Abstract
Emerging methods based on mass spectrometry (MS) can be used in the rapid identification of microorganisms. Thus far, these practical and rapidly evolving methods have mainly been applied to characterize prokaryotes. We applied matrix-assisted laser-desorption-ionization-time-of-flight mass spectrometry MALDI-TOF MS in the analysis of whole cells of 18 N. fowleri isolates belonging to three genotypes. Fourteen originated from the cerebrospinal fluid or brain tissue of primary amoebic meningoencephalitis patients and four originated from water samples of hot springs, rivers, lakes or municipal water supplies. Whole Naegleria trophozoites grown in axenic cultures were washed and mixed with MALDI matrix. Mass spectra were acquired with a 4700 TOF-TOF instrument. MALDI-TOF MS yielded consistent patterns for all isolates examined. Using a combination of novel data processing methods for visual peak comparison, statistical analysis and proteomics database searching we were able to detect several biomarkers that can differentiate all species and isolates studied, along with common biomarkers for all N. fowleri isolates. Naegleria fowleri could be easily separated from other species within the genus Naegleria. A number of peaks detected were tentatively identified. MALDI-TOF MS fingerprinting is a rapid, reproducible, high-throughput alternative method for identifying Naegleria isolates. This method has potential for studying eukaryotic agents.
Keywords: Free-living amoebae, matrix-assisted laser-desorption-ionization-time-of-flight mass spectrometry, Naegleria, protein fingerprints
NAEGLERIA fowleri is a free-living, amphiozoic, eukaryotic protist that occurs world-wide and can potentially infect humans and other animals (Visvesvara 2013; Visvesvara et al. 2007). Among at least 30 species described in the genus Naegleria, N. fowleri is the only species that can infect children and young adults causing an acute, fulminant, fatal brain disease known as primary amoebic meningoencephalitis. This protist can be acquired through the exposure to thermally polluted streams, ponds, lakes, or inadequately chlorinated swimming pools. There are well-established morphological, serologic and molecular methods to characterize different species within the genus Naegleria as well as intraspecies genetic diversity (Visvesvara et al. 2007; Zhou et al. 2003). However, there are only a few reports on the use of matrix-assisted laser-desorption-ionization-time-of-flight mass spectrometry (MALDI-TOF MS) to characterize this organism (Visvesvara et al. 2007).
Matrix-assisted laser-desorption-ionization-time-of-flight mass spectrometry is a practical and rapidly evolving application of MS for rapid identification of microorganisms and strain differentiation (Fenselau and Demirev 2001; Lay 2001; van Baar 2000). Spectra obtained by MALDI-TOF MS provide characteristic patterns of proteins (fingerprints composed of unique biomarkers) from whole organisms that can be used to identify bacteria, viruses, protozoa and fungi (Amiri-Eliasi and Fenselau 2001; Croxatto et al. 2012; Glassmeyer et al. 2007; Moura et al. 2003; Villegas et al. 2006; Wunschel et al. 2005). Improved algorithms have been developed to interpret MALDI-TOF MS data obtained from whole organisms. MALDI-TOF MS has recently caught the attention of clinical microbiologists as a fast and effective method in identifying microorganisms, and it is now considered a revolution in microbial routine identification (De Bruyne et al. 2011; Seng et al. 2010). There are dedicated instruments with improved databases and the method has been adapted to use in routine clinical microbiology laboratories (Clark et al. 2013; Patel 2013a).
For the past 10 yr, we have been using MALDI-TOF MS to characterize different genera of culture-derived bacteria including Bacillus, Coxiellla and Streptococcus (Moura et al. 2003, 2008; Pierce et al. 2007; Satten et al. 2004; Shaw et al. 2004; Williamson et al. 2008; Woolfitt et al. 2011). Consistent and unique spectral patterns were obtained for each organism examined. Using MALDI-TOF MS analysis coupled with statistical analysis we have been able to identify, characterize and differentiate isolates, species, and genera. Examples include discrimination of necrotizing fasciitis-causing invasive group A Streptococcus strains from noninvasive strains and identification of specific biomarkers associated with conjunctivitis Streptococcus pneumoniae outbreak isolates (Moura et al. 2003; Pierce et al. 2007; Shaw et al. 2004; Williamson et al. 2008; Woolfitt et al. 2011). Among the select agents characterized Coxiella burnetti prototype strains isolated from different geographical and/or historical origins were differentiated as well as numerous Bacillus anthracis strains (Pierce et al. 2007; Shaw et al. 2004; Woolfitt et al. 2011). Most organisms studied in our laboratory were bacterial species and only a few microsporidia among eukaryotic organisms have been analyzed and reported (Moura et al. 2003).
We report here the combined development and application of MALDI-TOF MS and statistical analysis as a potential complementary method for N. fowleri characterization and strain differentiation. We have applied MALDI-TOF MS with Random Forest analysis, hierarchical cluster analysis, and proteomic database searching to a number of N. fowleri isolates. Using a combination of novel data processing methods for visual peak comparison, statistical analysis, and proteomics database searching we were able to demonstrate the power of this combined approach on a number of well characterized N. fowleri human and environment isolates. We believe that the combined approach will strengthen the ability of MALDI-TOF MS to differentiate species and isolates of N. fowleri.
MATERIALS AND METHODS
Chemicals
All free-living amoebae (FLA) isolates were a gift from Dr. G.S. Visvesvara (DPD, CDC). All chemicals used during this study were purchased from Sigma-Aldrich (St. Louis, MO) except where indicated. Buffers and culture media were obtained from the Scientific Resources Program at the Centers for Disease Control & Prevention (CDC).
Free-living amoebae isolates
We included in this study a total of 24 Naegleria isolates, comprising seven species, including 18 N. fowleri genotypes I, II, and III from different sources listed in Table 1. These isolates were previously identified by using conventional phenotypic tests and further characterized using the internal transcriber spacer and mitochondrial small subunit rRNA gene (Zhou et al. 2003). For comparison, we included an additional six species described within the genus Naegleria. All Naegleria spp. used in this study were grown in double-modified Nelson’s medium. Growth temperature for N. jadini and N. gruberi was 25 °C; the other Naegleria species were grown at 37 °C (Zhou et al. 2003). All isolates were washed with amoeba saline as described before (Zhou et al. 2003), pelleted and stored at −80 °C. To account for growth variability, we grew, harvested and analyzed separately each organism on different occasions over a 2-wk (batch 2) and 4-wk (batch 3) interval.
Table 1.
Characteristics of the Naegleria spp. isolates included in the present
| Number | Isolate no. | State/or country/year | Sex/age/source | Species/genotype | References |
|---|---|---|---|---|---|
| 1 | CDC:V414 | FL, 1998 | M/16/CSF | N. fowleri, I | Zhou et al. (2003) |
| 2 | CDC:V511 | GA, 2002 | M/11/CSF | N. fowleri, I | Zhou et al. (2003) |
| 3 | CDC:V518 | AZ, 2002 | Water filter 2 | N. fowleri, I | Zhou et al. (2003) |
| 4 | CDC:V413 | TX, 1998 | M/17/CSF | N. fowleri, I | Zhou et al. (2003) |
| 5 | CDC:V212 | AL, 1990 | M/?/CSF | N. fowleri, I | Zhou et al. (2003), Herman et al. (2013) |
| 6 | CDC:V020 | TX, 1984 | M/12/CSF | N. fowleri, I | Zhou et al. (2003) |
| 7 | CAMP | CA, 1978 | F/8/CSF | N. fowleri, II | Zhou et al. (2003) |
| 8 | CDC:V236 | CA, 1991 | M/29/CSF | N. fowleri, II | Zhou et al. (2003) |
| 9 | CDC:V444 | CA, 1999 | F/Cow/Brain | N. fowleri, I | Visvesvara et al. (2005) |
| 10 | CDC:V513 | GA, 2002 | Water | N. fowleri, III | Zhou et al. (2003) |
| 11 | CDC:V019 | TX, 1984 | M/25/CSF | N. fowleri, III | Zhou et al. (2003) |
| 12 | CDC:V516 | AZ, 2002 | Water filter 4 | N. fowleri, I | Zhou et al. (2003) |
| 13 | CDC:V068 | AZ, 1987 | Water | N. fowleri, I | Zhou et al. (2003) |
| 14 | CDC:V067 | AZ, 1987 | M/30/CSF | N. fowleri, III | |
| 15 | CDC:V455 | NV, 2000 | M/??/CSF | N. fowleri, III | Zhou et al. (2003) |
| 16 | CDC:V457 | TX, 2000 | M/12/CSF | N. fowleri | Visvesvara, G.S. (unpubl. data) |
| 17 | CDC:V551 | Colombia, 2004 | M/??/CSF | N. fowleri | Visvesvara, G.S. (unpubl. data) |
| 18 | 76-15-250 | Belgium, 1976 | Water | N. lovaniensis | Stevens et al. (1980) AUTHOR: Stevens et al. (1980) has not been included in the Reference List, please supply full publication details. |
| 19 | ATCC30958 | Australia | Water | N. australiensis | De Jonckheere, (1981) |
| 20 | AB-T-F3 | Italy, 1982 | Thermal mud | N. italica | Scaglia et al. (1983) |
| 21 | CDC:419 | CA, 1999 | Water | N. dunnebackei | Visvesvara et al. (2005) |
| 22 | EGs | CA, 1964 | Soil | N. gruberi | Schuster (1964) |
| 23 | 400 | Belgium, 1973 | Water | N. jadini | Willaert et al. (1980) |
| 24 | HB3 | Czechoslovakia, 1969 | M/??/CSF | N. fowleri | Cerva et al. (1969) |
Sample preparation
We prepared protein extracts for MS analysis as described previously (Moura et al. 2003, 2008; Shaw et al. 2004; Williamson et al. 2012) with a few modifications. Briefly, extracts were obtained from 100 μl frozen amoeba pellets containing 108 trophozoites after several freeze-thaw cycles and extracted with a 10% formic acid-50% acetonitrile solution. Five microliters of each extract were aliquoted and then premixed with equal volumes of the respective matrix solutions just before spotting on the MALDI target. We followed the protocol described previously (Moura et al. 2008) employing MALDI matrices (saturated solutions of α-cyano-4-hydroxycinnamic acid, or 3,5-dimethoxy-4-hydroxycinnamic acid [sinapinic acid]), 192-well stainless steel sample target plates (Applied Biosystems [AB], Framingham, MA) and mass standards for calibration (Sequazyme Peptide Mass Standards Kit, AB) prior to MALDI-TOF MS analysis. Details of conditions and parameters for MALDI-TOF MS analysis using a MALDI-TOF/TOF mass spectrometer (AB 4700 Proteomics Analyzer) are those described before (Moura et al. 2008).
Data processing, peak matching and database searching
We processed mass spectra from three harvestings as described before with a few modifications (Moura et al. 2008; Satten et al. 2004). Briefly, we exported profile spectral data as text-format m/z-intensity lists. The text data were further processed and viewed by use of a suite of custom Microsoft Visual Basic.NET (VB.NET) programs, the VBA macros and VB.NET programs “MultiSpec Processor” and “MultiSpec Viewer”. We normalized these spectra to the base peak, smoothed and finally standardized and denoised by use of a custom Fortran program (Satten et al. 2004). We used PAST software v1.34 (http://folk.uio.no/ohammer/past/doc1.html) for hierarchical cluster analysis, with the single summed spectra (one spectrum representing each organism) for input. We used Random Forest (RF) v5.1 (http://www.stat.berkeley.edu/users/breiman/RandomForests/cc_home.htm) for classification and identification, in this case, with ~9 summed spectra from three separated harvestings (2 wk apart) of each organism as a training set and ~3 summed spectra as unknowns. The recompilation of the Fortran RF code for each experimental condition was automatically driven by VB.NET programs, and custom viewing applications were developed to aid in the interpretation of the RF results.
In addition, we imported the summed spectra into the BioNumerics software v7.1 (Applied Maths Inc., Sint-Martens-Latem, Belgium) that were preprocessed using default parameters. A similarity matrix was generated using Ranked Pearson Correlation and provided the basis for constructing Neighbor Joining trees. Next, we submitted the extracted peak lists from all studied Naegleria to an in-house script for tentative protein identification. We used a total of 31,833 Naegleria spp. proteins entries available at GenBank (http://www.ncbi.nlm.nih.gov/genbank/) as of January, 2014. The Pepstats software from the European Molecular Biology Open Software Suite (EMBOOS) (Olson 2002) was used to retrieve protein database information and to calculate statistics of protein properties. We subtracted the molecular mass of the N-terminal methionine (131.00 Da), as described by before (Moura et al. 2008) and subtracted the proton charge (1 Da) as well. We matched the observed peaks (m/z minus 1 Da) and the predicted protein MW proteins in the database using an in-house script considering ±3.00 Da and posttranslational modifications such as the oxidation of methionine (+16 Da). Finally, the list of possible proteins for each peak was manually curated in order to exclude partial or fragment proteins and hypothetical proteins.
RESULTS
Visual analysis revealed differences among species and isolates of Naegleria
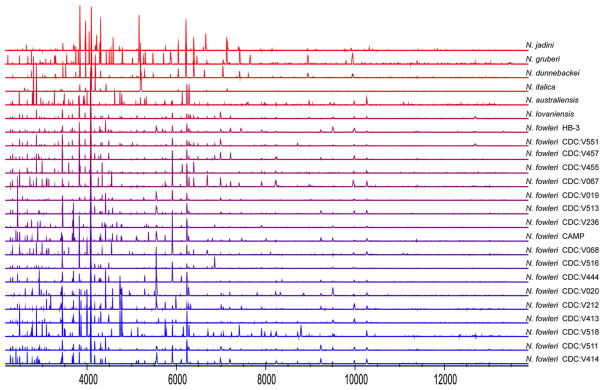
Extracts from three batches of the FLA species and isolates described in Table 1 were analyzed by MALDI-TOF MS. Spectra obtained revealed complex spectral patterns with 20–40 peaks in the m/z range corresponding to 1,000–6,000 Da or 2,000–14,000 Da. The mass range corresponding to 2,000–14,000 Da was preferred for this study because the peaks in this region were more abundant and more consistent. Spectra of amoeba protein extracts revealed reproducible patterns and differences among species could be detected visually after analyzing several runs. As expected, only minor variation from batch to batch and from well to well was detected in the analysis. Careful analysis of the spectra in Fig. 1 reveals that several peaks desorbed from extracts of Naegleria species and isolates are markedly similar to each other. However, there are specific peaks that can be used to easily differentiate among the different species although isolate differentiation would require more effort.
Figure 1.
MALDI-TOF analysis of different Naegleria species. MultiSpec Viewer depicting denoised peaks (m/z 2,000–14,000 Da) from extracts of vegetative forms that are consistently seen in all wells and reveal differences among species and isolates.
Statistical analyses improved interspecies and strain discrimination in Naegleria
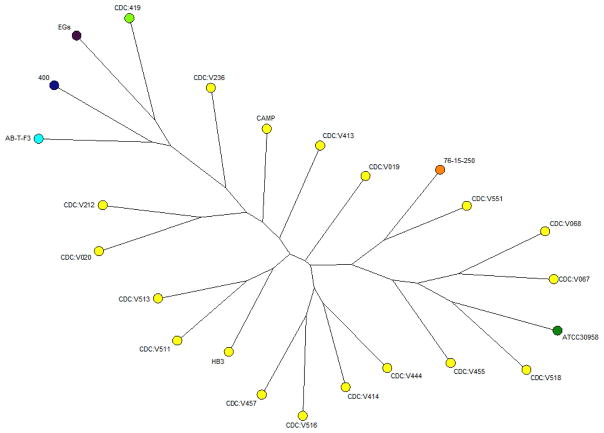
Because spectral visual analysis is not an easy task, we preferred to employ a statistical analysis of the MALDI-TOF MS data to quickly reveal significant differences among the strains analyzed. Naegleria fowleri could be discriminated from each other by Random Forest using summed spectra from three harvestings of each organism as a training set and ~3 summed spectra as unknowns. With low estimated classification error (0.75%) the RF algorithm successfully classified all the Naegleria species and the majority of the isolates in the training set (Fig. S1). Two isolates (CDC:V019 and CDC:V551) shared several peaks and the RF algorithm was unable to differentiate this subset. Results with three clustering algorithms within PAST, such as paired group, single linkage, and Ward’s method, using similarity measure methods such as Euclidean, Dice, and Jaccard, among others, were analogous and quite consistent regardless of the analysis method applied, so that the different Naegleria species and isolates studied could be reliably and reproducibly separated from each other (Fig. S2). As expected, N. fowleri isolates were clustered together, and even sub-clusters could be detected. Interestingly, two isolates (CDC:V020 and CDC:V518) were clustered with other species within the genus Naegleria. Naegleria lovaniensis was clustered with N. fowleri isolates CDC:V551 while N. australiensis is the closest species to the N. fowleri isolates. Even more, isolates were mainly clustered according to the species in Neighbor Joining trees constructed using BioNumerics (Fig. 2). Naegleria fowleri isolates could be distinguished from related species, such as N. dunnebackei, N. gruberi, N. jadini, and N. italica. In this analysis, N. australiensis was closely related to a N. fowleri isolate recovered from water (CDC:V518) and N. lovaniensis was closely related to a N. fowleri isolate recovered from CSF (CDC:V551). Among N. fowleri strains, various subgroups were observed.
Figure 2.
Neighbor joining tree obtained after BioNumerics analysis. The images are color-coded according to the Naegleria species analyzed. Naegleria fowleri isolates could be distinguished from the other species in the genus.
Tentative identification of peaks using an in-house script disclosed a large number of small proteins
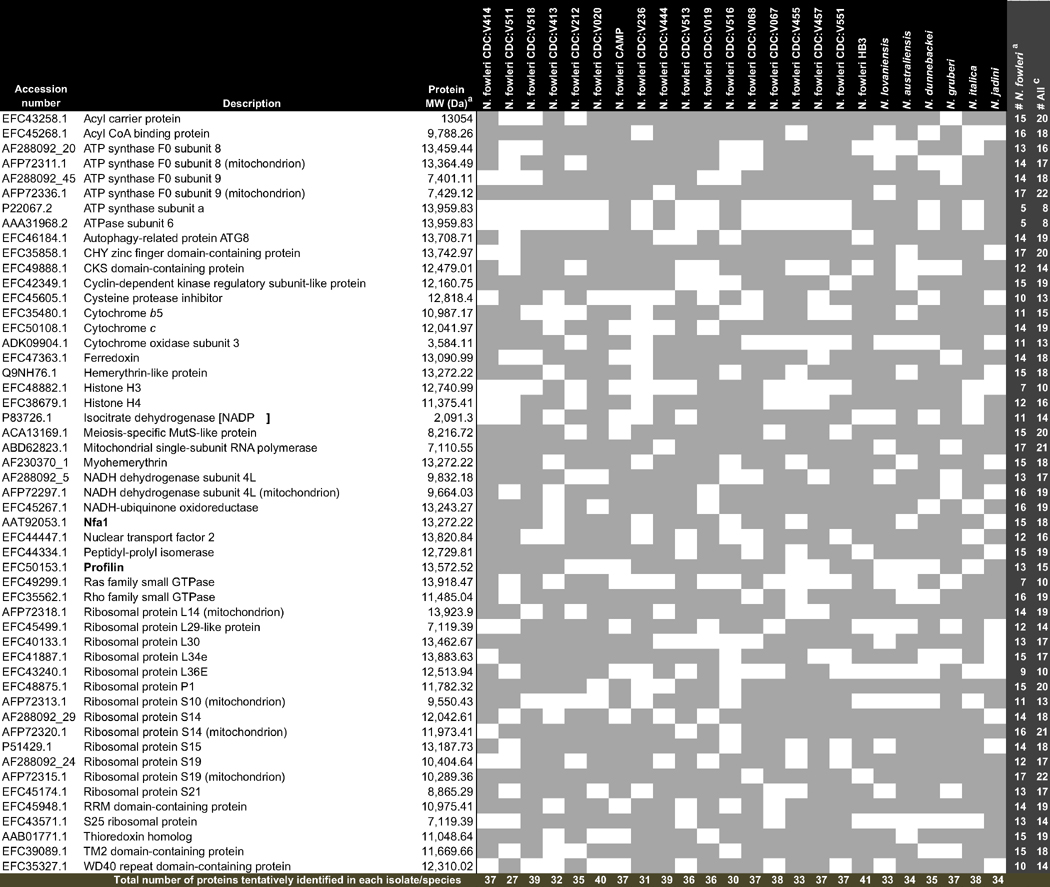
The peak lists compared with proteins of Naegleria spp. in the database revealed 51 distinct matches among all Naegleria, including peaks corresponding to 13 ribosomal proteins (Table 2). Considering the peaks observed among all 18 N. fowleri isolates studied plus the strain HB3, a mean of 37 proteins were identified. The isolate that generate the smaller number of peaks was CDC:V511 with only 27 proteins identified. A total of 42 of the 51 proteins identified were found in N. fowleri HB3. Nevertheless, none of the 19 N. fowleri studied presented all the 51 proteins identified, although 29 proteins were identified in 14 isolates.
Table 2.
Tentative identification of detected biomarkers of the studied Naegleria spp. isolates
Protein MW expected without N-terminal methionine (Da).
Number of N. fowlerid isolates where the tentatively identified proteins were found.
Number of Naegleri spp. where the tentatively identified proteins were found.
DISCUSSION
Emerging methods based on MS can be used to rapidly identify microorganisms. In particular, MALDI-TOF MS detects small acidic proteins and peptides desorbed from whole cells. Observed mass spectra are believed to consist primarily of protonated peptide and protein signals, although some signals below m/z 4,000 may represent other materials. The method is applicable to the identification of microorganisms at different levels (Clark et al. 2013; Sandrin et al. 2013). After spectra collection, multiple types of data analysis can be applied to discriminate genus, species, and even strains of the microorganisms. MALDI-TOF MS is making a fundamental shift in the routine practice of clinical microbiology (Patel 2013a,b). With over 500 publications each year in the past 5 yr, thus far these practical and rapidly evolving methods have been applied mostly to characterize and identify prokaryotes and clinically relevant fungi (Cayrou et al. 2010; Clark et al. 2013).
In this study, we used a high-resolution MALDI-TOF MS instrument and processing methods that we have developed (Moura et al. 2008; Satten et al. 2004; Woolfitt et al. 2011) to successfully detect differences and similarities among species and isolates of the genus Naegleria. Our work was expedited by use of improved in-house developed software that combined mass spectral visual analysis, statistical analysis using Random forest and tentative peak identification by database search and proteomics strategies (Moura et al. 2008; Pierce et al. 2007; Satten et al. 2004; Williamson et al. 2008; Woolfitt et al. 2011). In addition, we were able to apply commercial software (BioNumerics) that added another layer of certainty to the analysis. Concerns from early studies done with prokaryotes about MALDI-TOF reproducibility, the nature of the observed peaks and the best data analysis algorithms have been addressed in a number of publications (De Bruyne et al. 2011). However, there are still limitations of the MALDI method that need to be addressed. For example, the variability presented in MALDI chromatograms can be minimized by acquiring a number of spectra and can use the averaged spectrum for identification. In this study, complex eukaryotic organisms from the genus Naegleria were analyzed and different MALDI preparations varying solvents, matrices, plating techniques, and mass ranges were tested. Several factors can influence the composition of the MALDI-TOF spectra resulting from whole organism analysis. Although the metabolic state of the organism cannot be ruled out, MALDI identification is based largely on small conserved proteins such as the ribosomal proteins. In addition, due to the complexity of the material analyzed, which is composed of an expressive number of proteins with a large dynamic range, and without any analytical separation, it is not expected that all peaks be detected in each species in each analysis. The way to compensate for the variability presented in MALDI chromatograms, which may be a limitation of the MALDI analysis, is to run the samples several times acquiring a number of spectra and to apply statistical analysis. However, as even after these analyses the total number of expected peaks is generally not achieved for each species, a common criteria for microorganism identification by MALDI is to accept a percentage of representative peaks. For example, the two MALDI instruments in the market dedicated for bacterial identification (Bruker Biotyper and bioMerieuex Vitek-MS) assign species identification with high confidence when over 70% of the peaks of the unknown organism match the species in their database. Overall, in this study, the spectra of replicate samples were similar, with a common subset of specific biomarkers found consistently in all studied conditions. Only minor differences were detected from analyses on different days.
It is important to note that the spectra obtained from different Naegleria spp. could be easily set apart from each other by visual inspection of their spectra. Because visual analysis was not an ideal approach we applied the previously reported Random Forest method for spectral standardizing and denoising to differentiate species within the genus Naegleria (Satten et al. 2004). In addition, the combined algorithms for data processing applied in this study proved to be useful. Random Forest is a robust method which can be used to differentiate all Naegleria species and separate most of the strains tested. More importantly, results from Neighbor Joining trees constructed using a similarity matrix generated with Ranked Pearson Correlation revealed similar results to outcomes using Random Forest. In concordance with PAST analysis, N. australiensis was closely related to a N. fowleri isolate recovered from water. Interestingly, N. australiensis can cause infection in mice after intranasal or intracerebral inoculation. Likewise, N. lovaniensis was closely related to a N. fowleri isolate (CDC:V551) recovered from CSF; the close relationship between these two species was previously reported (Zhou et al. 2003). Among N. fowleri strains, the various subgroups observed may reflect either genotype distribution or epidemiological characteristics of the strains, including year of isolation and geographical origin. Moreover, while some genotype I and III isolates were gathered in the same N. fowleri subgroup, strains of genotype II were more distantly related, in agreement with previous results of mtSSU-rRNA gene and ITS analyses (Zhou et al. 2003). Still, discrimination among isolates belonging to the same genotype was also possible. Within three N. fowleri branches, isolates from environmental sources were clustered together with those from human sources. When considered collectively, these data confirmed the potential of MALDI-TOF MS for discrimination among species, and strains.
Challenges in tentative peak identification in this study exist because Naegleria gruberi, a nonpathogenic species is the only representative within the genus that had its genome completely sequenced (Fritz-Laylin et al. 2010). Only the mitochondrial genome and a 60-kb segment of nuclear genome from N. fowleri have been sequenced and assembled (Herman et al. 2013). However, we were able to retrieve 31,833 Naegleria spp. protein sequences available in GenBank that were tentatively matched to our results using a combination of an in-house script with the EMBOOS software. The complex procedure was fruitful since we have been able to tentatively identify over 50 small proteins or peptides occurring in the different Nagleria species studied. As expected, a number of them corresponded to ribosomal proteins which are extremely ancient molecules, and consequently one-third of ribosomal protein families are conserved among Bacteria, Archae, and Eucharia (Lecompte et al. 2002). These proteins are used to identify distinct microorganisms using MALDI-TOF spectra and these approaches are largely in current used (Hamprecht et al. 2014; Moura et al. 2008; Seng et al. 2010).
Matrix-assisted laser-desorption-ionization-time-of-flight mass spectrometry-based methods have been generally used for genus and species differentiation, and in a few cases for strain characterization (Sandrin et al. 2013). We report here one of the first uses of MALDI-TOF MS for analyzing Naegleria species and N. fowleri strains. Species within the genus and most of the strains were discriminated by MALDI-TOF MS. Although MS-based methods are sensitive and specific the present MALDI-TOF MS application was designed for microbial identification and not for microorganism detection, as there is a requirement of ~5,000 organisms per well for the observation of a clean and meaningful spectra.
In conclusion, we have successfully applied MALDI-TOF MS with statistical analysis to differentiate among different Naegleria species and isolates. Visual analysis of multiple spectra was sufficient to differentiate among the different species analyzed. However, the combined MS visual analysis, fingerprinting and proteomics approach yielded more robust biomarker identification by use of a database search, which permitted isolate differentiation among N. fowleri isolates. Small ribosomal proteins were consistently found among biomarkers detected in this study. We believe that this method is a rapid, reproducible, high-throughput alternative for the characterization of microorganisms.
Supplementary Material
Figure S1. Random Forest analysis of the studied Naegleria species and isolates. Datasets consisted of summed spectra from three harvestings of each organism as a training set (columns) and ~3 summed spectra as unknowns (rows). The estimated classification error was 0.75%, and the RF algorithm successfully classified all the Naegleria species and the majority of the isolates in the training set.
Figure S2. Dendrogram obtained with PAST (http://folk.uio.no/ohammer/past/index.html). For this analysis, all spectra for each strain were summed to give one representative spectrum per organism.
Acknowledgments
We thank Dr. Govinda S. Visvesvara for many helpful discussions and his valuable advice during the course of this investigation. We also thank Mrs. Rama Sriram for technical support in the cultivation and maintenance of the isolates.
Footnotes
DISCLAIMER
References in this article to any specific commercial products, process, service, manufacturer, or company do not constitute an endorsement or a recommendation by the U.S. Government or the Centers for Disease Control and Prevention. The findings and conclusions in this report are those of the authors and do not necessarily represent the views of CDC.
Additional Supporting Information may be found in the online version of this article:
LITERATURE CITED
- Amiri-Eliasi B, Fenselau C. Characterization of protein biomarkers desorbed by MALDI from whole fungal cells. Anal Chem. 2001;73:5228–5231. doi: 10.1021/ac010651t. [DOI] [PubMed] [Google Scholar]
- Cayrou C, Raoult D, Drancourt M. Matrix-assisted laser desorption/ionization time-of-flight mass spectrometry for the identification of environmental organisms: the Planctomycetes paradigm. Environ Microbiol Rep. 2010;2:752–760. doi: 10.1111/j.1758-2229.2010.00176.x. [DOI] [PubMed] [Google Scholar]
- Cerva L, Zimak V, Novak K. Amoebic meningoencephalitis: a new amoeba isolate. Science. 1969;163:575–576. doi: 10.1126/science.163.3867.575. [DOI] [PubMed] [Google Scholar]
- Clark AE, Kaleta EJ, Arora A, Wolk DM. Matrix-assisted laser desorption ionization-time of flight mass spectrometry: a fundamental shift in the routine practice of clinical microbiology. Clin Microbiol Rev. 2013;26:547–603. doi: 10.1128/CMR.00072-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croxatto A, Prod’hom G, Greub G. Applications of MALDI-TOF mass spectrometry in clinical diagnostic microbiology. FEMS Microbiol Rev. 2012;36:380–407. doi: 10.1111/j.1574-6976.2011.00298.x. [DOI] [PubMed] [Google Scholar]
- De Bruyne K, Slabbinck B, Waegeman W, Vauterin P, De Baets B, Vandamme P. Bacterial species identification from MALDI-TOF mass spectra through data analysis and machine learning. Syst Appl Microbiol. 2011;34:20–29. doi: 10.1016/j.syapm.2010.11.003. [DOI] [PubMed] [Google Scholar]
- De Jonckheere JF. Naegleria australiensis sp Nov., another pathogenic Naegleria from water. Protistologica. 1981;17:423–429. [Google Scholar]
- Fenselau C, Demirev PA. Characterization of intact microorganisms by MALDI mass spectrometry. Mass Spectrom Rev. 2001;20:157–171. doi: 10.1002/mas.10004. [DOI] [PubMed] [Google Scholar]
- Fritz-Laylin LK, Prochnik SE, Ginger ML, Dacks JB, Carpenter ML, Field MC, Kuo A, Paredez A, Chapman J, Pham J, Shu S, Neupane R, Cipriano M, Mancuso J, Tu H, Salamov A, Lindquist E, Shapiro H, Lucas S, Grigoriev IV, Cande WZ, Fulton C, Rokhsar DS, Dawson SC. The genome of Naegleria gruberi illuminates early eukaryotic versatility. Cell. 2010;140:631–642. doi: 10.1016/j.cell.2010.01.032. [DOI] [PubMed] [Google Scholar]
- Glassmeyer ST, Ware MW, Schaefer FW, III, Shoemaker JA, Kryak DD. An improved method for the analysis of Cryptosporidium parvum oocysts by matrix-assisted laser desorption/ionization time of flight mass spectrometry. J Eukaryot Microbiol. 2007;54:479–481. doi: 10.1111/j.1550-7408.2007.00287.x. [DOI] [PubMed] [Google Scholar]
- Hamprecht A, Christ S, Oestreicher T, Plum G, Kempf VA, Gottig S. Performance of two MALDI-TOF MS systems for the identification of yeasts isolated from bloodstream infections and cerebrospinal fluids using a time-saving direct transfer protocol. Med Microbiol Immunol. 2014;203:93–99. doi: 10.1007/s00430-013-0319-9. [DOI] [PubMed] [Google Scholar]
- Herman EK, Greninger AL, Visvesvara GS, Marciano-Cabral F, Dacks JB, Chiu CY. The mitochondrial genome and a 60-kb nuclear DNA segment from Naegleria fowleri, the causative agent of primary amoebic meningoencephalitis. J Eukaryot Microbiol. 2013;60:179–191. doi: 10.1111/jeu.12022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lay JO., Jr MALDI-TOF mass spectrometry of bacteria. Mass Spectrom Rev. 2001;20:172–194. doi: 10.1002/mas.10003. [DOI] [PubMed] [Google Scholar]
- Lecompte O, Ripp R, Thierry JC, Moras D, Poch O. Comparative analysis of ribosomal proteins in complete genomes: an example of reductive evolution at the domain scale. Nucleic Acids Res. 2002;30:5382–5390. doi: 10.1093/nar/gkf693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moura H, Ospina M, Woolfitt AR, Barr JR, Visvesvara GS. Analysis of four human microsporidian isolates by MALDI-TOF mass spectrometry. J Eukaryot Microbiol. 2003;50:156–163. doi: 10.1111/j.1550-7408.2003.tb00110.x. [DOI] [PubMed] [Google Scholar]
- Moura H, Woolfitt AR, Carvalho MG, Pavlopoulos A, Teixeira LM, Satten GA, Barr JR. MALDI-TOF mass spectrometry as a tool for differentiation of invasive and noninvasive Streptococcus pyogenes isolates. FEMS Immunol Med Microbiol. 2008;53:333–342. doi: 10.1111/j.1574-695X.2008.00428.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson SA. EMBOSS opens up sequence analysis. European Molecular Biology Open Software Suite. Brief Bioinform. 2002;3:87–91. doi: 10.1093/bib/3.1.87. [DOI] [PubMed] [Google Scholar]
- Patel R. Matrix-assisted laser desorption ionization-time of flight mass spectrometry in clinical microbiology. Clin Infect Dis. 2013a;57:564–572. doi: 10.1093/cid/cit247. [DOI] [PubMed] [Google Scholar]
- Patel R. MALDI-TOF mass spectrometry: transformative proteomics for clinical microbiology. Clin Chem. 2013b;59:340–342. doi: 10.1373/clinchem.2012.183558. [DOI] [PubMed] [Google Scholar]
- Pierce CY, Barr JR, Woolfitt AR, Moura H, Shaw EI, Thompson HA, Massung RF, Fernandez FM. Strain and phase identification of the U.S. category B agent Coxiella burnetii by matrix assisted laser desorption/ionization time-of-flight mass spectrometry and multivariate pattern recognition. Anal Chim Acta. 2007;583:23–31. doi: 10.1016/j.aca.2006.09.065. [DOI] [PubMed] [Google Scholar]
- Sandrin TR, Goldstein JE, Schumaker S. MALDI TOF MS profiling of bacteria at the strain level: a review. Mass Spectrom Rev. 2013;32:188–217. doi: 10.1002/mas.21359. [DOI] [PubMed] [Google Scholar]
- Satten GA, Datta S, Moura H, Woolfitt AR, da Carvalho MG, Carlone GM, De BK, Pavlopoulos A, Barr JR. Standardization and denoising algorithms for mass spectra to classify whole-organism bacterial specimens. Bioinformatics. 2004;20:3128–3136. doi: 10.1093/bioinformatics/bth372. [DOI] [PubMed] [Google Scholar]
- Scaglia M, Strosselli M, Graziolo V, Gatti S, Bernuzzi AM, deJonckheere JF. Isolation and identification of pathogenic Naegleria australiensis (Amoebida, Vahlkampfiidae) from a spa in Northern Italy. Appl Environ Microbiol. 1983;46:1282–1285. doi: 10.1128/aem.46.6.1282-1285.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuster FL. An electron microscope study of the amoeb-o-flagellate, Naegleria gruberi (Schardinger). I The amoeboid and flagellate stages. J Protozool. 1963;10:293–313. doi: 10.1111/j.1550-7408.1963.tb01681.x. [DOI] [PubMed] [Google Scholar]
- Seng P, Rolain JM, Fournier PE, La Scola B, Drancourt M, Raoult D. MALDI-TOF-mass spectrometry applications in clinical microbiology. Future Microbiol. 2010;5:1733–1754. doi: 10.2217/fmb.10.127. [DOI] [PubMed] [Google Scholar]
- Shaw EI, Moura H, Woolfitt AR, Ospina M, Thompson HA, Barr JR. Identification of biomarkers of whole Coxiella burnetii phase I by MALDI-TOF mass spectrometry. Anal Chem. 2004;76:4017–4022. doi: 10.1021/ac030364k. [DOI] [PubMed] [Google Scholar]
- van Baar BL. Characterisation of bacteria by matrix-assisted laser desorption/ionisation and electrospray mass spectrometry. FEMS Microbiol Rev. 2000;24:193–219. doi: 10.1016/S0168-6445(99)00036-4. [DOI] [PubMed] [Google Scholar]
- Villegas EN, Glassmeyer ST, Ware MW, Hayes SL, Schaefer FW., III Matrix-assisted laser desorption/ionization time-of-flight mass spectrometry-based analysis of Giardia lamblia and Giardia muris. J Eukaryot Microbiol. 2006;53(Suppl 1):S179–S181. doi: 10.1111/j.1550-7408.2006.00223.x. [DOI] [PubMed] [Google Scholar]
- Visvesvara GS. Infections with free-living amebae. Handb Clin Neurol. 2013;114:153–168. doi: 10.1016/B978-0-444-53490-3.00010-8. [DOI] [PubMed] [Google Scholar]
- Visvesvara GS, Moura H, Schuster FL. Pathogenic and opportunistic free-living amoebae: Acanthamoeba spp. Balamuthia mandrillaris, Naegleria fowleri, and Sappinia diploidea. FEMS Immunol Med Microbiol. 2007;50:1–26. doi: 10.1111/j.1574-695X.2007.00232.x. [DOI] [PubMed] [Google Scholar]
- Visvesvara GS, De Jonckheere JF, Sriram R, Daft B. Isolation and molecular analysis of Naegleria fowleri from a cow brain that died of primary amebic meningoencephalitis. J Clin Microbiol. 2005;43:4203–4204. doi: 10.1128/JCM.43.8.4203-4204.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willaert E, Stevens AR. Experimental pneumonitis induced by Naegleria fowleri in mice. Trans Roy Soc Trop Med Hyg. 1980;74:779–783. doi: 10.1016/0035-9203(80)90199-6. [DOI] [PubMed] [Google Scholar]
- Williamson YM, Moura H, Woolfitt AR, Pirkle JL, Barr JR, da Carvalho MG, Ades EP, Carlone GM, Sampson JS. Differentiation of Streptococcus pneumoniae conjunctivitis outbreak isolates by matrix-assisted laser desorption ionization-time of flight mass spectrometry. Appl Environ Microbiol. 2008;74:5891–5897. doi: 10.1128/AEM.00791-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williamson YM, Moura H, Simmons K, Whitmon J, Melnick N, Rees J, Woolfitt A, Schieltz DM, Tondella ML, Ades E, Sampson J, Carlone G, Barr JR. A gel-free proteomic-based method for the characterization of Bordetella pertussis clinical isolates. J Microbiol Methods. 2012;90:119–133. doi: 10.1016/j.mimet.2012.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woolfitt AR, Boyer AE, Quinn CP, Hoffmaster AR, Kozel TR, De BK, Gallegos M, Moura H, Pirkle JL, Barr JR. Matrix assisted laser desorption ionization mass spectrometric analysis of Bacillus anthracis: from finger-print analysis of the bacterium to quantification of its toxins in clinical samples. In: Banoub J, editor. Detection of Biological Agents for the Prevention of Bioterrorism, NATO Science for the Peace and Security. Vol. 1. Springer Science + Business Media B.V; Dordecht, The Netherlands: 2011. pp. 83–98. [Google Scholar]
- Wunschel DS, Hill EA, McLean JS, Jarman K, Gorby YA, Valentine N, Wahl K. Effects of varied pH, growth rate and temperature using controlled fermentation and batch culture on matrix assisted laser desorption/ionization whole cell protein fingerprints. J Microbiol Methods. 2005;62:259–271. doi: 10.1016/j.mimet.2005.04.033. [DOI] [PubMed] [Google Scholar]
- Zhou L, Sriram R, Visvesvara GS, Xiao L. Genetic variations in the internal transcribed spacer and mitochondrial small subunit rRNA gene of Naegleria spp. J Eukaryot Microbiol. 2003;50(Suppl):522–526. doi: 10.1111/j.1550-7408.2003.tb00617.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Random Forest analysis of the studied Naegleria species and isolates. Datasets consisted of summed spectra from three harvestings of each organism as a training set (columns) and ~3 summed spectra as unknowns (rows). The estimated classification error was 0.75%, and the RF algorithm successfully classified all the Naegleria species and the majority of the isolates in the training set.
Figure S2. Dendrogram obtained with PAST (http://folk.uio.no/ohammer/past/index.html). For this analysis, all spectra for each strain were summed to give one representative spectrum per organism.