Abstract
The TET enzymes oxidize 5-methylcytosine to 5-hydroxymethylcytosine, which can lead to DNA demethylation. However, direct connections between TET-mediated DNA demethylation and transcriptional output are difficult to establish due to challenges of distinguishing global versus locus-specific effects. Here we show that TET1/2/3 triple knockout (TKO) human embryonic stem cells (hESCs) exhibit prominent bivalent promoter hypermethylation without an overall corresponding gene expression decrease in the undifferentiated state. Focusing on the bivalent PAX6 locus, we find increased DNMT3B binding is associated with promoter hypermethylation, which precipitates a neural differentiation defect and failure of PAX6 induction during differentiation. dCas9-mediated locus-specific demethylation and global inactivation of DNMT3B in TKO hESCs partially reverses the hypermethylation at the PAX6 promoter and improves differentiation to neuroectoderm. Taken together with further genome-wide methylation and TET1 and DNMT3B ChIP-Seq analysis, we conclude that the TET proteins safeguard bivalent promoters from de novo methylation to ensure robust lineage-specific transcription upon differentiation.
DNA methylation is a key mechanism for transcriptional regulation, and dramatic changes in DNA methylation of regulatory regions occur during normal development and in pathological conditions1–2. Deletion of the DNA methyltransferases (Dnmt1, Dnmt3a and Dnmt3b) in mice leads to embryonic or postnatal lethality3–4. Proper development also requires active and passive DNA demethylation 5. The three mammalian TET proteins (TET1, TET2 and TET3) catalyze the oxidation of 5-methylcytosine (5mC) to 5-hydroxymethylcytosine (5hmC), which can serve as an intermediate in active DNA demethylation6–8. Loss of the TET proteins leads to hypermethylation of the promoters and enhancers of developmental genes in mouse embryos and mouse embryonic stem cells (mESCs)7, 9–11. Although TET inactivation clearly impairs embryonic development and cellular differentiation, it is difficult to establish direct causal relationships between TET-mediated DNA demethylation, transcriptional output and developmental or pathological phenotypes due to the general challenge of distinguishing global versus locus-specific effects for epigenetic regulators12–14.
In order to link transcriptional outcomes to TET-mediated demethylation, previous studies have focused on TET actions at enhancers and found examples in which loss of the TET proteins causes hypermethylation and decreased gene expression11,15. However, the consequences of TET activities at promoters remain ambiguous. In particular, bivalent promoters, which have the H3K4me3 and H3K27me3 marks on the same or adjacent nucleosomes, are hypomethylated like active promoters. Yet unlike active promoters, which support productive transcription, bivalent promoters are associated with negligible transcription similar to silent promoters that have high levels of DNA methylation16–19. Thus at bivalent promoters the importance of DNA methylation for gene expression regulation is not readily apparent.
Human embryonic stem cells (hESCs) reflect a later developmental stage than mESCs. Here we have generated viable hESC lines with mutations in all 3 TET genes (TKO hESCs). Although hESCs have greater global CpG methylation than mESCs20, inactivating the TET genes still produces hypermethylation in a locus-specific manner. This hypermethylation is observed among enhancers and other regulatory regions, and is particularly prominent at bivalent promoters. In the absence of the TET proteins, the de novo methyltransferase DNMT3B causes aberrant hypermethylation at bivalent promoters, which leads to impaired gene activation upon differentiation. Thus the TET proteins are necessary to maintain hypomethylation at bivalent promoters, which is critical for proper cellular differentiation during early human development.
Bivalent promoter hypermethylation in TKO hESCs
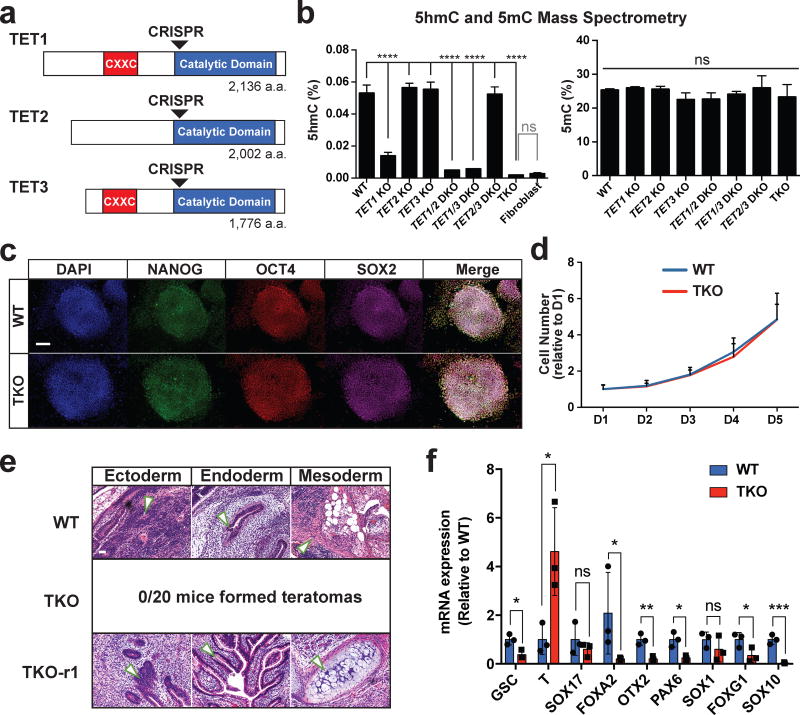
Since all three TET genes are expressed in hESCs (Supplementary Fig. 1a), and none has been genetically deleted previously, we used the iCRISPR platform developed in our lab21 to generate a panel of TET1/2/3 knockout lines in the HUES8 and MEL-1 hESC backgrounds (Supplementary Tables 1, 2, Fig. 1a). hESCs in which all 3 TET genes have been inactivated (TKO hESCs) had no detectable 5hmC signal by mass spectrometry or 5hmC dot blot (Fig. 1b, Supplementary Fig. 1b, 1c) but showed no difference in morphology, self-renewal capacity or pluripotency marker expression when compared to wild-type (WT) hESCs (Fig. 1c–d, Supplementary Fig. 1d). However, TKO hESCs showed a complete inability to form teratomas and impaired induction of key early differentiation genes upon spontaneous embryoid body differentiation (Fig. 1e–f), suggesting that the TET proteins may be particularly important for the regulation of cellular differentiation.
Figure 1. TET TKO hESCs exhibit differentiation defects.
a, TET knockout mutants were generated using CRISPR gRNAs (arrowheads) that target the beginning of the catalytic domain of TET1, TET2 and TET3. b, Analysis of 5hmC (left) and 5mC (right) in HUES8 WT and TET knockout hESCs by mass spectrometry. For all mass spectrometry analysis, 2 mutant lines were used for all KO genotypes except for TKO. For TKO lines, 2 different passages of the same line were used for mass spectrometry measurements. Human fibroblasts were used as a negative control for mass spectrometry of 5hmC. Data presented are mean ± STD. Statistical analysis: black lines indicate comparisons to WT, one-way ANOVA, ****P<0.0001. c, Pluripotency markers NANOG, OCT4 and SOX2, were detected by immunofluorescence. Scale bar indicates 100 µm. d, Growth curves for WT and TKO hESCs, n= 3 independent experiments. Data presented are mean ± STD. e, Hematoxylin and eosin staining of teratoma sections from WT teratomas. Arrows point to representative tissues for the respective germ layers. No teratomas were obtained 6 months after injection of TKO hESCs. f, Expression for markers of mesoderm (GSC T), endoderm (SOX17 FOXA2), neuroectoderm (OTX2 PAX6 SOX1 FOXG1) and neural crest (SOX10) at D12 of spontaneous embryoid body differentiation, n= 3 independent experiments. Data presented are mean ± STD. Statistical analysis: Student’s t test (two sided), *P<0.05,**P<0.01,***P<0.001.
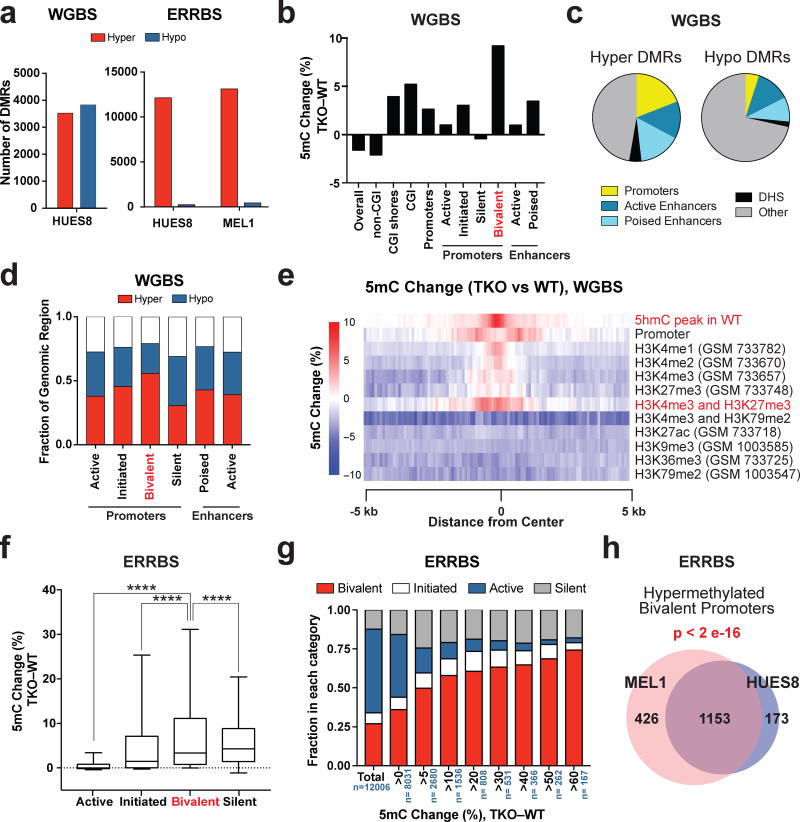
Loss of the TET genes results in locus-specific hypermethylation rather than a global gain of methylation. Mass spectrometry analysis did not show a difference in 5mC levels between TKO and WT hESCs (Fig. 1b, Supplementary Fig. 1c) similar to previous findings in mESCs11. Instead, whole genome bisulfite sequencing (WGBS) of the HUES8 WT and TKO hESCs identified 3,523 hypermethylated differentially methylated regions (hyper-DMRs) with at least 5 hypermethylated CpGs and ≥10% methylation difference when comparing TKO to WT hESCs (Supplementary Data Set 1). Using the same criterion, we also observed 3,832 hypomethylated differentially methylated regions (hypo-DMRs) (Fig. 2a). These hypo-DMRs primarily occurred at CpGs outside of CpG islands (CGI) (Fig. 2b). They could be a direct result of TET inactivation or a secondary effect, possibly due to the redirection of the DNMT proteins to novel sites in TKO hESCs. Importantly, CGIs are enriched in regulatory regions, and they showed increased methylation in TKO hESCs (Fig. 2b). Furthermore, hyper-DMRs were enriched for regulatory regions such as promoters and enhancers (Fig. 2c). Compared to other promoter types, bivalent promoters had the greatest magnitude of methylation increase (Fig. 2b) and a relatively high proportion of bivalent promoters gained methylation (Fig. 2d), as exemplified by the HOXA7 and HOXA9 loci21 (Supplementary Fig. 1e, Supplementary Data Set 2). Indeed significant gain of methylation in TKO hESCs was found to center around genomic sites with both H3K4me3 and H3K27me3 marks (Fig. 2e). Finally, we performed genome wide 5hmC profiling through 5hmC-Seal22 and found that DNA hypermethylation in TKO hESCs was detected most strongly in regions that had 5hmC peaks in WT hESCs (Fig. 2e), supporting our hypothesis that the loss of the TET proteins is directly responsible for the gain of methylation. Gene Ontology (GO) analysis revealed that hypermethylated bivalent promoters in particular, but also hypermethylated poised enhancers (marked by H3K4me1 but not H3K27ac), were associated with developmental categories, suggesting that these methylation aberrations could be responsible for the differentiation defects of TKO hESCs (Supplementary Fig. 2a).
Figure 2. Hypermethylation of bivalent promoters in TET TKO hESCs.
a, Total number of hyper-DMRs (TKO vs. WT) for HUES8 and MEL1 TKO lines compared to HUES8 and MEL1 WT lines by whole genome bisulfite sequencing (WGBS, left) and enhanced reduced representation bisulfite sequencing (ERRBS, right). b, Average percent DNA methylation change between HUES8 TKO and WT hESCs by WGBS at different genomic regions. The definitions for the different genomic regions can be found in the Methods section. c, Enrichment of various regulatory regions in hyper and hypo-DMRs by WGBS. (DHS) DNase I Hypersensitive sites. The definitions for the different genomic regions can be found in the Methods section. d, Fraction of genomic regions that show >5% increase in 5mC (Hyper) or a >5% decrease in 5mC (Hypo) in methylation between TKO and WT hESCs by WGBS of HUES8 WT and TKO hESCs. e,, Heat map of the average 5mC level differences between HUES8 TKO and WT hESCs at the center of the annotated histone modifications by WGBS. f, DNA methylation change between HUES8 TKO and WT hESCs by ERRBS at different promoter types. Box and whisker plots were generated using the methylation change at individual promoters. The error bars show 10 and 90 percent confidence intervals and the bar at the center of the box and whisker plot indicates the median. The promoters are divided into four groups based on histone modification patterns. The details of promoter definitions can be found in the Methods section, n= 2 independent experiments. Statistical analysis: one-way ANOVA, ****P<0.0001. g, Representation of active, initiated, bivalent and silent promoters among promoters that show different degrees of methylation change between HUES8 TKO and HUES8 WT hESCs by ERRBS, n indicates the total number of promoters in each DNA methylation change group, n= 2 independent experiments. h, Overlap of the bivalent promoters that show greater than 5% methylation increase in HUES8 and MEL1 TKO lines compared to HUES8 and MEL1 WT lines by ERRBS. The p-value for the overlap between HUES8 and MEL1 hypermethylation at bivalent promoters is given (Fisher’s exact test). WGBS: n = 1 independent experiment, ERRBS: n = 2 independent experiments (2 independent experiments for HUES8 WT and HUES8 TKO, 2 independent experiments for MEL1 WT and MEL1 TKO).
To further investigate the methylation changes at bivalent promoters we performed enhanced reduced representation bisulfite sequencing (ERRBS), which focuses on CGIs and thus increases sequencing coverage of promoter regions23. By ERRBS, we observed similar methylation changes in the MEL1 TKO line as the HUES8 TKO line. ERRBS analysis showed ~12,000 hyper-DMRs and an insignificant number of hypo-DMRs in TKO lines compared to WT hESCs (Fig. 2a, Supplementary Data Sets 3, 4). We further analyzed the methylation change based on promoter types: bivalent promoters (marked by H3K4me3 and H3K27me3), active promoters (marked by H3K4me3 and H3K79me2), initiated promoters (marked by H3K4me3 only) and silent promoters (based on the absence of H3K4me3)24. Overall about half of bivalent promoters showed hypermethylation by ERRBS and WGBS analysis (Fig. 2d, Supplementary Fig. 2b). Although WGBS also showed that a fraction of promoters became hypomethylated after TET inactivation, this was not observed by ERRBS. The hypomethylation detected by WGBS is likely due to hypomethylation of non-CGI CpGs that are frequently found within or near the promoter regions.
Compared to other promoter types, bivalent and silent promoters showed a greater magnitude of methylation increase in HUES8 and MEL1 TKO lines (Fig. 2f, Supplementary Fig. 2c). Importantly we observed increasing fractions of bivalent promoters among promoters with greater methylation changes (Fig. 1g, Supplementary Fig. 2d). Bivalent promoters constituted 60–74% of all promoters with >60% increase in DNA methylation, greatly exceeding the overall ~27% composition of bivalent promoters evaluated by ERRBS. We observed similar results in both HUES8 and MEL1 TKO hESCs, with 1,326 and 1,579 bivalent promoters gaining methylation, respectively (Supplementary Data Sets 5, 6). Individual bivalent promoters showed similar methylation changes between these two lines (Supplementary Fig. 2e) and 87% of the 1,326 hypermethylated bivalent promoters in HUES8 TKO hESCs also gained methylation in MEL1 TKO hESCs (Fig.2h). Overall these results indicate that the TET proteins are critical to preserve hypomethylation at bivalent promoters and in their absence a reproducible subset of bivalent promoters becomes aberrantly hypermethylated.
We also compared our data with previous mESC TKO data11. Out of the 732 bivalent promoters that become hypermethylated (>20% increase) in TKO mESCs, 517 are also bivalent in hESCs, out of which 289 (~56%) were associated with hyper-DMRs in TKO hESCs. The remaining 215 promoters are not bivalent in hESCs, and only 55 (~25.6%) were associated with hyper-DMRs in TKO hESCs (Supplementary Fig. 2f). Thus bivalent promoters found in both mESCs and hESCs tend to show similar methylation changes after TET inactivation.
Hypermethylation of the PAX6 promoter
TKO hESCs showed relatively few transcriptional changes compared to WT cells (Supplementary Fig. 3a). We found that hypermethylation of active, initiated and silent promoters was associated with a decrease in gene expression. In contrast, genes associated with hypermethylated bivalent promoters and poised enhancers did not show an overall change in expression in TKO hESCs (Supplementary Fig. 3b). 5mC MassArray Epityper analysis confirmed that the bivalent promoters of selected developmental genes (FOXA2, GATA2, PAX6, SOX10, and SOX17) showed significant hypermethylation in TKO hESCs, whereas the active promoters for housekeeping and pluripotency genes did not (Supplementary Fig. 3c). The hypermethylation of the LEFTY2 enhancer (an active enhancer marked by H3K4me1 and H3K27ac) in TKO hESCs was associated with a significant decrease in gene expression, as has been previously described in TKO mouse embryos10 (Supplementary Fig. 3c–d); whereas no expression change was detected for housekeeping and pluripotency genes. Notably hypermethylation of the bivalent promoters was not associated with a consistent decrease in gene expression. For instance no change in PAX6 expression was observed. A few bivalent promoter genes showed up- or downregulation, but the expression levels were generally low (Supplementary Fig. 3d, Supplementary Data Set 7).
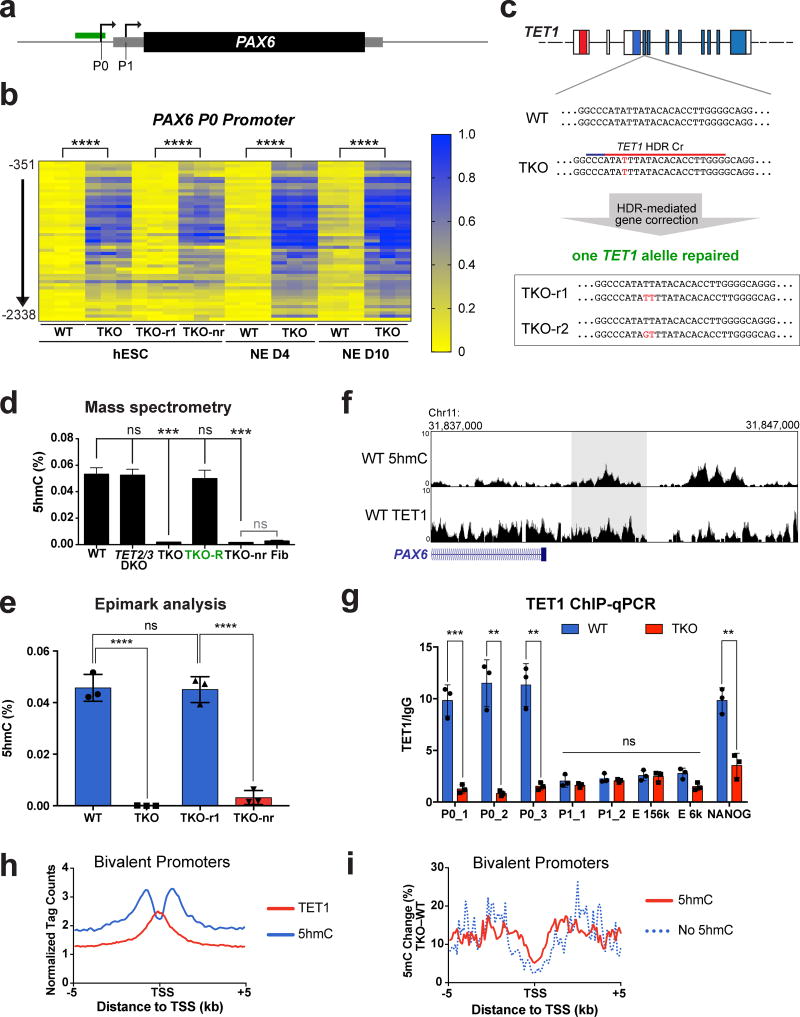
We were intrigued that despite promoter hypermethylation, bivalent promoter genes such as PAX6 did not show a change in expression at the hESC stage (Supplementary Fig 3d). Noticing that PAX6 (as well as FOXA2 and SOX10) failed to be upregulated upon spontaneous embryoid body differentiation (Fig. 1f), we speculated that the hypermethylation at bivalent promoters could affect activation of gene expression following exposure to differentiation signals. We chose to focus on PAX6 as it is a critical gene for human neural development and is highly expressed during in vitro differentiation of hESCs into the neural lineage25–27, thus allowing us to use neural differentiation to track the effects of TET deletion on PAX6 expression as well as cellular differentiation. PAX6 has well annotated promoters and enhancers (Fig. 3a, Supplementary Fig. 4a)28–32. 5mC MassArray analysis of these regions revealed that only the bivalent P0 promoter of PAX6 showed hypermethylation (Fig. 3b, Supplementary Fig. 4b), which was recapitulated in the MEL1 TKO line (Supplementary Fig. 4c). We further generated two “TKO-repaired” lines through CRISPR/Cas-mediated homology directed repair for comparison with isogenic WT and TKO hESCs to rule out potential CRISPR off-target effects (Fig. 3c). Repair of one TET1 allele to the WT sequence in TKO hESCs was sufficient to restore 5hmC to near WT levels by mass spectrometry (Fig. 3d), and importantly, it also reversed the PAX6 P0 promoter hypermethylation (Fig. 3b). Notably the TKO-repaired line was able to form teratomas, indicating a rescue in their differentiation capacity (Fig. 1e).
Figure 3. TKO hESCs show hypermethylation at the PAX6 P0 bivalent promoter.
a, Schematic for analysis of 5mC, 5hmC and TET1 binding at the PAX6 locus. Arrows represent the P0 and P1 promoter, the grey box represents the PAX6 mRNA transcript and the black box represents the PAX6 protein. The region analyzed for 5mC using MassArray and for 5hmC using hMe-Seal profiling is shown by a green box. b, Heat map of MassArray analysis of 5mC at the PAX6 P0 promoter. The location of each row of CpGs with respect to the P0 TSS is shown to the left of the heat map. For each cell line three independent experiments are shown as three columns. (NE D4) Neuroectoderm differentiation day 4; (NE D10) Neuroectoderm Differentiation day 10. Statistical analysis: Student’s t test (two sided), ****P<0.0001. c, Diagram of homology-directed repair (HDR) of the TET1 locus in TKO hESCs. Red letters indicate mutations of the WT sequence. The sequences of the two repaired lines (TKO-r1, TKO-r2) are shown below. d, Mass spectrometry analysis of 5hmC levels in WT, TET KO mutant lines, lines in which one allele of TET1 has been repaired (TKO-R) and lines which underwent HDR targeting but retained the TKO mutations in the TET1 locus (TKO-nr). For all mass spectrometry analysis, 2 mutant lines were used for all genotypes except for TKO. For TKO lines, 2 different passages of the same line were used for mass spectrometry measurements. Data presented are mean ± STD. Statistical analysis: black lines indicate comparisons to WT, one-way ANOVA, ***P<0.001. e, Analysis of percent 5hmC at the PAX6 P0 promoter by Epimark, n= 3 independent experiments. Data presented are mean ± STD. Statistical analysis: Student’s t test (two sided), ****P<0.0001. f, Top panel: Analysis of 5hmC peak at the PAX6 P0 promoter by hMe-Seal in WT hESCs. Bottom panel: Analysis of TET1 peak at the PAX6 P0 promoter by TET1 ChIP-Seq in WT hESCs. Shaded area represents the region of the PAX6 P0 promoter assayed for 5hmC by Epimark (e) and TET1 binding by ChIP-qPCR (g). g, ChIP-qPCR for TET1 in WT and TKO hESCs, n= 3 independent experiments. Data presented are mean ± STD. Statistical analysis: Student’s t test (two sided), *P<0.05, **P<0.01, ***P<0.001. h, Analysis of 5hmC and TET1 peaks at bivalent promoters in WT hESCs. The height above the x-axis reflects the normalized tag count. i, Percent DNA methylation change (TKO – WT) in bivalent promoters that have 5hmC peaks compared to bivalent promoters that don’t have 5hmC peaks.
To confirm that the hypermethylation of the PAX6 P0 promoter is a direct consequence of losing the TET proteins, we performed 5hmC analysis by the Epimark 5hmC Analysis Kit33 and genome-wide hMe-Seal22: both showed 5hmC enrichment at the PAX6 P0 promoter in WT hESCs (Fig. 3e,f). We also detected TET1 binding at the PAX6 P0 promoter in WT hESCs by ChIP-Seq and ChIP-qPCR (Fig. 3f,g). Approximately 51% of 5hmC peaks at promoters and 75% of 5hmC peaks globally overlapped with TET1 peaks (Supplementary Fig. 4d–e). Incomplete overlap between 5hmC and TET1 could be due to 5hmC production by TET2 or TET3. Globally, TET1 binds to bivalent, active and initiated promoters at the transcription start site. Binding of TET1 overlaps with 5hmC signals, which extends into the promoter and gene body (Fig. 3h, Supplementary Fig. 4f). The functional relevance of TET binding and 5hmC signal is supported by a greater methylation increase in TKO hESCs associated with bivalent promoters that have 5hmC peaks compared to bivalent promoters that don’t have 5hmC peaks (Fig. 3i, Supplementary Fig. 4g). Overall ~92% and ~50% of hyper-DMRs at bivalent promoters overlapped with 5hmC or TET1 peaks, respectively (Supplementary Fig. 4h). The greater overlap between hyper-DMRs and 5hmC peaks is likely due to the production of 5hmC by TET2 and TET3. Overall these findings support the conclusion that TET1 binding at bivalent promoters (such as the PAX6 P0 promoter) leads to 5hmC production, and TET inactivation causes 5hmC depletion and aberrant promoter hypermethylation.
Impaired neural differentiation of TKO hESCs
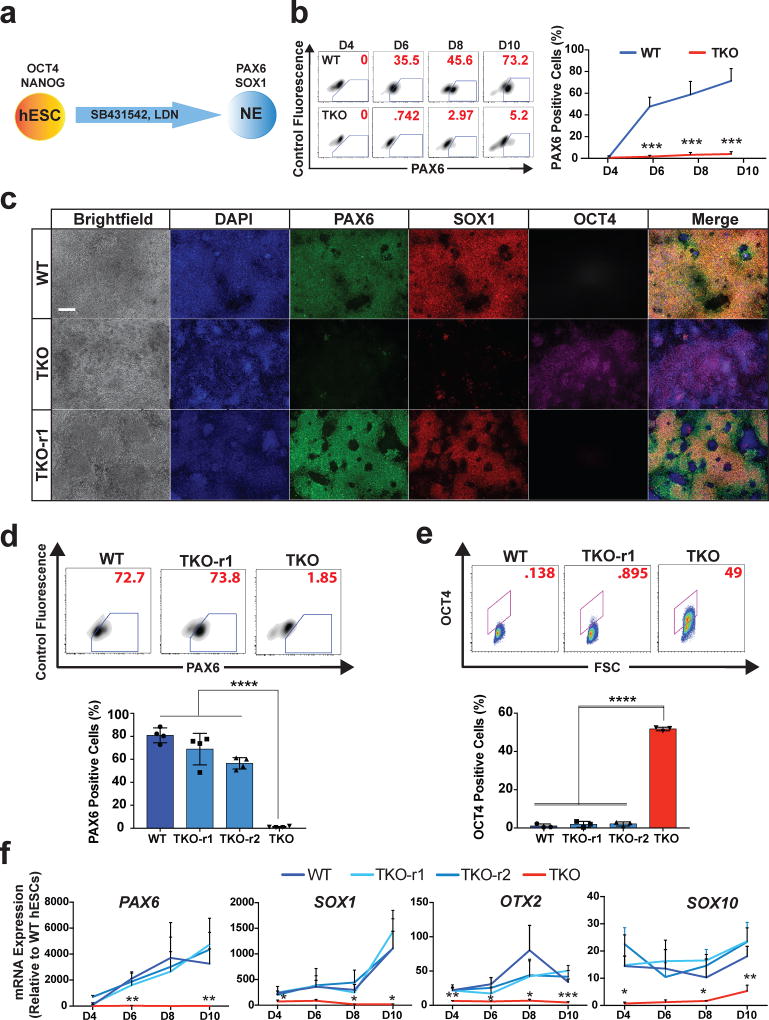
Since PAX6 is expressed in early neuroectoderm (NE) derived from hESCs26–27, and it is shown to be both necessary and sufficient for NE formation from hESCs25, we speculate that hypermethylation of the PAX6 P0 promoter in TKO hESCs may impede hESC differentiation into the neural lineage as suggested by the embryoid body differentiation results (Fig. 1f).
We performed NE differentiation using the dual SMAD inhibition protocol34 (Fig. 4a, Supplementary Fig. 5a). In contrast to WT and TKO-repaired hESCs, TKO hESCs failed to form significant numbers of PAX6-positive NE cells at any point during the differentiation (Fig. 4b–d), suggesting a defect in the acquisition rather than the maintenance of the NE fate. Notably with differentiation of TKO hESCs, ~40% of cells remained OCT4-positive after 10 days of differentiation as shown by immunostaining and FACS analysis (Fig. 4c, e). Analysis of additional markers as well as the MEL1 TKO line confirmed the NE differentiation defect of TKO hESCs (Fig. 4f, Supplementary Fig. 5b–d). Notably, comparison of TKO and TET single and double knockout lines showed that the severity of the NE differentiation defect depended on the TET1/2/3 gene dosage. Loss of TET1 had the largest effect on bulk 5hmC levels (Fig. 1b) as well as NE differentiation as determined by FACS, immunostaining, and RT-qPCR analysis for PAX6 and SOX1 expression (Supplementary Fig. 6a–c).
Figure 4. TKO hESCs show a defect in neuroectoderm differentiation.
a, Schematic for neuroectoderm (NE) differentiation. b, Representative FACS plots of PAX6 staining at D4, D6, D8 and D10 of WT and TKO cells (left panel). On the right is the quantification of PAX6-positive cells at D4, D6, D8 and D10 of NE differentiation, n= 3 independent experiments. Data presented are mean ± STD. Statistical analysis: Student’s t test (two sided), ***P<0.001. c, Immunofluorescence of PAX6, SOX1 and OCT4 at the endpoint of differentiation (D10) of WT, TKO and TKO-r1 cells. Scale bar indicates 100 µm. d, Representative FACS plots (top) and quantification (bottom) of PAX6 staining at D10 of NE Differentiation, n= 3 independent experiments. Data presented are mean ± STD. Statistical analysis: one-way ANOVA, ****P<0.0001. e, Representative FACS plots (top) and quantification (bottom) of OCT4 staining at D10 of NE differentiation, n= 3 independent experiments. Data presented are mean ± STD. Statistical analysis: one-way ANOVA, ****P<0.0001. f, qPCR analysis for epiblast (OTX2), neuroectoderm (PAX6 SOX1 and OTX2) and neural crest (SOX10) markers during NE differentiation, n= 3 independent experiments. Data presented are mean ± STD. Statistical analysis: one-way ANOVA, *P<0.05, **P<0.01, ***P<0.001.
Promoter hypermethylation hinders PAX6 expression upon differentiation
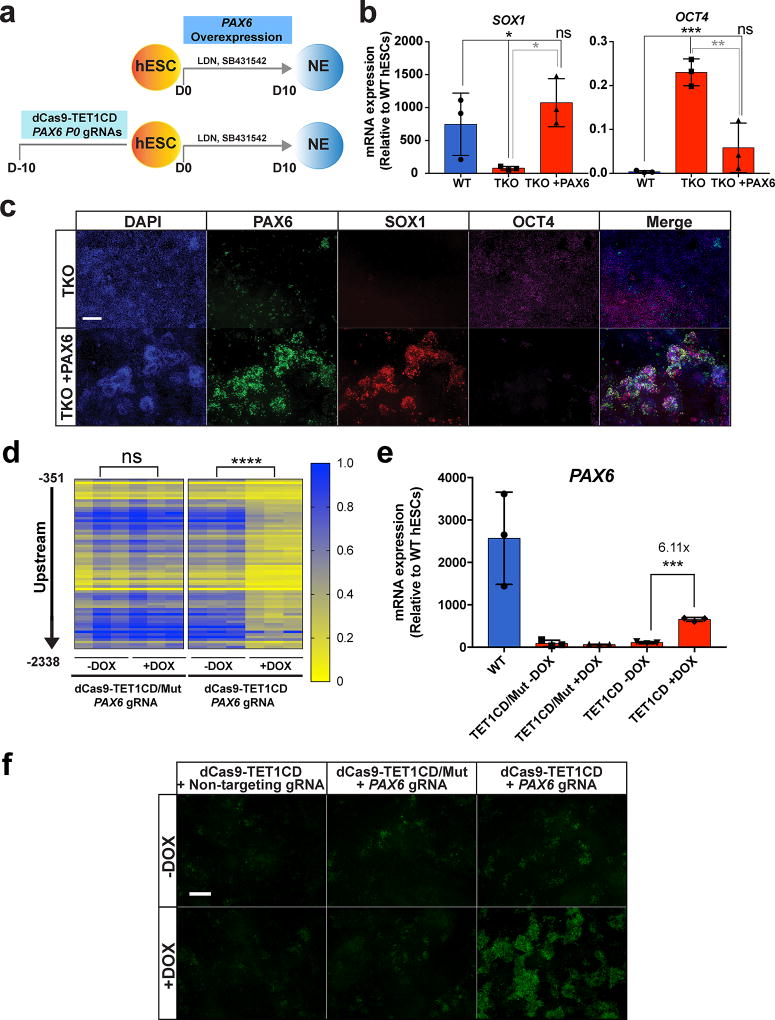
PAX6 is expressed at a very low level in hESCs, and the P0 promoter hypermethylation in TKO hESCs had no effect on PAX6 gene expression in hESCs (Supplementary Fig. 3d). We hypothesized that the hypermethylation of the PAX6 P0 promoter prevents activation of PAX6 expression upon differentiation and leads to the NE differentiation defect in TKO hESCs. Supporting this hypothesis, 5mC MassArray analysis showed aberrant hypermethylation at the PAX6 P0 promoter in TKO hESCs both before and during NE differentiation (Fig. 3b). To establish direct causality we needed to determine whether loss of PAX6 expression accounts for the NE differentiation defect of TKO hESCs and then investigate whether the P0 promoter hypermethylation is responsible for the loss of PAX6 expression. We first performed a rescue experiment in which we expressed the PAX6 transgene under the control of a doxycycline inducible promoter in TKO cells during NE differentiation (Fig. 5a). TKO cells exposed to doxycycline were able to upregulate NE markers SOX1 and FOXG1 and downregulate the pluripotency markers OCT4 and NANOG (Fig. 5b–c, Supplementary Fig. 7a). However, SOX10 and endogenous PAX6 expression were not restored (Supplementary Fig. 7a). Compared to the promoters of SOX1 and FOXG1, the SOX10 and PAX6 promoters showed a much greater methylation increase in TKO hESCs (Supplementary Data Set 5), which may prevent their expression even when the PAX6 transgene is overexpressed.
Figure 5. Hypermethylation of the PAX6 P0 bivalent promoter in TKO hESCs leads to a failure of PAX6 induction upon neuroectoderm differentiation.
a, Schema for rescue of the NE differentiation defect in TKO hESCs using PAX6 overexpression or targeted demethylation of the PAX6 P0 promoter. b, qPCR analysis of neuroectoderm (SOX1) and pluripotency (OCT4) markers in WT and TKO cells without doxycycline (TKO) and with doxycycline treatment (TKO +PAX6) at D10 of NE differentiation, n= 3 independent experiments. Data presented are mean ± STD. Statistical analysis: one-way ANOVA, *P<0.05, **P<0.01, ***P<0.001. c, Immunofluorescence of PAX6, SOX1 and OCT4 at the endpoint of differentiation (D10) of TKO cells without doxycycline treatment (TKO) and with doxycycline treatment (TKO + PAX6). Scale bar indicates 100 µm. d, Heat map of MassArray analysis of 5mC at the PAX6 P0 promoter for TKO hESCs that express PAX6 targeting gRNAs with either a dCas9-TET1CD/Mut (left) or a dCas9-TET1CD (right) fusion protein. The location of each row of CpGs with respect to the P0 TSS is shown to the left of the heat map. Methylation analysis at the PAX6 P0 promoter was analyzed for these cell lines with and without doxycycline treatment, n= 3 independent experiments. Statistical analysis: Student’s t test (two sided), ****P<0.0001. e, qPCR of PAX6 expression on D10 of NE differentiation for TKO hESCs that express PAX6 targeting gRNAs with either a dCas9-TET1CD/Mut or a dCas9-TET1CD fusion protein. PAX6 expression was analyzed for these cell lines with and without doxycycline treatment prior to differentiation, n= 3 independent experiments. Data presented are mean ± STD. Statistical analysis: Student’s t test (two sided), ***P<0.001. f, Immunofluorescence of PAX6 on D10 of NE differentiation for TKO hESCs that express PAX6 targeting gRNAs with either a dCas9-TET1CD/Mut or a dCas9-TET1CD fusion protein. TKO hESCs expressing the dCas9-TET1CD fusion and a non-targeting gRNA were also used as a control. PAX6 immunofluorscence was analyzed for these cell lines with and without doxycycline treatment prior to differentiation. Scale bar indicates 100 µm.
The above findings suggest that failure of PAX6 induction is largely responsible for the impaired NE differentiation observed in TKO hESCs. The failure of PAX6 induction could be due to hypermethylation of the PAX6 P0 promoter. Alternatively, it could result from other causes, such as the failure to induce upstream transcription factors that activate PAX6 expression or perhaps a failure of exiting the pluripotency network. Thus we investigated whether reversing hypermethylation specifically at the PAX6 P0 promoter could rescue PAX6 induction during NE differentiation. We developed a targeted demethylation strategy by first fusing the TET1 catalytic domain (TET1CD) to a nuclease “dead” Cas9 (dCas9, Supplementary Fig. 7b). With the appropriate gRNAs this dCas9-TET1CD fusion protein can be recruited to target genomic sites where the TET1 catalytic domain can oxidize 5mC to 5hmC and induce DNA demethylation as shown in recent studies using similar strategies35–38. We transiently transfected 3 individual CRISPR gRNAs targeting the PAX6 P0 promoter region with TET1 enrichment and a 5hmC peak in WT hESCs (Supplementary Fig. 7c). All 3 gRNAs, Cr6, Cr7 and Cr9, caused a significant decrease in methylation by MassArray analysis at the PAX6 P0 promoter in TKO hESCs compared to the non-transfected controls (Supplementary Fig. 7d).
To enable persistent demethylation, we used a lentiviral vector to constitutively express the Cr6, Cr7 and Cr9 gRNAs in TKO hESCs with the doxycycline inducible dCas9-TET1CD fusion protein. A number of controls were performed, including a no doxycycline control, a non-targeting gRNA control, and a control dCas9-TET1CD mutated fusion protein (dCas9-TET1CD/Mut), in which the TET1 catalytic domain has been inactivated by targeted mutagenesis (Supplementary Fig. 7b). After 10 days of doxycycline treatment we performed 5mC MassArray analysis at the PAX6 P0 promoter (Fig. 5a), and observed greater demethylation in TKO hESCs that expressed both the dCas9-TET1CD and the 3 PAX6 P0 gRNAs than in the controls (Fig. 5d, Supplementary Fig. 7e). NE differentiation was then performed without further doxycycline treatment. dCas9-TET1CD TKO hESCs expressing PAX6 P0 gRNAs and treated with doxycycline prior to NE differentiation showed upregulation of PAX6 expression after NE differentiation, while the controls did not (Fig. 5e–f, Supplementary Fig. 7e). Thus the PAX6 promoter hypermethylation observed in undifferentiated TKO hESCs is directly responsible for the failure of PAX6 induction following NE differentiation.
We further tested dCas9-TET1CD mediated targeted demethylation on the hypermethylated bivalent promoter of SOX10, and the hypermethylated enhancer of LEFTY2 in TKO hESCs. We found that targeted demethylation of the hypermethylated LEFTY2 enhancer was able to increase expression of LEFTY2 in TKO hESCs. In contrast targeted demethylation of the hypermethylated SOX10 bivalent promoter had no effect on SOX10 expression at the hESC stage but increased SOX10 expression after differentiation, as seen with PAX6 (Supplementary Fig. 7e). These results support the conclusion that hypermethylation of a bivalent promoter impairs gene expression upon differentiation. Although the targeted demethylation induces a significant increase in gene expression that is comparable to previous reports35–38, the expression did not reach WT levels. This may be due to incomplete demethylation. Additional chromatin changes at the target locus or elsewhere in TKO hESCs could also affect the target gene expression directly or indirectly.
De novo methylation causes PAX6 promoter hypermethylation
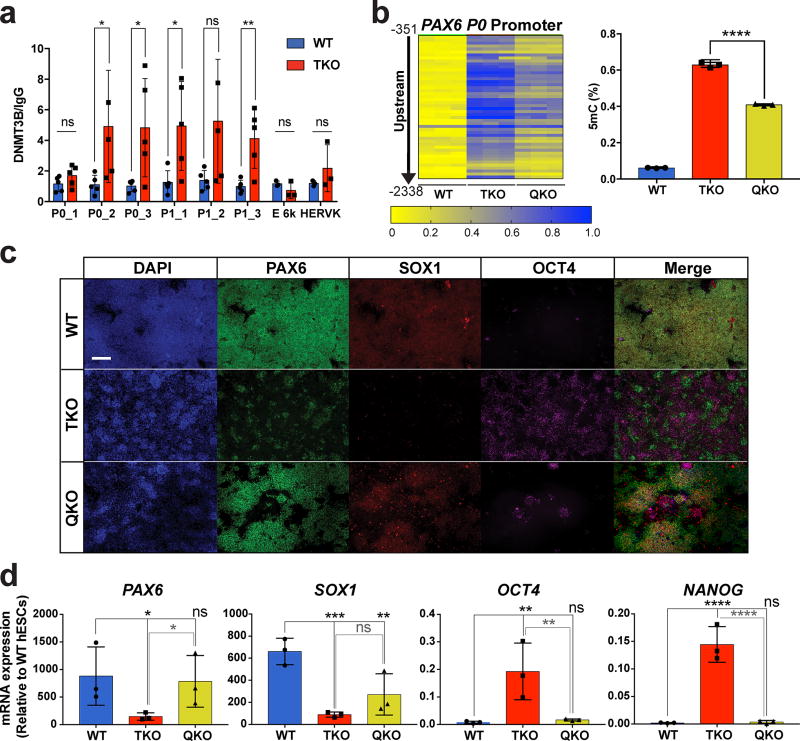
The DNA methyltransferases, DNMT1, DNMT3A and DNMT3B, are responsible for cytosine methylation. We speculated that the hypermethylation of the PAX6 P0 promoter and the resulting NE differentiation defect could be due to increased expression or activity of the DNMTs in TKO hESCs. There were no differences in the expression of the DNMT genes between WT and TKO hESCs (Supplementary Fig. 8a); however, ChIP-qPCR analysis showed increased binding of DNMT3B, but not DNMT1 or DNMT3A, at the PAX6 P0 promoter in TKO hESCs compared to WT hESCs (Fig. 6a, Supplementary Fig. 8b). Thus we further investigated whether DNMT3B is responsible for the hypermethylation at the PAX6 P0 promoter in TKO hESCs through genetic deletion. By applying a CRISPR gRNA that targets the sequence corresponding to the cytosine C5-methyltransferase domain of DNMT3B in TKO hESCs, we generated a quadruple TET/DNMT3B knockout (QKO) line. QKO hESCs had a ~35% reduction in the methylation at the PAX6 P0 promoter compared to passage matched TKO hESCs (Fig. 6b). Furthermore, there was a significant rescue of the NE differentiation phenotype compared to TKO hESCs: QKO cells formed PAX6 and SOX1 double positive cells (Fig. 6c) and few cells remained OCT4-positive after 10 days of NE differentiation. RT-qPCR analysis also showed rescue of NE markers PAX6 and SOX1 and proper downregulation of pluripotency markers OCT4 and NANOG (Fig. 6d). Our results suggest that DNMT3B activity at the PAX6 P0 promoter is responsible for the hypermethylation and NE differentiation phenotypes observed in TKO hESCs. In WT hESCs the TET proteins or the resulting 5hmC marks may actively block de novo methylation by DNMT3B to maintain a hypomethylated PAX6 P0 promoter and enable NE differentiation (Fig. 7).
Figure 6. Genetic inactivation of DNMT3B partially rescues the neuroectoderm differentiation defect of TKO hESCs.
a, ChIP-qPCR for DNMT3B at the PAX6 locus in WT and TKO hESCs, n= 3 independent experiments. Data presented are mean ± STD. Statistical analysis: Student’s t test (two sided), *P<0.05, **P<0.01. b, Heat map of MassArray analysis of 5mC at the PAX6 P0 promoter in WT, TKO and QKO hESCs. WT and TKO hESCs are passage-matched with the QKO hESCs. The location of each row of CpGs with respect to the TSS is shown to the left of the heat map. For each cell line three independent experiments are shown as three columns. Quantification of the percent methylation is shown on the right, n= 3 independent experiments. Data presented are mean ± STD. Statistical analysis: Student’s t test (two sided), ****P<0.0001. c, Immunofluorescence of PAX6, SOX1 and OCT4 at D10 of NE differentiation in WT, TKO and QKO cells. Scale bar indicates 100 µm. d, qPCR analysis of neuroectoderm (PAX6 and SOX1) and pluripotency (OCT4 and NANOG) markers in WT, TKO and QKO cells at D10 of NE differentiation, n= 3 independent experiments. Data presented are mean ± STD. Statistical analysis: black lines indicate comparisons to WT, one-way ANOVA, *P<0.05,**P<0.01,***P<0.001, ****P<0.0001.
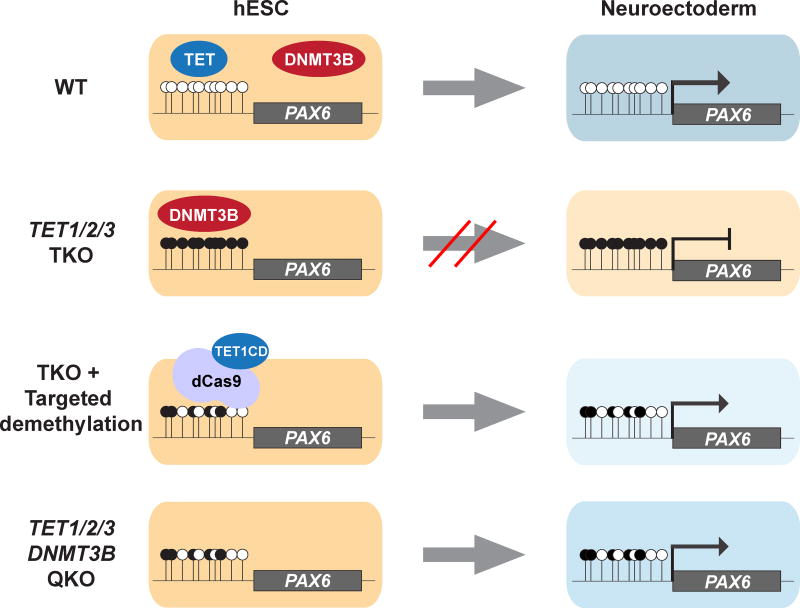
Figure 7. TET1 and DNMT3B compete to regulate the methylation of the PAX6 P0 bivalent promoter.
In WT hESCs TET1 or 5hmC functions to antagonize DNMT3B at the PAX6 bivalent promoter and prevents DNA hypermethylation. Upon NE differentiation PAX6 expression is activated and leads to the production of PAX6 and SOX1 double positive cells that are negative for the pluripotency marker OCT4. In TKO hESCs increased DNMT3B binding at the PAX6 bivalent promoter leads to increased DNA methylation. As a result PAX6 expression is not activated and ultimately few PAX6 or SOX1-positive cells are produced and a large number of cells still express the pluripotency marker OCT4. Targeted demethylation of the PAX6 P0 bivalent promoter or inactivation of DNMT3B in the TKO background (QKO) reduces methylation at the PAX6 bivalent promoter, allowing activation of PAX6 expression upon differentiation.
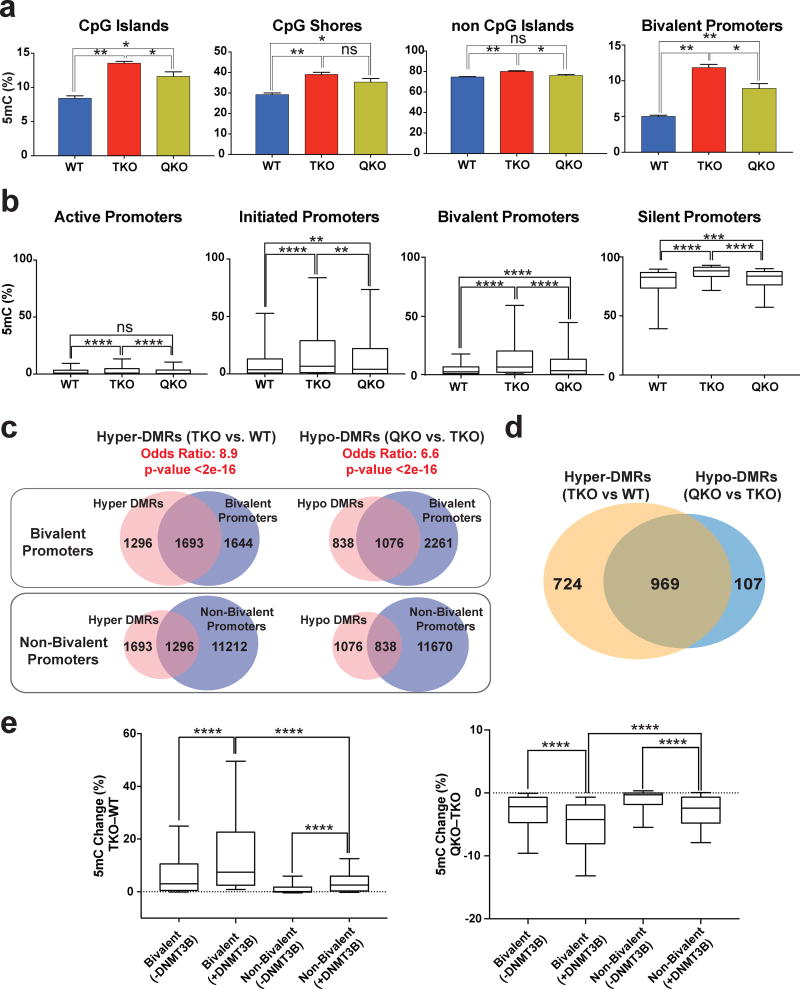
We performed ERRBS for passage-matched WT, TKO and QKO lines to investigate whether our findings at the PAX6 P0 promoter applies to other bivalent promoters. We found that QKO hESCs showed a global decrease in methylation, both within and outside of CpG islands, and in all promoter types. Bivalent promoters showed a larger decrease in methylation between QKO and TKO hESCs than active and initiated promoter types, but less than silent promoters (Supplementary Data Set 8, Fig. 8a–b, Supplementary Fig. 8c–d). Furthermore bivalent promoters, in comparison to non-bivalent promoters, had a greater overlap with hyper-DMRs that occur after TET inactivation as well as a higher overlap with hypo-DMRs that occur after DNMT3B inactivation in the TKO background (Fig. 8c, Supplementary Fig. 8e). This suggests that the TET and DNMT proteins dynamically regulate the methylation state of bivalent promoters. ~90% of the bivalent promoters that lost methylation in the QKO hESCs had previously gained methylation in the TKO hESCs (compared to WT hESCs). Conversely, ~57% of the bivalent promoters that gained methylation after TET inactivation lost methylation after DNMT3B was mutated (Fig. 8d). Thus at many bivalent promoters continuous DNMT3B activity is needed for the hypermethylation phenotype in TKO hESCs.
Figure 8. DNMT3B regulates the methylation level at bivalent promoters.
a, Average methylation at different genomic regions and bivalent promoters for WT, TKO and QKO hESCs by enhanced reduced representation bisulfite sequencing (ERRBS), n = 2 independent experiments. Data presented are mean ± STD. Statistical analysis: one-way ANOVA, *P<0.05,**P<0.01. b, Percent methylation in WT, TKO and QKO hESCs for active, initiated, bivalent and silent promoters. Error bars show 10 and 90 percent confidence intervals and the bar at the center of the box and whisker plot indicates the median, n = 2 independent experiments. Statistical analysis: one-way ANOVA, **P<0.01,***P<0.001, ****P<0.0001. c, Top: Overlap of bivalent promoters with hyper-DMRs at promoter regions (TKO vs. WT) and hypo-DMRs at promoter regions (QKO vs. TKO). Bottom: Overlap of non-bivalent promoters with hyper-DMRs at promoter regions (TKO vs. WT) and hypo-DMRs at promoter regions (QKO vs. TKO). The odd’s ratio and p-value for a comparison between bivalent and non-bivalent promoters is provided (Fisher’s exact test). d, The overlap between hyper-DMR (TKO vs. WT) associated bivalent promoters and hypo-DMR (QKO vs. TKO) associated bivalent promoters. e, Left: Methylation change (TKO – WT) for bivalent and non-bivalent promoters that either have DNMT3B peaks in TKO hESCs (+DNMT3B) or do not have DNMT3B peaks in TKO hESCs (−DNMT3B). Right: Methylation change (QKO – TKO) for bivalent and non-bivalent promoters that either have DNMT3B peaks in TKO hESCs (+DNMT3B) or do not have DNMT3B peaks in TKO hESCs (−DNMT3B). Error bars show 10 and 90 percent confidence intervals and the bar at the center of the box and whisker plot indicates the median, n = 2 independent experiments. Statistical analysis: one-way ANOVA, ****P<0.0001.
Our results thus far indicate that DNMT3B is responsible for the majority of bivalent promoter hypermethylation that occurs after TET inactivation. We performed DNMT3B ChIP-Seq to investigate whether bivalent promoters are directly targeted by DNMT3B. DNMT3B shows relatively insignificant overall binding to promoter regions (Supplementary Fig. 8e) in WT and TKO hESCs, which is similar to previous results of DNMT3B ChIP-Seq in WT mESCs39. This may be due to weak DNMT3B binding, or technical difficulties of DNMT3B ChIP. Nevertheless, among promoters with DNMT3B peaks in TKO hESCs, 74% of the 293 bivalent promoters and 21% of the 1,017 non-bivalent promoters had gained methylation after TET inactivation. Furthermore bivalent promoters with DNMT3B peaks in TKO hESCs showed a greater methylation increase (TKO versus WT) and a greater methylation decrease (QKO versus TKO) than bivalent promoters that lacked DNMT3B peaks in TKO hESCs or non-bivalent promoters with DNMT3B peaks in TKO hESCs (Fig. 8e). Thus DNMT3B binding correlates with more dynamic changes in DNA methylation at bivalent promoters and suggests that at these promoters the TET and DNMT3B proteins function in a competitive manner.
DISCUSSION
In mouse and human ESCs40,41 the promoter regions of differentiation-associated genes are enriched for the bivalent marks. Previous studies have focused on the establishment and maintenance of the bivalent histone marks. However, it was unclear whether DNA methylation is also actively regulated at bivalent promoters and if it has functional relevance for cell differentiation. Here we show that the TET proteins are critical for maintaining a hypomethylated state at bivalent promoters in hESCs. Although we focused on lineage regulators, such as PAX6 and SOX10, bivalent promoters may also regulate signaling pathways during development, which, along with the regulation of enhancers and other regulatory regions, contributes to proper embryonic development and cellular differentiation. Importantly, the alteration of DNA methylation at bivalent promoters does not cause immediate changes in transcription but impairs hESC differentiation. We therefore propose to revise the predominant approach for studying epigenetic regulators, which focuses on epigenetic changes (including changes in DNA methylation) that have an immediate impact on gene expression, by expanding the analysis to additional epigenetic changes that do not immediately affect gene expression but may influence future cell behavior during embryonic development or adult stem/progenitor cell differentiation.
Previous studies have indicated that genomic regions marked by the H3K4me3 modification are refractory to de novo DNA methylation42,43. TET deletion causes bivalent promoter hypermethylation without causing significant changes in H3K4me3 occupancy (Supplementary Fig. 8f). One possibility is that some of the effects associated with the H3K4me3 mark could be due to TET or TET-dependent 5hmC or other associated proteins, which often co-occur with H3K4me344. A second, non-mutually exclusive possibility is that de novo methyltransferases may be actively recruited to bivalent promoters in the absence of TET, overcoming the repulsion by H3K4me3. In fact it was recently shown that the PRC2 complex recruits DNMT3L, DNMT3A and DNMT3B to the bivalent promoters of genes involved in germ cell differentiation45.
It was also interesting that this aberrant hypermethylation at bivalent promoters, once established in TKO hESCs, is not adequately maintained by DNMT1. Based on the >99.7% overall fidelity of DNMT1 in preserving methylation46, DNMT1 would be expected to largely preserve the hypermethylation seen in TKO hESCs during the approximately 6 passages it took to generate and expand the QKO cells for analysis. Yet we observed a global reduction of DNA methylation at bivalent promoters including PAX6 in QKO cells, indicating that the hypermethylation phenotype requires continuous DNMT3B activity. It also suggests that additional mechanisms, such as transient transcription47, may inhibit DNMT1 activity at these loci, which may be an additional protective measure against hypermethylation of bivalent promoters even in TKO hESCs. Alternatively, though not mutually exclusively, DNMT3B may function as both a de novo and maintenance methyltransferase, as previously suggested48,49. It must be noted, however, that the maintenance function of DNMT3A and DNMT3B has only been observed for limited genomic regions in mESCs, such as repetitive sequences48,49, and inactivation of both DNMT3A and DNMT3B in hESCs only causes gradual DNA demethylation46.
Previous studies have shown that DNMT3A and DNMT3B share largely overlapping targets46, yet deletion of DNMT3B alone in TKO hESCs was sufficient to partially reverse bivalent promoter hypermethylation along with the associated NE differentiation defect. DNMT3B may have stronger preference or activity, compared to DNMT3A, at bivalent promoters such as PAX6, and such loci have been identified previously in hESCs46. Perhaps more likely, the rescue by DNMT3B deletion alone could be due to the relatively low DNMT3A expression compared to DNMT3B (Supplementary Fig. 8a) and the lack of compensatory increase of DNMT3A expression upon DNMT3B deletion (Supplementary Fig. 8g). Similarly a recent paper found that inactivating either DNMT3A or DNMT3B in Tet TKO mouse embryos was able to rescue the gastrulation phenotype10. It is possible that in other cell types DNMT3A, along with or instead of DNMT3B, may counteract the TET proteins to regulate methylation50,51.
Our work highlights the utility of locus-specific epigenome editing tools to directly probe the functional consequences of epigenetic changes and to distinguish direct, locus-specific effects from indirect effects52. The competitive balance between the TET proteins and de novo methyltransferases at bivalent promoters and other genomic loci could facilitate rapid changes of their methylation state to either activate or silence transcription in a cell-lineage and locus-specific manner. Further work could investigate the factors that influence whether the methylation state of a genomic region is dynamically regulated and ultimately how methylation states predict (in the context of cell differentiation) cell-type specific transcriptional programs.
ONLINE METHODS
hESC Culture
HUES8 (NIHhESC-09–0021) and MEL1 (NIHhESC-11–0139) hESCs were cultured on irradiated mouse embryonic fibroblasts (iMEFs) feeder layers in DMEM/F12 medium (Life Technologies, 12500-062) supplemented with 20% KnockOut Serum Replacement (Life Technologies, 10828028), 1X MEM Non-Essential Amino Acids (Life Technologies, 11140050), 1X GlutaMAX (Life Technologies, 35050079), 100U/ml Penicillin and 100 µg/ml Streptomycin (Gemini, 15070063), 0.055 mM 2-mercaptoethanol (Life Technologies, 21985023) and 10 ng/ml recombinant human bFGF (EMD Millipore, GF003AF). Cells were incubated at 37 °C with 5% (vol/vol) CO2, and media was changed daily. Cultures were passaged at a 1:6 – 1:12 split ratio every 4 – 6 days using TrypLE (Life Technologies, 12563-029). 5 µM Rho-associated protein kinase (ROCK) inhibitor Y-27632 (Selleck Chemicals, S1049) was added into the culture medium when passaging or thawing cells. Cells are regularly confirmed to be mycoplasma-free by the MSKCC Antibody & Bioresource Core Facility.
Generation of mutant lines
In vitro transcription of gRNAs
CRISPR gRNAs were designed to target the genomic sequence corresponding to the beginning of the catalytic domain for TET1, TET2, and TET3. For TET1 and TET3 the two gRNAs most efficient at producing indel mutations were used for the targeting experiments. For TET2 only one gRNA was found to be efficient at producing frameshift indel mutations. A CRISPR gRNA used in a previous study was used to target the C5-methyltransferase domain of DNMT3B1. The procedure to generate mutants has been previously described2. For each CRISPR a 20 bp T7 promoter was added to the 20 bp gRNA target sequence (Supplementary Table 1) followed by a 80 bp constant gRNA backbone to form a 120 bp oligonucleotide. The T7-gRNA oligo nucleotide was amplified by PCR and the T7-gRNA PCR products were used as templates for in vitro transcription using the MEGAshortscript T7 kit (Life Technologies, AM1354M). The resulting gRNAs were purified using the MEGAclear kit (Life Technologies, AM1908M), eluted in RNase-free water and stored at −80°C until use.
Transfection of gRNAs or gRNA + ssDNA
HUES8 and MEL1 iCas9 hESCs3 or HUES8 TKO hESCs were treated with ROCK inhibitor and doxycycline one day before transfection. For transfection, confluent cells were dissociated using TrypLE (Life Technologies, 12563-029), replated at a 1:6 ratio in iMEF-coated 24-well plates and transfected in suspension with gRNAs or a mixture of gRNA and ssDNA. A second transfection was performed 24 hrs later. Transfection was performed using Lipofectamine RNAiMAX (Life Technologies, 13778-150) following the manufacturer’s guidelines. For each targeting, TET1, TET2, TET3, DNMT3B gRNAs, each at a 10 nM final concentration, were used. For repair of the TET1 allele in TKO hESCs, gRNAs and ssDNA (of TET1 WT sequence) were transfected at a 10nM and 20nM concentration, respectively. Lipofectamine RNAiMAX and gRNA or gRNA + ssDNA were diluted separately in Opti-MEM (Life Technologies, 31985070), then mixed together, incubated for 5 min, and added drop-wise to cultured hESCs.
Isolation, expansion and identification of mutant clones
Two days after the last gRNA transfection, hESCs were dissociated into single cells and replated at ~ 2,000 cells per 10-cm dish. Cells were allowed to grow until colonies from single cells became visible (~10 days). At this stage, single colonies were manually picked, mechanically disaggregated and replated individually into 96-well plates. Colonies were amplified and analyzed by Sanger sequencing at the 3 TET and the DNMT3B loci for the presence of mutations. Clonal cell lines carrying the desired mutations or TKO hESCs in which one TET1 allele was repaired were amplified and frozen down. For all targeting experiments (TET, DNMT3B and HDR repair of TET1) we also isolated and froze down lines that had undergone the targeting procedure, but whose genomic sequence was not changed. These lines were used as passage-matched controls for methylation analysis and differentiation.
hESC Neuroectoderm Differentiation
90% confluent hESC cultures were disaggregated using TrypLE (Life Technologies, 12563-029) for 5 minutes and washed using hESC media. The cells were plated on Matrigel (BD, 354234) coated dishes in hESC media with ROCK-inhibitor at a density of 180,000–200,000 cells/cm2. After 12 hrs, differentiation into neuroectoderm was initiated by switching to knockout serum replacement (KSR) media with 10 µM TGF-β inhibitor SB431542 (Tocris 161410) and 100 nM BMP inhibitor LDN193189 (Axonmedchem, 1509). On day 1 and day 2 of differentiation, the media was removed and fresh KSR with 10 µM SB431542 and 100 nM LDN193189 was added. Starting on day 4 of differentiation an increasing amount of N2 media was added to the KSR media every two days, while maintaining 10 µM SB431542 and 100 nM LDN193189. On day 4 a 3:1 mixture of KSR/N2 media was added. On day 6 a 1:1 mixture of KSR/N2 media was added and on day 8, a 1:3 mixture of KSR/N2 media was added. The cells were isolated for analysis on day 10 of differentiation. KSR media contains Knockout DMEM (Life Technologies, 10829018), Knockout Serum Replacement (Life Technologies, 10828028), 1X MEM Non-Essential Amino Acids (Life Technologies, 11140050), 1X GlutaMAX (Life Technologies, 35050079), and 2-mercaptoethanol (Life Technologies, 21985023). N2 media contains DMEM/F12 medium (Life Technologies, 12500-062), glucose (Sigma, G8270), sodium bicarbonate (Sigma, S5761), putrescine (Sigma, P5780), progesterone (Sigma, P8783), sodium selenite (Sigma, S5261), apo-transferrin (Sigma, T1147), and insulin (Sigma, I2643).
RNA-Seq
For RNA-Seq total RNA was isolated with the RNeasy Mini Kit (Qiagen, 74136) from HUES8 WT and TKO hESCs (n=2 each). RNA samples were submitted to the MSKCC Integrated Genomics Core for library prep and sequencing.
RNA-seq data was aligned to the hg19 reference genome using Bowtie4 Read counts were derived from HTSeq.scripts.count module in HTSeq-0.6.0 with default parameters in a non-directional model5. Differentially expressed genes were generated by DES eq2 -1.4.5 in R6. Upregulated genes were decided by logFC > 2 and p-adj < 0.1 (n=134). Downregulated genes were decided by logFC < −2 and p-adj < 0.1 (n=233).
5hmC Profiling
5hmC-Seal was performed as previously described7. 40 µg of genomic DNA was sonicated to ~200–400bp fragments using a Diagenode Bioruptor Sonicator. Sonicated DNA was then labeled with azide glucose in a 1 hr reaction at 37°C catalyzed by recombinant -GT utilizing UDP-6-N3-glucose as the sugar donor. The reactions were cleaned up using a Zymo DNA Clean & Concentrator kit (Zymo, D4003), then a biotin moiety was added to the azide-labeled DNA via a copper-free click chemistry reaction with DBCO-S-S-PEG3-Biotin in water at 37°C for 1 hr. Reactions were once again cleaned with the Zymo kit and then bound to Dynabeads MyOne Streptavidin C1 beads (Life Technologies, 65001) for 15 minutes at RT and washed 5 times with binding buffer (5mM Tris-HCl, pH7.5, 0.5mM EDTA, 1M NaCl, 0.01% Tween 20). Bound DNA was eluted by reducing the disulfide in the biotin linker with 100mM DTT for 2h at RT with gentle rotation. Eluted DNA was cleaned on a Micro Bio-spin Column (BioRad, 7326204) to remove DNA and then purified by the Zymo kit. Libraries were constructed from eluted DNA by end repair, A-tailing, and adapter ligation, followed by 4 cycles of PCR, size selection via agarose gel electrophoresis, 12 additional PCR cycles, and a final size selection via agarose gel electrophoresis. Library quantity and quality were analyzed by Bioanalyzer prior to sequencing on an Illumina HiSeq 2500. Sequence alignment and peak identification were performed as previously described7. Peak calling was performed by macs14 1.4.2 with default parameters8
WGBS and ERRBS
For WGBS, genomic DNA was isolated from HUES8 WT and HUES8 TKO hESCs. Genomic DNA was sheared using E220 focused-ultrasonicator (Covaris) to 250–350bp fragments. After end-repair and TruSeq adapter (Illumina) ligation the DNA-libraries were denatured followed by treatment with bisulfite (Zymo Research, D5020) for 30 min at 65C. Single stranded DNA was purified using silica gel columns and amplified using HiFi Uracil+ polymerase (Kapa biosciences, KK2802). The amplified libraries were quantified and mixed equimolarly for sequencing on Illumina HiSeq2500 by the MSKCC Integrated Genomics Core. One replicate for HUES8 WT and HUES8 TKO was submitted for sequencing.
For ERRBS, genomic DNA was isolated from HUES8 WT, HUES8 TKO and HUES8 QKO hESCs. Genomic DNA was submitted to the Weill Cornell Medical College Epigenomics Core for ERRBS. Two independent replicates each for HUES8 WT, TKO and QKO cells were submitted for sequencing.
WGBS data were aligned to the bisulfite-converted hg19 reference genome using Bismark v0.13.09. We extracted the methylation status with the bismark_methylation_extractor script in Bismark9. Only CpGs with at least 3 covered reads were used for downstream analysis. The WCMC Computational Genomics Core Facility supported alignment and methylation extraction for ERRBS data10. DMRs were defined as regions containing at least five differentially methylated CpGs (DMCs, false discover rate=20%, Chi square test) and whose total methylation difference was more than 10%11. For Figures 1h and Extended Data Fig. 3b–c hypermethylation was defined as a 5% increase in methylation in order to set a uniform cutoff for comparison between different promoters or cell lines. For Extended Data Fig. 3b–c hypomethylation was defined as a 5% decrease in methylation. For whole genome bisulfite sequencing only one replicate was used but the use of five or more DMCs partially overcomes the statistical limitation of individual Chi square tests based on a n=1 sample, as the latter should be cautiously interpreted in the absence of multiple measurements from independent samples. DMRs calling was performed with RRBSeeqer with default parameters11. DMRs were annotated using ChIPseeqerAnnotate from ChIPseeqer package12. Methylation of a specific region was calculated by averaging the methylation levels of all covered CpGs in that region.
Genomic regions for CpGs are defined as the following definitions. CpG islands (CGI) are defined as annotations from refSeq. CGI shores are defined as the region encompassing 1 kb upstream and downstream of known CGIs. Non-CGIs are defined as at least 10 kb away from known CGIs. Promoters are defined as the region encompassing 2 kilobases (kb) upstream and downstream from the transcription start site (TSS) from refSeq. The following characteristics were used to classify promoters as active, initiated, bivalent and silent. Active promoters are associated with H3K4me3 (in 1.5 kb region flanking the TSS) and H3K79me2 (in 5 kb downstream from the TSS). Initiated promoters are associated with H3K4me3 (in 1.5 kb region flanking the TSS). Bivalent promoters are associated with H3K4me3 and H3K27me3 (in 1.5 kb region flanking the TSS). Silent promoters were not associated with H3K4me3 (in 1.5 kb region flanking the TSS). With WGBS (HUES8 TKO vs. HUES8 WT) 6,695 active promoters, 989 initiated promoters, 3,327 bivalent promoters and 4,707 silent promoters were analyzed. With ERRBS (HUES8 TKO vs. HUES8 WT) 6,450 active promoters, 846 initiated promoters, 3,234 bivalent promoters and 1,476 silent promoters were analyzed. With ERRBS (MEL1 TKO vs. MEL1 WT) 6,420 active promoters, 3,222 bivalent promoters, 835 initiated promoters and 1,413 silent promoters were analyzed.
Enhancers were defined as outside of promoters and exons (RefSeq). Poised enhancers were identified as regions overlapping with H3K4me1 peaks only. Active enhancers were identified as regions overlapping with H3K4me1 and H3K27ac peaks.
Overlap of 5mC change with histone marks was determined using previous ChIP-Seq datasets for histone marks (H3K4me1: GSM 733782; H3K4me2: GSM 733670; H3K4me3: GSM 733657; H3K27me3: GSM 733748; H3K27ac: GSM 733718; H3K9me3: GSM 1003585; H3K36me3: GSM 1003585; H3K79me2: GSM 1003547). Peak calling was performed by ChIPseeqer-2.1 with default parameters12.
ChIP-Seq
ChIP-Seq was performed for TET1 (WT hESCs) and DNMT3B (WT and TKO hESCs). WT and TKO hESCs were cultured in standard hESC media, as described above. ~5 × 107 cells were fixed, washed and snap-frozen according to the Cell Fixation protocol from Active Motif (http://www.activemotif.com/documents/1848.pdf). Chromatin immunoprecipitation and DNA sequencing was performed by Active Motif.
ChIP-seq data was aligned to the hg19 genomes using bowtie-0.12.9 with default parameters except --n 2 and --best4. Peak calling and read density in peak regions were performed by macs14 1.4.2 with default parameters8. The technical success of the ChIP-Seq was confirmed by using standard quality control measures and determines the overlap between TET1 binding and the presence of 5hmC. We observed that 52.6 % of 5hmC peaks associated with gene promoters overlapped with TET1 peaks (Supplementary Fig. 5f), which is similar to previous results obtained from TET1 ChIP-Seq in mouse embryonic stem cells13,14. Approximately 75% of total TET1 peaks overlapped with total 5hmC peaks present in HUES8 WT hESCs, in contrast only 25% of TET1 peaks overlapped with randomly generated 5hmC peaks (Supplementary Fig. 5g). Random peaks were generated with ChIPseeqerCreateRandomRegions in ChIPseeqer-2.1 with default parameters12. The lack of a complete overlap between 5hmC and TET1 could be due to a number of factors including: 5hmC production by TET2 and TET3, rapid turnover of 5hmC and reduced binding of TET1 to 5hmC. Peaks from ChIP-seq were annotated using ChIPseeqerAnnotate from ChIPseeqer package12.
ChIP-qPCR
Confluent hESCs from a 100 mm dish were used for 2 immunoprecipitations. ChIP was performed using the SimpleChIP Plus Enzymatic Chromatin IP kit (Cell Signaling Technology, 9003S) according to the manufacturer protocols. The antibodies used for ChIP were: H3 (Cell Signaling Technology, 9003S), IgG (Cell Signaling Technology, 9003S), TET1 (Genetex, GTX627420), DNMT1 (Active Motif, 39204). DNMT3A (Abcam, ab2850), and DNMT3B (Novus Biologicals, NB100–56514). Primers for ChIP-qPCR are provided in Supplementary Table 3.
5mC methylation analysis by MassArray
hESCs were disaggregated using TrypLE and genomic DNA was extracted using the DNeasy Blood & Tissue Kit (Qiagen, 69504) following manufacturer’s guidelines. MassArray Epityper analysis was performed using the WCMC Epigenomics core. MassArray Epityper analysis consists of bisulfite treatment of genomic DNA, followed by PCR for the specific region of analysis, base specific cleavage that differentiates between previously methylated and unmethylated DNA and finally MALDI-TOF mass spectrometry15. Primers used for mass array 5mC methylation analysis are found in Supplementary Table 4. Genomic coordinates of regions assayed by MassArray are provided in Supplementary Table 5.
PAX6 overexpression lentivirus construct and generation
To generate the PAX6 overexpression construct PAX6 cDNA from D10 of WT NE differentiation was tagged with a 2A–GFP sequence and inserted into a lentiviral backbone. The 2A–GFP sequence was amplified from the OCT4-eGFP-PGK-Puro plasmid (Addgene, plasmid # 31937) using primers that contain BamHI (Forward) and AscI sites (Reverse). The PAX6 cDNA PCR product was cloned into the pENTR-dTOPO vector (Thermo Fisher Scientific, K240020). Digestion of the PAX6-pENTR-dTOPO vector and the 2A–GFP PCR product with BamHI and Asc1 was followed with ligation to form the PAX6-2A–GFP-pENTR-dTOPO vector. The PAX6-2A–GFP insert was then transferred into a doxycycline inducible lentiviral backbone through a LR reaction. Primers used for cloning are listed in Supplementary Table 6.
Cloning dCas9 TET1 catalytic domain fusion and site-directed mutagenesis
The TET1 catalytic domain was amplified using forward and reverse primers that contain the Fse1 restriction site from the pJFA274 plasmid (Addgene, plasmid # 49239). This PCR product was then digested with Fse1. The dCas9-krab construct was cloned from the pHR-SFFV-dCas9-BFP-KRAB plasmid (Addgene, Plasmid # 46911) into the pENTR-dTOPO vector (Thermo Fisher Scientific, K240020). The KRAB protein was removed by Fse1 digestion and the Fse1-digested TET1 catalytic domain PCR product was ligated to produce the dCas9-TET1 catalytic domain fusion vector (dCas9-TET1CD). Primers used for cloning are listed in Supplementary Table 6.
The TET1 catalytic domain, from the dCas9 TET1 catalytic domain fusion vector (described above), was mutated at two sites (H1671A and D1673A) to produce a catalytic null control (dCas9-TET1CDmut). Mutagenesis was performed using the QuikChange II XL Site-Directed Mutagenesis kit (Agilent Technologies, 200521) according to the manufacturer’s protocol. The primers used for mutagenesis are in Supplementary Table 6.
Targeting dCas9-TET1CD into TKO hESCs
The dCas9-TET1CD was cloned into the pINDUCER lentiviral backbone (Addgene, Plasmid # 46948) under the control of doxycycline inducible promoter. This lentiviral backbone also constitutively expresses GFP, which allows us to isolate infected cells. Prior to infection of TKO hESCs with pINDUCER-dCas9-TET1CD, the already AAVS1-integrated Cas9 was inactivated through CRISPR targeting of the beginning of the Cas9 protein. The gRNA used to inactivate Cas9 is listed in Supplementary Table 1. After knocking out Cas9, clones were isolated and infected with virus containing the pINDUCER-dCas9-TET1CD construct. Infected cells were isolated by FACS sorting of GFP-positive cells. These cells were expanded and frozen down.
After identifying CRISPR gRNAs that produce demethylation at the SOX10 promoter and LEFTY2 enhancer we cloned these CRISPR gRNAs into the piCRg Entry vector (Addgene, Plasmid # 58904). A gRNA that targets the HBB promoter was cloned into the piCRg Entry vector to be used as a non-targeting control. For the PAX6 P0 promoter we identified 3 gRNAs (Cr6, Cr7, Cr9) that were able to produce demethylation. We performed golden gate cloning to assemble these 3 gRNAs in tandem. An LR reaction (Thermo Fisher Scientific, 11791100) was then performed to transfer the U6 promoter and either the tandem array of Cr6, Cr7 and Cr9 gRNAs for the PAX6 P0 promoter or the individual gRNAs for SOX10, LEFTY2 and HBB (non-targeting control) into a lentiviral backbone containing a hygromycin selection cassette. TKO hESCs containing the dCas9-TET1CD were then infected by the lentivirus. Infected cells were isolated by 4 days of hygromycin selection (40 µg/mL) and then amplified and frozen down. As described in the text targeted clones were treated with doxycycline for 10 days prior to methylation analysis and RNA expression analysis at the hESC stage. For PAX6 and SOX10 targeted demethylation the cells were also differentiated using the standard NE differentiation protocol and then RNA expression was analyzed by qPCR.
Statistical analysis
Data are presented as mean ± STD (unless otherwise noted) and were derived from at least three independent experiments. Data on replicates (n) is given in figure legends. Statistical analysis was performed using the two sided Student’s t-test (comparing two groups) or one-way comparison ANOVA (comparing multiple groups against one group). Variance was similar between the groups that are being compared. Distribution of the raw data approximated normal distribution (Kolmogorov–Smirnov normality test) for data with sufficient number of replicates to test for normality. No method of randomization was performed and investigators were not blinded to the genotypes of cell lines. Statistical analysis for whole genome bisulfite sequencing (WGBS), enhanced reduced representation bisulfite sequencing (ERRBS) and RNA-Seq data are described in those separate methods section. Additional methodological details are provided in the Supplementary Note.
Data Availability
All sequencing datasets are available under the Gene Expression Omnibus accession GSE89728.
URLs
R language, http://www.r-project.org/.
DAVID Bioinformatics Resources 6.8, https://david.ncifcrf.gov
ENCODE: Encyclopedia of DNA Elements, https://www.encodeproject.org.
Supplementary Material
Acknowledgments
We thank the WCMC Epigenomics core for ERRBS and 5mC MassArray analysis and the MSKCC integrated genomics core for performing RNA-seq and WGBS. We also thank L. Studer and J. Tchieu for advice on neuroectoderm differentiation, E. Apostolou and M. Donohoe for comments on the manuscript, and M. Goll and members of the Huangfu laboratory for insightful discussions and critical reading of the manuscript. This study was funded in part by Tri-Institutional Stem Cell Initiative (#2016-032), New York State Stem Cell Science (NYSTEM C029156) and a MSKCC Cancer Center Support grant (P30 CA008748). N.V. is supported by Weill Graduate School of Medical Sciences at Cornell University/The Rockefeller University/Sloan-Kettering Institute Tri-Institutional M.D.-Ph.D. Program.
Footnotes
Author Contributions
N.V. and D.H. devised experiments and interpreted results. N.V. performed most experiments and collected data. H.P. and O.E. performed computational analysis on WGBS, ERRBS, RNA-seq, ChIP-Seq and 5hmC profiling. L.D. and C.H. performed 5hmC profiling, sequencing and analysis and mass spectrometry. A.K. generated libraries for WGBS. A.S., Q.V.L., B.P., V.T., F.G. and C.C. assisted with additional experiments. N.V. and D.H. wrote the manuscript; all other authors provided editorial advice.
Competing financial interests
5hmC-Seal has been licensed to Active Motif and Epican by the University of Chicago.
References
- 1.Ziller MJ, Gu H, Muller F, Donaghey J, Tsai LT, Kohlbacher O, De Jager PL, Rosen ED, Bennett DA, Bernstein BE, et al. Charting a dynamic DNA methylation landscape of the human genome. Nature. 2013;500:477–481. doi: 10.1038/nature12433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Robertson KD. DNA methylation and human disease. Nature Reviews Genetics. 2005;6:597–610. doi: 10.1038/nrg1655. [DOI] [PubMed] [Google Scholar]
- 3.Li E, Bestor TH, Jaenisch R. Targeted mutation of the DNA methyltransferase gene results in embryonic lethality. Cell. 1992;69:915–926. doi: 10.1016/0092-8674(92)90611-f. [DOI] [PubMed] [Google Scholar]
- 4.Okano M, Bell DW, Haber DA, Li E. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell. 1999;99:247–257. doi: 10.1016/s0092-8674(00)81656-6. [DOI] [PubMed] [Google Scholar]
- 5.Smith ZD, Meissner A. DNA methylation: roles in mammalian development. Nature Reviews Genetics. 2013;14:204–220. doi: 10.1038/nrg3354. [DOI] [PubMed] [Google Scholar]
- 6.Tahiliani M, Koh KP, Shen Y, Pastor WA, Bandukwala H, Brudno Y, Agarwal S, Iyer LM, Liu DR, Aravind L, et al. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science. 2009;324:930–935. doi: 10.1126/science.1170116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ito S, D’Alessio AC, Taranova OV, Hong K, Sowers LC, Zhang Y. Role of Tet proteins in 5mC to 5hmC conversion, ES-cell self-renewal and inner cell mass specification. Nature. 2010;466:1129–1133. doi: 10.1038/nature09303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Guo JU, Su Y, Zhong C, Ming GL, Song H. Hydroxylation of 5-methylcytosine by TET1 promotes active DNA demethylation in the adult brain. Cell. 2011;145:423–434. doi: 10.1016/j.cell.2011.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dawlaty MM, Breiling A, Le T, Barrasa MI, Raddatz G, Gao Q, Powell BE, Cheng AW, Faull KF, Lyko F. Loss of tet enzymes compromises proper differentiation of embryonic stem cells. Dev Cell. 2014;29:102–111. doi: 10.1016/j.devcel.2014.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dai HQ, Wang BA, Yang L, Chen JJ, Zhu GC, Sun ML, Ge H, Wang R, Chapman DL, Tang F, et al. TET-mediated DNA demethylation controls gastrulation by regulating Lefty-Nodal signalling. Nature. 2016;538:528–532. doi: 10.1038/nature20095. [DOI] [PubMed] [Google Scholar]
- 11.Lu F, Liu Y, Jiang L, Yamaguchi S, Zhang Y. Role of Tet proteins in enhancer activity and telomere elongation. Genes Dev. 2014;28:2103–2119. doi: 10.1101/gad.248005.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jaenisch R, Bird A. Epigenetic regulation of gene expression: how the genome integrates intrinsic and environmental signals. Nature Genetics. 2003;33(Suppl):245–254. doi: 10.1038/ng1089. [DOI] [PubMed] [Google Scholar]
- 13.Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 14.Rasmussen KD, Helin K. Role of TET enzymes in DNA methylation, development, and cancer. Genes & Development. 2016;30:733–750. doi: 10.1101/gad.276568.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bogdanovic O, Smits AH, de la Calle Mustienes E, Tena JJ, Ford E, Williams R, Senanayake U, Schultz MD, Hontelez S, van Kruijsbergen I, et al. Active DNA demethylation at enhancers during the vertebrate phylotypic period. Nature Genetics. 2016;48:417–426. doi: 10.1038/ng.3522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mikkelsen TS, Ku M, Jaffe DB, Issac B, Lieberman E, Giannoukos G, Alvarez P, Brockman W, Kim TK, Koche RP, et al. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007;448:553–560. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fouse SD, Shen Y, Pellegrini M, Cole S, Meissner A, Van Neste L, Jaenisch R, Fan G. Promoter CpG methylation contributes to ES cell gene regulation in parallel with Oct4/Nanog, PcG complex, and histone H3 K4/K27 trimethylation. Cell Stem Cell. 2008;2:160–169. doi: 10.1016/j.stem.2007.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lister R, Pelizzola M, Dowen RH, Hawkins RD, Hon G, Tonti-Filippini J, Nery JR, Lee L, Ye Z, Ngo QM, et al. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature. 2009;462:315–322. doi: 10.1038/nature08514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Meissner A, Mikkelsen TS, Gu H, Wernig M, Hanna J, Sivachenko A, Zhang X, Bernstein BE, Nusbaum C, Jaffe DB, et al. Genome-scale DNA methylation maps of pluripotent and differentiated cells. Nature. 2008;454:766–770. doi: 10.1038/nature07107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.von Meyenn F, Berrens RV, Andrews S, Santos F, Collier AJ, Krueger F, Osorno R, Dean W, Rugg-Gunn PJ, Reik W. Comparative Principles of DNA Methylation Reprogramming during Human and Mouse In Vitro Primordial Germ Cell Specification. Developmental Cell. 2016;39:104–115. doi: 10.1016/j.devcel.2016.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gonzalez F, Zhu Z, Shi ZD, Lelli K, Verma N, Li QV, Huangfu D. An iCRISPR platform for rapid, multiplexable, and inducible genome editing in human pluripotent stem cells. Cell Stem Cell. 2014;15:215–226. doi: 10.1016/j.stem.2014.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Song CX, Szulwach KE, Fu Y, Dai Q, Yi C, Li X, Li Y, Chen CH, Zhang W, Jian X, et al. Selective chemical labeling reveals the genome-wide distribution of 5-hydroxymethylcytosine. Nat Biotechnol. 2011;29:68–72. doi: 10.1038/nbt.1732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gu H, Smith ZD, Bock C, Boyle P, Gnirke A, Meissner A. Preparation of reduced representation bisulfite sequencing libraries for genome-scale DNA methylation profiling. Nature Protocols. 2011;6:468–481. doi: 10.1038/nprot.2010.190. [DOI] [PubMed] [Google Scholar]
- 24.Zhou V, Goren A, Bernstein B. Charting histone modifications and the functional organization of mammalian genomes. Nature Reviews Genetics. 2011;12:7–18. doi: 10.1038/nrg2905. [DOI] [PubMed] [Google Scholar]
- 25.Zhang X, Huang CT, Chen J, Pankratz MT, Xi J, Li J, Yang Y, Lavaute TM, Li XJ, Ayala M, et al. Pax6 is a human neuroectoderm cell fate determinant. Cell Stem Cell. 2010;7:90–100. doi: 10.1016/j.stem.2010.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li XJ, Du ZW, Zarnowska ED, Pankratz M, Hansen LO, Pearce RA, Zhang SC. Specification of motoneurons from human embryonic stem cells. Nat Biotechnol. 2005;23:215–221. doi: 10.1038/nbt1063. [DOI] [PubMed] [Google Scholar]
- 27.Pankratz MT, Li XJ, Lavaute TM, Lyons EA, Chen X, Zhang SC. Directed neural differentiation of human embryonic stem cells via an obligated primitive anterior stage. Stem Cells. 2007;25:1511–1520. doi: 10.1634/stemcells.2006-0707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Anderson TR, Hedlund E, Carpenter EM. Differential Pax6 promoter activity and transcript expression during forebrain development. Mechanisms of Development. 2002;114:171–175. doi: 10.1016/s0925-4773(02)00051-5. [DOI] [PubMed] [Google Scholar]
- 29.Kammandel B, Chowdhury K, Stoykova A, Aparicio S, Brenner S, Gruss P. Distinct cis-essential modules direct the time-space pattern of the Pax6 gene activity. Developmental Biology. 1999;205:79–97. doi: 10.1006/dbio.1998.9128. [DOI] [PubMed] [Google Scholar]
- 30.Plaza S, Saule S, Dozier C. High conservation of cis-regulatory elements between quail and human for the Pax-6 gene. Development Genes and Evolution. 1999;209:165–173. doi: 10.1007/s004270050240. [DOI] [PubMed] [Google Scholar]
- 31.Xu ZP, Saunders GF. PAX6 intronic sequence targets expression to the spinal cord. Developmental Genetics. 1998;23:259–263. doi: 10.1002/(SICI)1520-6408(1998)23:4<259::AID-DVG1>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- 32.Zheng JB, Zhou YH, Maity T, Liao WS, Saunders GF. Activation of the human PAX6 gene through the exon 1 enhancer by transcription factors SEF and Sp1. Nucleic Acids Research. 2001;29:4070–4078. doi: 10.1093/nar/29.19.4070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stroud HFS, Morey Kinney S, Pradhan S, Jacobsen SE. Hydroxymethylcytosine is associated with enhancers and gene bodies in human embryonic stem cells. Genome Biology. 2011:R54. doi: 10.1186/gb-2011-12-6-r54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chambers SM, Fasano CA, Papapetrou EP, Tomishima M, Sadelain M, Studer L. Highly efficient neural conversion of human ES and iPS cells by dual inhibition of SMAD signaling. Nat Biotechnol. 2009;27:275–280. doi: 10.1038/nbt.1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xu X, Tao Y, Gao X, Zhang L, Li X, Zou W, Ruan K, Wang F, Xu GL, Hu R. A CRISPR-based approach for targeted DNA demethylation. Cell Discovery. 2016;2:16009. doi: 10.1038/celldisc.2016.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu XS, Wu H, Ji X, Stelzer Y, Wu X, Czauderna S, Shu J, Dadon D, Young RA, Jaenisch R. Editing DNA Methylation in the Mammalian Genome. Cell. 2016;167:233–247. e217. doi: 10.1016/j.cell.2016.08.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Choudhury SR, Cui Y, Lubecka K, Stefanska B, Irudayaraj J. CRISPR-dCas9 mediated TET1 targeting for selective DNA demethylation at BRCA1 promoter. Oncotarget. 2016;29:46545–46556. doi: 10.18632/oncotarget.10234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Morita S, Noguchi H, Horii T, Nakabayashi K, Kimura M, Okamura K, Sakai A, Nakashima H, Hata K, Nakashima K, et al. Targeted DNA demethylation in vivo using dCas9-peptide repeat and scFv-TET1 catalytic domain fusions. Nature biotechnology. 2016;34:1060–1065. doi: 10.1038/nbt.3658. [DOI] [PubMed] [Google Scholar]
- 39.Baubec T, Colombo DF, Wirbelauer C, Schmidt J, Burger L, Krebs AR, Akalin A, Schubeler D. Genomic profiling of DNA methyltransferases reveals a role for DNMT3B in genic methylation. Nature. 2015;520:243–247. doi: 10.1038/nature14176. [DOI] [PubMed] [Google Scholar]
- 40.Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, Fry B, Meissner A, Wernig M, Plath K, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- 41.Pan G, Tian S, Nie J, Yang C, Ruotti V, Wei H, Jonsdottir GA, Stewart R, Thomson JA. Whole-genome analysis of histone H3 lysine 4 and lysine 27 methylation in human embryonic stem cells. Cell Stem Cell. 2007;1:299–312. doi: 10.1016/j.stem.2007.08.003. [DOI] [PubMed] [Google Scholar]
- 42.Guo H, Zhu P, Yan L, Li R, Hu B, Lian Y, Yan J, Ren X, Lin S, Li J, et al. The DNA methylation landscape of human early embryos. Nature. 2014;511:606–610. doi: 10.1038/nature13544. [DOI] [PubMed] [Google Scholar]
- 43.Ooi SK, Qiu C, Bernstein E, Li K, Jia D, Yang Z, Erdjument-Bromage H, Tempst P, Lin SP, Allis CD, et al. DNMT3L connects unmethylated lysine 4 of histone H3 to de novo methylation of DNA. Nature. 2007;448:714–717. doi: 10.1038/nature05987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Xu Y, Wu F, Tan L, Kong L, Xiong L, Deng J, Barbera AJ, Zheng L, Zhang H, Huang S, et al. Genome-wide regulation of 5hmC, 5mC, and gene expression by Tet1 hydroxylase in mouse embryonic stem cells. Molecular Cell. 2011;42:451–464. doi: 10.1016/j.molcel.2011.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Neri F, Krepelova A, Incarnato D, Maldotti M, Parlato C, Galvagni F, Matarese F, Stunnenberg H, Oliviero S. DNMT3L antagonizes DNA methylation at bivalent promoters and favors DNA methylation at gene bodies in ESCs. Cell. 2013;155:121–134. doi: 10.1016/j.cell.2013.08.056. [DOI] [PubMed] [Google Scholar]
- 46.Liao J, Karnik R, Gu H, Ziller MJ, Clement K, Tsankov AM, Akopian V, Gifford CA, Donaghey J, Galonska C, et al. Targeted disruption of DNMT1, DNMT3A and DNMT3B in human embryonic stem cells. Nature Genetics. 2015;47:469–478. doi: 10.1038/ng.3258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Di Ruscio A, Ebralidze AK, Benoukraf T, Amabile G, Goff LA, Terragni J, Figueroa ME, De Figueiredo Pontes LL, Alberich-Jorda M, Zhang P, et al. DNMT1-interacting RNAs block gene-specific DNA methylation. Nature. 2013;503:371–376. doi: 10.1038/nature12598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chen T, Ueda Y, Dodge JE, Wang Z, Li E. Establishment and maintenance of genomic methylation patterns in mouse embryonic stem cells by Dnmt3a and Dnmt3b. Molecular and Cellular Biology. 2003;23:5594–5605. doi: 10.1128/MCB.23.16.5594-5605.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Liang G, Chan MF, Tomigahara Y, Tsai YC, Gonzales FA, Li E, Laird PW, Jones PA. Cooperativity between DNA methyltransferases in the maintenance methylation of repetitive elements. Molecular and Cellular Biology. 2002;22:480–491. doi: 10.1128/MCB.22.2.480-491.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang X, Su J, Jeong M, Ko M, Huang Y, Park HJ, Guzman A, Lei Y, Huang YH, Rao A, et al. DNMT3A and TET2 compete and cooperate to repress lineage-specific transcription factors in hematopoietic stem cells. Nature Genetics. 2016;48:1014–1023. doi: 10.1038/ng.3610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rinaldi L, Datta D, Serrat J, Morey L, Solanas G, Avgustinova A, Blanco E, Pons JI, Matallanas D, Von Kriegsheim A, et al. Dnmt3a and Dnmt3b Associate with Enhancers to Regulate Human Epidermal Stem Cell Homeostasis. Cell Stem Cell. 2016;19:491–501. doi: 10.1016/j.stem.2016.06.020. [DOI] [PubMed] [Google Scholar]
- 52.Pulecio J, Verma N, Meija-Ramirez E, Huangfu D, Raya A. CRISPR/Cas9 for epigenome engineering and investigation. Cell Stem Cell. 2017;21:431–447. doi: 10.1016/j.stem.2017.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Liao J, Karnik R, Gu H, Ziller MJ, Clement K, Tsankov AM, Akopian V, Gifford CA, Donaghey J, Galonska C, et al. Targeted disruption of DNMT1, DNMT3A and DNMT3B in human embryonic stem cells. Nature Genetics. 2015;47:469–478. doi: 10.1038/ng.3258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhu Z, Gonzalez F, Huangfu D. The iCRISPR platform for rapid genome editing in human pluripotent stem cells. Methods in Enzymology. 2014;546:215–250. doi: 10.1016/B978-0-12-801185-0.00011-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gonzalez F, Zhu Z, Shi ZD, Lelli K, Verma N, Li QV, Huangfu D. An iCRISPR platform for rapid, multiplexable, and inducible genome editing in human pluripotent stem cells. Cell Stem Cell. 2014;15:215–226. doi: 10.1016/j.stem.2014.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Langmead B, Trapnell C, Pop M, Salzberg S. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Anders S, Pyl PT, Huber W. HTSeq--a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31:166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biology. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Song CX, Szulwach KE, Fu Y, Dai Q, Yi C, Li X, Li Y, Chen CH, Zhang W, Jian X, et al. Selective chemical labeling reveals the genome-wide distribution of 5-hydroxymethylcytosine. Nat Biotechnol. 2011;29:68–72. doi: 10.1038/nbt.1732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhang Y, Liu T, Meyer C, Eeckhoute J, Johnson D, Bernstein B, Nusbaum C, Myers R, Brown M, Li W, Liu X. Model-based analysis of ChIP-Seq (MACS) Genome Biol. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Krueger F, Andrews SR. Bismark: a flexible aligner and methylation caller for Bisulfite-Seq applications. Bioinformatics. 2011;27:1571–1572. doi: 10.1093/bioinformatics/btr167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Akalin A, Garrett-Bakelman FE, Kormaksson M, Busuttil J, Zhang L, Khrebtukova I, Milne TA, Huang Y, Biswas D, Hess JL, et al. Base-pair resolution DNA methylation sequencing reveals profoundly divergent epigenetic landscapes in acute myeloid leukemia. PLoS Genetics. 2012;8:e1002781. doi: 10.1371/journal.pgen.1002781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Pan H, Jiang Y, Boi M, Tabbo F, Redmond D, Nie K, Ladetto M, Chiappella A, Cerchietti L, Shaknovich R, et al. Epigenomic evolution in diffuse large B-cell lymphomas. Nature Communications. 2015;6:6921. doi: 10.1038/ncomms7921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Giannopoulou EG, Elemento O. An integrated ChIP-seq analysis platform with customizable workflows. BMC Bioinformatics. 2011;12:277. doi: 10.1186/1471-2105-12-277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Williams K, Christensen J, Pedersen MT, Johansen JV, Cloos PA, Rappsilber J, Helin K. TET1 and hydroxymethylcytosine in transcription and DNA methylation fidelity. Nature. 2011;473:343–348. doi: 10.1038/nature10066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Xu Y, Wu F, Tan L, Kong L, Xiong L, Deng J, Barbera AJ, Zheng L, Zhang H, Huang S, et al. Genome-wide regulation of 5hmC, 5mC, and gene expression by Tet1 hydroxylase in mouse embryonic stem cells. Molecular Cell. 2011;42:451–464. doi: 10.1016/j.molcel.2011.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ehrich M, Nelson MR, Stanssens P, Zabeau M, Liloglou T, Xinarianos G, Cantor CR, Field JK, van den Boom D. Quantitative high-throughput analysis of DNA methylation patterns by base-specific cleavage and mass spectrometry. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:15785–15790. doi: 10.1073/pnas.0507816102. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All sequencing datasets are available under the Gene Expression Omnibus accession GSE89728.