Abstract
The viral protein R (Vpr) is an accessory virulence factor of HIV-1 that facilitates infection in immune cells. Cellular functions of Vpr are tied to its interaction with DCAF1, a substrate receptor component of the CRL4 E3 ubiquitin ligase. Recent proteomic approaches suggested that Vpr degrades helicase-like transcription factor (HLTF) DNA helicase in a proteasome-dependent manner by redirecting the CRL4-DCAF1 E3 ligase. However, the precise molecular mechanism of Vpr-dependent HLTF depletion is not known. Here, using in vitro reconstitution assays, we show that Vpr mediates polyubiquitination of HLTF, by directly loading it onto the C-terminal WD40 domain of DCAF1 in complex with the CRL4 E3 ubiquitin ligase. Mutational analyses suggest that Vpr interacts with DNA-binding residues in the N-terminal HIRAN domain of HLTF in a manner similar to the recruitment of another target, uracil DNA glycosylase (UNG2), to the CRL4-DCAF1 E3 by Vpr. Strikingly, Vpr also engages a second, adjacent region, which connects the HIRAN and ATPase/helicase domains. Thus, our findings reveal that Vpr utilizes common as well as distinctive interfaces to recruit multiple postreplication DNA repair proteins to the CRL4-DCAF1 E3 ligase for ubiquitin-dependent proteasomal degradation.
Keywords: human immunodeficiency virus (HIV), polyubiquitin chain, protein degradation, ubiquitin, ubiquitin ligase, virulence factor, Vpr
Introduction
CD4+ T lymphocytes and macrophages are preferentially targeted by human immunodeficiency virus-1 (HIV-1) infection. HIV-1 replication in these cells and disease progression are enhanced by accessory virulence proteins, which counteract potent intrinsic and innate antiviral mechanisms (1, 2). These virulence factors often hijack specific E3 ubiquitin ligases and direct them to degrade antiviral proteins via the ubiquitin-proteasome system (3).
The viral protein R (Vpr)2, an accessory virulence protein found in all primate lentiviruses, facilitates HIV-1 replication in infected cells (4–6). Although the precise molecular mechanism by which Vpr enhances HIV-1 infection remains elusive, a direct interaction with DDB1 and CUL4-associated factor 1 (DCAF1), a substrate receptor of the cullin4-RING E3 ubiquitin ligase (CRL4) comprising DDB1, CUL4, and RBX1, mediates Vpr's cellular functions (7–9). In particular, Vpr was proposed to positively and/or negatively modulate, in a DCAF1-dependent manner, innate response to HIV-1 infection in target cell types (10, 11).
HIV-1 Vpr induces cell-cycle arrest in G2 phase by activating the DNA damage response pathway associated with ataxia telangiectasia–mutated (ATM) and Rad3-related (ATR) serine/threonine kinase (12–14). Further, HIV-1 Vpr expression induces nuclear foci formations similar to those observed following activation of DNA damage repair by genotoxic agents (15). Significantly, Vpr was localized to the DNA repair foci and proposed to recruit DCAF1 to these sites (16). Although the exact mechanism of Vpr-mediated DNA damage response pathway activation is not known, the interaction between Vpr and DCAF1 in complex with the CRL4 E3 ubiquitin ligase was linked to Vpr's ability to induce arrest cells in G2 phase of the cell cycle (17–22). Thus, one plausible hypothesis is that Vpr activates ATR and induces DNA repair foci by interfering with postreplication DNA repair and loading DNA repair proteins onto the CRL4-DCAF1 for proteasome-dependent degradation. Indeed, uracil DNA glycosylase 2 (UNG2) and the structure-selective endonuclease complex of MUS81-EME1 are direct substrates, targeted by Vpr-bound CRL4-DCAF1 for polyubiquitination and proteasome-dependent degradation (23, 24). However, depletion of these DNA repair enzymes appears insufficient to induce DNA damage checkpoint and arrest cells in G2 phase (23–26), suggesting that additional targets of Vpr, that remain to be identified, mediate these effects.
Helicase-like transcription factor (HLTF) is a multi-functional protein that contributes to the maintenance of genome stability by mediating postreplication DNA repair. In response to DNA damage, HLTF, which possesses a RING E3 ligase domain, loads a nonproteolytic Lys-63 polyubiquitin chain onto proliferating cell nuclear antigen (PCNA) (27, 28). This enables replication fork reversal, i.e. switching leading strand replication from the damaged template to the nascent lagging strand, thus allowing error-free bypass of the lesion that blocks progression of the replication fork (29). Replication fork reversal requires both the 3′-end single-strand DNA binding activity of its N-terminal HIRAN domain and ATP hydrolysis–driven double-stranded DNA translocase activity (30–33). HLTF also possesses ATP hydrolysis–dependent protein remodeling activity, which allows it to dislodge DNA-binding proteins, possibly to clear the stalled replication fork (34).
Recently, we and others found that HLTF is depleted by Vpr in HIV-1 infected cells in a ubiquitin-proteasome–dependent manner (26, 35). However, the precise molecular mechanism by which Vpr depletes HLTF is not known. Here, using in vitro reconstitution assays with homogeneously purified recombinant protein complexes, we show that Vpr directly loads HLTF onto the DCAF1 substrate receptor subunit in complex with the CRL4 E3 ubiquitin ligase. Strikingly, Vpr engages two separable N-terminal regions of HLTF to target it for proteasome-dependent degradation. These, together with the earlier findings, reveal that Vpr has evolved the ability to specifically recruit multiple DNA repair proteins to the ubiquitin-proteasome degradation pathway, via the CRL4-DCAF1 E3 ligase.
Results
HIV-1 Vpr recruits HLTF to CRL4-DCAF1 to mediate its proteasome-dependent degradation
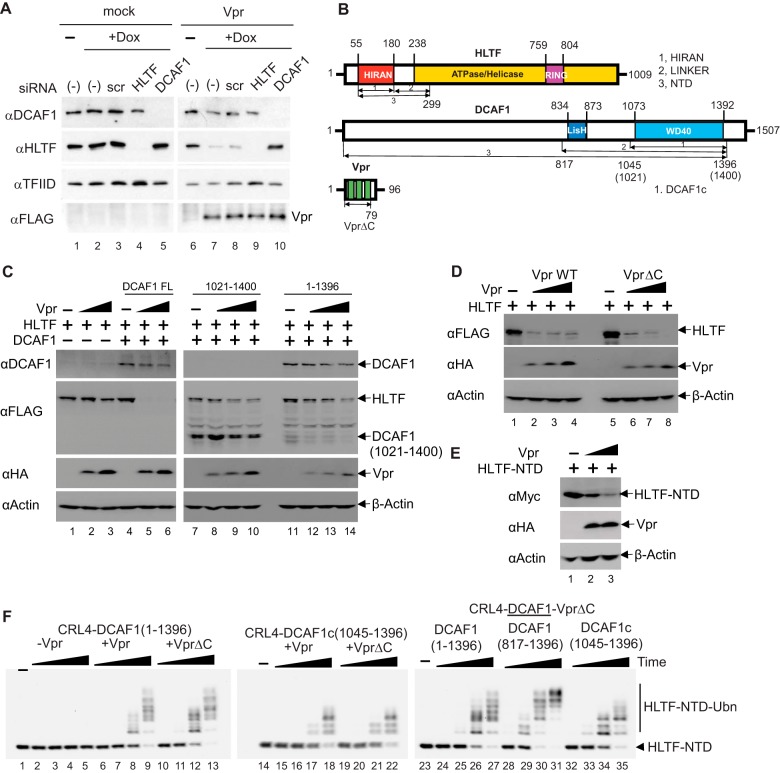
Proteomic screens for cellular targets of Vpr suggested that Vpr usurps the CRL4-DCAF1 E3 ubiquitin ligase to down-regulate HLTF in a proteasome-dependent manner (26, 35). To validate these findings, we interrogated the endogenous level of HLTF in U2OS cell lines engineered to express HIV-1 NL4-3 Vpr in a doxycycline-dependent manner (Fig. 1A). Prior to the induction of Vpr expression, mock and Vpr-inducible cell lines were transiently transfected with a scrambled control siRNA or HLTF- or DCAF1-targeting siRNAs. In control U2OS cells, the level of HLTF was significantly reduced only when siRNA targeting HLTF was transfected. In Vpr-inducible cell lines, the level of HLTF was dramatically reduced upon induction of Vpr. Importantly, HLTF levels were restored when siRNA targeting DCAF1 was transfected into these Vpr expressing cells. These data reveal that Vpr down-regulates HLTF in a DCAF1-dependent manner.
Figure 1.
HIV-1 Vpr directly loads HLTF onto CRL4-DCAF1 to mediate its proteasome-dependent degradation. A, U2OS cells harboring doxycycline (Dox) inducible HIV-1 Vpr transgene were transfected with scrambled (scr) siRNA or siRNA targeting HLTF or DCAF1 and treated with doxycycline for 48 h. Cell extracts were analyzed by immunoblotting with antibodies reacting with DCAF1, HLTF, and FLAG-tagged Vpr. TFIID was used as a loading control. B, domain organization of HLTF, DCAF1, and Vpr. HLTF comprises a single-stranded DNA–binding HIRAN domain, an ATPase domain, and a RING domain. Recombinant HLTF-HIRAN (residues 55–180), LINKER (residues 181–299), and NTD (residues 55–299) constructs were prepared for in vitro reconstitution assays. DCAF1 contains a LisH dimerization domain and a WD40 domain followed by a stretch of acidic amino acid residues. Recombinant DCAF1 constructs (residues 1045–1396, henceforth DCAF1c; 817–1396; 1–1396) were prepared for in vitro assays. Alternatively, a DCAF1 construct of residues 1021–1400 was transiently transfected into HEK 293T cells. Vpr comprises three α helices within residues 16–32, 36–51, and 53–74 (39). Recombinant WT and VprΔC (residues 1–79) constructs were prepared. C, constant amounts of plasmids expressing HLTF and the indicated DCAF1 constructs (full-length (FL), residues 1021–1400, and residues1–1396) were transiently co-transfected into HEK 293T cells together with increasing amounts of HIV-1 Vpr expression plasmid. At 48 h post transfection, cells were harvested and analyzed by Western blotting. D, HEK 293T cells were transfected with a constant amount of plasmids expressing full-length HLTF and DCAF1, and increasing amounts of a plasmid expressing Vpr WT or VprΔC. E, constant amounts of HLTF-NTD (residues 55–299) expression plasmid were transiently co-transfected with increasing amounts of Vpr expression plasmid into HEK 293T cells. F, in vitro ubiquitination assays of HLTF-NTD with CRL4-DCAF1, CRL4-DCAF1-Vpr, or CRL4-DCAF1-VprΔC, where DCAF1 constructs vary (residues 1–1396, 816–1396, or 1045–1396) as indicated. Polyubiquitinated HLTF species are indicated (HLTF-NTD-Ubn). All experiments were repeated three times with equivalent results.
To map the minimal DCAF1 domain required for Vpr-dependent HLTF down-regulation, full-length DCAF1 or two WD domain constructs (residues 1–1396 and residues 1021–1400) (Fig. 1B) were transiently co-transfected with a constant amount of HLTF and increasing amounts of Vpr expression plasmids into HEK 293T cells (Fig. 1C). The WD40 domain alone (residues 1021–1400) was sufficient to induce down-regulation of HLTF in a Vpr-dependent manner (Fig. 1C), although not to the same degree as full-length DCAF1. The C terminus of Vpr, divergent among Vpr proteins representative of main groups of HIV-1 and SIVcpz viruses isolated from human and chimpanzee, respectively (26), was not required for HLTF down-regulation (Fig. 1D). Consistent with previous findings (26), the N-terminal region of HLTF (residues 55–299) was targeted by Vpr (Fig. 1E). Of note, the endogenous DCAF1 levels in HEK 293T cells are much lower than those of the ectopically expressed DCAF1, so the presence of the endogenous protein is not likely to confound the results from transient assays of DCAF1 mutants. Similarly, the ectopically expressed HLTF levels in HEK 293T cells were much higher than those of endogenously expressed HLTF in U2OS cells, hence probably overwhelming the endogenously expressed CRL4-DCAF1, which explains why not all HLTF was depleted by Vpr in transient overexpression assays in HEK 293T cells.
To ascertain that no other host factors are required for Vpr-dependent HLTF down-regulation, we performed in vitro ubiquitination assays with reconstituted CRL4-DCAF1 E3 ubiquitin ligase (23, 36, 37). Polyubiquitination of HLTF-NTD (residues 55–299) was apparent only in the presence of Vpr (Fig. 1F; compare lanes 2–5 versus lanes 6–13). Consistent with our observations from transient transfection assays (Fig. 1, C–E), the WD40 domain of DCAF1 (residues 1045–1396, henceforth abbreviated as DCAF1c) and VprΔC were sufficient to mediate polyubiquitination of HLTF-NTD in vitro. In addition, the ubiquitin ligase activity of Vpr-bound CRL4-DCAF1 (817–1396) toward HLTF substrate appeared to be more potent than that of Vpr-bound CRL4-DCAF1c. This is likely because of DCAF1 dimerization via its LisH domain; such dimerization is thought to confer conformational flexibility to permit productive loading of substrates and to accommodate increasing numbers of ubiquitin during polyubiquitination (38).
HIV-1 Vpr binds two separable regions of HLTF
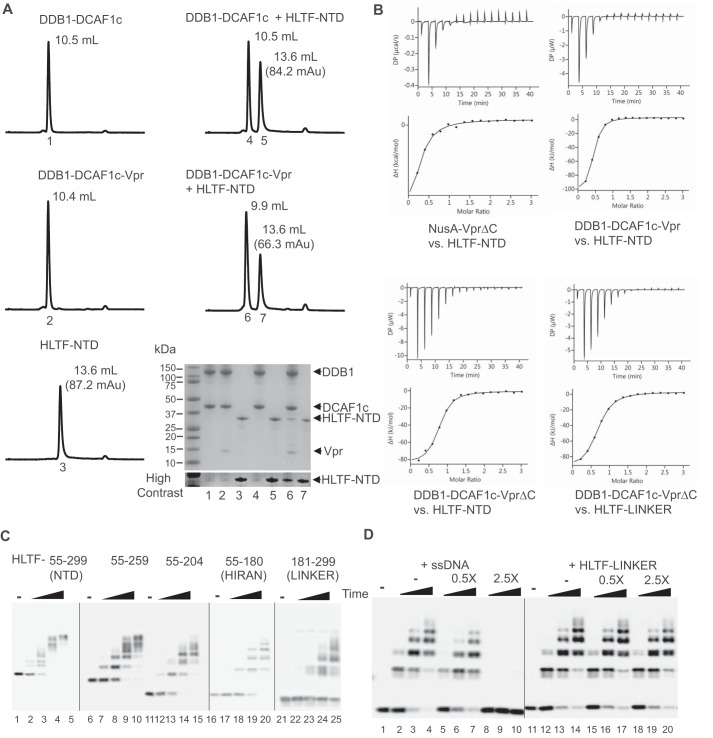
To investigate direct loading of HLTF onto DCAF1 by Vpr, we carried out analytical gel filtration column chromatography. DDB1-DCAF1c, DDB1-DCAF1c-Vpr complexes, and HLTF-NTD were individually injected into a Superdex200 column, and their retention volumes were determined (Fig. 2A, left panels). Mixtures of HLTF-NTD with DDB1-DCAF1c or DDB1-DCAF1c-Vpr were then analyzed for co-elution (Fig. 2A, right panels). Elution volumes of DDB1-DCAF1c and HLTF-NTD were essentially the same for individual component and mixtures (10.5 ml and 13.6 ml, respectively). On the other hand, preincubation of the DDB1-DCAF1c-Vpr complex with the HLTF-NTD led to a shift of the first peak in complex elution compared with that in absence of HLTF-NTD (9.9 ml versus 10.4 ml), and the second elution peak height corresponding to HLTF-NTD (13.6 ml) was reduced (66.3 versus 87.2 milli absorbance units). SDS-PAGE analysis of the elution peak collected after separation of protein mixtures with analytical gel filtration column revealed co-elution of HLTF with DDB1-DCAF1c-Vpr (Fig. 2A, lane 6, bottom right panel).
Figure 2.
Two separate N-terminal regions of HLTF are targeted by Vpr. A, DDB1-DCAF1c, DDB1-DCAF1c-Vpr, both at 12.5 μm, or HLTF-NTD at 62.5 μm was injected into an analytical Superdex200 Increase gel filtration column at a flow rate of 0.5 ml/min (left panels). Absorbance at UV 280 nm was recorded. Protein mixtures of DDB1-DCAF1c or DDB1-DCAF1c-Vpr, with HLTF-NTD were also analyzed (right panels). The peak elution volume (ml) is labeled in each panel. The peak absorbance for HLTF-NTD is also indicated. Elution peaks were collected, analyzed by SDS-PAGE, and visualized by Coomassie Brilliant Blue staining (bottom right panel). The number under each peak refers to the lane number in the SDS-PAGE analysis panel. B, isothermal titration trace for NusA-VprΔC binding to HLTF-NTD is shown (left top panel). A single-site binding isotherm curve fitting yielded a Kd of 5.8 ± 2.2 μm. Isothermal titration traces for HLTF-NTD binding to DDB1-DCAF1c-Vpr (right top panel) and DDB1-DCAF1c-VprΔC (left bottom panel) are also shown. The data were best fitted with a single-site binding isotherm yielding a Kd of 1.1 ± 0.2 μm and 1.3 ± 0.2 μm, respectively. Isothermal titration traces for HTLF-LINKER binding to DDB1-DCAF1c-VprΔC (right bottom panel) yielded a Kd of 2.1 ± 0.1 μm. C, in vitro ubiquitination assays of various truncation constructs of HLTF-NTD, including residues 55–259, 55–204, 55–180 (HIRAN), and 181–299 (LINKER), with CRL4-DCAF1c-Vpr E3 ligase. D, in vitro ubiquitination assays of HLTF-HIRAN with CRL4-DCAF1c-Vpr E3 ligase. The reaction mixtures contained increasing concentrations of single-stranded DNA (ssDNA) or HLTF-LINKER at molar ratios of 0-, 0.5-, and 2.5-fold over HLTF-HIRAN. The sequence of oligonucleotide used in the assay is 5′-AGCTACCATGCCTGCCTCAAGAATTCGTAA-3′. All experiments were repeated two to three times with similar results.
To quantitatively determine the binding affinity between Vpr, or DDB1-DCAF1c-Vpr, and HLTF-NTD or HLTF-LINKER, we performed isothermal titration calorimetry (ITC) (Fig. 2B). HLTF-NTD bound to NusA-VprΔC and DDB1-DCAF1c-Vpr with Kd value of 5.8 ± 2.2 μm and 1.1 ± 0.2 μm, respectively (Fig. 2B, upper panels). HLTF-NTD bound DDB1-DCAF1c-VprΔC with Kd of 1.3 ± 0.2 μm (Fig. 2B, left bottom panel). That HLTF-NTD bound the DDB1-DCAF1c-VprΔC and DDB1-DCAF1c-Vpr complexes with similar affinities is not surprising, because both E3 complexes mediated polyubiquitination of HLTF-NTD to similar degrees (Fig. 1F). Of note, neither NusA nor DDB1-DCAF1c showed a measurable binding affinity by ITC (data not shown), indicating that DDB1-DCAF1 and HLTF are bridged by Vpr. On the other hand, HLTF-LINKER (residues 181–299) bound DDB1-DCAF1c-VprΔC with an affinity of 2.1 ± 0.1 μm, suggesting that the HIRAN domain contributes to the interaction with DDB1-DCAF1c-VprΔC (Fig. 2B, right bottom panel).
To further investigate the interaction between HLTF and DDB1-DCAF1-Vpr, we performed in vitro ubiquitination assays with several deletion constructs of the HLTF-NTD (Fig. 2C). Both HLTF-HIRAN and HLTF-LINKER were efficiently polyubiquitinated by CRL4-DCAF1c-Vpr. The HIRAN domain specifically binds to the 3′-end of single-stranded DNA (ssDNA) (33). Surprisingly, the degree of HLTF-HIRAN polyubiquitination was effectively reduced when increasing amounts of ssDNA were added to the reaction (Fig. 2D). Further, polyubiquitination of HLTF-NTD was moderately inhibited by addition of ssDNA into the reaction mixtures, whereas polyubiquitination of HLTF-LINKER was not affected by ssDNA (supplemental Fig. 1). Moreover, addition of increasing amounts of HLTF-LINKER to in vitro ubiquitination assays did not inhibit HIRAN domain polyubiquitination (Fig. 2D). Taken altogether, these data suggest that the HIRAN and LINKER domains independently interact with Vpr and that the DNA-binding surface of the HIRAN domain may overlap with the Vpr-binding interface.
The interaction with Vpr reveals two separate recognition sequences in the N-terminal region of HLTF
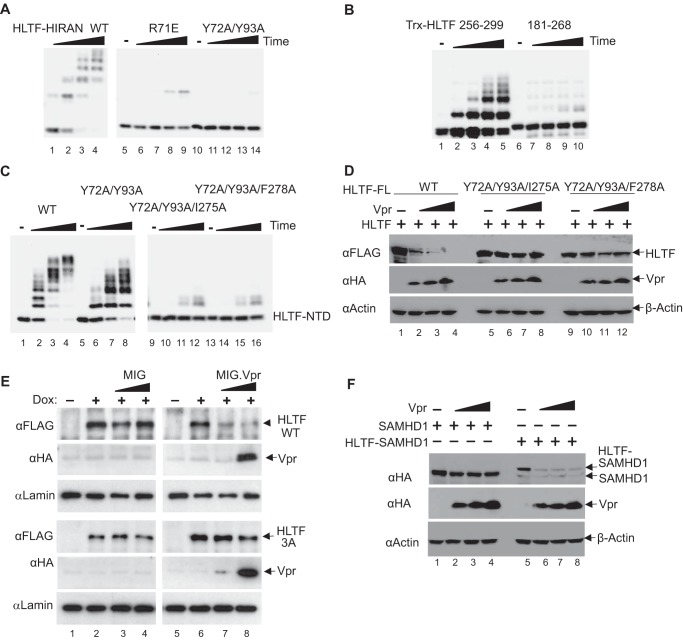
To address the molecular basis for the recruitment of HLTF onto the CRL4-DCAF1 E3 ligase by Vpr, we performed mutagenesis of HLTF-HIRAN and HLTF-LINKER domains separately. The crystal structure of the DDB1-DCAF1-Vpr-UNG2 complex, that we solved previously, suggested that Vpr mimics DNA and interacts with DNA interfacing residues in UNG2 (39). Because the HIRAN domain binds specifically to the 3′-end of single-stranded DNA, we hypothesized that the previously identified DNA binding residues in the HIRAN domain, including Arg-71, Tyr-72, and Tyr-93 (33), may interface with Vpr. As shown in Fig. 3A, R71E and Y72A/Y93A mutations indeed prevented polyubiquitination of the HIRAN domain mediated by the CRL4-DCAF1c-Vpr E3 complex. Further, as expected, these mutations significantly reduced single-stranded DNA–binding activity of the HIRAN domain without causing significant secondary structure changes, as evidence by circular dichroism (supplemental Fig. 2, A and B). Consistent with the observation from the in vitro ubiquitination assays, levels of HLTF-NTD with Y72A/Y93A mutation were higher than those of HLTF-NTD WT, when ectopically co-expressed with Vpr (supplemental Fig. 2C). Note, the Y72A/Y93A mutated HLTF-NTD was still down-regulated by Vpr; this was expected, because the LINKER region is also recognized by Vpr (see Fig. 2, B and C).
Figure 3.
Mapping of HLTF residues interfacing Vpr. A, in vitro ubiquitination assays of HLTF-HIRAN WT, R71E, or Y72A/Y93A mutant with CRL4-DCAF1c-Vpr E3 ligase. B, in vitro ubiquitination assays of thioredoxin-fused HLTF constructs (residues 256–299 and 181–268) with CRL4-DCAF1c-Vpr. C, in vitro ubiquitination assays of HLTF-NTD WT, Y72A/Y93A, Y72A/Y93A/I275A, or Y72A/Y93A/F278A with CRL4-DCAF1c-Vpr. D, a constant amount of plasmid expressing full-length (FL) HLTF WT or with Y72A/Y93A/I275A or Y72A/Y93A/F278A mutations was co-transfected with a constant amount of full-length DCAF1 and increasing amounts of Vpr expression plasmids, into HEK 293T cells. E, in Vpr resistance assays, U2OS cells expressing doxycycline (Dox) inducible full-length HLTF WT or 3A mutant (Y72A/Y93A/I275A) were transduced with MIG virus expressing HIV-1 Vpr, or not. F, constant amounts of plasmid expressing SAMHD1, or a fusion construct comprising HLTF residues 256–299 and SAMHD1 (HLTF-SAMHD1), were co-transfected with a constant amounts of full-length DCAF1 and increasing amounts of Vpr expression plasmids. All experiments were repeated three times with similar results.
To identify HLTF-LINKER residues that mediate specific recruitment by Vpr to the CRL4-DCAF1 E3, we first carried out in vitro ubiquitination assays with two HLTF polypeptides fused with the thioredoxin (Trx) moiety (Fig. 3B). The Trx fused with HLTF residues 256–299 showed robust polyubiquitination, whereas that fused to the N-terminal part of the LINKER comprising residues 181–268 did not. We then performed Ala scanning mutagenesis of HLTF-NTD containing Y72A and Y93A mutation in the HIRAN domain. As shown in Fig. 3C, additional I275A or F278A mutation within the LINKER region disrupted polyubiquitination of HLTF-NTD by the CRL4-DCAF1c-Vpr. Those mutations did not affect the overall secondary structure of HLTF-NTD (supplemental Fig. 2D).
To validate our in vitro results, we transiently co-expressed full-length HLTF WT, Y72A/Y93A/I275A, and Y72A/Y93A/F278A with increasing amounts of Vpr (Fig. 3D). Both triple mutants resisted Vpr-dependent down-regulation. To further corroborate our findings, we generated U2OS cell lines harboring doxycycline-inducible WT or Y72A/Y93A/I275A HLTF transgenes and transduced the cells with retroviral vector expressing Vpr, or a control empty vector (Fig. 3E). The levels of HLTF WT were effectively down-regulated in cells expressing Vpr, compared with those infected with the empty vector. In contrast, HLTF Y72A/Y93A/I275A appeared resistant to depletion by Vpr.
To assess the individual contribution of HIRAN and LINKER domain to Vpr-dependent down-regulation of the full-length HLTF protein, we transiently co-expressed full-length HLTF proteins with Y72A/Y93A, I275A, or F278A mutation with increasing amounts of Vpr. Although Y72A/Y93A mutant showed some resistance to Vpr-dependent down-regulation (supplemental Fig. 3A), WT HLTF, the I275A and F278A mutants were efficiently down-regulated by Vpr (supplemental Fig. 3B).
The observation that I275A and F278A mutants are more sensitive to Vpr expression than Y72A/Y93A was somewhat surprising. However, it should be noted that Ile-275 and Phe-278 are located at the N-terminal region of ATPase/helicase domain (Fig. 2B) and, hence, accessibility of these residues to Vpr could be different in the context of full-length HLTF versus individual domains such as NTD or LINKER. Consistent with this notion, fusing the C-terminal half of the HLTF-LINKER (residues 256–299), spanning residues Ile-275 and Phe-278, to SAMHD1, rendered the fusion protein sensitive to down-regulation by Vpr (Fig. 3F). Taken together, these findings support the model in which Vpr targets two separate regions in the HLTF-NTD.
Vpr substrate recruitment interface mediates polyubiquitination of HLTF by the CRL4-DCAF1 E3 complex
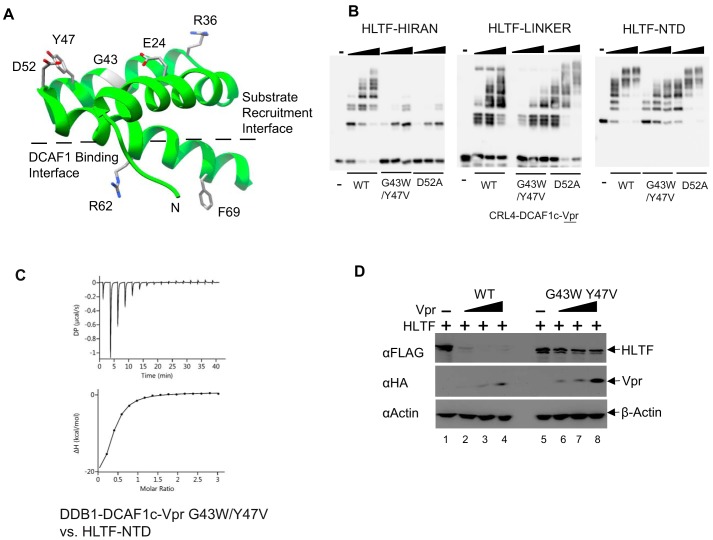
The crystal structure of DDB1-DCAF1-Vpr-UNG2 (39) revealed two distinct Vpr interfaces: A DCAF1-binding interface and a substrate recruitment interface (Fig. 4A). To determine Vpr residues that mediate HLTF recruitment to the CRL4-DCAF1, we began by mutating the substrate-binding interface of Vpr. In particular, we mutated Asp-52, a residue that engages the active site of UNG2 (39). We also mutated Gly-43 and Tyr-47 residues, which also interface with UNG2 (39). The DDB1-DCAF1c proteins in complex with Vpr D52A, or G43W/Y47V mutant were prepared (supplemental Fig. 4) and then tested for their ability to mediate polyubiquitination of the HLTF-HIRAN, -LINKER, and -NTD fragments, respectively. The E3 ligase in complex with either of the two Vpr mutants failed to efficiently polyubiquitinate HLTF-HIRAN (Fig. 4B, left panel). This suggested that both the Asp-52 and the Gly-43 to Tyr-47 residues interface with the HIRAN domain of HLTF. In contrast, the D52A Vpr mutant did not affect polyubiquitination of HLTF-LINKER and HLTF-NTD by the ligase, whereas the G43W/Y47V Vpr mutant reduced the ligase's activity toward both HLTF constructs (Fig. 4B, right panels). ITC confirmed that G43W/Y47V mutations reduced DDB1-DCAF1c-Vpr binding HLTF-NTD by 4-fold (Fig. 4C). These observations suggest a model in which the Asp-52 residue exclusively interacts with the HIRAN domain, whereas the region encompassing Gly-43 and Tyr-47 interfaces with both the HIRAN and LINKER domains. This model is also supported by results from transient co-expression assays of HLTF and Vpr in HEK 293T cells. In particular, the G43W/Y47V mutation disrupted Vpr's ability to down-regulate full-length HLTF WT (Fig. 4D). Of note, other elements of the substrate interface, Glu-24 and Arg-36 in particular (Fig. 4A), were previously shown to mediate HLTF depletion in a cell-based assay (26). In sum, the above observations suggest that Vpr's substrate recruitment interface, which is located opposite to the DCAF1-binding interface, mediates direct loading of HLTF onto the CRL4-DCAF1 E3 ligase complex.
Figure 4.
Vpr residues that mediate HLTF degradation. A, ribbon and stick representation of Vpr (green, taken from PDB ID, 5JK7) illustrating two separate interfaces: The substrate recruitment and DCAF1 binding interfaces. Critical residues of Vpr interfacing with DCAF1 (Arg-62 and Phe-69) or substrates, including UNG2 and HLTF (Glu-24, Arg-36, Gly-43, Tyr-47, and Asp-52) identified in this and previous reports (26, 39) are shown in stick representation. B, in vitro ubiquitination assays of HLTF-HIRAN, HLTF-LINKER and HLTF-NTD with Vpr WT, G43W/Y47V or D52A in complex with CRL4-DCAF1c. C. isothermal titration trace for DDB1-DCAF1c-Vpr with G43W/Y47V mutation binding to HLTF-NTD yielded a Kd of 4.6 ± 0.4 μm. D, HEK 293T cells were co-transfected with a constant amount of full-length HLTF and DCAF1 and increasing amounts of Vpr WT or G43W/Y47V expression plasmids. All experiments were performed three times with equivalent results.
Discussion
Reprogramming of the CRL4-DCAF1 E3 ubiquitin ligase by Vpr toward host cellular factors likely facilitates HIV-1 replication (17–22). Recent proteomic studies identified HLTF, a DNA helicase involved in postreplication DNA repair, as a target of the Vpr-bound CRL4-DCAF1 E3 (26, 35). Here, we provide extensive biochemical evidence for a direct role of Vpr in proteasome-dependent degradation of HLTF mediated by the CRL4-DCAF1 E3 ubiquitin ligase. In particular, results from size exclusion column chromatography, ITC, and in vitro ubiquitination assays performed with purified recombinant proteins unequivocally support the model in which Vpr directly loads HLTF onto DCAF1, a substrate receptor module of the CRL4 E3 (Fig. 5). This model is further supported by the results from our structure-function studies of the Vpr-HLTF interfaces, using biochemical and cell-based assays. Specifically, Vpr, in complex with the WD40 domain of DCAF1, directly interacts with N-terminal region of HLTF, including the HIRAN domain possessing the 3′-end, the single-stranded DNA–binding activity, and a LINKER region connecting the HIRAN and ATPase/helicase domains. Whereas other DCAF1 regions, such as the LisH dimerization domain and sequences C proximal to the WD40 domain, may contribute to efficient processing of HLTF by Vpr-bound CRL4-DCAF1 E3 in cells (Fig. 1, B, C, and F), our results demonstrate that the WD40 domain of DCAF1 is the minimal region required for Vpr-dependent HLTF polyubiquitination in vitro and depletion in vivo.
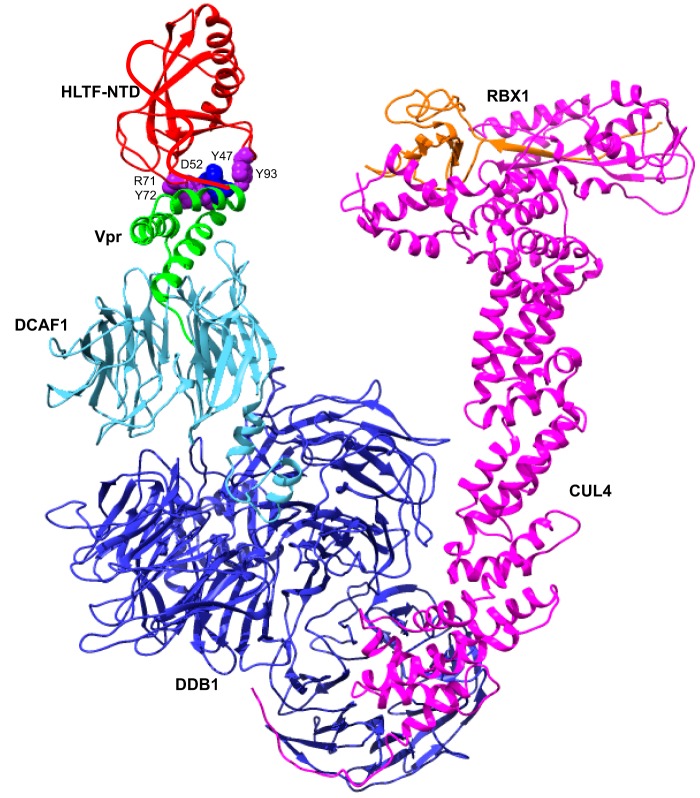
Figure 5.
Model of Vpr loading HLTF onto the CRL4-DCAF1-Vpr. Vpr (green) directly loads HLTF-NTD (red) onto the DCAF1 (cyan) substrate receptor of the CRL4 E3 ubiquitin ligase comprising DDB1 (blue), CUL4 (magenta), and RBX1 (orange). RBX1 mediates a direct transfer of ubiquitin from the E2 ubiquitin-conjugating enzyme, which is loaded with activated ubiquitin by E1 ubiquitin-activating enzyme, to substrates bound to a substrate recognition module (43, 44). Side chains of Arg-71, Tyr-72, and Tyr-93 in HLTF are shown in purple sphere and Vpr side chains of Asp-52 and Tyr-47 are shown in blue. A structure with the best HADDOCK score is shown. The trace of HLTF residues 181–299 is hand-drawn in red.
Because HLTF-mediated lesion bypass at damaged replication fork requires its HIRAN and ATPase/helicase domains (30–33), our results suggest that direct binding of both HIRAN and LINKER located at the ATPase/helicase domains with Vpr alone or in the context of the CRL4-DCAF1 complex may impinge on HLTF function during DNA replication. Interestingly, our data reveal that the effect of LINKER region mutations is more pronounced in the context of HLTF-NTD and HLTF-LINKER fragments, than in the context of the full-length HLTF protein, suggesting that LINKER accessibility to Vpr varies between these constructs. This in turn raises the possibility that the LINKER domain accessibility can be modulated by HLTF conformational states, for example upon HIRAN binding to DNA. This possibility as well as whether and how Vpr-mediated down-modulation of HLTF function and/or level facilitates HIV-1 replication in target cells will be addressed in future studies.
The structure of DDB1-DCAF1-Vpr in complex with UNG2 revealed that one surface of Vpr's three-helix bundle interfaces with DCAF1, whereas the opposite surface is utilized for recruitment of UNG2 (39). Our findings from mutagenesis studies suggest that the latter surface of Vpr is also utilized for recruitment of HLTF (Fig. 4). For interaction with UNG2, Vpr employs structural mimicry of DNA and engages the DNA-binding region of UNG2 (39). In a similar fashion, Vpr targets the DNA-binding interfaces in the HLTF HIRAN domain (Fig. 3A and supplemental Fig. 2A). The finding that Vpr targets both UNG2 and HLTF via their DNA-binding domains is intriguing. One possible advantage of such a strategy is that these surfaces are constrained by structures of their DNA ligands and hence not free to generate escape variants. Another is that Vpr binding to these surfaces competes with their binding to DNA and thus likely inhibits their functions even prior to their processing by the CRL4-DCAF1 E3 ligase.
Although Vpr targets UNG2 and HLTF DNA-binding surfaces in similar modes, some differences in the recruitment of the two substrates are apparent. Besides engaging the DNA-binding interface in the HLTF HIRAN domain, Vpr also binds hydrophobic residues in an adjacent region that connects the HIRAN and ATPase/helicase domains. In particular, mutations of Glu-24 and Arg-36 in the Vpr substrate recruitment interface (Fig. 4A) abrogated Vpr's ability to deplete HLTF without impinging on UNG2 degradation (19). Thus, the above observations suggest that the Vpr-HLTF interface is more extensive than that of Vpr-UNG2. Despite this, the affinity of the Vpr-HLTF binding appears to be weaker than that of Vpr-UNG2, at least under experimental conditions employed in our study. For example, although analytical size exclusion column chromatography analyses of the mixtures of Vpr, DDB1, DCAF1, and UNG2 revealed that they form a stable near stoichiometric complex (23, 39), similar analysis of HLTF showed a sub-stoichiometric amount of HLTF in the complex with DDB1, DCAF1, Vpr, and HLTF-NTD (Fig. 2A). Further, endogenous UNG2 appears to be depleted by Vpr more rapidly than HLTF (26). Future structural studies will provide a more detailed understanding of how Vpr engages HLTF for its degradation.
Recent studies have begun to reveal that Vpr targets multiple DNA repair pathways. In addition to HLTF, direct interactions between DCAF1-Vpr and UNG2 and MUS81-EME1 were reported (23, 24, 39). These data suggest that Vpr disrupts cellular DNA repair machinery to facilitate efficient HIV-1 replication/persistence in the host. Importantly, Vpr forms nuclear foci in cycling cells and in CD4+ T lymphocytes and recruits DCAF1 to sites, where DNA repair proteins are also observed (16). Thus, it is of interest to test whether down-regulation of these cellular factors facilitates HIV-1 infection. Further, searches for additional Vpr targets in the DNA repair pathways are also warranted.
Experimental procedures
Mammalian expression plasmids
Human HLTF cDNA point mutants were constructed using standard techniques and subcloned into pCG plasmid encoding triple FLAG epitope tag (26). The N terminus of HLTF, residues 55–299, was cloned into the pcDNA 3.1 (Thermo Fisher Scientific) with a Myc epitope tag at the N terminus. The pcDNA 3.1 plasmids expressing DCAF1, SAMHD1, and HIV-1 NL4-3 Vpr were described previously (24, 37, 39). A fusion construct of HLTF residues 256–299 and SAMHD1 was cloned in pcDNA 3.1 plasmid with an HA epitope tag at the N terminus. Triple FLAG epitope–tagged WT and mutant HLTF cDNAs were amplified by PCR and subcloned into retroviral tetracycline-inducible vector pRetroX-TetOne-Puro (Clontech), using standard molecular biology techniques.
Mammalian cell lines and transfection
Human embryonic kidney cells (HEK 293T) (ATCC, catalog no. CRL-3216) were cultured in advanced DMEM, supplemented with 1% (v/v) 100× glutamine and 10% (v/v) fetal bovine serum, at 37 °C and with 5% CO2. Typically, 1 × 105 cells were plated in each well of a 12-well culture plate 24 h before transfection. Transfection of HEK 293T cells with appropriate DNA mixtures using Lipofectamine 2000 (Life Technologies, Inc.) was performed according to the manufacturer's protocol. U2OS cell lines harboring doxycycline-inducible HIV-1 NL4-3 Vpr or control lacking Vpr were used as described previously (26). U2OS cells were transfected with siRNA targeting DCAF1, HLTF, or nontargeting scrambled siRNA (26, 40) using Lipofectamine RNAiMAX (Life Technologies, Inc.) according to the manufacturer's instructions. 24 h later, cells were treated with doxycycline. To generate U2OS cell lines harboring doxycycline-inducible FLAG epitope–tagged HLTF, cells were transduced with pRetroX-TetOne-Puro viruses expressing full-length HLTF WT or the 3A (Y72A/Y93A/I275A) mutant. After 2 days, cells were selected with puromycin (2 μg/ml) for 3 days and then expanded for experiments.
Western blotting
Cells were collected 48 h post transfection. After washing twice with PBS, cells were lysed with sonication. Cell lysate was mixed with loading buffer and heated at 95 °C for 5 min. Cell extracts were separated by SDS-PAGE and analyzed by Western blotting with appropriate antibodies. The antibodies used in this study included anti-FLAG (catalog no. F1804, Sigma), anti-HA (catalog no. MMS-101P, Covance), anti-actin (catalog no. sc-47778, Santa Cruz Biotechnology), anti-Myc (catalog no.MMS-150P, Covance), anti-DCAF1 (catalog no. sc-376850, Santa Cruz Biotechnology), anti-HLTF (catalog no. PA5–30178, Thermo Fisher Scientific), anti-TFIID (catalog no., sc-204, Santa Cruz Biotechnology), anti-Lamin B1 (catalog no. ab16048, Abcam), anti-rabbit IgG (catalog no. A8275, Sigma), and anti-mouse IgG (catalog no. A2304, Sigma).
Retroviral vectors and infections
HIV-1 NL4-3 Vpr gene was subcloned into a retroviral bi-cistronic murine stem cell virus-based vector MIG co-expressing GFP from an internal ribosome entry site element. Viruses were produced from HEK 293T cells, as described previously (19). For Vpr-mediated HLTF degradation assays, U2OS cells (1 × 105) harboring doxycycline inducible HLTF transgenes were treated with doxycycline (100 ng/ml) and then transduced with a MIG retroviral vector expressing FLAG-HA epitope–tagged Vpr, or with a control MIG vector lacking Vpr cDNA, at multiplicity of infection of 2:1, or 6:1. Cells were harvested 48 h post infection. Total cell extracts, prepared as previously described, were analyzed by immunoblotting for HLTF with anti-FLAG, or Vpr with anti-HA antibodies, and enhanced chemiluminescence (26).
Escherichia coli expression plasmids
The cDNA encoding HLTF residues 55–299, 55–259, 55–204, 55–180 (HIRAN), or 181–299 (LINKER) were cloned into the pET21 vector (EMD Biosciences) with Myc at the N terminus and His6 at the C terminus, respectively. Additionally, the LINKER was cloned into the pET21 vector only with His6 at the C terminus. The thioredoxin (Trx) fusion constructs with HLTF residues 256–299 or 181–266 were cloned in the pET32 vector (EMD Biosciences) with His6 tag connecting Trx and HLTF. The plasmid expressing Vpr was described previously (23). Site-specific mutants of Vpr and HLTF were prepared with a QuikChange mutagenesis kit (Agilent Technologies).
Protein expression and purification
The various HLTF constructs cloned in pET21 and pET32 vectors were expressed in E. coli Rosetta 2 (DE3) in autoinduction medium at 18 °C for 16 h. All proteins were first purified using a 5-ml nickel–nitrilotriacetic acid column (GE Healthcare), then aggregates were removed by gel-filtration column chromatography (Hi-Load Superdex200 16/60, GE Healthcare). Protein complexes including various DDB1-DCAF1, DDB1-DCAF1-Vpr, and CUL4-RBX1 were prepared as described previously (23, 38). The proteins were stored in a buffer containing 25 mm sodium phosphate, pH 7.5, 150 mm NaCl, 1 mm DTT, 5% glycerol, and 0.02% sodium azide.
In vitro ubiquitination assays
Typically, E1 (UBA1, 0.8 μm), E2 (Ubc-5Hb, 5 μm), and appropriate E3 ubiquitin ligase complexes (mixtures of CUL4A-RBX1 and DDB1-DCAF1 or DDB1-DCAF1-Vpr, 0.6 μm) were incubated with 3 μm HLTF and 5 μm ubiquitin in a buffer containing 10 mm Tris-HCl, pH 7.5, 150 mm NaCl, 5% glycerol, 20 units/ml pyrophosphatase, 1 mm tris(2-carboxyethyl)phosphine, and 5 mm ATP. The reactions were terminated with SDS-PAGE sample loading buffer at 2-, 5-, 10-, and 15-min intervals or 5-, 10-, and 15-min intervals. The extent of ubiquitination was revealed by immunoblotting with anti-Myc or anti-HLTF antibodies.
Size exclusion column chromatography
Proteins or protein mixtures (50 μl) at a concentration of 12.5 μm or 62.5 μm in a buffer containing 25 mm Tris-HCl, pH 7.5, 5% glycerol, and 0.02% sodium azide were incubated at 4 °C for 30 min prior to separation with a 24-ml analytical Superdex 200 Increase gel filtration column at a flow rate of 0.5 ml/min. Peak fractions were collected at 0.5 ml volume, concentrated with Amicon concentrator (EMD Millipore), separated by 12% SDS-PAGE, and stained with Coomassie Brilliant Blue.
Isothermal titration calorimetry (ITC)
Protein complexes, NusA-VprΔC, DDB1-DCAF1c-Vpr, or DDB1-DCAF1c-VprΔC (30 μm) were placed in the sample cell of the PEAQ-ITC Calorimeter (Malvern Instruments Ltd.), and aliquots of HLTF-NTD or HLTF-LINKER (450 μm) were titrated at 20 °C. Titration was carried out in a buffer containing 20 mm sodium phosphate, pH 7.4, and 100 mm NaCl. Samples were dialyzed against the same buffer and concentrated using Amicon concentrator (EMD Millipore). Data analyses were performed with the PEAQ-ITC analysis software.
Molecular modeling
The structure of CRL4 E3 ligase comprising CUL4, RBX1, and DDB1 (PDB ID, 2HYE) was superimposed on the DDB1-DCAF1-Vpr component of DDB1-DCAF1-Vpr-UNG2 structure (PDB ID, 5JK7). Binding of HLTF HIRAN domain (PDB ID, 4XZG) to Vpr was modeled with HADDOCK using Vpr residues Gly-43, Tyr-47, and Asp-52, and HLTF residues Arg-71, Tyr-72, and Tyr-93 as active constraints (41). The structure was rendered with Chimera (42).
Author contributions
J. A. and J. S. conceived the project. X. Z., M. D., C. H., K. H., C. M., J. S., and J. A. designed the experiments and analyzed the data. X. Z., J. S., and J. A. wrote the paper.
Supplementary Material
Acknowledgments
We thank Dr. Teresa Brosenitsch for critical reading of the manuscript and editorial help. We thank Juan Qian for expert assistance and Dr. Andy Hinck for sharing the PEAQ-ITC calorimeter.
This work was supported by National Institute of Health Grants P50GM082251 (J. A. and J. S.), R01GM116642 (J. A.), R01AI100673 (J. S.), and R01GM123973 (J. S.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
This article contains supplemental Figs. 1–4.
- Vpr
- viral protein R
- DCAF1
- DDB1 and CUL4-associated factor 1
- CRL4
- cullin4-RING ligase
- HLTF
- helicase-like transcription factor
- UNG2
- uracil DNA glycosylase 2
- SIV
- simian immunodeficiency virus
- ITC
- isothermal titration calorimetry
- Trx
- thioredoxin.
References
- 1. Malim M. H., and Emerman M. (2008) HIV-1 accessory proteins—ensuring viral survival in a hostile environment. Cell Host Microbe 3, 388–398 [DOI] [PubMed] [Google Scholar]
- 2. Collins D. R., and Collins K. L. (2014) HIV-1 accessory proteins adapt cellular adaptors to facilitate immune evasion. PLoS Pathog. 10, e1003851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Simon V., Bloch N., and Landau N. R. (2015) Intrinsic host restrictions to HIV-1 and mechanisms of viral escape. Nat. Immunol. 16, 546–553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Connor R. I., Chen B. K., Choe S., and Landau N. R. (1995) Vpr is required for efficient replication of human immunodeficiency virus type-1 in mononuclear phagocytes. Virology 206, 935–944 [DOI] [PubMed] [Google Scholar]
- 5. Goh W. C., Rogel M. E., Kinsey C. M., Michael S. F., Fultz P. N., Nowak M. A., Hahn B. H., and Emerman M. (1998) HIV-1 Vpr increases viral expression by manipulation of the cell cycle: A mechanism for selection of Vpr in vivo. Nat. Med. 4, 65–71 [DOI] [PubMed] [Google Scholar]
- 6. Subbramanian R. A., Kessous-Elbaz A., Lodge R., Forget J., Yao X. J., Bergeron D., and Cohen E. A. (1998) Human immunodeficiency virus type 1 Vpr is a positive regulator of viral transcription and infectivity in primary human macrophages. J. Exp. Med. 187, 1103–1111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Dehart J. L., and Planelles V. (2008) Human immunodeficiency virus type 1 Vpr links proteasomal degradation and checkpoint activation. J. Virol. 82, 1066–1072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Romani B., and Cohen E. A. (2012) Lentivirus Vpr and Vpx accessory proteins usurp the cullin4-DDB1 (DCAF1) E3 ubiquitin ligase. Curr. Opin. Virol. 2, 755–763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Nakagawa T., Mondal K., and Swanson P. C. (2013) VprBP (DCAF1): A promiscuous substrate recognition subunit that incorporates into both RING-family CRL4 and HECT-family EDD/UBR5 E3 ubiquitin ligases. BMC Mol. Biol. 14, 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Laguette N., Brégnard C., Hue P., Basbous J., Yatim A., Larroque M., Kirchhoff F., Constantinou A., Sobhian B., and Benkirane M. (2014) Premature activation of the SLX4 complex by Vpr promotes G2/M arrest and escape from innate immune sensing. Cell 156, 134–145 [DOI] [PubMed] [Google Scholar]
- 11. Vermeire J., Roesch F., Sauter D., Rua R., Hotter D., Van Nuffel A., Vanderstraeten H., Naessens E., Iannucci V., Landi A., Witkowski W., Baeyens A., Kirchhoff F., and Verhasselt B. (2016) HIV triggers a cGAS-dependent, Vpu- and Vpr-regulated type I interferon response in CD4+ T cells. Cell Rep. 17, 413–424 [DOI] [PubMed] [Google Scholar]
- 12. Poon B., Jowett J. B., Stewart S. A., Armstrong R. W., Rishton G. M., and Chen I. S. (1997) Human immunodeficiency virus type 1 vpr gene induces phenotypic effects similar to those of the DNA alkylating agent, nitrogen mustard. J. Virol. 71, 3961–3971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Roshal M., Kim B., Zhu Y., Nghiem P., and Planelles V. (2003) Activation of the ATR-mediated DNA damage response by the HIV-1 viral protein R. J. Biol. Chem. 278, 25879–25886 [DOI] [PubMed] [Google Scholar]
- 14. Lai M., Zimmerman E. S., Planelles V., and Chen J. (2005) Activation of the ATR pathway by human immunodeficiency virus type 1 Vpr involves its direct binding to chromatin in vivo. J. Virol. 79, 15443–15451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Zimmerman E. S., Chen J., Andersen J. L., Ardon O., Dehart J. L., Blackett J., Choudhary S. K., Camerini D., Nghiem P., and Planelles V. (2004) Human immunodeficiency virus type 1 Vpr-mediated G2 arrest requires Rad17 and Hus1 and induces nuclear BRCA1 and γ-H2AX focus formation. Mol. Cell Biol. 24, 9286–9294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Belzile J. P., Abrahamyan L. G., Gérard F. C., Rougeau N., and Cohen E. A. (2010) Formation of mobile chromatin-associated nuclear foci containing HIV-1 Vpr and VPRBP is critical for the induction of G2 cell cycle arrest. PLoS Pathog. 6, e1001080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Belzile J. P., Duisit G., Rougeau N., Mercier J., Finzi A., and Cohen E. A. (2007) HIV-1 Vpr-mediated G2 arrest involves the DDB1-CUL4AVPRBP E3 ubiquitin ligase. PLoS Pathog. 3, e85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. DeHart J. L., Zimmerman E. S., Ardon O., Monteiro-Filho C. M., Argañaraz E. R., and Planelles V. (2007) HIV-1 Vpr activates the G2 checkpoint through manipulation of the ubiquitin proteasome system. Virol. J. 4, 57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hrecka K., Gierszewska M., Srivastava S., Kozaczkiewicz L., Swanson S. K., Florens L., Washburn M. P., and Skowronski J. (2007) Lentiviral Vpr usurps Cul4-DDB1[VprBP] E3 ubiquitin ligase to modulate cell cycle. Proc. Natl. Acad. Sci. U.S.A. 104, 11778–11783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Le Rouzic E., Belaïdouni N., Estrabaud E., Morel M., Rain J. C., Transy C., and Margottin-Goguet F. (2007) HIV1 Vpr arrests the cell cycle by recruiting DCAF1/VprBP, a receptor of the Cul4-DDB1 ubiquitin ligase. Cell Cycle 6, 182–188 [DOI] [PubMed] [Google Scholar]
- 21. Tan L., Ehrlich E., and Yu X. F. (2007) DDB1 and Cul4A are required for human immunodeficiency virus type 1 Vpr-induced G2 arrest. J. Virol. 81, 10822–10830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Wen X., Duus K. M., Friedrich T. D., and de Noronha C. M. (2007) The HIV1 protein Vpr acts to promote G2 cell cycle arrest by engaging a DDB1 and Cullin4A-containing ubiquitin ligase complex using VprBP/DCAF1 as an adaptor. J. Biol. Chem. 282, 27046–27057 [DOI] [PubMed] [Google Scholar]
- 23. Ahn J., Vu T., Novince Z., Guerrero-Santoro J., Rapic-Otrin V., and Gronenborn A. M. (2010) HIV-1 Vpr loads uracil DNA glycosylase-2 onto DCAF1, a substrate recognition subunit of a cullin 4A-ring E3 ubiquitin ligase for proteasome-dependent degradation. J. Biol. Chem. 285, 37333–37341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Zhou X., DeLucia M., and Ahn J. (2016) SLX4-SLX1 protein-independent down-regulation of MUS81-EME1 protein by HIV-1 viral protein R (Vpr). J. Biol. Chem. 291, 16936–16947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. DePaula-Silva A. B., Cassiday P. A., Chumley J., Bosque A., Monteiro-Filho C. M., Mahon C. S., Cone K. R., Krogan N., Elde N. C., and Planelles V. (2015) Determinants for degradation of SAMHD1, Mus81 and induction of G2 arrest in HIV-1 Vpr and SIVagm Vpr. Virology 477, 10–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Hrecka K., Hao C., Shun M. C., Kaur S., Swanson S. K., Florens L., Washburn M. P., and Skowronski J. (2016) HIV-1 and HIV-2 exhibit divergent interactions with HLTF and UNG2 DNA repair proteins. Proc. Natl. Acad. Sci. U.S.A. 113, E3921–E3930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Motegi A., Liaw H. J., Lee K. Y., Roest H. P., Maas A., Wu X., Moinova H., Markowitz S. D., Ding H., Hoeijmakers J. H., and Myung K. (2008) Polyubiquitination of proliferating cell nuclear antigen by HLTF and SHPRH prevents genomic instability from stalled replication forks. Proc. Natl. Acad. Sci. U.S.A. 105, 12411–12416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Unk I., Hajdú I., Fátyol K., Hurwitz J., Yoon J. H., Prakash L., Prakash S., and Haracska L. (2008) Human HLTF functions as a ubiquitin ligase for proliferating cell nuclear antigen polyubiquitination. Proc. Natl. Acad. Sci. U.S.A. 105, 3768–3773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Chiu R. K., Brun J., Ramaekers C., Theys J., Weng L., Lambin P., Gray D. A., and Wouters B. G. (2006) Lysine 63-polyubiquitination guards against translesion synthesis-induced mutations. PLoS Genet. 2, e116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Blastyák A., Hajdú I., Unk I., and Haracska L. (2010) Role of double-stranded DNA translocase activity of human HLTF in replication of damaged DNA. Mol. Cell Biol. 30, 684–693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Achar Y. J., Balogh D., Neculai D., Juhasz S., Morocz M., Gali H., Dhe-Paganon S., Venclovas, ., and Haracska L. (2015) Human HLTF mediates postreplication repair by its HIRAN domain-dependent replication fork remodelling. Nucleic Acids Res. 43, 10277–10291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Hishiki A., Hara K., Ikegaya Y., Yokoyama H., Shimizu T., Sato M., and Hashimoto H. (2015) Structure of a novel DNA-binding domain of helicase-like transcription factor (HLTF) and its functional implication in DNA damage tolerance. J. Biol. Chem. 290, 13215–13223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kile A. C., Chavez D. A., Bacal J., Eldirany S., Korzhnev D. M., Bezsonova I., Eichman B. F., and Cimprich K. A. (2015) HLTF's ancient HIRAN domain binds 3′ DNA ends to drive replication fork reversal. Mol. Cell 58, 1090–1100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Achar Y. J., Balogh D., and Haracska L. (2011) Coordinated protein and DNA remodeling by human HLTF on stalled replication fork. Proc. Natl. Acad. Sci. U.S.A. 108, 14073–14078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Lahouassa H., Blondot M. L., Chauveau L., Chougui G., Morel M., Leduc M., Guillonneau F., Ramirez B. C., Schwartz O., and Margottin-Goguet F. (2016) HIV-1 Vpr degrades the HLTF DNA translocase in T cells and macrophages. Proc. Natl. Acad. Sci. U.S.A. 113, 5311–5316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Ahn J., Hao C., Yan J., DeLucia M., Mehrens J., Wang C., Gronenborn A. M., and Skowronski J. (2012) HIV/simian immunodeficiency virus (SIV) accessory virulence factor Vpx loads the host cell restriction factor SAMHD1 onto the E3 ubiquitin ligase complex CRL4DCAF1. J. Biol. Chem. 287, 12550–12558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. DeLucia M., Mehrens J., Wu Y., and Ahn J. (2013) HIV-2 and SIVmac accessory virulence factor Vpx down-regulates SAMHD1 enzyme catalysis prior to proteasome-dependent degradation. J. Biol. Chem. 288, 19116–19126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Ahn J., Novince Z., Concel J., Byeon C. H., Makhov A. M., Byeon I. J., Zhang P., and Gronenborn A. M. (2011) The cullin-RING E3 ubiquitin ligase CRL4-DCAF1 complex dimerizes via a short helical region in DCAF1. Biochemistry 50, 1359–1367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Wu Y., Zhou X., Barnes C. O., DeLucia M., Cohen A. E., Gronenborn A. M., Ahn J., and Calero G. (2016) The DDB1-DCAF1-Vpr-UNG2 crystal structure reveals how HIV-1 Vpr steers human UNG2 toward destruction. Nat. Struct. Mol. Biol. 23, 933–940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Srivastava S., Swanson S. K., Manel N., Florens L., Washburn M. P., and Skowronski J. (2008) Lentiviral Vpx accessory factor targets VprBP/DCAF1 substrate adaptor for cullin 4 E3 ubiquitin ligase to enable macrophage infection. PLoS Pathog. 4, e1000059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. van Zundert G. C. P., Rodrigues J. P. G. L. M., Trellet M., Schmitz C., Kastritis P. L., Karaca E., Melquiond A. S. J., van Dijk M., de Vries S. J., and Bonvin A. M. J. J. (2016) The HADDOCK2.2 web server: User-friendly integrative modeling of biomolecular complexes. J. Mol. Biol. 428, 720–725 [DOI] [PubMed] [Google Scholar]
- 42. Pettersen E. F., Goddard T. D., Huang C. C., Couch G. S., Greenblatt D. M., Meng E. C., and Ferrin T. E. (2004) UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 [DOI] [PubMed] [Google Scholar]
- 43. Petroski M. D., and Deshaies R. J. (2005) Mechanism of lysine 48-linked ubiquitin-chain synthesis by the cullin-RING ubiquitin-ligase complex SCF-Cdc34. Cell 123, 1107–1120 [DOI] [PubMed] [Google Scholar]
- 44. Pickart C. M. (2001) Mechanisms underlying ubiquitination. Ann. Rev. Biochem. 70, 503–533 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.