Abstract
Spatiotemporal interrogation of signal transduction at the single cell level is necessary to answer a host of important biological questions. This protocol describes a nanotechnology-based single-cell and single-molecule perturbation tool, termed mechanogenetics, that enables precise spatial and mechanical control over genetically encoded cell-surface receptors in live cells. The key components of this tool are a magnetoplasmonic nanoparticle (MPN) actuator that delivers defined spatial and mechanical cues to receptors through target-specific one-to-one engagement, and a micromagnetic tweezers (µMT) that remotely controls the magnitude of force exerted on a single MPN. This protocol consists of three major parts: 1) Preparation of small (40–50 nm), monovalent, and modular MPN probes, including chemical synthesis of MPN nanoparticles, conjugation with DNA, and purification of monovalent MPNs, 2) µMT setup and single particle force calibration as a function of µMT-to-MPN distance, and 3) control of spatial and mechanical properties of targeted mechanosensitive receptors in live cells by adjusting the µMT-to-MPN distance. Using benzylguanine-functionalized MPNs and model cell lines expressing either SNAP-tagged Notch or vascular endothelial cadherin (VE-cadherin), we provide stepwise instructions for mechanogenetic control of receptor clustering or mechanical activation. The ability of this method to differentially control spatial and mechanical inputs to targeted receptors makes it particularly useful for interrogating the differential contributions of each individual cue on cell signaling. The entire procedure takes up to 1 week. This tool has versatile applications spanning the basic to applied biological sciences.
Keywords: Mechanogenetics, Magnetoplasmonic nanoparticles, Monovalent conjugation, Mechanical signaling, Biophysical methods, Single cell perturbation, Magnetic nanoparticles, Magnetic tweezers
INTRODUCTION
Tools for analyzing and perturbing selected biochemical processes at the single cell and single molecule levels allow for interrogation of the mechanisms underlying complex cellular signaling activities. Recent advances in this area have contributed to a paradigm shift in cell biology, allowing the properties of biomolecules to be studied in the context of a larger and more integrated cellular signaling system1–5. Using techniques such as optogenetics, molecules or cells can be perturbed selectively and systematically, at a desired location and time1–5. By measuring the cellular response to these controlled perturbations, existing models can be tested and new biological principles can be revealed at both the molecular and systems-level. This allows the investigator to deconstruct and decode the working mechanisms of complex receptor signaling processes. However, tools for manipulating mechanical inputs to cell receptors have, until recently, remained elusive.
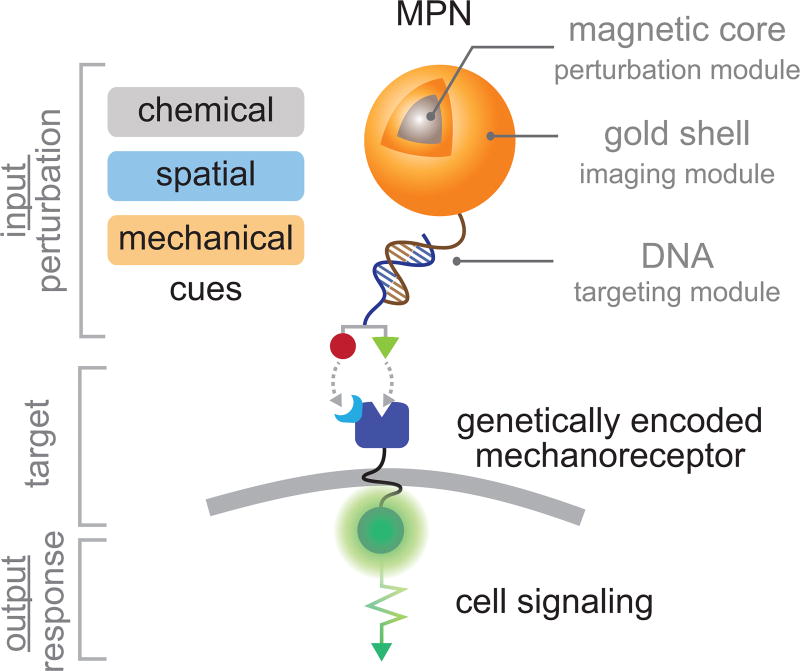
Colloidal nanoparticles have recently emerged as probes for the selective perturbation of biomolecular processes6–16. Their size is comparable to that of cell surface receptors, they are easily conjugated with targeting domains, and they can uniquely deliver a variety of physical perturbations to targeted receptors or cells not accessible to small molecules or most genetically encoded probes. For example, magnetic and metal nanoparticles have provided an unprecedented means of selectively and quantitatively delivering spatial7–10, mechanical11,12, or thermal13–17 perturbation to cell surface receptors and ion channels. Additionally, these perturbation components can be further assembled with other functional modules for chemical targeting and imaging into a single integrated platform, providing a complete tool for perturbation biology with single-cell and single-molecule resolution. We recently developed such a modular nanoprobe based on magnetoplasmonic nanoparticles (MPNs), comprising a Zn-doped ferrite magnetic-core, a plasmonic-shell, and an oligonucleotide-based targeting module (Fig. 1)12. Importantly, each module of the MPN can be modified to alter the magnetic, light scattering, or targeting properties. We further demonstrated cell labeling via Watson-Crick hybridization between the monovalent MPN probes and genetically encoded mechanoreceptors, and mechanogenetic control of the targeted receptors to promote their subcellular spatial localization or mechanical activation12. By measuring cellular responses to these controlled stimuli using fluorescence proteins and reporter genes, we also identified the differential roles of receptor clustering and mechanical force in Notch and VE-cadherin signaling12.
Figure 1. Schematic illustration of magnetoplasmonic nanoparticles (MPNs).
A MPN is comprised of a magnetic-core (perturbation module), a plasmonic-shell (imaging and conjugation module), and a functionalized oligonucleotide (chemical targeting module).
These mechanogenetic tools can be generalized to tackle many outstanding questions related to the dynamics of signal transduction. These include elucidating receptor activation mechanisms, interrogating the spatiotemporal dynamics of cell signaling, and single cell transcriptional control. Here, we present a detailed and optimized protocol for synthesis, bioconjugation, and mechanogenetic applications of MPN probes so as to open their application to researchers from diverse backgrounds.
Development and overview of the protocol
An ideal mechanogenetic probe selectively targets and quantitatively perturbs cell surface receptors at a desired location and time. To meet these requirements, the following critical considerations in probe design should be taken into account:
Probe size and quality
Probes that are too large (> 100 nm) limit target accessibility, perturb receptor diffusion, and alter the mechanical properties of the receptor microenvironment (e.g. membrane)12. On the other hand, too small of a probe (< 10 nm) cannot provide pico Newton (pN)-forces or single particle light scattering signals18. Probes should also be of high quality, crystallinity, and monodisperse in size for generating strong and homogeneous mechanical and optical signals.
Monovalency
Multivalent probes (e.g. probes containing multiple targeting ligands) result in uncontrolled receptor crosslinking, leading to non-specific change in receptor diffusion dynamics19,20, translocation21, and signal transduction12. Additionally, such probes are unable to deliver a defined force to single receptors, as the force generated by a single probe is distributed across multiple receptors. Monovalent probes ensure one-to-one probe-receptor engagement, eliminating problems associated with probe-multivalency12.
Functional Modularity
Targeting of different sites on the same protein or multiple different proteins can be achieved using modular probes bearing interchangeable targeting domains. This allows for chemical control of receptor activity (e.g. agonistic, antagonistic, or non-stimulating) and provides more general applicability of the probes to a wider range of cell surface receptors.
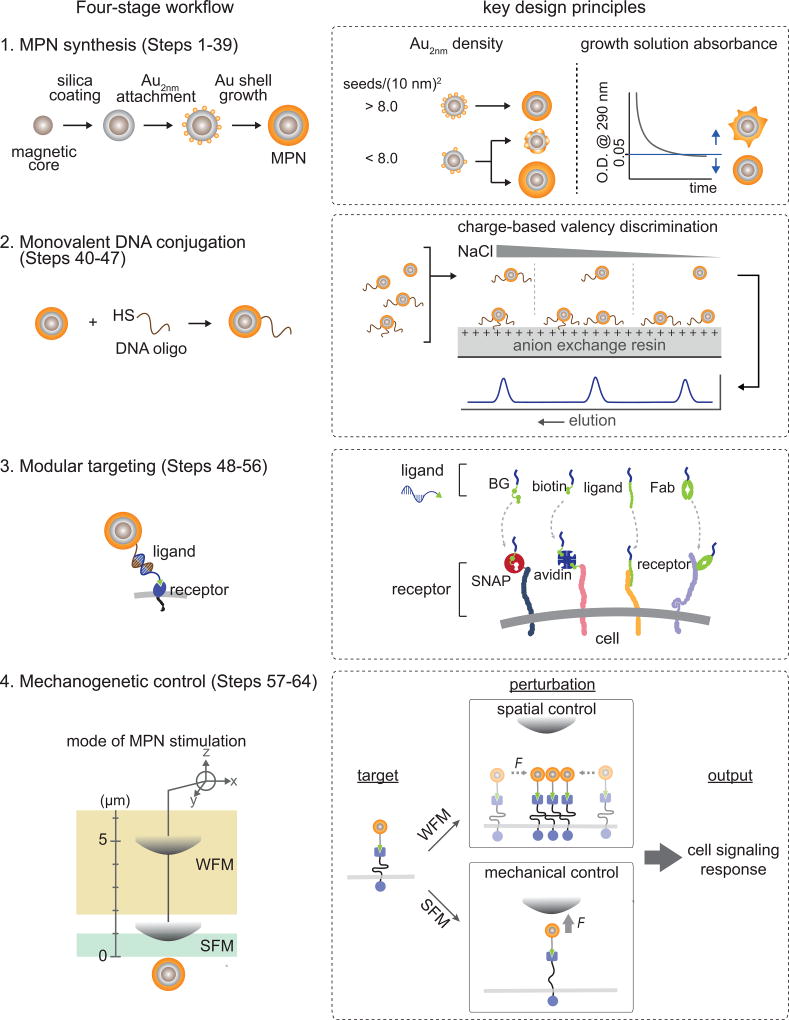
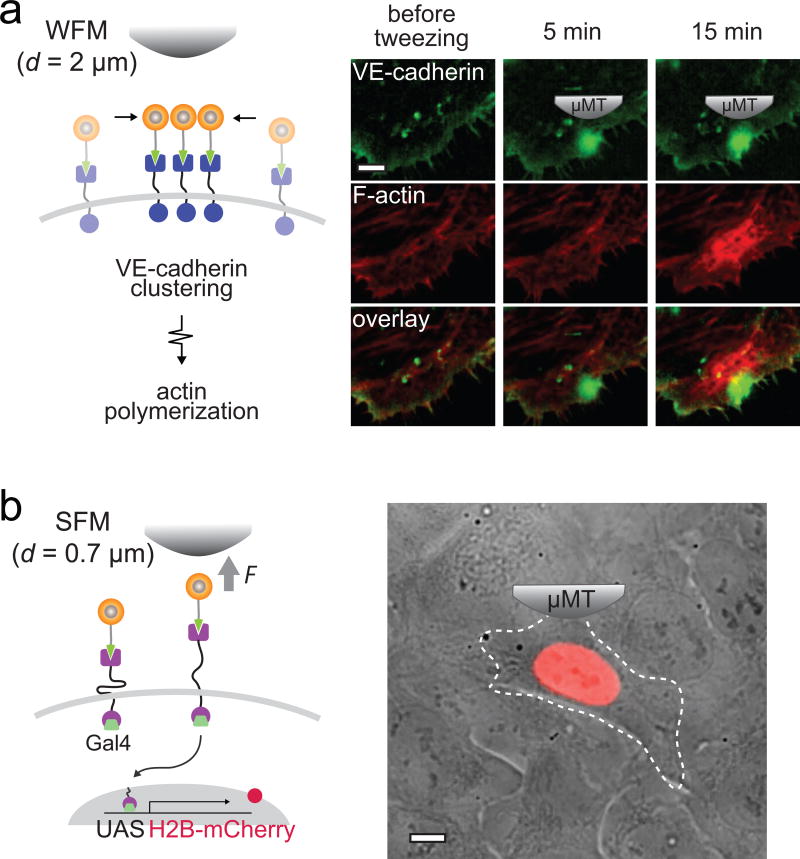
The concept and methodology elaborated here is built upon our previous efforts to develop MPN-based mechanogenetic tools12. The mechanogenetic tool leverages nanofabrication techniques to synthesize multifunctional MPN probes that integrate the perturbation, imaging, and targeting modules without altering probe size, monovalent targeting, and force generating profiles. Key technical challenges have been addressed through sequential nanocrystal growth strategy, modular oligonucleotide conjugation and DNA base-pairing, and charge-based valency discrimination. We have characterized MPN growth kinetics, optical properties and force-generation as a function of MPN size, and the extent to which MPN perturb cells. In this protocol we present a comprehensive stepwise procedure to replicate our MPN-based mechanogenetic system for spatial and mechanical control of SNAP-tagged cell surface proteins. The experimental workflow is outlined in Figure 2, which includes MPN synthesis, monovalent DNA conjugation, and modular targeting. By controlling force application as a function of µMT-to-MPN distance (d, Box 1), MPNs can provide spatial (with a weak force mode, WFM; force exertion on a single MPN < 1 pN) and mechanical (with a strong force mode, SFM; force exertion on a single MPN > 5 pN) perturbation of the targeted receptors.
Figure 2. Schematic workflow of the protocol (left panel), key design principles (right panel).
Synthesis of small and monodisperse MPNs is achieved by sequential nanoparticle growth of a magnetic core, dielectric inner shell, and plasmonic outer shell. Dense loading of Au2nm seeds and sufficiently long incubation of the shell growth are essential for the uniform shell formation (Stage 1, Steps 1–39). A targeting functionality is introduced by conjugation of MPNs with thiolated DNA via Au-thiol chemistry. Monovalently conjugated MPNs are selectively isolated via charge-based valency discrimination with anion exchange HPLC (Stage 2, Steps 40–47). The MPNs can target cell surface receptors with a diverse set of receptors via modular targeting and Watson-Crick hybridization (Stage 3, Steps 48–56). Coupled with an external micro magnetic tweezers (µMT), the targeted MPNs can provide two different modes of perturbation to the receptors as a function of distance between the MPN and the µMT: The SFM and WFM modes can be used for spatial and mechanical control, respectively (Stage 4, Steps 57–64).
Box 1 | Calibration of force exertion on a single MPN ● TIMING ~6 h.
We suggest two approaches to measure force intensities of single MPNs. To determine the force generated by an MPN located at > 1 µm distance from µMT, the viscous drag of the nanoparticles can be utilized (Option A). The velocity of magnetic nanoparticles should be experimentally determined. Because velocity of nanoparticles is too fast for optical imaging once the particles are within 1 µm of the magnet, TGT force sensors with defined rupture forces can be utilized (Option B). In this case, the µMT gradually approaches a MPN-TGT duplex immobilized on a coverslip until the initial detachment event is observed.
Advantages, limitations, and potential improvements to the protocol
A key advantage of the MPN-based mechanogenetic tool is the capacity to provide both spatial and mechanical perturbations to targeted receptors, while effectively decoupling each cue by using different modes of stimulation. There are several techniques that provide single modes of perturbation of targeted receptors2,7–11,13–17,41,42, but no technologies exist allowing both the spatial and mechanical control, particularly on the single molecule and single cell levels. This unique ability of MPNs enables interrogation of the differential contributions of, for example, ligand binding, receptor clustering, receptor location, and force on downstream signaling. Importantly, each of these variables can be modified independently, while keeping other experimental conditions identical, hence minimizing experimental artifacts associated with use of different methodologies or batch-to-batch heterogeneity. For example, this allows direct comparison of receptor responses to two different stimuli on a single cell or on two neighboring cells in a cell population.
Another critical advantage of MPNs is their negligible nonspecific perturbation of cells when compared to traditional force-microscopy tools or multivalent nanoparticle probes. Microprobe-based force microscopy tools – including atomic force microscopy (AFM), magnetic tweezers, and optical tweezers – can deliver a defined force to a single, specific biomolecule22,23. However, due to the large size and multivalent character of microprobes, these tools frequently influence the dynamic properties of targeted receptors (e.g. immobilization of receptors due to probe viscous drag) and mechanical environments surrounding the probe (e.g. cell membrane tension), leading to nonspecific cell signal activation or changes in cell behavior12. Multivalent magnetic nanoparticles that have been previously used for receptor spatial control, also significantly alter the dynamic properties of the targeted receptors12,21. These probes crosslink receptors nonspecifically, slowing receptor diffusion and enhancing receptor endocytosis24,25. More significantly, the poorly defined number of proteins linked to single particles prevents the delivery a defined force to the targeted proteins. The protocol presented here produces small (40–50 nm) and monovalent MPNs, eliminating the nonspecific perturbation associated with probe size and valency.
Rapid spatial control with the MPN-based tool is another advantage. Upon application of a focused magnetic field gradient, protein translocation is induced by force-induced convection rather than by passive diffusion followed by the trapping of proteins, as in optogenetic methods26. This property of MPNs is advantageous for directing the reorganization of confined or slowly diffusing membrane proteins such as Notch and Cadherin.
A major drawback of the current MPN-based mechanogenetic tool is the limited accessibility of particles to intracellular targets. Unlike optogenetic methods that only require stimulus-sensitive proteins, this tool additionally requires targeted delivery of MPNs to mechanosensitive proteins. Traditional intracellular delivery methods of nanoparticles through transfection and electroporation are not ideal because of low delivery efficiency, poor endosomal escape, and particle aggregation27,28. Microinjection offers efficient intracellular delivery of particles, and has been successfully applied for localizing soluble proteins9,10,29. However, removal of excess particles from the cytosol is difficult with microinjection methods, which is an additional requirement of the MPN-mechanogenetic tool. Genetic engineering approaches that endogenously expresses magnetic particles inside a protein complex can be a promising alternative approach15,16,30, but significant improvements in magnetic properties of the small or poorly crystalline particles expressed endogenously will be necessary for mechanogenetic applications18. Controlling proteins presenting at the basal cell surface is also difficult with the current version of the µMT in the MPN-based mechanogenetic tool. Implementing micropatterned superparamagnets on a cell culture substrate can potentially resolve the problem of tweezer accessibility29,31.
Another drawback of the MPN-based mechanogenetics is its limited force resolution. The MPN exerts forces of up to tens of pN to target receptors. However, under the µMT-SFM (d ≤ 1 µm), slight changes of the µMT-to-MPN distance caused by experimental noise can lead to significant changes in MPN-force exertion, because the force-exertion changes steeply as a function of d. Hence, precise force control within the pN range is limited. Use of MPNs with higher magnetic moments can shift the operating distance toward the long-distance range and thus significantly improve force control. One strategy is to replace the Zn-doped ferrite core with other magnetic materials with higher susceptibility (e.g. Fe, Co)32–34. Alternatively, larger Zn-doped ferrite cores can increase the magnetic moment without significantly increasing the total MPN size if the thickness of the silica and gold shells are reduced. For example, use of a 30 nm magnetic core may generate a 10-times stronger force than our current version of the MPN with a 13 nm core35.
The spatial resolution of the µMT may also be improved further. The current µMT design allows focused pico-newton force generation with a resolution of a few-micrometers, providing subcellular activation of multiple receptors in a densely labeled cell or single-molecule perturbation in a sparsely labeled cell12. Higher resolution spatial control of mechanical force generation may provide additional utility of the tool, such as single-molecule and quantitative activation of multiple different receptors in live cells. Cutting-edge nanofabrication technologies will facilitate the development of more precise magnetic tweezers and the realization of these new applications36.
To summarize the advantages and limitations of the mechanogenetic tool presented here, we provide a direct comparison with other tools in Table 1, based on our studies and others.
Table 1.
Comparison of MPN-based mechanogenetics with existing techniques.
| this protocol | existing techniques | |||
|---|---|---|---|---|
|
|
|
|||
| MPN-based mechanogenetcs | optogenetics | microprobe-based force microscopy (magnetic, optical tweezers) | substrate based mechanical stimulation | |
|
| ||||
| input | gradient magnetic field | light | gradient magnetic field/ light/ mechanical force | mechanical force, magnetic field |
|
| ||||
| applicable target proteins | mechanosensitive proteins | light-sensitive proteins | mechanosensitive proteins | mechanosensitive proteins |
|
| ||||
| advantages |
|
|
|
|
|
| ||||
| limitations |
|
|
low spatiotemporal resolution | |
Applications of the method
One of the primary applications of the MPN-based mechanogenetic tool is to interrogate the chemical, spatial, and mechanical responses of receptors during signal transduction. We previously applied this tool for manipulating the spatial and mechanical properties of two mechanoreceptors (i.e. Notch and Cadherin) at single molecule and single cell resolution, where we clarified the activation mechanism of Notch and described the differential roles of spatial and mechanical cues on Cadherin junction formation, respectively12. Use of the MPN-based mechanogenetic tool is not limited to these specific applications but has numerous applications across diverse biological systems. In principle, any cell surface receptor can be targeted with MPNs to investigate the differential roles of spatial and mechanical cues on their signaling properties. Receptors and channels exhibiting mechanosensitivity such as NOMPC37, PIEZO38,39, T cell receptors (TCRs)40–42 integrin43, selectin44,45, and CAM46 are potential future applications.
MPNs provide an alternative platform for single molecule force microscopy and for investigating ligand-receptor complexes in live cells47. For example, dissociation rate constants (koff) of a respective ligand-receptor pair can be determined by simply measuring bond lifetime (1/koff) of a monovalent MPN-ligand conjugate bound to a target receptor on the cell surface. Analysis of single molecule detachment kinetics as a function of force applied will characterize the nature of ligand-receptor binding, i.e. slip vs. catch bond45,48. Again, small and monovalent MPNs effectively eliminate probe-induced modification of the ligand-receptor properties (e.g. receptor crosslinking and nonspecific interaction between a microbead and the cell surface)12.
This tool will also be useful for systems biology applications by providing the ability to perturb transcriptional programs of target cells in a population selectively and quantitatively. We previously showed that, in a model UAS-Gal4 system, transcriptional profiles of targeted cells can be systematically tuned with MPNs at any desired location, time, and quantity with single cell resolution12. Thus, this MPN-based mechanogenetic tool can provide an alternative and complementary tool to other the state-of-the-art single cell perturbation tools (e.g. optogenetics), for probing the logic of cellular cognition in space and time.
Experimental design
Optimization of MPN synthesis
We employ a sequential nanoparticle growth strategy to fabricate MPNs that includes i) magnetic core synthesis, ii) dielectric inner shell growth, and plasmonic outer shell growth. We use a superparamagnetic Zn-doped ferrite (ZnxFe3-xO4) core and a plasmonic Au shell geometry to maximize the magnetic and optical properties of MPNs49–51. The dielectric silicon dioxide (SiO2) inner shell serves as a buffer layer to prevent plasmonic damping by the ferrite core.
We first synthesize the ZnxFe3-xO4 magnetic core through thermal decomposition of iron (III) acetylacetonate and zinc (II) chloride in hot surfactant solution containing oleic acid and oleylamine49. The composition of Zn-dopants (x) in the ferrite matrix can be controlled by simply changing the stoichiometric ratio between iron and zinc precursors. We recommend x of 0.4 to provide a maximal magnetic moment of the core where zinc dopants mainly occupy tetrahedral sites of the ferrite matrix eliminating antiparallel magnetic spins52,53. At higher x, the dopants fill the octahedral sites as well, reducing net magnetic moments54. Varying the precursor concentration can control the size of the magnetic core. In this protocol, we used 13 nm or 30 nm superparamagnetic ferrite particles. Larger magnetic cores may lead to residual remanence magnetization, causing irreversible aggregation of particles after magnetization. The detailed synthesis of the magnetic core (M) is presented in the PROCEDURES, Steps 1–11.
To add a SiO2 inner shell layer onto the magnetic core, we use a standard water-in-oil microemulsion sol-gel approach55. Base-catalyzed hydrolysis and polycondensation of tetraethoxy silane (TEOS) with surface hydroxyl groups on the ferrite core facilitates the homogeneous and selective dielectric shell coating. Amine surface functionalization of the silica shell is subsequently followed by addition of amine-containing silica precursors. We recommend aminoethylaminopropyltrimethoxysilane (AEAPTMS) as the precursor instead of a standard aminopropyltrimetoxysilane (APTMS) for higher amine contents. The silica shell thickness can be tuned from 3.5–40 nm with nanometer resolution by varying TEOS concentration (Table 2). The detailed silica shell coating (M-SiO2) is presented in the PROCEDURES, Steps 12–22.
Table 2.
Relationship between amounts of TEOS and silica shell thickness
| TEOS (µl) | 2 | 4 | 30 | 135 | 1080 |
| silica shell thickness (nm) | 3.5 | 6.0 | 8.5 | 18.5 | 40 |
| total size (nm) | 20 | 25 | 30 | 50 | 93 |
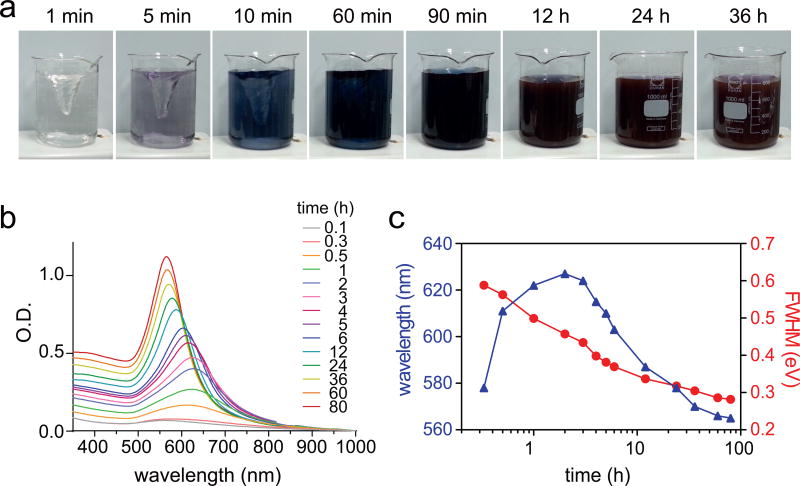
We employ a seed-mediated growth method for gold shell formation56,57. Charge interaction between a positively charged M-SiO2 core and multiple negatively charged Au2nm seeds forms an assembly with a core-satellite geometry (M-SiO2@(Au2nm)n)58. Dense loading of satellite Au2nm seeds on the core is essential for uniform shell formation, whereas sparse loading results in non-uniform shell growth (Fig.2, stage 1). To maximize the seed loading, we recommend performing reactions between Au2nm seeds and AEAPTMS-functionalized M-SiO2 at high nanoparticle concentration (Supplementary Fig. 1). Removal of unbound Au2nm seeds should be performed with a magnetic column, instead of traditional purification methods using centrifugation. Pellet formation of nanoparticles by centrifugation results in irreversible micro-aggregations of nanoparticles. Finally, mild reduction of the gold precursor (HAuCl4) with hydroxylamine hydrochloride (NH2OH·HCl) in the presence of M-SiO2@(Au2nm)n forms MPNs. The shell growth can be monitored by gradual changes in both color and absorption spectra (Fig. 3). Initial redshifts followed by blue shifts at later growth stages in plasmon resonance indicate gradual formation of continuous shell followed by the shell thickening, respectively (Fig. 3b,c). Varying either the M-SiO2 concentration or reaction time can control the gold shell thickness. Detailed procedures for gold shell coating are presented in the PROCEDURES, Steps 23–39.
Figure 3. Growth kinetics of MPNs.
(a) Photographs and (b) absorption spectra of reaction solution as a function of time. (c) Changes in plasmon resonance and measurements of full width at half maximum (FWHM) scattering as a function of time.
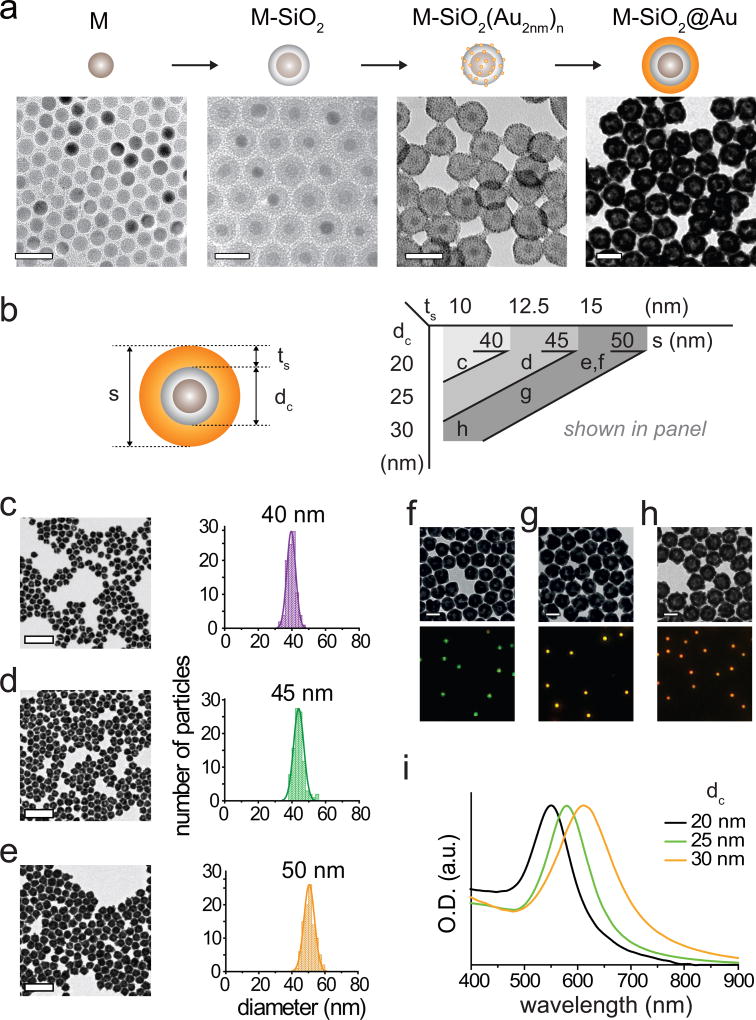
Synthetic protocols presented in this section provide the capacity to tune structure, magnetism, and optical properties of MPNs. The final particle size as well as the thickness of individual components can be tuned with nanometer resolution, and each of these structural features contribute to the magnetic and optical properties of MPNs (Fig. 4).
Figure 4. Controlled synthesis of MPNs.
(a) TEM images of 13 nm Zn-doped iron oxide nanoparticles (M), silica-coated Zn-doped iron oxide magnetic nanoparticles (M-SiO2), Au2nm seed bound M-SiO2, and gold coated M-SiO2. Scale bars, 50 nm. (b) Structural parameters of MPNs shown in panel c–h. (c–e) Control of gold shell thickness; TEM images of MPNs (left) with 10 nm (c), 12.5 nm (d), and 15 nm (e) shell thicknesses and their size distribution (right). The 96, 64, and 24 pmol of Au2nm seed bound 20 nm M-SiO2 are used in Step 34, respectively. Scale bars, 200 nm. (f–h) M-SiO2 size dependent plasmonic absorption shift of 50 nm MPNs. TEM images (top) of 50 nm MPNs with 20 nm (f), 25 nm (g), and 30 nm (h) M-SiO2 core and their corresponding scattering images (bottom). Scale bars, 50 nm. (i) Absorption spectra of 50 nm MPNs with 20 nm (black), 25 nm (green), and 30 nm (orange) M-SiO2 core.
Monovalent and modular functionalization
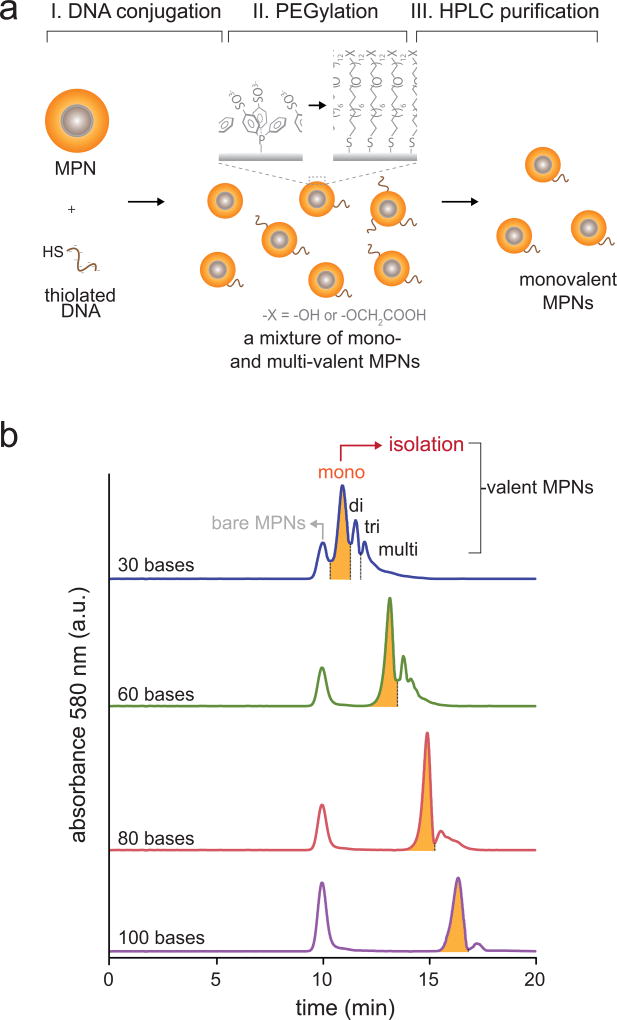
We introduce targeting functionality to MPNs by conjugating them with a thiolated biomolecule via Au-thiol chemistry. Specifically, we use a DNA oligonucleotide for tethering because of two characteristic features facilitating monovalent and modular MPN functionalization. First, polyanionic phosphate groups of DNA allow post-synthetic isolation of monovalent MPNs because of their high charge density12,59,60. As-synthesized MPN-DNA conjugates with Au-thiol conjugation chemistry have a mixed valency and traditional electrophoretic isolation methods based on size-exclusion are restricted to very small particles (≤ 10 nm). On the other hand, DNA conjugation drastically increases anionic charge density of the MPNs as a function of particle valency. This enables purification of monovalent MPNs from non-conjugated or multivalent species via charge-based valency discrimination (Fig. 2, stage 2). We provide examples in Figure 5 of anion-exchange high performance liquid chromatography (HPLC) elution profiles of MPNs bearing different oligonucleotide lengths. While all MPN-DNA conjugation reactions show discrete peaks for MPNs with different valencies, longer oligonucleotides provide more resolved peak separations. We recommend 30-mer or longer DNA oligonucleotides for efficient separation of monovalent products. To maximize the production of MPNs, typically a 5–20 times excess of thiolated DNA is used. Because the reactivity of thiolated DNA is sequence-and length-dependent, a series of small-scale test experiments with varying DNA/MPN ratios is recommended to determine optimal condition for each DNA sequences.
Figure 5. Monovalent DNA conjugation of MPNs.
(a) Schematic illustration of oligonucleotide conjugation and purification of monovalent MPNs. Monovalent MPNs can be isolated via charge-based valency discrimination in an anion exchange HPLC column. To enhance colloidal stability and increase separation yield, thiolated PEG is used as an additional coating. (b) Oligo-length dependent HPLC elution profiles of MPNs. 30, 60, 80 and 100 bases DNA oligos are conjugated to 40 nm MPNs.
Second, the DNA oligonucleotide tether provides a simple, selective, and modular handle for functionalization. A number of companies sell synthetic oligonucleotides with diverse options for 3’ and 5’-functionalization useful for direct targeting (e.g. biotin) or which can be used as substrates for further bioconjugation (e.g. NH2, SH, alkyne/azide). These modified oligonucleotides then react with monovalent MPNs via Watson-Crick hybridization – a highly specific and rapid non-covalent reaction – providing a highly modular strategy for functionalizing MPNs. The detailed protocol for MPN functionalization is presented in the PROCEDURES, Steps 40–47.
Optimization of cell labeling
We use a two-step targeting and hybridization procedure for efficient and specific labeling of cells with MPNs. First, cells expressing receptors are targeted with an oligonucleotide bearing the receptor’s ligand conjugated at its 5’-end. Second, the DNA-conjugated receptors hybridize with monovalent MPNs bearing complementary sequences. DNA-mediated targeting has the advantage of high specificity, modularity, and a rapid on-rate. Cells can be labeled with the same batch of MPNs using the endogenous ligand or the ligand for a recombinant protein tag by simply replacing the targeting DNA strand (Fig. 2, stage 3). The DNA tether length is critical for improving target accessibility of MPNs and hence the cell labeling efficiency. The optimal length depends on the cell type (e.g. thickness of glycocalyx), accessibility of the receptor above the glycocalyx (e.g. receptor height), and steric environment of the receptors (e.g. glycosylation level and conformation). We recommend an oligonucleotide with linker lengths of at least 60 bases or longer for efficient MPN labeling. Cells can be labeled with MPNs either sparsely or densely depending on desired applications by simply adjusting particle concentrations. Sparse labeling (<10 particles per 100 µm2) is preferred for single-molecule perturbation studies to avoid activation of multiple receptors. While, dense labeling (> 100 particles per 100 µm2) is required for activating downstream signals.
Another important consideration in cell labeling is optimization of blocking conditions, a step necessary to minimize nonspecific cell surface interactions. We recommend screening various blocking reagents, since optimal blocking reagents may vary between cell types. For guidance, we provide optimal labeling conditions of U2OS cells expressing either SNAP-tagged VE-cadherin or SNAP-tagged Notch with benzylguanine (BG) functionalized MPNs in the PROCEDURES, Steps 48–56.
Optimization of µMT configuration for spatial and mechanical control of targeted receptors
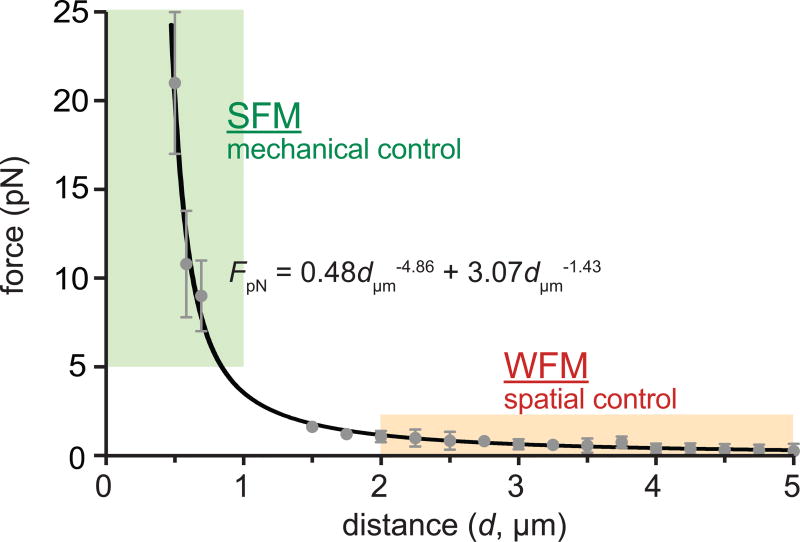
Quantitative and defined force application to an MPN-labeled receptor is required for spatial and mechanical control of receptors with desired modes. To deliver a controlled force, we take advantage of the distance-dependent force generation capability of MPNs coupled with a custom-built µMT. The µMT configurations are described in the Equipment Setup section and Figure 6. We calibrate force delivery per MPN as a function of distance (d) between the MPN and the magnetic tip end. Two force calibration methods are useful for “weak-force” or “strong-force” regimes where the d is ≥ 2 µm or < 2 µm, respectively: viscous drag force calibration experiments61,62 for weak force calibration and DNA rupture experiments using reference tension-gauge tethers (TGTs)63–65 for strong force calibration. A calibration curve can be obtained by fitting the data obtained by these two calibration experiments with a power-law function66. For example, we obtained the calibration curve for MPNs with a 13 nm Zn0.4Fe2.6O4 core, FpN = 0.48dµm−4.86 + 3.07dµm−1.43 (Box 1, Fig. 7), where a single MPN was able to generate forces from 0.1 to 50 pN by controlling d. Use of a larger magnetic core (e.g. 30 nm) is recommended for applications that require higher force ranges (Supplementary Fig. 2). The force ranges exerted by a single MPN are ideally suited for spatial and mechanical studies (Fig. 2, stage 4). Typically, spatial control of receptors can be induced with sub-piconewton force, while mechanical activation of receptors requires a few to tens of piconewton force per receptor12,22. Hence, different modes of perturbation to MPN-labeled receptors can be achieved by simply changing d. We provide an example, spatial and mechanical control of Notch and VE-cadherin on U2OS cell membranes with controlled modes of perturbation, in the PROCEDURES, Steps 57–64.
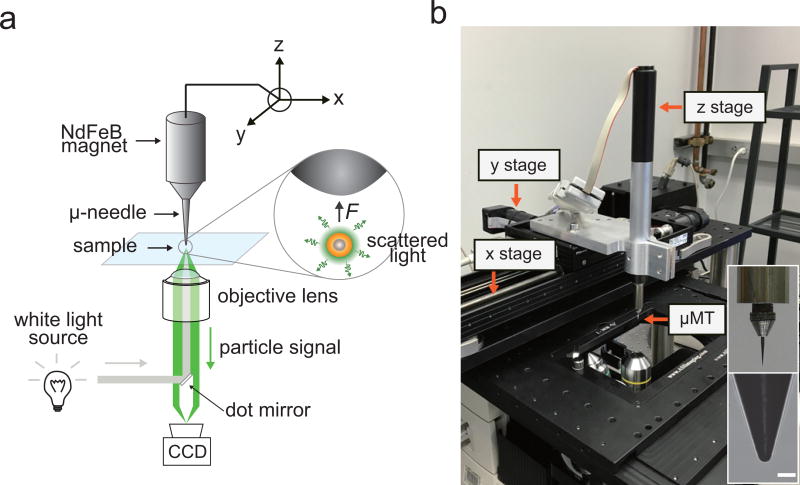
Figure 6. Dark-field microscope and micro-magnetic tweezer (µMT) setup.
(a) A schematic illustration of µMT and dark-field microscopy setup. Steel needle (tip radius: 5 µm) - NdFeB magnet (diameter 1 cm) assembly is mounted to the xyz translation stage. A 4 mm ellipsoidal dot mirror is installed at a filter cube set for a dark-field illumination. (b) A photograph of experimental setup. Scale bar, 20 µm.
Figure 7. Distance dependent force generation of MPNs.
Force acting on particles was plotted (gray dots) and was fitted with a power law (black solid line). TGT DNA force sensor and Stokes’ drag force are used to measure the applied force at SFM and WFM, respectively Each data point represents the mean value of multiple repetitions (n= 10 for SFM, n=30 for WFM). Error bars represent ±s.d.
MATERIALS
REAGENTS
! CAUTION All organic solvents and chemicals should be handled inside a chemical fume hood with the personal protective equipment.
Iron (III) acetylacetonate (Strem Chemicals, cat. no. 26–2300)
Zinc chloride (Sigma-Aldrich, cat. no. 208086)
Oleic acid (Sigma-Aldrich, cat. no. 364525)
Oleylamine (Sigma-Aldrich, cat. no. O7805)
Trioctylamine (Sigma-Aldrich, cat. no. T81000)
Igepal® CO-520 (Sigma-Aldrich, cat. no. 238643)
Ammonium hydroxide solution 28.0–30.0 % (NH4OH; Sigma-Aldrich, cat. no. 221228) ! CAUTION This reagent is a hazardous and corrosive to skin and eye.
Tetraethyl orthosilicate (TEOS; Sigma-Aldrich, cat.no. 333859) ▲ CRITICAL This reagent is sensitive to moisture.
(3-Aminopropyl)trimethoxysilane (APTMS; Sigma-Aldrich, cat. no. 281778) ▲ CRITICAL This reagent is sensitive to moisture.
[3-(2-Aminoethylamino)propyl]trimethoxysilane (AEAPTMS; Sigma-Aldrich, cat. no. 440302) ▲ CRITICAL This reagent is sensitive to moisture.
Tetramethylammonium hydroxide in methanol solution (25 % (wt/vol), TMAOH in methanol solution; Sigma-Aldrich, cat. no. 334901) ! CAUTION This reagent is harmful on ingestion, inhalation and skin contact.
Au(III) chloride trihydrate (HAuCl4·3H2O; Sigma-Aldrich, cat. no. 520918) ! CAUTION This reagent is a hazardous and corrosive to skin and eye.
Sodium hydroxide (NaOH; Sigma-Aldrich, cat.no. 221465) ! CAUTION This reagent is a hazardous and corrosive.
Tetrakis(hydroxymethyl)phosphonium chloride solution (THPC; Sigma-Aldrich, cat. no. 404861) ! CAUTION This reagent is a hazardous and corrosive to skin and eye.
Potassium carbonate (K2CO3; Sigma Aldrich, cat. no. 347825) ! CAUTION This reagent is harmful on ingestion, inhalation and skin contact.
Bis(p-sulfonatophenyl)phenylphosphine dihydrate dipotassium salt (BSPP; Strem Chemicals, cat. no. 15-0463)
Hydroxylamine hydrochloride (NH2OH·HCl; Sigma-Aldrich, cat. no. 159417) ! CAUTION This reagent is a hazardous and corrosive to skin and eye.
Hydrogen chloride (HCl; Sigma-Aldrich, cat. no. 258148) ! CAUTION This reagent is a hazardous and corrosive to skin and eye.
30 nm iron oxide nanoparticle (Ocean Nanotech, cat. no. SOR-30-50)
Tris base (Thermo Fisher Scientific, cat. no. BP152-1)
Sodium Chloride (NaCl; Thermo Fisher Scientific, cat. no. BP358-1)
Toluene (≥99.9% (vol/vol); Sigma-Aldrich, cat. no. 650579) ! CAUTION This reagent is flammable and toxic.
Ethanol (≥99.9% (vol/vol); Sigma-Aldrich, cat. no. 362808) ! CAUTION This reagent is highly volatile and flammable, and it may cause skin irritation, serious eye damage and respiratory irritation.
Cyclohexane (≥99.7% (vol/vol); Sigma-Aldrich, cat. no. 34855) ! CAUTION This reagent is flammable and toxic.
Methanol (≥99.8% (vol/vol); Sigma-Aldrich, cat. no. 179337) ! CAUTION This reagent is a toxic and highly flammable solvent, and it is harmful on inhalation, ingestion or on skin and eye contact.
Dimethylsulfoxide (DMSO; Sigma-Aldrich, cat.no. D5879) ! CAUTION This reagent is hazardous on inhalation, ingestion, skin and eye contact.
Thiol-DNA1: 5’-Thiol- CTC TCT CTC TCT CTC TCT CTC TCT CTC TCT CTC TCT CTC TCA GTC AGT CAG TCA GTC AGT CAG TCA GTC AGT CAG TCA GT -3’ (Integrated DNA Technologies, Customized)
Amine-DNA2: 5’-Amine- CTC TCT CTC TCT CTC TCT CTC TCT CTC TCT CTC TCT CTC TAC TGA CTG ACT GAC TGA CTG ACT GAC TGA CTG ACT GAC TG -3’ (Integrated DNA Technologies, Customized)
BG-GLA-NHS (New England BioLabs, cat. no. S9151S) ▲ CRITICAL This reagent is highly sensitive to moisture. It should be stored at −20 °C.
Tris(2-carboxyethyl)phosphine hydrochloride (TCEP·HCl; TCI, cat. no. T1656) ▲ CRITICAL This reagent should be stored at 4 °C and protected from moisture
HS-(CH2)11-EG12-OH (Prochimia Surfaces, Customized) ▲ CRITICAL This reagent is sensitive to moisture. It should be stored at −20 °C.
HS-(CH2)11-EG12-OCH2-COOH (Prochimia Surfaces, Customized) ▲ CRITICAL This reagent is sensitive to moisture. It should be stored at −20 °C.
HEPES (Thermo Fisher Scientific, cat. no. BP310-1)
U2OS cells (ATCC, cat. no. HTB-96) ! CAUTION The cell lines should be regularly checked to ensure that they are authentic and not infected with mycoplasma.
McCoy’s 5A (modified) medium (Thermo Fisher Scientific, cat.no. 16600082)
HyClone™ McCoy’s 5A medium; without phenol red (GE Healthcare Life Sciences, cat. no. SH30270.01)
FBS (heat-inactivated FBS; Gibco, cat. no. 10500)
Trypsin-EDTA solution (0.25% (wt/vol) trypsin/ 1 mM EDTA-4Na (1×), liquid; Thermo Fisher Scientific, cat. no. 25200-072)
Bovine serum albumin (BSA; Thermo Fisher Scientific, cat. no. BP9703100)
5% (wt/vol) alkali-soluble casein (Merck Millipore, cat. no. 70955)
Collagen type I, rat tail (Corning, cat. no. 354236)
Doxycycline hydrochloride (Sigma-Aldrich, cat. no. D3447)
Plasmid encoding SNAP-Human Notch 1 (original pcDNA5-hNotch1-Gal4 plasmid was a gift from Stephen Blacklow)(SNAP-hNotch1-mCherry and SNAP-hNotch1-GAL4, available upon request)
Plasmid encoding VE-cadherin-mEmerald (mEmerald-VE-Cadherin-N-10, Michael Davidson Fluorescent Protein Collection, Nikon Imaging Center at UCSF, https://nic.ucsf.edu/resources/, SNAP-VEcadherin-mEmerald, available upon request, Full sequence information: https://benchling.com/s/seq-e4p28LgRtyZLdHQMpnFl)
Plasmid encoding Lifeact7-mCherry (mCherry-Lifeact-7, Addgene cat. No. 54491)
Polycaprolactone (Sigma-Aldrich, cat. no. 440744)
EQUIPMENT
50-ml three-neck round bottom flask (Pyrex)
Reflux condenser (Pyrex)
Vacuum pump (EDWARDS, RV8)
Schlenk line connected to a vacuum pump and supply of Argon
Top-loading balance (0.0001 g readability, METTLER, AE240)
20, 50-ml vial (VWR)
Glass jars (100 ml, 1 l, Pyrex)
Thermo controller (POONG LIM, MC-NP200))
Hot plate (Corning, cat. no. PC420D) with magnetic stir bar
Bench top centrifuge (Eppendorf, Model 5424)
Centrifuge (Beckman, Model J2-21M)
Shaker (Wisd laboratory instruments, SHO-1D)
MidiMACS™ Separator (Miltenyl Biotec, cat. no. 130-042-302)
MACS separation columns (25 LS columns, Miltenyl Biotec, cat. no. 130-042-401)
Amicon® ultra -15 centrifugal filter (10K, Merck Millipore, cat. no. UFC801008)
Illustra™ NAP™-10 column (GE Healthcare, cat. no. 17-0854-02)
Transmission electron microscope (TEM; JEOL, JEM-2100)
TEM grid (Carbon on 400 mesh, Tedpella, 01754-F)
Plasma cleaner (Harrick Plasma, PDC-32G)
UV-Vis absorption spectrophotometer (Shimazu corporation, UV-1650(PC)s)
Zetasizer (Malvern instruments, ZEN3600)
Inverted microscope (Nikon, ECLIPSE Ti-E)
White light source (NKT Photonics, SuperK COMPACT supercontinuum lasers) (Other collimated white light source such as mercury, xenon and LED lamps may be used instead)
Dot mirror (Chroma Technology, 4 mm ellipsoid silver coated, customized)
EM CCD camera (Princeton Instruments, ProEM 512)
Color CCD (Nikon, DS-Ri2)
Oil immersion dark-field condenser (Nikon, Oil type, N.A. 1.43–1.20)
pH meter (Fisher Scientific, AB15)
Zeba™ spin desalting columns, 7K MWCO, 0.5 mL (Thermo Fisher Scientific, cat. no. 89882)
High Performance Liquid Chromatography System (HPLC; Agilent Technologies)
DNAPac™ PA100 Oligonucleotide Column 4×250 mm (Thermo Fisher Scientific, cat. no. 043010)
Disposable smartSpatula® (anti-static; Sigma-Aldrich, cat. no. Z561762)
35 mm dish with 7 mm glass bottom dish (MatTeK Corporation, cat. no. P35G-1.5-7-C)
Neon® transfection system (Thermo Fisher Scientific, cat. no. MPK5000)
Steel needle (Semprex, customized; length, 15 mm; diameter, 0.8 mm; tapered angle, 15°)
NdFeB permanent magnet (K&J magnetics, N52; length, 25 mm; diameter, 10 mm)
Magnet adaptor (Customized, cone shape; length, 5 mm; upper diameter, 1 mm; bottom diameter, 5 mm; hole size, 0.8 mm)
xyz translation stage (Physik Instrument, cat. no. VT-80 250-DC, VT-80 25-DC, M-227.50 for x, y, and z translation stage, respectively)
µManager (https://micro-manager.org/)
ImageJ (https://micro-manager.org/)
REAGENT SETUP
50 mM TMAOH in methanol solution Dilute 2.1 ml TMAOH in methanol solution with 97.9 ml methanol. This solution is stable for 2 months at room temperature (RT, 15–25 °C).
10 M NaOH solution Dissolve 400 g of NaOH into 800 ml deionized water. Add deionized water to a total volume of 1.0 l. This solution can be stored at RT for several months.
1.0% (wt/vol) HAuCl4 solution Dissolve 24 mg of HAuCl4·3H2O in 1.5 ml deionized water. ! CAUTION HAuCl4 can rust stainless steel spatulas. It should be weighed with disposable plastic spatulas. Make up volume to 2.4 ml with deionized water. This solution should be freshly prepared before use.
1.0 M Tris·HCl buffer (pH 8.0) Dissolve 121.1 g of Tris base in 800 ml of deionized water. Adjust the pH to 8.0 with concentrated HCl. Make up volume to 1.0 l with deionized water. This solution can be stored at RT for several months.
5.0 M NaCl solution Dissolve 292.2 g of NaCl in 800 ml of deionized water. Make up volume to 1.0 l with deionized water. This solution can be stored at RT for several months.
1.0 M HEPES buffer (pH 8.5) Dissolve 238.3 g of HEPES into 800 ml of deionized water. Adjust pH to 8.5 with 10 M NaOH. Make up volume to 1.0 l with deionized water. This solution can be stored at 4 °C for several months.
BG-DNA2 preparation Dissolve 2 mg of BG-GLA-NHS in 200 µl of anhydrous DMSO. Add 20 µl of 1 mM amine-DNA2 into 100 µl of 1.0 M HEPES buffer (pH 8.5). Mix both solution and make up volume to 1 ml with deionized water. Incubate reaction solution at RT for 1 h. Purify BG-DNA2 with illustra™ NAP™-10 columns. Follow manufacturer provided protocol and elute with deionized water. Concentrate the solution with ethanol precipitation method and dissolve the product in 10 mM Tris-HCl butter (pH 8.0). This product is stable for 6 months at −80 °C.
10 mM HS-C11-EG12-OH solution Dissolve 7.33 mg of HS-(CH2)11-EG12-OH in 1 ml deionized water. This solution can be stored at −20 °C for > 1 month.
10 mM HS-C11-EG12-COOH solution Dissolve 7.91 mg of HS-(CH2)11-EG12-OCH2-COOH in 1 ml deionized water. This solution can be stored at −20 °C for > 1 month.
Collagen solution Dilute concentrated collagen solution to 50 µg/ml using 0.02 M acetic acid, and filter the solution with 0.22 µm syringe filter. This solution can be stored at 4 °C for 1 month.
McCoy’s 5A culture medium (with or without phenol red) To prepare a total 100 ml of McCoy’s 5A culture medium, mix 90 ml of McCoy’s 5A medium with 10 ml of FBS. This medium can be stored at 4 °C up to 1 week.
McCoy’s 5A medium containing 10 % (wt/vol) BSA Dissolve 1 g BSA in 10 ml McCoy’s 5A medium. Filter the solution with 0.22 µm syringe filter. This solution should be freshly prepared before use.
0.5% (wt/vol) casein solution Add 1 ml of 5% (wt/vol) alkali-soluble casein into 9 ml McCoy’s 5A medium. Filter the solution with 0.22 µm syringe filter. This solution should be freshly prepared before use.
200 nM thiol-DNA1 solution Dissolve 24.5 µg of thiol-DNA1 into 100 µl of 500 mM Tris-HCl buffer (pH 8.0). Dilute 50 times and store at −20 °C for up to 6 months.
EQUIPMENT SETUP
Transmission Electron Microscopy (TEM) TEM analyses are conducted on a JEOL JEM-2100 electron microscope at an acceleration voltage of 200 kV.
UV-Vis spectrophotometer setup UV-Vis absorption spectroscopy was conducted on an Agilent 8453 UV-Visible spectroscopy system.
HPLC The Agilent high performance liquid chromatography system (1220 Infinity II LC system) equipped with a DNAPac™ PA100 Oligonucleotide Column (4 × 250 mm) is used.
Collagen coating on to glass bottom dish Spread 50 µl of 50 µg/ml collagen solution to 7 mm glass bottom dish. Incubate at 37 °C for 30 min and wash with 10 ml cell culture medium. This collagen coated glass bottom dish can be stored at 4 °C for several days.
Transfection of U2OS cells by electroporation Neon® transfection system from Thermo Fisher Scientifics is used for electroporation of U2OS cells. Electroporation parameters are 1,230V, 10 ms, 4 pulse number with 10 µl tip type at recommended cell density and DNA concentration (e.g. 5–10 × 106 cells/ml, 0.5–1 µg total). Right after electroporation, cells are transferred to 35 mm cell culture dish with appropriate media.
Human Notch 1 Expressing Stable Cell Line Generation. SNAP-human Notch1-mCherry (SNAP-hN1-mC), and SNAP-human Notch1-Gal4 (SNAP-hN1-Gal4) constructs are stably incorporated into a parental U2OS T-rex cell line, and U2OS T-rex cell line containing a gene encoding a H2B-mCherry reporter sequence, respectively. The transfected cells were selected in 400 µg/mL of hygromycin for 2 weeks and the selective media were replaced every 2–4 days.
µMT setup The µMT is comprised of three key components: a microneedle, a permanent magnet, and xyz-controller. The first two components can be purchased from multiple providers (e.g. Semprex Inc, K&J Magnetics) and can be easily assembled through a needle holder (can be fabricated through commercial 3D printing or machine shop services). We integrated a xyz controller system by assembling three single-axis motorized translation stages, but a microinjection controller system that is routinely used for gene-transfection may be used instead. In order to prepare the µMT setup, (1) install a xyz translation stage on the inverted microscope. (2) Assemble a steel needle onto a magnet adaptor and (3) place the steel needle-magnet adaptor assembly on a hot-plate and heat for 100 °C for 3 min. (4) Apply 50 mg of polycaprolactone on the top of needle tip using a tweezers to coat and prevent oxidation of the steel needle. Polycaprolactone will quickly melt and cover the entire needle-magnet adaptor assembly. Apply heat until polycaprolactone has spread along the entire surface of steel probe. (6) Cool the polycaprolactone coated needle-magnet adaptor assembly at RT for 10 min. (7) Magnetically attach the steel needle to the NdFeB magnet and mount it to the z translation stage (Fig. 6b). The horizontal position of the needle can be controlled by linking the xy-stage to a computer control program, such as µManager.
PROCEDURE
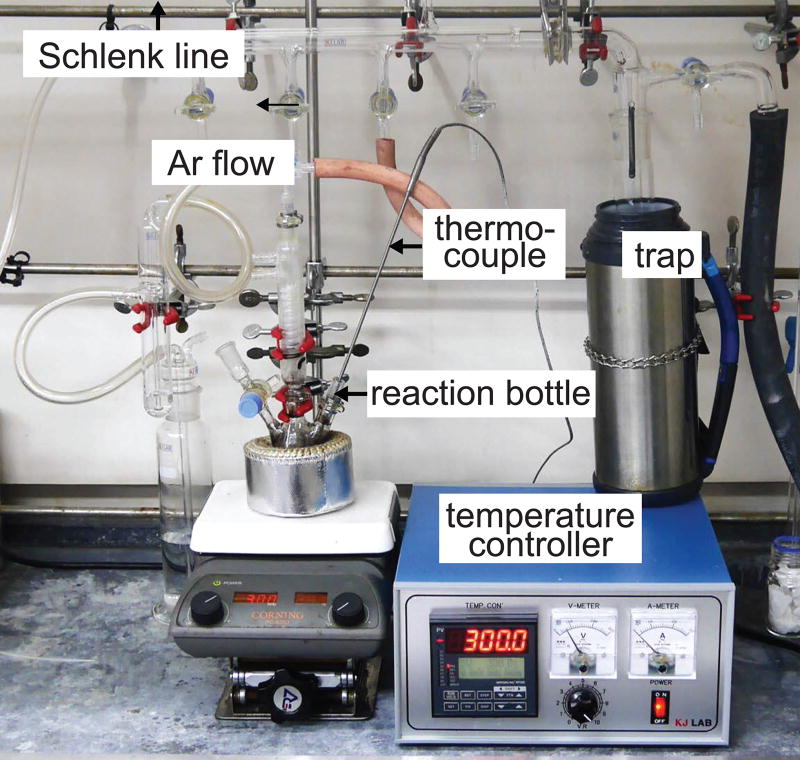
Synthesis of 13 nm Zn-doped ferrite (Zn0.4Fe2.6O4) core nanoparticles ● TIMING 6 hours (h)
-
1|
Add 423.6 mg of iron(III) acetylacetonate and 120 mg of zinc chloride into a 50-ml three-neck round bottom flask.
-
2|
Add 1.2 ml of oleic acid, 4.8 ml of oleylamine, and 4.8 ml of trioctylamine into the flask.
-
3|
Connect the flask to a Schlenk line and stir the mixture solution for 3 min at 300 rpm under vacuum conditions. Experimental setup is depicted in Figure 8.
-
4|Heat the mixture solution using a temperature controller under argon gas flow at 90 ml/min. Stir the mixture solution at 300 rpm during the reaction followings the three-step heating processes shown below.
Time (hh:mm) Temperature Process 00:00 – 00:25 RT – 200 °C Gradual heating 00:25 – 01:25 200 °C Constant temperature 01:25 – 02:25 200 °C – 330 °C Gradual heating 02:25 – 330 °C – RT Gradual cooling -
5|
Add 1.92 ml of toluene and 30 ml of ethanol to the solution. Transfer the solution to 50-ml centrifuge tube.
-
6|
Centrifuge the solution at 1,600g for 5 min at RT.
-
7|
Discard the supernatant, redisperse the black precipitate in 8 ml of toluene and add 30 µl of oleylamine.
-
8|
Centrifuge the solution at 650g for 3 min at RT and collect the supernatant.
-
9|
Add 4 ml of ethanol to the supernatant and centrifuge again at 1,600g for 5 min at RT.
-
10|
Discard the supernatant and redisperse the precipitate (final product, 13 nm Zn0.4Fe2.6O4 nanoparticles) in 4 ml of toluene. Perform TEM analysis under an acceleration voltage of 200 kV.
-
11|
Measure absorption of the nanoparticle solution at 400 nm using a UV-Vis absorption spectrophotometer and determine the nanoparticle concentration using the extinction coefficient of 13 nm Zn0.4Fe2.6O4 nanoparticles of 5.6 ×107 M−1 cm−1 at 400 nm.
■PAUSE POINT The product can be stored at RT; The nanoparticles are stable for at least 2 months. For long-term storage, determine the nanoparticle concentration again before use.
Figure 8. Experimental setup for colloidal synthesis of magnetic nanoparticles.
Preparation of silica coated Zn0.4Fe2.6O4 nanoparticles (M-SiO2) ● TIMING 24 h
-
12|
Add 12.6 ml of cyclohexane into a 50-ml vial.
-
13|
Add 770 mg of Igepal® CO-520 and shake until it is completely dissolved.
-
14|
Add 100 µl of the 2.75 µM Zn0.4Fe2.6O4 nanoparticle solution from Step 10.
-
15|
Add 105 µl of NH4OH solution and shake for 1 min in a fume hood. Upon addition of NH4OH, the solution becomes turbid and soon turns to transparent under to shaking.
? TROUBLESHOOTING
-
16|
Add 30 µl of TEOS to produce a 7.5 nm thick silica shell. Other thicknesses within a range between 3.5 and 40 nm can be created based on amounts of TEOS. Table 2 represents relationship between amounts of TEOS and silica shell thickness.
-
17|
Close the cap and shake the vial for 1 min. Incubate the mixture solution for 48 h at RT.
■ PAUSE POINT The product can be stored at RT for several weeks.
-
18|
Add 2 µl of AEAPTMS and shake for 90 min at 100 rpm. For the M-SiO2 with thin silica layer (< 4 nm), add 0.8 µl of AEAPTMS.
? TROUBLESHOOTING
-
19|
Add 4.0 ml of 50 mM TMAOH in methanol solution. Shaking the vial for 5 s. In 30 s, phase separation between the methanol solution containing nanoparticles (dark brown color) and cyclohexane becomes evident. Carefully remove the dark brown layer (approximately 4.5 ml) using a pipet and transfer to a new vial.
-
20|
Mount a MACS® LS column to a MidiMACS™ separator and load 3.0 ml of solution from step 19. Allow the nanoparticle solution to enter the column completely. Then, apply 8 ml of 50 mM TMAOH in methanol solution to washout byproducts.
-
21|
Allow 1 ml of DMSO to enter to the column completely. Apply an additional 1.5 ml of DMSO and elute the M-SiO2 by dismounting the column from the MidiMACS™ separator.
-
22|
Determine the particle solution concentration using a UV-Vis absorption spectrophotometer. Dilute the particle solution to 400 nM using DMSO.
Synthesis of Au2nm seeds ● TIMING 20 min
-
23|
To a 100-ml glass jar containing 60 ml of deionized water, add 664 µl of 1.0 M NaOH solution and 16 µl of THPC sequentially.
-
24|
Stir the reaction mixture for 10 s at 500 rpm.
-
25|
Inject 2.4 ml of 1.0% (wt/wt) HAuCl4 solution, this results in a brown color.
Attachment of Au2nm seeds to M-SiO2 (M-SiO2 (Au2nm)n) ● TIMING 8 h
-
26|
Concentrate the Au2nm seed solution from step 25 to approximately 1 ml with centrifugal filter unit, Amicon® Ultra-15 (10K) at 3,000g for 30 min at RT.
-
27|
Desalt the mixture solution from step 26 with an illustra™ NAP™-10 column pre-equilibrated with deionized water. Dilute the solution to a final volume of 2.0 ml. Incubate the solution for 1 h.
-
28|
Add 100 µl of M-SiO2 solution from step 21 in 1.9 ml of Au2nm seed solution from step 27 while shaking.
-
29|
Cover the reaction bottle with aluminum foil and shake the mixture for 5 h at 200 rpm.
-
30|
Load the mixture solution onto a MACS® LS column. Wash column with 5.0 ml of 1.0 mM Tris-HCl buffer (pH 8.0). Apply an additional 3.0 ml of 1.0 mM Tris-HCl buffer and collect the dark-red Au2nm seeds bound M-SiO2 (M-SiO2(Au2nm)n) solution.
-
31|
Determine the particle concentration.
▲ CRITICAL STEP The extinction coefficient of M-SiO2(Au2nm)n can be estimated as a simple summation of extinction coefficients for each component. This can be stated as:
where, n is number of Au2nm seeds on the surface of single M-SiO2. εM-SiO2 (Au2nm), εM-SiO2, and εAu2nm are extinction coefficients of M-SiO2(Au2nm)n, M-SiO2, and Au2nm, respectively. To determine the seed density per M-SiO2 particle, perform TEM analysis under an acceleration voltage of 200 kV and take the image at 80,000× magnification. The density of Au2nm seeds on M-SiO2 is approximately 8.16 particles/(10 nm)2. We found that the seed density is consistent within the range of 8–9 particles/(10 nm)2 for all M-SiO2 size. Extinction coefficient of Au2nm seed is 4.71 × 105 M−1cm−1. In case of 30 nm M-SiO2 the number of Au2nm seeds bound single M-SiO2 is 230 and estimated extinction coefficient is 1.64 × 108 M−1cm−1.? TROUBLESHOOTING
Formation of Au shell (M-SiO2@Au) ● TIMING 3 days
-
32|
To a 1-l glass jar containing 800 ml deionized water, add 100 mg of K2CO3 and 60 mg of HAuCl4.
-
33|
Stir the mixture at 400 rpm for 24 h.
▲ CRITICAL STEP During the aging step, the color of the mixture slowly changes from yellow to colorless. Monitor the progression of the reaction by measuring absorbance of the gold chloride solution at λ = 290 nm using a UV-Vis spectrophotometer. The absorbance gradually decreases from 0.5 to 0.05 (Supplementary Fig. 3). Move to the next step when the absorbance of the growth solution reaches 0.05. Avoid direct sunlight.
-
34|
Add 16 pmol of 30 nm M-SiO2(Au2nm)n and 20 ml of 1.0 mg/ml BSPP aqueous solution in order. Vary the M-SiO2(Au2nm)n concentration to obtain MPNs having different core/shell ratios. For example, 96, 64, and 24 pmol of 20 nm M-SiO2(Au2nm)n are used to produce 40 nm, 45 nm, and 50 nm MPNs, respectively (Fig. 4, c–e).
-
35|
Add 22 ml of 130 µg/ml NH2OH·HCl aqueous solution.
-
36|
Monitor the color change of the solution during the reaction. The color of the solution changes continuously during the reaction due to the change of Au shell thickness (Fig. 3a). Initial redshifts (e.g. from λmax = 550 nm to λmax = 630 nm for 30 nm M-SiO2) represent a gold shell formation (Fig. 3b). Subsequent blue shifts in plasmon resonance (e.g. from λmax = 630 nm to λmax = 560 nm for 30 nm M-SiO2) indicate thickening of the gold shell (Fig. 3b). Stop stirring at the desired λmax. For example, to achieve a 10 nm thick gold shell on the 30 nm M-SiO2, stop the reaction at λmax = 620 nm (Fig. 4a). Before measuring the absorbance, remove the byproduct using MACS® LS column.
-
37|
Transfer the product solution into a 50-ml tube and centrifuge for 60 min at 1,500g at RT.
-
38|
Discard the supernatant carefully, and add 1.0 mg/ml of BSPP solution to each tube.
-
39|
Collect the gold coated M-SiO2 nanoparticles using a MACS® LS column. Use the 1.0 mg/ml BSPP solution as an eluent. Verify the presence of the reaction product using TEM at an acceleration voltage of 200 kV (Fig. 4 c–h). Measure absorbance of the MPN solution at λmax using a UV-Vis spectrophotometer and determine the MPN concentration using the extinction coefficient of the respective MPN at λmax. The extinction coefficients of 40 nm and 50 nm MPNs (independent on the core-shell ratio67) are 9.0 × 109 M−1 cm−1 and 1.9 × 1010 M−1 cm−1 at λmax, respectively.
? TROUBLESHOOTING
Monovalent conjugation of MPNs ● TIMING 12 h
-
40|
Add 10 µl of 250 mg/ml TCEP to 100 µl of 200 nM thiol-DNA1 to reduce disulfide bonds. Incubate for 30 min at RT.
-
41|
Remove excess TCEP and byproducts with a Zeba™ spin desalting column. Measure absorbance of the eluted fraction at λ = 260 nm using a UV-Vis spectrophotometer and determine the DNA concentration with the extinction coefficient of thiol-DNA1 of 699,900 M−1cm−1.
-
42|
Add 100 µl of reduced 100 nM thiol-DNA1 solution to 50 µl of 20 nM MPN (40 nm) solution.
-
43|
Add 50 µl of 120 mM NaCl solution (in 40 mM Tris-HCl pH 8.0) carefully into the mixture solution and shake for 6 h.
▲ CRITICAL STEP High salt concentration increases the binding efficiency between negatively charged MPNs and oligonucleotides. However, an abrupt increment of salt concentration causes aggregation between the nanoparticles. Add the NaCl solution dropwise, 5 µl increments every 30 s while vortexing the solution.
-
44|
Add 20 µl of 5 mM HS-C11-EG12-OH and 500 µM HS-C11-EG12-COOH to the mixture solution and shake an additional 4 h.
-
45|
Inject 100 µl of the reaction solution to an HPLC system to separate the monovalent MPNs. (DNAPac™ PA100 Oligonucleotide Column 4×250 mm2, salt concentration gradient: 40 – 1200 mM NaCl in 25 mM Tris-HCl buffer, pH 8.0 for 30 min at 1 ml/min of flow rate; detection wavelength 580 nm). Collect a fraction containing monovalent MPNs (1 ml, Fig. 5b).
? TROUBLESHOOTING
-
46|
Dilute the eluent 10 times with deionized water and centrifuge at 1500g for 10 min at RT. Carefully remove supernatant and redisperse the bottom nanoparticle layer containing monovalent MPNs in 200 µl of deionized water.
-
47|
Measure absorbance of the MPN solution at λmax (e.g. λmax= 620 nm for 50 nm MPNs having a 30 nm M-SiO2 core, λmax= 560 nm for 40 nm MPNs having a 20 nm M-SiO2 core) using a UV-Vis spectrophotometer and determine the concentration using the extinction coefficients denoted in step 39.
■ PAUSE POINT The product can be stored at 4 °C for several days.
Cell culture and transfection of SNAP-tagged receptors ● TIMING 40–50 h
-
48|
Detach 5 × 105 U2OS cells from a cell culture plate (70–100% confluency on 60 mm dish or T25 flask) with 1 ml of 0.05% Trypsin-EDTA solution. Neutralize the lifted cells with 2 ml McCoy’s 5A media with 10% FBS and transfer the cells into a 35 mm dish.
-
49|
Transfect SNAP-tagged receptor constructs transiently to U2OS cells with Neon transfection system (see Equipment Setup). In this procedure, as an example, SNAP-VEcadherin-mEmerald (SNAP-VEcad-mEm) and Lifeact7-mCherry constructs are co-transfected, and SNAP-human Notch1-mCherry (SNAP-hN1-mC) construct are transfected for mechanical control of VE-cadherin and Notch receptors.
-
50|
Following transfection, wash the cells 3 times with 15 ml of PBS after 6 h of incubation to remove cellular debris and inclusion bodies.
▲ CRITICAL STEP Cellular debris and inclusion bodies remaining on glass and/or cell surface induce nonspecific binding of DNA and MPNs causing nonspecific perturbation of receptor signaling.
-
51|
After 15 h, detach the U2OS cells transfected with SNAP-tagged receptors using 1 ml of 0.05% Trypsin-EDTA solution and transfer the cells to collagen coated glass bottom dishes. Incubate another 36–48 h.
Labeling of MPNs to SNAP-tagged receptors ● TIMING 1 h
-
52|
Treat the U2OS cells expressing SNAP-tagged receptors with 1 µM BG-DNA2 in McCoy’s 5A culture medium without FBS. Incubate for 30 min.
-
53|
Wash the cells five times with 5 ml of McCoy’s 5A culture medium containing 10% (wt/vol) BSA. Alexa 647-(ACTG)5 (10 nM) can be used to confirm the binding of BG-DNA2. A non-transfected control cell line can be used as a negative control.
▲ CRITICAL STEP Remaining free BG-DNA2 can bind to MPNs and decrease the labeling efficiency. Wash out the unbound BG-DNA2 with plenty of culture media.
-
54|
Treat the cells with 1 nM of MPN-DNA1 conjugates dispersed in McCoy’s 5A culture medium containing 0.5% (wt/vol) of alkaline casein. Incubate for 10 min.
▲ CRITICAL STEP For single-molecule perturbation studies (i.e. sparse labeling), use 0.1 pM MPN-DNA solution. This typically resulted in 1–2 MPNs per cell. To test non-specific binding, MPNs conjugated with a non-complementary DNA sequence (e.g. polyT), and/or only PEG molecules can be used as negative controls
? TROUBLESHOOTING
-
55|
Wash the cells three times with 10 ml of McCoy’s 5A culture medium containing 10% FBS.
-
56|
Add phenol red free McCoy’s 5A culture medium with 10% FBS for imaging.
Preparations for mechanogenetic control of cells ● TIMING 1 h
-
57|
Equilibrate the temperature of a microscope cell culture chamber to 37 °C before starting the experiment.
▲ CRITICAL STEP Always keep the microscope cell culture chamber at 37 °C with a humidified atmosphere (80%) of 5% CO2 for long term time-lapse experiments. In this procedure, a stage-top incubation system was used for all steps. Direct contact between the oil immersion objective and culture dish may cause rapid temperature changes within the culture dish depending on changes in room temperature. To solve this problem, use a closed-loop temperature regulating system to maintain the temperature of the cell culture dish.
-
58|
Align the µMT with the microscope objective lens. Position the end of the µMT tip at the center of the objective oculus with bright field illumination. Perform this step sequentially with a 20×, 40×, and 100× objective lens and set the height of the µMT tip ~5 cm above the 100× objective lens.
-
59|
To prevent contamination, dip the µMT briefly in 70% ethanol and dry it before use.
-
60|
Place a small drop of immersion oil onto the 100× objective lens, and place the bottom dish containing the cells on the microscope stage.
-
61|
Find and center a subcellular region of interest by visualizing the MPNs using dark-field microscopy.
? TROUBLESHOOTING
-
62|
Lower the µMT into the medium and place the end of the tip of the µMT above the target cell.
▲ CRITICAL STEP Focus above the cells. Carefully adjust the µMT z-position using the z translation stage until scattering from the tip is centered in the field of view.
-
63|
Determine the distance (d) between the tip of µMT and the MPNs on a cell membrane and adjust the d to 40 µm.
▲ CRITICAL STEP To determine the d, focus at the sample plane and the µMT tip sequentially by using Nikon perfect focus system (axial resolution: 25 nm) and then measuring the focal height difference.
▲ CRITICAL STEP The magnetic force between the µMT and the MPN is negligible when d > 20 µm. Do not place the µMT closer than 20 µm during preparation.
Probing single cell mechanogenetics
-
64|
To control the spatial distribution of receptors with the weak force mode (WFM), perform the procedures described in option A; to induce a conformational change of receptors with the strong force mode (SFM), perform the procedures described in option B; to apply single-molecule perturbations, perform the procedures described in option C. In order to study MPN-induced single cell transcription, perform the procedures described in option D. For all options, 40 nm gold nanoparticles without a magnetic core can be used as negative controls.
(A) Spatial control of receptors ● TIMING 1 h
CRITICAL In this procedure, cells expressing both SNAP-VEcad-mEm and Lifeact7-mCherry are used, but similar procedures can be followed for cells expressing tagged versions of other types of receptors.
Choose a dense MPN labeled cell, and place the µMT 2 µm (WFM) above a target subcellular location for 5–15 min.
During stimulation, monitor the spatial localization of the MPNs at the µMT by imaging nanoparticle scattering in dark-field microscopy.
Monitor the fluorescence signals of SNAP-VEcad-mEm (λem = 509 nm) and Lifeact7-mCherry (λem = 610 nm) within the same region. If desired, time-lapse fluorescence imaging can also be performed.
(B) Mechanical control of receptors ● TIMING 1 h
CRITICAL In this procedure, cells expressing SNAP-hN1-mC are used, but similar procedures can be followed for cells expressing tagged versions of other types of receptors. For these experiments, 40 nm gold nanoparticles without a magnetic core or 5 µM of DAPT and/or 50 µM of TAPI-2 to inhibit initiation of Notch signaling may be used as negative controls.
Choose a dense MPN labeled cell, and place the µMT 0.7 µm (SFM) above a target subcellular location for 5–20 min.
-
Monitor the spatial distribution and activation of the receptors by imaging fluorescence signals of mCherry (λem = 610 nm) during stimulation. If desired, time-lapse fluorescence imaging can also be performed.
▲ CRITICAL STEP Be careful not to touch the cells with the tweezers tip. Touching the cell induces a rapid membrane rupture due to the strong magnetic force between concentrated MPNs and µMT.
(C) Single receptor perturbation. ● TIMING 1 h
CRITICAL In this procedure, sparsely labeled cells (1–2 MPNs per cell, from step 54) expressing SNAP-hN1-mCh are used, but similar procedures can be followed for cells expressing tagged versions of other types of receptors. For these experiments, cells can be treated with 5 µM of DAPT and/or 50 µM of TAPI-2 to inhibit initiation of Notch signaling as negative controls. P
Place the µMT 0.7 µm (SFM) above a target receptor conjugated with a MPN. Single receptors are identifiable as discrete particles diffusing within the cell membrane.
Monitor the MPN detachment (disappearance of the MPN) as a result of Notch extracellular shedding by imaging the scattering with dark-field microscopy during stimulation.
(D) MPN-induced single cell gene transcription ● TIMING 40–50 h
CRITICAL 40 nm non-magnetic nanoparticle (e.g. gold nanoparticle)-DNA1 conjugates may be used for a negative control of force induced signaling. For Notch receptor activation, cells may be treated with 5 µM of DAPT and/or 50 µM of TAPI-2 as a control for specific activation of Notch signaling.
Prepare a target transcription system (see EQUIPMENT SETUP). In this procedure, U2OS cells expressing both SNAP-hN1-Gal4 and UAS driven H2B-mCherry are used, but similar procedures can be followed for cells expressing tagged versions of other types of receptors.
To activate Notch receptors mechanically with SFM, place the µMT 0.7 µm above a target subcellular location for 5–20 min.
-
Remove the µMT probe tip from the microscope and monitor nuclear mCherry fluorescence signals (λem = 610 nm) of the cell for the next 20 hours using a 40× oil immersion objective. Place a small drop of immersion oil onto the 40× objective lens before changing objective, and gently align the 40× objective under the cell culture dish.
▲ CRITICAL STEP Be careful not to move the xy stage while changing objectives. Digitally storing the xy position using the controller of the motorized xy stage is helpful for this step.
Find the stimulated cell with the 40× objective lens, and center the cell in the field of view using the xy stage.
-
Image nuclear mCherry expression in the target cell. Image in the bright field for cell tracking. Acquire bright-field and fluorescence image sequences every 30 min for 20 h.
▲ CRITICAL STEP To reduce photobleaching, minimize exposure time, and use neutral density filters.
? TROUBLESHOOTING
TIMING
Setting up the µ-magnetic tweezers (µMT) (Equipment Setup): ~1 h
Steps 1–11, Synthesis of 13 nm Zn0.4Fe2.6O4 nanoparticles: 6 h
Steps 12–22, Preparation of silica coated 13 nm Zn0.4Fe2.6O4 nanoparticles: 48 h
Steps 23–25, Synthesis of Au2nm seeds: 20 min
Steps 26–31, Attachment of Au2nm seeds to M-SiO2: 5 h
Steps 32–39, Formation of Au shell: 3 days
Steps 40–47, Monovalent conjugation of MPNs with oligonucleotide: 12 h
Steps 48–51, Cell culture and transfection of SNAP tagged receptor: 40–50 h
Steps 52–56, The SNAP tagged receptor labeling with MPNs: 1 h
Steps 57–63, Preparations for mechanogenetic control of cells: 1 h
Step 64 Option A, Spatial distribution control of receptors: 1 h
Step 64 Option B, Mechanical control of receptors: 1 h
Step 64 Option C, Single receptor perturbation: 1 h
Step 64 Option D, MPN induced single cell gene transcription: 40–50 h
Box 1, Calibration of force exertion on a single MPN: ~6 h
?TROBLESHOOTING
Troubleshooting advice can be found in Table 3.
TABLE 3.
Troubleshooting table
| Step | Problem | Possible reason | Solution |
|---|---|---|---|
| 15 | Aggregate present in the bottom | Igepal® CO-520 may not be completely dissolved | Before adding the Zn0.4Fe2.6O4 solution, shake the Igepal® CO-520 and cyclohexane mixture until solution turns to completely transparent (Step 13) |
| 18 | Silica shell thickness changes | AEAPTMS may form multiple layers | Decrease reaction time. Do not exceed 2 h |
| 26–31 | Low binding density of Au2nm seeds | AEAPTMS may not appropriately coat the silica surface | Use fresh AEAPTMS. Avoid moisture. |
| Au2nm seeds may grow slowly by Ostwald ripening, especially at high concentrations, due to their active surface energy. | Use freshly prepared Au2nm seeds. Keep recommended reaction time, and avoid intermittence among steps | ||
| 39 | MPNs have irregular shape | The activity of gold chloride growth solution is too high | Check absorbance of the growth solution at 290 nm before starting the experiment. |
| To smooth the surface of synthesized nanoparticles, disperse them in 800 ml of deionized water before adding BSPP solution (Step 38 and incubate 1 day with 150 r.p.m. shaking | |||
| A large amount of MPNs form dimers (peanut shape) | Inappropriate volume ratio between DMSO and deionized water in step 28 | Final solution should be 5% DMSO in deionized water. | |
| To separate out the peanut shape particles, use 1–2 % agarose gel electrophoresis (120V, 90 min) and recover the first band | |||
| Broad size distribution | AEAPTMS treated M-SiO2 may aggregate before gold shell formation reaction. Aggregation occurs easily in Au2nm seed solution with neutral pH | After the 1h incubation of Au2nm seed solution, pH decreases from 7 to 5 (Step 27). Add AEAPTMS treated M-SiO2 at this pH to prevent aggregation | |
| 45 | Low yield in HPLC separation | MPNs can aggregate at high salt concentration. | Ratio between carboxyl-PEG-thiol and hydroxyl-PEG-thiol in Step 44 is 1:10. Increase the carboxyl-PEG-thiol ratio up to 1:3 |
| 54 | Cellular uptake of MPNs (MPNs located at cytosolic area, and show slow and directional movements.) | MPN labeled receptor recycled and endocytozed within 30–60 min at 37 °C | Incubate MPN less than 10 min. |
| To increase the labeling yield, use higher concentration of MPN rather than using longer incubation time. | |||
| 61 | The MPNs are immobile | MPNs may aggregate and non-specifically bind to the cell membrane due to poor long-term colloidal stability of MPNs | Use fresh MPNs (Step 40–46) |
| Cellular debris and inclusion bodies remaining on cell surface induce nonspecific binding of MPNs | Wash the cells more extensively with PBS or media before MPN labeling (Step 48) | ||
| 64 | Cells become unhealthy during time-lapse imaging: usually showing large vacuoles and cell detaching | Cell damage from phototoxicity and/or environmental shock | To reduce phototoxicity, minimize the exposure time, and light intensity. |
| Check the temperature, CO2, and humidity of microscope cell culture chamber. |
Materials
Alexa Fluor 594 Carboxylic acid, Succinimidyl Ester, mixed isomers (ThermoFisher scientific, cat. no. A20004)
NHS-biotin (ThermoFisher scientific, cat. no. 20217)
NHS-PEG-COOH (NANOCS, cat. no. PG2-CANS-2k)
Glycerol (C3H8O3, Sigma-Aldrich, cat. no. G9012)
10× PBS (Sigma-Aldrich, cat. no. P7059)
TGT-thiol: 5’- CAC AGC ACG GAG GCA CGA CAC (ACTG)5 thiol-3’
TGT-biotin-1: 5’- biotin (ACTG)10 GTG TCG TGC CTC CGT GCT GTG-3’
TGT-biotin-2: 5’- GTG TBiotinCG TGC CTC CGT GCT GTG-3’
TGT-biotin-3: 5’- GTG TCG TBiotinGC CTC CGT GCT GTG-3’
TGT-biotin-4: 5’- GTG TCG TGC CTC CGT GCT GTG (ACTG)10 biotin-3’
(TBiotin : biotin group is placed on the thymine),
(All DNAs are custom-ordered from Integrated DNA Technologies)
M-SiO2 (diameter of Zn0.4Fe2.6O4 nanoparticle, 13 nm; thickness of silica layer, 40 nm)
AEAPTMS coated MPN with 50 nm in diameter
Equipment
Coverslip (Fisher Scientific, cat. No.12-544-G/22X60-1.5)
Streptavidin coated cover slip (NANOCS, cat. no. CS-SV-5)
Dark field microscope with 100× objective lens (N.A. 1.49)
552 nm solid-state laser (Applied Research Inc., Spectral)
µMT
Procedure
A. Force calibration using the viscous drag of the nanoparticle
Add 16.4 µg of NHS-Alexa Fluor® 594 and 160 µg NHS-PEG-COOH into 100 µl of 400 nM AEAPTMS coated M-SiO2 dispersed in DMSO of Step 22. Incubate for 8 h at RT.
Load the mixture solution onto a MACS® LS column and wash with 5 ml deionized water. Apply an additional 1.5 ml of deionized water and elute the Alexa 594 conjugated M-SiO2.
Determine the particle solution concentration using UV-Vis absorption spectrophotometer. Use same extinction coefficient with step 11. Dilute the particle solution to 400 nM using deionized water.
Add 10 µl of 400 nM Alexa 594 conjugated M-SiO2 to 990 µl glycerol.
Apply 30 µl of M-SiO2 dispersed in glycerol on the surface of a cover slip.
Immerse the µMT into the solution containing nanoparticles with an immersion angle of 30 degrees toward the cover glass surface.
Image particle movement with an EM CCD camera at 100 fps for 10 sec.
Analyze the velocity of the particles using the ImageJ particle tracker plugin (http://mosaic.mpi-cbg.de/ParticleTracker/).
Calculate the force acting on a single particle using Stokes’ law (F⃑ = 6πηrv⃗, where η is the viscosity of solvent, r is the radius of particles, v is the velocity of particles. As specified, the fluid viscosity is 1.1908 Ns/m2 and the Stokes radius of the particle is 50 nm.
B. Force calibration using TGT force sensors
Prepare 100 µl of 1 nM monovalently conjugated MPNs with TGT-thiol (See steps 40–47) in 200 mM NaCl, 10 mM tris buffer.
Add 1 µl of 100 µM TGT-biotin-1 oligonucleotides bearing complementary sequences to form a TGT-duplex force sensor and incubate for 4 h at RT.
Add 900 µl of 30 mM NaCl, 10 mM tris buffer and centrifuge at 1,500g for 10 min at RT. Discard the supernatant. Repeat 3 times. Dilute the bottom nanoparticle layer containing MPNs to 1 ml.
Measure absorbance of the solution at λ = 620 nm using a UV-Vis spectrophotometer. Determine the nanoparticle concentration using the extinction coefficient of 1.9 × 1010 M−1 cm−1 of 50 nm MPNs at λ = 620 nm.
Place a streptavidin-coated coverslip onto a dark-field microscope. Apply 30 µl of 1 pM TGT-sensor conjugated MPNs to the coverslip and monitor the particle immobilization.
When the desired nanoparticle density is reached (e.g. 1–2 particles/100 µm2), wash the coverslip 5 times with 200 µL with 30 mM NaCl, 10 mM Tris-HCl buffer (pH 8.0), and then 3 times with 200 µL PBS.
Gradually approach the µMT in the z-direction with a 50 nm-step size at an approach rate of 100 nm/s while monitoring detachment events of the particles from the surface (i.e. disappearance of particles from the imaging area) using dark-field microscope. Repeat steps 1 to 5 with TGT-biotin-2, TGT-biotin-3, and TGT-biotin-4.
Measure the rupture distance (d) of MPN-TGT sensor, and calculate the loading rate from a power law fit taking into account the loading rate-force relationship and the most probable rupture force of each dsDNA with unzipping geometry (Fig. 7).
ANTICIPATED RESULTS
Synthesis of MPNs
This protocol reproducibly produces high quality and monodisperse MPNs with respect to size, shape, and crystallinity, which together provide for uniform optical and magnetic properties. Fabrication of the magnetic core coated with a silica shell is relatively straight forward, whereas the gold shell coating steps on the M-SiO2 often results in imperfect and/or irregular particles when critical steps in the procedure section are improperly performed. For guidance, we show representative reaction kinetics for MPN shell formation, monitored by changes in solution color and plasmon resonance, in Figure 3. Characteristic initial redshifts are followed by blue shifts in plasmon resonance that confirm uniform shell coating (Fig. 3b). Gradual narrowing in FWHM of plasmon peaks as time proceeds additionally indicates homogenous MPN formation (Fig. 3c). Observation of continuous red shifts with no blue shift or broad plasmon peaks indicates one of the following two representative cases: 1) Incomplete shell formation due to low Au2nm seed density on the M-SiO2 core (< 8.2 seeds/(10 nm)2, Supplementary Fig. 1) and/or 2) irregular shell formation that results from excessive growth kinetics in the reactive precursor solution (OD290 nm > 0.05).
Once all reaction conditions are met, the size of MPNs can be easily tuned by controlling the reaction time, the concentration of M-SiO2@Au2nm, and the thickness of the silica shells (Fig. 4); Longer reaction times and decreased M-SiO2@Au2nm concentration increase gold shell thickness. The plasmon excitation wavelength and the scattering color of the MPNs can be tuned depending on the thickness of silica and gold shells (Fig. 4f–i). For example, MPNs with green (Fig. 4f, λmax = 550 nm), yellow (Fig. 4g, λmax = 570 nm), and orange (Fig. 4h, λmax = 620 nm) single particle scattering colors are obtained from the 50 nm MPNs with 20 nm, 25 nm, and 30 nm M-SiO2, respectively.
Monovalent DNA conjugation
Conjugation of MPNs with thiolated DNA generates products with mixed valencies. Monovalent MPNs can be isolated via anion exchange HPLC, providing discrete peaks corresponding to MPNs with different DNA valencies. Figure 5b shows example elution profiles of MPN-DNA conjugates bearing different oligonucleotide lengths, where the monovalent MPNs are then selectively collected. With optimal conditions, isolation yield of monovalent MPNs is approximately 40%. Lower isolation yield could result from several possible reasons: 1) sub-optimal MPN/DNA reaction ratio, 2) colloidal instability of MPN-DNA conjugates against high salt environment, and 3) poor peak separation between monovalent and multivalent MPNs. Improvement in the product yield can be achieved by revisiting the critical steps described above.
Mechanogenetic control of cell signaling
Cell labeling of MPNs via BG-SNAP chemistry typically provides excellent target-specificity ratio, that is, the ratio between the number of particles per cell expressing SNAP-tagged proteins and the number of particles per cell without SNAP expression, is usually > 100. Comparing the diffusivity of MPN-labeled receptors with small fluorescent dye-labeled receptors can validate the one-to-one engagement of MPNs with target proteins. In our previous report, we showed that the monovalent MPN minimally influences membrane diffusion dynamics of targeted Notch receptors12.
Application of µMT with differential modes of stimulation (WFM vs. SFM) allows us to investigate the differential roles of spatial and mechanical cues in cell signaling. Figure 9 shows examples of receptor activation through either spatial segregation or mechanical activation. Subcellular spatial segregation of MPN-labeled VE-cadherin recruits F-actin (Fig. 9a), whereas mechanical loading of Notch-Gal4 receptors with MPNs activates transcription programs (i.e. UAS-H2B-mCherry) within a single cell (Fig. 9b).
Figure 9. Mechanogenetic control.
(a) Spatial control of actin polymerization using WFM mode of MPNs. (left) Schematic illustration of MPN induced spatial segregation of VE-cadherin and actin polymerization. (right) Spatial segregation of VE-cadherin receptors (green) and resulting actin polymerization (red) are observed by time-lapse fluorescence imaging of SNAP-VE-cadherin-mEmerald and Lifeact7-mCherry signals, respectively. Scale bars, 4 µm. (b) Mechanical control of single cell gene transcription using SFM mode of MPNs (9 pN). (left) Schematic illustration of UAS-Gal4 fluorescence reporter system and transcriptional activation by MPNs. (right) A representative optical microscope image of single cell mCherry expression. Scale bars, 4 µm.
Supplementary Material
Acknowledgments
The authors thank S. Blacklow (Harvard Univ.) for the generous gift of Notch plasmids. This work was supported by IBS-R026-D1 from IBS (Y.J., J.C.), HI08C2149 from Korea Healthcare Technology R&D Project (J.C.), 2017R1C1B2010945 from National Research Foundation of Korea (NRF) grant funded by the Korea government (MSIP, D.S.), 1R01GM112081-01 from the National Institute of General Medical Science (NIGMS) and the National Institute of Health (NIH) (Y.J.), the UCSF Program for Breakthrough Biomedical Research-Sandler Foundation (Y.J.), 1R21HL123329-01 from the National Heart, Lung, and Blood Institute (NHLBI) and NIH (Y.J.), DP2 HD080351-01 from the NIH common fund (Z.J.G.), and P50 GM081879 from the NIGMS UCSF Center for Synthetic and Systems Biology (Z.J.G.).
Footnotes
AUTHOR CONTRIBUTIONS
J.K., D.S., Y.J. and J.C. conceived and designed the project; D.S., J.K., J.L., Y.L., D.K., Y.J. and J.C. contributed nanoparticle synthesis; D.S., K.S., Z.J.G., and Y.J. designed biological experiments; J.K., D.S., J.L., K.S., Y.L., and D.K. performed experiments and analyzed the data; and J.K., D.S., J.L., Y.J. and J.C. wrote the manuscript. All authors discussed and commented on the manuscript.
COMPETING FINANCIAL INTEREST The authors declare no competing financial interests.
Supplementary Information is available in the online version of the paper.
References
- 1.Banghart M, Borges K, Isacoff E, Trauner D, Kramer RH. Light-activated ion channels for remote control of neuronal firing. Nat. Neurosci. 2004;7:1381–1386. doi: 10.1038/nn1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Deisseroth K. Optogenetics. Nat. Methods. 2011;8:26–29. doi: 10.1038/nmeth.f.324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Miesenbock G. The optogenetic catechism. Science. 2009;326:395–399. doi: 10.1126/science.1174520. [DOI] [PubMed] [Google Scholar]
- 4.Toettcher JE, Voigt CA, Weiner OD, Lim WA. The promise of optogenetics in cell biology: interrogating molecular circuits in space and time. Nat. Methods. 2011;8:35–38. doi: 10.1038/nmeth.f.326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wu YI, et al. A genetically encoded photoactivatable Rac controls the motility of living cells. Nature. 2009;461:104–108. doi: 10.1038/nature08241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mannix RJ, et al. Nanomagnetic actuation of receptor-mediated signal transduction. Nat. Nanotechnol. 2008;3:36–40. doi: 10.1038/nnano.2007.418. [DOI] [PubMed] [Google Scholar]
- 7.Lee JH, et al. Artificial control of cell signaling and growth by magnetic nanoparticles. Angew. Chem. Int. Ed. 2010;49:5698–5702. doi: 10.1002/anie.201001149. [DOI] [PubMed] [Google Scholar]
- 8.Cho MH, et al. A magnetic switch for the control of cell death signalling in in vitro and in vivo systems. Nat. Mater. 2012;11:1038–1043. doi: 10.1038/nmat3430. [DOI] [PubMed] [Google Scholar]
- 9.Etoc F, et al. Subcellular control of Rac-GTPase signalling by magnetogenetic manipulation inside living cells. Nat. Nanotechnol. 2013;8:193–198. doi: 10.1038/nnano.2013.23. [DOI] [PubMed] [Google Scholar]
- 10.Hoffmann C, et al. Spatiotemporal control of microtubule nucleation and assembly using magnetic nanoparticles. Nat. Nanotechnol. 2013;8:199–205. doi: 10.1038/nnano.2012.246. [DOI] [PubMed] [Google Scholar]
- 11.Liu Z, et al. Nanoscale optomechanical actuators for controlling mechanotransduction in living cells. Nat Methods. 2016;13:143–146. doi: 10.1038/nmeth.3689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Seo D, et al. A Mechanogenetic Toolkit for Interrogating Cell Signaling in Space and Time. Cell. 2016;165:1507–1518. doi: 10.1016/j.cell.2016.04.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huang H, Delikanli S, Zeng H, Ferkey DM, Pralle A. Remote control of ion channels and neurons through magnetic-field heating of nanoparticles. Nat. Nanotechnol. 2010;5:602–606. doi: 10.1038/nnano.2010.125. [DOI] [PubMed] [Google Scholar]
- 14.Chen R, Romero G, Christiansen MG, Mohr A, Anikeeva P. Wireless magnetothermal deep brain stimulation. Science. 2015;347:1477–1480. doi: 10.1126/science.1261821. [DOI] [PubMed] [Google Scholar]
- 15.Stanley SA, et al. Radio-wave heating of iron oxide nanoparticles can regulate plasma glucose in mice. Science. 2012;336:604–608. doi: 10.1126/science.1216753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stanley SA, Sauer J, Kane RS, Dordick JS, Friedman JM. Remote regulation of glucose homeostasis in mice using genetically encoded nanoparticles. Nat. Med. 2015;21:92–98. doi: 10.1038/nm.3730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Carvalho-de-Souza JL, et al. Photosensitivity of neurons enabled by cell-targeted gold nanoparticles. Neuron. 2015;86:207–217. doi: 10.1016/j.neuron.2015.02.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Meister M. Physical limits to magnetogenetics. Elife. 2016;5 doi: 10.7554/eLife.17210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Howarth M, et al. Monovalent, reduced-size quantum dots for imaging receptors on living cells. Nat. Methods. 2008;5:397–399. doi: 10.1038/nmeth.1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Farlow J, et al. Formation of targeted monovalent quantum dots by steric exclusion. Nat Methods. 2013;10:1203–1205. doi: 10.1038/nmeth.2682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Banerjee D, Liu AP, Voss NR, Schmid SL, Finn MG. Multivalent display and receptor-mediated endocytosis of transferrin on virus-like particles. Chembiochem. 2010;11:1273–1279. doi: 10.1002/cbic.201000125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dufrene YF, et al. Five challenges to bringing single-molecule force spectroscopy into living cells. Nat. Methods. 2011;8:123–127. doi: 10.1038/nmeth0211-123. [DOI] [PubMed] [Google Scholar]
- 23.Neuman KC, Nagy A. Single-molecule force spectroscopy: optical tweezers, magnetic tweezers and atomic force microscopy. Nat. Methods. 2008;5:491–505. doi: 10.1038/nmeth.1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jiang W, Kim BY, Rutka JT, Chan WC. Nanoparticle-mediated cellular response is size-dependent. Nat. Nanotechnol. 2008;3:145–150. doi: 10.1038/nnano.2008.30. [DOI] [PubMed] [Google Scholar]
- 25.Xu S, Olenyuk BZ, Okamoto CT, Hamm-Alvarez SF. Targeting receptor-mediated endocytotic pathways with nanoparticles: rationale and advances. Adv. Drug Deliv. Rev. 2013;65:121–138. doi: 10.1016/j.addr.2012.09.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Levskaya A, Weiner OD, Lim WA, Voigt CA. Spatiotemporal control of cell signalling using a light-switchable protein interaction. Nature. 2009;461:997–1001. doi: 10.1038/nature08446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shete HK, Prabhu RH, Patravale VB. Endosomal escape: a bottleneck in intracellular delivery. J. Nanosci. Nanotechnol. 2014;14:460–474. doi: 10.1166/jnn.2014.9082. [DOI] [PubMed] [Google Scholar]
- 28.Martens TF, Remaut K, Demeester J, Smedt SCD, Braeckmans K. Intracellular delivery of nanomaterials: How to catch endosomal escape in the act. Nano Today. 2014;9:344–364. [Google Scholar]
- 29.Tseng P, Judy JW, Di Carlo D. Magnetic nanoparticle-mediated massively parallel mechanical modulation of single-cell behavior. Nat. Methods. 2012;9:1113–1119. doi: 10.1038/nmeth.2210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wheeler MA, et al. Genetically targeted magnetic control of the nervous system. Nat. Neurosci. 2016;19:756–761. doi: 10.1038/nn.4265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tseng P, et al. Flexible and stretchable micromagnet arrays for tunable biointerfacing. Adv. Mater. 2015;27:1083–1089. doi: 10.1002/adma.201404849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Farrell D, Majetich SA, Wilcoxon JP. Preparation and Characterization of Monodisperse Fe Nanoparticles. J. Phys. Chem. B. 2003;107:11022–11030. [Google Scholar]
- 33.Zhang S, et al. Halide ion-mediated growth of single crystalline Fe nanoparticles. Nanoscale. 2014;6:4852–4856. doi: 10.1039/c4nr00193a. [DOI] [PubMed] [Google Scholar]
- 34.Zalich MA, Baranauskas VV, Riffle JS, Saunders M, Pierre TGS. Structural and Magnetic Properties of Oxidatively Stable Cobalt Nanoparticles Encapsulated in Graphite Shells. Chem. Mater. 2006;18:2648–2655. [Google Scholar]
- 35.Pankhurst QA, Connolly J, Jones SK, Dobson J. Applications of magnetic nanoparticles in biomedicine. J. Phys. D. 2003;36:167–181. [Google Scholar]
- 36.Matthews BD, LaVan DA, Overby DR, Karavitis J, Ingber DE. Electromagnetic needles with submicron pole tip radii for nanomanipulation of biomolecules and living cells. Appl Phys Lett. 2004;85:2968–2970. [Google Scholar]
- 37.Yan Z, et al. Drosophila NOMPC is a mechanotransduction channel subunit for gentle-touch sensation. Nature. 2013;493:221–225. doi: 10.1038/nature11685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Coste B, et al. Piezo1 and Piezo2 are essential components of distinct mechanically activated cation channels. Science. 2010;330:55–60. doi: 10.1126/science.1193270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kim SE, Coste B, Chadha A, Cook B, Patapoutian A. The role of Drosophila Piezo in mechanical nociception. Nature. 2012;483:209–212. doi: 10.1038/nature10801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.van der Merwe PA. The TCR triggering puzzle. Immunity. 2001;14:665–668. doi: 10.1016/s1074-7613(01)00155-8. [DOI] [PubMed] [Google Scholar]
- 41.Dustin ML, Zhu C. T cells like a firm molecular handshake. Proc. Natl. Acad. Sci. USA. 2006;103:4335–4336. doi: 10.1073/pnas.0600899103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Das DK, et al. Force-dependent transition in the T-cell receptor beta-subunit allosterically regulates peptide discrimination and pMHC bond lifetime. Proc. Natl. Acad. Sci. U. S. A. 2015;112:1517–1522. doi: 10.1073/pnas.1424829112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wichert Gv, et al. RPTP-α acts as a transducer of mechanical force on αv/β3-integrin–cytoskeleton linkages. J. Cell Biol. 2003;161 doi: 10.1083/jcb.200211061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Huang RB, Eniola-Adefeso O. Shear stress modulation of IL-1beta-induced E-selectin expression in human endothelial cells. PLoS One. 2012;7:e31874. doi: 10.1371/journal.pone.0031874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Marshall BT, et al. Direct observation of catch bonds involving cell-adhesion molecules. Nature. 2003;423:190–193. doi: 10.1038/nature01605. [DOI] [PubMed] [Google Scholar]
- 46.Tzima E, et al. A mechanosensory complex that mediates the endothelial cell response to fluid shear stress. Nature. 2005;437:426–431. doi: 10.1038/nature03952. [DOI] [PubMed] [Google Scholar]
- 47.Chen Y, et al. Fluorescence Biomembrane Force Probe: Concurrent Quantitation of Receptor-ligand Kinetics and Binding-induced Intracellular Signaling on a Single Cell. J Vis Exp. 2015:e52975. doi: 10.3791/52975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Luca VC, et al. Notch-Jagged complex structure implicates a catch bond in tuning ligand sensitivity. Science. 2017;355:1320–1324. doi: 10.1126/science.aaf9739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jang JT, et al. Critical enhancements of MRI contrast and hyperthermic effects by dopant-controlled magnetic nanoparticles. Angew. Chem. Int. Ed. 2009;48:1234–1238. doi: 10.1002/anie.200805149. [DOI] [PubMed] [Google Scholar]
- 50.Prodan E, Radloff C, Halas NJ, Nordlander P. A hybridization model for the plasmon response of complex nanostructures. Science. 2003;302:419–422. doi: 10.1126/science.1089171. [DOI] [PubMed] [Google Scholar]
- 51.Jain PK, Lee KS, El-Sayed IH, El-Sayed MA. Calculated absorption and scattering properties of gold nanoparticles of different size, shape, and composition: applications in biological imaging and biomedicine. J. Phys. Chem. B. 2006;110:7238–7248. doi: 10.1021/jp057170o. [DOI] [PubMed] [Google Scholar]
- 52.Cullity BD. Introduction to magnetic materials. Addison-Wesley Pub. Co., Reading, Mass; 1972. [Google Scholar]
- 53.Bercoff PG, Bertorello HR. Exchange constants and transfer integrals of spinel ferrites. J. Magn. Magn. Mater. 1997;169:314–322. [Google Scholar]
- 54.Schiessl W, et al. Magnetic properties of the ZnFe2O4 spinel. Phys. Rev. B Condens Matter. 1996;53:9143–9152. doi: 10.1103/physrevb.53.9143. [DOI] [PubMed] [Google Scholar]
- 55.Yi DK, Lee SS, Papaefthymiou GC, Ying JY. Nanoparticle Architectures Templated by SiO2/Fe2O3 Nanocomposites. Chem. Mater. 2006;18:614–619. [Google Scholar]
- 56.Oldenburg SJ, Averitt RD, Westcott SL, Halas NJ. Nanoengineering of optical resonances. Chem. Phys. Lett. 1998;288:243–247. [Google Scholar]
- 57.Brinson BE, et al. Nanoshells made easy: improving Au layer growth on nanoparticle surfaces. Langmuir. 2008;24:14166–14171. doi: 10.1021/la802049p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sarah LW, Oldenburg SJ, Lee TR, Halas NJ. Formation and Adsorption of Clusters of Gold Nanoparticles onto Functionalized Silica Nanoparticle Surfaces. Langmuir. 1998;14:5396–5401. [Google Scholar]
- 59.Claridge SA, Liang HW, Basu SR, Frechet JM, Alivisatos AP. Isolation of discrete nanoparticle-DNA conjugates for plasmonic applications. Nano Lett. 2008;8:1202–1206. doi: 10.1021/nl0802032. [DOI] [PubMed] [Google Scholar]
- 60.Seo D, Lee H, Lee J-u, Haas TJ, Jun Y-w. Monovalent plasmonic nanoparticles for biological applications. Proc. of SPIE. 2016;9722:97220I–97221. [Google Scholar]
- 61.Batchelor GK. An introduction to fluid dynamics. U.P., Cambridge: 1967. [Google Scholar]
- 62.Rogers HB, Anani T, Choi YS, Beyers RJ, David AE. Exploiting Size-Dependent Drag and Magnetic Forces for Size-Specific Separation of Magnetic Nanoparticles. Int. J. Mol. Sci. 2015;16:20001–20019. doi: 10.3390/ijms160820001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wang X, Ha T. Defining single molecular forces required to activate integrin and notch signaling. Science. 2013;340:991–994. doi: 10.1126/science.1231041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ho D, et al. Force-driven separation of short double-stranded DNA. Biophys. J. 2009;97:3158–3167. doi: 10.1016/j.bpj.2009.09.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kuhner F, Morfill J, Neher RA, Blank K, Gaub HE. Force-induced DNA slippage. Biophys. J. 2007;92:2491–2497. doi: 10.1529/biophysj.106.095836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Cocco S, Monasson R, Marko JF. Force and kinetic barriers to unzipping of the DNA double helix. Proc. Natl. Acad. Sci. USA. 2001;98:8608–8613. doi: 10.1073/pnas.151257598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Jain PK, El-Sayed MA. Universal scaling of plasmon coupling in metal nanostructures: extension from particle pairs to nanoshells. Nano Lett. 2007;7:2854–2858. doi: 10.1021/nl071496m. [DOI] [PubMed] [Google Scholar]
- 68.Wang Y, et al. Visualizing the mechanical activation of Src. Nature. 2005;434:1040–1045. doi: 10.1038/nature03469. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.