Abstract
The heme-based oxygen sensor histidine kinase AfGcHK is part of a two-component signal transduction system in bacteria. O2 binding to the Fe(II) heme complex of its N-terminal globin domain strongly stimulates autophosphorylation at His183 in its C-terminal kinase domain. The 6-coordinate heme Fe(III)-OH− and -CN− complexes of AfGcHK are also active, but the 5-coordinate heme Fe(II) complex and the heme-free apo-form are inactive. Here, we determined the crystal structures of the isolated dimeric globin domains of the active Fe(III)-CN− and inactive 5-coordinate Fe(II) forms, revealing striking structural differences on the heme-proximal side of the globin domain. Using hydrogen/deuterium exchange coupled with mass spectrometry to characterize the conformations of the active and inactive forms of full-length AfGcHK in solution, we investigated the intramolecular signal transduction mechanisms. Major differences between the active and inactive forms were observed on the heme-proximal side (helix H5), at the dimerization interface (helices H6 and H7 and loop L7) of the globin domain and in the ATP-binding site (helices H9 and H11) of the kinase domain. Moreover, separation of the sensor and kinase domains, which deactivates catalysis, increased the solvent exposure of the globin domain-dimerization interface (helix H6) as well as the flexibility and solvent exposure of helix H11. Together, these results suggest that structural changes at the heme-proximal side, the globin domain-dimerization interface, and the ATP-binding site are important in the signal transduction mechanism of AfGcHK. We conclude that AfGcHK functions as an ensemble of molecules sampling at least two conformational states.
Keywords: bacterial protein kinase, crystal structure, histidine kinase, hydrogen-deuterium exchange, signal transduction, globin, heme-containing oxygen sensor, two component signal transduction system
Introduction
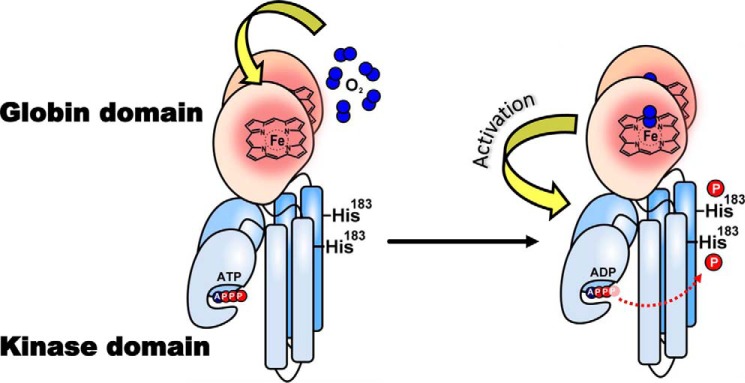
The heme-based oxygen sensor kinase from the soil bacterium Anaeromyxobacter sp. Fw109-5, AfGcHK is a globin-coupled oxygen sensor (GCS)3 consisting of an N-terminal heme-binding globin domain and a C-terminal histidine kinase domain (1, 2). AfGcHK is an element of a two-component signal transduction system (3–6) in which the binding of O2 to the heme Fe(II) complex of its oxygen-sensing globin domain significantly enhances the autophosphorylation activity of its kinase domain via intramolecular signal transduction (Fig. 1). The phosphate group is subsequently transferred from His183 of the AfGcHK to the Asp52 and Asp169 residues of its cognate response regulator protein (1, 2, 7) (supplemental Fig. S1). Like the 6-coordinate heme Fe(II)-O2 complex, the 6-coordinate heme Fe(III)-OH− and CN− complexes are also active, despite having a different redox state at the iron center (8). However, AfGcHK containing a globin domain with a 5-coordinate Fe(II) heme complex or a heme-free apo-globin domain exhibits low kinase activity (8). This strongly suggests that structural changes near the heme-binding site in the globin domain are tightly linked to the catalytic regulation of the kinase domain via intraprotein signal transduction.
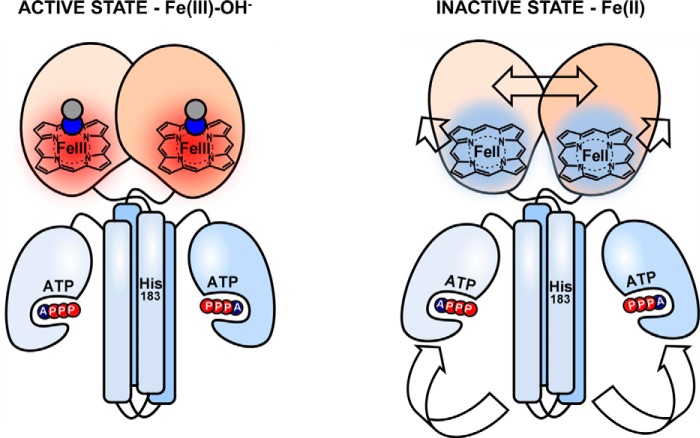
Figure 1.
Intramolecular signal transduction of homodimeric AfGcHK. O2 binds to the heme Fe(II) complex in the globin domain at the N terminus, changing the structure of the protein surrounding the heme. These structural changes function as a signal that is transduced to the kinase domain (at the C terminus), activating the kinase reaction and inducing autophosphorylation at His183.
To understand the molecular mechanism of this intramolecular signal transduction, it is important to know 1) how the structure of the globin domain differs between the Fe(III) (active) and Fe(II) (inactive) heme complex–bound forms and between forms with and without axial ligands; 2) how structural changes in the globin domain propagate to the kinase domain; and 3) how the transition from the inactive to the active state in response to the transduction of the globin domain signal affects the kinase domain.
The structure–function relationships of oxygen sensors containing a heme-bound globin, PAS, and GAF folds have been investigated in various proteins (2). Both heme iron reduction and the binding of external axial ligands such as O2 can induce global structural changes (affecting both the heme surroundings and more distant regions) in the isolated heme-bound PAS domains of oxygen sensors, such as FixL (9, 10) and EcDOS (11, 12). However, these effects were found to be less pronounced in the isolated heme-bound GAF domains of oxygen sensors, such as DosS/DosT, than those observed for the PAS domain (13–15). Additionally, changes in heme redox state or external ligand binding had no significant impact on the structure of the isolated heme-bound globin domain of GCS (16–18). In this work, we present the first X-ray crystal structures of heme Fe(III) and Fe(II) complexes of the isolated dimeric globin domain of AfGcHK, revealing large structural differences between the active and inactive forms in the vicinity of the heme and at the globin dimerization interface.
Importantly, no X-ray crystal structure of any full-length heme-based oxygen sensor has yet been reported. We previously studied the full-length AfGcHK protein in solution by combining hydrogen/deuterium exchange coupled with mass spectrometry (HDX–MS) with homology modeling (7). However, the structural changes in this protein caused by heme redox state changes and the binding of different axial ligands (both of which are closely linked to catalytic regulation) have not been characterized.
This work uses HDX–MS to describe the structural differences between the active and inactive forms of full-length AfGcHK and its isolated globin and kinase domains in solution and analyzes structural differences between the active and inactive forms of the AfGcHK globin domain revealed by X-ray crystallography. It was found that changes in the heme redox state and axial ligand binding to the heme iron complex lead to structural alterations in the heme-proximal side of the globin domain and affect domain-domain interactions within the dimeric globin domains. Differences were also observed around the two helices forming the ATP-binding site necessary for autophosphorylation in the kinase domain. These structural changes associated with catalytic regulation revealed by HDX–MS corroborated the conclusions drawn by analyzing the X-ray crystal structures of the isolated globin domain in two redox states.
Results
We first determined the X-ray crystal structures of isolated AfGcHK globin domains bearing heme in the Fe(III)-CN− form (active state) and containing heme Fe(II) (inactive state) complexes and then compared them with HDX–MS data for the full-length AfGcHK protein.
Crystal structure of the isolated heme Fe(III)-CN− globin domain (PDB code 5OHE)
The crystal structure of the isolated globin domain in the presence of KCN was solved at a resolution of 1.85 Å. Relevant data processing parameters and structure characteristics are presented in Table 1.
Table 1.
Data processing statistics and structure refinement parameters
Values in parentheses refer to the highest-resolution shell.
| Globin domain of AfGcHK with heme Fe(III)-CN− | Globin domain of AfGcHK with cyanide, partially reduced (the dithionite-soaked crystal) | |
|---|---|---|
| PDB code | 5OHE | 5OHF |
| Data processing statistics | ||
| Space group | P41212 | P41212 |
| Unit-cell parameters | ||
| a, b, c (Å) | 78.1, 78.1, 441.8 | 77.7, 77.7, 441.2 |
| α, β, γ (degrees) | 90.0, 90.0, 90.0 | 90.0, 90.0, 90.0 |
| Resolution range (Å) | 46.82–1.85 (1.88–1.85) | 49.17–1.8 (1.83–1.80) |
| No. of observations | 1,413,447 (62,729) | 931,937 (47,063) |
| No. of unique reflections | 118,539 (5739) | 127,077 (6192) |
| Data completeness (%) | 100 (100) | 100 (100) |
| Average redundancy | 11.9 (10.9) | 7.3 (7.6) |
| Mosaicity (degrees) | 0.118 | 0.07 |
| Average I/σ(I) | 11.1 (1.9) | 16.7 (2.7) |
| Solvent content (%) | 46 | 46 |
| Matthews coefficient (Å3/Da) | 2.27 | 2.27 |
| Rmergea | 0.139 (1.157) | 0.059 (0.761) |
| Rp.i.m.a | 0.059 (0.517) | 0.034 (0.442) |
| CC1/2 | 0.998 (0.804) | 0.993 (0.844) |
| Structure refinement parameters | ||
| Rworka | 0.192 | 0.190 |
| Rfree | 0.241 | 0.239 |
| Rall | 0.194 | 0.192 |
| Average B-factor (Å2) | 24 | 28 |
| RMSD bond lengths from ideal (Å) | 0.015 | 0.009 |
| RMSD bond angles from ideal (degrees) | 1.653 | 1.267 |
| Number of non-hydrogen atoms | 11,456 | 12,325 |
| Dimers per asymmetric unit (chains) | AB, CD, EF, GH | AB, CD, EF, GH |
| No. of water molecules | 1117 | 1017 |
| Ramachandran statistics | ||
| Residues in favored regions (%) | 100 | 99.9 |
| No. of outliers | 0 | 0 |
a Rmerge = ΣhΣi|Ihi − 〈Ih〉|/ΣhΣiIhi, Rp.i.m. = ΣhΣi(nh − 1)−½|Ihi − 〈Ih〉|/ΣhΣiIhi, and R = Σh‖Fh,obs| − | Fh,calc‖/Σh|Fh,obs|, where Ihi is the observed intensity, 〈Ih〉 is the mean intensity of multiple observations of symmetry-related reflections, and Fh,obs and Fh,calc are the observed and calculated structure factor amplitudes. Rwork is the R factor calculated on 95% of reflections excluding a random subset of 5% of reflections marked as “free.” The final structure refinement was performed on all observed structure factors.
Overall structure
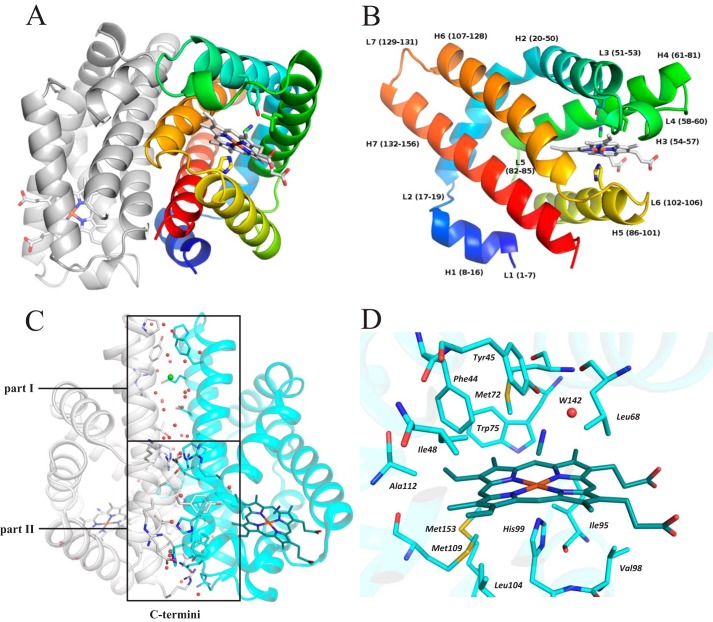
The asymmetric unit of the crystal consists of four dimers (Fig. 2A) formed by eight protein chains (AB, CD, EF, and GH) positioned in a disphenoidal arrangement. The crystal structure confirms that the domain adopts the characteristic globin fold of GCSs (Fig. 2B). Each monomer consists of seven helices, H1–H7 (also known as helices A–G (2)), and seven loops, L1–L7. RMSD values of coordinates of Cα atoms of the aligned protein chains (monomers) are in the range of 0.2–0.5 Å with no significant difference between the structures of the monomers. The fold of the dimer is most similar to that of the globin domain of Geobacter sulfurreducens GCS (PDB code 2W31 (17), 34% sequence identity); the RMSD between the Cα atom coordinates of the aligned dimers is 1.4 Å (PDBeFold (19)).
Figure 2.
Crystal structure of a dimer of the globin domain of AfGcHK with an Fe(III)-CN− heme complex (active state, PDB code 5OHE) and a close-up view of the heme surroundings. A, secondary structure representation of the dimer with the N terminus of chain A in blue, the C terminus of chain A in red, and chain B in gray. The heme, selected neighboring residues, and bound cyanide molecule are shown using a stick representation. B, close-up view of the molecular architecture of protein chain A, using the same color coding and representations as in A; individual α-helices, loops, and the corresponding residue numbers are marked. C, dimerization interface of the isolated globin domain dimer, showing its division into parts I and II; part I is situated opposite the kinase domain in the full-length protein and relies only on van der Waals interactions and water-mediated hydrogen bonds, with no direct protein–protein hydrogen bonds, whereas part II is near the C terminus (close to the kinase domain in the full-length protein) and features 19 interchain protein–protein hydrogen bonds, hydrophobic interactions, and several water-mediated hydrogen bonds. Red dots, water molecules; green sphere, a chloride ion. D, the heme pocket residues (whose carbon atoms are shown in cyan), the heme group (carbon atoms shown in dark cyan), and the cyanide ligand (carbon atom shown in dark cyan) are shown in a stick representation with color coding according to atom type. The graphics were generated using PyMOL (Schrödinger, LLC, New York).
Crystal contacts, electron density, and B-factors; differences among the chains
All four dimers in the asymmetric unit have approximately the same contact surface area at the dimer interface (as determined by a PISA assembly analysis (20)) except in the region of residues Arg55–Val65. This region is very well localized in the electron density map in chains A and D due to stabilizing interchain contacts within the asymmetric unit. The remaining chains form weaker water-mediated contacts with symmetry-related chains. In chains C and G, this region exhibits higher disorder and weaker electron density. This is also reflected in B-factors of the protein chains; the C and G chains of the dimer had higher chain-averaged B-factors than the other chains (28 and 31 Å2, respectively, compared with 21–23 Å2 for the others). The higher average B-factors of both chains are primarily due to B-factor “hot spots” around residues 55–65, which are close to the heme-binding pocket. These observations are in agreement with the greater susceptibility of chains C and G to structural changes and in part explain why the effects of the dithionite soaking were observed only in chain G (see below).
Dimer interface
Interestingly, the dimer interface largely consists of water-mediated hydrogen bonds. The interface between protein chains A and B features 28 water molecules involved in water-mediated hydrogen bonds, of which 12 participate in residue-water-residue and four participate in residue-water-water-residue bonds (Fig. 2C). Another eight water molecules together with a chloride ion fill a large cavity in the upper part of the dimerization interface (the area labeled part I of the dimerization interface in Fig. 2C), which is located opposite the kinase domain in the full-length protein. The total interface area varies between 1722 and 1800 Å2, depending on the pair of chains analyzed, and accounts for around 21% of the total monomer surface area of 8500 Å2. The interface can be divided into two regions (Fig. 2C): part I and part II. Part I features mainly van der Waals interactions and water-mediated hydrogen bonds and has no direct protein–protein hydrogen bonds. Part II is close to the C-terminus (and thus close to the kinase domain in the full-length protein) and features 19 interchain protein–protein hydrogen bonds, hydrophobic interactions, and several water-mediated hydrogen bonds. Part I of the interface therefore allows for more variability in the relative arrangement of the two monomers, whereas part II functions as a more tightly tethered hinge. Together, the two parts allow the monomers' relative positions and orientations to be quite well defined and controlled near the heme sites while also permitting a highly dynamic arrangement, especially in the part I region (Fig. 2C).
Heme surroundings
The heme is bound in a hydrophobic pocket formed by residues from helices H2–H7. The 6-coordinate Fe(III) ion is stabilized by direct interactions with His99 (belonging to H5) at the proximal side and Tyr45 (belonging to H2) at the distal side, with the latter interaction being mediated by a small ligand (Fig. 2D). In most chains, the small ligand can be interpreted as a CN− ion, but the electron density maps for chains D and F lack the diatomic character and were interpreted as water molecules.
The distal side of the heme-binding pocket is formed from eight residues located within 4 Å of the heme: Phe44, Tyr45, Ile48, Leu68, Met72, Trp75, Ala112, and Met113. Additionally, Tyr15 from the neighboring chain completes the binding pocket. The Leu68 residue is located at the entrance to the heme-binding pocket and represents a “gate” for incoming ligand molecules. The proximal side is formed from residues Ile95, Val98, His99 (which binds directly to the Fe(III) center), Ile102, Leu104, Tyr108, Met109, and Met153. As observed in our crystal structure, the distance between the Fe(III) center and the Nϵ2 center of His99 is 2.1 Å.
The observed position of Tyr45 is consistent with resonance Raman data on its interaction with O2 bound to Fe(II) (1). In addition, mutation of Tyr45 destabilizes the Fe(II)-O2 complex, increasing its autoxidation rate several hundredfold (supplemental Table S1 and Figs. S2 and S3). The corresponding Tyr residue (Tyr43) of the homologous GCS sensor protein, YddV, is also assumed to interact with O2 bound to the heme Fe(II) center. Accordingly, mutation of this residue significantly changes the O2-binding behavior of YddV: the Y43A and Y43L mutants do not bind O2, and the autoxidation rates of the Y43F and Y43W mutants are more than 10 times higher than that of the wild type (supplemental Table S1) (1, 21–24). Leu68, which is located between the small heme ligand and the bulk solvent, also has important effects on the activity of AfGcHK. Its role in stabilizing the Fe(II)-O2 complex has been confirmed using site-directed mutagenesis; the L68G, L68N, and L68V mutations all significantly increased the heme iron autoxidation rate (supplemental Table S1 and Figs. S2 and S3).
Crystal structure of the AfGcHK globin domain after reduction with sodium dithionite (PDB code 5OHF)
To obtain a crystal structure of the AfGcHK globin domain with an Fe(II) heme complex, the crystal of the Fe(III)-CN−-containing globin domain was soaked in a solution of the strong reductant sodium dithionite. The resulting diffraction data were processed at a resolution of 1.8 Å. The data processing parameters and structure characteristics for the reduced crystal are presented in Table 1.
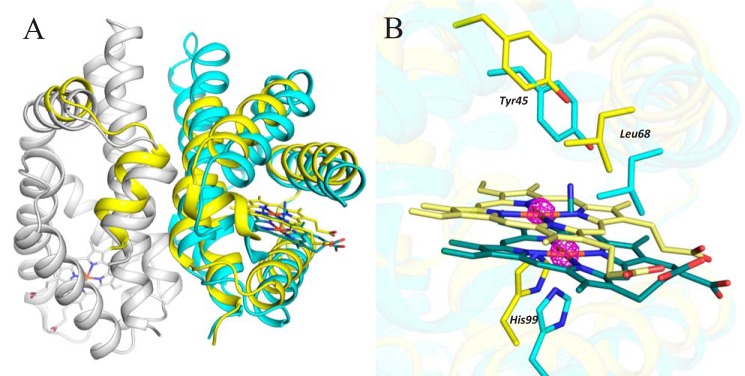
The reduced crystal retains the unit cell and asymmetric unit ordering of the parent (non-soaked) crystal. However, the structure of protein chain G (which had the highest B-factors in the parent crystal) differs sharply from that in the parent crystal (Fig. 3A). The iron center of the heme group from chain G was found in two distinct positions, each having an occupancy of 0.5 (confirmed by anomalous scattering; see Fig. 3B). Similarly, the heme group and protein chain were also observed in two alternative positions designated A and B (supplemental Fig. S4). In alternative A, the structure of the G chain resembles that of the other chains, and the sixth ligand is visible above the heme; if the sixth ligand is not modeled, there is a positive difference peak in the position it would occupy. Alternative A thus represents an Fe(III)-CN−-bound form. Omit electron density does not contain any significant positive difference peak that would justify building of a small ligand in the expected position above heme in alternative B. Therefore, alternative B contains a heme with no sixth ligand and thus represents a reduced heme iron complex formed by the action of dithionite. Based on the experimental conditions and the lack of a small axial ligand, we assume that the iron center in this alternative is 5-coordinate and exists in the Fe(II) oxidation state (Fig. 3B). In alternative A, the atomic coordinates for all amino acid residues in chain G and all of the other protein chains are consistent with those for the corresponding amino acids in the original 6-coordinate Fe(III)-CN− structure. In alternative B, the positions of most residues in chain G and some in chain H (its dimerization partner protein chain) differed significantly from those in alternative A. In total, 93 residues in chain G were modeled in alternative positions: Pro38–Leu58, Gln64–Leu120, and Gly143–Arg158 at the end of the chain (residues Val59–Ser63 could not be localized and therefore were not modeled). Residues in chain H that required interpretation by two alternative positions were Thr7–Ala25, located at the beginning of the chain (Fig. 3A). All residues built in two alternatives belong to helices H2–H7 of chain G and helices H1 and H2 of its partner chain H. The positional changes in chain H are clearly due to those in chain G rather than any redox process affecting the chain H heme complex. The heme and heme pocket in alternative B are shifted away from the globin dimer interface by 2.5 Å relative to their position in alternative A, and much of the positional difference between the protein chains in two alternatives is due to the displacement of residues to follow and accommodate this shift. The residue exhibiting the greatest displacement from its position in alternative A (3.6 Å) is Ile102, which forms van der Waals interactions with the heme.
Figure 3.
Structural changes induced by sodium dithionite soaking (PDB code 5OHF). A, chains G and H are shown using a secondary structure representation (in cyan and gray, respectively) except for their heme and CN− moieties, which are shown using a stick representation (colored in dark cyan for chain G and dark gray for chain H). Alternative B is shown in yellow, and the position of its heme group is shown in light yellow using a stick representation. Most of the residues adopting different positions in alternative B are in chain G. B, detailed view of the shift of the heme group and the surrounding heme pocket upon dithionite soaking. The active Fe(III)-CN− form is shown in cyan, and the inactive Fe(II) form is shown in yellow. The anomalous difference map (magenta, contoured at 5σ) confirms the presence of iron in two positions (observed in chain G only). The graphics were generated using PyMOL (Schrödinger, LLC).
The crystal structure presented in Fig. 3 is the best from a series of eight diffraction data sets obtained by soaking of the original crystals of the AfGcHK globin domain in sodium dithionite for various periods. All eight data sets were checked, and similar trends of structural changes were observed. The data set allowing the most accurate interpretation of the data in terms of structural changes was selected. For details, see the supplemental materials.
Determination of the oligomeric state of the isolated globin domain and the full-length protein in solution by analytical ultracentrifugation
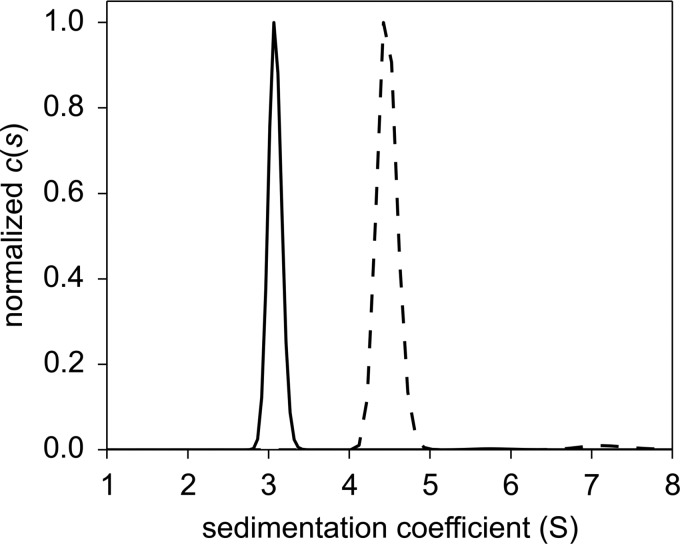
The full-length AfGcHK protein forms a dimer in solution (Fig. 4) because its sedimentation coefficient of s20,w = 4.74 S corresponds to a molecular mass of 82 kDa, but its molecular mass as computed from its amino acid sequence is 43 kDa. The isolated globin domain of AfGcHK also forms a dimer in solution (Fig. 4) because its sedimentation coefficient of s20,w = 3.20 S corresponds to a molecular mass of 39 kDa, but its molecular mass as computed from its amino acid sequence is 19 kDa.
Figure 4.
Sedimentation coefficient distributions for the isolated globin domain of AfGcHK (15 μm; black solid line) and the full-length AfGcHK (15 μm; black dashed line). The weight average sedimentation coefficients were calculated from the absorbance data. For further details, see “Experimental procedures.” c(s) denotes the continuous sedimentation coefficient distribution.
Similarities and differences between the active Fe(III)-OH− and inactive Fe(II) forms of the full-length AfGcHK protein revealed by HDX–MS data
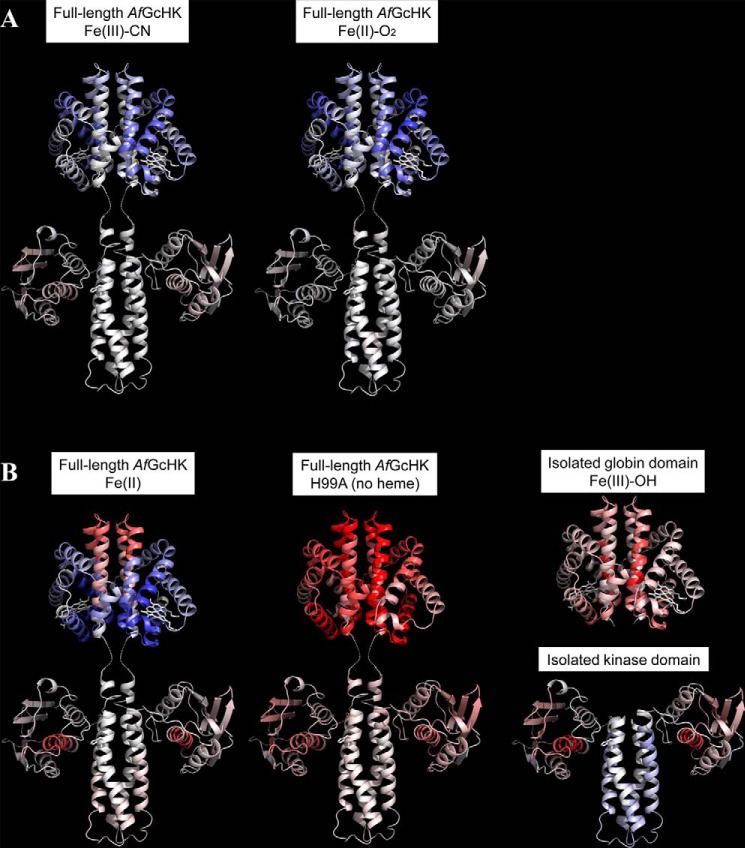
The structure of the isolated globin domain of AfGcHK containing Fe(III)-CN− determined by X-ray crystallography in this work and our previously reported model structure of the isolated kinase domain (7) were used to visualize the changes in the functional states of AfGcHK that were observed by HDX–MS, which are summarized in Fig. 5.
Figure 5.
Conformational changes revealed by HDX–MS after 60 min of deuteration of the full-length AfGcHK proteins and the separated domains, visualized on the protein's structure. Differences between the Fe(III)-OH− form and other active or inactive forms are color-coded; gray indicates no difference, whereas higher and lower levels of deuteration in the specified protein form are indicated by increasingly intense red and blue coloration, respectively. A, active full-length AfGcHK proteins with Fe(III)-CN− (left) and Fe(II)-O2 (center) heme complexes. B, inactive structures, including the full-length AfGcHK protein with an Fe(II) heme complex (left), the full-length heme-free (apo) H99A mutant (center), the isolated globin domain (top right), and the isolated kinase domain (bottom right). The globin domain structure is based on the X-ray crystal data (PDB code 5OHE) presented in this work, and the kinase domain was modeled as an asymmetric homodimer using subunits A and C of the sensor histidine kinase (HK853) from Thermotoga maritima (PDB code 3DGE). This approach was validated by experiments presented elsewhere (7).
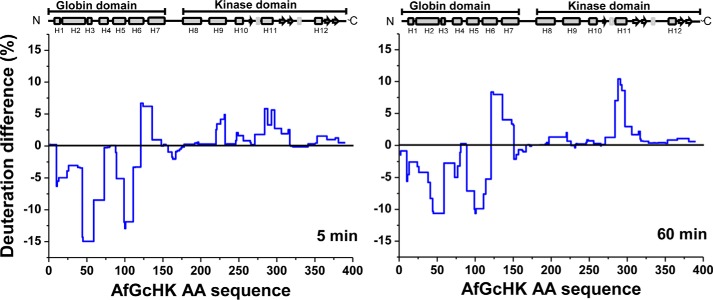
Reduction of the Fe(III)-OH− complex in AfGcHK changes the iron coordination number from 6 to 5 and is accompanied by changes in deuteration across the entire protein. This is shown in Fig. 6, which presents the relative differences in deuteration between the active full-length AfGcHK protein containing the Fe(III)-OH− complex and the inactive 5-coordinate Fe(II) complex with no sixth ligand. The greatest decrease in deuteration was observed in the globin domain, and the magnitude and extent of the decrease observed 5 min after reduction were similar to those seen after 60 min (Fig. 6). Regions of the protein that were stabilized or more shielded from deuteration included helices H2 and H3 (residues 44–72), and H5 and H6 (residues 89–120); these helices are shown in blue in the top left structure in Fig. 5B. On the other hand, heme iron reduction increased deuteration in part I of the globin dimerization interface, which is distant from the connection to the kinase domain. Regions exhibiting pronounced increases in deuteration included the end of H6, loop L7, and the beginning of H7 (residues 121–150); see the red areas in the top left structure in Fig. 5B. The HDX–MS results are consistent with the globin domain structure determined by X-ray crystallography (Fig. 2C); part I shows greater dynamics, whereas part II functions as a more tightly tethered hinge. This behavior was observed for the isolated globin domain with an Fe(III)-CN− heme complex and is readily apparent in the HDX–MS results for the active and inactive forms of the full-length AfGcHK protein (compare the top left structures shown in Fig. 5, A and B). The structural data suggest that the globin dimerization interface undergoes a sort of scissor motion as the protein transitions from the active to the inactive state.
Figure 6.
HDX–MS protection plots showing the differences in deuteration after 5 min (left) and 60 min (right) of exchange for the full-length Fe(III)-OH−bound (active) and Fe(II)-bound (inactive) forms of AfGcHK. The deuteration levels of the active form were subtracted from those of the inactive form and plotted against the protein's sequence (x axis). The blue line thus represents the structural changes caused by reducing the heme iron center in the globin domain of the full-length protein.
Interestingly, reduction of the heme iron center has little effect on the deuteration profile of the kinase domain outside two distinct regions: residues 221–231 of H9 and residues 284–296 of H11 (Figs. 5B (bottom left) and 6). The largest changes occur in the part of the kinase domain (helix H11) that forms the ATP binding site, which is more flexible and solvent-exposed in the inactive Fe(II)-bound form. It may be that heme iron reduction increases the separation of H9 and H11 (the ATP binding site), indirectly hindering His183 (at H8) phosphorylation.
The differences in deuteration between the active Fe(III)-OH− and inactive Fe(II) forms of the full-length AfGcHK protein were analogous in all tested incubation times. Data for the 5- and 60-min time points were selected as representative results (Fig. 6); the measurements for the other time points (0.5, 20, and 180 min) showed similar trends (supplemental Fig. S5).
Similarities and differences between the other active and inactive forms of the full-length AfGcHK protein revealed by HDX–MS data
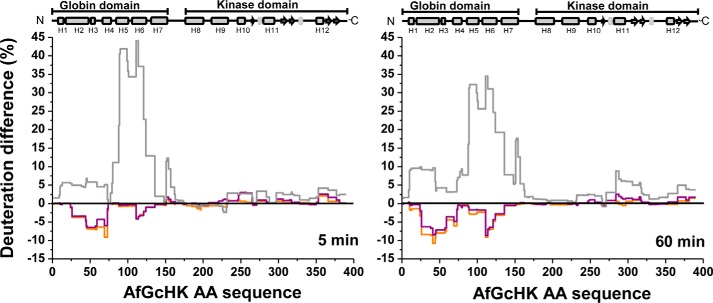
To determine whether the changes in the deuteration profile of AfGcHK upon reduction of the active Fe(III)-OH− globin domain heme complex to the inactive Fe(II) form are consistent with those observed for other active and inactive forms (supplemental Fig. S6), we performed HDX–MS experiments with three other full-length variants: the inactive heme-free apoprotein (the H99A mutant), the active Fe(III)-CN− complex, and the active Fe(II)-O2 complex (Fig. 7).
Figure 7.
HDX–MS deuteration differences for the full-length active Fe(II)-O2 (orange) and Fe(III)-CN (purple) forms of AfGcHK and the inactive full-length heme-free H99A mutant (gray) after 5 and 60 min of exchange. The deuteration differences for each form are computed relative to the deuteration of the full-length AfGcHK protein with the Fe(III)-OH− heme complex.
The differences in deuteration between the Fe(III)-OH− form and the other active forms containing CN− and O2 were rather small and confined to the heme-distal side of the globin domain, specifically residues 30–60 and 120–130 in helices H2, H3, and H6 (represented as blue ribbons in the top left and central structures in Fig. 5A). It thus seems that the nature of the sixth heme ligand affects the protein's dynamics and the solvent accessibility in the vicinity of the heme prosthetic group on the heme-distal side but has little effect on the transduction of dynamic effects between the globin and kinase domains (represented as gray ribbons in the bottom left and central structures of Fig. 5A).
The HDX–MS patterns for the two inactive forms, the 5-coordinate Fe(II) complex and the H99A apo-mutant, differ significantly from those for the active forms. In particular, their globin domains have deuteration patterns very different from those of the active Fe(III)-OH− form; the 5-coordinate Fe(II) complex is much less extensively deuterated on the heme-proximal side (helices H2, H3, and H5) but more strongly deuterated around the dimerization interface (helices H6 and H7) (see the top part of the left structure in Fig. 5B). On the other hand, the entire globin domain of the apo-form is completely deprotected (Fig. 5B, top part of the central structure). Interestingly, despite these differences in the globin domains, the kinase domains of the two inactive forms exhibit rather similar deuteration patterns, with sharply increased deuteration in helix H11(284–296) (see the bottom parts of the left and central structures in Fig. 5B). Increases in the solvent accessibility and dynamic behavior of helix H11 were also identified as the major changes associated with domain separation and kinase inactivation (Fig. 5B, bottom right). This may indicate that this area is involved in intramolecular signal transduction. The deuteration patterns of the active and inactive forms of AfGcHK did not vary appreciably over the incubation times considered in this work.
Comparison of the full-length AfGcHK protein with its isolated globin and kinase domains based on HDX–MS data
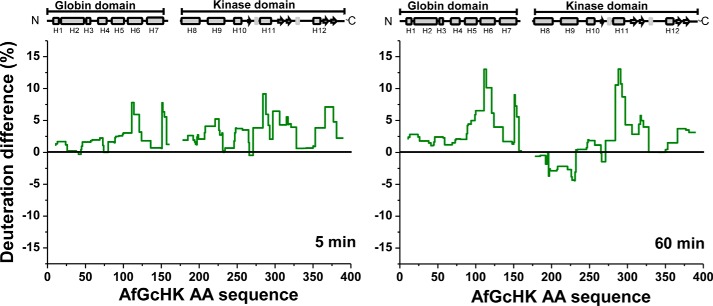
We also compared the HDX–MS exchange profile of the Fe(III)-OH− complex of the full-length AfGcHK protein with those of the isolated Fe(III)-OH− globin domain and the isolated kinase domain (Fig. 8).
Figure 8.
HDX–MS deuteration differences between the active full-length Fe(III)-OH− form of AfGcHK and a mixture of its isolated Fe(III)-OH−-bound globin domain with its isolated inactive kinase domain. Deuteration differences are shown for incubation times of 5 and 60 min lined in dark green, and the plotted values were obtained by subtracting the deuteration levels of the active full-length Fe(III)-OH− form from those for the isolated domains.
The globin domain of the full-length protein is dimeric, and ultracentrifugation experiments showed that the isolated globin domain is too (Fig. 4). However, the absence of the covalently attached kinase domain increases the solvent accessibility of the globin domain contact interface (H6 and L7) in the isolated domain (Fig. 5B, top right structure). Interestingly, another region showing increased deuteration upon isolation of the globin domain was helix H5 on the heme-proximal side (shown in pink in the bottom parts of the top right structure in Fig. 5B; see also Fig. 8). This helix is in direct contact with both the heme and the C terminus of the globin domain, the latter of which is attached to the kinase domain via a short linker. Therefore, H5 might be important in intraprotein signal transduction.
The separation of the kinase domain from the globin domain eliminates its autokinase activity, suggesting that the attachment of the globin domain in the full-length AfGcHK protein is required for the functioning of the kinase domain (supplemental Fig. S7). HDX–MS experiments indicated that the isolated kinase domain is more solvent-accessible than that in the full-length protein. The dark red ribbon in the bottom part of the right structure in Fig. 5B shows the significant structural destabilization of helix H11 (residues 284–292) associated with the isolation of the kinase domain in the full-length protein. The isolation of the kinase domain, which inactivates it, also affects helix H9, which is slightly deprotected over short deuteration times (Fig. 8) but moderately protected over longer deuteration times (see the light blue ribbons in the bottom right structure in Fig. 5B). Conversely, both incubation times make helix H11 more solvent-accessible. A slight increase in deuteration was also observed for the large β-sheet–forming solvent-exposed part of the ATP-binding subdomain (pink arrows in the bottom right structure in Fig. 5B).
The deuteration profiles of the isolated globin (Fig. 5B, top right) and kinase (Fig. 5B, bottom right) domains show several similarities to the inactive forms of the full-length protein (i.e. the 5-coordinate heme Fe(II) form and the H99A apo-form) (Figs. 5–8). Specifically, helices H6 and H11 behave similarly when the heme iron is reduced and when the domains are separated.
Characterization of the contact interface between the globin and kinase domains based on HDX–MS data for isolated domains and mixtures of isolated domains
HDX–MS can be used to map interaction interfaces between proteins or domains if they interact in a way that affects solvent accessibility and/or hydrogen bonding. However, as shown in supplemental Fig. S8, the deuteration profiles of the isolated globin and kinase domains when measured in isolation (dark green lines) did not differ from those seen with an equimolar mixture of the two (light green lines). This implies that the domains are not in direct close contact with a clearly defined interaction interface. The absence of protection in a HDX–MS experiment does not necessarily mean the absence of a direct interaction; weak and/or transient interactions are not captured by this method (25–27). However, such weak interactions would probably not explain the efficient signal transduction between the sensor and kinase domains.
Discussion
Full-length heme-based gas sensor proteins are very difficult to crystallize. X-ray structures have been reported for a small full-length heme-based CO-sensing transcription activator, CooA (28, 29), but not for any full-length heme-based oxygen sensors. HDX–MS is a powerful complementary tool to crystallography for mapping the folding, interactions, and conformational changes of proteins in solution, including those that are not readily crystallized. For example, it was used to characterize significant conformational changes in the heme-based NO sensor soluble guanylate kinase induced by NO binding (30). In addition, we have previously studied the folding, interactions, and conformational changes of heme Fe(III) complexes of AfGcHK in the presence and absence of its response regulator partner protein (7). In this work, we determined the X-ray crystal structures of the isolated globin domain of AfGcHK with 6-coordinate Fe(III)-CN− and 5-coordinate Fe(II) complexes and compared these structures with conformational changes observed by HDX–MS.
X-ray structures of the AfGcHK globin domain in various forms
The X-ray structure of AfGcHK globin domain containing the Fe(III)-CN− complex (Fig. 2) revealed slight differences between the individual chains, mainly in the long loop (L3) connecting helices H3 and H4. This segment, which consists of residues Ala57–Met71, has the highest absolute hydrogen/deuterium exchange level in both the isolated globin domain and the full-length AfGcHK protein, suggesting that it is very flexible and solvent-accessible. The large contact area observed between the two chains of the AfGcHK dimer supports the proposed physiological relevance of the dimerization of its globin domain, in keeping with the results of ultracentrifugation experiments (Fig. 4) and predictions based on earlier HDX–MS experiments (7). Moreover, all globin domains from homologous GCS proteins with known structures form analogous dimers (16–18). The unique organization of the globin domain dimerization interface, with its flexible part I and tightly bound part II (see Fig. 2C), implies a high capacity for dynamic rearrangement (31). This is consistent with the HDX–MS data for active and inactive forms of the full-length AfGcHK protein (Figs. 5 (A and B), top parts in left structures), which suggest that the globin domain dimerization interface undergoes a “scissor” motion as the protein transitions between active and inactive forms (Fig. 9). A scissor-like dynamic motion was inferred for another oxygen sensor, EcDOS, based on the crystal structure of its dimeric isolated heme-bound PAS domain (11). A structural comparison of the isolated globin domains of AfGcHK with other GCSs is presented in the supplemental material.
Figure 9.
Proposed intramolecular signal transduction mechanism in the full-length AfGcHK protein that explains its catalytic inactivation upon heme reduction. Heme reduction induces conformational changes in the globin domain, simultaneously widening and splitting its internal dimeric interface, and making the initial residues of H7 accessible to the solvent. This signal is propagated down to the kinase domain via the linker, causing the ATP binding site to be separated from His183, hindering the autophosphorylation of the latter residue. Data previously obtained by our group indicated that heme reduction increases the enzyme's KmATP value and thus reduces the ATP affinity of the kinase domain (8).
Interestingly, reducing the heme iron in the crystallized Fe(III)-CN− form of the isolated globin domain induced a structural change (Fig. 3). The most significant changes were observed in chain G, in the chain with elevated temperature factors and weaker protein–protein contacts of the H3–L3 segment in the crystal. Surprisingly, only some heme iron molecules populating chain G were reduced upon soaking the original crystal in the dithionite solution, resulting in the observation of two different alternatives, A and B. Observations of alternative localization of protein atoms in crystal structures are common and can provide vital insights into how conformational changes enable proteins to function (32). Alternative A is consistent with the original atomic coordinates of the chains in the 6-coordinate Fe(III)-CN− structure, whereas alternative B represents the reduced state of the heme complex. Because this change occurred under strongly reducing conditions and the heme of the B alternative lacks a distal ligand, we conclude that the heme in chain G was partially reduced to form a 5-coordinate Fe(II) complex. Therefore, the two alternatives represent globin domain structures associated with one active form (Fe(III)-CN− complex) and one inactive form (Fe(II) complex).
The most important result is the observation that the heme and a significant part of the protein chain G and minor part of the protein chain H are shifted in the case of the heme iron reduction. There is thus a clear structural difference between the globin domains associated with the active and inactive forms. Conversely, Tarnawski et al. (16) found that heme iron reduction had no significant structural effect on the isolated globin domain of YddV from Escherichia coli (PDB entries 4ZVA and 4ZVB). They reached this conclusion by preparing two different crystals and superimposing their structures, whereas we prepared a single protein crystal containing AfGcHK in a mixture of two states. In our case, the four dimers in the asymmetric unit formed luckily a scaffold in which it is possible to see larger changes in one of the dimers without destroying the crystal. If all of the protein chains of this asymmetric crystal underwent similar reduction-induced changes, it would probably cause major structural changes in the crystal lattice; indeed, prolonged soaking in the dithionite solution led to macroscopic crystal reshaping. Based on the consistent experimental results for crystals soaked in the strong reducing agent and the adverse effects of long soaking times, we suggest that the concurrent observation of both states in the crystal was only possible because the heme of chain G in the asymmetric crystal reacts more rapidly than the hemes of the other chains.
Signal transduction in the full-length AfGcHK protein observed by HDX–MS
For the first time, we have observed conformational changes associated with signal transduction in a full-length heme-containing sensor protein. All known active forms of the full-length AfGcHK protein, namely the Fe(III)-CN−-, Fe(III)-OH−- and Fe(II)-O2-containing forms, exhibit very similar dynamic behavior and solvent accessibility, unlike the inactive Fe(II)- and heme-free forms (Fig. 5). The most important differences between the active and inactive forms were found 1) near the heme in the globin domain; 2) in the dimerization interface (helices H6 and H7) of the globin domain, where the end of helix H6, loop L7, and the beginning of helix H7 are more solvent-accessible in the inactive forms; and 3) in helices H11 and (to a lesser extent) H9 in the kinase domain, which undergo faster hydrogen/deuterium exchange after heme iron reduction or in the absence of heme. The faster hydrogen/deuterium exchange in H11 and H9 could be caused by their opening or an increase in the mobility of this region, which is consistent with the hypothesis that the distance between the helices increases in the inactive states. These changes are probably significant because the HDX–MS results for the three active forms were similar, as were those for the two inactive forms, especially with respect to the kinase domain. The HDX–MS data thus highlighted the key regions involved in the molecular signal transduction mechanism and important structural differences between the active and inactive forms.
We have previously shown that the heme Fe(III)-OH−-, Fe(III)-CN−-, and Fe(II)-O2-bound forms of full-length AfGcHK are active, whereas heme-free and heme Fe(II)-bound forms are inactive (1, 8). Here we reported for the first time that the isolated kinase domain is inactive, and its His183 residue is not autophosphorylated (supplemental Fig. S7). Importantly, the HDX–MS profile of the isolated kinase domain (and especially that of H11) is similar to that of the kinase domain joined to the “inactive” Fe(II)-bound and heme-free globin domains in full-length AfGcHK (Fig. 5B).
Mechanism of signal transduction in AfGcHK
Two mechanisms have been proposed to explain how the kinase domain's autophosphorylation activity could be altered in response to signaling from the globin domain. The first is based on steric hindrance, with autophosphorylation being suppressed by physically blocking the autophosphorylation site (33). A similar mechanism was proposed to explain signal transduction in gas sensors; the protein surface of the heme-bound globin domain was suggested to directly contact the protein surface of the kinase domain such that signals from the globin domain could directly modulate the kinase domain's activity (34). The second mechanism involves indirect regulation of the autophosphorylation activity, with no direct blocking (“touching”) of the phosphorylation site in the kinase domain (35). The data presented here for AfGcHK strongly support this mechanism; there does not seem to be any direct interaction between the globin and kinase domains, which are only connected by a 15-residue-long covalent linker peptide in the full-length protein (supplemental Fig. S8).
The indirect mechanism of AfGcHK signal transduction loosely resembles that proposed for transmembrane signal propagation in membrane-bound two-component systems (3–6), although AfGcHK is not a membrane-bound protein.
In terms of dynamics, AfGcHK can be regarded as an ensemble of molecules sampling at least two conformational states. The kinase domains in these states feature different mutual arrangements of segments, including helices H11 (the ATP-binding site) and H9. In one state, the orientation of the H11 and H9 segments supports autophosphorylation; in the other, these segments occupy catalytically unfavorable positions. We suggest that the oxygen-binding signal is propagated from the heme to the kinase domain via the globin dimerization interface and the long helices (H8 and H9) that form the dimerization interface of the kinase domain. The short and structurally undetermined linker sequence connecting the globin and kinase domains may also play some role. On the contrary, there was no difference in its behavior under the studied conditions; therefore, its functional significance is currently unknown. Finally, the signal is propagated throughout the kinase domain, reorienting helices H11 and H9 in a way that impairs autophosphorylation and possibly ATP binding, as suggested by an increase in the domain's KmATP value (8). This signal transduction process may involve a scissor-like motion as was discussed above or a larger-scale dynamic rearrangement of the globin domain that transfers the signal to the kinase domain (Fig. 9). The X-ray crystallography and HDX–MS experiments performed in this study provided complementary information on the functional and structural behavior of AfGcHK. For the first time, the conformational changes associated with signal transduction were studied in a full-length globin-coupled oxygen sensor protein and linked to directly observed structural changes in the globin domain. As such, this work represents an important step toward a deeper understanding of the mechanism of heme-containing sensor regulation.
Experimental procedures
Materials
Ampicillin was obtained from P-lab (Prague, Czech Republic). Isopropyl β-d-thiogalactopyranoside, hemin, and acrylamide were obtained from Sigma-Aldrich. Water, doubly distilled over quartz, was purified using a Milli-Q Plus system (EMD Millipore, Billerica, MA). All glassware used for sample preparation was conditioned in advance by standing for 24 h in 10% (v/v) HCl Suprapur (Merck, Darmstadt, Germany). Phos-tag was from the Phos-tag consortium, Wako Pure Chemical Industries (Osaka, Japan). All chemicals used were of the highest purity grade available from commercial sources and used without further purification.
Protein production and purification
Cloning, overexpression in E. coli, and purification of the full-length wild-type, Y45F, Y45L, Y45W, L68N, L68F, L68G, L68W, and H99A AfGcHK proteins were performed as described previously (1, 7, 8). Briefly, His-tagged AfGcHK was expressed in BL21(DE3) (Novagen, Madison, WI) harboring the pET21c(+) plasmid. Cell lysates containing the His-tagged AfGcHK were isolated by affinity chromatography on a TALON® metal affinity resin column (Clontech), followed by size-exclusion chromatography on a Superdex Increase 200 GL 10/300 column (GE Healthcare). Protein and heme concentrations were determined using the BCA assay (Sunrise Absorbance Reader, TECAN, Männedorf, Switzerland) and the pyridine hemochromogen assay, respectively (36). Purified proteins were >90% homogeneous, as confirmed by SDS-PAGE.
The isolated globin domain of AfGcHK was prepared as follows. 500 ml of TB medium containing 100 μg/ml ampicillin was inoculated with 500 μl of O/N BL21(DE3) cells containing the appropriate pET21c(+) plasmid. Cells were grown at 37 °C for 5 h (220 rpm), and then the temperature was lowered to 18 °C (160 rpm), and protein expression was initiated by adding 0.1 mm isopropyl 1-thio-β-d-galactopyranoside followed by further shaking for 18 h. Cells were then harvested by centrifugation at 4 °C and 5000 × g, and stored at −80 °C. The pellets were resuspended in 50 mm Tris, 150 mm NaCl, pH 8, containing 1 mm PMSF, 1 mm EDTA, and 0.2 mg/ml lysozyme. After sonication (10 × 1 min), reconstitution with 300 μm hemin for 20 min, and 70 min of centrifugation at 50,000 × g, the supernatant was applied to a TALON® metal affinity resin column (Clontech), and the protein was eluted using 200 mm imidazole. The eluted protein was concentrated to a volume of 2 ml and desalted on Sephadex G10 column equilibrated with 20 mm Tris, 150 mm NaCl, pH 8. The protein was then diluted to 0.5 mg/ml, and the His tag was cleaved with the tobacco etch virus protease (final concentration 0.05 mg/ml, 48 h at 10 °C). The tobacco etch virus protease and uncleaved protein were removed using a TALON® metal affinity resin column, leaving the isolated protein in the flow-through fraction. Finally, size-exclusion chromatography was performed using a Superdex Increase 200 GL 10/300 column (GE Healthcare), and the final protein preparation was frozen in liquid nitrogen for further use. SDS-PAGE experiments showed that the purified isolated globin domain of AfGcHK was >99% homogeneous.
The isolated kinase domain (Δ161) of AfGcHK was prepared as follows. 500 ml of TB medium containing 100 μg/ml ampicillin was inoculated with 500 μl of O/N BL21(DE3) cells containing the appropriate pET21c(+) plasmid. Cells were grown at 37 °C for 5 h (220 rpm), and then the temperature was lowered to 18 °C (160 rpm), and protein expression was initiated by adding 0.1 mm isopropyl 1-thio-β-d-galactopyranoside followed by further shaking for 18 h. Cells were harvested by centrifugation at 4 °C and 5000 × g and stored at −80 °C. Pellets were resuspended in 50 mm Tris, 150 mm NaCl, pH 8, containing 1 mm PMSF, 1 mm EDTA, and 0.2 mg/ml lysozyme. After sonication (5 × 1 min) and 70 min of centrifugation at 50,000 × g, the supernatant was applied to a TALON® metal affinity resin column (Clontech), and the protein was eluted using 200 mm imidazole. The protein was concentrated to a volume of 0.5 ml, size-exclusion chromatography was performed on a Superdex 200 GL 10/300 column (GE Healthcare), and the final protein preparation was frozen in liquid nitrogen for further use. SDS-PAGE experiments showed that the purified isolated kinase domain of AfGcHK was >70% homogeneous.
Crystallization of the isolated AfGcHK globin domain
A 5.4 mg/ml solution of the AfGcHK globin domain with the Fe(III)-OH− complex in 150 mm NaCl, 20 mm Tris, pH 8.0, was crystallized using the hanging-drop vapor diffusion method at 25 °C. The drops contained 2 μl of protein and 1 μl of reservoir. The reservoir contained 20.7% (w/v) PEG 3350, 0.1 m MMT buffer (dl-malic acid, MES, and Tris base in a molar ratio of 1:2:2), pH 6.7, 0.2 m MgCl2, 10 mm KCN, and 7.5% (v/v) glycerol. This yielded red wedge block–shaped crystals with dimensions of up to 80 × 80 × 300 μm. The crystals were vitrified in liquid nitrogen without any cryoprotection. Before vitrification, the dithionite-soaked crystal was soaked for 15 min in a maternal drop containing 0.2 μl of added 100 mm sodium dithionite (giving a final dithionite concentration of ∼10 mm). Soaking did not cause any visible changes in the crystal.
Diffraction data collection, structure determination, and refinement
Data for the crystal with the Fe(III)-CN− complex were collected at the Helmholtz-Zentrum Berlin (Bessy II) on beamline 14.1 equipped with a Dectris Pilatus 6M detector at 100 K, using wavelength 0.918409 Å, detector distance 0.375 m, Δϕ 0.1° per image, and exposure time 0.5 s. Data for the sodium dithionite–soaked crystal were collected at the synchrotron light source Petra III in Hamburg on beamline PX13 equipped with a Dectris Pilatus 6M detector at 100 K using wavelength 0.92010 Å, detector distance 0.369 m, Δϕ 0.05° per image, and exposure time 0.1 s.
The diffraction images were processed using XDS (37) and scaled using AIMLESS from the CCP4 program package (38). Data processing statistics are presented in Table 1. The phase problem was solved by molecular replacement using the MORDA program (39) and the structure of the globin domain of the globin-coupled sensor from G. sulfurreducens (PDB code 2W31 (17)). Structures were refined using REFMAC5 with manual editing using COOT (40), with 5% of reflections used as the testing set (the Rfree set). The last cycle of the refinement was performed using all reflections. Structure quality was checked using the validation tools implemented in MOLPROBITY (41) and COOT.
The atomic coordinates and structure factors have been deposited in the Protein Data Bank under the codes 5OHE (globin domain of AfGcHK with cyanide) and 5OHF (globin domain of AfGcHK with cyanide, partially reduced).
HDX–MS
HDX–MS experiments using samples of full-length wild type AfGcHK (80 μm), the globin domain of AfGcHK (80 μm), the kinase domain of AfGcHK (80 μm), and a 1:1 mixture of the globin and kinase domains with a total protein concentration of 160 μm were initiated by 10-fold dilution in a deuterated buffer containing 20 mm Tris-HCl (pD 7.6), 150 mm NaCl, 50 mm KCl, and 5 mm MgCl2 at 20 °C. Solutions of AfGcHK with different heme iron ligands in the globin domain were prepared as follows: Fe(III)-CN−, 1-h incubation of the protein in 10 mm KCN solution and use of a deuterated buffer containing 10 mm KCN; Fe(II)-O2, 2-min incubation of the protein in 10 mm sodium dithionite followed by desalting on a Sephadex G10 column equilibrated with 20 mm Tris and 150 mm NaCl, pH 8; Fe(II), prepared in an anaerobic glove box (Jacomex GP-Concept; oxygen concentration <1 ppm) by 2-min incubation of the protein in 10 mm sodium dithionite followed by desalting on a Sephadex G10 column equilibrated with degassed 20 mm Tris, 150 mm NaCl, pH 8, buffer. All solutions used in this experiment were thoroughly degassed and equilibrated in the anaerobic box, and UV-visible spectra were recorded throughout the procedure to ensure the exclusive presence of the Fe(II) form.
Aliquots (25 μl) were taken after 0.5, 5, 20, 60, and 180 min of exchange. The exchange reaction in the aliquots was immediately quenched by adding 25 μl of 1 m glycine (pH 2.3), and the samples were then rapidly frozen in liquid nitrogen. HPLC-MS analysis of deuterated samples was performed using a high-performance liquid chromatograph (1200 Agilent Technologies, Waldbronn, Germany) connected to an ESI-FT-ICR mass spectrometer (15T solariX XR, Bruker Daltonics, Billerica, MA). The analysis was initiated by rapidly thawing the sample, followed by digestion on a pepsin column (66-μl bed volume, flow rate 100 μl min−1). The resulting peptides were desalted online using a Peptide MicroTrap (MichromBioresources, Auburn, CA) and separated on a C18 reversed phase column (0.5 × 50 mm; Jupiter, Phenomenex, Torrance, CA) using a linear gradient of 10–45% (v/v) solvent B over 7 min, where solvent A was 2% (v/v) acetonitrile with 0.4% (v/v) formic acid in water and solvent B was 95% (v/v) acetonitrile, 5% (v/v) water with 0.4% (v/v) formic acid. The injector and switching valve, pepsin column, peptide trap, and analytical column were placed in an ice box to minimize back-exchange. Peptide identification (mapping, HPLC-MS/MS) was done using the system described above, and MASCOT was used to compare the obtained MS/MS spectra with a database containing a sequence for AfGcHK. Deuterated data were processed using an in-house program called DeutEx,4 which records the average mass for each peptide at each time point and each condition studied and calculates the percentage of deuteration relative to the maximum achievable deuteration based on the number of exchangeable amide hydrogens in each peptide. The deuterium content of each peptide was reported as a percentage of the maximum achievable deuteration based on the number of exchangeable amide hydrogens in each peptide (42).
Mass spectrometric analysis of AfGcHK phosphorylation at His183
The above-mentioned LC-MS setup designed for HDX–MS samples was also used to analyze the phosphorylation status of His183 in the full-length AfGcHK and the isolated kinase domain. Extracted ion chromatograms for non-phosphorylated and phosphorylated peptides including His183 were plotted in Data Analysis (Bruker Daltonics).
UV-visible spectroscopy and autoxidation rate estimation
Optical absorption spectral data were obtained using an HP 8453 UV-visible spectrophotometer (Agilent Technologies) at 20 °C under aerobic conditions as described previously (1). Proteins were diluted to 300 μl in 20 mm Tris/Cl, 150 mm NaCl, pH 8. A dithiothreitol solution (2 m) was then added to a final concentration of 0.5 m, and the protein sample was reduced for 60 min; the reduction was monitored by UV-visible spectrophotometry. Excess dithiothreitol was removed using a Sephadex G-25 desalting chromatography column, and the eluate was placed in a cuvette and diluted such that its absorbance at 413 nm was around 0.8. Spectra were collected every 15 or 30 min (depending on the protein) for 48 h at 25 °C (supplemental Fig. S2). The kox value was calculated under the assumption of first order kinetics based on the time-dependent decrease of the α band (577–580 nm) absorbance.
Enzyme activity of the isolated kinase domain of AfGcHK
A reaction mixture containing 10 μm full-length wild-type AfGcHK or its isolated kinase domain, 50 mm Tris-HCl, pH 8.0, 50 mm KCl, and 5 mm MgCl2 was preincubated for 5 min at 20 °C, and then the reaction was initiated by adding 1 mm ATP at 20 °C. At designated times, the autophosphorylation reaction was terminated by adding 100-μl aliquots of termination buffer (125 mm Tris-HCl, pH 6.8, 4% (w/v) SDS, 10% (v/v) 2-mercaptoethanol, 20% (v/v) glycerol, 0.004% (w/v) bromphenol blue). Samples of the quenched reaction mixtures were then loaded on a 10% (w/v) SDS-polyacrylamide gel containing 75 μm Phos-tag acrylamide and 0.2 mm MnCl2. Each lane was loaded with a quantity of quenched reaction mixture containing 0.5 μg of AfGcHK. Phosphorylated proteins in the sample interacted with the Phos-tag manganese complex in the gel, reducing their mobility relative to phosphate-free proteins (1, 43, 44). After electrophoresis, the proteins were visualized by staining with Coomassie Brilliant Blue R350, and the stained gels were imaged using a Scanjet G3010 (HP) scanner. The protein loadings were then quantified by analyzing the scanned images using ImageJ.
Analytical ultracentrifugation
Sedimentation velocity experiments were performed using a ProteomeLab XL-I (Beckman Coulter, Brea, CA) analytical ultracentrifuge at loading concentrations of 6–15 μm for the isolated globin domain of AfGcHK and the full-length protein, at 20 °C and a rotor speed of 36,000 rpm. All data were collected using absorbance optics at 411 nm (selective for hemoproteins). Data analysis was performed with the SEDFIT, SEDPHAT, and Gussi packages (45–47). Sedimentation velocity data were analyzed using a continuous sedimentation coefficient distribution model, c(s). The resulting apparent sedimentation coefficients were used to compute sedimentation coefficients in water at 20 °C (s20,w).
Author contributions
M. S. planned experiments, performed experiments and analyzed data; P. M. planned, performed and interpreted MS experiments; T. S., P. K., and J. B. designed, performed, and analyzed the crystallization, diffraction data collection, structure determination, and refinement experiments; V. F. performed experiments; V. M. performed and interpreted protein models and discussed and interpreted data; J. D. performed interpretation and validation of structures and contributed to the manuscript; A. L. provided technical assistance and contributed to the preparation of the figures; M. R. provided technical assistance; T. S. discussed and interpreted data; M. M. discussed and interpreted data and wrote the paper. All authors reviewed the results and approved the final version of the manuscript.
Supplementary Material
Acknowledgments
We are grateful to Dr. Kenichi Kitanishi and Dr. Ondrej Vanek for technical assistance during the early stages of this work.
This work was supported in part by Charles University Grant UNCE 204025/2012; Grant Agency of the Czech Republic Grant 15-19883S; Grant Agency of Charles University Grants 362115 and 704217; Ministry of Education, Youth, and Sports of the Czech Republic Grant LM2015043 CIISB, Biocev - Crystallization, Diffraction and LTC17065 in frame of COST Action CA15126 MOBIEU; ERDF fund Grants CZ.02.1.01/0.0/0.0/16_013/0001776 and CZ.1.05/1.1.00/02.0109; and Grant Agency of the Czech Technical University in Prague Grant SGS16/246/OHK4/3T/14. This work was also partly funded by Instruct, part of the European Strategy Forum on Research Infrastructures (ESFRI) and supported by national member subscriptions. The authors declare that they have no conflicts of interest with the contents of this article.
This article contains supplemental information on the crystal structure of AfGcHK globin domain, a structural comparison of the isolated globin domains of AfGcHK and other GCSs, supplemental Table S1, and supplemental Figs. S1–S8.
The atomic coordinates and structure factors (codes 5OHE and 5OHF) have been deposited in the Protein Data Bank (http://wwpdb.org/).
D. Kavan, P. Man, and G. Kruppa, unpublished data.
- GCS
- globin-coupled oxygen sensor
- AfGcHK
- a globin-coupled histidine kinase from Anaeromyxobacter sp. Fw109-5
- EcDOS
- E. coli direct oxygen sensor or heme-regulated phosphodiesterase from E. coli or EcDosP
- Fe(III)
- Fe(III)-protoporphyrin IX complex, or hemin
- Fe(II)
- Fe(II)-protoporphyrin IX complex
- FixL
- an oxygen-sensor histidine kinase with the heme-bound PAS domain that regulates nitrogen fixation in Rhizobium meliloti or Bradyrhizobium japonicum
- GAF
- domain conserved in cyclic GMP-specific and stimulated phosphodiesterases, adenylate cyclases, and E. coli formate hydrogenlyase transcriptional activator
- GsGCS
- globin-coupled sensor from G. sulfurreducens
- HDX–MS
- hydrogen/deuterium exchange coupled with mass spectrometry
- HK853
- the sensor histidine kinase from T. maritima
- PAS
- Per (Drosophila period clock protein)-Arnt (vertebrate aryl hydrocarbon receptor nuclear translocator)-Sim (Drosophila single-minded protein)
- YddV
- heme-bound diguanylate cyclase from E. coli or EcDosC
- PDB
- Protein Data Bank
- RMSD
- root mean square deviation.
References
- 1. Kitanishi K., Kobayashi K., Uchida T., Ishimori K., Igarashi J., and Shimizu T. (2011) Identification and functional and spectral characterization of a globin-coupled histidine kinase from Anaeromyxobacter sp. Fw109-5. J. Biol. Chem. 286, 35522–35534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Martínková M., Kitanishi K., and Shimizu T. (2013) Heme-based globin-coupled oxygen sensors: linking oxygen binding to functional regulation of diguanylate cyclase, histidine kinase, and methyl-accepting chemotaxis. J. Biol. Chem. 288, 27702–27711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Gushchin I., Melnikov I., Polovinkin V., Ishchenko A., Yuzhakova A., Buslaev P., Bourenkov G., Grudinin S., Round E., Balandin T., Borshchevskiy V., Willbold D., Leonard G., Büldt G., Popov A., and Gordeliy V. (2017) Mechanism of transmembrane signaling by sensor histidine kinases. Science 10.1126/science.aah6345 [DOI] [PubMed] [Google Scholar]
- 4. Abriata L. A., Albanesi D., Dal Peraro M., and de Mendoza D. (2017) Signal sensing and transduction by histidine kinases as unveiled through studies on a temperature sensor. Acc. Chem. Res. 50, 1359–1366 [DOI] [PubMed] [Google Scholar]
- 5. Willett J. W., and Crosson S. (2017) Atypical modes of bacterial histidine kinase signaling. Mol. Microbiol. 103, 197–202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Zschiedrich C. P., Keidel V., and Szurmant H. (2016) Molecular mechanisms of two-component signal transduction. J. Mol. Biol. 428, 3752–3775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Stranava M., Martínek V., Man P., Fojtikova V., Kavan D., Vaněk O., Shimizu T., and Martinkova M. (2016) Structural characterization of the heme-based oxygen sensor, AfGcHK, its interactions with the cognate response regulator, and their combined mechanism of action in a bacterial two-component signaling system. Proteins 84, 1375–1389 [DOI] [PubMed] [Google Scholar]
- 8. Fojtikova V., Stranava M., Vos M. H., Liebl U., Hranicek J., Kitanishi K., Shimizu T., and Martinkova M. (2015) Kinetic analysis of a globin-coupled histidine kinase, AfGcHK: effects of the heme iron complex, response regulator, and metal cations on autophosphorylation activity. Biochemistry 54, 5017–5029 [DOI] [PubMed] [Google Scholar]
- 9. Gong W., Hao B., and Chan M. K. (2000) New mechanistic insights from structural studies of the oxygen-sensing domain of Bradyrhizobium japonicum FixL. Biochemistry 39, 3955–3962 [DOI] [PubMed] [Google Scholar]
- 10. Hao B., Isaza C., Arndt J., Soltis M., and Chan M. K. (2002) Structure-based mechanism of O2 sensing and ligand discrimination by the FixL heme domain of Bradyrhizobium japonicum. Biochemistry 41, 12952–12958 [DOI] [PubMed] [Google Scholar]
- 11. Kurokawa H., Lee D.-S., Watanabe M., Sagami I., Mikami B., Raman C. S., and Shimizu T. (2004) A redox-controlled molecular switch revealed by the crystal structure of a bacterial heme PAS sensor. J. Biol. Chem. 279, 20186–20193 [DOI] [PubMed] [Google Scholar]
- 12. Park H., Suquet C., Satterlee J. D., and Kang C. (2004) Insights into signal transduction involving PAS domain oxygen-sensing heme proteins from the X-ray crystal structure of Escherichia coli Dos heme domain (Ec DosH). Biochemistry 43, 2738–2746 [DOI] [PubMed] [Google Scholar]
- 13. Cho H. Y., Cho H. J., Kim Y. M., Oh J. I., and Kang B. S. (2009) Structural insight into the heme-based redox sensing by DosS from Mycobacterium tuberculosis. J. Biol. Chem. 284, 13057–13067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Podust L. M., Ioanoviciu A., and Ortiz de Montellano P. R. (2008) 2.3 Å X-ray structure of the heme-bound GAF domain of sensory histidine kinase DosT of Mycobacterium tuberculosis. Biochemistry 47, 12523–12531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Yukl E. T., Ioanoviciu A., Nakano M. M., de Montellano P. R., and Moënne-Loccoz P. (2008) A distal tyrosine residue is required for ligand discrimination in DevS from Mycobacterium tuberculosis. Biochemistry 47, 12532–12539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Tarnawski M., Barends T. R. M., and Schlichting I. (2015) Structural analysis of an oxygen-regulated diguanylate cyclase. Acta Crystallogr. D 71, 2158–2177 [DOI] [PubMed] [Google Scholar]
- 17. Pesce A., Thijs L., Nardini M., Desmet F., Sisinni L., Gourlay L., Bolli A., Coletta M., Van Doorslaer S., Wan X., Alam M., Ascenzi P., Moens L., Bolognesi M., and Dewilde S. (2009) HisE11 and HisF8 provide bis-histidyl heme hexa-coordination in the globin domain of Geobacter sulfurreducens globin-coupled sensor. J. Mol. Biol. 386, 246–260 [DOI] [PubMed] [Google Scholar]
- 18. Zhang W., and Phillips G. N. (2003) Structure of the oxygen sensor in Bacillus subtilis: signal transduction of chemotaxis by control of symmetry. Structure 11, 1097–1110 [DOI] [PubMed] [Google Scholar]
- 19. Krissinel E., and Henrick K. (2004) Secondary-structure matching (SSM), a new tool for fast protein structure alignment in three dimensions. Acta Crystallogr. D 60, 2256–2268 [DOI] [PubMed] [Google Scholar]
- 20. Krissinel E., and Henrick K. (2007) Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 372, 774–797 [DOI] [PubMed] [Google Scholar]
- 21. Nakajima K., Kitanishi K., Kobayashi K., Kobayashi N., Igarashi J., and Shimizu T. (2012) Leu65 in the heme distal side is critical for the stability of the Fe(II)-O2 complex of YddV, a globin-coupled oxygen sensor diguanylate cyclase. J. Inorg. Biochem. 108, 163–170 [DOI] [PubMed] [Google Scholar]
- 22. Zhang W., Olson J. S., and Phillips G. N. (2005) Biophysical and kinetic characterization of HemAT, an aerotaxis receptor from Bacillus subtilis. Biophys. J. 88, 2801–2814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Aono S., Kato T., Matsuki M., Nakajima H., Ohta T., Uchida T., and Kitagawa T. (2002) Resonance Raman and ligand binding studies of the oxygen-sensing signal transducer protein HemAT from Bacillus subtilis. J. Biol. Chem. 277, 13528–13538 [DOI] [PubMed] [Google Scholar]
- 24. Springer B. A., Sligar S. G., Olson J. S., and Phillips G. N. J. (1994) Mechanisms of ligand recognition in myoglobin. Chem. Rev. 94, 699–714 [Google Scholar]
- 25. Engen J. R. (2003) Analysis of protein complexes with hydrogen exchange and mass spectrometry. Analyst 128, 623–628 [DOI] [PubMed] [Google Scholar]
- 26. Kadek A., Kavan D., Marcoux J., Stojko J., Felice A. K. G., Cianférani S., Ludwig R., Halada P., and Man P. (2017) Interdomain electron transfer in cellobiose dehydrogenase is governed by surface electrostatics. Biochim. Biophys. Acta 1861, 157–167 [DOI] [PubMed] [Google Scholar]
- 27. Rezabkova L., Man P., Novak P., Herman P., Vecer J., Obsilova V., and Obsil T. (2011) Structural basis for the 14-3-3 protein-dependent inhibition of the regulator of G protein signaling 3 (RGS3) function. J. Biol. Chem. 286, 43527–43536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lanzilotta W. N., Schuller D. J., Thorsteinsson M. V., Kerby R. L., Roberts G. P., and Poulos T. L. (2000) Structure of the CO sensing transcription activator CooA. Nat. Struct. Biol. 7, 876–880 [DOI] [PubMed] [Google Scholar]
- 29. Komori H., Inagaki S., Yoshioka S., Aono S., and Higuchi Y. (2007) Crystal structure of CO-sensing transcription activator CooA bound to exogenous ligand imidazole. J. Mol. Biol. 367, 864–871 [DOI] [PubMed] [Google Scholar]
- 30. Underbakke E. S., Iavarone A. T., Chalmers M. J., Pascal B. D., Novick S., Griffin P. R., and Marletta M. A. (2014) Nitric oxide-induced conformational changes in soluble guanylate cyclase. Structure 22, 602–611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Skálová T., Dohnálek J., Spiwok V., Lipovová P., Vondrácková E., Petroková H., Dusková J., Strnad H., Králová B., and Hasek J. (2005) Cold-active β-galactosidase from Arthrobacter sp. C2-2 forms compact 660 kDa hexamers: crystal structure at 1.9 Å resolution. J. Mol. Biol. 353, 282–294 [DOI] [PubMed] [Google Scholar]
- 32. Keedy D. A., Fraser J. S., and van den Bedem H. (2015) Exposing hidden alternative backbone conformations in X-ray crystallography using qFit. PLoS Comput. Biol. 11, e1004507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Cunningham K. A., and Burkholder W. F. (2009) The histidine kinase inhibitor Sda binds near the site of autophosphorylation and may sterically hinder autophosphorylation and phosphotransfer to Spo0F. Mol. Microbiol. 71, 659–677 [DOI] [PubMed] [Google Scholar]
- 34. Shimizu T., Huang D., Yan F., Stranava M., Bartosova M., Fojtíková V., and Martínková M. (2015) Gaseous O2, NO, and CO in signal transduction: structure and function relationships of heme-based gas sensors and heme-redox sensors. Chem. Rev. 115, 6491–6533 [DOI] [PubMed] [Google Scholar]
- 35. Rao M., Herzik M. A. Jr., Iavarone A. T., and Marletta M. A. (2017) Nitric oxide-induced conformational changes govern H-NOX and histidine kinase interaction and regulation in Shewanella oneidensis. Biochemistry 56, 1274–1284 [DOI] [PubMed] [Google Scholar]
- 36. Antonini E., and Brunori M. (1971) Hemoglobin and myoglobin in their reactions with ligands, North-Holland Publishing Co., Amsterdam, The Netherlands [Google Scholar]
- 37. Kabsch W. (2010) XDS. Acta Crystallogr. D 66, 125–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Winn M. D., Ballard C. C., Cowtan K. D., Dodson E. J., Emsley P., Evans P. R., Keegan R. M., Krissinel E. B., Leslie A. G. W., McCoy A., McNicholas S. J., Murshudov G. N., Pannu N. S., Potterton E. A., Powell H. R., et al. (2011) Overview of the CCP4 suite and current developments. Acta Crystallogr. D 67, 235–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Vagin A., and Lebedev A. (2015) MoRDa, an automatic molecular replacement pipeline. Acta Crystallogr. A 71, s19 [Google Scholar]
- 40. Emsley P., Lohkamp B., Scott W. G., and Cowtan K. (2010) Features and development of Coot. Acta Crystallogr. D 66, 486–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Chen V. B., Arendall W. B. 3rd, Headd J. J., Keedy D. A., Immormino R. M., Kapral G. J., Murray L. W., Richardson J. S., and Richardson D. C. (2010) MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D 66, 12–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Trcka F., Durech M., Man P., Hernychova L., Muller P., and Vojtesek B. (2014) The assembly and intermolecular properties of the Hsp70-Tomm34-Hsp90 molecular chaperone complex. J. Biol. Chem. 289, 9887–9901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Yamada S., Nakamura H., Kinoshita E., Kinoshita-Kikuta E., Koike T., and Shiro Y. (2007) Separation of a phosphorylated histidine protein using phosphate affinity polyacrylamide gel electrophoresis. Anal. Biochem. 360, 160–162 [DOI] [PubMed] [Google Scholar]
- 44. Igarashi J., Murase M., Iizuka A., Pichierri F., Martinkova M., and Shimizu T. (2008) Elucidation of the heme binding site of heme-regulated eukaryotic initiation factor 2α kinase and the role of the regulatory motif in heme sensing by spectroscopic and catalytic studies of mutant proteins. J. Biol. Chem. 283, 18782–18791 [DOI] [PubMed] [Google Scholar]
- 45. Houtman J. C. D., Brown P. H., Bowden B., Yamaguchi H., Appella E., Samelson L. E., and Schuck P. (2007) Studying multisite binary and ternary protein interactions by global analysis of isothermal titration calorimetry data in SEDPHAT: application to adaptor protein complexes in cell signaling. Protein Sci. 16, 30–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Schuck P. (2000) Size-distribution analysis of macromolecules by sedimentation velocity ultracentrifugation and Lamm equation modeling. Biophys. J. 78, 1606–1619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Brautigam C. A. (2015) Calculations and publication-quality illustrations for analytical ultracentrifugation data. Methods Enzymol. 562, 109–133 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.