Abstract
The human cytomegalovirus opening reading frame UL144 is an ortholog of the TNF receptor superfamily member, herpesvirus entry mediator (HVEM; TNFRSF14). HVEM binds the TNF ligands, LIGHT and LTa; the immunoglobulin inhibitory receptor, B and T lymphocyte attenuator (BTLA); and the natural killer cell–activating receptor CD160. However, UL144 selectively binds BTLA, avoiding activation of inflammatory signaling initiated by CD160 in natural killer cells. BTLA and CD160 cross-compete for binding HVEM, but the structural basis for the ligand selectivity by UL144 and how it acts as an anti-inflammatory agonist remains unclear. Here, we modeled the UL144 structure and characterized its binding with BTLA. The UL144 structure was predicted to closely mimic the surface of HVEM, and we also found that both HVEM and UL144 bind a common epitope of BTLA, whether engaged in trans or in cis, that is shared with a BTLA antibody agonist. On the basis of the UL144 selectivity, we engineered a BTLA-selective HVEM protein to understand the basis for ligand selectivity and BTLA agonism to develop novel anti-inflammatory agonists. This HVEM mutein did not bind CD160 or TNF ligands but did bind BTLA with 10-fold stronger affinity than wild-type HVEM and retained potent inhibitory activity that reduced T-cell receptor, B-cell receptor, and interferon signaling in B cells. In conclusion, using a viral immune evasion strategy that shows broad immune-ablating activity, we have identified a novel anti-inflammatory BTLA-selective agonist.
Keywords: cytokine, herpesvirus, immunology, lymphocyte, receptor structure-function, signal transduction, structural biology, structural model, viral immunology
Introduction
The human β-herpesvirus cytomegalovirus (CMV)4 devotes much of its genome to evasion of the host immune response, and some of these genes aid in the virus establishing persistent and latent infection (1, 2). The TNF receptor superfamily member HVEM is a focal point for manipulation by viral pathogens including CMV and mutation in cancers and autoimmunity (3–8). HVEM interacts with multiple ligands expressed in the immune system including the TNF superfamily cytokines, LIGHT and lymphotoxin-α, and the immunoglobulin superfamily members BTLA and CD160 that serve as checkpoint regulators in T cells (9). Indeed, human CMV evolved a mutated form of HVEM that specifically engages the immune inhibitor, BTLA. The mimic of HVEM encoded by CMV ORF UL144 binds the HVEM ligand BTLA and inhibits T cell proliferation to a greater extent than HVEM (10). We recently showed that UL144 does not bind the HVEM receptor CD160, avoiding natural killer (NK) cell activation (11).
In humans, membrane associated LIGHT, BTLA, and CD160, all activate NF-κB signaling downstream of HVEM following receptor engagement, whereas HVEM activates the receptors BTLA and CD160 resulting in bidirectional signaling between neighbor cells (12–14). Cells co-expressing HVEM and BTLA or CD160 can also form cell-intrinsic complexes of these proteins that prevent accessibility of these receptors to extracellular ligands because of steric hindrance (15). In T cells, BTLA was first shown to engage inhibitory signaling pathways including the activation of SH2 domain–containing protein tyrosine phosphatases (SHP)-1 and 2. Recently BTLA has been shown to regulate Toll-like receptor signaling in dendritic cells and IL-7 signaling in γδ T cells (16, 17). However, in human NK cells, HVEM can also promote cytolytic and pro-inflammatory pathways through CD160 as a host counter measure to human CMV (11). Recent work describing CD160 deficiency in mice confirms its pro-inflammatory function in NK cells (18).
Ligand-blocking biologics directed at checkpoint regulators such as CTLA4 and PD1 have proven successful in releasing the immune system to treat of a variety of cancers (19). However, agonists that activate the inhibitory checkpoint regulators to limit inflammation have resisted development. Here, we define the biophysical and functional interactions between HVEM and UL144 network proteins as a blueprint to bioengineer an HVEM mutein as a selective agonist for BTLA. We show that similar to the viral UL144 protein, a bioengineered HVEM mutein exhibits ligand selection for BTLA, without binding CD160. This HVEM mutein additionally activates broad inhibition of T cell receptor, B cell receptor, and cytokine signaling. Together, these data illustrate the successful design of a selective BTLA agonist to be used as an anti-inflammatory biologic that is based on selectivity observed in a viral protein and potentially useful as a human therapeutic.
Results
Characterization of the molecular mechanism of CMV UL144 selection for BTLA agonism
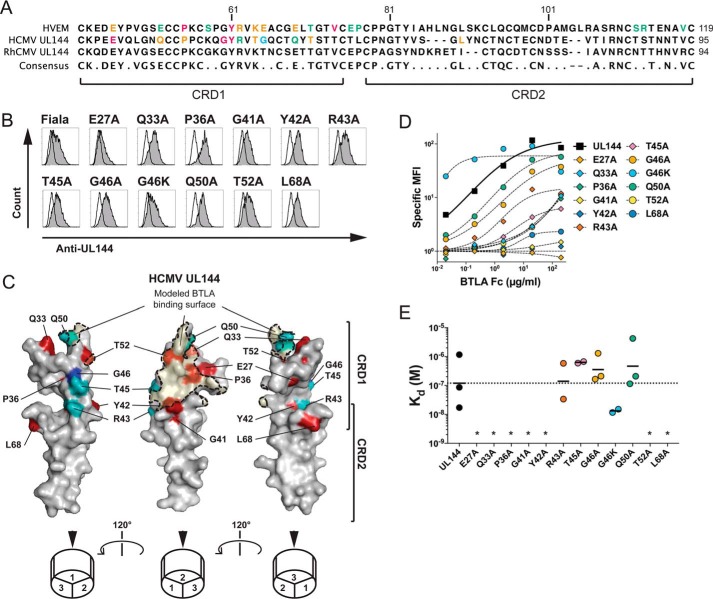
We previously showed that the HVEM ortholog in primate CMV ORF UL144 shows selective affinity for BTLA and not CD160 and that this protein can inhibit T-cell proliferation (10, 11). To confirm the agonistic epitope of UL144, we first modeled the UL144 structure and a hypothetical BTLA contact surface based on the HVEM-BTLA co-crystal structure. We next targeted proposed UL144 surface residues divergent from HVEM by alanine mutagenesis (Fig. 1, A and B) (20). Among UL144 alanine substitution mutants, for which we confirmed expression, we found several residues critical for BTLA binding including Glu27, Gln33, Pro36, Gly41, Tyr42, Thr52, and Leu68 (Fig. 1, C–E). These residues defines a surface on the modeled UL144 structure centered on one face of its CRD1 region similar to the BTLA binding surface of the solved HVEM structure. Notably, residues Gly41 and Tyr42 are homologous to Gly60 and Tyr61 in HVEM that were predicted to impact BTLA. However, one residue in UL144, Leu68 homologous to Leu90 in HVEM, is positioned outside of the hypothetical BTLA binding surface in the UL144 model (Fig. 1C). Mutation at this residue may alter the global UL144 structure, preventing BTLA engagement. Alanine substitutions at Arg43, Thr45, Gly46, and Gln50 did not significantly alter BTLA binding, whereas the previously described G46K mutant again showed enhanced affinity for BTLA (Fig. 1, C–E) (11). Together, the mutations describe a BTLA-binding region on the UL144 protein similar but not identical to the BTLA binding surface in HVEM.
Figure 1.
Modeling BTLA binding epitope in ligand selective CMV UL144. A, the extracellular domains of human HVEM and human CMV UL144 were aligned to show the positions of residues mutated in UL144 here and in HVEM from previous studies (10, 20). B, 293T cells transduced with wild-type or mutated human CMV UL144 were stained with anti-UL144 (clone 2F11). Histograms show UL144 expression (gray) overlaid onto cells transfected with control vector. C, three images of human CMV UL144 rotated 120° about the y axis showing mutated surface residues. Structure was predicted using HVEM as a model and with residue dihedral angles φ and ψ conforming to allowed ranges within the Ramachandran plot. Human CMV UL144 surface is shown in gray with CRD1 and CRD2 indicated. Residues required for BTLA binding are colored as follows: Glu27, Gln33, Pro36, Gly41, Tyr42, Thr52, and Leu68 in red (required) and Arg43, Thr45, and Gln50 in teal (no significant effect). Gly46 in blue showed increased binding when mutated to Lys. The BTLA binding surface was predicted using the structure of HVEM and BTLA together and shaded in orange. D and E, 293T cells transduced with wild-type or mutated human CMV UL144 (group C strain) were stained with the indicated concentrations of BTLA-Fc. Graphs in D show binding curves for the indicated UL144 mutants from a representative experiment. Kd of BTLA-Fc binding to UL144 proteins were plotted in E (geometric mean, at least three experiments per mutation). *, not determined.
Analysis of BTLA binding epitopes by diverse agonists
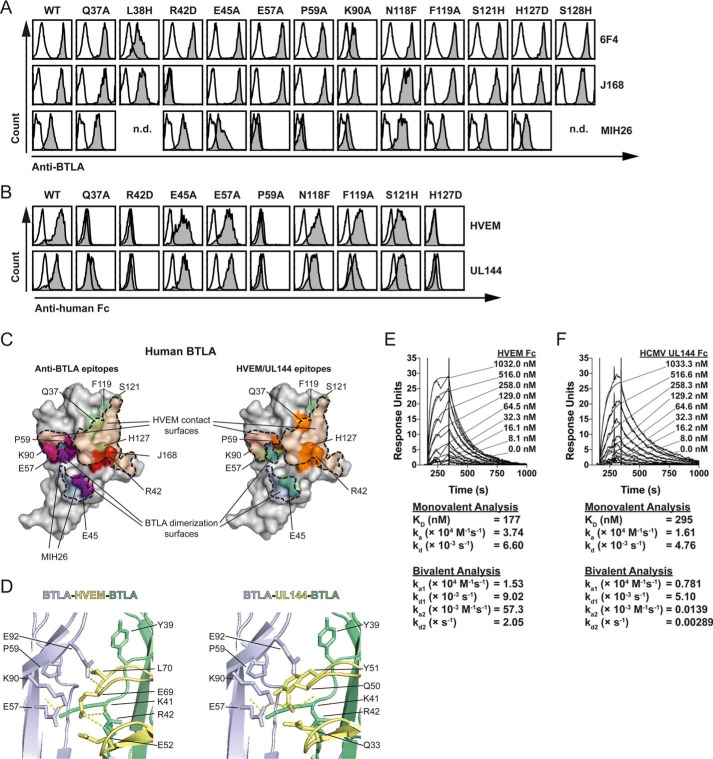
We next compared how HVEM, UL144, or anti-BTLA mAb could each bind BTLA to determine how diverse molecular agonists activate BTLA signaling. Using a panel of human BTLA mutants, we found that binding of the agonistic anti-BTLA mAb (clone MIH26) was disrupted by mutation at either Glu57 (homologous to Gln63 in mouse BTLA that binds the agonistic clone 6A6) or Pro59, whereas binding of the competitive anti-BTLA mAb (clone J168) was disrupted by mutation at Arg42 (Fig. 2, A and C) (21–23). In comparison, we observed similar requirements for binding of both HVEM-Fc and UL144-Fc to Gln37, Arg42, Pro59, and His127, consistent with previous studies in human and mouse BTLA (Fig. 2, B and C) (20, 24).
Figure 2.
Identification of BTLA binding epitopes for HVEM, UL144, and antibodies. A and B, EL4 cells transduced with wild-type or mutant human BTLA were stained with anti-human BTLA polyclonal or monoclonal antibodies or with 10 μg/ml of HVEM-Fc or human CMV UL144-Fc, followed by species specific secondary. A and B, from top to bottom, the graphs show specific MFI staining of 6F4, J168, or MIH26 anti-BTLA (A), or HVEM-Fc or UL144-Fc (B) staining on cells gated on GFP expression. C, two images of BTLA are shown in gray. Binding surfaces of HVEM (orange shade) and a BTLA dimer (blue shade) are predicted using the structure of HVEM and BTLA. Residues required for binding are colored as follows: left, Glu45, Glu57, and Pro59 in blue (required for MIH26 binding) and Arg42 in red (required for J168 binding); right, Gln37, Arg42, Pro59, and His127 in orange (required for HVEM/UL144 binding), Glu45, Glu57, Phe119, and Ser121 in teal (not required for HVEM/UL144 binding). D, the contact region between two BTLA monomers (blue, green) and HVEM (right, Protein Data Bank code 2AW2, yellow) or UL144 (left, modeled as in Fig. 1, yellow) are shown with interacting side chains and hydrogen bonds illustrated. E and F, 2-fold dilutions of human HVEM-Fc (E) or human CMV UL144-Fc (F) were injected over human BTLA-Fc immobilized to dextran sulfate and a control channel in replicate at the indicated concentrations. Traces of 3 min following injection and 15 min of dissociation of the analyte are shown, representative of three experiments. KD and rate constants were calculated by kinetics from modeling a 1:1 monovalent and 1:2 bivalent fit.
The common requirement for Pro59 by endogenous BTLA ligands and the MIH26 antibody indicates that this residue may be required for structural integrity of the BTLA immunoglobulin domain or for direct interactions with ligands. Interestingly, in the BTLA-HVEM co-crystal structure, both Glu57 and Pro59 are located on the distal surface to the HVEM domain where Glu57 shows hydrogen bonds with Lys90 (Fig. 2D) (20). However, in the tetrameric HVEM-BTLA complex, the pyrrolidine ring of Pro59 lies in apposition to the hydrophobic side chain of Leu70 in a secondary HVEM domain. In UL144 this position is replaced by Tyr51, which may favor greater hydrophobic interactions through its aromatic ring to Pro59 in the dimeric BTLA domain (Figs. 1A and 2D). Arg42 of BTLA shows polar contacts with HVEM Glu69 in the canonical BTLA interacting surface. However, in UL144, this position is replaced by Gln50 that is predicted to have polar contacts instead with Lys90 in the dimeric BTLA domain, again potentially strengthening interactions to a BTLA dimer. Thus, although the HVEM-BTLA tetramer was previously reported to be a weak interaction, UL144 may promote stronger higher order complexes (20). Additionally, whereas competition with HVEM is accomplished through blockade of the canonical surface between BTLA and HVEM monomers (J168: Arg42), the agonistic antibody binds the BTLA surface engaged in the HVEM-BTLA tetramer (MIH26: Glu57/Pro59).
We next confirmed similar affinities for BTLA to HVEM- and UL144-Fc using surface plasmon resonance (Fig. 2, E and F). Interestingly, rate constants calculated using a 1:2 model of ligand to analyte binding showed that ka2 and kd2 for UL144-Fc is much lower than for HVEM-Fc binding to BTLA, raising the possibility that UL144-Fc preferentially binds as a monomer to BTLA proteins compared with HVEM-Fc. Because HVEM oligomerization is not changed in the presence of BTLA (25), these affinity measurements indicate that oligomerization of soluble UL144 is also not changed in the presence of BTLA. Together, these data illustrate a common surface of BTLA that is engaged to form BTLA dimers and that may be activated to initiate receptor signaling events.
Comparison of HVEM and UL144 binding to BTLA in trans and in cis
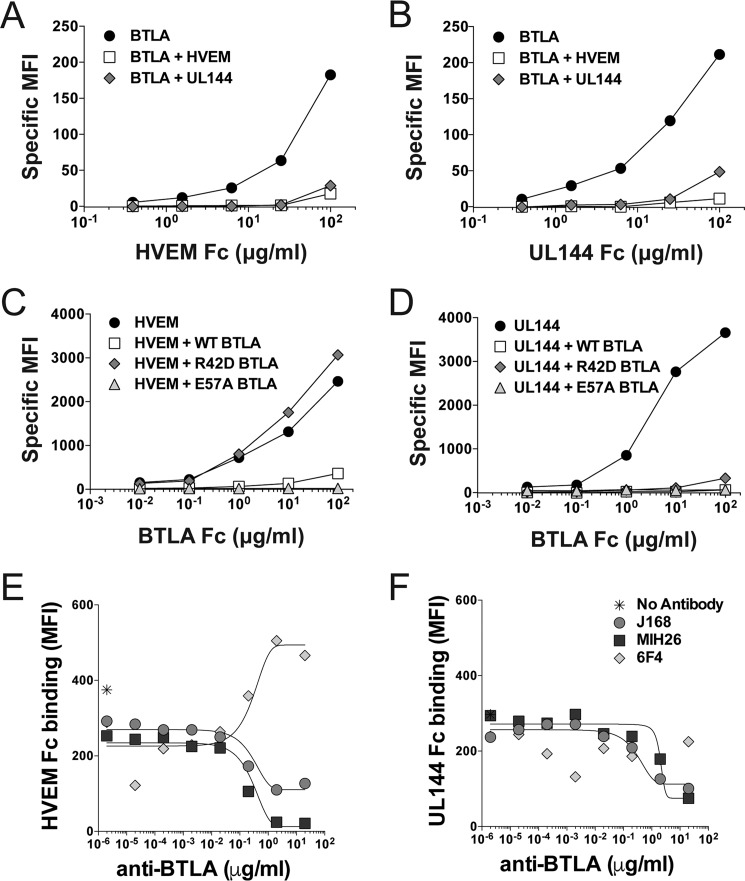
T and B lymphocytes co-express HVEM and BTLA resulting in formation of an intrinsic complex in cis located at the cell surface that competitively blocks agonistic activation by all extrinsic ligands in trans (9). We determined whether UL144 co-expressed with BTLA also formed complexes in cis, similarly engaging the BTLA agonistic surface. HVEM co-expressed with BTLA and UL144 co-expressed with BTLA could not bind soluble HVEM-Fc or UL144-Fc, demonstrating that soluble UL144 could not outcompete a preformed BTLA cis complex at cell surfaces and that HVEM and UL144 form similar complexes with BTLA in cis (Fig. 3, A–D). Interestingly, whereas the BTLA R42D mutant could not bind cis-expressed HVEM as expected, it could complex with UL144, preventing BTLA-Fc binding, indicating that the R42D mutation may not be sufficient to disrupt the BTLA-UL144 cis-complex. These data are consistent with the UL144-BTLA modeled structure in which UL144 does not have direct polar contacts with BTLA Arg42, but instead shows increased polar and hydrophobic interactions with a BTLA dimer domain, resulting in a more stable interaction in cis than between BTLA and HVEM (Fig. 2D).
Figure 3.
BTLA binds HVEM and UL144 in trans and in cis using an overlapping epitope. A and B, 293T cells transfected with BTLA alone or together with HVEM or human CMV UL144 were stained with the indicated concentrations of HVEM-Fc (A) or human CMV UL144-Fc (B). C and D, 293T cells transfected with HVEM (C) or human CMV UL144 (D) alone or together with wild-type, R42D, or E57A BTLA were stained with the indicated concentrations of BTLA-Fc. Graphs show specific MFI for Fc binding to co-expressed cells. E and F, BJAB cells transduced with BTLA were incubated with the indicated concentrations of anti-BTLA prior to staining with 20 μg/ml of HVEM-Fc (E) or human CMV UL144-Fc (F). Graphs show curves of antibody blockade of HVEM and UL144-Fc cell binding.
To confirm our prediction that the binding surfaces between agonistic antibodies and cellular and viral BTLA ligands overlap, we determined whether these antibodies competed with HVEM and UL144 (20). Titration of both J168 and MIH26 anti-BTLA clones interfered with HVEM-Fc and UL144-Fc binding to BTLA, whereas a third anti-BTLA mAb (clone 6F4) enhanced binding of HVEM-Fc to BTLA but did not affect binding of UL144-Fc (Fig. 3, E and F) (15). We could not identify the 6F4 epitope using these BTLA mutants, and this clone does not show any reactivity to HVEM (Fig. 2A; data not shown). Together, these data indicate that a core agonistic surface of BTLA binds HVEM, UL144, and agonistic antibodies. However, there may be additional structural elements that contribute to ligand binding and receptor activation.
Bioengineered HVEM selectively activates BTLA signaling
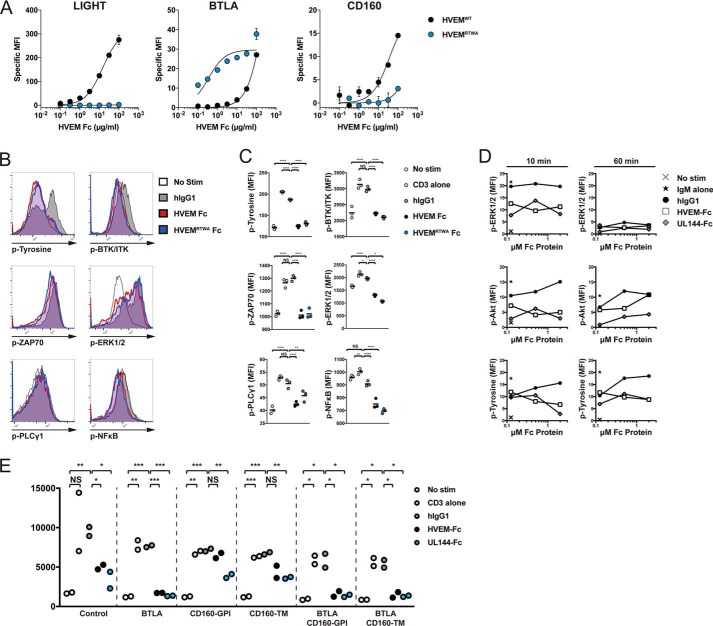
The ligand binding selectivity of UL144 suggested that de novo engineering of HVEM should yield a BTLA specific agonist. Bioengineered HVEM-Fc muteins were created through alanine scanning, saturation, and combinatorial mutagenesis. We found that HVEM-Fc muteins containing S58R and L90A conferred selectivity for BTLA, whereas additional changes at G68T and L70W enhanced BTLA affinity 10-fold, and together in a tetra-mutant (HVEMRTWA), we combined strong affinity for BTLA with loss of binding to LIGHT and CD160 in one protein (Fig. 4A). Interestingly, the L70W mutation introduces an aromatic ring into the BTLA dimer interface similar to Tyr51 in UL144. We next compared inhibitory signaling engaged by the wild-type HVEM-Fc, the engineered HVEM-Fc (HVEMRTWA), and UL144-Fc in a multiparameter phosphoflow assay across activated lymphocyte cell lines. Jurkat cells transduced with ectopic BTLA and CD160 isoforms were activated with immobilized anti-CD3 induced phosphorylation of interleukin-2-inducible T-cell kinase (ITK)/Bruton's tyrosine kinase (BTK), phospholipase C-γ1 (PLCγ1), ζ-chain associated protein kinase 70 kDa (ZAP70)/Syk, ERK1/2, NF-κB p65, and total cellular tyrosines (Fig. 4, B and C). In cells activated with co-immobilized anti-CD3 and a BTLA agonist, phosphorylation of each of these proteins was reduced. We further confirmed BTLA agonistic function in BJAB cells that lack CD160 and that activate Syk-dependent ERK and Akt phosphorylation in response to IgM stimulation (Fig. 4D) (26). These data confirm functional BTLA agonistic activity for each of the HVEM variants.
Figure 4.
A BTLA-selective HVEM mutein inhibits TCR and BCR signaling. A, 293 cells transduced with LIGHT (left panel), CD160 (right panel), or Jurkat cells transduced with BTLA (middle panel) were stained with the indicated concentrations of HVEM-Fc muteins. B, C, and E, JTAg cells expressing BTLA were cultured with microspheres coupled to anti-CD3 with or without Fc proteins for 20 (B and C) or 5 (E) min prior to intracellular staining for phosphotyrosine, phospho-BTK/ITK (Tyr551/Tyr511), phospho-PLCγ1 (Tyr783), phospho-ZAP70/Syk (Tyr319/Tyr352), phospho-ERK1/2 (Thr202/Tyr204), and phospho-NF-κB p65 (Ser529). The histograms in (B) show representative staining for each of the phosphoproteins. The graphs in C show the MFI of the phosphosignals for each of the treatments (geometric mean, representative of at least two experiments). D, BJAB cells were cultured with microspheres coupled to anti-IgM with or without titrated Fc proteins for the indicated times prior to intracellular staining. Graphs show the percentage of cells positive for phosphotyrosine, phospho-ERK1/2 (Thr(P)202/Tyr(P)204), or phospho Akt (Ser473). E, JTag expressing the indicated HVEM ligands cells were stimulated with microspheres as above for 5 min prior to intracellular staining for phospho-ERK1/2 (T202/Y204). Graphs show the phospho-ERK1/2 MFI for each of the treatments (geometric mean, representative of at least two experiments). *, p < 0.05; **, p < 0.01; ***, p < 0.001;****, p < 0.0001.
The expression of CD160 and LIGHT in diverse lymphocyte subsets may influence the capacity of HVEM to engage inhibitory signaling through BTLA. To assess whether altering the expression of HVEM ligands altered inhibitory function, we determined their impact on T cell receptor signaling (Fig. 4E). Ectopic BTLA expression enhanced the ability of HVEM and UL144 to inhibit anti-CD3–activated ERK1/2 phosphorylation to background levels. Importantly, in cells expressing ectopic CD160 (glycophosphoinositide or transmembrane isoforms), HVEM was unable to inhibit ERK1/2 phosphorylation unless BTLA was additionally present. In contrast, UL144 inhibited ERK1/2 phosphorylation regardless of CD160 isoform expression. Together, these data confirm that BTLA engagement by selective agonists potently inhibits antigen receptor signaling proximal to receptor stimulation.
BTLA regulates cytokine signaling in B cells and NK cells
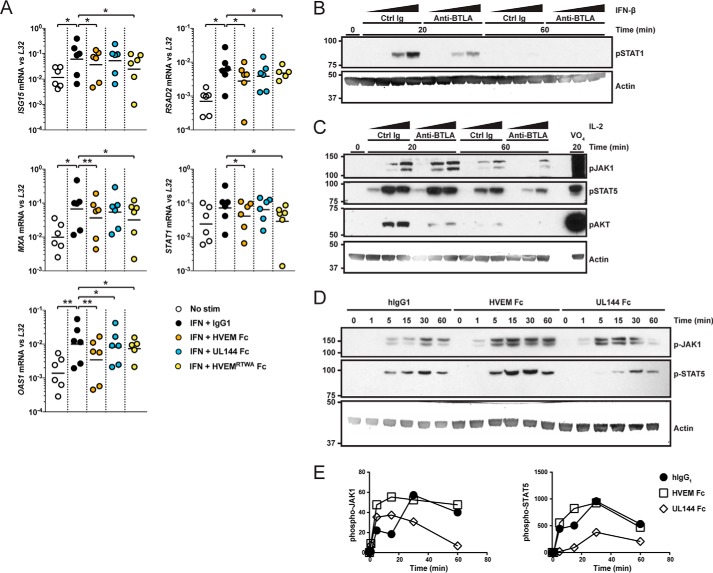
We previously found that BTLA regulated IL-7 signaling in homeostasis and activation of γδ T cells and innate lymphoid cells (17). We next assessed whether BTLA could similarly regulate additional cytokine pathways, such as type I interferon, that can promote adaptive responses and priming of innate cells such as NK, NKT, and γδ T cells (27). In human B cells treated with IFN-β, we found both HVEM-Fc and the HVEMRTWA-Fc protein significantly reduced expression of the type I signature genes ISG15, MXA, OAS1, RSAD2, and STAT1 (Fig. 5A). We observed less significant inhibition using UL144-Fc likely because of reduced affinity of this protein. We confirmed BTLA inhibition of interferon-induced STAT1 phosphorylation in NK92 cells activated with IFN-β using anti-BTLA mAb (clone MIH26) at early and late times (Fig. 5B). We additionally found that this anti-BTLA mAb and the BTLA–specific UL144-Fc protein could reduce IL-2–activated phosphorylation of JAK1, STAT5, and Akt proteins (Fig. 5, C–E). Together, these data demonstrate that BTLA exhibits significant inhibitory function in the SHP-1-sensitive type I interferon signaling pathway in B cells and NK cells. Thus, in addition to regulating T cell receptor signaling, BTLA agonism limits type I interferon activation in lymphocytes including innate cells.
Figure 5.
BTLA agonists inhibit IFN-β and IL-2 signaling. A, purified human B cells were stimulated with human IFN-β and the indicated microspheres coupled to Fc proteins for 6 h prior to harvest. Graphs show the mean relative log-transformed levels of the indicated interferon stimulated genes measured by quantitative real-time PCR from each donor (mean of log-transformed values pooled from three experiments, paired analysis). *, p < 0.05; **, p < 0.01. B and C, NK92 cells were stimulated with titrated human IFN-β (B) and IL-2 (C) at the indicated times after pretreating with anti-BTLA (MIH26) or control (Ctrl) Ig. Western blots show whole cell extracts of phospho-STAT1 to monitor IFN-β signaling, or phospho-JAK1, STAT5, and Akt (Ser473) to monitor IL-2 signaling and actin to control for total protein levels. D and E, NK92 cells were stimulated with IL-2 at the indicated times after pretreating with the indicated Fc and antibodies. Western blots show whole cell extracts of phospho-JAK1, STAT5, and Akt (Ser473) to monitor IL-2 signaling, and actin to control for total protein levels (D). Graphs show quantitation of band intensity normalized to actin (E).
Discussion
We report and compare the structural interactions of HVEM and the CMV UL144 proteins with BTLA and how these interactions activate similar inhibitory signaling pathways. UL144 functions as a structural mimic of HVEM in its interactions with BTLA, including binding an epitope in trans and in cis shared with HVEM. We show that these BTLA agonists inhibit T cell receptor activation affecting proximal (ITK, PLCγ1, ZAP70) and distal (ERK1/2, NFκB) signaling nodes. Importantly, we confirm that through these agonists BTLA inhibits type I interferon and IL-2 signaling in B cells and NK cells, illustrating significant inhibitory function of BTLA in several signaling pathways in lymphocytes. Together, these data illustrate how the potential to limit inflammatory signaling by inhibitory receptors can provide a selective advantage for intracellular pathogens such as viruses. The structural knowledge base of UL144 agonism prompted bioengineering of HVEM to achieve selectivity and high affinity for BTLA, which may show utility in altering inflammatory and proliferative processes.
We confirmed the role of BTLA as an immune checkpoint inhibitor regulating T and B cell receptor signaling (13, 28–30). The mechanism of inhibitory signaling in lymphocytes downstream of BTLA is thought to include activation of SHP-1 or 2 and likely involves additional pathways (29). The inhibitory effect of BTLA agonists on early events in antigen receptor signaling is consistent with previous studies demonstrating that CD3ζ and Syk are direct targets of BTLA activity in T and B cells (28, 30). Although direct targets of BTLA signaling remain challenging to identify, we found the greatest inhibitory effect in the reduction of BTK/ITK phosphorylation by all BTLA agonists. It remains to be determined whether this protein is directly inactivated by SHP-1/2 or through the engagement of unknown inhibitory pathways. The broad inhibitory effect of these HVEM muteins and of our bioengineered mutant in antigen receptor and cytokine signaling highlights their potential as novel anti-inflammatory therapeutics. These endogenously derived proteins may have the additional benefit of reduced antigenicity compared with alternative therapeutic modes.
In activated T cells, HVEM interacts with BTLA, LIGHT, and CD160, whereas in NK cells the abundant expression of CD160 sequesters HVEM, potentially activating pro-inflammatory signaling. In contrast, BTLA-selective UL144 proteins cannot bind CD160 and cannot activate NK cells, and potentially cytolytic CD8+ T cells, whereas retention of BTLA binding activates inhibitory signaling in neighboring cells (11). UL144 was shown to be expressed within an intracellular compartment of latent CMV-infected monocytes (31). However, in human colorectal cancer decreased disease-free survival was associated with CMV infection–expressing group B UL144 proteins, potentially linking UL144 agonism of inhibitory receptors to anti-inflammatory activity (32).
The expression of BTLA, LIGHT, and CD160 varies greatly between different cell types, activation, and differentiation states (11, 33). Thus, dynamic regulation of HVEM ligands provides a mechanism for control of activating and inhibitory signals depending on cellular context. The determination of factors regulating receptor and ligand expression will allow for a better understanding of the role of these proteins in immune responses and how these pathways can be manipulated for therapeutic intervention. Additionally, knowledge of tissue-specific regulation of immune responses by inhibitory receptors will allow for tailored design of anti-inflammatory therapeutics. The development of targeted agonists to BTLA or other inhibitory receptors may thus serve to increase the repertoire of treatments for inflammatory disease.
Materials and methods
Cells and surface protein expression
EL4 and 293T cells were maintained in DMEM with 10% heat-inactivated FBS, antibiotics, l-glutamine, and 50 μm 2-ME. BJAB, Jurkat TAg (JTAg) cells, and their derivatives were maintained in RPMI with 10% heat-inactivated FBS, antibiotics, l-glutamine, and 50 μm 2-ME. EL4 cells and BJAB cells transduced with retroviruses containing wild-type and mutant human BTLA (34) were sorted for GFP expression to increase the frequency of transduced cells. Pseudotyped single infection retrovirus was produced by co-transfection of retroviral plasmid, pCG VSVg envelope protein, and Hit60 gag-pol as previously described (13). UL144 and BTLA mutants were produced by round-the-world PCR mutagenesis. 293T cells transduced by calcium phosphate transfection with pND vector containing mutant or wild-type UL144 were used for Fc binding studies. All oligonucleotides used for PCR amplification and site-directed mutagenesis are listed in supplemental Table S1. For transient expression of HVEM ligands in JTAgs, cells were electroporated with 10 μg of the indicated DNA constructs with control vector at 230 V for 65 ms using a T820 square wave electroporator (BTX®; Harvard Apparatus, Inc., Hollison, MA).
Antibodies and Fc fusion protein production
Anti-human HVEM was from BD Biosciences (San Diego, CA). Anti-human BTLA antibodies clones MIH26 and J168 were obtained from eBioscience (San Diego, CA) and BD Biosciences (San Diego, CA). Anti-human BTLA clone 6F4 and anti-UL144 clone 2F11 were produced as previously described (10, 15). Anti-human CD160 was obtained from R&D Systems (Minneapolis, MN). Donkey anti-human Fcγ and anti-Fcμ F(ab′)2 were from Jackson Immunoresearch (West Grove, PA). Phospho-specific antibodies include phosphotyrosine PE, phospho-Akt (Ser473) PE, and phospho-PLCγ1 (Y783) Alexa-647 (BD Bioscience), phospho-ERK1/2 (Thr(P)202/Tyr(P)204) PerCP-e710, phospho-BTK/ITK (Tyr551/Tyr511) PerCP-e710, and phospho-ZAP70/Syk (Tyr319/Tyr352) APC (eBioscience).
The Fc fusion proteins were designed and produced as follow. The ectodomain (residues 1–184) including the native signal sequence of human HVEM was fused at the 3′ end to the human IgG Fc sequence. For human BTLA and CMV UL144, residues 26–150 and 19–132, respectively, were fused at the 5′ end to the human Ig signal sequence and the 3′ end to the human IgG Fc regions. The Fc fusion proteins were produced in transfected 293T cells grown in CellGro serum-free medium and purified by affinity chromatography on protein A columns. Control human IgG1 protein was from Sigma–Aldrich.
Binding assays
Flow cytometric binding assays were performed as previously described (11). Briefly, cells transduced with the indicated HVEM mutants, UL144 mutants, or HVEM ligands were incubated with Fc ligands for 30 min on ice in buffer (PBS with 2% FBS), washed twice, incubated with anti-Fc secondary for 15 min on ice in buffer, washed twice, and analyzed for ligand binding using flow cytometry. Specific mean fluorescence intensity (MFI) was calculated by subtracting experimental cellular MFI from the cellular MFI of control transduced cells. Binding curves were analyzed using the one site-specific binding module within Prism 7.0a to determine Kd.
Modeling of UL144 structure
The initial model was generated using a combination of homology modeling with Swiss-Model workspace v8.05 and MODELLER v9.18 for the present study (35, 36). The UL144 sequence was aligned to Protein Data Bank template structures (code 2AW2 42% sequence homology selected), and 3D models were generated using Swiss-Model. The best model was identified through evaluation of both overall model quality and maximization of global model quality estimation. Model, template, and sequence alignment were used with MODELLER to optimize flexible non-hydrogen atoms. The model was further improved by refinement cycles and energy minimization in Coot v0.8.8 EL (37). The refined structural model was further optimized using standard methods that control the residue dihedral angles and overall conformation and validated through Ramachandran plot visualization using RAMPAGE (38). The final model was rendered using a PyMOL Molecular Graphics System (version 0.99rc6; Schrödinger).
Surface plasmon resonance kinetic affinity measurement
Human BTLA-Fc ligand was immobilized onto a CM5 sensor chip to 150 response units using amine coupling. Sensograms were collected at a flow rate of 30 μl/min at 25 °C. Specific binding was determined by subtraction of control from ligand channels. Indicated concentrations of analyte were injected from vials cooled to 7 °C. Data collection includes 3 min of 90 μl of analyte followed by 15 min of disassociation. Regeneration after each cycle was with a 30-s pulse of 15 μl of 10 mm glycine (pH 2.5). The first 10 s following analyte injection and disassociation were applied for affinity measurements with both the Langmuir and the Bivalent fit models of BIAevaluation software (version 4.1) kinetic analysis module.
Directed mutagenesis of HVEM
BTLA selective HVEM mutant proteins were engineered de novo through three rounds of mutagenesis as follows: first, alanine mutagenesis of surface exposed residues to identify ligand-binding hot spots; second, saturation mutagenesis at hot spots to optimize targeting of ligand binding; and third, combinatorial mutagenesis to achieve BTLA selectivity and enhance affinity. Candidate mutant HVEM Fc proteins were transiently expressed in 293T cells and first assayed for binding to BTLA, CD160, LIGHT, and Lymphotoxin-α proteins using the Octet Red 384 System (Pall ForteBio, Fremont, CA). HVEM mutations exhibiting BTLA selection were confirmed using BIAcore analysis.
Fc protein activation and phosphoflow analysis
Aldehyde/sulfate latex microspheres (5 μm; Life Technologies) were covalently coupled to 100 μg/ml of anti-CD3 (OKT3; eBioscience) or anti-Fcμ F(ab')2 (Jackson Immunoresearch) alone or with the indicated concentrations of human IgG1, HVEM-Fc, or UL144-Fc as previously described (39). Briefly, microspheres were washed in PBS, incubated with the indicated proteins at 37 °C for 90 min, blocked in buffer (PBS with 1% BSA and 0.1% glycine), washed twice, resuspended in media, and counted. Microspheres were used to stimulate JTag and BJAB cells at a 3:1 ratio. Microspheres were added first to 96-well flat-bottomed plates followed by cells washed in PBS and resuspended to a concentration of 1.5 × 106 cells/ml. The plates were briefly spun and incubated for the indicated times at 37 °C followed by fixation with 2% paraformaldehyde in PBS and additional incubation at 37 °C for 10 min. The cells were washed in buffer (PBS with 2% FBS) and permeabilized in Perm III buffer (BD Bioscience) for 30 min on ice, washed twice, incubated with phospho-specific antibodies for 30 min on ice, washed twice, and analyzed.
Human B cell purification and interferon-stimulated gene expression analysis
Fresh human blood was collected and from healthy donors giving written informed consent at the Scripps Research Institute Normal Blood Donor Service, and all handling was approved by the Sanford Burnham Prebys Medical Discovery Institute Internal Review Board. Samples were mixed 1:1 with PBS and overlaid onto Ficoll (GE Healthcare) for density gradient centrifugation. PBMC were isolated from buffy coats and washed twice with PBS. B cells were further purified using an EasySep human B-cell enrichment kit (STEMCELL Technologies, Vancouver, Canada). The cells resuspended to 106 cells/ml in RPMI 1640 supplemented with 10% heat-inactivated FBS, antibiotics, l-glutamine, and 50 μm 2-ME were co-cultured with Fc protein-coupled microspheres produced as described above and with 1 unit/ml of recombinant human interferon-β (R&D Systems, Minneapolis, MN) for 6 h at 37 °C. Total RNA was prepared using the Qiagen RNeasy mini kit (Qiagen), and RNA was reverse-transcribed into cDNA using the iScriptTM cDNA synthesis kit (Bio-Rad) according to the manufacturer's instructions. Relative expression of Interferon stimulated genes was detected using intron-spanning primer pairs ISG15 (TTTGCCAGTACAGGAGCTTGTG, GGGTGATCTGCGCCTTCA), MXA (TTCAGCACCTGATGGCCTATC, GTACGTCTGGAGCATGAAGAACTG), OAS1 (TGCGCTCAGCTTCGTACTGA, GGTGGAGAACTCGCCCTCTT), RSAD2 (GTGGTTCCAGAATTATGGTGAGTATTT, CCACGGCCAATAAGGACATT), STAT1 (TGCATCATGGGCTTCATCAGC, GAAGTCAGGTTCGCCTCCGTTC), IRF7 (GCTCCCCACGCTATACCATCTAC, GCCAGGGTTCCAGCTTCAC), and IFNB (ACAGACTTACAGGTTACCTCCGAAAC, CTCCTAGCCTGTCCCTCTGGGACTGG) compared with L32 (GGATCTGGCCCTTGAACCTT, GAAACTGGCGGAAACCCA), and relative expression was measured using SYBR Green Master Mix (Bio-Rad). Reactions were carried out in clear 384 plates using an ABI® 7900HT real-time PCR system (Thermo Fisher Scientific, Waltham, MA). Relative expression was calculated as follows: 2−ΔCt, where Ct = cycle number, ΔCt = Ct(target gene) − Ct(L32).
Western analysis
The IL-2-dependent NK92 model cell line was activated using the indicated concentrations of interferon-β or recombinant human IL-2 (Miltenyi, San Diego, CA). In experiments testing IL-2 responses, NK92 cells were cultured overnight without IL-2 to re-establish basal signaling. In experiments using fusion proteins and antibodies, NK92 cells were coated with control human IgG1, HVEM-Fc, or UL144-Fc or control mouse IgG2a or anti-BTLA for at least 15 min on ice prior to activation. NK92 cells were aliquoted to 2 × 106 cells per condition in 100 μl and activated at 37 °C for the indicated times, quenched with ice-cold PBS, and lysed in radioimmune precipitation assay buffer at 4 °C for 20 min followed by centrifugation at 14,000 rpm, 4 °C. Extracts were boiled in SDS loading buffer containing 1% β-mercaptoethanol for 5 min and resolved by SDS-PAGE on 10% Bis-Tris gels (Bio-Rad). Proteins were transferred using a tank method to PVDF membrane and blocked with 1% ovalbumin in TBS-T buffer, and blotted with antibodies against phospho-JAK1, phospho-STAT5, phospho-STAT1, phospho-Akt (Ser473) (Cell Signaling, Danvers, MA), and total actin (Sigma–Aldrich), followed by anti-rabbit HRP (GE Healthcare) or anti-mouse HRP and visualized by enhanced chemiluminescence (Thermo Scientific, Rockford, IL).
Statistical analysis
HVEM ligand-binding curves, Kd values, and standard error were calculated from specific MFI using one site-specific binding module in Prism 7 for Mac OS X v7.0a. HVEM and UL144 Fc BIAcore affinity measurements were made during the first 10 s following analyte injection and disassociation using both the Langmuir fit model of BIAevaluation software (version 4.1) kinetic analysis module. In phosphoflow assays conditions, containing triplicate measures were compared against each other using unpaired Student's t test with equal S.D.
Author contributions
J. R. S. designed and performed the experiments and wrote the manuscript. M. O. B., B. C. W., and W. S. performed experiments. P. S. N., B. R. M., and D. A. designed and produced protein constructs. I. N. modeled the human CMV UL144 structure. C. F. W. designed the experiments and wrote the manuscript.
Supplementary Material
Acknowledgments
We thank Yoav Altman and Amy Cortez of the Flow Cytometry Shared Resources facility at Sanford Burnham Prebys Medical Discovery Institute and Rachel Roach, Jackie Vekich, and Mike Favis at Pfizer Centers for Therapeutic Innovation for technical assistance.
This work was supported by National Institutes of Health Grants AI048073, AI067890, CA164679, and CA177322 (to C. F. W.), a gift from the Perkins Family Foundation (to C. F. W.), American Heart Association Grant AHA-14BGIA20380277 (to J. R. S.), National Psoriasis Foundation Grant NPF2015 (to J. R. S.), Slovak Research and Development Agency Grant APVV-14-0839 (to I. N.), the Scientific Grant Agency of the Slovak Republic Grant 2/0103/15 (to I. N.), and a research contract from Pfizer Centers for Therapeutic Innovation. The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
This article contains supplemental Table S1.
- CMV
- cytomegalovirus
- HVEM
- herpesvirus entry mediator
- BTLA
- B and T lymphocyte attenuator
- NK
- natural killer
- SHP
- SH2 domain–containing protein tyrosine phosphatase
- ITK
- interleukin-2-inducible T-cell kinase
- BTK
- Bruton's tyrosine kinase
- PLCγ1
- phospholipase C-γ1
- ZAP70
- ζ-chain associated protein kinase 70 kDa
- 2-ME
- 2-mercaptoethanol
- MFI
- mean fluorescence intensity.
References
- 1. Reddehase M. J. (2002) Antigens and immunoevasins: opponents in cytomegalovirus immune surveillance. Nat. Rev. Immunol. 2, 831–844 [DOI] [PubMed] [Google Scholar]
- 2. Sedý J. R., Spear P. G., and Ware C. F. (2008) Cross-regulation between herpesviruses and the TNF superfamily members. Nat. Rev. Immunol. 8, 861–873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Alcami A. (2003) Viral mimicry of cytokines, chemokines and their receptors. Nat. Rev. Immunol. 3, 36–50 [DOI] [PubMed] [Google Scholar]
- 4. Murphy T. L., and Murphy K. M. (2010) Slow down and survive: enigmatic immunoregulation by BTLA and HVEM. Annu. Rev. Immunol. 28, 389–411 [DOI] [PubMed] [Google Scholar]
- 5. Pardoll D. M. (2012) The blockade of immune checkpoints in cancer immunotherapy. Nat. Rev. Cancer 12, 252–264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Callahan M. K., Wolchok J. D., Allison J. P., and Sharma P. (2013) Immune co-signaling to treat cancer. In Cancer Immunotherapy, pp. 211–280, Springer-Verlag, New York Inc., New York [Google Scholar]
- 7. Šedý J., Bekiaris V., and Ware C. F. (2014) Tumor necrosis factor superfamily in innate immunity and inflammation. Cold Spring Harb. Perspect. Biol. 7, a016279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Basso K., and Dalla-Favera R. (2015) Germinal centres and B cell lymphomagenesis. Nat. Rev. Immunol. 15, 172–184 [DOI] [PubMed] [Google Scholar]
- 9. Ware C. F., and Sedý J. R. (2011) TNF superfamily networks: bidirectional and interference pathways of the herpesvirus entry mediator (TNFSF14). Curr. Opin. Immunol. 23, 627–631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Cheung T. C., Humphreys I. R., Potter K. G., Norris P. S., Shumway H. M., Tran B. R., Patterson G., Jean-Jacques R., Yoon M., Spear P. G., Murphy K. M., Lurain N. S., Benedict C. A., and Ware C. F. (2005) Evolutionarily divergent herpesviruses modulate T cell activation by targeting the herpesvirus entry mediator cosignaling pathway. Proc. Natl. Acad. Sci. U.S.A. 102, 13218–13223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Šedý J. R., Bjordahl R. L., Bekiaris V., Macauley M. G., Ware B. C., Norris P. S., Lurain N. S., Benedict C. A., and Ware C. F. (2013) CD160 activation by herpesvirus entry mediator augments inflammatory cytokine production and cytolytic function by NK cells. J. Immunol. 191, 828–836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Cheung T. C., Steinberg M. W., Oborne L. M., Macauley M. G., Fukuyama S., Sanjo H., D'Souza C., Norris P. S., Pfeffer K., Murphy K. M., Kronenberg M., Spear P. G., and Ware C. F. (2009) Unconventional ligand activation of herpesvirus entry mediator signals cell survival. Proc. Natl. Acad. Sci. U.S.A. 106, 6244–6249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Sedy J. R., Gavrieli M., Potter K. G., Hurchla M. A., Lindsley R. C., Hildner K., Scheu S., Pfeffer K., Ware C. F., Murphy T. L., and Murphy K. M. (2005) B and T lymphocyte attenuator regulates T cell activation through interaction with herpesvirus entry mediator. Nat. Immunol. 6, 90–98 [DOI] [PubMed] [Google Scholar]
- 14. Cai G., Anumanthan A., Brown J. A., Greenfield E. A., Zhu B., and Freeman G. J. (2008) CD160 inhibits activation of human CD4+ T cells through interaction with herpesvirus entry mediator. Nat. Immunol. 9, 176–185 [DOI] [PubMed] [Google Scholar]
- 15. Cheung T. C., Oborne L. M., Steinberg M. W., Macauley M. G., Fukuyama S., Sanjo H., D'Souza C., Norris P. S., Pfeffer K., Murphy K. M., Kronenberg M., Spear P. G., and Ware C. F. (2009) T cell intrinsic heterodimeric complexes between HVEM and BTLA determine receptivity to the surrounding microenvironment. J. Immunol. 183, 7286–7296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kobayashi Y., Iwata A., Suzuki K., Suto A., Kawashima S., Saito Y., Owada T., Kobayashi M., Watanabe N., and Nakajima H. (2013) B and T lymphocyte attenuator inhibits LPS-induced endotoxic shock by suppressing Toll-like receptor 4 signaling in innate immune cells. Proc. Natl. Acad. Sci. U.S.A. 110, 5121–5126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Bekiaris V., Šedý J. R., Macauley M. G., Rhode-Kurnow A., and Ware C. F. (2013) The inhibitory receptor BTLA controls γδ T cell homeostasis and inflammatory responses. Immunity 39, 1082–1094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Tu T. C., Brown N. K., Kim T. J., Wroblewska J., Yang X., Guo X., Lee S. H., Kumar V., Lee K. M., and Fu Y. X. (2015) CD160 is essential for NK-mediated IFN-γ production. J. Exp. Med. 212, 415–429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Sharma P., and Allison J. P. (2015) The future of immune checkpoint therapy. Science 348, 56–61 [DOI] [PubMed] [Google Scholar]
- 20. Compaan D. M., Gonzalez L. C., Tom I., Loyet K. M., Eaton D., and Hymowitz S. G. (2005) Attenuating lymphocyte activity: the crystal structure of the BTLA-HVEM complex. J. Biol. Chem. 280, 39553–39561 [DOI] [PubMed] [Google Scholar]
- 21. Otsuki N., Kamimura Y., Hashiguchi M., and Azuma M. (2006) Expression and function of the B and T lymphocyte attenuator (BTLA/CD272) on human T cells. Biochem. Biophys. Res. Commun. 344, 1121–1127 [DOI] [PubMed] [Google Scholar]
- 22. Albring J. C., Sandau M. M., Rapaport A. S., Edelson B. T., Satpathy A., Mashayekhi M., Lathrop S. K., Hsieh C. S., Stelljes M., Colonna M., Murphy T. L., and Murphy K. M. (2010) Targeting of B and T lymphocyte associated (BTLA) prevents graft-versus-host disease without global immunosuppression. J. Exp. Med. 207, 2551–2559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hurchla M. A., Sedy J. R., Gavrieli M., Drake C. G., Murphy T. L., and Murphy K. M. (2005) B and T lymphocyte attenuator exhibits structural and expression polymorphisms and is highly Induced in anergic CD4+ T cells. J. Immunol. 174, 3377–3385 [DOI] [PubMed] [Google Scholar]
- 24. Nelson C. A., Fremont M. D., Sedy J. R., Norris P. S., Ware C. F., Murphy K. M., and Fremont D. H. (2008) Structural determinants of herpesvirus entry mediator recognition by murine B and T lymphocyte attenuator. J. Immunol. 180, 940–947 [DOI] [PubMed] [Google Scholar]
- 25. Liu W., Vigdorovich V., Zhan C., Patskovsky Y., Bonanno J. B., Nathenson S. G., and Almo S. C. (2015) Increased heterologous protein expression in Drosophila S2 cells for massive production of immune ligands/receptors and structural analysis of human HVEM. Mol. Biotechnol. 57, 914–922 [DOI] [PubMed] [Google Scholar]
- 26. Gobessi S., Laurenti L., Longo P. G., Sica S., Leone G., and Efremov D. G. (2007) ZAP-70 enhances B-cell-receptor signaling despite absent or inefficient tyrosine kinase activation in chronic lymphocytic leukemia and lymphoma B cells. Blood 109, 2032–2039 [DOI] [PubMed] [Google Scholar]
- 27. Gajewski T. F., Schreiber H., and Fu Y. X. (2013) Innate and adaptive immune cells in the tumor microenvironment. Nat. Immunol. 14, 1014–1022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Vendel A. C., Calemine-Fenaux J., Izrael-Tomasevic A., Chauhan V., Arnott D., and Eaton D. L. (2009) B and T lymphocyte attenuator regulates B cell receptor signaling by targeting Syk and BLNK. J. Immunol. 182, 1509–1517 [DOI] [PubMed] [Google Scholar]
- 29. Chen L., and Flies D. B. (2013) Molecular mechanisms of T cell co-stimulation and co-inhibition. Nat. Rev. Immunol. 13, 227–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Wu T. H., Zhen Y., Zeng C., Yi H. F., and Zhao Y. (2007) B and T lymphocyte attenuator interacts with CD3zeta and inhibits tyrosine phosphorylation of TCRzeta complex during T-cell activation. Immunol. Cell Biol. 85, 590–595 [DOI] [PubMed] [Google Scholar]
- 31. Poole E., Walther A., Raven K., Benedict C. A., Mason G. M., and Sinclair J. (2013) The myeloid transcription factor GATA-2 regulates the viral UL144 gene during human cytomegalovirus latency in an isolate-specific manner. J. Virol. 87, 4261–4271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Chen H. P., Jiang J. K., Chan C. H., Teo W. H., Yang C. Y., Chen Y. C., Chou T. Y., Lin C. H., and Chan Y. J. (2015) Genetic polymorphisms of the human cytomegalovirus UL144 gene in colorectal cancer and its association with clinical outcome. J. Gen. Virol. 96, 3613–3623 [DOI] [PubMed] [Google Scholar]
- 33. Murphy K. M., Nelson C. A., and Sedý J. R. (2006) Balancing co-stimulation and inhibition with BTLA and HVEM. Nat. Rev. Immunol. 6, 671–681 [DOI] [PubMed] [Google Scholar]
- 34. Watanabe N., Gavrieli M., Sedy J. R., Yang J., Fallarino F., Loftin S. K., Hurchla M. A., Zimmerman N., Sim J., Zang X., Murphy T. L., Russell J. H., Allison J. P., and Murphy K. M. (2003) BTLA is a lymphocyte inhibitory receptor with similarities to CTLA-4 and PD-1. Nat. Immunol. 4, 670–679 [DOI] [PubMed] [Google Scholar]
- 35. Arnold K., Bordoli L., Kopp J., and Schwede T. (2006) The SWISS-MODEL workspace: a web-based environment for protein structure homology modelling. Bioinformatics 22, 195–201 [DOI] [PubMed] [Google Scholar]
- 36. Webb B., and Sali A. (2014) Comparative protein structure modeling using MODELLER. Curr. Protoc. Bioinformatics 47, 5.6.1–32 [DOI] [PubMed] [Google Scholar]
- 37. Emsley P., Lohkamp B., Scott W. G., and Cowtan K. (2010) Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr 66, 486–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Lovell S. C., Davis I. W., Arendall W. B. 3rd, de Bakker P. I., Word J. M., Prisant M. G., Richardson J. S., and Richardson D. C. (2003) Structure validation by Cα geometry: φ, ψ and Cβ deviation. Proteins 50, 437–450 [DOI] [PubMed] [Google Scholar]
- 39. Sperling A. I., Sedy J. R., Manjunath N., Kupfer A., Ardman B., and Burkhardt J. K. (1998) TCR signaling induces selective exclusion of CD43 from the T cell-antigen-presenting cell contact site. J. Immunol. 161, 6459–6462 [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.