Abstract
DNA secondary structures and methylation are two well-known mechanisms that regulate gene expression. The catalytic subunit of telomerase, human telomerase reverse transcriptase (hTERT), is overexpressed in ∼90% of human cancers to maintain telomere length for cell immortalization. Binding of CCCTC-binding factor (CTCF) to the first exon of the hTERT gene can down-regulate its expression. However, DNA methylation in the first exon can prevent CTCF binding in most cancers, but the molecular mechanism is unknown. The NMR analysis showed that a stretch of guanine-rich sequence in the first exon of hTERT and located within the CTCF-binding region can form two secondary structures, a hairpin and a quadruplex. A key finding was that the methylation of cytosine at the specific CpG dinucleotides will participate in quartet formation, causing the shift of the equilibrium from the hairpin structure to the quadruplex structure. Of further importance was the finding that the quadruplex formation disrupts CTCF protein binding, which results in an increase in hTERT gene expression. Our results not only identify quadruplex formation in the first exon promoted by CpG dinucleotide methylation as a regulator of hTERT expression but also provide a possible mechanistic insight into the regulation of gene expression via secondary DNA structures.
Keywords: 5-methylcytosine, DNA methylation, DNA structure, G-quadruplex, telomerase reverse transcriptase (TERT)
Introduction
In mammals, methylation of cytosine in CpG dinucleotides of the promoter regions is a well-characterized epigenetic modification of DNA, which plays an important role in regulating many cellular processes, including development and tumorigenesis (1, 2). Previous studies have shown that the CpG dinucleotide often occurs within the potential G-quadruplex-forming sequences (PG4s)4 (3, 4). The folding of G-rich DNA or RNA sequences into a four-stranded conformation is named G-quadruplex (G4). Bioinformatics search found over 370,000 PG4s in the human genome on the basis of the sequence motif, G3N(x)G3N(y)G3N(z)G3 (x, y, z = 1–7) (5, 6). However, the high-throughput sequencing-based method detected 716,310 non-canonical G4 motifs, such as two G-quartet, bulge, and long loop fashion in the human genomic DNA, implying the propensity of G4 formation from diverse G-rich motifs (7). G4s may play a crucial role in gene transcription and regulation (8, 9). Many therapeutically relevant promoter G4s have been shown to have the ability to suppress their transcriptional activity, such as c-MYC (10), KRAS (11), BCL-2 (12), VEGF (13), c-KIT (14), hTERT (15), and WNT1 (16). Therefore, G4s have become an attractive target for drug development (8, 17). Although methylation of cytosine in CpG dinucleotides may enhance the stability of G4 structures (18–21), to the best of our knowledge the interplay between DNA methylation and DNA secondary structure and its following effect in regulating gene expression have not been addressed.
Cell immortalization is a key event in carcinogenesis, which requires activation of telomerase, a large ribonucleoprotein complex, to maintain the telomere length for unlimited proliferation in the majority of cancer cells (22). Human telomerase reverse transcriptase (hTERT) is the major component of the catalytic subunit of telomerase and plays a rate-limiting factor for telomerase activity. Telomerase activity is found in ∼90% of human cancers, whereas most somatic normal tissues are negative for hTERT expression (23). The aberrant hTERT gene expression may cause aging, cancer, and other diseases (24, 25). Several transcription factors on the promoter region have been documented to directly or indirectly regulate the hTERT gene expression, such as the activators, MYC and SP1, as well as the repressors, such as p53 (22). Many recurrent mutations were identified within the promoter region of hTERT in various cancers, which correlate with the increased hTERT transcriptional activity (26, 27), and the promoting potential of immortalization and tumorigenesis (28). Hurley and co-workers (29) found that in the core promoter of hTERT, located between −22 and −90, contains an end-to-end stacked pair of G-quadruplexes. Recently, they (30) showed that hTERT expression was regulated by G4 folding of the long G-tract within the hTERT promoter modulated by sequence mutation. They further found that aberrant G4 formation in the core promoter region disrupted repressor binding and drove hTERT transcription activation.
Meanwhile, it has been shown that binding of CCCTC-binding factor (CTCF) to the first exon of hTERT gene could suppress its transcription in telomerase-negative cells. Presently, there is no mutation or single nucleotide polymorphism identified within the CTCF-binding site (+4 to +39 from the ATG start codon (31)) in the first exon of hTERT. Of interest is that methylation of the first exon of hTERT prevents CTCF binding and allows hTERT gene expression in telomerase-positive cells (32). However, the underlying mechanism of the hTERT methylation in regulating its gene expression remains unclear. In this study, we found that two secondary structures, hairpin and quadruplex, could form within the CTCF-binding region of the first exon of hTERT. Further studies demonstrated that methylation of cytosine in a specific CpG in the first exon of the hTERT gene promotes quadruplex formation, which prevents CTCF binding and results in hTERT gene expression.
Results
Cytosine methylation and quadruplex formation of hT25
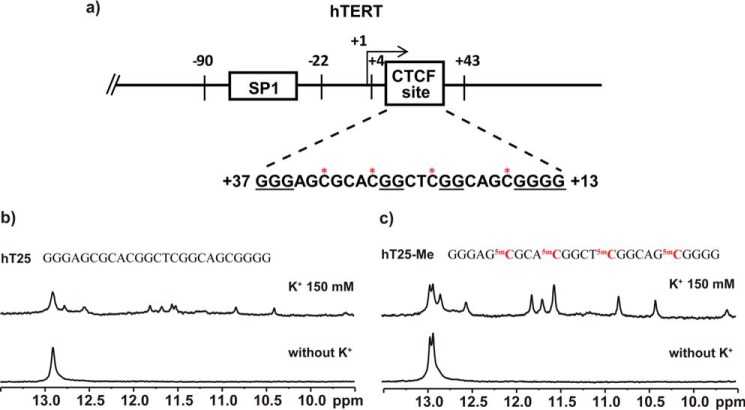
Within the CTCF-binding region of the first exon of the hTERT gene (32), we found a G-rich sequence, GGGAGCGCACGGCTCGGCAGCGGGG (Fig. 1a, named hT25) localized at +13 to +37 (at antisense strand), containing four CpG dinucleotides. To assess whether this sequence is involved in DNA secondary structure formation, we conducted the 1D imino proton NMR experiment to characterize the possible hydrogen-bonding formation of DNA secondary structure. The results showed some imino proton signals around 13 ppm in the absence of K+, implying the formation of Watson-Crick hydrogen bonding for hairpin structure (33), whereas the spectrum showed several imino proton signals at 10–12 ppm after addition of K+, suggesting the formation of Hoogsteen hydrogen bonding for the quadruplex structure (Fig. 1b). Of particular interest is that the imino proton signals at 10–12 ppm were more pronounced after CpG methylation (Fig. 1c), i.e. the population of quadruplex structures changes from less than 30% to over 60% (Table 1). It appears that methylation of CpG dinucleotides of this hTERT DNA sequence could enhance the formation of quadruplex structures.
Figure 1.
Characterization of DNA secondary structures of a G-rich sequence in the first exon of hTERT gene. a, identification of a potential G4-forming sequence, hT25, d(G3AGCGCACG2CTCG2CAGCG4), at the antisense (from +37 to +13) in the first exon of the hTERT gene, which is located within the CTCF-binding region. b and c, the imino proton NMR spectra of hT25 (b) and hT25-Me (c) in Tris-HCl buffer without (bottom panel) and with (top panel) 150 mm KCl. The methylated sequence, hT25-Me, was synthetically modified cytosine to 5-methylcytosine at the four CpG dinucleotides of hT25.
Table 1.
The population of hairpin and quadruplex structures of hT25 and its mutants
The listed populations of hairpin (H) and quadruplex (Q) structures were estimated by integrating the peak volume of hairpin and quadruplex structures. NA means not available.
| Name | Population |
|
|---|---|---|
| H | Q | |
| hT25 | >70% | <30% |
| hT25-m1 | >95% | <5% |
| hT25-m2 | <10% | >90% |
| hT25-m3 | >90% | <10% |
| hT25-m4 | <10% | >90% |
| hT25-ss1 | NA | NA |
| hT25-ss2 | NA | NA |
| hT25-ss3 | NA | NA |
| hT25-Me | <40% | >60% |
| hT25-Me(2,3) | >75% | <25% |
| hT25-Me(1,4) | <40% | >60% |
| hT25-Me(1) | >70% | <30% |
| hT25-Me(4) | <30% | >70% |
| hT25-m1-Me(4) | >95% | <5% |
NMR study of hT25 via mutation and site-specific methylation
To examine the formation of this wild-type sequence into a G4 structure, a number of mutants were designed for structural characterization (supplemental Table S1). Fig. 2a showed the imino proton NMR spectra of four hT25 mutants in 150 mm K+ solution. These mutations were designed not only to disrupt either hairpin or quadruplex formation but also to identify the bases involved in the quadruplex formation. The hT25-m1 was designed to disrupt the quadruplex formation by the change of the G2 and G24 in the first and last G-tracts of the hT25 sequence to T2 and T24. The imino proton NMR results of hT25-m1 showed distinct signals of Watson-Crick base pairing and no appreciable signals of quadruplex structure, implying that the first and last G-tracts of the hT25 sequence were involved in the quartet formation. The hT25-m2 was designed to disrupt the quadruplex formation by the change of the G12 and G17 in the second and third G-tracts to T12 and T17. Surprisingly, the imino proton NMR spectrum of hT25-m2 showed distinct signals of quadruplex structure at the 10–12-ppm region, implying that the middle two G-tracts, G11G12 and G16G17, were not involved in the quartet formation of quadruplex structure. Moreover, the imino proton signal pattern of hT25-m2 at the 10–12-ppm region was very similar to that observed for the hT25 and the methylated hT25 sequences.
Figure 2.
NMR study of hT25 DNA sequence by mutation and methylation. a, imino proton NMR spectra of four mutants, hT25-m1, hT25-m2, hT25-m3, and hT25-m4, in the presence of 150 mm K+, as shown in descending order. The quadruplex characters were detected in the spectra of hT25-m2 and hT25-m4. b, imino proton NMR spectra of hT25-Me, hT25-Me(2,3), hT25-Me(1,4), hT25-Me(1), and hT25-Me(4) for hT25 and hT25-m1-Me(4) for hT25-m1 in the presence of 150 mm K+, as shown in descending order. Their sequences are listed in supplemental Table S1.
We further investigated the effect of a single G-base mutation to the quadruplex formation. The hT25-m3 was designed by the change of G11 to T11, which may enhance the Watson-Crick base pairs in hairpin structure. Of interest is that the imino proton NMR spectrum of hT25-m3 showed distinct signals of Watson-Crick base pairing at 12.5–13 ppm and no appreciable signals of the quadruplex structure at the 10–12-ppm region. Given that two middle G-tracts were not involved in quadruplex formation and the CpG methylation could enhance quadruplex formation, the design of hT25-m4 was to disrupt quadruplex formation by replacing G20 with T20. In contrast to hT25, the imino proton NMR spectrum of the hT25-m4 showed a different pattern not only at 13.0 ppm for hairpin formation but also at 10–12 ppm quadruplex formation. It is noteworthy that G-rich sequences with only a single base difference could form not only various quadruplex structures but also different secondary structures. Such different conformational changes caused by a single base mutation deserve further study.
Considering that there are four CpG dinucleotides in hT25, it is important to determine which methylated CpG dinucleotide plays a critical role for shifting the equilibrium to quadruplex formation. A number of methylated hT25 sequences based on simple step by step screening are listed in supplemental Table S1. NMR spectra of hT25-Me and hT25-Me(1,4) showed almost identical imino proton signals in the region of 9.5–12.0 ppm (Fig. 2b). Their quadruplex formation is >60% population of quadruplex (Table 1). Further study of the methylation at the first (hT25-Me(1)) and fourth (hT25-Me(4)) CpG dinucleotides showed that the population of quadruplex in hT25-Me(1) (<30%) is lower than in hT25-Me(4) (>70%), suggesting that methylation of the fourth CpG dinucleotide (C21G22) plays the key role to enhance quadruplex formation.
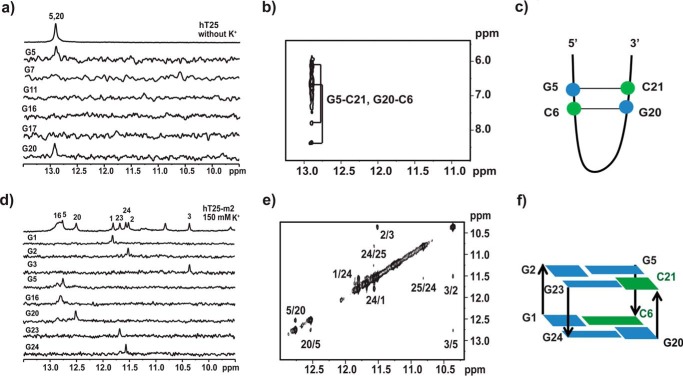
Topology study of hT25-m2 by site-specific 15N-labeled 1H NMR spectra
To examine the transition from hairpin to quadruplex topologies, we first used site-specific 15N isotope labeling to examine which G-bases are involved in hairpin and quadruplex formation of hT25. Six hT25 samples were synthesized, each of which was site-specifically labeled with 15N-enriched guanine. Two of the 15N-edited imino proton resonances of the guanines of G5 and G20 showed the contribution to the signal at around 13 ppm (Fig. 3a). Further 2D-NOESY spectra confirmed these two Watson-Crick signals (Fig. 3b). Considering all possible combinations of Watson-Crick hydrogen bonds involved G5 and G20, Fig. 3c shows the proposed hairpin structure of hT25 in Tris buffer. After addition of 150 mm K+ overnight, we were unable to detect the 15N-edited imino proton resonances at the 10–12-ppm region for the quadruplex formation because the signal was too weak (supplemental Fig. S1a). Of importance is the same detection of the 15N-edited imino proton resonances of G5 and G20 at around 13 ppm before and after addition of 150 mm K+, suggesting that the transition from hairpin to quadruplex topologies does not necessarily involve the opening of the hairpin formation.
Figure 3.
Characterization of secondary structure by NMR with site-specific labeling. a, site-specific assignments of imino proton resonances of hT25 hairpin structure. The 1D 15N-edited HMQC spectra of 8% 15N-enriched oligonucleotide samples were shown with assignments to the labeling sites. b, 2D NMR spectra of hT25 hairpin form. The recorded NOESY spectrum with mixing time of 300 ms showed two major GC correlations. c, proposed scheme of hairpin orientation of the hT25 with two G-C hydrogen-bonding pairs. d, site-specific assignments of imino proton resonances of hT25-m2 quadruplex structure. The 1D 15N-edited HMQC spectra of 8% 15N-enriched oligonucleotide samples are shown with assignments to the labeling sites. e, 2D NMR spectra of hT25-m2 quadruplex form. The recorded NOESY spectrum with mixing time of 250 ms showed the H1-H1 correlations. f, proposed scheme of quartet orientation of the hT25-m2 with two quartets of G-G-G-C configurations.
At present, we are unable to prepare the sample of site-specific 15N isotope labeling to methylated hT25. Therefore, we used hT25-m2 to study the quadruplex formation because similar imino proton NMR signals at 10–12 ppm were detected more prominently in hT25-m2 than in hT25. Using site-specific 15N isotope labeling, we found that three guanine residues, G5, G16, and G20, are involved in the initial hairpin formation of hT25-m2 (supplemental Fig. S1b), particularly G5 and G20, also detected in hT25. After addition of 150 mm K+ overnight, the 15N-edited imino proton spectra showed six signals of five G-bases of G1, G2, G3, G23, G24, and an unlabeled G base at the 10–12-ppm region for the quadruplex formation together with three signals of G5, G16, and G20 at 12.5–13 ppm region for the Watson-Crick base pairing (Fig. 3d). Further studies suggested that the unlabeled peak was G25 (data not shown). Again, the finding of the same three Watson-Crick signals in the hairpin and quadruplex structures implied that the K+-induced topological transition of hT25-m2 does not necessarily involve the opening of hairpin formation.
Considering that several G-rich sequences form the quadruplex structure containing a G·C·G·C quartet characterized by two imino proton signals at the 12.5-13-ppm region (20, 34–36), we anticipated that the quadruplex formation of hT25-m2 could also involve cytosine without using the two middle G-tracts. Further study of mutants (supplemental Table S1) was conducted to examine which cytosine residue is possibly involved in the quartet formation of hT25-m2. The imino proton NMR spectra showed different patterns of hT25-m5 and hT25-m6 from hT25-m2 and hT25-m7, suggesting that C6 and C21 are involved in the quartet formation (supplemental Fig. S2). In addition, several H1-H1 NOEs were analyzed to predict the possible topology between neighboring guanines, i.e. G5/G20, G1/G24, and G3/G5 (Fig. 3e). Although the precise quadruplex structure of hT25-m2 has not been determined, it is very likely that the quadruplex formation of hT25-m2 involves two G·G·G·C quartets (Fig. 3f). To our knowledge, such type of quadruplex structure containing two G·G·G·C quartets has not been reported yet.
Investigation of melting temperature and folding kinetics of hT25-Me(4)
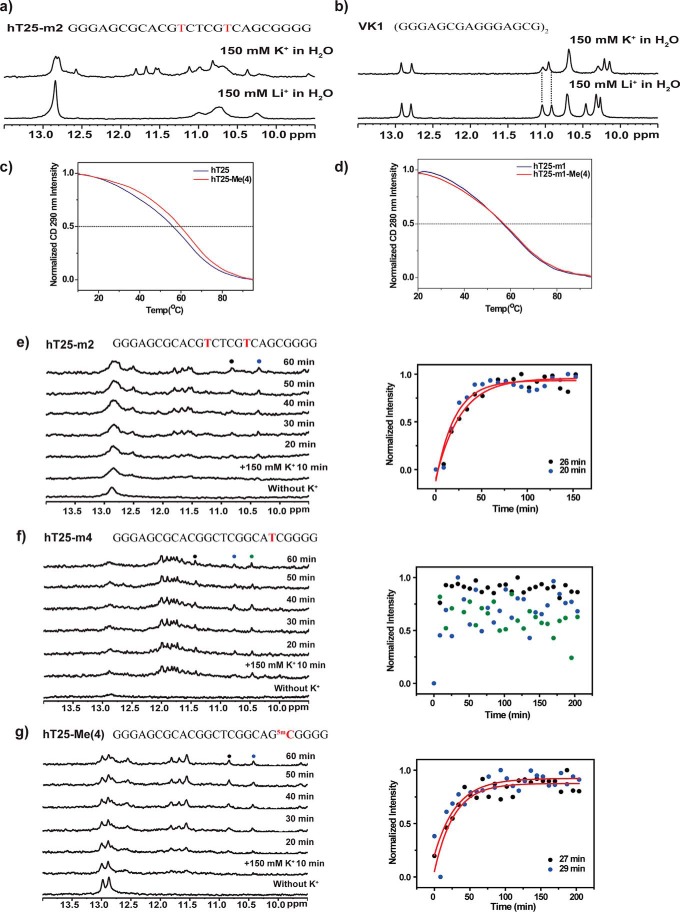
CD spectroscopy has been extensively used to characterize different types of G4 structures. In general, parallel G4 structures show a positive band at 265 nm and a negative band at 240 nm, whereas antiparallel G4 structures show two positive bands at 295 and 240 nm together with a negative band at 265 nm. Here, the CD spectra of hT25 and hT25-Me(4) showed no appreciable difference, even in the absence and presence of 150 mm K+ (supplemental Fig. S3). Recently, Kocman and Plavec (37) reported that a d(GGGAGCGAGGGAGCG) sequence, VK1, could form an unusual dimeric tetraplex structure stabilized by four G-C base pairs in Watson-Crick geometry. Noteworthy, their CD spectra also ruled out the formation of parallel and non-parallel G4 structures. Moreover, the CD spectra of VK1 are similar to the CD spectra of hT25. Of interest is the similarity found in the folding motif of hT25 (GGGAGCGCACGGCTCGGCAGCGGGG). The two G-C base pairs formed in the hT25 quadruplex structure is similar to the finding in the VK1 dimeric tetraplex structure, which may play a major contribution to CD spectra. However, NMR spectra showed that the cation-dependent quadruplex formation of hT25-m2 is very different from the cation-independent quadruplex formation of VK1 in the presence of 150 mm Li+ and 150 mm K+ (Fig. 4, a and b).
Figure 4.
CD melting and NMR spectra of quadruplex formation. a and b, the imino proton NMR spectra of hT25-m2 (a) and VK1 (b) under 150 mm Li+ or 150 mm K+ conditions in H2O at 10 °C. c, CD melting curves of hT25 (blue line) and hT25-Me(4) (red line) in 150 mm K+ monitored at 290 nm CD signal. d, CD melting curves of hT25-m1 (blue line) and hT25-m1-Me(4) (red line) in 150 mm K+ monitored at 280 nm CD signal. e–g, the time-dependent imino proton NMR spectra were recorded at 0, 10, 20, 30, 40, 50, and 60 min after addition of 150 mm K+ (e–g, left) together with the rise of quadruplex signals fitted with a single exponential (e–g, right) of hT25-m2 (e), hT25-m4 (f), and hT25-Me(4) (g). Since the average time for collecting NMR spectra is ∼8 min, the very fast rise of quadruplex signals of hT25-m4 cannot be fitted.
In addition, we measured the CD melting curves of hT25 and hT25-Me(4) at 290 nm in 150 mm K+ to examine the effect of methylation in quadruplex stability. The melting temperature (Tm) is 56.2 °C for hT25 and 59.8 °C for hT25-Me(4) (Fig. 4c), indicating that there is ∼3.6 °C increase on ΔTm upon methylation. However, the Tm is 58 °C for both h25-m1 and hT25-m1-Me(4) (Fig. 4d), which formed the hairpin structure, suggesting that methylation favors stabilizing the quadruplex formation. The slight increase of ΔTm could shift the equilibrium from the hairpin state to the quadruplex state. Previous studies have shown that the CpG methylation could stabilize G4 structure by favorable stacking interaction between the methyl group of 5-methylcytosine and guanine residues (18–20).
We then used real time imino proton NMR spectra to study the folding kinetics of hT25-m2, hT25-m4, and hT25-Me(4) quadruplexes after addition of 150 mm K+ (Fig. 4, e–g). The results showed that the transition time from hairpin to quadruplex structures of hT25-m2 is ∼25 min at 25 °C. Surprisingly, the folding transition is much faster for hT25-m4 than for hT25-m2, indicating that different transition pathways are involved. Of importance is that the same time scale for the folding transition of hT25-m2 is also measured for hT25-Me(4), indicating that similar transition pathways are involved.
Quadruplex formation mediated by DNA methylation modulates hTERT gene expression via CTCF binding
It has been shown that CpGs around the CTCF-binding site in the first exon of hTERT are highly methylated in the majority of tumor tissues and cancer cell lines (32). Here, we verified whether methylation of CpG dinucleotides could perturb CTCF binding and further regulate hTERT gene expression in telomerase-positive human melanoma A375 cells. The methylated reporter constructs (WT-Me) were generated by methylation from −36 to +110 via incubated wild-type plasmids (WT) with M.SssI CpG methyltransferase, which was confirmed by using bisulfite sequencing (Fig. 5a). The reporter assay showed that the expression level of WT-Me was markedly higher than WT (Fig. 5b). Consistent with the finding of reporter assay, the chromatin immunoprecipitation (ChIP) results showed a significant reduction of CTCF binding after methylation (WT-Me) (Fig. 5c). Although methylation of CpG dinucleotides indeed inhibits CTCF binding and results in gene expression, it is not clear whether the quadruplex structure promoted by methylation has a major effect on CTCF binding and gene regulation.
Figure 5.
Transcriptional effect of quadruplex formation promoted by DNA methylation within the CTCF-binding region of hTERT. a, schematic diagrams of the CpG free reporters containing the hTERT promoter without (WT) and with CpG methylation (WT-Me) from −36 to +110. The methylation patterns of reporters from four different clones were verified by bisulfite sequencing. Each square represents one CpG site. Filled squares, methylated; open squares, unmethylated. b, WT and WT-methyl plasmids were transiently transfected into A375 cells. Luciferase activities of methylated reporters were compared with that of cells transfected with WT reporters. c, binding of CTCF to the first exon of hTERT of reporters in A375 cell lines was analyzed by ChIP assay with antibody against CTCF. The semi-quantitative real-time PCR with primers that are complementary to the CTCF-binding site of hTERT and pCpG free-basic-Lucia vector backbone was used to analyze the level of CTCF binding. Representative results were obtained from three independent experiments. The results represent the mean ± S.D. for each group. **, p < 0.01; ***, p < 0.001 compared with WT.
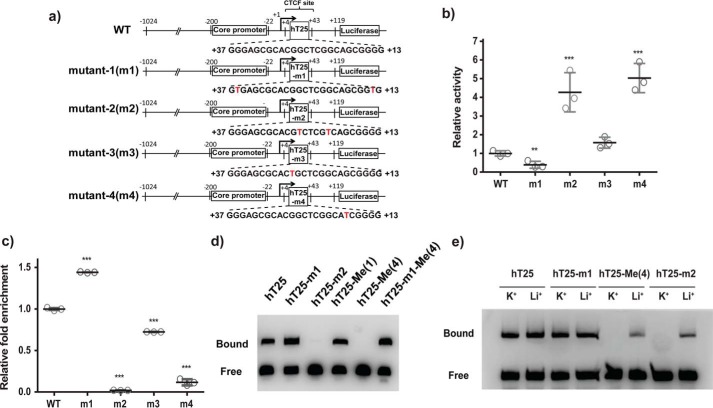
We therefore constructed different reporter plasmids to examine whether quadruplex formation is important in regulating CTCF binding. These reporter constructs were generated by employing the wild-type and mutated hTERT promoter region based on the previous NMR results (supplemental Table S1 and Fig. 6a). The luciferase expression level in A375 cells transfected with m2 and m4 plasmids with the preference to form quadruplex structure was much higher than wild type and transfected with m1 and m3 plasmids with the preference to form hairpin structure (Fig. 6b). In addition, ChIP analysis showed that the level of CTCF binding is much lower in cells transfected with m2 and m4 plasmids than WT and m1 plasmid (Fig. 6c). These results indicated that the hairpin and quadruplex structures play an important role for both CTCF binding and gene expression on hTERT.
Figure 6.
Effect of DNA secondary structures in the CTCF-binding site on hTERT gene expression. a, schematic diagrams of the CpG free reporters containing WT promoter and mutants. b, transcriptional activities of reporters containing the WT or mutated hTERT promoters with hairpin structure (m1 and m3) or quadruplex structure (m2 and m4) were analyzed in A375 cells. The luciferase activities of reporters with mutated promoters were compared with that of WT. c, binding of CTCF to the reporters with WT or mutated promoters was further analyzed by ChIP assay using antibody against CTCF. The results represent the mean ± S.D. from three independent experiments. **, p < 0.01; ***, p < 0.001 compared with WT. d, EMSA experiments were performed to verify the binding of CTCF to biotinylated hT25, hT25-m1, hT25-m2, hT25-Me(1), hT25-Me(4), and hT25-m1-Me incubated with CTCF in vitro. Representative results from three independent experiments are shown. e, EMSA experiments were performed in the presence of KCl or LiCl condition for biotinylated hT25, hT25-Me(4), hT25-m1, and hT25-m2 incubated with CTCF in vitro.
Electrophoretic mobility shift assay (EMSA) using recombinant full-length CTCF protein was conducted to examine the binding preference of CTCF protein, which is the secondary structure (hairpin or quadruplex) of hT25. The gel results showed that a large amount of the bound form of CTCF binds to hT25 and hT25-m1, suggesting that the CTCF protein favors the binding of hairpin structure (Fig. 6d). In contrast, the bound form of CTCF binding to hT25-m2 was hardly detected, implying that quadruplex formation prevents CTCF binding.
EMSA was also performed to verify how the methylated CpG dinucleotides of the hT25 structures perturb CTCF binding. The binding complex of hT25-Me(1) with CTCF was clearly detected, whereas the bound form of the CTCF binding to hT25-Me(4) was hardly observed (Fig. 6d). This difference is because the hairpin structure of hT25-Me(1) favors CTCF binding, and the quadruplex structure of hT25-Me(4) impedes CTCF binding. To test whether the inhibition of CTCF binding is simply due to methylation of CpG dinucleotides, the methylation of hT25-m1 (hT25-m1-Me(4)) was examined because of its hairpin structure. The detection of the bound form of the CTCF binding to hT25-m1-Me(4) (Fig. 6d) suggested that the DNA secondary structure may be more critical for CTCF binding.
We further conducted EMSA to examine the cation effect on the CTCF binding (Fig. 6e). The EMSA results of hT25 and hT25-m1 showed no appreciable difference in the presence of K+ and Li+. On the contrary, the binding of CTCF to hT25-Me(4) and hT25-m2 was markedly increased in the presence of Li+ than in the presence of K+, suggesting that the K+-induced quadruplex formation could prevent the binding of CTCF protein as well (Fig. 6e). In addition, ChIP analysis showed that methylated m1 plasmids (m1-Me) at the first exon present no appreciable effect on CTCF binding (supplemental Fig. S4), suggesting that CTCF protein favors the binding of hairpin structure.
CTCF prefers binding to hairpin structure
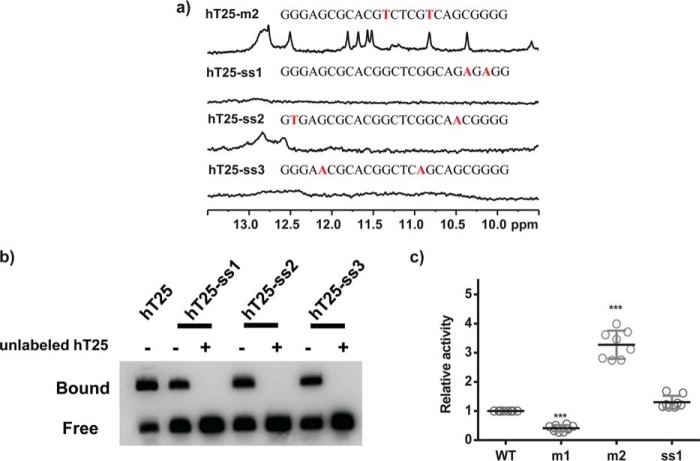
Next, we examined whether CTCF has a better binding preference to the single strand than to the hairpin DNA. Two mutants (hT25-ss1 and hT25-ss3) showed no distinct imino proton NMR signal in the region of 9.5–13.5 ppm, whereas hT25-ss2 mutant showed weak signals in the region of 12.5–13.0 ppm (Fig. 7a). The sequences of these mutants are listed in supplemental Table S1. Although EMSA results show that CTCF can bind these single-strand sequences, the competition between these single-strand sequences and the hT25 indicated that CTCF has a higher binding preference to the hairpin structure of hT25 (Fig. 7b). Consistently, reporter assay showed that the expression level of ss1 is higher than m1, similar to WT, and lower than m2 (Fig. 7c). These findings suggested that the hairpin structure is the major target in the first exon of hTERT gene for CTCF binding.
Figure 7.
Imino proton NMR spectra and the EMSA of single-strand mutants of hT25. a, imino proton NMR spectra of hT25-ss1, hT25-ss2, and hT25-ss3 in 150 mm K+ solution. b, EMSA experiments were performed to verify the binding of CTCF to biotinylated hT25 and single-strand mutants hT25-ss1, hT25-ss2, and hT25-ss3 incubated with CTCF in vitro. 1st lane showed the interaction of hT25 and CTCF, which was used as a positive control. For competition study, the interactions of hT25-ss1, hT25-ss2, and hT25-ss3 with CTCF were incubated with or without 25-fold molar of unlabeled hT25. c, reporter assays were performed in A375 cells transfected with WT, m1, m2, and ss1 mutant reporters. Representative results were obtained from three independent experiments. The results represent the mean ± S.D. for each group. ***, p < 0.001 compared with WT.
Because previous reports suggested that CTCF binds to double strands of promoter DNA (31, 38), EMSA was conducted to verify the binding of CTCF to the sense and antisense strands of the first exon of hTERT gene in vitro (supplemental Fig. S5). The gel results showed a large contrast in the detection of the appreciable amount of CTCF binding to the methylated sense strand (chT25-Me) and the absence of CTCF binding to the methylated wild-type antisense strand (hT25-Me), indicating that CTCF protein is capable of binding to the methylated sense strand. This finding also suggested that only methylation is not sufficient to inhibit CTCF binding. We anticipated that quadruplex formation enhanced by methylation plays a major role for the inhibition of CTCF binding. In addition, our results suggested that methylated antisense strand is mainly responsible for the inhibition of CTCF binding to the first exon of hTERT gene and the regulation of hTERT gene expression.
DNA methylation at the 5-position of cytosine is associated with multiple cellular processes that regulate gene expression in the mammalian genome. In addition to the methylation of CpG dinucleotides, demethylation enzyme, such as ten-eleven translocation (TET) 5mC-hydroxylases, could convert the 5-position of cytosine to 5-hydroxymethylcytosine (5hmC), which offers a means of dynamic regulation of DNA methylation (39, 40). It has been shown that 5hmC plays an important role for the regulation of genes involved in embryonic development, cellular differentiation, and stem cell programming (41, 42). By incorporating 5hmC into the hT25, we further examined whether the hydroxymethylation could influence the CTCF binding onto the hT25. The imino proton NMR spectrum of hT25-hydroxymethyl (hT25-hMe(4)) with modified 5hmC at the fourth CpG dinucleotide suggested that hT25-hMe(4) can form a hairpin structure (supplemental Fig. S6). The EMSA results showed the binding of CTCF to the hT25-hMe(4) but not hT25-Me (supplemental Fig. S6b) and hT25-Me(4) (Fig. 5d). These findings indicated that hydroxymethylation at hT25 does not influence the formation of hairpin structure and the following CTCF binding.
Discussion
Here, we demonstrated that G-rich sequences with only a single base difference could shift base pairs for different hairpin formation, which might lead to different secondary structures. In contrast to the transition from hairpin to quadruplex structures of hT25 after addition of K+, no appreciable imino proton NMR signal detected in a single G-base mutation by replacing G11 to T11 of hT25-m3 indicated the unfavorable transition from hairpin to quadruplex structures. It is likely due to the more stable hairpin structure of hT25-m3 because of the formation of four consecutive Watson-Crick base pairs between C10T11G12C13 and G17C18A19G20. In contrast, the single G-base mutation by replacing G20 to T20 of hT25-m4 disrupted the original G20-C6 base pairs of hT25 hairpin structure. Of interest is that an imino proton NMR signal located around 13 ppm shows no appreciable change, and the folding transition to quadruplex structure is fast after addition of 150 mm K+. In comparison, Gray and Chaires (43) used a stopped-flow method to obtain a single folding time of 20–60 ms for K+-induced quadruplex formation of single-stranded human telomeres. In addition, several mutants of WT22 (GGGCCACCGGGCAGGGGGCGGG) in the WNT1 gene promoter without formation of hairpin structures showed fast quadruplex formation within 2 min at 37 °C (33). The fast folding kinetics of hT25-m4 is similar to the transition from a single-stranded DNA. Nevertheless, verification of hT25-m4 structures with and without K+ for elucidating the transition pathway deserves more study.
Considering that two G-C base pairs of G5-C21 and G20-C6 are involved in both the hairpin formation and quadruplex formation, a possible scenario is proposed for the K+-induced quadruplex formation of hT25-m2 that results from a simple flip back of a hairpin form to fold into a quadruplex form with two G·G·G·C quartets. The proposed model is supported by the transition kinetics. The transition time from hairpin to quadruplex structures of hT25-m2 via a simple flip (∼25 min) is much faster than the transition time from hairpin to quadruplex structures of WT22 via unfolding–refolding (∼550 min) at 25 °C (33). To our knowledge, such a simple transition from hairpin to quadruplex structures for a genomic G-rich sequence has not been previously documented.
DNA methylation plays an important role in regulating hTERT expression. Benhattar et al. (32) found that partial hypomethylation in the core promoter region together with the hypermethylation in the CTCF-binding site of the first exon can lead to hTERT expression in telomerase-positive tumor cells. This finding was further supported by those studies in telomerase-positive cancer cells treated with 5-aza-2′-deoxycytidine or trichostatin A in which DNMT1 down-regulation correlates with the CpG islands demethylation, CTCF binding, and the repression of hTERT transcription (32, 44). By using the dimethyl sulfate–methylation interference assay, Renaud et al. (31) demonstrated that G20, G23 G24, and G25 were recognized by CTCF binding to the first exon of hTERT. Here, we showed that two secondary structures, hairpin and quadruplex, could form within the CTCF-binding region of hTERT first exon. The CTCF-contacting guanines are involved in the hairpin formation. Particularly, CpG methylation in the CTCF-binding site promotes quadruplex formation, which prevents CTCF binding and leads to hTERT expression. In addition, a transition pathway from hairpin to quadruplex topologies of hT25-m2 was proposed. Similar imino proton NMR spectra of hT25-Me suggested that hT25-Me could also adopt a similar quadruplex structure. It is feasible that such simple transition from hairpin to quadruplex topologies for a genomic G-rich sequence could perturb DNA–protein interaction.
G-quadruplex structures found in promoter regions are generally considered as negative regulators of gene expression (8). Zakian and co-workers (9) reported the potential function of quadruplex in gene regulation via stimulating activator binding or inhibiting repressor binding. The recent findings of Hurley and co-workers (30) showed that an aberrant G4 formation of a long G-tract mutation within hTERT promoter region could disrupt repressor binding and result in overexpression of hTERT. Here, our findings demonstrated that quadruplex formation enhanced by CpG dinucleotide methylation in the first exon of hTERT could impede CTCF binding and lead to hTERT expression. Therefore, in addition to transcriptional inhibition, quadruplex formation could also act as a stimulatory factor in modulating gene expression (9, 45).
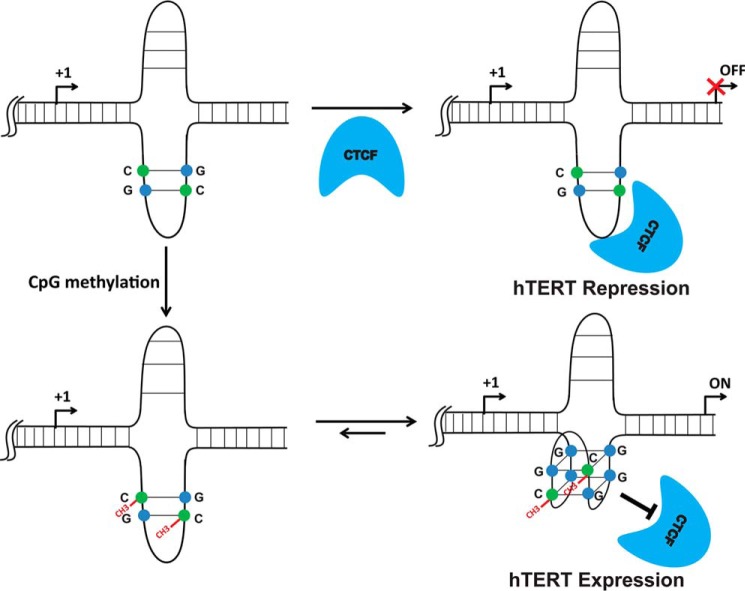
In summary, we demonstrated that methylated cytosine may directly participate in “quartet” formation. Here, methylation of a single cytosine at the specific CpG dinucleotide of the hTERT gene is capable of shifting the equilibrium from hairpin structure to quadruplex structure via a simple flipping process (Scheme 1). Our results showed that DNA methylation alone is not sufficient to inhibit CTCF binding to the first exon of hTERT, suggesting that quadruplex formation promoted by CpG methylation plays a major role in preventing CTCF binding and further regulating gene expression. These findings provided mechanistic insight to explain how the hypermethylated hTERT promoter can lead to its expression in most telomerase-positive tumors.
Scheme 1.
Proposed mechanism of CTCF binding to the first exon of hTERT gene for transcriptional regulation. CTCF favors binding hairpin structure, whereas quadruplex formation enhanced by CpG methylation impedes CTCF binding and further leads to gene expression.
Experimental procedures
DNA preparation
All unlabeled oligonucleotides were purchased from Bio Basic (Ontario, Canada). The DNA concentrations were determined by the absorption at 260-nm peaks using a UV-visible absorption spectrometer. The oligonucleotides were dissolved in 10 mm Tris-HCl (pH 7.5) without and with 150 mm KCl, followed by heat denaturation at 95 °C for 5 min and slowly annealed to room temperature.
NMR spectroscopy
All NMR experiments were performed on a Bruker AVIII 500 MHz NMR spectrometer equipped with a prodigy probe head and on a Bruker AVIII 800 MHz NMR spectrometer equipped with a cryoprobe. The 1D imino proton NMR spectra were recorded using a WATERGATE (46) or a jump-return pulse sequence (47) for water suppression. The 1D 15N-1H SOFAST-HMQC spectra were used for unambiguous assignments of individual imino proton resonances using a series of site-specifically 15N-labeled NMR samples, where 8% of 15N-labeled guanine was introduced into one of the 11 G-quartet-forming guanine residues as described previously (48, 49). The strand concentrations of the NMR samples were typically at 100–200 μm with specific salt conditions and an internal reference of 0.1 mm 4,4-dimethyl-4-silapentane-1-sulfonic acid.
Cell culture
Human melanoma A375 cells (American Type Culture Collection, Manassas, VA), telomerase-positive cells, were grown in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum. Cells were cultured at 37 °C in an incubator supplemented with 5% CO2. A375 cell line was authenticated by using the PromegaGenePrint®10 system (Promega, Madison, WI) and analyzed by ABI PRISM 3730 GENETIC ANALYZER and GeneMapper® software version 3.7. (Applied Biosystems, Carlsbad, CA). The DNA-methylated pattern of the promoter and the first exon region of the hTERT gene in these cells is similar to the pattern in telomerase-positive cells (32).
Promoter reporter assay
To verify the transcriptional activity, the hTERT sequence, including −1024 to +119 (translation start site as +1), was cloned into the pCpGfree-basic-Lucia vector (Invivogen, San Diego). To verify the effect of the DNA methylation in the first exon on the hTERT gene expression, methylated and mutant reporters were also generated and transfected into telomerase-positive A375 cells. Luciferase activity of these constructs was determined by using the QUANTI-Luc luciferase system (Invivogen). Details are provided in the supplemental Materials and methods.
Bisulfite genomic sequencing
To determine the methylation status of the reporter plasmid, genomic DNAs of A375 cells transfected with reporter plasmids were extracted. The bisulfite modification method and following sequences were employed for determining the methylation status of cytosine residues in DNA (50). Details are provided in supplemental Materials and methods.
Chromatin immunoprecipitation
Chromatin immunoprecipitation (ChIP) assays were performed as described previously with minor modifications (31). Briefly, A375 cells transfected with reporter plasmids were treated with 1% formaldehyde in phosphate-buffered saline. The cross-linked nuclei were sonicated to yield DNA fragments in the range of 200–1000 bp. ChIP grade polyclonal antibody against CTCF (Product code: ab70303, Abcam, Cambridge, MA) was incubated overnight with the nuclear lysates. Immune complexes were then collected with protein A magnetic beads (Millipore, Temecula, CA). Using the forward primer (5′-TGCGCACGTGGGAAGCCCTG-3′, complementary of nucleotides −38 to −1 of hTERT gene) and the reverse primer (5′-TGAGGGCAAACAGCACCTTGATTTCC-3′, complementary of nucleotides of pCpGfree-basic-Lucia vector backbone), the semi-quantitative real-time PCR was performed to analyze the CTCF-binding efficiency.
Electrophoretic mobility shift assay
EMSA was performed as described previously with minor modifications (51, 52). 5′-Biotin-labeled oligonucleotides (supplemental Table S1) were incubated with 0.2 μg of CTCF full-length recombinant protein (Abnova, Jhongli City, Taiwan) in binding buffer containing 50 mm KCl, 10 mm Tris, 5 mm MgCl2, 0.1 mm ZnSO4, 2 mm DTT, 0.05% Nonidet P-40, 2.5% glycerol, and 50 ng/μl of double-strand competitor DNA poly(dI-dC) (Thermo Fisher Scientific, San Jose, CA). After incubation for 20 min at room temperature, samples were analyzed on a 5% non-denaturing polyacrylamide gel in 0.5× TBE buffer at 100 V for 1 h. Gels were transferred to positively charged nylon membrane in 0.5× TBE at 380 mA for 40 min. Nylon membranes were immunoblotted and followed by a CCD camera for detecting biotin signals (Thermo Fisher Scientific).
Author contributions
P.-T. L. and Z.-F. W. participated in the design of the experiment and carried out the work and manuscript preparation. I.-T. C. and M.-H. L. participated in the work of analyzing the DNA secondary structure. Y.-M. K., M.-C. H., and P.-C. C. participated in performing the work of reporter construction and assays as well as the EMSA experiments. T.-C. C. and C.-T. C. conceived the study, participated in its design and coordination, and finalized the draft of the manuscript.
Supplementary Material
Acknowledgments
We thank Dr. Shing-Jong Huang and Shou-Ling Huang (Instrumentation Center, National Taiwan University) for assistance in obtaining the Bruker AVIII 500 and 800 MHz FT-NMR data.
This work was supported by Academia Sinica Grant AS-102-TPA07, Ministry of Science and Technology of the Republic of China Grants MOST-103-2113-M-001-017-MY3 (to T. C. C.) and MOST 105-2320-B-002-056 (to C. T. C.), and National Taiwan University and Academia Sinica Innovative Joint Program Grant NTU-SINICA-105R104510. The authors declare that they have no conflicts of interest with the contents of this article.
This article contains supplemental Figs. S1–S6, Tables S1–S2, and Materials and methods.
- PG4
- potential G-quadruplex-forming sequence
- hTERT
- human telomerase reverse transcriptase
- CTCF
- CCCTC-binding factor
- 5hmC
- 5-hydroxymethylcytosine
- HMQC
- heteronuclear multiple quantum coherence.
References
- 1. Feinberg A. P., and Tycko B. (2004) The history of cancer epigenetics. Nat. Rev. Cancer 4, 143–153 [DOI] [PubMed] [Google Scholar]
- 2. Smith Z. D., and Meissner A. (2013) DNA methylation: roles in mammalian development. Nat. Rev. Genet. 14, 204–220 [DOI] [PubMed] [Google Scholar]
- 3. Halder R., Halder K., Sharma P., Garg G., Sengupta S., and Chowdhury S. (2010) Guanine quadruplex DNA structure restricts methylation of CpG dinucleotides genome-wide. Mol. Biosyst. 6, 2439–2447 [DOI] [PubMed] [Google Scholar]
- 4. De S., and Michor F. (2011) DNA secondary structures and epigenetic determinants of cancer genome evolution. Nat. Struct. Mol. Biol. 18, 950–955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Maizels N., and Gray L. T. (2013) The G4 genome. PLoS Genet. 9, e1003468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Huppert J. L., and Balasubramanian S. (2005) Prevalence of quadruplexes in the human genome. Nucleic Acids Res. 33, 2908–2916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Chambers V. S., Marsico G., Boutell J. M., Di Antonio M., Smith G. P., and Balasubramanian S. (2015) High-throughput sequencing of DNA G-quadruplex structures in the human genome. Nat. Biotechnol. 33, 877–881 [DOI] [PubMed] [Google Scholar]
- 8. Balasubramanian S., Hurley L. H., and Neidle S. (2011) Targeting G-quadruplexes in gene promoters: a novel anticancer strategy? Nat. Rev. Drug Discov. 10, 261–275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Bochman M. L., Paeschke K., and Zakian V. A. (2012) DNA secondary structures: stability and function of G-quadruplex structures. Nat. Rev. Genet. 13, 770–780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Siddiqui-Jain A., Grand C. L., Bearss D. J., and Hurley L. H. (2002) Direct evidence for a G-quadruplex in a promoter region and its targeting with a small molecule to repress c-MYC transcription. Proc. Natl. Acad. Sci. U.S.A. 99, 11593–11598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Cogoi S., and Xodo L. E. (2006) G-quadruplex formation within the promoter of the KRAS proto-oncogene and its effect on transcription. Nucleic Acids Res. 34, 2536–2549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Dai J., Chen D., Jones R. A., Hurley L. H., and Yang D. (2006) NMR solution structure of the major G-quadruplex structure formed in the human BCL2 promoter region. Nucleic Acids Res. 34, 5133–5144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Guo K., Gokhale V., Hurley L. H., and Sun D. (2008) Intramolecularly folded G-quadruplex and i-motif structures in the proximal promoter of the vascular endothelial growth factor gene. Nucleic Acids Res. 36, 4598–4608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Rankin S., Reszka A. P., Huppert J., Zloh M., Parkinson G. N., Todd A. K., Ladame S., Balasubramanian S., and Neidle S. (2005) Putative DNA quadruplex formation within the human c-kit oncogene. J. Am. Chem. Soc. 127, 10584–10589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Lim K. W., Lacroix L., Yue D. J., Lim J. K., Lim J. M., and Phan A. T. (2010) Coexistence of two distinct G-quadruplex conformations in the hTERT promoter. J. Am. Chem. Soc. 132, 12331–12342 [DOI] [PubMed] [Google Scholar]
- 16. Wang J. M., Huang F. C., Kuo M. H., Wang Z. F., Tseng T. Y., Chang L. C., Yen S. J., Chang T. C., and Lin J. J. (2014) Inhibition of cancer cell migration and invasion through suppressing the Wnt1-mediating signal pathway by G-quadruplex structure stabilizers. J. Biol. Chem. 289, 14612–14623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Huang W. C., Tseng T. Y., Chen Y. T., Chang C. C., Wang Z. F., Wang C. L., Hsu T. N., Li P. T., Chen C. T., Lin J. J., Lou P. J., and Chang T. C. (2015) Direct evidence of mitochondrial G-quadruplex DNA by using fluorescent anti-cancer agents. Nucleic Acids Res. 43, 10102–10113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hardin C. C., Corregan M., Brown B. A. 2nd, and Frederick L. N. (1993) Cytosine-cytosine + base pairing stabilizes DNA quadruplexes and cytosine methylation greatly enhances the effect. Biochemistry 32, 5870–5880 [DOI] [PubMed] [Google Scholar]
- 19. Fry M., and Loeb L. A. (1994) The fragile X syndrome d(CGG)n nucleotide repeats form a stable tetrahelical structure. Proc. Natl. Acad. Sci. U.S.A. 91, 4950–4954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kettani A., Kumar R. A., and Patel D. J. (1995) Solution structure of a DNA quadruplex containing the fragile X syndrome triplet repeat. J. Mol. Biol. 254, 638–656 [DOI] [PubMed] [Google Scholar]
- 21. Lin J., Hou J. Q., Xiang H. D., Yan Y. Y., Gu Y. C., Tan J. H., Li D., Gu L. Q., Ou T. M., and Huang Z. S. (2013) Stabilization of G-quadruplex DNA by C-5-methyl-cytosine in bcl-2 promoter: implications for epigenetic regulation. Biochem. Biophys. Res. Commun. 433, 368–373 [DOI] [PubMed] [Google Scholar]
- 22. Kyo S., Takakura M., Fujiwara T., and Inoue M. (2008) Understanding and exploiting hTERT promoter regulation for diagnosis and treatment of human cancers. Cancer Sci. 99, 1528–1538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kim N. W., Piatyszek M. A., Prowse K. R., Harley C. B., West M. D., Ho P. L., Coviello G. M., Wright W. E., Weinrich S. L., and Shay J. W. (1994) Specific association of human telomerase activity with immortal cells and cancer. Science 266, 2011–2015 [DOI] [PubMed] [Google Scholar]
- 24. Bernardes de Jesus B., and Blasco M. A. (2013) Telomerase at the intersection of cancer and aging. Trends Genet. 29, 513–520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Armanios M., and Blackburn E. H. (2012) The telomere syndromes. Nat. Rev. Genet. 13, 693–704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Huang F. W., Hodis E., Xu M. J., Kryukov G. V., Chin L., and Garraway L. A. (2013) Highly recurrent TERT promoter mutations in human melanoma. Science 339, 957–959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Vinagre J., Almeida A., Pópulo H., Batista R., Lyra J., Pinto V., Coelho R., Celestino R., Prazeres H., Lima L., Melo M., da Rocha A. G., Preto A., Castro P., Castro L., et al. (2013) Frequency of TERT promoter mutations in human cancers. Nat. Commun. 4, 2185. [DOI] [PubMed] [Google Scholar]
- 28. Chiba K., Johnson J. Z., Vogan J. M., Wagner T., Boyle J. M., and Hockemeyer D. (2015) Cancer-associated TERT promoter mutations abrogate telomerase silencing. Elife 4, e07918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Palumbo S. L., Ebbinghaus S. W., and Hurley L. H. (2009) Formation of a unique end-to-end stacked pair of G-quadruplexes in the hTERT core promoter with implications for inhibition of telomerase by G-quadruplex-interactive ligands. J. Am. Chem. Soc. 131, 10878–10891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kang H. J., Cui Y., Yin H., Scheid A., Hendricks W. P., Schmidt J., Sekulic A., Kong D., Trent J. M., Gokhale V., Mao H., and Hurley L. H. (2016) A pharmacological chaperone molecule induces cancer cell death by restoring tertiary DNA structures in mutant hTERT promoters. J. Am. Chem. Soc. 138, 13673–13692 [DOI] [PubMed] [Google Scholar]
- 31. Renaud S., Loukinov D., Bosman F. T., Lobanenkov V., and Benhattar J. (2005) CTCF binds the proximal exonic region of hTERT and inhibits its transcription. Nucleic Acids Res. 33, 6850–6860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Renaud S., Loukinov D., Abdullaev Z., Guilleret I., Bosman F. T., Lobanenkov V., and Benhattar J. (2007) Dual role of DNA methylation inside and outside of CTCF-binding regions in the transcriptional regulation of the telomerase hTERT gene. Nucleic Acids Res. 35, 1245–1256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kuo M. H., Wang Z. F., Tseng T. Y., Li M. H., Hsu S. T., Lin J. J., and Chang T. C. (2015) Conformational transition of a hairpin structure to G-quadruplex within the WNT1 gene promoter. J. Am. Chem. Soc. 137, 210–218 [DOI] [PubMed] [Google Scholar]
- 34. Kettani A., Bouaziz S., Gorin A., Zhao H., Jones R. A., and Patel D. J. (1998) Solution structure of a Na cation stabilized DNA quadruplex containing G.G.G.G and G.C.G.C tetrads formed by G-G-G-C repeats observed in adeno-associated viral DNA. J. Mol. Biol. 282, 619–636 [DOI] [PubMed] [Google Scholar]
- 35. Bouaziz S., Kettani A., and Patel D. J. (1998) A K cation-induced conformational switch within a loop spanning segment of a DNA quadruplex containing G-G-G-C repeats. J. Mol. Biol. 282, 637–652 [DOI] [PubMed] [Google Scholar]
- 36. Lim K. W., Alberti P., Guédin A., Lacroix L., Riou J. F., Royle N. J., Mergny J. L., and Phan A. T. (2009) Sequence variant (CTAGGG)n in the human telomere favors a G-quadruplex structure containing a G.C.G.C tetrad. Nucleic Acids Res. 37, 6239–6248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Kocman V., and Plavec J. (2014) A tetrahelical DNA fold adopted by tandem repeats of alternating GGG and GCG tracts. Nat. Commun. 5, 5831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Klenova E. M., Nicolas R. H., Paterson H. F., Carne A. F., Heath C. M., Goodwin G. H., Neiman P. E., and Lobanenkov V. V. (1993) CTCF, a conserved nuclear factor required for optimal transcriptional activity of the chicken c-myc gene, is an 11-Zn-finger protein differentially expressed in multiple forms. Mol. Cell. Biol. 13, 7612–7624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Vincent J. J., Huang Y., Chen P. Y., Feng S., Calvopiña J. H., Nee K., Lee S. A., Le T., Yoon A. J., Faull K., Fan G., Rao A., Jacobsen S. E., Pellegrini M., and Clark A. T. (2013) Stage-specific roles for tet1 and tet2 in DNA demethylation in primordial germ cells. Cell Stem Cell 12, 470–478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Kaas G. A., Zhong C., Eason D. E., Ross D. L., Vachhani R. V., Ming G. L., King J. R., Song H., and Sweatt J. D. (2013) TET1 controls CNS 5-methylcytosine hydroxylation, active DNA demethylation, gene transcription, and memory formation. Neuron 79, 1086–1093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Gao Y., Chen J., Li K., Wu T., Huang B., Liu W., Kou X., Zhang Y., Huang H., Jiang Y., Yao C., Liu X., Lu Z., Xu Z., Kang L., et al. (2013) Replacement of Oct4 by Tet1 during iPSC induction reveals an important role of DNA methylation and hydroxymethylation in reprogramming. Cell Stem Cell 12, 453–469 [DOI] [PubMed] [Google Scholar]
- 42. Hackett J. A., Sengupta R., Zylicz J. J., Murakami K., Lee C., Down T. A., and Surani M. A. (2013) Germline DNA demethylation dynamics and imprint erasure through 5-hydroxymethylcytosine. Science 339, 448–452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Gray R. D., and Chaires J. B. (2008) Kinetics and mechanism of K+- and Na+-induced folding of models of human telomeric DNA into G-quadruplex structures. Nucleic Acids Res. 36, 4191–4203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Choi J. H., Min N. Y., Park J., Kim J. H., Park S. H., Ko Y. J., Kang Y., Moon Y. J., Rhee S., Ham S. W., Park A. J., and Lee K. H. (2010) TSA-induced DNMT1 down-regulation represses hTERT expression via recruiting CTCF into demethylated core promoter region of hTERT in HCT116. Biochem. Biophys. Res. Commun. 391, 449–454 [DOI] [PubMed] [Google Scholar]
- 45. Rigo R., Palumbo M., and Sissi C. (2017) G-quadruplexes in human promoters: a challenge for therapeutic applications. Biochim. Biophys. Acta 1861, 1399–1413 [DOI] [PubMed] [Google Scholar]
- 46. Piotto M., Saudek V., and Sklenár V. (1992) Gradient-tailored excitation for single-quantum NMR spectroscopy of aqueous solutions. J. Biomol. NMR 2, 661–665 [DOI] [PubMed] [Google Scholar]
- 47. Plateau P., and Gueron M. (1982) Exchangeable proton NMR without base-line distortion, using new strong-pulse sequences. J. Am. Chem. Soc. 104, 7310–7311 [Google Scholar]
- 48. Phan A. T., and Patel D. J. (2002) A site-specific low-enrichment 15N,13C isotope-labeling approach to unambiguous NMR spectral assignments in nucleic acids. J. Am. Chem. Soc. 124, 1160–1161 [DOI] [PubMed] [Google Scholar]
- 49. Wang Z. F., Li M. H., Hsu S. T., and Chang T. C. (2014) Structural basis of sodium-potassium exchange of a human telomeric DNA quadruplex without topological conversion. Nucleic Acids Res. 42, 4723–4733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Frommer M., McDonald L. E., Millar D. S., Collis C. M., Watt F., Grigg G. W., Molloy P. L., and Paul C. L. (1992) A genomic sequencing protocol that yields a positive display of 5-methylcytosine residues in individual DNA strands. Proc. Natl. Acad. Sci. U.S.A. 89, 1827–1831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Shukla S., Kavak E., Gregory M., Imashimizu M., Shutinoski B., Kashlev M., Oberdoerffer P., Sandberg R., and Oberdoerffer S. (2011) CTCF-promoted RNA polymerase II pausing links DNA methylation to splicing. Nature 479, 74–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Li T., Hu J. F., Qiu X., Ling J., Chen H., Wang S., Hou A., Vu T. H., and Hoffman A. R. (2008) CTCF regulates allelic expression of Igf2 by orchestrating a promoter-polycomb repressive complex 2 intrachromosomal loop. Mol. Cell. Biol. 28, 6473–6482 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.