Abstract
RUN and FYVE domain–containing 3 (Rufy3) is an adapter protein for small GTPase proteins and is bound to activated Rap2, a Ras family protein in the developing neuron. Previously, we reported the presence of a rapid cell polarity determination mechanism involving Rufy3, which is likely required for in vivo neuronal development. However, the molecular details of this mechanism are unclear. To this end, here we produced Rufy3 knock-out (Rufy3-KO) mice to study the role of Rufy3 in more detail. Examining Rufy3-KO neurons, we found that Rufy3 is recruited via glycoprotein M6A to detergent-resistant membrane domains, which are biochemically similar to lipid rafts. We also clarified that Rufy3, as a component of a ternary complex, induces the assembly of Rap2 in the axonal growth cone, whereas in the absence of Rufy3, the accumulation of a Rac guanine nucleotide exchange factor, T-cell lymphoma invasion and metastasis 2 (Tiam2/STEF), is inhibited downstream of Rap2. We also found that Rufy3 regulates the cellular localization of Rap2 and Tiam2/STEF. Taken together, we conclude that Rufy3 is a physiological adapter for Rap2 and activates Tiam2/STEF in glycoprotein M6A-regulated neuronal polarity and axon growth.
Keywords: cell polarity, lipid raft, neurite outgrowth, neurodevelopment, small GTPase
Introduction
Small GTP-binding proteins (also known as small GTPases) form a central large group that act as transducers in various cellular events (1). GTPases are classified into the Ras, Rho, and Rab subfamilies with each playing a specific role in signaling pathways, cytoskeletal rearrangement, and intracellular trafficking. These proteins share several common biochemical properties; for example, their activated (GTP-bound) form is selectively bound to specific adapter proteins for signal transduction (1). A group of such adapter proteins was recently shown to contain a RUN domain which is involved in binding small G proteins (2) of which one, RUN and FYVE domain–containing 3 (Rufy3;2 also known as Singar1/Rap2-interacting protein X), lacks an FYVE domain, which interacts with phosphatidylinositol 4,5-bisphosphate, and therefore cannot directly associate with the cell membrane (2). The analysis of Rufy3 using X-ray crystallography has shown that Rap2, a Ras family member, is tightly and stably bound to Rufy3 in a GTP- and RUN domain–dependent manner (3).
Small G proteins are critical in various steps of neuronal development in the mammalian brain (4–7). For example, accurate synaptogenesis requires formation of growth cones with highly motile activity at the tips of the extending axon. We previously conducted proteomic analysis of rodent growth cone proteins and showed that these proteins from more than 30 rodent species are small G proteins (4, 8). Of these, Rap2 was the most abundant membrane-bound Ras family protein, and Rufy3 was the most abundant adapter protein specific to small G proteins (4, 8). We recently confirmed that these two proteins form a complex and are colocalized in the growth cone (9). This complex is formed prior to axon growth in the first step of the determination of neuronal polarity (9) during which time a single axon grows in a neuron (10, 11). Complex formation during this step is dependent upon an abundant membrane protein, glycoprotein M6A (GPM6a) (4, 8). Using GPM6a knock-out (KO) mice, we recently demonstrated that GPM6a is responsible for neuronal polarity via the Rufy3-Rap2 complex (9). This protein complex accumulates in lipid rafts and activates T-cell lymphoma invasion and metastasis 2 (Tiam2/STEF), a Rac1 activator (9). Rufy3 and Tiam2/STEF were previously known as components of cell-autonomous determinants for neuronal polarity (12, 13), and more than 40 of these components have been characterized to date (11). Our findings regarding GPM6a and Rap2 demonstrated the presence of a rapid polarity determination mechanism that involves Rufy3 and is probably involved in in vivo neuronal development.
Here, we further investigated the mechanism underlying Rufy3-dependent processes by producing Rufy3-KO mice and examining the properties of Rufy3-deficient neurons. We found that Rufy3 is recruited via GPM6a to detergent-resistant membrane domains, which are biochemically equivalent to lipid rafts. We also showed that Rufy3 is a component of a ternary complex that induces the assembly of Rap2 in the growth cone. In the absence of Rufy3, the accumulation of a Rac guanine nucleotide exchange factor, Tiam2/STEF, is inhibited downstream of Rap2. In addition, Rufy3 regulates the localization of Rap2 and Tiam2/STEF. Taking these findings together, we conclude that Rufy3 is a physiological adapter for Rap2 and activates Tiam2/STEF for polarity determination under the regulation of GPM6a.
Results
Rufy3 deficiency leads to multiple-axon formation irrespective of the localization of GPM6a in cortical neurons
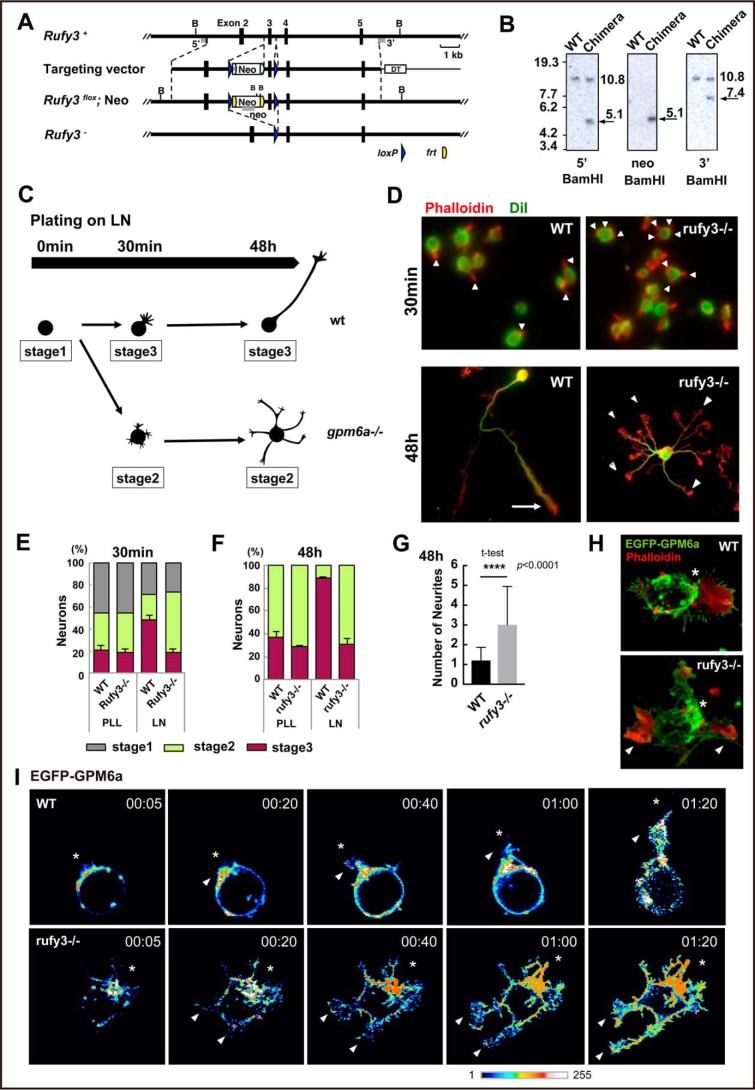
To further examine the physiological functions of Rufy3 in the determination of polarity, we generated KO mice deficient in Rufy3 using C57BL/6N strain mice (Fig. 1A; also see Fig. S1, A and B). Deficiency in Rufy3 was confirmed using Southern and Western blot analyses (Fig. 1B; also see Fig. S1, A and B). Rufy3-KO mice died just after birth. These mice did not show macroscopic brain abnormalities or microscopic defects in major neural networks, and therefore we first verified the formation of neuronal polarity in their embryonic cortical neurons. Consistent with the effects of Rufy3 RNAi (13), significant multiple-axon formation was observed in the rufy3−/− neurons at 2 days in vitro (Fig. 1, C–G). Overexpression of EGFP-Rufy3 in KO neurons suppressed excessive axon formation (Fig. S2, A and B), indicating that Rufy3 is involved in the determination of neuronal polarity. We previously reported that the determination of neuronal polarity is induced within a few hours of plating neurons on laminin (LN) (9) (summarized in Fig. 1C). To examine the effects of Rufy3 deficiency on GPM6a-regulated signaling, we confirmed neuronal polarity in wild type (WT) and rufy3−/− neurons 30 min after plating on LN (Fig. 1D). Multiple neurites protruded from the rufy3−/− neurons, whereas single protrusions were observed for WT neurons (Fig. 1, E and F). As we demonstrated previously (9), this neurite protruded from the GPM6a-assembled membrane area in the WT neuron, whereas the multiple neurites from rufy3−/− neurons and gpm6a−/− neurons (9) were positioned irrespective of GPM6a localization (Fig. 1, H–J). These results suggested that Rufy3 is involved in the regulation of neuronal polarity determination by GPM6a.
Figure 1.
Generation of Rufy3-KO mice and abnormalities in the polarity of their neurons. A, strategy for conditional inactivation of the mouse Rufy3 gene. B, BamHI site; Neo, neomycin resistance gene; DT, diphtheria toxin gene. B, Southern blot analysis of genomic DNA isolated from WT ES cells and chimeric clones. The DNA was digested with BamHI and hybridized with 5′ and 3′ probes. Each band was detected at the expected size. C, schematic models of the time course in neuronal polarity determination of the cultured neurons plated on LN. The schematics show the representative neuronal shapes at each time point. Developmental stages of the neurons were morphologically defined according to the definition of Dotti et al. (19); i.e. stage 1, cells with no neurite (gray); stage 2, multipolar cells (green); and stage 3, polarized cells (pink). In the WT, the neurons dramatically changed from stage 1 to 3 within 1–4 h after plating on LN. In contrast, the gpm6a−/− neurons still remained at stages 1 to 2 at the same time course (see Ref. 9). D–G, cortical neurons derived from WT or rufy3−/− mice were cultured for 30 min or 48 h on LN. D, the plasma membrane (green) and F-actin (red) were stained using DiI and phalloidin, respectively. Scale bars, 50 (upper) and 20 μm (lower). E and F, percentage of neurons at stage 1, 2, or 3 from WT or rufy3−/− mice plated for 30 min (E) or 48 h (F) on LN (see also Ref. 9) (one-way analysis of variance with Tukey's multiple comparisons test; n 500 cells (E) and n 300 cells (F) for each group; error bars represent S.D. of the percentage of the stage 3 neurons in each group). G, the numbers of neurites of WT or rufy3−/− mice after 48-h culture (two-tailed t test (means ± S.D.); WT (1.203 ± 0.661) versus rufy3−/− (3.00 ± 1.944); ****, p < 0.0001; n = 300 from five distinct preparations; error bars represent S.D.). H, in LN-dependent cultures, growth cones (arrowheads) protruded from a GPM6a (green)-enriched membrane area (asterisk) in the cortical neurons from WT but not from rufy3−/− mice. Scale bar, 20 μm. I, time-lapse imaging of EGFP-GPM6a overexpressed in the cortical neurons of WT and rufy3−/− mice for 80 min after plating on LN. The fluorescence intensities of EGFP-GPM6a are shown as pseudocolor images. Neurites (arrowheads) protruded from the GPM6a-enriched membrane area (asterisk) in WT but not in rufy3−/−. Scale bar, 20 μm. PLL, poly-l-lysine.
Rufy3 is recruited to detergent-resistant membrane domains via GPM6a
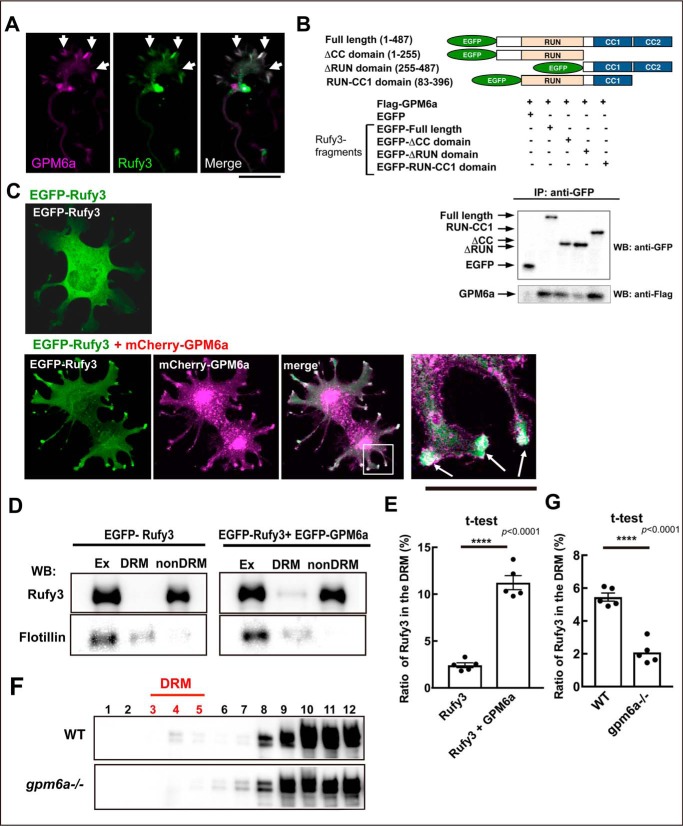
We recently reported that Rufy3 directly binds to GPM6a (9), and the complex colocalizes in the peripheral (P-) domain of the axonal growth cone (Fig. 2A; also see Fig. S3A). Previously, Rufy3 was reported to interact with fascin, an actin-bundling protein (14); however, our immunoanalysis showed that Rufy3 is localized at the edge of the growth cone rather than colocalized with F-actin bundles such as fascin (Fig. S3) (15). We further examined the interactions between Rufy3 and GPM6a in pulldown experiments using EGFP-tagged Rufy3 fragments and FLAG-GPM6a. The binding of Rufy3 to GPM6a required the Rufy3 RUN domain (Fig. 2B) as previously observed for the binding of Rufy3 to Rap2 (3, 9).
Figure 2.
Rufy3 is translocated to detergent-resistant membrane fractions through its interaction with GPM6a. A, immunofluorescence of endogenous GPM6a (magenta) and Rufy3 (green) in the growth cone of neurons plated on LN for 72 h. The proteins colocalized in the P-domain of the axonal growth cone (arrow). Scale bar, 5 μm. B, GPM6a-binding domain of Rufy3, visualized using a pulldown assay of FLAG-GPM6a with EGFP-Rufy3. EGFP-tagged full length (aa 1–487), N terminus + RUN domain (aa 1–255), two coiled-coil domains (CC1 + CC2 domains; aa 255–487), and RUN-CC1 (aa 83–396) of Rufy3 were coexpressed with FLAG-GPM6a in COS-7 cells. EGFP-Rufy3 fragments and coimmunoprecipitation (IP) of GPM6a were detected with both anti-GFP and anti-FLAG antibodies using Western blotting (WB), respectively. C, EGFP-Rufy3 (green) changes its localization upon coexpression with mCherry-GPM6a (magenta) in COS-7 cells. The translocated EGFP-GPM6a was colocalized with mCherry-GPM6a at the tips of the protrusions (arrow). A high magnification image of the protrusions in the inset is shown in the left panel. Scale bar, 10 μm. D and E, translocation of EGFP-Rufy3 to DRM fractions, induced by coexpression of FLAG-GPM6a in COS-7 cells, with or without FLAG-GPM6a. D, translocation of EGFP-Rufy3, induced by coexpression of FLAG-GPM6a in COS-7 cells, with or without FLAG-GPM6a. The cells were lysed using 1% Brij98, and the whole-cell extract (Ex) was fractionated into DRM and non-DRM fractions. The cells were lysed using 1% Brij 98 and fractionated into DRM and non-DRM fractions. The distribution of EGFP-Rufy3 was analyzed using EGFP antibody. Endogenous flotillin 1 was used as a lipid raft marker. EGFP-Rufy3 was partially translocated from the non-DRM to the DRM fractions. E, ratio of immunofluorescence of EGFP-Rufy3 in the DRM fractions to that in total EGFP-Rufy3 (two-tailed t test (means ± S.E.); Rufy3 (2.41 ± 0.25) versus Rufy3 + GPM6a (11.22 ± 0.74); ****, p < 0.0001; n = 5; error bars represent S.E.). F, differential localization of endogenous Rufy3 between WT and GPM6a-KO brains. Homogenized brains (E14.5 WT or gpm6a−/− embryos) were treated with 1% Brij 98, DRMs were fractionated, and the distribution of Rufy3 was analyzed as described above. In WT but not in gpm6a−/−, Rufy3 was partially distributed in the DRM fractions. G, ratio of Rufy3 in the DRM fractions to its total amount (two-tailed t test (means ± S.E.); WT (5.45 ± 0.2452) versus gpm6a−/− (2.085 ± 0.3087); ****, p < 0.0001; n = 5; error bars represent S.E.).
According to our proteomics data, Rufy3 is an abundant cytosolic protein in the axonal growth cone (8), and thus we investigated the effects of Rufy3 binding to GPM6a on the localization of the complex using a reconstitution assay. The results showed that EGFP-Rufy3 was originally distributed in the cytosol and then was transferred to the tips of the protrusions by colocalization with mCherry-GPM6a (Fig. 2C). As shown previously (9), we confirmed that detergent-resistant membrane (DRM) even prepared from embryonic brains represents the lipid rafts as described previously (16). DRM analysis showed that EGFP-Rufy3 coexpressed with GPM6a was partially distributed in the DRM fractions, although proteins expressed individually were exclusively localized in non-DRM fractions (Fig. 2, D and E). Similarly, endogenous Rufy3 was distributed in the DRM fractions of embryonic WT brains but not in brains from gpm6a−/− embryos (Fig. 2, F and G), indicating that Rufy3 is captured by GPM6a and accumulates in lipid rafts through Rufy3-GPM6a interactions.
Rufy3 mediates between GPM6a and GTP-bound Rap2
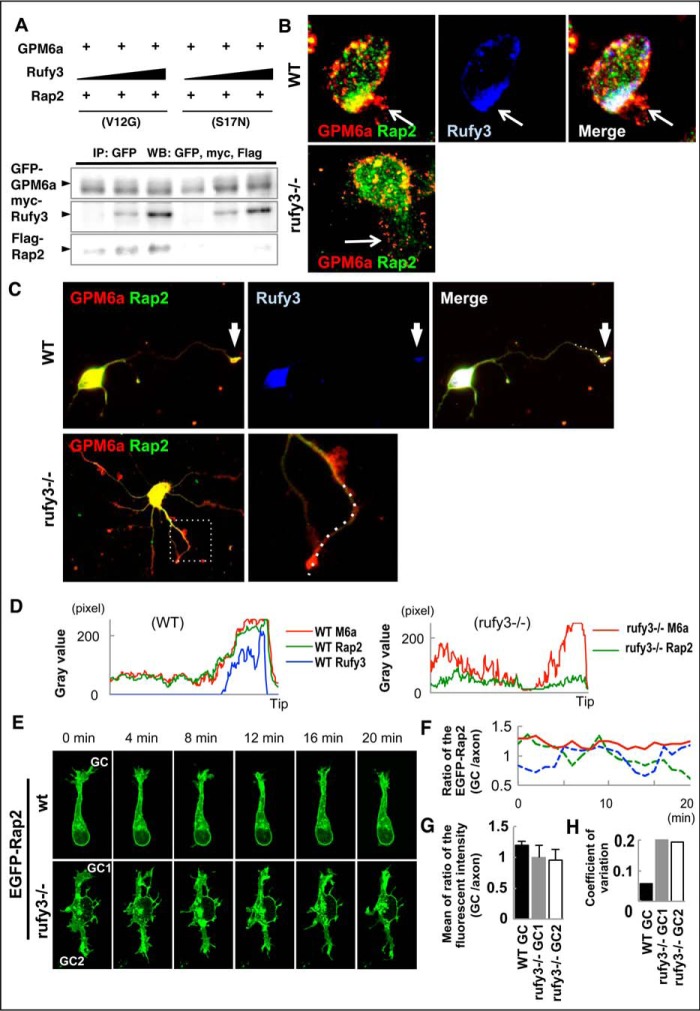
We previously reported that the active form (GTP-bound; i.e. V12G constitutively active mutant) of Rap2 specifically binds to Rufy3, and GPM6a forms a ternary complex with Rufy2 and Rufy3 (9). In coimmunoprecipitation experiments, the active form (V12G), but not the inactive form (S17N), of Rap2 was selectively coprecipitated with GPM6a via Myc-Rufy3 in a dose-dependent manner (Fig. 3A). Because the ternary complex asymmetrically accumulates at the origin sites of neurites (9), we hypothesized that Rufy3 intermediated between GPM6a and Rap2 to assemble the ternary complex and that this complex participates in the downstream signaling of GPM6a. To examine this possibility, we compared the localization of Rap2 at the growth cone in both WT and rufy3−/− neurons.
Figure 3.
Rufy3 mediates Rap2 accumulation in the growth cone through a GPM6a-Rufy3-Rap2 ternary complex. A, Rufy3 induced the formation of a GPM6a-Rufy3-Rap2 ternary complex. EGFP-GPM6a and Myc-Rufy3 were incubated with either FLAG-Rap2V12G or FLAG-Rap2S17N. The concentration of Myc-Rufy3 changed according to the concentration of FLAG-Rap2V12G or FLAG-Rap2S17N. EGFP-GPM6a and its binding proteins were immunoprecipitated (IP) with anti-EGFP antibody (arrows). B and C, immunofluorescence staining of cortical neurons at 30 min (B) and 48 h (C) and after plating on LN using anti-GPM6a (red), anti-Rufy3 (blue), and anti-Rap2 (green) antibodies. In WT neurons, GPM6a was assembled at the protruding site (arrow) and colocalized with Rufy3 and Rap2. In contrast, in rufy3−/− neurons, GPM6a and Rap2 were not assembled (arrow). In rufy3−/− neurons with multiple axons (C), the highlighted region (left) is shown enlarged (right). D, immunofluorescence intensity profiles of anti-GPM6a (red), -Rufy3 (green), and -Rap2 (blue) from the axon shaft to the growth cone (tip) shown as dotted lines in the photograph in C. E–H, in rufy3−/− neurons, the accumulation of Rap2 is not sustained in the growth cone. E, six sequential images (4 min per frame) of EGFP-Rap2 expressed in WT or rufy3−/− neurons at 24 h after plating on LN. The analyzed axonal growth cones (GC) are highlighted. F and G, chronological changes (F) and means (G) in the ratio of EGFP-Rap2 fluorescence intensity in the growth cone to that in the axon (mean ± S.D.; WT GC/axon, 1.20 ± 0.07; rufy3−/− GC1/axon, 1.00 ± 0.20; rufy3−/− GC2/axon, 0.96 ± 0.19; error bars represent S.D.). Coefficients of variation in G are shown in H. Scale bar, 20 μm (B, C, and E). WB, Western blotting.
Immunofluorescence analysis indicated that Rap2 is not specifically localized at the growth cone and is independent of the developmental stage of rufy3−/− neurons (Fig. 3, B–D). In contrast, GPM6a accumulated at the growth cone despite the deficiency of Rufy3, indicating that Rufy3 is required for Rap2, its downstream molecule, but not for its upstream molecule, GPM6a. Taken together with our previous report (9), we conclude that Rufy3 is required for the recruitment of Rap2 to form a ternary complex downstream of GPM6a-dependent signaling. Live imaging analysis of EGFP-Rap2 revealed that Rap2 stably remained in the growth cone in WT but not rufy3−/− neurons (Fig. 3, E–H). These results indicate that Rufy3 induces formation of a GPM6a-Rufy3-Rap2 ternary complex, acts as an adapter between GPM6a and Rap2, and assembles active GTP-bound Rap2 in the axonal growth cone.
Deficiency of Rufy3 inhibits the accumulation of Tiam2/STEF downstream of Rap2
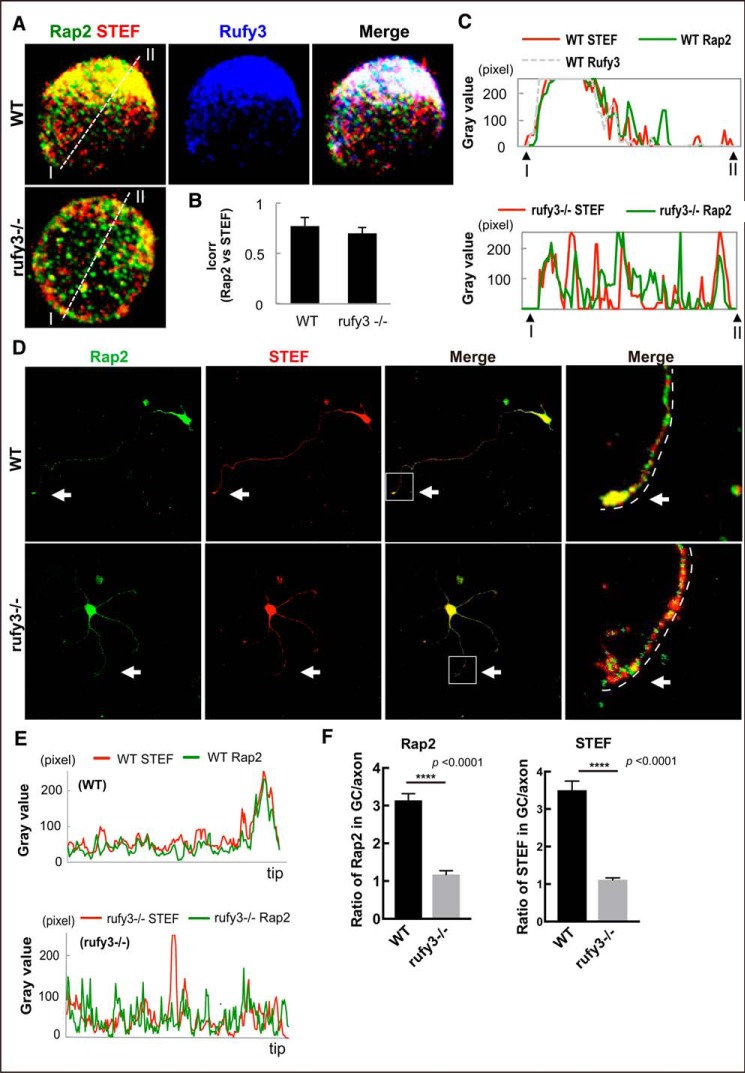
We previously identified Tiam2/STEF as an active Rap2 partner and demonstrated its colocalization with GPM6a-Rufy3-Rap2 ternary complex in the cortical neuron (9). Tiam2/STEF, a guanine exchange factor of Rac1, acts as a positive regulator for neuronal polarity by activating Rac1 (17, 18). The GEF activity of Tiam2/STEF is activated by its interaction with active Rap2 (9). To examine whether suppression of Rap2 accumulation in Rufy3-KO mice affected Tiam2/STEF localization, we analyzed the immunofluorescence of Rap2 and Tiam2/STEF in cortical neurons at different stages of development. In neurons plated on LN for 30 min (neurons at stage 1) (11, 19), the spatial interaction of Rap2 with Tiam2/STEF was not significantly different in WT and rufy3−/− neurons (Fig. 4, A and B). Rap2 and Tiam2/STEF were asymmetrically colocalized in WT but symmetrically colocalized in rufy3−/− neurons at stage 1 (Fig. 4C). In cortical neurons plated for 72 h, Rap2 and Tiam2/STEF specifically accumulated in the axonal growth cone of WT but not of rufy3−/− neurons (Fig. 4, D–F), suggesting that Rufy3 regulates the localization of Rap2 and of STEF in the axonal growth cone. Furthermore, these proteins were particularly accumulated in the P-domain of the axonal growth cone in WT neurons but dispersed in rufy3−/− neurons (Fig. 4F), suggesting that Rufy3 regulates the localization of Rap2 and STEF. A competition assay between Rufy3 and Tiam2/STEF for the binding of Rap2 showed that Rufy3 is released from Rap2 by the binding of STEF to Rap2 (Fig. S4). These results conclusively showed that Rufy3 serves as a transient adapter between Rap2 and GPM6a to recruit active Rap2 in GPM6a-enriched lipid rafts, resulting in the accumulation of Tiam2/STEF.
Figure 4.
Loss of Rufy3 disrupts the assembly of both proteins downstream of Rufy3. A, immunofluorescence staining of endogenous STEF (red), Rap2 (green), and Rufy3 (blue) in WT and rufy3−/− cortical neurons at 30 min after plating on LN. Scale bar, 20 μm. B, colocalization of endogenous Rap2 and STEF in WT and rufy3−/− cortical neurons is shown as the mean correlation index (Icorr). C, detailed fluorescence plot profiles of each fluorescence image along the I–II lines are shown. D, immunofluorescence staining of STEF (red), Rap2 (green), and Rufy3 (blue) in WT and rufy3−/− cortical neurons at 48 h after plating on LN. The highlighted regions (left panels) are shown enlarged (right). Arrows, growth cones. Scale bars, 30 (left panels) and 15 μm (right panels). E, immunofluorescence intensity profiles of anti-STEF (red) and -Rap2 (green) from the axon shaft to the growth cone (GC) shown as dotted lines in the photograph in D. F, ratio of the immunofluorescence intensity of Rap2 or STEF in the growth cone versus that of the axon (two-tailed t test (means ± S.E.); Rap2, WT (3.141 ± 0.178) versus rufy3−/− (1.175 ± 0.101); ****, p < 0.0001; STEF, WT (3.506 ± 0.244) versus rufy3−/− (1.115 ± 0.049); ****, p < 0.0001; n = 30 for each sample; error bars represent S.E.).
Rufy3 regulates the localization of neuronal polarity determinants through the Rap2-STEF signaling pathway
To confirm that Rap2 and Tiam2/STEF affect the same pathway controlling polarity downstream of Rufy3, we performed an inhibition assay with rufy3−/− neurons using dominant-negative mutations of Rap2 or Tiam2/STEF. The ratio of rufy3−/− polarized neurons was significantly decreased compared with control (WT) neurons on LN (Fig. S5, A–D), and neither overexpression of dominant-negative Rap2 nor overexpression of STEF significantly affected the reduced ratio of polarized rufy3−/− neurons (Fig. S5, A–D). These results are consistent with the idea that Rap2 and STEF are downstream of the Rufy3 in the signaling pathway for polarity determination.
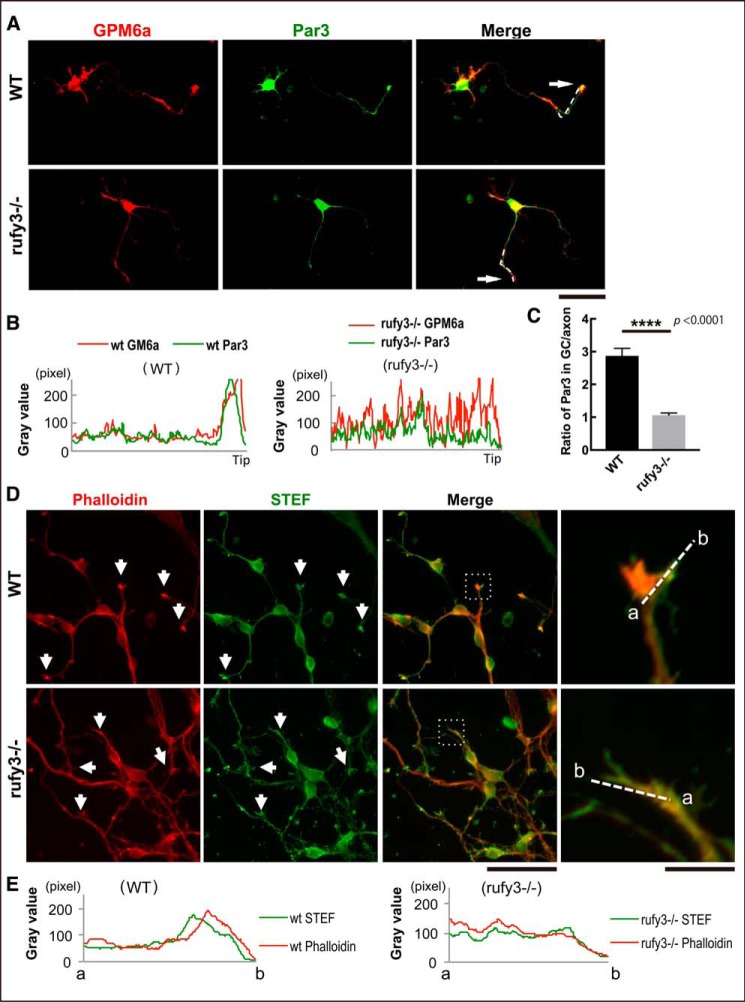
STEF was reported to be associated with the Par6-Par3-atypical PKC polarity complex through its direct interaction with Par3 (12). We examined whether localization of the Par complex in the growth cone was also regulated downstream of Rufy3 signaling. Par3 and GPM6a both assembled in the growth cone, but the localization of Par3 was significantly inhibited in rufy3−/− neurons (Fig. 5, A–C), suggesting that Rufy3 is required for polarity complex localization in the growth cone. Similar to Rap2, the polarity complex activates the guanine nucleotide exchange factor activity of STEF, resulting in Rac activation, which is crucial for actin organization. Because actin dynamics in the growth cone are thought to establish and maintain neuronal polarity, we examined actin distribution in rufy3−/− neurons. Only faint localization of F-actin enrichment was detected compared with WT neurons (Fig. 5, D and E), indicating that actin organization in the P-domain is changed through the inhibition of STEF accumulation in rufy3−/− neurons.
Figure 5.
Loss of Rufy3 disturbs the accumulation of neuronal polarity determinants in the growth cone. A, immunofluorescence staining of GPM6a (red) and Par3 (green) in WT and rufy3−/− cortical neurons (48 h after plating on LN). Arrows, growth cones. Scale bar, 50 μm. B, immunofluorescence intensity plot profiles of anti-GPM6a (red) and anti-Rufy3 (green) from the axon shaft to the growth cone (tip) are shown as dotted lines in A. C, ratio of the immunofluorescence intensity of Par3 in the growth cone versus that of the axon (two-tailed t test (means ± S.E.); WT (2.87 ± 0.23) versus rufy3−/− (1.06 ± 0.07); ****, p < 0.0001; n = 25; error bars represent S.E.). D, immunofluorescence staining of phalloidin (red) and anti-STEF antibody (green) in WT and rufy3−/− cortical neurons (48 h after plating on LN). High magnification images of insets are shown in the left panels. Arrows, growth cones. Scale bars, 100 (left) and 20 μm (right). E, detailed fluorescence plot profiles of each fluorescence image along the a–b lines in D are shown.
Discussion
Among a number of neuronal proteins involved in polarity determination (20), Rufy3 is already known as one of the cell-autonomous polarity determinants; however, using Rufy3-KO mice, we confirmed that Rufy3 is also physiologically involved in the determination of neuronal polarity (Figs. 1, C–I; 3, C–H; and 5) and axon growth at a later stage (Fig. 4, D–F). Consistent with other molecules involved in neuronal polarity, deficiency of Rufy3 alone in vivo did not result in large defects in the network, similar to several other cell-autonomous polarity determinants (10). We also demonstrated the importance of Rufy3 in its role in connecting its upstream partner protein, GPM6a, with its downstream partner, Rap2, in lipid rafts. With Rufy3, GPM6a signaling was effectively transduced to activate STEF/Tiam2 as demonstrated previously (9), whereas in the absence of Rufy3, Rac activation by STEF/Tiam2 was not effective (Figs. 3E and 4, D–F, and Figs. S3 and S5).
Because Rufy3-KO mice immediately died after birth with severe cyanosis but id not show marked developmental defects of the nervous system, we suspect that this death is probably due to a circulation problem. RPIP8, another binding partner of Rap2 enriched in brain that also has a RUN domain, may partially and functionally rescue the Rufy3 deficiency (21). Rufy3 was previously characterized as a regulator of neuronal cell polarity and to act in a cell-autonomous manner (14) as we confirmed using rufy3−/− neurons (Fig. 1, C and D). However, polarity determination was delayed in rufy3−/− neurons more markedly than in LN-dependent neurons (Fig. 1, D–F), consistent with our recent results showing that GPM6a, Rap2, and Tiam2/STEF are highly concentrated in lipid rafts and that the presence of Rap2 and Tiam2/STEF are GPM6a-dependent (9).
Rufy3 was previously reported to interact with fascin and control axon growth (14); however, our results did not show its colocalization with F-actin (Fig. S3). Because fascin is specifically localized in the F-actin bundles of the P-domain (Fig. S3; also see Ref. 20) and Rufy3 was mainly localized in the membrane area (Fig. S3), we could not confirm colocalization of these two proteins as reported previously (14) using our antibody (Fig. S3).
Previous in vitro studies showed that Rufy3 interacts with other small G proteins, including Rab family members such as Rab33 (22) reported to be involved in axon growth (23). However, we observed that Rufy3 was mainly distributed to the plasma membrane or the cytosol (Fig. 2A and Fig. S3A) and did not show the vesicular distribution along the cytoskeleton. These results indicate that Rufy3 is physiologically bound to Rap2 in the developmental stages of neurons unlike Rab33.
In conclusion, Rufy3 is believed to be an adapter of Rap2 as an upstream protein for the determination of neuronal polarity. We recently visualized the recycling of lipid raft domains in the growth cone using super-resolution microscopy (24) and will apply this new methodology to study the dynamics of Rufy3 using Rufy3-KO mouse neurons.
Experimental procedures
Animals
We obtained C57BL/6 BAC genomic clone RP24-132B11 mice, which contain the Rufy3 gene, from the BACPAC Resources Center. To construct the Rufy3-targeting vector, a 0.64-kb DNA fragment carrying exon 3 of Rufy3 was amplified by PCR and inserted into the KpnI-SacI sites of a middle entry clone plasmid (pDME-1). In this plasmid, a DNA fragment of phosphoglycerate kinase promoter–driven neomycin resistance gene-poly(A) (pgk-Neo) is flanked by two frt sites and a loxP sequence located 249 bp upstream of exon 3, and a second loxP sequence is placed 269 bp downstream of exon 3. The 3.02-kb upstream and 5.45-kb downstream homologous genomic DNA fragments were subcloned into the 5′ entry clone (pD3UE-2) and 3′ entry clone (pD5DE-2), respectively, using a Quick and Easy BAC modification kit (Gene Bridges, Dresden, Germany). To target vector assembly, the three entry clones were recombined to a destination vector plasmid (pDEST-DT; containing a CAG promoter-driven diphtheria toxin gene) using a MultiSite Gateway Three-fragment Vector construction kit (Invitrogen). The Rufy3-flox mouse line was established by introducing the linearized targeting vector into the C57BL/6N-derived embryonic stem cell line RENKA, and then recombinant clones were selected using medium containing 175 μg/ml G418. ES cells were cultured as described previously (23). The targeted clones were confirmed by Southern blot analysis using 5′, 3′, and neomycin resistance gene probes. Chimeric mice were generated as described previously (25). Briefly, targeted clones were microinjected into eight-cell-stage embryos of the CD-1 mouse strain. The resulting chimeric embryos were developed to the blastocyst stage by incubation for more than 24 h and then transferred to pseudopregnant CD-1 mouse uteruses. Germ line chimeras were crossed with C57BL/6N female mice, and the heterozygous offspring were crossed with a TLCN-Cre deleter mouse line (26) to establish the Rufy3 knock-out mouse line.
Neuronal cell culture
Dissociated neurons were cultured as described previously (9). The cortices of E14.5 embryonic mice were dissected in cold DMEM (Wako) and incubated in AccuMax (Innovative Cell Technologies) in Hanks' balanced salt solution (Wako) for 10 min at 37 °C. Neurons were suspended in 10% FBS (Gibco) in DMEM, filtered through a 40-μm Cell Strainer (BD Falcon), then counted, and plated on coverslips coated with LN (2 μg/ml; Invitrogen) using Neurobasal medium (Gibco) supplemented with B27, GlutaMAX (×1 final concentration; Gibco), and penicillin/streptomycin. For transfection, dissociated neurons (1 × 106) or COS-7 cells (1 × 106) were placed in 2-mm gap cuvettes (BEX Co., Ltd., Tokyo, Japan) containing 100 μl of Opti-MEM (Gibco) with 2–5 μg of plasmid DNA and then transfected using 10 direct-current 20-V electrical pulses at intervals of 50 ms using an electroporator (CUY21 Vitro-EX, BEX Co., Ltd.).
Plasmid construction
cDNA fragments of GPM6a and Rufy3 were amplified from a mouse brain cDNA library by PCR and subcloned into pmCherry-C1 (Clontech), pEGFP-C1 (Clontech), or pEF1-Myc plasmid vector (Clontech). DNA fragments encoding the RUN domain and coiled-coil domain of Rufy3 were amplified by PCR and subcloned into pEGFP-C1 vector. A constitutively active and a dominant-negative mutant of mouse Rap2 was designed by analogy to Ras: Gly-12 was substituted by Val in Rap2V12G, and Asn-17 was substituted by Ser in Rap2S17N by PCR-mediated mutagenesis. Wild-type Rap2A, Rap2V12G, and Rap2S17N subcloned into pFLAG-CMV-C1 (Sigma-Aldrich) were obtained from M. Matsuda (Kyoto University, Japan). Full-length STEF subcloned into pcDNA3-HA vector was obtained from M. Hoshino (National Center for Neurology and Psychiatry, Kodaira, Japan) (16).
Transfection of neurons
Dissociated neurons (1 × 106) or COS-7 cells (1 × 106) were placed in 2-mm gap cuvettes containing 100 μl of Opti-MEM with 2–5 μg of plasmid DNA and then transfected as described under “Neuronal cell culture.”
Immunofluorescence labeling
Indirect immunofluorescence labeling was performed as described previously (9) using the antibodies shown in Table S1. The cells were fixed for 30 min with 4% paraformaldehyde (PFA) in PBS (pH 7.4), and then the cells were permeabilized for 5 min with 0.1% Triton X-100 in PBS and incubated in 2% BSA in PBS for 1 h at room temperature, in 2% BSA in PBS for 1 h, and with 2% BSA in PBS containing primary antibodies overnight at 4 °C. Subsequently, the cells were rinsed and incubated with secondary antibody for 3 h three times with PBS and then mounted in fluorescent mounting medium (Dako).
Coronal slices (50 μm thick) were prepared from fixed brains using a Leica VT1000S Vibratome. The sections were permeabilized with 1% Triton X-100 in PBS for 1 h at room temperature and then blocked with 5% BSA in PBS for 1 h. Slices were incubated with 5% BSA in PBS containing primary antibodies overnight at 4 °C, rinsed with PBS, incubated with secondary antibody for 3 h, then washed with PBS, and mounted in fluorescent mounting medium. Preparations were examined under Olympus BX63 optical, Olympus FV2000 confocal, and Zeiss LSM510 confocal microscopes.
E16.5 mice were fixed by cardiac perfusion with 4% PFA in PBS (pH 7.4). The brains were dissected and soaked in 4% PFA in PBS (pH 7.4) at 4 °C overnight, and then the fixed brains were stored in 30% sucrose in PBS at 4 °C. The brains were frozen in powdered dry ice and embedded in OCT compound (Tissue-Tek, Sakura Finetek USA Inc.), and 30-μm-thick coronal sections were sliced using a sliding cryotome (Leica CM1850). Coronal sections were rinsed with PBS and incubated in 0.1% Triton X-100 and 3% H2O2 in PBS for 15 min at room temperature. After rinsing with PBS, the sections were incubated overnight at 4 °C with anti-Rufy3 antibody (1:1000) in 5% BSA in PBS, then rinsed with PBS, and incubated with biotinylated anti-rabbit IgG antibody (1:200) for 30 min at 37 °C. After rinsing with PBS, the sections were incubated in avidin-biotin-peroxidase complex (Vectastain ABC kit, Vector Laboratories) for 30 min at 37 °C, rinsed with PBS, and then incubated in 50 mm Tris-HCl (pH7.4) containing 0.01% H2O2 and 0.01% diaminobenzidine tetrahydrochloride at 37 °C for 10 min. The stained sections were mounted on MAS-coated glass slides (Matsunami Glass, Osaka, Japan) using fluorescent mounting medium. Bright-field images were taken with an Olympus BX63 microscope and DP-72 charge-coupled device camera.
Time-lapse experiment
Time-lapse recording of EGFP-GPM6a or EGFP-Rap2 was analyzed as described previously (9). pEGFP-C1-GPM6a or pEGFP-C1-Rap2 plasmid was electroporated into dissociated E14.5 mouse cortical neurons, and the transfected cells were precultured in L-15 medium (Gibco) supplemented with 10% FBS, B27, GlutaMAX, 0.45% glucose, and penicillin/streptomycin in a Lipidure-coated 96-well plate (NOF Corp., Japan) at 37 °C without CO2 for 24 h. The cells were dissociated with AccuMax and plated on a glass-bottom dish coated with LN. EGFP-positive neurons were observed in a temperature-controlled incubation chamber (at 37 °C; Tokai Hit) fitted onto a confocal microscope (Olympus FV2000). Fluorescence images were obtained with a cooled charge-coupled device camera and processed using image analysis software (Metamorph, Molecular Devices, CA).
Polarity assay
Neuronal polarity was analyzed as described previously (9). Briefly, at 30 min after plating, neurons with a single short filopodium or lamellipodium protruding within a 90° angle from the center were considered as monopolar, i.e. stage 3 neurons. Neurons with two or more filopodia or lamellipodia protruding within a 90° angle were considered as bi- or multipolar, i.e. stage 2 neurons. Neurons with a lamellipodium that formed at an angle over 180° or those with no filopodia or lamellipodia were defined as neutral or intact, i.e. stage 1 neurons.
At 18–72 h after plating, neurons projecting single major neurites that were at least twice as long as other protrusions were counted as polarized, i.e. stage 3 neurons. Neurons with minor and no major neurites were counted as unpolarized, i.e. stage 2 neurons. Neurite tracing and measurement of neurite length were conducted using ImageJ with the NeuronJ plugin module.
Pulldown assay and immunoprecipitation assay
COS-7 cells transfected by electroporation were washed with cold PBS and lysed with lysis buffer (50 mm Tris-HCl (pH 7.4), 150 mm NaCl, 1% Nonidet P-40, 1 mm EDTA). The homogenate was sonicated and centrifuged at 10,000 × g at 4 °C. The antibodies against the recombinant proteins were cross-linked to Protein G-Sepharose 4 Fast Flow (GE Healthcare) with disuccinimidyl suberate (Thermo Fisher Scientific Pierce) or to AminoLink Plus Coupling Resin (Thermo Fisher Scientific Pierce) (1 μg of antibody/μl of resin) and incubated with the cell extracts at 4 °C for 2 h. The resin was washed three times and eluted with 1× sample buffer for SDS-PAGE.
Preparation of DRM fractions
DRM fractions were prepared as described by Simons and Ikonen (27) with some modifications. For preparation of DRM from mouse brain, embryonic brains (E14.5) were dissected in PBS on ice. Lysis buffer (50 mm Tris-HCl (pH 7.4), 150 mm NaCl, 1 mm EDTA, phosphatase inhibitor mixture, 4-amidinobenzylsulfonyl fluoride hydrochloride, leupeptin, pepstatin) with 1% Brij 98 and 5% glycerol was added to the brain (brain:1% Brij 98 and 5% glycerol lysis buffer (v/w) = 8:1) and homogenized at 1000 rpm using 10 strokes. For preparation of DRMs from the transfected cells, cells were scraped in 1% Brij 98 and 5% glycerol lysis buffer after rinsing with cold PBS and homogenized at 1000 rpm using 10 strokes. The homogenates were mixed well and placed on ice for 1 h. After centrifugation, supernatants were gently mixed with an equal volume of 80% sucrose lysis buffer. The sample was overlaid with a discontinuous gradient of 35% sucrose lysis buffer (4 ml) to 5% sucrose lysis buffer (4 ml). The gradients were subjected to ultracentrifugation at 33,600 rpm (20,000 × g) for 18 h in a Hitachi ST40 Ti rotor. Twelve fractions (1 ml/fraction) were collected from the top of a centrifugation tube by a Hitachi gradient generator. The DRM fraction was positioned at the interface between 35 and 5% sucrose (fractions 3–5).
Colocalization assay
We used the ImageJ plugin Colocalization Colormap to determine protein colocalization to automatically quantify the correlation between a pair of pixels. Distributions of the normalized mean deviation product value shown with a color scale and average of normalized mean deviation products were compared.
In utero electroporation
In utero electroporation was performed on E14.5 embryos using pregnant ICR (Japan SLC, Inc.) mice as described previously (9, 28) with some modifications. In brief, pregnant mice were deeply anesthetized by intraperitoneal administration of pentobarbital (48.6 mg/kg of body weight), and their uterine horns were exposed. Plasmid DNA purified using a Qiagen Plasmid Maxi kit was dissolved in HBS buffer solution (10 mm Hepes (pH 7.4), 150 mm NaCl) with 0.01% Fast Green solution. Approximately 1 μl of plasmid solution was injected into the lateral ventricle of each embryo using a processed microcapillary made from a glass tube (number 3-000-105-G, Drummond, GD-1, Narishige). Electroporation was performed using a disc electrode (CUY650P3, BEX Co., Ltd.) and electroporator (CUY21 Edit II, BEX Co., Ltd.; Nepa21; NEPA gene) using the following conditions: voltage, 35 V; pulse-on, 50 ms; pulse-off, 450 ms; pulse time, 5 ms. The uterine horns were placed back into the abdominal cavity to allow the embryos to continue normal development.
Author contributions
M. I. and A. H. designed the experiments. A. H. performed the cellular experiments. H. U. and K. S. produced the Rufy3-KO mice. M. I. and A. H. wrote the paper.
Supplementary Material
Acknowledgments
We thank Dr. Yasuyuki Ito, Ph.D. (Igarashi laboratory) and the Animal Resource Department of Niigata University for the generation and maintenance of Rufy3-KO mice.
This work was supported in part by TOGO-NO and HOKATSU-NO (Comprehensive Brain Science Network) on Innovative Areas (to M. I.); project-promoting grants from Niigata University; Uehara Memorial Science Promoting Foundation (to M. I.); RIKEN BioResource Center grants for gene targeting in production of mice (to M. I.); KAKENHI Grants 17023019, 22240040, and 24111515 (to M. I.) and 15K06770 and 24700319 (to A. H.) from the Ministry of Education, Culture, Sports, Science and Technology (MEXT) of Japan and Japan Society for the Promotion of Science; the Platform of Advanced Animal Model Support from MEXT of Japan (to M. I. and K. S.); grants from Tsukada Milk Co. and Novartis Pharma Ltd. (to A. H.); and a Takeda Foundation research grant (to Y. I.). The authors declare that they have no conflicts of interest with the contents of this article.
This article contains Figs. S1–S5 and Table S1.
- Rufy
- RUN and FYVE domain–containing
- GPM6a
- glycoprotein M6A
- Tiam2
- T-cell lymphoma invasion and metastasis 2
- STEF
- SIF and Tiam 1-like exchange factor
- EGFP
- enhanced GFP
- LN
- laminin
- P
- peripheral
- DRM
- detergent-resistant membrane
- BAC
- bacterial artificial chromosome
- PFA
- paraformaldehyde
- E
- embryonic day
- aa
- amino acids
- CC
- coiled-coil domain.
References
- 1. Cromm P. M., Spiegel J., Grossmann T. N., and Waldmann H. (2015) Direct modulation of small GTPase activity and function. Angew. Chem. Int. Ed. Engl. 54, 13516–13537 [DOI] [PubMed] [Google Scholar]
- 2. Kitagishi Y., and Matsuda S. (2013) RUFY, Rab and Rap family proteins involved in a regulation of cell polarity and membrane trafficking. Int. J. Mol. Sci. 14, 6487–6498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Kukimoto-Niino M., Takagi T., Akasaka R., Murayama K., Uchikubo-Kamo T., Terada T., Inoue M., Watanabe S., Tanaka A., Hayashizaki Y., Kigawa T., Shirouzu M., and Yokoyama S. (2006) Crystal structure of the RUN domain of the RAP2-interacting protein x. J. Biol. Chem. 281, 31843–31853 [DOI] [PubMed] [Google Scholar]
- 4. Igarashi M. (2014) Proteomic identification of the molecular basis of mammalian CNS growth cones. Neurosci. Res. 88, 1–15 [DOI] [PubMed] [Google Scholar]
- 5. Stankiewicz T. R., and Linseman D. A. (2014) Rho family GTPases: key players in neuronal development, neuronal survival, and neurodegeneration. Front. Cell. Neurosci. 8, 314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Villarroel-Campos D., Bronfman F. C., and Gonzalez-Billault C. (2016) Rab GTPase signaling in neurite outgrowth and axon specification. Cytoskeleton 73, 498–507 [DOI] [PubMed] [Google Scholar]
- 7. Shah B., and Püschel A. W. (2016) Regulation of Rap GTPases in mammalian neurons. Biol. Chem. 397, 1055–1069 [DOI] [PubMed] [Google Scholar]
- 8. Nozumi M., Togano T., Takahashi-Niki K., Lu J., Honda A., Taoka M., Shinkawa T., Koga H., Takeuchi K., Isobe T., and Igarashi M. (2009) Identification of functional marker proteins in the mammalian growth cone. Proc. Natl. Acad. Sci. U.S.A. 106, 17211–17216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Honda A., Ito Y., Takahashi-Niki K., Matsushita N., Nozumi M., Tabata H., Takeuchi K., and Igarashi M. (2017) Extracellular signals induce glycoprotein M6a clustering of lipid rafts and associated signaling molecules. J. Neurosci. 37, 4046–4064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Cáceres A., Ye B., and Dotti C. G. (2012) Neuronal polarity: demarcation, growth and commitment. Curr. Opin. Cell Biol. 24, 547–553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Namba T., Kibe Y., Funahashi Y., Nakamuta S., Takano T., Ueno T., Shimada A., Kozawa S., Okamoto M., Shimoda Y., Oda K., Wada Y., Masuda T., Sakakibara A., Igarashi M., et al. (2014) Pioneering axons regulate neuronal polarization in the developing cerebral cortex. Neuron 81, 814–829 [DOI] [PubMed] [Google Scholar]
- 12. Nishimura T., Yamaguchi T., Kato K., Yoshizawa M., Nabeshima Y., Ohno S., Hoshino M., and Kaibuchi K. (2005) PAR-6-PAR-3 mediates Cdc42-induced Rac activation through the Rac GEFs STEF/Tiam1. Nat. Cell Biol. 7, 270–277 [DOI] [PubMed] [Google Scholar]
- 13. Mori T., Wada T., Suzuki T., Kubota Y., and Inagaki N. (2007) Singar1, a novel RUN domain-containing protein, suppresses formation of surplus axons for neuronal polarity. J. Biol. Chem. 282, 19884–19893 [DOI] [PubMed] [Google Scholar]
- 14. Wei Z., Sun M., Liu X., Zhang J., and Jin Y. (2014) Rufy3, a protein specifically expressed in neurons, interacts with actin-bundling protein fascin to control the growth of axons. J. Neurochem. 130, 678–692 [DOI] [PubMed] [Google Scholar]
- 15. Sasaki Y., Hayashi K., Shirao T., Ishikawa R., and Kohama K. (1996) Inhibition by drebrin of the actin-bundling activity of brain fascin, a protein localized in filopodia of growth cones. J. Neurochem. 66, 980–988 [DOI] [PubMed] [Google Scholar]
- 16. Hoshino M., Sone M., Fukata M., Kuroda S., Kaibuchi K., Nabeshima Y., and Hama C. (1999) Identification of the stef gene that encodes a novel guanine nucleotide exchange factor specific for Rac1. J. Biol. Chem. 274, 17837–17844 [DOI] [PubMed] [Google Scholar]
- 17. Matsuo N., Hoshino M., Yoshizawa M., and Nabeshima Y. (2002) Characterization of STEF, a guanine nucleotide exchange factor for Rac1, required for neurite growth. J. Biol. Chem. 277, 2860–2868 [DOI] [PubMed] [Google Scholar]
- 18. Miyamoto Y., Yamauchi J., Tanoue A., Wu C., and Mobley W. C. (2006) TrkB binds and tyrosine-phosphorylates Tiam1, leading to activation of Rac1 and induction of changes in cellular morphology. Proc. Natl. Acad. Sci. U.S.A. 103, 10444–10449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Dotti C. G., Sullivan C. A., and Banker G. A. (1988) The establishment of polarity by hippocampal neurons in culture. J. Neurosci. 8, 1454–1468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Namba T., Funahashi Y., Nakamuta S., Xu C., Takano T., and Kaibuchi K. (2015) Extracellular and intracellular signaling for neuronal polarity. Physiol. Rev. 95, 995–1024 [DOI] [PubMed] [Google Scholar]
- 21. Janoueix-Lerosey I., Pasheva E., de Tand M. F., Tavitian A., and de Gunzburg J. (1998) Identification of a specific effector of the small GTP-binding protein Rap2. Eur. J. Biochem. 252, 290–298 [DOI] [PubMed] [Google Scholar]
- 22. Fukuda M., Kobayashi H., Ishibashi K., and Ohbayashi N. (2011) Genome-wide investigation of the Rab binding activity of RUN domains: development of a novel tool that specifically traps GTP-Rab35. Cell Struct. Funct. 36, 155–170 [DOI] [PubMed] [Google Scholar]
- 23. Nakazawa H., Sada T., Toriyama M., Tago K., Sugiura T., Fukuda M., and Inagaki N. (2012) Rab33a mediates anterograde vesicular transport for membrane exocytosis and axon outgrowth. J. Neurosci. 32, 12712–12725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Nozumi M., Nakatsu F., Katoh K., and Igarashi M. (2017) Coordinated movement of vesicles and actin bundles during nerve growth revealed by superresolution microscopy. Cell Rep. 18, 2203–2216 [DOI] [PubMed] [Google Scholar]
- 25. Mishina M., and Sakimura K. (2007) Conditional gene targeting on the pure C57BL/6 genetic background. Neurosci. Res. 58, 105–112 [DOI] [PubMed] [Google Scholar]
- 26. Fuse T., Kanai Y., Kanai-Azuma M., Suzuki M., Nakamura K., Mori H., Hayashi Y., and Mishina M. (2004) Conditional activation of RhoA suppresses the epithelial to mesenchymal transition at the primitive streak during mouse gastrulation. Biochem. Biophys. Res. Commun. 318, 665–672 [DOI] [PubMed] [Google Scholar]
- 27. Simons K., and Ikonen E. (1997) Functional rafts in cell membranes. Nature 387, 569–572 [DOI] [PubMed] [Google Scholar]
- 28. Tabata H., and Nakajima K. (2001) Efficient in utero gene transfer system to the developing mouse brain using electroporation: visualization of neuronal migration in the developing cortex. Neuroscience 103, 865–872 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.