Abstract
Objective
A locally disturbed commensal microbiome might be an etiological factor in chronic rhinosinusitis (CRS) in general and in CRS without nasal polyps (CRSsNP) in particular. Lactic acid bacteria (LAB) have been suggested to restore commensal microbiomes. A honeybee LAB microbiome consisting of various lactobacilli and bifidobacteria have been found potent against CRS pathogens in vitro. Recently, we examined effects of single nasal administrations of this microbiome in healthy subjects and found it inert. In this study, we examined effects of repeated such administrations in patients with CRSsNP.
Study Design
The study was of a randomized, double‐blinded, crossover, and sham‐controlled design.
Methods
Twenty patients received 2 weeks' treatment administered using a nasal spray‐device. The subjects were monitored with regard to symptoms (SNOT‐22 questionnaire, i.e., the primary efficacy variable), changes to their microbiome, and inflammatory products (IL‐6, IL‐8, TNF‐, IL‐8,a, and MPO) in nasal lavage fluids.
Results
Neither symptom scores, microbiological explorations, nor levels of inflammatory products in nasal lavage fluids were affected by LAB (c.f. sham).
Conclusion
Two weeks' nasal administration of a honeybee LAB microbiome to patients with CRSsNP is well tolerated but affects neither symptom severity nor the microbiological flora/local inflammatory activity.
Level of Evidence
1b
Keywords: bacterial interference, probiotics, Lactobacillus, Bifidobacterium, LAB, paranasal sinuses, SNOT‐22, therapeutics, honey, bacteriology, rhinosinusitis, CRSsNP
INTRODUCTION
Chronic or recurrent rhinosinusitis (CRS) is a disease associated with impaired quality of life and substantial societal costs.1 While sometimes associated with asthma, allergy, or nasal polyposis1, many cases present without apparent underlying cause. Despite lack of etiology, and reflecting the inflammatory nature of the condition1, treatment protocols dictate intervention with antibiotics, anti‐inflammatory drugs, or even surgery1, 2, often with poor long‐term effects.
Recent studies indicate that commensal bacteria colonize the nasal and paranasal cavities of healthy individuals3, while damage to this natural microbiome, by either pathogens or antibiotics, may cause an imbalance that promotes CRS.4, 5 Arguably, treatments eliminating pathogens without damaging the commensal microbiome, or ideally helping restoring it6, may therefore offer an alternative to current protocols. Probiotic lactic acid bacteria (LAB) has been forwarded as such a measure.6
LAB, specifically a mixture of a synergistic LAB microbiota obtained from the honeybee Apis mellifera, can be administered safely to the human nasal airway7: single doses are well tolerated and does neither affect commensal bacteria nor produce any inflammatory response: as assessed by type‐1 cytokines in nasal lavages including interleukin‐1ß (IL‐1ß), interferon‐α (IFN‐α), interleukin‐6 (IL‐6), macrophage inflammatory proteins (MIP‐1α), interferon‐γ (IFN‐γ), and tumor necrosis factor‐α (TNF‐α). In vitro studies demonstrate that the honeybee LAB have potent antimicrobial effects against bacteria associated with CRS.8, 9
In this study including patients with CRS without nasal polyps (CRSsNP), we examine whether or not repeated administration of a honeybee specific LAB microbiota to the nasal cavity for 2 weeks affects symptoms of the condition as assessed by a questionnaire (SNOT‐22). In addition, we study if the intervention affects commensal bacteria or the inflammatory milieu in the nasal cavity (as assessed by nasal lavage fluid analyses of select cytokines/mediators).
MATERIALS AND METHODS
Study Design
The study was of a randomized, double‐blinded, crossover, and sham‐controlled design in which a mixture of 9 lactobacilli and 4 bifidobacteria (LAB) was examined against sham as a topical (nasal spray) 2 weeks' treatment for CRSsNP focusing on patient‐assessment (SNOT‐22 questionnaire, i.e., the primary efficacy variable), microbiology, and inflammatory indices. The study was approved by the regional ethics committee (2013/487) and informed consent was obtained.
Patients
Twenty‐one patients suffering from CRSsNP aged 21–80 years (mean age 58) were recruited (10 women and 11 men). Inclusion criteria were two nasal symptoms for more than 12 weeks, one of which was nasal obstruction or discoloured discharge, in addition to disease verification by endoscopy or CT scan according to the definition determined in the 2012 EPOS guidelines.1 Specific exclusion criteria were nasal polyposis and treatment with antibiotics within a period of 14 days prior to inclusion. Individuals with asthma and allergy were not excluded. At inclusion, an ENT surgeon (AM) examined all the subjects and a skin prick test was performed. Fourteen of the 20 patients who completed the study had undergone previous sinus surgery due to CRS.
Study Visits
Visit 1: The subjects completed a Sino‐Nasal Outcome Test (SNOT‐22) questionnaire to assess current upper respiratory tract health. An ENT surgeon (AM) conducted an examination and took a sample from the middle meatus using an E‐Swab (Copan, Murrieta, CA) for microbiological testing. Also, an assessment was performed using a fibre optic endoscope (Storz, Tuttlingen, Germany). A nasal lavage was performed.
The subjects were then given two bottles of nasal spray (marked 1 and 2; same content in both bottles) containing either LAB or sham solution, instructed on how to store and administer the spray, and administered the first dose under supervision of study personnel. The patients were also given a diary card focusing on symptoms from the upper and lower airways, the gastrointestinal tract, and other symptoms and they were instructed to note symptoms experienced during the following two weeks, i.e., the first treatment period.
Visit 2: Fourteen days after study visit 1 (i.e., after 14‐days' treatment), the patients were again asked to complete a SNOT‐22 questionnaire and the diary cards were collected. Again, AM repeated the examination, the nasal endoscopy, and the sampling as in visit 1. Finally, a nasal lavage was performed.
After a washout period of at least 4 weeks, the patients returned to the clinic and the abovementioned procedures and interventions were repeated in the same order, thereby constituting Visits 3–4. This time subjects who received nasal spray containing LAB in the first run now were subjected to sham and vice versa.
LAB Formulation and Administration
The formulation comprised 13 honeybee LAB species: Lactobacillus apinorum Fhon13N, Lactobacillus mellifer Bin4N, Lactobacillus mellis Hon2N, Lactobacillus kimbladii Hma2N, Lactobacillus melliventris Hma8N, Lactobacillus helsingborgensis Bma5N, Lactobacillus kullabergensis Biut2N, Lactobacillus kunkeei Fhon2N, Lactobacillus apis Hma11N, Bifidobacterium asteroides Bin2N, Bifidobacterium coryneforme Bma6N, Bifidobacterium Bin7N, and Bifidobacterium Hma3N.10, 11, 12
The initial concentration of each LAB was 108 CFU/mL and the matrix was Swedish sterilized heather honey (93%) and bee‐pollen in water (7%). Honey and bee pollen were used as nutrients and to stimulate the microbiota to produce bioactive metabolites.9, 13 A spray solution was obtained by mixing the abovementioned solution with sterile water (5 g/10 mL). The mixture was incubated for 48 hours at 35°C to allow the bacteria to become active. The total cell count after incubation was approximately 1011 CFU/mL. The same solution as described above, but without any bacteria, was used as sham provocation.
The spray‐device delivered 100 μL per actuation (Aptar Pharma, Crystal Lake, IL). The subjects were instructed to store the spray bottles in a fridge and to administer two spray doses to each nostril twice daily using the bottle marked 1 for the first week and the one marked 2 for the second week.
The bottles were weighed before being given to the subjects, who were instructed to bring them back at visits 2 and 4 when they were weighed again to check for compliance, which was found to be good (data not shown). Furthermore, the content of five of the spray bottles returned from different subjects were tested for viability of the LAB by comparing their bacterial growth when sprayed on de Man, Rogosa & Sharpe (MRS) (Oxoid, Hampshire, UK) agar plates supplemented with 2.0% fructose (Merck, Sollentuna, Sweden) and 0.1% L‐cysteine (Sigma‐Aldrich, Stockholm, Sweden) to a similar plate sprayed with a fresh spray bottle. Bacterial activity was found to be slightly lower than the fresh bottle yet still very high.
SNOT‐22 Questionnaire
The SNOT‐22 questionnaire comprises 22 questions relevant to the symptomatology and morbidity of upper respiratory tract conditions.14 Individual scores are produced and added to a total score (range 0–110). Eight specific questions make up a rhinology domain of the test (range 0–40). These questions are of relevance to need to blow the nose, sneezes, running nose, cough, nasal secretion going into the throat, thick nasal secretions, difficulty to sense smells/tastes, and stuffed nose, and are thought to relate directly to nasal symptoms.
E‐swab Handling and Microbiological Analysis
E‐swab samples from Visit 1, 2, 3, and 4 were diluted 1:10 and 1:100 with phosphate buffered saline (pH 7.2) and cultured on blood, chocolate, fastidious anaerobe agar (FAA), and supplemented MRS. Blood and chocolate agar plates were incubated aerobically for 24 hours at 37°C and FAA plates anaerobically for 48 hours at 37°C. The MRS plates were incubated anaerobically for 48 hours at 35°C. The colonies were counted and a selection giving representation of colonies from all different morphological appearances was selected for identification by mass spectrometry.
Matrix‐assisted laser desorption/ionization time of flight mass spectrometry (MALDI‐TOF MS) was performed as described previously.8, 15 Briefly, bacterial isolates were placed in duplicate as thin films onto a 96‐spot steel plate (Bruker Daltonics, Solna, Sweden) and dried at room temperature (i.e., direct colony technique). Each bacterial target was covered with 1.0 μL formic acid (Sigma‐Aldrich, Stockholm, Sweden), left to dry followed by 1.0 μL of a α‐cyano‐4‐hydroxycinnamic acid matrix solution (50% acetonitrile, 47.5% water, and 2.5% triflouroacetic acid) (Sigma‐Aldrich, Stockholm, Sweden). MALDI‐TOF MS was performed on an ultrafleXtreme MALDI‐TOF/TOF (Bruker Daltonics, Solna, Sweden) in linear positive mode in a mass range of 2–20 kDa. Mass spectra were analyzed using the FlexControl and MALDI Biotyper 3.1 software and reference database MBT‐BDAL‐5627. Identification criteria used in the analysis were as follows: a score of ≥2.000 indicated species level identification, a score of 1.700 to 1.999 indicated identification at the genus level, and a score of <1.700 was interpreted as no identification.
Nasal Lavages and Analysis
Nasal lavages were performed using the head‐back technique.16, 17 Briefly, using a pipette, 10 mL isotonic saline was instilled in the nasal cavity (5 mL per nostril) with the neck extended and the soft palate voluntarily closed. The head was then moved forward, allowing the lavage fluid to be blown out of the nose and into a container for further transfer to a test tube. The samples were centrifuged and frozen for later analysis as one batch.
Through magnetic Luminex assay, samples were analysed for interleukin‐6 (IL‐6), interleukin‐8 (IL‐8), interferon‐γ (IFN‐γ), tumor necrosis factor‐α (TNF‐α), myeloperoxidase (MPO) by a human premixed multi‐analyte kit (R&D Systems, Minneapolis, MN) and the Bio‐Plex 200 system (Bio‐Rad Lab, Hercules, CA). The analysis was performed in duplicates. To keep data quality high, coefficient of variance (CV) filtration was performed and duplicates exceeding a 20% CV‐cut off were omitted. Extrapolated values, as suggested by the machine, were used in case readouts were out of the detection range of the assay.
Statistics
Data are presented as individual values or as median values with interquartile ranges. The comparative analysis focused first on observation after LAB compared with sham and second on observation before and after LAB and sham, respectively, using the Friedman test and the Wilcoxon signed rank test. The statistical analysis was performed using SPSS Statistics: Version 24.0 (IBM, Armonk, NY) for all variables except for nasal lavage fluid indices that were analysed using GraphPad Prism: Version 7.01 (GraphPad, La Jolla, CA). p‐values <0.05 was considered statistically significant.
RESULTS
One of the 21 patients withdrew his consent without specifying any reason, but the remainder of the group completed the study. One patient was prescribed flucloxacillin for nasal symptoms 1 week into the LAB run and another one ciprofloxacin for pyelonephritis also 1 week into the LAB run. Clinical examinations of these individuals revealed nasal findings similar to those seen prior to administration of the antibiotics. Both patients were kept in the intention to treat analysis.
The median SNOT‐22 score for all observations prior to administration of LAB and sham was 45.5 (IQR 23.0–58.5). The scores after two weeks' administration of LAB and sham were 38.0 (IQR 28.0–68.5) and 34.0 (IQR 17–55), respectively. Individual data are indicated in Figure 1. Statistically significant differences were seen neither between LAB and sham (p = 0.082) nor between observations before and after LAB (p = 0.862) and sham (p = 0.577), respectively.
Figure 1.
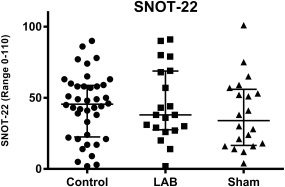
SNOT‐22 scores before treatment with LAB and sham (pooled data) and after each treatment (individual values, medians, and interquartile ranges). Statistically significant differences were neither seen between LAB and sham (p = 0.082) nor between observations before and after LAB (p = 0.862) and sham (p = 0.577), respectively. LAB = lactic acid bacteria.
For the SNOT‐22 rhinology domain, the median score for all observations prior to administration of LAB and sham was 18.0 (IQR 12.5–24.0). The scores after 2 weeks' administration of LAB and sham were 19.0 (IQR 15–28,5) and 17.5 (IQR 9.0–23), respectively. Individual data are indicated in Figure 2. Statistically significant differences were seen neither between LAB and sham (p = 0.061) nor between observations before and after LAB (p = 0.471) and sham (p = 0.992), respectively.
Figure 2.
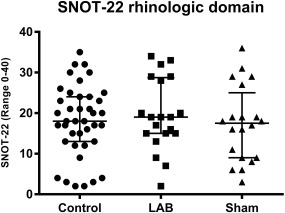
SNOT‐22 scores of the rhinology domain before treatment with LAB and sham (pooled data) and after each treatment (individual values, medians, and interquartile ranges). Statistically significant differences were neither seen between LAB and sham (p = 0.061) nor between observations before and after LAB (p = 0.471) and sham (p = 0.992), respectively. LAB = lactic acid bacteria.
Fifty‐two bacterial species were identified in the study. Of these, 13 were only found in samples before treatment and eleven only after treatment. Furthermore, some of the bacteria observed were found in separate individuals before and after treatment. Twenty‐three species were only found in one sample. In four samples, no bacteria could be identified, two before the LAB run and two after the sham run. Staphylococcus epidermidis was found in all samples after treatment with LAB. Results are indicated in Table 1. The differences in bacterial composition between observations before and after LAB treatment and sham were not significant: p = 0.219 and p = 0.263, respectively.
Table 1.
Number of patients with CRSsNP that were culture positive for specific bacterial species before and after treatment with LAB or sham. Statistically significant differences were neither seen between LAB and sham (p = 0.097) nor between observations before and after LAB (p = 0.219) and sham (p = 0.263), respectively. CRSsNP = chronic rhinosinusitis without nasal polyps; LAB = lactic acid bacteria
| Before sham | After sham | Before LAB | After LAB | |
|---|---|---|---|---|
| Actinomyces odontolyticus | 0 | 0 | 0 | 1 |
| Bacillus sonorensis | 0 | 0 | 1 | 0 |
| Brevibacterium casei | 0 | 0 | 0 | 1 |
| Citrobacter freundii | 0 | 0 | 1 | 0 |
| Clavispora lusitaniae | 0 | 1 | 0 | 0 |
| Corynebacterium propinquum/pseudodiphtheriticum/simulans/species | 5 | 1 | 6 | 3 |
| Enterobacter aerogenesis | 0 | 0 | 1 | 0 |
| Enterococcus faecalis | 0 | 1 | 1 | 2 |
| Escherichia coli | 0 | 0 | 0 | 1 |
| Gemella haemolysans | 0 | 1 | 0 | 0 |
| Haemophilus haemolyticus | 0 | 1 | 0 | 0 |
| Haemophilus influenzae | 1 | 2 | 2 | 0 |
| Klebsiella oxytoca | 0 | 1 | 0 | 0 |
| Klebsiella pneumoniae | 0 | 0 | 1 | 0 |
| Kocuria kristinae | 0 | 0 | 1 | 0 |
| Lactobacillus kunkeei | 0 | 0 | 0 | 2 |
| Micrococcus luteus | 2 | 0 | 0 | 0 |
| Moraxella_sg_Branhamella catarrhalis | 1 | 0 | 1 | 1 |
| Moraxella_sg_Moraxella lincolnii | 0 | 0 | 1 | 0 |
| Moraxella_sg_Moraxella nonliquefaciens | 1 | 1 | 3 | 1 |
| Neisseria perflava | 1 | 0 | 0 | 0 |
| Neisseria subflava | 2 | 0 | 0 | 0 |
| Propionibacterium acnes | 3 | 2 | 2 | 3 |
| Propionibacterium avidum | 0 | 0 | 1 | 0 |
| Propionibacterium species | 1 | 0 | 0 | 0 |
| Pseudomonas aeruginosa | 1 | 0 | 3 | 1 |
| Rothia amarae | 0 | 1 | 0 | 0 |
| Rothia mucilaginosa | 1 | 1 | 2 | 1 |
| Serratia marcescens | 0 | 1 | 1 | 0 |
| Serratia ureilytica | 0 | 0 | 1 | 0 |
| Staphylococcus aureus | 4 | 7 | 8 | 9 |
| Staphylococcus capitis | 5 | 2 | 7 | 2 |
| Staphylococcus epidermidis | 17 | 15 | 16 | 20 |
| Staphylococcus haemolyticus | 0 | 0 | 3 | 1 |
| Staphylococcus hominis | 1 | 2 | 1 | 2 |
| Staphylococcus intermedius | 0 | 1 | 0 | 0 |
| Staphylococcus lugdunensis | 6 | 7 | 8 | 7 |
| Staphylococcus pasteuri | 2 | 1 | 2 | 3 |
| Staphylococcus succinus | 1 | 0 | 0 | 0 |
| Staphylococcus warneri | 1 | 3 | 4 | 6 |
| Streptococcus gordonii, | 1 | 0 | 0 | 1 |
| Streptococcus infantis | 1 | 0 | 0 | 0 |
| Streptococcus oralis/mitis/peroris/parasanguinis/pneumoniae/pseudopneumoniae group | 5 | 3 | 5 | 2 |
| Streptococcus salivarius | 3 | 0 | 2 | 1 |
| Streptococcus sanguinis | 0 | 0 | 1 | 0 |
| Streptococcus urinalis | 0 | 0 | 0 | 1 |
| Streptococcus vestibularis | 0 | 0 | 0 | 1 |
IL‐6, IL‐8, IFN‐γ, TNF‐α, and MPO were measured in the nasal lavage fluid samples before and after the LAB and sham runs. Individual data are indicated in Figure 3. Statistically significant differences were not seen between LAB or sham: p‐values for the LAB versus sham calculations are indicated in Figure 3. Neither were any statistically significant differences seen between observations before and after LAB and sham runs, respectively.
Figure 3.
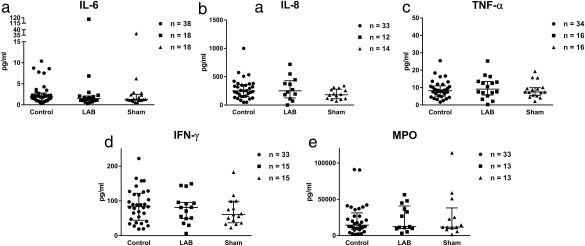
Levels of inflammatory analytes detected in collected nasal lavage fluid before treatment with LAB and sham (pooled data) and after each treatment (individual values with medians and interquartile ranges). No statistically significant differences were observed between LAB and sham: IL‐6 (p = 0.890), IL‐8 (p = 0.074), IFN‐γ (p = 0.391), TNF‐α (p = 0.380), and MPO (p = 0.966). Number of samples that passed CV filtration noted in each graph. CV = coefficient of variance; IL = interleukin; LAB = lactic acid bacteria; MPO = myeloperoxidase; TNF = tumor necrosis factor.
The patients' general experience of the treatments (LAB and sham) was as follows: 5 patients listed decreased nasal problems when using the nasal sprays, whereof 3 were in the LAB run and 2 in the sham run; 3 recorded a burning sensation in the nose after LAB/sham administration, 1 in the LAB run and 2 for both the LAB and the sham runs; 4 reported diffuse stomach problems, 3 in the sham run and 1 for both the LAB and the sham runs; 3 reported coughing, 1 in the LAB and 1 in the sham run and 1 for both the LAB and the sham runs; 2 patients reported a minor nose‐bleed, 1 in the sham run and 1 in both the sham and the LAB runs. Finally, 1 patient in the LAB run reported an unpleasant smell.
DISCUSSION
In this study, involving patients with well‐defined CRSsNP, we demonstrate that repeated nasal administration of a LAB microbiota composed of several species of lactobacilli and bifidobacteria over 2 weeks neither affects symptoms as assessed by SNOT‐22 questionnaire nor the bacterial composition or the inflammatory activity in the nasal cavity. The observations are of relevance to the evaluation of topical LAB treatment in the management of upper respiratory tract conditions such as CRS.
The patients included in this study had a history of CRSsNP treated at the regional ENT departments. In agreement, they all showed endoscopic findings of crusting, oedema, or purulent discharge at inclusion. Furthermore, the median SNOT‐22 score before any intervention was 45.5, which was high and in line with mean scores reported by Hopkins et al. for CRS with a symptom duration of more than 1 year (mean SNOT‐22 score of 42.6) and for patients eligible for revision sinonasal surgery (mean score of 44.8).14 Also, e.g., in contrast to healthy individuals included in our previous study7, the present microbiological examination at inclusion showed prevalence of CRS pathogens such as Haemophilus influenzae, Pseudomonas aeruginosa, Staphylococcus aureus, and various Moraxella species. Taken together, the abovementioned findings are in agreement with a diagnosis of CRSsNP and supports the criteria set by the EPOS 2012 guidelines used in this study.
Two hundred microliters per nostril of a 1011 CFU/mL solution of the honeybee LAB microbiome was administered twice a day for 14 days in this study. The dose is the same as used in our previous study on the effects of the honeybee LAB in healthy subjects7, but here used repeatedly, and higher than that used by Skovbjerg et al. in their study on effects of a nasal spray containing Lactobacillus rhamnosus in children suffering from secretory otitis media (100 μL per nostril twice a day of a 5 x 109 CFU/mL solution).18 While no adverse effects were observed in this study, the dose had no effect on the outcome of the parameters monitored (discussed below). Whether or not a higher dose of the honeybee LAB microbiota or a longer treatment period would have produced a different result seems unlikely but may be considered.
In this study, no statistically significant change in symptom scores, i.e., the primary efficacy variable, was observed for SNOT‐22 scores after treatment with LAB (c.f. sham). A problem studying patients suffering from CRS is the fluctuating symptomatology of the disease, which in turn makes crossover assessments difficult. Also, the minimal clinically important change of SNOT‐22 has been found to be 8.9 points.14 To study this, the data was also reviewed by exploring change of SNOT‐22 scores during each treatment. A decrease in SNOT‐22 score of 9 points or more was regarded as improvement, a change of less than 9 points in either direction was regarded as unchanged, and an increase by 9 points or more was regarded as deterioration. Depending on the results the subjects were graded 1 to 3 (deterioration to improvement) and then re‐analysed using the Wilcoxon signed rank test (p = 0.8). This strengthens our formal conclusion (from this study) that honeybee LAB had no effect on symptoms of CRSsNP.
With regard to the microbiological identification part of this study, The MALDI‐TOF method was chosen over 16S rRNA gene sequencing as it gives sufficient information on the entire microbiome at the strain level.19 No change to the nasal microbiota was observed for LAB compared to sham, neither for commensals nor CRS pathogens. This observation is relevant in relation to the proposed mechanisms that probiotics may affect diseases featuring altered commensals, pathogen colonisation, or recurrent infections as part of their pathophysiology, i.e., bacterial interference through, e.g., “quorum sensing”6, 20, including effector mechanisms such as production of antimicrobial peptides/proteins, altered pH (through production of organic acids) and competition for nutrients,2, 6 Based on this study, the LAB preparation, in its employed dose administered twice daily for 2 weeks, did not exert any detectable bacterial interference. It thereby extends our previous observation on commensals in health7 to include pathogens in CRS. Whether or not other LAB preparations or dosage schemes may possess such effects may be examined in future studies.
In order to explore the inflammatory load of the sinonasal airway, five different analytes associated with type‐1 inflammation were examined in this study (IL‐6, IL‐8, IFN‐γ, TNF‐α, MPO). These markers represent a range of biological events involving macrophages, neutrophil granulocytes (such as the neutrophil activation marker myeloperoxidase: MPO) etc.21, 22, 23 While no control subjects were included in the present study, a positive correlation between levels of different inflammatory markers were observed (using Kendall's Tau test), supporting the view that the present CRSsNP patients were indeed characterised by a sinonasal type‐1 inflammation. No differences were observed when comparing the level of these markers between LAB and sham, or when comparing levels before and after treatment with LAB and sham, respectively, suggesting that the treatment had no effect on the process of inflammation that characterises CRS/CRSsNP.
The strength of this study lies foremost in the crossover design that allows for paired comparisons. Also, the range of outcome measures, from SNOT‐22 assessment, to a microbiological assessment and to indices of inflammation, provide a global view of the disease. Main disadvantages may be a limited number of study subjects and the inherent fluctuating symptomatology of CRS. However, while we believe that the employed dosage and its mode of administration is relevant and our overall conclusion sound, the negative outcome may be discussed further. One consideration is nevertheless the mode of administration, where techniques for high‐volume delivery may be considered over the nasal spray‐device used in this study. Another may be the timing of the intervention in relation to other treatments. Specifically, would pretreatment with a course of antibiotics before the start of the topical LAB intervention, thereby reducing the bacterial load of the nasal airway, have produce different results? A final consideration is whether or not the honey bee LAB, even as a multispecies consortium of lactobacilli/bifidobacteria where individual species act synergistically and exert effects against bacteria associated with CRS in vitro 8, 9, is at all suitable for use in the human nasal airway. Alternatively, candidate bacteria may be sought after amongst commensal bacteria of the nasal airway itself. The above queries may be tested in future studies.
We conclude that 2 weeks' nasal administration of a honeybee LAB microbiome to patients diagnosed with CRSsNP is well tolerated, but neither affects symptom severity nor the microbiological flora/local inflammatory activity. Further studies are warranted to explore whether other tentative probiotic assemblages can confer positive health effects to patients suffering from inflammatory conditions of the upper airways.
ACKNOWLEDGMENTS
Lena Glantz‐Larsson and Charlotte Cervin‐Hoberg for study management assistance. Stig and Ragna Gorthons Foundation for funding the study. AV and TCO are grateful for contribution by Dr. P. Håkansson's foundation.
Funding information: The study was funded by Stig and Ragna Gorthons Foundation.
Conflict of interest (financial disclosure):
AV and TO: shareholders in ConCellae (the honeybee LAB is patented by ConCellae). LG: shareholder in Nares (active in the field of allergic rhinitis). AC: previous financial support from AstraZeneca and Mediplast. Except for what is stated above, all authors declare no conflict of interest.
BIBLIOGRAPHY
- 1. Fokkens WJ, Lund VJ, Mullol J, et al. European Position Paper on Rhinosinusitis and Nasal Polyps 2012. Rhinol Suppl 2012;3:1–298. [PubMed] [Google Scholar]
- 2. Cope EK, Lynch SV. Novel microbiome‐based therapeutics for chronic rhinosinusitis. Curr Allergy Asthma Rep 2015;15:504. [DOI] [PubMed] [Google Scholar]
- 3. Human Microbiome Project C. Structure, function and diversity of the healthy human microbiome. Nature 2012;486:207–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Abreu NA, Nagalingam NA, Song Y, et al. Sinus microbiome diversity depletion and Corynebacterium tuberculostearicum enrichment mediates rhinosinusitis. Sci Trans Med 2012;4:151ra124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Feazel LM, Robertson CE, Ramakrishnan VR, Frank DN. Microbiome complexity and Staphylococcus aureus in chronic rhinosinusitis. Laryngoscope 2012;122:467–472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Nagalingam NA, Cope EK, Lynch SV. Probiotic strategies for treatment of respiratory diseases. Trends Microbiol 2013;21:485–492. [DOI] [PubMed] [Google Scholar]
- 7. Mårtensson A, Greiff L, Lamei SS, et al. Effects of a honeybee lactic acid bacterial microbiome on human nasal symptoms, commensals, and biomarkers. Int Forum Allergy Rhinol 2016;6:956–963. [DOI] [PubMed] [Google Scholar]
- 8. Butler E, Oien RF, Lindholm C, Olofsson TC, Nilson B, Vasquez A. A pilot study investigating lactic acid bacterial symbionts from the honeybee in inhibiting human chronic wound pathogens. Int Wound J 2016;13:729–737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Olofsson TC, Butler E, Markowicz P, Lindholm C, Larsson L, Vasquez A. Lactic acid bacterial symbionts in honeybees ‐ an unknown key to honey's antimicrobial and therapeutic activities. Int Wound J 2016;13:668–679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Olofsson TC, Vasquez A. Detection and identification of a novel lactic acid bacterial flora within the honey stomach of the honeybee Apis mellifera. Curr Microbiol 2008;57:356–363. [DOI] [PubMed] [Google Scholar]
- 11. Vasquez A, Forsgren E, Fries I, et al. Symbionts as major modulators of insect health: lactic acid bacteria and honeybees. PloS one 2012;7:e33188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Olofsson TC, Alsterfjord M, Nilson B, Butler E, Vasquez A. Lactobacillus apinorum sp. nov., Lactobacillus mellifer sp. nov., Lactobacillus mellis sp. nov., Lactobacillus melliventris sp. nov., Lactobacillus kimbladii sp. nov., Lactobacillus helsingborgensis sp. nov. and Lactobacillus kullabergensis sp. nov., isolated from the honey stomach of the honeybee Apis mellifera. Int J Syst Evol Microbiol 2014;64:3109–3119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Butler E, Alsterfjord M, Olofsson TC, Karlsson C, Malmström J, Vasquez A. Proteins of novel lactic acid bacteria from Apis mellifera mellifera: an insight into the production of known extra‐cellular proteins during microbial stress. BMC Microbiol 2013;13:235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hopkins C, Gillett S, Slack R, Lund VJ, Browne JP. Psychometric validity of the 22‐item Sinonasal Outcome Test. Clin Otolaryngol 2009;34:447–454. [DOI] [PubMed] [Google Scholar]
- 15. Saffert RT, Cunningham SA, Ihde SM, Jobe KE, Mandrekar J, Patel R. Comparison of Bruker Biotyper matrix‐assisted laser desorption ionization‐time of flight mass spectrometer to BD Phoenix automated microbiology system for identification of gram‐negative bacilli. J Clin Microbiology 2011;49:887–892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Naclerio RM, Meier HL, Adkinson NF, Jr , et al. In vivo demonstration of inflammatory mediator release following nasal challenge with antigen. Eur J Respir Dis Suppl 1983;128:26–32. [PubMed] [Google Scholar]
- 17. Naclerio RM, Meier HL, Kagey‐Sobotka A, et al. Mediator release after nasal airway challenge with allergen. Am Rev Respir Dis 1983;128:597–602. [DOI] [PubMed] [Google Scholar]
- 18. Skovbjerg S, Roos K, Holm SE, et al. Spray bacteriotherapy decreases middle ear fluid in children with secretory otitis media. Arch Dis Child 2009;94:92–98. [DOI] [PubMed] [Google Scholar]
- 19. Lamei S, Hu YO, Olofsson TC, Andersson AF, Forsgren E, Vasquez A. Improvement of identification methods for honeybee specific Lactic Acid Bacteria; future approaches. PloS One 2017;12:e0174614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Gueimonde M, Salminen S. New methods for selecting and evaluating probiotics. Dig Liver Dis 2006;38:S242–247. [DOI] [PubMed] [Google Scholar]
- 21. Scheckenbach K, Wagenmann M. Cytokine patterns and endotypes in acute and chronic rhinosinusitis. Curr Allergy Asthma Rep 2016;16:3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Bachert C, Akdis CA. Phenotypes and emerging endotypes of chronic rhinosinusitis. J Allergy Clin Immunol Pract 2016;4:621–628. [DOI] [PubMed] [Google Scholar]
- 23. Bachert C, van Kempen MJ, Hopken K, Holtappels G, Wagenmann M. Elevated levels of myeloperoxidase, pro‐inflammatory cytokines and chemokines in naturally acquired upper respiratory tract infections. Eur Arch Otorhinolaryngol 2001;258:406–412. [DOI] [PMC free article] [PubMed] [Google Scholar]


