Abstract
The excision of introns from pre-mRNA is an essential step in mRNA processing. We developed LeafCutter to study sample and population variation in intron splicing. LeafCutter identifies variable splicing events from short-read RNA-seq data and finds events of high complexity. Our approach obviates the need for transcript annotations and circumvents the challenges in estimating relative isoform or exon usage in complex splicing events. LeafCutter can be used both for detecting differential splicing between sample groups, and for mapping splicing quantitative trait loci (sQTLs). Compared to contemporary methods, we find 1.4–2.1 times more sQTLs, many of which help us ascribe molecular effects to disease-associated variants. Strikingly, transcriptome-wide associations between LeafCutter intron quantifications and 40 complex traits increased the number of associated disease genes at 5% FDR by an average of 2.1-fold as compared to using gene expression levels alone. LeafCutter is fast, scalable, easy to use, and available online.
Introduction
The alternative removal of introns during mRNA maturation is essential for major biological processes in eukaryotes including cellular differentiation, response to environmental stress, and proper gene regulation1,2,3,4. Nevertheless, our ability to draw novel insights into the regulation and function of splicing is hindered by the challenge of estimating transcript abundances from short-read RNA-seq data.
Popular approaches for studying alternative splicing from RNA-seq estimate isoform ratios5,6,7,8 or exon inclusion levels9,10. Quantification of isoforms or exons is intuitive because RNA-seq reads generally represent mature mRNA molecules from which introns have already been removed. However, estimation of isoform abundance from conventional short-read data is statistically challenging, as each read samples only a small part of the transcript, and alternative transcripts often have substantial overlap11. Similarly, when estimating exon expression levels, read depths are often overdispersed due to technical effects, and there may be ambiguity about which version of an exon is supported by a read if there are alternative 5′ or 3′ splice sites.
Further, both isoform- and exon-quantification approaches rely on transcript models, or pre-defined splicing events, both of which may be inaccurate or incomplete12. Predefined transcript models are particularly limiting when comparing splicing profiles of healthy versus disease samples, as aberrant transcripts may be disease-specific; or when studying genetic variants that generate splicing events in a subset of individuals only13. Even when transcript models are complete, it is difficult to estimate isoform or exon usage of complex alternative splicing events12.
An alternative perspective is to focus on what is removed in each splicing event. Excised introns may be inferred directly from reads that span exon-exon junctions. Thus, there is little ambiguity about the precise intron that is cut out, and quantification of usage ratios is very accurate12. A recent method, MAJIQ12, also proposed to estimate local splicing variation using split-reads and identified complex splicing events, however it does not scale well above 30 samples and has not been adapted to map splicing QTLs (sQTLs). At present, there are several software programs for sQTL mapping: GLiMMPS14, sQTLseekeR15 and Altrans16. However, all three rely on existing isoform annotations and both GLiMMPS and sQTLseeker reported modest numbers of sQTLs in their analyses.
Here we describe LeafCutter, a suite of novel methods that allow identification and quantification of novel and known alternative splicing events by focusing on intron excisions. We show LeafCutter’s utility by applying it to three important problems: (1) identification of differential splicing across conditions, (2) identification of sQTLs in multiple tissues or cell types, and (3) ascribing molecular effects to disease-associated GWAS loci. Using an early version of LeafCutter, we found that alternative splicing is an important mechanism through which genetic variants contribute to disease risk17. We now show that LeafCutter dramatically increases the number of detectable associations between genetic variation and pre-mRNA splicing, thus enhancing our understanding of disease-associated loci.
Results
Overview of LeafCutter
LeafCutter uses short-read RNA-seq data to detect intron excision events at base-pair precision by analyzing split-mapped reads (Figure 1). LeafCutter focuses on alternative splicing events including skipped exons, 5′ and 3′ alternative splice site usage and additional complex events that can be summarized by differences in intron excision12 (Supplementary Figure 1). LeafCutter’s intron-centric view of splicing is motivated by the observation that mRNA splicing predominantly occurs through the step-wise removal of introns from nascent pre-mRNA19. (Unlike isoform quantification methods such as Cufflinks25, alternative transcription start sites, and alternative polyadenylation are not directly measured by LeafCutter as they are not generally captured by intron excision events.) The major advantage of this representation is that LeafCutter does not require read assembly or inference on which isoform is supported by ambiguous reads, both of which are computationally and statistically difficult. As a result we were able to improve speed and memory requirements by an order of magnitude or more as compared to similar methods such as MAJIQ12.
Figure 1.
Overview of LeafCutter. (a) LeafCutter uses split reads to uncover alternative choices of intron excision by finding introns that share splice sites. In this example, LeafCutter identifies two clusters of variably excised introns. (b) LeafCutter workflow. First, short reads are mapped to the genome. When SNP data are available, WASP18 should be used to filter allele-specific reads that map with a bias. Next, LeafCutter extracts junction reads from .bam files, identifies alternatively excised intron clusters, and summarizes intron usage as counts or proportions. Lastly, LeafCutter identifies intron clusters with differentially excised introns between two user-defined groups using a Dirichlet-multinomial model or maps genetic variants associated with intron excision levels using a linear model. (c) Visualization of differential splicing between 10 GTEx heart and brain samples using LeafViz. LeafViz is an interactive browser-based application that allows users to visualize results from LeafCutter differential splicing analyses. In this example, we observed that Rbfox1 shows differential usage of a mutually exclusive exon in heart compared to brain. For all examples, see URLs.
To identify alternatively-excised introns, LeafCutter pools all mapped reads from a study and finds overlapping introns demarcated by split reads. LeafCutter then constructs a graph that connects all overlapping introns that share a donor or an acceptor splice site. The connected components of this graph form clusters, which represent alternative intron excision events. Finally, LeafCutter iteratively applies a filtering step to remove rarely used introns, which are defined based on the proportion of reads supporting an intron compared to other introns in the same cluster, and re-clusters leftover introns (Methods, Supplementary Note). In practice, we found that this filtering is important to avoid arbitrarily large clusters when read depth increases to a level at which noisy splicing events are supported by multiple reads.
De novo identification of RNA splicing in mammalian organs
We tested LeafCutter’s novel intron detection method by analyzing mapped RNA-seq20 data from 2,192 samples (Supplementary Note) across 14 tissues from the GTEx consortium21. We then searched for introns predicted to be alternatively excised by LeafCutter, but that were missing in three commonly-used annotation databases (GENCODE v19, Ensembl, and UCSC). For this analysis, we ensured that the identified introns were indeed alternatively excised by only considering introns that were excised at least 20% of the time as compared to other overlapping introns, in at least one fourth of the samples, analyzing each tissue separately. We found that between 10.8% and 19.3% (Pancreas and Spleen, respectively) of alternatively spliced introns are unannotated – excluding testis which is the major outlier, in which 48.5% of alternatively spliced introns are novel (Figure 2a). The latter observation is compatible with the “out-of-testis” hypothesis, which proposes that transcription is more permissive in testis and allows novel genes or isoforms to be selected for if beneficial22,23. Thus 31.5% of the alternatively excised introns we detected are unannotated (Supplementary Note), consistent with a recent study that identified a similar proportion of novel splicing events in 12 mouse tissues12. To further confirm that these findings were not merely mapping or GTEx-specific artefacts, we searched for junction reads in 21,504 human RNA-seq samples from the Sequence Read Archive (SRA) obtained from Intropolis24. We found that most (86%, Figure 2b and Supplementary Figure 2) novel junctions identified in our study were also present in at least one RNA-seq sample from the corresponding tissue as identified in Intropolis. Furthermore, we found that, as expected, unannotated junctions tend to be tissue-specific, and often involve complex splicing patterns (Supplementary Figure 3 and Supplementary Note).
Figure 2.
LeafCutter discovers reproducible unannotated introns. (a) Using LeafCutter to discover novel introns, we find that for any given tissue, over 10% of alternatively excised introns are unannotated. Remarkably, 48.5% of testis alternatively excised introns are unannotated. Different colors denote the proportion of introns when one or more splice sites are unannotated “(ss absent)”, both splice sites are annotated but the intron is not part of any transcript “(ss present)”, or when the intron is annotated in some but not all databases. (b) Barplots showing the numbers of unannotated and annotated junctions discovered using LeafCutter that are also found in samples from the short read archive (SRA) using Intropolis24. Phenopredict25 was used to predict the tissue type corresponding to the SRA samples analyzed in Intropolis. (c) The unannotated splice sites of novel introns show moderate signature of sequence conservation as determined by vertebrate phastCons scores. Miss one: conservation of the unannotated splice site of an intron for which the cognate splice site is annotated. Miss both: conservation of splice sites of introns with both splice sites unannotated.
We next asked whether these novel introns show evidence of functionality as determined by sequence conservation. When we averaged phastCons scores over unannotated splice sites of introns that were absent in annotation databases, we found a moderate, but significant, signature of sequence conservation (Figure 2c). In particular, we found that a significant number (4,616 or 15–25%) of novel splice sites are conserved across vertebrates (ave. phastCons ≥ 0.6, Supplementary Figure 4), indicating that the alternative excision of thousands of introns may be functional (Supplementary Note).
Fast and robust identification of differential splicing
LeafCutter uses counts from the clustering step to identify introns with differential splicing between user-defined groups. Read counts in an intron cluster are jointly modeled using a Dirichlet-multinomial generalized linear model (GLM), which we found offers superior sensitivity relative to a beta-binomial GLM that tests each intron independently (Supplementary Figure 5). The implicit normalization of the multinomial likelihood avoids the estimation of library size parameters required by methods such as DEXSEQ10.
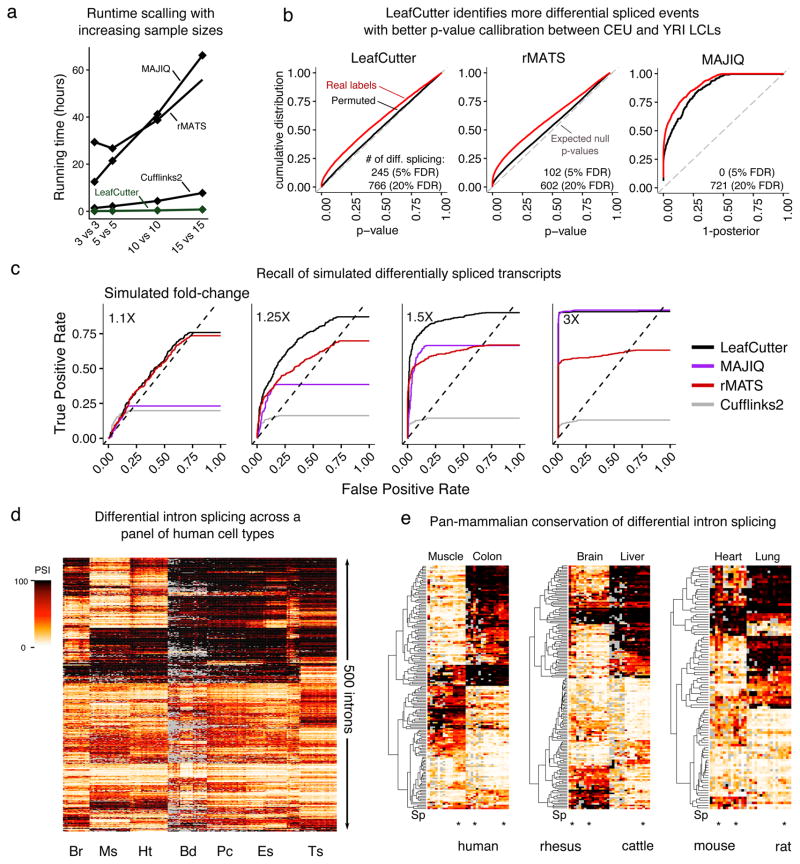
We compared LeafCutter against other methods for differential splicing detection including Cufflinks25, MAJIQ12, and rMATS26. We note that comparisons between algorithms have the complication that they there is typically no one-to-one mapping between the splicing events quantified by different methods. We discuss this issue and our solution in the Supplementary Note. For comparison, we applied each method to identify splicing differences between 3, 5, 10, and 15 Yoruba (YRI) versus European (CEU) LCL RNA-seq samples. In terms of runtime, we observed a large difference in scalability (Figure 3a). In our hands, only LeafCutter completed all comparisons within an hour, while Cufflinks2, rMATS, and MAJIQ took as long as 7.8, 55.7, and 66.2 hours to complete the largest comparison, respectively. In terms of memory usage, we also found that LeafCutter greatly outperforms the other software, using less than 400Mb of RAM for all comparisons, while MAJIQ required over 50Gb to perform the larger comparisons (Supplementary Figure 6). Although this range of sample sizes is representative for most biological studies, identifying differential splicing across groups in large studies such as GTEx would be impractically slow using rMATS or MAJIQ.
Figure 3.
Comparison of methods for detecting differential splicing. (a) Running time of differential splicing methods applied to comparisons between YRI and CEU LCLs RNA-seq samples. (b) Cumulative distributions of differential splicing test p-values (1-posterior for MAJIQ) for the 15 YRI versus 15 CEU LCLs comparison (red). The distribution of test p-values for a comparison with permuted labels is also shown (black). Cufflinks2 (not shown) detected 0 significantly differentially spliced genes (Supplementary Figure 8). (c) Receiver operating characteristic (ROC) curves of LeafCutter, Cufflinks2, rMATS and MAJIQ when evaluating differential splicing of genes with transcripts simulated to have varying levels of differential expression. ROC curves that do not reach 1.00 True Positive Rates reflect genes simulated to be differentially spliced that were not tested. (d) LeafCutter identifies tissue-regulated intron splicing events from GTEx organ samples. Heatmap of the intron excision ratios of the top 500 introns that were found to be differentially spliced between at least one tissue pair. Tissues include brain (Br), muscle (Ms), heart (Ht), blood (Bd), pancreas (Pc), esophagus (Eg), and testis (Ts). (e) Heatmap showing intron exclusion ratios of introns differentially spliced between pairs of tissues (Muscle vs Colon, Brain vs Liver, and heart vs Lung). Heatmap shows 100 random introns (97 for the heart vs lung comparison) that were predicted to be differentially excised in human with p-value < 10−10 (LR-test) and no more than 5 samples with missing data. Heatmap of all introns that pass our criteria can be found in Supplementary Figure 11.
To compare their ability to detect differential splicing, we reasoned that the p-values or posterior probabilities of the tests computed by each method are not directly comparable. We therefore computed an empirical FDR from the p-values of real comparisons between biologically distinct sample groups (i.e. YRI versus CEU here) and the p-values of permuted comparisons between samples with permuted labels (i.e. both groups contain YRI and CEU samples). If the p-values are well-calibrated, the p-value distribution of the permuted comparisons are expected to be uniform. Indeed, we observed that the distributions of LeafCutter and rMATS p-values for the permutations were close to uniform (Figure 3b and Supplementary Figure 7). However, the Cufflinks2 p-values were overly conservative (Supplementary Note) and the posterior probabilities P reported by MAJIQ for the permuted comparisons did not track the expected false discovery rate (FDR) of 1 - P (Supplementary Note). Altogether, we found that LeafCutter p-values showed better callibration compared to other methods, and that LeafCutter detected more differentially spliced events at all reasonable FDRs (≤ 0.2). Importantly, not only did LeafCutter detect more differentially spliced events at fixed FDRs, but it also achieved lower false negative rates when we evaluated the four methods on artificial data in which we simulated various levels of fold-changes in isoform levels (Figure 3c, Supplementary Note, Supplementary Figure 8). These comparisons show LeafCutter is a robust and highly scalable method for differential splicing analysis.
To evaluate LeafCutter’s suitability to detect differential splicing in a biological setting, we searched for intron clusters that show differential splicing between tissue pairs collected by the GTEx consortium, using all tissues to identify intron clusters. Combining all pairwise comparisons, we found 5,070 tissue-regulated splicing clusters at 10% FDR and with an estimated absolute effect size greater than 1.5 (Methods). As expected, GTEx samples mostly grouped by organ/tissue when hierarchically clustered according to the excision ratios of the five hundred most differentially spliced introns among all tissue pairs (Figure 3d, Supplementary Note).
To assess LeafCutter’s applicability to studies with smaller sample sizes, we used a small subset of GTEx samples and then evaluated replication using a larger subset. Using 220 samples (110 brain versus 110 muscle samples) we identified 1,906 differentially spliced clusters with estimated effect sizes greater than 1.5 at 10% FDR, compared to 885 when using only 8 samples (4 brain versus 4 muscle samples). Importantly, the strengths of associations (−log 10p-values) were highly correlated between our two analyses (Pearson R2 = 0.72, Supplementary Note, Supplementary Figure 9), and 98% of alternatively spliced clustered identified at 10% FDR in the analysis using 8 total samples replicated in the analysis using 220 samples, also at 10% FDR. These observations indicate that LeafCutter can detect differentially spliced introns even when the number of biological replicates is small.
We investigated whether the differentially spliced clusters identified using LeafCutter are likely to be functional by assessing the pan-mammalian conservation of their splicing patterns across multiple organs. Two previous studies analyzed the evolution of alternative splicing in mammals. When using gene expression levels, they saw clustering by organ as expected, however when using exon-skipping levels, they instead saw a clustering by species27,28. These observations indicate that a large number of alternative skipping events may lack function or undergo rapid turnover.
We initially clustered using all splicing events and confirmed the previous findings27,28 that the samples mostly clustered by species (Supplementary Figure 10). We then focused on a subset of introns that LeafCutter identified as differentially excised across tissue pairs in human and found that this subset shows splicing patterns that are broadly conserved across mammalian organs (Figure 3e, Supplementary Figure 11). To do this, we hierarchically clustered samples from eight organs in human and four mammals27 according to the orthologous intron excision proportions of differentially excised introns (p-value < 10−10 and β > 1.5) from our pairwise analyses of human GTEx samples (Supplementary Note). Unlike in the previous analyses, this revealed a striking clustering of the samples by organ, implying that hundreds of tissue-biased intron excisions events are conserved across mammals and likely have organ-specific functional roles29. Thus, while the majority of alternative splicing events likely undergo rapid turnover, events that show organ-specificity are much more often conserved across mammals and, therefore, are more likely to be functionally important.
Mapping splicing QTLs using LeafCutter
To evaluate LeafCutter’s ability to map splicing QTLs, we applied LeafCutter to 372 EU lymphoblastoid cell line (LCL) RNA-seq samples from GEUVADIS, and identified 42,716 clusters of alternatively excised introns. We used the proportion of reads supporting each alternatively excised intron identified by LeafCutter and a linear model30 to map sQTLs (Supplementary Note). We found 5,774 sQTLs at 5% FDR (compared to 620 trQTLs in the original study at 5% FDR, i.e., one ninth as many) and 4,543 at 1% FDR. To perform a controlled comparison, we also processed 85 YRI GEUVADIS LCLs RNA-seq samples and quantified RNA splicing events using LeafCutter, Altrans16, and Cufflinks25. We then uniformly standardized and normalized the estimates and used them as input to fastQTL30 to identify sQTLs (Supplementary Note). At a similar FDR, LeafCutter identifies 1.36X–1.46X and 1.83X–2.06X more sQTLs than Cufflinks2 and Altrans, respectively (Table 1). The rate of sQTL discoveries shared between methods is generally high (Storey’s π1 ranging from 0.53 to 0.72 for sQTLs identified at 10% FDR, Supplementary Note, Supplementary Figure 12), with LeafCutter sQTLs showing higher estimates of sharing (π1 = 0.70 and 0.72 with Cufflinks2 and Altrans, respectively) than Cufflinks2 sQTLs (0.52 with Altrans) or Altrans sQTLs (0.66 with Cufflinks2).
Table 1.
Summary of sQTLs identified in GEUVADIS samples using LeafCutter, Altrans16, and Cufflinks25. The numbers of transcript ratio QTLs (trQTLs) identified in the orginal GEUVADIS study31 are also listed in the sQTL columns. N/A: Not available.
| Method | YRI sQTLs (1% FDR) | YRI sQTLs (5% FDR) | CEU sQTLs (5% FDR) |
|---|---|---|---|
| LeafCutter | 1,294 | 1,982 | 5,775 |
| Altrans | 624 | 1,083 | N/A |
| Cufflinks2 | 888 | 1,459 | N/A |
| GEUVADIS study | N/A | 83 | 620 |
To further ensure that our sQTLs are not simply false positives, we verified that LeafCutter finds stronger associations between intronic splicing levels and SNPs previously identified as exon eQTLs and transcript ratio QTLs in GEUVADIS31 when compared to genome-wide SNPs (Figure 4a). Importantly, 399 (81.3%) of the 491 top trQTLs tested are significantly associated to intron splicing variation, as identified by LeafCutter (compared to 4.7% when our samples are permuted, Supplementary Note). Furthermore, we confirmed that the sQTLs we identified are located near splice sites, are close to the introns they affect (Figure 4b, Supplementary Figure 13), and are enriched in expected functional annotations such as “splice regions” and DNaseI hypersensitivity regions (Supplementary Figure 14).
Figure 4.
LeafCutter sQTLs augment interpretation of GWAS hits. (a) QQ-plot showing genome-wide sQTL signal in LCLs (black), sQTL signal conditioned on exon eQTLs (purple) and conditioned on transcript ratio QTLs (dark purple) from31. Signal from permuted data in light grey shows that the test is well-calibrated. (b) Positional distribution of sQTLs across LeafCutter-defined intron clusters. 1,421 of 4,543 sQTLs lie outside the boundaries (Supplementary Figure 13 for all sQTLs). (c) High proportion of shared sQTLs across four tissues from21. (d) Example of a SNP associated to the excision level of an intron in blood but not in other tissues. Boxplot center line: median, box: interquartile range (IQR), whiskers: range of data, excluding outliers beyond 1.5x IQR.
We used LeafCutter to identify sQTLs in four tissues from the GTEx consortium. Overall, we found 442, 1,058, 1,047, and 692 sQTLs at 1% FDR in heart, lung, thyroid gland, and whole blood, respectively (Supplementary Note). Using these, we estimated that 75–93% of sQTLs replicate across tissue pairs (Figure 4c, Supplementary Figure 15, Supplementary Note). This agrees with a high proportion of sharing of sQTLs across tissues32; and contrasts with much lower pairwise sharing reported for these data previously (9–48%)21. The high level of replication is likely due to LeafCutter’s increased power in detecting genetic associations with specific splicing events. Nevertheless, this leaves 7–25% of sQTLs that show tissue-specificity in our analysis. As expected we found that a large proportion of tissue-specific sQTLs arose from trivial cases where the intron is only alternatively excised, and therefore variable, in one tissue (Supplementary Figure 16). However, we also found cases in which the introns were alternatively excised in all tissues, yet show tissue-specific association with genotype (Figure 4d).
LeafCutter sQTLs link disease variants to mechanism
Finally, we asked whether sQTLs identified using LeafCutter could be used to ascribe molecular effects to disease-associated variants as determined by genome-wide association studies. eQTLs are enriched for disease-associated variants, and disease-associated variants that are eQTLs likely function by modulating gene expression31,21. We recently showed that sQTLs identified in LCLs are also enriched among autoimmune-disease-associated variants17. LeafCutter sQTLs can therefore help us characterize the functional effects of variants associated with complex diseases. Indeed, when we looked at the association signals of the top eQTLs and LeafCutter sQTLs from GEUVADIS to multiple sclerosis and rheumatoid arthritis (Supplementary Note), we found that both QTL types were enriched for stronger associations (Figure 5a) compared to genome-wide variants. Consistent with recent findings17, SNPs associated with multiple sclerosis are more highly enriched among sQTLs than eQTLs, while both eQTLs and sQTLs are similarly enriched among SNPs associated with rheumatoid arthritis (Figure 5a).
Figure 5.
LeafCutter sQTLs enable interpret disease-variants. (a) Enrichment of low p-value associations to multiple sclerosis and rheumatoid arthritis among LeafCutter sQTL and GEUVADIS eQTL SNPs. The numbers of top sQTLs and eQTLs that are tested in each GWAS are shown in parentheses. (b) Manhattan plot of S-PrediXcan association p-values from prediction models for intron quantification (LeafCutter; top) and gene expression (GEUVADIS; bottom). Genes that were found to be associated through RNA splicing are highlighted in orange, those associated through gene expression in purple, and those associated through both in black. The names of associated genes from the extended MHC region are not shown.
To further explore the utility of LeafCutter sQTLs for understanding GWAS signals, we applied S-PrediXcan33 to compute the association between predicted splicing quantification and 40 complex trait GWASs using models trained on GEUVADIS data (Methods and Supplementary Note). When applied to a rheumatoid arthritis (RA) GWAS, we found that considering intronic splicing allowed us to identify 18 putative disease genes (excluding genes in the extended MHC region), of which 13 were not associated using gene expression level measurements (Figure 5b). Novel putative disease genes associated through intronic splicing include CD40, a gene previously found to affect susceptibility to RA34. However, we found no overall enrichment of functional categories among the 18 or 13 putative disease genes. Overall, using LeafCutter splicing quantifications allowed us to increase the number of putative disease genes by an average of 2.1-fold as compared to using gene expression alone (Supplementary Data 1). These results demonstrate that by dramatically increasing the number of detected sQTLs, LeafCutter significantly enhances our ability to predict the molecular effects of disease-associated variants.
Discussion
While we applied LeafCutter to short-read RNA-seq data, the principles of LeafCutter could also be applied to long-read technologies. Long-read technologies may be particularly helpful in gene families where it is currently difficult to resolve splicing clusters with short reads due to multiple mapping.
In conclusion, our analyses show that LeafCutter is a powerful approach to study variation in alternative splicing. By focusing on intron removal rather than exon inclusion rates, we can accurately measure the step-wise intron-excision process orchestrated by the splicing machinery. Our count based statistical modeling, accounting for overdispersion, allows identification of robust variation in intron excision across conditions. Most importantly, LeafCutter allows the discovery of far more sQTLs than other contemporary methods, which improves our interpretation of disease-associated variants.
Online Methods
Identifying alternatively excised introns
To identify clusters of alternatively excised intron, split-reads that map with minimum 6nt into each exon are extracted from aligned .bam files. Overlapping introns defined by split-reads are then grouped together. For each of these groups LeafCutter constructs a graph where nodes are introns and edges represent shared splice junctions between two introns. The connected components of this graph define intron clusters. Singleton nodes (introns) are discarded. For each intron cluster, LeafCutter iteratively (1) removes introns that are supported by fewer than a number of (default 30) reads across all samples or fewer than a proportion (default 0.1%) of the total number of intronic read counts for the entire cluster, and (2) re-clustered introns according to the procedure above.
Dirichlet-multinomial generalized linear model
Intron clusters identified from LeafCutter comprise of two or more introns. More specifically, each intron clusters C identified using LeafCutter consists of J possible introns, which have counts yij for sample i and intron j (and cluster total niC = Σj′yij′)=, and N covariate column vectors xi of length P. LeafCutter uses a Dirichlet-Multinomial (𝒟ℳ) generalized linear model (GLM) to test for changes in intron usage across the entire cluster, instead of testing differential excision of each intron separately across conditions or genotypes.
where the softmax transform is used to ensure Σjpij=1. We perform maximum likelihood estimation for the outputs: the J coefficient row vectors βj of length P, the intercepts μj and concentration parameters αj. We use the following regularization to stabilize the optimization:
The Dirichlet-Multinomial likelihood is derived by integrating over a latent probability vector π in the hierarchy
where a = Σjaj, to give
In the limit πj = eaj/Σj′eaj′, aj → ∞ for all j, we have 𝒟ℳ(n,a) → Multinomial(n,π). For the GLM this means that as αj → ∞ we recover a multinomial model with no overdispersion. Smaller values of αj correspond to more overdispersion.
Differential intron excision across conditions
To test differential intron excision between two groups of samples, we encode xi = 0 for one group and xi = 1 for the other in the Dirichlet-Multinomial generalized linear model. For each cluster we compare the null model with only the intercept term to an alternative model including x using a likelihood ratio test with K-1 degrees of freedom, where K is the number of introns in the cluster.
We apply two filters to ensure we only perform reasonable tests:
Only introns which are detected (i.e. have at least one corresponding spliced read) in at least five samples are tested.
A cluster is only tested if each group includes at least 4 individuals with 20 spliced reads supporting introns in the cluster.
The thresholds in these filters are easily customizable as optional parameters.
Mapping splicing QTLs
To identify splicing QTLs, RNA-seq reads are mapped onto the genome using a RNA-aligner such as STAR35 or OLego20. Because LeafCutter only uses reads that map across junctions to estimate intron excision rates, it is essential to remove read-mapping biases caused by allele-specific reads. This is particularly significant when a variant is covered by reads that also span intron junctions as it can lead to spurious association between the variant and intron excision level estimates. Subsequent to mapping, LeafCutter finds alternatively excised intron clusters and quantifies intron excision levels in all samples. LeafCutter outputs intron excision proportions, which are used as input for standard QTL mapping tools such as MatrixEQTL or fastQTL (Supplementary Note).
S-PrediXcan analyses
Prediction models for intron quantification (LeafCutter) and gene expression (GEUVADIS) were trained using Elastic Net on GEUVADIS data. A value of α = 0.5 was chosen for the mixing parameter. Prediction performance for gene expression remains stable for a wide range of mixing parameters when α does not approach 0.0 (Ridge Regression)36,37. For each gene, we used SNPs within 1Mb upstream of the TSS and 1Mb downstream of the TTS. Similar windows around each splicing clusters were chosen.
We downloaded a genome-wide association meta-analysis summary statistics for Rheumatoid Arthritis (see URLs), and ran S-PrediXcan using these models. A total of 4,625 gene associations were obtained for the genetic expression model, and 41,196 intron quantification cluster associations for the splicing model, that had a model prediction False Discovery Ratio under 5%.
Visualizing LeafCutter differential splicing output
Using the R Shiny framework and ggplot2, we created an interactive browser-based application, LeafViz, that allows users to visualize LeafCutter differential splicing analyses. LeafViz generates LeafCutter cluster plots with information on the significance of the detected differential splicing and the estimated differences of the splicing changes. All significant clusters are labelled as “annotated” or “cryptic” by intersecting junctions with a user-defined set of transcripts (e.g. gencode v19). Users can directly download plots from the website in PDF format, which can be easily edited for publication. An example of LeafViz applied to a differential splicing analysis between 10 brain and 10 heart samples from GTEx is available online (see URLs).
Data Availability Statement
The datasets analyzed during the current study are available through dbGaP accession phs000424.v6.p1 (GTEx), GEO accession GSE41637 (RNA-seq data from mammalian organs), and ENA accession PRJEB3366 (Geuvadis).
Supplementary Material
Acknowledgments
We thank Xun Lan and other members of the Pritchard Lab for helpful discussions and comments. This work was supported by a CEHG Fellowship, the Howard Hughes Medical Institute, and the US National Institutes of Health (NIH grants HG007036, HG008140, HG009431 and R01MH107666).
Footnotes
Author Contributions
Y.I.L., D.A.K. and J.K.P. conceived of the project. Y.I.L. and D.A.K. performed the analyses and implemented the software. D.A.K. developed and performed the statistical tests and modeling. J.H. implemented the visualization application. A.N.B., S.P.D., and H.K.I. performed the S-PrediXcan analyses. Y.I.L. and J.K.P. wrote the manuscript.
Competing Financial Interests Statement
The authors declare no competing financial interests.
URLs: LeafCutter software: https://github.com/davidaknowles/leafcutter.
LeafViz visualizations: https://leafcutter.shinyapps.io/leafviz/.
RA summary statistics: http://plaza.umin.ac.jp/yokada/datasource/software.htm
References
- 1.Han H, et al. MBNL proteins repress ES-cell-specific alternative splicing and reprogramming. Nature. 2013;498:241–245. doi: 10.1038/nature12270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Calarco JA, et al. Regulation of vertebrate nervous system alternative splicing and development by an SR-related protein. Cell. 2009;138:898–910. doi: 10.1016/j.cell.2009.06.012. [DOI] [PubMed] [Google Scholar]
- 3.Brett D, Pospisil H, Valcarcel J, Reich J, Bork P. Alternative splicing and genome complexity. Nat Genet. 2002;30:29–30. doi: 10.1038/ng803. [DOI] [PubMed] [Google Scholar]
- 4.Pai AA, et al. Widespread Shortening of 3′ Untranslated Regions and Increased Exon Inclusion Are Evolutionarily Conserved Features of Innate Immune Responses to Infection. PLoS Genet. 2016;12:e1006338. doi: 10.1371/journal.pgen.1006338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Trapnell C, et al. Differential analysis of gene regulation at transcript resolution with RNA-seq. Nat Biotechnol. 2013;31:46–53. doi: 10.1038/nbt.2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Leng N, et al. EBSeq: an empirical Bayes hierarchical model for inference in RNA-seq experiments. Bioinformatics. 2013;29:1035–1043. doi: 10.1093/bioinformatics/btt087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Patro R, Mount SM, Kingsford C. Sailfish enables alignment-free isoform quantification from RNA-seq reads using lightweight algorithms. Nat Biotechnol. 2014;32:462–464. doi: 10.1038/nbt.2862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bray N, Pimentel H, Melsted P, Pachter L. Near-optimal rna-seq quantification. arxiv. 2015;1:1. [Google Scholar]
- 9.Katz Y, Wang ET, Airoldi EM, Burge CB. Analysis and design of RNA sequencing experiments for identifying isoform regulation. Nat Methods. 2010;7:1009–1015. doi: 10.1038/nmeth.1528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Anders S, Reyes A, Huber W. Detecting differential usage of exons from RNA-seq data. Genome Res. 2012;22:2008–2017. doi: 10.1101/gr.133744.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lacroix V, Sammeth M, Guigo R, Bergeron A. Exact Transcriptome Reconstruction from Short Sequence Reads. Springer Berlin Heidelberg; Berlin, Heidelberg: 2008. pp. 50–63. URL http://dx.doi.org/10.1007/978-3-540-87361-7_5. [Google Scholar]
- 12.Vaquero-Garcia J, et al. A new view of transcriptome complexity and regulation through the lens of local splicing variations. Elife. 2016;5 doi: 10.7554/eLife.11752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stein S, Lu ZX, Bahrami-Samani E, Park JW, Xing Y. Discover hidden splicing variations by mapping personal transcriptomes to personal genomes. Nucleic Acids Res. 2015;43:10612–10622. doi: 10.1093/nar/gkv1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhao K, Lu ZX, Park JW, Zhou Q, Xing Y. GLiMMPS: robust statistical model for regulatory variation of alternative splicing using RNA-seq data. Genome Biol. 2013;14:R74. doi: 10.1186/gb-2013-14-7-r74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Monlong J, Calvo M, Ferreira PG, Guigo R. Identification of genetic variants associated with alternative splicing using sQTLseekeR. Nat Commun. 2014;5:4698. doi: 10.1038/ncomms5698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ongen H, Dermitzakis ET. Alternative Splicing QTLs in European and African Populations. Am J Hum Genet. 2015;97:567–575. doi: 10.1016/j.ajhg.2015.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li YI, et al. RNA splicing is a primary link between genetic variation and disease. Science. 2016;352:600–604. doi: 10.1126/science.aad9417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.van de Geijn B, McVicker G, Gilad Y, Pritchard JK. WASP: allele-specific software for robust molecular quantitative trait locus discovery. Nat Methods. 2015;12:1061–1063. doi: 10.1038/nmeth.3582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tilgner H, et al. Deep sequencing of subcellular RNA fractions shows splicing to be predominantly co-transcriptional in the human genome but inefficient for lncRNAs. Genome Res. 2012;22:1616–1625. doi: 10.1101/gr.134445.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wu J, Anczukow O, Krainer AR, Zhang MQ, Zhang C. OLego: fast and sensitive mapping of spliced mRNA-Seq reads using small seeds. Nucleic Acids Res. 2013;41:5149–5163. doi: 10.1093/nar/gkt216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ardlie KG, et al. The Genotype-Tissue Expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science. 2015;348:648–660. doi: 10.1126/science.1262110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Soumillon M, et al. Cellular source and mechanisms of high transcriptome complexity in the mammalian testis. Cell Rep. 2013;3:2179–2190. doi: 10.1016/j.celrep.2013.05.031. [DOI] [PubMed] [Google Scholar]
- 23.Kaessmann H. Origins, evolution, and phenotypic impact of new genes. Genome Res. 2010;20:1313–1326. doi: 10.1101/gr.101386.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nellore A, et al. Human splicing diversity and the extent of unannotated splice junctions across human RNA-seq samples on the Sequence Read Archive. Genome Biol. 2016;17:266. doi: 10.1186/s13059-016-1118-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ellis SE, Collado Torres L, Leek J. Improving the value of public rna-seq expression data by phenotype prediction. bioRxiv. 2017 doi: 10.1093/nar/gky102. arXiv: http://www.biorxiv.org/content/early/2017/06/03/145656.full.pdf. [DOI] [PMC free article] [PubMed]
- 26.Shen S, et al. rMATS: robust and flexible detection of differential alternative splicing from replicate RNA-Seq data. Proc Natl Acad Sci USA. 2014;111:E5593–5601. doi: 10.1073/pnas.1419161111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Merkin J, Russell C, Chen P, Burge CB. Evolutionary dynamics of gene and isoform regulation in Mammalian tissues. Science. 2012;338:1593–1599. doi: 10.1126/science.1228186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Barbosa-Morais NL, et al. The evolutionary landscape of alternative splicing in vertebrate species. Science. 2012;338:1587–1593. doi: 10.1126/science.1230612. [DOI] [PubMed] [Google Scholar]
- 29.Reyes A, et al. Drift and conservation of differential exon usage across tissues in primate species. Proc Natl Acad Sci USA. 2013;110:15377–15382. doi: 10.1073/pnas.1307202110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ongen H, Buil A, Brown AA, Dermitzakis ET, Delaneau O. Fast and efficient QTL mapper for thousands of molecular phenotypes. Bioinformatics. 2016;32:1479–1485. doi: 10.1093/bioinformatics/btv722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lappalainen T, et al. Transcriptome and genome sequencing uncovers functional variation in humans. Nature. 2013;501:506–511. doi: 10.1038/nature12531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hsiao YE, et al. Alternative splicing modulated by genetic variants demonstrates accelerated evolution regulated by highly conserved proteins. Genome Res. 2016 doi: 10.1101/gr.193359.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Barbeira A, et al. Exploring the phenotypic consequences of tissue specific gene expression variation inferred from GWAS summary statistics. bioRxiv. 2017:045260. doi: 10.1038/s41467-018-03621-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Orozco G, et al. Association of CD40 with rheumatoid arthritis confirmed in a large UK case-control study. Ann Rheum Dis. 2010;69:813–816. doi: 10.1136/ard.2009.109579. [DOI] [PMC free article] [PubMed] [Google Scholar]
Methods-only References
- 35.Dobin A, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gamazon ER, et al. A gene-based association method for mapping traits using reference transcriptome data. Nature genetics. 2015;47:1091–1098. doi: 10.1038/ng.3367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wheeler HE, et al. Survey of the Heritability and Sparse Architecture of Gene Expression Traits across Human Tissues. PLoS Genet. 2016;12:e1006423. doi: 10.1371/journal.pgen.1006423. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets analyzed during the current study are available through dbGaP accession phs000424.v6.p1 (GTEx), GEO accession GSE41637 (RNA-seq data from mammalian organs), and ENA accession PRJEB3366 (Geuvadis).