Abstract
Bacterial phytopathogen type III secreted (T3S) effectors have been strongly implicated in altering the interaction of pathogens with host plants. Therefore, it is useful to characterize the whole effector repertoire of a pathogen to understand the interplay of effectors in plants. Pseudomonas syringae pv. actinidiae is a causal agent of kiwifruit canker disease. In this study, we generated an Agrobacterium-mediated transient expression library of YFP-tagged T3S effectors from two strains of Psa, Psa-NZ V13 and Psa-NZ LV5, in order to gain insight into their mode of action in Nicotiana tabacum and N. benthamiana. Determining the subcellular localization of effectors gives an indication of the possible host targets of effectors. A confocal microscopy assay detecting YFP-tagged Psa effectors revealed that the nucleus, cytoplasm and cell periphery are major targets of Psa effectors. Agrobacterium-mediated transient expression of multiple Psa effectors induced HR-like cell death (HCD) in Nicotiana spp., suggesting that multiple Psa effectors may be recognized by Nicotiana spp.. Virus-induced gene silencing (VIGS) of several known plant immune regulators, EDS1, NDR1, or SGT1 specified the requirement of SGT1 in HCD induced by several Psa effectors in N. benthamiana. In addition, the suppression activity of Psa effectors on HCD-inducing proteins and PTI was assessed. Psa effectors showed differential suppression activities on each HCD inducer or PTI. Taken together, our Psa effector repertoire analysis highlights the great diversity of T3S effector functions in planta.
Keywords: effector screening, avirulence, Nicotiana benthamiana, Nicotiana tabacum, hypersensitive response, virus-induced gene silencing, subcellular localization
Introduction
Plants have developed two layers of defense to protect themselves from invading pathogens. The first layer, PAMP-triggered immunity (PTI), is facilitated by pattern recognition receptors (PRRs) that recognize conserved microbial molecules termed pathogen associated molecular patterns (PAMPs). Recognition of PAMPs such as bacterial flagellin, lipopolysaccharide or elongation factor Tu (EF-Tu) allows plants to mount immunity against a broad range of pathogens (Bigeard et al., 2015). Nevertheless, successful bacterial pathogens can deliver a suite of effector proteins via a type III secretion system (T3S) into host cells to dampen PTI. In turn, plants have evolved a second layer of defense to recognize specific effectors and induce effector-triggered immunity (ETI). This is often accompanied by rapid programmed cell death termed a hypersensitive response (HR) at the site of infection. As a result, in most of cases, plants can survive despite constant exposure to a wide range of pathogens.
Bacterial effectors, which primarily function to suppress immunity, have diverse biochemical functions and host virulence targets. Appropriate subcellular localization within the host cell is required for effectors to properly reach their targets to function (Hicks and Galán, 2013). Many studies have demonstrated the vast array of effector biochemical functions and subcellular localizations. For example, the type III effector AvrPto from P. syringae pv. tomato (Pto) is localized to the plasma membrane where it targets the membrane-associated PRRs FLS2 (FLAGELLIN SENSING 2) and EFR (ELONGATION FACTOR Tu RECEPTOR) thus inhibiting their phosphorylation, leading to reduced PTI responses (Xiang et al., 2008). The plasma membrane localized effectors AvrRpm1 from P. syringae pv. maculicola and AvrB from P. syringae pv. glycinea both require membrane localization by myristoylation for targeting the host protein RIN4 as well as avirulence functions (Nimchuk et al., 2000; Mackey et al., 2002). Another Pto effector, HopAI1, localizes to the cytoplasm where it inactivates MPK3 and MPK6, that play a key role in PTI signaling, via its phosphothreonine lyase activity (Zhang et al., 2007). A number of P. syringae effectors including HopI1, HopN1, HopK1, and AvrRps4 target the chloroplast to suppress immunity (Jelenska et al., 2007; Rodriguez-Herva et al., 2012; Li et al., 2014). Moreover, the Pto DC3000 effector HopM1 localizes to the trans-Golgi network and interacts with host ADP-ribosylation factor guanine nucleotide exchange factor, AtMIN7, to suppress vesicle-trafficking during immune responses (Nomura et al., 2011). Bacterial effectors do not interfere solely with PTI; some bacterial effectors have been shown to suppress ETI responses. Recently, the Pto DC3000 effector HopD1 was reported to suppress ETI by localizing to the endoplasmic reticulum to interact with the host membrane-tethered transcription factor NTL9 (Block et al., 2014). Some fascinating mechanisms of effector activity within the nucleus have been demonstrated, such as the Ralstonia effector PopP2 that contains a nuclear localization signal (NLS) and targets WRKY transcription factors in order to disable defense signaling (Sarris et al., 2015). Interestingly, Arabidopsis has evolved the paired immune receptors, RRS1 and RPS4, that interact with PopP2 via the RRS1 C-terminal decoy WRKY domain to trap PopP2 and activate ETI responses (Williams et al., 2014). The host nucleus is a key effector target for the disruption of immune responses. A recent study of the localization of ∼50 RxLR effectors from the Arabidopsis downy mildew oomycete pathogen, Hyaloperonospora arabidopsidis (Hpa) Emoy2, revealed that the majority of these effectors were found to be localized at membranes or in the nucleus (Caillaud et al., 2012a,b). Furthermore, Caillaud et al. (2012a) identified that the tonoplast-localized Hpa effector HaRxL17 functions as a virulence effector during infection. It is clear that understanding the subcellular localization of pathogen-derived effectors is of great importance to better understand their virulence or avirulence mechanisms.
The co-evolutionary arms race between pathogen effectors and their corresponding plant intracellular immune receptors has shaped the highly diversified repertoire of both (Jones and Dangl, 2006). The majority of characterized plant intracellular immune receptors are nucleotide-binding and leucine-rich repeat receptors (NLRs). NLR proteins carry a variable amino-terminal region with either a CC (coiled-coil) or TIR (toll-interleukin1 receptor-like) domain that is generally involved in activating downstream defense signaling (Meyers et al., 2003; Cui et al., 2015). Only a few immune regulators have been characterized for their role in NLR-mediated signaling. Typically, the lipase-like protein EDS1 (ENHANCED DISEASE SUSCEPTIBILITY 1) is required for immune signaling pathways initiated by TIR-NLRs (TNLs), whereas integrin protein NDR1 (NON-RACE SPECIFIC DISEASE RESISTANCE 1) is required downstream of CC-NLRs (CNLs) (Aarts et al., 1998). Moreover, SGT1 (SUPPRESSOR OF THE G2 ALLELE OF SKP1) forms a complex with the molecular chaperone HSP90 (HEAT SHOCK PROTEIN 90) to maintain proper folding of NLR proteins and are often required for NLR functions (Austin, 2002; Takahashi et al., 2003). Therefore, investigating the requirement of these regulators in effector-triggered immunity would help better understand in planta functions of effectors.
A few effectors have been shown to have diverse and complex functions in planta. Previous studies have shown that the Pto DC3000 effector HopAB3 is able to suppress ETI and PTI (Abramovitch et al., 2006; Goehre et al., 2008; Guo et al., 2009). Moreover, suppression of P. syringae pv. syringae effector HopA1-triggered HR in Arabidopsis by co-delivered effectors identified many ETI-suppressing effectors from Pto DC3000 that were also able to suppress P. syringae pv. maculicola effector AvrRpm1-triggered ETI (Guo et al., 2009). In the same study, some of these ETI-suppressing effectors also suppressed PTI responses. Similarly, a large number of effectors from the oomycete pathogen Phytophthora sojae were found to suppress PTI and ETI (Wang et al., 2011). Upon infection, these PTI/ETI-suppressing effectors would greatly affect the plant–pathogen interaction.
Pseudomonas syringae pv. actinidiae (Psa) causes bacterial canker disease in kiwifruit. Since the 2008 outbreak of highly virulent Psa (Psa-V) in Italy, Psa-V has spread worldwide, including Chile (2010) and New Zealand (2010). Psa-V is phylogenetically distinct from the low virulent strain (Psa-LV) (McCann et al., 2013). These authors suggested that these distinctive clades are the result of various gene-shifting processes derived from different source populations, hinting at the likely probability of new virulence emergence in Psa populations. In fact, evidence suggests selection driving horizontal gene transfer in a Psa-V strain to gain copper resistance from a local Psa-LV strain (Colombi et al., 2017). Considering the active transfer and conversion events of virulence effector genes in Psa source populations, research efforts on the effector gene pools in geographically co-existing strains such as Psa-LV and Psa-V would be useful.
In this study, we sought to characterize the effector repertoire of one representative strain from each clade present in New Zealand, namely the virulent strain Psa NZ V-13 and the low virulence strain Psa NZ LV-5 (hereafter, Psa V13 and Psa LV5, respectively), both isolated from diseased orchards in the Bay of Plenty region, New Zealand (Chapman et al., 2012). We used Agrobacterium-mediated transient transformation to test subcellular localization and cell death-inducing activity of Psa effectors in non-host plants Nicotiana benthamiana and N. tabacum. We showed that Psa effectors localized to different plant cell compartments, presumably to interfere with multiple plant defense-related processes. We also found that multiple Psa effectors induced HR-like cell death and, using virus-induced gene silencing (VIGS), identified the requirement of known plant immunity regulators for effector-induced cell death. In addition, we demonstrated that several Psa effectors suppressed cell death triggered by other Psa effectors. We expect that the various effector characteristics found in this study associated with their putative biological functions will help to predict roles of these effectors in planta and aid in the development of bacterial canker-resistant kiwifruit.
Results
Construction of Pseudomonas syringae pv. actinidiae Type III Effector Libraries
To characterize the type-III secreted effector (T3E) repertoire of Psa, we cloned effectors from two Psa strains, V13 and LV5. Psa V13 and LV5 strains carry 38 and 26 T3Es, respectively, based on the computational prediction using their genome sequences (McCann et al., 2013; Templeton et al., 2015). Based on this, we selected a total of 48 T3Es for further study using the following criteria: (i) all V13 or LV5 specific T3Es, (ii) only V13 alleles for T3Es in both V13 and LV5 that shared more than 90% amino acid identity, or (iii) both V13 and LV5 alleles for T3Es that shared less than 90% amino acid identity (Table 1). Based on the protein sequence identity to P. syringae homologs, Psa V13 effectors that are predicted to be significantly truncated (hopA1, hopW1, hopAA1-1, and hopAA1-2) were excluded (Table 1). Only one allele of Psa V13 for duplicated effectors hopBB1 (hopBB1-1/hopBB1-2, 93.6% identity) and hopAM1 (hopAM1-1/hopAM1-2, 100% identity) were functionally analyzed. Additionally, both alleles of hopAY1 were analyzed because the predicted peptide sequence of Psa V13 allele was only 77 amino acids shorter than that of LV5. In order to generate broad host-range plasmid (pBBR 1MCS-5) constructs for Pseudomonas-delivery and binary plasmid constructs for in planta transient expression of the 48 selected Psa T3Es, we used the Golden Gate cloning method (Engler et al., 2008). Briefly, each T3E sequence was divided into several modules roughly of 1 kb size and each module was amplified by polymerase chain reaction (PCR) using primers with flanking BsaI restriction enzyme sites. These amplicons were cloned into the Golden Gate compatible entry vector pICH41021. The number of modules for each effector and their 4 bp BsaI overhangs are listed in Supplementary Table S1. All modules for a given effector were then assembled with a C-terminal 6xHA-tag module into the Golden Gate-compatible derivative of the broad host-range vector pBBR1MCS-5, which carries the bacterial avrRps4 promoter (Jayaraman et al., 2017); or with a C-terminal YFP-tag module into the binary vector pICH86988 under control of a cauliflower mosaic virus (CaMV) 35S promoter for functional analysis.
Table 1.
Type III secreted effectors from Psa V13 and LV5 used in this study.
| Effectors | Psa- | Psa- | AA | Predicted |
|---|---|---|---|---|
| V13 | LV5 | identity | protein size | |
| (%) | (kDa) | |||
| AvrB4-1 | o | NA | 36.0 | |
| AvrD1 | o | 34.6 | ||
| AvrE1 | o | o | 97.1 | 195.3 |
| AvrPto5 | o | 17.2 | ||
| AvrRpm1 | o | 24.9 | ||
| HopA1 | o§ | o | 22 (V13)/42.2 (LV5) | |
| HopD1 | o | 34.6 | ||
| HopE1 | o | 24.1 | ||
| HopF1 | o | 21.9 | ||
| HopF4b (HopF2) | o | 22.0 | ||
| HopH1 | o | 24.3 | ||
| HopI1 | o | 49.3 | ||
| HopM1 | o | 75.9 | ||
| HopN1 | o | o | 99.6 | 38.7 |
| HopO1 | o | 32.6 | ||
| HopQ1 | o | 48.8 | ||
| HopR1 | o | o | 99.0 | 210.3 |
| HopS1 | o | 19.9 | ||
| HopS2 | o | o | 94.9 | 18.7 |
| HopT1 | o | 41.5 | ||
| HopW1 | o§ | o | 40.8 (V13)/82.8 (LV5) | |
| HopX1 | o | 40.6 | ||
| HopX2 | o | 38.0 | ||
| HopX3 | o | 40.9 | ||
| HopY1 | o | o | 30.9 | |
| HopZ3 | o | 45.1 | ||
| HopAA1 | o§ | o | 21.8 (V13)/50.4 (LV5) | |
| HopAB3 | o | 62.6 | ||
| HopAE1 | o | o | 99.8 | 126.6 |
| HopAF1 | o | o | 23.4 | |
| HopAF1-2 | o | 30.9 | ||
| HopAG1 | o | 77.6 | ||
| HopAH1 | o | o | 91.2 | 42.8 |
| HopAI1† | o | o | 81.6 | 29.7 |
| HopAM1-1 | o | 31.3 | ||
| HopAO2 | o | 38.1 | ||
| HopAR1 | o | 28.5 | ||
| HopAS1 | o | o | 98.9 | 149.0 |
| HopAU1 | o | 88.2 | ||
| HopAV1 | o | 123.5 | ||
| HopAW1 | o | 24.9 | ||
| HopAY1‡ | o | o | 91.9 | 27.1(V13)/35.4(LV5) |
| HopAZ1 | o | o | 99.5 | 24.7 |
| HopBB1-1(2) | o | 30.7 | ||
| HopBN1 | o | 56.8 | ||
§Effectors highly truncated in Psa V13 indicating a pseudogene.
†Amino acid identity between alleles from Psa V13 and LV5 lower than 90%.
‡Psa V13 version has shorter amino acid sequence.
Subcellular Localization of Psa Effectors in Nicotiana benthamiana Leaf Cells
Determining the subcellular localization of effectors can suggest clues about their mode of action during infection. Several studies have highlighted the importance of localization of effectors in a particular cellular compartment for their in planta function (Lewis et al., 2008; Schornack et al., 2009; Sohn et al., 2014). To investigate the subcellular localization of Psa effectors, we expressed YFP-tagged Psa effectors in N. benthamiana leaf cells using Agrobacterium-mediated transient transformation (hereafter agroinfiltration). To visualize effector-YFP proteins, confocal laser scanning microscopy was undertaken at 48 h after agroinfiltration. The subcellular localization of all tested Psa effectors are shown in Figure 1 (larger images for the localization representatives are in Supplementary Figure S2). Interestingly, approximately half of the Psa effectors (23 effectors) were localized in the nucleus and cytoplasm. Only one effector, HopBN1, exclusively localized in the nucleus (Figure 1 and Supplementary Figure S2). HopBB1-2, localized in the nucleus and cytoplasm as well as subnuclear foci (Figure 1 and Supplementary Figure S2). We identified seven effectors (HopN1, HopR1, HopAB3, HopAG1, HopAH1, HopAM1-1, and HopAU1) that localized to the cytoplasm but were excluded from the nucleus (Figure 1 and Supplementary Figure S2). Among these, HopAG1 showed punctate localization. HopM1 localized to the chloroplasts, while HopAV1 and HopAZ1 localized to the cytoplasm but in strands resembling the cytoskeleton (Figure 1 and Supplementary Figure S2). Eleven effectors localized to the cell periphery (largely absent from cytoplasmic strands), and two of them (HopT1 and AvrE1) showed punctate localization (Figure 1) (Jayaraman et al., 2017). However, we could not determine the subcellular localization of HopAS1, HopW1, and HopX1 in our experimental conditions. Notably, these results suggest that Psa bacterial effectors are localized and, therefore, function at several distinct cellular compartments.
FIGURE 1.
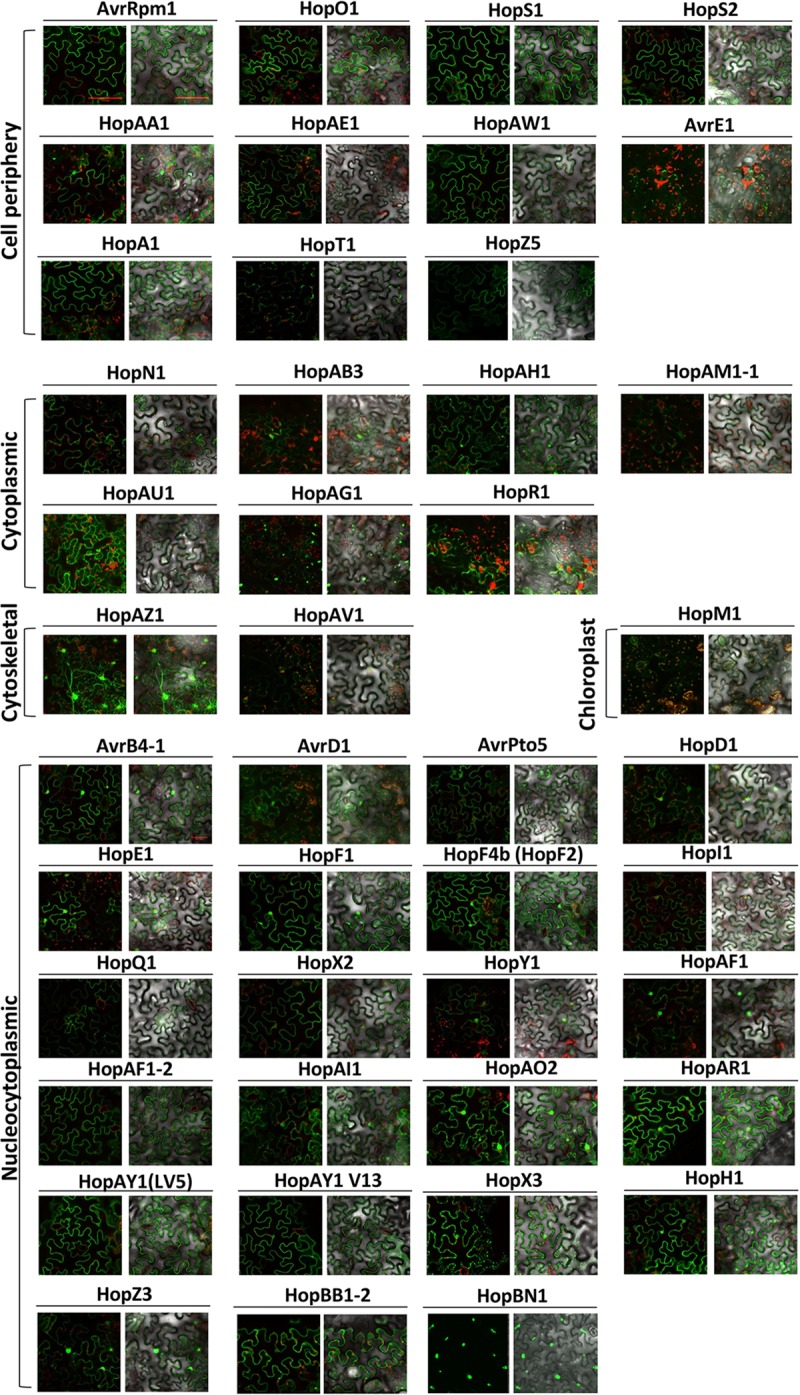
Subcellular localization of Psa effectors in Nicotiana benthamiana. Four–five week-old N. benthamiana leaf cells were infected with Agrobacterium AGL1 carrying C-terminally YFP-tagged Psa effectors for transient protein expression. At 2 days post infection (dpi), 8 mm diameter leaf disks were sampled and viewed using confocal microscopy. YFP fluorescence was excited at 488 nm with a 20 mW Argon laser and captured in the emission range between 500 and 530 nm. HopM1 YFP signal was determined by a sum-of-squares Z-projection. Chloroplast auto-fluorescence was detected between 600 and 680 nm. This experiment was repeated twice with similar results.
In order to validate the subcellular localization of effector-YFP fusion proteins, immunoblot analysis was conducted using total protein extracts from agroinfiltrated N. benthamiana leaves (Supplementary Figure S1). Out of 48 effectors, 38 were confirmed for protein expression by anti-GFP immunoblots (Supplementary Figure S1) (Jayaraman et al., 2017). Of the 10 effectors that could not be detected, six (AvrE1, HopR1, HopT1, HopW1, HopAM1-1, and HopAS1) triggered cell death in N. benthamiana (Figure 2). The remaining 4 effectors (HopI1, HopBN1, AvrD1, and HopAV1) could not be detected despite the lack of strong cell death response. In contrast, HopX1, which also triggered cell death, was detected in the immunoblot but could not be localized via confocal microscopy. The subcellular localization of AvrE1, HopR1, HopBN1, HopI1, HopAM1-1, HopAV1, and HopT1 was detected by confocal microscopy but protein expression was not validated by immunoblot analysis.
FIGURE 2.
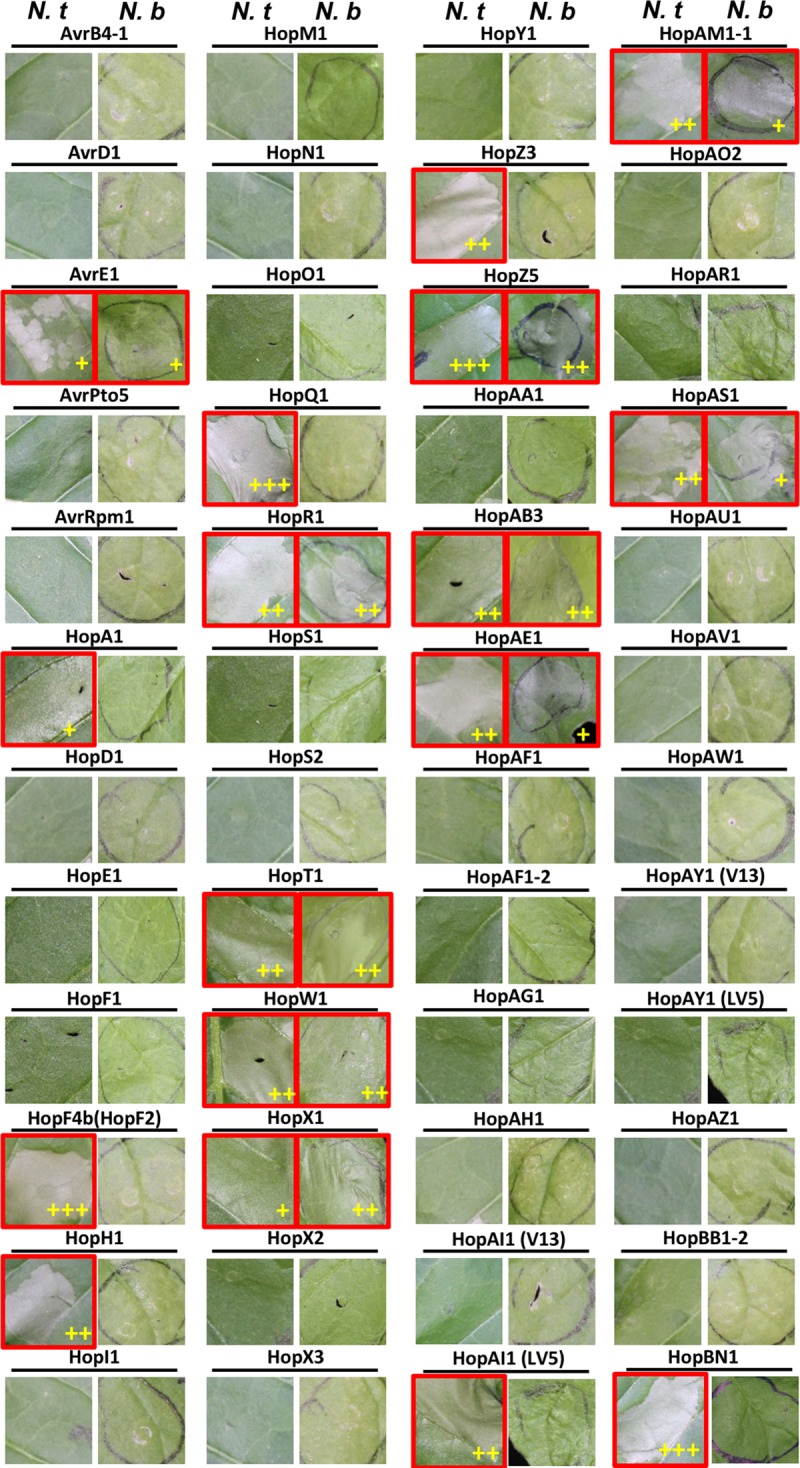
Multiple Psa effectors trigger HR-like cell death in Nicotiana spp. Four–five weeks-old N. benthamiana and N. tabacum plants were infiltrated with Agrobacterium tumefaciens AGL1 strains carrying C-terminally YFP-tagged Psa effector constructs. Photographs of the leaves showing HR-like cell death (HCD) were taken at 3 dpi. Leaf images showing HCD are indicated with a red border. (+), (++), or (+++) indicate the strength of the HCD phenotype as measured by ion conductivity (Supplementary Figures S3, S4). This experiment was conducted at least three times with similar results.
Multiple Psa Effectors Induce HR-Like Cell Death in Nicotiana spp.
Multiple P. syringae effectors have been shown to induce HR-like cell death (HCD) in Nicotiana (Vinatzer et al., 2006; Wei et al., 2007; Wroblewski et al., 2009). To identify Psa T3Es that induce HCD in N. benthamiana or N. tabacum, we transiently expressed each Psa effector using agroinfiltration. Surprisingly, 17 Psa T3Es induced HCD in N. tabacum. Of these, 10 Psa T3Es also triggered HCD in N. benthamiana (Figure 2 and Supplementary Figures S3, S4) (Jayaraman et al., 2017). Interestingly, none of the tested Psa T3Es triggered N. benthamiana-specific HCD. In N. tabacum, 17 T3Es [AvrE1, HopA1, HopF4b, HopH1, HopQ1, HopR1, HopT1, HopW1, HopX1, HopZ3, HopAB3, HopAE1, HopAI1 (LV5), HopBN1, HopAM1-1, HopZ5 and HopAS1] induced HCD in 3–4 days. The intensity of HCD was quantified by ion conductivity measurement at different time points (Supplementary Figure S3). Fourteen Psa T3Es triggered a full collapse of infiltrated leaf tissue while 3 effectors (AvrE1, HopX1, and HopA1) triggered a slower/weaker HCD (Figure 2 and Supplementary Figure S3). Psa-effector triggered HCD was assessed similarly for N. benthamiana, and 6 T3Es (HopR1, HopW1, HopX1, HopAB3, HopZ5, and HopT1) induced strong HCD while 4 T3Es (AvrE1, HopAM1-1, HopAE1, and HopAS1) induced a slower/weaker response (Figure 2 and Supplementary Figure S4).
Natural sequence diversity in the T3E repertoire of P. syringae strains can cause variation in their ability to induce HCD (Ma et al., 2006). In order to identify if allelic sequence variation between Psa T3Es and their homologs found in other P. syringae strains caused an altered HCD phenotype in Nicotiana, we compared their amino acid sequence identity and HCD-inducing ability based on published literature (Table 2). In total, 15 P. syringae homologs of Psa T3Es that were previously tested for HCD induction in N. tabacum and/or N. benthamiana were included in our comparison. Phylogenetic analysis showed that Pto DC3000 is closely related to Psa and 23 T3Es are conserved between these strains (Butler et al., 2013). We selected 10 Pto DC3000 T3Es that triggered HCD, or had a Psa homolog that triggered HCD in Nicotiana spp., for comparison (Table 2) (Wei et al., 2007; Wroblewski et al., 2009). In addition to Pto DC3000, 3 T3E homologs from P. syringae pv. syringae (Psy) B728a, one from P. cannabina pv. alisalensis ES4326 and one from P. syringae pv. tomato (Pto) T1 were analyzed (Lin et al., 2006; Robert-Seilaniantz et al., 2006; Vinatzer et al., 2006; Wroblewski et al., 2009).
Table 2.
Comparison of HR-like cell death (HCD) phenotypes induced by Psa effectors and their Pseudomonas syringae homologs.
| Psa effector | N. benthamiana/N. tabacum | Homologs | Source strain | N. benthamiana/N. tabacum | AA identity (%) | Reference |
|---|---|---|---|---|---|---|
| AvrE1 | +/+ | AvrE1 | Pto DC3000 | +/NA | 94.1 | 1∗ |
| HopA1 | -/+ | HopA1 | Pto DC3000 | -/NA | 93.9 | 1 |
| HopF4b | -/+ | HopF2 | Pto DC3000 | -/+ | 57.6 | 1,2 |
| HopF1 | -/- | 47.8 | ||||
| HopH1 | -/+ | HopH1 | Pto DC3000 | -/NA | 97.7 | 1 |
| HopM1 | -/- | HopM1 | Psy B728a | +/+ | 71.9 | 3 |
| HopQ1 | -/+ | HopQ1-1a | Pto DC3000 | -/NA | 99.1 | 1,4 |
| HopR1 | +/+ | HopR1 | Pto DC3000 | -/NA | 95.9 | 1 |
| HopT1 | +/+ | HopT1-1b | Pto DC3000 | +/NA | 70.4 | 1,4 |
| HopW1 | +/+ | HopW1-1 | Pcal ES4326 | +/NA | 77.8 | 4 |
| HopX1 | +/+ | HopX1 | Pto DC3000 | +/NA | 73.7 | 1 |
| HopZ3 | -/+ | HopZ3 | Psy B728a | -/+ | 72.1 | 3 |
| HopAA1 | -/+ | HopAA1-1 | Pto DC3000 | +/NA | 93.8 | 1 |
| HopAB3 | +/+ | AvrPtoB (HopAB3) | Pto T1 | -/NA | 83.5 | 5 |
| HopAE1 | +/+ | HopAE1 | Psy B728a | +/+ | 73.6 | 3 |
| HopAM1-1 | +/+ | HopAM1-1c | Pto DC3000 | -/NA | 98.9 | 1 |
aPto DC3000 HopQ1-2 is highly truncated (Buell et al., 2003).
bPsa HopT1 shares 96.9% of amino acid sequence identity with Pto DC3000 HopT1-2.
cBoth Psa and Pto DC3000 have two identical copies of HopAM1 (HopAM1-1/HopAM1-2).
NA, not available; Pto, Pseudomonas syringae pv. tomato; Pcal, Pseudomonas cannabina pv. alisalensis; Psy, Pseudomonas syringae pv. syringae.
1(Wei et al., 2007), ∗Effector-triggered cell death was assessed by Pseudomonas type III secretion in this literature.
2(Robert-Seilaniantz et al., 2006).
5(Lin et al., 2006).
Many P. syringae homologs showed similar phenotypes to their Psa counterparts in triggering HCD in N. benthamiana such as AvrE1, HopF4b, HopT1, HopW1, HopX1, HopAA1, and HopAE1 (Table 2). In Pto DC3000, there are two copies of hopT1 (indicated as hopT1-1 and hopT1-2). Psa HopT1 shares the highest amino acid sequence identity with Pto DC3000 HopT1-2 (96.9%). Despite this, we compared Psa HopT1 to Pto DC3000 HopT1-1 (70.4%) because the HCD phenotype was only tested for HopT1-1 (Wei et al., 2007; Wroblewski et al., 2009). Regardless, both Psa HopT1 and Pto DC3000 HopT1-1 triggered HCD in N. benthamiana. Conversely, there are several T3E homolog pairs that show differing phenotypes despite high amino acid sequence identity. For example, Pto T1 HopAB3 shares high amino acid identity with Psa HopAB3 (84.5%) (Lin et al., 2006). However, unlike HopAB3 from Pto T1, Psa HopAB3 induced HCD in both N. benthamiana and N. tabacum. In addition, Pto DC3000 T3Es HopAA1-1 and HopR1 are highly similar to Psa HopAA1 and HopR1 (93.8 and 95.9% amino acid identity, respectively) but only Pto DC3000 HopAA1-1 and Psa HopR1 induced HCD in N. benthamiana (Figure 3). Pto DC3000 HopQ1-1 and Psa HopQ1 share high amino acid sequence identity (99.1%) and they both trigger strong HCD in N. tabacum (Table 2). In contrast, despite relatively low amino acid sequence identity (57.6%), Pto DC3000 HopF2 and its Psa homolog HopF4b triggered HCD in N. tabacum while Psa HopF1 did not (Table 2). Taken together, our data indicate that natural sequence variation might have caused loss or gain of HCD-inducing activity of Psa effectors.
FIGURE 3.
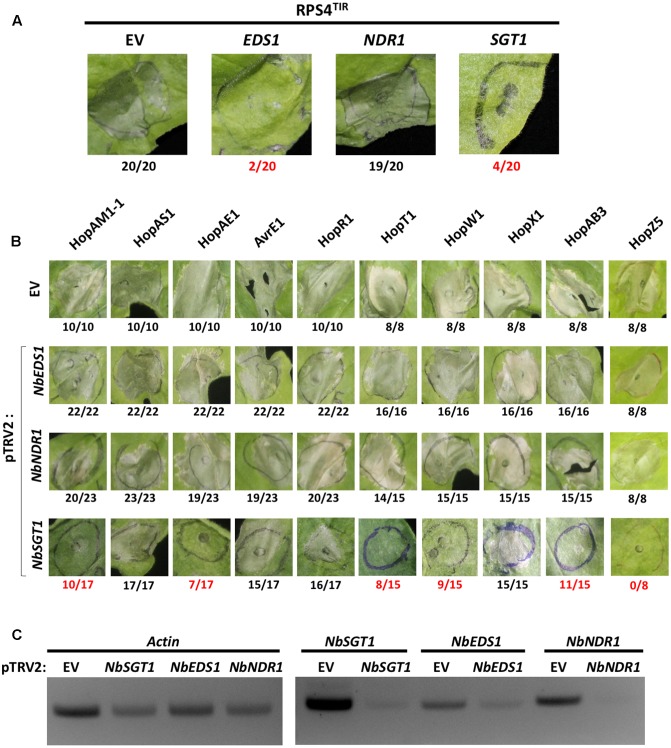
NbSGT1 is partially required for HR triggered by several Psa effectors. (A) RPS4TIR-triggered HCD in EV–, NbEDS1–, NbNDR1–, or NbSGT1–silenced N. benthamiana plants. Two-week old N. benthamiana plants were infiltrated with mixture of A. tumefaciens AGL1 strains carrying pTRV1 or pTRV2 construct as indicated in section “Materials and Methods.” Four weeks later, plants were agroinfiltrated to express the RPS4 TIR domain (RPS4TIR). RPS4TIR-induced HCD was scored at 4 dpi. The number of HCD-showing patches out of total infiltrated patches are indicated under each panel. Numbers labeled in red indicate a significant reduction of HCD. (B) Psa effector-triggered HCD in NbEDS1–, NbNDR1–, and NbSGT1–silenced N. benthamiana plants. VIGS and agroinfiltration to induce HCD are indicated in (A). Psa effector-induced HCD was scored at 4 dpi. The number of HCD-showing patches out of total infiltrated patches are indicated under each panel. Numbers labeled in red indicate a significant reduction of HCD. (C) Semi-quantitative PCR amplification of silenced genes in VIGS plants. 8 mm diameter leaf disks were harvested from VIGSed plants, total RNA was extracted by Trizol RNA method and cDNA was synthesized. Using cDNA as PCR template, partial fragments of Actin (control) and VIGSed genes were amplified with gene-specific primers.
HR-Like Cell Death Induced by Several Psa Effectors Requires SGT1 in Nicotiana benthamiana
Knocking down gene expression of key components of plant immunity by VIGS has been widely used to investigate the involvement of defense components in plant immunity (Liu et al., 2002; Peart et al., 2002; Oh et al., 2014). To test the requirement of general immunity regulators for Psa effector-triggered HCD, we applied VIGS to generate N. benthamiana plants silenced for EDS1 (pTRV2:NbEDS1), NDR1 (pTRV2:NbNDR1), or SGT1 (pTRV2:NbSGT1) together with control plants (pTRV2:EV). Two alleles of NbEDS1 have been reported, NbEDS1a and NbEDS1b, and our VIGS construct was designed to target NbEDS1a as NbEDS1b is likely a pseudogene (Adlung et al., 2016; Ordon et al., 2017). Hereafter, NbEDS1a is referred to as NbEDS1. First, in order to validate our experimental conditions, HCD induced by agroinfiltration of the RPS4 TIR domain (RPS4TIR) was tested in N. benthamiana plants VIGSed for EV, NbEDS1, NbNDR1, or NbSGT1. RPS4TIR induced HCD in EV– or NbNDR1–silenced but not in NbEDS1– or NbSGT1–silenced plants, consistent with previous results (Figure 3A and Table 3) (Zhang et al., 2004; Swiderski et al., 2009). Next, 10 T3Es (HopAM1-1, HopAS1, HopAE1, AvrE1, HopR1, HopT1, HopW1, HopX1, HopZ5, and HopAB3) that triggered HCD in N. benthamiana were agroinfiltrated in NbEDS1–, NbNDR1– or NbSGT1–silenced plants. Interestingly, Psa effector-triggered HCD in NbEDS1– or NbNDR1–silenced plants was comparable to EV-silenced plants (Figure 3B). However, in NbSGT1–silenced plants, 6 T3Es, HopAM1-1, HopAE1, HopT1, HopW1, HopZ5, and HopAB3 were significantly affected in their ability to trigger HCD (Figure 3B and Table 3) (Jayaraman et al., 2017). In contrast, HopAS1-, HopX1-, AvrE1-, and HopR1-triggered HCD was not affected by NbSGT1–silencing. To further analyze the efficiency of VIGS, semi-quantitative RT-PCR was conducted. As a result, the level of NbEDS1, NbNDR1, and NbSGT1 transcripts were significantly reduced in the respective silenced plants compared to EV-silenced control plants indicating that these genes were sufficiently silenced (Figure 3C). However, we noticed that unlike NbNDR1, NbEDS1-, or NbSGT1-silenced plants showed significantly reduced but not completely eliminated transcript levels of silenced genes. Taken together, these results suggest that SGT1 plays a key role in HCD induced by several Psa effectors.
Table 3.
Summary of Nicotiana HCD-triggering Psa effectors.
| Psa effector | In vivo localization | HR-like cell death in Nicotiana | Requirement of defense regulators HR-like cell death in N. benthamiana | |
|---|---|---|---|---|
| N. tabacum | N. benthamiana | |||
| AvrE1 | Cell periphery | + | + | - |
| HopA1 | Cell periphery | + | - | NA |
| HopF4b | Nucleus and cytoplasm | + | - | NA |
| HopH1 | Nucleus and cytoplasm | + | - | NA |
| HopQ1 | Nucleus and cytoplasm | + | - | NA |
| HopR1 | Cytoplasm | + | + | - |
| HopT1 | Cell periphery | + | + | SGT1 |
| HopW1 | Not localized | + | + | SGT1 |
| HopX1 | Not localized | + | + | - |
| HopZ3 | Nucleus and cytoplasm | + | - | NA |
| HopAB3 | Cytoplasm | + | + | SGT1 |
| HopAE1 | Cell periphery | + | + | SGT1 |
| HopAI1 (LV5) | Nucleus and cytoplasm | + | - | NA |
| HopAM1-1 | Cytoplasm | + | + | SGT1 |
| HopAS1 | Not localized | + | + | - |
| HopBN1 | Nucleus | + | - | NA |
+, cell death.
-, no cell death.
NA, not available.
Multiple Psa Effectors Suppress Effector-Triggered Cell Death in N. benthamiana
Some pathogen effectors suppress ETI to enable pathogen proliferation. In order to investigate if Psa effectors can suppress ETI, we conducted an agroinfiltration assay of HCD-inducing proteins with or without Psa effectors in N. benthamiana. Bcl-2-associated X (BAX) is an animal pro-apoptotic regulator that induces HCD in plant cells when overexpressed (Lacomme and Santa Cruz, 1999). As previously shown, agroinfiltration of BAX with a GFP control induced strong HCD in N. benthamiana leaf cells (Figure 4A). Interestingly, several Psa effectors, HopF4b, HopQ1, HopF1, and HopAR1, suppressed BAX-induced HCD when coexpressed in N. benthamiana, indicating that these effectors may interfere with cell death signaling. The P. syringae effector AvrPto is recognized by tomato kinase Pto and its cognate R protein Prf (Tobias et al., 1999). Transient expression of AvrPto and Pto trigger HCD in N. benthamiana, which carries a Prf homolog (Scofield et al., 1996). Another P. syringae effector AvrPtoB interferes with Pto-mediated recognition of AvrPto due to its E3-ligase activity (Abramovitch, 2003; Abramovitch et al., 2006). As expected, agroinfiltration of AvrPto and Pto induced strong HCD whereas coexpression of AvrPtoB did not (Figure 4B). We identified only one Psa effector, HopQ1, that suppressed AvrPto/Pto-induced HCD. In addition, since some Psa effectors induced HCD in N. benthamiana (Figure 2), we conducted an agroinfiltration assay to test if other Psa effectors have the ability to suppress Psa effector-induced HCD. Among 8 tested, 5 effectors showed suppression activity on Psa effector-induced HCD (Figure 4C). Interestingly, HopQ1 suppressed HCD triggered by multiple Psa effectors whereas AvrB4-1, HopF4b, HopAR1, and HopA1 suppressed HCD induced by one or two effectors. None of the Psa effectors were able to suppress HopAM1-1 or HopZ5-triggered HCD (Figure 4C). Overall, these results illustrate the specificity of HCD suppression by multiple Psa effectors.
FIGURE 4.

Multiple Psa effectors suppress HCD triggered by different effectors in N. benthamiana. (A) Psa effectors suppress BAX-induced HCD in N. benthamiana. Agrobacterium strains were mixed in a 1:4 ratio (BAX:Psa effector or GFP). (B) Psa effectors suppress AvrPto/Pto-induced HCD in N. benthamiana. Agrobacterium strains were mixed in 1:1:1 ratio (AvrPto:Pto:Psa effector or GFP). (C) Suppression of Psa effector-induced HCD in N. benthamiana. Agrobacterium strains were mixed in a 1:4 ratio (HCD-inducing effector:HCD-suppressing effector). Four–five week-old N. benthamiana leaves were agroinfiltrated and HCD was photographed at 3 (A,B) or 5 (C) dpi. This experiment was repeated more than three times with similar results.
HopD1 Suppresses PAMP-Induced Inhibition of Pto DC3000-Induced Hypersensitive Response in N. benthamiana
Many bacterial effectors were shown to interfere with PTI to enhance bacterial virulence. PTI triggered by a non-pathogenic bacterial strain can suppress ETI-associated HR elicited by a subsequent infiltration of another bacterial strain (Oh and Collmer, 2005; Crabill et al., 2010). The presence of a PTI-suppressing effector in the non-pathogenic first strain can suppress PTI sufficiently to allow the second ETI-triggering strain to trigger an HR (Le Roux et al., 2015). Pto DC3000 delivers effectors via the T3S and induces HCD in N. benthamiana. Activation of PTI prior to Pto DC3000 infection inhibits effector secretion by the T3S, resulting in significantly reduced HR triggered by Pto DC3000. We performed a PTI inhibition assay by delivery of each Psa effector from P. fluorescens Pf0-1(T3S) prior to Pto DC3000 infection in N. benthamiana (Le Roux et al., 2015). As shown previously, delivery of the wild-type Ralstonia solanacearum effector PopP2 from Pf0-1(T3S) interfered with Pf0-1(T3S)-mediated inhibition of Pto DC3000-induced HCD (Figure 5 and Supplementary Figure S5) (Le Roux et al., 2015). In contrast, the enzymatically inactive PopP2-C321A variant did not affect Pf0-1(T3S)-mediated inhibition of Pto DC3000-induced HCD. Among 35 Psa effectors tested, we found that only HopD1 showed robust PTI suppression activity in our assay (Figure 5 and Supplementary Figure S5). However, this result does not exclude the possibility that other Psa effectors carry PTI-suppression activity.
FIGURE 5.
The Psa effector HopD1 interferes with Pf Pf0-1(T3S)-induced suppression of Pto DC3000-triggered HCD. Leaves from 4 to 5 week-old N. benthamiana plants were infiltrated with Pf Pf0-1(T3S) carrying popP2, popP2-C321A or hopD1 (2 × 107 CFU/mL) 8 h prior to Pto DC3000 (3 × 108 CFU/mL) infection. Pto DC3000-triggered cell death was scored and photographed at 48 h post Pto DC3000 infection.
Discussion
In our study, we aimed to investigate functions of T3S effectors from two economically important strains of P. syringae pv. actinidiae, a causal agent of bacterial canker in kiwifruit. To better understand the functions of Psa T3Es, we generated a library comprising 48 cloned effectors from Psa V13 and LV5 strains, characterized their subcellular localization and showed that Psa effectors are localized at a diverse range of cellular compartments. By using agroinfiltration, we identified an unusually large number of effectors that induce HCD in Nicotiana spp. The requirement of SGT1 for HCD induced by some Psa effectors was demonstrated using a VIGS assay. Moreover, we showed that multiple Psa effectors suppress HCD. Finally, it was demonstrated that HopD1 interferes with PTI. Taken together, we conclude that multiple Psa effectors modify host immune responses via various mechanisms.
Diverse Localization of Psa T3Es Implicates Distinct Functions within the Host Cell
Localization of a T3E in planta provides an indication of the host target protein location. For example, several plasma membrane-localized T3Es including AvrRpm1, AvrB, and AvrRpt2 target membrane-associated RIN4 while nuclear-localized T3E PopP2 binds its corresponding NLR, RRS1, inside the nucleus (Nimchuk et al., 2000; Axtell and Staskawicz, 2003; Deslandes et al., 2003). Through our large-scale screening for the sub-cellular localization of transiently expressed Psa T3Es, we concluded that the nucleus, cytoplasm and plasma membrane are major target compartments for Psa T3Es. Psa T3Es of a multitude of sizes that localize to the nucleus make up a large proportion of the T3Es (23/44). Proteins between 90 and 110 kDa were found to passively diffuse through nuclear pores (Wang and Brattain, 2007). This may be one of the reasons for nucleocytoplasmic localization of a large number of effectors. Notably, since the in planta subcellular localization was assessed by transient expression of effector-YFP proteins, we cannot rule out the possibility that effector localization may be affected by over-accumulation of proteins or C-terminally tagged YFP. Nonetheless, it appears that Psa effectors localized in diverse cell compartments.
Previously, HopZ3 from P. syringae pv. syringae B728a (approximately 76 kDa including the YFP tag; 72% amino acid identity to Psa HopZ3) showed a nucleocytoplasmic localization to target MPK3 and MPK6, while HopBB1 from P. syringae pv. mori 301020 (approximately 60 kDa including the YFP tag; 93% amino acid identity to Psa HopBB1-2) was localized in the nucleus to target TCP14 and JAZ3 for degradation, despite not possessing a specific nuclear localization signal (Lewis et al., 2014; Yang et al., 2017). However, a nuclear localization may be critical to effector function with studies showing that several nucleus-localized T3Es induce transcriptional reprogramming in the host cell (Nissan et al., 2006; Kay and Bonas, 2009). On the other hand, many PRRs and their associated proteins involved in perception of PAMPs or activation of PTI are localized at the plasma membrane. Thus, many T3Es target these PTI components at the plasma membrane once delivered into the host cell (Kim et al., 2005; Zhou et al., 2014). This is consistent with our finding that some Psa T3Es are localized at the cell periphery. It would be interesting to further investigate if the Psa T3Es localized at the cell periphery enhance bacterial virulence by targeting novel PTI components.
A future application of findings from our study will be to examine Psa T3E function in relation to localization. Promising candidates for this could be HopAZ1 or HopAV1 that both appear to target the host cell cytoskeleton. HopZ1a and HopW1 are two known T3Es from P. syringae that target the cytoskeleton to disrupt PTI (Lee et al., 2012; Kang et al., 2014). Notably, a HopAZ1 homolog from P. savastanoi was recently reported to suppress both ROS production and callose deposition, critical markers of PTI (Matas et al., 2014). hopAZ1 is present in all sequenced strains of Psa and appears to be a ‘Psa core effector’ (McCann et al., 2013). Interestingly, hopAZ1 was reported to have been independently acquired multiple times into the T3E repertoire of pathogens of hazelnut, P. syringae pv. avellenae (O’Brien et al., 2012). These strains that cause hazelnut decline disease are from two different phylogroups of P. syringae strains, suggesting that hopAZ1 is closely associated with a gain of virulence in this woody host. Considering the subcellular localization of HopAZ1, identification of its interacting proteins in planta that also localize to the cytoskeleton could help reveal the role that HopAZ1 plays in promoting Psa virulence.
We note several differences for subcellular localization between Psa effectors and other known Pseudomonas effector homologs. HopM1 from Pto DC3000 is known to target AtMIN7 in Arabidopsis and destabilize AtMIN7 within the trans-golgi network and early endosome (Nomura et al., 2011). However, Psa HopM1 localized to the chloroplast in N. benthamiana. As the Pto DC3000 and Psa homologs of HopM1 share only 66.9% amino acid sequence identity, it is plausible that they target different host proteins in planta. However, previous findings have highlighted an alternative possibility since HopM1 was found to suppress SA-mediated immunity (DebRoy et al., 2004) as well as interact with 14-3-3 proteins to mediate pathogen virulence in planta (Lozano-Duran et al., 2014). This is particularly interesting since SA synthesis is primarily localized within the chloroplast and exported out by the multidrug and toxin extrusion-like transporter, EDS5, and 14-3-3 proteins have been associated with proteins targeted to the chloroplast (Sehnke et al., 2000; Serrano et al., 2013). Taken together, different localization of T3E homologs suggests that diversity of effector functions and targets may exist even in situations where effectors share significant homology.
Evidence of HCD-Inducing and -Suppressing Psa Effectors Support Pathogen Effector Interplay and Evolutionary Diversification
Multiple Psa effectors and their homologs from other P. syringae strains were compared for their HCD-inducing activity and, despite high amino acid sequence identity, a few effector homologs (HopAA1 and HopR1) showed different activities in Nicotiana spp. (Table 2). On the other hand, a HopF homolog from Psa V13 and Pto DC3000 showed similar HCD-inducing activity in N. tabacum despite relatively low amino acid identity. An R protein-mediated recognition for an effector can occur by direct interaction or indirectly through ‘guardee’ or ‘decoy’ molecules which interact with the effector (Jones and Dangl, 2006; van der Hoorn and Kamoun, 2008). Loss of interaction between the effector and its host target (R protein or guardee/decoy) may cause failure of effector recognition and associated cell death (Jones and Dangl, 2006; van der Hoorn and Kamoun, 2008). The HCD-inducing activities for HopAA1 (Pto DC3000) and HopR1 (Psa V13) may be lost in their corresponding homologs due to evolutionary pressure on the effector active site or recognition determinant to evade recognition while retaining virulence contribution. HopF4b (Psa HopF2), meanwhile, is a member of a wide-spread family of ADP-ribosyltransferases present in P. syringae strains with extensive genetic diversity (Lo et al., 2016). Psa possesses several effectors which share significant amino acid similarity with HopF2: HopF1, HopF4b, HopX3 (a novel member of the HopF family), HopBB1-1, and HopBB1-2. Among these, HopX3, HopBB1-1, and HopBB1-2 are located on the same pathogenicity island, termed the exchangeable effector locus (EEL), sites of significant genome variation between different Psa strains (McCann et al., 2013). This suggests that this family of effectors is under evolutionary pressure and may be a case study in evolutionary functional diversification. Unfortunately, mechanisms of cell death triggered by these effectors are still poorly understood. It would be interesting for future work to identify the recognition mechanisms and to search for the sequence variation in the interacting motifs responsible for variation in the HCD phenotype.
Several Psa effectors were shown to suppress effector-triggered HCD in this study (Figure 4). There are a few hypotheses on how effectors suppress ETI: (1) they share a common target with HCD-eliciting effectors and undergo biochemical processes to antagonistically interfere with the recognition, e.g., AvrRpm1/AvrRpt2/HopF2 (Kim et al., 2005; Wilton et al., 2010) and AvrPto/AvrPtoB (Abramovitch, 2003); (2) they may interfere with the downstream signaling of defense responses triggered by the effectors, e.g., HopF2 (Wang et al., 2010) and HopE1 (Guo et al., 2016); (3) they can interfere with the HCD-eliciting effectors through direct interaction, e.g., HopZ3/AvrB (Lee et al., 2015); or (4) they may suppress Agrobacterium-mediated transient protein expression in N. benthamiana, e.g., HopQ1 (Adlung and Bonas, 2017). Indeed, Psa HopQ1 suppressed most effector-triggered HCD tested in this study but notably failed to suppress HCD triggered by HopZ5, HopT1 and HopAM1-1. It would be interesting if HopZ5, HopT1 and/or HopAM1-1 affect HopQ1-mediated cell death suppression activity. Apart from HopQ1, other Psa effector-mediated HCD suppression activities were specific to one or two HCD-eliciting effectors (Figure 4). Nevertheless, several Psa effectors could suppress Psa effector-triggered HCD in N. benthamiana. Further research on these specific suppression mechanisms would help reveal the mechanism of ETI/HCD-suppression specificity.
From HCD-Triggering Effectors to Developing Resistance in Kiwifruit
Not all HCD triggered by agroinfiltration of effectors may be due to activation of immune responses (Vinatzer et al., 2006; Wroblewski et al., 2009). For instance, HopT1-1 triggered HCD but does not appear to be associated with significant growth restriction of virulent bacteria in N. benthamiana (Wei et al., 2007; Wroblewski et al., 2009). Conversely, P. syringae pv. tomato DC3000 lacking hopQ1-1 can cause disease symptoms in the non-host N. benthamiana but agroinfiltration of HopQ1-1 does not result in significant cell death (Wei et al., 2007; Wroblewski et al., 2009; Adlung and Bonas, 2017). Utilization of VIGS to suppress T3E-triggered cell death can offer clues about the downstream mechanism of effector-triggered HCD. The requirement of known immunity-regulator genes would suggest that the effector-triggered HCD is an immune response (Vinatzer et al., 2006). Wei et al. (2007), using VIGS, confirmed that SGT1 was required for cell death triggered by Pto-delivered HopQ1-1. Similarly, the PsyB728a effector, HopAA1, partially contributes to growth restriction of this strain on N. benthamiana and requires EDS1 for its HCD development (Vinatzer et al., 2006).
In addition to HopZ5 (Jayaraman et al., 2017), SGT1 is at least partially required for 5 Psa T3Es: HopAM1-1, HopAE1, HopT1, HopW1, and HopAB3. hopAM1-1 is unique to Psa V13 while hopT1, hopW1, and hopAB3 are unique to Psa LV5. hopAE1 alleles are present in both Psa strains. Interestingly, HopAM1 from Pto DC3000 is associated with resistance in Arabidopsis accession Bur-0 (Iakovidis et al., 2016), while HopAB3, a homolog of AvrPtoB from PtoDC3000, is an avirulence effector in tomato (Lin et al., 2006). Cell death triggered by HopX1, AvrE1, and HopR1 were largely unaffected by SGT1-silencing suggesting limited involvement in immune responses. Support for this notion comes from previous observations suggesting that HopX1, AvrE1, and HopR1 trigger necrosis rather than HR (Lorang and Keen, 1995; Badel et al., 2006; Nimchuk et al., 2007; Kvitko et al., 2009). Further characterization of Psa T3Es in N. benthamiana through gene knockouts in Psa would be useful for identification of avirulence effectors with a view to subsequently identify cognate R genes. Based on our results, the strongest avirulence gene candidates would be HopW1, HopAB3, HopZ5, HopAM1-1, and HopAE1.
Identification of R genes involved in the recognition of effectors is challenging and laborious. Recently, however, two advances in the field have facilitated this process. Firstly, Brendolise et al. (2017) have developed a hairpin-RNAi library targeting NLRs in N. benthamiana that allows for rapid identification of NLRs required for the recognition of an effector. They have demonstrated that the hairpin library is not only efficient at identifying the sensor NLR involved (Prf in the case of AvrPto) but also helper NLRs as well. This system is highly amenable for our purposes and multiple NLRs may be identified for cloning and downstream analyses without first mapping the resistance loci involved. The second technology is the development of RenSeq (Resistance gene enrichment sequencing) – a tool that capitalizes on the enrichment of specific NLR sequence(s) in a screened population of disease resistant plants (Jupe et al., 2013). This technology uses NLR enrichment and resequencing/reannotation to identify resistant alleles of NLRs even in crop plants that are still in the early stages of study. RenSeq and its derivative technologies have been used to identify candidate NLRs for multiple crop plants, including potato, tomato, and wheat (Jupe et al., 2013; Steuernagel et al., 2016; Witek et al., 2016). Application of RenSeq to identify candidate NLRs in tobacco and N. benthamiana would accelerate the identification of NLRs that recognize Psa effectors and, ultimately, the development of Psa-resistant kiwifruit.
Materials and Methods
Bacterial Materials
Escherichia coli DH5α was used to clone and maintain effector constructs. Agrobacterium tumefaciens AGL1 was used for transient transformation of N. benthamiana and N. tabacum leaf cells. E. coli DH5α and A. tumefaciens AGL1 were cultured in low salt L-media with appropriate antibiotics at 37 and 28°, respectively. The final concentrations of antibiotics used for bacterial cultures were 100 μg/ml for ampicillin and 50 μg/ml for kanamycin.
Plant Materials
Nicotiana tabacum Wisconsin 38 (W38) and N. benthamiana were grown for 4–5 weeks in a controlled plant growth room in long-day conditions (24°C, 16 h light/8 h dark).
Psa Effector Library Cloning Using Golden Gate Assembly
Psa strains genomic DNA was extracted using Thermo gDNA extraction kit (GeneJETTM, Thermo). All effector sequences were extracted from the published Psa genome database (McCann et al., 2013). Briefly, each effector sequence was divided into 1–1.5 kb modules based on length and presence of internal BsaI or SmaI restriction enzyme recognition sequences. Psa effector modules were PCR-amplified with flanking BsaI site-containing primers using high-fidelity polymerase (Phusion HiFi, Thermo). Amplified PCR products were ligated with SmaI-digested entry vector pICH41021. Ligated constructs were verified by BsaI digestion and Sanger sequencing. If present, internal BsaI sites in effector modules were removed by site-directed mutagenesis, as per instructions for the QuickChange II site-directed mutagenesis kit (Agilent, New Zealand). Chaperones for effectors, where present, were amplified with and cloned into the first module and were used exclusively for Pseudomonas-delivery constructs. Modules starting with the first codon of the effector coding sequence alone were used for Agrobacterium constructs. For Pseudomonas-delivery, effectors we cloned into the broad host-range vector pBBR1MCS5B:avrRps4pro with a C-terminal 6xHA tag using the Golden Gate cloning method (Engler et al., 2008) as described previously (Jayaraman et al., 2017). For agroinfiltration and construction of the Agrobacterium-expression library, effectors were cloned into the Golden Gate compatible binary vector pICH86988 under the CaMV 35S promoter and TMV Ω leader and with a C-terminal YFP tag using the Golden Gate assembly method (Engler et al., 2008).
Psa-Triggered HR and Pseudomonas-Delivery of Psa Effectors
Broad host-range plasmid constructs of each Psa effector were mobilized from E. coli DH5α to P. fluorescens Pf0-1(T3S) strains (Thomas et al., 2009) by triparental mating using E. coli HB101 (pRK2013) as a helper strain. Psa V13, Psa LV5, or Pf0-1(T3S) carrying empty vector or Psa effector constructs, were streaked from glycerol stocks onto King’s B plates with antibiotic selection and grown for 2 days at 28°C. Bacteria were then harvested from plates, resuspended in 10 mM MgCl2 and diluted to required OD600. Infiltrations were carried out on fully expanded leaves of 4- to 5-week-old N. benthamiana with a blunt end syringe. For assessing HR, Psa V13, Psa LV5 or Pto DC3000 (OD600 = 0.2; 1 × 108 CFU/mL), or Pf0-1(T3S) (OD600 = 0.6; 3 × 108 CFU/mL) were infiltrated in patches on three N. benthamiana leaves (replicates). HR was assayed visually at 1 day post infiltration (dpi) (Psa/Pto) or 2–3 dpi (Pf0-1). For assessing PTI suppression, the method was adapted from that described previously (Le Roux et al., 2015). Briefly, 12 h after Pf0-1 infiltration (for non-HR-triggering Psa effectors), Pto DC3000 (OD600 = 0.03; 2 × 107 CFU/mL) was infiltrated in an overlapping area of the leaves. Pto DC3000-triggered tissue collapse was scored at 3 dpi. PTI suppression experiments were conducted four times with effectors demonstrating full suppression of Pto DC3000-triggered tissue collapse in all three reps in at least three out of four experiments scored as strong suppressors of PTI.
Agrobacterium-Mediated Transient Expression
Agrobacterium tumefaciens AGL1 strain was transformed with binary vector effector constructs by electroporation (2.2 kV/6ms/Bio-Rad). Transformed cells were grown on L-agar plates with selective antibiotics for 2 days then inoculated and grown in liquid L-media with selective antibiotics. Overnight grown cultures were centrifuged at 5000 rpm for 4 min, resuspended in agroinfiltration solution (10 mM MgCl2/10 mM MES) and adjusted at OD600 = 0.4 before leaf infiltration with a blunt end syringe.
Immunoblot Analysis
Nicotiana benthamiana leaves were infiltrated with a mixture of Agrobacterium AGL1 harboring the binary vector effector construct (OD600 = 0.4) and the silencing suppressor P19 (OD600 = 0.1). Five leaf disks (8 mm in diameter) were harvested from the infiltrated patch at 2 days post infiltration and snap frozen in liquid nitrogen. Samples were ground in Laemmli protein loading buffer (Tris-Cl pH 6.8 250 mM, SDS 8%, Bromophenol blue 0.1%, Glycerol 40% and DTT 100 mM) and boiled for 10 min. Total protein extracts were separated by SDS-PAGE, transferred into PVDF membrane (Sigma–Aldrich) and probed with anti-GFP-HRP conjugated antibodies (Santa Cruz Biotech). Super signal West Pico Chemiluminescent sensitivity substrate (Thermo Fisher) and Super signal West Pico Maximum sensitivity substrate (Thermo Fisher) were used for detection.
Virus-Induced Gene Silencing (VIGS)
Short specific fragments (∼400 bp) from NbSGT1, NbNDR1, and NbEDS1 were designed using the Solgenomics VIGS tool (vigs.solgenomics.net). These fragments were PCR-amplified with primers introducing 5′ end EcoRI and 3′ end XhoI sites and were cloned into the pTRV2 vector between EcoRI and XhoI sites (Liu et al., 2002). Two week-old N. benthamiana seedlings were infiltrated with a mixture of A. tumefaciens AGL1 harboring pTRV1 (OD600 = 0.5) and pTRV2 : gene to be silenced (OD600 = 0.5) into cotyledons and grown for a further 4–5 weeks (Liu et al., 2002) in short day conditions (22°C, 11 h light/13 h dark). Gene silencing was confirmed by semi-quantitative amplification of the silencing target gene. Total RNA of N. benthamiana was extracted from one leaf disk (8 mm in diameter) using the Tri-reagent RNA extraction method (Ambion). cDNA was synthesized with Maxima kit (Thermo) following the manufacturer’s instruction and was used as template to amplify silenced genes with specific primers and Prime Taq polymerase (GenetBio).
Confocal Microscopy Analysis
Agrobacterium tumefaciens AGL1 harboring each Psa effector C-terminally tagged with YFP was infiltrated in 4-week-old N. benthamiana leaves and YFP signal was detected with a Carl Zeiss LSM700 confocal microscope at 2 days post infiltration. YFP fluorescence was excited at 488 nm with a 20 mW Argon laser and captured by the confocal channel in the emission range 500–530 nm. Images were processed using ImageJ software.
Author Contributions
SC, JJ, CS, and KS conceived and designed the study. SC, JJ, CS, and KS carried out the experiments. H-JP, HP, and S-WH provided materials and related information. SC, JJ, and KS analyzed and interpreted the data. SC, JJ, and KS prepared the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Matthew Templeton and Erik Rikkerink (New Zealand Institute for Plant & Food Research Limited) for providing Psa strains, and Toby Newman for critical reading of the manuscript.
Footnotes
Funding. This work was carried out with the support of Cooperative Research Program for Agriculture Science & Technology Development (Project No. PJ011913) from the Rural Development Administration, Basic Science Research Program (NRF-2016R1D1A1B03934707) from the National Research Foundation, South Korea, and Bioprotection Centre of Research Excellence, New Zealand. SC and JJ are the recipients of Ph.D. scholarships from Zespri and Plant & Food Research, respectively, in New Zealand.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2017.02157/full#supplementary-material
References
- Aarts N., Metz M., Holub E., Staskawicz B. J., Daniels M. J., Parker J. E. (1998). Different requirements for EDS1 and NDR1 by disease resistance genes define at least two R gene-mediated signaling pathways in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 95 10306–10311. 10.1073/pnas.95.17.10306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abramovitch R. B. (2003). Pseudomonas type III effector AvrPtoB induces plant disease susceptibility by inhibition of host programmed cell death. EMBO J. 22 60–69. 10.1093/emboj/cdg006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abramovitch R. B., Janjusevic R., Stebbins C. E., Martin G. B. (2006). Type III effector AvrPtoB requires intrinsic E3 ubiquitin ligase activity to suppress plant cell death and immunity. Proc. Natl. Acad. Sci. U.S.A. 103 2851–2856. 10.1073/pnas.0507892103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adlung N., Bonas U. (2017). Dissecting virulence function from recognition: cell death suppression in Nicotiana benthamiana by XopQ/HopQ1-family effectors relies on EDS1-dependent immunity. Plant J. 91 430–442. 10.1111/tpj.13578 [DOI] [PubMed] [Google Scholar]
- Adlung N., Prochaska H., Thieme S., Banik A., Blüher D., John P., et al. (2016). Non-host resistance induced by the Xanthomonas effector XopQ is widespread within the genus Nicotiana and functionally depends on EDS1. Front. Plant Sci. 7:1796. 10.3389/fpls.2016.01796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Austin M. J. (2002). Regulatory role of SGT1 in early R gene-mediated plant defenses. Science 295 2077–2080. 10.1126/science.1067747 [DOI] [PubMed] [Google Scholar]
- Axtell M. J., Staskawicz B. J. (2003). Initiation of RPS2-specified disease resistance in Arabidopsis is coupled to the AvrRpt2-directed elimination of RIN4. Cell 112 369–377. 10.1016/S0092-8674(03)00036-9 [DOI] [PubMed] [Google Scholar]
- Badel J. L., Shimizu R., Oh H.-S., Collmer A. (2006). A Pseudomonas syringae pv. tomato avrE1/hopM1 mutant is severely reduced in growth and lesion formation in tomato. Mol. Plant Microbe Interact. 19 99–111. 10.1094/MPMI-19-0099 [DOI] [PubMed] [Google Scholar]
- Bigeard J., Colcombet J., Hirt H. (2015). Signaling mechanisms in pattern-triggered immunity (PTI). Mol. Plant 8 521–539. 10.1016/j.molp.2014.12.022 [DOI] [PubMed] [Google Scholar]
- Block A., Toruño T. Y., Elowsky C. G., Zhang C., Steinbrenner J., Beynon J., et al. (2014). The Pseudomonas syringae type III effector HopD1 suppresses effector-triggered immunity, localizes to the endoplasmic reticulum, and targets the Arabidopsis transcription factor NTL9. New Phytol. 201 1358–1370. 10.1111/nph.12626 [DOI] [PubMed] [Google Scholar]
- Brendolise C., Montefiori M., Dinis R., Peeters N., Storey R. D., Rikkerink E. H. (2017). A novel hairpin library-based approach to identify NBS-LRR genes required for effector-triggered hypersensitive response in Nicotiana benthamiana. Plant Methods 13:32. 10.1186/s13007-017-0181-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buell C. R., Joardar V., Lindeberg M., Selengut J., Paulsen I. T., Gwinn M. L., et al. (2003). The complete genome sequence of the Arabidopsis and tomato pathogen Pseudomonas syringae pv. tomato DC3000. Proc. Natl. Acad. Sci. U.S.A. 100 10181–10186. 10.1073/pnas.1731982100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler M. I., Stockwell P. A., Black M. A., Day R. C., Lamont I. L., Poulter R. T. M. (2013). Pseudomonas syringae pv. actinidiae from recent outbreaks of kiwifruit bacterial canker belong to different clones that originated in China. PLOS ONE 8:e57464. 10.1371/journal.pone.0057464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caillaud M.-C., Piquerez S. J. M., Fabro G., Steinbrenner J., Ishaque N., Beynon J., et al. (2012a). Subcellular localization of the Hpa RxLR effector repertoire identifies a tonoplast-associated protein HaRxL17 that confers enhanced plant susceptibility. Plant J. 69 252–265. 10.1111/j.1365-313X.2011.04787.x [DOI] [PubMed] [Google Scholar]
- Caillaud M.-C., Wirthmueller L., Fabro G., Piquerez S. J. M., Asai S., Ishaque N., et al. (2012b). Mechanisms of nuclear suppression of host immunity by effectors from the Arabidopsis downy mildew pathogen Hyaloperonospora arabidopsidis (Hpa). Cold Spring Harb. Symp. Quant. Biol. 77 285–293. 10.1101/sqb.2012.77.015115 [DOI] [PubMed] [Google Scholar]
- Chapman J. R., Taylor R. K., Weir B. S., Romberg M. K., Vanneste J. L., Luck J., et al. (2012). Phylogenetic relationships among global populations of Pseudomonas syringae pv. actinidiae. Phytopathology 102 1034–1044. 10.1094/PHYTO-03-12-0064-R [DOI] [PubMed] [Google Scholar]
- Colombi E., Straub C., Künzel S., Templeton M. D., McCann H. C., Rainey P. B. (2017). Evolution of copper resistance in the kiwifruit pathogen Pseudomonas syringae pv. actinidiae through acquisition of integrative conjugative elements and plasmids: evolution of copper resistance. Environ. Microbiol. 19 819–832. 10.1111/1462-2920.13662 [DOI] [PubMed] [Google Scholar]
- Crabill E., Joe A., Block A., van Rooyen J. M., Alfano J. R. (2010). Plant immunity directly or indirectly restricts the injection of type III effectors by the Pseudomonas syringae type III secretion system. Plant Physiol. 154 233–244. 10.1104/pp.110.159723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui H., Tsuda K., Parker J. E. (2015). Effector-triggered immunity: from pathogen perception to robust defense. Annu. Rev. Plant Biol. 66 487–511. 10.1146/annurev-arplant-050213-040012 [DOI] [PubMed] [Google Scholar]
- DebRoy S., Thilmony R., Kwack Y. B., Nomura K., He S. Y. (2004). A family of conserved bacterial effectors inhibits salicylic acid-mediated basal immunity and promotes disease necrosis in plants. Proc. Natl. Acad. Sci. U.S.A. 101 9927–9932. 10.1073/pnas.0401601101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deslandes L., Olivier J., Peeters N., Feng D. X., Khounlotham M., Boucher C., et al. (2003). Physical interaction between RRS1-R, a protein conferring resistance to bacterial wilt, and PopP2, a type III effector targeted to the plant nucleus. Proc. Natl. Acad. Sci. U.S.A. 100 8024–8029. 10.1073/pnas.1230660100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engler C., Kandzia R., Marillonnet S. (2008). A one pot, one step, precision cloning method with high throughput capability. PLOS ONE 3:e3647. 10.1371/journal.pone.0003647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goehre V., Spallek T., Haeweker H., Mersmann S., Mentzel T., Boller T., et al. (2008). Plant Pattern-recognition receptor FLS2 is directed for degradation by the bacterial ubiquitin ligase AvrPtoB. Curr. Biol. 18 1824–1832. 10.1016/j.cub.2008.10.063 [DOI] [PubMed] [Google Scholar]
- Guo M., Kim P., Li G., Elowsky C. G., Alfano J. R. (2016). A bacterial effector co-opts calmodulin to target the plant microtubule network. Cell Host Microbe 19 67–78. 10.1016/j.chom.2015.12.007 [DOI] [PubMed] [Google Scholar]
- Guo M., Tian F., Wamboldt Y., Alfano J. R. (2009). The majority of the type III effector inventory of Pseudomonas syringae pv. tomato DC3000 can suppress plant immunity. Mol. Plant Microbe Interact. 22 1069–1080. 10.1094/mpmi-22-9-1069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hicks S. W., Galán J. E. (2013). Exploitation of eukaryotic subcellular targeting mechanisms by bacterial effectors. Nat. Rev. Microbiol. 11 316–326. 10.1038/nrmicro3009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iakovidis M., Teixeira P. J. P. L., Exposito-Alonso M., Cowper M. G., Law T. F., Liu Q., et al. (2016). Effector-triggered immune response in Arabidopsis thaliana is a quantitative trait. Genetics 204 337–353. 10.1534/genetics.116.190678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jayaraman J., Choi S., Prokchorchik M., Choi D. S., Spiandore A., Templeton M. D., et al. (2017). A bacterial acetyltransferase triggers immunity in Arabidopsis thaliana independent of hypersensitive response. Sci. Rep. 7:3557. 10.1038/s41598-017-03704-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jelenska J., Yao N., Vinatzer B. A., Wright C. M., Brodsky J. L., Greenberg J. T. (2007). A J domain virulence effector of Pseudomonas syringae remodels host chloroplasts and suppresses defenses. Curr. Biol. 17 499–508. 10.1016/j.cub.2007.02.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones J. D. G., Dangl J. L. (2006). The plant immune system. Nature 444 323–329. 10.1038/nature05286 [DOI] [PubMed] [Google Scholar]
- Jupe F., Witek K., Verweij W., Sliwka J., Pritchard L., Etherington G. J., et al. (2013). Resistance gene enrichment sequencing (RenSeq) enables reannotation of the NB-LRR gene family from sequenced plant genomes and rapid mapping of resistance loci in segregating populations. Plant J. 76 530–544. 10.1111/tpj.12307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang Y., Jelenska J., Cecchini N. M., Li Y., Lee M. W., Kovar D. R., et al. (2014). HopW1 from Pseudomonas syringae disrupts the actin cytoskeleton to promote virulence in Arabidopsis. PLOS Pathog. 10:e1004232. 10.1371/journal.ppat.1004232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kay S., Bonas U. (2009). How Xanthomonas type III effectors manipulate the host plant. Curr. Opin. Microbiol. 12 37–43. 10.1016/j.mib.2008.12.006 [DOI] [PubMed] [Google Scholar]
- Kim M. G., da Cunha L., McFall A. J., Belkhadir Y., DebRoy S., Dangl J. L., et al. (2005). Two Pseudomonas syringae type III effectors inhibit RIN4-regulated basal defense in Arabidopsis. Cell 121 749–759. 10.1016/j.cell.2005.03.025 [DOI] [PubMed] [Google Scholar]
- Kvitko B. H., Park D. H., Velasquez A. C., Wei C. F., Russell A. B., Martin G. B., et al. (2009). Deletions in the repertoire of Pseudomonas syringae pv. tomato DC3000 type III secretion effector genes reveal functional overlap among effectors. PLOS Pathog. 5:e1000388. 10.1371/journal.ppat.1000388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacomme C., Santa Cruz S. (1999). Bax-induced cell death in tobacco is similar to the hypersensitive response. Proc. Natl. Acad. Sci. U.S.A. 96 7956–7961. 10.1073/pnas.96.14.7956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Roux C., Huet G., Jauneau A., Camborde L., Trémousaygue D., Kraut A., et al. (2015). A receptor pair with an integrated decoy converts pathogen disabling of transcription factors to immunity. Cell 161 1074–1088. 10.1016/j.cell.2015.04.025 [DOI] [PubMed] [Google Scholar]
- Lee A. H.-Y., Hurley B., Felsensteiner C., Yea C., Ckurshumova W., Bartetzko V., et al. (2012). A bacterial acetyltransferase destroys plant microtubule networks and blocks secretion. PLOS Pathog. 8:e1002523. 10.1371/journal.ppat.1002523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J., Manning A. J., Wolfgeher D., Jelenska J., Cavanaugh K. A., Xu H., et al. (2015). Acetylation of an NB-LRR plant immune-effector complex suppresses immunity. Cell Rep. 13 1670–1682. 10.1016/j.celrep.2015.10.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis J. D., Abada W., Ma W., Guttman D. S., Desveaux D. (2008). The HopZ family of Pseudomonas syringae type III effectors require myristoylation for virulence and avirulence functions in Arabidopsis thaliana. J. Bacteriol. 190 2880–2891. 10.1128/JB.01702-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis J. D., Wilton M., Mott G. A., Lu W., Hassan J. A., Guttman D. S., et al. (2014). Immunomodulation by the Pseudomonas syringae HopZ type III effector family in Arabidopsis. PLOS ONE 9:e116152. 10.1371/journal.pone.0116152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G., Froehlich J. E., Elowsky C., Msanne J., Ostosh A. C., Zhang C., et al. (2014). Distinct Pseudomonas type-III effectors use a cleavable transit peptide to target chloroplasts. Plant J. 77 310–321. 10.1111/tpj.12396 [DOI] [PubMed] [Google Scholar]
- Lin N.-C., Abramovitch R. B., Kim Y. J., Martin G. B. (2006). Diverse AvrPtoB homologs from several Pseudomonas syringae pathovars elicit pto-dependent resistance and have similar virulence activities. Appl. Environ. Microbiol. 72 702–712. 10.1128/AEM.72.1.702-712.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Schiff M., Marathe R., Dinesh-Kumar S. P. (2002). Tobacco Rar1, EDS1 and NPR1/NIM1 like genes are required for N-mediated resistance to tobacco mosaic virus. Plant J. 30 415–429. 10.1046/j.1365-313X.2002.01297.x [DOI] [PubMed] [Google Scholar]
- Lo T., Koulena N., Seto D., Guttman D. S., Desveaux D. (2016). The HopF family of Pseudomonas syringae type III secreted effectors. Mol. Plant Pathol. 18 457–468. 10.1111/mpp.12412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorang J. M., Keen N. T. (1995). Characterization of avrE from Pseudomonas syringae pv. tomato: a hrp-linked avirulence locus consisting of at least two transcriptional units. Mol. Plant Microbe Interact. 8 49–57. 10.1094/MPMI-8-0049 [DOI] [PubMed] [Google Scholar]
- Lozano-Duran R., Bourdais G., He S. Y., Robatzek S. (2014). The bacterial effector HopM1 suppresses PAMP-triggered oxidative burst and stomatal immunity. New Phytol. 202 259–269. 10.1111/nph.12651 [DOI] [PubMed] [Google Scholar]
- Ma W., Dong F. F. T., Stavrinides J., Guttman D. S. (2006). Type III effector diversification via both pathoadaptation and horizontal transfer in response to a coevolutionary arms race. PLOS Genet. 2:e209. 10.1371/journal.pgen.0020209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackey D., Holt B. F., Wiig A., Dangl J. L. (2002). RIN4 interacts with Pseudomonas syringae type III effector molecules and is required for RPM1-mediated resistance in Arabidopsis. Cell 108 743–754. 10.1016/S0092-8674(02)00661-X [DOI] [PubMed] [Google Scholar]
- Matas I. M., Castañeda-Ojeda M. P., Aragón I. M., Antúnez-Lamas M., Murillo J., Rodríguez-Palenzuela P., et al. (2014). Translocation and functional analysis of Pseudomonas savastanoi pv. savastanoi NCPPB 3335 type III secretion system effectors reveals two novel effector families of the Pseudomonas syringae complex. Mol. Plant Microbe Interact. 27 424–436. 10.1094/MPMI-07-13-0206-R [DOI] [PubMed] [Google Scholar]
- McCann H. C., Rikkerink E. H. A., Bertels F., Fiers M., Lu A., Rees-George J., et al. (2013). Genomic analysis of the kiwifruit pathogen Pseudomonas syringae pv. actinidiae provides insight into the origins of an emergent plant disease. PLOS Pathog. 9:e1003503. 10.1371/journal.ppat.1003503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyers B. C., Kozik A., Griego A., Kuang H., Michelmore R. W. (2003). Genome-wide analysis of NBS-LRR-encoding genes in Arabidopsis. Plant Cell 15 809–834. 10.1105/tpc.009308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nimchuk Z., Marois E., Kjemtrup S., Leister R. T., Katagiri F., Dangl J. L. (2000). Eukaryotic fatty acylation drives plasma membrane targeting and enhances function of several type III effector proteins from Pseudomonas syringae. Cell 101 353–363. 10.1016/S0092-8674(00)80846-6 [DOI] [PubMed] [Google Scholar]
- Nimchuk Z. L., Fisher E. J., Desveaux D., Chang J. H., Dangl J. L. (2007). The HopX (AvrPphE) family of Pseudomonas syringae type III effectors require a catalytic triad and a novel N-terminal domain for function. Mol. Plant Microbe Interact. 20 346–357. 10.1094/MPMI-20-4-0346 [DOI] [PubMed] [Google Scholar]
- Nissan G., Manulis-Sasson S., Weinthal D., Mor H., Sessa G., Barash I. (2006). The type III effectors HsvG and HsvB of gall-forming Pantoea agglomerans determine host specificity and function as transcriptional activators. Mol. Microbiol. 61 1118–1131. 10.1111/j.1365-2958.2006.05301.x [DOI] [PubMed] [Google Scholar]
- Nomura K., Mecey C., Lee Y.-N., Imboden L. A., Chang J. H., He S. Y. (2011). Effector-triggered immunity blocks pathogen degradation of an immunity-associated vesicle traffic regulator in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 108 10774–10779. 10.1073/pnas.1103338108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Brien H. E., Thakur S., Gong Y., Fung P., Zhang J., Yuan L., et al. (2012). Extensive remodeling of the Pseudomonas syringae pv. avellanae type III secretome associated with two independent host shifts onto hazelnut. BMC Microbiol. 12:141. 10.1186/1471-2180-12-141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh H.-S., Collmer A. (2005). Basal resistance against bacteria in Nicotiana benthamiana leaves is accompanied by reduced vascular staining and suppressed by multiple Pseudomonas syringae type III secretion system effector proteins: vascular staining assay for basal resistance in Nicotiana benthamiana. Plant J. 44 348–359. 10.1111/j.1365-313X.2005.02529.x [DOI] [PubMed] [Google Scholar]
- Oh S. K., Kwon S. Y., Choi D. (2014). Rpi-blb2-mediated hypersensitive cell death caused by Phytophthora infestans AVRblb2 requires SGT1, but not EDS1, NDR1, salicylic acid-, jasmonic acid-, or ethylene-mediated signaling. Plant Pathol. J. 30 254–260. 10.5423/ppj.oa.2014.0027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ordon J., Gantner J., Kemna J., Schwalgun L., Reschke M., Streubel J., et al. (2017). Generation of chromosomal deletions in dicotyledonous plants employing a user-friendly genome editing toolkit. Plant J. 89 155–168. 10.1111/tpj.13319 [DOI] [PubMed] [Google Scholar]
- Peart J. R., Lu R., Sadanandom A., Malcuit I., Moffett P., Brice D. C., et al. (2002). Ubiquitin ligase-associated protein SGT1 is required for host and nonhost disease resistance in plants. Proc. Natl. Acad. Sci. U.S.A. 99 10865–10869. 10.1073/pnas.152330599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robert-Seilaniantz A., Shan L. B., Zhou J. M., Tang X. Y. (2006). The Pseudomonas syringae pv. tomato DC3000 type III effector HopF2 has a putative myristoylation site required for its avirulence and virulence functions. Mol. Plant Microbe Interact. 19 130–138. 10.1094/mpmi-19-0130 [DOI] [PubMed] [Google Scholar]
- Rodriguez-Herva J. J., Gonzalez-Melendi P., Cuartas-Lanza R., Antunez-Lamas M., Rio-Alvarez I., Li Z., et al. (2012). A bacterial cysteine protease effector protein interferes with photosynthesis to suppress plant innate immune responses. Cell. Microbiol. 14 669–681. 10.1111/j.1462-5822.2012.01749.x [DOI] [PubMed] [Google Scholar]
- Sarris P. F., Duxbury Z., Huh S. U., Ma Y., Segonzac C., Sklenar J., et al. (2015). A plant immune receptor detects pathogen effectors that target WRKY transcription factors. Cell 161 1089–1100. 10.1016/j.cell.2015.04.024 [DOI] [PubMed] [Google Scholar]
- Schornack S., Fuchs R., Huitema E., Rothbauer U., Lipka V., Kamoun S. (2009). Protein mislocalization in plant cells using a GFP-binding chromobody. Plant J. 60 744–754. 10.1111/j.1365-313X.2009.03982.x [DOI] [PubMed] [Google Scholar]
- Scofield S. R., Tobias C. M., Rathjen J. P., Chang J. H., Lavelle D. T., Michelmore R. T., et al. (1996). Molecular basis of gene-for-gene specificity in bacterial speck disease of tomato. Science 274 2063–2065. 10.1126/science.274.5295.2063 [DOI] [PubMed] [Google Scholar]
- Sehnke P. C., Henry R., Cline K., Ferl R. J. (2000). Interaction of a plant 14-3-3 protein with the signal peptide of a thylakoid-targeted chloroplast precursor protein and the presence of 14-3-3 isoforms in the chloroplast stroma. Plant Physiol. 122 235–242. 10.1104/pp.122.1.235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serrano M., Wang B., Aryal B., Garcion C., Abou-Mansour E., Heck S., et al. (2013). Export of salicylic acid from the chloroplast requires the multidrug and toxin extrusion-like transporter EDS5. Plant Physiol. 162 1815–1821. 10.1104/pp.113.218156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sohn K. H., Segonzac C., Rallapalli G., Sarris P. F., Woo J. Y., Williams S. J., et al. (2014). The nuclear immune receptor RPS4 is required for RRS1SLH1-dependent constitutive defense activation in Arabidopsis thaliana. PLOS Genet. 10:e1004655. 10.1371/journal.pgen.1004655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steuernagel B., Periyannan S. K., Hernández-Pinzón I., Witek K., Rouse M. N., Yu G., et al. (2016). Rapid cloning of disease-resistance genes in plants using mutagenesis and sequence capture. Nat. Biotechnol. 4 652–655. 10.1038/nbt.3543 [DOI] [PubMed] [Google Scholar]
- Swiderski M. R., Birker D., Jones J. D. G. (2009). The TIR domain of TIR-NB-LRR resistance proteins is a signaling domain involved in cell death induction. Mol. Plant Microbe Interact. 22 157–165. 10.1094/MPMI-22-2-0157 [DOI] [PubMed] [Google Scholar]
- Takahashi A., Casais C., Ichimura K., Shirasu K. (2003). HSP90 interacts with RAR1 and SGT1 and is essential for RPS2-mediated disease resistance in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 100 11777–11782. 10.1073/pnas.2033934100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Templeton M. D., Warren B. A., Andersen M. T., Rikkerink E. H. A., Fineran P. C. (2015). Complete DNA sequence of Pseudomonas syringae pv. actinidiae, the causal agent of kiwifruit canker disease. Genome Announc. 3:e01054–15. 10.1128/genomeA.01054-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas W. J., Thireault C. A., Kimbrel J. A., Chang J. H. (2009). Recombineering and stable integration of the Pseudomonas syringae pv. syringae 61 hrp/hrc cluster into the genome of the soil bacterium Pseudomonas fluorescens Pf0-1. Plant J. Cell Mol. Biol. 60 919–928. 10.1111/j.1365-313X.2009.03998.x [DOI] [PubMed] [Google Scholar]
- Tobias C. M., Oldroyd G. E., Chang J. H., Staskawicz B. J. (1999). Plants expressing the Pto disease resistance gene confer resistance to recombinant PVX containing the avirulence gene AvrPto. Plant J. Cell Mol. Biol. 17 41–50. 10.1046/j.1365-313X.1999.00350.x [DOI] [PubMed] [Google Scholar]
- van der Hoorn R. A. L., Kamoun S. (2008). From Guard to Decoy: a new model for perception of plant pathogen effectors. Plant Cell 20 2009–2017. 10.1105/tpc.108.060194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinatzer B. A., Teitzel G. M., Lee M.-W., Jelenska J., Hotton S., Fairfax K., et al. (2006). The type III effector repertoire of Pseudomonas syringae pv. syringae B728a and its role in survival and disease on host and non-host plants. Mol. Microbiol. 62 26–44. 10.1111/j.1365-2958.2006.05350.x [DOI] [PubMed] [Google Scholar]
- Wang Q., Han C., Ferreira A. O., Yu X., Ye W., Tripathy S., et al. (2011). Transcriptional programming and functional interactions within the Phytophthora sojae RXLR effector repertoire. Plant Cell 23 2064–2086. 10.1105/tpc.111.086082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R., Brattain M. G. (2007). The maximal size of protein to diffuse through the nuclear pore is larger than 60 kDa. FEBS Lett. 581 3164–3170. 10.1016/j.febslet.2007.05.082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Li J., Hou S., Wang X., Li Y., Ren D., et al. (2010). A Pseudomonas syringae ADP-ribosyltransferase inhibits Arabidopsis mitogen-activated protein kinase kinases. Plant Cell 22 2033–2044. 10.1105/tpc.110.075697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei C.-F., Kvitko B. H., Shimizu R., Crabill E., Alfano J. R., Lin N.-C., et al. (2007). A Pseudomonas syringae pv. tomato DC3000 mutant lacking the type III effector HopQ1-1 is able to cause disease in the model plant Nicotiana benthamiana. Plant J. Cell Mol. Biol. 51 32–46. 10.1111/j.1365-313X.2007.03126.x [DOI] [PubMed] [Google Scholar]
- Williams S. J., Sohn K. H., Wan L., Bernoux M., Sarris P. F., Segonzac C., et al. (2014). Structural basis for assembly and function of a heterodimeric plant immune receptor. Science 344 299–303. 10.1126/science.1247357 [DOI] [PubMed] [Google Scholar]
- Wilton M., Subramaniam R., Elmore J., Felsensteiner C., Coaker G., Desveaux D. (2010). The type III effector HopF2Pto targets Arabidopsis RIN4 protein to promote Pseudomonas syringae virulence. Proc. Natl. Acad. Sci. U.S.A. 107 2349–2354. 10.1073/pnas.0904739107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witek K., Jupe F., Witek A. I., Baker D., Clark M. D., Jones J. D. G. (2016). Accelerated cloning of a potato late blight–resistance gene using RenSeq and SMRT sequencing. Nat. Biotechnol. 34 656–660. 10.1038/nbt.3540 [DOI] [PubMed] [Google Scholar]
- Wroblewski T., Caldwell K. S., Piskurewicz U., Cavanaugh K. A., Xu H., Kozik A., et al. (2009). Comparative large-scale analysis of interactions between several crop species and the effector repertoires from multiple pathovars of Pseudomonas and Ralstonia. Plant Physiol. 150 1733–1749. 10.1104/pp.109.140251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang T., Zong N., Zou Y., Wu Y., Zhang J., Xing W., et al. (2008). Pseudomonas syringae effector AvrPto blocks innate immunity by targeting receptor kinases. Curr. Biol. 18 74–80. 10.1016/j.cub.2007.12.020 [DOI] [PubMed] [Google Scholar]
- Yang L., Teixeira P. J. P. L., Biswas S., Finkel O. M., He Y., Salas-Gonzalez I., et al. (2017). Pseudomonas syringae type III effector HopBB1 promotes host transcriptional repressor degradation to regulate phytohormone responses and virulence. Cell Host Microbe 21 156–168. 10.1016/j.chom.2017.01.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Shao F., Li Y., Cui H., Chen L., Li H., et al. (2007). A Pseudomonas syringae effector inactivates MAPKs to suppress PAMP-induced immunity in plants. Cell Host Microbe 1 175–185. 10.1016/j.chom.2007.03.006 [DOI] [PubMed] [Google Scholar]
- Zhang Y., Dorey S., Swiderski M., Jones J. D. G. (2004). Expression of RPS4 in tobacco induces an AvrRps4-independent HR that requires EDS1, SGT1 and HSP90. Plant J. 40 213–224. 10.1111/j.1365-313X.2004.02201.x [DOI] [PubMed] [Google Scholar]
- Zhou J. G., Wu S. J., Chen X., Liu C. L., Sheen J., Shan L. B., et al. (2014). The Pseudomonas syringae effector HopF2 suppresses Arabidopsis immunity by targeting BAK1. Plant J. 77 235–245. 10.1111/tpj.12381 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.