Abstract
Uncovering the genetic factors that correlate with a clinical deviation of previously unknown etiology helps to diminish the unknown variation influencing the phenotype. Clinical studies, particularly those that consider the effects of an appliance or treatment regimen on growth, need to be a part of these types of genetic investigations in the future. While the day-to-day utilization of “testing” for genetic factors is not ready for practice yet, genetic testing for monogenic traits such as Primary Failure of Eruption (PFE) and Class III malocclusion is showing more promise as knowledge and technology advances. Although the heterogeneous complexity of such things as facial and dental development, the physiology of tooth movement, and the occurrence of External Apical Root Resorption (EARR) make their precise prediction untenable, investigations into the genetic factors that influence different phenotypes, and how these factors may relate to or impact environmental factors (including orthodontic treatment) are becoming better understood. The most important “genetic test” the practitioner can do today is to gather the patient’s individual and family history. This would greatly benefit the patient, and augment the usefulness of these families in future clinical research in which clinical findings, environmental, and genetic factors can be studied.
Introduction
Many orthodontic clinicians think of genetics in terms of controlling and therefore predicting facial growth, and/or in terms of a patients’ genetics defining the limits of what can be changed with treatment. Knowing whether “genetics” can cause a phenotype has been cited as a factor in a patients’ eventual outcome; that is, if the phenotype is genetically programmed, then orthodontists may be limited in what they can do to change it.1 However, this concept has often been misapplied. Inappropriate use of heritability estimates in the orthodontic literature have occurred when authors misinterpret these estimates to be a proxy for determining whether a phenotype is of “genetic origin”; particularly when a malocclusion or other anatomic morphology (e.g., arch width) exhibits a complex inheritance pattern, or etiology, as most phenotypes do.2
The most practical and significant way that genetics will help in clinical practice is through the identification of specific genetic factors and factor variations that can influence the craniofacial traits that are identified within an individual; not by using heritability estimates. However, since there is practically no aspect of orthodontic practice that can be precisely predicted or explained by only one mutation in one gene, the expectation that genetics will be a crystal ball that tells all is unfounded. To paraphrase the writer HL Mencken, “For every complex problem there is an answer that is clear, simple, and wrong.”3 Hence, it most likely will be a combination of genetic factor effects and variations that will be central to enhancing our understanding of the genetic influence(s) that act in on numerous complex oral-facial phenotypes and responses to treatment. An overview of several studies pertinent to the orthodontist are presented below which have examined different genetic factors in the attempt to explain a portion of the individual variation observed in facial growth and response to treatment.
Facial Growth
Investigations into the influence of genetic factors on facial growth have incorporated measurements on lateral cephalometric radiographs to explore: (1) whether genetic variation within candidate genes are associated with measurable differences in the rate of annualized sagittal growth of the jaws in males and females, and (2) assess how genetic variations correlate with the size and/or morphology of skeletal variation and malocclusions.
Differences in the Rate of Annualized Sagittal Growth of the Jaws
While approximate facial growth predictions based upon expected growth curves can be useful for the average patient, more precise and personalized predictions would incorporate and account for the growth potential associated with an individual’s inherited genetic factors; particularly those factors that are highly pertinent to the pubertal growth spurt (PGS). In addition, as orthodontists we regularly observe a great deal of variation between the facial growth responses of different patients to various appliances such as those used for mandibular protrusion. It would be great valuable if we as clinicians could explain these growth response differences in a more concrete manner than simply saying some individuals are “better responders” than others. After all, how does a clinician know that the patients are not “better growers” to begin with? How do they know if certain patients will respond better than they would have without the appliance intervention? Hence, we need a definable reference frame with which to address these questions, and many of these questions may best be answered in the context of defining and understanding a patient’s growth potential due to their inherited genetic variation.
Based on years of investigation, it is understood that the PGS response is mediated by the combination of sex steroids, growth hormone, insulin-like growth factor (IGF-I) and other endocrine, paracrine and autocrine factors. Administration of low doses of testosterone in boys with delayed puberty not only accelerates their statural growth rate, but their craniofacial growth rate as well.4 In addition to testosterone, estrogen-based hormones are also involved in growth and development.5 Aromatase (also known as estrogen synthetase) is a key cytochrome P450 enzyme involved in estrogen biosynthesis which catalyzes the final rate limiting step in the conversion of testosterone and androstenedione to estradiol and estrone, respectively.6 CYP19A1 is the gene that encodes aromatase; therefore regulation of the transcription of this gene is critical for the testosterone/estrogen (T/E) ratio in the body. The T/E ratio is vital in the development of sex-indexed (male) facial characteristics such as the growth of cheekbones, the mandible and chin, the prominence of eyebrow ridges and the lengthening of the lower face.7
A significant difference in the average sagittal jaw growth has been observed when comparing groups of Caucasian males who inherited different genetic variations within or near the CYP19A1 gene (i.e., different CYP19A1 alleles); with the greatest observed differences in the annualized growth per year during orthodontic treatment shown to be over 1.5 mm/yr for the maxilla and 2.5 mm/yr for the mandible.8 By comparison, no significant differences were detected in female jaw growth based on the inherited CYP19A1 alleles.8 This was particularly impressive in the males studied because no statistical differences in jaw size were observed at the start of treatment based upon the CYP19A1 allele they inherited. The significant difference in jaw size was only expressed over the course of orthodontic treatment in patients who were transitioning through the cervical vertebral stages (CVS) that are known to be associated with increased growth velocity during puberty.8 Interestingly, the same results were also demonstrated in a study of Chinese males and females, strongly suggesting that this variation in the CYP19A1 gene may be a multi-ethnic marker for male sagittal jaw growth.9
Variations in other genetics factors, such as the Pro561Thr (P561T) variant in the growth hormone receptor gene (GHR) has been associated with a difference in adult mandibular ramus length (condylion-gonion); with those who had the GHR P56IT allele having a significantly shorter mandibular ramus length than did those who did not have the GHR P561T allele in a “normal” Japanese sample of 50 men and 50 women. The average mandibular ramus height in those with the GHR P561T allele was 4.65 mm shorter than the average for those without the GHR P561T allele. This significant correlation between the GHR P561T allele and shorter mandibular ramus height was confirmed in an additional 80 women.10 Hence variation in Growth Hormone Receptor and other genes associated with variation in adult morphology are also likely to play a role in the variation in skeletal growth velocity to achieve that adult morphology and should be evaluated in future investigations.11 It is important to note that even by examining multiple genetic factors that influence a complex trait (jaw growth) may only account for a portion of the overall variation in growth observed, and therefore these factors alone may only offer a projected, and not precise, prediction of a patient’s growth. Further investigation of these and other genetic factors, as well as their interactions with each other and with environmental factors, will help to explain what has up to now been an unknown component of individual variations in pubertal facial growth.
Recent studies related to the treatment of Class II malocclusion have indicated that when a removable mandibular protrusion appliance is used specifically during the PGS, an average increase in growth may occur consisting mainly of mandibular elongation and increase in ramus height. This contrasts with using the same appliances for treatment prior to the onset of puberty, or used postpubertally.12 However, the control group may be another group of patients who may be matched for gender, age, and other variables, but not for polymorphisms in genes that affect facial growth, particularly during puberty. Not all patients, even during the PGS, are “good” growers. Therefore, genetic studies that can identify and better define “good” growers (or late growers) are needed, and would aid not only in selecting the optimum treatment timing, but the use of the proper appliance in the appropriate patient.
Skeletal Variation and Malocclusions
Class III “skeletal” malocclusion (often referred to as mandibular prognathism) may be due to a short maxilla, long mandible, or both when examined in the sagittal plane. Studies have shown that this phenotype can occur in families with an autosomal dominant mode of inheritance, variable expressivity, and incomplete penetrance.13 It has also been said to occur due to a major gene effect and multifactorial influence.14 In other words, the phenotype runs strongly in families, but can vary in how severely it affects members of the same family (variable expressivity), and the appearance of the phenotype can even “skip a generation” (incomplete penetrance). Prior to genetic analysis of individuals and families with Class III skeletal malocclusion, the question was whether there was one, or only a few, genetic markers that were the primary driving factors. It was hypothesized that resolving this question and determining which factors were involved could make it more likely to forecast the Class III skeletal growth pattern and growth timing of a patient. For example, would knowing the gene(s) involved in “late” growth help to predict who would be more likely to grow out of a Phase I reverse headgear negative overjet correction? Would it help to determine if a further sagittal (or also vertical) growth discrepancy would occur in the mid or late teens, making a camouflage treatment plan less advisable versus waiting for orthognathic surgery?
In Genome Wide Association Studies (GWAS) of the Class III phenotype, the subjects chosen for analysis are not related to each other to not confound the results of this type of analysis. One problem with many of the GWAS studies, however, has been that if Class III malocclusion is as heterogeneous genetically as it seems to be clinically, this type of study involving many affected and unaffected who all come from different families will be less likely to pinpoint a contributing factor, since it is likely that many contributing factors may be present amongst all the represented families.15 In contrast, Genetic Linkage Analysis studies have been done on large families and require a different type of statistical approach than GWAS in order to account for the shared DNA in common among the family members that is not linked to the phenotype being studied. To date, family linkage analyses in combination with DNA sequencing technologies have been more effective in the identification of genetic mutations that are thought to cause Class III malocclusion than the association studies of Class III. For those who are interested, more information on these different types of studies maybe found elsewhere.16
Genetic linkage and association studies have identified multiple loci and candidate genes connected to the Class III phenotype (Figures 1a and 1b).14,17–26 While numerous genetic loci have been associated with Class III, so far causal genetic mutations have been identified within four unique genes in five families with Class III malocclusions (the genes with an asterisk in Figure 1a).27–31 In future studies of large numbers of unrelated individuals diagnosed with a Class III malocclusion, it will become important to morphologically sub-classify patients with a combination of cephalometric and/or geometric morphometric information to better study the genetics of the predominant sub-type(s) of skeletal and dental Class III across families.32 This type of approach would answer the question of whether Class III patients with a similar subtype will also have similar genetic factors in common, or if this will primarily be the case among affected members of a family.
Figure 1.
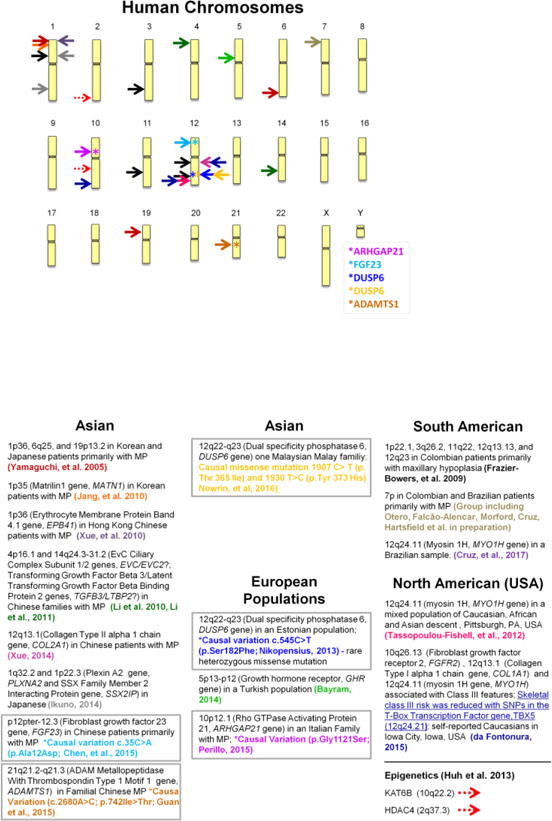
1A: Genetic Loci Identified in Linkage and Association Studies of Class III Malocclusion Ideogram of human chromosomes showing the locations of statistically significant genetic markers in mixed groups; and genes (ARHGAP21, FGF23, DUSP6, and ADAMTS1) where an actual causative mutation has been found in one or more families.
1B: Brief summaries of the genetic finding noted in Figure 1A matching by arrow color and reference with groupings by continental population. MP=mandibular prognathism
There have not been as many genetic studies published on Class II malocclusion as have been published on Class III malocclusion. In the past, some clinical studies supported a genetic basis for Class II division 2 malocclusion,33,34 although so far, no genome wide analysis of this type of malocclusion has been published. Recent studies on muscle fiber type and genetic factors for Class II and other malocclusions features will be reviewed in the next section.
Genetic Variation in Muscle and its Influence on Malocclusion
The Functional Matrix Theory of craniofacial growth emphasizes that skeletal development is secondary to muscle function, airway requirements, and other causes extrinsic to the bone.35 However, what about genetic and epigenetic effects on muscle that then affect skeletal development? Recent research by J. Sciote and collaborators has shown that variations in masseter muscle fiber type, gene expression in masseter muscle, and epigenetic changes that alter gene expression are associated with anterior open versus deep bites, mandibular retrognathism versus prognathism, and mandibular asymmetry.36–39
Skeletal muscle cells produce many proteins that, when in combination, define the unique characteristics and function of the muscle fiber tissue. The myosin heavy chain (MHC) is primarily responsible for the contraction velocity of a muscle fiber.40 In studies of muscle composition, at least four unique muscle fiber types can be described including: type I, IIA, IIX and hybrid type I/II MHC-protein containing muscle fibers. Additional muscle proteins may include cytoskeletal muscle proteins such as α-actinin-2 and -3. While α-actinin-2 appears to be found in all types of skeletal muscle fibers, α-actinin-3 is restricted to fast-contracting type II muscle fibers41 where it enhances muscle force.42
Interestingly, differences in muscle fiber composition have been noted in masseter muscle tissue obtained from patients with a mandibular asymmetry.37 Significant increases in type II muscle fiber area and frequency on the same side as the deviation were discovered when compared to muscle fibers on the side opposite the deviation. Moreover, no significant differences were noted when comparing the muscle composition on the right and left sides of symmetrical patients.37 Additional studies have shown that greater human facial height (i.e., vertical dimension) is inversely related to the size and proportion of masseter muscle fast, type II muscle fibers. Accordingly, short-faced, deep bite phenotypes correlated with increased type II fiber area and frequency, while long faced, open bite phenotypes showed increased type I fiber area and frequency.36,38,39
In contrast to the muscle fiber variations observed in vertical dimension malocclusions, muscle fiber compositions varied to a lesser degree in malocclusions affecting the sagittal dimension, although type IIA and IIX MHC proteins were expressed in the masseter muscles of individuals with mandibular prognathism.38,43 This observation may help to explain the relative difficulty in maintaining an overbite correction, particularly of a deep bite. Among Class III cases, differences were observed in the average fiber area when comparing normal, open bite and deep bite cases. Class III deep bite cases showed an increased amount of type I and hybrid type I/II muscle fiber areas in the masseter muscle, compared to normal and open bite cases. Sex differences have also been observed between masseter muscle fiber type and size, along with muscle growth factor-related gene expression levels.38 Interestingly, when the α-actinin-3 gene (ACTN3) alleles were homozygous for the polypeptide to stop being made at the 577th amino acid (577Stop/Stop), the patient was more likely to have a Class II malocclusion, while this genetic variation was less likely to be found in patients with a deep bite.42
Epigenetic factors may also influence muscle fiber types, vertical and/or sagittal dimension variations and malocclusion types. Masseter muscle from patients with deep bite malocclusions had a higher level of gene expression (i.e., greater mRNA production) for both histone deacetylase 4 (HDAC4) and lysine acetyltransferase 6B (KAT6B; OMIM 605880) compared to masseter muscle from patients with an open bite phenotype. The enzymes encoded by these two genes play an important role in regulating histone acetylation patterns (a form of epigenetic regulation on gene expression), which may affect the development of the malocclusion by increased expression of the genes for fast (type II) muscle fiber types; combined with decreased expression of the gene for the slow-contracting (type I) muscle fiber types.
In support of this hypothesis, increased gene expression of HDAC4 was associated with increased gene expression of the fast type IIX MHC (MYH1), and decreased gene expression of slow type I MHC (MYH7), in subjects diagnosed with Class II. Increased gene expression from the KAT6B locus, in contrast, correlated negatively with type IIX MHC (MYH1) gene expression in Class III malocclusions.36 Overall, gene expression at both KAT6B and HDAC4 loci were elevated in masseter muscle from patients with Class III malocclusions compared to individuals diagnosed with Class II.36,44 It has been proposed that the KAT6B protein could play a potential role in mandibular prognathism through its ability to activate the runt-related transcription factor 2 gene (RUNX2), which encodes an osteogenic transcription factor.44 These studies of how the genetic variation can affect muscle variation, which in turn can affect variation in morphology from Class II to Class III, and from deep to open bites, are areas for research in the correction and stability of correction of facial morphology variation and skeletal malocclusion. Better understanding of the genetics of muscle composition, and how muscle can be re-programmed prior to surgical correction of either Class II or III surgical cases, may greatly aid in reducing the number of surgical relapse cases (and may also aid in the identification of late growers and/or slow growers).
External Apical Root Resorption (EARR) Concurrent with Orthodontics and Rate of Tooth Movement
EARR occurring during orthodontic treatment has been attributed to the use of excessive forces on the teeth, and therefore may be seen by the patient, dentist, or other orthodontists as the fault of the treating practitioner. However, evidence does exists for the occurrence of EARR in some patients who have not received orthodontic treatment; such as in a number of patients diagnosed with missing teeth, increased periodontal probing depths, reduced crestal bone heights, bruxism, chronic nail-biting, and/or anterior open bites with a concomitant tongue thrust.45 It is true that EARR can be increased as a pathologic consequence of orthodontic mechanical loading in some patients.46,47 In addition, the amount of orthodontic movement (and not necessarily the amount of force applied) may be positively, but not absolutely, associated with the extent of EARR.48 There is a considerable amount of variation in the severity of EARR associated with an individual patients’ customized orthodontic treatment.49 Orthodontic tooth movement or “biomechanics”, however, appears to account for less than half (approximately one-tenth to one-third) of the total variation observed clinically for EARR.50,51 So, what accounts for the remaining variation in EARR concurrent with orthodontia?
Tooth movement studies using a panel of different inbred mice have supported the notion that there is a genetic component involving multiple genes in histological root resorption.52,53 In the patient populations examined, heritability estimates have suggested that approximately half of the EARR variation observed concurrent with orthodontics, and almost two-thirds of maxillary central incisor EARR specifically, may be attributed to genetic variation within the constraints of heritability measures already mentioned in this chapter.54 In a retrospective twin study on EARR, examiners found supporting evidence for both genetic and environmental factors influencing EARR.55 In addition, it is noteworthy to add that although EARR was not specifically examined, studies of the reaction to orthodontic force application in humans, including measurements of the rate of tooth movement, have shown differences depending on an individual’s genetic background.56
Since the initial explorative investigations were published in 2003,53,57 looking for possible genetic factors associated with EARR concurrent with orthodontia, many additional candidate gene studies have been conducted using a number of different populations (Figure 2).57–67 Even though heritability studies had hinted that genetic influences on EARR concurrent with orthodontics might account for a large portion of the total phenotypic variation observed, the contribution of the individual genes investigated to date appear to be relatively small and inconsistent, as would be expected with a complex trait; particularly in studies with fairly small sample sizes. While some of these genetic markers are associated with the occurrence of EARR most of the time, there are patients who have inherited the genetic marker that usually accompanies EARR but do not develop EARR, and there are some patients with EARR who do not have the marker. In this light, the “predictive” value of a single marker appears to be limited when used alone (e.g., without information concerning other DNA (gene) markers and additional variables that may be involved).16
Figure 2.
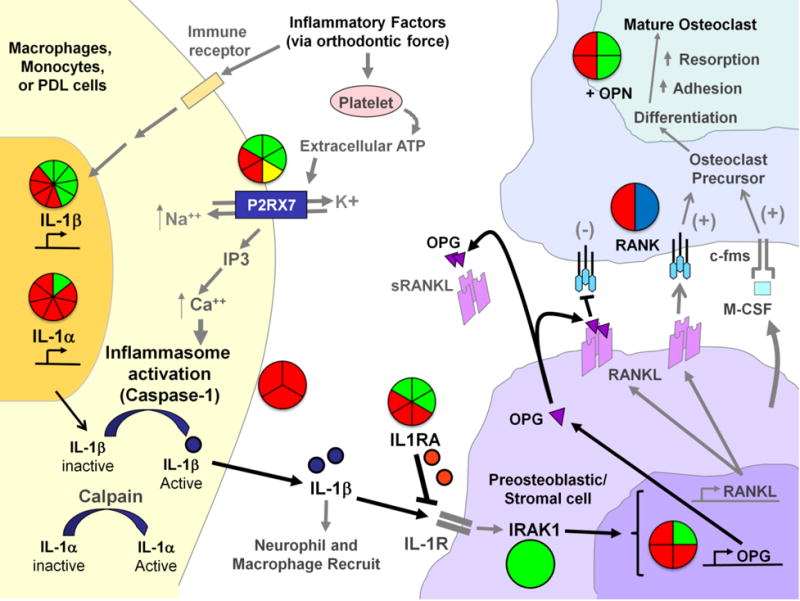
Genetic Associations to EARR within the ATP/P2RX7/IL-1β and RANK/RANKL/OPN signaling pathways-The pie-charts located throughout the diagram summarize the genetic association and linkage findings connected to different factors within the pathways and EARR. The number of triangles within each pie shape summarize the number of markers or independent tests that were examined for each factor; where (green) indicates genetic association, (red) indicates no association, (yellow) indicates a haplotype association, and (blue) indicates genetic linkage.
As orthodontic force is placed on the teeth and the neighboring periodontal ligament (PDL) is compressed, the immune system responds to the site in order to relieve the tissue stress and damage. As part of the stress response, ATP is released from platelets and binds to the purinergic receptor P2X, ligand-gated ion channel 7 (P2RX7) membrane channel protein located on the surface of immune cells and/or cells of the PDL. Upon binding ATP, the P2RX7 ion channel is opened, allowing the exchange of intracellular potassium (K+) and extracellular sodium (Na++), and triggering the elevation of calcium (Ca++) from intracellular stores. Elevation of intracellular Ca++ activates caspase-1 (also termed IL-1β converting enzyme or ICE) which is located in inflammasome complexes within the cell. Caspase-1 cleaves the pro-IL-1β molecule, releasing active mature interleukin-1beta (IL-1β) for biological function. IL-1β can recruit other inflammatory cells to the site of tissue damage, and can bind to interleukin-1 receptors (IL-1R) on the surface of pro-osteoblastic cells. The IL-1 receptor antagonist (IL1RA) may interrupt IL-1 signaling. Once IL-1β is bound to the IL-1Rs, a signaling cascade involving the Interleukin-1 Receptor Associated Kinase 1 (IRAK1) and other molecules (not shown) lead to the activation of such genes as the receptor activator of nuclear factor kappa-B ligand (RANKL) and Osteoprotegerin (OPG). When RANKL is synthesized, and expressed on the surface of the osteoblastic cells it can bind RANK on preosteoclasts. This interaction, in concert with M-CSF production from osteoblasts and binding to the c-fms receptor on the surface of pre-osteoclastic cells signals the osteoclast precursor cells to mature into functional osteoclasts. OPG and soluble RANKL (sRANKL) can act to dampen the maturation signal to pro-osteoclast cells by interfering with RANKL:RANK interactions. Osteopontin (OPN) plays an important role in bone remodeling by increasing osteoclast anchoring. The action of both osteoblasts and osteoclasts are needed to resolve the tissue stresses within the PDL from orthodontic force application. Additional associations with EARR that not depicted in this figure include the vitamin D receptor (VDR) on osteoblasts and Interleukin-6 (IL-6).
IL-1a
(Al-Qawasmi, et.al., 2003) Am J Orthod Dentofacial Orthop 123:242–52
(Gülden, et al., 2009) J Orofac Orthop 70: 20–18
(Iglesias-Linares, et. al., 2012) Oral Diseases 18:198–205
(Iglesias-Linares, et. al., 2012) International Endodontic Journal 45: 1018–1026
(Iglesias-Linares, et. al., 2012) Journal of Endodontics, 2012, Vol.38(3), p.283–287
(Linhartova, et. al., 2013) Oral Diseases 19:262–70
(Sharab, et al., 2015) Orthod Craniofac Res. 2015 Apr;18 Suppl 1:71–82. doi: 10.1111/ocr.12078
IL-1β
(Al-Qawasmi, et al., 2003) Am J Orthod Dentofacial Orthop 123:242–52
(Bastos Lages, et al., 2009) Am J Orthod Dentofacial Orthop 136:542–6
(Gülden, et al., 2009) J Orofac Orthop 70: 20–18
(Iglesias-Linares, et al., 2012) Oral Diseases 18:198–205
(Iglesias-Linares, 2012) International Endodontic Journal 45: 1018–1026
(Iglesias-Linares, et. al., 2012) Journal of Endodontics,38: 283–287
(Linhartova, et. al., 2013) Oral Diseases 19:262–70
Pereira, et. al., 2014) Oral Dis 20: 659–667
(Pereira, et. al., 2016) Oral Diseases (2016) doi: 10.1111/odi.12514
(Sharab, et al., 2015) Orthod Craniofac Res. 2015 Apr;18 Suppl 1:71–82. doi: 10.1111/ocr.12078.
IL-1RA
Pereira, et. al., 2016) Oral Diseases (2016) doi: 10.1111/odi.12514
(Iglesias-Linares, et. al., 2012) Oral Diseases 18:198–205
(Iglesias-Linares, et. al., 2013) Histol Histopathol. 28:767–73.
(Sharab, et al., 2015) Orthod Craniofac Res. 2015 Apr;18 Suppl 1:71–82. doi: 10.1111/ocr.12078
(Linhartova, et. al., 2013) Oral Diseases 19:262–70
(Gua, et al., 2016) Am J Orthod Dentofacial Orthop. 150:283–9
IRAK1
(Pereira, et. al., 2016) Oral Diseases (2016) doi: 10.1111/odi.12514
Caspase-1
(Sharab, et al., 2015) Orthod Craniofac Res. 2015 Apr;18 Suppl 1:71–82. doi:10.1111/ocr.12078. → (3 different SNPs examined)
P2RX7
(Sharab, et al., 2015) Orthod Craniofac Res. 2015 Apr;18 Suppl 1:71–82. doi:10.1111/ocr.12078. → 3 different SNPs examined
(Linhartova, et. al., 2016) Oral Dis. 2016 Aug 5. doi: 10.1111/odi.12564. [Epub ahead of print]
(Pereira, et. al., 2014) Oral Dis 20: 659–667
(Linhartova, et. l., 2016) Oral Dis. 2016 Aug 5. doi: 10.1111/odi.12564. [Epub ahead of print]
RANK
(Al-Qawasmi, et. al., 2003) J Dent Res. 2003 May;82(5):356–60
(Pereira, et. al., 2014) Oral Dis 20: 659–667
OPG
(Hartsfield, 2009) Orthod Craniofac Res 12: 236–242
(Pereira, et. al., 2014) Oral Dis 20: 659–667
(Linhartova, et. al., 2016) Oral Dis. 2016 Aug 5. doi: 10.1111/odi.12564. [Epub ahead of print] → 2 SNPs examined
OPN
(Iglesias-Linares, et. al., 2014) Oral Diseases 20: 307–312
(Linhartova, et. al., 2016) Oral Dis. 2016 Aug 5. doi: 10.1111/odi.12564. [Epub ahead of print] → 2 SNPs examined
(Iglesias-Linares, 2014) Oral Diseases 20: 307–312
VDR
(Fontana, et al., 2012) Am J Orthod Dentofacial Orthop. 142:339–47
IL-6 (Gua, et al., 2016) Am J Orthod Dentofacial Orthop. 150:283–9
More recent studies have incorporated multiple treatment and genetic factors in models to explain the occurrence of EARR concurrent with orthodontics. For example, 30% of the EARR variability in one study was explained by a combination of the treatment duration, the use of a Hyrax appliance, premolar extractions, sex and/or variation in the purinergic receptor P2X, ligand-gated ion channel 7 (P2RX7) gene-associated single nucleotide polymorphism (SNP) rs1718119; while age, pre-treatment overjet, a tongue thrust habit, a skeletal class II diagnosis and other genetic polymorphisms provided minor contributions.68 Similarly, a second modeling study examined the relative influence of multiple parameters on the occurrence of EARR including (a) treatment duration, (b) extraction of maxillary premolars, (c) numerous cephalometric measurements (pretreatment values and/or overall post-treatment change in the values), and (d) nine unique genetic variations found within the Interleukin (IL) -1α, -1β, IL1-RA, P2RX7 and Caspase-1 genes. This study found that a longer length of treatment, together with specific genotypes for the P2RX7-SNP rs208294 explained 25% of the total variation associated with EARR concurrent with orthodontia in the sample tested.67
Based on cumulative research to date, the molecular pathways influencing the development of EARR appear to involve: (1) clastic cell adhesion with specific roles for the α/β-integrins, osteopontin (OPN) and other related extracellular matrix proteins, (2) clastic cell fusion and activation via the Receptor Activator of Nuclear Factor κ B (RANK)/Receptor for RANK-Ligand (RANKL)/osteoprotegerin (OPG) and ATP/P2RX7/-IL1β pathways, and (3) regulatory mechanisms of root resorption repair by cementum at the proteomic and transcriptomic levels.69 In addition, the relationships between bone density, the rate of tooth movement and root resorption have been studied in lactating calcium-deficient female Sprague-Dawley rats. These studies showed that decreased alveolar bone density facilitated rapid orthodontic tooth movement, and reduced cratering of the root outer cementum, implying that there was less strain on the dental root as the tooth moved faster, with differential resorption of alveolar bone versus root structure.70 Gene-specific knockout mouse models and human genetic association studies have supported the “Root Strain Mechanism” as one model for root resorption.57,67,68,71,72 Future estimation of susceptibility to EARR likely will require the analysis of several genes as mentioned previously, root morphology, skeletal-dental values, and the treatment method to be used, or essentially the amount of tooth movement planned for treatment.54,73
Primary Failure of Eruption (PFE)
This condition appears to have an autosomal dominant inheritance with variable expressivity.74 It is characterized by two phenotypic types: 1) having all teeth distal to the most mesial involved tooth partly or completely unable to erupted, or 2) some of the teeth have no apparent reason for failure to eruption, but they do not follow the pattern that all teeth distal to the most mesial involved tooth are also affected. Patients or family members may also have a history of primary tooth ankylosis. So far as a constant, attempts to apply orthodontic force generally lead to ankylosis.75 It may affect only one or up to all four quadrants and while it is principally observed in the permanent dentition, examples affecting the primary dentition have also been noted.76
The familial occurrence of this phenotype in approximately one-quarter of all individuals in an affected family facilitated the investigation and discovery of mutations within the parathyroid hormone 1 receptor (PTHR1) gene being involved.77 The PTH1R gene is located on human chromosome 3p21.31 (OMIM *168468) comprised of 16 exons. To date, over 25 unique mutations within the PTH1R gene appear to be associated with PFE; with some mutations introducing an immediate stop code or frameshifts in the code leading to the premature truncation of the protein and haploinsufficiency, while other mutations lead to aberrant RNA splicing of the PTH1R messenger RNA.74,78–82 Advancements in this area could not only help to define patients who are likely to develop or have PFE, but also potentially result in the molecular manipulation of selective tooth eruption rates to enhance treatment protocols on an individual basis.83
Additional Areas
Numerous investigators are starting to look at several additional types of cases to better identify and understand how genetic influences direct aspects of growth and development, and treatment of malocclusion and associated anomalies. These include with only one or few selected references such as facial scan morphology,84 dental crowding in Class I malocclusion,85 arch form,86 facial morphology associated with obstructive sleep apnea,87,88 dental agenesis,89 palatally displaced canines (PDC),90 canine impaction,91 pain perception,92 and multiple (cluster phenomenon) dental implant failure.93,94 As mentioned previously, genetic studies are needed which examine the contribution of genetics in slow craniofacial growers and/or late growers (especially growth within the jaws) with and without the use of specific appliances such as the Herbst, MARA, and use of TAD-assisted mandibular growth restriction. These types of studies would help to when and on whom a specific growth modifying appliance should be utilized to synergistically enhance the growth or changes with the patients’ craniofacial features.
Discussion
While the field of oral and craniofacial genetics expands to learn more about the genetic factors that would help to better treat individual patients, it should not be overlooked that today the practitioner in their practice could start to take and consider family history in the diagnosis and treatment planning of malocclusion.16 This can be used to help understand the approximate likelihood that the patient or a sibling may also develop the same trait, which still may vary in its severity even within the same family. This can be particularly useful for monogenic traits including Class III malocclusion, hypodontia, PFE, and developmental dental dysplasias such as types of dentinogenesis and amelogenesis imperfecta. A family history may also be useful for complex traits such as skeletal Class III and Class II/division 2 malocclusions, EARR, PDC, or any trait that occurs in more than one member of the family.
Summary
Due to the heterogeneous complexity of facial and dental development, the physiology of tooth movement, and the occurrence of EARR, the precise prediction of outcomes is not tenable. Many genetic factors and how they may relate to environmental (including treatment) factors are being investigated and are starting to be better understood. While precise prediction is not at hand, the influence of genetic factors for example in EARR is clear. The uncovering of genetic factors that correlate with clinical variation of previously unknown etiology helps to diminish the unknown variation that is present in clinical studies, particularly those that consider the effect of appliances or treatment regiments on growth, and should in the future be a part of these investigations. The day-to-day utilization of the “testing” for these factors is not yet ready for practice, although monogenic traits such as PFE, and Class III malocclusion, show promise as knowledge and technology advances.
As the recognition of specific genetic factors that affect our patients are discovered, the three-generation (including parents and grandparents with aunts, uncles and patient siblings asked about) family history is something the practitioner can implement in their practice today. This would greatly benefit the patient at that time, and augment the usefulness of these families in future clinical research in which clinical findings, environmental, and genetic factors can be studied.
Acknowledgments
Supported in part by NIH P30GM110788 for the Center for the Biologic Basis of Oral/Systemic Disease (COBRE): Phase III (JKH, LAM), and the E. Preston Hicks Professor Endowment (JKH)
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
James K. Hartsfield, Jr., E. Preston Hicks Professor of Orthodontics and Oral Health Research, University of Kentucky Center for the Biologic Basis of Oral/Systemic Diseases, Hereditary Genetics/Genomics Core.
George Jeryn Jacob, Craniofacial Genetics Fellow, University of Kentucky Center for the Biologic Basis of Oral/Systemic Diseases, Hereditary Genetics/Genomics Core.
Lorri Ann Morford, Research Assistant Professor, University of Kentucky Center for the Biologic Basis of Oral/Systemic Diseases, Hereditary Genetics/Genomics CoreThe University of Kentucky College of Dentistry, 800 Rose Street, Lexington, Kentucky USA 40536-0297.
References
- 1.Manfredi C, Martina R, Grossi GB, Giuliani M. Heritability of 39 orthodontic cephalometric parameters on MZ, DZ twins and MN-paired singletons. Am J Orthod Dentofacial Orthop. 1997;111:44–51. doi: 10.1016/s0889-5406(97)70301-9. [DOI] [PubMed] [Google Scholar]
- 2.Hartsfield JK, Jr, Morford LA. Genetic Implications in Orthodontic Tooth Movement. In: Shroff B, editor. Biology of Orthodontic Tooth Movement - Current Concepts and Applications in Orthodontic Practice. Springer; 2016. pp. 103–132. [Google Scholar]
- 3.Mencken HL. A Mencken chrestomathy. New York: A. A. Knopf; 1949. [Google Scholar]
- 4.Verdonck A, Gaethofs M, Carels C, de Zegher F. Effect of low-dose testosterone treatment on craniofacial growth in boys with delayed puberty. Eur J Orthod. 1999;21:137–143. doi: 10.1093/ejo/21.2.137. [DOI] [PubMed] [Google Scholar]
- 5.Moss ML. The regulation of skeletal growth Regulation of Organ and Tissue Growth. Acad Press NY: 1972. pp. 127–142. [Google Scholar]
- 6.Guo Y, Xiong D-H, Yang T-L, Guo Y-F, Recker RR, Deng H-W. Polymorphisms of estrogen-biosynthesis genes CYP17 and CYP19 may influence age at menarche: a genetic association study in Caucasian females. Hum Mol Genet. 2006;15:2401–2408. doi: 10.1093/hmg/ddl155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schaefer K, Fink B, Mitteroecker P, Neave N, Bookstein FL. Visualizing facial shape regression upon 2nd to 4th digit ratio and testosterone. Collegium antropologicum. 2005;29:415–419. [PubMed] [Google Scholar]
- 8.Hartsfield JK, Jr, Zhou J, Chen S. The Importance of Analyzing Specific Genetic Factors in Facial Growth for Diagnosis and Treatment Planning. In: McNamara JA Jr, Kapila SD, editors. Surgical Enhancement of Orthodontic Treatment. Ann Arbor: University of Michigan; 2010. pp. 267–281. [Google Scholar]
- 9.He S, Hartsfield JK, Jr, Guo Y, Cao Y, Wang S, Chen S. Association between< i> CYP19A1</i> genotype and pubertal sagittal jaw growth. Am J Orthod Dentofacial Orthop. 2012;142:662–670. doi: 10.1016/j.ajodo.2012.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yamaguchi T, Maki K, Shibasaki Y. Growth hormone receptor gene variant and mandibular height in the normal Japanese population. Am J Orthod Dentofacial Orthop. 2001;119:650–653. doi: 10.1067/mod.2001.114536. [DOI] [PubMed] [Google Scholar]
- 11.Cruz CV, Mattos CT, Maia JC, Granjeiro JM, Reis MF, Mucha JN, et al. Genetic polymorphisms underlying the skeletal Class III phenotype. American Journal of Orthodontics and Dentofacial Orthopedics. 151:700–707. doi: 10.1016/j.ajodo.2016.09.013. [DOI] [PubMed] [Google Scholar]
- 12.Perinetti G, Primožič J, Franchi L, Contardo L. Treatment Effects of Removable Functional Appliances in Pre-Pubertal and Pubertal Class II Patients: A Systematic Review and Meta-Analysis of Controlled Studies. PLOS ONE. 2015;10:e0141198. doi: 10.1371/journal.pone.0141198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.El-Gheriani AA, Maher BS, El-Gheriani AS, Sciote JJ, Abu-Shahba FA, Al-Azemi R, et al. Segregation analysis of mandibular prognathism in Libya. J Dent Res. 2003;82:523–527. doi: 10.1177/154405910308200707. [DOI] [PubMed] [Google Scholar]
- 14.Cruz RM, Krieger H, Ferreira R, Mah J, Hartsfield J, Jr, Oliveira S. Major gene and multifactorial inheritance of mandibular prognathism. Am J Med Genet A. 2008;146A:71–77. doi: 10.1002/ajmg.a.32062. [DOI] [PubMed] [Google Scholar]
- 15.King RA, Rotter JI, Motulsky AG. Approach to genetic basis of common diseases. Oxford: Oxford University Press; 2002. [Google Scholar]
- 16.Hartsfield JK, Jr, Morford LA. Genetics and Orthodontics. In: Graber LW, Vanarsdall RL Jr, Vig KWL, Huang GJ, editors. Orthodontics Current Principles and Techniques. St. Louis, MO: Elsevier; 2017. pp. 31–50. [Google Scholar]
- 17.Falcão-Alencar G, Otero L, Cruz R, Foroud T, Dongbing L, Koller D, et al. Evidence for genetic linkage of the class III craniofacial phenotype with human chromosome 7 in 36 South American families 60th Annual Meeting of The American Society of Human Genetics. Washington, DC: 2010. [Google Scholar]
- 18.Frazier-Bowers S, Rincon-Rodriguez R, Zhou J, Alexander K, Lange E. Evidence of linkage in a Hispanic cohort with a Class III dentofacial phenotype. J Dent Res. 2009;88:56–60. doi: 10.1177/0022034508327817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ikuno K, Kajii TS, Oka A, Inoko H, Ishikawa H, Iida J. Microsatellite genome-wide association study for mandibular prognathism. Am J Orthod Dentofacial Orthop. 2014;145:757–762. doi: 10.1016/j.ajodo.2014.01.022. [DOI] [PubMed] [Google Scholar]
- 20.Jang JY, Park EK, Ryoo HM, Shin HI, Kim TH, Jang JS, et al. Polymorphisms in the Matrilin-1 gene and risk of mandibular prognathism in Koreans. J Dent Res. 2010;89:1203–1207. doi: 10.1177/0022034510375962. [DOI] [PubMed] [Google Scholar]
- 21.Li Q, Li X, Zhang F, Chen F. The identification of a novel locus for mandibular prognathism in the Han Chinese population. J Dent Res. 2011;90:53–57. doi: 10.1177/0022034510382546. [DOI] [PubMed] [Google Scholar]
- 22.Li Q, Zhang F, Li X, Chen F. Genome scan for locus involved in mandibular prognathism in pedigrees from China. PLoS One. 2010;5 doi: 10.1371/journal.pone.0012678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tassopoulou-Fishell M, Deeley K, Harvey EM, Sciote J, Vieira AR. Genetic variation in myosin 1H contributes to mandibular prognathism. Am J Orthod Dentofacial Orthop. 2012;141:51–59. doi: 10.1016/j.ajodo.2011.06.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xue F, Rabie AB, Luo G. Analysis of the association of COL2A1 and IGF-1 with mandibular prognathism in a Chinese population. Orthod Craniofac Res. 2014;17:144–149. doi: 10.1111/ocr.12038. [DOI] [PubMed] [Google Scholar]
- 25.Xue F, Wong RW, Rabie AB. Genes, genetics, and Class III malocclusion. Orthod Craniofac Res. 2010;13:69–74. doi: 10.1111/j.1601-6343.2010.01485.x. [DOI] [PubMed] [Google Scholar]
- 26.Yamaguchi T, Park SB, Narita A, Maki K, Inoue I. Genome-wide linkage analysis of mandibular prognathism in Korean and Japanese patients. J Dent Res. 2005;84:255–259. doi: 10.1177/154405910508400309. [DOI] [PubMed] [Google Scholar]
- 27.Chen F, Li Q, Gu M, Li X, Yu J, Zhang YB. Identification of a mutation in FGF23 Involved in mandibular prognathism. Scientific reports. 2015:5. doi: 10.1038/srep11250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Guan X, Song Y, Ott J, Zhang Y, Li C, Xin T, et al. The adamts1 gene is associated with familial mandibular prognathism. Journal of Dental Research. 2015;94:1196–1201. doi: 10.1177/0022034515589957. [DOI] [PubMed] [Google Scholar]
- 29.Nikopensius T, Saag M, Jagomägi T, Annilo T, Kals M, Kivistik PA, et al. A Missense Mutation in DUSP6 is Associated with Class III Malocclusion. J Dent Res. 2013;92:893–898. doi: 10.1177/0022034513502790. [DOI] [PubMed] [Google Scholar]
- 30.Nowrin SA, Basri R, Alam MK, Yusa T, Nakano J, Jaafar S, et al. Craniofacial morphology of class III malocclusion with DUSP6 gene: Mutation and non-mutation groups. Journal of Hard Tissue Biology. 2016;25:247–256. [Google Scholar]
- 31.Perillo L, Monsurrò A, Bonci E, Torella A, Mutarelli M, Nigro V. Genetic Association of ARHGAP21 Gene Variant with Mandibular Prognathism. Journal of Dental Research. 2015 doi: 10.1177/0022034515572190. [DOI] [PubMed] [Google Scholar]
- 32.Moreno Uribe LM, Vela KC, Kummet C, Dawson DV, Southard TE. Phenotypic diversity in white adults with moderate to severe Class III malocclusion. Am J Orthod Dentofacial Orthop. 2013;144:32–42. doi: 10.1016/j.ajodo.2013.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Basdra EK, Kiokpasoglou M, Stellzig A. The Class II Division 2 craniofacial type is associated with numerous congenital tooth anomalies. Eur J Orthod. 2000;22:529–535. doi: 10.1093/ejo/22.5.529. [DOI] [PubMed] [Google Scholar]
- 34.Morrison MW. Relative Risk of Class II division 2 Malocclusion in First-Degree Relatives of Probands with Class II division 2 Malocclusion Department of Orthodontics and Oral Facial Genetics. Indiana University School of Dentistry; 2008. [Google Scholar]
- 35.Moss ML. The functional matrix hypothesis revisited. 4. The epigenetic antithesis and the resolving synthesis. American Journal of Orthodontics and Dentofacial Orthopedics. 1997;112:410–417. doi: 10.1016/s0889-5406(97)70049-0. [DOI] [PubMed] [Google Scholar]
- 36.Huh A, Horton MJ, Cuenco KT, Raoul G, Rowlerson AM, Ferri J, et al. Epigenetic influence of KAT6B and HDAC4 in the development of skeletal malocclusion. Am J Orthod Dentofacial Orthop. 2013;144:568–576. doi: 10.1016/j.ajodo.2013.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Raoul G, Rowlerson A, Sciote J, Codaccioni E, Stevens L, Maurage CA, et al. Masseter myosin heavy chain composition varies with mandibular asymmetry. J Craniofac Surg. 2011;22:1093–1098. doi: 10.1097/SCS.0b013e3182107766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rowlerson A, Raoul G, Daniel Y, Close J, Maurage CA, Ferri J, et al. Fiber-type differences in masseter muscle associated with different facial morphologies. Am J Orthod Dentofacial Orthop. 2005;127:37–46. doi: 10.1016/j.ajodo.2004.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sciote JJ, Raoul G, Ferri J, Close J, Horton MJ, Rowlerson A. Masseter function and skeletal malocclusion. Rev Stomatol Chir Maxillofac Chir Orale. 2013;114:79–85. doi: 10.1016/j.revsto.2013.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Staron R. Correlation between myofibrillar ATPase activity and myosin heavy chain composition in single human muscle fibers. Histochemistry and Cell Biology. 1991;96:21–24. doi: 10.1007/BF00266756. [DOI] [PubMed] [Google Scholar]
- 41.North KN, Beggs AH. Deficiency of a skeletal muscle isoform of alpha-actinin (alpha-actinin-3) in merosin-positive congenital muscular dystrophy. Neuromuscul Disord. 1996;6:229–235. doi: 10.1016/0960-8966(96)00361-6. [DOI] [PubMed] [Google Scholar]
- 42.Zebrick B, Teeramongkolgul T, Nicot R, Horton MJ, Raoul G, Ferri J, et al. ACTN3 R577X genotypes associate with Class II and deepbite malocclusions. Am J Orthod Dentofacial Orthop. 2014;146:603–611. doi: 10.1016/j.ajodo.2014.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sciote JJ, Horton MJ, Rowlerson AM, Ferri J, Close JM, Raoul G. Human masseter muscle fiber type properties, skeletal malocclusions, and muscle growth factor expression. J Oral Maxillofac Surg. 2012;70:440–448. doi: 10.1016/j.joms.2011.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Desh H, Gray SL, Horton MJ, Raoul G, Rowlerson AM, Ferri J, et al. Molecular motor MYO1C, acetyltransferase KAT6B and osteogenetic transcription factor RUNX2 expression in human masseter muscle contributes to development of malocclusion. Arch Oral Biol. 2014;59:601–607. doi: 10.1016/j.archoralbio.2014.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Harris EF, Robinson QC, Woods MA. An analysis of causes of apical root resorption in patients not treated orthodontically. Quintessence Int. 1993;24:417–428. [PubMed] [Google Scholar]
- 46.Brezniak N, Wasserstein A. Root resorption after orthodontic treatment: Part 2. Literature review Am J Orthod Dentofacial Orthop. 1993;103:138–146. doi: 10.1016/S0889-5406(05)81763-9. [DOI] [PubMed] [Google Scholar]
- 47.Brezniak N, Wasserstein A. Root resorption after orthodontic treatment: Part 1. Literature review Am J Orthod Dentofacial Orthop. 1993;103:62–66. doi: 10.1016/0889-5406(93)70106-X. [DOI] [PubMed] [Google Scholar]
- 48.Parker RJ, Harris EF. Directions of orthodontic tooth movements associated with external apical root resorption of the maxillary central incisor. Am J Orthod Dentofacial Orthop. 1998;114:677–683. doi: 10.1016/s0889-5406(98)70200-8. [DOI] [PubMed] [Google Scholar]
- 49.Sameshima GT, Sinclair PM. Predicting and preventing root resorption: Part I. Diagnostic factors. Am J Orthod Dentofacial Orthop. 2001;119:505–510. doi: 10.1067/mod.2001.113409. [DOI] [PubMed] [Google Scholar]
- 50.Baumrind S, Korn EL, Boyd RL. Apical root resorption in orthodontically treated adults. Am J Orthod Dentofacial Orthop. 1996;110:311–320. doi: 10.1016/s0889-5406(96)80016-3. [DOI] [PubMed] [Google Scholar]
- 51.Horiuchi A, Hotokezaka H, Kobayashi K. Correlation between cortical plate proximity and apical root resorption. Am J Orthod Dentofacial Orthop. 1998;114:311–318. doi: 10.1016/s0889-5406(98)70214-8. [DOI] [PubMed] [Google Scholar]
- 52.Al-Qawasmi RA, Hartsfield JK, Jr, Everett ET, Weaver MR, Foroud TM, Faust DM, et al. Root resorption associated with orthodontic force in inbred mice: genetic contributions. Eur J Orthod. 2006;28:13–19. doi: 10.1093/ejo/cji090. [DOI] [PubMed] [Google Scholar]
- 53.Abass SK, Hartsfield JK, Jr, Al-Qawasmi RA, Everett ET, Foroud TM, Roberts WE. Inheritance of susceptibility to root resorption associated with orthodontic force in mice. Am J Orthod Dentofacial Orthop. 2008;134:742–750. doi: 10.1016/j.ajodo.2007.04.035. [DOI] [PubMed] [Google Scholar]
- 54.Hartsfield JK, Jr, Everett ET, Al-Qawasmi RA. Genetic Factors in External Apical Root Resorption and Orthodontic Treatment. Crit Rev Oral Biol Med. 2004;15:115–122. doi: 10.1177/154411130401500205. [DOI] [PubMed] [Google Scholar]
- 55.Ngan DC, Kharbanda OP, Byloff FK, Darendeliler MA. The genetic contribution to orthodontic root resorption: a retrospective twin study. Aust Orthod J. 2004;20:1–9. [PubMed] [Google Scholar]
- 56.Iwasaki LR, Crouch LD, Nickel JC. Genetic Factors and Tooth Movement. Semin Orthod. 2008;14:135–145. [Google Scholar]
- 57.Al-Qawasmi RA, Hartsfield JK, Jr, Everett ET, Flury L, Liu L, Foroud TM, et al. Genetic predisposition to external apical root resorption in orthodontic patients: linkage of chromosome-18 marker. J Dent Res. 2003;82:356–360. doi: 10.1177/154405910308200506. [DOI] [PubMed] [Google Scholar]
- 58.Al-Qawasmi RA, Hartsfield JK, Jr, Everett ET, Flury L, Liu L, Foroud TM, et al. Genetic predisposition to external apical root resorption. Am J Orthod Dentofacial Orthop. 2003;123:242–252. doi: 10.1067/mod.2003.42. [DOI] [PubMed] [Google Scholar]
- 59.Fontana MLSSN. de Souza CM, Bernardino JF, Hoette F, Hoette ML, Thum L, et al. Association analysis of clinical aspects and vitamin D receptor gene polymorphism with external apical root resorption in orthodontic patients. Am J Orthod Dentofacial Orthop. 2012;142:339–347. doi: 10.1016/j.ajodo.2012.04.013. [DOI] [PubMed] [Google Scholar]
- 60.Gülden N, Eggermann T, Zerres K, Beer M, Meinelt A, Diedrich P. Interleukin-1 polymorphisms in relation to externa] apical root resorption (EARR) J Orofac Orthop. 2009;70:20–38. doi: 10.1007/s00056-009-8808-6. [DOI] [PubMed] [Google Scholar]
- 61.Iglesias-Linares A, Yanez-Vico R, Ballesta-Mudarra S, Ortiz-Ariza E, Ortega-Rivera H, Mendoza-Mendoza A, et al. Postorthodontic external root resorption is associated with IL1 receptor antagonist gene variations. Oral Dis. 2012;18:198–205. doi: 10.1111/j.1601-0825.2011.01865.x. [DOI] [PubMed] [Google Scholar]
- 62.Iglesias-Linares A, Yanez-Vico RM, Ballesta S, Ortiz-Ariza E, Mendoza-Mendoza A, Perea E, et al. Interleukin 1 gene cluster SNPs (rs1800587, rs1143634) influences post-orthodontic root resorption in endodontic and their contralateral vital control teeth differently. Int Endod J. 2012;45:1018–1026. doi: 10.1111/j.1365-2591.2012.02065.x. [DOI] [PubMed] [Google Scholar]
- 63.Iglesias-Linares A, Yanez-Vico RM, Ballesta-Mudarra S, Ortiz-Ariza E, Mendoza-Mendoza A, Perea-Perez E, et al. Interleukin 1 receptor antagonist (IL1RN) genetic variations condition post-orthodontic external root resorption in endodontically-treated teeth. Histol Histopathol. 2013;28:767–773. doi: 10.14670/HH-28.767. [DOI] [PubMed] [Google Scholar]
- 64.Iglesias-Linares A, Yanez-Vico RM, Moreno-Fernandez AM, Mendoza-Mendoza A, Orce-Romero A, Solano-Reina E. Osteopontin gene SNPs (rs9138, rs11730582) mediate susceptibility to external root resorption in orthodontic patients. Oral Dis. 2014;20:307–312. doi: 10.1111/odi.12114. [DOI] [PubMed] [Google Scholar]
- 65.Iglesias-Linares A, Yanez-Vico RM, Ortiz-Ariza E, Ballesta S, Mendoza-Mendoza A, Perea E, et al. Postorthodontic external root resorption in root-filled teeth is influenced by interleukin-1beta polymorphism. J Endod. 2012;38:283–287. doi: 10.1016/j.joen.2011.12.022. [DOI] [PubMed] [Google Scholar]
- 66.Linhartova P, Cernochova P, Izakovicova Holla L. IL1 gene polymorphisms in relation to external apical root resorption concurrent with orthodontia. Oral Dis. 2013;19:262–270. doi: 10.1111/j.1601-0825.2012.01973.x. [DOI] [PubMed] [Google Scholar]
- 67.Sharab LY, Morford LA, Dempsey J, Falcão-Alencar G, Mason A, Jacobson E, et al. Genetic and treatment-related risk factors associated with external apical root resorption (EARR) concurrent with orthodontia. Orthod Craniofac Res. 2015;18:71–82. doi: 10.1111/ocr.12078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Pereira S, Lavado N, Nogueira L, Lopez M, Abreu J, Silva H. Polymorphisms of genes encoding P2X7R, IL-1B, OPG and RANK in orthodontic- induced apical root resorption. Oral diseases. 2014;20:659–667. doi: 10.1111/odi.12185. [DOI] [PubMed] [Google Scholar]
- 69.Iglesias-Linares A, Hartsfield J. Cellular and Molecular Pathways Leading to External Root Resorption. Journal of Dental Research. 2016 doi: 10.1177/0022034516677539. 0022034516677539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Goldie RS, King GJ. Root resorption and tooth movement in orthodontically treated, calcium-deficient, and lactating rats. American Journal of Orthodontics. 1984;85:424–430. doi: 10.1016/0002-9416(84)90163-5. [DOI] [PubMed] [Google Scholar]
- 71.Iglesias-Linares A, Morford LA, Hartsfield JK. Bone Density and Dental External Apical Root Resorption. Current osteoporosis reports. 2016;14:292–309. doi: 10.1007/s11914-016-0340-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Viecilli RF, Katona TR, Chen J, Hartsfield JK, Roberts WE. Orthodontic mechanotransduction and the role of the P2X7 receptor. American Journal of Orthodontics and Dentofacial Orthopedics. 2009;135:694. e691–694. e616. doi: 10.1016/j.ajodo.2008.10.018. [DOI] [PubMed] [Google Scholar]
- 73.Hartsfield JK., Jr Pathways in external apical root resorption associated with orthodontia. Orthod Craniofac Res. 2009;12:236–242. doi: 10.1111/j.1601-6343.2009.01458.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Stellzig-Eisenhauer A, Decker E, Meyer-Marcotty P, Rau C, Fiebig BS, Kress W, et al. Primary failure of eruption (PFE)–clinical and molecular genetics analysis. Journal of Orofacial Orthopedics/Fortschritte der Kieferorthopädie. 2010;71:6–16. doi: 10.1007/s00056-010-0908-9. [DOI] [PubMed] [Google Scholar]
- 75.Frazier-Bowers SA, Long S, Tucker M. Primary failure of eruption and other eruption disorders—Considerations for management by the orthodontist and oral surgeon Seminars in Orthodontics. Elsevier; 2016. pp. 34–44. [Google Scholar]
- 76.Ahmad S, Bister D, Cobourne MT. The clinical features and aetiological basis of primary eruption failure. The European Journal of Orthodontics. 2006;28:535–540. doi: 10.1093/ejo/cjl033. [DOI] [PubMed] [Google Scholar]
- 77.Decker E, Stellzig-Eisenhauer A, Fiebig BS, Rau C, Kress W, Saar K, et al. PTHR1 loss-of-function mutations in familial, nonsyndromic primary failure of tooth eruption. Am J Hum Genet. 2008;83:781–786. doi: 10.1016/j.ajhg.2008.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Frazier-Bowers S, Hendricks H, Wright J, Lee J, Long K, Dibble C, et al. Novel mutations in PTH1R associated with primary failure of eruption and osteoarthritis. Journal of dental research. 2014;93:134–139. doi: 10.1177/0022034513513588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Roth H, Fritsche LG, Meier C, Pilz P, Eigenthaler M, Meyer-Marcotty P, et al. Expanding the spectrum of PTH1R mutations in patients with primary failure of tooth eruption. Clinical oral investigations. 2014;18:377–384. doi: 10.1007/s00784-013-1014-3. [DOI] [PubMed] [Google Scholar]
- 80.Risom L, Christoffersen L, Daugaard-Jensen J, Hove HD, Andersen HS, Andresen BS, et al. Identification of six novel PTH1R mutations in families with a history of primary failure of tooth eruption. PloS one. 2013;8:e74601. doi: 10.1371/journal.pone.0074601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Yamaguchi T, Hosomichi K, Narita A, Shirota T, Tomoyasu Y, Maki K, et al. Exome resequencing combined with linkage analysis identifies novel PTH1R variants in primary failure of tooth eruption in Japanese. Journal of Bone and Mineral Research. 2011;26:1655–1661. doi: 10.1002/jbmr.385. [DOI] [PubMed] [Google Scholar]
- 82.Pilz P, Meyer-Marcotty P, Eigenthaler M, Roth H, Weber BH, Stellzig-Eisenhauer A. Differential diagnosis of primary failure of eruption (PFE) with and without evidence of pathogenic mutations in the PTHR1 gene. Journal of Orofacial Orthopedics/Fortschritte der Kieferorthopädie. 2014;75:226–239. doi: 10.1007/s00056-014-0215-y. [DOI] [PubMed] [Google Scholar]
- 83.Wise GE, Frazier-Bowers S, D’Souza RN. Cellular, molecular, and genetic determinants of tooth eruption. Crit Rev Oral Biol Med. 2002;13:323–334. doi: 10.1177/154411130201300403. [DOI] [PubMed] [Google Scholar]
- 84.Shaffer JR, Orlova E, Lee MK, Leslie EJ, Raffensperger ZD, Heike CL, et al. Genome-Wide Association Study Reveals Multiple Loci Influencing Normal Human Facial Morphology. PLOS Genetics. 2016;12:e1006149. doi: 10.1371/journal.pgen.1006149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Ting TY, Wong RWK, Rabie ABM. Analysis of genetic polymorphisms in skeletal Class I crowding. Am J Orthod Dentofacial Orthop. 2011;140:e9–e15. doi: 10.1016/j.ajodo.2010.12.015. [DOI] [PubMed] [Google Scholar]
- 86.Miller SF, Shaffer JR, Marazita ML, Levy SM, Warren JJ, Hartsfield JK, et al. Exploratory Genome-Wide Association Study of Dental Arch Shape IADR/AADR/CADR General Session & Exhibition. Boston, MA. J Dent Res. 2015;94(Spec Iss A) abstract number 3982. [Google Scholar]
- 87.Wachs L, Elmahdy MN, Morford LA, Hartsfield JK, Jr, Roedig J, Phillips B, et al. AHI, BMI, TNFa-rs1800629 Comparison Among Obstructive Sleep Apnea Patients with Different Facial Profiles IADR/AADR/CADR General Session & Exhibition. Boston, MA. J Dent Res. 2015;94(Spec Iss A) abstract number 4377. [Google Scholar]
- 88.Lavezzi AM, Casale V, Oneda R, Gioventù S, Matturri L, Farronato G. Obstructive sleep apnea syndrome (OSAS) in children with Class III malocclusion: involvement of the PHOX2B gene. Sleep and Breathing. 2013;17:1275–1280. doi: 10.1007/s11325-013-0833-4. [DOI] [PubMed] [Google Scholar]
- 89.Chhabra N, Goswami M, Chhabra A. Genetic basis of dental agenesis-molecular genetics patterning clinical dentistry. 2014 doi: 10.4317/medoral.19158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Rutledge MS, Hartsfield JK., Jr Genetic Factors in the Etiology of Palatally Displaced Canines. Semin Orthod. 2010;16:165–171. [Google Scholar]
- 91.Chung DD, Weisberg M, Pagala M. Incidence and effects of genetic factors on canine impaction in an isolated Jewish population. American Journal of Orthodontics and Dentofacial Orthopedics. 2011;139:e331–e335. doi: 10.1016/j.ajodo.2010.06.023. [DOI] [PubMed] [Google Scholar]
- 92.Hartsfield J, Jr, Ponce C, Mofid A, Morford L, Kluemper G. Pain Perception Following Orthodontic Separator Placement and COMT Haplotype. J Dent Res. 2014;93(Spec Iss B) abstract #1321. [Google Scholar]
- 93.Dirschnabel AJ, Alvim- Pereira F, Alvim- Pereira CC, Bernardino JF, Rosa EAR, Trevilatto PC. Analysis of the association of IL1B (C-511T) polymorphism with dental implant loss and the clusterization phenomenon. Clinical oral implants research. 2011;22:1235–1241. doi: 10.1111/j.1600-0501.2010.02080.x. [DOI] [PubMed] [Google Scholar]
- 94.Doetzer AD, Schlipf N, Alvim-Pereira F, Alvim-Pereira CC, Werneck R, Riess O, et al. Lactotransferrin Gene (LTF) Polymorphisms and Dental Implant Loss: A Case-Control Association Study. Clinical implant dentistry and related research. 2015;17:e550–e561. doi: 10.1111/cid.12284. [DOI] [PubMed] [Google Scholar]


