Abstract
Enzymes of the glycoside hydrolase family 42 (GH42) are widespread in bacteria of the human gut microbiome and play fundamental roles in the decomposition of both milk and plant oligosaccharides. All GH42 enzymes characterized so far have β-galactosidase activity. Here, we report the existence of a GH42 subfamily that is exclusively specific for α-l-arabinopyranoside and describe the first representative of this subfamily. We found that this enzyme (BlArap42B) from a probiotic Bifidobacterium species cannot hydrolyze β-galactosides. However, BlArap42B effectively hydrolyzed paeonolide and ginsenoside Rb2, plant glycosides containing an aromatic aglycone conjugated to α-l-arabinopyranosyl-(1,6)-β-d-glucopyranoside. Paeonolide, a natural glycoside from the roots of the plant genus Paeonia, is not hydrolyzed by classical GH42 β-galactosidases. X-ray crystallography revealed a unique Trp345-X12-Trp358 sequence motif at the BlArap42B active site, as compared with a Phe-X12-His motif in classical GH42 β-galactosidases. This analysis also indicated that the C6 position of galactose is blocked by the aromatic side chains, hence allowing accommodation only of Arap lacking this carbon. Automated docking of paeonolide revealed that it can fit into the BlArap42B active site. The Glcp moiety of paeonolide stacks onto the aromatic ring of the Trp252 at subsite +1 and C4-OH is hydrogen bonded with Asp249. Moreover, the aglycone stacks against Phe421 from the neighboring monomer in the BlArap42B trimer, forming a proposed subsite +2. These results further support the notion that evolution of metabolic specialization can be tracked at the structural level in key enzymes facilitating degradation of specific glycans in an ecological niche.
Keywords: carbohydrate metabolism, crystallography, enzyme structure, glycobiology, glycoside hydrolase, CAZyme, beta-galactosidase, bifidobacterium, microbiota
Introduction
The complex microbial consortium in the human gastrointestinal tract, referred to as the gut microbiota, has an increasingly recognized vital role in human health (1). Human diet is rich in a wide variety of non-digestible saccharides from plants and animals, and glycan metabolism is a pivotal factor shaping the dynamics and the evolution of gut microbiota (2). Specific taxa gain a competitive edge in fitness in this highly competitive niche through metabolic specialization, which is reflected by their enzymatic machinery being fine-tuned for specific glycans (3). Glycans containing β-galactoside are abundant in human infant and adult nutrition (4–6), and hydrolysis of the β-galactosidic bond by β-galactosidases is a prerequisite for their utilization.
Key enzymes in the hydrolysis and utilization of β-galactosides in gut microbes assign into glycoside hydrolase (GH)3 family 42 in the sequence-based classification system of the Carbohydrate Active enZymes (CAZy) database (7). Genomes of probiotic strains from the Bifidobacterium genus often encode several GH42 enzymes (8) with distinct subspecificities matching diversity and abundance of β-galactosides. For example, Bifidobacterium longum subsp. infantis encodes three GH42 β-galactosidases related to the utilization of the human milk oligosaccharide lacto-N-tetraose (LNT), β-galactosides from bovine milk, and arabinogalactan, respectively (9). A similar specialization exists in Bifidobacterium breve that encodes two GH42 β-galactosidases, one targeting LNT (10) and the other plant galactan (11). Notably, B. bifidum utilizes mucin-derived β-galactosides from the glycoprotein layer coating the human host in epithelial colonocytes (12), which is proposed to be possible through a key GH42 β-galactosidase (13).
The genome of Bifidobacterium animalis subsp. lactis Bl-04 encodes two GH42 enzymes (14). Galacto-oligosaccharides (GOS) were shown to up-regulate gene locus balac_0848 (15), experimentally verified to be a β(1,6)/β(1,3)-galactosidase (BlGal42A) (13). The second GH42 (BlArap42B) gene with locus tag balac_0053 was not differentially up-regulated by GOS (15), and belongs to a distinct, not previously characterized clade of bifidobacterial GH42 enzymes (16).
This distinct clade is here shown to form a novel GH42 subfamily present throughout the bacterial kingdom, which seems to have exclusive α-l-arabinopyranoside specificity as evaluated for the soluble intracellular BlArap42B using a panel of p-nitrophenyl (pNP)-substrates and oligosaccharides. BlArap42B effectively hydrolyzed paeonolide (Fig. 1A), a plant glycoside that contains a non-reducing end α-l-arabinopyranoside and is found in the roots of the widespread plant genus Paeonia (17). This apparent metabolic specialization could be tracked at the structural level, as the crystal structure of BlArap42B suggested GH42 members with α-l-arabinopyranosidase activity recognize their substrates through an active site Trp345–Trp358 motif as compared with a Phe–His motif in classical GH42 β-galactosidases.
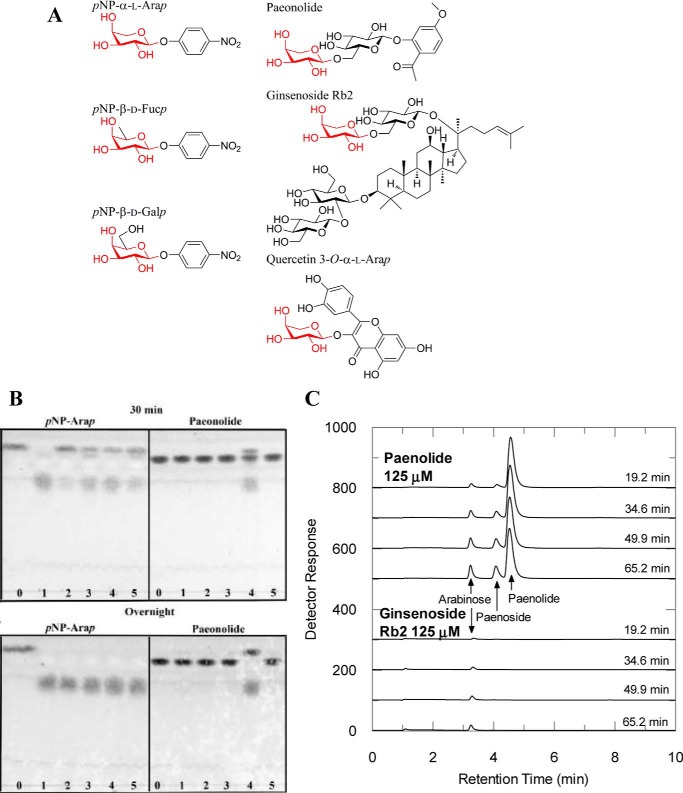
Figure 1.
A, chemical structures of substrates; B, TLC analysis on the hydrolysis of pNP-α-l-Arap and paeonolide by BlArap42B and other GH42 enzymes; and C, HPAEC-PAD analysis on the hydrolysis of plant glycosides containing a non-reducing end α-l-arabinopyranoside by BlArap42B. A, structures of pNP-α-l-Arap, pNP-β-d-Fucp, pNP-β-d-Galp, paeonolide, ginsenoside Rb2, and quercetin 3-O-α-l-Arap are shown. pNP-α-l-Arap and pNP-β-d-Fucp differ from pNP-β-d-Galp by lacking C6 and C6-OH, respectively. Non-reducing end α-l-arabinopyranose is found in 2-acetyl-5-methoxyphenyl-α-l-Arap-(1,6)-β-d-Glcp (paeonolide) from Paeonia, ginsenoside Rb2 from P. ginseng, and quercetin 3-O-α-l-Arap from P. guajava Linn. B, substrate activity screening using TLC of BlArap42B (lane 4) and previously characterized bifidobacterial GH42 β-galactosidases, Bga42A (lane 1), Bga42B (lane 2), and Bga42C (lane 3) from B. longum subsp. Infantis, ATCC 15697 as well as BlGal42A (lane 5) from B. animalis subsp. lactis Bl-04 (lane 0) are controls without enzyme. BlArap42B (land 4) displayed significant activity toward paeonolide by 30 min incubation, with complete hydrolysis overnight. Bga42A, Bga42B, Bga42C, and BlGal42A did not show significant activity toward paeonolide after 24 h incubation, despite their ability to hydrolyze pNP-α-l-Arap. C, HPAEC-PAD chromatograms showing activity toward paeonolide and ginsenoside Rb2. Paeonoside is the de-arabinosylated product of paeonolide. Activity was not detected toward quercetin 3-O-α-l-Arap. Peaks corresponding to the substrate ginsenoside Rb2 and its de-arabinosylated product (= ginsenoside Rd) could not be detected in the elution range under the conditions employed (see “Experimental procedures”).
Results
Biochemical properties
The recombinant BlArap42B was produced and purified to electrophoretic homogeneity. BlArap42B migrated as a single band in SDS-PAGE with the expected theoretically calculated molecular mass of 78.3 kDa. Analytical gel filtration data showed that the protein eluted as a single peak corresponding to 201 kDa. Although this is smaller than the theoretically calculated size for the trimer (235 kDa), GH42 enzymes, which are generally trimeric, usually exhibit similar discrepancy between their size estimations using gel filtration chromatography and crystallography (18). BlArap42B showed significant activity toward pNP-α-l-arabinopyranoside (pNP-α-l-Arap), very low activity toward pNP-β-d-galactopyranoside (pNP-β-d-Galp) and pNP-β-d-fucopyranoside (pNP-β-d-Fucp), and no detected activity on pNP-α-l-arabinofuranoside (pNP-α-l-Araf), pNP-β-d-xylopyranoside (pNP-β-d-Xylp), and pNP-β-d-glucopyranoside (pNP-β-d-Glcp) (Table 1). The kinetics of hydrolysis of pNP-α-l-Arap were best described by Michaelis-Menten kinetics with kcat = 160 ± 10 s−1 and Km = 3.4 ± 0.4 mm. RiArap42B, a homologue from Roseburia intestinalis M50/1 (Firmicutes isolated from human gut), sharing 42% sequence identity with BlArap42B (Actinobacteria), has a similar activity profile on aryl substrates (Table 1). BlArap42B displays significant activity toward 2-acetyl-5-methoxyphenyl-α-l-Arap-(1,6)- β-d-Glcp (paeonolide, Fig. 1A) in a 30-min assay, with complete hydrolysis by overnight incubation as verified by TLC (Fig. 1B). We also tested the activity of BlArap42B on plant glycosides containing a terminal α-l-arabinopyranoside group, paeonolide, ginsenoside Rb2, and quercetin 3-O-α-l-Arap (Fig. 1A), by high-performance anion-exchange chromatography with pulsed amperometric detection (HPAEC-PAD) (Fig. 1C). Ginsenoside Rb2 is a triterpene saponin isolated from the root of Panax ginseng (19), whereas quercetin 3-O-α-l-Arap is a flavonoid isolated from the aerial parts (leaves) of Alchemilla xanthochlora (Lady's mantle) (20) and Psidium guajava L. (guava) (21). BlArap42B hydrolyzed paeonolide and ginsenoside Rb2 (Fig. 1C) but not quercetin 3-O-α-l-Arap (data not shown). Kinetic parameters toward the plant glycosides indicate that these substrates exhibit significantly lower Km (<0.1 mm) and higher catalytic efficiencies (>1500 mm−1 s−1) compared with those of pNP-α-l-Arap (Table 2). By contrast, no activity was observed on an extensive series of β-galactosides from plants and mammals, including human milk (the substrates are listed under “Experimental procedures”). Bga42A (Blon_2016), Bga42B (Blon_2123), and Bga42C (Blon_2416) from B. longum subsp. infantis ATCC 15697, and BlGal42A from B. animalis subsp. lactis Bl-04, which are distantly related to BlArap42B (see Fig. 4), did not cleave paeonolide, although they were able to slowly hydrolyze pNP-α-l-Arap (Fig. 1B).
Table 1.
Specific activity (units mg−1 = μmol min−1 mg−1 mg−1) of BlArap42B from B. animalis subsp. lactis BL-04 (wild-type and mutants) and RiArap42B from R. intestinalis M50/1 toward 5 mmpNP-substrates
The activity was measured at 37 °C in 40 mm sodium citrate (pH 6.5) and 0.005% Triton X-100. All enzymes including the mutants showed no detectable activity toward pNP-β-d-Glcp, pNP-α-l-Araf, and pNP-β-d-Xylp.
| Substrate |
BlArap42B |
RiArap42B | |||
|---|---|---|---|---|---|
| Wild-type | W345F | W358H | W345F/W358H | ||
| pNP-α-l-Arap | 79.0 ± 0.7 | 2.7 ± 0.1 | 5.9 ± 0.1 | 0.1 ± 0.0 | 55.2 ± 2.0 |
| pNP-β-d-Galp | 0.1 ± 0.0 | NDa | NDa | NDa | 0.1 ± 0.0 |
| pNP-β-d-Fucp | 1.9 ± 0.0 | NDa | 0.7 ± 0.0 | NDa | 2.6 ± 0.1 |
a ND, not detected (<0.05 units mg−1).
Table 2.
Kinetic parameters of BlArap42B on plant glycosides and pNP-αl-Arap
| Substrate | Km | kcat | kcat/Km |
|---|---|---|---|
| mm | s−1 | mm−1 s−1 | |
| Paeonolidea | 0.074 ± 0.007 | 240 ± 10 | 3,200 ± 200 |
| Ginsenoside Rb2a | 0.051 ± 0.018 | 79 ± 8 | 1,500 ± 400 |
| pNP-α-l-Arapb | 3.4 ± 0.4 | 160 ± 10 | 46 ± 0.3 |
a Measured at 30 °C in 40 mm sodium citrate (pH 6.5).
b Measured at 37 °C in 40 mm sodium citrate (pH 6.5) and 0.005% Triton X-100.
Figure 4.
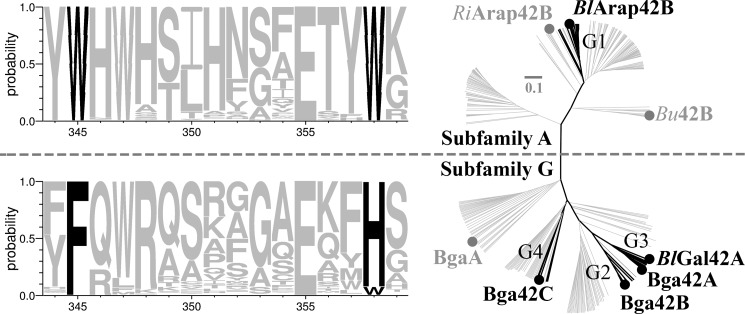
BlArap42B defines a novel GH42 α-l-arabinopyranosidase subfamily. The phylogenetic tree of GH42 enzymes, divides the family into the two distinct α-l-arabinopyranosidases (subfamily A) and β-galactosidases (subfamily G) subfamilies, respectively. Bifidobacterial GH42 enzymes (black) from the CAZy database reveals four distinct groups, with functional assignment for groups G2, G3, and G4. Group G1 was previously uncharacterized and multiple sequence alignment shows this group is diverse and comprises a unique Trp-X12-Trp sequence motif as compared with the Phe-X12-His sequence motif of groups G2, G3, and G4. Approximately 200 non-identical sequences (gray) found throughout the bacterial kingdom from each of the two Trp-X12-Trp and Phe-X12-His sequence motifs were merged with the bifidobacterial GH42 sequences and previously characterized GH42 enzymes (gray) to yield the phylogenetic tree.
Three-dimensional structure of BlArap42B
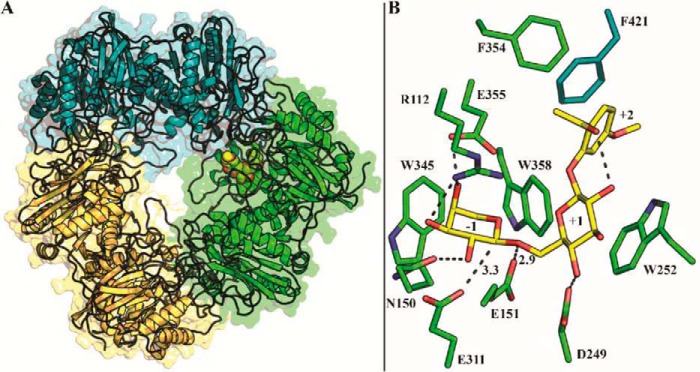
The three-dimensional structure of BlArap42B in its ligand-free form was determined by X-ray crystallography at 2.0-Å resolution with six chains (two trimers) in the asymmetric unit. Data collection, refinement, and stereochemical statistics are summarized in Table 3. BlArap42B is a homotrimer of a 701-residues subunit, with three subunits related by a 3-fold axis (Fig. 2A). Each subunit consists of three domains. Domain A is a (β/α)8 barrel containing the catalytic residues. Domain B is a structural domain that packs onto Domain A of the adjacent monomer. Domain C, whose role is unknown, adopts an anti-parallel β-sandwich fold. The first seven residues (Met1–Asp7) and the region Pro663–Thr672 in all six chains, as well as a variable number of residues in the C terminus of each chain (Gly695–Asn701), were disordered and not included in the final model.
Table 3.
Data collection and refinement statistics for BlArap42B
Numbers in parentheses are for the highest resolution shell.
| Data collection | BlArap42B |
|---|---|
| Beamline | KEK-PF BL5A |
| Resolution (Å) | 50.00–2.00 (2.03–2.00) |
| Space group | P41212 |
| Unit cell parameters (Å) | a = b = 177.95, c = 375.70 |
| No. of reflections | 5,987,058 |
| No. of unique reflections | 400,874 (19,842) |
| Mean I/σ (I) | 27.7 (3.8) |
| R-sym (%) | 12.7 (50.2) |
| Completeness (%) | 100.0 (100.0) |
| Redundancy | 14.9 (14.6) |
| Refinement | |
| Resolution (Å) | 50.00–2.00 |
| R-factor (%) | 15.5 |
| R-free (%) | 19.1 |
| No. of atoms | |
| Protein atoms (non-hydrogen) | 32,478 |
| Water molecules | 3,330 |
| R.m.s. deviation bond lengths (Å) | 0.022 |
| R.m.s. deviation bond angles (°) | 1.972 |
| Average B-factors (Å2) | |
| Protein atoms | 26.3 |
| Water | 47.9 |
| Ramachandran plot (%) | |
| Favored regions | 96.0 |
| Allowed regions | 3.8 |
| Disallowed regions | 0.2 |
| PDB code | 5XB7 |
Figure 2.
Overall structure and docking of paeonolide into BlArap42B. A, top view along the axis of the BlArap42B trimer with docked paeonolide shown as spheres in one of the three active sites. B, active site residues of BlArap42B shown with docked paeonolide. The Arap moiety binds at subsite −1 with distances of 3.3 Å between the catalytic nucleophile Glu311 and the anomeric carbon, and 2.9 Å between the catalytic acid/base Glu151 and the glycosidic oxygen. Glcp participates in an aromatic stacking interaction with the proposed subsite +1 platform Trp252 and C4-OH hydrogen bonds to Asp249. Paeonolide in this conformation can form an intramolecular hydrogen bond between its acetyl oxygen and C2-OH of the Glcp. The neighboring monomer participates in determining the substrate specificity by Phe421 engaged in aromatic stacking with the phenyl group of paeonolide at a possible subsite +2.
BlArap42B active-site architecture and ligand docking
Similar to structures of GH42 β-galactosidases, the active sites and the vicinity encompassing the proposed subsites +1 and +2 are formed at the interface of two adjacent monomers in the BlArap42B trimer (18). It was possible to dock an energetically preferred conformation of paeonolide, the preferred substrate for BlArap42B, into the active site with an estimated affinity of −8.8 kcal mol−1 (Fig. 2B). The docked paeonolide molecule makes an intramolecular hydrogen bond between the acetyl oxygen of the aglycon (Fig. 1A) and C2-OH of the Glcp. The Arap ring binds at subsite −1, with distances of 3.3 Å between the catalytic nucleophile Glu311 and the anomeric carbon and 2.9 Å between the catalytic acid/base Glu151 and the glycosidic oxygen, respectively. The Glcp ring bound at subsite +1, is stacking onto Trp252, and C4-OH is hydrogen bonded with Asp249. Furthermore, the aromatic ring of the aglycone and Phe421 from the neighboring monomer are almost parallel, making an aromatic stacking at this position, indicative of the presence of a subsite +2.
Comparison with the BlGal42A β-galactosidase structure
The structure of BlArap42B was compared with that of the other GH42 enzyme from the same organism (BlGal42A) that has specificity toward β(1,6)/β(1,3)-galactoside linkages and is one of the best characterized enzymes (both biochemically and structurally) among GH42 β-galactosidases (13). Fig. 3A shows superimposition of the structures of BlArap42B and BlGal42A in complex with galactose (PDB code 4UNI). From the structural comparison, Glu151 and Glu311 of BlArap42B were inferred as the acid/base catalyst and the catalytic nucleophile, respectively. Although BlArap42B and BlGal42A only share 25% sequence identity their overall structures are very similar as reflected by the root mean square deviation (r.m.s. deviation) for Cα atoms of 1.75 Å when chain A (689/695 residues) of BlGal42A is superimposed on chain A (682/701 residues) of BlArap42B.
Figure 3.
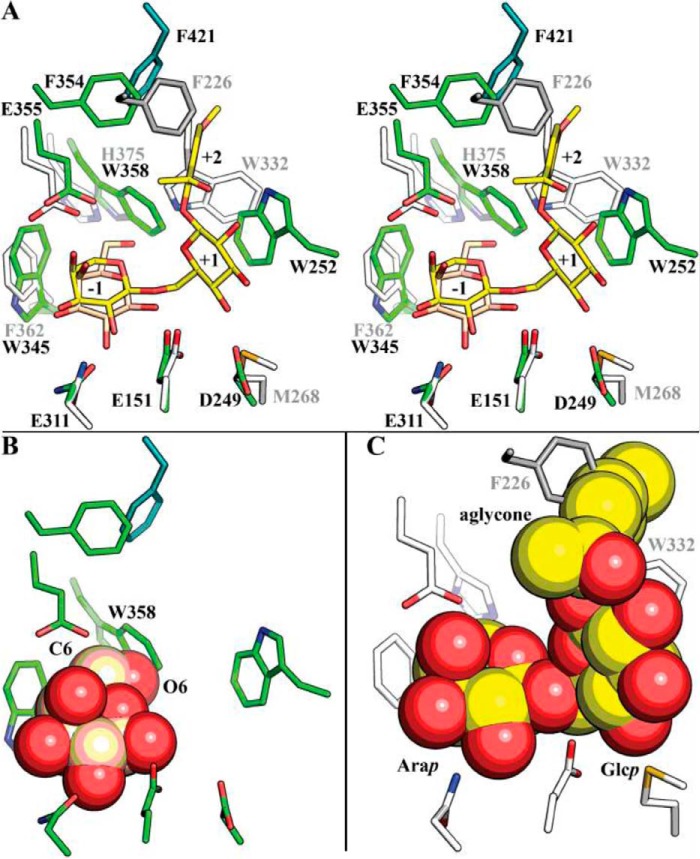
Structural comparison of BlArap42B α-l-arabinopyranosidase and BlGal42A β-galactosidase. A, stereo view of the comparison of the active site with subsites −1 through +2 of BlArap42B (green) and the BlGal42A β-galactosidase (white and gray) in complex with galactose from B. animalis subsp. lactis Bl-04 (PDB code 4UNI). Trp345 and Trp358 occupy the active site environment in BlArap42B, as compared with the smaller side chains of Phe362 and His375 in BlGal42A. The subsite +1 aromatic stacking platform is composed of Trp residues from different loops (Trp252 in BlArap42B and Trp332 in BlGal42A) and are spatially differently located in the two enzymes. B, the residues of subsite −1 of BlArap42B were overlaid with galactose from BlGal42A (sphere). The Trp358 of BlArap42B, which is the smaller His in BlGal42A, is clashing with the C6-OH of galactose in complex with BlGal42A, explaining the lack of activity toward β-galactosides of BlArap42B. C, active site residues of BlGal42A with paeonolide docked from BlArap42B are shown as spheres. Both the side chains of Trp332 and Phe226 at subsite +1 and +2 sterically hinder the docking of paeonolide, in agreement with the lack of activity for BlGal42A.
Remarkably, three residues at the substrate-binding site are either variant or spatially differently located in BlArap42B as compared with BlGal42A (Fig. 3A). Trp345 is phenylalanine in BlGal42A (Phe362) and the aromatic side chain is involved in substrate recognition in subsite −1 by forming a hydrophobic platform for the C4 side of the arabino- or galactopyranoside. The change of a histidine in BlGal42A (His375) to Trp358 restricts the space in subsite −1 around the C6–O6 hydroxymethyl group of galactoside, which is not present in arabinopyranoside (Fig. 3B). Moreover, Trp332 of BlGal42A makes a hydrogen bond to the C6–OH of galactose, and is also proposed to mediate aromatic stacking to substrate at subsite +1 in GH42 β-galactosidases. In contrast, the conformation of the loop containing Trp332 is different in BlArap42B and locates a tryptophan at the corresponding position in the sequence (Trp320, not shown in Fig. 3A), distantly from the active site in space. Instead, a differently located Trp252, which is not conserved in previously characterized GH42 enzymes, is taking the role of stacking platform at subsite +1 in BlArap42B. Additionally, Met268 in BlGal42A replaces Asp249 that in the docked structure of BlAra42B forms a hydrogen bond to the Glcp C4-OH in paeonolide.
Mutational analysis
From the structural comparison with BlGal42A, two non-conserved residues (Trp345 and Trp358) are identified in subsite −1. To examine the role of these residues in relationship to substrate specificity, these single residues were replaced using site-directed mutagenesis with those found in the β-galactosidase (Phe or His). Table 1 shows specific activities of the single (W345F and W358H) and double (W345F/W358H) mutants toward pNP-substrates, the single mutants having more than 10-fold reduced activity on pNP-α-l-Arap and the double mutant being almost inactive. W358H was slightly active on pNP-β-d-Fucp, but none of the mutants showed detectable activity on pNP-β-d-Galp or any of the other pNP-substrates. Clearly, the substrate specificity of the GH42 α-l-arabinopyranosidase was not changed to β-galactosidase by these simple mutations.
Phylogeny and active site motifs of GH42
A phylogenetic and active site sequence motif analysis of GH42 α-l-arabinopyranosidases (subfamily A) and β-galactosidases (subfamily G), divides GH42 into two subfamilies, respectively (Fig. 4). BlArap42B assigns into a distinct uncharacterized clade, group one (G1) in the phylogenetic tree. Based on sequence analysis, it is evident that the two tryptophan residues (Trp345 and Trp358) in BlArap42B are conserved in G1 and subfamily A, and comprise a unique Trp-X12-Trp sequence motif through the active site as compared with subfamily G that includes G2–G4. All of the G2–G4 groups of subfamily G contain at least one characterized β-galactosidase that has a Phe-X12-His motif. Trp252 of BlArap42B in subsite +1 is conserved in G1 but not in G2–G4 (data not shown).
Discussion
BlArap42B is a novel GH42 α-l-arabinopyranosidase
The GH42 family has been assumed very homogenous because all hitherto characterized members exhibited β-galactosidase activity despite representing broad diverse taxa. This further indicated active site conservation within the family, where evolution of the molecular machinery was driven toward subtle changes in the plus subsites for fine-tuning subspecificities, e.g. for β(1,6)-, β(1,4)-, or β(1,3)-linked galactosides or N-acetylglucosides (22–25).
Although GH42 BlArap42B from B. animalis subsp. lactis Bl-04 was unable to hydrolyze synthetic β-galactosides and those from plants, mammalian milk, and host mucin, so far demonstrated as substrates for GH42 β-galactosidases, it was able to hydrolyze, the different yet structurally similar, α-l-arabinopyranosides (Tables 1 and 2 and Fig. 1). BlArap42B effectively released α-l-arabinopyranose from paeonolide and ginsenoside Rb2, both containing an aglycone conjugated to α-l-Arap-(1,6)-β-d-Glcp, but was unable to hydrolyze quercetin 3-O-α-l-Arap. The subsite +1 is apparently specific for the β(1,6)-linked glucoside with Trp252 as a sugar-stacking residue (Fig. 3A), and aromatic residues surrounding this site (Trp358, Phe354, and Phe421) probably make steric clash with the large flavonoid group of quercetin 3-O-α-l-Arap. Thus, BlArap42B have the capabilities to discriminate substrates beyond subsite −1 and demonstrate the potential existence of different α-l-arabinopyranoside subspecificities. Previously characterized bifidobacterial GH42 β-galactosidases tested here did not cleave paeonolide, although they were able to hydrolyze pNP-α-l-Arap (Fig. 1B). BlArap42B is clearly a GH42 α-l-arabinopyranosidase, distinctly different from previously characterized GH42 enzymes. The kcat (240 s−1) and Km (0.074 mm) values of BlArap42B toward its preferred substrate, paeonolide, are in the same range as those of previously characterized GH42 enzymes toward their preferred substrates (13, 24).
Structural specificity determinants in GH42 α-l-arabinopyranosidases
The residues creating the spatial and chemical environment at subsite −1 are invariant in characterized GH42 β-galactosidases (13, 18, 22, 26, 27). However, the change from a histidine residue in classical GH42 β-galactosidases to Trp358 in BlArap42B limits the space at subsite −1, which would cause clashing with C6 of galactose in accordance with BlArap42B being unable to hydrolyze β-galactosides (Fig. 3B). A similar structural change is observed between GH27 α-d-galactosidases and β-l-arabinopyranosidases, the only other similar described activity. In GH27 β-l-arabinopyranosidases, a mutagenesis study revealed that the single substitution from an aspartic acid to the slightly larger glutamic acid in the catalytic pocket around the C-6 of galactose as compared with GH27 α-d-galactosidases is critical for modulating the enzyme activity (28). However, a similar attempt to change the specificity of BlArap42B from α-l-arabinopyranosidase to β-galactosidase through the single and double mutations of Trp345 and Trp358 failed (Table 1). This may suggest that the active site of this enzyme is optimized to α-l-arabinopyranoside through accumulation of other substitutions around subsite −1 during the molecular evolution after it diverged from the β-galactosidases.
Notably, a major change is observed in the positioning of the subsite +1 stacking platform and potential recognition of hydroxyl groups between BlArap42B and BlGal42A (Fig. 3A). Superimposing the docked paeonolide from BlArap42B into BlGal42A shows the phenyl aglycone clashes with the backbone of Trp332, which is invariant and similarly spatially located in structurally characterized GH42 enzymes (13, 18, 22, 26, 27), and Phe226 of the neighboring monomer in BlGal42A, which can explain its lack of activity for paeonolide (Fig. 3C). The present findings indicate that GH42 α-l-arabinopyranosidases have evolved from a common scaffold to target specific α-l-arabinopyranosides present in plant oligosaccharides.
BlArap42B defines a novel GH42 α-l-arabinopyranosidase subfamily
Applying a phylogenetic and sequence analysis approach links the α-l-arabinopyranoside specificity of BlArap42B with a unique Trp-X12-Trp sequence motif at the active site, as opposed to Phe-X12-His in classical bifidobacterial GH42 β-galactosidases (Fig. 4). The Trp-X12-Trp sequence motif is present in enzymes with a GH42 catalytic domain from more than 150 different bacterial species and subspecies of various phyla, and uncovers a novel α-l-arabinopyranosidase subfamily (subfamily A) distinct from GH42 β-galactosidases (subfamily G). As a confirmation that the subfamily division reflects specificity, we have shown that RiArap42B, a homologue and subfamily A member from a human gut bacterium, R. intestinalis M50/1 (29), shows the same activity profile on pNP-derived substrates, hydrolyzing pNP-α-l-Arap, but not pNP-β-d-Galp (Table 1), supporting the exclusive activity toward α-l-arabinopyranoside of this subfamily. Noticeably, BgaA from Clostridium cellulovorans (30), belonging to subfamily G, contains a Phe-X12-Trp-mixed sequence motif and is the only α-l-arabinopyranosidase currently assigned to GH42 in the CAZy database. C. cellulovorans BgaA, however, hydrolyzes pNP-β-d-Galp with about 10% activity of pNP-α-l-Arap (30) as opposed to BlArap42B that shows <0.15% activity of pNP-α-l-Arap on pNP-β-d-Galp.
Subspecificities within the α-l-arabinopyranosidase subfamily A
Distinct groups clearly exist within subfamily A (Fig. 4), with members of different groups organized in different gene landscapes. The gene encoding BlArap42B is located adjacent to a gene encoding a putative GH30 enzyme, with no close characterized homologues based on sequence similarity, as well as in the near vicinity of a putative GH3 enzyme (BlGH3). The closest characterized homologue to BlGH3 is the GH3 Apy-H1 from B. longum H-1, showing 61% sequence identity and 74% similarity, and dual α-l-arabinopyranosidase/β-galactosidase specificity, releasing α-l-arabinopyranoside from ginsenoside Rb2 found in Panax ginseng (31). Interestingly, BlArap42B is also able to release α-l-arabinopyranoside from ginsenoside Rb2 with high efficiency. This gene cluster in B. animalis subsp. lactis Bl-04 likely targets α-l-arabinopyranosides found in plants of especially Asian origin (17, 32). Similarly, a GH3 member is encoded adjacent to the gene of RiArap42B in R. intestinalis M50/1, but it clusters with characterized xylan β(1,4)-xylosidases based on a phylogenetic analysis of this family (data not shown).
The genes surrounding the gene encoding a putative GH42 α-l-arabinopyranosidase from Bacteroides uniformis ATCC 8492 (Bu42B) of subfamily A (Fig. 4) contains all glycoside hydrolase-encoding genes necessary for complete utilization of xyloglucan oligosaccharides (33), suggesting that α-l-arabinopyranosides exist in xyloglucan, but this has yet to be identified. Generally, α-l-arabinopyranoside is sparsely reported in plant oligosaccharides, which is in sharp contrast to the evident appearance of BlArap42B homologues in different gene landscapes found throughout the bacterial kingdom. GH42 α-l-arabinopyranosidases therefore may prove useful in the identification of α-l-arabinopyranosides in plant glycans, oligosaccharides, and glycoconjugates.
α-l-Arabinopyranosidase activity in the human gut niche
Recently, the human gut microbe Bacteroides thetaiotaomicron was shown to be capable to completely depolymerize the highly complex pectic Rhamnogalacturonan II, containing buried α-l-Arap-(1,4)-β-d-Galp, through the novel BT0983 GH2 α-l-arabinopyranosidase (34). Additionally, the human gut microbe B. breve K-110, isolated from healthy adults, can utilize ginsenoside Rb2 (35). However, the internal amino acid sequence (VIYLTDA) of the purified α-l-arabinopyranosidase from B. breve K-110 match with an AAA+ family ATPase but not with any GHs in the genome sequences, suggesting that the sequence was derived from a contaminated protein. Because ginsenoside Rb2 was effectively hydrolyzed by BlArap42B, B. animalis subsp. lactis Bl-04 is potentially conferred with the ability to utilize this compound. The present study reports the enzymology and structure of a GH42 α-l-arabinopyranosidase from the probiotic B. animalis subsp. lactis Bl-04, uncovering the existence of a distinct GH42 subfamily, with novel specificity toward α-l-arabinopyranosides and identifying a unique associated active site sequence motif. The molecular insights together with a deep phylogenetic analysis identify key structural elements discriminating the GH42 α-l-arabinopyranosidases from the classical GH42 β-galactosidases. The phylogenetic analysis uncovered the existence of an unexpectedly large number (>150) of potential GH42 α-l-arabinopyranosidases in bacterial genomes. It is noteworthy that the plant glycosides effectively hydrolyzed by BlArap42B, paeonolide, and ginsenoside Rb2, are contained in traditional herbal medicines in East Asian countries, such as Cortex moutan (Mu Dan Pi in Chinese and Botanpi in Japanese) and Asian ginseng, suggesting that other edible root vegetables may also contain certain amounts of glycosides with an α-l-arabinopyranoside. Altogether, these results indicate that α-l-arabinopyranoside metabolism evolved in different ecological niches and probably is widespread in important probiotic members of the human gut microbiota.
Experimental procedures
Substrates
pNP-β-d-galactopyranoside (pNP-β-d-Galp), pNP-β-d-glucopyranoside (pNP-β-d-Glcp), pNP-α-l-arabinofuranoside (pNP-α-l-Araf), pNP-α-l-arabinopyranoside (pNP-α-l-Arap), pNP-β-d-fucopyranoside (pNP-β-d-Fucp), and pNP-β-d-xylopyranoside (pNP-β-d-Xylp) were purchased from Sigma. 4-β-Galactobiose, 6-β-galactobiose, and LacNAc were purchased from Dextra Laboratories (Readings, UK). Lacto-N-biose I and galacto-N-biose were synthesized as described previously (36, 37). LNT and lacto-N-neotetraose (LNnT) were purchased from Elicityl (Crolles, France). 2-Acetyl-5-methoxyphenyl α-l-Arap-(1,6)-β-d-Glcp (paeonolide), ginsenoside Rb2, and quercetin 3-O-α-l-Arap were purchased from Tokyo Chemical Industry (Tokyo, Japan), Carbosynth (Berkshire, UK), and eNovation Chemicals LLC (Bridgewater, NJ), respectively.
Expression and purification
The gene encoding a putative GH42 enzyme (locus tag: balac_0053, GenBankTM accession number ACS45449.1) was amplified from B. animalis subsp. lactis Bl-04 genomic DNA by PCR using High-fidelity DNA polymerase (Fermentas, St. Leon-Rot, Germany) with the following primers: 5′-CTA GCT AGC GCC CGC GCA TAC ACC-3′; reverse: 5′-ATA AGA ATG CGG CCG CAT TCA TTG TGG GTT GC-3′. Amplified DNA was cloned into pET21(a)+ (Novagen, Darmstadt, Germany) using the NheI and NotI restriction sites (underlined) resulting in BlArap42B with a C-terminal His tag. Escherichia coli BL21(DE3) ΔlacZ(β-Gal) containing the above expression plasmid was grown at 20 °C in 1 liter of Luria-Bertani medium containing 50 μg ml−1 of ampicillin and 34 μg ml−1 of chloramphenicol to an A600 of 0.5, and expression was induced by 100 μm isopropyl β-thiogalactopyranoside for 36 h at 18 °C. The recombinant enzyme (BlArap42B) was purified using a 5-ml HisTrap HP column (GE Healthcare) with a linear gradient 2.5–100% 400 mm imidazole (20 mm HEPES buffer, 0.5 m NaCl, pH 7.5). The His-purified fractions were pooled and purified by gel filtration (HiLoad 26/60 Superdex G200; GE Healthcare) in 10 mm MES-NaOH, 150 mm NaCl (pH 6.5) eluted by 1.2 column volumes of this buffer at a flow rate of 1 ml min−1. The purity of the enzyme was analyzed by SDS-PAGE. The enzyme concentration was determined spectrophotometrically (ϵ280 = 152,890 m−1 cm−1). The W345F and W358H and double W345F/W358H mutants were constructed using the QuikChange® Site-directed Mutagenesis Kit (Stratagene), and the recombinant proteins were prepared and purified as described for the wild-type. The gene with locus tag Roi_37780 from R. intestinalis M50/1 (GenBank accession number CBL10553.1) encoding a homologue of BlArap42B was synthesized with a C-terminal His tag and cloned into pET21(a)+ (GenScript), expressed, and the resulting recombinant protein (RiArap42B) was purified as described above for BlArap42B. Bga42A, Bga42B, and Bga42C from B. longum subsp. infantis ATCC 15697, and BlGal42A from B. animalis subsp. lactis Bl-04 were prepared as previously described (13, 24).
Biochemical characterization
Activity was assayed toward pNP-β-d-Galp, pNP-α-l-Arap, pNP-β-d-Fucp, pNP-α-l-Araf, pNP-β-d-Glcp, and pNP-β-d-Xylp (final concentration 5 mm) at 37 °C in 40 mm sodium citrate, 0.005% Triton X-100, pH 6.5 (50 μl) by addition of enzyme (2–2,000 nm) and stopping the reaction after 10 min by 1 m Na2CO3 (200 μl) for BlArap42B wild-type and mutants, and RiAra42B. The amount of released pNP was measured spectrophotometrically at A410 using pNP (0–2 mm) as standard. One unit of activity was defined as the amount of enzyme that released 1 μmol of pNP min−1. The kinetic parameters kcat and Km were determined from initial rates of pNP-α-l-Arap (0.5–7.5 mm) hydrolysis in the above buffer by non-linear regression fit of the Michaelis-Menten model: v = kcat × [E] × [S]/(Km + [S]) to data from triplicate experiments (GraphPad Prism 6, La Jolla, CA). Activity of BlArap42B as well as Bga42A, Bga42B, and Bga42C from B. longum subsp. infantis ATCC 15697, and BlGal42A from B. animalis subsp. lactis Bl-04 (75–125 nm) were measured toward 4 mm pNP-α-l-Arap and 4 mm paeonolide in 50 mm sodium phosphate (pH 6.5) at 30 °C for 30 min and 24 h and monitored by TLC. HPAEC-PAD experiments were carried out using a Dionex ICS-3000 HPLC system (Chromeleon software version 6.80, Dionex) with a Dionex Carbopac PA20 column. The analytes were separated using 40 mm NaOH in isocratic mode at 0.5 ml min−1. Initial hydrolysis activities were measured using 20 nm BlArap42B at 30 °C in 40 mm sodium citrate (pH 6.5) toward paeonolide, ginsenoside Rb2, and quercetin 3-O-α-l-Arap (100 μm) in 19.2–65.2 min assays. The kinetic parameters kcat and Km were determined from initial rates of paeonolide, and ginsenoside Rb2 (7.812–1000 μm) using 40 pm BlArap42B at 30 °C in the above buffer at 4 time points by integrating the area of the peaks corresponding to released l-arabinose.
Crystallization, data collection, and structure determination of BlArap42B
BlArap42B was concentrated to 12.0 mg ml−1 in 10 mm MES-NaOH (pH 6.5), 150 mm NaCl and screened for initial crystallization conditions at 20 °C with the JCSG core I-IV screens (Qiagen). Crystals were observed in the JCSG core IV screen (1.5 m ammonium sulfate, 12% glycerol (v/v), 0.1 m Tris, pH 8.5). The final crystallization condition was 12% glycerol, 1.6 m ammonium sulfate, 0.1 m MES-NaOH buffer (pH 6.5) obtained from optimization in 24-well VDX trays (Hampton Research) in sitting drops containing 1 μl of protein stock and 1 μl of reservoir at 20 °C. Glycerol (20% final concentration) was added as cryoprotectant before harvesting.
A data set to 2.0-Å resolution was obtained at the BL5A beamline, Photon Factory, Tsukuba, Japan. Processing and scaling of the data were done with HKL2000 (38). The space group was determined to be P41212 with six molecules in the asymmetric unit. Molecular replacement was performed using Balbes (39) with a chain of the homotrimer of the PDB entry code 1KWG (18) as the initial search model. ARP/wARP (40) was used to partially build the structure, and refinement was completed using Coot (41) and Refmac5 (42). The quality of the structures, including Ramachandran statistics, was verified by MolProbity (43), and PyMOL version 1.8.5 (Schrödinger, LLC, New York) was used for structural analysis and rendering of molecular graphics.
Ligand preparation and docking
Paeonolide was prepared from the PubChem project (CID: 92043525) and PCModel version 9.20 (Serena Software) using energy minimization with MMX force field. The energy minimized molecule was docked into the active site of BlArap42B using AutoDock Vina 1.1.2 (44) with the grid box (16 Å × 16 Å × 16 Å) centered on the scissile glycosidic bond oxygen. The ligand structure was docked with flexible torsion angles, whereas the protein structure was fixed.
Bioinformatics analysis
The amino acid sequences of bifidobacterial GH42 members were extracted from the CAZy database (7) using CAZy Tools and aligned using MUSCLE version 3.8 with default settings (45). The active site sequence motif (WHWHSIHNSFETYW) for group one (G1) and (FQWRQSRGGAEKFH) for group two, three, and four (G2, G3, and G4) were queried against the NCBI nr-database using BLASTP. Non-identical sequences were extracted and the redundancy was decreased by USEARCH (46) to yield ∼200 sequences from each of the two sequence motifs, which were merged with the bifidobacterial entries and previously characterized GH42 enzymes. Sequence motif logos for the subfamilies were rendered by WebLogo 3 (47). ClustalW2 was used to render a phylogenetic tree with default settings, which was visualized in Dendroscope 3.0 (48).
Author contributions
A. H. V., M. A. H., and B. S. conceived the study. A. H. V. conducted most of the experiments and analyzed the results. A. H. V. wrote the manuscript with contributions from S. F. T. K. conducted TLC experiments on the hydrolysis of paeonolide. T. A. and S. F. assisted in X-ray data collection, structure determination and analysis. S. F. performed molecular replacement and ligand docking. L. L. L. assisted in preliminary crystallization trials, structural analysis and verification. T. K., M. A. H., M. K., and B. S. assisted in experimental design and analysis of the data. All authors reviewed the results and approved the final version of the manuscript.
Acknowledgments
We thank Dr. M. Hidaka and the staff of the Photon Factory and SPring-8 for the X-ray data collection. We thank Prof. Harry Brumer for laboratory facilities, as well as Dr. Shaheen Shojania and Nicholas McGregor for laboratory assistance.
This work was supported by a Grant-in-Aid for Japan Society for the Promotion of Science (JSPS) Research Fellowship (to A. H. V.), and JSPS KAKENHI Grant Numbers 15H02443 and 26660083 (to S. F.), the Platform for Drug Discovery, Informatics, and Structural Life Science funded by MEXT, and a Technical University of Denmark Ph.D. fellowship (to A. H. V.). The authors declare that they have no conflicts of interest with the contents of this article.
The atomic coordinates and structure factors (code 5XB7) have been deposited in the Protein Data Bank (http://wwpdb.org/).
- GH
- glycoside hydrolase
- CAZy
- Carbohydrate Active enZymes
- GNB
- galacto-N-biose
- GOS
- galacto-oligosaccharide
- HPAEC-PAD
- high-performance anion-exchange chromatography with pulsed amperometric detection
- LNnT
- lacto-N-neotetraose
- LNT
- lacto-N-tetraose
- pNP
- p-nitrophenyl
- r.m.s. deviation
- root mean square deviation
- PDB
- Protein Data Bank.
References
- 1. Nicholson J. K., Holmes E., Kinross J., Burcelin R., Gibson G., Jia W., and Pettersson S. (2012) Host-gut microbiota metabolic interactions. Science 336, 1262–1267 [DOI] [PubMed] [Google Scholar]
- 2. Koropatkin N. M., Cameron E. A., and Martens E. C. (2012) How glycan metabolism shapes the human gut microbiota. Nat. Rev. Microbiol. 10, 323–335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Peacock K. S., Ruhaak L. R., Tsui M. K., Mills D. A., and Lebrilla C. B. (2013) Isomer-specific consumption of galactooligosaccharides by bifidobacterial species. J. Agric. Food Chem. 61, 12612–12619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Han N. S., Kim T.-J., Park Y.-C., Kim J., and Seo J.-H. (2012) Biotechnological production of human milk oligosaccharides. Biotechnol. Adv. 30, 1268–1278 [DOI] [PubMed] [Google Scholar]
- 5. Ridley B. L., O'Neill M. A., and Mohnen D. (2001) Pectins: structure, biosynthesis, and oligogalacturonide-related signaling. Phytochemistry 57, 929–967 [DOI] [PubMed] [Google Scholar]
- 6. Vincken J. P., Schols H. A., Oomen R. J., McCann M. C., Ulvskov P., Voragen A. G., and Visser R. G. (2003) If homogalacturonan were a side chain of rhamnogalacturonan I. Implications for cell wall architecture. Plant Physiol. 132, 1781–1789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Lombard V., Golaconda Ramulu H., Drula E., Coutinho P. M., and Henrissat B. (2014) The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res. 42, D490–D495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Sela D. A., Chapman J., Adeuya A., Kim J. H., Chen F., Whitehead T. R., Lapidus A., Rokhsar D. S., Lebrilla C. B., German J. B., Price N. P., Richardson P. M., and Mills D. A. (2008) The genome sequence of Bifidobacterium longum subsp. infantis reveals adaptations for milk utilization within the infant microbiome. Proc. Natl. Acad. Sci. U.S.A. 105, 18964–18969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Viborg A. H., Katayama T., Abou Hachem M., Andersen M. C., Nishimoto M., Clausen M. H., Urashima T., Svensson B., and Kitaoka M. (2014) Distinct substrate specificities of three glycoside hydrolase family 42 β-galactosidases from Bifidobacterium longum subsp. infantis ATCC 15697. Glycobiology 24, 208–216 [DOI] [PubMed] [Google Scholar]
- 10. James K., Motherway M. O., Bottacini F., and van Sinderen D. (2016) Bifidobacterium breve UCC2003 metabolises the human milk oligosaccharides lacto-N-tetraose and lacto-N-neo-tetraose through overlapping, yet distinct pathways. Sci. Rep. 6, 38560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. O'Connell Motherway M., Fitzgerald G. F., and van Sinderen D. (2011) Metabolism of a plant derived galactose-containing polysaccharide by Bifidobacterium breve UCC2003. Microb. Biotechnol. 4, 403–416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Turroni F., Bottacini F., Foroni E., Mulder I., Kim J.-H., Zomer A., Sánchez B., Bidossi A., Ferrarini A., Giubellini V., Delledonne M., Henrissat B., Coutinho P., Oggioni M., Fitzgerald G. F., et al. (2010) Genome analysis of Bifidobacterium bifidum PRL2010 reveals metabolic pathways for host-derived glycan foraging. Proc. Natl. Acad. Sci. U.S.A. 107, 19514–19519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Viborg A. H., Fredslund F., Katayama T., Nielsen S. K., Svensson B., Kitaoka M., Lo Leggio L., and Abou Hachem M. (2014) A β1–6/β1–3 galactosidase from Bifidobacterium animalis subsp. lactis Bl-04 gives insight into sub-specificities of β-galactoside catabolism within Bifidobacterium. Mol. Microbiol. 94, 1024–1040 [DOI] [PubMed] [Google Scholar]
- 14. Barrangou R., Briczinski E. P., Traeger L. L., Loquasto J. R., Richards M., Horvath P., Coûté-Monvoisin A.-C., Leyer G., Rendulic S., Steele J. L., Broadbent J. R., Oberg T., Dudley E. G., Schuster S., Romero D. A., and Roberts R. F. (2009) Comparison of the complete genome sequences of Bifidobacterium animalis subsp. lactis DSM 10140 and Bl-04. J. Bacteriol. 191, 4144–4151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Andersen J. M., Barrangou R., Abou Hachem M., Lahtinen S. J., Goh Y. J., Svensson B., and Klaenhammer T. R. (2013) Transcriptional analysis of oligosaccharide utilization by Bifidobacterium lactis Bl-04. BMC Genomics 14, 312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Viborg A. H. (2015) Diversity in β-galactosidase specificities within Bifidobacterium: towards an understanding of β-galactoside metabolism in the gut niche. Trends Glycosci. Glycotech. 27, E9–E12 [Google Scholar]
- 17. Ding L., Zuo Q., Li D., Feng X., Gao X., Zhao F., and Qiu F. (2017) A new phenone from the roots of Paeonia suffruticosa Andrews. Nat. Prod. Res. 31, 253–260 [DOI] [PubMed] [Google Scholar]
- 18. Hidaka M., Fushinobu S., Ohtsu N., Motoshima H., Matsuzawa H., Shoun H., and Wakagi T. (2002) Trimeric crystal structure of the glycoside hydrolase family 42 β-galactosidase from Thermus thermophilus A4 and the structure of its complex with galactose. J. Mol. Biol. 322, 79–91 [DOI] [PubMed] [Google Scholar]
- 19. Oura H., Hiai S., Odaka Y., and Yokozawa T. (1975) Studies on the biochemical ation of ginseng saponin. J. Biochem. 77, 1057–1065 [DOI] [PubMed] [Google Scholar]
- 20. Fraisse D., Heitz A., Carnat A., Carnat A. P., and Lamaison J. L. (2000) Quercetin 3-arabinopyranoside, a major flavonoid compound from Alchemilla xanthochlora. Fitoterapia 71, 463–464 [DOI] [PubMed] [Google Scholar]
- 21. Metwally A. M., Omar A. A., Harraz F. M., and El Sohafy S. M. (2010) Phytochemical investigation and antimicrobial activity of Psidium guajava L. leaves. Pharmacogn. Mag. 6, 212–218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Godoy A. S., Camilo C. M., Kadowaki M. A., Muniz H. D., Espirito Santo M., Murakami M. T., Nascimento A. S., and Polikarpov I. (2016) Crystal structure of β1→6-galactosidase from Bifidobacterium bifidum S17: trimeric architecture, molecular determinants of the enzymatic activity and its inhibition by α-galactose. FEBS J. 283, 4097–4112 [DOI] [PubMed] [Google Scholar]
- 23. Tabachnikov O., and Shoham Y. (2013) Functional characterization of the galactan utilization system of Geobacillus stearothermophilus. FEBS J. 280, 950–964 [DOI] [PubMed] [Google Scholar]
- 24. Yoshida E., Sakurama H., Kiyohara M., Nakajima M., Kitaoka M., Ashida H., Hirose J., Katayama T., Yamamoto K., and Kumagai H. (2012) Bifidobacterium longum subsp. infantis uses two different β-galactosidases for selectively degrading type-1 and type-2 human milk oligosaccharides. Glycobiology 22, 361–368 [DOI] [PubMed] [Google Scholar]
- 25. Van Laere K. M., Abee T., Schols H. A., Beldman G., and Voragen A. G. (2000) Characterization of a novel β-galactosidase from Bifidobacterium adolescentis DSM 20083 active towards transgalactooligosaccharides. Appl. Environ. Microbiol. 66, 1379–1384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Maksimainen M., Paavilainen S., Hakulinen N., and Rouvinen J. (2012) Structural analysis, enzymatic characterization, and catalytic mechanisms of β-galactosidase from Bacillus circulans sp. alkalophilus. FEBS J. 279, 1788–1798 [DOI] [PubMed] [Google Scholar]
- 27. Solomon H. V., Tabachnikov O., Lansky, Salama R., Feinberg H., Shoham Y., and Shoham G. (2015) Structure-function relationships in Gan42B, an intracellular GH42 β-galactosidase from Geobacillus stearothermophilus. Acta Crystallogr. D Biol. Crystallogr. 71, 2433–2448 [DOI] [PubMed] [Google Scholar]
- 28. Ichinose H., Fujimoto Z., Honda M., Harazono K., Nishimoto Y., Uzura A., and Kaneko S. (2009) A β-l-arabinopyranosidase from Streptomyces avermitilis is a novel member of glycoside hydrolase family 27. J. Biol. Chem. 284, 25097–25106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Duncan S. H., Hold G. L., Barcenilla A., Stewart C. S., and Flint H. J. (2002) Roseburia intestinalis sp. nov., a novel saccharolytic, butyrate-producing bacterium from human faeces. Int. J. Syst. Evol. Microbiol. 52, 1615–1620 [DOI] [PubMed] [Google Scholar]
- 30. Kosugi A., Murashima K., and Doi R. (2002) Characterization of two noncellulosomal subunits, ArfA and BgaA, from Clostridium cellulovorans that cooperate with the cellulosome in plant cell wall degradation. J. Bacteriol. 184, 6859–6865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Lee J. H., Hyun Y.-J., and Kim D.-H. (2011) Cloning and characterization of α-l-arabinofuranosidase and bifunctional α-l-arabinopyranosidase/β-d-galactopyranosidase from Bifidobacterium longum H-1. J. Appl. Microbiol. 111, 1097–1107 [DOI] [PubMed] [Google Scholar]
- 32. Lee D. Y., Jeong Y. T., Jeong S. C., Lee M. K., Min J. W., Lee J. W., Kim G. S., Lee S. E., Ahn Y. S., Kang H. C., and Kim J. H. (2015) Melanin biosynthesis inhibition effects of ginsenoside Rb2 isolated from panax ginseng berry. J. Microbiol. Biotechnol. 25, 2011–2015 [DOI] [PubMed] [Google Scholar]
- 33. Larsbrink J., Rogers T. E., Hemsworth G. R., McKee L. S., Tauzin A. S., Spadiut O., Klinter S., Pudlo N. A., Urs K., Koropatkin N. M., Creagh A. L., Haynes C. A., Kelly A. G., Cederholm S. N., Davies G. J., Martens E. C., and Brumer H. (2014) A discrete genetic locus confers xyloglucan metabolism in select human gut Bacteroidetes. Nature 506, 498–502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Ndeh D., Rogowski A., Cartmell A., Luis A. S., Baslé A., Gray J., Venditto I., Briggs J., Zhang X., Labourel A., Terrapon N., Buffetto F., Nepogodiev S., Xiao Y., Field R. A., et al. (2017) Complex pectin metabolism by gut bacteria reveals novel catalytic functions. Nature 10.1038/nature21725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Shin H. Y., Park S. Y., Sung J. H., and Kim D. H. (2003) Purification and characterization of α-l-arabinopyranosidase and α-l-arabinofuranosidase from Bifidobacterium breve K-110, a human intestinal anaerobic bacterium metabolizing ginsenoside Rb2 and Rc. Appl. Environ. Microbiol. 69, 7116–7123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Nishimoto M., and Kitaoka M. (2007) Practical preparation of lacto-N-biose I, a candidate for the bifidus factor in human milk. Biosci. Biotechnol. Biochem. 71, 2101–2104 [DOI] [PubMed] [Google Scholar]
- 37. Nishimoto M., and Kitaoka M. (2009) One-pot enzymatic production of β-d-galactopyranosyl-(1→3)-2-acetamido-2-deoxy-d-galactose (galacto-N-biose) from sucrose and 2-acetamido-2-deoxy-d-galactose (N-acetylgalactosamine). Carbohydr. Res. 344, 2573–2576 [DOI] [PubMed] [Google Scholar]
- 38. Otwinowski Z., and Minor W. (1997) Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 [DOI] [PubMed] [Google Scholar]
- 39. Long F., Vagin A. A., Young P., and Murshudov G. N. (2008) BALBES: A molecular-replacement pipeline. Acta Crystallogr. D Biol. Crystallogr. 64, 125–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Perrakis A., Morris R., and Lamzin V. S. (1999) Automated protein model building combined with iterative structure refinement. Nat. Struct. Biol. 6, 458–463 [DOI] [PubMed] [Google Scholar]
- 41. Emsley P., Lohkamp B., Scott W. G., and Cowtan K. (2010) Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Nicholls R. A., Long F., and Murshudov G. N. (2012) Low-resolution refinement tools in REFMAC5 research papers. Acta Crystallogr. D Biol. Crystallogr. 68, 404–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Chen V. B., Arendall W. B. 3rd, Headd J. J., Keedy D. A., Immormino R. M., Kapral G. J., Murray L. W., Richardson J. S., and Richardson D. C. (2010) MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 66, 12–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Trott O., and Olson A. (2010) AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 31, 455–461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Edgar R. C. (2004) MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Edgar R. C. (2010) Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26, 2460–2461 [DOI] [PubMed] [Google Scholar]
- 47. Crooks G. E., Hon G., Chandonia J. M., and Brenner S. E. (2004) WebLogo: a sequence logo generator. Genome Res. 14, 1188–1190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Huson D. H., and Scornavacca C. (2012) Dendroscope 3: an interactive tool for rooted phylogenetic trees and networks. Syst. Biol. 61, 1061–1067 [DOI] [PubMed] [Google Scholar]