Abstract
Complex and dynamic networks of molecules are involved in human diseases. High-throughput technologies enable omics studies interrogating thousands to millions of makers with similar biochemical properties (e.g. transcriptomics for RNA transcripts). However, a single layer of ‘omics’ can only provide limited insights into the biological mechanisms of a disease. In the case of GWAS, although thousands of SNPs have been identified for complex diseases and traits, the functional implications and mechanisms of the associated loci are largely unknown. Additionally, the genomic variants alone are not able to explain the changing disease risk across the life span. DNA, RNA, protein, and metabolite often have complementary roles to jointly perform a certain biological function. Such complementary effects and synergistic interactions between omic layers in the life-course can only be captured by integrative study of multiple molecular layers. Building upon the success in single-omics discovery research, population studies started adopting the multi-omics approach to better understanding the molecular function and disease etiology. Multi-omics approaches integrate data obtained from different omic levels to understand their interrelation and combined influence on the disease processes. Here, we summarize major omics approaches available in population research, and review integrative approaches and methodologies interrogating multiple omic layers, which enhance the gene discovery and functional analysis of human diseases. We seek to provide analytical recommendations for different types of multi-omics data and study designs to guide the emerging multi-omic research, and to suggest improvement of the existing analytical methods.
Keywords: Integrative Genomics, GWAS, Transcriptome, Epigenome, Proteome, Metabolome, DNA Methylation, Gene Expression, Quantitative Trait Loci, Genomic Epidemiology
1. Introduction
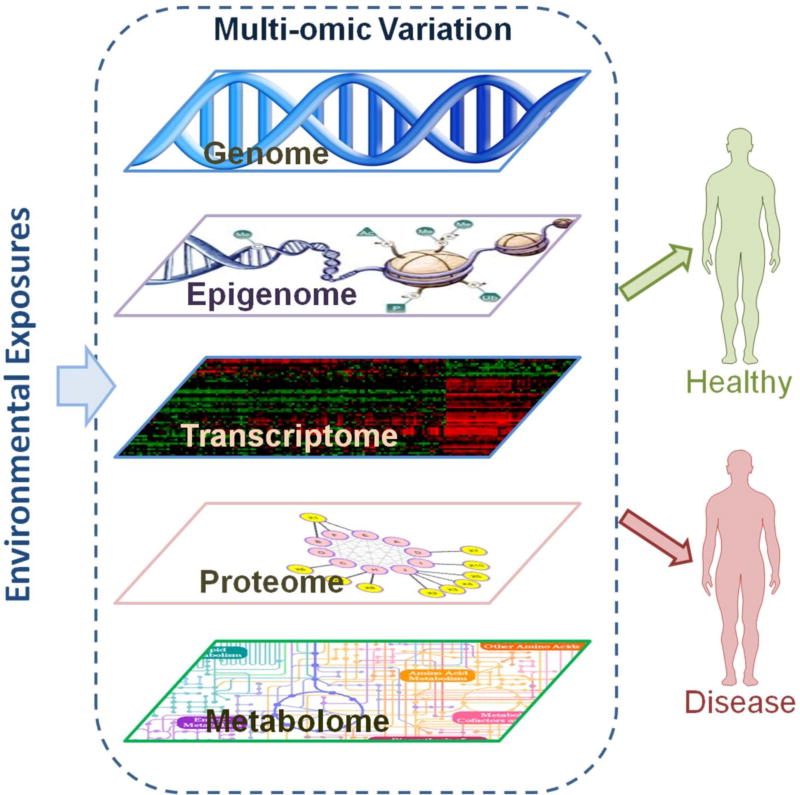
Biological processes, such as the development of human diseases, involve a highly dynamic and interactive system of molecular layers (e.g. genetics, epigenetics, mRNA transcripts, proteins and metabolites) and are influenced by many environmental factors. Recent technological advancement has permitted high-throughput measurement of human genome, epigenome, metabolome, transcriptome and proteome at the population level (G. T. Consortium, 2015; Kim et al., 2014; Roadmap Epigenomics et al., 2015; Shin et al., 2014; Wellcome Trust Case Control, 2007; Ziller et al., 2013). Although each layer of the omic profile allows a comprehensive survey for that particular type of disease associations, the cross-talk between multiple molecular layers cannot be assessed by such simplified reduction approach (Chen et al., 2012). A critical but challenging next step is to obtain a holistic understanding of the molecular information flow and the interactive molecular system, which can only be achieved by studying multiple layers of omic data simultaneously. Incorporating multi-omic measures of population samples into multidimensional network and system analyses (Figure 1), will address the gaps in our current knowledge of molecular mediation mechanisms, gene-environment interactions, and longitudinal effects during the development of chronic diseases (Civelek & Lusis, 2014; Ritchie, Holzinger, Li, Pendergrass, & Kim, 2015). The integrative approach of multi-omic data may enhance the understanding of the molecular dynamics underlying the pathophysiology of diseases, and may lead to novel strategies for early detection, prevention and treatment of human diseases. Given the increasing number of population studies collecting multi-omic data but limited overview of the methodological framework for integrative analyses (Liu, Ding, et al., 2013; Petersen et al., 2014; Shah et al., 2015), we summarize the analytical methods for high-throughput multi-omic data, and provide an updated analytical framework to incorporate genomic, epigenomic, transcriptomic, proteomic, and metabolomics data for the emerging field of multi-omic association study of human diseases. In this article, we do not cover the topic of disease classification and prediction, which does not aim to understand the biological and functional roles of omic markers (Wan & Pal, 2014).
Figure 1. Conceptual model of multi-omics and human disease.
2. Overview of omics technologies and association studies
Transcriptomic and genomic association approaches have been widely adopted in biomedical research and have successfully identified genes and genetic loci involved in the development of human diseases. These findings revealed the complexity of biological systems, and provided insights for new approaches to disease diagnosis, treatment and prevention. Additionally, other high-throughput omics technologies have been developed to measure other importance biomolecules such as epigenomics for epigenetic markers, proteomics for proteins and peptides and metabolomics for low-molecular-weight metabolites. In many ways, omic association studies are similar in that they search for omic biomarkers connected with phenotype by unbiased genome-wide screening. The high-throughput experimental methods allow us to study a large number of omic markers simultaneously. As a result, such omics studies emphasize the role of corresponding molecules of their kind. Although each omics technology is capable of measuring one type of biomolecule accurately and comprehensively, by themselves, they are all limited by the functional roles of each type of molecule in a biological system. From a typical protein-coding DNA sequence to its functional protein product in cell, multiple molecular machineries are involved, such as transcriptional regulation, translational regulation, RNA/peptide degradation, post-translational modification and transportation. In addition, different type of molecules may function together to play a joint role in a system. Thus, emphasizing only one omic layer can miss important information, particularly the complementary effects and interactions between omic layers. For example, GWAS have successfully identified the genetic susceptibility of human diseases, however, it cannot capture the intra-individual changes over time and how such changes relate to the disease risk; Proteomics measures all proteins and peptides in biological samples, and offers highly complementary information to genomics. As many biological functions are transmitted through proteins, proteomics can yield new biology and insights into disease. The population level omics studies have highlighted the robust association with disease traits as well as the inter-individual variation. As we summarize in the following sections, recent population studies started incorporating high-throughput omics data beyond a single type of molecules. The new multi-omic studies will provide a combined view of multiple functional layer at a system level, and pave the road to precision medicine, which emphasizes the tailored medical practice to optimize the clinical outcome given the unique individual profile comparing to the population average.
2.1 Genome-wide association study (GWAS) and transcriptomics studies of human diseases
Genome-wide profiling of genetic variants (e.g. SNPs and copy number variations) and RNA transcripts have been used to study human disease in populations for over a decade (Alizadeh et al., 2000; Klein et al., 2005). The earlier genome-wide scans rely on microarray technologies with a large number of pre-selected probes targeting the corresponding genetic variants or RNA transcripts. In the case of GWAS, thousands of genetic variants have been linked with hundreds of human traits and diseases (Welter et al., 2014). Given the extensive coverage of the study designs and methods for GWAS and transcriptomic studies from existing literatures (Cookson, Liang, Abecasis, Moffatt, & Lathrop, 2009; Manolio, 2010; Ziegler & Sun, 2012), we focus on the most recent technological development in the context of multi-omic studies. For both GWAS and transcriptomic studies, the next generation sequencing (NGS) technologies provide the most complete genome-wide coverage (Metzker, 2010).
Transcriptomic association studies of human diseases have been used to identify cellular pathways important for disease pathology, distinguish between healthy and diseased individuals, and identify changes during disease progression. The newest transcriptomics approaches are based on the simultaneous deep sequencing of all RNA transcripts expression (i.e., RNA-Seq) in biological samples across the genome (Ozsolak & Milos, 2011; Z. Wang, Gerstein, & Snyder, 2009). RNA-Seq provides comprehensive information on mRNA abundance, alternative splicing, nucleotide variation, and structure alteration. The resulting short sequence reads need to be processed following an extensive bioinformatics work flow including the evaluation of the data quality, the alignment to a reference genome, identification and annotation of variants, and quantification of the transcript levels (Trapnell et al., 2012). Similarly, NGS technology has been used to call SNP genotypes and structural variations and provides the ultimate coverage and resolution of genetic variations including private mutations of individuals. Bioinformatic procedures transform the raw data from NGS technology into aligned reads, call genotypes from samples (single or multiple), and perform quality control of individual genotype calls (Nielsen, Paul, Albrechtsen, & Song, 2011). A large number of human samples have been or are being sequenced to fully understand the role of genetics in human health and disease (Marx, 2015; The 1000 Genomes Project Consortium et al., 2012).
2.2 Epigenome-wide association studies
Epigenetics usually refers to the heritable molecular modifications that have effects independent of the primary DNA sequence that can be modified by environmental exposures at various developmental stages throughout the lifespan (Bird, 2007; Foley et al.). Epigenetic modifications, through DNA methylation (DNAm) and other mechanisms, can regulate gene expression and exert a long-term impact on the development of chronic diseases. Most human diseases are thought to be due to both genetic and environmental factors, and the interplay between genes and environment. Epigenetic modifications are shaped by environmental (e.g. smoking (Breitling, Yang, Korn, Burwinkel, & Brenner, 2011; Shenker et al.; Sun, 2014; Sun, Smith, et al., 2013), poor nutrition(Waterland & Jirtle, 2003), genetic factors (Sun, 2014; D. Zhang et al., 2010), and age – all well known risk factors human diseases (Bocklandt et al., 2011; Teschendorff et al., 2010). Additionally, inflammatory markers have been associated with epigenetics at the gene level in peripheral leukocytes (Sun, Lazarus, et al., 2013; Uddin et al., 2011), which play a critical role in acute and chronic inflammation related to pathophysiology of many human diseases. Studying the epigenome in the well-characterized samples may enable us to discover novel genes and pathways through which genetic factors and environmental exposures influence disease initiation and development, and thereby provide new targets for prevention and treatment (Foley et al., 2009; Jablonka, 2004; Relton & Davey Smith, 2010). We focus on DNA methylome in this article, because it is the only epigenomic mechanism being robustly measured in population samples.
Technologies and resources
The most recent population level studies of human epigenome rely on high-density microarrays such as the Illumina Infinium HumanMethylation450 (450K) BeadChip (Dedeurwaerder et al., 2011) or sequencing-based methods (McClay et al., 2014) following biochemical modifications or enrichments of genomic DNA (Rakyan, Down, Balding, & Beck). The Illumina 450K platform allows simultaneous assessment of the DNAm levels of over 480,000 sites across the genome. The 450K chip offers accurate and reproducible performance (Bibikova et al., 2011; Sandoval et al., 2011) with known but adjustable bias between Infinium I and II assays (Dedeurwaerder et al., 2011). On the down side, it only represents ~2% of all DNAm sites of the human genome, and over-represents the genic regions (75% of sites) including promoter, gene body and 3’-UTR (Sandoval et al., 2011). Because the 450K platform provides a balanced package of per sample cost, assay throughput, accuracy and coverage, it has become the most popular technology in human epigenomics research.
Several sequencing-base techniques, including methyl-CpG binding domain protein sequencing (MBD-seq) (Serre, Lee, & Ting, 2010), reduced-representation bisulfite sequencing (RRBS) (Meissner et al., 2008), and whole genome bisulfite sequencing (Y. Li, Zhu, et al., 2010) can also profile the DNA methylome. They provide better genomic coverage than 450K array, and can assess the genetic variants and allele-specific methylation beyond the preselected microarray probes. For example, MBD-seq enriches for the methylated DNA fraction, followed by the next-generation sequencing. MBD-seq was successfully used to screen over 10 million DNAm sites in a population study of schizophrenia (Aberg et al., 2014). But these sequencing-based methods suffer from unique technical biases and variations in sample preparation, processing and experimental procedures, which may not be fully addressable in the statistical analyses or require intensive bioinformatic/biostatistical analyses (Michels et al., 2013).
Most epigenome-wide profiling methods use bisulfite conversion of genomic DNA in order to distinguish methylated from unmethylated cytosines. This chemical conversion does not distinguish 5-methylcytosine (5mC) from 5-hydroxymethylcytosine (5hmC) (Jin, Kadam, & Pfeifer, 2010), another type of cytosine modification with potentially different function in cellular processes (Ito et al., 2010), particular in tissues with higher content of 5hmC. Alternative methylation forms, such as 5hmC, require modified experimental protocol to complete our understating of the genomic DNA methylation.
Depending on the epityping protocol, a corresponding data quality control, processing and analysis pipeline need to be appropriately carried out (D. Li, Xie, Pape, & Dye, 2015; Michels et al., 2013). These omics-specific procedures for obtaining high-quality data are essential for the success of the multi-omics study. They have been thoroughly investigated within each omics research community. Thus, we only focus on the common themes across omics and the approaches to multi-omics analysis.
As the HapMap Project and the 1000 Genome Project to GWAS, the success of EWAS relies on comprehensive reference panels of human epigenome. National Institutes of Health (NIH) Roadmap Epigenomics project aims to produce a public resource of human epigenomic data to catalyze basic biology and human disease research (Roadmap Epigenomics et al., 2015). The International Human Epigenome Consortium (IHEC) is a global consortium with the primary goal of establishing high-resolution reference human epigenome maps for normal and disease cell types to the research community (Adams et al., 2012). ENCyclopedia Of DNA Elements (ENCODE) targeted the identification of all functional DNA elements in the human genome including some epigenetic modifications (Bernstein et al., 2012; E. P. Consortium, 2004). These consortial efforts have provided important insight of molecular mechanism linking epigenetic variants and functional outcomes, and resulted in increasing knowledge of the important roles of epigenetics in both normal development and the disease process.
Studies of human diseases
Epigenome-wide association study (EWAS) is an examination of epigenome-wide markers in many individuals to scan for epigenetic markers associated with a trait (Sun, 2014). Current EWAS in human populations are all based on genome-wide measurement of DNA methylation of cytosine. EWAS has emerged as a valuable approach to searching for molecular mediators of genetic and environmental factors, and for unexplained disease risks. Although epigenomics techniques has only been available and affordable recently, numerous studies have successfully identified leukocyte-based DNAm markers associated with disease traits (Demerath et al., 2015; Dick et al., 2014; Hidalgo et al., 2014; Irvin et al., 2014; Liang et al., 2015; Liu, Aryee, et al., 2013). Using hundreds to thousands of subjects, these EWAS identified DNAm sites with much larger effect sizes compared to a typical GWAS of the same trait (Demerath et al., 2015; Dick et al., 2014). Despite these encouraging findings from the first wave of EWAS of disease traits, numerous issues such as imperfect epityping technologies, limited types of specimens from human subjects, cell-type specificity, sample size and data analysis framework need to be further addressed and improved (Heijmans & Mill, 2012). The field has rapidly evolved and offered improved methods and tools for the design, analysis and interpretation of EWAS of human diseases.(Cortessis et al., 2012; Michels et al., 2013; Mill & Heijmans, 2013; Rakyan et al., 2011).
Cross-talks with other omic layers
In addition to the genetic association analysis of DNA methylation levels (see section 3.1), the relationship between DNA methylation and Gene expression levels have been integrated at the genome-wide scale (Gibbs et al., 2010; Liu, Ding, et al., 2013). Although epigenetic markers can induce the transcriptional regulation, the epigenetic modification may not be sufficient to cause changes of gene expression levels. In other words, epigenetic modifications potentiate the gene expression changes conditional on other co-regulators. Therefore, lack of correlation between epigenetic variation and gene expression levels does not mean that the epigenetic modifications have no functional consequence in gene expression. In a recent study, the associations between 649 metabolic traits and over 400,000 DNA methylation markers were examined using blood samples from 1,814 European participants. The epigenome-wide association approach revealed strong associations with metabolic traits driven by either genetic effects or possible environmental factors (Petersen et al., 2014). The group of correlated epigenetic and metabolic markers influenced by common environmental exposures suggest their potential utility in studying the environmental risk and gene-environmental interaction for human diseases.
2.3 Proteomic studies of human biology and diseases
The proteome includes the entire set of proteins expressed by a genome (Wilkins et al., 1996). Proteomics can measure amino-acid mutations, peptide isoforms, and post-translational modifications that may influence cellular functions and physiology. Thus, proteomics is positioned to define the functional roles of proteins in normal and disease-related cellular processes, and to enable hypothesis-driven and discovery research of human diseases (Nilsson et al., 2010). Proteomics can also measure where and when proteins localize in the cell or tissue, that is important to understand the disease process but cannot be captured by other genomic technologies.
Proteomic technologies
Since 1990’s, numerous biochemical approaches have been developed to target the large-scale proteome-wide study. The technological advance in mass spectrometry (MS) and protein separation now allows rapid and accurate detection of hundreds of human proteins and peptides from a small amount of body fluid or tissue (Kraemer et al., 2011). Proper procedures in sample collection, sample preparation, MS experiments and data analysis are all critical to obtain high-quality data for hypothesis-driven or proteome-wide discovery research (Nilsson et al., 2010). Recently, the protein products of 17,294 genes were identified and mapped in the draft of human proteome using 30 human samples of tissues and cells (Kim et al., 2014). This draft proteome demonstrated the feasibility of building a complete human proteome encompassing over 200 cell types and all body fluids. Individual proteins can span 10 orders of magnitude in abundance (e.g. serum albumin and interleukin 6) (Anderson & Anderson, 2002). Current MS-based technology can detect and quantify proteins with at least six-fold difference in dynamic range and is still improving. Although, highly abundant “housekeeping” proteins from 2,350 genes constitute approximately 75% of total protein mass, low-abundance proteins constitute the majority of the protein species in human (Kim et al., 2014).
Proteomic studies of human diseases
Proteomics approach has been widely adopted in studies of human diseases including cancer (Frantzi et al., 2015; S. Pan, Brentnall, & Chen, 2015; Petricoin et al., 2002; Tsai et al., 2015), multiple sclerosis (Farias, Pradella, Schmitt, Santos, & Martins-de-Souza, 2014) and schizophrenia (Al Awam et al., 2015; Nascimento & Martins-de-Souza, 2015). In an early study of ovarian cancer, an independently trained proteomic profile of serum samples was able to prospectively classify ovarian cancer cases and controls (Petricoin et al., 2002). On the contrary, proteomic studies of multiple sclerosis and schizophrenia have not established any reliable protein biomarkers to classify patients after years of investigation of different biological samples (e.g. CNS tissue, cerebrospinal fluid, peripheral blood, plasma and serum) (Farias et al., 2014; Nascimento & Martins-de-Souza, 2015). However, these studies have found many protein candidates, which can potentially improve prognosis, diagnosis, and effectiveness of treatment. More recent proteomic studies also used body fluid samples to identify protein markers for bladder cancer (Frantzi et al., 2015), pancreatic cancer (S. Pan et al., 2015) and hepatocellular carcinoma (Tsai et al., 2015). These proteomic studies of human diseases provided integrated pictures of the protein networks involved in the pathophysiology, and might eventually lead to the development of novel and more efficient treatment therapies.
Cross-talks with other omic layers
Proteins function together with other genomic features in complex biological pathways and networks. They are products of genes and RNA transcripts, and play critical roles in cellular structure, transportation, transcriptional and translational regulation. A natural extension of proteomics is to understand the interplay with other genomic layers such as DNA variation and gene expression levels. The studies of protein quantitative trait loci are summarized in the later section along with other omic studies of quantitative trait loci.
Transcription and translation are two key biological processes underlying gene expression and disease etiology. To understand the relationship between the corresponding levels of mRNAs and proteins expressed from the genome, the proteome and transcriptome of the same samples need to be measured and analyzed in a single study. High-throughput transcriptomic and proteomic technologies identify and quantify RNA transcripts and proteins to achieve more comprehensive understanding of gene expression in biological systems.
Schwanhäusser et al. conducted a joint proteome-transcriptome study of mouse fibroblasts (Schwanhausser et al., 2011). Overall, the corresponding mRNA and protein levels from the same genes were moderately correlated with global R2 of 0.41. However, the corresponding half-lives of the mRNAs and proteins showed virtually no correlation. Another joint proteome-transcriptome study of mouse liver samples observed lower level of protein-mRNA correlation (Ghazalpour et al., 2011). The levels of transcripts and proteins correlated significantly for only about half of the genes tested. Employing a genome-wide association approach to map loci affecting mRNA and protein levels, little overlap was found between the protein- and transcript-associated loci. In association analyses of numerous clinically relevant metabolic traits, they found that the majority of associations with metabolic traits were specific to either the protein levels or transcript levels, and only a small number of clinical traits were correlated with both protein and mRNA products of the same gene. Surprisingly, these metabolic traits correlated better to RNA levels than to protein levels, which could be caused by less robust quantification method of the protein abundance. Using genetic data of the same mouse strains, more genetic loci were associated with the mRNA levels than of its corresponding protein levels (Ghazalpour et al., 2011).
Wang et al. recently reviewed studies integrating RNA-Seq with LC-MS/MS-based shotgun proteomics data to enhance protein identification (X. Wang, Liu, & Zhang, 2014). These studies showed how to effectively leverage the complementary information from RNA-Seq and proteomic data in understanding gene expression. Meanwhile, proteomic data provide confirmation of gene product and functional relevance of novel transcriptomic findings. Current studies highlighted that a comprehensive understanding of the control of proteome will require precise quantitative information at all levels, including DNA variants, mRNAs and proteins at a genome-wide scale.
2.4 Metabolome-wide association studies
Metabolome and metabolomics
The metabolome is the global collection of all low-molecular-weight metabolites that are produced by cells during metabolism, and provides a direct functional readout of cellular activity and physiological status. It reflects the combined exogenous effects of lifestyle and environmental factors, as well as the endogenous effects of genetic, developmental and pathological factors. Metabolomics is an emerging discipline that aims to profile all low-molecular-weight metabolites present in biological samples. It provides a tool for interrogating how mechanistic biochemistry links to cellular phenotypes. Compared to human genome, epigenome, transcriptome and proteome, metabolome is not directly involved in the information flow of the central dogma. However, metabolomics measures both upstream and downstream changes that are close to environmental exposures and phenotypic changes.
Metabolomics technologies
The field of metabolomics has made remarkable advance in the past decade. It is now possible to perform metabolome-wide association studies as a powerful way to address complex biological questions (Jones, Park, & Ziegler, 2012; Kaddurah-Daouk, Kristal, & Weinshilboum, 2008; Patti, Yanes, & Siuzdak, 2012). Current metabolomic technologies with computational methods for chemical identities and abundance allow for simultaneous measurements of hundreds to thousands of metabolites from minimal amounts of biological samples. The metabolome-wide study has become possible with recent developments in instrumentation, bioinformatics tools and software.
Nuclear magnetic resonance (NMR) is a common metabolomics method, that measures the molecules’ responses to radiofrequency stimuli by chemically distinct atomic nuclei in a magnetic field to provide information about the structure and dynamics of molecules (Patti et al., 2012). NMR spectroscopy can provide detailed information on the molecular structure of compounds found in complex mixtures, and a wide range of small molecule metabolites in a sample can be detected simultaneously. It usually requires minimal sample separation and preparation. The two-dimensional (2D) NMR spectra can reliably detect and quantify individual metabolites for metabolomic profiling.
Mass spectrometry is chemical method that can determine the type and abundance of chemicals through the accurate measurements of their mass-to-charge ratios (m/z). Tandem mass spectrometry (MS/MS) is type of mass spectrometry in which ions are selectively isolated and then fragmented. Recent technology of MS profiling involves the use of liquid chromatography (LC) to separate analytes and high-resolution MS to accurately measure mass and abundance (Jones et al., 2012; Patti et al., 2012). In complex biological samples, high-resolution detection allows quantification based on accurate m/z. This minimizes the need for separation by traditional LC-MS and does not require a priori knowledge of MS/MS spectra (Jones et al., 2012). In metabolomic applications, typical MS data consist of lists of metabolomic features characterized by their mass-to-charge ratios from the MS spectra. The mass-to-charge ratio of each feature is measured and used for structural characterization.
Depending on the scope of metabolites to be determined in a single analysis, a metabolomics study can use a targeted or an untargeted approach. Each approach has distinct capacity in addressing research questions, and requires unique experimental design including sample preparation, instrumentation, and data analysis pipeline. Targeted metabolomics measures a predefined set of metabolites, typically focusing on one or more types of chemicals or related pathways of interest (Dudley, Yousef, Wang, & Griffiths, 2010). Targeted metabolomic approaches are driven by a specific hypothesis about a particular biochemical pathway (e.g. lipid profile, carbon metabolism, amino acids and nucleotides). Although other analytical methods are available for targeted study, MS and NMR methods have been widely adopted in targeted metabolomics research, because they offer superior analytical specificity, reproducibility and accurate quantification (Astarita, Ahmed, & Piomelli, 2009; Dudley et al., 2010).
Untargeted metabolomic methods aim to simultaneously measure as many low-molecular-weight metabolites as possible without bias. Between NMR and MS technologies, LC-MS detects the most metabolites and has been the choice of global untargeted metabolomic profiling. Thus, we focus on LC-MS-based technology of metabolomics in this article. Using LC-MS-based high-resolution metabolomic (HRM) methods, thousands of m/z features (i.e. peaks of MS spectrum) can be consistently detected and quantified from biological samples (Hoffman et al., 2014; Wikoff et al., 2009). Each metabolomic feature represents a detected ion with a unique combination of m/z ratio and retention time. Due to the complexity in samples and instruments as well as variations in experimental conditions, the large output files from the high-resolution metabolomic method require automated computational algorithms to adjust these non-biological variations for downstream analyses. Untargeted HRM data are challenging to annotate because each metabolite may have multiple m/z peaks and multiple chemicals may have identical m/z value. This potential many-to-many relationship between metabolomic features and known chemicals requires comprehensive approach in annotation. To enhance the accuracy of metabolite annotation, the metabolomic features and metabolic modules need to be matched to reference databases such as the human metabolome database (HMDB) (Wishart et al., 2009), KEGG (Kanehisa & Goto, 2000), the Madison Metabolomics Consortium Database (MMCD) (Cui et al., 2008), Metlin (C. A. Smith et al., 2005), and chemical databases (Baker, 2011). The enriched metabolic modules obtained by these methods can then be mapped to metabolic pathways using online metabolomics tools such Metscape and MetaCore. There are more advanced methods that directly select sub-regions from the metabolome-scale network without limiting analyses to curated pathways, including network-based penalized regression (W. Pan, Xie, & Shen, 2010) and the Markov Random Fields model (Wei & Li, 2007). New pathway-based methods can incorporate the uncertainty of the metabolite annotation into the statistical analysis to identify the pathway modules associated with a complex disease (S. Li et al., 2013). Using the untargeted approach, recent studies are able to assess the global metabolomic profile involving over 20,000 metabolomic features from many metabolic pathways in human samples (Zhao et al., 2015). The untargeted HRM has demonstrated its unique contribution to understanding fundamental biological processes of human diseases, and identification of uncharacterized chemicals linked to human health and disease (Baker, 2011).
Metabolome-wide association studies (MWAS) of human diseases
The accomplishments in technology/instrumentation, data processing/analysis and feature annotation have already revealed that numerous metabolites correlate with complex human traits and diseases. Efforts have also been made to catalogue the thousands of metabolites present in the human samples to enable systematic discovery, curation and interpretation of metabolomic findings from human disease research.
Targeted metabolomics approaches have played an important role in understanding human diseases. A recent targeted study of 295 metabolites revealed serum effects of antihypertensives and lipid-lowering drugs in an European population (Altmaier et al., 2014). Significant associations with beta-blockers, angiotensin-converting enzyme (ACE) inhibitors, diuretics, statins, and fenofibrates were identified in 1,762 participants. The metabolic changes supported known pathways directly targeted by these drugs, and identified novel metabolites included by the drugs. For instance, the intake of statins resulted in changes of serum metabolites of both the biosynthesis and the degradation of cholesterol. These results provide a basis for a deeper functional understanding of the action and side effects of commonly-used drugs. Another targeted study of 68 metabolites aimed to identify the biomarkers for incident cardiovascular disease (CVD) during more than a decade follow-up in a Finnish population (Wurtz et al., 2015). Replication of metabolite associations with CVD were examined in two population-based studies from the United Kingdom. Four metabolites were associated with incident cardiovascular events after adjusting for established CVD risk factors in the meta-analysis. Higher serum levels of phenylalanine and monounsaturated fatty acid were associated with increased cardiovascular risk, while higher serum levels of omega-6 fatty acids and docosahexaenoic acid decreased cardiovascular risk. This study supported the value of high-throughput metabolomics in biomarker discovery and risk assessment, which may lead to improved disease diagnosis and prevention. Targeted metabolomic analyses also revealed citric acid metabolites and essential amino acids as metabolic signatures of myocardial ischemia (Sabatine et al., 2005) and diabetes (T. J. Wang et al., 2011), respectively.
Recent advances make untargeted studies (i.e. metabolome-wide association studies) now possible, as a powerful way to investigate complex biological questions (Jones et al., 2012; Kaddurah-Daouk et al., 2008; Patti et al., 2012). Using an untargeted HRM approach, Zhao et al. identified five known and two unknown metabolites significantly predict the incidence of type 2 diabetes (T2D) among 2,117 normoglycemic American Indians followed for an average of 5.5 years (Zhao et al., 2015). A multi-metabolite score significantly improved risk prediction beyond established diabetes risk factors. The findings demonstrated the utility of metabolomics in the discovery of novel prognostic markers of T2D in population studies.
Metabolomics has also been applied in studies of infectious disease. An exploratory study of over 400 metabolites identified 6 metabolites that differentiated latent tuberculosis (TB) infection from healthy uninfected patients (Weiner et al.). A recent metabolomic study of over 23,000 metabolites identified pathophysiologic pathways distinguishing 17 active TB patients from 17 asymptomatic household contacts (Frediani et al., 2014). Analysis revealed a metabolite profile including specific resolvins, glutamate, and trehalose-6-mycolate, as well as other Mycobacterium tuberculosis cell wall metabolites, that could distinguish those with active TB disease.
2.5 Common issues and limitations of omics approaches
Experimental and technical variation
The systematic differences in high-throughput omics data between laboratories, operators and batches of products have been well documented. Although standardized experimental protocol may reduce the so-called “batch effects”, they can hardly be eliminated in studies with large sample sizes involving multiple collaborative sites. Therefore, the “batch effect” can be an important confounder in association studies, and potentially causes spurious associations unrelated to the outcome of interests. Additionally, multiple technical platforms are usually available for the same type of omics profiling. For example, multiple versions of microarray and sequencing platforms have been available from various manufactures for transcriptomic and epigenomic association studies. They usually have different coverage of the genomic regions and features. The evolution of mass spectrometry has also introduced several generations of proteomic and metabolomic platforms detecting various ranges of chemicals in terms of identity and abundance. Such technical heterogeneity often makes the replication, validation and joint analysis of different omics studies very challenging. Using standard control samples to harmonize these measurement variations, and applying appropriate statistical models (e.g. mixed effect model) to adjust for batch effect can address some issues of technical variation. More importantly, recognizing and considering these issues in the early design stage is the most effective way to minimize the negative impacts.
Biological Variation
Another source of omics data variation is from the biological samples and specimens being measured. Except for genetic profile being identical across tissues and cell types, all other omic profiles (e.g. transcriptome, epigenome, proteome and metabolome) vary across tissues and cell types. The tissue and cell type specificity leads to two important issues in multi-omics studies, selection of tissues and cell types, and heterogeneity of tissues.
The most accessible specimen in human samples is peripheral blood. The blood based specimens such as plasma, serum and leukocytes are often used in omic association studies of human diseases due to the limited access to other disease-relevant tissues (e.g. brain for neurological disorders). Although the use of blood as surrogate tissue is sometimes relevant (e.g. autoimmune diseases and inflammatory processes), the biological relevance of blood-based omic profiles may not be apparent for many human diseases. There are reports showing that some blood-based omic makers share similar association as in other tissues (e.g. smoking-related DNA methylation (Sun, 2014)), but there is no clear evidence linking global omic mechanisms to disease development and environmental exposures across tissues. On the contrary, consortia studies have demonstrated distinct patterns of omic profiles across tissues and cell types (Bernstein et al., 2012; Roadmap Epigenomics et al., 2015). Using blood-based specimens is a convenient start of searching novel disease-related biomarkers, however, using blood as a surrogate tissue requires cautious validation and interpretation when the study aims to unravel disease mechanism.
A tissue sample always involves several cell types, each having a unique omic profile. Depending on the location of a tissue sample (e.g. micro-dissections of brain), or an individual’s physiological condition (e.g. peripheral leukocytes after acute infection), the proportions of multiple cell types of a tissue sample can change substantially, causing the heterogeneity of cell population. Such heterogeneity can shift the summary level of omic markers which are cell-type specific, and leads to associations unrelated to the direct omic changes (e.g. modifications of RNA and peptide expression levels). Statistical methods have been developed to adjust for potential confounding effects due to cell type heterogeneity. The contributing cell type(s) of the associated markers among the mixture remain unidentified (Abbas, Wolslegel, Seshasayee, Modrusan, & Clark, 2009; Houseman et al., 2012; Houseman, Molitor, & Marsit, 2014). Measuring the omic profile in each purified cell types is an ideal solution but often unrealistic due to folds of increase of measurement cost associated with all cell types. An alternative approach is to conduct initial association study using a large sample of mixed cell types adjusted for the effect of heterogeneity. Once an averaged effect is identified, a follow up study of the sorted cells can focus on a specific omic maker to identify the most relevant cell types. The analysis in the second stage does not require expensive omic profiling or large amount of samples, but provides direct inference on the molecular mechanism of a disease.
3. Genetic and environmental determinants of epigenomic, transcriptomic, metabolomic and proteomic markers
3.1 Genetic determinants of omic markers - studies of quantitative trait loci (QTL)
Given the mature analysis pipeline and the large amount of genome-wide SNP data available in population studies, one natural extension of the single layer omics study is to unravel the genetic loci affecting other omic markers (e.g. transcriptomic, epigenomic, proteomic and metabolomic markers) through a genome-wide QTL analysis. Disease-associated genetic variants do not directly cause disease at the molecular level. They affect intermediate phenotypes that in turn induce molecular and physiological changes. Thus, identifying the intermediate phenotypes of gene expression, DNA methylation, protein and metabolite levels that directly influence by genetic variants has the potential to provide the functional information of disease-causing genes and pathways, and to unravel the genetic-controlled molecular systems underlying the changes of health and disease status. Therefore, the QTLs of each omic layer hold the key functional information linking the observed genetic association and the disease phenotypes. In the context of multi-omic study, QTL analysis targets a set of similarly measured quantitative traits to screen for their associated genetic loci. The exhaustive pair-wise omic search demands computational resources to run thousands to millions of GWAS depending on each omic technology.
Studies of gene expression QTL (eQTLs)
Ten years ago, Schadt et al. demonstrated that integration of genetic variation and gene expression data with phenotypic data may identify key genes of complex traits in segregating mouse populations. Mapping eQTLs, particularly cis-eQTLs (i.e. eQTLs colocated with the gene encoding the RNA transcript), facilitated the understanding of functional linkage between disease-associated loci and the disease phenotype via gene expression regulation (Schadt et al., 2005). Since then, numerous human studies have generated eQTLs maps in multiple tissues such as brain, liver, skin, immune cells, and lymphoblastoid cell lines (Ding et al., 2010; Fairfax et al., 2012; Myers et al., 2007; Pickrell et al., 2010; Schadt et al., 2008; Veyrieras et al., 2008; W. Zhang et al., 2008). Studies have revealed that over 30% of gene transcripts are substantially influenced by eQTLs (Romanoski et al., 2010). The overlap between eQTL and disease-associated loci may indicate the putative functional role. Therefore, the eQTLs databases have been routinely queried to infer potential functional roles of disease-associated loci from GWAS. Most GWAS findings map to non-coding regions, but a large proportion map to ENCODE regulatory elements (Schaub, Boyle, Kundaje, Batzoglou, & Snyder, 2012), suggesting the important role of transcriptional regulation underlying human diseases.
The most recent and significant development of human eQTL research is from the Genotype-Tissue Expression (GTEx) Consortium (G. T. Consortium, 2013, 2015). GTEx collects and analyzes the genome-wide genetic variation and tissue-specific gene expression data to understand the genetic basis for gene expression variation across multiple human tissues (43 up to date). Using RNA-seq method, GTEx interrogates all RNA molecules, including messenger RNA, ribosomal RNA, transfer RNA, and other long noncoding RNA transcripts expression in all collected tissues. Through the online portal, researchers can view and download tissue-specific eQTL results. GTEx also provide a controlled access system for de-identified individual-level genotype, expression, and clinical data to support the broader research of human diseases.
Studies of DNA methylation QTLs (meQTLs)
Genetics is one of the primary determinants of epigenetic variation including DNA methylation (Bjornsson, Fallin, & Feinberg, 2004). Key genes such as DNA methyltransferases (DNMTs) directly control the DNA methylation profile. Other genetic variants may also influence the patterns of DNA methylation by modifying the accessibility or binding affinity of enzymes. Over a dozen of meQTL studies of numerous tissues (e.g. peripheral blood, lung, brain, adipose tissue and tumor tissues) have been reported by correlating the genome-wide SNP data with tissue-specific DNA methylome data (Bell et al., 2011; Drong et al., 2013; Gibbs et al., 2010; Grundberg et al., 2013; Gutierrez-Arcelus et al., 2013; Heyn et al., 2013; Heyn et al., 2014; Shi et al., 2014; A. K. Smith et al., 2014; Sun, 2014; Teh et al., 2014; D. Zhang et al., 2010; X. Zhang et al., 2014; Zhi et al., 2013). Many meQTL studies prioritized on the local genetic associations with DNA methylation sites (i.e. cis-meQTLs), which typically had larger effects and required less number of pair-wise tests than the trans-meQTL analysis (Sun, 2014). Because of the large number of SNP-DNA methylation site pairs to test, the trans-meQTL analysis is more computational intensive and requires a more stringent multiple-testing correction than the cis-meQTL analysis. The details of these methylome-wide meQTL studies have been recently reviewed by Sun (Sun, 2014). Because of tissue and cell type specificity of DNA methylation, the genome-wide meQTLs can distribute differently from tissue to tissue (Grundberg et al., 2013; A. K. Smith et al., 2014), as well as from cell type to cell type (Gutierrez-Arcelus et al., 2013). The functional impact of these tissue-specific meQTLs may be important to understand the pathophysiology of diseases in target tissues. The national institute of health Roadmap Epigenomics Consortium generated 111 human reference epigenomes for primary cells and tissues, the largest collection so far (Roadmap Epigenomics et al., 2015). These reference epigenomes, combined with other genomics data, have provided functional and causal insights about several human disease (De Jager et al., 2014; Farh et al., 2015; Yao, Tak, Berman, & Farnham, 2014). The maps of meQTL and other epigenetics QTLs of many targeted tissues and cell types from sizeable samples will continue to assist in the functional understanding of disease processes.
Studies of metabolite QTLs (mQTLs)
Metabolites play critical roles in biological pathways, and are partially controlled by genetic regulations. Several systems genetics studies of metabolites in human plasma or serum have been reported (Gieger et al., 2008; Kettunen et al., 2012; Shin et al., 2014; Suhre et al., 2011; Yu et al., 2013). The levels of a set of metabolites are strongly associated with genetic loci, and some of these loci overlapped with GWAS loci for disease traits. A non-targeted genome-metabolome-wide study analyzed more than 250 metabolites from 60 known pathways in human serum samples. Combined with genome-wide SNP data from the same 2,820 individuals with metabolomic data, 37 mQTLs were significant at a stringent genome-wide threshold. Comparing to most GWAS findings, these associations showed much larger effect sizes than those for disease traits (Suhre et al., 2011) Another population study of 8,330 European individuals analyzed 216 serum metabolites using NMR (Kettunen et al., 2012). The GWAS of these metabolites identified 31 mQTLs including 11 novel loci without known association with any traits or diseases. The most recent large scale mQTL study investigated 529 metabolites of plasma or serum samples from 7,824 adult Europeans using MS technology. A total of 299 SNP-metabolite associations (i.e. mQTLs) at 145 independent loci were genome-wide significant after correction for the number of SNP-metabolite association tests (Shin et al., 2014). The web-based database provides comprehensive genetic information about circulating metabolites in human body.
Studies of Protein QTLs (pQTLs)
aim to assess the correlation between genetic variation and protein abundance. Thus, they require precise measurements of both genotypes and protein abundance in high-throughput mode. The advance in quantitative proteomics allows a genome-wide map of pQTL model organisms (Foss et al., 2007; Picotti et al., 2013). Early pQTL studies suffered from an inconsistent detection of proteins across samples and a limited dynamic range for low abundance proteins (Foss et al., 2007). A GWAS of 42 proteins identified eight strong pQTLs with rather large effect sizes (0.19 to 0.69 standard deviations per allele). However, the panel of proteins only covered a small fraction of the proteome (Melzer et al., 2008). Using precise MS measurement of peptides over a large number of samples, recent pQTL analyses revealed complex genetic influence on the levels of proteins in yeast (Picotti et al., 2013) and mouse (Holdt et al., 2013). Picotti et al. used a nearly complete map of yeast proteome and genetic variation data to identify strong genetic effects of protein abundance, and to demonstrate epistatic interactions affecting protein levels (Picotti et al., 2013). The pQTL analysis of mouse plasma identified strong genetic determinants for approximately 40% of tested proteins, and suggested causal genetic variants affecting abundance of plasma proteins (Holdt et al., 2013).
Utilities of omic QTLs
Overall, these studies catalogued a large number of QTLs across multiple omic layers in multiple tissues and cell types, and provide a rich resource not only to understand the genetic regulation of intermediate phenotypes, but also to illustrate important molecular networks mediating the interaction between genetic variants and environment for human diseases. Recent studies have demonstrated the utilities of the QTL data including eQTLs and meQTLs. First, these QTLs have been used to infer the functional link between genetic variants and disease traits in recent GWAS, and lead to follow-up studies uncovering the biological functions of disease-associated loci. Significant GWAS loci are enriched for e QTLs (Nicolae et al., 2010) and meQTLs (X. Zhang et al., 2014). In numerous examples, the disease-associated loci were hinted to function via the regulation of gene expression or the modification of epigenetic markers (Liu, Aryee, et al.; Shi et al., 2014). Secondly, these QTLs can be used as the instrumental variables in Mendelian Randomization (MR) study of the omic markers for their potential causal roles (Relton & Davey Smith, 2010, 2012). MR was initially proposed as an epidemiologic method to obtain unbiased estimates of the putative casual effects without conducting a randomized trial (Gray & Wheatley, 1991; Katan, 1986). The MR approach uses the genetic variant mimicking the biological effects of a modifiable exposure. If the exposure truly alters the disease risk, the genetic variant should also be associated with the disease through the causal pathway. Because the genetic variant is randomly assigned to the offspring during meiosis in a population, the genotype distribution should not be biased by confounding. Only the genetic variant in the causal pathway should be associated with disease outcome by carrying the association through the causal exposure. MR approach can be applied to study the causal omic risk factor for human diseases. The study design and analytical issues have been recently discussed for epigenomic study (Relton & Davey Smith, 2010, 2012). but the same principles can be applied to other omic layers.
3.2 Environmental influences on multi-omic markers
Both genetic and environmental factors contribute to the development of human diseases. The genetic causes have been demonstrated by decades of research, and have been further endorsed by recent findings of thousands of genetic associations with disease traits. However, the static genome has its limitation to capture the time-varying changes caused by environmental factors or physiological conditions. The non-genetic factors can cause important changes in proteins, nucleic acids, lipids and other biomolecules, which have direct roles in biochemical and cellular functions. Recent advances of technology enabled robust and cost-effective measurement of genomic variants and identified thousands of genetic factors associated with human diseases. On the other hand, a comparable high-throughput approach to studying the environmental causes of human diseases remains unavailable, which leaves a large proportion of the phenotypic variance unexplained. Exposome refers to the totality of human environmental exposures encompassing both exogenous and endogenous exposures (Rappaport, Barupal, Wishart, Vineis, & Scalbert, 2014), and measures the accumulation of environmental influences and associated biological responses throughout the lifespan, including exposures from the environment, diet, behavior, and physiological processes (Miller & Jones, 2014). Although we are not able to fully measure or model the exposome, improved omics technologies including metabolomics and epigenomics provide promising methods to partially investigate the human exposome (Miller & Jones, 2014; Rappaport et al., 2014). Metabolomics, epigenomics and other omics approaches can complement genomics research by identifying time-varying omic markers in pathways and networks associated with a particular environmental exposure and a disease state. The multi-omic studies have the potential to transform not only the research of genetic variants, but also the research of the environmental exposure and the biological responses to the environment underlying disease development.
4. Analytical approaches and methods for multi-omic association studies
Existing studies in human and model organisms highlighted the complexity of genomic information flow and the interactive networks in biological processes and disease development. The multi-omics approach thus holds the promises to further advance human disease research. However, such enthusiasm can only be translated into scientific discoveries with sound study designs and solid analytical strategies.
The ideal datasets for such an integrative analysis are multi-omics data all collected on the same set of samples. However, this is often not possible because of the cost or because the control samples simply do not have the appropriate tissues to study. Another type of datasets is multi-omics data collected on different sets of individuals from different studies. Different research questions can be answered for each type of multi-omic dataset using corresponding statistical approaches.
4.1 Regression-based joint modeling
The regression-based approach jointly models multi-omics data, using the framework of mediation analysis. These data are typically collected on the same subjects. Throughout this section, we let Y be the dichotomous disease outcome, G be a SNP or a set of SNPs depending on the specific method, E be the mRNA expression of a gene or a set of genes, and X be all non-genomic covariates (such as clinical or environmental measurements) with the first covariate being 1. In the following, we review four methods in this category.
Huang, Vanderweele, and Lin (2014) developed a method that integrates SNP and gene expression data, treating gene expression as the mediator in the causal mechanism from SNPs to the disease outcome (Figure 2). They used a logistic regression model
| (1) |
to characterize the dependence of the disease outcome on a set of SNPs G, the expression E of a gene, and other covariates X. A SNP-expression pair can be defined in two ways. First, one can choose the SNPs mapped to a gene and the expression of the gene. Second, one can choose the eQTL SNPs and the corresponding gene expression based on an eQTL study. The dependence of the gene expression on the set of SNPs and other covariates is formulated through a linear regression model
| (2) |
The goal is to test the hypothesis
| (3) |
This null hypothesis can be interpreted within the framework of causal mediation analysis based on the causal diagram in Figure 2. Define the total effect (TE) of the set of SNPs on the disease outcome as
in which both probabilities are marginalized over E. The TE of SNPs can be decomposed into the direct effect (DE) and the indirect effect (IE). The DE is the effect of the SNPs on the disease outcome that is not through gene expression, whereas the IE is the effect of the SNPs that is mediated through the gene expression. When the SNPs are associated with the gene expression (i.e., eQTL SNPs; αG ≠ 0), the null hypothesis (3) is equivalent to the null hypothesis of DE = 0 and IE = 0 (i.e., no TE of the SNPs). When the SNPs have no effect on the gene expression (i.e., not eQTL SNPs; αG = 0), then there is no IE of the SNPs on Y, so that the null hypothesis (3) is not equivalent to testing for no TE, but simply whether there exists a joint effect of the SNPs, the gene expression, and possibly their interactive effect on the disease risk. This causal interpretation is helpful for understanding genetic etiology of diseases as well as for applications in pharmaceutical research (Y. Li, Tesson, Churchill, & Jansen, 2010).
Figure 2. Causal diagram of SNP (G), gene expression (E), and disease outcome (Y).
Gene expression is a potential mediator of genetic effects on the disease outcome.
As the number of SNPs in G may be large and some SNPs may be highly correlated with each other due to linkage disequilibrium (LD), the standard likelihood ratio test (LRT) or multivariate Wald test for the null hypothesis (3) would use a large number of degrees of freedom and would thus have limited power. To overcome this problem, Huang et al. (2014) proposed a variance component test. They assumed that the components in the vector βG are independent and follow an arbitrary distribution with mean 0 and variance τG, and that the components in βGE are independent and follow an arbitrary distribution with mean 0 and variance GE. The disease model (1) hence becomes a logistic mixed-effect model, and the test of hypothesis (3) becomes a joint test of variance components and a scalar regression coefficient:
Therefore, the proposed variance component test is insensitive to the number of SNPs in G. As the true disease model is unknown and can be different from (1), e.g., without the interaction term, Huang et al. (2014) further proposed an omnibus test that accommodates different possible disease models.
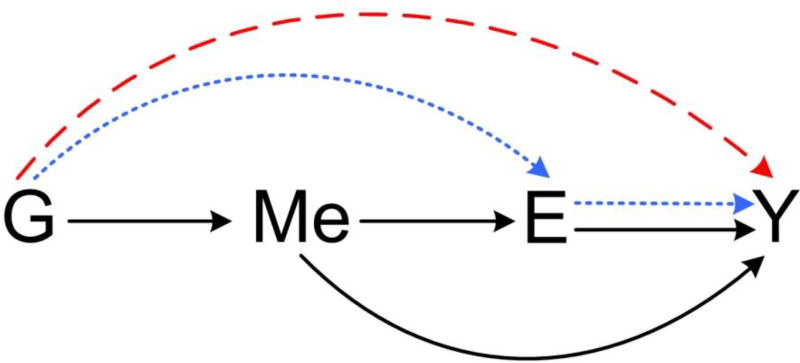
Later, Huang (2015) extended the work of Huang et al. (2014) to jointly analyze SNP, DNA methylation, and gene expression data with respective to a disease outcome, adding the layer of DNA methylation data to the existing framework. In addition, the earlier work only focused on testing the overall effect of a set of SNPs and the expression of a gene, without distinguishing the mechanisms of DE of the SNPs on the disease and IE of the SNPs mediated by the expression. In the later work, Huang (2015) studied path-specific effects, as depicted in the causal diagram (Figure 3), by jointly modeling a set of SNPs within a gene, the DNA methylation and expression of the gene, and the disease outcome as a biological process. Let M denote the DNA methylation measurement at a CpG site. Then the logistic model in (1) is expanded as
| (4) |
The dependence of the DNA methylation on the set of SNPs and other covariates and the dependence of the gene expression on the SNPs, DNA methylation, and other covariates are specified in the linear regression models
| (5) |
and
| (6) |
respectively, where , and FM and FE|M are any arbitrary distributions.
Figure 3. Causal diagram of SNP (G), DNA methylation (Me), gene expression (E), and disease outcome (Y).
Three path-specific effects are 1) Direct effect of SNPs on outcome (dashed red line), 2). Indirect effect of SNP mediated through gene expression but not through methylation (dotted blue lines), and 3). Indirect effect of SNP mediated through methylation (solid black lines).
An arbitrary set of regression coefficients in model (4) can be tested. For example,
| (7) |
can be assessed by a variance component test as proposed in Huang et al. (2014). To provide a mechanistic interpretation of the hypothesis (7), Huang (2015) first decomposed the overall genetic effect into three path-specific effects: 1) the DE of the SNPs on the outcome, not through the DNA methylation or the expression (denoted by ΔG→Y), 2) the IE of the SNPs on the outcome that is mediated through the gene expression but not through the DNA methylation (ΔG→E→Y), and 3) another IE of the SNPs on the outcome that is mediated through the DNA methylation (ΔG→MY). Within the causal mediation framework, the correspondence of a path-specific effect and a set of regression coefficients in the disease model (4) can be established. For example, the DE ΔG→Y corresponds to βG, βGM, βGE, and βGME, which is not influenced by the relationship among G, M, and E. By contrast, the IE ΔG→E→Y of SNPs mediated through expression is affected by the G-M-E relationship. Evidently, if there does not exist an effect of G on E, ΔG→E→Y is zero. If there exists an effect of G on E, ΔG→E→Y corresponds to βE, βME, βGE, and βGME; it means that the test of the hypothesis (7) is equivalent to the test of the IE of SNPs mediated through gene expression. To determine the relationship among G, M, and E, one can rely on prior knowledge of existing biological evidence, or statistical analyses that estimate the relationship, or model selection criteria such as Akaike information criterion (AIC) (Akaike, 1974) and Bayesian information criterion (BIC) (Schwarz, 1978).
To apply this method to the genome-wide data, it is unclear how to select the DNA methylation measurement for a gene. It is possible to consider each of the CpG sites that map to the gene including the upstream and downstream of the gene, but this strategy will result in too many tests. The data application of Huang (2015) does not illustrate this point. Instead, the application concerns 12 methylation loci, a micro-RNA expression, and a gene expression, substituting a set of methylation loci for the set of SNPs in the methodology and substituting a micro-RNA expression for the DNA methylation.
While Huang et al. (2014) and Huang (2015) jointly analyze multi-omics data from the same subjects, Huang (2014) extended the methodologies to analyze the data from different subjects. This is motivated by the fact that the GWAS and QTL studies are likely to be conducted in different subjects due to the availability of tissue samples and the tissue specificity of expression and DNA methylation. Specifically, in GWAS, SNPs and the disease outcomes are collected, but not gene expression/methylation; in QTL studies, SNPs, gene expression and methylation are collected, but not the disease outcome. Define μM = E(M | X, G), μE = E(E | X, G) and μME = E(ME | X, G). From expression (5), we have μM = GTδG + XTδX. The μE and μME can be obtained by marginalizing (6) over M. With different omics data on different subjects, the only testable effect is the overall SNP effect on the disease outcome, not any of the path-specific effects. In the statistic of the corresponding variance component test developed in Huang (2015), the M, E and ME terms should be replaced by the estimated μM, μE and μME, respectively. Thus, the testing procedure in Huang (2015) can be applied in settings where methylation and/or expression data are not collected in the subjects of GWAS but their associations with SNPs (i.e., μM, μE and μME) can be consistently estimated from external meQTL and eQTL studies. Note that the meQTL and eQTL studies should be conducted on the same subjects in order to calculate μME.
Zhao et al. (2014) considered the same omic datasets that Huang et al. (2014) have dealt with, i.e., SNP, gene expression, and disease data collected on the same set of subjects. However, Zhao et al. (2014) focused on testing the IE of the SNPs on the disease outcome that is regulated by gene expression. They proposed the following two-stage model for each SNP G,
| (8) |
| (9) |
where E may include the expression for a set of genes. Model (9) is significantly different from model (2) in that the former does not consider the regulation of the SNP on the expression of an individual gene, but on one particular linear combination of them; it hence requires estimating fewer parameters. Note that this is the same linear combination of gene expression in the disease model (8). Based on the two-stage model, one can test for SNP-disease association by testing H0: αG = 0, assuming that the SNP affects disease risk through affecting the gene expression levels. This work is analogous to the work by Huang et al. (2014) but focuses solely on increasing the power of SNP association testing, rather than on assigning causal interpretations to any of the parameters. When a particular set of genes or a pathway is of interest such that the number of genes in E does not exceed the number of subjects, Zhao et al. (2014) proposed to use the standard estimating equation theory for inference. To apply their method in an agnostic, genome-wide manner, they proposed to consider one gene in E at a time; to reduce the multiple testing burden imposed by the huge number of pairwise tests they proposed to restrict to testing only those SNPs located cis to each gene. This method works best when there is no DE of the SNPs on the disease outcome, such that the SNPs act only through regulating gene expression. In this case, the gene expression can help explain the variability of the SNP effect on disease and thus increases the power of detecting the overall effect of SNPs on disease. Indeed, Kenny and Judd (2014) noted that in the absence of a DE, testing the IE in a mediation analysis can be dramatically more powerful than the standard method testing SNP-disease associations directly. Even in the presence of a DE so that model (8) mis-specifies the true disease risk, Zhao et al. (2014) showed, both analytically and numerically, that their method is still more powerful than the standard method when the magnitude of DE is lower than the magnitude of IE.
4.2 Matching Patterns of eQTL and GWAS
He et al. (2013) developed a method to detect disease-associated genes (i.e., genes whose expression level influences the disease risk) by matching the eQTL patterns of each gene with the patterns of disease-associated SNPs. This method is especially useful when eQTL and GWAS studies were conducted on different samples. The rationale is that, for a disease-associated gene, any genetic variation that perturbs its expression is also likely to influence the disease risk (Figure 4). Thus, the eQTLs of the gene, which constitute a unique “genetic signature” of this gene, should overlap significantly with the set of loci associated with the disease. Because many eQTLs act in trans, this approach can identify important genes that are distal to any GWAS association signals and thus impossible to be detected with GWAS alone.
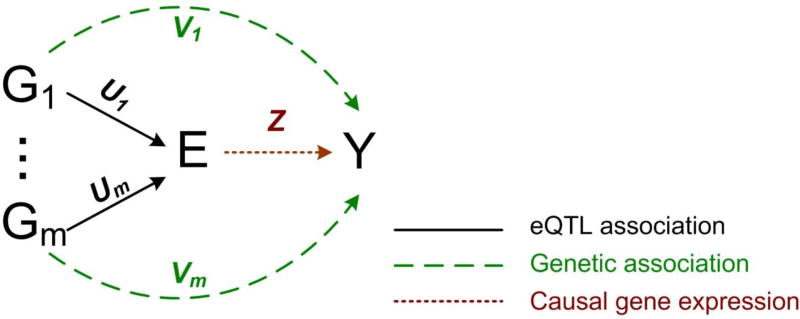
Figure 4. Matching the genetic signatures of gene expression traits (eQTLs) to that of the disease trait to identify gene expression-disease associations.
Ui: binary indicator variables to represent the true SNP-gene expression causal relationship;, Vi: binary indicator variables for the true SNP-disease relationship. Z is a binary variable indicating whether the gene expression trait influences the disease.
He et al. (2013) implemented the above idea of genetic signature matching by a Bayesian framework. Suppose that, given a gene, there are m putative eQTLs that pass some low, less stringent significance threshold in the eQTL study. Let Uj and Vj be binary indicators to represent whether the ith SNP is associated with the expression and the disease outcome, respectively. Let Z be a binary indicator that represents whether the expression of the gene is associated with the disease. If, for a significant number of SNPs, Uj = 1 and Vj = 1, then it is likely that Z = 1. The available data consist of the p-values of SNPs relative to the gene expression from an eQTL study, denoted by the vector peQTL, and the p-values of the SNPs relative to the disease outcome from a GWAS, denoted by the vector pGWAS. Although Uj and Vj are not observed, they are related to peQTL,j and pGWAS,j: when peQTL,j (pGWAS,j) is small, it is likely that Uj (Vj) = 1. Thus, the data peQTL and pGWAS can be used to test the hypothesis H0: Z = 0 that the gene is not associated with the disease. The inference of Z is based on the Bayes factor (BF):
which is the ratio of the probabilities of data under H1 and H0. When all SNPs are unlinked, the BF of the gene is the product of the BFs of all SNPs:
When there is LD among SNPs, He et al. (2013) proposed to use a block-level BF, which is the mean of the BFs of all SNPs in that block (Servin and Stephens, 2007). The probability P(peQTL,j, pGWAS,j | Z) is computed by summing over the hidden variables Uj and Vj:
The components on the right hand side are specified as follows. Uj is a Bernoulli variable with the success probability α, which is the prior probability of a SNP being associated with the gene expression. He et al. (2013) chose α = 1.0 × 10−3 for cis-eQTLs (within 1Mb of the gene) and α = 5.0 × 10−5 for trans-eQTLs. When Z = 0, the gene is irrelevant to the disease and thus Uj and Vj are independent. When Z = 1 and Uj =0, this SNP is not an eQTL and thus Uj and Vj are also independent. In both cases, Vj is a Bernoulli variable with the success probability β, which is the prior probability of a SNP being associated the disease. He et al. (2013) chosen β = 1.0 × 10−3. When Z = 1 and Uj = 1, Vj should always be 1, as a true eQTL of the gene is expected to be associated with the disease. The probabilities P(peQTL,j | Uj) and P(pGWAS,j | Vj) reflect the distributions of p-values under the null or alternative hypothesis. Let TeQTL,j and TGWAS,j be the test statistics corresponding to peQTL,j and pGWAS,j, respectively. Under the null, P(TeQTL,j | Uj = 0) and P(TGWAS,j | Vj = 0) follow the standard normal distribution. Under the alternative, P(TeQTL,j | Uj = 1) and P(TGWAS,j | Vj = 1) depend on the tests through which the test statistics are derived and the effect size of the SNP. Finally, the BF of the jth SNP, Bj, can be expressed as
where
are BFs measuring the association of the jth SNP with the expression and the disease, respectively. Thus the BF of the gene being tested depends only on α, β, and SNP-level BFs. (If Bayesian inference has been performed in both the eQTL and GWAS analysis, it is straightforward to combine the resulting BFs to obtain the BF for the gene.) To assess the statistical significance of BF, a simulation approach was proposed to compute the p-value of the BF for a gene.
Because this method does not directly test the relationships between genotypes, gene expression, and disease outcomes, but only requires p-values, the eQTL and GWAS data do not have to come from the same subjects. This method is also generalizable to molecular traits other than gene expression, such as metabolites, non-coding RNAs, and epigenetic modifications. It has been implemented in a software program called Sherlock. The name implies that the method works as a detective, comparing the fingerprint from a crime scene (the GWAS signature) against a database of fingerprints (the eQTL signature of all genes) to determine the real culprit (disease-associated genes).
4.3 Aggregating evidence of multi-omics data over gene set/pathway
Xiong et al. (2012) developed a statistical framework, called Gene Set Association Analysis (GSAA), that aggregates genetic and gene expression evidence in terms of “association scores” at the level of gene sets or pathways for genome-wide association analysis of gene sets or pathways. The gene expression data and the SNP genotype data are allowed to be collected on the same samples or different samples. The dashed box in Figure 5 illustrates the three-step aggregation procedure of GSAA without consideration of DNA methylation sites, proteins and metabolites.
Figure 5. Aggregation model of multi-omics evidence.
An omnibus test of pathways enriched for trait-associated SNPs, gene expressions, CpG sites, proteins and metabolomic features. This multi-layer approach allows aggregation of single association signals from individual markers to genes to pathways. The original aggregation model limited to SNPs and gene expression levels within the dashed box.
First, the SNP set association score and the gene expression association score are calculated respectively. The gene expression association score that reflects the degree to which a gene is differential expressed between cases and controls is calculated as the difference of the group means scaled by the standard deviation. The SNP set association score is the maximum of the single-SNP score over all the SNPs mapped to the gene region, where the single-SNP score is calculated as the genotype- or allele-based χ2 statistic and the gene region is a pre-defined genomic interval encompassing the gene and the upstream of and downstream from the transcribed region.
Second, the SNP set association score and the gene expression association score are combined to generate a gene association score. This step integrates evidence for association across the gene expression and SNP data. Before the integration of expression and SNP data, the absolute values of the gene expression scores are taken in order to capture both up-regulation and down-regulation in pathways and to be consistent with the magnitude of the SNP set association scores. Both gene expression score and SNP set score are also standardized by the mean and standard deviation of its respective null distributions, which are generated by a phenotype-based permutation procedure, so that the scores from different statistical tests or on different scales are brought on a common scale and thus directly comparable with each other. The gene association score is the sum of the two standardized scores.
Third, the gene set is evaluated by a weighted Kolmogorov-Smirnov (K-S) statistic (i.e., gene set association score) to determine whether the genes belonging to this gene set are preferentially near the top of the ranked ordered list based on gene association scores. Based on a phenotype-based permutation procedure that preserves LD structure in SNP data and gene-gene correlation structure in gene expression data, the false discovery rate (FDR) or the family-wise error rate (FWER) can be calculated and the significant gene sets are declared controlling for FDR or FWER below a certain threshold.
Although Xiong et al. (2012) only focused on integrating the gene expression and SNP genotype data, the flexibility of this framework allows integration of other omic data such as DNA methylation, proteomics, and metabolomics data (Figure 5). Analogous to the SNP set association score, we can first calculate the χ2 statistic at single CpG sites based on the beta values (measuring DNA methylation level) and then obtain a CpG-set association score for the gene using a maximum statistic. We can also calculate the χ2 statistic at each protein. These statistics are aggregated into the gene association score after proper standardization, along with those for SNP sets and gene expression. Finally, we perform a weighted K-S test for metabolites within each pathway to obtain a metabolite-set association score. The pathway association scores are the sums of the gene- and metabolite-set association scores.
5. Discussion
The thorough scan of a single type of biologically function features (e.g., GWAS and EWAS) continues to provide insights into the mechanism and etiology of human diseases. To fully elucidate the dynamic molecular system and understand the biological processes involved in disease development, we need to not only look through the right lens (i.e. omic layer) at the right time, but also to capture multiple dimensions of biological information flow together. Despite of its appealing scientific gain, the materialization of the multi-omics study has been hampered mainly due to the lack of feasible technologies for large-scale population studies, availability of biospecimens and the high cost attached to them. In recent years, the core technologies behind the high-throughput assays, such as sequencing and mass spectrometry, have become more and more sensitive, accurate, and affordable. Interrogation of complementary omics beyond a single omic layer has been explored for several diseases and model systems to demonstrate the utility and feasibility of multi-omics research. Multi-omics approach has emerged as a promising and power tool to comprehensive study human diseases at a system level across several types of functional layers over time.
Because of the known issues of experimental and biological variations, each layer of omic data needs to be cautiously processed, controlled for data quality, corrected for technical bias and properly adjusted in the analysis. To conduct a meaningful multi-omic study, the collection and production of a high-quality data set is an essential step and should never be underestimated. Although the technical and analytical details for preparing each type of omic data are beyond the scope of this article, we want to emphasize the data quality issues of multi-omic analysis. A close collaboration and continuous communication between scientists with different expertise in each related subject area (e.g., laboratory science, genomics, epidemiology, statistics and bioinformatics) is essential to fully address these issues.
Current multi-omics research of human diseases demands a large amount of resources to carry out a population study. The biomedical research community should invest not only in the technological innovation and data generation, but also in the design and development of analytical strategies to fully use of the big data from these new technologies. The consideration of multi-omic analysis should be integrated into the early phase of study design, rather than a post-hoc process after the data production. Given the rapid development in both laboratory assays and analytical capabilities, we anticipate that the multi-omics approach will grow rapidly to provide novel means to study human health and diseases. Such approach can be extremely effective to investigate understudied and rare diseases where the biological and pathophysiological mechanisms are largely unknown. With available clinical and epidemiologic samples, the multi-omics approach can facilitate a giant leap in understanding these disease conditions, and to offer effective treatment and prevention strategies in the era of precision medicine. In not too distant future, one or several of the omics technologies can be part of the standard precision medicine panel, in addition to the genomic data, which will accurately profile an individual’s genetic predisposition and environmental risk to guide diagnosis, and to optimize treatment and prevention of varies human diseases.
Table 1.
Summary of multi-omics data analysis approaches
| Approach | Reference | Omics- Data types |
On same samples? |
Effects that can be tested | Software |
|---|---|---|---|---|---|
| Regression-based joint modeling | Huang et al. 2014 (Ann App Stat) | G, E, Y | Yes | (for eQTL) TE of G on Y (for non-eQTL) joint effect of G, E, and G × E | |
| Huang 2015 (Stat Med) | G, M, E, Y | Yes | Three path-specific effects:
|
||
| Huang 2014 (Biostatistics) | G, M, E, Y | No | TE of G on Y | ||
| Zhao et al., 2014 (Biometrics) | G, E, Y | Yes | IE of G on Y | Integration | |
| Matching patterns of eQTL and GWAS | He et al. 2013 (Am J Hum Gen) | G, E, Y | No | Effect of E on Y | Sherlock |
| Aggregating evidence of multi-omics data | Xiong et al. 2012 (Genome Res) | G, E, Y | No | Gene set or pathway association | GSAA |
G: SNP genotype; E: Gene expression; M: Methylation; Y: disease outcome. TE: total effect of SNPs on disease; DE: direct effect of SNPs on disease; IE: indirect effect of SNPs on disease.
Acknowledgments
This study was supported by R01 NR013520, P30ES019776, R01GM116065, and R03AI111396 from the National Institute of Health; and by grants 13GRNT17060002 from the American Heart Association.
References
- Abbas AR, Wolslegel K, Seshasayee D, Modrusan Z, Clark HF. Deconvolution of blood microarray data identifies cellular activation patterns in systemic lupus erythematosus. PloS one. 2009;4(7):e6098. doi: 10.1371/journal.pone.0006098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aberg KA, McClay JL, Nerella S, Clark S, Kumar G, Chen W, van den Oord EJ. Methylome-wide association study of schizophrenia: identifying blood biomarker signatures of environmental insults. JAMA Psychiatry. 2014;71(3):255–264. doi: 10.1001/jamapsychiatry.2013.3730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams D, Altucci L, Antonarakis SE, Ballesteros J, Beck S, Bird A, Willcocks S. BLUEPRINT to decode the epigenetic signature written in blood. Nat Biotechnol. 2012;30(3):224–226. doi: 10.1038/nbt.2153. [DOI] [PubMed] [Google Scholar]
- Akaike H. A new look at the statistical model identification. Automatic Control, IEEE Transactions on. 1974;19(6):716–723. [Google Scholar]
- Al Awam K, Haussleiter IS, Dudley E, Donev R, Brune M, Juckel G, Thome J. Multiplatform metabolome and proteome profiling identifies serum metabolite and protein signatures as prospective biomarkers for schizophrenia. J Neural Transm. 2015;122(Suppl 1):111–122. doi: 10.1007/s00702-014-1224-0. [DOI] [PubMed] [Google Scholar]
- Alizadeh AA, Eisen MB, Davis RE, Ma C, Lossos IS, Rosenwald A, Staudt LM. Distinct types of diffuse large B-cell lymphoma identified by gene expression profiling. Nature. 2000;403(6769):503–511. doi: 10.1038/35000501. [DOI] [PubMed] [Google Scholar]
- Altmaier E, Fobo G, Heier M, Thorand B, Meisinger C, Romisch-Margl W, Kastenmuller G. Metabolomics approach reveals effects of antihypertensives and lipid-lowering drugs on the human metabolism. Eur J Epidemiol. 2014;29(5):325–336. doi: 10.1007/s10654-014-9910-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson NL, Anderson NG. The human plasma proteome: history, character, and diagnostic prospects. Mol Cell Proteomics. 2002;1(11):845–867. doi: 10.1074/mcp.r200007-mcp200. [DOI] [PubMed] [Google Scholar]
- Astarita G, Ahmed F, Piomelli D. Lipidomic analysis of biological samples by liquid chromatography coupled to mass spectrometry. Methods Mol Biol. 2009;579:201–219. doi: 10.1007/978-1-60761-322-0_10. [DOI] [PubMed] [Google Scholar]
- Baker M. Metabolomics: from small molecules to big ideas. nature methods. 2011;8(2):117. [Google Scholar]
- Bell JT, Pai AA, Pickrell JK, Gaffney DJ, Pique-Regi R, Degner JF, Pritchard JK. DNA methylation patterns associate with genetic and gene expression variation in HapMap cell lines. Genome Biol. 2011;12(1):R10. doi: 10.1186/gb-2011-12-1-r10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein BE, Birney E, Dunham I, Green ED, Gunter C, Snyder M. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489(7414):57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bibikova M, Barnes B, Tsan C, Ho V, Klotzle B, Le JM, Shen R. High density DNA methylation array with single CpG site resolution. Genomics. 2011;98(4):288–295. doi: 10.1016/j.ygeno.2011.07.007. [DOI] [PubMed] [Google Scholar]
- Bird A. Perceptions of epigenetics. Nature. 2007;447(7143):396–398. doi: 10.1038/nature05913. [DOI] [PubMed] [Google Scholar]
- Bjornsson HT, Fallin MD, Feinberg AP. An integrated epigenetic and genetic approach to common human disease. Trends Genet. 2004;20(8):350–358. doi: 10.1016/j.tig.2004.06.009. [DOI] [PubMed] [Google Scholar]
- Bocklandt S, Lin W, Sehl ME, Sanchez FJ, Sinsheimer JS, Horvath S, Vilain E. Epigenetic predictor of age. PLoS One. 2011;6(6):e14821. doi: 10.1371/journal.pone.0014821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breitling LP, Yang R, Korn B, Burwinkel B, Brenner H. Tobacco-smoking-related differential DNA methylation: 27K discovery and replication. Am J Hum Genet. 2011;88(4):450–457. doi: 10.1016/j.ajhg.2011.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen R, Mias GI, Li-Pook-Than J, Jiang L, Lam HY, Chen R, Snyder M. Personal omics profiling reveals dynamic molecular and medical phenotypes. Cell. 2012;148(6):1293–1307. doi: 10.1016/j.cell.2012.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Civelek M, Lusis AJ. Systems genetics approaches to understand complex traits. Nat Rev Genet. 2014;15(1):34–48. doi: 10.1038/nrg3575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Consortium, ENCODE Project. The ENCODE (ENCyclopedia Of DNA Elements) Project. Science. 2004;306(5696):636–640. doi: 10.1126/science.1105136. [DOI] [PubMed] [Google Scholar]
- Consortium, GTEx. The Genotype-Tissue Expression (GTEx) project. Nat Genet. 2013;45(6):580–585. doi: 10.1038/ng.2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Consortium, GTEx. Human genomics. The Genotype-Tissue Expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science. 2015;348(6235):648–660. doi: 10.1126/science.1262110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cookson W, Liang L, Abecasis G, Moffatt M, Lathrop M. Mapping complex disease traits with global gene expression. Nat Rev Genet. 2009;10(3):184–194. doi: 10.1038/nrg2537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortessis VK, Thomas DC, Levine AJ, Breton CV, Mack TM, Siegmund KD, Laird PW. Environmental epigenetics: prospects for studying epigenetic mediation of exposure-response relationships. Hum Genet. 2012;131(10):1565–1589. doi: 10.1007/s00439-012-1189-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui Q, Lewis IA, Hegeman AD, Anderson ME, Li J, Schulte CF, Markley JL. Metabolite identification via the Madison Metabolomics Consortium Database. Nature biotechnology. 2008;26(2):162–164. doi: 10.1038/nbt0208-162. [DOI] [PubMed] [Google Scholar]
- De Jager PL, Srivastava G, Lunnon K, Burgess J, Schalkwyk LC, Yu L, Bennett DA. Alzheimer's disease: early alterations in brain DNA methylation at ANK1, BIN1, RHBDF2 and other loci. Nat Neurosci. 2014;17(9):1156–1163. doi: 10.1038/nn.3786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dedeurwaerder S, Defrance M, Calonne E, Denis H, Sotiriou C, Fuks F. Evaluation of the Infinium Methylation 450K technology. Epigenomics. 2011;3(6):771–784. doi: 10.2217/epi.11.105. [DOI] [PubMed] [Google Scholar]
- Demerath EW, Guan W, Grove ML, Aslibekyan S, Mendelson M, Zhou YH, Boerwinkle E. Epigenome-wide association study (EWAS) of BMI, BMI change and waist circumference in African American adults identifies multiple replicated loci. Hum Mol Genet. 2015;24(15):4464–4479. doi: 10.1093/hmg/ddv161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dick KJ, Nelson CP, Tsaprouni L, Sandling JK, Aissi D, Wahl S, Samani NJ. DNA methylation and body-mass index: a genome-wide analysis. Lancet. 2014 doi: 10.1016/S0140-6736(13)62674-4. [DOI] [PubMed] [Google Scholar]
- Ding J, Gudjonsson JE, Liang L, Stuart PE, Li Y, Chen W, Abecasis GR. Gene expression in skin and lymphoblastoid cells: Refined statistical method reveals extensive overlap in cis-eQTL signals. Am J Hum Genet. 2010;87(6):779–789. doi: 10.1016/j.ajhg.2010.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drong AW, Nicholson G, Hedman AK, Meduri E, Grundberg E, Small KS, Mol PC. The presence of methylation quantitative trait loci indicates a direct genetic influence on the level of DNA methylation in adipose tissue. PloS one. 2013;8(2):e55923. doi: 10.1371/journal.pone.0055923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudley E, Yousef M, Wang Y, Griffiths WJ. Targeted metabolomics and mass spectrometry. Adv Protein Chem Struct Biol. 2010;80:45–83. doi: 10.1016/B978-0-12-381264-3.00002-3. [DOI] [PubMed] [Google Scholar]
- Fairfax BP, Makino S, Radhakrishnan J, Plant K, Leslie S, Dilthey A, Knight JC. Genetics of gene expression in primary immune cells identifies cell type-specific master regulators and roles of HLA alleles. Nat Genet. 2012;44(5):502–510. doi: 10.1038/ng.2205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farh KK, Marson A, Zhu J, Kleinewietfeld M, Housley WJ, Beik S, Bernstein BE. Genetic and epigenetic fine mapping of causal autoimmune disease variants. Nature. 2015;518(7539):337–343. doi: 10.1038/nature13835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farias AS, Pradella F, Schmitt A, Santos LM, Martins-de-Souza D. Ten years of proteomics in multiple sclerosis. Proteomics. 2014;14(4–5):467–480. doi: 10.1002/pmic.201300268. [DOI] [PubMed] [Google Scholar]
- Foley DL, Craig JM, Morley R, Olsson CA, Dwyer T, Smith K, Saffery R. Prospects for epigenetic epidemiology. Am J Epidemiol. 2009;169(4):389–400. doi: 10.1093/aje/kwn380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foss EJ, Radulovic D, Shaffer SA, Ruderfer DM, Bedalov A, Goodlett DR, Kruglyak L. Genetic basis of proteome variation in yeast. Nat Genet. 2007;39(11):1369–1375. doi: 10.1038/ng.2007.22. [DOI] [PubMed] [Google Scholar]
- Frantzi M, Latosinska A, Fluhe L, Hupe MC, Critselis E, Kramer MW, Vlahou A. Developing proteomic biomarkers for bladder cancer: towards clinical application. Nat Rev Urol. 2015;12(6):317–330. doi: 10.1038/nrurol.2015.100. [DOI] [PubMed] [Google Scholar]
- Frediani JK, Jones DP, Tukvadze N, Uppal K, Sanikidze E, Kipiani M, Ziegler TR. Plasma metabolomics in human pulmonary tuberculosis disease: a pilot study. PloS one. 2014;9(10):e108854. doi: 10.1371/journal.pone.0108854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghazalpour A, Bennett B, Petyuk VA, Orozco L, Hagopian R, Mungrue IN, Lusis AJ. Comparative analysis of proteome and transcriptome variation in mouse. PLoS Genet. 2011;7(6):e1001393. doi: 10.1371/journal.pgen.1001393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbs JR, van der Brug MP, Hernandez DG, Traynor BJ, Nalls MA, Lai SL, Singleton AB. Abundant quantitative trait loci exist for DNA methylation and gene expression in human brain. PLoS Genet. 2010;6(5):e1000952. doi: 10.1371/journal.pgen.1000952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gieger C, Geistlinger L, Altmaier E, Hrabe de Angelis M, Kronenberg F, Meitinger T, Suhre K. Genetics meets metabolomics: a genome-wide association study of metabolite profiles in human serum. PLoS Genet. 2008;4(11):e1000282. doi: 10.1371/journal.pgen.1000282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray R, Wheatley K. How to avoid bias when comparing bone marrow transplantation with chemotherapy. Bone marrow transplantation. 1991;7(Suppl 3):9–12. [PubMed] [Google Scholar]
- Grundberg E, Meduri E, Sandling JK, Hedman AK, Keildson S, Buil A, Deloukas P. Global analysis of DNA methylation variation in adipose tissue from twins reveals links to disease-associated variants in distal regulatory elements. Am J Hum Genet. 2013;93(5):876–890. doi: 10.1016/j.ajhg.2013.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez-Arcelus M, Lappalainen T, Montgomery SB, Buil A, Ongen H, Yurovsky A, Dermitzakis ET. Passive and active DNA methylation and the interplay with genetic variation in gene regulation. Elife. 2013;2:e00523. doi: 10.7554/eLife.00523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heijmans BT, Mill J. Commentary: The seven plagues of epigenetic epidemiology. Int J Epidemiol. 2012;41(1):74–78. doi: 10.1093/ije/dyr225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heyn H, Moran S, Hernando-Herraez I, Sayols S, Gomez A, Sandoval J, Esteller M. DNA methylation contributes to natural human variation. Genome Res. 2013;23(9):1363–1372. doi: 10.1101/gr.154187.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heyn H, Sayols S, Moutinho C, Vidal E, Sanchez-Mut JV, Stefansson OA, Esteller M. Linkage of DNA methylation quantitative trait loci to human cancer risk. Cell Rep. 2014;7(2):331–338. doi: 10.1016/j.celrep.2014.03.016. [DOI] [PubMed] [Google Scholar]
- Hidalgo B, Irvin MR, Sha J, Zhi D, Aslibekyan S, Absher D, Arnett DK. Epigenome-wide association study of fasting measures of glucose, insulin, and HOMA-IR in the Genetics of Lipid Lowering Drugs and Diet Network study. Diabetes. 2014;63(2):801–807. doi: 10.2337/db13-1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman JM, Soltow QA, Li S, Sidik A, Jones DP, Promislow DE. Effects of age, sex, and genotype on high-sensitivity metabolomic profiles in the fruit fly, Drosophila melanogaster. Aging Cell. 2014;13(4):596–604. doi: 10.1111/acel.12215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holdt LM, von Delft A, Nicolaou A, Baumann S, Kostrzewa M, Thiery J, Teupser D. Quantitative trait loci mapping of the mouse plasma proteome (pQTL) Genetics. 2013;193(2):601–608. doi: 10.1534/genetics.112.143354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houseman EA, Accomando WP, Koestler DC, Christensen BC, Marsit CJ, Nelson HH, Kelsey KT. DNA methylation arrays as surrogate measures of cell mixture distribution. BMC bioinformatics. 2012;13(1):86. doi: 10.1186/1471-2105-13-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houseman EA, Molitor J, Marsit CJ. Reference-free cell mixture adjustments in analysis of DNA methylation data. Bioinformatics. 2014;30(10):1431–1439. doi: 10.1093/bioinformatics/btu029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang YT. Integrative modeling of multiple genomic data from different types of genetic association studies. Biostatistics. 2014;15(4):587–602. doi: 10.1093/biostatistics/kxu014. [DOI] [PubMed] [Google Scholar]
- Huang YT. Integrative modeling of multi-platform genomic data under the framework of mediation analysis. Stat Med. 2015;34(1):162–178. doi: 10.1002/sim.6326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang YT, Vanderweele TJ, Lin X. Joint Analysis of Snp and Gene Expression Data in Genetic Association Studies of Complex Diseases. Ann Appl Stat. 2014;8(1):352–376. doi: 10.1214/13-AOAS690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irvin MR, Zhi D, Joehanes R, Mendelson M, Aslibekyan S, Claas SA, Arnett DK. Epigenome-Wide Association Study of Fasting Blood Lipids in the Genetics of Lipid Lowering Drugs and Diet Network Study. Circulation. 2014 doi: 10.1161/CIRCULATIONAHA.114.009158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito S, D'Alessio AC, Taranova OV, Hong K, Sowers LC, Zhang Y. Role of Tet proteins in 5mC to 5hmC conversion, ES-cell self-renewal and inner cell mass specification. Nature. 2010;466(7310):1129–1133. doi: 10.1038/nature09303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jablonka E. Epigenetic epidemiology. Int J Epidemiol. 2004;33(5):929–935. doi: 10.1093/ije/dyh231. [DOI] [PubMed] [Google Scholar]
- Jin SG, Kadam S, Pfeifer GP. Examination of the specificity of DNA methylation profiling techniques towards 5-methylcytosine and 5-hydroxymethylcytosine. Nucleic Acids Res. 2010;38(11):e125. doi: 10.1093/nar/gkq223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones DP, Park Y, Ziegler TR. Nutritional metabolomics: progress in addressing complexity in diet and health. Annu Rev Nutr. 2012;32:183–202. doi: 10.1146/annurev-nutr-072610-145159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaddurah-Daouk R, Kristal BS, Weinshilboum RM. Metabolomics: a global biochemical approach to drug response and disease. Annu Rev Pharmacol Toxicol. 2008;48:653–683. doi: 10.1146/annurev.pharmtox.48.113006.094715. [DOI] [PubMed] [Google Scholar]
- Kanehisa M, Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28(1):27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katan MB. Apolipoprotein E isoforms, serum cholesterol, and cancer. Lancet. 1986;1(8479):507–508. doi: 10.1016/s0140-6736(86)92972-7. [DOI] [PubMed] [Google Scholar]
- Kettunen J, Tukiainen T, Sarin AP, Ortega-Alonso A, Tikkanen E, Lyytikainen LP, Ripatti S. Genome-wide association study identifies multiple loci influencing human serum metabolite levels. Nat Genet. 2012;44(3):269–276. doi: 10.1038/ng.1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim MS, Pinto SM, Getnet D, Nirujogi RS, Manda SS, Chaerkady R, Pandey A. A draft map of the human proteome. Nature. 2014;509(7502):575–581. doi: 10.1038/nature13302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein RJ, Zeiss C, Chew EY, Tsai JY, Sackler RS, Haynes C, Hoh J. Complement factor H polymorphism in age-related macular degeneration. Science. 2005;308(5720):385–389. doi: 10.1126/science.1109557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraemer S, Vaught JD, Bock C, Gold L, Katilius E, Keeney TR, Sanders GM. From SOMAmer-based biomarker discovery to diagnostic and clinical applications: a SOMAmer-based, streamlined multiplex proteomic assay. PloS one. 2011;6(10):e26332. doi: 10.1371/journal.pone.0026332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D, Xie Z, Pape ML, Dye T. An evaluation of statistical methods for DNA methylation microarray data analysis. BMC bioinformatics. 2015;16:217. doi: 10.1186/s12859-015-0641-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Park Y, Duraisingham S, Strobel FH, Khan N, Soltow QA, Pulendran B. Predicting network activity from high throughput metabolomics. PLoS Comput Biol. 2013;9(7):e1003123. doi: 10.1371/journal.pcbi.1003123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Tesson BM, Churchill GA, Jansen RC. Critical reasoning on causal inference in genome-wide linkage and association studies. Trends Genet. 2010;26(12):493–498. doi: 10.1016/j.tig.2010.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Zhu J, Tian G, Li N, Li Q, Ye M, Zhang X. The DNA methylome of human peripheral blood mononuclear cells. PLoS Biol. 2010;8(11):e1000533. doi: 10.1371/journal.pbio.1000533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang L, Willis-Owen SA, Laprise C, Wong KC, Davies GA, Hudson TJ, Cookson WO. An epigenome-wide association study of total serum immunoglobulin E concentration. Nature. 2015 doi: 10.1038/nature14125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Aryee MJ, Padyukov L, Fallin MD, Hesselberg E, Runarsson A, Feinberg AP. Epigenome-wide association data implicate DNA methylation as an intermediary of genetic risk in rheumatoid arthritis. Nat Biotechnol. 2013;31(2):142–147. doi: 10.1038/nbt.2487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Ding J, Reynolds LM, Lohman K, Register TC, De La Fuente A, Hoeschele I. Methylomics of gene expression in human monocytes. Hum Mol Genet. 2013;22(24):5065–5074. doi: 10.1093/hmg/ddt356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manolio TA. Genomewide association studies and assessment of the risk of disease. N Engl J Med. 2010;363(2):166–176. doi: 10.1056/NEJMra0905980. [DOI] [PubMed] [Google Scholar]
- Marx V. The DNA of a nation. Nature. 2015;524(7566):503–505. doi: 10.1038/524503a. [DOI] [PubMed] [Google Scholar]
- McClay JL, Aberg KA, Clark SL, Nerella S, Kumar G, Xie LY, Van Den Oord EJ. A methylome-wide study of aging using massively parallel sequencing of the methyl-CpG-enriched genomic fraction from blood in over 700 subjects. Hum Mol Genet. 2014;23(5):1175–1185. doi: 10.1093/hmg/ddt511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meissner A, Mikkelsen TS, Gu H, Wernig M, Hanna J, Sivachenko A, Lander ES. Genome-scale DNA methylation maps of pluripotent and differentiated cells. Nature. 2008;454(7205):766–770. doi: 10.1038/nature07107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melzer D, Perry JR, Hernandez D, Corsi AM, Stevens K, Rafferty I, Ferrucci L. A genome-wide association study identifies protein quantitative trait loci (pQTLs) PLoS Genet. 2008;4(5):e1000072. doi: 10.1371/journal.pgen.1000072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metzker ML. Sequencing technologies - the next generation. Nat Rev Genet. 2010;11(1):31–46. doi: 10.1038/nrg2626. [DOI] [PubMed] [Google Scholar]
- Michels KB, Binder AM, Dedeurwaerder S, Epstein CB, Greally JM, Gut I, Irizarry RA. Recommendations for the design and analysis of epigenome-wide association studies. Nat Methods. 2013;10(10):949–955. doi: 10.1038/nmeth.2632. [DOI] [PubMed] [Google Scholar]
- Mill J, Heijmans BT. From promises to practical strategies in epigenetic epidemiology. Nat Rev Genet. 2013;14(8):585–594. doi: 10.1038/nrg3405. [DOI] [PubMed] [Google Scholar]
- Miller GW, Jones DP. The nature of nurture: refining the definition of the exposome. Toxicol Sci. 2014;137(1):1–2. doi: 10.1093/toxsci/kft251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers AJ, Gibbs JR, Webster JA, Rohrer K, Zhao A, Marlowe L, Hardy J. A survey of genetic human cortical gene expression. Nat Genet. 2007;39(12):1494–1499. doi: 10.1038/ng.2007.16. [DOI] [PubMed] [Google Scholar]
- Nascimento JM, Martins-de-Souza D. The proteome of schizophrenia. npj Schizophrenia. 2015;1 doi: 10.1038/npjschz.2014.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicolae DL, Gamazon E, Zhang W, Duan S, Dolan ME, Cox NJ. Trait-associated SNPs are more likely to be eQTLs: annotation to enhance discovery from GWAS. PLoS Genet. 2010;6(4):e1000888. doi: 10.1371/journal.pgen.1000888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen R, Paul JS, Albrechtsen A, Song YS. Genotype and SNP calling from next-generation sequencing data. Nat Rev Genet. 2011;12(6):443–451. doi: 10.1038/nrg2986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson T, Mann M, Aebersold R, Yates JR, 3rd, Bairoch A, Bergeron JJ. Mass spectrometry in high-throughput proteomics: ready for the big time. Nat Methods. 2010;7(9):681–685. doi: 10.1038/nmeth0910-681. [DOI] [PubMed] [Google Scholar]
- Ozsolak F, Milos PM. RNA sequencing: advances, challenges and opportunities. Nat Rev Genet. 2011;12(2):87–98. doi: 10.1038/nrg2934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan S, Brentnall TA, Chen R. Proteomics analysis of bodily fluids in pancreatic cancer. Proteomics. 2015;15(15):2705–2715. doi: 10.1002/pmic.201400476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan W, Xie B, Shen X. Incorporating predictor network in penalized regression with application to microarray data. Biometrics. 2010;66(2):474–484. doi: 10.1111/j.1541-0420.2009.01296.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patti GJ, Yanes O, Siuzdak G. Innovation: Metabolomics: the apogee of the omics trilogy. Nat Rev Mol Cell Biol. 2012;13(4):263–269. doi: 10.1038/nrm3314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen AK, Zeilinger S, Kastenmuller G, Romisch-Margl W, Brugger M, Peters A, Suhre K. Epigenetics meets metabolomics: an epigenome-wide association study with blood serum metabolic traits. Hum Mol Genet. 2014;23(2):534–545. doi: 10.1093/hmg/ddt430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petricoin EF, Ardekani AM, Hitt BA, Levine PJ, Fusaro VA, Steinberg SM, Liotta LA. Use of proteomic patterns in serum to identify ovarian cancer. Lancet. 2002;359(9306):572–577. doi: 10.1016/S0140-6736(02)07746-2. [DOI] [PubMed] [Google Scholar]
- Pickrell JK, Marioni JC, Pai AA, Degner JF, Engelhardt BE, Nkadori E, Pritchard JK. Understanding mechanisms underlying human gene expression variation with RNA sequencing. Nature. 2010;464(7289):768–772. doi: 10.1038/nature08872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picotti P, Clement-Ziza M, Lam H, Campbell DS, Schmidt A, Deutsch EW, Aebersold R. A complete mass-spectrometric map of the yeast proteome applied to quantitative trait analysis. Nature. 2013;494(7436):266–270. doi: 10.1038/nature11835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rakyan VK, Down TA, Balding DJ, Beck S. Epigenome-wide association studies for common human diseases. Nat Rev Genet. 2011;12(8):529–541. doi: 10.1038/nrg3000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rappaport SM, Barupal DK, Wishart D, Vineis P, Scalbert A. The Blood Exposome and Its Role in Discovering Causes of Disease. Environ Health Perspect. 2014 doi: 10.1289/ehp.1308015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Relton CL, Davey Smith G. Epigenetic epidemiology of common complex disease: prospects for prediction, prevention, and treatment. PLoS Med. 2010;7(10):e1000356. doi: 10.1371/journal.pmed.1000356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Relton CL, Davey Smith G. Two-step epigenetic Mendelian randomization: a strategy for establishing the causal role of epigenetic processes in pathways to disease. Int J Epidemiol. 2012;41(1):161–176. doi: 10.1093/ije/dyr233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritchie MD, Holzinger ER, Li R, Pendergrass SA, Kim D. Methods of integrating data to uncover genotype-phenotype interactions. Nat Rev Genet. 2015;16(2):85–97. doi: 10.1038/nrg3868. [DOI] [PubMed] [Google Scholar]
- Roadmap Epigenomics C, Kundaje A, Meuleman W, Ernst J, Bilenky M, Yen A, Kellis M. Integrative analysis of 111 reference human epigenomes. Nature. 2015;518(7539):317–330. doi: 10.1038/nature14248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romanoski CE, Lee S, Kim MJ, Ingram-Drake L, Plaisier CL, Yordanova R, Lusis AJ. Systems genetics analysis of gene-by-environment interactions in human cells. Am J Hum Genet. 2010;86(3):399–410. doi: 10.1016/j.ajhg.2010.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabatine MS, Liu E, Morrow DA, Heller E, McCarroll R, Wiegand R, Gerszten RE. Metabolomic identification of novel biomarkers of myocardial ischemia. Circulation. 2005;112(25):3868–3875. doi: 10.1161/CIRCULATIONAHA.105.569137. [DOI] [PubMed] [Google Scholar]
- Sandoval J, Heyn H, Moran S, Serra-Musach J, Pujana MA, Bibikova M, Esteller M. Validation of a DNA methylation microarray for 450,000 CpG sites in the human genome. Epigenetics. 2011;6(6):692–702. doi: 10.4161/epi.6.6.16196. [DOI] [PubMed] [Google Scholar]
- Schadt EE, Lamb J, Yang X, Zhu J, Edwards S, Guhathakurta D, Lusis AJ. An integrative genomics approach to infer causal associations between gene expression and disease. Nat Genet. 2005;37(7):710–717. doi: 10.1038/ng1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schadt EE, Molony C, Chudin E, Hao K, Yang X, Lum PY, Ulrich R. Mapping the genetic architecture of gene expression in human liver. PLoS Biol. 2008;6(5):e107. doi: 10.1371/journal.pbio.0060107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaub MA, Boyle AP, Kundaje A, Batzoglou S, Snyder M. Linking disease associations with regulatory information in the human genome. Genome Res. 2012;22(9):1748–1759. doi: 10.1101/gr.136127.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwanhausser B, Busse D, Li N, Dittmar G, Schuchhardt J, Wolf J, Selbach M. Global quantification of mammalian gene expression control. Nature. 2011;473(7347):337–342. doi: 10.1038/nature10098. [DOI] [PubMed] [Google Scholar]
- Schwarz G. Estimating the dimension of a model. The annals of statistics. 1978;6(2):461–464. [Google Scholar]
- Serre D, Lee BH, Ting AH. MBD-isolated Genome Sequencing provides a high-throughput and comprehensive survey of DNA methylation in the human genome. Nucleic Acids Res. 2010;38(2):391–399. doi: 10.1093/nar/gkp992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah S, Bonder MJ, Marioni RE, Zhu Z, McRae AF, Zhernakova A, Visscher PM. Improving Phenotypic Prediction by Combining Genetic and Epigenetic Associations. Am J Hum Genet. 2015;97(1):75–85. doi: 10.1016/j.ajhg.2015.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shenker NS, Polidoro S, van Veldhoven K, Sacerdote C, Ricceri F, Birrell MA, Flanagan JM. Epigenome-wide association study in the European Prospective Investigation into Cancer and Nutrition (EPIC-Turin) identifies novel genetic loci associated with smoking. Hum Mol Genet. 2013;22(5):843–851. doi: 10.1093/hmg/dds488. [DOI] [PubMed] [Google Scholar]
- Shi J, Marconett CN, Duan J, Hyland PL, Li P, Wang Z, Landi MT. Characterizing the genetic basis of methylome diversity in histologically normal human lung tissue. Nat Commun. 2014;5:3365. doi: 10.1038/ncomms4365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin SY, Fauman EB, Petersen AK, Krumsiek J, Santos R, Huang J, Soranzo N. An atlas of genetic influences on human blood metabolites. Nat Genet. 2014;46(6):543–550. doi: 10.1038/ng.2982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith AK, Kilaru V, Kocak M, Almli LM, Mercer KB, Ressler KJ, Conneely KN. Methylation quantitative trait loci (meQTLs) are consistently detected across ancestry, developmental stage, and tissue type. BMC Genomics. 2014;15:145. doi: 10.1186/1471-2164-15-145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith CA, O'Maille G, Want EJ, Qin C, Trauger SA, Brandon TR, Siuzdak G. METLIN: a metabolite mass spectral database. Ther Drug Monit. 2005;27(6):747–751. doi: 10.1097/01.ftd.0000179845.53213.39. [DOI] [PubMed] [Google Scholar]
- Suhre K, Shin SY, Petersen AK, Mohney RP, Meredith D, Wagele B, Gieger C. Human metabolic individuality in biomedical and pharmaceutical research. Nature. 2011;477(7362):54–60. doi: 10.1038/nature10354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun YV. The Influences of Genetic and Environmental Factors on Methylome-wide Association Studies for Human Diseases. Curr Genet Med Rep. 2014;2(4):261–270. doi: 10.1007/s40142-014-0058-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun YV, Lazarus A, Smith JA, Chuang YH, Zhao W, Turner ST, Kardia SL. Gene-specific DNA methylation association with serum levels of C-reactive protein in African Americans. PLoS One. 2013;8(8):e73480. doi: 10.1371/journal.pone.0073480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun YV, Smith AK, Conneely KN, Chang Q, Li W, Lazarus A, Kardia SL. Epigenomic association analysis identifies smoking-related DNA methylation sites in African Americans. Hum Genet. 2013;132(9):1027–1037. doi: 10.1007/s00439-013-1311-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teh AL, Pan H, Chen L, Ong ML, Dogra S, Wong J, Holbrook JD. The effect of genotype and in utero environment on interindividual variation in neonate DNA methylomes. Genome Res. 2014;24(7):1064–1074. doi: 10.1101/gr.171439.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teschendorff AE, Menon U, Gentry-Maharaj A, Ramus SJ, Weisenberger DJ, Shen H, Widschwendter M. Age-dependent DNA methylation of genes that are suppressed in stem cells is a hallmark of cancer. Genome research. 2010;20(4):440–446. doi: 10.1101/gr.103606.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The 1000 Genomes Project Consortium. Abecasis GR, Auton A, Brooks LD, DePristo MA, Durbin RM, McVean GA. An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491(7422):56–65. doi: 10.1038/nature11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, Pachter L. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc. 2012;7(3):562–578. doi: 10.1038/nprot.2012.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai TH, Song E, Zhu R, Di Poto C, Wang M, Luo Y, Ressom HW. LC-MS/MS-based serum proteomics for identification of candidate biomarkers for hepatocellular carcinoma. Proteomics. 2015;15(13):2369–2381. doi: 10.1002/pmic.201400364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uddin M, Koenen KC, Aiello AE, Wildman DE, de los Santos R, Galea S. Epigenetic and inflammatory marker profiles associated with depression in a community-based epidemiologic sample. Psychol Med. 2011;41(5):997–1007. doi: 10.1017/S0033291710001674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veyrieras JB, Kudaravalli S, Kim SY, Dermitzakis ET, Gilad Y, Stephens M, Pritchard JK. High-resolution mapping of expression-QTLs yields insight into human gene regulation. PLoS Genet. 2008;4(10):e1000214. doi: 10.1371/journal.pgen.1000214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan Q, Pal R. An ensemble based top performing approach for NCI-DREAM drug sensitivity prediction challenge. PloS one. 2014;9(6):e101183. doi: 10.1371/journal.pone.0101183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang TJ, Larson MG, Vasan RS, Cheng S, Rhee EP, McCabe E, Gerszten RE. Metabolite profiles and the risk of developing diabetes. Nat Med. 2011;17(4):448–453. doi: 10.1038/nm.2307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Liu Q, Zhang B. Leveraging the complementary nature of RNA-Seq and shotgun proteomics data. Proteomics. 2014;14(23–24):2676–2687. doi: 10.1002/pmic.201400184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Gerstein M, Snyder M. RNA-Seq: a revolutionary tool for transcriptomics. Nat Rev Genet. 2009;10(1):57–63. doi: 10.1038/nrg2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterland RA, Jirtle RL. Transposable elements: targets for early nutritional effects on epigenetic gene regulation. Mol Cell Biol. 2003;23(15):5293–5300. doi: 10.1128/MCB.23.15.5293-5300.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei Z, Li H. A Markov random field model for network-based analysis of genomic data. Bioinformatics. 2007;23(12):1537–1544. doi: 10.1093/bioinformatics/btm129. [DOI] [PubMed] [Google Scholar]
- Weiner J, 3rd, Parida SK, Maertzdorf J, Black GF, Repsilber D, Telaar A, Kaufmann SH. Biomarkers of inflammation, immunosuppression and stress with active disease are revealed by metabolomic profiling of tuberculosis patients. PloS one. 2012;7(7):e40221. doi: 10.1371/journal.pone.0040221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wellcome Trust Case Control, C. Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447(7145):661–678. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welter D, MacArthur J, Morales J, Burdett T, Hall P, Junkins H, Parkinson H. The NHGRI GWAS Catalog, a curated resource of SNP-trait associations. Nucleic Acids Res. 2014;42(Database issue):D1001–1006. doi: 10.1093/nar/gkt1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wikoff WR, Anfora AT, Liu J, Schultz PG, Lesley SA, Peters EC, Siuzdak G. Metabolomics analysis reveals large effects of gut microflora on mammalian blood metabolites. Proc Natl Acad Sci U S A. 2009;106(10):3698–3703. doi: 10.1073/pnas.0812874106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkins MR, Pasquali C, Appel RD, Ou K, Golaz O, Sanchez JC, Hochstrasser DF. From proteins to proteomes: large scale protein identification by two-dimensional electrophoresis and amino acid analysis. Biotechnology (N Y) 1996;14(1):61–65. doi: 10.1038/nbt0196-61. [DOI] [PubMed] [Google Scholar]
- Wishart DS, Knox C, Guo AC, Eisner R, Young N, Gautam B, Forsythe I. HMDB: a knowledgebase for the human metabolome. Nucleic Acids Res. 2009;37(Database issue):D603–610. doi: 10.1093/nar/gkn810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wurtz P, Havulinna AS, Soininen P, Tynkkynen T, Prieto-Merino D, Tillin T, Salomaa V. Metabolite profiling and cardiovascular event risk: a prospective study of 3 population-based cohorts. Circulation. 2015;131(9):774–785. doi: 10.1161/CIRCULATIONAHA.114.013116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong Q, Ancona N, Hauser ER, Mukherjee S, Furey TS. Integrating genetic and gene expression evidence into genome-wide association analysis of gene sets. Genome Res. 2012;22(2):386–397. doi: 10.1101/gr.124370.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao L, Tak YG, Berman BP, Farnham PJ. Functional annotation of colon cancer risk SNPs. Nat Commun. 2014;5:5114. doi: 10.1038/ncomms6114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu B, Zheng Y, Alexander D, Manolio TA, Alonso A, Nettleton JA, Boerwinkle E. Genome-wide association study of a heart failure related metabolomic profile among African Americans in the Atherosclerosis Risk in Communities (ARIC) study. Genet Epidemiol. 2013;37(8):840–845. doi: 10.1002/gepi.21752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D, Cheng L, Badner JA, Chen C, Chen Q, Luo W, Liu C. Genetic control of individual differences in gene-specific methylation in human brain. Am J Hum Genet. 2010;86(3):411–419. doi: 10.1016/j.ajhg.2010.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Duan S, Kistner EO, Bleibel WK, Huang RS, Clark TA, Dolan ME. Evaluation of genetic variation contributing to differences in gene expression between populations. Am J Hum Genet. 2008;82(3):631–640. doi: 10.1016/j.ajhg.2007.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Moen EL, Liu C, Mu W, Gamazon ER, Delaney SM, Zhang W. Linking the genetic architecture of cytosine modifications with human complex traits. Hum Mol Genet. 2014 doi: 10.1093/hmg/ddu313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J, Zhu Y, Hyun N, Zeng D, Uppal K, Tran VT, Howard BV. Novel metabolic markers for the risk of diabetes development in American Indians. Diabetes Care. 2015;38(2):220–227. doi: 10.2337/dc14-2033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhi D, Aslibekyan S, Irvin MR, Claas SA, Borecki IB, Ordovas JM, Arnett DK. SNPs located at CpG sites modulate genome-epigenome interaction. Epigenetics. 2013;8(8):802–806. doi: 10.4161/epi.25501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziegler A, Sun YV. Study designs and methods post genome-wide association studies. Hum Genet. 2012;131(10):1525–1531. doi: 10.1007/s00439-012-1209-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziller MJ, Gu H, Muller F, Donaghey J, Tsai LT, Kohlbacher O, Meissner A. Charting a dynamic DNA methylation landscape of the human genome. Nature. 2013;500(7463):477–481. doi: 10.1038/nature12433. [DOI] [PMC free article] [PubMed] [Google Scholar]