Abstract
The small heme-containing protein cytochrome b5 can facilitate, inhibit, or have no effect on cytochrome P450 catalysis, often in a P450-dependent and substrate-dependent manner that is not well understood. Herein, solution NMR was used to identify b5 residues interacting with different human drug-metabolizing P450 enzymes. NMR results revealed that P450 enzymes bound to either b5 α4-5 (CYP2A6 and CYP2E1) or this region and α2-3 (CYP2D6 and CYP3A4) and suggested variation in the affinity for b5. Mutations of key b5 residues suggest not only that different b5 surfaces are responsible for binding different P450 enzymes, but that these different complexes are relevant to the observed effects on P450 catalysis.
Keywords: cytochrome P450, cytochrome, protein-protein interaction, nuclear magnetic resonance (NMR), membrane protein
Introduction
Cytochrome P450 (P450)2 monooxygenases have vital roles both in the metabolism of xenobiotics, including drugs, and in the biosynthesis of endogenous compounds, such as steroids, vitamins, fatty acids, and eicosanoids. Although the ability of P450 enzymes to oxidize such substrates requires interaction with a redox partner protein, catalysis can additionally be modulated by interactions with the membrane-bound heme protein cytochrome b5 (b5). Cytochrome b5 has a complex influence, such that P450 catalysis can be stimulated, not affected, or even inhibited, prompting numerous investigations into the mechanism(s) by which these varied responses are elicited (1, 2). Proposals have largely focused on b5 functioning 1) in a purely redox role of electron delivery or 2) as an allosteric modulator of P450 conformation. Metal-substituted, redox-silent forms of b5 still stimulate certain P450 reactions (4, 5), supporting a purely allosteric role for b5, at least in some cases. Allosteric modulation of P450 conformation could result in alterations in substrate binding, electron or proton delivery, or coupling of NADPH consumption to metabolite formation (1–3).
Regardless of the mechanism, the capability of b5 to modulate P450 catalysis relies on direct binding between b5 and a P450. Insights into individual P450/b5 complexes gained from mutagenesis (3, 4), chemical modification (5), and cross-linking studies (6, 7) are consistent with the convex, negatively charged, heme-exposed face of b5 (Fig. 1A) transiently interacting with the concave, largely positively charged, proximal face of P450 enzymes (Fig. 1B). This proximal P450 face is the same surface to which the required redox partners bind (NADPH-cytochrome P450 oxidoreductase (CPR) for microsomal P450 enzymes), underscoring the necessity of transient interactions between P450 and its protein partners (3, 8).
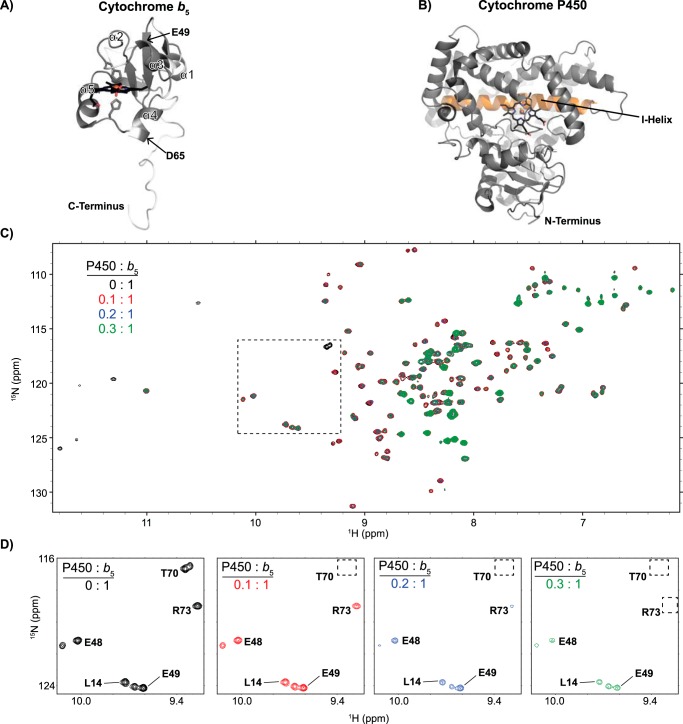
Figure 1.
General structures of proteins in this study and overview of their interaction as monitored by NMR. A, structure of the human cytochrome b5 soluble domain (residues 1–107), as determined by NMR (PDB entry 2I96). The heme and coordinating histidine residues are shown as sticks. α-Helices are labeled 1–5. The locations of Glu-49 and Asp-65 in helices 3 and 4 are indicated. B, P450 structure (CYP2A6, PDB 1Z10) viewed from the proximal face proposed to interact with b5. The heme is again shown as sticks. As a reference, the I helix is highlighted (orange) on the distal side of the heme where the active site is located. C, the 1H-15N HSQC NMR spectra of the soluble domain of human cytochrome b5 titrated with increasing amounts of an unlabeled CYP2A6 saturated with coumarin are representative of the type of changes seen for all P450 enzymes examined herein. Progressive signal loss or line-broadening of most b5 resonances occurs with increasing concentrations of P450, indicating progressive complex formation between the two proteins. D, a selected region of the NMR spectra (the region indicated as a dashed box in C) is shown with increasing P450 to demonstrate differential line broadening effects. In this example with CYP2A6, some resonances are broadened to a greater extent (Thr-70 and Arg-73) than others (Leu-14, Glu-48, and Glu-49).
Fundamental questions persist with regard to P450/b5 interactions. How variable is the binding interface between b5 and different P450 enzymes? Do the affinities vary? Could differences in the b5/P450 interaction underlie the different effects b5 has on P450 catalysis? To complicate matters, the effects that b5 has on catalysis can depend not only on the P450 enzyme, but also the P450 substrate being metabolized (9–11) and experimental parameters, including the ratio of P450 to b5 (10). Differences in b5 effects with different substrates have fueled interest in whether conformational linking exists between the P450 active site cavity and the P450 proximal face interacting with b5 and CPR (8, 12, 13).
Because there are no X-ray structures available of b5 interacting with any P450, solution NMR is an attractive high-resolution approach to decoding these transient binding interactions. Previous studies of 15N-labeled b5 binding to increasing concentrations of unlabeled human steroidogenic CYP17A1 enzyme or rabbit CYP2B4 revealed only small chemical shift perturbations, but more pronounced decreases in the intensity of b5 resonances (8, 14). These line-broadening effects may result from a number of potential sources. Binding of the smaller ∼16-kDa b5 to a larger P450 enzyme (∼50–55 kDa) would enhance the transverse relaxation rate, but could also result in paramagnetic effects from the P450 heme and/or alter the protein-protein interaction to fall within the intermediate-exchange time scale (8, 14). Thus, whereas progressive line broadening occurs upon formation of increasing amounts of the reversible P450:b5 complex, there is not necessarily a 1:1 correspondence between signal loss and amount of complex formed, which prevents the determination of meaningful Kd values. Regardless, the degree of broadening has varied between these two different P450 enzymes, the P450 ligands present, and experimental conditions, such as the presence of lipid mimic systems (8, 14, 15), providing various insights into these P450/b5 interactions. Both CYP17A1 and CYP2B4 studies reported that certain b5 resonances are broadened substantially more than the average. Because most of the b5 amino acids have been assigned to specific resonances (16), such differential line broadening has been used to successfully map out specific b5 residues that transiently interact with CYP17A1 and CYP2B4 (8, 14).
The current study employed such solution NMR experiments to identify and compare b5 residues interacting with the human xenobiotic cytochrome P450 enzymes 2A6, 2D6, 2E1, and 3A4. Results indicate that both shared and distinct interaction surfaces on b5 are present in these respective complexes. Additionally, substantial differences between CYP2D6 and the other P450 enzymes examined are consistent with different relative affinities of the complex with b5. Key b5 residues identified using this approach were subsequently validated by performing mutagenesis and evaluating both complex formation and effects on catalysis. This comparison of P450:b5 complexes provides a physical basis to begin understanding the ability of b5 to differentially modulate P450 catalytic activities.
Results and discussion
Rationale for experimental scope
The specific human P450 enzymes included in this study were selected for their importance in clinical drug metabolism, their diversity across the xenobiotic P450 subfamily, and the range of b5 effects on metabolite production as reported in the literature. Cytochrome b5 reportedly stimulates CYP2E1-mediated oxidation of a number of different substrates, including acetaminophen, aniline, N-nitrosodimethylamine, and chlorzoxazone (17, 18), but may play a redox role, as apo-b5 does not increase product formation (19, 20). The influence of b5 on the major drug-metabolizing P450 enzyme CYP3A4 has been widely documented, with numerous CYP3A4-mediated reactions showing stimulation in the presence of both holo- and apo-b5 (21). CYP2A6 coumarin 7-hydroxylation has also been reported to be stimulated 1.5–2.5-fold by b5 (19, 22–24). CYP2D6 is a clinically significant P450 enzyme because of its ability to metabolize a wide array of pharmaceuticals and its high degree of polymorphism (25), but its metabolite production is reportedly less influenced by b5, at least with in vitro experiments (19, 20, 26, 27). All of the human P450 enzymes in this study were the N-terminally truncated, C-terminally His-tagged forms used to determine X-ray crystallographic structures. They are expressed recombinantly in the significant quantities required to make multiple NMR samples, are highly purified, and are functionally active without lipid addition. The latter aspect provides a common environment across NMR and catalytic assays and avoids variation due to variability in P450 incorporation into lipid.
Each P450 enzyme was saturated with a substrate to additionally stabilize the P450 proteins under NMR-compatible conditions. From significant previous experience with P450 enzymes produced for crystallographic studies, it is clear that high-affinity ligands often greatly improve the stability of these flexible, promiscuous enzymes. They also did so under conditions where the NMR experiments could be collected: 10–100 μm P450 in glycerol-free, low-ionic-strength buffer at 25 °C for several hours. Additionally, some previous reports on P450/b5 interactions have indicated a greater affinity of the complex between the two proteins when P450 substrate is present (5). In this study, classical substrates with a single or at least one major metabolite were preferred, as this might be expected to simplify interaction modes by promoting a more homogeneous P450 conformation as well as permitting quantification of a single product in catalytic assays. Thus, CYP2E1 was saturated with the muscle relaxant chlorzoxazone (denoted as CYP2E1(CZN)), CYP3A4 with the hypotensive drug nifedipine (denoted as CYP3A4(NFP)), CYP2D6 with the cough suppressant dextromethorphan (denoted as CYP2D6(DXM)), and CYP2A6 with coumarin and p-nitrophenol (denoted as CYP2A6(pNP)). Of these substrates, the addition of coumarin, chlorzoxazone, and dextromethorphan to CYP2A6, CYP2E1, and CYP2D6, respectively, yielded shifts in the difference spectra indicating a transition from low spin to high spin, facilitating the ability to monitor protein saturation. The intrinsic absorbance of nifedipine and p-nitrophenol impaired such observations for CYP3A4 and CYP2A6, respectively. Under these conditions, after collection of NMR spectra none of the samples resulted in visible precipitation, and the reduced-carbon monoxide difference spectra were unchanged from the freshly purified P450 protein.
Whereas NMR-observed titrations necessarily involved various concentrations of P450, other experiments were designed to reflect ratios of P450:CPR:b5 often used in P450 functional assays in the literature. This is important, as thoroughly demonstrated in studies on the rabbit CYP2B4 by Waskell and co-workers (10). Herein, all catalytic assays used 1:2:2 ratios of P450 to CPR to b5, rather than optimizing these ratios for maximal turnover with individual P450/substrate combinations as is frequently done. In NMR experiments, this same ratio of P450 to b5 was used to evaluate the b5 mutants (0.5:1 P450:b5, which is the same as 1:2 in the assays). To ensure that these ratios are accurate, special precautions were taken to avoid frequent problems with the quality and quantification of reductase and b5. Cytochrome b5 was reconstituted with heme during purification to avoid large amounts of apo-b5 that can otherwise occur when polypeptide production exceeds heme production and that could also possibly bind P450. Reductase was purified to remove as much as possible of the proteolyzed form that can reduce cytochrome c and might be able to bind P450 but is not able to reduce P450 and promote catalysis. Thus, reductase was quantitated by flavin content instead of cytochrome c reduction. Reductase was also quantitated by total protein from the bicinchoninic acid assay using bovine serum albumin as a standard and was no more than 30% higher than quantitation by flavin absorbance. Both b5 and reductase were the human forms to match the human P450 enzymes, rather than the rat versions often employed. Finally, because electrostatics are probably involved in both steering and binding between b5 and P450 enzymes, to provide additional consistency, the same buffer was used in NMR experiments and all catalytic assays, with the exception of the CYP3A4 nifedipine metabolism assay, which required slightly different conditions (see below).
Finally, although the effects of b5 on P450 catalysis are often simply evaluated by monitoring differences in metabolite formation in the presence and absence of b5, b5 may also have other effects not readily detected by this approach. It has been reported that b5 can change the amount of NADPH consumed per amount of product formation (percent coupling) (1), which would not necessarily alter the amount of product formed. For this reason, NADPH consumption and its coupling to product formation were also evaluated in the absence and presence of wild-type and mutant b5 proteins.
Interaction and catalytic effects of b5 on CYP2A6
When the 2D HSQC spectrum of 15N-labeled b5 alone is collected (e.g. see Fig. 1C (black)), the intensity of each resonance is by definition 100%. As reported previously for CYP17A1 and CYP2B4, titrations of [15N]b5 with increasing concentrations of 2A6(coumarin) result in few chemical shifts, but line broadening occurs and the intensity of most resonances decreases (Fig. 1C), with more marked reductions for specific residues (Fig. 1D). Because the dominant features are changes in intensity and because such overlaid spectra are difficult to comprehend visually, each resonance in each series of spectra was analyzed, and the percentage of intensity remaining was plotted graphically (e.g. Fig. 2A).
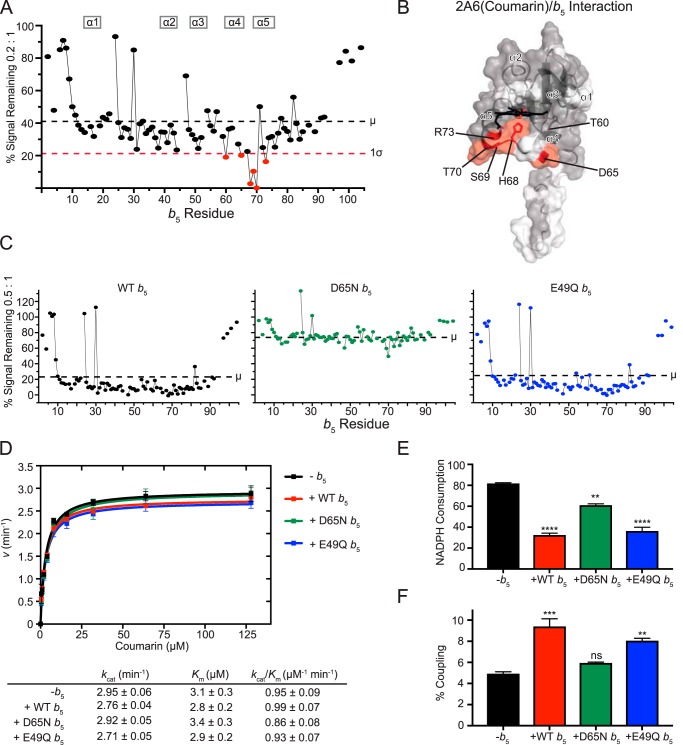
Figure 2.
Interaction of CYP2A6(coumarin) with [15N]b5 by NMR and b5 modulation of CYP2A6 catalysis. A, a single NMR spectrum of 0.2:1 CYP2A6(coumarin):[15N]b5 is shown graphically. The intensity of each b5 resonance (circle) corresponding to an individual b5 residue (x axis) is plotted as a percentage of that resonance's intensity in the absence of P450 (y axis). Fine lines between circles indicate continuous assignments for sequential b5 amino acids. Gaps in the line between circles indicate that one or more intervening b5 amino acids have not been assigned to an individual resonance in the spectrum (also shown in white in B). The average (μ) and average minus one S.D. (1σ) are indicated on the plot by black and red dashed lines, respectively, and constitute the selection criteria of b5 residues involved in P450 binding (red circles). B, on the surface of the human soluble domain b5 structure (PDB entry 2I96), residues displaying differential broadening effects are colored red, whereas residues that have assigned NMR resonances that are not differentially affected by P450 addition are colored gray, and unassigned residues are colored white. C, resonance intensity plots as in A, but comparing the effects of line broadening between WT b5 and b5 mutants D65N and E49Q at a fixed 0.5:1 CYP2A6(coumarin):[15N]b5 ratio. D, effects of wild-type and mutant b5 proteins on Michaelis–Menten kinetic parameters of CYP2A6-mediated coumarin 7-hydroxylation. Each sample was generated in duplicate with S.D. illustrated by error bars. Steady-state kinetic constants below are shown with S.D. E, measurement of NADPH consumed (nmol of NADPH/min/nmol of CYP2A6) for the CYP2A6 coumarin reaction at a fixed coumarin concentration of 128 μm. Samples were generated in duplicate with the S.D. illustrated by error bars. F, percent coupling of CYP2A6-mediated coumarin 7-hydroxylation of coumarin. Samples were generated in duplicate with the S.D. illustrated by error bars. ns, p >0.05; **, p ≤ 0.01; ***, p ≤ 0.001; ****, p ≤ 0.0001.
For CYP2A6(coumarin), reductions in the average b5 signal intensity occur even at very low P450:b5 ratios. At a CYP2A6(coumarin):b5 ratio of 0.1:1, an average of ∼68% of the original signal remained. As expected with increasing formation of the complex, the average intensities continued to decrease as more and more CYP2A6(coumarin) was added. At 0.2:1, on average, ∼41% of the original signal intensity remained (Fig. 2A, black dashed line). At 0.3:1, 0.4:1, and 0.5:1, ∼35%, 26%, and 22% of the original signal remained.
At titration points with ∼40–60% reduction in intensity, there is enough P450:b5 complex formed to clearly identify specific b5 amino acids whose resonances are differentially broadened compared with the average. For CYP2A6(coumarin), this corresponds to the 0.2:1 ratio, which reveals that a number of b5 resonances were broadened more than one S.D. value (Fig. 2A, red dashed line) from the average (Fig. 2A, black dashed line), consistent with the involvement of these b5 residues in binding CYP2A6(coumarin). These b5 resonances correspond to residues Thr-60, Asp-65, His-68, Ser-69, Thr-70, and Arg-73, which cluster on b5 helices 4 and 5 and the loop connecting them (Fig. 2B, red).
To further examine the role of specific b5 residues interacting with CYP2A6(coumarin), a series of mutations were examined. Residues Asp-65 and Glu-49 were selected based on 1) their location on the b5 surface, 2) previous evidence that electrostatic pairing is important in b5/P450 complex formation (4, 8), 3) locations on distinct b5 faces (Fig. 1A), and 4) their location in or near binding interfaces identified herein (see below). Asp-65 is located in b5 α4, whereas Glu-49 is in b5 α3. The effects of the charge-neutralizing E49Q and D65N mutations on P450 interactions with b5 were evaluated at a P450:b5 ratio of 0.5:1 for all P450 enzymes. As indicated above, under these conditions with wild-type b5, the intensity for the average b5 resonance decreases to ∼22% of the original signal (Fig. 2C, left). However, when the b5 mutant D65N was mixed with CYP2A6(coumarin) at the same ratio, the overall line broadening was much less, to only 76% of the original (Fig. 2C, middle), consistent with decreased complex formation between b5 D65N and CYP2A6 (coumarin) compared with wild-type b5. Conversely, mixing the b5 mutant E49Q with CYP2A6(coumarin) yielded average line broadening (to ∼25%; Fig. 2C, right) similar to wild-type b5 (22%; Fig. 2C, left), suggesting that this mutation did not adversely affect formation of the b5/CYP2A6(courmarin) complex. These results suggest that whereas the anionic charge on Asp-65 in b5 helix 4 has a significant role in b5 binding to CYP2A6(coumarin), Glu-49 in b5 helix 3 has little to no effect on complex formation.
To relate these structural observations to enzymatic function, coumarin metabolism assays were subsequently employed. Under our conditions, there was no significant effect of b5 on CYP2A6 7-hydroxycoumarin formation. Neither kcat nor Km was substantially altered in the presence of b5 (Fig. 2D). Thus, it was not surprising that neither the D65N nor E49Q b5 mutants altered coumarin metabolism (Fig. 2D).
Although the presence of wild-type b5 did not alter formation of the 7-hydroxycoumarin product, coupling studies revealed that NADPH consumption was reduced by ∼60% (Fig. 2E, red). Thus, wild-type b5 increased productive coupling of the reaction by ∼1.9-fold (Fig. 2F, red). The b5 D65N mutant, which appeared to decrease b5 binding to CYP2A6(coumarin) in the NMR experiments, yielded a much smaller decrease in NADPH consumption (Fig. 2E, green) and had coupling similar to that when b5 was not present (Fig. 2F, green versus black). The E49Q b5, which did not appear to alter b5 binding to CYP2A6(coumarin) in the NMR experiments, functioned more like wild-type b5, with a similar NADPH consumption (Fig. 2E, blue versus red) and productive coupling (Fig. 2F, blue versus red). Overall, wild-type and E49Q b5 acted similarly, with 1.9- and 1.6-fold increases in coupling, respectively, whereas D65N has little effect on coupling (a 1.2-fold increase in coupling over reactions without b5).
Whereas the effects of b5 on NADPH consumption observed herein are consistent with the ∼50% reduction previously reported for CYP2A6 coumarin 7-hydroxylation (22, 24), there have also been reports that b5 can increase product formation in the range of 1.5–2.5-fold (19, 22–24). However, no such increase in 7-hydroxycoumarin was observed herein. This apparent discrepancy could be due to other differences in experimental conditions. Of particular importance are the protein ratios used for P450:CPR:b5 in catalytic assays. Experimental evidence to date strongly supports mutually exclusive, overlapping binding sites for CPR and b5 on the proximal face of P450 enzymes. As a result, the ratio and relative affinities of reductase and b5 for a particular P450 would dictate the observed effects (1). At high relative ratios and/or affinities, b5 could inhibit required reduction of ferric P450 by reductase, whereas at lower relative ratios and/or affinities, b5 may exert stimulatory effects on other steps of the P450 catalytic cycle to increase coupling (13). That b5 significantly inhibits NADPH consumption without changes in product formation suggests that for this enzyme(substrate) combination, the 1:2:2 ratio may balance the inhibitory and stimulatory effects of b5. However, other significant differences, for example in the species and quantitation of b5 and reductase, the presence of lipid, etc., may also contribute to different observations across the literature with respect to b5 effects on product formation.
Regardless, NMR and functional assays herein consistently suggest that D65N in b5 helix 4 is important in both binding to CYP2A6(coumarin) and in improving coupling of NADPH consumption to product formation. More broadly, the NMR data suggest that b5 surface residues in both helix 4 and helix 5 are involved in binding to CYP2A6(coumarin).
Interaction and catalytic effects of b5 on CYP2E1
CYP2E1-mediated oxidation of many different substrates is reportedly stimulated by b5. These include chlorzoxazone, acetaminophen, aniline, and N-nitrosodimethylamine (17, 18). Studies suggest that b5 may play a redox role with CYP2E1, as apo-b5 does not increase product formation (19, 20).
NMR experiments performed in this study revealed that titration of b5 with CYP2E1(chlorzoxazone), like CYP2A6(coumarin), also caused fairly substantial overall broadening at moderate P450:b5 ratios. At 0.25:1, for example, an average of ∼55% of signal remained (Fig. 3A), and by 0.5:1, only ∼28% of the original signal was present (Fig. 3C). The resonances for b5 residues Val-34, His-68, Ser-69, Thr-70, and Ser-76 had signal losses more than one S.D. below this average (Fig. 3A). Each of these residues except for Val-34 reside on α5 or the preceding loop (Fig. 3B). This result is consistent with a monofacial interaction between b5 and CYP2E1(CZN) involving α5 and nearby residues.
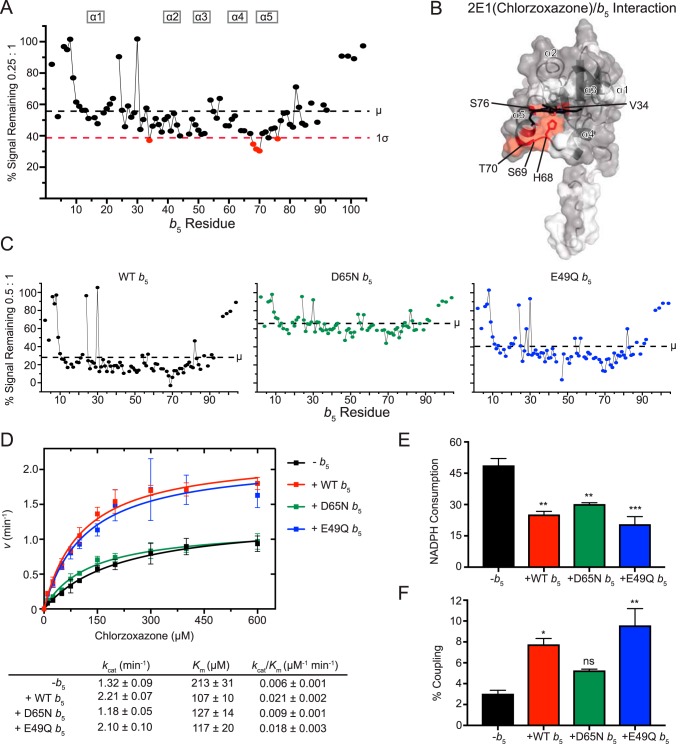
Figure 3.
Interaction of CYP2E1(chlorzoxazone) with [15N]b5 by NMR and b5 modulation of CYP2E1 catalysis. A, a single NMR spectrum of 0.25:1 CYP2E1(CZN):[15N]b5 is shown graphically as described in Fig. 2A. B, mapping of differentially affected residues (red) on human b5. The color code is as described in the legend to Fig. 2B. C, resonance intensity plots comparing line broadening for wild-type b5 and b5 mutants at a 0.5:1 CYP2E1(CZN):[15N]b5 ratio. D, effect of wild-type and mutated b5 on Michaelis–Menten kinetic parameters of CYP2E1-mediated chlorzoxazone 6-hydroxylation. Each sample was generated in triplicate with S.D. illustrated by error bars. Steady-state kinetic constants below are shown with S.D. E, NADPH consumed (nmol of NADPH/min/nmol of CYP2E1) for CYP2E1-mediated chlorzoxazone metabolism at a fixed chlorzoxazone concentration of 600 μm. Samples were generated in duplicate with the S.D. illustrated by error bars. F, percent coupling of CYP2E1-mediated chlorzoxazone 6-hydroxylation. Samples were generated in duplicate with the S.D. illustrated by error bars. ns, p > 0.05; **, p ≤ 0.01; ***, p ≤ 0.001.
Consistent with the above idea, when evaluated at a consistent P450:b5 ratio of 0.5:1, the b5 D65N mutant had less loss of average signal intensity (to 66%; Fig. 3C, middle) than wild-type b5 (Fig. 3C, left). This suggests that the D65N b5 mutant reduces binding to CYP2E1(CZN). Notably, the E49Q mutant on the opposite b5 surface had an intermediate effect on average signal intensity to ∼41% of the original signal.
Comparisons of the mutational effects are clearer when evaluating chlorzoxaone 6-hydroxylation. Consistent with previous literature reports (28, 29), the effect of wild-type b5 on 2E1 chlorzoxazone 6-hydroxylation was pronounced, with both an increased kcat and decreased Km (Fig. 3D). Specifically, experiments herein revealed an ∼1.7-fold increase in kcat and a 2-fold decrease in Km, yielding an ∼3.5-fold increase in kcat/Km. The E49Q b5 mutant performed very similarly to wild-type b5 (Fig. 3D), suggesting that this residue is not critical. However, using the b5 mutant D65N in these reactions yielded substrate metabolism and steady-state kinetic parameters more similar to when b5 was not present at all (Fig. 3D). Thus, the surface near Asp-65 is important in both b5 binding to CYP2E1(CZN) and stimulation of product formation.
Coupling between NADPH consumption and product formation is typically very poor for CYP2E1 in general. Under the current conditions in the absence of b5, CYP2E1 coupling was only ∼3%. The addition of wild-type b5 significantly decreased the NADPH consumption by 48% (Fig. 3E) while increasing product formation, resulting in increased coupling of the reaction by ∼2.5-fold (Fig. 3F). The b5 mutant E49Q behaved similarly to wild-type b5 in that NADPH consumption was decreased to a similar extent (Fig. 3E), and combined with increased product formation, the coupling increased ∼3.1-fold (Fig. 3F), slightly more than in the presence of wild-type b5. The b5 mutant D65N also reduced NADPH consumption (∼38%; Fig. 3E) but had little effect on product formation, thus increasing coupling by ∼1.7-fold when compared with reactions in the absence of b5 (Fig. 3F).
Interactions between b5 and CYP2E1 have previously been studied using cross-linking of the complex coupled with mass spectrometry. Those studies also identified residues on the same surface of b5: Asp-58 and Glu-61 in one study (6) and Asp-58 and Asp-65 in another study (30). In the latter study, these interactions were further tested for their functional role, and both residues appeared to be important in b5 stimulation of the 6-hydroxylation of CZN (30). Unfortunately, the resonance for Asp-58 in b5 has not been assigned, but it would be predicted to undergo differential line broadening. Overall, the current experiments and previous studies support the concept that the region surrounding Asp-65 is involved in both physical interaction between b5 and CYP2E1 and increases observed in product formation.
Interaction and catalytic effects of b5 on CYP3A4
Influence of b5 on CYP3A4 has been reported broadly. A number of CYP3A4-mediated reactions are stimulated by both holo- and apo-b5 (21), which may support a more allosteric role for b5. The current studies evaluated interactions and the effects of b5 on the hypotensive drug NFP.
Using solution NMR to visualize the interaction between CYP3A4(NFP) and b5, it was clear that considerable broadening occurred for the average b5 resonance with low concentrations of CYP3A4(NFP), suggesting substantial formation of the P450:b5 complex. With a CYP3A4(NFP):b5 ratio of 0.1:1, 85% of the average b5 signal remained. Additional CYP3A4(NFP) resulted in further decreases in the average resonance signal (0.2:1, ∼59%; 0.3:1, ∼59%; 0.4:1, ∼41%; 0.5:1, ∼29%).
Although the trends were similar throughout the titration range, differential line broadening was most distinct at a 0.2:1 ratio of CYP3A4(NFP):b5 (Fig. 4A). Residues of b5 differentially broadened more than one S.D. below the average are Leu-41, Glu-42, His-44, Gly-47, Glu-48, Glu-49, Leu-51, Arg-52, His-68, Ser-69, Ala-72, and Arg-73 (Fig. 4A). These residues cluster on helices 2, 3, and 5, as well as the loop between helices 2 and 3, and the loop preceding helix 5 (Fig. 4B). This suggests that these two faces of b5 are likely to interact with CYP3A4(NFP).
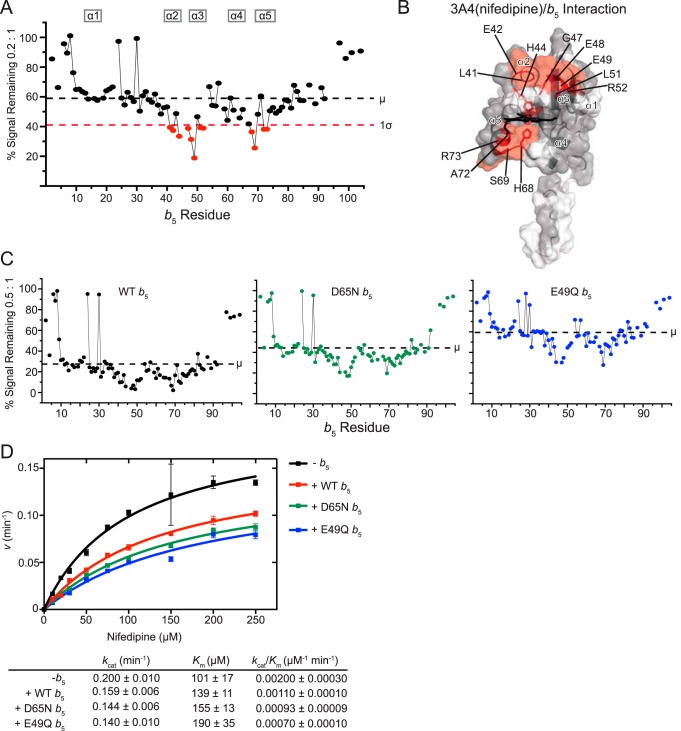
Figure 4.
Interaction of CYP3A4(nifedipine) with [15N]b5 as determined by NMR and catalytic modulation of b5 on CYP3A4-mediated metabolism of nifedipine. A, b5 resonance intensity plot at a 0.2:1 (CYP3A4(nifedipine):[15N]b5) ratio normalized to the free b5 resonance intensity. The color code is as described in the legend to Fig. 2B. B, human soluble domain b5 structure (PDB entry 2I96) with residues displaying differential broadening effects colored red, residues that are assigned in the NMR spectrum colored gray, and unassigned residues colored white. C, b5 resonance intensity plots comparing the effects of line broadening between WT b5 and b5 mutants D65N and E49Q at a fixed 0.5:1 (CYP3A4(nifedipine):[15N]b5) ratio. D, effect of WT b5 and mutants on Michaelis–Menten kinetic parameters of CYP3A4-mediated oxidation of nifedipine. Each sample was generated in duplicate with S.D. illustrated by error bars. Steady-state kinetic constants below are shown with S.D.
To evaluate the significance of these two surfaces, the b5 mutants D65N and E49Q were used to further evaluate the physical interaction between b5 and CYP3A4(NFP). As with the other enzymes, this was conducted at the P450:b5 ratio of 0.5:1. Whereas CYP3A4(NFP) resulted in a signal reduction to an average of 29% for wild-type b5 (Fig. 4C, left), this effect was dampened for both b5 mutants. At this same ratio, ∼45% of the average signal remained for D65N (Fig. 4C, middle) and ∼60% for E49Q (Fig. 4C, right). Thus, both mutations appeared to reduce CYP3A4(NFP):b5 complex formation. Notably, although the overall average intensities were higher for each b5 mutant compared with wild type, distinctive differential broadening still occurred (Fig. 4C, middle and right), suggesting that specific interactions between b5 and 3A4(nifedipine) were not completely disrupted by either single point mutation.
CYP3A4 converts nifedipine to dehydronifedipine. The addition of wild-type b5 to such reactions resulted in inhibition of this reaction (Fig. 4D). The addition of b5 both decreased the kcat and increased the Km such that the kcat/Km was almost half that when b5 was not present. Literature has reported varied stimulation of nifedipine by b5, typically ∼1.3–3-fold increased product formation (19, 31). The differences in these observations may lie in the protein ratios used to perform the experiments. Peng and Auchus (31) used a 1:2:4 ratio in their assays, whereas Yamazaki et al. (19) used a 1:4:1 ratio. The overlapping b5 and reductase binding sites and the variability in these results under different conditions may suggest that at higher concentrations of b5 relative to reductase, b5 may begin to outcompete reductase binding, resulting in an overall reduction in product formation, whereas lower relative concentrations of b5 could be stimulatory. However, there were also other potentially significant differences between how the assays were accomplished in different reports, including the presence of lipid or the constructs or buffer used. CYP3A4-mediated metabolism seems to be more sensitive to these environmental parameters than many other P450 enzymes. Notably, the single b5 mutants D65N and E49Q had effects on nifedipine oxidation most similar to wild-type b5. Thus, the interactions between CYP3A4(NFP) and b5 are not disrupted enough by either single-point mutation to ameliorate the effect of b5 on product formation. Unfortunately, NADPH consumption and coupling of the 3A4(nifedipine) reaction were not successful due to low turnover rates of the reaction under the conditions employed for all of the assays herein.
Overall, the differential line broadening of NMR signal suggests that CYP3A4(NFP) interacts with b5 over the widest surface area for any of the P450/b5 complexes in this study. Interactions between CYP3A4 and b5 have been reported previously in cross-linking and mutational studies. Zhao et al. (7) used mass spectrometry to identify cross-links between the two proteins; residues on human b5 identified were Glu-42, Glu-48, and Glu-61, which are located on α2, α3, and the start of α4. Another study reported similar residues, where mutations in human b5 residues Glu-48, Glu-49, Asp-58, and Asp-65 had significant decreases in the ability of b5 to enhance CYP3A4-mediated testosterone and nifedipine oxidation (31). Moreover, the b5 double mutant D58G/D65G significantly impaired the ability to form cross-links with CYP3A4, whereas the double mutant E48G/E49G had reduced stimulation but was highly dependent on the type of phospholipid in the assay (31). Overall, the current NMR data of the CYP3A4/b5 complex are consistent with reported interactions in these previous studies but highlight additional residues.
Interaction and catalytic effects of b5 on CYP2D6
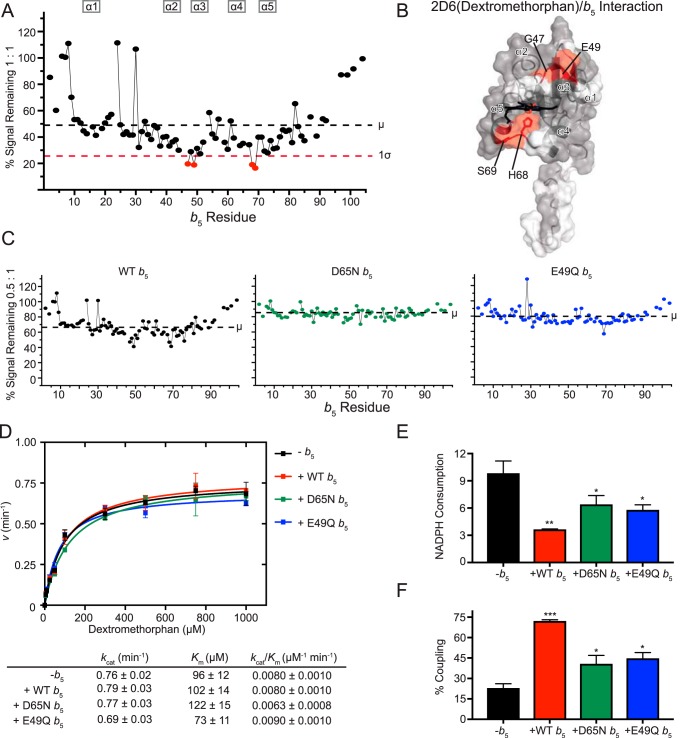
CYP2D6 metabolizes numerous pharmaceuticals, including many antidepressants and antipsychotics. It exhibits substantial polymorphism in humans (25). To assess whether 2D6 forms a complex with b5, NMR-monitored titrations of [15N]b5 with CYP2D6 were carried out using a saturating amount of its common antitussive substrate DXM. The titrations resulted in line broadening, consistent with complex formation between the two proteins, but required much higher concentrations of CYP2D6 than any of the other drug-metabolizing enzymes in this study. For example, it takes a 1:1 CYP2D6(DXM):b5 ratio before the average signal intensity drops to about half (48%; Fig. 5A). In comparison, CYP2A6(coumarin) had more signal reduction by a 0.2:1 ratio.
Figure 5.
Interaction of CYP2D6(dextromethorphan) with [15N]b5 as determined by NMR and catalytic modulation of b5 on CYP2D6-mediated metabolism of dextromethorphan. A, b5 resonance intensity plot at a 1:1 (CYP2D6(DXM):[15N]b5) ratio normalized to the free b5 resonance intensity. The color code is as described in the legend to Fig. 2B. B, human soluble domain b5 structure (PDB entry 2I96) with residues displaying differential broadening effects colored red, residues that are assigned in the NMR spectrum colored gray, and unassigned residues colored white. C, b5 resonance intensity plots comparing the effects of line broadening between WT b5 and b5 mutants D65N and E49Q at a fixed 0.5:1 (CYP2D6(DXM):[15N]b5) ratio. D, effect of WT b5 and mutants on Michaelis–Menten kinetic parameters of CYP2D6-mediated O-demethylation of dextromethorphan. Each sample was generated in triplicate with S.D. illustrated by error bars. Steady-state kinetic constants below are shown with S.D. E, measurement of NADPH consumed (nmol of NADPH/min/nmol of CYP2D6) for CYP2D6 dextromethorphan reaction at a fixed dextromethorphan concentration of 1 mm. Samples were generated in duplicate with the S.D. illustrated by error bars. F, percent coupling of CYP2D6-mediated O-demethylation of dextromethorphan. Samples were generated in duplicate with the S.D. illustrated by error bars. *, p ≤ 0.05; **, p ≤ 0.01; ***, p ≤ 0.001.
Within these data, the resonances for four residues are broadened more than one S.D. from the average. Gly-47, Glu-49, His-68, and Ser-69 are all differentially broadened (Fig. 5A). On the b5 surface, these residues fall into two disconnected regions of b5: the start of α3 (Gly-47 and Glu-49) and the loop between α4 and α5 (His-68 and Ser-69) (Fig. 5B). These data suggest that when a complex is formed at higher concentrations of CYP2D6(DXM), both faces of b5 are involved.
To probe the importance of these distinct faces, the E49Q and D65N b5 mutants were also evaluated for their ability to bind CYP2D6(DXM). At the uniform ratio of 0.5:1, wild-type b5 retained an average signal intensity of 69% (Fig. 5C, left), but both mutants retained even more signal intensity. E49Q and D65N retained ∼80 and ∼84% of the signal intensity, respectively (Fig. 5C, right and middle). This is consistent with both charges contributing to the CYP2D6(DXM):b5 complex when it does occur.
To link these structural observations with the effects of b5 on CYP2D6 dextromethorphan metabolism, both wild-type and mutant b5 proteins were employed in steady-state turnover assays. Similar to other in vitro reports on CYP2D6 reactions (19, 32), the presence of wild-type b5 had no significant effects on kinetics of metabolite formation from dextromethorphan (Fig. 5D). Thus, not surprisingly, neither b5 mutant altered product formation.
When NADPH consumption was measured, however, the presence of b5 did result in a two-thirds reduction (Fig. 5E) and therefore an increase in coupling (from ∼23 to ∼72%; Fig. 5F). Each of the single E49Q and D65N b5 mutants also reduced NADPH consumption (to ∼59 and ∼65%, respectively; Fig. 5E), but not as effectively as wild-type b5. Thus, the ∼2.0-fold increases in coupling with E49Q and D65N b5 were less than the ∼3-fold increase in coupling observed for wild-type b5 (Fig. 5F).
CYP2D6(DXM) interaction with b5 appeared to be similar to CYP3A4(NFP) in that opposite surfaces of b5 were implicated, but extensive broadening of b5 resonances did not occur until an equal molar (1:1) ratio of 2D6(DXM):b5, a feature that would be consistent with a weaker interaction with CYP2D6(DXM) compared with the other P450:b5 pairs studied. Most previous studies have reported no b5 stimulation of CYP2D6-mediated in vitro bufuralol 1′-hydroxylation, tamoxifen 4-hydroxylation, or acetaminophen conversion to its toxic metabolite N-acetyl-p-benzoquinone imine (19, 20, 26, 27). There is one conflicting report indicating that b5 could modulate CYP2D6 metabolite formation in vivo, as mice humanized for CYP2D6 have decreased bufuralol and debrisoquine turnover upon hepatic deletion of the b5 gene. This could be ameliorated by the addition of membranes containing b5 (33).
Interaction and catalytic effects of b5 on CYP2A6 saturated with p-nitrophenol
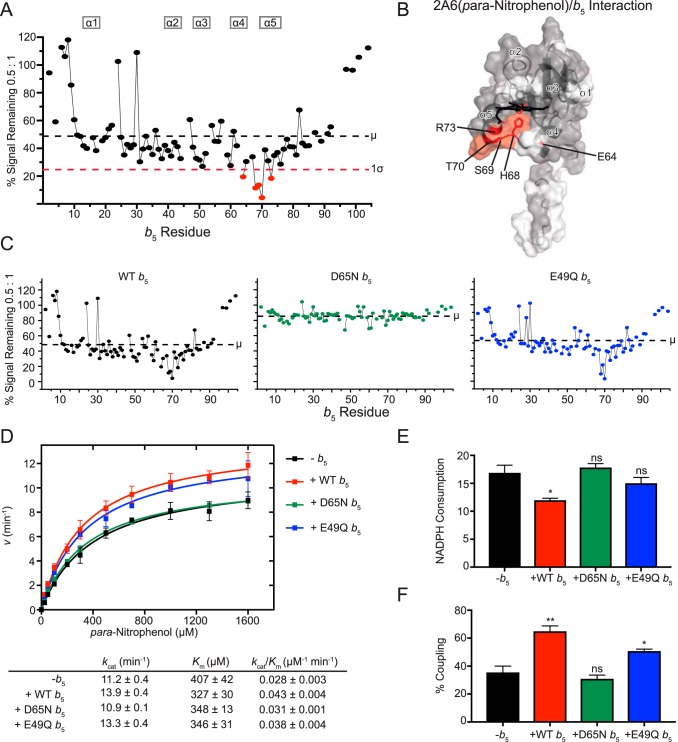
To begin to probe the effects that the identity of a given P450 substrate might have on interactions with b5, the interactions of CYP2A6 saturated with pNP were also investigated using NMR and functional assays and compared with earlier results for CYP2A6(coumarin).
When [15N]b5 was titrated with CYP2A6(pNP), differential line broadening was observed for very similar residues observed with 2A6(coumarin). Specifically, b5 residues significantly affected were Glu-64, His-68, Ser-69, Thr-70, and Arg-73 (Fig. 6A), which comprise b5 α4, α5, and the intervening loop (Fig. 6B). However, the average reduction in signal was not as pronounced when CYP2A6 was saturated with pNP as it was when CYP2A6 was saturated with coumarin. For example, at a P450:b5 ratio of 1:0.5, ∼49% of the average b5 signal remained for the CYP2A6(pNP)/b5 mixture (Fig. 6C, left), compared with ∼22% remaining signal for the CYP2A6(coumarin)/b5 mixture (Fig. 2C, left).
Figure 6.
Interaction of CYP2A6(pNP) with [15N]b5 as determined by NMR and catalytic modulation of b5 on CYP2A6 mediated metabolism of para-nitrophenol. A, b5 resonance intensity plot at a 0.5:1 (CYP2A6(pNP):[15N]b5) ratio normalized to the free b5 resonance intensity. The color code is as described in the legend to Fig. 2B. B, human soluble domain b5 structure (PDB entry 2I96) with residues displaying differential broadening effects colored red, residues that are assigned in the NMR spectrum colored gray, and unassigned residues colored white. C, b5 resonance intensity plots comparing the effects of line broadening between WT b5 and b5 mutants D65N and E49Q at a fixed 0.5:1 (CYP2A6(pNP):[15N]b5) ratio. D, effect of WT b5 and mutants on Michaelis–Menten kinetic parameters of CYP2A6-mediated 2-hydroxylation of pNP. Each sample was generated in duplicate with S.D. illustrated by error bars. Steady-state kinetic constants below are shown with S.D. E, measurement of NADPH consumed (nmol of NADPH/min/nmol of CYP2A6) for CYP2A6 pNP reaction at a fixed pNP concentration of 1.6 mm. Samples were generated in duplicate with the S.D. illustrated by error bars. F, percent coupling of CYP2A6-mediated 2-hydroxylation of pNP. Samples were generated in duplicate with the S.D. illustrated by error bars. ns, p > 0.05; *, p ≤ 0.05; **, p ≤ 0.01.
The importance of Asp-65 in forming the complex between b5 and CYP2A6(pNP) was similar to that observed with the substrate coumarin. At a uniform 0.5:1 P450:b5 ratio, the b5 D65N mutation retained more signal average intensity (∼86%; Fig. 6C, middle) than wild-type b5 (49% signal remaining; Fig. 6C, left), consistent with reduced complex formation for the mutant. By comparison, under the same conditions, the b5 E49Q mutant had an average intensity (54%; Fig. 6C, right) much more similar to that of wild-type b5, suggesting that this b5 residue does not play a significant role in binding CYP2A6(pNP).
The effects of these b5 mutants on formation of a CYP2A6(pNP)/b5 complex correlated with observations of the kinetics of CYP2A6 para-nitrophenol metabolism to 4-nitrocatechol. Whereas wild-type b5 modestly stimulated the reaction with both an increase in kcat and decrease in Km, resulting in an overall ∼1.5-fold increase in catalytic efficiency (Fig. 6D), the b5 mutant E49Q performed very similarly (Fig. 6D), suggesting that this residue is not critical for b5 simulation of CYP2A6-mediated pNP metabolism. In contrast, reactions substituting the b5 mutant D65N resulted in steady-state kinetic parameters more similar to reactions containing no b5 at all (Fig. 6D), suggesting that Asp-65 is critical for the b5 stimulation of CYP2A6-mediated pNP oxidation. It has previously been reported that CYP2A6-mediated pNP oxidation is not significantly altered by the presence of b5 (34). However, these results were obtained at a single pNP concentration that was relatively low (5 μm) and at a CYP2A6:CPR ratio of 1:5.8 (34). The CYP2A6 pNP assays performed in the current study used a range of pNP concentrations (25–1600 μm) and a CYP2A6:CPR ratio of 1:2. Thus, differences in experimental parameters could contribute to observed differences in b5 stimulation and again suggest that the ratio of b5 to CPR is likely to determine the outcome because these two proteins both bind, and probably compete for, the P450 proximal surface.
Evaluation of NADPH coupling herein revealed that wild-type b5 decreased NADPH consumption by ∼30% (Fig. 6E) and increased coupling of NADPH consumption to formation of the 4-nitrocatechol product formation 1.8-fold (Fig. 6F). By comparison, neither the E49Q or D65N mutation significantly altered NADPH consumption (Fig. 6E). However, the increased product formation observed for the b5 E49Q results in a ∼1.4-fold increase in coupling, whereas the absence of effect of D65N b5 on product formation and NADPH consumption meant coupling was the same as in the absence of b5 (Fig. 6F). Thus, these studies also support a significant role for Asp-65, but less so for Glu-49.
Overall, the NMR and mutation data herein suggest that the b5 surface involved in binding to CYP2A6(pNP) (Fig. 6B) is very similar to the b5 surface binding CYP2A6(coumarin) (Fig. 2B). Mutation studies confirmed that Asp-65 on one face of b5 plays a significant role in both CYP2A6/b5 interactions, whereas Glu-49 on the opposing b5 face had relatively little contribution. The greater degree of line broadening occurring with the substrate coumarin compared with pNP is potentially consistent with a stronger b5 interaction with CYP2A6(coumarin) compared with CYP2A6(pNP). However, we cannot exclude effects due to possible differences in CYP2A6 paramagnetism when bound to different ligands. CYP2A6(coumarin) results in almost complete conversion to high spin, whereas the CYP2A6(pNP) spin state cannot be readily assessed because of significant pNP absorbance in the region of interest. Using immobilized CYP2A6 and biotinylated b5, Guengerich and co-workers (27) found no difference in binding for unliganded CYP2A6 versus CYP2A6 bound to coumarin. Like the structural data, the effects of b5 mutants in turnover assays support an important role for D65N in coupling and metabolite formation. In contrast, E49Q performed more similarly to wild-type enzyme, suggesting that this residue is not critical for changes in CYP2A6-mediated pNP metabolism. In both CYP2A6-mediated coumarin and pNP metabolism, b5 increased coupling ∼2-fold. However, this mostly stemmed from a decrease in NADPH utilization for coumarin hydroxylation, whereas for pNP oxidation, both decreased NADPH consumption and increased metabolite formation contributed. Thus, although the b5 residues binding CYP2A6 are conserved across these two substrates, the effects on CYP2A6 catalysis vary.
Comparison of P450/b5 interactions across P450 enzymes and substrates
Of the four human P450 enzymes surveyed, all of them were found to interact with cytochrome b5 and to do so predominately on at least one of the surfaces surrounding the heme-exposed face of b5. This is in general agreement with studies assessing b5 interactions with CYP17A1 (4, 8), CYP2B4 (14), CYP3A4 (7, 31), and CYP2E1 (6, 30). However, the use of NMR allows one to simultaneously probe all possible b5 residues involved in the interaction without modifying either interacting partner aside from isotopic labeling. As a result, this study identified two b5 surfaces differentially interacting with different xenobiotic P450 enzymes. All four P450 enzymes in this study interacted with His-68 and Ser-69 on the loop between α4 and α5. Three of them (CYP2A6, CYP2E1, and CYP3A4) additionally interacted with adjacent residues in b5 α5. In addition to this patch on one side of the heme, CYP2D6 and CYP3A4 can also bind to the opposite surface of b5, α3, with CYP3A4 having the broadest interaction surface for both regions. Notably, CYP2A6 saturated with either coumarin or pNP interacted with the same face of b5, but it remains to be seen whether this is true for other P450/substrate pairs. It is clear that across these P450/substrate pairs and across the different binding interfaces observed, b5 consistently increases coupling but may or may not alter product formation, as was also seen for rabbit CYP2B4 (10).
Although the current NMR studies provide detailed structural information about the P450/b5 interaction, they are not well suited to the determination of dissociation constants. However, it is notable that CYP2D6(DXM) was a very distinct outlier in terms of the amount of line broadening observed for b5 resonances compared with CYP3A4(NFP), CYP2A6(coumarin or pNP), CYP2E1(CZN), and even CYP17A1 in a previous parallel study (8). Although other explanations are possible, the simplest explanation is that this observation is consistent with less complex formation. A study evaluating the physical interaction between P450 enzymes immobilized on a plastic plate and biotinylated b5 ranked the affinities as unliganded CYP3A4 > CYP2A6 ∼ CYP2D6 > CYP2E1 but noted that much less CYP2D6/b5 complex was formed in these experimental conditions as well (27).
In conclusion, it appears that cytochrome b5 interacts with different drug-metabolizing P450 enzymes with both shared and distinct surfaces. Disruption of these surfaces correlates with functional effects on metabolite production and/or NADPH consumption. Thus, in the absence of X-ray structures, solution NMR is a high-resolution technique to examine these transient P450 interactions with other proteins. Further work remains to map the P450 residues involved in binding b5, to compare b5 and reductase competition for binding different P450 enzymes, and to determine the mechanism(s) by which b5 modulates P450 catalysis.
Experimental procedures
Generation of the soluble domain of human cytochrome b5 with 15N-labeling for NMR experiments
A synthetic, codon-optimized gene encoding the soluble domain (residues 1–108) of human microsomal cytochrome b5 with a C-terminal His6 tag (Genewiz) was cloned into the NcoI and BamHI restriction sites of pET15b and transformed into E. coli C41 (DE3) cells already containing the pGro7 plasmid (Takara Bio) for expression of GroEL/GroES chaperones. Transformed cells were selected by growing cells for ∼18 h at 37 °C on a non-inducing minimal medium plate (MDAG-11) (35) supplemented with carbenicillin (100 μg/ml) and chloramphenicol (20 μg/ml) to select for the b5 and chaperone plasmids, respectively. All subsequent cultures contained these antibiotics. A single colony was picked and grown for ∼16 h at 37 °C with shaking (250 rpm) in a 50-ml liquid culture of non-inducing minimal medium MDAG-135 (35). Expression cultures consisted of 1 liter of a defined minimal medium (48 mm Na2HPO4, 22 mm KH2PO4, 9 mm NaCl, 19 mm 15N NH4Cl, 53 mm glucose, 4 mm MgSO4, and trace metals) inoculated with 5 ml of the liquid starter culture. Cells were grown with shaking (250 rpm) at 37 °C, to an optical density at 600 nm (A600) of 0.3, at which point the heme precursor δ-aminolevulinic acid (to 1 mm) and the chaperone inducer l-arabinose (to 13 mm) were added. After A600 reached 0.7–0.8, b5 expression was induced by adding 0.4 mm isopropyl 1-thio-β-d-galactopyranoside, and the temperature and shaking were reduced to 30 °C and 225 rpm, respectively. Cultures were subsequently grown for 20 h before harvesting and freezing the cell pellet at −80 °C. Purification was initiated by resuspending cells in lysis buffer (500 mm potassium phosphate, 100 mm NaCl, 15% glycerol, 1 mm EDTA, pH 7.4) with the addition of 1 mm PMSF. Resuspended cells were lysed by a French press, and heme reconstitution was performed as described (36). Lysed cells were clarified by ultracentrifugation at 140,000 × g and loaded onto a pre-equilibrated 25-ml Ni-NTA column (Qiagen) with loading buffer (100 mm potassium phosphate, 20% glycerol, 200 mm NaCl, 20 mm imidazole, pH 7.4), washed with an additional 8 column volumes of loading buffer, and eluted using a 6 column volumes of elution buffer (loading buffer with 200 mm imidazole). Eluted fractions with A413/A280 > 4.0 were pooled and concentrated before loading on a Superdex 200 Increase 10/300 GL column (GE Healthcare) and run with gel filtration buffer (50 mm potassium phosphate, 20% glycerol, 100 mm NaCl, pH 7.4). The protein sample was then exchanged into a storage buffer (50 mm potassium phosphate, 10% glycerol, pH 7.4) using a HiTrap desalting column (GE Healthcare). Final protein preparation was evaluated by SDS-PAGE and UV-visible spectroscopy (A413/A280 > 6.0) and quantified using an extinction coefficient of 117 mm−1 cm−1 at 413 nm (37). Aliquots were stored at −80 °C until use. Samples for NMR were created by exchanging labeled b5 into NMR buffer (50 mm potassium phosphate, 50 mm NaCl, 10% D2O, pH 6.5) through dilution and centrifugation.
Generation of full-length human cytochrome b5 for catalytic assays
A synthetic, codon-optimized DNA sequence encoding full-length human cytochrome b5 plus a C-terminal His6 tag was generated (GenScript) and cloned into pET-15b vector using restriction enzymes NcoI and BamHI. Expression of full-length b5 was similar to the soluble domain of b5, except 1) expression cultures were grown in Terrific broth, 2) induction was performed with 0.1 mm isopropyl 1-thio-β-d-galactopyranoside at an A600 of 1.1, and 3) induced cultures were grown at 20 °C with 250 rpm shaking for 44 h. Purification of full-length b5 was initiated by resuspending cells in resuspension buffer (100 mm potassium phosphate, 20% glycerol, 1 mm EDTA, pH 7.4) supplemented with 1× HALT protease inhibitor mixture (Thermo Fisher) and 1 mm PMSF. Cells were lysed and reconstituted with heme as described above for the soluble domain. Membrane fractions were isolated by ultracentrifugation at 140,000 × g for 30 min, and b5 was extracted using resuspension buffer with the addition of 0.2 m NaCl and 1% (w/v) CHAPS. Extracted membranes were pelleted by a second ultracentrifuge step at 140,000 × g for 30 min, and the b5-containing supernatant was loaded onto a Ni-NTA column (Qiagen) and washed as described for the soluble domain of b5 but with the addition of 0.5% (w/v) CHAPS. Eluted fractions with A413/A280 > 2.0 were pooled, concentrated, and loaded on a Sephacryl S-200 HR column (GE Healthcare) equilibrated and run with gel filtration buffer (50 mm potassium phosphate, 20% glycerol, 100 mm NaCl, 0.05% (w/v) CHAPS, pH 7.4) and exchanged into storage buffer as described above. Protein purity was assessed on SDS-PAGE, and UV-visible spectroscopy was used to confirm degree of heme incorporation (A413/A280 = 3.0) and quantify dithionite-reduced b5 (36).
Generation of full-length human NADPH-cytochrome P450 reductase for catalytic assays
A synthetic, codon-optimized gene for full-length human NADPH-cytochrome P450 reductase (Integrated DNA Technologies) preceded by the ompA signal peptide and followed by a His6 tag was cloned into pET-29a(+) using NdeI and HindIII. Expression of reductase was performed as described previously for a truncated CPR construct (38) with modifications. Modifications to the expression included 1) transformation and generation of the starter culture as described for soluble b5, 2) supplementation of expression cultures with 2 mg/liter riboflavin, and 3) shaking at 200 rpm after induction. Full-length CPR was also purified in a manner similar to a previously described method (38), with modifications: 1) membrane fractions after cell lysis were isolated by ultracentrifugation at 140,000 × g for 25 min before detergent extraction, 2) omission of the ammonium sulfate precipitation step, and 3) the addition of a second nickel affinity column after elution from Octyl-Sepharose 4 Fast Flow (GE Healthcare) resin to deplete detergent. Final reductase samples were evaluated on SDS-PAGE and by UV-visible spectroscopy. CPR was quantitated by flavin absorbance of the fully oxidized protein at 454 nm with an extinction coefficient of 21.4 mm−1 cm−1 (39).
Generation of P450 enzymes for NMR and catalytic assays
All cytochrome P450 enzymes were the forms used previously to generate crystallographic structures and resulted in catalytically active protein omitting the N-terminal transmembrane helix and adding a C-terminal His tag. Constructs and expression and purification of human CYP2A6 (40) and CYP2E1 (41) were reported previously. Synthetic, codon-optimized genes encoding CYP2D6 and CYP3A4 were generated (GenScript) to match the reported constructs (42, 43), cloned into the pCWori vector, and expressed and purified as described in these same publications with some modifications. Briefly, E. coli DH5α containing the pGro7 plasmid (Takara Bio) was used to express the P450 enzymes. Transformation and starter cultures were performed as described for soluble b5. Proteins were purified by isolating spheroplasts (44), lysis by French press, and detergent extraction using 4.8 mm CYMAL-5 (CYP2A6 and CYP2E1) or 14 mm CHAPS (CYP2D6 and CYP3A4). After extraction, lysate was clarified by ultracentrifugation at 104,000 × g and purified through Ni-NTA, carboxymethyl cellulose ion-exchange, and size-exclusion chromatography. All proteins were depleted of detergent during ion-exchange chromatography, exchanged into storage buffer (50 mm potassium phosphate, 10% glycerol, pH 7.4), and frozen as aliquots at −80 °C. P450 samples were evaluated by SDS-PAGE and UV-visible spectroscopy. The reduced-carbon monoxide difference spectra revealed, at most, only trace amounts of P420.
Mutagenesis
Single-amino acid mutants of cytochrome b5 E49Q and D65N were produced by the QuikChange Lightning site-directed mutagenesis kit (Stratagene) for the full-length construct and commercially (GENEWIZ) for the soluble-domain construct. The complete genes were verified by sequencing, and protein was expressed and purified as described for the respective wild-type constructs.
NMR spectroscopy
NMR experiments were acquired at 298 K using a Bruker Avance III HD 800 MHz with a QCI cryogenic probe. Data were processed using NMRPipe (45) and visualized and analyzed using Sparky version 3 (T. D. Goddard and D. G. Kneller, University of California, San Francisco). Assignments for the backbone amide signals for human soluble domain of b5 were transferred from the deposited chemical shifts in the Biological Magnetic Resonance Data Bank (accession number 6921) (16). All of the 2D 15N HSQC experiments were obtained on 15N-labeled, soluble domain b5 (0.1 mm) in NMR buffer (50 mm potassium phosphate, 50 mm NaCl, 10% D2O, pH 6.5) acquired with 64 scans and 128 increments, which took about 2.5 h. The signal for each individual resonance was measured as peak height, with the value at 0:1 P450:b5 defined as 100% signal. For each titration point within a series, a new sample of labeled b5 was prepared, and a defined molar amount of an unlabeled P450 enzyme was added to the sample. For samples including P450 enzyme, the peak height for each resonance was expressed as a percentage of the resonance peak height in the absence of the P450. Concentration of the P450 substrate was saturating and constant between samples (500 μm coumarin for 2A6, 500 μm CZN for 2E1, 2 mm DXM for 2D6, 5 mm pNP for 2A6, or 200 μm NFP for 3A4). NMR experiments were performed with both b5 and P450 in their oxidized states. At the conclusion of each NMR experiment, samples were collected from the NMR tube, centrifuged to determine whether any precipitation had occurred, and evaluated in terms of the reduced-carbon monoxide difference spectrum.
Cytochrome P450 catalytic assays
All catalytic assays were performed by incubating a P450 with full-length human P450 reductase and full-length human b5 (when applicable) at a 1:2:0 ratio for minus b5 reactions or a 1:2:2 ratio for b5 containing reactions for 20 min at room temperature. The amounts of P450 enzyme were 10 pmol (coumarin assay) or 50 pmol (pNP assay) of CYP2A6, 50 pmol of CYP2D6 (DXM assay), 50 pmol of CYP2E1 (CZN assay), and 50 pmol of CYP3A4 (NFP assay). This protein mixture was added to the same buffer used for NMR experiments, except for the nifedipine assay, which used an assay buffer consisting of 40 mm HEPES, 30 mm MgCl2, pH 7.4. The reactions also contained the respective P450 substrate (0–128 μm coumarin, 0–600 μm CZN, 0–1600 μm pNP, 0–1000 μm DXM, 0–250 μm nifedipine). For the nifedipine assay, all reactions were performed in amber vials. The reactions were preincubated at 37 °C for 3 min and then initiated with 1 mm NADPH and allowed to proceed at 37 °C for 10 min (coumarin, CZN, and pNP assays), 15 min (DXM assay), or 20 min (nifedipine assay). Reactions were terminated by the addition of diluted perchloric acid (acetonitrile for the nifedipine assay), placed on ice, and centrifuged at 5000 × g for 5 min before injection onto a Luna C18 (5 μm, 150 × 4.60 mm; Phenomenex) column at a flow rate of 1 ml/min (0.75 ml/min for the nifedipine assay). Separation on HPLC was obtained using the following mobile phase solutions for each of the respective assays. For separation of coumarin and its 7-OH metabolite, the mobile phase consisted of 50%/50% 20 mm potassium phosphate, pH 2.8/methanol. For CZN and its 6-OH metabolite, the mobile phase consisted of 75%/25% 20 mm potassium phosphate, pH 2.8/acetonitrile to elute product and then a sharp gradient to 40%/60% to elute substrate, followed by reequilibration to 75%/25%. For pNP and its 4-nitrocatechol metabolite, the mobile phase consisted of 73%/27% 10 mm potassium phosphate, pH 3.5/acetonitrile. For DXM and its O-demethylated metabolite dextrorphan, the mobile phase consisted of 50%/50% 10 mm potassium phosphate, pH 3.5/50% acetonitrile and 100% methanol (250:200, v/v). For nifedipine and its metabolite dehydronifedipine, the mobile phase consisted of 45%/55% water/methanol. Metabolite detection occurred by fluorescence for coumarin (355-nm excitation, 460-nm emission) and DXM (280-nm excitation, 310-nm emission) assays and by UV absorbance for CZN (287 nm), pNP (345 nm), and NFP (270 nm) assays. The amounts of the different metabolites produced were calculated using authentic standards prepared by the same method as the samples. Each reaction at each substrate concentration was performed at least in duplicate. Data were fit to the Michaelis–Menten equation using GraphPad Prism.
Measurement of NADPH consumption
NADPH consumption during the various P450 reactions was measured by creating reaction samples similar to the catalytic assays except for the following: 1) 100 pmol P450 was used to increase signal/noise, 2) reactions were scaled up to 1 ml in the respective assay buffer containing the maximum substrate concentration used in turnover reactions, 3) NADPH concentration was reduced to 0.5 mm to reduce background, and 4) reactions were allowed to proceed for 10 min. CYP2A6(pNP) samples required the use of 2-mm path length cuvettes due to high substrate absorbance, so these reactions used 50 pmol of P450 and a final volume of 500 μl. Rates of NADPH consumption were measured by monitoring linear decreases in absorbance at 340 nm. The amount of NADPH consumed was calculated using an extinction coefficient of 6.22 mm−1 cm−1. Control reactions omitting P450 provided the background NADPH consumption, which was subtracted. The amount of product at the end of each reaction was measured using the same methods described for each catalytic assay. Coupling efficiencies were calculated by dividing metabolite product formation in these experiments by their respective NADPH consumption. All measurements were performed in duplicate.
Author contributions
A. G. B. and E. E. S. conceptualization; A. G. B. data curation; A. G. B. and E. E. S. formal analysis; A. G. B. and E. E. S. investigation; A. G. B. visualization; A. G. B. and E. E. S. methodology; A. G. B. and E. E. S. writing-original draft; A. G. B. and E. E. S. writing-review and editing; E. E. S. supervision; E. E. S. funding acquisition; E. E. S. project administration.
Acknowledgments
NMR data were collected at the Ohio State University Campus Chemical Instrument Center. Sarah D. Burris did site-directed mutagenesis to make the full-length cytochrome b5 mutants E49Q and D65N and performed expression and purification. She also expressed and purified CYP3A4 and provided feedback on the manuscript.
This work was supported by National Institutes of Health Grants F37 GM076343 (to E. E. S.) and T32 GM008545. The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
- P450
- cytochrome P450
- CPR
- NADPH-cytochrome P450 oxidoreductase
- CYP
- cytochrome P450
- b5
- cytochrome b5
- CZN
- chlorzoxazone
- pNP
- para-nitrophenol
- DXM
- dextromethorphan
- NFP
- nifedipine
- Ni-NTA
- nickel-nitrilotriacetic acid
- HSQC
- heteronuclear single quantum coherence
- PDB
- Protein Data Bank.
References
- 1. Waskell L., and Kim J.-J. P. (2015) Electron transfer partners of cytochrome P450. In Cytochrome P450: Structure, Mechanism, and Biochemistry, 4th Ed (Ortiz de Montellano P. R., ed) pp. 33–68, Springer International Publishing, Cham, Switzerland [Google Scholar]
- 2. Schenkman J. B., and Jansson I. (2003) The many roles of cytochrome b5. Pharmacol. Ther. 97, 139–152 [DOI] [PubMed] [Google Scholar]
- 3. Bridges A., Gruenke L., Chang Y. T., Vakser I. A., Loew G., and Waskell L. (1998) Identification of the binding site on cytochrome P450 2B4 for cytochrome b5 and cytochrome P450 reductase. J. Biol. Chem. 273, 17036–17049 [DOI] [PubMed] [Google Scholar]
- 4. Naffin-Olivos J. L., and Auchus R. J. (2006) Human cytochrome b5 requires residues E48 and E49 to stimulate the 17,20-lyase activity of cytochrome P450c17. Biochemistry 45, 755–762 [DOI] [PubMed] [Google Scholar]
- 5. Tamburini P. P., White R. E., and Schenkman J. B. (1985) Chemical characterization of protein-protein interactions between cytochrome P-450 and cytochrome b5. J. Biol. Chem. 260, 4007–4015 [PubMed] [Google Scholar]
- 6. Gao Q., Doneanu C. E., Shaffer S. A., Adman E. T., Goodlett D. R., and Nelson S. D. (2006) Identification of the interactions between cytochrome P450 2E1 and cytochrome b5 by mass spectrometry and site-directed mutagenesis. J. Biol. Chem. 281, 20404–20417 [DOI] [PubMed] [Google Scholar]
- 7. Zhao C., Gao Q., Roberts A. G., Shaffer S. A., Doneanu C. E., Xue S., Goodlett D. R., Nelson S. D., and Atkins W. M. (2012) Cross-linking mass spectrometry and mutagenesis confirm the functional importance of surface interactions between CYP3A4 and holo/apo cytochrome b(5). Biochemistry 51, 9488–9500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Estrada D. F., Laurence J. S., and Scott E. E. (2013) Substrate-modulated cytochrome P450 17A1 and cytochrome b5 interactions revealed by NMR. J. Biol. Chem. 288, 17008–17018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Gorsky L. D., and Coon M. J. (1986) Effects of conditions for reconstitution with cytochrome b5 on the formation of products in cytochrome P-450-catalyzed reactions. Drug Metab. Dispos. 14, 89–96 [PubMed] [Google Scholar]
- 10. Gruenke L. D., Konopka K., Cadieu M., and Waskell L. (1995) The stoichiometry of the cytochrome P-450-catalyzed metabolism of methoxyflurane and benzphetamine in the presence and absence of cytochrome b(5). J. Biol. Chem. 270, 24707–24718 [DOI] [PubMed] [Google Scholar]
- 11. Katagiri M., Kagawa N., and Waterman M. R. (1995) The role of cytochrome b5 in the biosynthesis of androgens by human P450c17. Arch. Biochem. Biophys. 317, 343–347 [DOI] [PubMed] [Google Scholar]
- 12. Estrada D. F., Skinner A. L., Laurence J. S., and Scott E. E. (2014) Human cytochrome P450 17A1 conformational selection: modulation by ligand and cytochrome b5. J. Biol. Chem. 289, 14310–14320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Zhang H., Im S. C., and Waskell L. (2007) Cytochrome b5 increases the rate of product formation by cytochrome P450 2B4 and competes with cytochrome P450 reductase for a binding site on cytochrome P450 2B4. J. Biol. Chem. 282, 29766–29776 [DOI] [PubMed] [Google Scholar]
- 14. Ahuja S., Jahr N., Im S. C., Vivekanandan S., Popovych N., Le Clair S. V., Huang R., Soong R., Xu J., Yamamoto K., Nanga R. P., Bridges A., Waskell L., and Ramamoorthy A. (2013) A model of the membrane-bound cytochrome b5-cytochrome P450 complex from NMR and mutagenesis data. J. Biol. Chem. 288, 22080–22095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Zhang M., Le Clair S. V., Huang R., Ahuja S., Im S. C., Waskell L., and Ramamoorthy A. (2015) Insights into the role of substrates on the interaction between cytochrome b(5) and cytochrome P450 2B4 by NMR. Sci. Rep. 5, 8392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Nunez M., Guittet E., Pompon D., van Heijenoort C., and Truan G. (2010) NMR structure note: oxidized microsomal human cytochrome b5. J. Biomol. NMR 47, 289–295 [DOI] [PubMed] [Google Scholar]
- 17. Chen W., Koenigs L. L., Thompson S. J., Peter R. M., Rettie A. E., Trager W. F., and Nelson S. D. (1998) Oxidation of acetaminophen to its toxic quinone imine and nontoxic catechol metabolites by baculovirus-expressed and purified human cytochromes P450 2E1 and 2A6. Chem. Res. Toxicol. 11, 295–301 [DOI] [PubMed] [Google Scholar]
- 18. Yamazaki H., Nakano M., Gillam E. M., Bell L. C., Guengerich F. P., and Shimada T. (1996) Requirements for cytochrome b5 in the oxidation of 7-ethoxycoumarin, chlorzoxazone, aniline, and N-nitrosodimethylamine by recombinant cytochrome P450 2E1 and by human liver microsomes. Biochem. Pharmacol. 52, 301–309 [DOI] [PubMed] [Google Scholar]
- 19. Yamazaki H., Nakamura M., Komatsu T., Ohyama K., Hatanaka N., Asahi S., Shimada N., Guengerich F. P., Shimada T., Nakajima M., and Yokoi T. (2002) Roles of NADPH-P450 reductase and apo- and holo-cytochrome b5 on xenobiotic oxidations catalyzed by 12 recombinant human cytochrome P450s expressed in membranes of Escherichia coli. Protein Expr. Purif. 24, 329–337 [DOI] [PubMed] [Google Scholar]
- 20. Yamazaki H., Gillam E. M., Dong M. S., Johnson W. W., Guengerich F. P., and Shimada T. (1997) Reconstitution of recombinant cytochrome P450 2C10(2C9) and comparison with cytochrome P450 3A4 and other forms: effects of cytochrome P450-P450 and cytochrome P450-b5 interactions. Arch. Biochem. Biophys. 342, 329–337 [DOI] [PubMed] [Google Scholar]
- 21. Yamazaki H., Shimada T., Martin M. V., and Guengerich F. P. (2001) Stimulation of cytochrome P450 reactions by apo-cytochrome b(5): evidence against transfer of heme from cytochrome P450 3A4 to apo-cytochrome b5, or heme oxygenase. J. Biol. Chem. 276, 30885–30891 [DOI] [PubMed] [Google Scholar]
- 22. Yun C. H., Kim K. H., Calcutt M. W., and Guengerich F. P. (2005) Kinetic analysis of oxidation of coumarins by human cytochrome P450 2A6. J. Biol. Chem. 280, 12279–12291 [DOI] [PubMed] [Google Scholar]
- 23. Soucek P. (1999) Expression of cytochrome P450 2A6 in Escherichia coli: purification, spectral and catalytic characterization, and preparation of polyclonal antibodies. Arch. Biochem. Biophys. 370, 190–200 [DOI] [PubMed] [Google Scholar]
- 24. Tan Y., Patten C. J., Smith T., and Yang C. S. (1997) Competitive interactions between cytochromes P450 2A6 and 2E1 for NADPH-cytochrome P450 oxidoreductase in the microsomal membranes produced by a baculovirus expression system. Arch. Biochem. Biophys. 342, 82–91 [DOI] [PubMed] [Google Scholar]
- 25. Guengerich F. P. (2015) Human cytochrome P450 enzymes. In Cytochrome P450: Structure, Mechanism, and Biochemistry, 4th Ed (Ortiz de Montellano P. R., ed) pp. 523–785, Springer International Publishing, Cham, Switzerland [Google Scholar]
- 26. Dehal S. S., and Kupfer D. (1997) CYP2D6 catalyzes tamoxifen 4-hydroxylation in human liver. Cancer Res. 57, 3402–3406 [PubMed] [Google Scholar]
- 27. Shimada T., Mernaugh R. L., and Guengerich F. P. (2005) Interactions of mammalian cytochrome P450, NADPH-cytochrome P450 reductase, and cytochrome b(5) enzymes. Arch. Biochem. Biophys. 435, 207–216 [DOI] [PubMed] [Google Scholar]
- 28. Gillam E. M., Guo Z., and Guengerich F. P. (1994) Expression of modified human cytochrome P450 2E1 in Escherichia coli, purification, and spectral and catalytic properties. Arch. Biochem. Biophys. 312, 59–66 [DOI] [PubMed] [Google Scholar]
- 29. Chen W., Peter R. M., McArdle S., Thummel K. E., Sigle R. O., and Nelson S. D. (1996) Baculovirus expression and purification of human and rat cytochrome P450 2E1. Arch. Biochem. Biophys. 335, 123–130 [DOI] [PubMed] [Google Scholar]
- 30. Peng H. M., and Auchus R. J. (2013) The action of cytochrome b(5) on CYP2E1 and CYP2C19 activities requires anionic residues D58 and D65. Biochemistry 52, 210–220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Peng H. M., and Auchus R. J. (2014) Two surfaces of cytochrome b5 with major and minor contributions to CYP3A4-catalyzed steroid and nifedipine oxygenation chemistries. Arch. Biochem. Biophys. 541, 53–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Yu A., Dong H., Lang D., and Haining R. L. (2001) Characterization of dextromethorphan O- and N-demethylation catalyzed by highly purified recombinant human CYP2D6. Drug Metab. Dispos. 29, 1362–1365 [PubMed] [Google Scholar]
- 33. Henderson C. J., McLaughlin L. A., Scheer N., Stanley L. A., and Wolf C. R. (2015) Cytochrome b5 is a major determinant of human cytochrome P450 CYP2D6 and CYP3A4 activity in vivo. Mol. Pharmacol. 87, 733–739 [DOI] [PubMed] [Google Scholar]
- 34. Fukami T., Katoh M., Yamazaki H., Yokoi T., and Nakajima M. (2008) Human cytochrome P450 2A13 efficiently metabolizes chemicals in air pollutants: naphthalene, styrene, and toluene. Chem. Res. Toxicol. 21, 720–725 [DOI] [PubMed] [Google Scholar]
- 35. Studier F. W. (2005) Protein production by auto-induction in high-density shaking cultures. Protein Expr. Purif. 41, 207–234 [DOI] [PubMed] [Google Scholar]
- 36. Mulrooney S. B., and Waskell L. (2000) High-level expression in Escherichia coli and purification of the membrane-bound form of cytochrome b(5). Protein Expr. Purif. 19, 173–178 [DOI] [PubMed] [Google Scholar]
- 37. Strittmatter P., and Velick S. F. (1956) The isolation and properties of microsomal cytochrome. J. Biol. Chem. 221, 253–264 [PubMed] [Google Scholar]
- 38. Peng H. M., Im S. C., Pearl N. M., Turcu A. F., Rege J., Waskell L., and Auchus R. J. (2016) Cytochrome b(5) activates the 17,20-lyase activity of human cytochrome P450 17A1 by increasing the coupling of NADPH consumption to androgen production. Biochemistry 55, 4356–4365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Oprian D. D., and Coon M. J. (1982) Oxidation-reduction states of FMN and FAD in NADPH-cytochrome-P-450 reductase during reduction by NADPH. J. Biol. Chem. 257, 8935–8944 [PubMed] [Google Scholar]
- 40. DeVore N. M., Smith B. D., Urban M. J., and Scott E. E. (2008) Key residues controlling phenacetin metabolism by human cytochrome P450 2A enzymes. Drug Metab. Dispos. 36, 2582–2590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Porubsky P. R., Meneely K. M., and Scott E. E. (2008) Structures of human cytochrome P-450 2E1: insights into the binding of inhibitors and both small molecular weight and fatty acid substrates. J. Biol. Chem. 283, 33698–33707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Wang A., Savas U., Hsu M. H., Stout C. D., and Johnson E. F. (2012) Crystal structure of human cytochrome P450 2D6 with prinomastat bound. J. Biol. Chem. 287, 10834–10843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Yano J. K., Wester M. R., Schoch G. A., Griffin K. J., Stout C. D., and Johnson E. F. (2004) The structure of human microsomal cytochrome P450 3A4 determined by X-ray crystallography to 2.05-Å resolution. J. Biol. Chem. 279, 38091–38094 [DOI] [PubMed] [Google Scholar]
- 44. Wester M. R., Stout C. D., and Johnson E. F. (2002) Purification and crystallization of N-terminally truncated forms of microsomal cytochrome P450 2C5. Methods Enzymol. 357, 73–79 [DOI] [PubMed] [Google Scholar]
- 45. Delaglio F., Grzesiek S., Vuister G. W., Zhu G., Pfeifer J., and Bax A. (1995) NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR 6, 277–293 [DOI] [PubMed] [Google Scholar]