Abstract
Scavenger receptor Class B type 1 (SR-B1) is a lipid transporter and sensor. In intestinal epithelial cells, SR-B1-dependent lipid sensing is associated with SR-B1 recruitment in raft-like/detergent-resistant membrane domains and interaction of its C-terminal transmembrane domain with plasma membrane cholesterol. To clarify the initiating events occurring during lipid sensing by SR-B1, we analyzed cholesterol trafficking and raft-like domain composition in intestinal epithelial cells expressing wild-type SR-B1 or the mutated form SR-B1-Q445A, defective in membrane cholesterol binding and signal initiation. These features of SR-B1 were found to influence both apical cholesterol efflux and intracellular cholesterol trafficking from plasma membrane to lipid droplets, and the lipid composition of raft-like domains. Lipidomic analysis revealed likely participation of d18:0/16:0 sphingomyelin and 16:0/0:0 lysophosphatidylethanolamine in lipid sensing by SR-B1. Proteomic analysis identified proteins, whose abundance changed in raft-like domains during lipid sensing, and these included molecules linked to lipid raft dynamics and signal transduction. These findings provide new insights into the role of SR-B1 in cellular cholesterol homeostasis and suggest molecular links between SR-B1-dependent lipid sensing and cell cholesterol and lipid droplet dynamics.
Keywords: scavenger receptor, cholesterol, lipid raft, sphingolipid, lipid trafficking
1. Introduction
Cellular transport and trafficking of cholesterol, as well as the molecular composition of the plasma membrane, are critically involved in the governance of cell fate and cell metabolism. The scavenger receptor Class B type 1 (SR-B1) is primarily known for its function as a cholesterol transporter, particularly in the liver, where it is responsible for the last step in the reverse cholesterol transport pathway, which entails the selective uptake of cholesterol ester from High Density Lipoprotein (HDL). SR-B1 also mediates the bidirectional transport of cholesterol and phospholipids from peripheral tissues or cells to lipoproteins [1]. In the intestine, the functions of SR-B1 regarding cholesterol and lipid metabolism remain unclear. Although SR-B1 is expressed at the apical membrane of enterocytes [2–4], which are the absorptive cells of the intestinal epithelium, its role in intestinal cholesterol uptake has been questioned since NPC1L1 has been demonstrated to be responsible for 70% of cholesterol entry in enterocytes [5–7]. Moreover SR-B1-null mice [8] and mice overexpressing SR-B1 in the proximal intestine [9] do not display alterations in intestinal cholesterol absorption or in trans-intestinal cholesterol efflux [10]. However, other reports showed that SR-B1 is involved in the secretion of triglyceride-rich lipoproteins by enterocytes upon lipid supply [9, 11, 12].
In addition to serving in cholesterol movement, SR-B1 functions as a signal transducer in certain cell types, particularly in endothelial cells and in intestinal epithelium. In endothelial cells, the binding of HDL to SR-B1 triggers the sequential activation of protein kinases, among which the activating phosphorylation of Src kinase represents the earliest signaling event identified [13]. This signaling cascade leads to the production of nitric oxide and it promotes endothelial cell migration, which both contribute to the maintenance of endothelial monolayer integrity by HDL and SR-B1 [13]. In Caco-2/TC7 enterocyte-like cells, SR-B1 triggers intracellular signaling in response to the supply, at the apical pole of the cells, of postprandial lipid micelles (PPM), which mimic the form and composition of the dietary lipids that are present in the gut lumen following a meal. The sensing of PPM by SR-B1 activates MAP kinases and the trafficking of apolipoprotein B, the structural apolipoprotein required for triglyceride-rich lipoprotein assembly and secretion, from the apical region of the cell to basolateral secretory domains [14, 15]. These intracellular events are associated with the recruitment of SR-B1 into raft-like membrane domains [14, 15], which are known to be signaling platforms [16].
Studies have been conducted to analyze the molecular mechanisms involved in SR-B1-dependent lipid sensing. It has been established that the C-terminal transmembrane domain (CTTM) and the C-terminal cytosolic domain of SR-B1 are required for its property to sense lipid movement and initiate intracellular signaling [17]. The discovery of the effects of a single point mutation in the SR-B1 CTTM domain, which represents the unique domain of SR-B1 able to interact with plasma membrane cholesterol [17, 18], highlighted the importance of plasma membrane cholesterol for SR-B1-dependent lipid sensing and resulting signaling [15]. This mutated form of SR-B1, created by the replacement of the glutamine residue in position 445 of SR-B1 amino acid sequence by an alanine residue (SR-B1-Q445A), provoked a marked decrease in the capacity of SR-B1 to bind plasma membrane cholesterol, whereas the capacities of the receptor to bind HDL and to transport cholesterol were unmodified [15]. In intestinal epithelial cells and endothelial cells expressing SR-B1-Q445A, the addition of ligand (PPM and HDL respectively) failed to activate signaling and subsequent cellular events [15]. Moreover, it was shown that SR-B1-Q445A is not recruited in raft-like membrane domains of intestinal epithelial cells after a PPM supply, contrary to what is observed for wild-type SR-B1 [14, 15]. The initiating cellular events involved in SR-B1-dependent lipid sensing remain unknown. However, plasma membrane cholesterol movement, through cyclodextrin treatment, is sufficient to reproduce, in intestinal epithelial cells and endothelial cells, all the signaling cascades and subcellular events in wild-type SR-B1-expressing cells while SR-B1-Q445A transfected cells are unresponsive [15]. Altogether these results emphasize the critical importance of plasma membrane cholesterol in SR-B1-dependent intracellular signaling, and they suggest that the initiating events in SR-B1-related lipid sensing take place at the plasma membrane and may involve cholesterol trafficking. Although it has been shown that SR-B1 reorganizes the free cholesterol pool in the plasma membrane [19–21], the modifications in the plasma membrane microenvironment that occur upon SR-B1 lipid sensing remain largely unknown.
The purpose of the present study was to analyze the early cellular events involved in intestinal epithelial SR-B1-dependent lipid sensing by analyzing cholesterol trafficking and the remodeling of raft-like membrane domains upon supply of lipid micelles.
2. Materials and Methods
2.1 Cell culture and lipid micelle supply
Caco-2/TC7 cell line is a clonal population of the human colon carcinoma-derived Caco-2 cells, which reproduces to a high degree most of the morphological and functional characteristics of enterocytes [22, 23]. These cells were stably transfected with constructs expressing mouse WT SR-B1 or SR-B1-Q445A as previously described [15]. The presence of SR-B1 at plasma membrane (Suppl. Fig. 1) was shown by FACS analysis (BD LSRII FACS system and FlowJo software) performed on unpermeabilized cells as described in [24], using an anti-SR-B1 antibody (NB400-113 Novus Biologicals, Lille, France) and an anti-rabbit antibody conjugated to phycoerythrin (111-116-144 Jackson ImmunoResearch Laboratories Inc. West Grove, PA, USA). As previously described [15], SR-B1-Q445A has a dominant negative effect on signaling triggered by endogenous SR-B1 in both endothelial and Caco-2/TC7 cells. Cells were tested for mycoplasma contamination and found clean. In all experiments, cells were cultured on filter supports for 3 weeks to obtain fully differentiated enterocyte-like cells. Lipid micelles (2 mM sodium taurocholate, 0.6 mM oleic acid, 0.2 mM lysophosphatidylcholine, 0.05 mM cholesterol, and 0.2 mM monoacylglycerol) were prepared in serum-free medium as previously described [14, 15, 25] and added to the upper compartment for the indicated times. When appropriate, lipid micelles were supplemented with 5μM BODIPY-cholesterol (Avanti polar lipids, Alabaster, AL, USA) or with 5μCi/ml [1,2-3H(N)]-cholesterol (57,6Ci/mmol, PerkinElmer, Villebon-sur-Yvette, France). Transfection of Caco-2/TC7 cells with control RNAi and SR-B1 RNAi were performed as previously described [26].
2.2 Cholesterol labelling, trafficking and distribution
The intracellular distribution of cholesterol was analyzed in pulse-chase experiments. Caco-2/TC7 cells were incubated with BODIPY-cholesterol-containing lipid micelles for 1h hour, washed twice with warm phosphate-buffered saline (PBS), and then incubated with non-fluorescent lipid micelles for one more hour. At the end of the experiment cells were rinsed with cold PBS and prepared for confocal microscopy analysis.
The trafficking of cholesterol from the plasma membrane towards intracellular domains was analyzed as described previously [27]. Briefly, BODIPY-cholesterol was combined with 370 mM methyl- β-cyclodextrin (MβCD, Sigma, Saint-Quentin Fallavier, France) in a 100:1 molar ratio (MβCD/BODIPY-chol). Caco-2/TC7 cells were incubated at their apical pole with MβCD/BODIPY-chol complex in serum-free medium for 1 min at 37°C. The final concentration of MβCD and BODIPY-cholesterol were 0.185 mM and ~1μM, respectively. Cells were rinsed three times in warm PBS and then incubated with serum-free medium for the indicated times and analyzed by confocal microscopy.
Free cholesterol was visualized by filipin staining. Caco-2/TC7 cells were incubated or not with 3μg/ml U-18666A (Sigma, Saint-Quentin Fallavier, France) added in both compartments of the filter support for 18h. Cells were then rinsed three times with PBS, fixed with 4% paraformaldehyde (PFA), and incubated with 0.05mg/ml filipin (Sigma, Saint-Quentin Fallavier, France) in PBS/10%serum for 15 min. After extensive washing with PBS, cells were analyzed by confocal microscopy using a UV filter set.
2.3 Cholesterol efflux analysis
Caco-2/TC7 cells were cultured on filter supports and incubated with [1,2-3H(N)]-cholesterol containing lipid micelles in the apical compartment for 48h. Apical and basal compartments of the filter were then rinsed three times with serum-free medium. Cells were then collected (T0) or incubated with fresh serum-free medium in the basal compartment and with unlabelled lipid micelles devoid of cholesterol in the apical compartment for 2h. Total radioactivity was measured in cells (T0) and in apical medium (T2h) by liquid-scintillation counting after lipid extraction with chloroform/methanol (2:1, v:v).
2.4 Confocal microscopy analysis
Confocal microscopy analyses were performed as described [26]. Briefly, cells were fixed with 4% PAF, permeabilized by 0.05% saponin in PBS/10% serum and, when appropriate, stained for neutral lipids with LD540 dye [28]. After nuclear staining by DAPI and postfixation with 4% PFA, images were acquired by laser scanning confocal microscopy (LSM 710 microscope; Carl Zeiss) and analyzed with ZEN software (Carl Zeiss).
2.5 RT-PCR analysis
Total RNAs were extracted from differentiated Caco-2/TC7 cells and used for real-time PCR analysis as previously described [29]. Oligonucleotide sequences were as follows: forward, 5′-GTGAGATGGCAGAGAACGGTGTG-3′ and reverse, 5′-TGCCCCTTTGGTCTTGTCCA-3′ for human PLIN2 mRNA; forward 5′-GCCTTAGCTACAGGAGAGAA-3′ and reverse, 5′-TTTCCTCCTGTGCCATCTC-3′ for human PPIB mRNA (reference gene).
2.6 Western blot analysis
Cell lysates and western blots were performed as previously described [14]. Primary antibodies were used against human SR-B1 (BD 610882, BD Biosciences, Le Pont de Claix, France) and human ANXA2 (BD 610068, BD Biosciences, Le pont de Claix, France).
2.7 Preparation of raft-like/detergent-resistant membranes
Detergent-resistant membranes (DRM) were prepared from a sucrose gradient as previously described [14]. Briefly, 108 Caco-2/TC7 cells were homogenized on ice in 2 ml of TRIS-buffered saline (TBS) (10 mM Tris-HCl, pH 8, 150 mM NaCl) containing 1% Triton X-100 and protease inhibitors. Cell homogenates were adjusted to 40% sucrose with 2 ml of 80% sucrose/TBS. The resulting 4 ml were covered with 4 ml of 30% sucrose and 4 ml of 5% sucrose and centrifuged (SW41, L8 Beckman, 18h, 39,000 rpm, 4°C). Sequential 1 ml fractions were collected from the top of the tube and fractions 3 to 5, corresponding to DRM were pooled.
2.8 Lipidomic analysis
Detergent-resistant membranes collected from the sucrose gradient were kept at −80°C until use. Lipid standards di-myristoyl phosphatidylcholine (DMPC), 19:0-lysophosphatidylcholine (19:0-LPC), di-myristoyl phosphatidylethanolamine (DPME), myristoyl-lysophosphatidylethanolamine (MLPE), di-myristoyl phosphatidylserine (DMPS), d18:1/17:0 sphingomyelin (17:0-SM) and d18:1/17:0 ceramide were used; all of them were purchased from Avanti Polar Lipids (Coger, Paris, France). LC-MS/MS quality grade solvents were purchased from Fischer Scientific (Illkirch, France). Other chemicals of the highest grade available were purchased from Sigma Aldrich (Saint-Quentin Fallavier, France). For quantification of total cholesterol by GS-MS analysis, DRM fractions (20 μl) were spiked with epicoprostanol (2 μg) used as internal standard. Total cholesterol was quantified by GC-MS as previously described [30]. A calibration curve was generated with cholesterol standards processed as DRM fraction samples. For quantification of the different classes of lipids by LC-MS/MS, the DRM fraction (180 μl) was spiked with 20 μl of an internal standard mix containing 1000 ng DMPC, 250 ng 17:0-SM, 500 ng 19:0LPC, 200 ng DMPE, 400 ng DMPS, 100 ng MLPE and 100 ng d18:1/17:0 Cer. Total lipids were further extracted according to the method of Folch et al. [31]. Targeted lipidomic analysis by LC-MS/MS was conducted in Mutiple Reaction Monitoring mode as previously described [32].
2.9 Proteomic analysis
Detergent-resistant membranes collected from sucrose gradient were dialyzed (Flot-A-lyser G2 3.5–5kDa, Spectrumlabs, Breda, The Netherlands) for 3h at 4°C and then concentrated using an amicon Ultra-4 filter (ultracel-3 membrane 3kDa, Millipore, Fontenay-sous-Bois, France) according to the manufacturer’s instructions. DRM were then centrifuged (20 min, 12,000g, 4°C) and kept at −80°C until use. Proteins from DRM samples were processed as previously described [33]. Briefly, proteins were first extracted with a Urea-containing buffer (6M urea, 2.2M thiourea, 5mM EDTA, 0.1%SDS, 5% N-octyl glucoside, 50mM Tris-HCl) and separated shortly (1 cm) on a 1D 4–12 % acrylamide gel electrophoresis (ThermoScientific, Villebon sur Yvette, France). Proteins were excised from gel, reduced, alkylated and digested by Trypsin Gold Mass Spectrometry Grade, (Promega, Charbonnières-les-Bains, France). Peptides (2 μg) were separated with the nanoRSLC (ThermoScientific) fitted with a C18 trapping column (5 μm average particle diameter; 300 μm inner diameter × 5 mm length; ThermoScientific) and a C18 analytical column (2 μm average particle diameter; 75 μm inner diameter × 150 mm length; ThermoScientific). A 120-minute gradient was performed and peptides were analyzed by nanoLC-MS/MS using an LTQ-Orbitrap Elite mass spectrometer equipped with the Advion TriVersa NanoMate nanospray source. Full-scan spectra from a mass/charge ratio of 400 to one of 1,700 at a resolution of 120,000 full width at half maximum were acquired in the Orbitrap mass spectrometer. From each full-scan spectrum, the 20 ions with the highest intensity were selected for fragmentation in the ion trap.
The acquired data were searched against the International Protein Index/UniProt using Mascot and X!Tandem. Based on tandem mass spectometry data, peptides and proteins identifications were validated through Peptide- and ProteinProphet software [34]. At the end of the above steps, each injection is described by a list of validated proteins and peptides. Peptides that were not identified in at least 2 of the 3 technical replicates injected per sample were excluded [35]. Retention times were then aligned and peptide intensities extracted using the MASIC software [36]. Using this quantification, a new filter based on the coefficients of variation of each peptide in each sample is applied to discard poorly reproducible peptides [35]. A linear mixed model was used to select differential proteins in DRM from WT SR-B1- and SR-B1-Q445A-expressing cells (Clough et al., 2012). The FDR (false discovery rate) correction was applied to take into account the high dimension (far more tested variables than samples). Only proteins identified with at least 2 peptides fragments and with a fold change higher than 2, with a FDR at 5%, were kept.
2.10 Statistical analysis
For comparing two groups, unpaired Mann-Whitney test was used. For multiple comparisons, one-way ANOVA or two-way ANOVA were performed, followed by Dunnett’s multiple comparison tests or Bonferroni’s multiple comparison tests, depending on the combinations of comparisons. All analyses were performed using Prism software (GraphPad prism software, La Jolla, CA, USA).
3. Results
3.1 SR-B1-dependent lipid sensing modulates cholesterol distribution in intestinal epithelial cells
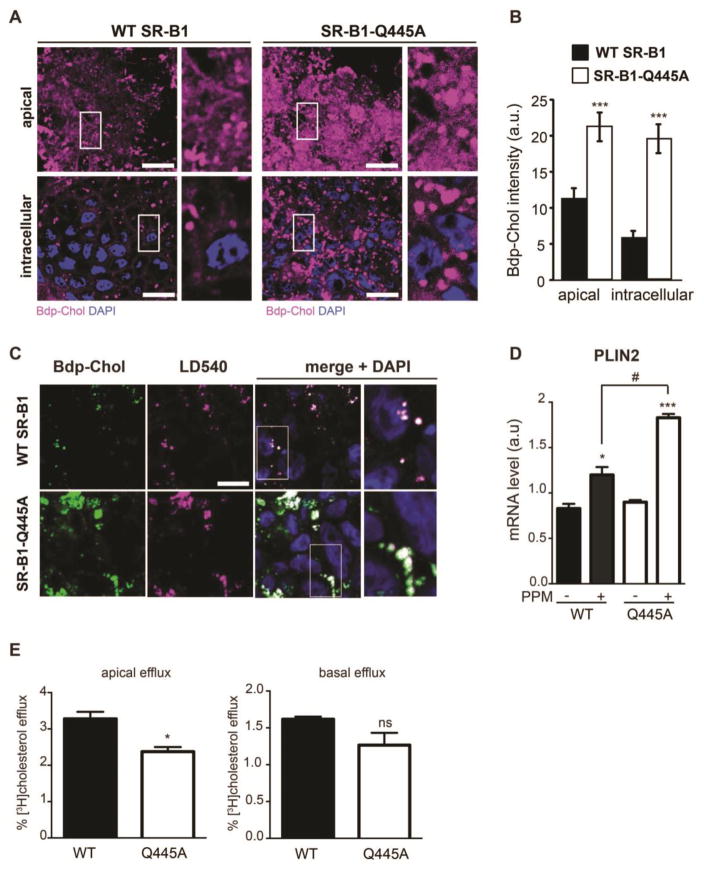
We first analyzed the link between SR-B1-dependent lipid sensing and intracellular cholesterol distribution using the incapacity of the SR-B1-Q445A mutant to bind plasma membrane cholesterol and trigger intracellular signaling [15]. The intracellular distribution of cholesterol was studied in WT SR-B1- or SR-B1-Q445A-expressing Caco-2/TC7 cells in a pulse-chase experiment using BODIPY-cholesterol (Fig. 1A and Fig. 1B). Cells were incubated for 1h with BODIPY-cholesterol-containing lipid micelles followed by 1h of chase with unlabelled cholesterol-containing lipid micelles. At the end of the experiment, we observed an increased amount of BODIPY-cholesterol in SR-B1-Q445A-expressing cells as compared to WT SR-B1-expressing cells, at both the apical and intracellular levels. These results revealed impaired cellular cholesterol trafficking in intestinal epithelial cells when SR-B1 cannot trigger lipid sensing.
Figure 1. SR-B1-dependent lipid sensing modulates cholesterol processing and lipid droplet dynamics.
(A) Pulse-chase experiments were conducted on Caco-2/TC7 cells stably transfected with WT SR-B1 or SR-B1-Q445A. Cells were cultured on semi-permeable filters in the presence of BODIPY-cholesterol-containing lipid micelles in the apical compartment of the filter for 1h (pulse), and then with unlabelled cholesterol-containing lipid micelles for one more hour (chase). At the end of the experiment, BODIPY-cholesterol (Bdp-Chol; purple) distribution was analyzed by confocal microscopy. Diamidino-2-phenylindole (DAPI; blue) is used for visualization of nuclei. Bar = 10μm. (B) Summary data (mean ± SEM) for Bdp-Chol fluorescence quantified at both apical and intracellular XY planes (5 fields per condition). ***p<0.001 versus WT SR-B1 expressing cells. (C) Transfected Caco-2/TC7 cells described in (A) were cultured for 1h with Bdp-Chol-containing lipid micelles. Neutral lipids of lipid droplets were stained with LD540 dye (purple). Co-localization of lipid droplets and Bdp-Chol (green) was analyzed by confocal microscopy. Enlarged views of the fields designated by white rectangles are displayed on the right. DAPI (blue) is used for nucleus staining. Bar = 10μm. (D) Cells expressing WT SR-B1 (WT) and SR-B1-Q445A (Q445A) were cultured in the presence (+) or absence (−) of PPM for 24h. The mRNA levels of PLIN2 were quantified by RT-PCR. Results are expressed as the ratio (mean ± SEM, n=6) of PLIN2 mRNA levels to cyclophilin mRNA level used as control. *p<0.05 and ***p<0.01 as compared to the same cell line in absence of PPM; #p<0.05, n=5. (E) Cells were cultured in the presence of [3H]-cholesterol-containing lipid micelles in the apical compartment for 48h. At the end of treatment (T0), cells were washed and then incubated for 2h with unlabelled lipid micelles devoid of cholesterol in the apical compartment. Radioactivity was then quantified in the apical (left panel) and basal (right panel) medium. Results are expressed as percentage of radioactivity counted in the corresponding medium to the radioactivity counted in cells at T0, mean ± SEM of 6 independent cultures. * p<0.05, ns: non-significant.
We then analyzed more precisely the intracellular localization of BODIPY-cholesterol in both WT and SR-B1-Q445A-expressing cells. In enterocytes as well as in Caco-2/TC7 cells, the supply of PPM is known to induce the formation of lipid droplets [26, 37], which store triglycerides and cholesterol esters. We analyzed the localization of BODIPY-cholesterol and of lipid droplets that were stained with the neutral lipid dye LD540. We observed that BODIPY-cholesterol co-localizes with lipid droplet staining (Fig 1C) in both WT and SR-B1-Q445A-expressing cells. However, SR-B1-Q445A cells display a more intense co-labeling (Fig. 1C, merge), which could be related to an increased accumulation of lipid droplets (Fig 1C, LD540 staining) in these cells as compared to WT SR-B1-expressing cells. Interestingly, such an accumulation of lipid droplets is also observed in Caco-2/TC7 cells transfected with RNAi for SR-B1 (Suppl. Fig. 2). We thus analyzed, in WT- and SR-B1-Q445A-expressing cells, the expression of PLIN-2 (Fig. 1D), which is known to be associated with lipid droplets and to modulate their formation and/or their stabilization [37, 38]. Before the addition of lipid micelles, we observed that the two cell lines express similar PLIN-2 mRNA levels. After a 24 hour incubation with PPM, an increase of PLIN-2 mRNA abundance is observed in both cell lines; however, the upregulation occurs to a greater extent in SR-B1-Q445A cells as compared to WT SR-B1 cells. Altogether these results indicate that SR-B1-dependent lipid sensing participates to the dynamics of lipid droplets.
We next studied whether the observed perturbation of cholesterol distribution in SR-B1-Q445A-expressing cells is associated with a modification of cholesterol efflux. Cells were incubated for 48h with [3H]-cholesterol-containing lipid micelles and then chased for 2h with lipid micelles devoid of cholesterol in the apical compartment (Fig. 1E). We found that the apical efflux of [3H]-cholesterol is lower in SR-B1-Q445A-expressing cells as compared to WT SR-B1-expressing cells, whereas basal efflux of [3H]-cholesterol is similar in both cell types. Thus, the modification of cholesterol distribution in SR-B1-Q445A cells is associated with attenuated apical cholesterol efflux.
3.2 SR-B1 is involved in cholesterol trafficking from plasma membrane
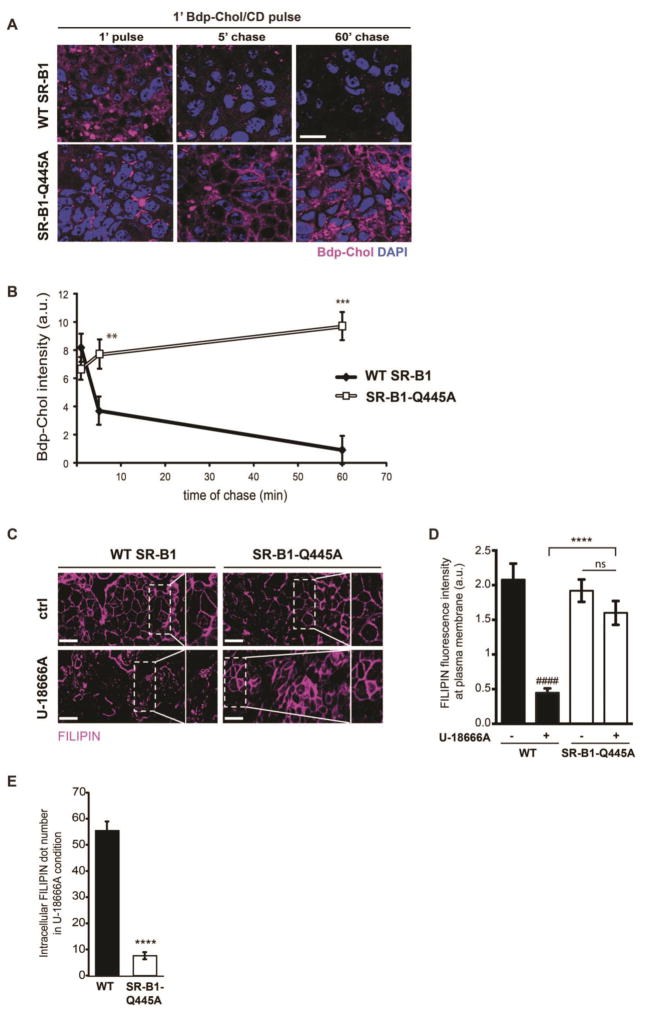
Having previously observed that plasma membrane cholesterol movement is required for SR-B1-dependent lipid sensing [15], we analyzed the cholesterol trafficking between plasma membrane and intracellular compartments in WT SR-B1- and SR-B1-Q445A-expressing cells (Fig. 2). Cells were pulse-labelled by the addition of BODIPY-cholesterol/methyl-β-cyclodextrin complexes (Bdp-Chol/CD) to the apical compartment for 1 min at 37°C, as previously described [27], and then chased with a medium without Bdp-Chol/CD for 5min or 60min. This short pulse allows limited incorporation of BODIPY-cholesterol into the cells, thus avoiding a saturation of the labelling and a better visualization of cholesterol trafficking from the plasma membrane [27]. After a 1min pulse, the intensity of sub-apical BODIPY-cholesterol labelling is similar in both cells (1′ pulse, Fig. 2A and quantification in Fig. 2B). During the pulse-chase, the intensity of sub-apical BODIPY-cholesterol labelling rapidly decreased in WT SR-B1-expressing cells, probably due to a rapid metabolism or efflux of BODIPY-cholesterol. In contrast, in SR-B1-Q445A cells BODIPY-cholesterol continued to label the plasma membrane after 5 min of chase, and accumulated in intracellular compartments after 60 min of chase.
Figure 2. SR-B1 is involved in cholesterol trafficking from plasma membrane.
(A) Pulse-chase experiments were conducted on Caco-2/TC7 cells stably transfected with WT SR-B1 or SR-B1-Q445A. Cells were cultured on semi-permeable filters and incubated in presence of BODIPY-cholesterol combined with methyl- β-cyclodextrin (Bdp-Chol/CD) in the apical compartment for 1 min (pulse), and then with culture medium without cholesterol for 5 min and 1 hour (chase). BODIPY-Cholesterol (purple) distribution was analyzed by confocal microscopy. DAPI (blue) is used for visualization of nucleus. Sub-apical XY planes are shown. Bar= 10μm. (B) Summary data (mean ± SEM) for Bdp-Chol fluorescence intensity quantified at sub-apical XY planes (5 fields per condition) are expressed as arbitrary units (a.u.). **p<0.01 and ***p<0.001 versus WT SR-B1 expressing cells. (C) Cells were incubated or not (ctrl) with U-18666A for 18h, then free cholesterol was visualized by filipin staining (purple). Rectangles and squares on the right of the XY planes represent enlargement of the selected area. Bar= 10μm (D) Filipin fluorescence intensity at plasma membrane reported to surface area was quantified in both cell lines with or without U18666A treatment. Results are expressed as arbitrary units (a.u.), mean ± SEM, (5 fields per condition). ####p<0.0001 as compared to WT SR-B1 in absence of U-18666A, ****p<0.0001 as compared to U-18666A-treated WT SR-B1 cells, ns: non-significant (E) Intracellular filipin dot numbers were quantified in both cell lines after U18666A treatment. Results are expressed as mean ± SEM (5 fields per condition). ****p<0.0001 as compared to WT SR-B1.
We next analyzed the capacity of plasma membrane cholesterol in WT SR-B1- and SR-B1-Q445A-expressing cells to be distributed in intracellular compartments. We visualized free cholesterol using filipin staining and studied free cholesterol movement using the properties of compound U-18666A to inhibit cholesterol transport leading to an intracellular accumulation of cholesterol [39]. Before U-18666A treatment, a similar localization of free cholesterol at the plasma membrane is observed in both WT and SR-B1-Q445A cell lines (Fig. 2C, ctrl panels). After U-18666A treatment, intracellular filipin signal is observed in WT SR-B1-expressing cells, indicating that free cholesterol is accumulating in vesicular structures (Fig. 2C, square showing an enlargement of the XY plan). By contrast, in SR-B1-Q445A-expressing cells treated with U-18666A, filipin staining is mainly observed at the plasma membrane, indicating that free cholesterol remains trapped at this level. The quantification of filipin fluorescence intensity showed a higher signal at plasma membrane (Fig. 2D) in SR-B1-Q445A cells treated with U-18666A (as compared to U-18666A-treated WT SR-B1 cells) that is associated with a decreased intracellular filipin dot numbers (Fig. 2E). Altogether these results reveal an impairment of cholesterol trafficking in cells expressing the SR-B1-Q445A mutant.
3.3 The expression of SR-B1-Q445A modifies the lipid composition of raft-like domain
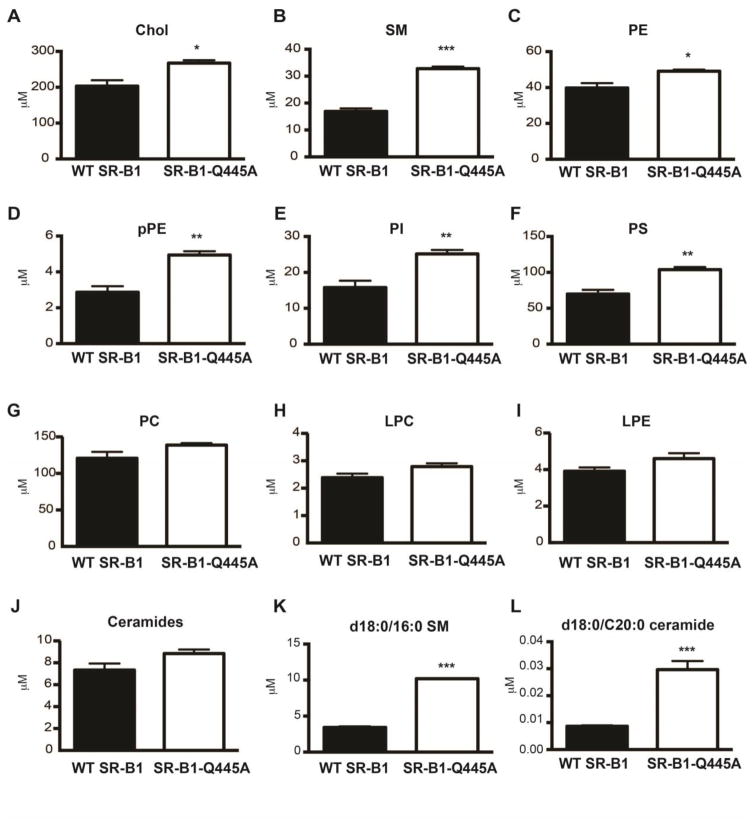
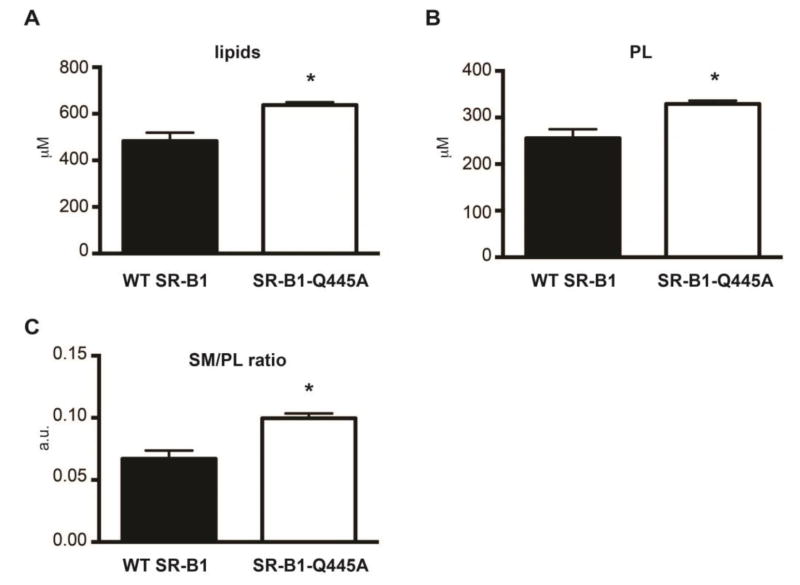
We showed previously, in intestinal epithelial cells, that SR-B1-dependent lipid sensing is associated with the recruitment of SR-B1 in raft-like membrane domains [14, 15]. Raft domains of the plasma membrane are signaling platforms characterized by a large amount of cholesterol and sphingolipids that can be isolated by their properties to be resistant to detergent action [40]. We thus determined whether the observed perturbations of cholesterol trafficking in SR-B1-Q445A cells are associated with alterations in raft lipid composition. We isolated detergent-resistant membranes (DRM) from WT SR-B1 and SR-B1-Q445A cells treated or not with PPM for 15min, which is a time compatible with the activation of lipid signaling [14]. We conducted LC-MS/MS studies to determine lipid composition of DRM and analyzed the total concentration of 10 lipid subclasses, cholesterol, ceramides, phosphatidylcholine (PC), lysopPC (LPC), phosphatidylethanolamine (PE), lysoPE (LPE), plasmalogen PE (pPE), phosphoinositide (PI), phosphatidylserine (PS) and sphingomyelin (SM), as well as several individual species in each subclass. In the absence of lipid micelle treatment (Fig. 3), we observed that DRM from SR-B1-Q445A-expressing cells display higher concentrations of cholesterol (+32%, Fig. 3A), SM (+94%, Fig. 3B), PE (+24%, Fig. 3C), pPE (+72%, Fig. 3D), PI (+59%, Fig. 3E) and PS (+51%, Fig. 3F), as compared to DRM from WT SR-B1-expressing cells. By contrast, no change in the total concentration of PC, LPC, LPE and ceramides is observed between the two cell lines (Fig. 3G, H, I, J). Among the individual lipid species analyzed in the different lipid subclasses, d18:0/16:0 SM (Fig. 3K), one of the most represented SM, and d18:0/C20:0 ceramide (Fig. 3L), a minor species in this subclass, displayed the greatest increase of concentration (approximately a 3 fold change) in DRM from SR-B1-Q445A cells as compared to WT cells. Overall, the lipidomic analysis revealed that DRM from SR-B1-Q445A cells display higher concentrations of lipids (Fig. 4A), in particular of phospholipids (PL) (Fig. 4B), than those from WT SR-B1 cells. The qualitative analysis of lipid composition of DRM, achieved by the calculation of SM to total PL ratio (Fig. 4C) revealed that DRM from SR-B1-Q445A cells are enriched in SM as compared to WT SR-B1 cells. Altogether, these results show that the presence of the mutated form of SR-B1 modifies the lipid composition of DRM.
Figure 3. SR-B1-Q445A-expressing cells display changes in detergent-resistant membrane lipid composition as compared to WT SR-B1 cells.
Detergent-resistant membranes were isolated from WT SR-B1- and SR-B1-Q44A-expressing cells. Concentrations of six lipid subclasses (A–J), of d18:0/16:0 SM (K) and d18:0/C20:0 ceramide (L) were determined by LC-MS/MS: cholesterol (chol), SM (sphingomyelin), PE (phosphatidylethanolamine), pPE (plasmalogen PE), PI (phosphoinositide) and PS (phosphoserine), PC (phosphatidylcholin), LPC (lysopPC), LPE (LysoPE) and ceramides. Results are expressed in μM as mean ± SEM from three independent experiments. *p<0.05,**p<0.01 and ***p<0.001 as compared to WT SR-B1-expressing cells.
Figure 4. Changes of total lipid and total phospholipid concentrations, as well as sphingomyelin to phospholipid ratio in detergent-resistant membranes of SR-B1-Q445A cells as compared to WT SR-B1 cells.
Concentration in DRM fractions of (A) total lipid calculated by summing all lipid subclasses and (B) total phospholipid calculated by summing the PC, PE, PS, PI, LPC, pPE, LPE obtained from LC-MS/MS experiments. Results are expressed in μM as mean ± SEM from three independent experiments. *p<0.05 as compared to WT SR-B1-expressing cells. (C) The SM/PL ratio was calculated from concentrations of total SM and total PL for each condition. Results are expressed as arbitrary units (a.u.) ± SEM from three independent experiments, *p<0.05 as compared to WT SR-B1-expressing cells.
After the addition of lipid micelles, changes in DRM lipid composition were observed. Lipid micelle treatment promotes a marked increase in LPC concentration, but this modification occurs to the same extent in both cell lines (Suppl Fig. 3). This dramatic increase in LPC concentration probably results from the incorporation of exogenous LPC, provided by PPM, within cell membranes. More interestingly, after the supply of lipid micelles, the alterations in the concentrations of certain lipids differed in DRM of WT SR-B1 as compared to SR-B1-Q445A cells (Table 1). In DRM from WT SR-B1-expressing cells, the concentration of d18:0/16:0 SM increased by 17% upon lipid micelle provision, and the concentration of 18:0/0:0 LPE increased by 43%. In contrast, no alterations were observed in these lipids in DRM from SR-B1-Q445A-expressing cells after PPM treatment. These results indicate that d18:0/16:0 SM and 18:0/0:0 LPE may be involved in SR-B1-dependent signaling in response to lipid micelles. On the other hand, several lipid concentrations in DRM were decreased after PPM supply in SR-B1-Q445A-expressing cells whereas they remained unchanged after such treatment in WT SR-B1 cells (Table 1). This included a decrease of total ceramide (−28%) and of total PE (-10%) as well as several individual lipid species in different subclasses (Table 1).
Table 1.
Lipid concentrations in DRM are modified differentially between WT SR-B1 and SR-B1-Q445A after PPM supply
| Lipids | WT SR-B1 (mM, mean ± SEM) | SR-B1-Q445A (mM, mean ± SEM) | WT vs WT PPM | Q445A vs Q445A PPM | |||||
|---|---|---|---|---|---|---|---|---|---|
|
| |||||||||
| subclass | species | - | PPM | - | PPM | variation | p value | variation | p value |
| SM | d18:0/16:0 | 3.44 ± 0.07 | 4.04 ± 0.17 | 10.21 ± 0.04 | 10.19 ± 0.42 | +17% | 0.03 | - | 0,96 |
| d18:1/18:0 | 0.38 ± 0.02 | 0.36 ± 0.01 | 0.58 ± 0.02 | 0.50 ± 0.01 | - | 0.4 | −14% | 0.02 | |
| d18:1/24:1 | 2.68 ± 0.35 | 2.42 ± 0.17 | 4.23 ± 0.15 | 3.58 ± 0.13 | - | 0.54 | −15% | 0.03 | |
|
| |||||||||
| PC | 16:0/16:1 | 13.9 ± 1.61 | 12.44 ± 2.13 | 20.37 ± 0.65 | 17.79 ± 0.64 | - | 0.61 | −12% | 0.047 |
|
| |||||||||
| LPE | 18:0/0:0 | 2.78 ± 0.27 | 3.98 ± 0.27 | 3.25 ± 0.2 | 3.79 ± 0.03 | +43% | 0.035 | - | 0.053 |
|
| |||||||||
| PE | Total | 39.78 ± 2.68 | 40.14 ± 3.53 | 49.14 ± 0.85 | 44.01 ± 0.98 | - | 0.94 | −10% | 0.017 |
|
| |||||||||
| 16:0/18:2 | 1.51 ± 0.22 | 1.37 ± 0.34 | 1.91 ± 0.08 | 1.55 ± 0.05 | - | 0.75 | −19% | 0.016 | |
| 16:0/18:1 | 6.47 ± 0.6 | 6.15 ± 0.92 | 8.89 ± 0.18 | 7.48 ± 0.17 | - | 0.78 | −16% | 0.005 | |
| 17:0/17:0 | 0.57 ± 0.05 | 0.53 ± 0.03 | 0.47 ± 0.004 | 0.41 ± 0.01 | - | 0.56 | −13% | 0.019 | |
| 18:1/18:2 | 0.84 ± 0.08 | 0.84 ± 0.1 | 1.03 ± 0.04 | 0.90 ± 0.03 | - | 0.99 | −12% | 0.038 | |
| 18:0/18:2 | 6.31 ± 0.56 | 6.41 ± 0.87 | 7.8 ± 0.22 | 6.98 ± 0.19 | - | 0.93 | −10% | 0.048 | |
| 18:0/18:1 | 12.32 ± 0.68 | 12.36 ± 0.92 | 14.69 ± 0.04 | 13.2 ± 0.23 | - | 0.97 | −10% | 0.0029 | |
| 16:0/22:6 | 0.59 ± 0.06 | 0.58 ± 0.06 | 0.78 ± 0.03 | 0.67 ± 0.02 | - | 0.84 | −14% | 0.045 | |
| 18:1/22:6 | 0.24 ± 0.02 | 0.24 ± 0.02 | 0.26 ± 0.01 | 0.23 ± 0.004 | - | 0.96 | −11% | 0.049 | |
|
| |||||||||
| pPE | 16:0p/18:1 | 0.32 ± 0.027 | 0.33 ± 0.026 | 0.57 ± 0.016 | 0.46 ± 0.009 | - | 0.8 | −19% | 0.0036 |
| 16:0p/20:4 | 0.11 ± 0.014 | 0.13 ± 0.011 | 0.18 ± 0.007 | 0.16 ± 0.001 | - | 0.33 | −11% | 0.030 | |
|
| |||||||||
| PS | 16:0/18:1 | 5.55 ± 0.29 | 5.14 ± 0.58 | 8.10 ± 0.17 | 7.10 ± 0.25 | - | 0.56 | −12% | 0.029 |
|
| |||||||||
| ceramide | Total | 7.33 ± 0.603 | 6.07 ± 0.242 | 8.84 ± 0.362 | 6.39 ± 0.223 | - | 0.12 | −28% | 0.0044 |
|
| |||||||||
| d18:1/14:0 | 0.24 ± 0.022 | 0.18 ± 0.015 | 0.3 ± 0.024 | 0.18 ± 0.005 | - | 0.09 | −38% | 0.0085 | |
| d18:1/16:0 | 1.62 ± 0.081 | 1.39 ± 0.076 | 1.85 ± 0.109 | 1.32 ± 0.039 | - | 0.11 | −29% | 0.01 | |
| d18:1/18:0 | 0.036 ± 0.004 | 0.028 ± 0.002 | 0.043 ± 0.002 | 0.028 ± 0.003 | - | 0.13 | −25% | 0.01 | |
| d18:1/20:0 | 0.027 ± 0.002 | 0.017 ± 0.003 | 0.043 ± 0.004 | 0.029 ± 0.002 | - | 0.061 | −35% | 0.025 | |
| d18:1/22:0 | 0.5 ± 0.055 | 0.39 ± 0.017 | 0.69 ± 0.013 | 0.48 ± 0.044 | - | 0.14 | −32% | 0.01 | |
| d18:1/24:1 | 2.33 ± 0.373 | 1.77 ± 0.226 | 2.56 ± 0.07 | 1.69 ± 0.092 | - | 0.27 | −33% | 0.0017 | |
| d18:1/24:0 | 1.88 ± 0.105 | 1.63 ± 0.109 | 2.31 ± 0.125 | 1.76 ± 0.067 | - | 0.16 | −24% | 0.018 | |
| d18:1/26:0 | 0.039 ± 0.005 | 0.034 ± 0.04 | 0.047 ± 0.001 | 0.037 ± 0.001 | - | 0.58 | −21% | 0.0011 | |
WT SR-B1- and SR-B1-Q445A-expressing cells were cultured in absence (−) or in presence of lipid micelles (PPM) for 15 min. Detergent-resistant membranes (DRM) were isolated through a sucrose gradient and lipid concentrations were determined by LC-MS/MC analysis. Results (mM) are expressed as mean ± SEM from three independent experiments. The species representing more than 10% of the total amount of species analyzed in their lipid subclass are underlined. Statistical changes (variation and p value) are in bold.
Altogether these results show that lipid composition of raft-like membrane domains is modified during SR-B1-dependent lipid sensing.
3.4 SR-B1-dependent lipid signaling modifies the protein composition of raft-like domains
In order to identify proteins in raft domains that could be involved in the SR-B1-dependent lipid sensing and resulting signaling in intestinal epithelial cells, we analyzed and compared changes in the amounts of DRM proteins after PPM supply in WT SR-B1- versus SR-B1-Q445A-expressing cells (Table 2). We isolated detergent-resistant membranes from both cell lines before and 15 min after PPM supply. The quantity of total protein was similar in all DRM (data not shown). Proteins were identified by mass spectrometry, and changes in protein amounts induced by PPM treatment for each cell line were determined.
Table 2.
List of proteins whose abundance in DRM changes after PPM supply in WT SR-B1- versus SR-B1-Q445A-expressing cells.
| WT SR-B1 | Q445A-SR-B1 | |||||
|---|---|---|---|---|---|---|
|
| ||||||
| fold change after PPM | p value | Uniprot | protein name | function | fold change after PPM | p value |
| 212.49 | 2.34E-05 | P00403 | COX2, Cytochrome C oxydase sub unit 2 | oxidation-reduction | 0.67 | 0.517 |
| 76.88 | 3.5E-07 | Q9UJU6 | DBNL, Drebin-like protein | cell adhesion/junction and cytoskeleton | 1.27 | 0.345 |
| 9.42 | 0.012 | P40227 | TCPZ, T-complex protein 1 subunit zeta | protein interaction/processing | 0.52 | 0.369 |
| 8.42 | 0.007 | P42167 | LAP2B, Lamina-associated polypeptide 2 isoforms b/g | nucleus/transcription/translation | 1.38 | 0.747 |
| 8.17 | 1.01E-05 | P46777 | RL560S, ribosomal protein L5 | protein interaction/processing | 1.09 | 0.715 |
| 7.13 | 0.014 | Q9Y5K8 | VATD, V-type proton ATPase subunit D | ion transport | 0.76 | 0.77 |
| 6.89 | 0.016 | P37059 | DHB2, Estradiol 17-beta-dehydrogenase | steroid, lipid metabolism | 0.85 | 0.864 |
| 6.29 | 1.4E-07 | O43169 | CYB5B, Cytochrome b5 | oxidation-reduction | 1.31 | 0.011 |
| 5.55 | 0.032 | P61978 | HNRPK, Heterogeneous nuclear ribonucleoprotein K | nucleus/transcription/translation | 1.73 | 0.524 |
| 4.10 | 0.009 | O75381 | PEX14, Peroxisomal membrane protein PEX14 | protein interaction/processing | 0.80 | 0.727 |
| 3.90 | 0.004 | P56134 | ATPK, ATP synthase subunit f | oxidation-reduction | 0.52 | 0.071 |
| 3.55 | 0.044 | Q9NRR3 | C42S2, Cdc42 small effector protein 2 | cell adhesion/junction and cytoskeleton | 0.89 | 0.851 |
| 3.36 | 0.011 | O95994 | AGR2, Anterior gradient protein 2 homolog | protein interaction/processing | 0.12 | 0.0006 |
| 3.13 | 0.004 | O43678 | NDUA2, NAH dehydrogenase (ubiquinone) 1 alpha subcomplex subunit 2 | oxidation-reduction | 1.09 | 0.801 |
| 3.12 | 0.047 | P50454 | SERPH, Serpin H | cell adhesion/junction and cytoskeleton | 0.46 | 0.175 |
| 3.01 | 0.002 | Q8NBX0 | SCPDL, Saccharopine dehydrogenase-like oxidoreductase | oxidation-reduction | 1.31 | 0.456 |
| 2.87 | 0.035 | Q12907 | LMAN2, Vesicular integral-membrane protein VIP36 | vesicular traficking/autophagy | 1.14 | 0.815 |
| 2.65 | 0.025 | P51114 | FXR1, Fragile X mental retardation syndrome-related protein 1 | nucleus/transcription/translation | 1.80 | 0.123 |
| 2.54 | 0.01 | P40879 | S26A3, Chloride-anion exchanger | ion transport | 0.98 | 0.956 |
| 2.46 | 0.003 | Q9H6A9 | PCX3, Pecanex-like protein 3 | unclassified | 2.36 | 0.0033 |
| 2.28 | 0.017 | P36578 | RL460S, ribosomal protein L4 | protein interaction/processing | 1.40 | 0.296 |
| 2.17 | 0.015 | Q9Y624 | JAM1, Junctional adhesion molecule A | cell adhesion/junction and cytoskeleton | 1.25 | 0.545 |
| 2.13 | 3.22E-06 | P35527 | K1C9, Keratin, type I cytoskeletal 9 | cell adhesion/junction and cytoskeleton | 0.60 | 0.0019 |
| 2.12 | 1.41E-06 | P27824 | CALX, Calnexin | protein interaction/processing | 1.01 | 0.974 |
| 2.06 | 5.95E-05 | Q16864 | VATFV-type proton ATPase subunit F | ion transport | 1.37 | 0.0061 |
|
| ||||||
| 0.50 | 0.031 | P13861 | KAP2, cAMP-dependent protein kinase type II-alpha regulatory subunit | signal transduction | 0.81 | 0.547 |
| 0.49 | 0.002 | Q15363 | TMED2, Transmembrane emp24 domain-containing protein 2 | vesicular traficking/autophagy | 0.66 | 0.0197 |
| 0.47 | 0.0036 | Q9Y2B5 | VP9D1, VPS9 domain-containing protein 1 | ion transport | 1.78 | 0.012 |
| 0.46 | 0.0067 | p07195 | LDHB, L-lactate dehydrogenase B chain | oxidation-reduction | 1.66 | 0.035 |
| 0.43 | 0.002 | Q14974 | IMB, 1Importin subunit beta-1 | protein interaction/processing | 1.16 | 0.394 |
| 0.41 | 0.012 | Q9BV40 | VAMP8, Vesicle-associated membrane protein 8 | vesicular traficking/autophagy | 0.97 | 0.956 |
| 0.41 | 0.044 | Q5T0W9 | FA83B, Protein FAM83B | signal transduction | 0.74 | 0.541 |
| 0.41 | 0.011 | Q8N3R9 | MPP5, MAGUK p55 subfamily member 5 | cell adhesion/junction and cytoskeleton | 0.76 | 0.515 |
| 0.40 | 0.005 | O14662 | STX16, Syntaxin-16 | vesicular traficking/autophagy | 2.88 | 0.002 |
| 0.38 | 0.030 | Q99536 | VAT1, Synaptic vesicule amine transport | oxidation-reduction | 1.02 | 0.974 |
| 0.38 | 0.016 | Q99623 | PHB2, Prohibitin-2 | nucleus/transcription/translation | 1.40 | 0.412 |
| 0.32 | 0.001 | P05362 | ICAM1, Intercellular adhesion molecule 1 | cell adhesion/junction and cytoskeleton | 0.77 | 0.411 |
| 0.29 | 9.3E-05 | Q5T0Z8 | CF132, Uncharacterized protein C6orf132 | unclassified | 1.22 | 0.224 |
| 0.25 | 0.0002 | P08574 | CY1, Ubiquinol-Cytochrome-C Reductase Complex Cytochrome C1 Subunit |
oxidation-reduction | 0.48 | 0.043 |
| 0.17 | 1.92E-06 | P31943 | HNRH1, Heterogeneous nuclear ribonucleoprotein H | nucleus/transcription/translation | 1.41 | 0.022 |
| 0.14 | 0.024 | O60658 | PDE8A, High affinity cAMP-specific and IBMX-insensitive 3′,5′-cyclic phosphodiesterase 8A | signal transduction | 0.92 | 0.950 |
WT SR-B1- and SR-B1-Q445A- expressing cells were cultured in absence (−) or in presence of lipid micelles (PPM) for 15 min. Detergent-resistant membranes (DRM) were isolated through sucrose gradient and protein enrichment was determined by tandem mass spectrometry analysis. Proteins with a fold change higher than 2 in DRM from WT SR-B1-expressing cells after PPM supply are listed. Fold change variation of these proteins in DRM from SR-B1-Q445A-expressing cells are reported. P values <0.05 are in bold. Proteins with similar direction change in their abundance in DRM after PPM in WT SR-B1- and SR-B1-Q445A-expressing cells are highlighted in green. Proteins with opposite variations induced by PPM in DRM in WT SR-B1- versus SR-B1-Q445A expressing cells are highlighted in red. Proteins unchanged in abundance in DRM after PPM supply in SR-B1-Q445A-expressing cells are indicated by a lack of highlighting.
We first analyzed modifications of protein amounts in DRM from WT SR-B1-expressing cells after PPM supply. We found that PPM supply promotes significant changes in the abundance of 41 proteins in DRM from WT SR-B1 cells, 25 of them being more represented and 16 of them less represented as compared to the condition without PPM treatment (Table 2). These proteins have been classified in several function categories according to UniProt Consortium annotations.
We then determined whether the amounts of these 41 identified proteins were also modified after PPM supply in DRM from SR-B1-Q445A cells. We observed that 5 of them (Table 2, green lines) displayed similar directional change in abundance in DRM from SR-B1-Q445A as compared to WT SR-B1 cells after PPM supply, suggesting that these proteins are not involved in SR-B1-dependent lipid sensing. More importantly, we found that 6 proteins, namely anterior gradient protein 2 homolog (AGR2), keratin, type I cytoskeletal 9 (K1C9), VPS9 domain-containing protein 1 (VP9D1), L-lactate dehydrogenase B chain (LDHB), Syntaxin-16 (STX16) and heterogeneous nuclear ribonucleoprotein H (HNRH1) displayed opposite variations in DRM from SR-B1-Q445A as compared to WT SR-B1 cells after PPM supply (Table 2, red lines). AGR2 is involved in protein processing, K1C9 belongs to an intermediate filament family, VP9D1 participates in ion transport, LDHB is involved in oxidation-reduction processes, STX16 plays a role in vesicular docking and fusion, and HNRH1 participates in the maturation of mRNA. Displaying disparate recruitment to DRM in WT versus SR-B1-Q445A cells, these six proteins are candidates for molecules involved in SR-B1-dependent signaling in response to lipid movement. Additional candidates are the 30 proteins whose abundance changed after PPM supply in DRM from WT SR-B1 but not in SR-B1-Q445A cells (Table 2, uncoloured lines). These proteins belong to functional categories related to 1) cell adhesion/junction and cytoskeleton, such as the junctional adhesion molecule A (JAM1) and the intercellular adhesion molecule 1 (ICAM1), which are involved in cell-cell contact; 2) protein interaction/processing, such as peroxisomal membrane protein PEX14 involved in protein-protein docking, and calnexin (CALX) involved in protein folding; 3) oxidation/reduction, such as the NADH dehydrogenase (ubiquinone) 1 alpha subcomplex subunit 2 (NDUA2); 4) ion transport, such as V-type proton ATPase subunit D (VATD); 5) pre-mRNA processing, such as Heterogeneous nuclear ribonucleoprotein K (HNRPK); and 6) signal transduction, such as the cAMP-dependent protein kinase type II-alpha regulatory subunit (KAP2), protein FA83B and the High affinity cAMP-specific and IBMX-insensitive 3′,5′-cyclic phosphodiesterase 8A (PDE8A). The vesicle-associated membrane protein 8 (VAMP8), which is involved in vesicular trafficking, is also present among the 30 proteins that remain unchanged after PPM supply in DRM from SR-B1-Q445A cells as compared to WT SR-B1 cells.
Altogether these results showed that SR-B1-dependent lipid sensing is associated with changes in the amount of several proteins in detergent-resistant membranes.
4. Discussion
SR-B1-dependent lipid sensing has been previously characterized in endothelial cells and in intestinal epithelial cells [14, 15, 41]. The discovery that the glutamine at the position 445 in the C-terminal transmembrane domain of SR-B1 was crucial in this process revealed that the lipid sensing is dependent on the interaction of SR-B1 with plasma membrane cholesterol [15]. The plasma membrane cholesterol sensing is also required for the recruitment of SR-B1 in raft-like membrane domains upon lipid micelle stimulation in intestinal epithelial cells. In the present study, we analyzed changes in the lipid and protein composition of raft-like domains occurring during SR-B1-dependent lipid sensing in intestinal epithelial cells, and also the impact of this sensing on cholesterol trafficking. We discovered that a defect in SR-B1-mediated lipid sensing leads to an alteration of cholesterol distribution and to a decreased apical cholesterol efflux. Associated with these events, we observed, using SR-B1-Q445A or RNA interference, an exaggerated accumulation of lipid droplets in the absence of efficient SR-B1-dependent lipid sensing. Such a link between SR-B1 and lipid droplet dynamics could be suggested from previous reports. Indeed, SR-B1 is found associated with lipid droplets during fat absorption in enterocytes [42, 43]. SR-B1 is also present in mature adipocytes, where it could be involved in lipid storage in adipose tissue [44, 45]. Interestingly, our proteomic analysis of DRM from WT SR-B1 cells revealed increased DHB2 protein abundance in DRM after PPM supply, and this did not occur in DRM from SR-B1-Q445A-expressing cells. Although DHB2, which belongs to the family of 17β-hydroxysteroid dehydrogenases, is mainly known for its role in steroid metabolism, it has been shown to be associated with lipid droplets in intestinal epithelial cells. Its depletion increased triglyceride secretion, suggesting a role for this enzyme in the control of the balance between lipid storage and lipid secretion in these cells [29].
The impact of SR-B1 on cellular cholesterol metabolism has been mainly understood in the context of selective cholesteryl ester uptake from HDL and in the bidirectional flux of free cholesterol between cells and HDL (for review [21]). SR-B1 is known to be localized to caveolae/lipid rafts and to interact with several phospholipids [46–48]. A number of these studies indicated that SR-B1-dependent cholesterol flux may be mediated by changes in the lipid organization of the plasma membrane and particularly that of the raft membrane domains [49–52]. Our lipidomic analysis revealed that the presence of SR-B1-Q445A per se modifies the lipid composition of DRM as compared to cells expressing the wild type form of SR-B1. These modifications, which are characterized by increases in cholesterol, sphingomyelins, and several phospholipids, result in a relative enrichment of sphingomyelins versus total phospholipids in DRM. It has been observed that sphingomyelins are critical for cholesterol sequestration in the plasma membrane [53] and for the regulation of cholesterol efflux [54]. Moreover, modulation of SM amount in HDL or in cell membrane modified SR-B1-dependent cholesterol flux [55, 56]. Interestingly, two point mutations in the extracellular loop of human SR-B1 (S112F and T175A) result in a reduced efflux of free cholesterol to HDL and in the impairment of the redistribution of free cholesterol pools at the plasma membrane [57]. Thus, our findings provide evidence that, through its capacity to bind plasma membrane cholesterol, SR-B1 governs the lipid composition of raft-like membrane domains, thereby preparing a favourable environment for lipid sensing processes.
We analyzed additionally whether SR-B1-dependent lipid sensing is associated with modifications in the lipid composition of DRM. Lipidomic analysis showed that PPM supply provokes an enrichment of d18:0/16:0 SM and 18:0/0:0 lysophosphatidylethanolamines in WT SR-B1 cells. This enrichment is not observed in DRM from SR-B1-Q445A after PPM supply, suggesting that these lipids may be involved in SR-B1-dependent lipid sensing and in the resulting intracellular signaling. Although these lipids belong to classes of lipids recognized to be involved in cell signaling, their specific roles in such processes are largely unknown. Sphingomyelins are important structural component of lipid rafts that can be metabolized into ceramides, which induce changes in membrane fluidity [58] and impairment of receptor-dependent signaling [59, 60]. Lysophosphatidylethanolamines are minor components of the cell membrane which are able to induce calcium-related and MAPK signaling in several cell lines [61–64].
In addition to the changes in the lipid composition of DRM that occurred during SR-B1-dependent lipid signaling, we compared alterations in protein abundance in DRM from WT SR-B1- and SR-B1-Q445A-expressing cells after PPM supply. Proteins present in DRM are generally hydrophobic and their identification by proteomic analysis is dependent on their intrinsic properties and on the detergents used to solubilize and extract them [65, 66]. Moreover, we selected only proteins that were covered by more than 2 peptides. SR-B1 itself was not reproducibly identified, in reproducible manner, by our proteomic analysis procedure, probably due to its low solubility. Nevertheless, our study revealed modifications in the abundance of a variety of proteins involved in several cellular processes (Table 2). Most of them are related to the cytoskeleton and oxidation/reduction metabolism. It is well known that the dynamics of plasma membrane organization and lipid raft assembly are dependent on the actin cytoskeleton and that they consume large amounts of ATP (for review [67]). Additionally, many proteins involved in oxidation/reduction process are located in DRM [68] and they were identified to be altered by PPM supply (such as COX2, VATD, ATPK, VATF). Moreover, this dynamic organization of the plasma membrane is linked to the endosomal/lysosomal system, where an ATP-dependent acidification of vesicles occurs [69, 70]. In our study, we also observed changes in the abundance of syntaxin-16 (STX16) and vesicle-associated membrane protein 8 (VAMP8). These proteins belong to the SNARE (soluble N-ethylmaleimide-sensitive factor attachment protein receptor) family, which is involved in membrane fusion and vesicular trafficking [71]. SNARE proteins participate also in cholesterol trafficking [72], and particularly in its movement from lipid droplets to mitochondria during steroidogenesis [73, 74]. SNARE proteins have been implicated additionally in autophagy [75], which is a cellular process that controls lipid droplet dynamics in intestinal epithelial cells [26]. Moreover, our proteomic analysis showed changes in the abundance of proteins involved directly in signal transduction. These included KAP2 (cAMP-dependent protein kinase type-II), and PDE8A (phosphodiesterase 8A), which are linked to cAMP-dependent signaling [76, 77], and FA83B (Protein FAM83B), which is involved in epidermal growth factor receptor activation and downstream RAS/MAPK and PI3K/AKT/TOR signaling cascades [78, 79]. Finally, we observed an increased abundance of PEX14 (Peroxisomal membrane protein PEX14) in DRM from WT SR-B1 cells after PPM supply that did not occur in DRM from SR-B1-Q445A cells. PEX14 is an important component of peroxisomal import machinery and it interacts directly with other peroxin proteins such as PEX5 [80, 81], which binds peroxisomal targeting signal type 1 (PTS1)–containing proteins such as SR-B1 [82]. Interestingly, PEX14 is also able to interact with LC3 protein involved in autophagy in a process dependent on nutrient availability [83]. Although the discrete functions of these proteins that are pertinent to their recruitment to DRM in response to PPM are yet to be elucidated, it is apparent that the dynamics of DRM composition are greatly influenced by the capacity of SR-B1 to bind plasma membrane-associated cholesterol and to initiate signaling in response to PPM.
In summary, our studies demonstrated that a point mutation in SR-B1 abolishing the property of the receptor to sense lipids induces modifications in lipid composition of raft-like membrane domains and cell cholesterol distribution in intestinal epithelial cells. The SR-B1-dependent lipid sensing influences both apical cholesterol efflux and intracellular cholesterol trafficking from the plasma membrane to lipid droplets. Two saturated lipids (d18:0/16:0 sphingomyelin and 16:0/0:0 lysophosphatidylethanolamine) and several proteins involved in lipid raft dynamics and signal transduction play an important role in lipid sensing by SR-B1 (Fig. 5). Our findings, which provide new insights into the role of SR-B1 in cellular cholesterol homeostasis, suggest molecular links between SR-B1-dependent lipid sensing and cell cholesterol and lipid droplet dynamics.
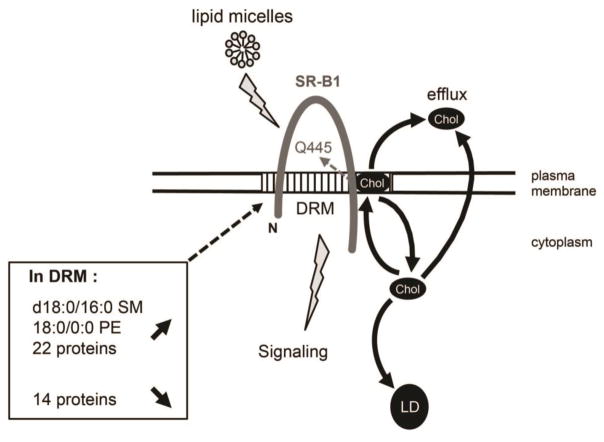
Figure 5. SR-B1-dependent lipid sensing is linked to cholesterol trafficking and modifications of lipid and protein composition of detergent-resistant membranes in intestinal epithelial cells.
In intestinal epithelial cells, SR-B1-dependent lipid sensing in response to lipid micelles requires plasma membrane cholesterol (Chol) interaction with the glutamine at position 445 (Q445) in the C-terminal transmembrane domain of SR-B1 [14, 15]. In this study, we identified that SR-B1-dependent lipid sensing is linked to intracellular cholesterol trafficking from plasma membrane and towards lipid droplet (LD) as well as cholesterol efflux. Lipidomic analysis of detergent-resistant membranes (DRM) revealed that the enrichment of d18:0/16:0 SM and 18:0/0:0 PE is associated with SR-B1-dependent lipid sensing. Moreover, we observed an increased abundance of 22 proteins and a decreased abundance of 14 proteins in DRM after lipid micelles supply, highlighting their potential role in SR-B1-dependent lipid signaling. SM: spingomyelin; PE: phosphatidylethanolamine; N: N-terminal region.
Supplementary Material
Highlights.
SR-B1-dependent lipid sensing is associated with cholesterol trafficking
Changes in raft-like lipid and protein composition occurs during this sensing
Identification of cellular processes driven by SR-B1
Acknowledgments
We thank Patrick Ducoroy and Delphine Pecqueur from CLIPP (Dijon, France) for their expertise in proteomic analysis. We thank Béatrice Riveau for the maintenance of Caco-2/TC7 cell culture. We thank Zeina Chamoun, Alexandra Grosfeld and Sophie Thenet for helpful discussion. We thank Maude Le Gall for helping us with FACS analysis. Confocal microscopy was performed at the Centre d’Imagerie Cellulaire et de Cytométrie (Centre de Recherche des Cordeliers, UMRS 1138, Paris, France).
This research was supported by Institut National de la Santé et de la Recherche Médicale (INSERM) and Université Pierre et Marie Curie (Paris 6), and also by National Institutes of Health grant HL131597 (PWS).
Abbreviations
- AGR2
anterior gradient protein 2 homolog
- CALX
calnexin
- CTTM
C-terminal transmembrane domain
- DRM
detergent-resistant membranes
- HDL
high density lipoprotein
- HNRH1
heterogeneous nuclear ribonucleoprotein H
- HNRPK
heterogeneous nuclear ribonucleoprotein K
- ICAM1
intercellular adhesion molecule 1
- JAM1
junctional adhesion molecule A
- K1C9
keratin, type I cytoskeletal 9
- KAP2
cAMP-dependent protein kinase type II-alpha regulatory subunit
- LDHB
L-lactate dehydrogenase B chain
- LPC
lysophosphatidylcholine
- LPE
lysophosphatidylethanolamine
- MβCD
methyl- β-cyclodextrin
- NDUA2
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex subunit 2
- PC
phosphatidylcholine
- PDE8A
high affinity cAMP-specific and IBMX-insensitive 3′,5′-cyclic phosphodiesterase 8A
- PE
phosphatidylethanolamine
- PI
phosphoinositide
- PL
phospholipids
- pPE
plasmalogen phosphatidylethanolamine
- PPM
postprandial lipid micelles
- PS
phosphatidylserine
- SM
sphingomyelin
- SR-B1
scavenger receptor Class B type 1
- STX16
syntaxin-16
- VAMP8
vesicle-associated membrane protein 8
- VATD
ion transport, such as V-type proton ATPase subunit D
- VP9D1
VPS9 domain-containing protein 1
Footnotes
6. Conflict of interest
No conflict of interest declared.
7. Authors contributions
E.M., S.G. G.L., C.T., J-P.B., S.D., M.R. and V.C. designed and conducted the experiments and analyzed the data. M.R. and V.C. wrote the paper. F.S-P. coordinated lipidomic and proteomic analyses. C.M. and P.W.S. created SR-B1-Q445A mutant. A.L., M.R., F.S-P, C.M. and P.W.S. revised the manuscript. All authors reviewed the results and approved the final version of the manuscript.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Hoekstra M, Van Berkel TJ, Van Eck M. Scavenger receptor BI: a multi-purpose player in cholesterol and steroid metabolism. World J Gastroenterol. 2010;16:5916–5924. doi: 10.3748/wjg.v16.i47.5916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Han CH, Lee MH. Topology of scavenger receptor class B type I (SR-BI) on brush border membrane. J Vet Sci. 2002;3:265–272. [PubMed] [Google Scholar]
- 3.Lobo MV, Huerta L, Ruiz-Velasco N, Teixeiro E, de la Cueva P, Celdran A, Martin-Hidalgo A, Vega MA, Bragado R. Localization of the lipid receptors CD36 and CLA-1/SR-BI in the human gastrointestinal tract: towards the identification of receptors mediating the intestinal absorption of dietary lipids. J Histochem Cytochem. 2001;49:1253–1260. doi: 10.1177/002215540104901007. [DOI] [PubMed] [Google Scholar]
- 4.Cai SF, Kirby RJ, Howles PN, Hui DY. Differentiation-dependent expression and localization of the class B type I scavenger receptor in intestine. Journal of lipid research. 2001;42:902–909. [PubMed] [Google Scholar]
- 5.Mardones P, Quinones V, Amigo L, Moreno M, Miquel JF, Schwarz M, Miettinen HE, Trigatti B, Krieger M, VanPatten S, Cohen DE, Rigotti A. Hepatic cholesterol and bile acid metabolism and intestinal cholesterol absorption in scavenger receptor class B type I-deficient mice. Journal of lipid research. 2001;42:170–180. [PubMed] [Google Scholar]
- 6.Altmann SW, Davis HR, Jr, Zhu LJ, Yao X, Hoos LM, Tetzloff G, Iyer SP, Maguire M, Golovko A, Zeng M, Wang L, Murgolo N, Graziano MP. Niemann-Pick C1 Like 1 protein is critical for intestinal cholesterol absorption. Science (New York, NY. 2004;303:1201–1204. doi: 10.1126/science.1093131. [DOI] [PubMed] [Google Scholar]
- 7.Altmann SW, Davis HR, Jr, Yao X, Laverty M, Compton DS, Zhu LJ, Crona JH, Caplen MA, Hoos LM, Tetzloff G, Priestley T, Burnett DA, Strader CD, Graziano MP. The identification of intestinal scavenger receptor class B, type I (SR-BI) by expression cloning and its role in cholesterol absorption. Biochimica et biophysica acta. 2002;1580:77–93. doi: 10.1016/s1388-1981(01)00190-1. [DOI] [PubMed] [Google Scholar]
- 8.Rigotti A, Trigatti BL, Penman M, Rayburn H, Herz J, Krieger M. A targeted mutation in the murine gene encoding the high density lipoprotein (HDL) receptor scavenger receptor class B type I reveals its key role in HDL metabolism. Proceedings of the National Academy of Sciences of the United States of America. 1997;94:12610–12615. doi: 10.1073/pnas.94.23.12610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bietrix F, Yan D, Nauze M, Rolland C, Bertrand-Michel J, Comera C, Schaak S, Barbaras R, Groen AK, Perret B, Terce F, Collet X. Accelerated lipid absorption in mice overexpressing intestinal SR-BI. The Journal of biological chemistry. 2006;281:7214–7219. doi: 10.1074/jbc.M508868200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bura KS, Lord C, Marshall S, McDaniel A, Thomas G, Warrier M, Zhang J, Davis MA, Sawyer JK, Shah R, Wilson MD, Dikkers A, Tietge UJ, Collet X, Rudel LL, Temel RE, Brown JM. Intestinal SR-BI does not impact cholesterol absorption or transintestinal cholesterol efflux in mice. Journal of lipid research. 2013;54:1567–1577. doi: 10.1194/jlr.M034454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hayashi AA, Webb J, Choi J, Baker C, Lino M, Trigatti B, Trajcevski KE, Hawke TJ, Adeli K. Intestinal SR-BI is upregulated in insulin-resistant states and is associated with overproduction of intestinal apoB48-containing lipoproteins. American journal of physiology. 2011;301:G326–337. doi: 10.1152/ajpgi.00425.2010. [DOI] [PubMed] [Google Scholar]
- 12.Lino M, Farr S, Baker C, Fuller M, Trigatti B, Adeli K. Intestinal scavenger receptor class B type I (SR-BI) as a novel regulator of chylomicron production in healthy and diet-induced obese states. American journal of physiology. 2015 doi: 10.1152/ajpgi.00086.2015. ajpgi 00086 02015. [DOI] [PubMed] [Google Scholar]
- 13.Mineo C, Shaul PW. Functions of scavenger receptor class B, type I in atherosclerosis. Current opinion in lipidology. 2012;23:487–493. doi: 10.1097/MOL.0b013e328357ba61. [DOI] [PubMed] [Google Scholar]
- 14.Beaslas O, Cueille C, Delers F, Chateau D, Chambaz J, Rousset M, Carriere V. Sensing of dietary lipids by enterocytes: a new role for SR-BI/CLA-1. PloS one. 2009;4:e4278. doi: 10.1371/journal.pone.0004278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Saddar S, Carriere V, Lee WR, Tanigaki K, Yuhanna IS, Parathath S, Morel E, Warrier M, Sawyer JK, Gerard RD, Temel RE, Brown JM, Connelly M, Mineo C, Shaul PW. Scavenger Receptor Class B Type I (SR-BI) is a Plasma Membrane Cholesterol Sensor. Circulation research. 2013;112:140–151. doi: 10.1161/CIRCRESAHA.112.280081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Levental I, Veatch SL. The Continuing Mystery of Lipid Rafts. J Mol Biol. 2016;428:4749–4764. doi: 10.1016/j.jmb.2016.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Saddar S, Mineo C, Shaul PW. Signaling by the high-affinity HDL receptor scavenger receptor B type I. Arteriosclerosis, thrombosis and vascular biology. 2010;30:144–150. doi: 10.1161/ATVBAHA.109.196170. [DOI] [PubMed] [Google Scholar]
- 18.Assanasen C, Mineo C, Seetharam D, Yuhanna IS, Marcel YL, Connelly MA, Williams DL, de la Llera-Moya M, Shaul PW, Silver DL. Cholesterol binding, efflux, and a PDZ-interacting domain of scavenger receptor-BI mediate HDL-initiated signaling. The Journal of clinical investigation. 2005;115:969–977. doi: 10.1172/JCI200523858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.de La Llera-Moya M, Connelly MA, Drazul D, Klein SM, Favari E, Yancey PG, Williams DL, Rothblat GH. Scavenger receptor class B type I affects cholesterol homeostasis by magnifying cholesterol flux between cells and HDL. Journal of lipid research. 2001;42:1969–1978. [PubMed] [Google Scholar]
- 20.de la Llera-Moya M, Rothblat GH, Connelly MA, Kellner-Weibel G, Sakr SW, Phillips MC, Williams DL. Scavenger receptor BI (SR-BI) mediates free cholesterol flux independently of HDL tethering to the cell surface. Journal of lipid research. 1999;40:575–580. [PubMed] [Google Scholar]
- 21.Phillips MC. Molecular mechanisms of cellular cholesterol efflux. The Journal of biological chemistry. 2014;289:24020–24029. doi: 10.1074/jbc.R114.583658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zucco F, Batto AF, Bises G, Chambaz J, Chiusolo A, Consalvo R, Cross H, Dal Negro G, de Angelis I, Fabre G, Guillou F, Hoffman S, Laplanche L, Morel E, Pincon-Raymond M, Prieto P, Turco L, Ranaldi G, Rousset M, Sambuy Y, Scarino ML, Torreilles F, Stammati A. An inter-laboratory study to evaluate the effects of medium composition on the differentiation and barrier function of Caco-2 cell lines. Altern Lab Anim. 2005;33:603–618. doi: 10.1177/026119290503300618. [DOI] [PubMed] [Google Scholar]
- 23.Chantret I, Rodolosse A, Barbat A, Dussaulx E, Brot-Laroche E, Zweibaum A, Rousset M. Differential expression of sucrase-isomaltase in clones isolated from early and late passages of the cell line Caco-2: evidence for glucose-dependent negative regulation. Journal of cell science. 1994;107(Pt 1):213–225. doi: 10.1242/jcs.107.1.213. [DOI] [PubMed] [Google Scholar]
- 24.Michau A, Guillemain G, Grosfeld A, Vuillaumier-Barrot S, Grand T, Keck M, L’Hoste S, Chateau D, Serradas P, Teulon J, De Lonlay P, Scharfmann R, Brot-Laroche E, Leturque A, Le Gall M. Mutations in SLC2A2 gene reveal hGLUT2 function in pancreatic beta cell development. The Journal of biological chemistry. 2013;288:31080–31092. doi: 10.1074/jbc.M113.469189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chateau D, Pauquai T, Delers F, Rousset M, Chambaz J, Demignot S. Lipid micelles stimulate the secretion of triglyceride-enriched apolipoprotein B48-containing lipoproteins by Caco-2 cells. Journal of cellular physiology. 2005;202:767–776. doi: 10.1002/jcp.20173. [DOI] [PubMed] [Google Scholar]
- 26.Khaldoun SA, Emond-Boisjoly MA, Chateau D, Carriere V, Lacasa M, Rousset M, Demignot S, Morel E. Autophagosomes contribute to intracellular lipid distribution in enterocytes. Molecular biology of the cell. 2014;25:118–132. doi: 10.1091/mbc.E13-06-0324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jansen M, Ohsaki Y, Rita Rega L, Bittman R, Olkkonen VM, Ikonen E. Role of ORPs in sterol transport from plasma membrane to ER and lipid droplets in mammalian cells. Traffic. 2011;12:218–231. doi: 10.1111/j.1600-0854.2010.01142.x. [DOI] [PubMed] [Google Scholar]
- 28.Spandl J, White DJ, Peychl J, Thiele C. Live cell multicolor imaging of lipid droplets with a new dye, LD540. Traffic. 2009;10:1579–1584. doi: 10.1111/j.1600-0854.2009.00980.x. [DOI] [PubMed] [Google Scholar]
- 29.Beilstein F, Bouchoux J, Rousset M, Demignot S. Proteomic analysis of lipid droplets from Caco-2/TC7 enterocytes identifies novel modulators of lipid secretion. PloS one. 2013;8:e53017. doi: 10.1371/journal.pone.0053017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Menegaut L, Masson D, Abello N, Denimal D, Truntzer C, Ducoroy P, Lagrost L, Pais de Barros JP, Athias A, Petit JM, Martin L, Steinmetz E, Kretz B. Specific enrichment of 2-arachidonoyl-lysophosphatidylcholine in carotid atheroma plaque from type 2 diabetic patients. Atherosclerosis. 2016;251:339–347. doi: 10.1016/j.atherosclerosis.2016.05.004. [DOI] [PubMed] [Google Scholar]
- 31.Folch J, Lees M, Sloane Stanley GH. A simple method for the isolation and purification of total lipides from animal tissues. The Journal of biological chemistry. 1957;226:497–509. [PubMed] [Google Scholar]
- 32.Vial G, Chauvin MA, Bendridi N, Durand A, Meugnier E, Madec AM, Bernoud-Hubac N, Pais de Barros JP, Fontaine E, Acquaviva C, Hallakou-Bozec S, Bolze S, Vidal H, Rieusset J. Imeglimin normalizes glucose tolerance and insulin sensitivity and improves mitochondrial function in liver of a high-fat, high-sucrose diet mice model. Diabetes. 2015;64:2254–2264. doi: 10.2337/db14-1220. [DOI] [PubMed] [Google Scholar]
- 33.Stanislas T, Bouyssie D, Rossignol M, Vesa S, Fromentin J, Morel J, Pichereaux C, Monsarrat B, Simon-Plas F. Quantitative proteomics reveals a dynamic association of proteins to detergent-resistant membranes upon elicitor signaling in tobacco. Mol Cell Proteomics. 2009;8:2186–2198. doi: 10.1074/mcp.M900090-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nesvizhskii AI, Keller A, Kolker E, Aebersold R. A statistical model for identifying proteins by tandem mass spectrometry. Anal Chem. 2003;75:4646–4658. doi: 10.1021/ac0341261. [DOI] [PubMed] [Google Scholar]
- 35.Lai X, Wang L, Tang H, Witzmann FA. A novel alignment method and multiple filters for exclusion of unqualified peptides to enhance label-free quantification using peptide intensity in LC-MS/MS. J Proteome Res. 2011;10:4799–4812. doi: 10.1021/pr2005633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Monroe ME, Shaw JL, Daly DS, Adkins JN, Smith RD. MASIC: a software program for fast quantitation and flexible visualization of chromatographic profiles from detected LC-MS(/MS) features. Comput Biol Chem. 2008;32:215–217. doi: 10.1016/j.compbiolchem.2008.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bouchoux J, Beilstein F, Pauquai T, Guerrera IC, Chateau D, Ly N, Alqub M, Klein C, Chambaz J, Rousset M, Lacorte JM, Morel E, Demignot S. The proteome of cytosolic lipid droplets isolated from differentiated Caco-2/TC7 enterocytes reveals cell-specific characteristics. Biol Cell. 2011;103:499–517. doi: 10.1042/BC20110024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Beilstein F, Carriere V, Leturque A, Demignot S. Characteristics and functions of lipid droplets and associated proteins in enterocytes. Exp Cell Res. 2016;340:172–179. doi: 10.1016/j.yexcr.2015.09.018. [DOI] [PubMed] [Google Scholar]
- 39.Cenedella RJ. Cholesterol synthesis inhibitor U18666A and the role of sterol metabolism and trafficking in numerous pathophysiological processes. Lipids. 2009;44:477–487. doi: 10.1007/s11745-009-3305-7. [DOI] [PubMed] [Google Scholar]
- 40.Brown DA, Rose JK. Sorting of GPI-anchored proteins to glycolipid-enriched membrane subdomains during transport to the apical cell surface. Cell. 1992;68:533–544. doi: 10.1016/0092-8674(92)90189-j. [DOI] [PubMed] [Google Scholar]
- 41.Mineo C, Shaul PW. Regulation of Signal Transduction by HDL. Journal of lipid research. 2013;54:2315–2324. doi: 10.1194/jlr.R039479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hansen GH, Niels-Christiansen LL, Immerdal L, Danielsen EM. Scavenger receptor class B type I (SR-BI) in pig enterocytes: trafficking from the brush border to lipid droplets during fat absorption. Gut. 2003;52:1424–1431. doi: 10.1136/gut.52.10.1424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Soayfane Z, Terce F, Cantiello M, Robenek H, Nauze M, Bezirard V, Allart S, Payre B, Capilla F, Cartier C, Peres C, Al Saati T, Theodorou V, Nelson DW, Yen CL, Collet X, Comera C. Exposure to dietary lipid leads to rapid production of cytosolic lipid droplets near the brush border membrane. Nutr Metab (Lond) 2016;13:48. doi: 10.1186/s12986-016-0107-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tondu AL, Robichon C, Yvan-Charvet L, Donne N, Le Liepvre X, Hajduch E, Ferre P, Dugail I, Dagher G. Insulin and angiotensin II induce the translocation of scavenger receptor class B, type I from intracellular sites to the plasma membrane of adipocytes. The Journal of biological chemistry. 2005;280:33536–33540. doi: 10.1074/jbc.M502392200. [DOI] [PubMed] [Google Scholar]
- 45.Yvan-Charvet L, Bobard A, Bossard P, Massiera F, Rousset X, Ailhaud G, Teboul M, Ferre P, Dagher G, Quignard-Boulange A. In vivo evidence for a role of adipose tissue SR-BI in the nutritional and hormonal regulation of adiposity and cholesterol homeostasis. Arteriosclerosis, thrombosis and vascular biology. 2007;27:1340–1345. doi: 10.1161/ATVBAHA.106.136382. [DOI] [PubMed] [Google Scholar]
- 46.Kawasaki Y, Nakagawa A, Nagaosa K, Shiratsuchi A, Nakanishi Y. Phosphatidylserine binding of class B scavenger receptor type I, a phagocytosis receptor of testicular sertoli cells. The Journal of biological chemistry. 2002;277:27559–27566. doi: 10.1074/jbc.M202879200. [DOI] [PubMed] [Google Scholar]
- 47.Orlowski S, Comera C, Terce F, Collet X. Lipid rafts: dream or reality for cholesterol transporters? Eur Biophys J. 2007;36:869–885. doi: 10.1007/s00249-007-0193-8. [DOI] [PubMed] [Google Scholar]
- 48.Rigotti A, Acton SL, Krieger M. The class B scavenger receptors SR-BI and CD36 are receptors for anionic phospholipids. The Journal of biological chemistry. 1995;270:16221–16224. doi: 10.1074/jbc.270.27.16221. [DOI] [PubMed] [Google Scholar]
- 49.Fielding CJ, Fielding PE. Cholesterol and caveolae: structural and functional relationships. Biochimica et biophysica acta. 2000;1529:210–222. doi: 10.1016/s1388-1981(00)00150-5. [DOI] [PubMed] [Google Scholar]
- 50.Hu J, Zhang Z, Shen WJ, Azhar S. Cellular cholesterol delivery, intracellular processing and utilization for biosynthesis of steroid hormones. Nutr Metab (Lond) 2010;7:47. doi: 10.1186/1743-7075-7-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kellner-Weibel G, de La Llera-Moya M, Connelly MA, Stoudt G, Christian AE, Haynes MP, Williams DL, Rothblat GH. Expression of scavenger receptor BI in COS-7 cells alters cholesterol content and distribution. Biochemistry. 2000;39:221–229. doi: 10.1021/bi991666c. [DOI] [PubMed] [Google Scholar]
- 52.Parathath S, Connelly MA, Rieger RA, Klein SM, Abumrad NA, De La Llera-Moya M, Iden CR, Rothblat GH, Williams DL. Changes in plasma membrane properties and phosphatidylcholine subspecies of insect Sf9 cells due to expression of scavenger receptor class B, type I, and CD36. The Journal of biological chemistry. 2004;279:41310–41318. doi: 10.1074/jbc.M404952200. [DOI] [PubMed] [Google Scholar]
- 53.Leventhal AR, Chen W, Tall AR, Tabas I. Acid sphingomyelinase-deficient macrophages have defective cholesterol trafficking and efflux. The Journal of biological chemistry. 2001;276:44976–44983. doi: 10.1074/jbc.M106455200. [DOI] [PubMed] [Google Scholar]
- 54.Nagao K, Takahashi K, Hanada K, Kioka N, Matsuo M, Ueda K. Enhanced apoA-I-dependent cholesterol efflux by ABCA1 from sphingomyelin-deficient Chinese hamster ovary cells. The Journal of biological chemistry. 2007;282:14868–14874. doi: 10.1074/jbc.M611230200. [DOI] [PubMed] [Google Scholar]
- 55.Yancey PG, de la Llera-Moya M, Swarnakar S, Monzo P, Klein SM, Connelly MA, Johnson WJ, Williams DL, Rothblat GH. High density lipoprotein phospholipid composition is a major determinant of the bi-directional flux and net movement of cellular free cholesterol mediated by scavenger receptor BI. The Journal of biological chemistry. 2000;275:36596–36604. doi: 10.1074/jbc.M006924200. [DOI] [PubMed] [Google Scholar]
- 56.Subbaiah PV, Gesquiere LR, Wang K. Regulation of the selective uptake of cholesteryl esters from high density lipoproteins by sphingomyelin. Journal of lipid research. 2005;46:2699–2705. doi: 10.1194/jlr.M500263-JLR200. [DOI] [PubMed] [Google Scholar]
- 57.Chadwick AC, Sahoo D. Functional Characterization of Newly-Discovered Mutations in Human SR-BI. PloS one. 2012;7:e45660. doi: 10.1371/journal.pone.0045660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kronke M. Biophysics of ceramide signaling: interaction with proteins and phase transition of membranes. Chem Phys Lipids. 1999;101:109–121. doi: 10.1016/s0009-3084(99)00059-6. [DOI] [PubMed] [Google Scholar]
- 59.Chakraborty M, Jiang XC. Sphingomyelin and its role in cellular signaling. Adv Exp Med Biol. 2013;991:1–14. doi: 10.1007/978-94-007-6331-9_1. [DOI] [PubMed] [Google Scholar]
- 60.Langeveld M, Aerts JM. Glycosphingolipids and insulin resistance. Progress in lipid research. 2009;48:196–205. doi: 10.1016/j.plipres.2009.03.002. [DOI] [PubMed] [Google Scholar]
- 61.Park SJ, Lee KP, Im DS. Action and Signaling of Lysophosphatidylethanolamine in MDA-MB-231 Breast Cancer Cells. Biomol Ther (Seoul) 2014;22:129–135. doi: 10.4062/biomolther.2013.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Park KS, Lee HY, Lee SY, Kim MK, Kim SD, Kim JM, Yun J, Im DS, Bae YS. Lysophosphatidylethanolamine stimulates chemotactic migration and cellular invasion in SK-OV3 human ovarian cancer cells: involvement of pertussis toxin-sensitive G-protein coupled receptor. FEBS letters. 2007;581:4411–4416. doi: 10.1016/j.febslet.2007.08.014. [DOI] [PubMed] [Google Scholar]
- 63.Nishina A, Kimura H, Sekiguchi A, Fukumoto RH, Nakajima S, Furukawa S. Lysophosphatidylethanolamine in Grifola frondosa as a neurotrophic activator via activation of MAPK. Journal of lipid research. 2006;47:1434–1443. doi: 10.1194/jlr.M600045-JLR200. [DOI] [PubMed] [Google Scholar]
- 64.Lee JM, Park SJ, Im DS. Calcium Signaling of Lysophosphatidylethanolamine through LPA1 in Human SH-SY5Y Neuroblastoma Cells. Biomol Ther (Seoul) 2016 doi: 10.4062/biomolther.2016.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Whitelegge JP. Integral membrane proteins and bilayer proteomics. Anal Chem. 2013;85:2558–2568. doi: 10.1021/ac303064a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tan S, Tan HT, Chung MC. Membrane proteins and membrane proteomics. Proteomics. 2008;8:3924–3932. doi: 10.1002/pmic.200800597. [DOI] [PubMed] [Google Scholar]
- 67.Simons K, Sampaio JL. Membrane organization and lipid rafts. Cold Spring Harb Perspect Biol. 2011;3:a004697. doi: 10.1101/cshperspect.a004697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kim KB, Lee JW, Lee CS, Kim BW, Choo HJ, Jung SY, Chi SG, Yoon YS, Yoon G, Ko YG. Oxidation-reduction respiratory chains and ATP synthase complex are localized in detergent-resistant lipid rafts. Proteomics. 2006;6:2444–2453. doi: 10.1002/pmic.200500574. [DOI] [PubMed] [Google Scholar]
- 69.Hu YB, Dammer EB, Ren RJ, Wang G. The endosomal-lysosomal system: from acidification and cargo sorting to neurodegeneration. Transl Neurodegener. 2015;4:18. doi: 10.1186/s40035-015-0041-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hsu VW, Prekeris R. Transport at the recycling endosome. Curr Opin Cell Biol. 2010;22:528–534. doi: 10.1016/j.ceb.2010.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lou X, Shin YK. SNARE zippering. Biosci Rep. 2016;36 doi: 10.1042/BSR20160004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Enrich C, Rentero C, Hierro A, Grewal T. Role of cholesterol in SNARE-mediated trafficking on intracellular membranes. Journal of cell science. 2015;128:1071–1081. doi: 10.1242/jcs.164459. [DOI] [PubMed] [Google Scholar]
- 73.Lin Y, Hou X, Shen WJ, Hanssen R, Khor VK, Cortez Y, Roseman AN, Azhar S, Kraemer FB. SNARE-Mediated Cholesterol Movement to Mitochondria Supports Steroidogenesis in Rodent Cells. Molecular endocrinology (Baltimore, Md. 2016;30:234–247. doi: 10.1210/me.2015-1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kraemer FB, Shen WJ, Azhar S. SNAREs and cholesterol movement for steroidogenesis. Mol Cell Endocrinol. 2017;441:17–21. doi: 10.1016/j.mce.2016.07.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wang Y, Li L, Hou C, Lai Y, Long J, Liu J, Zhong Q, Diao J. SNARE-mediated membrane fusion in autophagy. Semin Cell Dev Biol. 2016;60:97–104. doi: 10.1016/j.semcdb.2016.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Taylor SS, Buechler JA, Yonemoto W. cAMP-dependent protein kinase: framework for a diverse family of regulatory enzymes. Annu Rev Biochem. 1990;59:971–1005. doi: 10.1146/annurev.bi.59.070190.004543. [DOI] [PubMed] [Google Scholar]
- 77.Fisher DA, Smith JF, Pillar JS, St Denis SH, Cheng JB. Isolation and characterization of PDE8A, a novel human cAMP-specific phosphodiesterase. Biochemical and biophysical research communications. 1998;246:570–577. doi: 10.1006/bbrc.1998.8684. [DOI] [PubMed] [Google Scholar]
- 78.Cipriano R, Miskimen KL, Bryson BL, Foy CR, Bartel CA, Jackson MW. FAM83B-mediated activation of PI3K/AKT and MAPK signaling cooperates to promote epithelial cell transformation and resistance to targeted therapies. Oncotarget. 2013;4:729–738. doi: 10.18632/oncotarget.1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Cipriano R, Graham J, Miskimen KL, Bryson BL, Bruntz RC, Scott SA, Brown HA, Stark GR, Jackson MW. FAM83B mediates EGFR- and RAS-driven oncogenic transformation. The Journal of clinical investigation. 2012;122:3197–3210. doi: 10.1172/JCI60517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Azevedo JE, Schliebs W. Pex14p, more than just a docking protein. Biochimica et biophysica acta. 2006;1763:1574–1584. doi: 10.1016/j.bbamcr.2006.09.002. [DOI] [PubMed] [Google Scholar]
- 81.Neufeld C, Filipp FV, Simon B, Neuhaus A, Schuller N, David C, Kooshapur H, Madl T, Erdmann R, Schliebs W, Wilmanns M, Sattler M. Structural basis for competitive interactions of Pex14 with the import receptors Pex5 and Pex19. EMBO J. 2009;28:745–754. doi: 10.1038/emboj.2009.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Johnson MS, Johansson JM, Svensson PA, Aberg MA, Eriksson PS, Carlsson LM, Carlsson B. Interaction of scavenger receptor class B type I with peroxisomal targeting receptor Pex5p. Biochemical and biophysical research communications. 2003;312:1325–1334. doi: 10.1016/j.bbrc.2003.11.076. [DOI] [PubMed] [Google Scholar]
- 83.Hara-Kuge S, Fujiki Y. The peroxin Pex14p is involved in LC3-dependent degradation of mammalian peroxisomes. Exp Cell Res. 2008;314:3531–3541. doi: 10.1016/j.yexcr.2008.09.015. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.