Abstract
In this unit, protocols are provided for detection of disulfide bond formation in cultures of intact cells and in an in-vitro-translation system containing isolated microsomes or semi-permeabilized cells. First, the newly synthesized protein of interest is biosynthetically labeled with radioactive amino acids in a short pulse. The labeled protein then is chased with unlabeled amino acids. At different times during the chase, a sample is collected, membranes are lysed with detergent, and the protein is isolated by immunoprecipitation, as described. A support protocol is provided for analysis of disulfide bonds in the immunoprecipitates by SDS-PAGE with and without prior reduction. The difference in mobility observed between the gels with nonreduced and reduced samples is due to disulfide bonds in the nonreduced protein. An additional support protocol is included that uses PEG-maleimide to modify free thiols and follow disulfide-bond formation by SDS-PAGE.
Keywords: disulfide bonds, endoplasmic reticulum, protein folding, secretory pathway, pulse-chase, radiolabeling
INTRODUCTION
Most proteins synthesized in the endoplasmic reticulum (ER) in eukaryotic cells and in the periplasmic space in prokaryotes are stabilized by disulfide bonds. Disulfide bonds are of two types: intrachain (within a polypeptide chain) and interchain (between separate chains). Intrachain disulfide bonds are formed during cotranslational and post-translational folding of a newly synthesized protein (Braakman and Hebert, 2013). Most interchain disulfide bonds are formed at a later stage in the maturation process and establish covalent links between subunits in oligomeric proteins.
Disulfide bond formation can be followed in cultures of intact cells (Basic Protocol 1 and Alternate Protocols 1 and 2) or in an in-vitro-translation system containing isolated microsomes (Basic Protocol 2 and Alternate Protocol 3) or semi-permeabilzed cells (Alternate Protocol 4). First, the newly synthesized protein of interest is biosynthetically labeled with radioactive amino acids in a short pulse. The labeled protein is chased with unlabeled amino acids. At different times during the chase, a sample is collected, membranes are lysed with detergent, and the protein is isolated by immunoprecipitation (Support Protocol 1). The immunoprecipitates are analyzed for the presence of disulfide bonds by SDS-PAGE with and without prior reduction (Support Protocol 2). The difference in mobility observed between the gels with nonreduced and reduced samples is due to disulfide bonds in the nonreduced protein. The presence of disulfide bonds versus unpaired cysteines at steady state, in the total pool of a protein, can be monitored through electrophoretic mobility shifts upon modification by PEG-Maleimide (Support Protocol 3).
CAUTION: Radioactive materials require special handling. See APPENDIX 2B concerning safe use of radioisotopes.
Radiation safety
Allthough the [35S] methionine and cysteine-labeling mix is stabilized, some decomposition will occur, yielding volatile radioactive compounds. To protect experimentor and apparatus some precautions can be taken. Stock vials with [35S] methionine and cysteine should always be opened in a fumehood or under a local aspiration point, with a charcoal nursing mask. Known laboratory contamination spots are centrifuges, pipets, waterbaths, incubators, and shakers. Contamination is diminished by the use of charcoal pipet tips (solvent-safe tips, Molecular BioProducts), safe-seal microtubes (Sarstedt), acquarium charcoal sponges in waterbaths, activated charcoal filter papers in the pulse-chase dishes, and activated charcoal in the aspiration system, [35S] stock storage and incubators.
BASIC PROTOCOL 1. ANALYSIS OF DISULFIDE BOND FORMATION IN INTACT MONOLAYER CELLS
In vivo, disulfide bond formation can be examined in cells growing in monolayers on cell-culture dishes (Braakman et al., 1991; Jansens and Braakman, 2003). This allows multiple wash steps in a short period of time and makes it easy to use short pulse and chase times. In addition, adherent cells are suitable for studies requiring frequent changes of medium with very different composition. When cells do not adhere or when low volumes of medium are desirable (e.g., when expensive additives are needed), the analysis can be done in suspension (see Alternate Protocol 1). This method detects cotranslational and post-translational disulfide bond formation; it is also possible to analyze postponed post-translational disulfide bond formation (see Alternate Protocol 2).
Materials
Adherent cells
Cell culture medium containing methionine, 37°C
Wash buffer (see recipe), 37°C
Depletion medium (see recipe), 37°C
Labeling medium (containing 125 to 250 μCi/ml [35S]methionine; see recipe), 37°C
Chase medium (see recipe), 37°C
Stop buffer (see recipe), 0°C
Lysis buffer (see recipe), 0°C
60-mm cell culture dishes
37°C humidified 5% CO2 incubator
37°C water bath with rack to hold cell-culture dishes (e.g., Unwire racks for 15- and 50-ml tubes, Nalgene)
Liquid aspiration system for radioactive waste
Large laboratory ice pan with fitted metal plate (e.g. VWR International)
Cell scraper
Additional reagents and equipment for immunoprecipitation (see Support Protocol 1) and nonreducing and reducing SDS-PAGE (see Support Protocol 2)
NOTE: The volumes described here are for a 60-mm dish of cells. Volumes must be adjusted, based on the surface area of the dish, for other sizes (double volumes for 100-mm dishes, half-volumes for 35-mm dishes).
NOTE: All culture incubations are performed in a humidified 37°C, 5% CO2 incubator unless otherwise specified.
Perform pulse and chase
-
1
Set up cultures of adherent cells in cell culture medium in 60-mm tissue culture dishes so the cells will form a subconfluent monolayer on the day of the experiment.
The experiment requires at least 1 dish per time point, and each dish should contain sufficient cells to detect the radiolabeled protein (usually ≥106 cells).
-
2
Set up 37°C water bath with racks for cell culture dishes. Check that the water level is in contact with the bottom of the dish but does not allow it to float when the lid is removed. Arrange the aspiration flask for radioactive waste so the pipet easily reaches dishes in the water bath.
-
3
Rinse cells with 2 ml wash buffer. Aspirate wash buffer and add 2 ml depletion medium. Incubate 15 to 30 min at 37°C in an incubator.
This step depletes cells from methionine.
-
4
Remove dishes from the incubator and place on rack in 37°C water bath.
-
5
Pulse-label the cells, one dish at a time: aspirate depletion medium, add 400 μl labeling medium containing 50 to 100 μCi [35S]methionine, and incubate 1 to 5 min on rack in 37°C water bath.
The pulse time should be equal to or shorter than the time required to synthesize the protein of interest (assuming a rate of 4 to 5 amino acid residues/sec) and long enough to detect the protein. When longer labeling periods are needed, (methionine) starvation should be avoided. See for this Alternate protocol 3.
If the protein of interest does not contain many methionines, or if it contains several cysteines, it may be worthwhile to deplete cysteine (as well as methionine) and label with [35S]methionine + [35S]cysteine (50 to 100 μC) or with the same quantity of unpurified [35S]methionine, which usually contains ~15% (v/v) [35S]cysteine as well. The stabilized form of unpurified cysteine and methionine (e.g. EasyTag EXPRESS 35S Protein labeling mix, Perkin Elmer) is less volatile and should be used for short pulses in a water bath to minimize radioactive contamination of air, pipets, and equipment. In all protocols in this chapter methionine can be replaced with cysteine or a mixture of methionine and cysteine.
For 0-min chase interval
-
6a
Add 2 ml chase medium to stop the pulse at precisely the end of the labeling interval.
-
7a
Aspirate chase medium as quickly as possible. Transfer dish to aluminum plate on ice pan. Immediately add 2.5 ml cold stop buffer to end the chase.
For all other chase intervals
-
6b
At precisely the end of the pulse labeling interval, add 2 ml chase medium to start the chase. Rock gently to mix. Aspirate medium and add 2 ml chase medium again. Incubate in a 37°C incubator or 37°C water bath for the desired chase intervals.
Choice of chase times is determined by the time it takes a protein to fold and form disulfide bonds. Generally, the first chase interval is equal to pulse time, the next approximately double that, and so on (e.g. 2, 5, 10, 20, and 40 min).
For short chase intervals, dishes can be incubated in a water bath, but for chase intervals >15 to 20 min, pH is best maintained in a 5%CO2 incubator. Beware that HEPES in the medium may influence intracellular pH and that the HEPES buffer should not be stronger than the bicarbonate/CO2 buffer.
-
7b
At the end of the chase interval, aspirate chase medium and transfer dish to metal plate on ice pan. Add 2.5 ml cold stop buffer to end the chase.
Cold stop buffer is used to stop all cellular processes. Cells may be left ≤30 min on ice in stop buffer.
Prepare the lysate
-
8
Remove stop buffer and add fresh 2.5 ml cold stop buffer.
-
9
Aspirate dish as dry as possible. Add 600 μl cold lysis buffer.
No incubation with lysis buffer is necessary when the stop buffer contains an alkylating agent. If alkylating agent is omitted from the lysis buffer, incubate 10 min in the stop buffer with 20 mM N-ethylmaleimide (NEM) or longer with 20 mM iodoacetamide or 20 mM iodoacetic acid (typically 15 to 45 min).
-
10
Scrape dish and mix lysate well with cell scraper. Transfer the lysate to a labeled 1.5-ml microcentrifuge tube.
-
11
Microcentrifuge 5 min at 12,000 rpm, 0°C, to pellet nuclei. Transfer supernatant (postnuclear lysate) to a clean 1.5-ml microcentrifuge tube.
-
12
Immediately analyze postnuclear lysate by immunoprecipitation (see Support Protocol 1 and UNIT 13.2) and nonreducing and reducing SDS-PAGE (see Support Protocol 2) or freeze it rapidly in liquid nitrogen and store at –80°C.
Lysate with alkylating agent can be stored 1 to 2 h on ice; lysate without alkylating agent should not be stored on ice. but snap-frozen or immediately used for analysis.
ALTERNATE PROTOCOL 1. ANALYSIS OF DISULFIDE BOND FORMATION IN CELLS IN SUSPENSION
When cells do not adhere adequately to a solid support, or when it is desirable to use small volumes of reagents for analyses, cells in suspension may be used to analyze disulfide bond formation. One advantage to this approach is that samples at different chase intervals may be collected from a single tube of labeled cells. However, wash steps cannot be included during the pulse-chase incubations because the washes take too much time. Before starting the experiment, it is necessary to determine: (1) the minimum volume for incubating the cells (x μl); (2) the number of chase time points (y); and (3) the desired sample volume (z μl). Approximately 106 cells should be used for each time point.
For most cell types an x of 100 μl would suffice for 106 cells.
Additional Materials (also see Basic Protocol 1)
Culture of suspension cells
11 mCi/ml [35S]methionine (>1,000 Ci/mmol; Perkin Elmer or Amersham)
2× lysis buffer (see recipe)
Concentrated chase medium (see recipe)
50-ml polystyrene tube with cap, sterile
Cell centrifuge
Additional reagents and equipment for immunoprecipitation (see Support Protocol 1) and nonreducing SDS-PAGE (see Support Protocol 2)
NOTE: All culture incubations are performed in a humidified 37°C, 5% CO2 incubator unless otherwise specified.
NOTE: Keep cells in suspension during incubations by gently swirling the tube at regular intervals.
-
Transfer suspension cells for analysis to a sterile 50-ml polystyrene tube with cap.
Use ~106 cells per time point.
If it is necessary to minimize the volumes of reagents used for the experiment, adherent cells can be removed from the dish by treating them with 0.25% (w/v) trypsin/0.2% (w/v) EDTA (or a milder alternative if cells allow) and resuspending them in a low-calcium medium.
-
Centrifuge cells 4 min at 500 × g 20° to 37°C. Resuspend pellet in 2y ml depletion medium and centrifuge again. Resuspend in 2y ml depletion medium. Incubate 15 to 30 min at 37°C.
Pelleting conditions may vary with cell type.
Centrifuge cells 4 min at 500 × g, at 20° to 37°C. Resuspend cells in x μl depletion medium in appropriate tube and place in water bath.
At the start of pulse, add 50 to 100 μCi undiluted [35S]methionine per time point to the cell suspension and mix by swirling. Incubate for the pulse period.
-
At the end of the pulse, add ≥4 vol (4 × x μl) concentrated chase medium. Mix by swirling.
The total volume after addition of concentrated chase medium should be slightly more than y × z μl to allow for loss due to evaporation.
The final concentrations of chase medium components must be identical to those for the chase medium used in Basic Protocol 1 (see recipe for chase medium). Calculate the concentrations needed for the concentrated chase medium. A 1.25× solution is appropriate if 4 vol concentrated chase medium is added to the labeling mixture; for other added volumes, the concentrations must be adjusted accordingly.
Immediately collect the first sample of volume z. Add an equal volume of 2× lysis buffer, mix well, and place on ice.
-
At every chase time point, take a sample, add an equal volume of 2× lysis buffer, mix well, and place on ice.
After the sample for the last chase time point is collected, the tube of cells should be almost empty.
Immediately analyze samples by immunoprecipitation (see Support Protocol 1) and nonreducing and reducing SDS-PAGE (see Support Protocol 2) or freeze rapidly in liquid nitrogen and store at –80°C.
ALTERNATE PROTOCOL 2. ANALYSIS OF POSTPONED POST-TRANSLATIONAL DISULFIDE BOND FORMATION IN INTACT CELLS
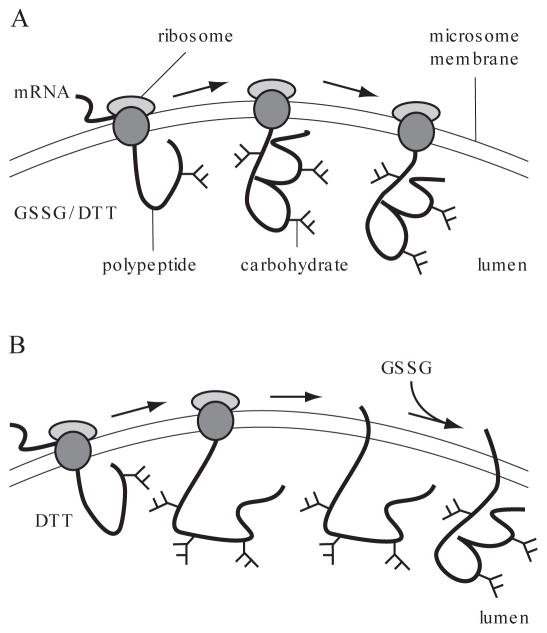
To determine the effect of certain conditions, such as ATP depletion, on folding, it may be necessary to separate the folding process from translation. This can be done by delaying folding until translation, glycosylation, and signal-peptide cleavage are complete. Folding of a disulfide-bonded protein may be prevented by incubating cells in reducing agent. Newly synthesized proteins cannot form disulfide bonds and often cannot fold when oxidation is prevented. When the reducing agent is removed after pulse-labeling, formation of disulfide bonds and folding may proceed (Braakman et al., 1992a,b; see Fig. 14.1.1).
Figure 14.1.1.
In vitro translation/rough endoplasmic reticulum–derived microsome translocation and folding system. (A) For cotranslational folding, the protein is translocated into microsomes containing an oxidizing environment (GSSG). This provides an opportunity for the protein to fold vectorially during the translation and translocation processes. (B) For post-translational folding, oxidizing agent is added after translation and translocation, permitting synchronization and isolation of the folding process.
Wash the cells and deplete the methionine (see Basic Protocol 1, steps 2 and 3). Add dithiothreitol (DTT; APPENDIX 3A) to labeling medium to a final concentration of 5 mM. Perform a pulse-chase experiment and prepare lysates (see Basic Protocol 1, steps 4 to 11).
It may be necessary to optimize the final concentration of DTT in labeling medium for each cell type. During longer incubations, DTT may affect cellular ATP levels, likely induces cell stress or may lead to rapid cell death, whereas some cells tolerate prolonged exposure to DTT without damage. Efficiency of incorporation may also be slightly diminished by the presence of DTT. The duration of pulse labeling may be increased to improve incorporation of label in protein.
Before the effects of different conditions on folding are tested, the efficiency, rate, and outcome of post-translational folding should be compared to those of cotranslational folding.
BASIC PROTOCOL 2. ANALYSIS OF DISULFIDE BOND FORMATION IN ROUGH ENDOPLASMIC RETICULUM–DERIVED MICROSOMES
Formation of disulfide bonds in proteins which possess signal sequences that target the protein to the endoplasmic reticulum can also be studied with an in vitro translation/rough endoplasmic reticulum–derived microsome translocation and folding system (Fig. 14.1.1). 35S-labeled proteins are generated by the translation of mRNA with a rabbit reticulocyte lysate in the presence of [35S]methionine. Translation is carried out in the presence of canine pancreas microsomes; this combination permits cotranslational insertion of 35S-labeled protein into microsomes. An oxidizing agent (i.e., oxidized glutathione, GSSG; APPENDIX 3A) is present during translation to allow monitoring of cotranslational or vectorial oxidation from the N- to the C-terminus of the protein, the mechanism by which proteins acquire their disulfide bonds under normal physiological conditions. Oxidation of proteins in microsomes provides a system where the experimental conditions can be more extensively controlled and manipulated. The oxidation state of the protein can be trapped by alkylation with N-ethylmaleimide (NEM) and monitored by immunoprecipitation (see Support Protocol 1) and nonreducing and reducing SDS-PAGE (see Support Protocol 2).
Materials
1 equivalent/μl nuclease-treated canine pancreas microsomes (Promega)
Rabbit reticulocyte lysate treated with ATP-regenerating system and nucleases (Promega)
1 mM amino acid mixture lacking methionine (Promega)
10 mCi/ml [35S]methionine (1,000 Ci/mmol, Amersham or Perkin Elmer)
100 mM dithiothreitol (DTT)
RNase-free H2O (e.g., DEPC-treated; see recipe)
23 U/ml RNase inhibitor (e.g., RNasin, Promega)
1 μg/μl mRNA for the protein of interest
100 mM oxidized glutathione (GSSG; APPENDIX 3A), titrated to neutrality with KOH
120 mM N-ethylmaleimide (NEM) in 100% ethanol (prepare from 1 M stock; see recipe)
Lysis buffer (see recipe)
2× SDS sample buffer (UNIT 10.1; optional)
27°C water bath
1.5-ml microcentrifuge tubes, RNase-free
Ice bath
Additional reagents and equipment for immunoprecipitation (see Support Protocol 1) and nonreducing SDS-PAGE (see Support Protocol 2)
NOTE: The major source of failure in the in-vitro translation/translocation of proteins is contamination with ribonucleases (RNases) that degrade mRNA. To avoid contamination, water and salt solutions should be treated with diethylpyrocarbonate (DEPC) to chemically inactivate RNases; glass and plasticware should be treated with DEPC-treated water or otherwise treated to remove RNase activity (see recipe for DEPC treatment). Freshly opened plasticware that has not been touched by unprotected hands is also acceptable. It is also helpful to keep a set of solutions for RNA work alone to ensure that “dirty” pipets do not contaminate them. In addition, gloves should be worn at all times.
-
Thaw reagents and solutions rapidly with agitation in 27°C water bath.
Rabbit reticulocyte lysate and canine pancreas microsomes should be stored in aliquots of working volumes (i.e., 6 μl and multiples of 6 μl). Repeated freeze-thaw cycles result in inefficient translation and translocation of protein. Freeze other reagents rapidly in liquid nitrogen after use.
-
Prepare reaction mix in an RNase-free 1.5-ml plastic microcentrifuge tube (96.7 μl total):
6 μl 1 equivalent/μl nuclease-treated canine pancreas microsomes
52 μl treated rabbit reticulocyte lysate
2 μl 1 mM amino acid mixture lacking methionine
8 μl 10 μCi/μl [35S]methionine
1 μl 100 mM DTT
16 μl RNase-free H2O
4 μl 23 U/μl RNase inhibitor
4 μl 1μg/μl mRNA for the protein of interest
-
3.7 μl 100 mM GSSG.
This reaction mix is enough for 10 samples. Adjust the volumes proportionately for more or fewer samples. Each sample represents a different time point.
mRNA for the protein of interest can be transcribed from cDNA for the protein of interest into a commercially available vector (e.g., pBluescript, Stratagene) which contains both T7 and T3 promoters for RNA polymerases.
-
Mix thoroughly with a pipet. Incubate at 27°C.
Set pipettor to less than the total volume of reaction mixture to avoid introducing air bubbles. Mix by repeatedly taking the mixture up and gently expelling it with the pipettor.
-
At the appropriate time (e.g., 0.5, 1, 1.5, 2, 3,…n h), transfer 9-μl samples to separate tubes and add 2.4 μl of 120 mM NEM in ethanol (25 mM final) to alkylate proteins. Incubate 10 min on ice.
In vitro translation of proteins is 3- to 5-fold slower than in vivo translation, so extended time points are required.
Add 600 μl lysis buffer to each alkylated sample or add 30 μl of 2× sample buffer for direct analysis of total proteins translated.
Analyze lysate by immunoprecipitation (see Support Protocol 1) and nonreducing SDS-PAGE (see Support Protocol 2).
ALTERNATE PROTOCOL 3. ANALYSIS OF DISULFIDE-BOND FORMATION IN INTACT CELLS WITHOUT STARVATION
Before the short pulse with [35S] methionine, cells are starved for methionine to deplete the endogenous pool of these amino acids and increase incorporation of the radiolabeled amino acids into newly synthesized proteins. During the starvation period however, cells perceive amino-acid starvation as stress and respond by, amongst others, activating the ubiquitin-proteasomal pathway, which leads to degradation of pre-existing proteins as well as growing nascent chains to ensure continuation of amino-acid supply for protein synthesis. Fast proteins may fold within 15 minutes, but others may take up to several hours. In case of the longer pulse-labeling and chase times the starvation, labeling and chase conditions need to be modified to maintain stress-free conditions for the cells.
Wash the cells and deplete cells from methionine for 10 – 15 min (see Basic Protocol 1, steps 2 and 3) with a modified depletion medium. Add at least 0.5 % (v/v) plain DMEM, containing 200 μM methionine and 200 μM cystine, to the DMEM depletion medium to yield 1 μM methionine and cystine. As MEM contains 100 μM methionine and 100 μM cystine, 1% (v/v) plain MEM should be added. This needs to be checked and calculated for each cell line and medium. Under these conditions translation is maintained at its original level in Hela, HEK293 cells (as example). Pulse label the cells (see Basic Protocol 1, steps 4 and 5). For labeling longer than 5 min methionine (and cystine) need to be added to the pulse medium as well.
To keep depletion as short as possible, steps 1, 2, and 3 of the pulse-chase protocol can be done at the water bath for exactly 15 minutes, scheduled precisely before the pulse labeling, step 4.
When labeling HeLa cells from 5 – 15 min, a concentration of 0.5 μM methionine is sufficient to maintain translation (and hence label incorporation). For 15 – 30 min labeling 1 μM methionine will suffice, and between 30 and 60 minutes 2 μM methionine is needed. In case of very long labeling (8–16 h), the pulse medium should be depletion medium mixed 1:1 with the plain medium, with all culture constituents present in their normal concentrations (FBS, glutathione, non-essential amino acids, sodium pyruvate, etc.). In this case, methionine depletion can be omitted.
The EasyTag Express protein labeling mix (Perkin Elmer) contains 8.0 mCi/ml methionine (specific activity 1175 Ci/mmol) and 2.4 mCi/ml cysteine (specific activity 1075 Ci/mmol), which is 6.8 μM methionine and 2.2 μM cysteine. If the labeling medium contains 250 μCi/ml [35S], the label will contribute 0.25 μM methionine and 0.08 μM cysteine. The EasyTag label contains the reduced form of cystine, because the oxidized cysteine form cannot enter all cell types.
After pulse labeling, samples can be chased (see Basic Protocol 1, steps 6 and 7). In case of chase times >15 min, optimal is a chase in the CO2 incubator with their normal growth medium.
There is no need to add extra methionine in the chase medium, because this already contains a >500× excess. In practice, it often is added fresh, as it is crucial to stop any label incorporation after the pulse-labeling.
Prepare lysates (see Basic Protocol 1, steps 8 to 11).
ALTERNATE PROTOCOL 4. ANALYSIS OF POST-TRANSLATIONAL DISULFIDE BOND FORMATION IN ROUGH ENDOPLASMIC RETICULUM–DERIVED MICROSOMES
Post-translational oxidation is performed by translating the protein under reducing conditions in the presence of microsomes to allow accumulation of translocated and glycosylated proteins with the signal sequence removed and lacking disulfide bonds. Oxidizing agents are added post-translationally to initiate oxidation (see Fig. 14.1.1).
Perform in-vitro translation and analysis as for the previous method (see Basic Protocol 2) with the following exceptions in the indicated steps.
Omit oxidized glutathione (GSSG) from the reaction mixture (to make 93 μl).
After the initial incubation of the reaction, add 4.2 μl of 100 mM GSSG and incubate 1 h more at 27°C before proceeding to alkylate and analyze proteins (Basic Protocol 2, steps 3 to 6)..
ALTERNATE PROTOCOL 5. IN-VITRO-TRANSLATION IN THE PRESENCE OF SEMI-PERMEABILIZED CELLS
The ER-derived microsomes can be substituted with semi-permeabilized cells to provide a more physiological and unperturbed ER microenvironment to follow disulfide bond formation (Wilson et al., 1995; Francis et al., 2003; Kleizen et al., 2005). Low levels of digitonin are used to partially permeabilize the cholesterol-rich plasma membrane of cells, while leaving the endomembranes, including the ER, intact. The preparation of ER-derived microsome involves rupturing of ER membranes during their production providing potential leakage of ER factors including oxidoreductases involved in disulfide bond formation. Using the intact ER is expected to support oxidative folding in an environment with physiologically relevant resident-ER proteins levels. An additional advantage is that in principle each cell can be used as membrane source. This allows not only use of the physiologically relevant cell type but also use of cells that contain mutations or changes in expression levels (overexpression, depletion or deletion) of select ER-resident proteins. For example, a CHO cell that has a defect in the alg6 gene that is involved in transferring glucose to the N-linked glycan dolichol precursor has been used to monitor reglucosylation of target substrates using semi-permeabilized cells (Pearse et al., 2008). Altogether, the coupling of in-vitro translations with semi-permeabilized cells provides a more physiological and manipulatable ER source.
Materials
MEF, COS7, HT1080, HEK293, Hela, B cells or CHO cell lines with appropriate media grown in 75 cm2 flask to near confluence (~5 × 107 cells)
Tissue culture grade Trypsin/EDTA (0.25%) solution
-
KHM buffer
110 mM KOAc, 2 mM MgOAc, 20 mM HEPES pH 8.2
-
Resuspension buffer
50 mM KOAc, 90 mM HEPES pH 7.2
Digitonin (20 mg/ml)
8 ml KHM buffer containing 16 μl STI (soybean trypsin inhibitor) at 50 mg/ml (KHM-STI)
0.1 M CaCl2
Micrococcal nuclease (15,000 Units/ml), thawed immediately before use
250 mM EGTA
Phosphate Buffered Saline (PBS buffer)
-
Wash flasks twice with PBS buffer and detach cells with 4 ml of trypsin.
To accelerate trypsin treatment place in 37°C incubator for 2 – 5 min.
Add 6 ml of KHM-STI to each flask and resuspend cells.
Transfer resuspended cells into sterile 15 ml conical tubes, and place on ice.
Rinse flask with 2 ml KHM-STI.
Transfer resuspended cells to the same 15 ml conical tubes (as described above in 3).
Count cells to obtain an average number of live cells.
Harvest cells by centrifugation at 250 × g for 7 min at 4°C.
Remove supernatant, and gently resuspend the cell pellet in 1 ml of KHM buffer, then add 5 ml of KHM buffer for a final volume of 6 ml.
Permeabilize cells by adding 6 μl of digitonin for a final concentration of 20 μg/ml, and incubate on ice for 5 min.
Stop permeabilization by adding 8 ml of KHM buffer to the tube.
Spin at 250 × g for 7 min at 4°C.
Remove supernatant and resuspend cell pellet in 14 ml of resuspension buffer, and incubate on ice for 10 min.
Spin cells at 250 × g for 7 min at 0°C.
Resuspend pellet in KHM buffer (~4 × 106 cells/100 μl KHM).
Transfer the resuspended cells to a sterile 1.5 ml microcentrifuge tube, and place on ice.
-
Treat permeabilized cells with micrococcal nuclease to remove endogenous RNAs, incubate at 25°C for 12 min.
For an average of 2 – 5 × 106 cells / 100 μl KHM buffer add 1 μl 0.1 M CaCl2, and 2 μl micrococcal nuclease (15 K units/ml).
Stop the reaction by adding EGTA (4 mM final concentration).
Spin at 6,500 g for 5 min at 4°C.
Resuspend the pellet in KHM buffer to a final concentration of 1 × 105 cells/μl.
Proceed to in-vitro-translation by substituting semipermeabilized cells (1.3 × 104 cells/μl) for rough ER-derived microsomes.
SUPPORT PROTOCOL 1. IMMUNOPRECIPITATION OF LYSATES
The protein of interest is precipitated from lysates of cells or microsomes using a specific antibody bound to Protein A–Sepharose beads. The immunoprecipitate is then analyzed by nonreducing and reducing SDS-PAGE (see Support Protocol 2).
Instead of Protein A-Sepharose beads, 10% (w/v) killed, fixed Staphylococcus aureus cells (Zymed) may be used. These are slightly more sticky than beads and thereby may lead to increased background. On the other hand, as they provide tighter pellets, they may allow increased speed of handling. If antibodies do not bind well to Protein A, Protein G-Sepharose beads may be used or a bridging antibody (such as Rabbit anti-mouse Ig bridging a mouse monoclonal antibody and Protein A). For higher-throughput screening, increase of efficiency and decrease of background and sample volumes, Protein-A magnetic beads are the optimal choice.
Materials
10% (w/v) pre-washed Protein-A-Sepharose beads
Antibody against protein of interest
Lysate from pulse-chase labeled cells or microsomes (see Basic Protocol 1 or 2 or Alternate Protocol 1, 2, or 3)
Immunoprecipitation wash buffer (see recipe), 37°C
TE buffer, pH 6.8 (see recipe)
2× nonreducing sample buffer (see recipe)
200 mM dithiothreitol (DTT)
Rotator and microcentrifuge-tube shaker
Additional reagents and equipment for nonreducing and reducing SDS-PAGE (see Support Protocol 2)
-
Add 50 μl of 10% pre-washed Protein-A-Sepharose beads to 0.5 to 500 μl antibody. Shake 1 h at 4°C.
The amount of antibody used depends on the antibody and its concentration (ranging from ascites to diluted cell-culture supernatant).
The time required to bind antibody to Protein A–Sepharose beads usually is less than 1 h.
Before use the Protein A–Sepharose beads are washed five times in lysis buffer without alkylating, reducing or protease inhibitor agents, containing 0.25% BSA.
-
Add 100 to 600 μl postnuclear lysate. Incubate 1 h at 4°C in a rotator.
If the beads tolerate it, shaking is advised. Some antibodies need significantly more time than the default 1 h. In this case rotating overnight at 4°C is an option.
If background is high, preincubate the lysate with S. aureus cells without antibody 1 h at 4°C. Microcentrifuge the cells 4 min at 2,000 × g, room temperature, and transfer the cleared lysate to antibody-coated S. aureus cells.
-
Pellet beads by centrifuging 1 min at 12,000 × g, room temperature. Remove supernatant and resuspend cells in 1 ml wash buffer. Shake 5–10 min. Repeat wash once and pellet beads.
The temperature and time of the wash can affect the level of background; generally, the higher the temperature, the lower the background. Typical wash temperatures range from 10°C to room temperature. Time may be extended to an hour of longer, as long as antibody and antigen remain stable for that long.
-
Aspirate the supernatant. Add 20 μl TE buffer and resuspend beads thoroughly.
If Staph aureus cells are used for immunoprecipitation, their resuspension takes longer and should be continued for 5 min after resuspension of the cell pellet.
Add 20 μl of 2× nonreducing sample buffer (without reducing agent!) and vortex. Heat 5 min at 95°C and vortex.
Pellet beads by centrifuging 1 min at 12,000 × g to give nonreduced sample in the supernatant.
Transfer 20 μl supernatant to a tube containing 2 μl of 200 mM DTT and vortex. Heat 5 min at 95°C. Centrifuge briefly at 12,000 × g to give reduced sample.
-
Analyze nonreduced and reduced samples by SDS-PAGE (see Support Protocol 2).
Samples can be rapidly frozen in liquid nitrogen and stored at –80°C, but storage may increase background.
SUPPORT PROTOCOL 2. NONREDUCING AND REDUCING SDS-PAGE
Immunoprecipitates (see Support Protocol 1) are analyzed by nonreducing SDS-PAGE to detect changes in mobility due to disulfide bond formation.
Materials
Samples in 1× sample buffer (see Support Protocol 1)
2× nonreducing sample buffer (see recipe)
Whatman 3MM filter paper
Additional reagents and equipment for SDS-PAGE minigel with Laemmli buffers (UNIT 10.1; Gallagher, 2012) and Coomassie blue staining and destaining (UNIT 10.5)
-
1
Prepare a 1- or 0.75-mm thick polyacrylamide separating and stacking minigel.
Percent acrylamide depends on molecular weight of the protein.
-
2
Load 8 μl of each sample. Load 1× nonreducing sample buffer in the two lanes next to the samples.
When nonreduced and reduced samples are to be loaded on the same gel, leave two empty lanes between the two sample types, and load empty lanes with 1× nonreducing sample buffer. If nonreduced and reduced samples need to be loaded next to each other, let the heated samples cool to room temperature and add NEM to a final concentration of 50 mM to both sets of samples. Mix well and shortly spin down the volume before loading the samples on the gel.
-
3
Run gel ~1 h at 20 to 25 mA until the dye front is close to or at the bottom of the gel. Monitor whether the gel warms up in the center. It should run at maximum speed (current) without heating up, to keep diffusion to a minimum.
-
4
Stain the gel, including the stacking gel, with Coomassie blue stain and destain (see UNIT 10.5).
Stacking gels often contain nonreduced or aggregated protein.
-
7
Dry gel onto Whatman 3MM filter paper. Autoradiograph/fluorograph, perhaps at –80°C.
SUPPORT PROTOCOL 3. ANALYSIS OF DISULFIDE AND FREE THIOL BY PEG-MALEIMIDE MODIFICATION
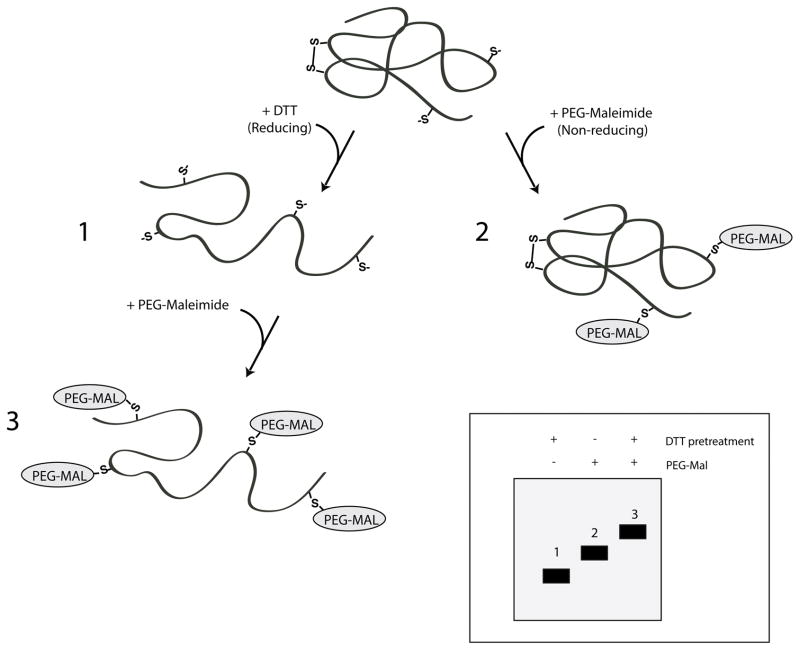
Monitoring disulfide bond formation as well as the presense of unpaired cysteines at steady state can be achieved in cell lysates. This method uses gel mobility shifts of the oxidized protein of interest modified with PEG-maleimide compared to the mobility of the modified or unmodified reduced protein sample. The gel shifts are caused by the addition of the bulky PEG group attached to a thiol reactive maleimide (see Fig. 14.X.X). An advantage of using this method to monitor the presence of disulfide bonds instead of conventional methods such as comparing mobility shifts between non-reducing and reducing gels, is that in some cases the formation of a disulfide bond may not sufficiently affect the compactness of the protein or its mobility on a non-reducing gel. Using a large group such as PEG-maleimide (MW 5,000) to modify unpaired cysteines will affect the mobility of the protein on a gel especially when multiple cysteines are present.
Figure 14.X.X.
Assessing oxidative state using PEG-Maleimide modification. Cartoon representation of a protein possessing two cysteines involved in an intramolecular disulfide bond and two unpaired cysteines. The presence of free thiols is analyzed using sulfhydryl-reactive maleimide linked to a polyethylene glycol group (PEG-Mal). Cells expressing the protein of interest are treated with or without DTT to assess reducing and non-reducing states, respectively (1 and 2). Cells are lysed in the presence and absence of PEG-maleimide and protein samples are resolved by SDS-PAGE. Insert, reduced and unlabeled proteins are expected to migrate fastest through the SDS-PA gel (1), followed by nonreduced proteins labeled with PEG-maleimide (2), with reduced and fully labeled proteins being slowest (3).
Materials
HEK293T cells with DMEM media
Methoxypolyethelyne glycol maleimide 5,000 (PEG-maleimide)
1 M Dithiothreitol (DTT)
PBS
-
Lysis buffer: 9% SDS, 30 mM Tris-HCl pH 6.8, 15% glycerol
Add 0.05% bromophenol Blue for gel loading purposes
-
Plate cells to a density of 1 × 106 cells on 6 cm dishes.
Calculate number of dishes for each condition −/+ DTT pretreatment, −/+ PEG-maleimide.
Cells should be treated with and without DTT to break and maintain disulfides, respectively, in the presence and absence of PEG-maleimide.
Pretreat appropriate dishes with DTT to reduce disulfides: Remove media and replace with fresh media containing 5 mM DTT. Remove and replace media for the minus DTT pretreatment dishes. Incubate for 1 h at 37°C, in a 5% CO2 incubator.
Remove media and wash off DTT with 2 ml 1× PBS.
Alkylate proteins by adding 300 μl of lysis buffer containing 5 mM PEG-maleimide to appropriate dishes, scrape off cellular lysates, and transfer to a 1.5 ml microcentrifuge tube.
Incubate for 2 h at room temperature in the dark.
Add 30 μl of 1 M DTT (for final concentration of 100 mM) to quench labeling reaction.
Sonicate samples using a probe sonicator set to 30 AMP for 30 sec, repeat for a total of three times to reduce sample viscosity.
-
Load 20 – 30 μl of each sample per lane on SDS-PAGE.
Amount loaded depends on signal intensity and should be determined on a protein-to-protein basis, and gel percentage depends on size and number of free thiols
Run gels at 20 mA per gel for about 1.5 – 2 h, and perform routine Western blot.
REAGENTS AND SOLUTIONS
Use Milli-Q-purified water or equivalent in all recipes and protocol steps. For common stock solutions, see APPENDIX 2E; for suppliers, see SUPPLIERS APPENDIX.
Chase medium
Complete tissue culture medium appropriate for the cells containing:
20 mM HEPES (sodium salt), pH 7.4 (see recipe, needs to be the same as the pH of the medium)
5 mM methionine (see recipe)
1 mM cycloheximide (optional; see recipe)
-
Prepare fresh
Add HEPES, methionine, and optional cycloheximide from concentrated stocks (see recipes).
The chase medium may contain fetal bovine serum or another protein source, especially when chase times are long. Protein should be omitted when more defined conditions are needed. When the reducing agent dithiothreitol (DTT) is added, proteins present in the medium will partly quench the reductant.
Concentrated chase medium
Adjust the concentration of chase medium components (see recipe) so the final concentrations are 20 mM HEPES, 5 mM methionine, and 5 mM cycloheximide after the concentrated chase medium has been added to the labeling medium for cells in suspension (see Alternate Protocol 1).
Cycloheximide, 500 mM stock
28.1 g cycloheximide
H2O to 100 ml
-
Store 1- to 2-ml aliquots ≤2 years at –20°C
Thaw and warm to room temperature and vortex to completely dissolve before using. Do not freeze and thaw more than three times.
Cysteine, 0.5 M stock
6.06 g cysteine
H2O to 100 ml
-
Store 1- to 2-ml aliquots ≤2 years at –20°C
Thaw and warm to room temperature and vortex to completely dissolve before using. Do not freeze and thaw more than three times.
Depletion medium
Methionine- (and cysteine-)free cell culture medium containing:
20 mM HEPES, pH 7.4 (from 1 M stock; see recipe, should be same pH as the medium)
-
Prepare fresh:
Methionine- (and cysteine-)free cell culture medium are commercially available as DMEM and MEM. All components present in the normal culture medium except methionine (and cysteine) should be present (glutamine, sodium pyruvate, non-essential amino acids)
Diethylpyrocarbonate (DEPC) treatment of solutions and labware
CAUTION: Wear gloves and use a fume hood when working with DEPC because it is a suspected carcinogen.
Solutions: Add 0.2 ml DEPC per 100 ml of the solution to be treated. Shake vigorously to dissolve DEPC. Autoclave the solution to inactivate remaining DEPC.
Any water or salt solutions used in RNA preparation should be treated with DEPC. Note that solutions containing Tris cannot be effectively treated with DEPC because Tris reacts with DEPC to inactivate it.
Labware: Rinse glass and plasticware thoroughly with DEPC solution. Alternatively, bake glassware 4 h at 300°C; rinse plasticware with chloroform, or use fresh plasticware straight from a package that has not been touched by unprotected hands. Wear gloves for all manipulations.
Note that autoclaving alone will not fully inactivate many RNases.
Ethylenediaminetetraacetic acid (EDTA), 200 mM stock
14.6 g EDTA
180 ml H2O
10 mM NaOH added dropwise with mixing just until EDTA dissolves
Adjust pH to 7.4
H2O to 250 ml
-
Store at 4°C
For EDTA, pH 6.8, adjust the pH to 6.8.
HEPES (N-2-hydroxyethylpiperidine-N′-ethanesulfonic acid), 1 M stock
119.15 g HEPES 400 ml H2O
Adjust pH to 7.4 with 10 M NaOH
H2O to 500 ml
Store at 4°C
Immunoprecipitation wash buffer
PBS (APPENDIX 2E)/0.5% (v/v) Triton X-100 or
PBS/150 mM NaCl
-
Store at 4°C or room temperature
For every protein-antibody combination, the optimal wash buffer needs to be determined. The two buffers listed are particularly mild wash buffers that will maintain most antigen-antibody interactions but may lead to high background.
To decrease background, other detergents or multiple detergents may be added, salt concentration may be increased, or a combination of the two may be used. SDS at a concentration ≥0.05% (w/v) may be especially helpful, with or without the quenching effect of added nondenaturing detergents.
Labeling medium
Methionine- (and cysteine-)free tissue culture medium containing:
125 to 250 μCi [35S]methionine (and cysteine)/ml
-
Prepare fresh for each experiment (400 μl per chase time point)
Addition of 50 to 100 μCi in 400 μl to a 60-mm-diameter dish (~3–5 × 106 cells) should be sufficient to visualize a 1- to 2-min-pulse-labeled protein that is expressed to a high level in the cell.
Lysis buffer
PBS (APPENDIX 2E) or similar buffer containing:
0.5% (v/v) Triton X-100
1 mM EDTA, pH 7.4 (optional, see recipe), added just before use
20 mM NEM (see recipe), added just before use
1 mM PMSF (see recipe), added just before use
Chymostatin, Leupeptin, Antipain, Pepstatin A, mixture (CLAP) (10 μg/ml each final; see recipe), added just before use
-
Use within 3 h
Add EDTA, NEM, and protease inhibitors (PMSF and CLAP mixture) from concentrated stocks (see recipes). Thaw and warm stocks to room temperature and vortex to completely dissolve before using.
For 2× lysis buffer, double the concentrations of reagents.
Depending on the protein analyzed and on the purpose of the experiment, a variety of detergents may be used. Triton X-100 may disrupt noncovalent interactions between proteins. Milder detergents that can be used include CHAPS (3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonate), deoxycholate, n-octylglucoside, or a mixture of detergent and lipid.
Methionine, 250 mM stock
3.73 g methionine
H2O to 100 ml
-
Store 1- to 2-ml aliquots ≤2 years at −20°C
Thaw and warm to room temperature and vortex to completely dissolve before using. Do not freeze and thaw more than three times.
N-ethylmaleimide (NEM), 1 M stock
12.5 g NEM
100 ml 100% ethanol
-
Store 1- to 2-ml aliquots ≤1 years at −20°C protected from light
Thaw and warm to room temperature and vortex to completely dissolve before using.
NEM is sensitive to light and to hydrolysis in water. If it is frozen and thawed too many times or exposed to too much light, the solution will turn yellow and the NEM will precipitate.
Nonreducing sample buffer, 2×
400 mM Tris·Cl, pH 6.8
6% (w/v) SDS or 6% (v/v) from a commercial 20% (w/v) SDS solution.
20% (v/v) glycerol
2 mM EDTA, pH 6.8 (from 200 mM stock; see recipe)
0.01% (w/v) bromphenol blue
Phenylmethylsulfonyl fluoride (PMSF), 200 mM stock
3.48 g PMSF
100 ml anhydrous isopropanol
-
Store in 500-μl aliquots ≤2 years at −20°C
PMSF is highly unstable in water (the half life is ~30 min at 37°C and a few hours on ice). Add this stock to solution immediately before use. Thaw and warm to room temperature and vortex to completely dissolve before using.
CLAP mixture stock
10 mg/ml chymostatin in dimethyl sulfoxide (DMSO)
10 mg/ml leupeptin in DMSO
10 mg/ml antipain in DMSO
10 mg/ml pepstatin A in DMSO
Store in 20-μl aliquots ≤2 years at −20°C
Use at a final concentration of 10 μg/ml each
Stop buffer
Hanks’ balanced salt solution containing calcium, magnesium and glucose
20 mM NEM (from 1 M stock; see recipe)
-
Prepare fresh and store on ice <5 h
NEM may be replaced by iodoacetamide or iodoacetic acid, which react more slowly with free –SH groups. They are less specific than NEM and penetrate cells more slowly than NEM, but they irreversibly carboxymethylate the –SH group, whereas NEM may dissociate slowly. Deblocking of –SH groups after NEM treatment is not a problem if cells are stored ≤2 h on ice.
TE buffer, pH 6.8
10 mM Tris·Cl, pH 6.8
1 mM EDTA, pH 6.8 (from 200 mM stock; see recipe)
Store at 4°C
Wash buffer
-
HBSS containing calcium, magnesium and glucose.
For most cells, calcium ions are crucial to maintain adherence to the plastic dishes, especially to withstand many washes.
COMMENTARY
Background Information
Folding of a newly synthesized protein can be followed by assaying for conformational changes in the molecule during and immediately after synthesis. For this type of analysis, a large number of antibodies that recognize specific epitopes are required to analyze the changes in the complete molecule. In a mature protein that contains intrachain disulfide bonds (which is true of most proteins synthesized in the endoplasmic reticulum), the formation of these cross-links is indicative of folding (Creighton, 1986). A disulfide bond can be formed only when the conformation allows the two participating cysteines to be in close proximity. With only few antibodies available, the rate and extent of folding of a protein can be examined grossly through detection of disulfide bond formation. Of course, disulfide bond formation is not identical to folding; therefore, a comparison of disulfide bond formation and conformational changes is required for every protein studied.
When a disulfide bond–containing protein is prepared for SDS-PAGE, it is routinely reduced. In nonreducing denaturing polyacrylamide gel electrophoresis, however, the protein is denatured with SDS, but the disulfide bonds remain intact. The consequence is a more compact conformation of the protein, resulting in a higher electrophoretic mobility. In some cases the protein may bind less SDS, resulting in a lower electrophoretic mobility. Most proteins will run faster in oxidized form than in reduced form.
The choice of radioactive labeling is dictated by the desire to follow a protein from the moment of synthesis. No other method permits following the life of a protein in a cell in such detail. Any other method of detection still would require bulk production of proteins, which would be difficult and nonphysiological.
The in-vitro-translation system coupled with canine pancreas microsomes traditionally has been employed to study the translocation of secretory, lysosomal, and many integral membrane proteins across the membrane of the endoplasmic reticulum (Blobel and Dobberstein, 1975). Since then, microsomes have been shown to possess a complete set of folding enzymes and chaperones that provide an environment for rapid and efficient oxidative protein folding (Scheele and Jacoby, 1982; Marquardt et al., 1993; Hebert et al., 1995). As microsome isolation involves fragmentation and resealing of ER membranes, different preparations of microsomes have retained variable quantities of ER-resident chaperones and folding enzymes. This disadvantage, paired with the inflexibility of ER source, has popularized the use of semi-permeabilized cells as a source of ER membranes (Alternate Protocol 5; Wilson et al., 1995). Any cell type can be used and the integrity of the ER membrane is maintained.
Microsomes and semi-permeabilized cells allow improved control and manipulation of the ER environment than intact cells. The reticulocyte lysate (equivalent to the cytosol in the cell) is directly accessible, and intralumenal components of the ER can be manipulated with ionophores, detergents, toxins, or alkaline pH (Bulleid and Freedman, 1988; Nicchitta and Blobel, 1993; Hebert et al., 1995). Post-translational oxidative foling of proteins (see Alternate Protocol 2 and Alternate Protocol 3) can be used in both intact cells and in-vitro translations to separate the oxidative folding process from other biosynthetic events such as translation. Together, analysis of cellular and microsomal folding allow effective dissection of the folding pathway of a protein.
Critical Parameters
Wash buffer for adherent cells should contain Ca2+and Mg2+ to maintain adherence to the plate, especially when there are several washes.
Labeling medium should contain 50 to 100 μCi 35S-labeled methionine for ~3–5 × 106 cells. If the protein of interest has few methionines, or if it contains several cysteines, a combination of [35S]methionine and [35S]cysteine, or unpurified labeled methionine and cysteine, may be used for labeling (see comments in protocols). When short pulses are done on a water bath, the stabilized version of unpurified methionine should be used (in e.g. EasyTag EXPRESS 35S Protein labeling mix, Perkin Elmer) to minimize contamination of air, pipets, and incubators.
Cycloheximide in the chase medium will stop elongation of unfinished nascent peptide chains. To study kinetics of disulfide bond formation, cycloheximide should be added to the chase medium. Because cycloheximide blocks completion of nascent chains, it will prevent the increase in amount of full-length labeled protein during the chase and hence will lead to lower signals; cycloheximide hence should be omitted when a maximum amount of incorporated label is required (Braakman et al., 1991).
An alkylating agent—e.g., N-ethylmaleimide, iodoacetamide, or iodoacetic acid—must be included in the stop buffer and lysis buffer to prevent artifactual formation and isomerization of disulfide bonds. N-ethylmaleimide reacts faster than the other two alkylating agents, but at least two alkylating agents should be tested and compared with the absence of alkylating agent. Ideally, there should be no difference in results obtained with the various alkylating agents, except possibly a change in electrophoretic mobility due to alkylation. The alkylating agent may allow trapping of an oxidative folding intermediate that would be unstable without it.
SDS-PAGE should be performed under nonreducing conditions; reducing agents should be completely absent. To analyze nonreducing and reducing samples side by side on the same gel, to all samples (nonreducing as well as reducing) the previously used alkylating agent should be added to at least a two-fold concentration compared to the reducing agent.
The translocation process is dependent upon free sulfhydryl groups, but oxidation (and oxidative folding) requires an oxidizing environment. To simultaneously create a reducing extralumenal compartment and an oxidizing intralumenal compartment, DTT should be omitted from the reticulocyte lysate and a glutathione buffer (GSSG/GSH) should be titrated in. The redox range over which both translocation and oxidative folding occur efficiently is small. As the redox potential is directly related to the pH, pH must be carefully controlled as well.
Troubleshooting
The presence of disulfide bonds in a protein does not guarantee an electrophoretic mobility difference between reduced and oxidized forms, especially when a protein is large and disulfide bonds form small peptide loops. Electrophoretic mobility is influenced most by SDS binding, which may be lower in acidic proteins and strongly hydrophobic proteins. The largest shift can be expected at a late chase time when all disulfide bonds have been formed. If the presence of disulfide bonds is not certain and needs to be tested, the radioactive cell lysate can be treated with GSSG, followed by immunoprecipitation and analysis by nonreducing and reducing SDS-PAGE as described (see Support Protocol 1 and Support Protocol 2). Alternatively, free sulfhydryl groups can be labeled with [14C]iodoacetamide before and after reduction or identified with the PEG-maleimide protocol (Support Protocol 3).
The mobility difference between reduced and oxidized protein may be minimal, but might be improved by using a lower percentage acrylamide gel, a different cross-linker instead of bisacrylamide, and/or changing the alkylating agent.
The treated reticulocyte lysate used here is optimized to efficiently translate a large range of mRNAs with an ATP-generating system, mixture of tRNAs, and salts. The concentration of salts greatly affects the translation efficiency. If the efficiency of translation is low, an untreated lysate should be used and optimized specifically for the mRNA for the protein of interest.
If there is difficulty in differentiating between translocated and untranslocated proteins in microsomes or semi-permeabilized cells, untranslocated protein can be removed by protease digestion or centrifugation. Translocation of protein across ER membrane protects the protein from digestion by added proteases. Alternatively, dense rough microsomes and semi-permeabilized cells can be pelleted easily by centrifugation, thereby isolating translocated proteins from extralumenal untranslocated proteins.
If the background is high in immunoprecipitation, try different immunoprecipitation wash buffers, using other detergents or multiple detergents, increased salt concentration, or a combination of both. SDS at concentrations ≥0.05% (w/v) may be especially helpful, with or without the quenching effect of added nondenaturing detergents. Antibodies targeting the disulfide-containing protein of interest need to be carefully selected as antibodies are commonly conformation dependent and frequently recognize mature protein more efficiently than folding intermediates or non-native products.
Anticipated Results
In most cases, formation of disulfide bonds creates a more compact structure that results in an increase in the mobility of a protein in nonreducing SDS-PAGE (for representative results, see Braakman et al., 1992a,b). The number of disulfide bonds and the distance between the cysteines in a sulfhydryl pair influences the mobility change. During the chase, the protein will move from the more reduced position in the gel to the more oxidized position. For some proteins, distinct oxidative intermediates may be detectable, but for others a fuzzy band may precede accumulation of native protein.
In reducing SDS-PAGE, ideally one band will be found in intact cells. Two bands will be found in the microsomal system—one for untranslocated protein and one for translocated protein. When the ER-targeting signal sequence (1.5 to 3.0 kDa) is cleaved during translocation, a small mobility shift may ensue, dependent upon the protein’s molecular weight. Differentiation of translocated and untranslocated proteins in reducing SDS-PAGE is most prominent for multiglycosylated proteins, because each glycosylation step adds ~2.5 kDa to the translocated protein. Modifications of oligosaccharides and other post-translational modifications can change electrophoretic mobility as well, which are distinguishable from disulfide-bond changes by comparison of reducing with nonreducing gels.
Time Considerations
Design and preparation of the experiment should take the most time. The very first time, this may take 1 or more days, depending on experience and available equipment. Preparation for routine experiments is around 1 h. The pulse-chase portion requires 1 h plus the maximum chase time. For rapidly folding proteins, folding can be over in 15 min; for others it may take hours. Cultures of cells should be started 1 or 2 days before the experiment so they will be subconfluent on the day of the experiment.
Pulse-chase, immunoprecipitation, and SDS-PAGE are optimally done in 1 day. It is possible to rapid-freeze lysates or immunoprecipitates in liquid nitrogen and store them at –80°C until further use, but freezing may lead to higher background and less reproducibility. Freezing of detergent cell lysates is highly preferred over freezing of immunoprecipitates, as SDS will precipitate in the cold.
Acknowledgments
This work was supported by the National Institutes of Health under award number GM086874 (to D.N.H.); and a Chemistry-Biology Interface program training grant (T32 GM08515 to L.L.). This work was also supported by the CF Foundation and NWO (Netherlands Organization for Scientific Research).
Footnotes
Key References
Braakman et al., 1991. See above.
Describes the protocol in intact cells, results with influenza hemagglutinin, and considerations for ultra-short pulse times.
Hebert et al., 1995. See above.
Describes the use of rough-ER derived microsomes to follow oxidation and chaperone binding of a maturing substrates.
Marquardt et al., 1993. See above.
Describes the protocol in microsomes.
Jansens and Braakman, 2003. See above.
Describes the pulse-chase laboratory set up and protocols.
Contributor Information
Ineke Braakman, Cellular Protein Chemistry, Bijvoet Center for Biomolecular Research, Faculty of Science, Utrecht University, The Netherlands.
Lydia Lamriben, Department of Biochemistry and Molecular Biology, Program in Molecular and Cellular Biology, University of Massachusetts, Amherst, MA.
Guus van Zadelhoff, Cellular Protein Chemistry, Bijvoet Center for Biomolecular Research, Faculty of Science, Utrecht University, The Netherlands.
Daniel N. Hebert, Department of Biochemistry and Molecular Biology, Program in Molecular and Cellular Biology, University of Massachusetts, Amherst, MA
Literature Cited
- Blobel G, Dobberstein B. Transfer of proteins across membranes. II. Reconstitution of functional rough microsomes from heterologous components. J Cell Biol. 1975;67:852–862. doi: 10.1083/jcb.67.3.852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braakman I, Hoover-Litty H, Wagner KR, Helenius A. Folding of influenza hemagglutinin in the endoplasmic reticulum. J Cell Biol. 1991;114:401–411. doi: 10.1083/jcb.114.3.401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braakman I, Helenius J, Helenius A. Manipulating disulfide bond formation and protein folding in the endoplasmic reticulum. EMBO J. 1992a;11:1717–1722. doi: 10.1002/j.1460-2075.1992.tb05223.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braakman I, Helenius J, Helenius A. Role of ATP and disulphide bonds during protein folding in the endoplasmic reticulum. Nature. 1992b;356:260–262. doi: 10.1038/356260a0. [DOI] [PubMed] [Google Scholar]
- Braakman I, Hebert DN. Protein folding in the endoplasmic reticulum. Cold Spring Harb Perspect Biol. 2013;5:a013201. doi: 10.1101/cshperspect.a013201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulleid NJ, Freedman R. Defective cotranslational formation of disulphide bonds in protein disulphide-isomerase-deficient microsomes. Nature. 1988;335:649–651. doi: 10.1038/335649a0. [DOI] [PubMed] [Google Scholar]
- Creighton TE. Disulfide bonds as probes of protein folding pathways. Methods Enzymol. 1986;131:83–106. doi: 10.1016/0076-6879(86)31036-x. [DOI] [PubMed] [Google Scholar]
- Francis E, Wang N, Parag H, Halaban R, Hebert DN. Tyrosinase maturation and oligomerization in the endoplasmic reticulum requires a melanocyte-specific factor. J Biol Chem. 2003;278:25607–25617. doi: 10.1074/jbc.M303411200. [DOI] [PubMed] [Google Scholar]
- Hebert DN, Foellmer B, Helenius A. Glucose trimming and reglycosylation determine glycoprotein association with calnexin in the endoplasmic reticulum. Cell. 1995;81:425–433. doi: 10.1016/0092-8674(95)90395-x. [DOI] [PubMed] [Google Scholar]
- Jansens A, Braakman I. Pulse-chase labeling techniques for the analysis of protein maturation and degradation. Methods Mol Biol. 2003;232:133–145. doi: 10.1385/1-59259-394-1:133. [DOI] [PubMed] [Google Scholar]
- Kleizen B, van Vlijmen T, de Jonge HR, Braakman I. Folding of CFTR is predominantly cotranslational. Mol Cell. 2005;20:277–287. doi: 10.1016/j.molcel.2005.09.007. [DOI] [PubMed] [Google Scholar]
- Marquardt T, Hebert DN, Helenius A. Post-translational folding of Influenza hemagglutinin in isolated endoplasmic reticulum–derived microsomes. J Biol Chem. 1993;268:19618–19625. [PubMed] [Google Scholar]
- Nicchitta CV, Blobel G. Lumenal proteins of the mammalian endoplasmic reticulum are required to complete protein translocation. Cell. 1993;73:989–998. doi: 10.1016/0092-8674(93)90276-v. [DOI] [PubMed] [Google Scholar]
- Pearse BR, Gabriel L, Wang N, Hebert DN. A cell-based reglucosylation assay demonstrates the role of GT1 in the quality control of a maturing glycoprotein. J Cell Biol. 2008;181:309–320. doi: 10.1083/jcb.200712068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheele G, Jacoby R. Conformational changes associated with proteolytic processing of presecretory proteins allow glutathione-catalyzed formation of native disulfide bonds. J Biol Chem. 1982;257:12277–12282. [PubMed] [Google Scholar]
- Wilson R, Allen AJ, Oliver J, Brookman JL, High S, Bulleid NJ. The translocation, folding, assembly and redoxdependent degradation of secretory and membrane proteins in semi-permeabilized mammalian cells. Biochem J. 1995;307:679–687. doi: 10.1042/bj3070679. [DOI] [PMC free article] [PubMed] [Google Scholar]