Abstract
HCM, the most common inherited cardiac disease, is mainly caused by mutations in sarcomeric genes. More than a third of the patients are heterozygous for mutations in the MYH7 gene encoding for the β-myosin heavy chain. In HCM-patients, expression of the mutant and the wildtype allele can be unequal, thus leading to fractions of mutant and wildtype mRNA and protein which deviate from 1:1. This so-called allelic imbalance was detected in whole tissue samples but also in individual cells. There is evidence that the severity of HCM not only depends on the functional effect of the mutation itself, but also on the fraction of mutant protein in the myocardial tissue. Allelic imbalance has been shown to occur in a broad range of genes. Therefore, we aimed to examine whether the MYH7-alleles are intrinsically expressed imbalanced or whether the allelic imbalance is solely associated with the disease. We compared the expression of MYH7-alleles in non-HCM donors and in HCM-patients with different MYH7-missense mutations. In the HCM-patients, we identified imbalanced as well as equal expression of both alleles. Also at the protein level, allelic imbalance was determined. Most interestingly, we also discovered allelic imbalance and balance in non-HCM donors. Our findings therefore strongly indicate that apart from mutation-specific mechanisms, also non-HCM associated allelic-mRNA expression regulation may account for the allelic imbalance of the MYH7 gene in HCM-patients. Since the relative amount of mutant mRNA and protein or the extent of allelic imbalance has been associated with the severity of HCM, individual analysis of the MYH7-allelic expression may provide valuable information for the prognosis of each patient.
Keywords: Hypertrophic cardiomyopathy, Allelic imbalance, MYH7, Beta-myosin
Introduction
HCM is the most common disease of the heart with an incidence of 1:500 (Maron et al. 2012). Mutations in different sarcomeric genes have been shown to account for most of the HCM-cases (Cecconi et al. 2016; Maron et al. 2012). One of the most affected genes is MYH7 (Marsiglia and Pereira 2014), encoding for the β-myosin heavy chain (β-MyHC), a central player in cardiac and slow muscle contraction.
Numerous mutations in the MYH7-gene have been associated with HCM (Walsh et al. 2009). Since mutations in the MYH7-gene are mostly missense mutations, a poison peptide effect has been postulated for the mutant proteins. With respect to disease, benign or malignant phenotypes were assigned. This assignment was then transmitted to the causative mutations, classifying mutations as benign or malignant (Maron 2002). However, this was contrasted by the finding that the severity of the disease can vary even between patients with the same mutation (Maron 2002; Maron et al. 2012). Therefore additional environmental and genetic factors must influence the disease phenotype (Lopes et al. 2013).
Unequal expression of the two alleles of a gene (allelic imbalance) has been reported for a broad range of genes and tissues and the imbalanced expression of disease associated variants can influence the progression of different diseases (Pastinen 2010). Also for HCM it was shown that the relative abundance of mutant and wildtype mRNA and protein varies between the HCM-mutations (Di Domenico et al. 2012; Helms et al. 2014; Tripathi et al. 2011; Witjas-Paalberends et al. 2013) and the ratio of mutant/wildtype β-MyHC seems to be associated with disease severity (Jiang et al. 2013; Tripathi et al. 2011). In addition to this tissue-level allelic imbalance, we have shown recently that the relative fraction of mutant vs. wildtype mRNA is also variable from cell to cell in the myocardium of HCM-patients (Kraft et al. 2016). We have evidence that this cell-to-cell allelic imbalance is caused by an independent and stochastic ON- and OFF-switching of the mutant and the wildtype MYH7 allele in each individual cell. In principle, such a stochastic expression should result in nearly equal fractions of mRNA from both alleles at the tissue level. Yet, on average we found the same deviation from a 1:1 relation of mutant and wildtype mRNA for all analyzed single cells as it was determined at the tissue level for the same patients (Kraft et al. 2016; Tripathi et al. 2011). Therefore, factors additional to the stochastic ON- and OFF-switch of the alleles must induce the allelic imbalance at the tissue level. The regulatory mechanisms of allelic expression imbalance encompass variants in different cis-regulatory elements such as RNA-stability and turnover, transcription factor binding or splicing regulators but also epigenetic regulators such as DNA methylation (Bjornsson et al. 2008; Milani et al. 2009). Therefore, the allelic imbalance of the MYH7 gene in HCM may either be affected by intrinsic sequence variations in regulatory regions of the HCM-associated alleles or be directly altered by the mutations.
We hypothesize that if not only the HCM-mutation but also intrinsic, non-HCM-related expression regulating factors on the alleles hold responsible for the MYH7 allelic expression, allelic imbalance will also be detected in non-HCM controls. To address this question we examined the relative expression of the MYH7 alleles based on single base substitutions in 11 non-HCM donors and in ten HCM-patients with heterozygous mutations in the MYH7-gene.
Materials and methods
RNA extraction and RT-qPCR
Muscle tissue of Musculus soleus and myocardial tissues were flash frozen directly after excision and stored under liquid nitrogen. RNA was extracted using the PeqGold Total RNA Kit (PeqLab, Erlangen, Germany) according to the supplier’s instructions. Total RNA was subjected to cDNA-synthesis using 1× reaction buffer, 0.125 mM dNTPs each, 0.4 µM MYH7 specific primers (Table 1), 1 U/µl RNase inhibitor (RiboSafe, Bioline, Luckenwalde, Germany), and 5 U/µl reverse transcriptase (Tetro RT, Bioline) and 1 µl RNA for 1 h at 42 °C. Unless stated otherwise in Table 1, for amplification 1 µl cDNA was mixed with 1x reaction buffer, 0.5 mM MgCl2, 0.2 mM of each dNTP, 0.2 µM of both forward and reverse Primers (Table 1), and 0.04 U/µl HotStarTaq (Qiagen, Hilden, Germany). Initial activation was performed for 15 min at 95 °C. Subsequently 45 cycles were applied with 95 °C for 30 s, 64 °C for 30 s, and 72 °C for 30 s. The final elongation was performed at 72 °C for 2 min. To minimize heteroduplexes a reconditioning PCR was performed. 2.5 µl PCR product were transferred to a final volume of 25 µl respective PCR reaction mix, and the respective PCR protocol was run for three successive cycles (Thompson et al. 2002).
Table 1.
Oligonucleotides and peptides used for allele specific quantification assays
| Variant | cDNA-primer | Primer forward | Primer reverse | Buffer | Annealing (°C) | endo-nuclease | Fragments WT | Fragments Mut | Peptide wildtype native | Peptide wildtype isotope | Peptide mutant native | Peptide mutant isotope |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A200V | GCCTTGCCTTTGCCCTTCTCA | AGGGTCATCCAGTACTTTGCTGTTATTAC | ATGTCTGCAGATGCCAACTTTCCT | Standard | 64 | Hpy188I | 103; 154 | 27; 103; 127 | NH2-VIQYFAVIAAIGDR-COOH | NH2-VIQYF*(9C13 + N15)AVIAAIGDR-COOH | NH2-VIQYFAVIAVIGDR-COOH | NH2-VIQYF*(9C13 + N15)AVIAVIGDR-COOH |
| R453C | GCCTTGCCTTTGCCCTTCTCA | GTTCAACTGGATGGTGACGCGCAT | TTGGCCAGTGCCCCAGTGGCATATAT | Standard | 68 | NlaIII | 48; 175 | 48; 50;120 | ||||
| V606M | TGCCAGGTTGTCTTGTTCCG | TCCCCAAGGCCACCGACATGACCTTCAAGGCCAAGC | CTTTGCCCTTCTCAATAGGCGCATCAG | Standard | 68 | NlaIII | 20; 268 | 20; 88; 180 | ||||
| G716R | TGCCAGGTTGTCTTGTTCCG | GTGCTGGAGGGCATCCGCATCT | CCATTCTGGCGAGCACACCT | 2,5 mM MgCl2 | 64 | BslI | 54; 115; 138 | 115; 192 | NH2-GFPNRILYGDFRQRYRILNPAAIPEGQFIDSRK-COOH | NH2-GFPNRIL*(6C13 + N15)YGDFRQRYRIL*(6C13 + N15)NPAAIPEGQFIDSRK-COOH | NH2-GFPNRILYRDFRQRYRILNPAAIPEGQFIDSRK-COOH | NH2-GFPNRIL*(6C13 + N15)YRDFRQRYRIL*(6C13 + N15)NPAAIPEGQFIDSRK-COOH |
| R719W | TGCCAGGTTGTCTTGTTCCG | ACGAAGTCTCCAGGCGTGATGGACAACC | CTTTTTGTACTCCATTCTGGCGAGCACA | 2,5 mM MgCl2 | 68 | MspI | 109; 123; 146 | 109; 269 | ||||
| G741R | TGCCAGGTTGTCTTGTTCCG | ATCCCTGAGGGACAGTTCATTGATAGCAGGACG | CTTTTTGTACTCCATTCTGGCGAGCACA | Standard | 67 | BstUI | 51; 171 | 33; 51; 138 | NH2-DSRKGAEKLLSSL-COO3 | NH2-DSRKGAEKL*(6C13 + N15)LSSL-COO3 | NH2-DSRKRAEKLLSSL-COO3 | NH2-DSRKRAEKL*(6C13 + N15)LSSL-COO3 |
| D752N | TGCCAGGTTGTCTTGTTCCG | GGACAGTTCATTGATAGCAGGAAGGGGGCA | CTCGGGACTGGGCCTGGATACGCGTGATCA | 2 mM MgCl2 | 68 | BclI | 29; 57; 98 | 29; 155 | ||||
| T1377M | TCATGGATAGTCTTTCCGCTGGA | GCGTCCTTTCCAAGGCCAACT | AGTTCCTCTGCTTCTTGTCCAGG | 65 | BccI | 67; 203 | 41; 67; 156 | |||||
| T63T | GGTTCTGCTGCATCACCTG | CGTGCCTGATGACAAACAGG | GGAGTTGGTGAGTGACAGG | 3 mM MgCl2 | 68 | DdeI | 125; 294 | 75. 125, 219 | ||||
| I989I | GATGCGTGCCTGAAGCTCCTT | CTCAAAAGGGACTCGATGAT | AAAAAAAAAAAAATTCATCCTCAATCATTGC | 2 mM MgCl2 | 58 | BsrDI | 24; 116; 318 | 24; 434 | ||||
| A1702A | TGTCCTCCTCCGTCTGGTAG | CCAACGACGACCTGGCGGAG | CTCAGCATTCCTGCACTCCT | Additional 1 mM MgCl2 and 4% DMSO | 58 | EciI | 29; 101; 139 | 29; 240 |
Allele specific restriction digest
12.5 µl of the reconditioned PCR products were treated with respective restriction enzymes (Table 1) in a final volume of 15 µl for at least 3 h to yield the allele specific fragments (Table 1). The fragments were separated on 3% sieving agarose gels stained with ethidiumbromide and mutant vs. wildtype MYH7 mRNA was quantified as described previously in detail (Tripathi et al. 2011). In brief, the restriction fragments were analysed densitometrically using the TotalLab (Newcastle upon Tyne, Great Britain) and Origin (OriginLab, Northampton, MA, USA) software, yielding the integrated optical density (IOD) of each band. The IOD was normalized against the number of base pairs. The fraction of mutant per wildtype MYH7 mRNA was calculated from the IOD/bp values of the respective bands.
Relative quantification of mutant and wildtype myosin
The quantification of mutant and wildtype β-MyHC protein was performed as described previously in detail (Becker et al. 2007). In brief, for each mutation a specific set of isotope labelled peptides (Table 1) was spiked in equal quantities to extracted myosin from tissue samples of the HCM-patients. The mixture was digested using trypsin (A200V), Lys-C (G716R) or Asp-N (G741R) and subjected to LC/ESI-based analysis of the ratio of WT- and mutant-specific peptides in the samples. The isotope labelled peptides were used as internal standards for the quantification to correct for sequence specific ionization. The assays were established using mutant and wildtype specific synthetic peptides (Table 1).
Statistics
For statistical analysis of the deviation from the 50:50 ratio, we used one way ANOVA test. We compared the fractions of all quantification experiments per mutation or variant, respectively, with the theoretically expected 50%. Analysis was performed using the GraphPad Prism software, significance was assigned for p < 0.0001.
Ethics statement
Informed consent was obtained from all individuals according to approved Ethics Committee protocols of the institutions involved. The study was approved by the Ethics Committee of Hannover Medical School (no. 2276-2014). The investigations conformed to the principles of the Declaration of Helsinki (1997).
Results
Patients and non-HCM donor genetics
We analyzed the allelic expression of the MYH7 gene in 21 individuals based on single base substitutions (Table 2). We analyzed flash frozen tissue from slow skeletal muscle (M. soleus) from one male HCM-patient with the mutation R453C and one male and two female patients with the mutation G741R. In addition, left ventricular/interventricular septum tissue from one HCM-patient each with the mutation A200V, V606M, G716R, R719W, D752N, and T1377M was analyzed. Slow skeletal muscle fibers express mainly the β-myosin isoform and as we have shown previously, the allelic imbalance of the MYH7 gene is comparable in slow skeletal muscle and heart tissue (Tripathi et al. 2011). The three patients with the mutation G741R were related, however showed distinct progression of HCM at biopsy ranging from asymptomatic (G741R-1, age at biopsy 45), dyspnea on effort (G741R-3, age at biopsy 39) to severe heart failure (G741R-2, age at biopsy 55). Exemplary analysis revealed that the patients A200V, R453C, G716R, G741R-1, G741R-2 and G741R-3 showed no variants in the in the 5′ and 3′-UTR. The patients G741R-1, G741R-2 and G741R-3 were further analyzed for SNPs in the promotor region and also showed no variants.
Table 2.
Relative expression of the MYH7 alleles in HCM-patients and non-HCM donors
| Variant | Individual | Gender | Tissue | Fraction of mutant allele (mean ± SEM) | |
|---|---|---|---|---|---|
| mRNA | Protein | ||||
| A200V | Female | Septum | 48.3 ± 1.9 | 49.1 ± 0.9 | |
| R453C | Male | Soleus | 37.0 ± 1.0* | ||
| V606M | Male | Septum | 37.5 ± 0.8* | ||
| G716R | Male | Left ventricle | 88.6 ± 0.2* | 29.9 ± 0.9* | |
| R719W | Male | Left ventricle | 52.2 ± 0.4 | ||
| G741R | 1 | Male | Soleus | 18.8 ± 0.4* | 21.4 ± 0.4* |
| 2 | Female | Soleus | 49.4 ± 1.0 | 26.9 ± 0.6* | |
| 3 | Female | Soleus | 44.8 ± 2.9 | 26.7 ± 0.6* | |
| D752N | Male | Septum | 7.9 ± 0.6* | ||
| T1377M | Female | Septum | 52.3 ± 1.1 | ||
| T63T | 1 | Male | Left ventricle | 51.1 ± 0.33 | |
| 2 | Female | Left ventricle | 49.2 ± 0.97 | ||
| 3 | Male | Left ventricle | 5.5 ± 0.15* | ||
| I989I | 1 | Female | Left ventricle | 51.2 ± 0.04 | |
| 2 | Female | Left ventricle | 51.3 ± 0.1 | ||
| 3 | Female | Left ventricle | 53.2 ± 0.1 | ||
| 4 | Male | Left ventricle | 52.6 ± 0.1 | ||
| A1702A | 1 | Female | Left ventricle | 60.2 ± 0.2* | |
| 2 | Male | Left ventricle | 60.3 ± 0.7* | ||
| 3 | Female | Left ventricle | 58.1 ± 0.2* | ||
| 4 | Male | Left ventricle | 50.1 ± 0.3 | ||
*Significant deviation from 50% of equal allelic expression (p > 0.0001)
The MYH7-variants R453C and V606M (Watkins et al. 1992), G716R and R719W (Anan et al. 1994), and G741R (Fananapazir et al. 1993) have been previously described as disease causing and are listed as “pathogenic” in the ClinVar database. In addition, they are reported with a very low allele frequency or are even not present in the Exac or GnomAD databases and at least three out of four in silico prediction softwares report the variants as pathogenic (Table 3). Therefore, these HCM-variants strongly fulfill the criteria of pathogenicity according to the ACMG guidelines (Richards et al. 2015). The mutation T1377M is listed as “likely pathogenic” in ClinVar, it has been described previously as HCM-mutation (Helms et al. 2014) and it is highly seldom in the reference population as determined by the GnomAD allele frequency of 4.06 × 10− 6. In addition, in silico prediction of pathogenicity by two out of four independent software tools suggests a deleterious effect on the mutant protein (Table 3). According to the ACMG guidelines (Richards et al. 2015) this indicates strongly to a pathogenic effect of the T1377M-variant. The variants A200V and D752N are not listed in ClinVar. However, the variant alleles are not present in the Exac/GnomAD database and both variants are predicted to be deleterious for protein function by the three in silico analysis tools. In addition, at position 200 another pathogenic variant has been determined (A200T) (Fujino et al. 2013). Taken together, the ACMG guidelines (Richards et al. 2015) strongly support a pathogenic effect also of these mutations (Table 3). Therefore, we assume that all MYH7-variants in the HCM patients as pathogenic under the light of current evidence.
Table 3.
Pathogenicity of MYH7-variants of the HCM-patients and donors
| Variant | Exac/GnomAD allele frequency | In silico predictions of pathogenicity | ClinVar classification | ACMG classificatione | Initial reference for pathogenic variant | |||
|---|---|---|---|---|---|---|---|---|
| Mutation tastera | PolyPhen2b | PhD-SNPc | PANTHERd | |||||
| A200V | Not present | Disease causing | Probably damaging | Disease | Neutral | No data | Pathogenic (PS1 + PM2 + PP3) | Pathogenic variant at same amino acid: Fujino et al. (2013) |
| R453C | Not present | Disease causing | Probably damaging | Disease | Disease | Pathogenic | Pathogenic (known disease causing, PS3 + PM2 + PP3) | Watkins et al. (1992) |
| V606M | 3.230 × 10−5 | Disease causing | Probably damaging | Neutral | Disease | Pathogenic | Pathogenic (known disease causing + PS3 + PM2 + PP3) | Watkins et al. (1992) |
| G716R | Not present | Disease causing | Probably damaging | Disease | Disease | Pathogenic | Pathogenic (known disease causing PS3 + PM2 + PP3) | Anan et al. (1994) |
| R719W | 3.231 × 10−5 | Disease causing | Probably damaging | Disease | Disease | Pathogenic | Pathogenic (known disease causing PS3 + PM2 + PP3) | Anan et al. (1994) |
| G741R | 3.232 × 10−5 | Disease causing | Probably damaging | Disease | Neutral | Pathogenic | Pathogenic (known disease causing PS3 + PM2 + PP3) | Fananapazir et al. (1993) |
| D752N | Not present | Disease causing | Probably damaging | Disease | Neutral | No data | Pathogenic (PS2 + PM2 + PP3) | Not described previously |
| T1377M | 4.06 × 10−6 | Disease causing | Probably damaging | Neutral | Neutral | Likely pathogenic | Pathogenic (known disease causing, PS2 + PM2 + PP3) | Helms et al. (2014) |
| T63T | 0.47960 | Polymorphism | Not applicable | Not applicable | Not applicable | Benign | Benign (BA1) | |
| I989I | 0.32119 | Polymorphism | Not applicable | Not applicable | Not applicable | Benign | Benign (BA1) | |
| A1702A | 0.10702 | Polymorphism | Not applicable | Not applicable | Not applicable | Benign | Benign (BA1) | |
In addition, interventricular septum samples from non-HCM donors were analyzed for single nucleotide polymorphisms (SNPs) in the MYH7 gene. We identified three donors heterozygous for the SNP T63T (rs2069540), four donors heterozygous for the SNP I928I (rs7157716) and four donors heterozygous for the SNP A1702A (rs3729830). Under the light of current evidence, the variants are not pathogenic (Table 3). To our knowledge, the donors were not related to each other. In total five male and six female donors were analyzed.
Allelic balance and imbalance of the MYH7 gene in HCM-patients
We have shown previously, that the mutant and wildtype MYH7 alleles are expressed imbalanced at the tissue level in HCM-patients with different missense mutations in this gene. To examine whether this phenomenon can be detected for a broader range of MYH7-mutations, we determined the ratio of mutant vs. wildtype transcript in samples of slow skeletal muscle tissue and cardiac tissue from HCM-patients with eight different MYH7-missense mutations.
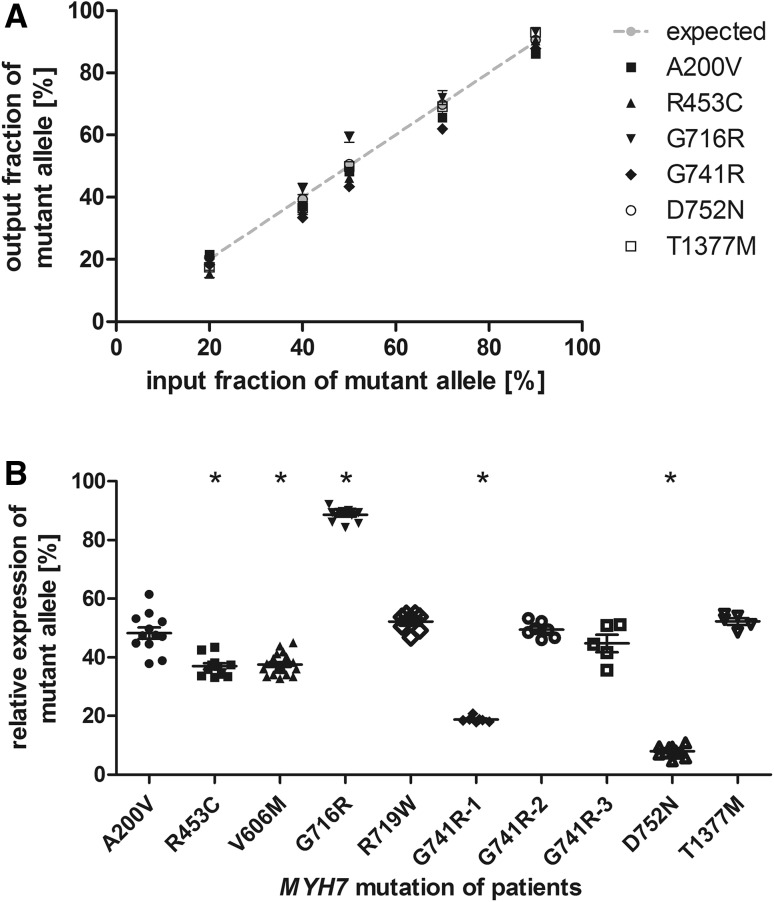
The analyses were designed as shown previously for HCM-associated MYH7-mutations (Tripathi et al. 2011). In brief, the PCR amplicons contained a restriction site that was either generated or disrupted by the respective base substitution. The region of interest was PCR-amplified, reconditioning PCR was performed to eliminate potential heteroduplexes originating from the end point PCR and subsequently subjected to restriction digest. The allele specific restriction fragments were separated by agarose gel electrophoresis, quantified densitometrically and the respective ratios of each allele were calculated. Each analysis was optimized and finally validated using defined mixtures of recombinant plasmids encoding either for the wildtype or the mutant PCR amplicon. Using the finalized protocols, we relatively quantified both alleles in the standard plasmid mixtures for ratios between 10:90 and 80:20 in at least three independent experiments (Fig. 1A). For each patient 2–4 independent RNA extractions were performed and used for at least two cDNA-syntheses. The expression of the mutant allele was quantified from each cDNA at least in duplicates and statistically compared to a theoretical value of 50% reflecting balanced allelic expression (one way ANOVA) (Fig. 1b).
Fig. 1.
Relative quantification of mutant and wildtype MYH7-mRNA in HCM-patients. a Defined mixtures of synthetic plasmids encoding for the wildtype and the respective mutated sequences were amplified by quantitative PCR and quantified densitometrically from allele specific restriction digests. Quantifications were performed in at least 4 independent experiments. The expected ratios are indicated by the grey line. b Relative quantification of mutant and wildtype MYH7-mRNA in M. soleus or myocardial tissue from HCM-patients. Quantification was performed on RNA extracted from at least two pieces of tissue at least in duplicate analysis. Each dot represents a single quantification analysis. The asterisks indicate significant deviation from 50% (one way ANOVA; p < 0.0001)
The mutations R435C, V606M, and D752N showed a significantly reduced expression of the mutated allele of on average 37.0, 37.5 and 7.9%, respectively (Fig. 1b). In the patients with the mutations A200V, R719W, and T1377M both alleles were expressed at a similar ratio of 48.3, 52.2, and 52.3%, respectively. The mutation G716R showed a high expression of the mutant allele with 88.6% (Table 2).
Interestingly, in skeletal muscle tissue from three related patients with the mutation G741R we found a varying expression pattern. Whereas one patient (G741R-1) showed a significantly reduced expression of the mutated allele of 18.8%, both other patients (G741R-2 and G741R-3) showed equal expression of both alleles of 45.6 and 46.6% of the mutant allele (Fig. 1b; Table 2). To test for differences between the G741R patients we compared SNPs in the promotor region and in exons and flanking intron sequences. No variant was identified in the promotor region or the 3′ UTR. However, the patients showed different exonic SNPs (Table 4).
Table 4.
Variants detected upon analysis of the MYH7 promoter, 5′-UTR, 3′-UTR, exons, and flanking intron sequences
| Patient | Location | Web Ref. | HGVS nomenclature | Effect | Genotype |
|---|---|---|---|---|---|
| G741-1 | Exon | rs2069540 | c.189C>T | SNP | Homozygous |
| Exon | c.2221G>C | Mutation | Heterozygous | ||
| Exon | rs3729830 | c.5106G>A | SNP | Heterozygous | |
| G741R-2 | Exon | rs2069540 | c.189C>T | SNP | Heterozygous |
| Exon | c.1095G>A | SNP | Heterozygous | ||
| Exon | c.2221G>C | Mutation | Heterozygous | ||
| Exon | rs7157716 | c.2967T>C | SNP | Heterozygous | |
| G741R-3 | Exon | rs2069540 | c.189C>T | SNP | Homozygous |
| Exon | c.2221G>C | Mutation | Heterozygous |
However, at the protein level the difference between patients with the mutation G741R was no longer detectable. The myocardial tissue of the three patients contained 21.4, 26.9, and 26.7%, respectively, of the mutant β-MyHC protein (Table 2). Also the patient with mutation G716R showed different levels of mutant mRNA and protein. At the mRNA level the mutant allele was predominant with 89%, while at the protein level only 29.9% mutant myosin was found. The patient with mutation A200V in contrast showed comparable levels of mutant mRNA and protein.
Allelic imbalance of the MYH7 gene in non-HCM individuals
To test whether the MYH7 alleles are generally expressed imbalanced and that this is not specific for HCM patients, we analyzed cardiac tissue from non-HCM control individuals. Three different single nucleotide polymorphisms within the MYH7 gene were used for allelic discrimination.
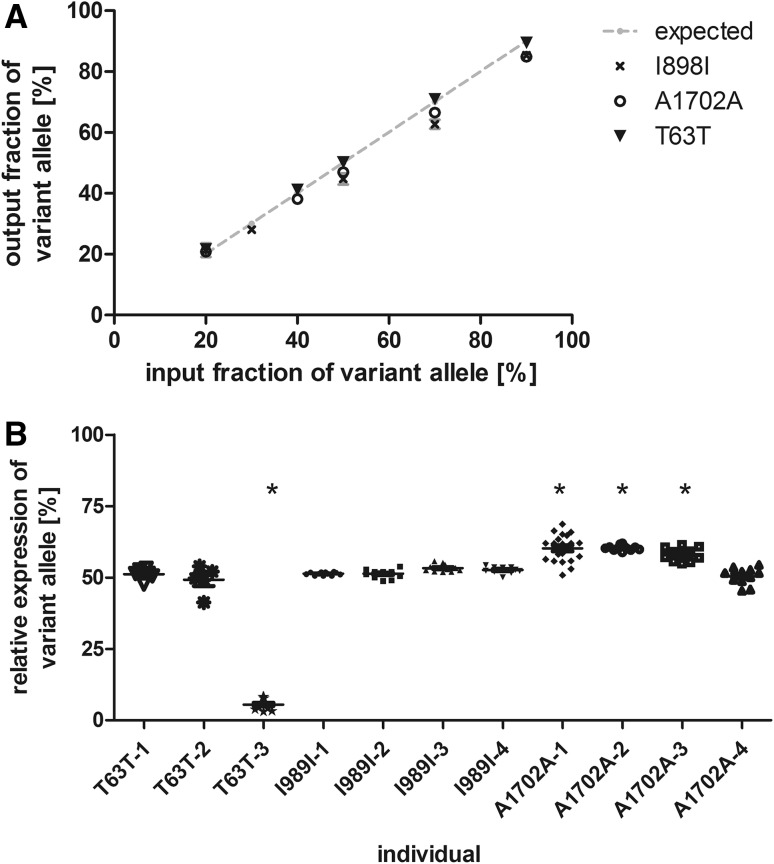
Specific qPCR and restriction digest analyses were established for each SNP and validated using standard plasmid mixtures as described for the HCM-mutations (Fig. 2a). For each individual, three independent RNA extractions were performed and analyzed as described for the HCM-patients. Most interestingly, we determined a statistically significant deviation from the 50:50 allelic expression ratio in four out of 11 non-HCM donors (Fig. 2b). For three control individuals we determined up to 60% of the variant allele, and one control individual showed a low fraction of the variant allele of on average 5.5% (Table 2). Therefore, the allelic expression ranges from equal to almost exclusively one allele. Nevertheless, both alleles were expressed in all individuals.
Fig. 2.
Relative quantification of the MYH7 alleles in myocardium of non-HCM control individuals using heterozygous single nucleotide polymorphisms. a Defined mixtures of synthetic plasmids encoding for the wildtype and T63T (triangle) I989I (cross) or A1702A (open circle) variant, respectively were amplified by quantitative PCR and quantified densitometrically from allele specific restriction digests. Quantifications were performed in at least four independent experiments. The expected ratios are indicated by the grey line. b Relative quantification of MYH7-mRNA in myocardial tissue from 11 non-HCM control individuals carrying the variants T636T, I989I, and A1702A, respectively. Quantification was performed on RNA extracted from at least two pieces of tissue at least in duplicate analysis. Each dot represents a single quantification analysis. The asterisks indicate significant deviation from 50% (one way ANOVA; p < 0.0001)
Discussion
Allelic imbalance is a general feature of MYH7 mRNA expression
Several studies on HCM-patients have shown unequal expression of the mutant and wildtype alleles of β-myosin (MYH7) (Di Domenico et al. 2012; Nier et al. 1999; Tripathi et al. 2011; Witjas-Paalberends et al. 2013) and myosin binding protein (MYBPC3) (Helms et al. 2014) at mRNA and protein level. Also balanced allelic expression of the mutant and wildtype mRNA associated with imbalanced expression of mutant and wildtype protein have been reported (Helms et al. 2014). Clinically, the severity of HCM has been linked to the fraction of mutant protein (Jiang et al. 2013; Tripathi et al. 2011) and to the degree of imbalance (Helms et al. 2014). In line with these findings our present study shows MYH7-allelic expression imbalance and balance in the HCM-patients, both at the mRNA and at the protein level. The allelic imbalance encompasses a broad range, from an almost exclusive expression of either the mutant or the wildtype allele, respectively, to a balanced expression of both alleles. However, allelic expression imbalance has also been shown for many different genes under healthy conditions (Jeffries et al. 2016). In accordance, also non-HCM control individuals showed imbalanced expression of the MYH7-alleles. Next to only slight deviations from the 50:50 ratio also a marked imbalance was detected. Our findings contradict the assumption that MYH7-missense mutations cannot be expressed imbalanced in HCM-patients at the mRNA or protein level (Helms et al. 2014). The authors assume, that only premature termination codons would cause allelic imbalance. In contrast, our study shows, in line with findings from other groups (Heap et al. 2009; McDaniell et al. 2010; Serre et al. 2008), that allelic imbalance is a common mechanism of gene expression; also under non-disease associated conditions. Therefore, our findings provide evidence that a general variation in MYH7-allelic expression exists and importantly, may also underlie at least part of the allelic imbalance experienced in HCM-patients.
At the mRNA-level, a marked allelic imbalance was only found in male individuals. However, in previous studies we have also determined ratios of 70/30 in a female patient (H29, Tripathi et al. 2011). Therefore, we assume that our cohort is not representative with regard to gender diversity. In addition, at the protein level both, male and female HCM-patients show significant allelic imbalance.
It should be noted that we used the different SNPs in the non-HCM individuals only as indicators for the differential expression of the MYH7-alleles. These SNPs are very common (Table 3) and are also found in some of the HCM-patients (Table 4). Since the allele specific expression levels vary largely between individuals with the same SNPs we do not assume that the examined SNPs dominate allelic imbalance establishment. However, it seems likely that at least some SNPs may contribute significantly to regulation of the allelic expression of the MYH7-alleles.
Our results from donor individuals show for the first time, that allelic imbalance is an intrinsic, non-HCM-dependent feature of MYH7-gene expression. Under healthy conditions—without disease causing mutations—the imbalanced allelic expression of genes will have no further effect on functional parameters. However, altered expression of variants that affect protein function or increase disease susceptibility may exhibit severe effects on the metabolism, e.g. cardiomyocyte and thus cardiac function. Finally, this may increase the severity or occurrence of the respective disorder as shown for multiple sclerosis (Keshari et al. 2016), autoimmune diseases (Ge et al. 2009), cancer (Chen et al. 2008; Wang et al. 2016) and HCM (Di Domenico et al. 2012; Helms et al. 2014; Tripathi et al. 2011). And vice-versa, in a mouse model of HCM, the targeted knock-down of the mutant allele has been shown to improve HCM-phenotype (Jiang et al. 2013). Therefore, next to the direct “poisoning” effect of the MYH7-mutations on β-MyHC function, the unequal expression of mutated and wildtype alleles may provide an essential player in HCM development (Brenner et al. 2014). Our results indicate that in patients with HCM-mutations, the intrinsic MYH7-allelic imbalance may be one additional factor that affects disease progression.
Mutation dependent and independent mechanisms regulate unequal allelic expression of MYH7
In a previous study we have shown that the MYH7 alleles are expressed at different ratios in neighboring cells in the myocardium, causing a functional imbalance between these cells that may finally trigger the progression of myofibrillar and myocardial disarray and hypertrophy (Kraft et al. 2016). We assume that this cell-to-cell allelic imbalance is caused by independent, stochastic ON- and OFF-switching of each MYH7-allele, so-called bursts-of-transcription. However, the stochastic expression pattern should result in equal levels of each allele averaged over the multitude of cells analyzed in our tissue samples. Nevertheless, the average of all single cells in patients with the mutation R723G show a comparable level of allelic imbalance as determined at the tissue level (Kraft et al. 2016). In consequence, additional mechanisms must cause an average drift of the allelic expression towards one allele. Our present study did not address the exact mechanisms of allelic expression establishment. However, our findings allow to draw some conclusions that may provide deeper insights into such potential mechanisms.
Exemplary analysis of the promotor region in three patients and the 5′- and the 3′-UTR in 6 patients revealed no variant. Variants in these regions can affect mRNA- and protein levels by altering transcription factor- or miRNA-binding, respectively. Based on the absence of variants in the analyzed patients with and without allelic imbalanced expression, we assume no influence on allelic expression regulation in these cases. Nevertheless, in other patients imbalance may be induced by variants in these regions.
The observation of unequal expression of the two MYH7-alleles in non-HCM control individuals suggests that allelic imbalance of MYH7 is an intrinsic feature of this gene, which may be modulated by HCM-associated missense mutations. Interestingly, in most cases the relative fraction of mutant alleles seems to be comparable between patients with identical mutations as we have shown previously for related patients (I736T) and unrelated patients with presumably a common founder mutation (R723G) (Enjuto et al. 2000; Tripathi et al. 2011). Even though this may be explained by identical inherited - but mutation independent - regulatory motifs, we have hypothesized that also a mutation induced alteration of the mRNA turnover may cause this imbalance (Tripathi et al. 2011). In line with this hypothesis, here we found comparable allelic imbalance in three patients unrelated to patients from previous studies: the patient with mutation V606M had 38% mutated mRNA in myocardial tissue, which is similar to the previously determined 28% mutated mRNA in M. soleus (Tripathi et al. 2011). In addition, patients with the mutations R719W and T1377M, respectively, show only a moderate increased expression of the mutant allele or a balanced expression of both alleles in this and previous studies (Helms et al. 2014; Tripathi et al. 2011). This supports the assumption that the mutations may also directly influence the allelic expression (Tripathi et al. 2011). E.g. we have preliminary evidence that the mutation R723G alters mRNA secondary structure (unpublished data), indicating that mutations can influence the mRNA stability. In addition point mutations can act as a cis-regulatory variant for methylation, alter the affinity of RNA-binding proteins that affect recruitment of DNA-methylation complexes or histone modifications (Zhang et al. 2009).
Next to mutation associated regulators, our results indicate that also mutation independent factors influence the allelic expression. One of the three patients with the mutation G741R has a significantly lower fraction of the mutant mRNA compared to the others. Analysis of the promotor and 3′-UTR (Table 4) did not reveal any difference between the patients, indicating that here either intragenic or epigenetic factors may also affect the allelic expression of MYH7. Regulation of allelic expression biases have been assumed to be genetically determined (Pastinen 2010) and also epigenetically regulated (Tycko 2010). Especially the allele specific methylation (ASM) and allele specific transcription factor binding (ASTF) have been determined as potent cis-acting regulators of allelic imbalance (Jeffries et al. 2016; Nakaoka et al. 2016; Tycko 2010). The expression of the MYH7 gene is regulated by DNMT3a dependent methylation of the promotor (Fang et al. 2016). Therefore, allele specific levels of methylation of the MYH7 promotor may also provide one mechanism for the establishment of allelic imbalance irrespective of the HCM mutations.
Taken together, our data indicate that the MYH7-alleles can be expressed imbalanced in healthy individuals, without causing cardiac pathologies. In patients with HCM-mutations, this intrinsic imbalance may then be one factor that determines disease progression. However, most patients with identical mutations show comparable levels of allelic imbalance. Therefore, in addition to the intrinsic allelic imbalance, also mutation-associated regulators may determine the allelic expression.
Transcriptional and post-transcriptional mechanisms induce allelic imbalance of mutant and wildtype β-MyHC at the protein level
In six out of six HCM-patients from a previous study (Tripathi et al. 2011) and in two out of five mutations in this study the allelic imbalance at the protein level is comparable to the mRNA allelic imbalance. This in accordance with the assumption that “Protein Levels at Steady State Are Primarily Determined by mRNA Levels” (Liu et al. 2016). Therefore, we assume that in most cases the MYH7-allelic imbalance is induced prior to translation of the MYH7-gene and thus mRNA imbalance is paralleled by protein level imbalance.
Most interestingly, the missense mutations G741R and G716R show lower fractions of mutated protein as compared to the respective mutated mRNA fractions. The examined mutations are missense mutations that do not cause frame shifts. Therefore, mechanisms such as nonsense mediated decay are highly unlikely (Helms et al. 2014). The disparity of mRNA and protein levels can be linked to different mechanisms as reviewed in (Maier et al. 2009): first, the RNA secondary structure can alter the translation efficacy. As stated above, we have preliminary evidence, that HCM-mutations may alter the secondary structure of the mRNA. Future analyses will address a possible effect of these specific mutations on the secondary structure. Second, regulatory RNAs such as miRNAs may inhibit mRNA translation. However, analysis of the 5′- and the 3′-UTR of the patients revealed no variant, therefore an aberrant miRNA-binding seems rather unlikely. Third, the ribosomal density can regulate the translation efficiency. Determinants of ribosomal density are located in the 5′-UTR (Dvir et al. 2013), which contains no variants in the analyzed patients. Therefore, the ribosomal density will most likely not affect the protein levels. Forth, a reduced protein half-life can lead to reduced levels of protein. The HCM-mutations G716R and G741R could lead to proteins that are degraded faster than the wildtype proteins. This would lead to the observed decreased mutant:wildtype ratio at the protein as compared to the mRNA level. Future experiments will have to show, whether the half-life of the β-MyHC protein is shortened by the mutations G716R and G741R. Last but not least, alternative splicing resulting in non-functional proteins may have caused the reduced mutant protein levels as compared to the mRNA. For myosin binding protein C (MYBPC3) mutations, differential splicing of wildtype and mutant mRNA is an important mechanism that affects protein level imbalance (Helms et al. 2014). For MYH7 mutations splice effects have not been experimentally determined so far, however, bioinformatically predicted (Tripathi et al. 2011). Also for the base substitution c.2146G>A leading to the mutation G716R the generation of an alternative splice site, however with a comparable activity score as the original splice site, is predicted (Desmet et al. 2009). Nevertheless, differential splicing may act as one mechanism underlying protein level allelic imbalance. Most interestingly, the three G741R patients with different fractions of mutant mRNA showed comparable fractions of mutant and wildtype protein for all three patients. Here a post-transcriptional mechanism seems to establish a constant mutant protein ratio, irrespective of the initial fraction of mutant mRNA. Further studies will have to clarify the post-transcriptional mechanisms that affect the expression of mutant and wildtype β-MyHC.
Conclusion
Our study provides evidence that the MYH7 alleles can be expressed balanced and highly imbalanced at the mRNA and at the protein level in both, HCM-patients and non-HCM controls. In non-HCM control individuals, the imbalanced expression will have no physiological impact. However, in HCM-patients one allele encodes for a functionally altered protein of the contractile apparatus. The influence of this alteration may increase with an increasing fraction of mutant protein. Our results suggest that diverse mechanisms must exist for the establishment of allelic imbalance not only between patients with different mutations but also between patients sharing identical HCM-mutations.
Acknowledgements
The funding was provided by the Deutsche Forschungsgemeinschaft (grant KR1187/22-1 to TK).
Footnotes
Deceased: Bernhard Brenner.
References
- Adzhubei I, Jordan DM, Sunyaev SR. Predicting functional effect of human missense mutations using PolyPhen-2. Curr Protoc Hum Genet Unit. 2013;7:20. doi: 10.1002/0471142905.hg0720s76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anan R, Greve G, Thierfelder L, Watkins H, McKenna WJ, Solomon S, Vecchio C, Shono H, Nakao S, Tanaka H, et al. Prognostic implications of novel beta cardiac myosin heavy chain gene mutations that cause familial hypertrophic cardiomyopathy. J Clin Invest. 1994;1:280–285. doi: 10.1172/JCI116957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker E, Navarro-Lopez F, Francino A, Brenner B, Kraft T. Quantification of mutant versus wild-type myosin in human muscle biopsies using nano-LC/ESI-MS. Anal Chem. 2007;24:9531–9538. doi: 10.1021/ac701711h. [DOI] [PubMed] [Google Scholar]
- Bjornsson HT, Albert TJ, Ladd-Acosta CM, Green RD, Rongione MA, Middle CM, Irizarry RA, Broman KW, Feinberg AP. SNP-specific array-based allele-specific expression analysis. Genome Res. 2008;5:771–779. doi: 10.1101/gr.073254.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner B, Seebohm B, Tripathi S, Montag J, Kraft T (2014) Familial hypertrophic cardiomyopathy: functional variance among individual cardiomyocytes as a trigger of FHC-phenotype development. Front Physiol 392:1–15 [DOI] [PMC free article] [PubMed]
- Capriotti E, Calabrese R, Casadio R. Predicting the insurgence of human genetic diseases associated to single point protein mutations with support vector machines and evolutionary information. Bioinformatics. 2006;22:2729–2734. doi: 10.1093/bioinformatics/btl423. [DOI] [PubMed] [Google Scholar]
- Cecconi M, Parodi MI, Formisano F, Spirito P, Autore C, Musumeci MB, Favale S, Forleo C, Rapezzi C, Biagini E, Davi S, Canepa E, Pennese L, Castagnetta M, Degiorgio D, Coviello DA. Targeted next-generation sequencing helps to decipher the genetic and phenotypic heterogeneity of hypertrophic cardiomyopathy. Int J Mol Med. 2016;4:1111–1124. doi: 10.3892/ijmm.2016.2732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Weaver J, Bove BA, Vanderveer LA, Weil SC, Miron A, Daly MB, Godwin AK. Allelic imbalance in BRCA1 and BRCA2 gene expression is associated with an increased breast cancer risk. Hum Mol Genet. 2008;9:1336–1348. doi: 10.1093/hmg/ddn022. [DOI] [PubMed] [Google Scholar]
- Desmet FO, Hamroun D, Lalande M, Collod-Beroud G, Claustres M, Beroud C. Human Splicing Finder: an online bioinformatics tool to predict splicing signals. Nucleic Acids Res. 2009;9:e67. doi: 10.1093/nar/gkp215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Domenico M, Casadonte R, Ricci P, Santini M, Frati G, Rizzo A, Carratelli CR, Lamberti M, Parrotta E, Quaresima B, Faniello CM, Costanzo F, Cuda G. Cardiac and skeletal muscle expression of mutant beta-myosin heavy chains, degree of functional impairment and phenotypic heterogeneity in hypertrophic cardiomyopathy. J Cell Physiol. 2012;10:3471–3476. doi: 10.1002/jcp.24047. [DOI] [PubMed] [Google Scholar]
- Dvir S, Velten L, Sharon E, Zeevi D, Carey LB, Weinberger A, Segal E. Deciphering the rules by which 5′-UTR sequences affect protein expression in yeast. Proc Natl Acad Sci USA. 2013;30:E2792–E801. doi: 10.1073/pnas.1222534110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enjuto M, Francino A, Navarro-Lopez F, Viles D, Pare JC, Ballesta AM. Malignant hypertrophic cardiomyopathy caused by the Arg723Gly mutation in beta-myosin heavy chain gene. J Mol Cell Cardiol. 2000;12:2307–2313. doi: 10.1006/jmcc.2000.1260. [DOI] [PubMed] [Google Scholar]
- Fananapazir L, Dalakas MC, Cyran F, Cohn G, Epstein ND. Missense mutations in the beta-myosin heavy-chain gene cause central core disease in hypertrophic cardiomyopathy. Proc Natl Acad Sci USA. 1993;9:3993–3997. doi: 10.1073/pnas.90.9.3993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang X, Poulsen RR, Wang-Hu J, Shi O, Calvo NS, Simmons CS, Rivkees SA, Wendler CC. Knockdown of DNA methyltransferase 3a alters gene expression and inhibits function of embryonic cardiomyocytes. Faseb J. 2016;9:3238–3255. doi: 10.1096/fj.201600346R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujino N, Konno T, Hayashi K, Hodatsu A, Fujita T, Tsuda T, Nagata Y, Kawashiri MA, Ino H, Yamagishi M. Impact of systolic dysfunction in genotyped hypertrophic cardiomyopathy. Clin Cardiol. 2013;3:160–165. doi: 10.1002/clc.22082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge B, Pokholok DK, Kwan T, Grundberg E, Morcos L, Verlaan DJ, Le J, Koka V, Lam KC, Gagne V, Dias J, Hoberman R, Montpetit A, Joly MM, Harvey EJ, Sinnett D, Beaulieu P, Hamon R, Graziani A, Dewar K, Harmsen E, Majewski J, Goring HH, Naumova AK, Blanchette M, Gunderson KL, Pastinen T. Global patterns of cis variation in human cells revealed by high-density allelic expression analysis. Nat Genet. 2009;11:1216–1222. doi: 10.1038/ng.473. [DOI] [PubMed] [Google Scholar]
- Heap GA, Trynka G, Jansen RC, Bruinenberg M, Swertz MA, Dinesen LC, Hunt KA, Wijmenga C, Vanheel DA, Franke L (2009) Complex nature of SNP genotype effects on gene expression in primary human leucocytes. BMC Med Genom 1 [DOI] [PMC free article] [PubMed]
- Helms AS, Davis FM, Coleman D, Bartolone SN, Glazier AA, Pagani F, Yob JM, Sadayappan S, Pedersen E, Lyons R, Westfall MV, Jones R, Russell MW, Day SM. Sarcomere mutation-specific expression patterns in human hypertrophic cardiomyopathy. Circ Cardiovasc Genet. 2014;4:434–443. doi: 10.1161/CIRCGENETICS.113.000448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeffries AR, Uwanogho DA, Cocks G, Perfect LW, Dempster E, Mill J, Price J (2016) Erasure and reestablishment of random allelic expression imbalance after epigenetic reprogramming. RNA [DOI] [PMC free article] [PubMed]
- Jiang J, Wakimoto H, Seidman JG, Seidman CE. Allele-specific silencing of mutant Myh6 transcripts in mice suppresses hypertrophic cardiomyopathy. Science. 2013;6154:111–114. doi: 10.1126/science.1236921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keshari PK, Harbo HF, Myhr KM, Aarseth JH, Bos SD, Berge T (2016) Allelic imbalance of multiple sclerosis susceptibility genes IKZF3 and IQGAP1 in human peripheral blood. BMC Genet 59 [DOI] [PMC free article] [PubMed]
- Kraft T, Montag J, Radocaj A, Brenner B. Hypertrophic cardiomyopathy: cell-to-cell imbalance in gene expression and contraction force as trigger for disease phenotype development. Circ Res. 2016;9:992–995. doi: 10.1161/CIRCRESAHA.116.309804. [DOI] [PubMed] [Google Scholar]
- Liu Y, Beyer A, Aebersold R. On the dependency of cellular protein levels on mRNA abundance. Cell. 2016;3:535–50. doi: 10.1016/j.cell.2016.03.014. [DOI] [PubMed] [Google Scholar]
- Lopes LR, Rahman MS, Elliott PM. A systematic review and meta-analysis of genotype–phenotype associations in patients with hypertrophic cardiomyopathy caused by sarcomeric protein mutations. Heart. 2013;24:1800–1811. doi: 10.1136/heartjnl-2013-303939. [DOI] [PubMed] [Google Scholar]
- Maier T, Guell M, Serrano L. Correlation of mRNA and protein in complex biological samples. FEBS Lett. 2009;24:3966–3973. doi: 10.1016/j.febslet.2009.10.036. [DOI] [PubMed] [Google Scholar]
- Maron BJ. Hypertrophic cardiomyopathy: a systematic review. JAMA. 2002;10:1308–1320. doi: 10.1001/jama.287.10.1308. [DOI] [PubMed] [Google Scholar]
- Maron BJ, Maron MS, Semsarian C. Genetics of hypertrophic cardiomyopathy after 20 years: clinical perspectives. J Am Coll Cardiol. 2012;8:705–715. doi: 10.1016/j.jacc.2012.02.068. [DOI] [PubMed] [Google Scholar]
- Marsiglia JD, Pereira AC. Hypertrophic cardiomyopathy: how do mutations lead to disease? Arq Bras Cardiol. 2014;3:295–304. doi: 10.5935/abc.20140022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDaniell R, Lee BK, Song L, Liu Z, Boyle AP, Erdos MR, Scott LJ, Morken MA, Kucera KS, Battenhouse A, Keefe D, Collins FS, Willard HF, Lieb JD, Furey TS, Crawford GE, Iyer VR, Birney E. Heritable individual-specific and allele-specific chromatin signatures in humans. Science. 2010;5975:235–239. doi: 10.1126/science.1184655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mi H, Huang X, Muruganujan A, Tang H, Mills C, Kang D, Thomas PD. PANTHER version 11: expanded annotation data from Gene Ontology and Reactome pathways, and data analysis tool enhancements. Nucleic Acids Res. 2016;D1:D183–D189. doi: 10.1093/nar/gkw1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milani L, Lundmark A, Nordlund J, Kiialainen A, Flaegstad T, Jonmundsson G, Kanerva J, Schmiegelow K, Gunderson KL, Lonnerholm G, Syvanen AC. Allele-specific gene expression patterns in primary leukemic cells reveal regulation of gene expression by CpG site methylation. Genome Res. 2009;1:1–11. doi: 10.1101/gr.083931.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakaoka H, Gurumurthy A, Hayano T, Ahmadloo S, Omer WH, Yoshihara K, Yamamoto A, Kurose K, Enomoto T, Akira S, Hosomichi K, Inoue I. Allelic imbalance in regulation of ANRIL through chromatin interaction at 9p21 endometriosis risk locus. PLoS Genet. 2016;4:e1005893. doi: 10.1371/journal.pgen.1005893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nier V, Schultz I, Brenner B, Forssmann W, Raida M. Variability in the ratio of mutant to wildtype myosin heavy chain present in the soleus muscle of patients with familial hypertrophic cardiomyopathy. A new approach for the quantification of mutant to wildtype protein. FEBS Lett. 1999;3:246–252. doi: 10.1016/S0014-5793(99)01433-7. [DOI] [PubMed] [Google Scholar]
- Pastinen T. Genome-wide allele-specific analysis: insights into regulatory variation. Nat Rev Genet. 2010;8:533–538. doi: 10.1038/nrg2815. [DOI] [PubMed] [Google Scholar]
- Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, Grody WW, Hegde M, Lyon E, Spector E, Voelkerding K, Rehm HL. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;5:405–424. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz JM, Cooper DN, Schuelke M, Seelow D. MutationTaster2: mutation prediction for the deep-sequencing age. Nat Methods. 2014;4:361–362. doi: 10.1038/nmeth.2890. [DOI] [PubMed] [Google Scholar]
- Serre D, Gurd S, Ge B, Sladek R, Sinnett D, Harmsen E, Bibikova M, Chudin E, Barker DL, Dickinson T, Fan JB, Hudson TJ. Differential allelic expression in the human genome: a robust approach to identify genetic and epigenetic cis-acting mechanisms regulating gene expression. PLoS Genet. 2008;2:e1000006. doi: 10.1371/journal.pgen.1000006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JR, Marcelino LA, Polz MF. Heteroduplexes in mixed-template amplifications: formation, consequence and elimination by ‘reconditioning PCR’. Nucleic Acids Res. 2002;9:2083–2088. doi: 10.1093/nar/30.9.2083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tripathi S, Schultz I, Becker E, Montag J, Borchert B, Francino A, Navarro-Lopez F, Perrot A, Ozcelik C, Osterziel KJ, McKenna WJ, Brenner B, Kraft T. Unequal allelic expression of wild-type and mutated beta-myosin in familial hypertrophic cardiomyopathy. Basic Res Cardiol. 2011;6:1041–1055. doi: 10.1007/s00395-011-0205-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tycko B. Allele-specific DNA methylation: beyond imprinting. Hum Mol Genet. 2010;R2:R210–R20. doi: 10.1093/hmg/ddq376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh R, Rutland C, Thomas R, Loughna S. Cardiomyopathy: a systematic review of disease-causing mutations in myosin heavy chain 7 and their phenotypic manifestations. Cardiology. 2009;1:49–60. doi: 10.1159/000252808. [DOI] [PubMed] [Google Scholar]
- Wang Q, An Y, Yuan Q, Qi Y, Ou Y, Chen J, Huang J. Identification of allelic expression imbalance genes in human hepatocellular carcinoma through massively parallel DNA and RNA sequencing. Med Oncol. 2016;4:38. doi: 10.1007/s12032-016-0751-y. [DOI] [PubMed] [Google Scholar]
- Watkins H, Rosenzweig A, Hwang DS, Levi T, McKenna W, Seidman CE, Seidman JG. Characteristics and prognostic implications of myosin missense mutations in familial hypertrophic cardiomyopathy. N Engl J Med. 1992;17:1108–1114. doi: 10.1056/NEJM199204233261703. [DOI] [PubMed] [Google Scholar]
- Witjas-Paalberends ER, Ferrara C, Scellini B, Piroddi N, Montag J, Tesi C, Stienen GJ, Michels M, Ho CY, Kraft T, Poggesi C, van der Velden J. Faster cross-bridge detachment and increased tension cost in human hypertrophic cardiomyopathy with the R403Q MYH7 mutation. J Physiol. 2013;15:3257–3272. doi: 10.1113/jphysiol.2014.274571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Medical Association Declaration of Helsinki Recommendations guiding physicians in biomedical research involving human subjects. Cardiovasc Res. 1997;1:2–3. [PubMed] [Google Scholar]
- Zhang K, Li JB, Gao Y, Egli D, Xie B, Deng J, Li Z, Lee JH, Aach J, Leproust EM, Eggan K, Church GM. Digital RNA allelotyping reveals tissue-specific and allele-specific gene expression in human. Nat Methods. 2009;8:613–618. doi: 10.1038/nmeth.1357. [DOI] [PMC free article] [PubMed] [Google Scholar]