Abstract
Aim: Stroke is associated closely with vascular homeostasis, and several complex processes and interacting pathways, which involve various genetic and environmental factors, contribute to the risk of stroke. Although adrenomedullin (ADM) has a number of physiological and vasoprotective functions, there are few studies of the ADM receptor system in humans. The ADM receptor comprises a calcitonin-receptor-like receptor (CLR) and receptor activity-modifying proteins (RAMPs). We analyzed single nucleotide polymorphisms (SNPs) in the RAMP2 and CLR genes to determine their association with stroke in the light of gene-environment interactions.
Methods: Using cross-sectional data from the Japan Multi-Institutional Collaborative Cohort Study in the baseline surveys, 14,087 participants from 12 research areas were genotyped. We conducted a hypothesis-based association between stroke prevalence and SNPs in the RAMP2 and CLR genes based on data abstracted from two SNPs in RAMP2 and 369 SNPs in CLR. We selected five SNPs from among the CLR variants (rs77035639, rs3815524, rs75380157, rs574603859, and rs147565266) and one RAMP2 SNP (rs753152), which were associated with stroke, for analysis.
Results: Five of the SNPs (rs77035639, rs3815524, rs75380157, rs147565266, and rs753152) showed no significant association with obesity, ischemic heart disease, hypertension, dyslipidemia, and diabetes. In the logistic regression analysis, rs574603859 had a lower odds ratio (0.238; 95% confidence interval, 0.076–0.745, adjusted for age, sex, and research area) and the other SNPs had higher odds ratios for association with stroke.
Conclusions: This was the first study to investigate the relationships between ADM receptor genes (RAMP2 and CLR) and stroke in the light of gene-environment interactions in human.
Keywords: Adrenomedullin, Receptor activity-modifying protein 2, Calcitonin-receptor-like receptor, Stroke
Introduction
The vascular system plays a crucial role in organ homeostasis, being essential for organ and tissue construction, the supply of oxygen and nutrients, and mobilization of inflammatory cells to regions of injury1, 2). Current and novel therapeutic approaches aimed at improving vascular function provide real benefits with respect to reducing cerebrovascular disease3). In addition, the vascular system can be considered the largest system in the body, given its length and area, and via its active secretion of bioactive molecules, plays a central role in vascular homeostasis4–6). Revealing the mechanisms underlying the functional integrity of the vascular system could lead to novel approaches to therapy and preventive medicine.
Strokes are associated closely with vascular homeostasis, and disruption of vascular function can also cause a stroke. A stroke is the clinical culmination of several complex processes and interacting pathways that involve various genetic and environmental factors7). Genetic contributions to strokes may result from common variants with small effect sizes, rare variants with large effect sizes, or their combination8). However, environmental risk factors are associated with the pathogenesis of stroke, and considerable evidence suggests that geneenvironment interactions are important9).
Adrenomedullin (ADM) is a vasoactive peptide first identified in human pheochromocytoma10). Although ADM is secreted by various organs and tissues, it is produced mainly by vascular endothelial cells and serves a number of physiological functions11–14). The ADM receptor is a seven transmembrane domain G protein-coupled receptor, called the calcitonin-receptor-like receptor (CLR)15, 16). The specificity of CLR for ADM is thought to be regulated by receptor activity-modifying proteins (RAMPs), which are membrane proteins having a single membrane-spanning domain. Analysis of genetically engineered knockout mice revealed that the ADM signal is indispensable for CLR and RAMP2 function17–20). By contrast, there have been few studies of the ADM receptor system in humans.
The present study aimed to analyze RAMP2 and CLR single nucleotide polymorphisms (SNPs) and evaluate their association with stroke in the light of gene-environment interactions. This analysis was conducted as a cross-sectional study using a large-scale pooled analysis of the Japanese general population.
Methods
Study Participants
In the present study, we evaluated participant data collected during the Japan Multi-Institutional Collaborative Cohort (J-MICC) Study from the baseline surveys using the cross-sectional data. That cohort study evaluated the general Japanese population in 12 research areas, using genetic and clinical data to detect and confirm gene-environment interactions related to lifestyle-associated diseases21). The study participants were 35–69 years old, and were enrolled after responding to study announcements in their specific research areas, attending health check-up examinations that were commissioned by their local governments, visiting local health check-up centers, or visiting a cancer hospital. A total of 14,539 participants were selected. We analyzed the data while minimizing the number of deleted participants. Each parameter was separated in the analysis because of missing data.
The J-MICC study participants included citizens, health check examiners, and first-visit patients to a cancer hospital. All participants in this study gave written informed consent. The study protocol was approved by the Ethics Committees at Aichi Cancer Center, the Nagoya University Graduate School of Medicine, and other institutions participating in the J-MICC study. The present study was conducted according to the principles expressed in the World Medical Association Declaration of Helsinki.
Lifestyle and Blood Biochemistry Data
In the present study, we evaluated the lifestyle and medical information obtained through self-administered questionnaires (alcohol consumption status, smoking habits, and physical exercise). The body mass index (BMI) was calculated as weight (kg) divided by the square of height (m2). Obesity was defined as a BMI ≥ 25.0 kg/m2. Alcohol consumption of each type of beverage was determined by the average number of drinks per day, and then converted into the Japanese sake unit, ‘gou’ (180 ml), which is equivalent to 23 g of ethanol (0, 0.1–22.9, 23.0–45.9, or ≥ 46.0 g ethanol/day). Regular physical activity was defined as three times a week and lasting over 30 minutes. Anamnesis and medication history were also assessed using a questionnaire. Information on stroke (n = 248) and ischemic heart disease (n = 403) was available from the self-administered questionnaires. Hypertension was defined as a systolic/diastolic blood pressure ≥ 140/90 mm Hg and/or current use of medication for hypertension. Dyslipidemia was defined as non-high density lipoprotein-C (HDL-C) ≥ 170 mg dl−1 and/or HDL-C < 40 mg dl−1 and/or current use of medication for dyslipidemia. Diabetes was defined as a glycated hemoglobin (HbA1c) level ≥ 6.5% and/or current use of medication for diabetes. The participants who have the absence of laboratory data and/or insufficient data were excluded in each analytic criterion.
In addition, blood chemistry data (serum levels of triglycerides, total cholesterol, HDL-C, non-HDL-C, creatinine, and HbA1c) and anthropometric data were obtained from health check-ups performed in the research areas. The estimated glomerular filtration rate (eGFR) was calculated using the following equation: eGFR (mL/min/1.73 m2) = 194 × creatinine−1.094 × age−0.287 (for men) and eGFR (mL/min/1.73 m2) = 194 × creatinine−1.094 × age−0.287 × 0.739 (for women)22). Each blood sample was centrifuged and the plasma was separated and stored at −80°C until analysis. Laboratories in each research area analyzed the serum samples.
Genotyping and Quality Control Filtering
In the study, buffy coat fractions and DNA were prepared from blood samples and stored at −80°C at the central J-MICC study office. DNA was extracted from all buffy coat fractions using a BioRobot M48 Workstation (Qiagen Group, Tokyo, Japan) at the central study office. For the samples from two areas (Fukuoka and Kyushu-KOPS), DNA was extracted locally from samples of whole blood, using an automatic nucleic acid isolation system (NA-3000, Kurabo, Co., Ltd, Osaka, Japan). The 14,539 study participants from the 12 areas of the J-MICC study were genotyped at RIKEN Center for Integrative Medicine using a Human-OmniExpressExome-8 v1.2 BeadChip array (Illumina Inc., San Diego, CA, USA). Twenty-six samples with inconsistent sex information between the questionnaire and the estimate from the genotyping results were excluded. The identity-by-descent method implemented in the PLINK 1.9 software23, 24) identified 388 closely related pairs (pi-hat > 0.1875) and one sample of each pair was excluded. Principal component analysis (PCA)25, 26) with the 1000 Genomes reference panel (phase 3)27) detected 34 subjects whose estimated ancestries were outside of the Japanese population28). These 34 samples were excluded. All the remaining 14,091 samples met a sample-wise genotype call rate criterion (≥ 0.99). SNPs with a genotype call rate < 0.98 and/or a Hardy-Weinberg equilibrium exact test P-value < 1 × 10−6 were removed, resulting in 873,254 autosomal variants. Among these, 298,644 variants with a low minor allele frequency (MAF) < 0.01 were excluded. This quality control filtering resulted in 14,091 participants and 570,162 SNPs. After genotyping, data from four participants who withdrew from follow-up were excluded from further analysis, resulting in 14,087 participants were included in the analyses.
Genotype Imputation
Genotype imputation was performed using SHA-PIT29) and Minimac330) software based on the 1000 Genomes Project cosmopolitan reference panel (phase 3)27). After the genotype imputation, variants with an R2 value < 0.3 were excluded, resulting in 12,617,547 variants. We identified variants in the RAMP2 and CLR loci, which identified two and 369 SNPs, respectively.
Statistical Analyses
We compared the associations between various genotypes and stroke, after combining the heterozygous and minor homozygous alleles because of the small number of minor homozygotes. We selected SNPs from the RAMP2 and CLR genes that exhibited a statistically significant association with stroke. Continuous variables are expressed as mean ± standard deviation (SD) and categorical data are expressed as sums and percentages. Inter-group comparisons were performed using Welch's t-tests for continuous variables, and the chi-square or Fisher's exact tests for categorical variables (sex, alcohol consumption, regular physical activity, smoking, stroke, obesity, ischemic heart disease, hypertension, dyslipidemia, and diabetes). The chisquare test was performed to examine the Hardy–Weinberg equilibrium for each locus studied. Odds ratios (OR) and 95% confidence intervals (CI) were calculated using logistic regression analyses, in which stroke was defined as the dependent variable, and age, sex, research area, alcohol intake, current smoking, regular physical activity, obesity, hypertension, diabetes, dyslipidemia, and ischemic heart disease, were included as independent variables. The haplotype frequency and linkage disequilibrium of the SNPs were estimated using Haploview31). All statistical tests were two-tailed, and differences with a p-value < 0.05 were considered statistically significant. SPSS software (version 18.0, SPSS, Japan, Inc.) was used for all statistical analyses.
Results
Among these 14,087 participants, the mean age of the included 6337 men was 55.4 years, compared to 54.3 years for the 7750 women.
We identified two and 369 SNPs among the genetic variants of RAMP2 and CLR, respectively (Supplementary Table 1, Chromosomal locations were described based on hg19/GRCh37 coordinates). Supplementary Fig. 1 shows the linkage disequilibrium analyses of 13 CLR SNPs associated with stroke identified using the chi-square test. The position of the studied SNPs in CLR is shown. Pair-wise SNP R-squared D' linkage values (multiplied by 100) are also shown. We then selected five SNPs from among the CLR variants (rs77035639, rs3815524, rs75380157, rs574603859, and rs147565266) to avoid similar haplotypes. Similarly, RAMP2 SNP (rs753152), which is associated with stroke, was selected for analysis. The distributions of genotypes and alleles of the evaluated SNPs are summarized in Supplementary Table 2. Supplementary Fig. 2 shows exons (shown as boxes) 1–4 for RAMP2, and exons 1–15 for CLR. For analysis, we compared the associations between genotypes and stroke, after combining the heterozygous and minor homozygous alleles because of the small number of minor homozygotes alleles.
Supplementary Table 1. Genetic variants of the RAMP2 and CLR loci.
| Gene | chromosome | Position | |||||||
|---|---|---|---|---|---|---|---|---|---|
| RAMP2 | 17 | 40913366 | 188216783 | 188225844 | 188236012 | 188252411 | 188269685 | 188287434 | 188298742 |
| 40913505 | 188216807 | 188225970 | 188236025 | 188252561 | 188269709 | 188287456 | 188298787 | ||
| CLR | 2 | 188206953 | 188217247 | 188226414 | 188236053 | 188252608 | 188270337 | 188288165 | 188300228 |
| 188207245 | 188217379 | 188226520 | 188236279 | 188253796 | 188270560 | 188288259 | 188300873 | ||
| 188207585 | 188217500 | 188226900 | 188236354 | 188253902 | 188270687 | 188288266 | 188301149 | ||
| 188207611 | 188217501 | 188227008 | 188236378 | 188254046 | 188270864 | 188288315 | 188301544 | ||
| 188208012 | 188217706 | 188227300 | 188236458 | 188254173 | 188270914 | 188289056 | 188302053 | ||
| 188208120 | 188217738 | 188227302 | 188236819 | 188254581 | 188270925 | 188289079 | 188302587 | ||
| 188208130 | 188218342 | 188227302 | 188237045 | 188255091 | 188271085 | 188289172 | 188302634 | ||
| 188208290 | 188218458 | 188227613 | 188237868 | 188255237 | 188271819 | 188289174 | 188302770 | ||
| 188208736 | 188218683 | 188227754 | 188238101 | 188255432 | 188272092 | 188289448 | 188303700 | ||
| 188209158 | 188218937 | 188227921 | 188238103 | 188255549 | 188272460 | 188289795 | 188303845 | ||
| 188209159 | 188218946 | 188228516 | 188238334 | 188255912 | 188272951 | 188289849 | 188303979 | ||
| 188209179 | 188219052 | 188228911 | 188239574 | 188256158 | 188273829 | 188289964 | 188304095 | ||
| 188209709 | 188219186 | 188229007 | 188239647 | 188256237 | 188275146 | 188289971 | 188304213 | ||
| 188210214 | 188219325 | 188229335 | 188239809 | 188256685 | 188275930 | 188290419 | 188304446 | ||
| 188210256 | 188219447 | 188229622 | 188240559 | 188256910 | 188275982 | 188290490 | 188304891 | ||
| 188210257 | 188219468 | 188229739 | 188241519 | 188257985 | 188276272 | 188290717 | 188305046 | ||
| 188210415 | 188219975 | 188229820 | 188241522 | 188258904 | 188276515 | 188290969 | 188305110 | ||
| 188210586 | 188220301 | 188230146 | 188241953 | 188259620 | 188276584 | 188290969 | 188305797 | ||
| 188210673 | 188220317 | 188230333 | 188242861 | 188260401 | 188276906 | 188291105 | 188306215 | ||
| 188210960 | 188220384 | 188230588 | 188243363 | 188260747 | 188276987 | 188291382 | 188306229 | ||
| 188211005 | 188220446 | 188230692 | 188243503 | 188260912 | 188277123 | 188291723 | 188306543 | ||
| 188211112 | 188220865 | 188230976 | 188243620 | 188260913 | 188277455 | 188291869 | 188306768 | ||
| 188211296 | 188221302 | 188231072 | 188243633 | 188260921 | 188277682 | 188292242 | 188306985 | ||
| 188211443 | 188221350 | 188231216 | 188243658 | 188261266 | 188278035 | 188292458 | 188307038 | ||
| 188211568 | 188221547 | 188231433 | 188243684 | 188261890 | 188278203 | 188293391 | 188307334 | ||
| 188211568 | 188221648 | 188231645 | 188243940 | 188261922 | 188278226 | 188293438 | 188307429 | ||
| 188211610 | 188221793 | 188231887 | 188244430 | 188262362 | 188278525 | 188293545 | 188307608 | ||
| 188211789 | 188221911 | 188232223 | 188245890 | 188262468 | 188278822 | 188293614 | 188307747 | ||
| 188212371 | 188221943 | 188232805 | 188246023 | 188263003 | 188279122 | 188293635 | 188307962 | ||
| 188212423 | 188222351 | 188233502 | 188247648 | 188263325 | 188279606 | 188293921 | 188308056 | ||
| 188213235 | 188222428 | 188233524 | 188247843 | 188264246 | 188280382 | 188294598 | 188308240 | ||
| 188213336 | 188222469 | 188233714 | 188248440 | 188264602 | 188280870 | 188294980 | 188308604 | ||
| 188213538 | 188222560 | 188234194 | 188248594 | 188264833 | 188280896 | 188295534 | 188308640 | ||
| 188213819 | 188222561 | 188234520 | 188248663 | 188265242 | 188282330 | 188295611 | 188308853 | ||
| 188214239 | 188222581 | 188234678 | 188248727 | 188265543 | 188282703 | 188296132 | 188309875 | ||
| 188214694 | 188222928 | 188234844 | 188249420 | 188266531 | 188283002 | 188296488 | 188310118 | ||
| 188214823 | 188222946 | 188234928 | 188250234 | 188266951 | 188283061 | 188297133 | 188310305 | ||
| 188214924 | 188223256 | 188235033 | 188250476 | 188267147 | 188283123 | 188297160 | 188311515 | ||
| 188215045 | 188223889 | 188235100 | 188250718 | 188267147 | 188284000 | 188297214 | 188311900 | ||
| 188215156 | 188224057 | 188235162 | 188250860 | 188267193 | 188284127 | 188297348 | 188311992 | ||
| 188215209 | 188224322 | 188235258 | 188251427 | 188267470 | 188284824 | 188297349 | |||
| 188215241 | 188224457 | 188235340 | 188251432 | 188268001 | 188285455 | 188297388 | |||
| 188215292 | 188224928 | 188235611 | 188251535 | 188268541 | 188285710 | 188298159 | |||
| 188215299 | 188225030 | 188235956 | 188251702 | 188268931 | 188286217 | 188298197 | |||
| 188216078 | 188225240 | 188236000 | 188251972 | 188269235 | 188286516 | 188298341 | |||
Supplementary Fig. 1.
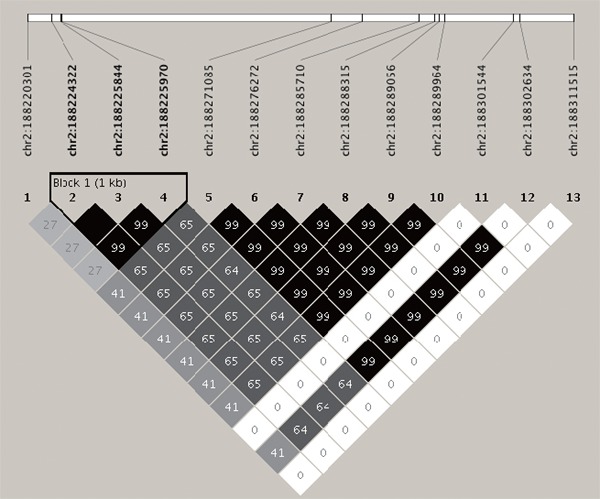
The linkage disequilibrium analyses of 13 CLR (calcitonin-receptor-like receptor) SNPs associated with stroke.
The haplotype structure and the position of the studied single nucleotide polymorphisms in the CLR gene exhibited a statistically significant association with stroke.
Supplementary Table 2. Allele and genotype frequencies of the RAMP2 and CLR genes in the participants.
| SNP | Allele frequency | Genotype frequency | n | P for Hardy-Weinberg equilibrium |
|---|---|---|---|---|
| rs753152 (T/G) | ||||
| TT | T = 0.965 | T/T = 0.933 | 13137 | |
| TG | T/G = 0.065 | 921 | 0.003 | |
| GG | G = 0.035 | G/G = 0.002 | 29 | |
| rs77035639 (A/G) | ||||
| AA | A = 0.983 | A/A = 0.965 | 13604 | |
| AG | A/G = 0.034 | 476 | 0.177 | |
| GG | G = 0.017 | G/G = 0.001 | 7 | |
| rs3815524 (G/C) | ||||
| GG | G = 0.940 | G/G = 0.885 | 12464 | |
| GC | G/C = 0.110 | 1552 | 0.003 | |
| CC | C = 0.060 | C/C = 0.005 | 71 | |
| rs75380157 (A/T) | ||||
| AA | A = 0.959 | A/A = 0.921 | 12978 | |
| AT | A/T = 0.076 | 1073 | 0.006 | |
| TT | T = 0.041 | T/T = 0.003 | 36 | |
| rs574603859 (A/T) | ||||
| AA | A = 0.975 | A/A = 0.952 | 13405 | |
| AT | A/T = 0.047 | 664 | 0.001 | |
| TT | T = 0.025 | T/T = 0.001 | 18 | |
| rs147565266 (T/A) | ||||
| TT | T = 0.998 | T/T = 0.996 | 14024 | |
| TA | T/A = 0.004 | 63 | 0.790 | |
| AA | A = 0.002 | A/A = 0.000 | 0 |
Supplementary Fig. 2.
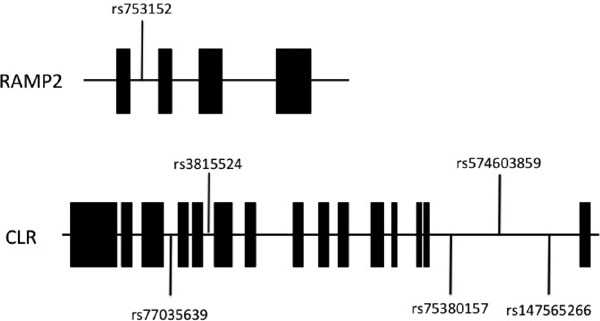
Organization of the RAMP2 (receptor activity-modifying protein 2) and CLR (calcitonin-receptor-like receptor) genes and locations of the SNPs used in the present study. Closed boxes indicate exons and lines represent introns.
Table 1 shows the distribution of stroke for each SNP. The major homozygotes had significantly higher incidences of stroke compared with the heterozygotes and minor homozygotes, except for rs574603859. SNP rs574603859 showed an inverse ratio between major homozygotes and the other genotypes. Table 2 summarizes the baseline characteristics of the participants divided into two groups, classified by major homozygous alleles versus heterozygous and minor homozygous alleles. None of the SNPs showed a constant tendency for these characteristics. Table 3 shows the distribution of obesity, ischemic heart disease, hypertension, dyslipidemia, and diabetes for each SNP. SNP rs574603859 was associated with a higher incidence of obesity in the heterozygotes and minor homozygotes. SNP rs753152 was associated with a higher incidence of diabetes in the heterozygotes and minor homozygotes. The other four SNPs showed no significant association with these diseases.
Table 1. Genotype and allele distributions in stroke.
| SNPs Chromosome: position | Genotype |
p value | ||
|---|---|---|---|---|
| Major Homo | Hetero+Minor Homo | |||
| rs753152 (T/G) chr17: 40913505 | control (n) | 12139 | 881 | 0.003 |
| (%) | 93.2% | 6.8% | ||
| Stroke (n) | 219 | 29 | ||
| (%) | 88.30% | 11.7% | ||
| rs77035639 (A/G) chr2: 188220301 | control (n) | 12579 | 441 | 0.020 |
| (%) | 96.6% | 3.4% | ||
| Stroke (n) | 232 | 16 | ||
| (%) | 93.5% | 6.5% | ||
| rs3815524 (G/C) chr2: 188224322 | control (n) | 11519 | 1501 | 0.041 |
| (%) | 88.5% | 11.5% | ||
| Stroke (n) | 209 | 39 | ||
| (%) | 84.3% | 15.7% | ||
| rs75380157 (A/T) chr2: 188271085 | control (n) | 12005 | 1015 | 0.002 |
| (%) | 92.2% | 7.8% | ||
| Stroke (n) | 215 | 33 | ||
| (%) | 86.7% | 13.3% | ||
| rs574603859 (A/T) chr2: 188301544 | control (n) | 12375 | 645 | 0.003 |
| (%) | 95.0% | 5.0% | ||
| Stroke (n) | 245 | 3 | ||
| (%) | 98.8% | 1.2% | ||
| rs147565266 (T/A) chr2: 188311515 | control (n) | 12967 | 53 | 0.022 |
| (%) | 99.6% | 0.4% | ||
| Stroke (n) | 244 | 4 | ||
| (%) | 98.4% | 1.6% | ||
Homo, homozygote; Hetero, heterozygote.
Table 2. Characteristics of study participants for each single nucleotide polymorphism (SNP).
| Genotype | rs753152 |
p value | |||
|---|---|---|---|---|---|
| Major Homo |
Hetero+Minor Homo |
||||
| n | mean ± SD (%) | n | mean ± SD (%) | ||
| Sex (male) | 5890 | 44.8% | 447 | 47.1% | 0.188 |
| Age (year) | 13137 | 54.8 ± 9.4 | 950 | 55.0 ± 9.5 | 0.462 |
| BMI (kg/m2) | 10578 | 23.2 ± 3.4 | 752 | 23.1 ± 3.5 | 0.809 |
| Systolic blood pressure (mmHg) | 10514 | 128 ± 20.2 | 747 | 128 ± 19.1 | 0.528 |
| Diastolic blood pressure (mmHg) | 10513 | 78.2 ± 12.3 | 747 | 77.9 ± 11.7 | 0.441 |
| Triglyceride (mg/dl) | 10861 | 128 ± 96.5 | 792 | 130 ± 94.0 | 0.585 |
| Total cholesterol (mg/dl) | 9947 | 211 ± 34.7 | 749 | 211 ± 36.0 | 0.821 |
| nonHDL-C (mg/dl) | 9946 | 148 ± 35.1 | 749 | 149 ± 36.1 | 0.781 |
| HDL-C (mg/dl) | 10863 | 62.7 ± 16.3 | 792 | 62.3 ± 15.8 | 0.458 |
| Hemoglobin A1C (%) | 8057 | 5.55 ± 0.73 | 581 | 5.61 ± 0.74 | 0.055 |
| eGFR (mL/min/1.73 m2) | 10509 | 78.8 ± 15.1 | 774 | 78.3 ± 14.9 | 0.374 |
| Alcohol drinking | |||||
| 0 g/d | 5912 | 45.8% | 433 | 46.6% | |
| 0.1–22.9 g/d | 4224 | 32.7% | 305 | 32.8% | |
| 23–45.9 g/d | 1412 | 10.9% | 101 | 10.9% | 0.870 |
| 46.0+ g/d | 1359 | 10.5% | 90 | 9.7% | |
| Smoking | 2471 | 18.8% | 171 | 18.0% | 0.574 |
| Regular physical activity | 3830 | 29.2% | 289 | 30.5% | 0.417 |
| Genotype | rs77035639 |
p value | |||
|---|---|---|---|---|---|
| Major Homo |
Hetero+Minor Homo |
||||
| n | mean ± SD (%) | n | mean ± SD (%) | ||
| Sex (male) | 6114 | 44.9% | 223 | 46.2% | 0.609 |
| Age (year) | 13604 | 54.8 ± 9.4 | 483 | 54.5 ± 9.3 | 0.587 |
| BMI (kg/m2) | 10937 | 23.2 ± 3.4 | 393 | 23.3 ± 3.2 | 0.562 |
| Systolic blood pressure (mmHg) | 10874 | 128 ± 20.1 | 387 | 128 ± 20.4 | 0.605 |
| Diastolic blood pressure (mmHg) | 10873 | 78.2 ± 12.2 | 387 | 77.6 ± 12.3 | 0.318 |
| Triglyceride (mg/dl) | 11246 | 128 ± 96.3 | 407 | 129 ± 96.0 | 0.868 |
| Total cholesterol (mg/dl) | 10326 | 211 ± 34.7 | 370 | 211 ± 36.1 | 0.966 |
| nonHDL-C (mg/dl) | 10325 | 148 ± 35.1 | 370 | 149 ± 36.5 | 0.879 |
| HDL-C (mg/dl) | 11248 | 62.7 ± 16.3 | 407 | 62.5 ± 15.6 | 0.795 |
| Hemoglobin A1C (%) | 8347 | 5.55 ± 0.73 | 291 | 5.63 ± 0.91 | 0.171 |
| eGFR (mL/min/1.73 m2) | 10899 | 78.7 ± 15.1 | 384 | 78.2 ± 13.8 | 0.527 |
| Alcohol drinking | |||||
| 0 g/d | 6136 | 45.9% | 209 | 44.5% | |
| 0.1–22.9 g/d | 4376 | 32.7% | 153 | 32.6% | |
| 23–45.9 g/d | 1458 | 10.9% | 55 | 11.7% | 0.856 |
| 46.0+ g/d | 1396 | 10.4% | 53 | 11.3% | |
| Smoking | 2548 | 18.7% | 94 | 19.5% | 0.682 |
| Regular physical activity | 3985 | 29.3% | 134 | 27.7% | 0.475 |
| Genotype | rs3815524 |
p value | |||
|---|---|---|---|---|---|
| Major Homo |
Hetero+Minor Homo |
||||
| n | mean ± SD (%) | n | mean ± SD (%) | ||
| Sex (male) | 5613 | 45.0% | 724 | 44.6% | 0.750 |
| Age (year) | 12464 | 54.8 ± 9.4 | 1623 | 54.7 ± 9.4 | 0.816 |
| BMI (kg/m2) | 10010 | 23.2 ± 3.4 | 1320 | 23.2 ± 3.3 | 0.950 |
| Systolic blood pressure (mmHg) | 9949 | 128 ± 20.1 | 1312 | 128 ± 20.1 | 0.868 |
| Diastolic blood pressure (mmHg) | 9948 | 78.3 ± 12.2 | 1312 | 77.9 ± 12.1 | 0.374 |
| Triglyceride (mg/dl) | 10288 | 128 ± 96.3 | 1365 | 130 ± 96.3 | 0.447 |
| Total cholesterol (mg/dl) | 9426 | 211 ± 34.8 | 1270 | 212 ± 34.2 | 0.703 |
| nonHDL-C (mg/dl) | 9425 | 148 ± 35.2 | 1270 | 149 ± 35.0 | 0.382 |
| HDL-C (mg/dl) | 10289 | 62.7 ± 16.3 | 1366 | 62.3 ± 15.8 | 0.372 |
| Hemoglobin A1C (%) | 7659 | 5.56 ± 0.74 | 979 | 5.55 ± 0.66 | 0.684 |
| eGFR (mL/min/1.73 m2) | 9966 | 78.8 ± 15.2 | 1317 | 78.3 ± 14.1 | 0.237 |
| Alcohol drinking | |||||
| 0 g/d | 5615 | 45.8% | 730 | 46.1% | |
| 0.1–22.9 g/d | 4007 | 32.7% | 522 | 32.9% | |
| 23–45.9 g/d | 1336 | 10.9% | 177 | 11.2% | 0.848 |
| 46.0+ g/d | 1293 | 10.6% | 156 | 9.8% | |
| Smoking | 2345 | 18.8% | 297 | 18.3% | 0.635 |
| Regular physical activity | 3655 | 29.4% | 464 | 28.6% | 0.542 |
| Genotype | rs75380157 |
p value | |||
|---|---|---|---|---|---|
| Major Homo |
Hetero+Minor Homo |
||||
| n | mean ± SD (%) | n | mean ± SD (%) | ||
| Sex (male) | 5843 | 45.5% | 494 | 44.5% | 0.777 |
| Age (year) | 12978 | 54.8 ± 9.4 | 1109 | 54.6 ± 9.4 | 0.629 |
| BMI (kg/m2) | 10439 | 23.2 ± 3.4 | 891 | 23.0 ± 3.2 | 0.280 |
| Systolic blood pressure (mmHg) | 10377 | 128 ± 20.1 | 884 | 128 ± 20.3 | 0.845 |
| Diastolic blood pressure (mmHg) | 10376 | 78.3 ± 12.2 | 884 | 77.9 ± 12.3 | 0.367 |
| Triglyceride (mg/dl) | 10734 | 128 ± 96.1 | 919 | 129 ± 98.9 | 0.663 |
| Total cholesterol (mg/dl) | 9829 | 211 ± 34.8 | 867 | 211 ± 34.4 | 0.888 |
| nonHDL-C (mg/dl) | 9829 | 148 ± 35.2 | 867 | 149 ± 34.7 | 0.930 |
| HDL-C (mg/dl) | 10735 | 62.7 ± 16.3 | 920 | 62.5 ± 15.6 | 0.723 |
| Hemoglobin A1C (%) | 7954 | 5.56 ± 0.74 | 684 | 5.56 ± 0.71 | 0.794 |
| eGFR (mL/min/1.73 m2) | 10393 | 78.8 ± 15.2 | 890 | 78.1 ± 14.0 | 0.190 |
| Alcohol drinking | |||||
| 0 g/d | 5841 | 45.8% | 504 | 46.5% | |
| 0.1–22.9 g/d | 4169 | 32.7% | 360 | 33.2% | |
| 23–45.9 g/d | 1393 | 10.9% | 120 | 11.1% | 0.637 |
| 46.0+ g/d | 1348 | 10.6% | 101 | 9.3% | |
| Smoking | 2424 | 18.7% | 218 | 19.7% | 0.424 |
| Regular physical activity | 3801 | 29.3% | 318 | 28.7% | 0.679 |
| Genotype | rs74603859 |
p value | |||
|---|---|---|---|---|---|
| Major Homo |
Hetero+Minor Homo |
||||
| n | mean ± SD (%) | n | mean ± SD (%) | ||
| Sex (male) | 6044 | 45.1% | 293 | 43.0% | 0.287 |
| Age (year) | 13405 | 54.8 ± 9.4 | 682 | 54.8 ± 9.2 | 0.889 |
| BMI (kg/m2) | 10778 | 23.1 ± 3.4 | 552 | 23.6 ± 3.5 | 0.001 |
| Systolic blood pressure (mmHg) | 10711 | 128 ± 20.1 | 550 | 130 ± 20.6 | 0.020 |
| Diastolic blood pressure (mmHg) | 10710 | 78.2 ± 12.2 | 550 | 79.0 ± 12.1 | 0.137 |
| Triglyceride (mg/dl) | 11082 | 128 ± 95.0 | 571 | 133 ± 118 | 0.192 |
| Total cholesterol (mg/dl) | 10183 | 211 ± 34.8 | 513 | 212 ± 34.4 | 0.871 |
| nonHDL-C (mg/dl) | 10182 | 148 ± 35.2 | 513 | 150 ± 34.8 | 0.393 |
| HDL-C (mg/dl) | 11084 | 62.7 ± 16.3 | 571 | 61.9 ± 15.9 | 0.213 |
| Hemoglobin A1C (%) | 8235 | 5.55 ± 0.72 | 403 | 5.59 ± 0.94 | 0.277 |
| eGFR (mL/min/1.73 m2) | 10732 | 78.7 ± 15.1 | 551 | 79.4 ± 14.7 | 0.302 |
| Alcohol drinking | |||||
| 0 g/d | 6033 | 45.8% | 312 | 46.7% | |
| 0.1–22.9 g/d | 4325 | 32.8% | 204 | 30.5% | |
| 23–45.9 g/d | 1432 | 10.9% | 81 | 12.1% | 0.558 |
| 46.0+ g/d | 1378 | 10.5% | 71 | 10.6% | |
| Smoking | 2523 | 18.8% | 119 | 17.5% | 0.392 |
| Regular physical activity | 3905 | 29.2% | 214 | 31.4% | 0.210 |
| Genotype | rs147565266 |
p value | |||
|---|---|---|---|---|---|
| Major Homo |
Hetero+Minor Homo |
||||
| n | mean ± SD (%) | n | mean ± SD (%) | ||
| Sex (male) | 6305 | 45.0% | 32 | 50.8% | 0.376 |
| Age (year) | 14024 | 54.8 ± 9.4 | 63 | 56.0 ± 8.3 | 0.312 |
| BMI (kg/m2) | 11277 | 23.2 ± 3.4 | 53 | 23.3 ± 3.2 | 0.739 |
| Systolic blood pressure (mmHg) | 11209 | 128 ± 20.1 | 52 | 133 ± 26.2 | 0.189 |
| Diastolic blood pressure (mmHg) | 11208 | 78.2 ± 12.2 | 52 | 79.6 ± 14.6 | 0.501 |
| Triglyceride (mg/dl) | 11599 | 128 ± 95.9 | 54 | 151 ± 161 | 0.293 |
| Total cholesterol (mg/dl) | 10644 | 211 ± 34.7 | 52 | 215 ± 36.6 | 0.446 |
| nonHDL-C (mg/dl) | 10643 | 148 ± 35.1 | 52 | 152 ± 39.1 | 0.488 |
| HDL-C (mg/dl) | 11601 | 62.7 ± 16.2 | 54 | 62.9 ± 21.2 | 0.950 |
| Hemoglobin A1C (%) | 8592 | 5.56 ± 0.73 | 46 | 5.63 ± 0.84 | 0.520 |
| eGFR (mL/min/1.73 m2) | 11230 | 78.7 ± 15.1 | 53 | 80.2 ± 14.2 | 0.482 |
| Alcohol drinking | |||||
| 0 g/d | 6318 | 45.9% | 27 | 43.5% | |
| 0.1–22.9 g/d | 4513 | 32.8% | 16 | 25.8% | |
| 23–45.9 g/d | 1505 | 10.9% | 8 | 12.9% | 0.226 |
| 46.0+ g/d | 1438 | 10.4% | 11 | 17.7% | |
| Smoking | 2632 | 18.8% | 10 | 15.9% | 0.631 |
| Regular physical activity | 4105 | 29.3% | 14 | 22.2% | 0.268 |
Homo, homozygote; Hetero, heterozygote.
Table 3. Genotype and allele distributions in obesity, ischemic heart disease, hypertension, dyslipidemia, and diabetes.
| SNPs | Genotype | Obesity |
Ischemic heart disease |
Hypertension |
Dyslipidemia |
Diabetes |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ≥ 25 | < 25 | p value | (−) | (+) | p value | (−) | (+) | p value | (−) | (+) | p value | (−) | (+) | p value | ||
| rs753152 | Major Homo | 7823 | 2755 | 0.729 | 12006 | 374 | 0.774 | 6624 | 3889 | 0.432 | 6396 | 3676 | 0.846 | 7496 | 561 | 0.030 |
| 74.0% | 26.0% | 97.0% | 3.0% | 63.0% | 37.0% | 63.5% | 36.5% | 93.0% | 7.0% | |||||||
| Hetero+ | 561 | 191 | 883 | 29 | 482 | 265 | 476 | 278 | 526 | 55 | ||||||
| Minor Homo | 74.6% | 25.4% | 96.8% | 3.2% | 64.5% | 35.5% | 63.1% | 36.9% | 90.5% | 9.5% | ||||||
| rs77035639 | Major Homo | 8097 | 2840 | 0.643 | 12447 | 388 | 0.678 | 6864 | 4009 | 0.832 | 6642 | 3810 | 0.414 | 7758 | 589 | 0.163 |
| 74.0% | 26.0% | 97.0% | 3.0% | 63.1% | 36.9% | 63.5% | 36.5% | 92.9% | 7.1% | |||||||
| Hetero+ | 287 | 106 | 442 | 15 | 242 | 145 | 230 | 144 | 264 | 27 | ||||||
| Minor Homo | 73.0% | 27.0% | 96.7% | 3.3% | 62.5% | 37.5% | 61.5% | 38.5% | 90.7% | 9.3% | ||||||
| rs3815524 | Major Homo | 7423 | 2587 | 0.301 | 11399 | 351 | 0.388 | 6275 | 3673 | 0.878 | 6067 | 3473 | 0.498 | 7105 | 554 | 0.323 |
| 74.2% | 25.8% | 97.0% | 3.0% | 63.1% | 36.9% | 63.6% | 36.4% | 92.8% | 7.2% | |||||||
| Hetero+ | 961 | 359 | 1490 | 52 | 831 | 481 | 805 | 481 | 917 | 62 | ||||||
| Minor Homo | 72.8% | 27.2% | 96.6% | 3.4% | 63.3% | 36.7% | 62.6% | 37.4% | 93.7% | 6.3% | ||||||
| rs75380157 | Major Homo | 7718 | 2721 | 0.632 | 11878 | 366 | 0.349 | 6541 | 3835 | 0.611 | 6316 | 3634 | 1.000 | 7381 | 573 | 0.395 |
| 73.9% | 26.1% | 97.0% | 3.0% | 63.0% | 37.0% | 63.5% | 36.5% | 92.8% | 7.2% | |||||||
| Hetero+ | 666 | 225 | 1011 | 37 | 565 | 319 | 556 | 320 | 641 | 43 | ||||||
| Minor Homo | 74.7% | 25.3% | 96.5% | 3.5% | 63.9% | 36.1% | 63.5% | 36.5% | 93.7% | 6.3% | ||||||
| rs574603859 | Major Homo | 8001 | 2777 | 0.012 | 12255 | 385 | 0.803 | 6778 | 3932 | 0.085 | 6550 | 3759 | 0.575 | 7648 | 587 | 0.921 |
| 74.2% | 25.8% | 97.0% | 3.0% | 63.3% | 36.7% | 63.5% | 36.5% | 92.9% | 7.1% | |||||||
| Hetero+ | 383 | 169 | 634 | 18 | 328 | 222 | 322 | 195 | 374 | 29 | ||||||
| Minor Homo | 69.4% | 30.6% | 97.2% | 2.8% | 59.6% | 40.4% | 62.3% | 37.7% | 92.8% | 7.2% | ||||||
| rs147565266 | Major Homo | 8345 | 2932 | 1.000 | 12833 | 402 | 1.000 | 7079 | 4129 | 0.111 | 6841 | 3933 | 0.570 | 7981 | 611 | 0.378 |
| 74.0% | 26.0% | 97.0% | 3.0% | 63.2% | 36.8% | 63.5% | 36.5% | 92.9% | 7.1% | |||||||
| Hetero+ | 39 | 14 | 56 | 1 | 27 | 25 | 31 | 21 | 41 | 5 | ||||||
| Minor Homo | 73.6% | 26.4% | 98.2% | 1.8% | 51.9% | 48.1% | 59.6% | 40.4% | 89.1% | 10.9% | ||||||
Homo, homozygote; Hetero, heterozygote.
To determine the relationship of the SNPs with stroke in consideration of environmental factors, a logistic regression analysis adjusted for age, sex, research area, alcohol intake, current smoking, regular physical activity, obesity, hypertension, diabetes, dyslipidemia, and ischemic heart disease was performed. For the logistic regression analysis, the major homozygous genotypes were used as the reference group and the heterozygous and minor homozygous genotypes were used as the exposed group in the dominant model. Table 4 shows model I adjusted for basic characteristics (age, sex, research area), model II adjusted for lifestyle, and model III adjusted for anamnesis. RAMP2 SNP rs753152 was associated with a significantly higher OR in model I (OR, 1.773; 95% CI, 1.194–2.634). The CLR SNPs were associated with a significantly higher OR in model I (OR, 1.448–3.735) in participants with stroke, excluding rs574603859. SNP rs574603859 had a lower OR in model I (OR, 0.238; 95% CI, 0.076–0.745) between major homozygotes and the others. The model II results were similar to those for model I. In model III, rs574603859 showed no significant OR. The lack of statistical significance when adjusting for anamnesis indicated that rs574603859 has no strong effect on the risk of stroke.
Table 4. Associations between the RAMP2 and CLR gene variants and stroke.
| SNPs | Model I |
Model II |
Model III |
||||||
|---|---|---|---|---|---|---|---|---|---|
| OR | 95% CI | p-value | OR | 95% CI | p-value | OR | 95% CI | p-value | |
| rs753152 | 1.773 | 1.194–2.634 | 0.005 | 1.784 | 1.200–2.652 | 0.004 | 1.767 | 1.059–2.947 | 0.029 |
| rs77035639 | 2.015 | 1.199–3.385 | 0.008 | 2.084 | 1.239–3.506 | 0.006 | 2.25 | 1.177–4.302 | 0.014 |
| rs3815524 | 1.448 | 1.023–2.051 | 0.037 | 1.482 | 1.046–2.099 | 0.027 | 1.908 | 1.259–2.891 | 0.002 |
| rs75380157 | 1.845 | 1.270–2.683 | 0.001 | 1.877 | 1.290–2.731 | 0.001 | 2.115 | 1.338–3.342 | 0.001 |
| rs574603859 | 0.238 | 0.076–0.745 | 0.014 | 0.245 | 0.078–0.768 | 0.016 | 0.276 | 0.068–1.126 | 0.073 |
| rs147565266 | 3.735 | 1.325–10.52 | 0.013 | 3.997 | 1.412–11.31 | 0.009 | 5.316 | 1.775–15.92 | 0.002 |
Model I: adjusted for age, sex, research area
Model II: adjusted for age, sex, research area, alcohol intake, current smoking, regular physical activity, obesity
Model III: adjusted for age, sex, research area, alcohol intake, current smoking, regular physical activity, obesity, hypertension, diabetes, dyslipidemia, ischemic heart disease
Discussion
There is considerable evidence to suggest that the pathogenesis of stroke is affected by not only genetic factors, but also environment interactions9). Previous studies showed that a history of hypertension, dyslipidemia, diabetes, physical inactivity, diet, waist-to-hip ratio, current smoking, cardiac causes, and alcohol consumption were associated with risk of stroke32, 33). There was a J-shaped association between high amounts of alcohol and increased risk of both ischemic and hemorrhagic stroke34). Therefore, we defined age, sex, research area, alcohol consumption status, current smoking, regular physical activity, obesity, hypertension, diabetes, dyslipidemia, and ischemic heart disease as independent variables in the logistic regression analyses. To the best of our knowledge, this is the first study to investigate the relationships between RAMP2 and CLR and stroke in the light of gene-environment interactions in humans.
The pathogenesis of stroke is very complex and is associated closely with vascular dysfunction and disruption. Indeed, similar to chronic obstructive pulmonary disease, systemic inflammation and oxidative stress might play important roles in increasing the risk of stroke by promoting vascular dysfunction and platelet hyperactivity35). A review study showed that ADM has strong anti-oxidation and anti-inflammation activities14). Moreover, ADM acts via CLR/RAMP2 to prevent brain injury in both acute and chronic cerebral ischemia36), and exerts crucial vasoprotective effects following vascular injury37). The vascular ADM-CLR/RAMP2 system is critical in the regulation of vascular integrity, including the maintenance of vascular structure, and the regulation of angiogenesis and vasoprotection against vascular injury14, 19). Studying the ADM-CLR/RAMP2 system should reveal the mechanisms underlying the functional integrity of the vascular system, and could serve as the basis for novel approaches to therapy and preventive medicine.
RAMP expression is modulated by various agents in cell culture and in animal models of human disease38). For example, marked changes induced in the cardiovascular and renal systems provided evidence of an important role for dynamic RAMP regulation in those systems. Studies suggest that regulation of RAMPs might modulate the pathophysiology of conditions linked to RAMP-interacting G protein-coupled receptors. For example, human SNP studies described the relationship between CLR and essential hypertension and primary angle closure glaucoma39, 40). Polymorphisms in the ADM gene have also been reported to have a possible association with essential hypertension, dysglycemia, and adrenomedullin levels41–45). Genetic variants of the ADM-CLR/RAMP2 system might affect vascular homeostasis and cerebrovascular/cardiovascular disease. Several studies have revealed interactions of SNPs with stroke46–49). In these studies, the functional genetic polymorphisms were located in the promoters, which could cause differences in the plasma levels of the encoded target protein; were located in coding exons, leading to amino acid changes; or were located in an intron. Although we demonstrated that an intronlocated SNP is not functional, it could have other effects, such as influencing splicing or regulatory processes; for example, the binding of transcription factors to the gene. Furthermore, as a tag SNP, the polymorphism might be representative of many other variants, which could regulate the function of the receptor.
The limitations of our study include its cross-sectional design. However, case-control studies can be used to assess previously identified candidate regions and to determine target selections more precisely. In general, strokes can be divided into three subtypes: ischemic, lacunar, and hemorrhagic. In this study, we did not investigate the subtypes of stroke because we used a self-administered questionnaire to judge anamnesis. By contrast, a previous study reported that stroke and myocardial infarct seem sensitive enough to use self-administered questionnaire for judgment at baseline in Japanese cohort studies50). In a future (follow-up) survey, we plan to assess the participants by looking up the actual medical records; therefore, we expect these additional data will lead to further detailed analysis of genetic variants of ADM receptor genes in accordance with the stroke subtypes and cardiovascular disease. In addition, we only assessed Japanese participants in the present study, and further studies in other ethnic groups are needed to validate our findings.
Conclusions
In conclusion, the association of the RAMP2 and CLR genes with stroke in a Japanese cohort implicates these genes in the pathogenesis of stroke, although further investigation is required to confirm their associations. It will be interesting to determine whether the polymorphisms of RAMP2 and CLR are responsible for functional changes, and to reveal the underlying mechanism, given the potentially important role that ADM receptor genes play in stroke and/or vascular fragility.
Acknowledgements
The authors would like to thank Mr. Kyota Ashikawa, Ms. Tomomi Aoi, and the other members of the Laboratory for Genotyping Development, Center for Genomic Medicine, RIKEN, for support with the genotyping; and Mses. Yoko Mitsuda, Keiko Shibata, and Etsuko Kimura at the Department of Preventive Medicine, Nagoya University Graduate School of Medicine for their co- operation, technical assistance, and valuable comments.
Conflict of Interest
There are no conflicts of interest.
Sources of Funding
Funding was provided by Grant-in-Aid for Scientific Research on Priority Areas of Cancer (No. 17015018) and Grant-in-Aid for Scientific Research on Innovative Areas (No. 221S0001) from the Japanese Ministry of Education, Culture, Sports, Science and Technology. Japan Society for the Promotion of Science KAKENHI Grant Number 16H06277 supported this work.
References
- 1). Cleaver O, Melton DA: Endothelial signaling during development. Nat Med, 2003; 9: 661-668 [DOI] [PubMed] [Google Scholar]
- 2). Carmeliet P: Angiogenesis in health and disease. Nat Med, 2003; 9: 653-660 [DOI] [PubMed] [Google Scholar]
- 3). Miller AA, Budzyn K, Sobey CG: Vascular dysfunction in cerebrovascular disease: mechanisms and therapeutic intervention. Clin Sci (Lond), 2010; 119: 1-17 [DOI] [PubMed] [Google Scholar]
- 4). Higashi Y, Noma K, Yoshizumi M, Kihara Y: Endothelial function and oxidative stress in cardiovascular diseases. Circ J, 2009; 73: 411-418 [DOI] [PubMed] [Google Scholar]
- 5). Mochizuki N: Vascular integrity mediated by vascular endothelial cadherin and regulated by sphingosine 1-phosphate and angiopoietin-1. Circ J, 2009; 73: 2183-2191 [DOI] [PubMed] [Google Scholar]
- 6). Dejana E: Endothelial cell-cell junctions: happy together. Nat Rev Mol Cell Biol, 2004; 5: 261-270 [DOI] [PubMed] [Google Scholar]
- 7). Lanktree MB, Dichgans M, Hegele RA: Advances in genomic analysis of stroke: what have we learned and where are we headed? Stroke, 2010; 41: 825-832 [DOI] [PubMed] [Google Scholar]
- 8). Bevan S, Markus HS: Genetics of common polygenic ischaemic stroke: current understanding and future challenges. Stroke Res Treat, 2011; 2011: 179061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9). Markus HS: Stroke genetics. Hum Mol Genet, 2011; 20: R124-131 [DOI] [PubMed] [Google Scholar]
- 10). Kitamura K, Kangawa K, Kawamoto M, Ichiki Y, Nakamura S, Matsuo H, Eto T: Adrenomedullin: a novel hypotensive peptide isolated from human pheochromocytoma. Biochem Biophys Res Commun, 1993; 192: 553-560 [DOI] [PubMed] [Google Scholar]
- 11). Abe M, Sata M, Nishimatsu H, Nagata D, Suzuki E, Terauchi Y, Kadowaki T, Minamino N, Kangawa K, Matsuo H, Hirata Y, Nagai R: Adrenomedullin augments collateral development in response to acute ischemia. Biochem Biophys Res Commun, 2003; 306: 10-15 [DOI] [PubMed] [Google Scholar]
- 12). Brain SD, Grant AD: Vascular actions of calcitonin gene-related peptide and adrenomedullin. Physiol Rev, 2004; 84: 903-934 [DOI] [PubMed] [Google Scholar]
- 13). Kato J, Tsuruda T, Kita T, Kitamura K, Eto T: Adrenomedullin: a protective factor for blood vessels. Arterioscler Thromb Vasc Biol, 2005; 25: 2480-2487 [DOI] [PubMed] [Google Scholar]
- 14). Koyama T, Sakurai T, Kamiyoshi A, Ichikawa-Shindo Y, Kawate H, Shindo T: Adrenomedullin-RAMP2 System in Vascular Endothelial Cells. J Atheroscler Thromb, 2015; 22: 647-653 [DOI] [PubMed] [Google Scholar]
- 15). McLatchie LM, Fraser NJ, Main MJ, Wise A, Brown J, Thompson N, Solari R, Lee MG, Foord SM: RAMPs regulate the transport and ligand specificity of the calcitonin- receptor-like receptor. Nature, 1998; 393: 333-339 [DOI] [PubMed] [Google Scholar]
- 16). Parameswaran N, Spielman WS: RAMPs: The past, present and future. Trends Biochem Sci, 2006; 31: 631-638 [DOI] [PubMed] [Google Scholar]
- 17). Shindo T, Kurihara Y, Nishimatsu H, Moriyama N, Kakoki M, Wang Y, Imai Y, Ebihara A, Kuwaki T, Ju KH, Minamino N, Kangawa K, Ishikawa T, Fukuda M, Akimoto Y, Kawakami H, Imai T, Morita H, Yazaki Y, Nagai R, Hirata Y, Kurihara H: Vascular abnormalities and elevated blood pressure in mice lacking adrenomedullin gene. Circulation, 2001; 104: 1964-1971 [DOI] [PubMed] [Google Scholar]
- 18). Ichikawa-Shindo Y, Sakurai T, Kamiyoshi A, Kawate H, Iinuma N, Yoshizawa T, Koyama T, Fukuchi J, Iimuro S, Moriyama N, Kawakami H, Murata T, Kangawa K, Nagai R, Shindo T: The GPCR modulator protein RAMP2 is essential for angiogenesis and vascular integrity. J Clin Invest, 2008; 118: 29-39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19). Koyama T, Ochoa-Callejero L, Sakurai T, Kamiyoshi A, Ichikawa-Shindo Y, Iinuma N, Arai T, Yoshizawa T, Iesato Y, Lei Y, Uetake R, Okimura A, Yamauchi A, Tanaka M, Igarashi K, Toriyama Y, Kawate H, Adams RH, Kawakami H, Mochizuki N, Martinez A, Shindo T: Vascular endothelial adrenomedullin-RAMP2 system is essential for vascular integrity and organ homeostasis. Circulation, 2013; 127: 842-853 [DOI] [PubMed] [Google Scholar]
- 20). Fritz-Six KL, Dunworth WP, Li M, Caron KM: Adrenomedullin signaling is necessary for murine lymphatic vascular development. J Clin Invest, 2008; 118: 40-50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21). Hamajima N: The Japan Multi-Institutional Collaborative Cohort Study (J-MICC Study) to detect gene-environment interactions for cancer. Asian Pacific journal of cancer prevention: Asian Pac J Cancer Prev, 2007; 8: 317-323 [PubMed] [Google Scholar]
- 22). Matsuo S, Imai E, Horio M, Yasuda Y, Tomita K, Nitta K, Yamagata K, Tomino Y, Yokoyama H, Hishida A: Revised equations for estimated GFR from serum creatinine in Japan. Am J Kidney Dis, 2009; 53: 982-992 [DOI] [PubMed] [Google Scholar]
- 23). Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, deBakker PI, Daly MJ, Sham PC: PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet, 2007; 81: 559-575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24). Chang CC, Chow CC, Tellier LC, Vattikuti S, Purcell SM, Lee JJ: Second-generation PLINK: rising to the challenge of larger and richer datasets. GigaScience, 2015; 4: 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25). Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D: Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet, 2006; 38: 904-909 [DOI] [PubMed] [Google Scholar]
- 26). Patterson N, Price AL, Reich D: Population structure and eigenanalysis. PLoS Genet, 2006; 2: e190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27). Auton A, Brooks LD, Durbin RM, Garrison EP, Kang HM, Korbel JO, Marchini JL, McCarthy S, McVean GA, Abecasis GR: A global reference for human genetic variation. Nature, 2015; 526: 68-74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28). Yamaguchi-Kabata Y, Nakazono K, Takahashi A, Saito S, Hosono N, Kubo M, Nakamura Y, Kamatani N: Japanese population structure, based on SNP genotypes from 7003 individuals compared to other ethnic groups: effects on population-based association studies. Am J Hum Genet, 2008; 83: 445-456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29). Delaneau O, Marchini J, Zagury JF: A linear complexity phasing method for thousands of genomes. Nat Methods, 2011; 9: 179-181 [DOI] [PubMed] [Google Scholar]
- 30). Das S, Forer L, Schonherr S, Sidore C, Locke AE, Kwong A, Vrieze SI, Chew EY, Levy S, McGue M, Schlessinger D, Stambolian D, Loh PR, Iacono WG, Swaroop A, Scott LJ, Cucca F, Kronenberg F, Boehnke M, Abecasis GR, Fuchsberger C: Next-generation genotype imputation service and methods. Nat Genet, 2016; 48: 1284-1287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31). Barrett JC, Fry B, Maller J, Daly MJ: Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics, 2005; 21: 263-265 [DOI] [PubMed] [Google Scholar]
- 32). O'Donnell MJ, Chin SL, Rangarajan S, Xavier D, Liu L, Zhang H, Rao-Melacini P, Zhang X, Pais P, Agapay S, Lopez-Jaramillo P, Damasceno A, Langhorne P, McQueen MJ, Rosengren A, Dehghan M, Hankey GJ, Dans AL, Elsayed A, Avezum A, Mondo C, Diener HC, Ryglewicz D, Czlonkowska A, Pogosova N, Weimar C, Iqbal R, Diaz R, Yusoff K, Yusufali A, Oguz A, Wang X, Penaherrera E, Lanas F, Ogah OS, Ogunniyi A, Iversen HK, Malaga G, Rumboldt Z, Oveisgharan S, Al Hussain F, Magazi D, Nilanont Y, Ferguson J, Pare G, Yusuf S: Global and regional effects of potentially modifiable risk factors associated with acute stroke in 32 countries (INTERSTROKE): a case-control study. Lancet, 2016; 388: 761-775 [DOI] [PubMed] [Google Scholar]
- 33). Isabel C, Calvet D, Mas JL: Stroke prevention. Press Med, 2016; 45: e457-e471 [DOI] [PubMed] [Google Scholar]
- 34). Ikehara S, Iso H, Yamagishi K, Kokubo Y, Saito I, Yatsuya H, Inoue M, Tsugane S: Alcohol consumption and risk of stroke and coronary heart disease among Japanese women: the Japan Public Health Center-based prospective study. Prev Med, 2013; 57: 505-510 [DOI] [PubMed] [Google Scholar]
- 35). Austin V, Crack PJ, Bozinovski S, Miller AA, Vlahos R: COPD and stroke: are systemic inflammation and oxidative stress the missing links? Clin Sci (Lond), 2016; 130: 1039-1050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36). Igarashi K, Sakurai T, Kamiyoshi A, Ichikawa-Shindo Y, Kawate H, Yamauchi A, Toriyama Y, Tanaka M, Liu T, Xian X, Imai A, Zhai L, Owa S, Koyama T, Uetake R, Ihara M, Shindo T: Pathophysiological roles of adrenomedullin- RAMP2 system in acute and chronic cerebral ischemia. Peptides, 2014; 62: 21-31 [DOI] [PubMed] [Google Scholar]
- 37). Xian X, Sakurai T, Kamiyoshi A, Ichikawa-Shindo Y, Tanaka M, Koyama T, Kawate H, Yang L, Liu T, Imai A, Zhai L, Hirabayashi K, Dai K, Tanimura K, Liu T, Cui N, Igarashi K, Yamauchi A, Shindo T: Vasoprotective activities of the adrenomedullin-RAMP2 system in endothelial cells. Endocrinology, 2017; 158: 1359-1372 [DOI] [PubMed] [Google Scholar]
- 38). Udawela M, Hay DL, Sexton PM: The receptor activity modifying protein family of G protein coupled receptor accessory proteins. Semin Cell Dev Biol, 2004; 15: 299-308 [DOI] [PubMed] [Google Scholar]
- 39). Sano M, Kuroi N, Nakayama T, Sato N, Izumi Y, Soma M, Kokubun S: Association study of calcitonin-receptor-like receptor gene in essential hypertension. Am J Hypertens, 2005; 18: 403-408 [DOI] [PubMed] [Google Scholar]
- 40). Awadalla MS, Burdon KP, Thapa SS, Hewitt AW, Craig JE: A cross-ethnicity investigation of genes previously implicated in primary angle closure glaucoma. Mol Vis, 2012; 18: 2247-2254 [PMC free article] [PubMed] [Google Scholar]
- 41). Verweij N, Mahmud H, Mateo Leach I, de Boer RA, Brouwers FP, Yu H, Asselbergs FW, Struck J, Bakker SJ, Gansevoort RT, Munroe PB, Hillege HL, van Veldhuisen DJ, van Gilst WH, Sillje HH, van der Harst P: Genome-wide association study on plasma levels of midregional-proadrenomedullin and C-terminal-pro-endothelin-1. Hypertension, 2013; 61: 602-608 [DOI] [PubMed] [Google Scholar]
- 42). Ong KL, Tso AW, Leung RY, Cherny SS, Sham PC, Lam TH, Cheung BM, Lam KS: A genetic variant in the gene encoding adrenomedullin predicts the development of dysglycemia over 6.4 years in Chinese. Clin Chim Acta, 2011; 412: 353-357 [DOI] [PubMed] [Google Scholar]
- 43). Glorioso N, Herrera VL, Didishvili T, Ortu MF, Zaninello R, Fresu G, Argiolas G, Troffa C, Ruiz-Opazo N: Sex-specific effects of NLRP6/AVR and ADM loci on susceptibility to essential hypertension in a Sardinian population. PLoS one, 2013; 8: e77562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44). Cheung BM, Ong KL, Tso AW, Leung RY, Cherny SS, Sham PC, Lam TH, Lam KS: Plasma adrenomedullin level is related to a single nucleotide polymorphism in the adrenomedullin gene. Eur J Endocrinol, 2011; 165: 571-577 [DOI] [PubMed] [Google Scholar]
- 45). Chen S, Lu X, Zhao Q, Wang L, Li H, Huang J: Association of adrenomedullin gene polymorphisms and blood pressure in a Chinese population. Hypertens Res, 2013; 36: 74-78 [DOI] [PubMed] [Google Scholar]
- 46). Yi X, Wu L, Liao D, Wang C, Zhang B: Interactions Among CYP2C8, EPHX2, and CYP4A11 Variants and CYP Plasma Metabolite Levels in Ischemic Stroke. J Atheroscler Thromb, 2016; 23: 1286-1293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47). Zhong H, Cai Y, Cheng J, Cai D, Chen L, Su C, Li K, Chen P, Xu J, Cui L: Apolipoprotein E Epsilon 4 Enhances the Association between the rs2910164 Polymorphism of miR-146a and Risk of Atherosclerotic Cerebral Infarction. J Atheroscler Thromb, 2016; 23: 819-829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48). Zhang L, Yang J, Xue Q, Yang D, Lu Y, Guang X, Zhang W, Ba R, Zhu H, Ma X: An rs13293512 polymorphism in the promoter of let-7 is associated with a reduced risk of ischemic stroke. J Thromb Thrombolysis, 2016; 42: 610-615 [DOI] [PubMed] [Google Scholar]
- 49). Yi X, Lin J, Wang Y, Zhou Q, Wang C, Cheng W, Chi L: Association of Cytochrome P450 Genetic Variants with Clopidogrel Resistance and Outcomes in Acute Ischemic Stroke. J Atheroscler Thromb, 2016; 23: 1188-1200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50). Yamagishi K, Ikeda A, Iso H, Inoue M, Tsugane S: Self-reported stroke and myocardial infarction had adequate sensitivity in a population-based prospective study JPHC (Japan Public Health Center)-based Prospective Study. J Clin Epidemiol, 2009; 62: 667-673 [DOI] [PubMed] [Google Scholar]


