Abstract
Gram-negative bacteria remodel their surfaces to interact with the environment, particularly to protect pathogens from immune surveillance and host defenses. The enzyme AlmG is known to be involved in remodeling the Vibrio cholerae surface, but its specific role was not clear. A new study characterizes AlmG at the molecular level, showing it defies phylogenetic expectations to add amino acids to lipopolysaccharide (LPS). This LPS modification plays a pivotal role in V. cholerae resistance to antimicrobial peptides, weapons of the innate immune system against infections.
Keywords: Vibrio cholera, lipopolysaccharide (LPS), antimicrobial peptide (AMP), lipid A, synthetic biology, aminoacylation, polymyxin B
Introduction
A defining feature of Gram-negative bacteria is the presence of an outer membrane, which is an asymmetrical bilayer with glycerophospholipids on the cytoplasmic face and LPS2 anchored to the outer face. The LPS is composed of three regions: the lipid A domain, the core oligosaccharide, and the O-antigen polysaccharide. The lipid A domain is recognized by the innate immune system, leading to the activation of signaling pathways governing host-defense responses, and is the target of antimicrobial peptides such as defensins and polymyxin B, which kill bacteria by affecting membrane integrity. Recognition and exploitation of the lipid A structure therefore relies on the inability of bacteria to alter this component dramatically. However, a wealth of evidence demonstrates that bacteria do modify their lipid A as a virulence strategy to survive the onslaught of host defenses. The canonical lipid A structure, lipid IVA, is found in Escherichia coli K-12 and consists of a glucosamine disaccharide modified with two phosphate groups and four R-3-hydroxymyristoyl acyl chains. Two of the hydroxymyristoyl chains are further acylated with laureate (containing a C12 backbone) and myristate (C14) through the action of the late acyltransferases LpxL and LpxM, respectively (1). Pioneering studies demonstrated that Salmonella typhimurium remodels its lipid A by adding 4-amino-4-deoxy-l-arabinose and phosphoethanolamine to mask lipid A's negative charges, limiting its interaction with positively-charged antimicrobial peptides (2, 3), whereas Klebsiella pneumoniae produces a distinct lipid A in vivo to limit inflammation and to resist antimicrobial peptides and polymyxins (4). Although these remodeling events are therefore critical to understanding a variety of bacterial infections, most of the studies on lipid A remodeling have focused primarily on just a few bacterial species.
V. cholerae is the causative agent responsible for the severe diarrheal disease cholera. The global disease burden of cholera is estimated to be between 1.3 and 4 million cases per year with 21,000 to 143,000 deaths. For decades, this pathogen has been used as a model to study the regulation of host–pathogen interactions and, more recently, has enabled investigations of the type VI secretion system that facilitates direct killing of competitors. However, until recently, there was a major gap in our understanding of V. cholerae LPS and its contribution to virulence, even though resistance to polymyxin B has been used as diagnostic test to differentiate the two V. cholerae O1 biotypes, El Tor and Classical. In a landmark work, Stephen Trent's team uncovered that V. cholerae O1 El Tor pandemic strains synthesize novel mono- or diglycine-modified lipid A species (Fig. 1) that confer resistance to polymyxin B and identified the proteins AlmE, AlmF, and AlmG as required for this modification (5). Moreover, they later showed that Classical V. cholerae strains lack a functional AlmEGF due to a mutation in AlmF, providing further evidence for this mechanism and explaining the mystery of why pandemic Classical strains are polymyxin B–susceptible (6). However, further insights are still needed, as the polymyxin B–resistant O1 El Tor strains are currently causing the seventh V. cholerae pandemic.
Figure 1.
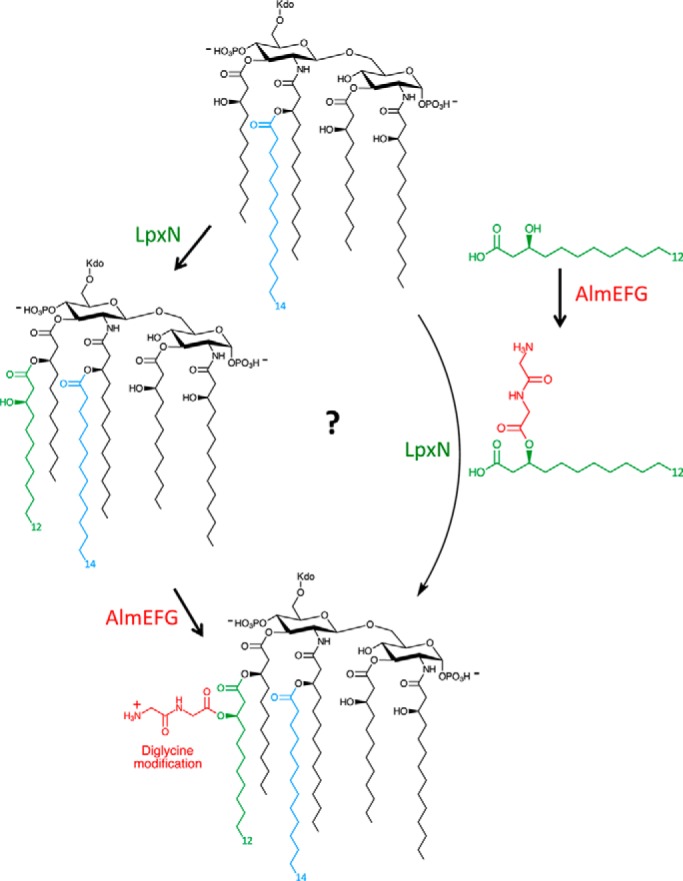
Synthesis of hexa-acylated lipid A in V. cholerae. The combined action of LpxN and AlmEFG lead to installation of a glycine-modified 3-hydroxylaurate group on the R-2′-hydroxymyristate acyl chain of Kdo-lipid IVA. Whether the process occurs in a stepwise manner (left) or, as Henderson et al. (7) speculate, whether AlmEFG might be creating a modified substrate for LpxN (right) remains to be seen.
Previous studies revealed that AlmF is an aminoacyl carrier protein and AlmE is the enzyme required to activate AlmF as a functional carrier protein (6). AlmG was suspected to be a glycyltransferase to complete the functional pathway, but its evolutionary context—it is only distantly related to enzymes of the lysophospholipid acyltransferase (LPLAT) superfamily, including LpxL, LpxM, and LpxN—provided no clear indication as to how catalysis might occur. Moreover, deciphering the function of an enzyme involved in lipid A modifications is technically challenging. In this issue of JBC, Jeremy Henderson and co-workers (7) present compelling biochemical evidence demonstrating that AlmG is the glycyltransferase in the AlmEFG pathway. To characterize the enzymatic activity of AlmG, the authors followed an elegant synthetic biology approach combining the power of bacterial engineering and biochemistry methods and exploiting E. coli as a workhorse. To define the minimum structural requirements required for the Kdo-lipid A glycine modification, the authors constructed an E. coli strain that produced a simplified Kdo-lipid A domain resembling V. cholerae lipid A. To do this, they generated an E. coli strain lacking lpxM to allow expression of Vibrio LpxN that transfers 3-hydroxylaurate to the Kdo-lipid A (8) (Fig. 1), lpxT, to facilitate the analysis of 32P-radiolabeled LPS by thin-layer chromatography, and the rfaDFC (also known as waaDFC) operon, to prevent addition of the inner core section of LPS and simplify the isolation of Kdo-lipid A material. Expression of LpxN in this background resulted in a hexa-acylated Kdo-lipid A containing a 3-hydroxylauoryl group at the 3′ position. However, mass spectrometry analysis also revealed the presence of lipid A species modified with phosphoethanolamine. It is not unprecedented that LPS truncations lead to changes in lipid A decorations most likely to maintain the overall outer membrane integrity; deletion of the phosphoethanolamine transferases eptA and eptB successfully blocked this decoration. With the resultant mutant strain in hand, the authors were ready to test AlmG function. As anticipated, AlmG was necessary and sufficient for the lipid A modification with glycine, with both mono- and diglycine-modified species observed. In agreement with the crucial role played by the lipid A–glycine modification in polymyxin B resistance, the V. cholerae almG mutant was 80-fold more susceptible to polymxyin B than the wild-type strain.
A HX4D/E catalytic dyad has been shown to be essential for the activity of the LPLAT family of proteins. Bioinformatic analysis showed that AlmG contains two putative catalytic dyads (H106X4D111 and H215X4E220). Site-directed mutagenesis experiments convincingly demonstrated that H215X4E220 was absolutely essential for the glycine modification, this being an unexpected result because the H106X4D111 dyad corresponds to the active site of the LPLAT family. Future structure–function studies are now warranted to explain AlmG's unique mechanism, including the authors' proposal that AlmG might be acting on the acyl precursor used as a substrate by LpxN rather than the intact lipid A domain (Fig. 1).
The work of Henderson et al. (7) makes a strong argument for exploiting synthetic biology approaches using E. coli to elucidate the activity of enzymes responsible for Kdo-lipid A biosynthesis and decoration from different bacteria. This strategy may also prove useful for purifying LPS of defined chemical structures to assess their potential as vaccine adjuvants and/or immunomodulators; Stephen Trent's group has published a proof-of-principle study showing the outstanding opportunities that await (9). In this context, the enzymes encoded by the alm operon represent a singular addition to the repertoire of proteins employed by Gram-negative bacteria to remodel their LPS. In a broader context, many questions remain to be investigated. Does the lipid A modification with glycine play any role in V. cholerae survival in the environment? Is there any connection between lipid A modifications and the expression/function of the Vibrio type VI secretion system? How does Vibrio coordinate the spatial–temporal expression of lipid A modifications with that of other virulence factors? How widespread is the modification of lipid A with amino acids? Answering these questions will not only advance our understanding of V. cholerae infection biology but also will provide further insights into the role of LPS in Gram-negative bacteria biology.
Acknowledgments
Research in my laboratory is supported by the Biotechnology and Biomedical Sciences Research Council (BBSRC, Grants BB/P006078/1, BB/P020194/1, and BB/N00700X/1) and the Medical Research Council (MRC, Grant MR/R005893/1).
The author declares that he has no conflicts of interests with the contents of this article.
- LPS
- lipopolysaccharide(s)
- LPLAT
- lysophospholipid acyltransferase.
References
- 1. Raetz C. R., and Whitfield C. (2002) Lipopolysaccharide endotoxins. Annu. Rev. Biochem. 71, 635–700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Lee H., Hsu F. F., Turk J., and Groisman E. A. (2004) The PmrA-regulated pmrC gene mediates phosphoethanolamine modification of lipid A and polymyxin resistance in Salmonella enterica. J. Bacteriol. 186, 4124–4133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Guo L., Lim K. B., Gunn J. S., Bainbridge B., Darveau R. P., Hackett M., and Miller S. I. (1997) Regulation of lipid A modifications by Salmonella typhimurium virulence genes phoP-phoQ. Science 276, 250–253 [DOI] [PubMed] [Google Scholar]
- 4. Llobet E., Martínez-Moliner V., Moranta D., Dahlström K. M., Regueiro V., Tomás A., Cano V., Pérez-Gutiérrez C., Frank C. G., Fernández-Carrasco H., Insua J. L., Salminen T. A., Garmendia J., and Bengoechea J. A. (2015) Deciphering tissue-induced Klebsiella pneumoniae lipid A structure. Proc. Natl. Acad. Sci. U.S.A. 112, E6369–E6378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Hankins J. V., Madsen J. A., Giles D. K., Brodbelt J. S., and Trent M. S. (2012) Amino acid addition to Vibrio cholerae LPS establishes a link between surface remodeling in Gram-positive and Gram-negative bacteria. Proc. Natl. Acad. Sci. U.S.A. 109, 8722–8727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Henderson J. C., Fage C. D., Cannon J. R., Brodbelt J. S., Keatinge-Clay A. T., and Trent M. S. (2014) Antimicrobial peptide resistance of Vibrio cholerae results from an LPS modification pathway related to nonribosomal peptide synthetases. ACS Chem.Biol. 9, 2382–2392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Henderson J. C., Herrera C. M., and Trent M. S. (2017) AlmG, responsible for polymyxin resistance in pandemic V. cholerae, is a glycyltransferase distantly related to lipid A late acyltransferases. J. Biol. Chem. 292, 21205–21215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hankins J. V., Madsen J. A., Giles D. K., Childers B. M., Klose K. E., Brodbelt J. S., and Trent M. S. (2011) Elucidation of a novel Vibrio cholerae lipid A secondary hydroxyacyltransferase and its role in innate immune recognition. Mol. Microbiol. 81, 1313–1329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Needham B. D., Carroll S. M., Giles D. K., Georgiou G., Whiteley M., and Trent M. S. (2013) Modulating the innate immune response by combinatorial engineering of endotoxin. Proc. Natl. Acad. Sci. U.S.A. 110, 1464–1469 [DOI] [PMC free article] [PubMed] [Google Scholar]


