highly enriched for genes associated with inducibly accessible loci in dentate gyrus
Induced gene sets from dentate gyrus comprising the genes with the most proximal TSS to inducibly accessible loci identified by Su et al. at 1 and 4 hours of stimulation (atac_1, atac_4) or the genes directly identified by Su et al. via bulk RNA-seq (rnaseq_1, rnaseq_4) were tested for enrichment among the induced genes identified in this study (Online Methods)48. The fractions of the visual cortex induced gene set identified in this study (611 genes) and its complement that were also induced in Su et al.’s paradigm are plotted. All gene sets show significant enrichment. Relative risk=1.84, 1.75, 3.67, 2.50; FDR-corrected q-value=3.2e-13, 3.4e-8, 3.4e-35, 2.7e-13 (in order shown in Figure). ***FDR-corrected q<1e-5.
Data Availability Statement
The data that support the findings of this study are available from the corresponding author. Raw and processed RNA-seq data for both single-cell and bulk experiments are available at GEO accession GSE102827.
To broadly share our data, we have also created an interactive website on which the gene expression of each of the genes in our dataset can be viewed. http://greenberg.hms.harvard.edu/project/gene-database/