Abstract
Late blight has been the most devastating potato disease worldwide. The causal agent, Phytophthora infestans, is notorious for its capability to rapidly overcome host resistance. Changes in the expression pattern and the encoded protein sequences of effector genes in the pathogen are responsible for the loss of host resistance. Among numerous effector genes, the class of RXLR effector genes is well-known in mediating host genotype-specific resistance. We therefore performed deep sequencing of five genetically diverse P. infestans strains using in planta materials infected with zoospores (12 h post inoculation) and focused on the identification of RXLR effector genes that are conserved in coding sequences, are highly expressed in early stages of plant infection, and have defense suppression activities. In all, 245 RXLR effector genes were expressed in five transcriptomes, with 108 being co-expressed in all five strains, 47 of them comparatively highly expressed. Taking sequence polymorphism into consideration, 18 candidate core RXLR effectors that were conserved in sequence and with higher in planta expression levels were selected for further study. Agrobacterium tumefaciens-mediated transient expression of the selected effector genes in Nicotiana benthamiana and potato demonstrated their potential virulence function, as shown by suppression of PAMP-triggered immunity (PTI) or/and effector-triggered immunity (ETI). The identified collection of core RXLR effectors will be useful in the search for potential durable late blight resistance genes. Analysis of 10 known Avr RXLR genes revealed that the resistance genes R2, Rpi-blb2, Rpi-vnt1, Rpi-Smira1, and Rpi-Smira2 may be effective in potato cultivars. Analysis of 8 SFI (Suppressor of early Flg22-induced Immune response) RXLR effector genes showed that SFI2, SFI3, and SFI4 were highly expressed in all examined strains, suggesting their potentially important function in early stages of pathogen infection.
Keywords: potato late blight, Phytophthora infestans, RXLR effectors, sequence polymorphism, durable resistance, resistance breeding
Introduction
Potato (Solanum tuberosum L.) is the world’s most important non-grain food crop and is central to global food security (Potato Genome Sequencing Consortium, 2011). However, its safe production is seriously threatened by late blight, a disease caused by Phytophthora infestans, the most destructive pathogen of potato (Yoshida et al., 2013). Historically, as the Irish potato famine agent, P. infestans has had a tremendous effect on human history, resulting in famine and population displacement (Haas et al., 2009). Nowadays, annual worldwide potato crop losses due to late blight are still huge, conservatively estimated at $6.7 billion (Nowicki et al., 2011). Although significant efforts have been made to control late blight disease, this pathogen is still a tremendous challenge for sustainable production of potato (Fry et al., 2015).
It is widely accepted that planting resistant cultivars is one of the most effective, economical, and environmentally friendly strategies to control P. infestans (Rodewald and Trognitz, 2013). Field-deployed resistant cultivars contain resistance (R) genes that can recognize P. infestans avirulence (Avr) RXLR effector genes and activate host defense responses to constrain disease development. However, some resistant cultivars have been defeated in a single season because the targets of potato R genes, Avr RXLR effector genes, evolve rapidly through present and absent variation (PAV), insertion and deletion (Indel), point mutations (SNPs), and gene silencing to avoid interaction with R genes (Raffaele et al., 2010; Vleeshouwers and Oliver, 2014). Due to the ability of P. infestans to rapidly overcome R genes, this pathogen is considered an “R gene destroyer” by phytopathologists and breeders (Haas et al., 2009; Fry et al., 2015). In order to effectively control late blight, plant breeders need to adopt new strategies and techniques in potato R gene identification, introgression, functional characterization, and field deployment (Vleeshouwers and Oliver, 2014).
Research over the last 15 years has led to an increasingly clear understanding of RXLR effector genes. Since Shan et al. (2004) cloned the first P. sojae Avr gene Avr1b, three other oomycete Avr genes have been cloned (Allen et al., 2004; Armstrong et al., 2005; Rehmany et al., 2005). Sequence alignment of the four AVR proteins found conservation motifs (RXLR and dEER) at their N terminals (Rehmany et al., 2005). These two motifs provide a powerful bioinformatic tool to identify the effector reservoir in oomycete pathogens, and has led to the prediction of up to 563 RXLR effector genes in the P. infestans genome (Haas et al., 2009) and 385 in the P. sojae genome (Jiang et al., 2008). Function analysis of those RXLR effectors indicated that they were secreted from the pathogen onto host cell surfaces or into host cells to defeat plant defenses by disturbing the host’s innate immune system (Wang et al., 2011). The precise virulence roles of RXLR effector genes such as PiAvrblb1 (Champouret et al., 2009), PiAvr3a (Bos et al., 2010), PiAvrblb2 (Bozkurt et al., 2011), and PiAvr2 (Saunders et al., 2012) have been dissected; the PTI supression function of eight SFI (Suppressor of early Flg22-induced Immune response) RXLR effectors, SFI1, SFI2, SFI3, SFI4, SFI5, SFI6, SFI7, and SFI8, have been preliminary analyzed (Zheng et al., 2014); they play critical role in host–pathogen interaction, especially during the infection and colonization stages (Cooke et al., 2012). However, apart from virulence roles, RXLR effectors also play avirulence roles when potato cultivars contain cognate R genes (Birch et al., 2008). The underlying reason for this is that co-evolution pushes the surviving hosts to evolve R genes to recognize cognate effectors and trigger the localized programed cell death called hypersensitive response (HR) (Oliva et al., 2015). To date, the ten known P. infestans Avr genes, Avr1, Avr2, Avr3a, Avr3b, Avr4, Avrblb1, Avrblb2, Avrvnt1, AvrSmira1, and AvrSmira2, are all RXLR effector genes (Armstrong et al., 2005; van Poppel et al., 2008; Champouret et al., 2009; Oh et al., 2009; Gilroy et al., 2011; Vleeshouwers et al., 2011). With our increasing understanding to the function of RXLR genes, those effectors are emerging as tools in modern potato resistance breeding to accelerate and improve the identification, functional characterization, and deployment of R genes (Vleeshouwers and Oliver, 2014).
Recently, Dangl et al. (2013) reviewed the principle of “core effectors” and proposed an improved practice to breed durable resistance using genomic strategies that start with identification of core effectors. In fact, identification of core effectors had already been conducted by Bart et al. (2012). They used a genome sequencing strategy to search for conserved effector genes in the bacterial pathogen Xanthomonas axonopodis pv. manihotis and found a set of conserved effectors (core effectors), which now serve as targets to define the R genes they activate in wild species of Manihot, and may potentially do the same in other related plants in the Euphorbiaceae. Although core effectors were considered ideal targets for deploy durable resistance, P. infestans core RXLR effectors are largely unknown currently, which limits our ability to deploy durable potato R genes. Thus, in the search for durable potato R genes and long-lasting control of late blight, it is urgent to identify the core RXLR effector set in P. infestans.
Considering that (1) effector genes with important functions in pathogen virulence are sequence-conserved and widely distributed in populations (Bart et al., 2012; Dangl et al., 2013) and, (2) all known P. infestans Avr effector genes are in planta induced RXLR effector genes (Cooke et al., 2012), in this study we utilized a next-generation transcriptome deep sequencing strategy to identify potentially conserved core RXLR effector genes in P. infestans, primarily based on their (1) conserved sequences among diverse strains, (2) high levels of expression at the early infection stage, and (3) potentially essential functions for pathogenesis. This resulted in the identification of 18 conserved candidate Core RXLR Effectors (CRE). Transient expression of selected CREs in Nicotiana benthamiana suggested that these effectors contribute to the virulence of P. infestans by enhancing pathogen colonization and expansion. Further analysis revealed that these CREs defeated the host defense response by suppressing plant PTI and ETI in both N. benthamiana and potato. The identified collection of CREs is a valuable resource and they can be used as tools to search for potential durable late blight R genes from potato germplasms and related Solanum species in modern breeding programs.
Materials and Methods
Phytophthora Infection Assays
Solanum tuberosum differential host ‘MaR3,’ a susceptible potato line, was used to prepare the infection materials. Potato seedlings were cultured in sterile Murashige and Skoog medium for a month, then transferred to vermiculite for another month, before being planted in pots containing a mix of peat moss and vermiculite (V/V = 2:1). After 5 weeks, the fully expanded leaves were used for inoculation. P. infestans strains were cultured and maintained on RSA (rye sucrose agar) medium plates. They were grown at 16°C in darkness for 2 weeks on RSA plates, then sporangia were harvested in cold sterile distilled water. The sporangial suspensions were adjusted to 4 × 104 spores mL-1 with distilled water and chilled at 4°C for 2 h to release motile zoospores (Tian et al., 2015a). The detached potato leaves were inoculated with the zoospore suspension, placed in a plastic tray, and maintained at 16°C and 100% relative humidity in the darkness to ensure infection. The infected leaves were observed under a microscope, pooled at 12 h post inoculation (hpi), frozen with liquid nitrogen, and stored at -80°C.
Trypan Blue Staining
Leaf inoculation sites were cut off and transferred into polypropylene tubes which were filled with diluted trypan blue solution (10 g phenol, 10 mL glycerol, 10 mL lactic acid, 10 mL water and 10 mg of trypan blue). The tubes (lid slightly unscrewed) were treated in a heated water bath and boiled for 2 min. After cooling to room temperature, tubes were boiled for another 2 min. Then, the samples were destained by replacing the staining solution with chloral hydrate solution (5 g chloral hydrate dissolving into 2 mL water) for 24 h. The samples were finally mounted in distilled water and viewed under an Olympus BX51 (Shinjuku-ku, Tokyo, Japan) microscope with differential interference contrast optics (Meng et al., 2015).
RNA Isolation and Sequencing
Total RNA was extracted from the infection tissues using TRIzol reagent (Invitrogen, Carlsbad, CA, United States) following the manufacturer’s protocol, then treated with DNase I (RNase free, TaKaRa, Japan) to remove genomic DNA contaminations. The quality, purity and concentration of each RNA sample were checked by using the 1% agarose gels, NanoDrop 2000c spectrophotometer (Thermo Scientific, Waltham, MA, United States) and Agilent 2100 bioanalyzer (Agilent Technologies, Santa Clara, CA, United States). Library preparation was performed from 1 μg of total RNA. Sequencing were conducted on Illumina HiSeq 2000 platform to produce100 bp paired-end reads (LC Company, Hangzhou, China). The sequencing results were deposited in the Sequence Read Archive (SRA) at the NCBI database (accession number: PRJNA415282).
RNA-Seq Data Analysis
The adapter sequence and low quality bases were trimmed from both ends of reads with the software HTQC (Yang et al., 2013). The command ‘ht-stat’ was used to assess the quality of reads. The command ‘ht-trim -S both -C 13 -W 5’ was used to trim both sides of reads. Then, the command ‘ht-filter –filter length -L 35’ was used to filter out short reads. The P. infestans reference genome of strain T30-4 was downloaded from the PID (Phytophthora infestans Database)1. TopHat2 was used to map the clean reads to the reference genome with the custom parameter setting ‘-r 0 –mate-std-dev 80’ (Trapnell et al., 2012). Cufflink was used to calculate the Fragments Per Kilobase of exon model per Million mapped reads (FPKM) of each sample. SAMtools was used to extract SNPs from the accepted.bam file, with parameters set as ‘DP = 4, QUAL = 20’, with other parameters set as default. MEGA6.0 was used to perform the sequences alignment analysis and construct the phylogenetic tree (Tamura et al., 2013).
Prediction of Novel RXLR Effector Genes
Pair-end reads were mapped to the genome of S. tuberosum using TopHat2 (Kim et al., 2013). Then, reads that could not be mapped to the potato genome were extracted and we performed de novo assembly with Trinity (Grabherr et al., 2011). The longest potential coding regions were identified by TransDecoder (Haas et al., 2013), then the sequence was trimmed before the first methionine. Sequences more than 70 bp long were analyzed with a python script to predict the RXLR proteins following the reported rules (Whisson et al., 2007).
qRT-PCR
For RNA-seq data validation, SYBR green qRT-PCR assays were performed. Primer pairs (Supplementary Table 1) were designed to anneal specifically to each of the selected genes. The housekeeping gene of P. infestans, PiUBC (PITG_08327), was used as endogenous control. Correlation analysis of RXLR genes were conducted by associating the ΔCt values calculated from the qRT-PCR assays with the log2-transformed expression levels from the RNA-seq data.
Transient Agro-Infiltration Assays
RXLR effector genes and GFP gene were amplified with primers containing ClaI and SalI restriction sites and ligated into PVX vector pGR106 (with 35S promoter) using standard molecular biology techniques. For in planta transient gene expression assays, A. tumefaciens strain GV3101, with the appropriate constructs, were grown in LB media to the late-log phase (supplemented with 50 μgcdotmL-1 of kanamycin and 20 μgcdotmL-1 of rifampicin). The cells were collected by centrifugation at room temperature (25°C, 3500 g, 5 min), resuspended in an infiltration medium (200 μM acetosyringone, 10 mM MES, pH 5.6, and 10 mM MgCl2), and then incubated for 1–3 h at room temperature before infiltration. A. tumefaciens suspensions were infiltrated at an OD600 value of 0.3–0.6. Agroinfiltration experiments were performed on leaves of 6- to 8-week-old N. benthamiana plants and 5-week-old S. tuberosum plants. After 24 h, infiltrated N. benthamiana leaves were detached and transferred into moist sealed plastic trays and inoculated with 10 μl P. infestans zoospore suspension (100 zoospores/μl) at the infiltration sites (Tian et al., 2016). P. infestans strain Pa21106 was used in the infection assay. Lesion diameters of the inoculated leaves were recorded and photographs were taken 5 days after infiltration (Gu et al., 2011).
To assay suppression of HR, A. tumefaciens cells carrying selected RXLR effector genes were infiltrated. About 24 h later, the same infiltration site was infiltrated with A. tumefaciens cells carrying the Bax gene. A. tumefaciens cells carrying Bax, or RXLR effector or GFP genes alone, were infiltrated in parallel as controls. Plants were grown and maintained in a cultivation room with an ambient temperature of 22–25°C and high light intensity throughout the experiments. Symptom development was observed from day 3 to day 8. Statistics were performed and photographs taken at day 5. Similar procedures were used for other cell death suppression assays, where, A. tumefaciens cells carrying Bax, NIP, Avh238, Avh241, or INF1 were infiltrated 24 h after the RXLR effector constructs were infiltrated (Wang et al., 2011). Each assay consisted of three biological replications (Gu et al., 2011).
Results
Five Diverse Strains Were Selected for Analysis of RXLR Effector Genes
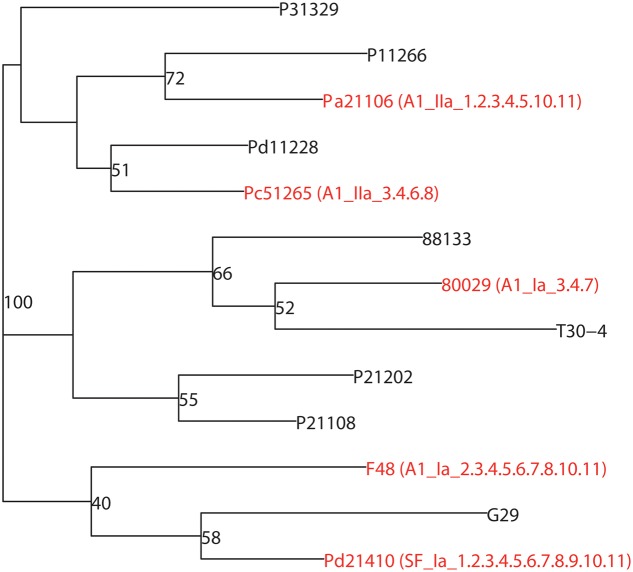
In our previous studies, more than 2,000 P. infestans isolates were collected and a population analysis was conducted (Tian et al., 2015a,b, 2016). Based on SSR genotype data, a phylogenetic tree containing thirteen representative strains was constructed (Supplementary Table 2). As shown in Figure 1, five diverse strains Pa21106, Pc51265 and Pd21410 (collected from a north-western China population), F48 (collected from a southern China population) and 80029 (collected from European population and used as reference strain) with different mating types, haplotypes, and pathotypes, were selected and used in the following work.
FIGURE 1.
Five diverse P. infestans strains with highly divergent genetic backgrounds were chosen for RNA-seq. Thirteen representative strains were used to build a phylogenetic tree according to the Bruvo distance using the R package “poppor” (Bruvo et al., 2004). The strains 80029, F48, Pa21106, Pc51265, and Pd21410 marked as red branches in the figure, were selected to conduct RNA-seq. Mating type: A1 and SF (self-fertilization); haplotype: Ia and IIa; pathotype: virulence race indicated by Black’s potato differentials.
Leaves 12 hpi Were Chosen to Prepare Early in Planta Materials
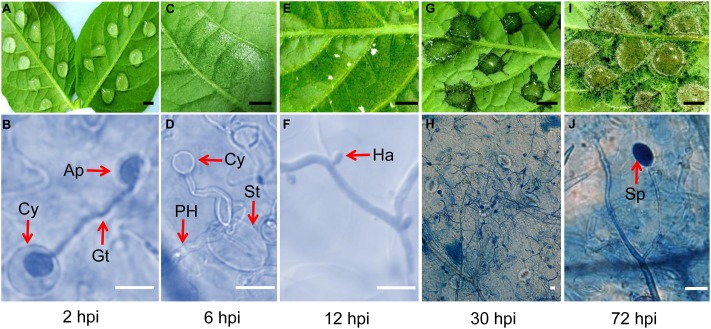
Inoculated potato leaf materials were continuously harvested at different time points. Macroscopic and microscopic observations were used to monitor the disease development process. At 2 hpi, the resting cysts of P. infestans germinated and extended geminating tubes. The appressoria were formed at the end of the tubes. No obvious leaf symptoms were observed (Figures 2A,B). At 6 hpi, the appressoria were observed to extend primary hypha and pass through the host stomata into the host tissues. Light water-soaked appearances were seen at the inoculation sites (Figures 2C,D). At 12 hpi, as in a previous report (Sivak and Shaw, 1969), haustoria were observed, which means that colonization had begun. Small dark spots were observed on leaves (Figures 2E,F). At 30 hpi, secondary mycelia were formed and widely expanded. The well-formed water-soaked lesions were observed at the infection sites (Figures 2G,H). At 72 hpi, aerial mycelia were observed surrounding the infection sites and young sporangia appeared in sporangiophores (Figures 2I,J). Based on these results, the time point of 12 hpi was chosen to prepare the early in planta materials to perform P. infestans deep sequencing.
FIGURE 2.
The ideal sampling time point of potato infection by P. infestans was confirmed by observation. (A,B) At 2 h post inoculation (hpi), the Cy (cyst) germinated and extended out Gt (germ tube) to form Ap (appressorium) and was ready to invade into host cells. (C,D) At 6 hpi, some pathogen PH (primary hyphae) invaded into host through the host St (stoma), and developed inside the host. (E,F) At 12 hpi, young hyphae broadly spread and haustoria formed, but only very small black spots could be macroscopically observed on the leaf. (G,H) At 30 hpi, the infection site turned to dark and obvious damage could be observed. (I,J) At 72 hpi, air hyphae appeared and young Sp (sporangium) was ready to form. (Black bar, 0.5 cm; white bar, 10 μm).
Detection of 245 RXLR Effector Genes Expressed at the Early Infection Stage
To understand genetic differences in RXLR effector genes, the five genetically diverse strains of P. infestans were used to prepare in planta infections. The inoculated leaf tissues were sampled at 12 hpi and total RNA was isolated for RNA sequencing. In total, 379 million raw pair-end reads were produced. After filtering, mapping, and gene assembling, 11,911 unique expressed genes were detected, including 243 RXLR effector genes (Supplementary Table 3). Recently, Cooke et al. (2012) and Martin et al. (2013) reported eleven additional RXLR genes. Therefore, sequencing reads were parallel-mapped to these eleven genes and one of them, PiRXLRe, was found to be expressed in Pa21106, as two unique reads of Pa21106 were successfully mapped to the gene. To confirm whether our strains had unique RXLR effector genes, RNA-seq reads were also used to carry out de novo assembly and RXLR effector gene prediction. One novel RXLR effector gene, comp858_c1_seq1 (GenBank accession number: MG269997), was identified in Pa21106 (Supplementary Table 4). In total, in this study, expression of 245 RXLR effector genes was detected in early infection stage in planta.
Expression and Sequence Analysis Uncovered 18 Candidate Core RLXR Effectors
RNA-seq data were partially validated by qRT-PCR assays. The transcript abundances determined by qRT-PCR were highly consistent with RNA-seq results, indicating that the RNA-seq data were reliable (R2 = 0.8783, p-value < 0.01) (Supplementary Figure 1). Among the detected 245 RXLR effector genes, 108 were expressed in all five strains, 34 in four strains, 29 in three strains, 30 in two strains, and 44 in one strain (Supplementary Figure 2). Meanwhile, SNPs extracted from our RNA-seq reads and from P. infestans strain 06_3928A resequencing reads (Cooke et al., 2012) were used to analyze RXLR effector gene sequence polymorphisms. In total, 468 SNPs were detected in 128 RXLR effector genes, resulting in the discovery of 115 potentially conserved RXLR effector genes, including 92 with no SNPs and 23 with synonymous substitutions (Supplementary Table 3). Furthermore, 23 RXLR effector genes that were highly expressed and potentially conserved were extracted; they were both conserved in protein coding sequences and expressed by all five strains at the early infection stage. Although 23 RXLR effector genes have been assigned different gene IDs by Haas et al. (2009), subsequent phylogenetic analysis of these genes showed that the genes PITG_14954, PITG_14959, PITG_14961, and PITG_14962 (CRE11), PITG_16233, and PITG_16240 (CRE13), PITG_23014, and PITG_23015 (CRE16) have consensus sequences. Since effector genes with same sequences can be considered one core RXLR effector, only 18 core RXLR effectors were uncovered (Table 2 and Supplementary Figure 3).
Table 2.
Identification of 18 conserved candidate core RXLR effector genes.
| Namea | Gene IDb | Familyc | Intergenici | Five strainsd |
06_3928Ae |
||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Snp | Nf | Sg | dN/dSh | Snp | N | S | dN/dS | ||||
| CRE1h | PITG_00821 | RxLRfam108 | InBtw | 0 | 0 | 0 | NA | 0 | 0 | 0 | NA |
| CRE2 | PITG_04196 | RxLRfam47 | GSR | 0 | 0 | 0 | NA | 0 | 0 | 0 | NA |
| CRE3 | PITG_05750 | RxLRfam29 | InBtw | 0 | 0 | 0 | NA | 0 | 0 | 0 | NA |
| CRE4 | PITG_05910 | RxLRfam52 | InBtw | 0 | 0 | 0 | NA | 0 | 0 | 0 | NA |
| CRE5 | PITG_06308 | RxLRfam23 | InBtw | 0 | 0 | 0 | NA | 0 | 0 | 0 | N/A |
| CRE6 | PITG_07451 | RxLRfam116 | InBtw | 1 | 0 | 1 | –1 | 0 | 0 | 0 | NA |
| CRE7 | PITG_08133 | RxLRsng158 | InBtw | 0 | 0 | 0 | NA | 0 | 0 | 0 | NA |
| CRE8 | PITG_09160 | RxLRfam42 | GSR | 0 | 0 | 0 | NA | 0 | 0 | 0 | N/A |
| CRE9 | PITG_09224 | RxLRfam55 | InBtw | 1 | 0 | 1 | –3.9521 | 0 | 0 | 0 | N/A |
| CRE10 | PITG_13452 | RxLRfam108 | InBtw | 0 | 0 | 0 | NA | 0 | 0 | 0 | NA |
| CRE11 | PITG_14954 | RxLRfam21 | GSR | 0 | 0 | 0 | NA | 0 | 0 | 0 | N/A |
| PITG_14959 | RxLRfam21 | GSR | 0 | 0 | 0 | NA | 0 | 0 | 0 | N/A | |
| PITG_14961 | RxLRfam21 | GSR | 0 | 0 | 0 | NA | 0 | 0 | 0 | N/A | |
| PITG_14962 | RxLRfam21 | GSR | 0 | 0 | 0 | NA | 0 | 0 | 0 | N/A | |
| CRE12 | PITG_14960 | RxLRfam21 | GSR | 0 | 0 | 0 | NA | 0 | 0 | 0 | NA |
| CRE13 | PITG_16233 | RxLRfam9 | InBtw | 0 | 0 | 0 | NA | 0 | 0 | 0 | NA |
| PITG_16240 | RxLRfam9 | InBtw | 0 | 0 | 0 | NA | 0 | 0 | 0 | N/A | |
| CRE14 | PITG_17063 | RxLRfam45 | Not | 0 | 0 | 0 | NA | 0 | 0 | 0 | NA |
| CRE15 | PITG_17316 | RxLRfam1 | InBtw | 1 | 0 | 1 | –2.6297 | 1 | 0 | 1 | 0 |
| CRE16 | PITG_23014 | RxLRfam100 | GSR | 0 | 0 | 0 | NA | 0 | 0 | 0 | N/A |
| PITG_23015 | RxLRfam100 | GSR | 0 | 0 | 0 | NA | 0 | 0 | 0 | NA | |
| CRE17 | PITG_23042 | RxLRfam25 | GDR | 0 | 0 | 0 | NA | 0 | 0 | 0 | N/A |
| CRE18 | PITG_23226 | RxLRfam100 | Not | 0 | 0 | 0 | NA | 0 | 0 | 0 | NA |
a,hDesignated names of core RXLR effector (CRE) genes.bGene ID of candidate conserved RXLR effectors in reference genome T30-4 (Haas et al., 2009).cRXLR effector gene families.dThe five diverse P. infestans strains used in our study (Pa21106, Pc51265, Pd21410, F48 and 80029).eP. infestans strains used in the study of Cooke et al. (2012).fNon-synonymous substitution.gSynonymous substitution.hThe ratio of non-synonymous substitution rate to synonymous substitution rate.iGSR (gene spare region), GDR (gene density region), and InBTw (gene in the region between GSR and GDR) are three types of intergenic regions.
Core RLXR Effectors Contribute to Virulence through Defense Suppression
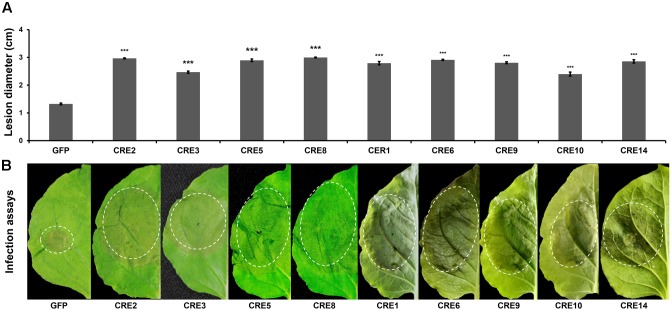
To examine whether the core RXLR effectors promote P. infestans virulence, the mature protein-coding regions of PITG_04196, PITG_05750, PITG_06308, PITG_09160, PITG_00821, PITG_07451, PITG_09224, PITG_13452, and PITG_17063 were cloned and transiently expressed in N. benthamiana using Agrobacterium-mediated expression followed by a P. infestans challenge. At 5 days post inoculation (dpi), significantly larger lesions were observed in areas expressing RXLR effectors compared to that of free GFP (Figure 3), suggesting that the core RLXR effectors confer a benefit to the pathogen. To examine how the core RXLR effectors promote virulence, constructs of PITG_04196, PITG_05750, PITG_06308 and PITG_09160 were randomly selected to test cell death suppressive activities triggered by PAMPs and effectors, using their Agrobacterium-mediated transient expression in leaves of N. benthamiana. None of the four genes induced cell death in N. benthamiana. In fact, comparing to negative control PITG_22804 and free GFP, they all suppressed the cell death caused by a range of cell death inducers, including BAX, INF1, NIP, Avh238 and Avh241 (Wang et al., 2011). These results indicated that these conserved core RXLR effectors contribute to virulence during the early infection stage (12 hpi), by inhibiting plant defense responses induced by both PTI and ETI (Figures 4A–F and Supplementary Figure 3). The functional characterization of these four RXLR effector genes was also conducted on leaves of potato cultivar ‘MaR3,’ S. tuberosum. Comparing to negative control Avr3aEM and free GFP, none of them induced cell death, but could suppress HR caused by transient expression of Avr3aKI, suggesting that these conserved core RXLR effectors that are expressed at the early infection stage are important in the suppression of ETI-induced host plant defense (Figures 4G–N).
FIGURE 3.
Transient overexpression of selected RXLR effector genes enhances P. infestans colonization. (A) Average lesion diameters of the inoculated leaves. Error bars represent standard errors calculated from at least 30 independent biological replicates. Asterisks indicate significant differences determined using Dunnett’s test (P < 0.005), (B) N. benthamiana leaf phenotypes upon P. infestans infection. Detached leaves of GFP- and RXLR effector-transgenic plants were inoculated with P. infestans zoospores. These representative photographs were taken at 120 hpi.
FIGURE 4.
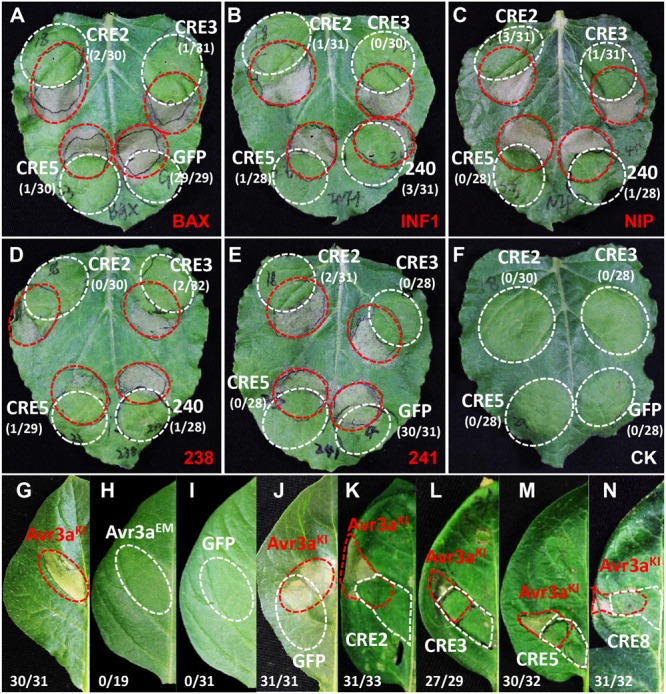
Plant defense suppression activities of selected P. infestans RXLR effector genes. Candidate RXLR effector genes that were highly expressed at the early stage of plant infection were selected and A. tumefaciens-mediated transient expression assays were performed on N. benthamiana (A–F) and host potato (G–N). In N. benthamiana, all examined RXLR effector genes suppressed HR mediated by a range of elicitors including: (A) BAX; (B) the P. infestans PAMP elicitor INF1; (C) NIP; the P. sojae RXLR effectors Avh238 (D,E) Avh241. The red circle indicates the area infiltrated with A. tumefaciens cells carrying elicitor constructs. The white circle indicates A. tumefaciens cells carrying the RXLR effector gene CRE2 (PITG_04196), CRE3 (PITG_05750), and CRE5 (PITG_06308), the negative control GFP (A,E), and the positive control Avh240 (B–D). In S. tuberosum, the P. infestans RXLR effector Avr3aKI induces HR in the differential host line ‘MaR3,’ which was suppressed by transient expression of RXLR effector genes (K) CRE2, (L) CRE3, (M) CRE5, and (N) CRE8 (PITG_09160); (G) the positive control Avr3aKI, and (H–J) the negative control Avr3aEM, GFP, and Avr3aKI plus GFP. The numbers show the ratio of infiltrated sites that developed cell death versus the total number of infiltrated sites.
Analysis of 10 Known Avr and 8 SFI RXLR Effector Genes
Previously, 10 known Avr RXLR effector genes had been reported (Vleeshouwers and Oliver, 2014). In this study, their expression at early stage of potato infection was detected. Avr1 was only found to be expressed in strain 80029. Avr2, Avr3a, Avrblb1, Avrvnt1, and AvrSmira1 were all expressed in all five tested P. infestans strains, while Avr3b, Avr4, Avrblb2 and AvrSmira2 were each expressed in two to four strains. Sequence polymorphism analyses showed that the Avr3aEM allele and the truncated Avr4 were expressed by the tested strains (Table 3 and Supplementary Figure 5).
Table 3.
Amino acid substitutions in the known RXLR effectors (AVR).
| Amino acid position | Strains |
|||||
|---|---|---|---|---|---|---|
| T30-4 | 80029 | F48 | Pa21106 | Pc51265 | Pd21410 | |
| AVR2 (PITG_22870) | ||||||
| 31 | N | K | K | K | K | N |
| AVR3a (PITG_14371) | ||||||
| 80 | E | E | E | E | E | E |
| 103 | M | M | M | M | M | M |
| 124 | R | G | R | R | R | G |
| AVR4 (PITG_07387) | ||||||
| 19 | T | -a | ∗b | ∗ | - | - |
| AVRblb1 (PITG_21388) | ||||||
| 122 | R | I | R | R | R | R |
| AVRblb2 (PITG_20300) | ||||||
| 79 | P | - | - | P | A | - |
| AVRvnt1 (PITG_16294) | ||||||
| 96 | A | A | V | A | - | A |
| AVRSmira1 (PITG_07550) | ||||||
| 131 | K | K | K | R | R | R |
| 170 | R | R | R | Q | Q | R |
| 199 | K | T | K | T | K | T |
| 209 | H | H | H | R | H | H |
| AVRSmira2 (PITG_07555) | ||||||
| 64 | M | V | – | M | M | M |
aRegions not covered by sequencing.bNucleotide sequence encodes a stop codon in this amino acid position.
Recently, Zheng et al. (2014) reported 8 SFI (Suppressor of early Flg22-induced Immune response) RXLR effector genes. Here, their expression was also detected, with SFI2, SFI3, and SFI4 being highly expressed in all five strains. SFI5 was expressed at a low level in four strains, SFI1 and SFI6 were not expressed in any strains, and SFI8 was only expressed at a low level in strain Pa21106. The expression of SFI7 was very distinct among individual strains, either highly expressed or completely undetectable in any given strains (Supplementary Figure 5).
Discussion
Core effectors were considered ideal target for deploying durable resistance, but P. infestans core RXLR effectors are largely unknown. In this study, to lay the foundation for searching for durable potato R genes and long-lasting controlling late blight disease, we utilized next-generation transcriptome deep sequencing strategy to identify potentially conserved core RXLR effector genes in P. infestans, primarily based on their (1) conserved sequences among diverse strains in the population, (2) high levels of expression in the early infection stage (12 hpi), and (3) potentially essential functions in pathogenesis.
In our previous studies, more than 2,000 P. infestans strains were collected from a broad potato cultivation area of China from 2008 to 2013 (Tian et al., 2015a,b, 2016). Their population structures were analyzed, which helped us to further identify core RXLR effectors at the population level. According to previous population analysis data (e.g., SSR genotype, mtDNA haplotype, mating type and pathotype), four representative strains Pa21106, Pc51265, Pd21410 (collected from northwestern China population), and F48 (collected from southern China population) were selected for further analysis. Meanwhile, since P. infestans populations in northwestern China were genetically distant from European lineages (Tian et al., 2016), a representative strain, named 80029, from European population was also used in our study. P. infestans RXLR effectors have been reported to be extremely up-regulated during infection and colonization (Cooke et al., 2012). Thus, in this study in planta infection stage materials of five diverse P. infestans strains were used to conduct transcriptome deep sequencing (Figure 1 and Supplementary Table 2). In addition, 12 hpi in-planta materials were collected for RNA-seq, because this is the time point at which the haustorium started to form (Figure 2), which means that colonization was established and the delivery of effectors into plant cells was underway (Sivak and Shaw, 1969; Petre and Kamoun, 2014).
In this study, deep-sequencing approach produced over four hundred million reads from five samples. Among them, more than 2 million reads were successfully mapped to the P. infestans reference genome, and 245 expressed RXLR effectors genes were detected (Table 1 and Supplementary Table 3), much more than that in previous studies in tomato-P. infestans interaction materials (79 effector genes) and potato-P. infestans interaction materials (31 effector genes) (Haas et al., 2009; Zuluaga et al., 2015). One reason for these results is the difference between the microarray and Illumina sequencing platforms (Haas et al., 2009). Another reason is the difference in the quantity of data between our sequencing and others (Zuluaga et al., 2015).
Table 1.
Deep sequencing data.
| Strain | Raw | Clean | Mapped | Ratio | No. of |
|---|---|---|---|---|---|
| reads | reads | reads | (%)a | RXLRb | |
| 80029 | 81,170,140 | 78,088,447 | 209,151 | 0.268 | 166 |
| F48 | 76,156,500 | 67,813,237 | 121,189 | 0.179 | 147 |
| Pa21106 | 71,809,644 | 62,860,454 | 1,577,134 | 2.509 | 226 |
| Pc51265 | 60,289,336 | 53,667,578 | 189,841 | 0.354 | 167 |
| Pd21410 | 89,624,368 | 79,813,868 | 147,278 | 0.185 | 162 |
| All | 379,049,988 | 342,243,584 | 2,244,593 | 0.656 | 245 |
aThe percentage of clean reads belonging to P. infestans in certain sample.
bNumber of RXLR effector genes detected in certain strain.
Expression of the 245 RXLR effector genes was comparably at high levels, consistent with the fact that RXLR effectors are in planta induced genes and are up-regulated during infection and colonization (Cooke et al., 2012) (Supplementary Figure 2A). Interestingly, among these expressed RXLR effector genes, most (108) were expressed by all strains and a large number (44) were specifically expressed by just one strain (Supplementary Figure 2B). Cooke et al. (2012) compared the RXLR expression profiles and aggressiveness of three P. infestans strains and found that highly aggressive strains expressed more RXLR effector genes. They, therefore, speculated that RXLR were likely virulence determinants that enhance aggressivenesses. Accordingly, we speculate that, as virulence factors, the expressed RXLR effectors may have partial contributions to fundamental pathogenesis and the uniquely expressed RXLR effectors may have partial contributions to divergence in virulence. However, additional work is required to determine exactly which genes contribute virulence to any particular strain.
In addition, to obtain conserved core RXLR effectors from P. infestans, 47 of the 108 RXLR effector genes with higher expression levels were selected for subsequent analysis. Bart et al. (2012) conducted SNP analysis to select conserved core effectors. Similarly, in this study, the sequence polymorphism analysis was performed by comparing SNPs and dN/dS ratios of 245 RXLR effector genes among strains. This led to the identification of 23 RXLR effector genes that are highly expressed at the early infection stage and conserved in protein coding sequences. Phylogenetic analysis of these 23 genes revealed that RXLR effector genes, PITG_14954, PITG_14959, PITG_14961, and PITG_14962 (CRE11), PITG_16233, and PITG_16240 (CRE13), PITG_23014, and PITG_23015 (CRE16), were designated different gene IDs yet shared consensus nucleotide sequences (Haas et al., 2009). Thus, these 23 genes were categorized as 18 conserved core RXLR effectors (Table 2 and Supplementary Figure 2).
Core effectors should make substantial contributions to pathogen virulence (Dangl et al., 2013). To validate the virulence contribution of the core RXLR effectors uncovered in this study, nine of the 18 candidate core effectors were selected and transiently expressed in N. benthamiana, then challenged with P. infestans. Results revealed that areas infiltrated with effectors showed significantly larger lesion comparing to free GFP control, which showed that those core effectors contribute to virulence during plant-pathogen interaction (Figure 3). BAX, INF1, NIP, Avh238 and Avh241 are all of cell death inducers that can activate plant PTI and ETI defenses and widely used in examining effector virulence functions (Wang et al., 2011). To further explore these virulence functions, the candidate core effectors were transiently expressed in N. benthamiana, then challenged by infiltrating cell death inducers at the same areas. Results showed that the core effectors could suppress the cell death caused by a range of inducers, suggesting that all four examined RXLR effector genes suppressed plant defenses mediated by both PTI and ETI (Figures 4A–F and Supplementary Figure 4). Meanwhile, cell death suppression assays were also tested on host potatoes. Potato differential host ‘MaR3’ contains resistance gene R3, which can recognize Avr3aKI to activate host defense response that results in cell death. Cell death suppression assays in ‘MaR3’ showed that the examined RXLR effector genes could suppress the HR caused by transient expression of Avr3aKI, suggesting that these conserved core RXLR effectors expressed at early infection stages are important in the suppression of host plant defenses induced by ETI (Figures 4G–N and Supplementary Figure 4).
Cooke et al. (2012) previously identified 45 core RXLR effectors showing in planta gene induction at 2 or 3 dpi in all three examined strains (06_3924A, NL07434, and T30-4), including 5 Avr genes with known gain-of-virulence variants (Vleeshouwers and Oliver, 2014). In this study, a collection of 18 core RXLR effectors, which are highly expressed at the early infection stage (12 hpi) and have sequences that are conserved among diverse strains, was identified. No previously known Avr RXLR effector genes were among these 18 core effectors. Meanwhile, transient expression assays in N. benthamiana and S. tuberosum showed that these tested candidate core RXLR effector genes have potentially important contributions to virulence (Figures 3, 4). These results are consistent with the fact that genes with important functions are evolutionary conserved and under negative selection (Dangl et al., 2013). More importantly, the 18 core effectors identified in this study are highly expressed at 12 hpi, suggesting that they could activate host defenses at the early infection stage, when plants contain cognate R genes, through which disease development can be quickly constrained and potential disease-related losses can be decreased.
In P. infestans, 10 Avr RXLR effector genes had been reported and their gain-of-virulence alleles were analyzed (Vleeshouwers and Oliver, 2014). However, less information is available about their polymorphisms in Chinese P. infestans populations. Thus, in the present study, sequence polymorphisms and detected expression of these 10 Avr effector genes were performed in five high divergence stains to reveal their gain-of-virulence alleles (Table 3 and Supplementary Figure 5). Avr1 was only detected in strain 80029. Previously, Cooke et al. (2012) found that Avr1 was not expressed in strain 06_3928A during the parasitic stage, because it was abandoned by the strain. Thus, we speculated that Avr1 might also be lost by Chinese P. infestans strains, or that it does not express in the early infection stage. Avr2 was highly expressed in all five examined strains. Sequence analyses showed an amino acid change (Asparagine to Lysine) at the 31st residue in strains 80029, F48, Pa21106, and Pc51265, but not in strain Pd21410. However, this change was reportedly not relevant to its Avr function (Gilroy et al., 2011). Avr3a was highly expressed in all five strains, and only the virulent form of E80M103 allele (80th residue Glutamic acid, 103rd residue Methionine) (Armstrong et al., 2005) was detected. Avr4 was expressed at a low level in strains Pa21106 and F48, and a stop codon was found at 196 nt. Although the Avr4 stop codon in strain 80029 was not detected in the present study due to its low level or complete lack of expression, it already has previously been found in this strain (van Poppel et al., 2008). Thus, the virulent truncated form of AVR4 is probably widely present in P. infestans populations. Avrblb1 was highly expressed in all five examined strains. However, ipiO4, the suppressor of Avrblb1, was also highly expressed in these strains; it functions in suppressing the defense response mediated by R gene Rpi-blb1 (Champouret et al., 2009; Chen et al., 2012). Avrvnt1, AvrSmira1, and AvrSmira2 were highly expressed in all tested strains. SNPs were detected in these Avr genes, but, according to previous reports, the substitutions did not influence their Avr function (Pel, 2010; Rietman et al., 2012). In summary, Chinese P. infestans populations may still contain functional Avr effector genes that could be recognized by the matching R genes R2, Rpi-blb2, Rpi-vnt1, Rpi-Smira1, and Rpi-Smira2.
The expression of 8 Suppressor of early Flg22-induced Immune response (SFI) RXLR effectors, which have been reported to function in suppressing host immune response (Zheng et al., 2014), were also examined (Supplementary Figure 5). Results revealed that SFI2, SFI3, and SFI4 were highly expressed in all five examined strains, suggesting their conserved function in interfering with plant immunity during the early infection stages. SF1, SF6, and SFI8 were not expressed, or expressed at a low level, in examined strains, indicating that they do not function in the early infection stage. SFI5 and SFI7 may have divergent functions in different strains, as their expression was different among strains. For example, SFI7 was highly expressed in strains 80029 and F48, but not expressed in strains Pa21106, Pc51265, or Pd21410. Although these 8 SFIs play virulent roles in suppressing PTI, they do not meet our standards for selecting core effectors (e.g., conserved protein coding sequences and/or being highly expressed at the early infection stage), suggesting that their virulence function may not be essential to P. infestans pathogenesis, at least not in the early infection stage.
The variation analysis of 10 avirulence and 8 virulence RXLR effectors suggests the possibility to monitor the pathogen virulence structure in population level and to trace down the virulence change based on the whole group of RXLR effector genes, which will help proper deployment of the resistance genes in the field to effectively control particular P. infestans population with cognate avirulence effectors. However, it should be noticed that, in our study, only one time point was used to profile the expression level of RXLR effector genes, while a time series sampling at early infection stage, such as 9, 12, and 15 hpi, would provide more information about the expression of RXLR effectors. In the future, we will expand the RNA-seq sequencing to more P. infestans strains and more sampling time points to reveal the expression pattern of RXLR effectors.
Conclusion
In this research, five genetically diverse P. infestans strains were selected to identify core P. infestans RXLR effectors, which resulted in the discovery of 18 RXLR effectors that are conserved in sequence and highly and widely expressed in the early stage of plant infection. Functional characterization by transient gene expression confirmed that these core effectors have critical functions in suppressing plant defenses mediated by PTI and ETI. These candidate core RXLR effectors are valuable in searching for cognate R genes for durable resistance breeding, and for function and virulence target studies. RXLR effectors are emerging as tools to accelerate and improve the identification, functional characterization, and deployment of resistance genes for modern potato breeding (Vleeshouwers and Oliver, 2014). Our results will speed up the usage of core RXLR effectors in potato resistance breeding.
Author Contributions
Conceived and designed the experiments: WS. Performed the experiments: JY, BG, GH, and YT. Analyzed the data: WS, JY, and HL-K. Contributed reagents/materials/analysis tools: JY, BG, GH, and JQ. Wrote the paper: WS and JY, with contributed from all authors.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Dr. Brett M. Tyler (Oregon State University, United States) for comments on the manuscript. We are grateful to Dr. Niklaus J. Grünwald (Oregon State University, United States) for helpful advice on genetic data analysis, Dr. Qinhu Wang and Dr. Jinbu Jia for advices on bioinformatics analysis.
Funding. This work was supported by National Natural Science Foundation of China (31561143007, 31301644) and China Agriculture Research System (CARS-10).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2017.02155/full#supplementary-material
References
- Allen R. L., Bittner-Eddy P. D., Grenville-Briggs L. J., Meitz J. C., Rehmany A. P., Rose L. E., et al. (2004). Host-parasite coevolutionary conflict between Arabidopsis and downy mildew. Science 306 1957–1960. 10.1126/science.1104022 [DOI] [PubMed] [Google Scholar]
- Armstrong M. R., Whisson S. C., Pritchard L., Bos J. I. B., Venter E., Avrova A. O., et al. (2005). An ancestral oomycete locus contains late blight avirulence gene Avr3a, encoding a protein that is recognized in the host cytoplasm. Proc. Natl. Acad. Sci. U.S.A. 102 7766–7771. 10.1073/pnas.0500113102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bart R., Cohn M., Kassen A., McCallum E. J., Shybut M., Petriello A., et al. (2012). High-throughput genomic sequencing of cassava bacterial blight strains identifies conserved effectors to target for durable resistance. Proc. Natl. Acad. Sci. U.S.A. 109 1972–1979. 10.1073/pnas.1208003109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birch P. R. J., Boevink P. C., Gilroy E. M., Hein I., Pritchard L., Whisson S. C. (2008). Oomycete RXLR effectors: delivery, functional redundancy and durable disease resistance. Curr. Opin. Plant Biol. 11 373–379. 10.1016/j.pbi.2008.04.005 [DOI] [PubMed] [Google Scholar]
- Bos J. I. B., Armstrong M. R., Gilroy E. M., Boevink P. C., Hein I., Taylor R. M., et al. (2010). Phytophthora infestans effector AVR3a is essential for virulence and manipulates plant immunity by stabilizing host E3 ligase CMPG1. Proc. Natl. Acad. Sci. U.S.A. 107 9909–9914. 10.1073/pnas.0914408107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruvo R., Michiels N. K., D’Souza T. G., Schulenburg H. (2004). A simple method for the calculation of microsatellite genotype distances irrespective of ploidy level. Mol. Ecol. 13 2101–2106. 10.1111/j.1365-294X.2004.02209.x [DOI] [PubMed] [Google Scholar]
- Bozkurt T. O., Schornack S., Win J., Shindo T., Ilyas M., Oliva R., et al. (2011). Phytophthora infestans effector AVRblb2 prevents secretion of a plant immune protease at the haustorial interface. Proc. Natl. Acad. Sci. U.S.A. 108 20832–20837. 10.1073/pnas.1112708109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Champouret N., Bouwmeester K., Rietman H., van der Lee T., Maliepaard C., Heupink A., et al. (2009). Phytophthora infestans isolates lacking class I ipiO variants are virulent on Rpi-blb1 potato. Mol. Plant Microbe Interact. 22 1535–1545. 10.1094/mpmi-22-12-1535 [DOI] [PubMed] [Google Scholar]
- Chen Y., Liu Z., Halterman D. A. (2012). Molecular determinants of resistance activation and suppression by Phytophthora infestans effector IPI-O. PLOS Pathog. 8:e1002595. 10.1371/journal.ppat.1002595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooke D. E. L., Cano L. M., Raffaele S., Bain R. A., Cooke L. R., Etherington G. J., et al. (2012). Genome analyses of an aggressive and invasive lineage of the Irish potato famine pathogen. PLOS Pathog. 8:e1002940. 10.1371/journal.ppat.1002940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dangl J. L., Horvath D. M., Staskawicz B. J. (2013). Pivoting the plant immune system from dissection to deployment. Science 341 746–751. 10.1126/science.1236011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fry W. E., Birch P. R. J., Judelson H. S., Grünwald N. J., Danies G., Everts K. L., et al. (2015). Five reasons to consider Phytophthora infestans a reemerging pathogen. Phytopathology 105 966–981. 10.1094/PHYTO-01-15-0005-FI [DOI] [PubMed] [Google Scholar]
- Gilroy E. M., Breen S., Whisson S. C., Squires J., Hein I., Kaczmarek M., et al. (2011). Presence/absence, differential expression and sequence polymorphisms between PiAVR2 and PiAVR2-like in Phytophthora infestans determine virulence on R2 plants. New Phytol. 191 763–776. 10.1111/j.1469-8137.2011.03736.x [DOI] [PubMed] [Google Scholar]
- Grabherr M. G., Haas B. J., Yassour M., Levin J. Z., Thompson D. A., Amit I., et al. (2011). Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 29 644–652. 10.1038/nbt.1883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu B., Kale S. D., Wang Q., Wang D., Pan Q., Cao H., et al. (2011). Rust secreted protein Ps87 is conserved in diverse fungal pathogens and contains a RXLR-like motif sufficient for translocation into plant cells. PLOS ONE 6:e27217. 10.1371/journal.pone.0027217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas B. J., Kamoun S., Zody M. C., Jiang R. H. Y., Handsaker R. E., Cano L. M., et al. (2009). Genome sequence and analysis of the Irish potato famine pathogen Phytophthora infestans. Nature 461 393–398. 10.1038/nature08358 [DOI] [PubMed] [Google Scholar]
- Haas B. J., Papanicolaou A., Yassour M., Grabherr M., Blood P. D., Bowden J., et al. (2013). De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat. Protoc. 8 1494–1512. 10.1038/nprot.2013.084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang R. H. Y., Tripathy S., Govers F., Tyler B. M. (2008). RXLR effector reservoir in two Phytophthora species is dominated by a single rapidly evolving superfamily with more than 700 members. Proc. Natl. Acad. Sci. U.S.A. 105 4874–4879. 10.1073/pnas.0709303105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D., Pertea G., Trapnell C., Pimentel H., Kelley R., Salzberg S. L. (2013). TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 14:R36. 10.1186/gb-2013-14-4-r36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin M. D., Cappellini E., Samaniego J. A., Zepeda M. L., Campos P. F., Seguin-Orlando A., et al. (2013). Reconstructing genome evolution in historic samples of the Irish potato famine pathogen. Nat. Commun. 4:2172. 10.1038/ncomms3172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng Y., Huang Y., Wang Q., Wen Q., Jia J., Zhang Q., et al. (2015). Phenotypic and genetic characterization of resistance in Arabidopsis thaliana to the oomycete pathogen Phytophthora parasitica. Front. Plant Sci. 6:378. 10.3389/fpls.2015.00378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowicki M., Foolad M. R., Nowakowska M., Kozik E. U. (2011). Potato and tomato late blight caused by Phytophthora infestans: an overview of pathology and resistance breeding. Plant Dis. 96 4–17. 10.1094/pdis-05-11-0458 [DOI] [PubMed] [Google Scholar]
- Oh S. K., Young C., Lee M., Oliva R., Bozkurt T. O., Cano L. M., et al. (2009). In planta expression screens of Phytophthora infestans RXLR effectors reveal diverse phenotypes, including activation of the Solanum bulbocastanum disease resistance protein Rpi-blb2. Plant Cell 21 2928–2947. 10.1105/tpc.109.068247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliva R. F., Cano L. M., Raffaele S., Win J., Bozkurt T. O., Belhaj K., et al. (2015). A recent expansion of the RXLR effector gene Avrblb2 is maintained in global populations of Phytophthora infestans indicating different contributions to virulence. Mol. Plant Microbe Interact. 28 901–912. 10.1094/MPMI-12-14-0393-R [DOI] [PubMed] [Google Scholar]
- Pel M. A. (2010). Mapping, Isolation and Characterization of Genes Responsible for Late Blight Resistance in Potato. Doctoral thesis, Wageningen University, Wageningen. [Google Scholar]
- Petre B., Kamoun S. (2014). How do filamentous pathogens deliver effector proteins into plant cells? PLOS Biol. 12:e1001801. 10.1371/journal.pbio.1001801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potato Genome Sequencing Consortium (2011). Genome sequence and analysis of the tuber crop potato. Nature 475 189–195. 10.1038/nature10158 [DOI] [PubMed] [Google Scholar]
- Raffaele S., Farrer R. A., Cano L. M., Studholme D. J., MacLean D., Thines M., et al. (2010). Genome evolution following host jumps in the Irish potato famine pathogen lineage. Science 330 1540–1543. 10.1126/science.1193070 [DOI] [PubMed] [Google Scholar]
- Rehmany A. P., Gordon A., Rose L. E., Allen R. L., Armstrong M. R., Whisson S. C., et al. (2005). Differential recognition of highly divergent Downy mildew avirulence gene alleles by RPP1 resistance genes from two Arabidopsis lines. Plant Cell 17 1839–1850. 10.1105/tpc.105.031807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rietman H., Bijsterbosch G., Cano L. M., Lee H.-R., Vossen J. H., Jacobsen E., et al. (2012). Qualitative and quantitative late blight resistance in the potato cultivar Sarpo Mira is determined by the perception of five distinct RXLR effectors. Mol. Plant Microbe Interact. 25 910–919. 10.1094/MPMI-01-12-0010-R [DOI] [PubMed] [Google Scholar]
- Rodewald J., Trognitz B. (2013). Solanum resistance genes against Phytophthora infestans and their corresponding avirulence genes. Mol. Plant Pathol. 14 740–757. 10.1111/mpp.12036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders D. G. O., Breen S., Win J., Schornack S., Hein I., Bozkurt T. O., et al. (2012). Host protein BSL1 associates with Phytophthora infestans RXLR effector AVR2 and the Solanum demissum immune receptor R2 to mediate disease resistance. Plant Cell 24 3420–3434. 10.1105/tpc.112.099861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan W., Cao M., Leung D., Tyler B. M. (2004). The Avr1b locus of Phytophthora sojae encodes an elicitor and a regulator required for avirulence on soybean plants carrying resistance gene Rps1b. Mol. Plant Microbe Interact. 17 394–403. 10.1094/mpmi.2004.17.4.394 [DOI] [PubMed] [Google Scholar]
- Sivak B., Shaw M. (1969). Nuclei in haustoria of Phytophthora infestans. Can. J. Bot. 47 1585–1587. 10.1139/b69-226 20847293 [DOI] [Google Scholar]
- Tamura K., Stecher G., Peterson D., Filipski A., Kumar S. (2013). MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 30 2725–2729. 10.1093/molbev/mst197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian Y., Sun J., Li H., Wang G., Ma Y., Liu D., et al. (2015a). Dominance of a single clonal lineage in the Phytophthora infestans population from northern Shaanxi, China revealed by genetic and phenotypic diversity analysis. Plant Pathol. 64 200–206. 10.1111/ppa.12251 [DOI] [Google Scholar]
- Tian Y., Yin J., Sun J., Ma H., Ma Y., Quan J., et al. (2015b). Population structure of the late blight pathogen Phytophthora infestans in a potato germplasm nursery in two consecutive years. Phytopathology 105 771–777. 10.1094/PHYTO-03-14-0073-R [DOI] [PubMed] [Google Scholar]
- Tian Y. E., Yin J. L., Sun J. P., Ma Y. F., Wang Q. H., Quan J. L., et al. (2016). Population genetic analysis of Phytophthora infestans in northwestern China. Plant Pathol. 65 17–25. 10.1111/ppa.12392 25738550 [DOI] [Google Scholar]
- Trapnell C., Roberts A., Goff L., Pertea G., Kim D., Kelley D. R., et al. (2012). Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 7 562–578. 10.1038/nprot.2012.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Poppel P. M. J. A., Guo J., van de Vondervoort P. J. I., Jung M. W. M., Birch P. R. J., Whisson S. C., et al. (2008). The Phytophthora infestans avirulence gene Avr4 encodes an RXLR-dEER effector. Mol. Plant Microbe Interact. 21 1460–1470. 10.1094/mpmi-21-11-1460 [DOI] [PubMed] [Google Scholar]
- Vleeshouwers V. G., Oliver R. P. (2014). Effectors as tools in disease resistance breeding against biotrophic, hemibiotrophic, and necrotrophic plant pathogens. Mol. Plant Microbe Interact. 27 196–206. 10.1094/mpmi-10-13-0313-ia [DOI] [PubMed] [Google Scholar]
- Vleeshouwers V. G., Raffaele S., Vossen J. H., Champouret N., Oliva R., Segretin M. E., et al. (2011). Understanding and exploiting late blight resistance in the age of effectors. Ann. Rev. Phytopathol. 49 507–531. 10.1146/annurev-phyto-072910-095326 [DOI] [PubMed] [Google Scholar]
- Wang Q., Han C., Ferreira A. O., Yu X., Ye W., Tripathy S., et al. (2011). Transcriptional programming and functional interactions within the Phytophthora sojae RXLR effector repertoire. Plant Cell 23 2064–2086. 10.1105/tpc.111.086082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whisson S. C., Boevink P. C., Moleleki L., Avrova A. O., Morales J. G., Gilroy E. M., et al. (2007). A translocation signal for delivery of oomycete effector proteins into host plant cells. Nature 450 115–118. 10.1038/nature06203 [DOI] [PubMed] [Google Scholar]
- Yang X., Liu D., Liu F., Wu J., Zou J., Xiao X., et al. (2013). HTQC: a fast quality control toolkit for Illumina sequencing data. BMC Bioinformatics 14:33. 10.1186/1471-2105-14-33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida K., Schuenemann V. J., Cano L. M., Pais M., Mishra B., Sharma R., et al. (2013). The rise and fall of the Phytophthora infestans lineage that triggered the Irish potato famine. eLife 2:e00731. 10.7554/eLife.00731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng X., McLellan H., Fraiture M., Liu X., Boevink P. C., Gilroy E. M., et al. (2014). Functionally redundant RXLR effectors from Phytophthora infestans act at different steps to suppress early flg22-triggered immunity. PLOS Pathog. 10:e1004057. 10.1371/journal.ppat.1004057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuluaga A. P., Vega-Arreguín J. C., Fei Z., Ponnala L., Lee S. J., Matas A. J., et al. (2015). Transcriptional dynamics of Phytophthora infestans during sequential stages of hemibiotrophic infection of tomato. Mol. Plant Pathol. 17 29–41. 10.1111/mpp.12263 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.