Abstract
Background and aims
Octamer-binding transcription factor 4 (OCT4) has been implicated in the development of hepatocellular carcinoma (HCC), although the findings are controversial. We conducted a meta-analysis to assess the correlation between OCT4 and the clinicopathological characteristics and the prognostic value in HCC.
Methods
An electronic search for relevant articles was conducted in PubMed, Cochrane Library, Web of Science, EMBASE database, Chinese CNKI, and Chinese WanFang database. Correlations between OCT4 expression and clinicopathological features and survival outcomes were analyzed. Pooled odds ratios and hazard ratios with 95% CIs were calculated using STATA 14.2 software.
Results
A total of 10 trials with 985 patients were included. Positive OCT4 expression was correlated with tumor size, tumor numbers, differentiation, and TNM stage. OCT4 expression was not correlated with gender, age, hepatitis B surface antigen, alfa-fetoprotein, liver cirrhosis, vascular invasion, or tumor encapsulation. OCT4 expression was associated with poor 3- and 5-year overall survival, and disease-free survival rate.
Conclusion
OCT4 expression was associated with tumor size, tumor numbers, differentiation, and TNM stage in HCC. OCT4 may be a useful prognostic biomarker for HCC.
Keywords: octamer-binding transcription factor 4, hepatocellular carcinoma, prognosis, meta-analysis
Introduction
Hepatocellular carcinoma (HCC) is one of the most common malignancies in the world, ranking fifth in the global incidence of malignant tumors. In 2012, 780,000 newly diagnosed HCC cases were reported.1–2 The mortality rate of HCC-related malignant tumors is second only to lung cancer. The highest incidence and mortality of HCC has been in China, with the incidence of liver cancer accounting for 55% of cases.3,4
HCC has a poor prognosis because of the difficulty of early diagnosis. The clinical features are not obvious, and the cancer has a high frequency of metastasis. The main treatments for HCC are surgical resection, transplantation, radiotherapy, chemotherapy, and radiofrequency. However, the benefits of treatments are poor, and recurrence is frequent.5,6 Although knowledge of the basic science and clinical treatment of liver cancer has increased markedly, there is still no effective means of early diagnosis, prevention, and treatment for HCC. Many recent studies have sought to identify effective methods of diagnosis and clarify the pathogenesis of HCC.
Transcription factor octamer-binding transcription factor 4 (OCT4) is a member of the POU transcription factor family,7 which is involved in the regulation of embryonic stem cells and self-renewal maintenance cells.8–9 The cells are over-expressed in a variety of tumors including esophageal cancer,10 gastric cancer,11 pancreatic cancer,12 breast cancer,13 and lung cancer.14 OCT4 expression is also associated with the clinicopathological features and prognosis of patients.15–17 OCT4 is highly expressed in HCC; however, the association between OCT4 expression and clinicopathological features of HCC is still controversial. Dong et al18 reported that the positive expression of OCT4 is related to differentiation of HCC, but not to tumor size, hepatitis B surface antigen (HBsAg), and alfa-fetoprotein (AFP). Interestingly, Huang et al17 found that OCT4 expression is associated with tumor size, vascular invasion, and TNM stage, but not with tumor differentiation. Zhao et al16 demonstrated that OCT4 expression is associated with tumor size and tumor differentiation grade.
A meta-analysis19 explored the relationship between OCT4 and the clinicopathological features and prognosis of gastrointestinal cancer. The analysis, which included five studies on the clinicopathological features and prognosis of HCC, did not correlate the positive expression of OCT4 with the clinicopathological features of HCC. However, the number of articles and patients included in the meta-analysis were limited, and the results were weakened by the lack of analysis of the relationship between the positive OCT4 expression and the 3- and 5-year survival rates. To provide more definitive clarity, we conducted this meta-analysis to determine the relationship between OCT4 expression and clinicopathological characteristics and the prognostic value in HCC.
Methods
Search strategy
A comprehensive literature search for articles published in any language up to September 1, 2017, was conducted in the Pubmed, Cochrane Library, Web of Science, EMBASE database, Chinese CNKI, and Chinese WanFang electronic databases. The keywords used were “Pou5f1 or OCT3 or OCT4” and “liver cancer” OR “hepatic carcinoma” OR “hepatocellular carcinoma” OR “cancer of the liver”. We also searched the references cited in the identified articles to identify other applicable studies.
Eligibility criteria
Studies were included if they met all of the following inclusion criteria: 1) the full text was available; 2) patients were clearly diagnosed as having HCC and directly examined for OCT4 expression status; 3) OCT4 expression was mainly tested by immunohistochemistry (IHC) or reverse transcription polymerase chain reaction (RT-PCR); 4) results included clinicopathological characteristics, disease-free (recurrence-free) survival (DFS), and overall survival (OS); and 5) hazard ratios (HRs) for OS were reported or could be calculated from the published data.
Studies were excluded if 1) they included ecological studies, case reports, reviews, editorials, letters, conference abstracts, or animal trials; 2) they were repetition studies based on the same database or patients; and 3) if samples were from lymph nodes or the peritoneal cavity.
Data extraction
Two investigators (Chaojie Liang and Hua Ge) independently screened all articles to determine if they met the inclusion criteria. Discrepancies were resolved by discussions, reextraction, or third-party adjudication. Extracted data included the name of the first author, publication year, number of patients, region of origin, patient characteristics, and HRs with 95% CIs for OS.
Quality assessment
The Newcastle–Ottawa Scale (NOS) was introduced to evaluate the quality of included studies. An NOS score ≥6 indicated good quality and ≤5 indicated poor quality. Studies considered to be of high quality were included.
Statistical analyses
Meta-analyses were conducted using STATA 14.2 software. Pooled odds ratios (ORs) with 95% CIs were used to quantitatively determine the association between positive OCT4 expression and clinicopathological features, including gender, age, HBsAg, AFP, liver cirrhosis, tumor size, vascular invasion, tumor encapsulation, tumor number, differentiation, and TNM stage. The heterogeneity between the studies was evaluated by the chi-squared-based Q and I2 tests. I2 values >50% and probability of hereogeneity ≥0.1 were considered to have a significant heterogeneity. According to the results of heterogeneity analysis, a random or fixed effects model was used, and subgroup analysis was conducted to explore the source of heterogeneity. Engauge Digitizer 10.0 software was used to extract the survival data from a Kaplan–Meier curve in the articles. An HR or OR >1 implies a worse prognosis for the group with positive OCT4 expression. The finding was considered statistically significant if the 95% CI did not overlap 1. Potential publication bias was examined by the Begg’s funnel plot test.
Results
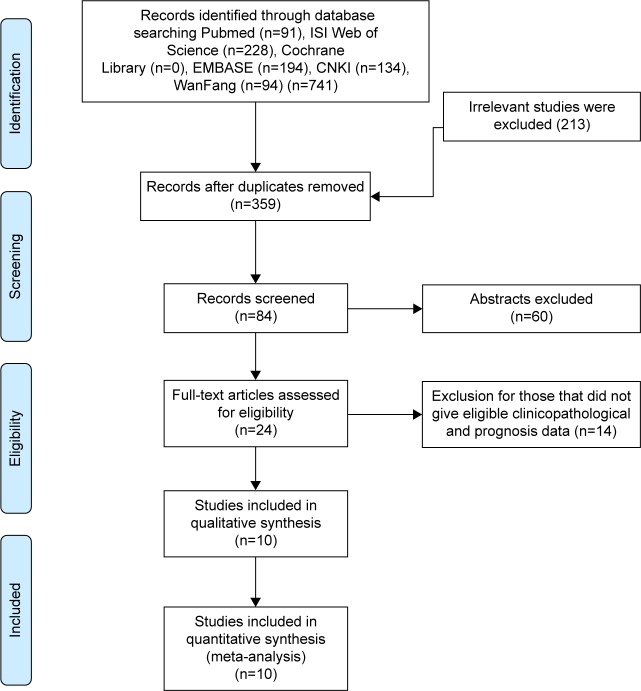
A total of 741 potential studies were initially identified on the basis of our defined criteria (Figure 1). Of these, 717 were excluded after reviewing the titles and abstracts, since these articles were duplicated, not related to OCT4, or did not involve tumor tissues. A total of 24 studies were assessed by reading the full text. Of these, 14 studies were excluded due to inefficient information and/or lack of cutoff value for OCT4 expression. Finally, 10 eligible articles15–18,20–25 were included in this meta-analysis. The characteristics of the included studies are summarized in Table 1. These studies were published from 2010 to 2016, with 985 HCC patients enrolled. Sample sizes ranged from 44 to 228 patients. Six studies16,20–24 included ≤100 patients, and four15,17,18,25 included >100 patients. Four17,21,24,25 of these studies utilized RT-PCR, while IHC was used in remaining six.15,16,18,20,22,23 All of these studies evaluated patients from China. Five studies were published in English and the others in Chinese. All of these studies scored ≥6 in methodological assessment, which implied they were of high quality.
Figure 1.
Flow diagram of study selection.
Table 1.
Characteristics of studies included in the meta-analysis
| Author | Year | Country | Type | No of patients | Experimental methods | Expression, cancer (+/−)/control (+/−) | Gender, male (+/−)/female (+/−) | Age, >50 (+/−)/≤50 (+/−) | HBsAg, positive (+/−)/negative (+/−) | AFP, positive (+/−)/negative (+/−) | Liver cirrhosis, Yes (+/−)/No (+/−) | Tumor size, >3 cm (+/−)/≤3 cm (+/−) | Vascular invasion, Yes (+/−)/No (+/−) | Tumor encapsulation, Yes (+/−)/No (+/−) | Tumor number, single (+/−)/multiple (+/−) | Differentiation, low (+/−)/moderate and high (+/−) | TNM stage, I (+/−)/II+III (+/−) | Survival information | Quality score (NOS) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Huang et al17 | 2011 | China | HCC | 136 | RT-PCR | 92/44 NA |
84/38 8/6 |
48/20 44/24 |
81/40 11/4 |
32/19 60/25 |
83/38 9/6 |
28/23 64/21 |
63/40 29/4 |
21/11 71/33 |
59/34 33/10 |
49/27 43/17 |
40/33 52/11 |
Y | 9 |
| Yin et al15 | 2012 | China | HCC | 228 | IHC | 88/140 NA |
78/116 10/24 |
40/69 48/71 |
75/120 13/20 |
64/100 24/40 |
74/125 14/15 |
40/77 48/63 |
46/56 42/84 |
37/76 51/64 |
75/124 13/16 |
56/104 32/36 |
42/79 46/61 |
Y | 8 |
| Zhao et al16 | 2016 | China | HCC | 86 | IHC | 52/34 NA |
42/25 10/9 |
10/7 42/27 |
46/27 6/7 |
31/13 21/21 |
28/17 24/17 |
19/20 33/14 |
19/11 33/23 |
28/20 24/14 |
39/29 13/5 |
14/3 38/31 |
NA | Y | 9 |
| Dong et al18 | 2012 | China | HCC | 152 | IHC | 103/49 1/13 |
92/41 11/8 |
50/15 53/34 |
88/43 15/6 |
76/34 27/15 |
49/23 54/26 |
31/14 72/35 |
NA | NA | NA | 37/29 60/20 |
NA | Y | 8 |
| Yin et al24 | 2013 | China | HCC | 57 | RT-PCR | 19/38 NA |
16/32 3/6 |
8/23 11/15 |
1/2 18/36 |
6/12 13/26 |
2/4 17/34 |
10/18 9/20 |
6/10 13/28 |
11/16 8/22 |
12/30 7/8 |
14/35 5/3 |
8/16 11/22 |
Y | 8 |
| Lin et al22 | 2015 | China | HCC | 60 | IHC | 41/29 | 19/14 14/13 |
18/8 23/11 |
27/19 9/5 |
22/6 27/5 |
NA | 13/10 31/6 |
15/2 26/17 |
NA | NA | NA | 15/17 21/7 |
NA | 6 |
| Zhang23 | 2012 | China | HCC | 50 | IHC | 38/12 25/33 |
31/7 7/5 |
23/6 15/6 |
NA | NA | NA | 10/4 28/8 |
NA | NA | NA | 13/10 25/2 |
11/9 27/3 |
NA | 6 |
| Xi et al20 | 2015 | China | HCC | 65 | IHC | 35/30 NA |
22/23 13/7 |
23/19 12/11 |
NA | 20/23 15/7 |
NA | 25/21 10/9 |
21/8 14/22 |
9/16 26/14 |
NA | 18/16 17/14 |
16/10 19/20 |
NA | 7 |
| Xu21 | 2016 | China | HCC | 44 | RT-PCR | 35/9 NA |
21/5 14/4 |
27/3 8/6 |
35/4 0/5 |
10/5 25/4 |
30/9 5/0 |
10/6 25/3 |
22/7 13/2 |
30/3 5/6 |
14/4 21/5 |
15/5 20/4 |
15/2 20/7 |
Y | 7 |
| Lu et al25 | 2010 | China | HCC | 107 | RT-PCR | 77/30 NA |
71/25 6/5 |
37/16 40/14 |
67/27 10/3 |
25/15 52/15 |
67/28 10/2 |
21/17 56/13 |
22/2 55/28 |
17/8 60/22 |
49/23 28/7 |
39/16 38/14 |
34/22 43/8 |
Y | 8 |
Notes: +, positive expression; −, negative expression.
Abbreviations: AFP, alfa-fetoprotein; HBsAg, hepatitis B surface antigen; HCC, hepatocellular carcinoma; IHC, immunohistochemistry; NA, not applicable; NOS, Newcastle–Ottawa Scale; RT-PCR, reverse transcription polymerase chain reaction.
Meta-analysis of clinicopathological characteristics
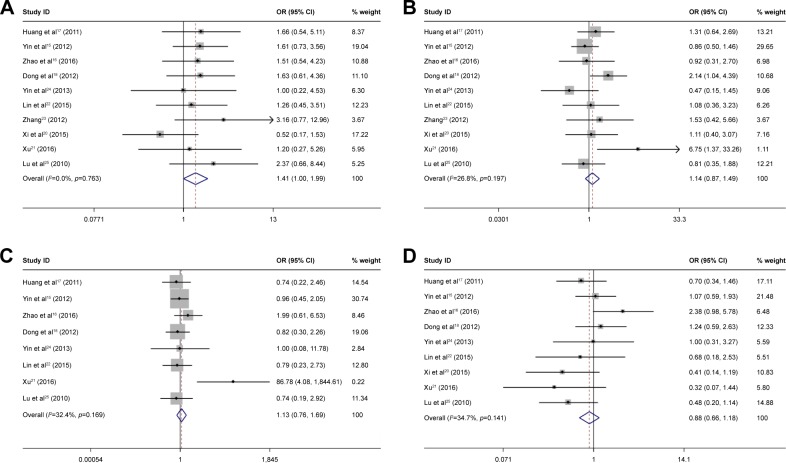
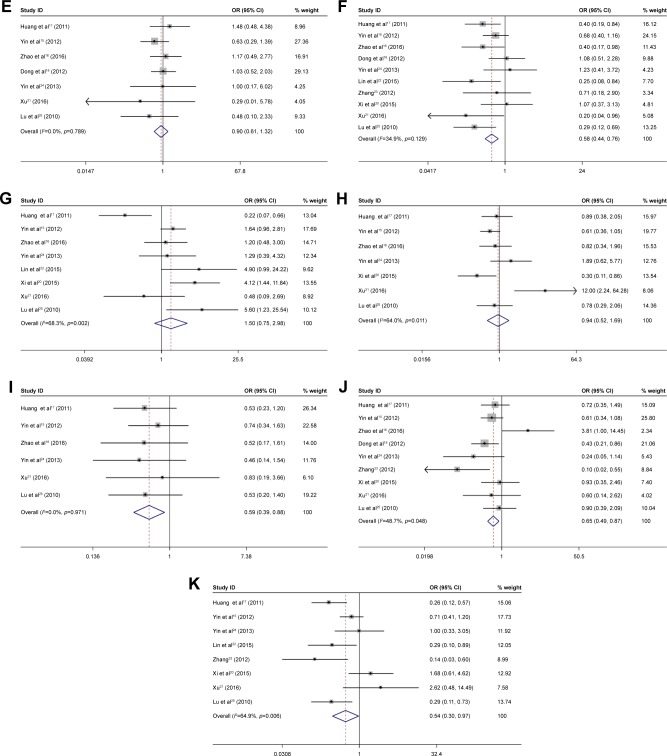
The correlation between OCT4 expression and clinicopathological features of HCC was assessed. Results are summarized in Table 2 and Figure 2. Positive OCT4 expression was associated with tumor size (OR =0.58, 95% CI =0.44–0.76, p=0.000, fixed effect), tumor number (OR =0.59, 95% CI =0.39–0.86, p=0.000, fixed effect), differentiation (OR =0.65, 95% CI =0.49–0.87, p=0.003, fixed effect), and TNM stage (OR =0.54, 95% CI =0.49–0.97, p=0.038, random effect). OCT4 expression was not significantly associated with the gender (OR =1.41, 95% CI =1.00–1.99, p=0.053, fixed effect), age (OR =2.02, 95% CI =0.97–4.21, p=0.344, fixed effect), HBsAg (OR =1.13, 95% CI =0.76–1.69, p=0.546, fixed effect), AFP (OR =0.86, 95% CI =0.66–1.18, p=0.403, fixed effect), liver cirrhosis (OR =0.9, 95% CI =0.61–1.32, p=0.584, fixed effect), vascular invasion (OR =1.50, 95% CI =0.75–2.98, p=0.253, random effect), and tumor encapsulation (OR =0.94, 95% CI =0.52–1.69, p=0.83, random effect).
Table 2.
OCT4 clinicopathological features for hepatocellular carcinoma
| Heterogeneity
| |||||||
|---|---|---|---|---|---|---|---|
| Clinicopathological features | No of studies | No of patients | Pooled OR (95% CI) | PHet | I2 (%) | p-value | Model used |
| Gender | 10 | 985 | 1.41 (1.00–1.99) | 0.053 | 0.0 | 0.763 | Fixed |
| Age | 10 | 985 | 1.14 (0.87–1.49) | 0.344 | 26.8 | 0.197 | Fixed |
| HBsAg | 8 | 870 | 1.13 (0.76–1.69) | 0.546 | 32.4 | 0.169 | Fixed |
| AFP | 9 | 935 | 0.86 (0.66–1.18) | 0.403 | 34.7 | 0.141 | Fixed |
| Liver cirrhosis | 7 | 810 | 0.90 (0.61–1.32) | 0.584 | 0.0 | 0.789 | Fixed |
| Tumor size | 10 | 985 | 0.58 (0.44–0.76) | 0.000 | 34.9 | 0.129 | Fixed |
| Vascular invasion | 8 | 783 | 1.50 (0.75–2.98) | 0.253 | 68.3 | 0.002 | Random |
| Tumor encapsulation | 7 | 723 | 0.94 (0.52–1.69) | 0.83 | 64 | 0.011 | Random |
| Tumor number | 6 | 658 | 0.59 (0.39–0.86) | 0.009 | 0.0 | 0.971 | Fixed |
| Differentiation | 9 | 919 | 0.65 (0.49–0.87) | 0.003 | 48.7 | 0.048 | Fixed |
| TNM stage | 8 | 747 | 0.54 (0.30–0.97) | 0.038 | 64.9 | 0.006 | Random |
Abbreviations: AFP, alfa-fetoprotein; Fixed, fixed-effects model; HBsAg, hepatitis B surface antigen; OCT4, octamer-binding transcription factor 4; OR, odds ratio; Random, random-effects model; PHet, probability of heterogeneity.
Figure 2.
Forest plot of studies evaluating the relationship between OCT4 expression and gender (A), age (B), HBsAg (C), AFP (D), liver cirrhosis (E), tumor size (F), vascular invasion (G), tumor encapsulation (H), tumor number (I), differentiation (J), and TNM stage (K).
Note: Weights are from random effects analysis.
Abbreviations: AFP, alfa-fetoprotein; HBsAg, hepatitis B surface antigen; OCT4, octamer-binding transcription factor 4.
Subgroup analysis was conducted to explore the potential source of heterogeneity. The results are summarized in Table 3. OCT4 expression correlated with TNM (OR =0.40, 95% CI =0.20–0.81, p=0.011, random effect) stage in the large sample size (>100) subgroup and with vascular invasion (OR =1.81, 95% CI =1.26–2.62, p=0.024, fixed effect) in the small sample size (<100) subgroup. TNM stage heterogeneity was evident in both the small sample size (I2=68.4%) and the large sample size (I2=63.7%) subgroups. Tumor encapsulation heterogeneity was evident in the small sample size group (I2=79.9%). Vascular invasion heterogeneity was evident in the large sample size group (I2=85.9%). In IHC analysis, OCT4 expression was correlated with vascular invasion (OR =1.95, 95% CI =1.30–2.92, p=0.013, fixed effect) and tumor encapsulation (OR =0.58, 95% CI =0.37–0.90, p=0.015, fixed effect). However, heterogeneity remained in the TNM stage and vascular invasion subgroups. The observations indicated that vascular invasion, tumor encapsulation, and TNM stage heterogeneity were most likely because of the sample size and methods.
Table 3.
Subgroup analysis of vascular invasion, tumor encapsulation, and TNM by methods or sample
| Subgroups | No of studies | No of patients | Pooled OR (95% CI) | PHet | I2 (%) | p-value |
|---|---|---|---|---|---|---|
| Vascular invasion | ||||||
| Methods | ||||||
| RT-PCR | 4 | 344 | 0.90 (0.23–3.64) | 0.040 | 76.0 | 0.888 |
| IHC | 4 | 439 | 1.95 (1.30–2.92) | 0.024 | 35.9 | 0.013 |
| Sample size | ||||||
| ≤100 | 5 | 412 | 1.81 (1.26–2.62) | 0.126 | 44.3 | 0.024 |
| >100 | 3 | 371 | 1.20 (0.25–5.76) | 0.001 | 85.9 | 0.819 |
| Tumor encapsulation | ||||||
| Methods | ||||||
| RT-PCR | 5 | 344 | 1.64 (0.63–4.24) | 0.029 | 68.8 | 0.311 |
| IHC | 3 | 379 | 0.58 (0.37–0.90) | 0.348 | 5.0 | 0.015 |
| Sample size | ||||||
| ≤100 | 4 | 471 | 0.94 (0.52–1.69) | 0.002 | 79.9 | 0.640 |
| >100 | 4 | 252 | 0.70 (0.46–1.05) | 0.741 | 0 | 0.086 |
| TNM stage | ||||||
| Methods | ||||||
| RT-PCR | 4 | 344 | 0.55 (0.22–1.38) | 0.031 | 66.1 | 0.201 |
| IHC | 4 | 403 | 0.53 (0.22–1.27) | 0.021 | 69.3 | 0.155 |
| Sample size | ||||||
| ≤100 | 5 | 276 | 2.91 (0.26–1.89) | 0.013 | 68.4 | 0.458 |
| >100 | 3 | 471 | 0.40 (0.20–0.81) | 0.064 | 63.7 | 0.011 |
Abbreviations: IHC, immunohistochemistry; OR, odds ratio; RT-PCR, reverse transcription polymerase chain reaction; PHet, probability of heterogeneity.
Meta-analysis of prognostic value
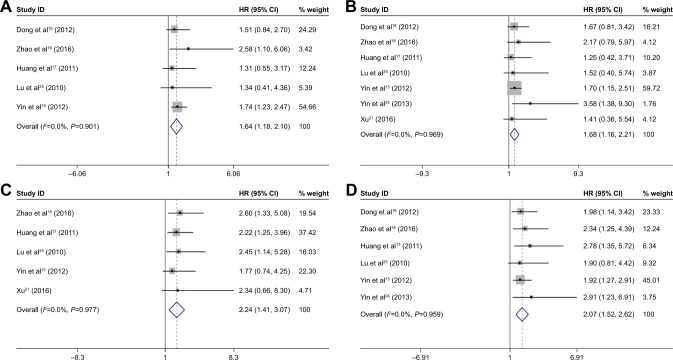
For HCC, positive OCT4 expression was associated with unfavorable 3-year OS (HR =1.68, 95% CI =1.16–2.21, p<0.001, fixed effect), 5-year OS (HR =1.64, 95% CI =1.18–2.10, p<0.001, fixed effect), and DFS (HR =2.24, 95% CI =1.41–3.07, p<0.001, fixed effect); see Figure 4. Cox multivariate analyses in six studies implicated OCT4 as an independent prognostic factor for OS of patients with HCC (HR =2.07, 95% CI =1.52–2.52, p<0.001, fixed effect).
Figure 4.
Pooled analysis for the association between OCT4 expression and 5-year survival rate (A: forest plot, E: Begg’s publication bias plot), 3-year survival rate (B: forest plot, F: Begg’s publication bias plot), disease-free survival rate (C: forest plot, G: Begg’s publication bias plot), and independent role for overall survival (D: forest plot, H: Begg’s publication bias plot).
Abbreviations: OCT4, octamer-binding transcription factor 4; SE, standard error; HR, hazard ratio.
Publication bias
No publication biases were evident for gender (p=0.721), age (p=0.592), HBsAg (p=0.536), AFP (p=0.602), liver cirrhosis (p=1), tumor size (p=0.721), vascular invasion (p=0.902), tumor encapsulation (p=0.368), tumor number (p=0.707), differentiation (p=0.754), TNM stage (p=0.711), 3-year OS (p=0.764), 5-year OS (p=0.806), DFS (p=0.806), and independent prognostic factor (p=0.296); see Figures 3 and 4.
Figure 3.
Begg’s publication bias plot for OCT4-related studies: (A) gender, (B) age, (C) HBsAg, (D) AFP, (E) liver cirrhosis, (F) tumor size, (G) vascular invasion, (H) tumor encapsulation, (I) tumor number, (J) differentiation, and (K) TNM stage.
Abbreviations: AFP, alfa-fetoprotein; HBsAg, hepatitis B surface antigen; OCT4, octamer-binding transcription factor 4; SE, standard error; OR, odds ratio.
Discussion
Recent discoveries concerning tumor biological characteristics have focused research attention on the cancer stem cell theory, which posits that tumor growth is the result of the proliferation of a small segment of tumor stem cells, which have tumor cell and stem cell biological characteristics, especially the capabilities of self-renewal, producing/maintaining tumor growth, and cell differentiation.26 Tumor cells likely originate from normal stem cells that experience a long-term cumulative mutation, and so finding the target gene of the mutation can lay the foundation for further study of tumor recurrence and metastasis.27 A current focus of oncology research is how to kill the entire tumor in order to obtain tumor stability, long-term tumor regression, and possible cure.28
Takahashi and Yamanaka29 reported the induction of fibroblasts to pluripotent stem cells by the simultaneous transfer of the genes encoding OCT4, SOX2, C-myc, and KLf4. The resulting cell lines displayed a similar appearance and behavior of embryonic stem cells. The transfer of OCT4 alone into mouse and human neural stem cells can produce these induced pluripotent cells,30 implicating OCT4 as a crucial prerequisite for cancer stem cells. OCT4 expression was subsequently demonstrated in a variety of tumors, including breast cancer,13 gastric cancer,11 and bladder cancer.31 OCT4-positive cells have been confirmed to have the characteristics of tumor stem cells, including self-renewal. Upregulation of OCT4 expression in HCC cell lines can promote the epithelial-to-mesenchymal transition of tumor cells through the STAT3/Snail signaling pathway and can increase cell proliferation, invasion, and migration.32 Silencing OCT4 in pancreatic cancer cells can inhibit the proliferation, invasion, and stem cell characteristics by inhibiting the AKT pathway,33 which suggests that OCT4 acts as a vital regulator of tumor stem cells and is likely to regulate the transformation of tumor stem cells and promote the occurrence and development of tumors.
OCT4 is over-expressed in HCC and is associated with clinicopathological features and prognosis of HCC. However, the results remain controversial. In this meta-analysis, we included a total of 985 patients from 10 studies. Positive OCT4 expression was associated with tumor size, tumor numbers, differentiation, and TNM stage, but not with gender, age, HBsAg, AFP, liver cirrhosis, tumor encapsulation, and vascular invasion. In addition, the positive expression of OCT4 predicted a poor 3- and 5-year OS as well as DFS, and OCT4 expression was shown to be an independent predictive factor for OS of HCC patients.
Some heterogeneity was evident in this study, despite subgroup and sensitivity analyses. While we cannot definitively explain the heterogeneity, suggested reasons include difference in laboratory methods, IHC and RT-PCR. Moreover, both methods were semiquantitative for IHC creating the possibility of nonspecific staining related to the quality and concentration of antibody, incubation time, and tissue embedding and slicing. Furthermore, some unavoidable error in the positive and negative judgment of OCT4 expression is likely given the mainly subjective approach used. For RT-PCR, errors may be related to primer design, annealing temperature, extension time, and number of cycles. Additionally, studies with fewer patients may have an impact on the outcome, so there is still a need for high-quality, large-scale studies to analyze the relationship between OCT4 and HCC.
Limitations
Limitations include the need for studies with larger sample sizes to provide confirmatory data, the potential contribution to bias by the extrapolation of HRs from the survival curves in articles, and the exclusive use of cohorts from China, which limit the generalization of the findings.
Conclusion
The meta-analysis indicates that OCT4 expression was associated with tumor size, tumor number, differentiation, and TNM stage in HCC. OCT4 may be a novel marker for prognosis of HCC. Well-designed studies with larger sample size are necessary to further confirm our results.
Acknowledgments
This study was funded by the Beijing Municipal Administration of Hospital Clinical Medicine Development of Special Funding Support (ZYLX201612), Capital Foundation of Medical Development (shoufa2016–2-2053), and Beijing Tongren Hospital Funds (TRYY-KYJJ-2015–032).
Footnotes
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Marquardt JU, Thorgeirsson SS. SnapShot: hepatocellular carcinoma. Cancer Cell. 2014;25(4):550.e1. doi: 10.1016/j.ccr.2014.04.002. [DOI] [PubMed] [Google Scholar]
- 2.Siegel R, Naishadham D, Jemal A. Cancer statistics, 2012. CA Cancer J Clin. 2012;62(1):10–29. doi: 10.3322/caac.20138. [DOI] [PubMed] [Google Scholar]
- 3.Forner A, Llovet JM, Bruix J. Hepatocellular carcinoma. Lancet. 2012;379(9822):1245–1255. doi: 10.1016/S0140-6736(11)61347-0. [DOI] [PubMed] [Google Scholar]
- 4.Niu J, Lin Y, Guo Z, Niu M, Su C. The epidemiological investigation on the risk factors of hepatocellular carcinoma: a case-control study in Southeast China. Medicine (Baltimore) 2016;95(6):e2758. doi: 10.1097/MD.0000000000002758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Avila MA, Berasain C, Sangro B, Prieto J. New therapies for hepatocellular carcinoma. Oncogene. 2006;25(27):3866–3884. doi: 10.1038/sj.onc.1209550. [DOI] [PubMed] [Google Scholar]
- 6.Nishikawa H, Kimura T, Kita R, Osaki Y. Treatment for hepatocellular carcinoma in elderly patients: a literature review. J Cancer. 2013;4(8):635–643. doi: 10.7150/jca.7279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nichols J, Zevnik B, Anastassiadis K, et al. Formation of pluripotent stem cells in the mammalian embryo depends on the POU transcription factor Oct4. Cell. 1998;95(3):379–391. doi: 10.1016/s0092-8674(00)81769-9. [DOI] [PubMed] [Google Scholar]
- 8.Radzisheuskaya A, Silva JC. Do all roads lead to Oct4? the emerging concepts of induced pluripotency. Trends Cell Biol. 2014;24(5):275–284. doi: 10.1016/j.tcb.2013.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kim JB, Sebastiano V, Wu G, et al. Oct4-induced pluripotency in adult neural stem cells. Cell. 2009;136(3):411–419. doi: 10.1016/j.cell.2009.01.023. [DOI] [PubMed] [Google Scholar]
- 10.Ge N, Lin HX, Xiao XS, et al. Prognostic significance of Oct4 and Sox2 expression in hypopharyngeal squamous cell carcinoma. J Transl Med. 2010;8:94. doi: 10.1186/1479-5876-8-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen Z, Xu WR, Qian H, et al. Oct4, a novel marker for human gastric cancer. J Surg Oncol. 2009;99(7):414–419. doi: 10.1002/jso.21270. [DOI] [PubMed] [Google Scholar]
- 12.Lu Y, Zhu H, Shan H, et al. Knockdown of Oct4 and Nanog expression inhibits the stemness of pancreatic cancer cells. Cancer Lett. 2013;340(1):113–123. doi: 10.1016/j.canlet.2013.07.009. [DOI] [PubMed] [Google Scholar]
- 13.Gwak JM, Kim M, Kim HJ, Jang MH, Park SY. Expression of embryonal stem cell transcription factors in breast cancer: Oct4 as an indicator for poor clinical outcome and tamoxifen resistance. Oncotarget. 2017;8(22):36305–36318. doi: 10.18632/oncotarget.16750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li B, Yao Z, Wan Y, Lin D. Overexpression of OCT4 is associated with gefitinib resistance in non-small cell lung cancer. Oncotarget. 2016;7(47):77342–77347. doi: 10.18632/oncotarget.12999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yin X, Li YW, Zhang BH, et al. Coexpression of stemness factors Oct4 and Nanog predict liver resection. Ann Surg Oncol. 2012;19(9):2877–2887. doi: 10.1245/s10434-012-2314-6. [DOI] [PubMed] [Google Scholar]
- 16.Zhao RC, Zhou J, Chen KF, et al. The prognostic value of combination of CD90 and OCT4 for hepatocellular carcinoma after curative resection. Neoplasma. 2016;63(2):288–298. doi: 10.4149/neo_2016_036. [DOI] [PubMed] [Google Scholar]
- 17.Huang P, Qiu J, Li B, et al. Role of Sox2 and Oct4 in predicting survival of hepatocellular carcinoma patients after hepatectomy. Clin Biochem. 2011;44(8–9):582–589. doi: 10.1016/j.clinbiochem.2011.02.012. [DOI] [PubMed] [Google Scholar]
- 18.Dong Z, Zeng Q, Luo H, et al. Increased expression of OCT4 is associated with low differentiation and tumor recurrence in human hepatocellular carcinoma. Pathol Res Pract. 2012;208(9):527–533. doi: 10.1016/j.prp.2012.05.019. [DOI] [PubMed] [Google Scholar]
- 19.Chen Z, Zhang L, Zhu Q, Wang X, Wu J, Wang X. Clinical value of octamer-binding transcription factor 4 as a prognostic marker in patients with digestive system cancers: a systematic review and meta-analysis. J Gastroenterol Hepatol. 2017;32(3):567–576. doi: 10.1111/jgh.13624. [DOI] [PubMed] [Google Scholar]
- 20.Xi Z, Guo Y, Wu Z, Pan Y. Expression and clinical significance of Epcam and OCT-4 in hepatocellular carcinoma. J Hepatobiliary Surg. 2015;23(1):389–391. Chinese. [Google Scholar]
- 21.Xu G. OCT-4 Expression in Hepatocellular Carcinoma Tissue and Its Role in the Malignant Biological Behavior. [master’s thesis] Nanjing Medical University; 2016. [Google Scholar]
- 22.Lin J, A S-L. Study on the relationship between expression of OCT4 protein with clinical pathogical factors in patients with hepatocellular carcinoma. J Clin Exp Med. 2015;14:1969–1971. Chinese. [Google Scholar]
- 23.Zhang M. Expression of OCT4, SOX2 and NANOG and its Clinical Significance in Hepatocellular Carcinoma. [master’s thesis] Hebei Medical University; 2012. [Google Scholar]
- 24.Yin X, Li YW, Jin JJ, et al. The clinical and prognostic implications of pluripotent stem cell gene expression in hepatocellular carcinoma. Oncol Lett. 2013;5(4):1155–1162. doi: 10.3892/ol.2013.1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lu C, Huang P, Li B, et al. OCT4 expression in hepatocellular carcinoma and its clinical significance. Chin J Cancer. 2010;29(1):111–116. doi: 10.5732/cjc.009.10232. Chinese. [DOI] [PubMed] [Google Scholar]
- 26.Beachy PA, Karhadkar SS, Berman DM. Tissue repair and stem cell renewal in carcinogenesis. Nature. 2004;432(7015):324–331. doi: 10.1038/nature03100. [DOI] [PubMed] [Google Scholar]
- 27.Korkaya H, Wicha MS. Cancer stem cells: nature versus nurture. Nat Cell Biol. 2010;12(5):419–421. doi: 10.1038/ncb0510-419. [DOI] [PubMed] [Google Scholar]
- 28.Shi Y, Du L, Lin L, Wang Y. Tumour-associated mesenchymal stem/stromal cells: emerging therapeutic targets. Nat Rev Drug Discov. 2017;16(1):35–52. doi: 10.1038/nrd.2016.193. [DOI] [PubMed] [Google Scholar]
- 29.Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126(4):663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 30.Bhartiya D, Kasiviswanathan S, Unni SK, et al. Newer insights into premeiotic development of germ cells in adult human testis using Oct-4 as a stem cell marker. J Histochem Cytochem. 2010;58(12):1093–1106. doi: 10.1369/jhc.2010.956870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jozwicki W, Brozyna AA, Siekiera J. Expression of OCT4A: the first step to the next stage of urothelial bladder cancer progression. Int J Mol Sci. 2014;15(9):16069–16082. doi: 10.3390/ijms150916069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yin X, Zhang BH, Zheng SS, et al. Coexpression of gene Oct4 and Nanog initiates stem cell characteristics in hepatocellular carcinoma and promotes epithelial-mesenchymal transition through activation of Stat3/Snail signaling. J Hematol Oncol. 2015;8:23. doi: 10.1186/s13045-015-0119-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lin H, Sun LH, Han W, et al. Knockdown of OCT4 suppresses the growth and invasion of pancreatic cancer cells through inhibition of the AKT pathway. Mol Med Rep. 2014;10(3):1335–1342. doi: 10.3892/mmr.2014.2367. [DOI] [PMC free article] [PubMed] [Google Scholar]