Abstract
Like Streptococcus mutans, lactobacilli are commonly isolated from carious sites, although their exact role in caries development remains unclear. This study used mixed-species models to analyze biofilm formation by major groups of oral lactobacilli, including L. casei, L. fermentum, L. rhamnosus, L. salivarius ssp. salivarius, and L. gasseri. The results showed that lactobacilli did not form good biofilms when grown alone, although differences existed between different species. When grown together with S. mutans, biofilm formation by L. gasseri and L. rhamnosus was increased by 2-log (P < 0.001), while biofilms by L. fermentum reduced by >1-log (P < 0.001). L. casei enhanced biofilm formation by ~2-log when grown with S. mutans wild-type, but no such effects were observed with S. mutans deficient of glucosyltransferase GtfB and adhesin P1. Both S. mutans and L. casei in dual-species enhanced resistance to acid killing with increases of survival rate by >1-log (P < 0.001), but drastically reduced the survival rates following exposure to hydrogen peroxide (P < 0.001), as compared to the respective mono-species cultures. When analyzed by RNA-seq, more than 134 genes were identified in S. mutans in dual-species with L. casei as either up- or down-regulated when compared to those grown alone. The up-regulated genes include those for superoxide dismutase, NADH oxidase, and members of the mutanobactin biosynthesis cluster. Among the down-regulated genes were those for GtfB and alternative sigma factor SigX. These results further suggest that interactions between S. mutans and oral lactobacilli are species-specific and may have significant impact on cariogenic potential of the community.
Keywords: S. mutans, oral lactobacilli, mixed-species biofilms, dental caries, RNA-seq
Introduction
The oral microbiome, represented by dental plaque, harbors diverse and abundant microbial communities consisting of over 700 different species or phylotypes (Jenkinson, 2011). Both intra- and inter-species interactions in the oral flora have been well documented, although the underlying mechanisms remain unclear (Kuramitsu et al., 2007). The oral cavity is featured with fluctuating and often unpredictable conditions, such as nutrient source and availability and pH. Both microbe-microbe and microbe-environment interactions can profoundly influence the composition and relative proportions of major groups in the dynamic communities, leading to dysbiosis and consequently, development of oral diseases such as dental caries and periodontitis (Kuramitsu et al., 2007; Jenkinson, 2011; Burne et al., 2012; Hajishengallis et al., 2017). Cariogenic plaque, for instance, is characterized by dramatic increases in the proportion of acidogenic and aciduric species, which include mutans streptococci and lactobacilli (Jenkinson, 2011).
As a major causative agent of dental caries, S. mutans possesses multiple mechanisms to colonize and persist on the tooth surface, and under certain conditions to become numerically significant, causing carious lesions (Hamada and Slade, 1980; Bowen et al., 1991; Bowen and Koo, 2011; Burne et al., 2011). Multi-functional adhesin P1 (also Antigen I/II, SpaP, or PAc) functions as the primary factor mediating early attachment to the tooth surface via interaction with salivary agglutinin-gp340 (Crowley et al., 1993). S. mutans also produces at least three glucosyltransferases (GtfB, -C, and -D) that polymerize the glucosyl moiety from sucrose, generating adhesive glucans (Bowen and Koo, 2011). The Gtfs and their glucan products, along with the glucan-binding proteins (Gbps), constitute the sucrose-dependent pathway central in plaque formation and caries development (Banas et al., 2007; Gregoire et al., 2011). In addition, multiple two-component signal transduction systems, molecular chaperones, and biofilm regulatory protein BrpA are shown to play an important role in S. mutans biofilm formation (Burne et al., 2011).
As the first microorganisms implicated in human dental caries (Owen, 1949), lactobacilli are frequently identified at carious sites, esp. in patients with advanced caries, with L. casei, L. fermentum, L. gasseri, L. salivarius, and L. rhamnosus among the most prevalent groups (Caufield et al., 2007; Badet and Thebaud, 2008; Gross et al., 2012). Lactobacilli can utilize various kinds of sugars, generating lactic acid and other organic acids, and are highly resistant to low pH. Previously, our in vitro model studies showed that L. casei alone did not colonize a surface efficiently, but drastically increased its ability to colonize and accumulate biofilms during growth with S. mutans (Wen et al., 2010). Similar results were also observed with Actinomyces spp. (Van Houte et al., 1991; Filoche et al., 2004; Wen et al., 2010). Our previous studies have shown that S. mutans in dual-species cultures with several other prominent bacterial species including L. casei had altered expression of several genes known to be critical to biofilm formation and cariogenicity (Wen et al., 2010). In this study, representatives of lactobacilli groups most frequently isolated from the carious sites were first analyzed using in vitro mixed-species models for their abilities to form biofilms with and without the presence of S. mutans. An in vitro continuous biofilm model, transcriptional profiling via RNA-seq and metabolite analysis via HPLC were then used to further investigate the interactions between S. mutans and L. casei and identify the factors that mediate these interspecies interactions. Results demonstrated that interactions between S. mutans and the major oral lactobacilli are species-specific and may have an impact on the pathogenicity of the community.
Materials and methods
Bacterial strains and cultivation
All bacterial strains used in this study are listed in Table 1. S. mutans strains were maintained in Brain Heart Infusion (BHI, Becton, Dickinson and Company, Sparks, MD). Lactobacilli were maintained in Lactobacillus MRS (Difco Laboratories, MI). For Veillonella dispar, tryptic soy broth (TSB, Becton, Dickinson and Company, Sparks, MD) plus lactic acid at 0.6% (v/v) was used (Liu et al., 2011). For S. mutans and lactobacilli dual-species biofilm formation, semi-defined biofilm medium (BM) with 18 mM glucose and 2 mM sucrose (BMGS) was used (Loo et al., 2000; Bitoun et al., 2012). For triple-species biofilms, S. mutans, lactobacilli and V. dispar were grown in TSB. For transcriptional profiling and metabolite analysis, chemically defined medium FMC was used with modifications (mFMC) in an effort to facilitate the growth of lactobacilli (Terleckyj et al., 1975; Elli et al., 2000; Savijoki et al., 2006; Wegkamp et al., 2009; Table S1). All solid media were prepared similarly with inclusion of Bacto agar (Difco Laboratories, Franklin Lakes, NJ) at the level of 1.5% (w/v). When needed, erythromycin (5 μg/ml), and/or kanamycin (1 mg/ml) were added to the proper medium. Unless otherwise stated, bacterial cells were grown at 37°C in an aerobic chamber with 5% CO2.
Table 1.
Bacterial strains used in this study.
| Strains /Plasmid | Relevant characteristics | References/Source |
|---|---|---|
| S. mutans UA159 | wild-type | Ajdic et al., 2002 |
| S. mutans SAN136 | ΔspaP, Kanr | Ahn et al., 2008 |
| S. mutans SAB106 | ΔgtfB, Kanr | The Burne lab |
| S. mutans SAB108 | ΔgtfC, Kanr | The Burne lab |
| S. mutans SAB109 | ΔgtfBC, Kanr | The Burne lab |
| S. mutans SAB110 | Δftf, Kanr | The Burne lab |
| S. milleri KSB8 | gtfB of S. mutans GS5, Emr | Vacca-Smith and Bowen, 1998 |
| L. casei 4646 | wild-type | Shu et al., 2003 |
| L. casei CB | derivative of 4646, Ermr | Wen et al., 2010 |
| L. fermentum ATCC 14931 | wild-type | ATCC# |
| L. rhamnosus ATCC7469 | wild-type | ATCC# |
| L. salivarius ssp. salivarius ATCC 11741 | wild-type | Caufield et al., 2015 |
| L. gasseri ATCC 33323 | Wild-type | Caufield et al., 2015 |
| L. gasseri M57-23 | mother-caries active* | Caufield et al., 2015 |
| L. fermentum M87-28 | mother-caries active* | Caufield et al., 2015 |
| L. salivarius M87-8 | mother-caries active* | Caufield et al., 2015 |
| L. rhamnosus M72-26 | mother-caries active* | Caufield et al., 2015 |
| V. dispar ATCC17745 | wild-type | Shu et al., 2003 |
Ermr and Kanr for erythromycin and kanamycin resistance, respectively.
strains purchased directly from American Type Culture Collection, Manassas, VA;
Clinical strains previously isolated from mothers of caries active kids.
Growth of mixed-species biofilms
For biofilm formation, 96-well plates and glass slides were used as substratum, and biofilms were grown by following protocols described previously (Loo et al., 2000; Wen and Burne, 2002; Wen et al., 2010). For the 96-well plate model, actively growing individual species were diluted by 1:100 (v/v) in proper growth medium, and 200 μL aliquots were transferred to the wells of the culture plates (Corning, New York) in triplicate. For dual- and triple-species biofilms, individual species were mixed proportionally and then diluted as described above. The plates were incubated at 37°C in an aerobic chamber with 5% CO2 or for biofilms involving veillonella, in an anaerobic box for 24 and 48 h. By the end of incubation, the adherent biofilms were stained with 0.1% crystal violet and quantified using a spectrophotometer at 575 nm (Loo et al., 2000; Wen and Burne, 2002; Wen et al., 2010). For the glass slides model, the biofilms were grown on the glass slides in 50 ml Falcon tubes, and the slides were aseptically transferred daily to fresh medium (Loo et al., 2000; Wen and Burne, 2002; Wen et al., 2010). For enumeration of the different species, aliquots of the mixed-species cultures were plated on BHI agar, where the two bacteria form colonies with distinctive morphological characteristics, and on Rogosa agar (Becton, Dickinson and Company, Sparks, MD), a selective medium for lactobacilli. For continuous flowing conditions, S. mutans and L. casei were grown in mFMC with glucose (18 mM) and sucrose (2 mM), and biofilms were grown on glass slides deposited in a Drip Flow Biofilm Reactor (BioSurface Technologies, Montana). The system was run with a medium flow rate of 60 ml per hour in a 37°C warm room for 48 and 120 h, respectively (Wen et al., 2010; Fan et al., 2012). By the end of the experiments, the biofilms were scratched off with a sterile spatula and suspended in 5 ml potassium phosphate buffer, 10 mM, pH 7.0, sonicated briefly to de-chain and separate the cells (Wen et al., 2010), and serial dilutions were then plated in triplicate on proper agar medium for enumeration of viable biofilm cells (Fan et al., 2012).
GtfB binding to L. casei surface and fluorescence imaging of glucan production on L. casei and S. mutans bound to L. casei
The GtfB enzyme was prepared from culture supernatant of S. milleri KSB8 harboring S. mutans gtfB and purified via hydroxyapatite column chromatography using our well-established protocol as detailed previously (Vacca-Smith and Bowen, 1998; Gregoire et al., 2011). The in vitro Gtf binding assay and fluorescence imaging were carried out similarly as described by Gregoire et al. (2011). Briefly, for in vitro binding assay, L. casei cells (~1 × 109 cfu/ml) were mixed with saturating amount of GtfB (25 μg/ml, 3 U), in adsorption buffer and incubated for 60 min at 37°C. For controls, L. casei cells were incubated in adsorption buffer alone. Following adsorption, the cells were stained with nucleic acid staining dye SYTO 9 (485/498 nm; Molecular Probes, Eugene, OR) that confers green fluorescence. Then, the cells were washed twice with adsorption buffer by centrifugation, and the pellets were resuspended in adsorption buffer. For glucan production, 1 μM Alexa Fluor 647-conjugated dextran (647/688 nm, Molecular Probes) was added to the reaction mixture containing L. casei (1 × 109 cells/ml) with and without surface-adsorbed GtfB and with and without sucrose (100 mM) for 1 h at 37°C. Following incubation, 20 μl of the mixture were placed on a glass slide and imaging was performed using an upright Olympus confocal microscope (Olympus Fluoview BX61). For L. casei-S. mutans interactions, S. mutans cells were washed with adsorption buffer similarly as described above, and stained with DAPI (4′,6-diamidino-2-phenylindole) blue fluorescence nucleic acid staining dye (358/461 nm; Molecular Probes). The fluorescently labeled cells (1 × 109 cells/ml) were mixed with L. casei with and without bound GtfB and L. casei with and without surface glucan coating, and incubated at 37°C for 1 h. After incubation, the suspension was immediately visualized under confocal microscope.
Acid and hydrogen peroxide killing assays
To evaluate the impact of growth in a consortium on stress tolerance response, the abilities of S. mutans and L. casei to withstand low pH and oxidative stress were evaluated using acid and hydrogen peroxide killing assays (Wen and Burne, 2004; Wen et al., 2006, 2010). For acid killing assays, 48-h biofilms of S. mutans and L. casei mono- and dual-species biofilms were grown on glass slides in BMGS. Biofilms were briefly sonicated to detach and de-chain S. mutans, washed once, and the acid killing assay was carried out by incubation of the bacterial cells in glycine buffer of pH 2.0, 0.1 M for L. casei and pH 2.8, 0.1 M for S. mutans for a period of 15, 30, 45 and 60 min (Wen and Burne, 2004; Wen et al., 2006, 2010). For hydrogen peroxide killing assay, the mono- and dual-species cultures were grown in BHI broth until early-exponential phase (OD600nm ≅0.4), and washed once as described above before subjecting to the killing assays by incubation of the bacterial cells in glycine buffer containing 0.2% hydrogen peroxide for period of 60, 90, 105, and 120 min as described elsewhere (Wen and Burne, 2004; Wen et al., 2006, 2010).
Competition assays on agar plates
Competitions between S. mutans and the lactobacilli were examined by following the method of Kreth et al. (2005).
Organic acid analysis
Organic acids were determined using UV-HPLC (Kara et al., 2006 88; Ausubel et al., 2010). For acidic metabolite analysis, mono- and dual species biofilms of S. mutans and L. casei were grown on glass slides in mFMC with 18 mM glucose and 2 mM sucrose for 24- and 48-h. At the end of incubation, the cultures were spun down, and supernates were collected and filtered via centrifugal units (Millipore, 10kDa MWCT). The supernates were then diluted 10-fold in 40 mM Na2SO4, pH 2.65, and passed over a Acclaim™ OA 3 μm column (DX070087, Dionex) optimized for organic acids, at 0.21 mL/min of 40 mM Na2SO4 mobile phase at 30°C on a Agilent 1100 Series UV/HPLC. Samples were detected at 210 nm. Sodium salts of fumaric, α-ketoglutaric, citric, proponic, butyric, succinic, and lactic acid (Sigma) were used to prepare single and mixed standard solutions in deionized water.
Transcriptomic analysis via RNA-Seq
For RNA extraction, early-exponential phase cultures, OD600nm ~0.3, were treated with RNAProtect (Qiagen Inc., CA), and total RNAs were extracted using a hot phenol method as detailed elsewhere (Wen and Burne, 2004; Wen et al., 2006). To remove all DNA, the purified RNAs were treated with DNase I (Ambion, Inc., TX) and RNA was retrieved with RNeasy Mini kit (Qiagen, Inc.), including an additional on-column DNase I treatment with RNase-free DNase I. The RNA samples were depleted of ribosomal RNA using Illumina's Ribo-Zero Bacterial Kit. 40 ng of RNA were then converted to cDNA, and libraries were generated using Illumina's ScriptSeq v2 Library Preparation Kit. Deep sequencing and post-sequencing analysis were carried out by following our established protocols (Robinson et al., 2010; Lo and Chain, 2014; Love et al., 2014; Maekawa et al., 2014; Kim et al., 2015).
Statistical analysis
Unless specified otherwise, all quantitative data were further analyzed by student's t-test or ANOVA and Tukey's pairwise comparison. A difference of P < 0.05 is considered as statistically significant.
Results
Lactobacilli differ in the ability to form biofilms
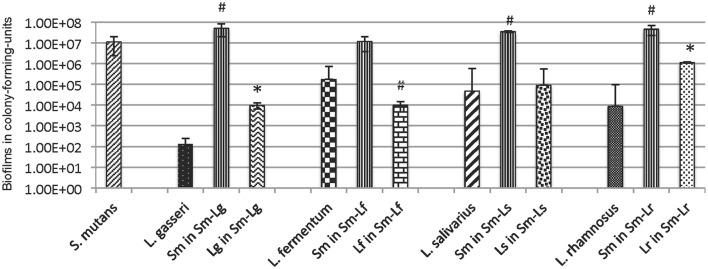
Five different species of lactobacilli that are most frequently isolated from carious sites were selected for the study (Table 1; Caufield et al., 2007, 2015; Badet and Thebaud, 2008; Gross et al., 2012). Like L. casei (Van Houte et al., 1972; Wen et al., 2010), all lactobacillus strains showed only limited abilities to form biofilms on glass, polystyrene and hydroxylapatite surfaces tested. When grown in BMGS, L. fermentum formed the best biofilms when grown alone over night, followed by L. salivarius ssp. salivarius, and L. rhamnosus, while L. gasseri possessed the least capacity to form biofilms under the conditions studied (Figure 1). When compared between 24- and 48-h incubation (without growth medium refreshment), 48-h incubation significantly reduced biofilm formation for all lactobacilli tested (Figure S1). When grown in TSB in 96-well plates, L. fermentum again accumulated the most biofilms overnight among the lactobacilli tested, while the least biofilms were observed with L. rhamnosus (Figure S2). Continuous incubation in TSB for 48 h moderately increased biofilms by L. casei and L. salivarius ssp. salivarius, while biofilms by L. gasseri and L. fermentum were reduced moderately. Interestingly, when grown in TSB on glass slides for 5 days, the highest biofilm was observed with L. rhamnosus, averaging just 8.7E7 ± 7.1E6 in colony-forming-units (CFU) (Figure S3). It was followed by L. fermentum, and L. salivarius ssp. salivarius, and the least was obtained with L. gasseri, with an average of 3.3E4 ± 4.4E3 in CFU. Selected clinical isolates of different major species, which were isolated from mothers of caries active kids from previous studies (Caufield et al., 2015), were also analyzed. Similar trends were also observed when they were grown in TSB, with the most biofilms produced by L. fermentum, followed by L. salivarius, L. gasseri, and the least was found with L. rhamnosus (Data not shown).
Figure 1.
Forty eight-hour biofilms of L. gasseri (Lg), L. fermentum (Lf), L. salivarius (Ls) and L. rhamnosus (Lr) in mono-species and dual-species with S. mutans (Sm) grown in BM plus glucose (18 mM) and sucrose (2 mM) on glass slides vertically deposited in 50 ml Falcon tubes. Data represent the average (±standard deviation in error bars) of at least three separate experiments, with * and # indicating statistical difference at P < 0.001 and 0.05, respectively, when compared to the respective mono-species biofilms.
Interactions between S. mutans and lactobacilli are species-specific
When compared to mono-species cultures grown in BMGS, L. gasseri and L. rhamnosus increased biofilm formation by ~2-log after 48 h when grown together with S. mutans (P < 0.001) (Figure 1, Figure S1). In contrast, biofilms of L. fermentum decreased by >1-log during growth with S. mutans (P < 0.05). No significant differences were measured with L. salivarius ssp. salivarius in dual-species with S. mutans in comparison to those grown alone. On the other hand, some significant increases were also observed with S. mutans when grown together with L. gasseri, L. salivarius ssp. salivarius, and L. rhamnosus (P < 0.05), while moderate decreases were measured after 24 h' growth with L. fermentum (Figure S1). When grown in a consortium in TSB in 96 well plates, the amount of total biofilms was decreased significantly when S. mutans was partnered with L. fermentum, L. salivarius ssp. salivarius, and L. rhamnosus, while moderate increases of biofilms were observed with S. mutans-L. gasseri biofilms (Figure S2). On glass slides (Figure S3), co-cultivation with S. mutans for 5 days significantly increased biofilms of L. salivarius ssp. salivarius (P < 0.05), while significantly reduced biofilms of L. fermentum (P < 0.05) and L. gasseri (P < 0.001). This is contrary to what were seen with 24- and 48-h biofilms in BMGS, when compared to the respective mono-species counter-parts. No significant differences were measured between S. mutans grown alone and those co-cultivated with the lactobacilli tested under these conditions (Figure S3).
GtfB and P1 play an important role in mixed-species biofilm formation
When grown in flow cells in mFMC for 5 days, L. casei alone accumulated a little over 1.3E5 ± 5.5E4 CFU per mL, but when grown together with S. mutans, its biofilm increased by 54-fold, averaging 7.0E6 ± 1.9E5 CFU (P < 0.001; Figure 2). No significant differences were observed between S. mutans grown alone and those in dual-species with L. casei. In an effort to find out if Gtf enzymes were involved in mixed-species biofilm formation, L. casei was grown in BMGS with S. mutans strains deficient of GtfB, GtfC, GtfB&C, Ftf (for fructosyltransferase, Ftf), and P1. The gtf and ftf mutants were generated by standard allelic replacement strategies and kindly provided by Robert A. Burne (Department of Oral Biology, University of Florida, Gainesville, FL). Consistently, co-cultivation of L. casei with the wild-type S. mutans UA159 increased biofilm formation by L. casei by >13-fold (P < 0.001), when compared to L. casei grown alone (Figure 3). Relatively, biofilm formation by L. casei increased by 8-fold when grown with a GtfC-deficient mutant (P < 0.001), by >9-fold (P < 0.001) when co-cultivated with an ftf-deficient mutant, but no significant differences were observed when incubated with GtfB- and GtfBC- and P1-deficient mutants (P > 0.05).
Figure 2.
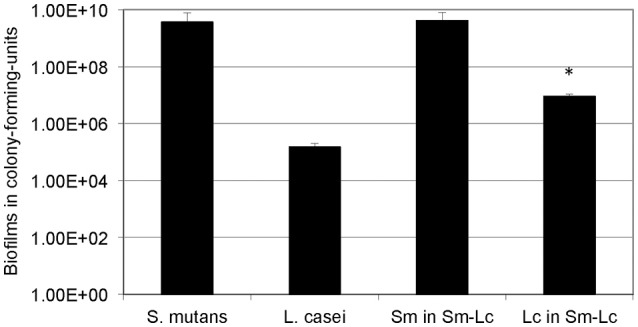
Five-day biofilm grown under continuous flowing conditions. S. mutans (Sm) and L. casei (Lc) mono- and dual-species biofilms were grown on glass slides deposited in a Drip Flow Biofilm Reactor in mFMC containing 18 mM glucose and 2 mM sucrose. Results represent the average (±standard deviation in error bars) of at least three separate experiments, with * indicating statistical difference at P < 0.001 relative to its mono-species biofilms.
Figure 3.
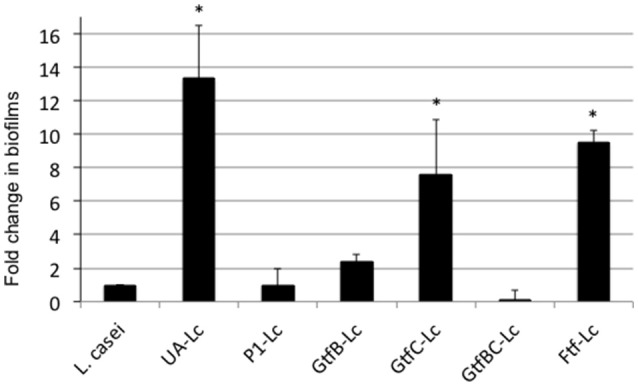
L. casei biofilm formation with S. mutans mutants. L. casei (Lc) was grown alone or together with S. mutans wild-type UA159 (UA) and its mutants deficient of P1, GtfB, GtfC, GtfBC, or Ftf in biofilm medium with glucose and sucrose for 48-h. Data are expressed as the ratio of L. casei biofilms (in colony-forming-units) in dual-species over those in mono-species, with * indicating significant differences at P < 0.001.
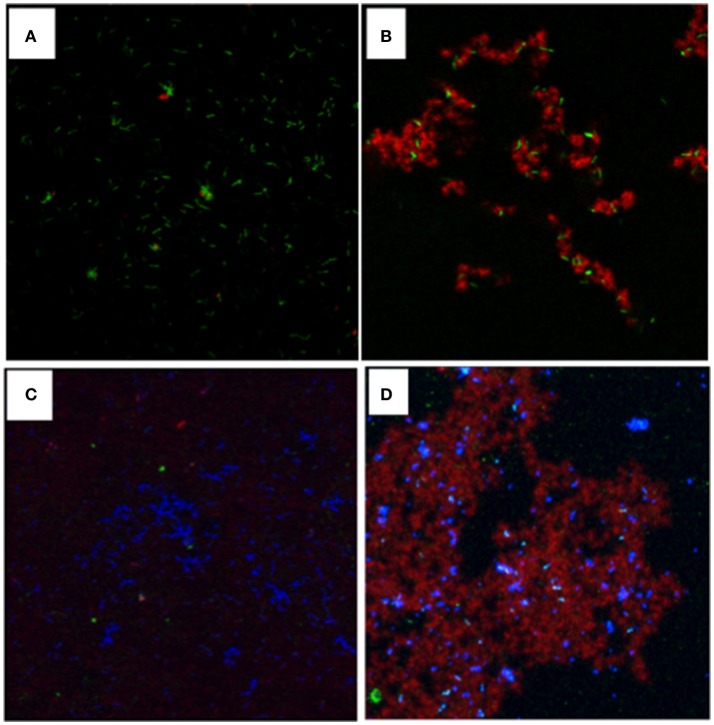
When analyzed using in vitro assays, the purified GtfB from S. mutans (Gregoire et al., 2011) was shown to readily bind to L. casei cells, and the bound GtfB was able to generate glucose polymers in the presence of sucrose (Figures 4A,B). When mixed together, S. mutans was shown to be able to bind to L. casei, forming microcolony-like structures with green L. casei (Data not shown). In contrast, more S. mutans cells were found on L. casei with GtfB-bound (data not shown) and especially, on the top of the glucan matrices surrounding the L. casei cells (Figures 4C,D), as compared to the controls with no GtfB glucans.
Figure 4.
Visualization of glucans synthesized in situ by GtfB adsorbed on L. casei and S. mutans binding to L. casei with and without glucans. For glucan visualization, L. casei was incubated with GtfB or buffer, and following washes, exposed to sucrose for 1 h, and glucans were imaged using a confocal microscope. (A) L. casei cells (in green) with GtfB incubated in buffer alone; (B) L. casei with bound-GtfB incubated with sucrose showing rich glucans (in red) engulfed in green L. casei cells. For interactions, S. mutans were mixed proportionally with L. casei with and without GtfB glucan-coating. (C) S. mutans (in blue) incubated with green L. casei with no glucans on surface; and (D) S. mutans (in blue) incubated with L. casei coated with GtfB glucans (in red). Images were obtained using an upright Olympus confocal microscope with a 100x oil objective.
V. dispar and S. mutans enhance biofilm formation when grown together
V. dispar did not form good biofilm when grown in TSB alone (Figure S2). When it was grown with S. mutans, the amount of mixed-species biofilms was significantly increased, especially for those grown continuously for 48 h (Figure S2). However, no such effects were observed when V. dispar was co-cultivated with the different species of lactobacilli tested. When S. mutans and V. dispar were grown together with different lactobacilli, the amounts of mixed-species biofilms were very similar to what were measured with the S. mutans-V. dispar dual-species biofilms, regardless of the lactobacilli in the consortium (Figure S2).
S. mutans and L. casei in consortium enhanced acid tolerance responses and reduced survival rates against hydrogen peroxide killing
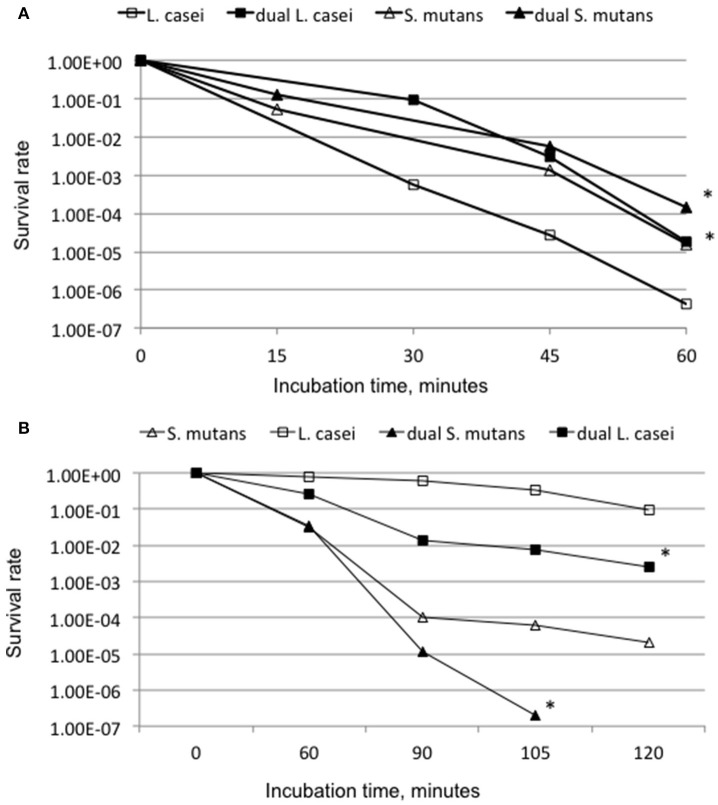
When compared to the respective mono-species cultures, both S. mutans and L. casei grown in dual-species consortium increased their survival rate by more than 1-log (P < 0.001) after incubation at pH 2.0 for L. casei and 2.8 for S. mutans for a period of 60 min (Figure 5A). When analyzed by hydrogen peroxide killing assay, S. mutans grown in dual-species reduced its survival rate by >1-log after 90 min, >2-log after 105 min and became undetectable after 120 min, as compared to those grown individually (P < 0.001; Figure 5B). Relatively, L. casei displayed higher resistance to hydrogen peroxide than S. mutans, although it also had a reduction of survival rate by >1-log for cells grown together with S. mutans (P < 0.05).
Figure 5.
Acid (A) and hydrogen peroxide (B) killing assays. (A) For acid killing, S. mutans, and L. casei biofilms were grown in biofilm medium with glucose and sucrose on glass slides individually or in dual-species for 48 h, and acid killing was carried out at pH 2.0 for L. casei and 2.8 for S. mutans. (B) For hydrogen peroxide killing assays planktonic cultures of S. mutans and L. casei were grown overnight in BHI broth, and killing assays were done in glycine buffer containing 0.2% hydrogen peroxide. Data presented here are representatives of more than three independent assays with * indicating significant difference at P < 0.001 in comparison to the respective mono-species cultures.
S. mutans produces antimicrobials against L. fermentum
Streptococcus mutans is known to produce mutacins that possess a broad spectrum of activities against closely associated bacterial species (Merritt and Qi, 2012). Several oral Lactobacillus spp. have also been shown to produce bacteriocins effective against S. mutans (Simark-Mattsson et al., 2007; Strahinic et al., 2007; Wu et al., 2015). When analyzed using a competition assay, S. mutans displayed inhibitory activity against L. fermentum when allowed to grow first on half-strength BHI agar plates. This was illustrated by a crescent zone of inhibition, but no such activity was detected when S. mutans and L. fermentum were spotted next to each other simultaneously nor was when L. fermentum was allowed to grow first (Figure S4). L. fermentum did not produce any visible inhibitory activity under the conditions studied. No apparent inhibitory activities were observed when the two bacteria were grown on regular BHI agar plates. No antimicrobial activity was detected from S. mutans against the other lactobacilli tested, or vice versa.
S. mutans and L. casei in dual-species had different metabolic profiles from those grown individually
For metabolite analysis, a chemically defined medium, termed mFMC, was developed using FMC as the basal medium and with defined lactobacillus medium as a reference (Table S1). This medium retains its strong support for S. mutans of FMC, while enhancing the growth of lactobacilli (Figure 2). When the dual-species overnight cultures of S. mutans and lactobacilli tested were grown in mFMC with 18 mM glucose and 2 mM sucrose and analyzed using a micro pH probe, all except L. fermentum, had a reduced cultural pH than the respective mono-species cultures (data not shown). Continuous incubation for 48 h led to pH elevation for both S. mutans and the lactobacilli strains, but again, the mixed-species cultures, including those with L. fermentum, had a significantly reduced pH than when they grew alone. When the organic acid profile was analyzed using HPLC, succinic acid was reduced by 4-fold in 24-h dual-species biofilms, as compared to the respective S. mutans and L. casei mono-species cultures (Figure S5). Other more subtle changes were also identified for lactic acid, α-ketoglutaric acid, fumaric acid, and citric acid. In addition, metabolites with major alterations with retention times of ~4 and ~8 min were also apparent under the conditions studied, although the exact identity of these metabolites currently remain unknown.
S. mutans in dual-species has an altered transcriptional profile
When analyzed by deep- sequencing, the transcriptional profile of S. mutans in dual-species with L. casei was shown to be significantly different from that in mono-species with >134 genes being identified as either up- or down regulated (P < 0.001; Table S2). Of the altered genes, six were down-regulated by more than 1.5-fold, and twelve were up-regulated by >1.5-fold. Among the down-regulated genes were those for GtfB, dextranase, and alternative sigma factor (SigX) (Table 2). The up-regulated genes include those for GtfD, Ftf, H2O-forming NADH oxidase (Nox1), Mn-dependent superoxide dismutase (SodA), lipid hydroperoxide peroxidase (Tpx), and members of the gene cluster for biosynthesis of mutanobactin (Wu et al., 2010).
Table 2.
Selected genes identified by RNA-Seq analysis.
| Locus | Description* | Ratio (D/M)# | P-value |
|---|---|---|---|
| SMU.265 | Putative amino acid kinase | 1.56 | 0.016654713 |
| SMU.270 | Unknown | 1.67 | 0.000835434 |
| SMU.629 | Mn-dependent Superoxide dismutase, SodA, | 1.60 | 2.48242E-08 |
| SMU.910 | Glucosyltransferase-S, GtfD | 1.50 | 1.98036E-14 |
| SMU.924 | Lipid hydroperoxide peroxidase, Tpx | 1.67 | 1.5412E-12 |
| SMU.1117 | NADH oxidase (H2O-forming), Nox1, Nox | 2.09 | 5.75842E-26 |
| SMU.1339 | Putative bacitracin synthetase | 1.65 | 6.9306E-08 |
| SMU.1340 | Putative surfactin synthetase | 1.72 | 2.61792E-10 |
| SMU.1342 | Putative bacitracin synthetase, BacA | 1.92 | 5.72903E-15 |
| SMU.1346 | Putative thioesterase, BacT | 2.15 | 2.98087E-12 |
| SMU.1425 | Putative Clp proteinase, ATP-binding subunit, ClpB | 1.53 | 0.008209022 |
| SMU.2028 | Levansucrase precursor, beta-D-fructosyltransferase, Ftf, SacB | 1.72 | 7.67161E-26 |
| SMU.20 | Unknown (MreC, putative cell shape determining protein) | −1.61 | 2.98587E-05 |
| SMU.183 | Putative Mn/Zn ABC transporter | −1.65 | 0.011069851 |
| SMU.184 | Putative ABC transporter metal binding lipoprotein | −1.69 | 7.99849E-07 |
| SMU.1004 | Gucosyltransferase-I, GtfB | −1.50 | 1.11398E-16 |
| SMU.1997 | Transcriptional regulator ComX, alternative sigma factor | −1.59 | 9.38051E-05 |
| SMU.2042 | Dextranase precursor | −1.66 | 3.37282E-19 |
Description and putative function of the identified genes are based on the published S. mutans database.
Defined as relative levels of expression in the dual-species over those in the mono-species with “–” indicating the down-regulated genes.
Discussions
While lactobacilli are frequently isolated from carious sites (Van Houte et al., 1972; Michalek et al., 1981), this study has generated evidence that none of the lactobacilli strains tested, including clinic isolates from mothers of caries active kids, possessed strong capacity to colonize and form biofilms in vitro consistent with our early studies with L. casei (Van Houte et al., 1972; Filoche et al., 2004; Wen et al., 2010). When grown together with S. mutans, however, all lactobacilli showed major increases in biofilm formation, except L. fermentum, which reduced biofilms significantly. These results further suggest differences exist in intercellular interactions between S. mutans and different Lactobacillus spp. as well as their impact on the mixed-species consortium. In an early animal model study by Fitzgerald et al. (Fitzgerald et al., 1980), it was found that clinic isolates of L. salivarius and L. fermentum were able to induce significant carious activity in conventional hamsters but not on hamsters whose regular oral flora was depressed via polyantibiotics. This also indicates results of likely interactions between members of the indigenous oral flora and the lactobacilli infected.
Inter-species interactions are well documented in the oral cavity, which include adhesin-receptor mediated coadhesion and coaggregation, nutrient coupling, and metabolite mediated inhibitions (Kuramitsu et al., 2007). S. mutans possesses at least three Gtf enzymes that produce glucose polymers (better known as glucans) from sucrose, playing a central role in bacterial adherence and biofilm accumulation (Bowen and Koo, 2011). These Gtf proteins can also function as adhesins that bind directly to their adhesive products, the glucans. In addition, GtfB can also avidly bind to the surfaces of other bacterial species, including Actinomyces vicosus, Candida albicans and L. casei (Vacca-Smith and Bowen, 1998; Gregoire et al., 2011). Using radiolabeled GtfB, -C and -D in an in vitro binding assay, GtfB was found to possess the best capacity to bind to other bacteria tested (Vacca-Smith and Bowen, 1998). Consistently, the results presented here have also shown that GtfB can readily bind to L. casei, converting the cells to de facto glucan producers. S. mutans can directly bind to the L. casei cells (see also below), but as expected, significantly more S. mutans were found on the glucans synthesized by the GtfB that were bound on the surface of L. casei cells. These results further suggest that as an adhesin and an enzyme, GtfB is a major contributing factor to the enhanced biofilm formation by L. casei when grown with S. mutans. This is similar to what have recently been reported in C. albicans (Gregoire et al., 2011; Ellepola et al., 2017; Hwang et al., 2017; Kim et al., 2017), but to the best of our knowledge, has never been reported in Lactobacillus spp. before. In C. albicans, presence of S. mutans' GtfB was also recently shown to modulate expression of genes critical to cell morphology and biofilm formation (Ellepola et al., 2017). Studies are underway to elucidate the receptors GtfB binds to and if GtfB binding also influences other aspects of the L. casei physiology.
P1 protein is a high-affinity adhesin that S. mutans utilizes to colonize the tooth's salivary pellicle via interactions with the host scavenger receptor glycoprotein called GP340 or DMBT-1 (Jakubovics et al., 2005; Brady et al., 2011; Larson et al., 2011). As multi-functional adhesins, the AgI/II family of proteins, including SspA/B in S. gordonii, also interact with host proteins such as fibronectin and collagen and other bacteria, such as Porphyromonas gingivalis and A. naeslundii (Brady et al., 2011; Sullan et al., 2015). The results that P1 deficiency significantly diminishes the ability of S. mutans to facilitate L. casei biofilm formation also suggests that P1 is part of the underlying factors in S. mutans-L. casei interactions. While not surprising, this, too, has not yet been reported before. What receptors on L. casei cells P1 binds to and what regions of P1 are involved in binding await further investigation.
Like S. mutans, Lactobacillus spp. utilize a variety of sugars, although the end products of sugar fermentation vary between different species in response to environmental conditions, such as oxygen tension and sugar availability (Van Houte et al., 1972; Iwami and Yamada, 1985; Yamada et al., 1985; Iwami et al., 1992). Consistent with our previous studies of L. casei and S. mutans (Wen et al., 2010), co-cultivation of S. mutans with all Lactobacillus spp. tested, except L. fermentum, resulted in a lower culture pH than the respective mono-species cultures (data not shown). The reduced culture pH is likely a major underlying factor as to why S. mutans and probably, L. casei in dual-species biofilms had a significantly enhanced resistance to low pH than the respective mono-species cultures (Lemos et al., 2013). Interestingly, however, both L. casei and especially, S. mutans in dual-species displayed a drastic reduction in resistance to hydrogen peroxide, when compared to respective mono-species cultures. Co-current with the reduced stress tolerance was also the up-regulation of several genes known to play major roles in oxidative stress tolerance responses, as revealed by RNA-seq analysis. These include sodA, nox1, and tpx, which are all known to be inducible in response to oxidative stresses (Lemos et al., 2013). Among the up-regulated genes were also those for biosynthesis of mutanobactin, whose deficiency was shown to lead to major defects in oxidative stress tolerance response (Wu et al., 2010), and clpB and clpX for subunits of the Clp proteinase, members of the stress tolerance response pathways (Lemos et al., 2013). These results all suggest that S. mutans in dual-species with L. casei underwent some major oxidative stresses.
Both S. mutans and lactobacilli produce reactive oxygen species from oxygen metabolism, such as hydrogen peroxide, which can trigger the expression of an array of cytoprotective enzymes, including SodA and Nox1 (Ajdic et al., 2002; Bitoun et al., 2011; Lemos et al., 2013; Baker et al., 2014). However, unlike S. mutans, L. casei possesses two copies of genes for pyruvate oxidase, which in S. gordonii and S. sanguinis are responsible for hydrogen peroxide production (Kreth et al., 2005; Zheng et al., 2011a,b). Both SpxB expression and hydrogen peroxide production are modulated in response to cellular metabolic status via catabolite repression protein CcpA (Zheng et al., 2011a). As revealed by RNA-seq analysis, S. mutans seemingly alters sugar metabolism during growth with L. casei (see more details below), but it is unclear if similar phenomenon took place in L. casei in dual-species. It is likely that production of hydrogen peroxide by L. casei in the mixed-species consortium is up-regulated. If proven to be true, the elevated hydrogen peroxide will certainly be part of the underlying factors for the up-regulation of the stress genes and the increased susceptibility to hydrogen peroxide killing. Several oral lactobacilli are capable of producing bacteriocins effective against S. mutans causing cell envelope damage and death (Simark-Mattsson et al., 2007; Strahinic et al., 2007; Wu et al., 2015). However, none of the lactobacilli tested were shown to produce any visible antimicrobial activity against S. mutans under the conditions studied. On the other hand, S. mutans UA159 did produce some antimicrobials effective against L. fermentum, but not the other lactobacilli tested (Figure S4). Interestingly, of the genes identified as down-regulated in S. mutans, comX encodes the alternative Sigma factor, SigX that is well documented for its role in regulation of bacteriocins production (Khan et al., 2016). However, the actual impact of the altered SigX expression on L. casei and the mixed-species consortium as well as the underlying mechanism awaits further investigation.
Veillonellae are closely associated with S. mutans and other lactic acid bacteria in a community due to their reliance on lactic acids for carbon and energy sources. Consistent with Kara et al. on V. parvula (Kara et al., 2006, 2007; Luppens et al., 2008), the results presented here also showed that V. dispar alone did not form good biofilms, but co-cultivation of S. mutans and V. dispar yielded significantly more biofilms than the respective mono-species. Likely, such an enhancement can be in part attributed to the metabolic coupling between the two bacteria when grown together. In addition, as shown with L. casei, S. mutans GtfB and/or its adhesive products could also play a role.
Recent studies in several oral pathogens have generated evidence that growth in a community not only causes changes in community behavior, but may also elevate the pathogenic potential (Kara et al., 2006; Kuboniwa et al., 2009; Wen et al., 2010; Whitmore and Lamont, 2011; Falsetta et al., 2014; Sztajer et al., 2014; He et al., 2017). As revealed by RNA-seq analysis, co-cultivation with L. casei causes S. mutans to alter its transcriptional profile, including those involved in biofilm formation and oxidative stress responses. Of note, the down-regulation of gtfB and spaP (Table S2) by S. mutans in dual-species with L. casei is consistent with the results of our previous studies by RealTime-PCR (Wen et al., 2010). Interestingly, among the up-regulated genes are also those for Ftf and GtfD, and the down-regulated include a dextranase precursor. Unlike GtfB, GtfD produces primarily α[1,6]-linked water soluble glucose polymers, while Ftf synthesizes β(2,1 & 2,6)-linked fructose polymers, both of which are believed to primarily serve as storage of carbon and energy sources (Burne, 1998; Moye et al., 2014). These alterations suggest that S. mutans may undergo some major shifts in metabolic pathways when grown in consortium with L. casei. In support of this notion, several genes were identified (in <1.5-fold but still statistically significant) to encode proteins with roles in sugar metabolism, including ccpA (for carbon catabolite protein CcpA, a global regulator), pfl (for pyruvate formate lyase), pflA (pyruvate formate lyase activating enzyme), msm operon (SMU.876/80, for members of the multiple sugar metabolism system), and loci SMU.270/2 for ascorbate-specific PTS system EIIABC (Ajdic et al., 2002). The significant reduction of succinic acid and the increase of lactic acid in the culture medium of 24-h S. mutans-L. casei co-cultures, as compared to the respective mono-species cultures, also support the concept that interactions between these two major oral bacteria may lead to changes in sugar fermentation and acid production. Similar observations have also been made with S. mutans grown with V. parvula and more recently in S. mutans grown with C. albicans (Kara et al., 2006; He et al., 2017). However, how the sugar fermentation is altered and what effects of such alterations may have on L. casei and others in the consortium and ultimately, the pathogenicity of the community await further investigation.
In summary, these results have shown that Lactobacillus ssp. do not form good biofilms, although differences exist between different species. In mixed-species consortium, S. mutans can drastically enhance biofilm formation by all Lactobacillus spp. tested except L. fermentum, and both GtfB and P1 are major contributors to the enhanced biofilm formation by L. casei. On the other hand, S. mutans in dual-species with L. casei also displayed major reductions in oxidative stress tolerance and altered the expression of a large number of genes including those involved in biofilm formation and oxidative stress responses. These results suggest that interactions between S. mutans and Lactobacillus spp. vary between different species, although further investigation on what effects of such interactions may have on the plaque community and its cariogenicity is needed.
Author contributions
SL, AD, and AJ performed most of the experiments and data analyses; JB and XX worked on metabolite analysis; SF and PSGC performed RNA-seq and post-sequencing analysis; PWC, YL, HK and ZW conceived the overall plan and interpreted the data; ZW wrote the draft; AJ help with revisions; YL and ZW wrote the final version.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest. The reviewer JK and handling Editor declared their shared affiliation.
Acknowledgments
We want to thank Dr. Gary G. Xie at the Los Alamos National Laboratory Division of Biology and Bioinformatics, NM, USA for his insights on small RNA and related aspects. We would like to thank Dr. Robert A. Burne at the University of Florida, Gainesville, FL for kindly providing us the gtf and the ftf mutants used for this study. We want also thank Dr. Jose Lemos at the University of Florida, Gainesville, FL for the discussions during the writing of the manuscript. This work is supported in part by NIDCR grants DE19452 and DE25348 to ZW.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2017.00524/full#supplementary-material
References
- Ahn S. J., Wen Z. T., Brady L. J., Burne R. A. (2008). Characteristics of biofilm formation by Streptococcus mutans in the presence of saliva. Infect. Immun. 76, 4259–4268. 10.1128/IAI.00422-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ajdic D., McShan W. M., McLaughlin R. E., Savic G., Chang J., Carson M. B., et al. (2002). Genome sequence of Streptococcus mutans UA159, a cariogenic dental pathogen. Proc. Natl. Acad. Sci. U.S.A. 99, 14434–14439. 10.1073/pnas.172501299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ausubel F. M., Brent R., Kingston R. E., Moore D. D., Seidman J. G., Smith J. A., et al. (eds.). (2010). Current Protocols in Molecular Biology. New York, NY: John Wiley and Sons, Inc. [Google Scholar]
- Badet C., Thebaud N. B. (2008). Ecology of lactobacilli in the oral cavity: a review of literature. Open Microbiol. J. 2, 38–48. 10.2174/1874285800802010038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker J. L., Derr A. M., Karuppaiah K., MacGilvray M. E., Kajfasz J. K., Faustoferri R. C., et al. (2014). Streptococcus mutans NADH oxidase lies at the intersection of overlapping regulons controlled by oxygen and NAD+ levels. J. Bacteriol. 196, 2166–2177. 10.1128/JB.01542-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banas J. A., Fountain T. L., Mazurkiewicz J. E., Sun K., Margaret Vickerman M. (2007). Streptococcus mutans glucan-binding protein-A affects Streptococcus gordonii biofilm architecture. FEMS Microbiol. Lett. 267, 80–88. 10.1111/j.1574-6968.2006.00557.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bitoun J. P., Liao S., Yao X., Xie G. G., Wen Z. T. (2012). The redox-sensing regulator rex modulates central carbon metabolism, stress tolerance response and biofilm formation by Streptococcus mutans. PLoS ONE 7:e44766. 10.1371/journal.pone.0044766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bitoun J. P., Nguyen A. H., Fan Y., Burne R. A., Wen Z. T. (2011). Transcriptional repressor Rex is involved in regulation of oxidative stress response and biofilm formation by Streptococcus mutans. FEMS Microbiol. Lett. 320, 110–117. 10.1111/j.1574-6968.2011.02293.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowen W. H., Koo H. (2011). Biology of Streptococcus mutans-derived glucosyltransferases: role in extracellular matrix formation of cariogenic biofilms. Caries Res. 45, 69–86. 10.1159/000324598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowen W. H., Schilling K., Giertsen E., Pearson S., Lee S. F., Bleiweis A., et al. (1991). Role of a cell surface-associated protein in adherence and dental caries. Infect. Immun. 59, 4604–4609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brady L. J., Maddocks S. E., Larson M. R., Forsgren N., Persson K., Deivanayagam C. C., et al. (2011). The changing faces of Streptococcus antigen I/II polypeptide family adhesins. Mol. Microbiol. 77, 276–286. 10.1111/j.1365-2958.2010.07212.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burne R. A. (1998). Oral streptococci…products of their environment. J. Dent. Res. 77, 445–452. 10.1177/00220345980770030301 [DOI] [PubMed] [Google Scholar]
- Burne R. A., Abranches J., Ahn S. J., Lemos J. A., Wen Z. T., Zeng L. (2011). Functional Genomics of Streptococcus mutans, in Oral Microbial Communities: Genomic Inquires and Interspecies Communication, ed Kolenbrander P. E. (Washington, DC: ASM Press; ), 185–204. [Google Scholar]
- Burne R. A., Zeng L., Ahn S. J., Palmer S. R., Liu Y., Lefebure T., et al. (2012). Progress dissecting the oral microbiome in caries and health. Adv. Dent. Res. 24, 77–80. 10.1177/0022034512449462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caufield P. W., Li Y., Dasanayake A., Saxena D. (2007). Diversity of lactobacilli in the oral cavities of young women with dental caries. Caries Res. 41, 2–8. 10.1159/000096099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caufield P. W., Schon C. N., Saraithong P., Li Y., Argimon S. (2015). Oral lactobacilli and dental caries: a model for niche adaptation in humans. J. Dent. Res. 94(9 Suppl.), 110S−118S. 10.1177/0022034515576052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crowley P. J., Brady L. J., Piacentini D. A., Bleiweis A. S. (1993). Identification of a salivary agglutinin-binding domain within cell surface adhesin P1 of Streptococcus mutans. Infect. Immun. 61, 1547–1552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellepola K., Liu Y., Cao T., Koo H., Seneviratne C. J. (2017). Bacterial GtfB augments Candida albicans accumulation in cross-kingdom biofilms. J. Dent. Res. 96, 1129–1135. 10.1177/0022034517714414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elli M., Zink R., Rytz A., Reniero R., Morelli L. (2000). Iron requirement of Lactobacillus spp. in completely chemically defined growth media. J. Appl. Microbiol. 88, 695–703. 10.1046/j.1365-2672.2000.01013.x [DOI] [PubMed] [Google Scholar]
- Falsetta M. L., Klein M. I., Colonne P. M., Scott-Anne K., Gregoire S., Pai C. H., et al. (2014). Symbiotic relationship between Streptococcus mutans and Candida albicans synergizes virulence of plaque biofilms in vivo. Infect. Immun. 82, 1968–1981. 10.1128/IAI.00087-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan Y., Wen Z. T., Liao S., Lallier T., Hagan J. L., Twomley J. T., et al. (2012). Novel amelogenin-releasing hydrogel for remineralization of enamel artificial caries. J. Bioact. Compat. Polym. 27, 585–603. 10.1177/0883911512458050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filoche S. K., Anderson S. A., Sissons C. H. (2004). Biofilm growth of Lactobacillus species is promoted by Actinomyces species and Streptococcus mutans. Oral Microbiol. Immunol. 19, 322–326. 10.1111/j.1399-302x.2004.00164.x [DOI] [PubMed] [Google Scholar]
- Fitzgerald R. J., Fitzgerald D. B., Adams B. O., Duany L. F. (1980). Cariogenicity of human oral lactobacilli in hamsters. J. Dent. Res. 59, 832–837. 10.1177/00220345800590051501 [DOI] [PubMed] [Google Scholar]
- Gregoire S., Xiao J., Silva B. B., Gonzalez I., Agidi P. S., Klein M. I., et al. (2011). Role of Glucosyltransferase B in Interactions of Candida albicans with Streptococcus mutans and with an Experimental Pellicle on Hydroxyapatite Surfaces. Appl. Environ. Microbiol. 77, 6357–6367. 10.1128/AEM.05203-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross E. L., Beall C. J., Kutsch S. R., Firestone N. D., Leys E. J., Griffen A. L. (2012). Beyond Streptococcus mutans: dental caries onset linked to multiple species by 16S rRNA community analysis. PLoS ONE 7:e47722. 10.1371/journal.pone.0047722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajishengallis E., Parsaei Y., Klein M. I., Koo H. (2017). Advances in the microbial etiology and pathogenesis of early childhood caries. Mol. Oral Microbiol. 32, 24–34. 10.1111/omi.12152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamada S., Slade H. D. (1980). Biology, immunology, and cariogenicity of Streptococcus mutans. Microbiol. Rev. 44, 331–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He J., Kim D., Zhou X., Ahn S. J., Burne R. A., Richards V. P., et al. (2017). RNA-Seq reveals enhanced sugar metabolism in Streptococcus mutans Co-cultured with Candida albicans within mixed-species biofilms. Front. Microbiol. 8:1036. 10.3389/fmicb.2017.01036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang G., Liu Y., Kim D., Li Y., Krysan D. J., Koo H. (2017). Candida albicans mannans mediate Streptococcus mutans exoenzyme GtfB binding to modulate cross-kingdom biofilm development in vivo. PLoS Pathog. 13:e1006407. 10.1371/journal.ppat.1006407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwami Y., Abbe K., Takahashi-Abbe S., Yamada T. (1992). Acid production by streptococci growing at low pH in a chemostat under anaerobic conditions. Oral Microbiol. Immunol. 7, 304–308. 10.1111/j.1399-302X.1992.tb00593.x [DOI] [PubMed] [Google Scholar]
- Iwami Y., Yamada T. (1985). Regulation of glycolytic rate in Streptococcus sanguis grown under glucose-limited and glucose-excess conditions in a chemostat. Infect. Immun. 50, 378–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakubovics N. S., Stromberg N., van Dolleweerd C. J., Kelly C. G., Jenkinson H. F. (2005). Differential binding specificities of oral streptococcal antigen I/II family adhesins for human or bacterial ligands. Mol. Microbiol. 55, 1591–1605. 10.1111/j.1365-2958.2005.04495.x [DOI] [PubMed] [Google Scholar]
- Jenkinson H. F. (2011). Beyond the oral microbiome. Environ. Microbiol. 13, 3077–3087. 10.1111/j.1462-2920.2011.02573.x [DOI] [PubMed] [Google Scholar]
- Kara D., Luppens S. B., Cate J. M. (2006). Differences between single- and dual-species biofilms of Streptococcus mutans and Veillonella parvula in growth, acidogenicity and susceptibility to chlorhexidine. Eur. J. Oral Sci. 114, 58–63. 10.1111/j.1600-0722.2006.00262.x [DOI] [PubMed] [Google Scholar]
- Kara D., Luppens S. B., van Marle J., Ozok R., ten Cate J. M. (2007). Microstructural differences between single-species and dual-species biofilms of Streptococcus mutans and Veillonella parvula, before and after exposure to chlorhexidine. FEMS Microbiol. Lett. 271, 90–97. 10.1111/j.1574-6968.2007.00701.x [DOI] [PubMed] [Google Scholar]
- Khan R., Rukke H. V., Hovik H., Amdal H. A., Chen T., Morrison D. A., et al. (2016). Comprehensive transcriptome profiles of Streptococcus mutans UA159 Map Core streptococcal competence genes. mSystems 1,e00038-15. 10.1128/mSystems.00038-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D., Langmead B., Salzberg S. L. (2015). HISAT: a fast spliced aligner with low memory requirements. Nat. Methods 12, 357–360. 10.1038/nmeth.3317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D., Sengupta A., Niepa T. H., Lee B. H., Weljie A., Freitas-Blanco V. S., et al. (2017). Candida albicans stimulates Streptococcus mutans microcolony development via cross-kingdom biofilm-derived metabolites. Sci. Rep. 7:41332. 10.1038/srep41332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreth J., Merritt J., Shi W., Qi F. (2005). Competition and coexistence between Streptococcus mutans and Streptococcus sanguinis in the dental biofilm. J. Bacteriol. 187, 7193–7203. 10.1128/JB.187.21.7193-7203.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuboniwa M., Hendrickson E. L., Xia Q., Wang T., Xie H., Hackett M., et al. (2009). Proteomics of Porphyromonas gingivalis within a model oral microbial community. BMC Microbiol. 9:98. 10.1186/1471-2180-9-98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuramitsu H. K., He X., Lux R., Anderson M. H., Shi W. (2007). Interspecies interactions within oral microbial communities. Microbiol. Mol. Biol. Rev. 71, 653–670. 10.1128/MMBR.00024-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larson M. R., Rajashankar K. R., Crowley P. J., Kelly C., Mitchell T. J., Brady L. J., et al. (2011). Crystal structure of the C-terminal region of Streptococcus mutans antigen I/II and characterization of salivary agglutinin adherence domains. J. Biol. Chem. 286, 21657–21666. 10.1074/jbc.M111.231100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemos J. A., Quivey R. G., Jr., Koo H., Abranches J. (2013). Streptococcus mutans: a new Gram-positive paradigm? Microbiology 159(Pt 3), 436–445. 10.1099/mic.0.066134-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Merritt J., Qi F. (2011). Genetic transformation of Veillonella parvula. FEMS Microbiol. Lett. 322, 138–144. 10.1111/j.1574-6968.2011.02344.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo C. C., Chain P. S. (2014). Rapid evaluation and quality control of next generation sequencing data with FaQCs. BMC Bioinformatics 15:366. 10.1186/s12859-014-0366-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loo C. Y., Corliss D. A., Ganeshkumar N. (2000). Streptococcus gordonii biofilm formation: identification of genes that code for biofilm phenotypes. J. Bacteriol. 182, 1374–1382. 10.1128/JB.182.5.1374-1382.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love M. I., Huber W., Anders S. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550. 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luppens S. B., Kara D., Bandounas L., Jonker M. J., Wittink F. R., Bruning O., et al. (2008). Effect of Veillonella parvula on the antimicrobial resistance and gene expression of Streptococcus mutans grown in a dual-species biofilm. Oral Microbiol. Immunol. 23, 183–189. 10.1111/j.1399-302X.2007.00409.x [DOI] [PubMed] [Google Scholar]
- Maekawa S., Suzuki A., Sugano S., Suzuki Y. (2014). RNA sequencing: from sample preparation to analysis. Methods Mol. Biol. 1164, 51–65. 10.1007/978-1-4939-0805-9_6 [DOI] [PubMed] [Google Scholar]
- Merritt J., Qi F. (2012). The mutacins of Streptococcus mutans: regulation and ecology. Mol. Oral Microbiol. 27, 57–69. 10.1111/j.2041-1014.2011.00634.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michalek S. M., Hirasawa M., Kiyono H., Ochiai K., McGhee J. R. (1981). Oral ecology and virulence of Lactobacillus casei and Streptococcus mutans in gnotobiotic rats. Infect. Immun. 33, 690–696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moye Z. D., Zeng L., Burne R. A. (2014). Fueling the caries process: carbohydrate metabolism and gene regulation by Streptococcus mutans. J. Oral Microbiol. 6, 1–15. 10.3402/jom.v6.24878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owen O. W. (1949). A study of bacterial counts (lactobacilli) in saliva related to orthodontic appliances; a preliminary report. Am. J. Orthod. 35, 672–678. 10.1016/0002-9416(49)90123-2 [DOI] [PubMed] [Google Scholar]
- Robinson M. D., McCarthy D. J., Smyth G. K. (2010). edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140. 10.1093/bioinformatics/btp616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savijoki K., Suokko A., Palva A., Varmanen P. (2006). New convenient defined media for [S]methionine labelling and proteomic analyses of probiotic lactobacilli. Lett. Appl. Microbiol. 42, 202–209. 10.1111/j.1472-765X.2005.01853.x [DOI] [PubMed] [Google Scholar]
- Shu M., Browngardt C. M., Chen Y. Y., Burne R. A. (2003). Role of urease enzymes in stability of a 10-species oral biofilm consortium cultivated in a constant-depth film fermenter. Infect. Immun. 71, 7188–7192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simark-Mattsson C., Emilson C. G., Hakansson E. G., Jacobsson C., Roos K., Holm S. (2007). Lactobacillus-mediated interference of mutans streptococci in caries-free vs. caries-active subjects. Eur. J. Oral Sci. 115, 308–314. 10.1111/j.1600-0722.2007.00458.x [DOI] [PubMed] [Google Scholar]
- Strahinic I., Busarcevic M., Pavlica D., Milasin J., Golic N., Topisirovic L. (2007). Molecular and biochemical characterizations of human oral lactobacilli as putative probiotic candidates. Oral Microbiol. Immunol. 22, 111–117. 10.1111/j.1399-302X.2007.00331.x [DOI] [PubMed] [Google Scholar]
- Sullan R. M., Li J. K., Crowley P. J., Brady L. J., Dufrene Y. F. (2015). Binding forces of Streptococcus mutans P1 adhesin. ACS Nano 9, 1448–1460. 10.1021/nn5058886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sztajer H., Szafranski S. P., Tomasch J., Reck M., Nimtz M., Rohde M., et al. (2014). Cross-feeding and interkingdom communication in dual-species biofilms of Streptococcus mutans and Candida albicans. ISME J. 8, 2256–2271. 10.1038/ismej.2014.73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terleckyj B., Willett N. P., Shockman G. D. (1975). Growth of several cariogenic strains of oral streptococci in a chemically defined medium. Infect. Immun. 11, 649–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vacca-Smith A. M., Bowen W. H. (1998). Binding properties of streptococcal glucosyltransferases for hydroxyapatite, saliva-coated hydroxyapatite, and bacterial surfaces. Arch. Oral Biol. 43, 103–110. 10.1016/S0003-9969(97)00111-8 [DOI] [PubMed] [Google Scholar]
- Van Houte J., Gibbons R. J., Pulkkinen A. J. (1972). Ecology of human oral lactobacilli. Infect. Immun. 6, 723–729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Houte J., Sansome C., Joshipura K., Kent R. (1991). Mutans streptococci and non-mutans streptococci acidogenic at low pH, and in vitro acidogenic potential of dental plaque in two different areas of human dentition. J. Dent. Res. 70, 1503–1507. 10.1177/00220345910700120601 [DOI] [PubMed] [Google Scholar]
- Wegkamp A., de Vos W. M., Smid E. J. (2009). Folate overproduction in Lactobacillus plantarum WCFS1 causes methotrexate resistance. FEMS Microbiol. Lett. 297, 261–265. 10.1111/j.1574-6968.2009.01690.x [DOI] [PubMed] [Google Scholar]
- Wen Z. T., Baker H. V., Burne R. A. (2006). Influence of BrpA on critical virulence attributes of Streptococcus mutans. J. Bacteriol. 188, 2983–2992. 10.1128/JB.188.8.2983-2992.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen Z. T., Burne R. A. (2002). Functional genomics approach to identifying genes required for biofilm development by Streptococcus mutans. Appl. Environ. Microbiol. 68, 1196–1203. 10.1128/AEM.68.3.1196-1203.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen Z. T., Burne R. A. (2004). LuxS-mediated signaling in Streptococcus mutans is involved in regulation of acid and oxidative stress tolerance and biofilm formation. J. Bacteriol. 186, 2682–2691. 10.1128/JB.186.9.2682-2691.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen Z. T., Yates D., Ahn S. J., Burne R. A. (2010). Biofilm formation and virulence expression by Streptococcus mutans are altered when grown in dual-species model. BMC Microbiol. 10, 111. 10.1186/1471-2180-10-111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitmore S. E., Lamont R. J. (2011). The pathogenic persona of community-associated oral streptococci. Mol. Microbiol. 81, 305–314. 10.1111/j.1365-2958.2011.07707.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C., Cichewicz R., Li Y., Liu J., Roe B., Ferretti J., et al. (2010). Genomic island TnSmu2 of Streptococcus mutans harbors a nonribosomal peptide synthetase-polyketide synthase gene cluster responsible for the biosynthesis of pigments involved in oxygen and H2O2 tolerance. Appl. Environ. Microbiol. 76, 5815–5826. 10.1128/AEM.03079-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C. C., Lin C. T., Wu C. Y., Peng W. S., Lee M. J., Tsai Y. C. (2015). Inhibitory effect of Lactobacillus salivarius on Streptococcus mutans biofilm formation. Mol. Oral Microbiol. 30, 16–26. 10.1111/omi.12063 [DOI] [PubMed] [Google Scholar]
- Yamada T., Takahashi-Abbe S., Abbe K. (1985). Effects of oxygen on pyruvate formate-lyase in situ and sugar metabolism of Streptococcus mutans and Streptococcus sanguis. Infect. Immun. 47, 129–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng L., Chen Z., Itzek A., Ashby M., Kreth J. (2011a). Catabolite control protein A controls hydrogen peroxide production and cell death in Streptococcus sanguinis. J. Bacteriol. 193, 516–526. 10.1128/JB.01131-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng L., Itzek A., Chen Z., Kreth J. (2011b). Environmental influences on competitive hydrogen peroxide production in Streptococcus gordonii. Appl. Environ. Microbiol. 77, 4318–4328. 10.1128/AEM.00309-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.