Abstract
Epithelial organs undergo steady-state turnover throughout adult life, with old cells being continually replaced by the progeny of stem cell divisions1. To avoid hyperplasia or atrophy, organ turnover demands strict equilibration of cell production and loss2–4. However, the mechanistic basis of this equilibrium is unknown. Using the adult Drosophila intestine5, we find that robustly precise turnover arises through a coupling mechanism in which enterocyte apoptosis breaks feedback inhibition of stem cell divisions. Healthy enterocytes inhibit stem cell division through E-cadherin, which prevents secretion of mitogenic EGFs by repressing transcription of the EGF maturation factor rhomboid. Individual apoptotic enterocytes promote divisions by loss of E-cadherin, which releases cadherin-associated β-catenin/Armadillo and p120-catenin to induce rhomboid. Induction of rhomboid in the dying enterocyte triggers EGFR activation in stem cells within a discrete radius. When we block apoptosis, E-cadherin-controlled feedback suppresses divisions, and the organ retains the same number of cells. When we disrupt feedback, apoptosis and divisions are uncoupled, and the organ develops either hyperplasia or atrophy. Altogether, our work demonstrates that robust cellular balance hinges on the obligate coupling of divisions to apoptosis, which limits the proliferative potential of a stem cell to the precise time and place that a replacement cell is needed. In this manner, localized cell-cell communication gives rise to tissue-level homeostatic equilibrium and constant organ size.
Main Text
Throughout an animal’s lifetime, mature organs undergo continuous cell turnover yet can maintain the same approximate size. This remarkable ability implies the existence of robust mechanisms to ensure that turnover is zero-sum, with cell production and loss held precisely equal1,2,4. In most organs, production of new cells ultimately depends on divisions of resident stem cells. Although much is understood about how excessive or insufficient divisions lead to disease, little is known about how equal rates of division and loss are sustained during the steady-state turnover of healthy tissues.
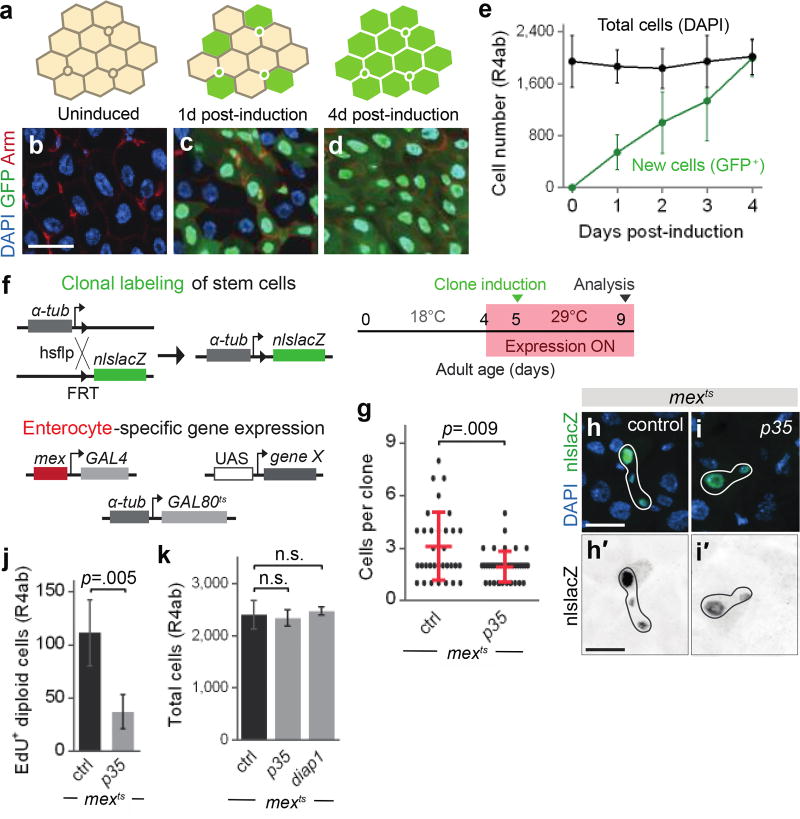
We investigated the regulation of turnover in the midgut epithelium of adult Drosophila5 (Extended Data Fig. 1a–e). To establish whether production of new cells equals loss of old cells, we measured kinetics of cell addition and loss in the R4ab region using escargot>GFP flp-out labeling (esgF/OGFP)6,7 (Fig. 1a–e, Extended Data Fig. 1f–g). Newly-added, GFP+ cells increased linearly over time and, after 4 days, comprised all cells in R4ab. Concomitantly, total cell number remained near-constant. We conclude that production of new cells quantitatively equals loss of old cells.
Fig. 1. Enterocyte apoptosis regulates the rate of stem cell division for homeostatic mainte-nance of total cell number.
a–e, Kinetics of midgut epithelial turnover: Cartoon (a), images (b–d) and quantitation (e) of esgF/O>GFP labeling. Progenitors (a, small circles) express GFP upon induction; new, but not old, enterocytes (a, large hexagons) inherit GFP from progenitors after induction. Quantitation of total and GFP+ cells over time shows complete replacement of unlabeled cells by GFP+ cells after 4 days (e, means ± S.D.; 3 midgut R4ab compartments per timepoint). See Extended Data Fig. 1. f, Genetic schema and experimental timeline for tracing stem cell divisions (split-lacZ clones) in a background of genetically manipulated enterocytes (mexts). See Extended Data Fig. 2. g–i, Sizes (g, means ± S.D; p values, Mann-Whitney test) and images (h, i) of stem cell clones following enterocyte inhibition of apoptosis (mexts>p35). Clone sizes are reduced by apoptotic inhibition. N=4–5 midguts per genotype. j, EdU incorporation in diploid cells is reduced by apoptotic inhibition. k, Total cell counts of the R4ab compartment with control or apoptosis-inhibited (p35, diap1) enterocytes. For j, k: Means ± S.D. shown; p values from unpaired t-test. N=4 midguts per genotype. One of three replicate experiments shown in each graph. All scale bars, 25 µm.
To probe the relationship between cell production and loss, we devised a system to manipulate mature enterocytes and simultaneously track divisions of stem cells by combining enterocyte-specific mexGAL4;GAL80ts (mexts) with split-nlsLacZ clonal labeling8 (Fig. 1f; Extended Data Fig. 2). Using this two-pronged system, we expressed the apoptotic inhibitor p35 in enterocytes and assessed the impact on stem cell divisions. Blocking enterocyte apoptosis resulted in fewer divisions, as indicated by smaller clones (Fig. 1g–i). Apoptotic inhibition also impeded S phase progression (Fig. 1j), consistent with a prior report9. Reduced divisions could be a compensatory means to keep a constant number of total cells. Indeed, total cell number, as well as physical size and morphology, of apoptosis-inhibited midguts remained normal (Fig. 1k, Extended Data Figs. 3a and 4a, b, d, e). These findings imply that enterocyte apoptosis and stem cell division are homeostatically coupled to maintain constant cell number and organ size.
How is coupling mediated? The cell-cell adhesion protein E-cadherin (E-cad) drew our attention because in the mouse intestine, enterocyte E-cad represses stem cell divisions10, and because in other epithelia, E-cad is degraded by caspases during apoptosis11. In the Drosophila midgut, we found that E-cad::mTomato was largely eliminated from the interfaces of dying, Sytox+ enterocytes (Fig. 2a), indicating that apoptotic enterocytes lose junctional E-cad.
Fig. 2. Homeostatic size control requires E-cad on enterocytes, but not on stem cells.
a, E-cad::m-Tomato (red-hot LUT) is absent from cell-cell junctions (arrowheads) of dying enterocytes (Sytox+, asterisks). Tracheal autofluorescence appears as bright, red-yellow lines (arrows). b–g, Sizes (b) and images (c–g) of stem cell clones following enterocyte manipulation of E-cad, with or without apoptotic inhibition (p35; experimental schema in Fig. 1f). Clone sizes are reduced by enterocyte E-cad. h–j. Total cell counts (h, means ± S.D. shown; p values, unpaired t-test) and whole-organ images (i, j; A, anterior; P, posterior). Midguts become hyperplastic following enterocyte co-expression of p35 and E-cadRNAi, but not expression of either p35 or E-cadRNAi alone, or co-expression of p35 and edRNAi. N=4 midguts per genotype. See Extended Data Figs. 3a, 5. k, Sizes of stem cell clones are unchanged by stem cell/enteroblast manipulation of E-cad. Experimental schema similar to Fig. 1f, but using stem/enteroblast esgtsGAL4. For b, k: Means ± S.D. shown; p values, Mann-Whitney test. N=4–5 midguts per genotype. One of three replicate experiments shown in each graph. Scale bars, 25 µm or as indicated.
To investigate whether E-cad couples divisions to apoptosis, we depleted E-cad in apoptosis-inhibited enterocytes and assessed stem cell divisions by measuring clones. Depletion of enterocyte E-cad did not disrupt epithelial architecture or integrity (Extended Data Fig. 4a, c, d, f–j). It did, however, prevent the reduction in stem cell divisions that would otherwise have occurred following apoptotic inhibition (Fig. 2b–e). Consequently, total cell number increased by 70%, and organs became markedly hyperplastic (Fig. 2h–j, Extended Data Fig 3a). These effects were specific to E-cad as depletion of another cell-cell adhesion protein, echinoid, did not affect cell number (Fig. 2h). Without apoptotic inhibition, E-cad depletion caused excess divisions but not hyperplasia (Fig. 2b), likely due to other, tissue-level effects (Extended Data Fig 5); conversely, E-cad overexpression suppressed divisions (Fig. 2f). These findings show that in apoptosis-inhibited midguts, homeostatic suppression of stem cell divisions requires enterocyte E-cad.
Because E-cad functions as an intercellular homodimer, we considered whether enterocyte E-cad acts by dimerizing with stem cell E-cad12,13. Surprisingly, we found that manipulation of E-cad in stem and enteroblast cells did not alter the rate of stem cell divisions, at least as measured by 4-day clones (Fig. 2k). Thus, enterocyte E-cad acts not through stem cell E-cad, but through a distinct intermediary.
We sought to identify this intermediary. Prime candidates included four E-cad-associated pathways: Wingless/Wnt, Hippo, JAK-STAT, and EGFR. To assess whether these pathways act downstream of enterocyte E-cad, we asked whether E-cad knockdown induced pathway-specific target genes and reporters (Extended Data Fig. 6a–j). E-cad knockdown did not induce Wingless or Hippo targets, JAK-STAT pathway components, or STAT signaling in stem cells, although STAT activity in enterocytes was mildly elevated. Strikingly by contrast, E-cad knockdown induced significant activation of EGFR targets in diploid cells, likely stem cells.
We therefore probed the functional relationship between enterocyte E-cad, stem cell EGFR, and organ size control. In the Drosophila midgut, a specific readout of EGFR activation is diphosphorylation of the effector kinase ERK (dpERK); endogenous diphosphorylation of ERK requires EGFR and occurs primarily in stem cells14–17 (Fig. 3d, e; Extended Data Figs. 3b and 6k). We found that enterocyte E-cad knockdown caused dpERK+ stem cells to increase, whereas overexpression caused them to decrease (Fig. 3a–c, Extended Data Fig. 3b). Furthermore, EGFR was necessary for the excess divisions and organ hyperplasia induced by knockdown of E-cad in apoptosis-inhibited enterocytes (Fig. 3f–g; Extended Data Fig. 3a). Thus, enterocyte E-cad inhibits stem cell EGFR to mediate homeostatic control of cell number and organ size.
Fig. 3. Enterocyte E-cad inhibits stem cell EGFR via a dispersed signal for homeostatic size control.
a–e, Immunostain for activated, diphospho-ERK (dpERK) following enterocyte manipulation of E-cad, without or with EGFR inhibition (AG1478, egfrtlsa). ERK activation in stem cells (Extended Data Fig. 6k) is repressed by E-cad and requires EGFR. See Extended Data Fig. 3b. f, Sizes of stem cell clones (means ± S.D; p-values, Mann-Whitney test) after induction of enterocyte p35 and E-cadRNAi, without or with AG1478. EGFR inhibition suppresses stem cell divisions. N=4–5 midguts per condition. g, Total cell counts (means ± S.D; p-values, unpaired t-test compared to control). Midgut hyperplasia (mexts>p35, E-cadRNAi) requires EGFR and enterocyte spi and krn. N=4 midguts per genotype or condition. See Extended Data Fig. 3a. h–k, The spatial distribution of dpERK cells around a single, E-cadRNAi enterocyte distinguishes direct and dispersed activation mechanisms (h). dpERK+ cells are both directly adjacent to and dispersed from GFP-marked, E-cadRNAi enterocytes (i), consistent with a dispersed mechanism. dpERK+ cells are infrequent near marked control enterocytes (j). Distribution of distances between dpERK+ cells and E-cadRNAi enterocytes (k, n=53 dpERK+ cells; N=4 midguts, 5 day induction). Green bar represents dpERK+ cells directly adjacent to enterocytes (0 µm). One of three replicate experiments shown in each graph. All scale bars, 25 µm.
How does E-cad on enterocytes control EGFR on stem cells? One possible mechanism involves direct, E-cad-EGFR interaction18; another involves a dispersed signal. To examine these possibilities, we generated isolated, GFP-marked enterocytes that were depleted of E-cad. Measuring the spatial distribution of dpERK+ stem cells surrounding each mutant enterocyte (Fig. 3h–j), we found a zone of strong EGFR activation within 25 µm and a second zone of weak activation from 25–50 µm (Fig. 3k). The spatial extent of these zones suggests the involvement of a dispersed signal.
Consistent with a dispersed signal, two enterocyte-derived EGFs, spitz (spi) and keren (krn), were necessary for organ hyperplasia caused by loss of E-cad in apoptosis-inhibited enterocytes (Fig. 3g, Extended Data Fig. 3a). Surprisingly, however, spi and krn were not induced by E-cad knockdown (Fig. 4a). The visceral muscle EGF vein, modulators of EGF star and argos, and egfr itself were also unaffected. In contrast, the obligate EGF protease rhomboid (rho) was markedly induced by E-cad knockdown and repressed by E-cad overexpression.
Fig. 4. Enterocyte apoptosis activates stem cell division by disrupting E-cad-controlled inhibition of rhomboid.
a, qPCR of whole midgut mRNAs following enterocyte-specific E-cad manipulation. E-cad represses rho but not EGFs (vn, spi, krn) or other EGF-related factors (egfr, aos, star). Red line indicates control. Means ± S.D. of 3 independent experiments. b, c, Enterocyte rho is required for ERK hyperactivation following depletion of enterocyte E-cad. See Extended Data Fig. 3b. d–f, rho-lacZ induction during physiological apoptosis. Immunostains for β-gal and activated Caspase-3 (Cas-3) mark identical enterocytes. Images show different fields in planar (d) and vertical (e) sections; dotted line in e indicates basal. Quantitation of Cas-3+, rho-lacZ+ enterocytes (f, means ± S.D). n=188 enterocytes from 3 experiments; N=3–4 midguts (6 days post-eclosion) per experiment. g–j, ERK activation is suppressed by apoptotic inhibition and depends on E-cad and rho. See Extended Data Fig. 3b. k, Total cell counts (means ± S.D; p values, unpaired t-test compared to control). Hyperplasia of mexts>p35, E-cadRNAi midguts requires rho, arm, and p120. Hyperplasia is induced by either rho or p120 alone. N=4 midguts per genotype. One of three replicate experiments shown. See Extended Data Fig. 3a. l, Model for steady-state equilibrium of division and death. All scale bars, 25 µm.
Rho operates within EGF-producing cells, where it cleaves EGF precursors for secretion19. Consistent with this function, we found that E-cad repressed rho-lacZ specifically in enterocytes (Extended Data Fig. 7c, d, f). Overexpression of rho in enterocytes caused dpERK+ stem cells to increase; conversely, rho depletion caused dpERK+ cells to virtually disappear (Extended Data Figs. 3b, 7m–o, s). Furthermore, combined depletion of rho and E-cad precluded the hyperactivation of ERK caused by depletion of E-cad alone (Fig. 4b, c). Altogether, these results imply that enterocyte E-cad inhibits stem cell EGFR by preventing EGF secretion through repression of rho.
Does this E-cad-Rho-EGFR relay couple stem cell divisions to enterocyte apoptosis? If so, then: (1) apoptotic enterocytes, which lose E-cad, should upregulate rho; (2) loss of E-cad in apoptotic enterocytes should underlie stem cell EGFR activation; and (3) exogenous manipulation of rho should alter organ size. We investigated each prediction. First, rho-lacZ predominantly marked apoptotic enterocytes during normal turnover (Fig. 4d–f). By comparison, the cardinal injury signal upd320,21 rarely marked apoptotic enterocytes; upd3 was also dispensable for EGFR activation (Extended Data Figs. 6l, 7i; Supplemental Discussion). Thus, rho is silenced in healthy enterocytes but upregulated in enterocytes undergoing physiological apoptosis.
To test the second prediction, we blocked enterocyte apoptosis and examined stem cell EGFR activation. ERK-activated stem cells were virtually absent following apoptotic inhibition but were restored, in a rho-dependent manner, by additional depletion of enterocyte E-cad (Fig. 4g–j, Extended Data Fig. 3b). These results demonstrate that the loss of E-cad in apoptotic enterocytes underlies EGFR activation in stem cells.
To investigate the third prediction, we manipulated enterocyte rho and measured cell number and organ size. Overexpression of rho in apoptosis-inhibited enterocytes led to organ hyperplasia (Fig. 4k, Extended Data Fig. 3a). Conversely, loss of rho in apoptosis-competent enterocytes resulted in organ atrophy (Extended Data Fig. 8). Moreover, combined loss of both rho and E-cad in apoptosis-inhibited enterocytes thwarted the hyperplasia that would have resulted from loss of E-cad alone (Fig. 4k, Extended Data Fig. 3a). These results show that downstream of E-cad, rho is the pivot point that balances division and death to sustain cellular equilibrium.
Finally, how does E-cad control rho expression? To address this, we examined three transcription factors whose nuclear localization is precluded by binding to junctional E-cad: β-catenin/Armadillo (Arm), p120-catenin (p120), and YAP/Yorkie (Yki)22. arm and p120, but not yki, were required in E-cad knockdown enterocytes for induction of rho and hyperactivation of stem cell EGFR (Extended Data Figs. 3b, 7a, g–l). In addition, arm and p120 were required for organ hyperplasia following combined E-cad knockdown and apoptotic inhibition (Fig 4k, Extended Data Fig. 3a). Conversely, overexpression of p120, but not activated armS10, was sufficient for rho induction, EGFR hyperactivation, and organ hyperplasia (Extended Data Figs. 3b, 7b–f, p–r). Thus, E-cad controls rho by inhibiting p120 and Arm, likely through physical sequestration at cell junctions.
Our results demonstrate that steady-state turnover is not driven by constitutive cycling of stem cells. Rather, healthy enterocytes enforce a default state of stem cell quiescence, while sporadic, apoptotic enterocytes trigger replacement divisions (Fig 4l). Because divisions are coupled to apoptosis, turnover remains zero-sum over time.
The molecular mechanism of coupling suggests a simple model for how, during continuous turnover, total cell number is held constant with such robust precision. Apoptotic enterocytes trigger stem cell divisions through loss of E-cad, which induces rho to permit secretion of mitogenic EGFs. Crucially, a single enterocyte can efficiently activate EGFR on stem cells within a ~25 µm radius (Fig. 3k). We propose that this local zone of activation enables organ size homeostasis. If, by chance, stem cells produce excess enterocytes, the stem cells’ physical spacing would increase; consequently, fewer stem cells would reside in the activation zone of the next dying enterocyte, and fewer divisions would result. Similarly, insufficient production of enterocytes would place more stem cells in the activation zone, and more divisions would result. We propose that the radii of individual activation zones, when integrated over the entire epithelium, sets total cell number and organ size. In this manner, localized cell-cell communication can give rise to tissue-level homeostatic equilibrium.
Methods
Drosophila Husbandry
Crosses utilizing the GAL4/GAL80ts system were performed at 18°C. Upon eclosion, adult animals remained at 18°C for 4 days, unless otherwise indicated. On adult day 4, animals were temperature shifted to 29°C to inactivate GAL80ts and induce GAL4-mediated expression. Midguts were harvested for immunostaining 4 days after induction, unless specified otherwise in the figure caption. All other crosses were performed at 25°C; refer to figure legends for individual timepoint information. Adult female flies were used in all experiments.
Fly Stocks
The following stocks were obtained from the Bloomington Stock Center: y, w; shg[mTomato]23, UAS-E-cadherinRNAi (TRiP.HMS00693, TRiP. JF02769, and TRiP.GL00646), UAS-echinoidRNAi (TRiP.GL00648), UAS-rhomboidRNAi (TRiP.JF03106), UAS-spitzRNAi (TRiP.HMS01120), UAS-armadilloRNAi (TRiP.JF01251), UAS-p120ctnRNAi (TRiP.HMC03276), UAS-yorkieRNAi (TRiP.JF03119), UAS-unpaired3RNAi (TRiP.HM05061), UAS-hisH2A:RFP, UAS-p35, UAS-diap1, UAS-rhomboid, UAS-armadilloS10, UAS-p120ctn, Egfrf24/T(2;3)TSTL, Egfrtsla/T(2;3)TSTL, cycE-lacZ, and 10xSTAT-GFP. UAS-kerenRNAi (KK104299) and UAS-armadilloRNAi (KK107344) were obtained from the Vienna Drosophila Resource Center. The following stocks were generous gifts: esg flp-out line6 (a gift from Bruce Edgar), mexGAL424 (a gift from Carl Thummel), UAS-E-cadherinDEFL (a gift from Margaret Fuller), rhoX81 (rho-lacZ) and Upd3.1-lacZ (gifts from Huaqi Jiang), UAS-groucho (a gift from Amir Orian). Other stocks used: esgGAL4, y w hsflp; X-15-29 w+ (‘split-lacZ’)8, y w; y+ X-15-33 (‘split-lacZ’)8, w UAS-CD8:GFP hsflp; tubGAL4; FRT82 tubGAL80, w; FRT82 (used in our prior study3). Detailed information on Drosophila genes and stocks is available from FlyBase (http://flybase.org/).
Immunohistochemistry and Microscopy
Samples were fixed, immunostained, and mounted as previously described3. Primary antibodies: mouse anti-β-galactosidase (1:400, Promega Z3781), mouse anti-Armadillo (1:100, DSHB N27A1), rabbit anti-cleaved caspase 3 (1:200, Cell Signaling, generous gift from D. Bilder3) rabbit anti-diphospho-ERK (1:400, Cell Signaling 4370P), goat anti-HRP-Cy3 (Cappel, 1:100) which stains stem cells and enteroblasts3, mouse anti-Coracle (1:50, DSHB C615.16), mouse anti-Discs large C615.16 (1:50, DSHB 4F3), and rabbit anti-phospho-histone H3, Ser 10 (1:1000, EMD Millipore). Secondary antibodies: Alexa Fluor 488-, 555- or 647-conjugated donkey anti-rabbit or anti-mouse IgGs (1:800, LifeTechnologies A31570, A11001, and A21244). Nuclei were stained with DAPI (LifeTechnologies, 1:1000). Actin was stained with SiR-Actin (Spiro-chrome, 1:500) or Alexa 647-conjugated phalloidin (1:100, LifeTechnologies). Samples were mounted in ProLong (LifeTechnologies). Imaging of samples was performed on a Leica SP8 confocal microscope, with serial optical sections taken at 3.5 µm intervals through the entirety of whole-mounted, immunostained midguts. Representative images are shown in all panels.
Regionalization of the Adult Midgut; Cell Counts and Size Measurements of the R4ab (P1–2) compartment
The Drosophila midgut is compartmentalized along its proximal-distal axis. Each compartment exhibits a characteristic digestive physiology, gene expression pattern, and stem cell division rate7,25. In general, stem cell clones do not cross compartment boundaries25. Our study focused specifically on two adjacent compartments, known alternatively as R4ab or P1–2, which comprise the major region of nutrient absorption. We observed that R4ab consistently exhibited complete cellular turnover between adult days 4–8, as indicated by esgF/O labeling (Fig. 1a–e, Extended Data Fig. 1f–g). Other midgut compartments exhibited variable, incomplete turnover during the same time period, consistent with prior reports; they were not analyzed in this study.
To perform total cell counts of R4ab, this region was first identified in confocal image stacks using morphological landmarks7 (Extended Data Fig 1b–e, g) and digitally isolated in Fiji. Bitplane Imaris software algorithms were applied to generate three-dimensional organ reconstructions and comprehensively count individual cell nuclei by mapping DAPI signals to Imaris surface objects. For analysis of esgF/O midguts, GFP+ cells were additionally counted by mapping DAPI/GFP colocalization signals to Imaris surface objects. R4ab lengths were measured by a spline through the center of individual midguts in Fiji.
Split-lacZ Clone Induction and Analysis
Animals were raised at 18°C and shifted to 29°C four days post-eclosion. Split-lacZ clone induction8 was performed by subjecting animals to two 30-min, 38.5°C heat shocks separated by a 5-min chill on ice. Four days after clone induction, midguts were immunostained and clones in the R4ab region were identified and analyzed by visual examination of serial confocal sections. Clones in regions outside R4ab were excluded from analysis. Clone size was measured as the number of contiguous cells in one discrete clone, as previously described3. No labeled cells were observed in the absence of 38.5°C heat shock.
To ensure that clone counts comprised exclusively stem cell clones and excluded any non-stem cell (transient) clones that were directly labeled by the heat shock, our split-lacZ clonal analyses incorporated two, redundant safeguards. First, a 4-day chase period was included between heat-shock induction and subsequent clonal analysis. Enteroblasts/enterocytes that were directly labeled by the heat shock would have been lost during the succeeding chase period. Confirming that transient clones were nearly absent, only 1–3 single labeled enterocytes were observed per midgut R4ab region after the 4-day chase. As a second safeguard, all single, labeled enterocytes were excluded from our clone counts. This induction protocol resulted in an average of 6–8 clones per midgut R4ab gut region, depending on experimental genotype.
EdU Labeling
Animals were fed yeast paste prepared with 1mg/mL EdU (5-ethynyl-2’-deoxyuridine, Invitrogen) dissolved in water. After 2 days, tissues were fixed as described above and stained for EdU using the Click-iT EdU kit (Invitrogen) based on manufacturer’s protocol.
Sytox Staining
Sytox Green (ThermoFisher, 5mM in DMSO) or Sytox Orange (ThermoFisher, 5mM in DMSO) was diluted 1:5,000 in 5% sucrose. Sytox solution was fed to animals in an empty vial for 5–6 hours26, after which midguts were dissected and mounted in ProLong (LifeTechnologies). Because Sytox is incompatible with fixation, live organs were imaged immediately after mounting.
MARCM Clone Induction
MARCM clone inductions27 were performed by subjecting animals to two 30-min, 38.5°C heat shocks separated by a 5-min chill on ice. For single-enterocyte MARCM clones, animals were dissected 5 days post-induction and terminal clones consisting of one GFP+ enterocyte (identified by its polyploid nucleus) were selected for analysis. GFP+ enterocytes were excluded from analysis if another GFP+ clone was present within an 80 µm radius. Fiji was used to measure the distance between the plasma membrane of the nearest GFP+ enterocyte and the center of dpERK+ stem cells within a 60 µm radius. For mosaic analyses of multicell MARCM clones, animals were fed Sytox three days post-induction and dissected. The proportion of labeled clone cells (GFP+) that were also Sytox+ was quantified.
AG1478 Drug Treatment
Stocks of AG1478 (Sigma) were dissolved in EtOH and subsequently diluted in dH2O to reach a working concentration of 100 µM AG1478 (in 0.02% EtOH). This 100 µM stock solution was used to prepare yeast paste, which was fed to animals as a supplement to their standard cornmeal-molasses diet for the duration of induced gene expression.
Smurf Assay
Smurf assays were conducted by feeding adult animals yeast paste containing 2.5% Brilliant Blue FCF (Sigma) and scoring animals for leakage of dye into the abdomen. Animals were scored as ‘non-Smurf’ if the blue dye was confined to the GI tract and ‘Smurf’ if blue dye leaked outside the GI tract. As a positive control, animals were fed dye in conjunction with 1% SDS.
qRT-PCR
mRNA was extracted from midguts (5 animals/experiment) followed by cDNA synthesis with Invitrogen SuperStrand III First Script Super Mix (Invitrogen). Real-time PCR was performed using the relative standard curve method with SYBR GreenER Supermix (Invitrogen) on a StepOnePlus ABI machine. Expression levels were normalized to mexGAL4ts>CD4-GFP midguts; mef2 transcripts were used as a reference3.
Statistical Analysis
All statistical analyses were performed using Graphpad Prism 6. For comparisons of clone size distributions, unpaired two-tailed Mann-Whitney tests were used to assess statistical significance. (Clone size distributions are non-normal, independent, and derived from a simple random sample.) For comparisons of cell numbers and gut length, unpaired two-tailed t-tests were used to assess statistical significance. (Organ cell number and size distributions are normal, independent, and derived from a simple random sample.) For comparisons of rho gene expression, unpaired two-tailed t-tests were used to assess statistical significance.
Study Design
Sample sizes were chosen based on our previous study3, which also characterized changes in organ cell number and clone sizes. In split-lacZ experiments, single enterocyte clones were excluded from analysis. No other exclusion criteria were applied. No sample randomization or blinding was performed, although automated, Imaris-based computer algorithms were used to analyze and quantify most data in this study.
Extended Data
Extended Data Fig. 1. Midgut lineage and morphology, esgF/O labeling system, and work-flow for semi-automated cell counting.
a, Lineage of the adult Drosophila midgut29,30. Stem cells are, in general, the only cells capable of division. Asymmetric stem cell divisions typically produce absorptive enterocytes; less frequently, they produce secretory enteroendocrine cells. Enterocytes arise through direct maturation of transient, post-mitotic intermediates called enteroblasts. Stem and enteroblast cells express the Snail-family transcription factor escargot (esg). b, Compartments of the female adult midgut7,25,31. The R4ab compartment (also known as P1–225) was used for all experiments in this study. Schematic adapted from Ref. 77. c–e, Identification of R4ab using morphological landmarks. As defined in Ref. 77, R4ab is bounded by: (1) the apex of the midgut tube’s most distal 180° turn (blue arrowheads in d) and (2) the first prominent muscle constriction distal to this 180° turn (red arrowheads in d). e, The R4ab distal muscle constriction (red arrowheads) is particularly apparent in confocal optical sections7. Visceral muscle stained with phalloidin. Midguts in panels c–d and e are two different samples. f, Genetic schema of the esgF/O system6. Stem and enteroblast cells are induced to express heritable GFP by temperature shift from 18°C to 29°C. The temperature shift inactivates GAL80ts, which allows esgGAL4 to drive expression of both UAS-GFP and UAS-flp in stem and enteroblast cells. In these cells, flp recombinase renders GFP expression permanent and heritable by excising a CD2 ‘flp-out’ cassette to generate a functional actGAL4; once generated, actGAL4 drives expression of UAS-GFP (and UAS-flp) irrespective of cell type. Thus, after temperature shift, all mature cells that arise from undifferentiated cells will express GFP. g, Pipeline for semi-automated, comprehensive cell counts of 3D, reconstructed midgut regions. (1) Confocal microscope z-stacks capturing the entire depth of the organ are visualized in Fiji. (2) The R4ab region of the midgut (yellow outline) is digitally isolated and exported to Imaris. (Only the top half of the gut tube is shown.) Note that different midgut regions have different rates of turnover: R4ab undergoes complete turnover between adult days 4–8 (at 29°C). However, other regions undergo slower turnover, as shown by large unlabeled regions outside of R4ab. See Methods for further discussion. (3) To quantify total cells, nuclei (DAPI) are mapped to surface objects using Imaris. To quantify newly-added cells in the esgF/O system, GFP+ cells are recognized in Imaris by co-localization of GFP and DAPI channels, and subsequently mapped to surface objects. Scale bars: 100µm.
Extended Data Fig. 2. Genetic schema of system to simultaneously manipulate enterocyte expression and trace stem cell divisions.
a, Detailed explanation of the genetic system in Fig. 1f. Animals are raised at 18°C; at this temperature, GAL80ts represses mex-driven GAL4 in enterocytes (mexts), and lacZ labeling of stem cells is not induced. At 4 days post-eclosion, animals are temperature-shifted to 29°C; consequent inactivation of GAL80ts allows mexGAL4 to express genes of interest (UAS-gene X) specifically in enterocytes. After 1 day of UAS gene expression (5 days post-eclosion), animals are shifted to 38.5°C for one hour to induce ubiquitous expression of flp recombinase, which is under control of a heat-shock promoter (hs-flp). Flp catalyzes trans-recombination of the two FRTs to place the α-tubulin promoter upstream of the promoter-less nls-lacZ cassette and, consequently, turn on permanent nls-lacZ expression. After heat shock, animals are returned to 29°C to maintain UAS-transgene expression. Midguts are harvested for clonal analysis 4 days after the 38.5°C heat shock (9 days post-eclosion). See also Methods. b, Validation of genetic system using mexts>his2av::RFP. β-galactosidase marks a stem cell clone (outlined) in a background of His2av::RFP+ enterocytes. Within the 5-cell clone, only the enterocyte (yellow asterisk, polyploid) expresses his2av::RFP.
Extended Data Fig. 3. Quantifications of organ size and EGFR activation in genetically manipulated midguts.
a, Lengths of the R4ab compartment. N=10–12 midguts per genotype or condition, analyzed after 4 days of UAS-transgene expression. b, dpERK+ cells in the R4ab compartment. N=4 midguts per genotype or condition, analyzed after 2 days of UAS-transgene expression. One of two replicate experiments was quantified. In a and b, values are means ± S.D. p values: unpaired t-test, compared to control.
Extended Data Fig. 4. Analysis of epithelial architecture, polarity, and barrier function.
a–f, Apoptotic inhibition (b, e) or E-cad depletion (c, f) in enterocytes does not disrupt either epithelial architecture or apical-basal polarity. Images show vertical sections through the midgut epithelium after 4 days of either mexts>p35 or mexts>E-cadRNAi expression. Enterocytes remain as a coherent monolayer. Apical-basal polarity is intact, as revealed by immunolocalization of apical, actin-rich microvilli (a–f; SiR-Actin, red) and of two apico-lateral septate junction proteins, Coracle (a–c, green) and Discs-large (d–f, green). At the basal surface of the epithelium (white dotted lines), midgut visceral muscle cells stain brightly for actin and Discs-large. Scale bars: 25µm. g–j, Depletion of E-cad in enterocytes does not compromise the intestinal barrier. To test the intestinal barrier, animals were subjected to Smurf assays in which a blue, non-absorbable food dye is administered by feeding. The dye remains within the midgut when the barrier is intact (g, non-Smurf) but leaks into the body cavity when the barrier is compromised, such as after consumption of 1% SDS (h, Smurf). After 10 days of mexts>E-cadRNAi expression, midguts still retain the blue dye; no Smurf phenotypes are observed (i–j).
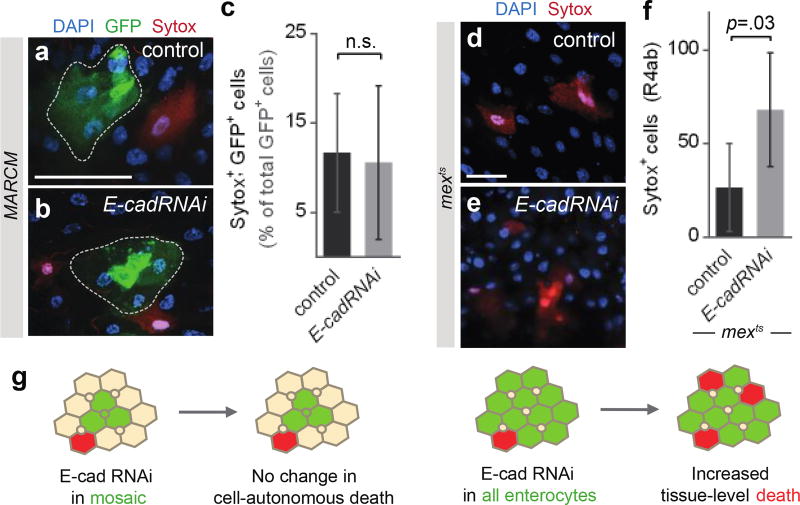
Extended Data Fig. 5. Depletion of E-cad has distinct cell-autonomous and tissue-level effects on cell death.
In Fig. 2h, total cell counts show that midguts accumulate excess cells when E-cad is depleted from apoptosis-inhibited enterocytes but not apoptosis-competent enterocytes. To shed light on this difference, we examined whether E-cad depletion itself promotes cell death. Two approaches, mosaic knockdown and pan-enterocyte knockdown, were used to distinguish direct, cell-autonomous effects from indirect, tissue-level effects. a–c, Mosaic knockdown of E-cad does not promote cell-autonomous death. To generate a mosaic epithelium, MARCM labeling27 was used to induce sparse, multicellular, GFP-marked clones in a background of unmarked, genetically unperturbed cells. a–b, Dotted outlines show representative control and E-cadRNAi clones (green). Sytox (red) identifies dying cells. c, Percentage of GFP+ cells that are also Sytox+. Dying cells occur with near-equal frequency within control and E-cadRNAi clones. N=5 midguts per genotype, analyzed 9 days after clone induction; n=873 cells in control clones and 698 cells in E-cadRNAi clones. d–f, Pan-enterocyte knockdown of E-cad promotes cell death, likely through a non-autonomous effect. d–e, Representative images of mexts control and mexts>E-cadRNAi epithelia. Sytox (red) identifies dying cells. f, Quantification of Sytox+ cells in the R4ab compartment. The number of dying cells increases ~2.5× in E-cadRNAi midguts compared to control. N=5 midguts per genotype, analyzed after 3 days of transgene induction. In both c and f, values are means ± S.D; comparisons by unpaired t-test. Scale bars are 25 µm. g, Summary. The unaltered frequency of dying cells in E-cadRNAi mosaic clones indicates that loss of E-cad does not cause cell-autonomous death. This result suggests that elevated death in mexts>E-cadRNAi guts is a non-autonomous, tissue-level effect, possibly due to excess divisions (Fig 2b) and consequent crowding. These findings may explain why p35, E-cadRNAi guts accumulate excess cells whereas E-cadRNAi guts retain a normal number of cells (Fig. 2h).
Extended Data Fig. 6. Loss of enterocyte E-cad activates EGFR, but not Wg, Hpo, or Upd/JAK/STAT.
a, Effect of enterocyte E-cad depletion on target mRNAs of known midgut regulatory pathways. mRNAs were measured by qPCR of mexts control or mexts>E-cadRNAi midguts. Relative to control (red line), mRNAs do not increase for: the Wg targets frizzled-3 (fz3) and senseless (sens), the Hpo/Yki targets expanded (ex) and diap1, the injury-associated cytokines upd and upd3, and the JAK/STAT target windpipe (wdp). The other JAK/STAT target, Socs36E, is elevated, likely reflecting its occasional activation in enterocytes (panel f). By comparison, the EGFR target pointed (pnt) is slightly increased, and the EGFR target cyclinE (cycE) is substantially increased. Values are means ± S.D. from 3 independent experiments. Midguts analyzed 4 days post-induction. b–d, The number of upd3-lacZ+ enterocytes in the R4ab compartment is unchanged by enterocyte E-cad depletion. e–g, The number of 10XSTAT-GFP+ diploid cells in R4ab is unchanged by enterocyte E-cad depletion. Occasional activation of 10XSTAT-GFP+ occurs in E-cad-depleted enterocytes (asterisk in f), consistent with elevated Socs36E (panel a). h–j, The number of cycE+ diploid cells in R4ab increases following enterocyte E-cad depletion. In panels d, g, and j, values are means ± S.D of 4 midguts, analyzed 2 days post-induction; comparisons by unpaired t-test. k, dpERK immunostaining is limited to stem cells (HRP+, Su(H)lacZ−; arrowheads in k' and k") and does not mark enteroblasts (HRP+, Su(H)lacZ+; asterisks in k' and k"), even in mexts>E-cadRNAi midguts14,16. l, Expression of upd3 is not associated with physiological apoptosis. Most enterocytes that express upd3-lacZ are non-apoptotic, as assessed by staining for cleaved Caspase-3. Values are means ± S.D of 4 midguts, analyzed 6 days post-eclosion. Representative images in all panels. Scale bars: 25 µm.
Extended Data Fig. 7. Two E-cad-associated transcription factors, Armadillo and p120-catenin, activate rho following loss of E-cad in enterocytes.
a, rho mRNA levels were measured by qPCR of either mexts control (red line) or mexts>E-cadRNAi midguts, the latter with additional manipulation of candidate rho regulators as indicated. Five candidates were examined: Yki, a transcriptional co-activator in the Hpo pathway; Groucho, a co-repressor known to target rho in some tissues32,33; puckered (puc), an inhibitor of Basket/JNK, the latter of which can enhance EGF signaling; and Arm and p120, co-activators that are inhibited by sequestration at E-cad adherens junctions. Knockdown of either arm or p120 significantly reduces rho activation. b, Overexpression of p120, but not armS10, in enterocytes is sufficient to increase rho mRNAs; control (red line). In a and b, values are means ± S.D. of 3 independent experiments; midguts were analyzed 4 days post-induction. c–f, Depletion of E-cad or overexpression of p120 induces rho-lacZ in enterocytes and not in diploid cells. f, Quantification. g–l, Enterocyte arm and p120, but not yki or upd3, are necessary for activation of stem cell EGFR (dpERK immunostain) following loss of E-cad. m–o, Enterocyte rho is necessary and sufficient for activation of stem cell EGFR. p–r, Enterocyte p120, but not arm, is sufficient to activate stem cell EGFR. See also Extended Data Fig. 3b. s, Overexpression of enterocyte rho increases the number of mitotic (phospho-histone H3+) stem cells. In f and s, values are means ± S.D of N=4 midguts, assessed after 2 days of transgene expression. p values: unpaired t-test, comparisons to control. Representative images shown in all panels. Scale bars: 25 µm.
Extended Data Fig. 8. Loss of rho, arm, or p120 in enterocytes results in organ atrophy.
a, Total cell counts. Depletion of rho, arm, or p120 in enterocytes reduces total cells compared to control. Values are means ± S.D from 1 of 3 replicate experiments. N=4 midguts per genotype, analyzed after 6 days of induction. b, Depletion of enterocyte rho, arm, or p120 reduces the length of the R4ab compartment compared to control. N=10–12 midguts per genotype, analyzed after 6 days of induction. In a and b, comparisons to control by unpaired t-test. c–d, Depletion of enterocyte rho leads to organ atrophy. Representative whole mount images. A, anterior; P posterior. Scale bar: 200µm.
Supplementary Material
Acknowledgments
J.L. was supported by NSF GRFP DGE-114747 and NIH T32GM007276. This work was supported by NIH R03DK104027 and R01GM116000-01A1 to L.E.O. Confocal microscopy was performed at the Stanford Beckman Cell Sciences Imaging Facility (NIH 1S10OD01058001A1). We thank D. Bilder for the gift of cCas-3 antibody; the Developmental Studies Hybridoma Bank for other antibodies; D. Bilder, B. Edgar, M. Fuller, H. Jiang, B. Ohlstein, C. Thummel, the Bloomington Drosophila Stock Center (NIH P40OD018537), the TRiP at Harvard Medical School (NIH/NIGMS R01-GM084947), and the Vienna Drosophila Resource Center for fly stocks; J. Axelrod, M. Goodman, M. Fuller, W.J. Nelson, R. Nusse, M. Krasnow, T. Nystul, and D. Fox for comments on the manuscript; and M. Mirvis, B. Benham-Pyle, N. Pierce, and D. Gordon for helpful discussions.
Footnotes
The authors declare no competing interests.
Author contributions: J.L. and L.E.O. designed the experiments and wrote the manuscript. J.L. and S.N. prepared microscopy specimens. J.L., S.B., and S.N. performed confocal microscopy. J.L. performed all other experiments, genetic crosses, data analysis, and statistical analysis.
References
- 1.Leblond CP, Stevens CE. The constant renewal of the intestinal epithelium in the albino rat. Anat Rec. 1948;100:357–377. doi: 10.1002/ar.1091000306. [DOI] [PubMed] [Google Scholar]
- 2.Pellettieri J, Sanchez Alvarado A. Cell turnover and adult tissue homeostasis: from humans to planarians. Annu Rev Genet. 2007;41:83–105. doi: 10.1146/annurev.genet.41.110306.130244. [DOI] [PubMed] [Google Scholar]
- 3.O'Brien LE, Soliman SS, Li X, Bilder D. Altered modes of stem cell division drive adaptive intestinal growth. Cell. 2011;147:603–614. doi: 10.1016/j.cell.2011.08.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.O'Brien LE, Bilder D. Beyond the niche: tissue-level coordination of stem cell dynamics. Annu Rev Cell Dev Biol. 2013;29:107–136. doi: 10.1146/annurev-cellbio-101512-122319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Apidianakis Y, Tamamouna V, Teloni S, Pitsouli C. Advances in Insect Physiology. Vol. 52. Ligoxygakis Petros: Academic Press; 2017. pp. 139–178. [Google Scholar]
- 6.Jiang H, et al. Cytokine/Jak/Stat signaling mediates regeneration and homeostasis in the Drosophila midgut. Cell. 2009;137:1343–1355. doi: 10.1016/j.cell.2009.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Buchon N, et al. Morphological and molecular characterization of adult midgut compartmentalization in Drosophila. Cell Rep. 2013;3:1725–1738. doi: 10.1016/j.celrep.2013.04.001. [DOI] [PubMed] [Google Scholar]
- 8.Harrison DA, Perrimon N. Simple and efficient generation of marked clones in Drosophila. Curr Biol. 1993;3:424–433. doi: 10.1016/0960-9822(93)90349-s. [DOI] [PubMed] [Google Scholar]
- 9.Takeishi A, et al. Homeostatic epithelial renewal in the gut is required for dampening a fatal systemic wound response in Drosophila. Cell Rep. 2013;3:919–930. doi: 10.1016/j.celrep.2013.02.022. [DOI] [PubMed] [Google Scholar]
- 10.Hermiston ML, Gordon JI. In vivo analysis of cadherin function in the mouse intestinal epithelium: essential roles in adhesion, maintenance of differentiation, and regulation of programmed cell death. J Cell Biol. 1995;129:489–506. doi: 10.1083/jcb.129.2.489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Steinhusen U, et al. Cleavage and shedding of E-cadherin after induction of apoptosis. J Biol Chem. 2001;276:4972–4980. doi: 10.1074/jbc.M006102200. [DOI] [PubMed] [Google Scholar]
- 12.Maeda K, Takemura M, Umemori M, Adachi-Yamada T. E-cadherin prolongs the moment for interaction between intestinal stem cell and its progenitor cell to ensure Notch signaling in adult Drosophila midgut. Genes Cells. 2008;13:1219–1227. doi: 10.1111/j.1365-2443.2008.01239.x. [DOI] [PubMed] [Google Scholar]
- 13.Choi NH, Lucchetta E, Ohlstein B. Nonautonomous regulation of Drosophila midgut stem cell proliferation by the insulin-signaling pathway. Proc Natl Acad Sci U S A. 2011;108:18702–18707. doi: 10.1073/pnas.1109348108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Biteau B, Jasper H. EGF signaling regulates the proliferation of intestinal stem cells in Drosophila. Development. 2011;138:1045–1055. doi: 10.1242/dev.056671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Buchon N, Broderick NA, Kuraishi T, Lemaitre B. Drosophila EGFR pathway coordinates stem cell proliferation and gut remodeling following infection. BMC Biol. 2010;8:152. doi: 10.1186/1741-7007-8-152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jiang H, Grenley MO, Bravo MJ, Blumhagen RZ, Edgar BA. EGFR/Ras/MAPK signaling mediates adult midgut epithelial homeostasis and regeneration in Drosophila. Cell Stem Cell. 2011;8:84–95. doi: 10.1016/j.stem.2010.11.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Strand M, Micchelli CA. Regional control of Drosophila gut stem cell proliferation: EGF establishes GSSC proliferative set point & controls emergence from quiescence. PLoS One. 2013;8:e80608. doi: 10.1371/journal.pone.0080608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Qian X, Karpova T, Sheppard AM, McNally J, Lowy DR. E-cadherin-mediated adhesion inhibits ligand-dependent activation of diverse receptor tyrosine kinases. EMBO J. 2004;23:1739–1748. doi: 10.1038/sj.emboj.7600136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shilo BZ. Developmental roles of Rhomboid proteases. Seminars in Cell & Developmental Biology. 2016;60:5–9. doi: 10.1016/j.semcdb.2016.07.014. [DOI] [PubMed] [Google Scholar]
- 20.Zhou F, Rasmussen A, Lee S, Agaisse H. The UPD3 cytokine couples environmental challenge and intestinal stem cell division through modulation of JAK/STAT signaling in the stem cell microenvironment. Dev Biol. 2013;373:383–393. doi: 10.1016/j.ydbio.2012.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Osman D, et al. Autocrine and paracrine unpaired signaling regulate intestinal stem cell maintenance and division. J Cell Sci. 2012;125:5944–5949. doi: 10.1242/jcs.113100. [DOI] [PubMed] [Google Scholar]
- 22.McCrea PD, Gottardi CJ. Beyond beta-catenin: prospects for a larger catenin network in the nucleus. Nat Rev Mol Cell Biol. 2016;17:55–64. doi: 10.1038/nrm.2015.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huang J, Zhou W, Dong W, Watson AM, Hong Y. From the Cover: Directed, efficient, and versatile modifications of the Drosophila genome by genomic engineering. Proc Natl Acad Sci U S A. 2009;106:8284–8289. doi: 10.1073/pnas.0900641106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen H, Zheng X, Zheng Y. Age-associated loss of lamin-B leads to systemic inflammation and gut hyperplasia. Cell. 2014;159:829–843. doi: 10.1016/j.cell.2014.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Marianes A, Spradling AC. Physiological and stem cell compartmentalization within the Drosophila midgut. Elife. 2013;2:e00886. doi: 10.7554/eLife.00886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kolahgar G, et al. Cell Competition Modifies Adult Stem Cell and Tissue Population Dynamics in a JAK-STAT-Dependent Manner. Dev Cell. 2015;34:297–309. doi: 10.1016/j.devcel.2015.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee T, Luo L. Mosaic analysis with a repressible cell marker for studies of gene function in neuronal morphogenesis. Neuron. 1999;22:451–461. doi: 10.1016/s0896-6273(00)80701-1. [DOI] [PubMed] [Google Scholar]
- 28.Shaw RL, et al. The Hippo pathway regulates intestinal stem cell proliferation during Drosophila adult midgut regeneration. Development. 2010;137:4147–4158. doi: 10.1242/dev.052506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Micchelli CA, Perrimon N. Evidence that stem cells reside in the adult Drosophila midgut epithelium. Nature. 2006;439:475–479. doi: 10.1038/nature04371. [DOI] [PubMed] [Google Scholar]
- 30.Ohlstein B, Spradling A. The adult Drosophila posterior midgut is maintained by pluripotent stem cells. Nature. 2006;439:470–474. doi: 10.1038/nature04333. [DOI] [PubMed] [Google Scholar]
- 31.O'Brien LE. Regional specificity in the Drosophila midgut: setting boundaries with stem cells. Cell Stem Cell. 2013;13:375–376. doi: 10.1016/j.stem.2013.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Daniels DL, Weis WI. Beta-catenin directly displaces Groucho/TLE repressors from Tcf/Lef in Wnt-mediated transcription activation. Nat Struct Mol Biol. 2005;12:364–371. doi: 10.1038/nsmb912. [DOI] [PubMed] [Google Scholar]
- 33.Zhang T, Du W. Groucho restricts rhomboid expression and couples EGFR activation with R8 selection during Drosophila photoreceptor differentiation. Dev Biol. 2015;407:246–255. doi: 10.1016/j.ydbio.2015.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.