Abstract
Curcumin has been shown to exert potential antitumor activity in vitro and in vivo involved in multiple signaling pathways. However, the application of curcumin is still limited because of its poor hydrophilicity and low bio-availability. In the present study, we investigated the therapeutic effects of a novel and water soluble bis(hydroxymethyl) alkanoate curcuminoid derivative, MTH-3, on human breast adenocarcinoma MDA-MB-231 cells. This study investigated the effect of MTH-3 on cell viability, cell cycle and induction of autophagy and apoptosis in MDA-MB-231 cells. After 24-h treatment with MTH-3, a concentration-dependent decrease in MDA-MB-231 cell viability was observed, and the IC50 value was 5.37±1.22 μM. MTH-3 significantly triggered G2/M phase arrest and apoptosis in MDA-MB-231 cells. Within a 24-h treatment, MTH-3 decreased the CDK1 activity by decreasing CDK1 and cyclin B1 protein levels. MTH-3-induced apoptosis was further confirmed by morphological assessment and Annexin V/PI staining assay. Induction of apoptosis caused by MTH-3 was accompanied by an apparent increase of DR3, DR5 and FADD and, as well as a marked decrease of Bcl-2 and Bcl-xL protein expression. MTH-3 also decreased the protein levels of Ero1, PDI, PERK and calnexin, as well as increased the expression of IRE1α, CHOP and Bip that consequently led to ER stress and MDA-MB-231 cell apoptosis. In addition, MTH-3-treated cells were involved in the autophagic process and cleavage of LC3B was observed. MTH-3 enhanced the protein levels of LC3B, Atg5, Atg7, Atg12, p62 and Beclin-1 in MDA-MB-231 cells. Finally, DNA microarray was carried out to investigate the level changes of gene expression modulated by MTH-3 in MDA-MB-231 cells. Taken together, our results suggest that MTH-3 might be a novel therapeutic agent for the treatment of triple-negative breast cancer in the near future.
Keywords: bis(hydroxymethyl) alkanoate curcuminoid derivative, MTH-3, apoptosis, autophagy, breast cancer MDA-MB-231 cells
Introduction
Breast cancer is the second leading cause of death in women and has approximately 1 million new cases per year worldwide (1,2). Breast cancer patients develop metastasis eventually leading to poor prognosis (3). Triple-negative breast cancer (TNBC) accounts for 12–20% of all breast cancer (4). It has more aggressive disease progress and worse prognosis (5). TNBC characteristics are the lack of expression of estrogen receptor (ER), progesterone receptor (PR) and the lack of overexpression of HER-2 (4,6). TNBC is resistance to anti-hormone therapies and HER-2-aiming target therapies (7,8). Treatment of TNBC remains a great clinical challenge because of the lack of targeting agents and limited therapeutic options (8,9).
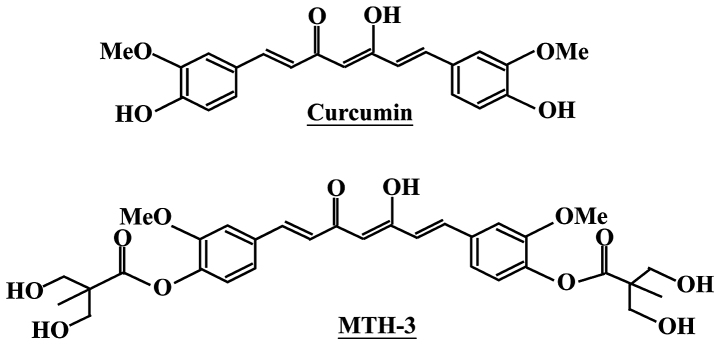
Curcumin has been used in traditional Chinese medicine for a long time in Taiwan, China and India (10). The pharmacological effects of curcumin include anti-amyloid (11), anti-bacterial (12), anti-depressant (13), anti-inflammatory (14), anti-oxidant (15), anti-diabetes (16) and anticancer properties (17,18). In addition, curcumin has been found to affect several anticancer signaling pathways such as inhibition of cancer cell proliferation (19,20) and induction of cell cycle arrest (21), apoptosis (22) or autophagy (23). Specifically, the phase II and III clinical trial of curcumin was advocated for use in patients with colon and pancreatic cancers (24,25), but its low water solubility exerts poor bioavailability and primary limiting factors (low efficacy and safety) (26,27). To improve these issues, we designed and developed a novel bis(hydroxymethyl) alkanoate curcuminoid derivative, MTH-3 (Fig. 1). In our previous studies, novel bis(hydroxymethyl) alkanoate curcuminoid derivatives were shown to exhibit antitumor effects on triple-negative breast cancer cells and in a xenograft animal experiment (28). The aim of the present study was to characterize the property of MTH-3 and to clarify the molecular mechanism of MTH-3 in human breast adenocarcinoma MDA-MB-231 cells in vitro.
Figure 1.
The chemical structures of curcumin (upper panel) and MTH-3 (bottom panel).
Materials and methods
Chemicals and reagents
MTH-3 was synthesized as previously described (28) (patent pending). The purity of MTH-3 is 98.7, and its molecular weight is 600.61. Leibovitz's L-15 medium, fetal bovine serum (FBS), penicillin-streptomycin, trypsin-EDTA, Premo Autophagy Sensor LC3B-GFP (BacMam 2.0) and 4′,6-diamidino-2-phenylindole (DAPI) were purchased from Thermo Fisher Scientific (Waltham, MA, USA). The antibodies were purchased from Cell Signaling Technology (Danvers, MA, USA). All chemicals were obtained from Sigma-Aldrich (St. Louis, MO, USA) unless otherwise stated.
Cell culture
The human breast adenocarcinoma cell line MDA-MB-231 was purchased from the Bioresource Collection and Research Center (BCRC; Hsinchu, Taiwan). Cells were cultured in Leibovitz's L-15 medium with 10% FBS and 1% penicillin-streptomycin (100 Units/ml penicillin and 100 μg/ml streptomycin) in an incubator under 95% air and 5% CO2 at 37°C.
Cell viability assay and morphologic changes
Cell viability was evaluated by the reduction in MTT to yield blue formazan. MDA-MB-231 cells (1×104 cells/well) in 96-well plates were allowed to attach overnight and then treated with different concentrations (1, 3, 5 and 10 μM) of MTH-3 for 24 h. After treatments, MTT solution was added to each well (a final concentration of 0.5 μg/ml), and then the plates were incubated for another 4 h. The medium was removed, blue formazan was dissolved in dimethyl sulfoxide (DMSO), and the absorbance was read at 570 nm as previously described (29). For trypan blue exclusion assay, cells were collected after 1, 3, 5 and 10 μM of MTH-3 exposure, stained with 0.4% trypan blue and then counted on a hemocytometer under a microscope. For morphological observation, cells were visualized and photographed using a phase-contrast microscope equipped with a digital camera (Leica Microsystems GmbH, Wetzlar, Germany) as in previous reports (26,30).
Distribution of cell cycle analysis
MDA-MB-231 cells (2×105 cells/well) in 12-well plates were exposed to 10 μM MTH-3. After a 24-h treatment, cells were harvested and fixed gently by putting 70% ethanol at 4°C overnight before being stained with PI solution (40 μg/ml PI and 0.1 mg/ml RNase and 0.1% Triton X-100) in the dark for 30 min as previously described (31). The cells were analyzed for the cell cycle distribution with a flow cytometer (FACSCalibur; BD Biosciences, San Jose, CA, USA).
CDK1 kinase assay
CDK1 kinase activity was analyzed according to the manufacturer's protocol (CycLex Cdc2-Cyclin B Kinase Assay kit; MBL International Corp., Woburn, MA, USA). The ability of CDK1 kinase from MDA-MB-231 cell extracts prepared from each treatment of 10 μM MTH-3 for 4, 8, 16 and 24 h was measured as previously described (32,33).
Apoptosis analysis
MDA-MB-231 cells (2×105 cells/well) into 12-well plates were incubated in the presence and absence of 10 μM MTH-3 for 24 and 48 h. Subsequently, cells were harvested and stained with Annexin V and propidium iodide (PI) using the Annexin V-FITC apoptosis detection kit (BD Biosciences, San Diego, CA, USA) and subjected to flow cytometry (BD FACSCalibur; BD Biosciences). The percentage of apoptotic cells were quantified with BD CellQuest Pro software (BD Biosciences) (34,35).
Cells lysate preparation and western blot analysis
After 10 μM MTH-3 treatments at indicated intervals of time, MDA-MB-231 cells were harvested, washed and suspended in the PRO-PREP Protein Extraction Solution (iNtRON Biotechnology, Gyeonggi-do, Korea). Protein concentrations were estimated using the Protein Assay kit (Bio-Rad Laboratories, Hercules, CA, USA). The samples were resolved with SDS-PAGE and transferred to a polyvinylidene difluo-ride membrane (PVDF) (EMD Millipore, Billerica, MA, USA). Each membrane was blocked in 5% non-fat milk in Tris-buffered saline with 0.1% Tween-20 for 1 h followed by individual incubation with specific primary antibodies [cyclin B1 (cat. no. 4138, 1:1,000), CDK1/Cdc2 (cat. no. 9116, 1:1,000), DR3 (cat. no. 4758, 1:1,000), DR5 (cat. no. 8074, 1:1,000), FADD (cat. no. 2782, 1:1,000), Bcl-2 (cat. no. 4223, 1:1,000), Bcl-xL (cat. no. 2764, 1:1,000), Ero1 (cat. no. 3264, 1:1,000), PDI (cat. no. 3501, 1:1,000), PERK (cat. no. 5683, 1:1,000), calnexin (cat. no. 2679, 1:1,000), IRE1α (cat. no. 3294, 1:1,000), CHOP (cat. no. 2895, 1:1,000), Bip (cat. no. 3177, 1:1,000), Atg5 (cat. no. 12994, 1:1,000), Atg7 (cat. no. 8558, 1:1,000), Atg12 (cat. no. 4180, 1:1,000), Beclin-1 (cat. no. 3495, 1:1,000), p62 (cat. no. 88588, 1:1,000), LC3A/B (cat. no. 12741, 1:1,000) and β-actin (cat. no. 3700, 1:5,000) (Cell Signaling Technology, Danvers, MA, USA)] at 4°C overnight. Each membrane was then incubated with anti-rabbit IgG (cat. no. 7074, 1:10,000) or anti-mouse IgG (cat. no. 7076, 1:10,000) horseradish peroxidase (HRP)-linked antibodies (Cell Signaling Technology) at room temperature for 1 h. The signal was detected with the Immobilon Western Chemiluminescent HRP substrate (EMD Millipore) and visualized using the LAS 4000 imaging system (Fuji, Tokyo, Japan) as previously described (36–38). The quantitative densitometric analysis of immunoreactive band was employed by ImageJ bundled with 64-bit Java 1.6.0_24 program for Windows from the National Institutes of Health (NIH; Bethesda, MD, USA).
Immunofluorescence staining
MDA-MB-231 cells (2×106 cells/dish) were grown on sterile coverslips placed in a 10-cm dish. After 10 μM MTH-3 treatment, cells were fixed with 4% paraformaldehyde and permeabilized with 0.2% Triton X-100 in phosphate-buffered saline (PBS). After blocking with 2% bovine serum albumin (BSA) in PBS, LC3B and p62 were detected using anti-LC3B and anti-p62 antibody followed by reaction with FITC- or PE-conjugated secondary antibody (BD Biosciences). Coverslips were mounted on glass slides with ProLong Gold Antifade reagents (Thermo Fisher Scientific) containing DAPI, and fluorescent image was taken on a Leica Microsystems TCS SP2 Confocal Spectral microscope as detailed by Lu et al (39).
cDNA microarray analysis
MDA-MB-231 cells were incubated with or without 10 μM MTH-3 for 24 h. After exposure, cell pellets were collected, and the total RNA from each treatment was purified using the Qiagen RNeasy Mini kit (Qiagen, Valencia, CA, USA). RNA purity was determined to check the quality at 260/280 nm using a NanoDrop 1000 spectrophotometer (Thermo Fisher Scientific). mRNA was amplified and labeled using the GeneChip WT Sense Target Labeling and Control Reagents kit (Affymetrix, Santa Clara, CA, USA) for expression analysis. The synthesized cDNA was labeled with fluorescence and then hybridized for 17 h using GeneChip Human Gene 1.0 ST array (Affymetrix) to determine microarray hybridization following the manufacturer's protocols. The arrays were subsequently washed using GeneChip Fluidics Station 450 (Affymetrix), stained with streptavidin-phycoerythrin (GeneChip Hybridization, Wash and Stain kit; Affymetrix) and scanned on a GeneChip Scanner 3000 (Affymetrix). The localized concentrations of fluorescent molecules were quantitated and analyzed using Expression Console Software (Affymetrix) with default RMA parameters as previously described (40). The gene expression level of a 2.5-fold change (log2 ratio) was considered a difference in MTH-3-treated cells in vitro (41,42).
Statistical analysis
Data are presented as the mean ± SD for three separate experiment. Differences among the groups were considered to be significant at P<0.05 using ANOVA followed by the Duncan's test.
Results
MTH-3 inhibits cell proliferation of human breast adenocarcinoma MDA-MB-231 cells
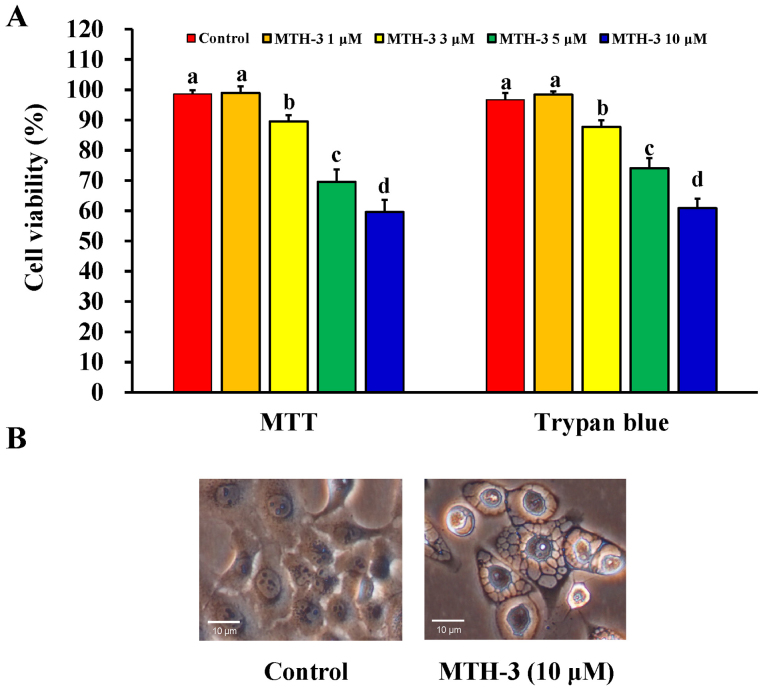
At first, the effect of MTH-3 on the viability of MDA-MB-231 cells was investigated using the MTT and trypan blue exclusion assays. MTH-3 at 1, 3, 5 and 10 μM significantly reduced the viability of MDA-MB-231 cells by 98.94±2.26, 89.57±2.07, 69.57±4.13 and 59.6±4.04%, respectively (Fig. 2A). Importantly, the cell viability reduction after 30 μM MTH-3 challenge is 34.23±3.31%. This effect is in a concentration-dependent manner. Data from morphological observation revealed that MTH-3 treatment at 10 μM caused obvious MDA-MB-231 cell apoptosis and autophagy with characteristics, including cytoplasmic membrane blebbing, cell shrinkage and autophagic vacuoles (Fig. 2B). Based on these findings and gaining effective evidence of cell death, we selected MTH-3 at 10 μM for the majority of the experiments in MDA-MB-231 cells.
Figure 2.
MTH-3 reduces cell viability and affects cell morphology in MDA-MB-231 cells. (A) Cells were incubated with or without various concentrations (1, 3, 5 and 10 μM) of MTH-3 for 24 h, and the viable cells were monitored using an MTT and trypan blue exclusion methods. Data are presented as the mean ± SD of three independent experiments. The different letters (a–d) show statistically significant differences (P<0.05) in each group by the Duncan's test. (B) Cells were examined after with or without 10 μM MTH-3 for 24 h to photograph the changes in cell morphology using a phase-contrast microscope as described in Materials and methods. Scale bar, 10 μm.
MTH-3 triggers G2/M phase arrest and reduces CDK1 activity in MDA-MB-231 cells
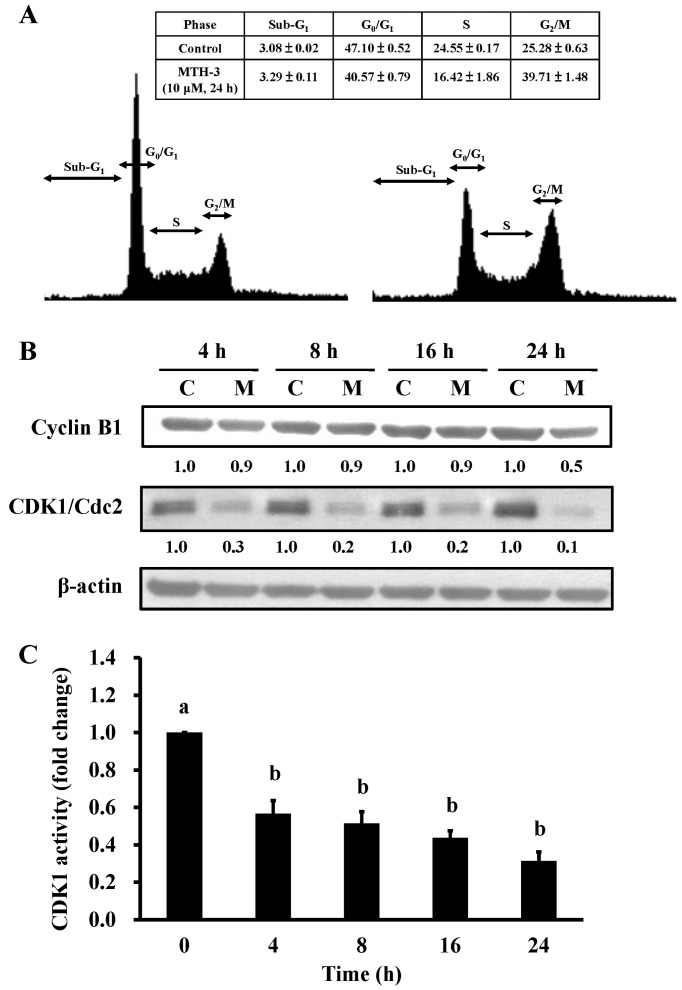
To investigate the cell cycle distribution of treated and untreated MDA-MB-231 cells, cells were monitored after 10 μM MTH-3 challenge. Results from flow cytometric analysis showed that MTH-3 treatment of MDA-MB-231 cells significantly increased G2/M phase cell population at 24 h (Fig. 3A). The effects of MTH-3 on G2/M phase-related proteins in MDA-MB-231 cells were investigated. Our results showed that MTH-3 effectively down-regulated the levels of cyclin B1 and CDK1 (Fig. 3B). We also tested the CDK1 kinase activity in MDA-MB-231 cells prior to MTH-3 treatment. MTH-3 markedly reduced CDK1 kinase activity at 4, 8, 12 and 24 h of treatment, respectively (Fig. 3C). Therefore, the finding showed that downregulation of CDK1 activity contributed to G2/M phase arrest caused by MTH-3 in MDA-MB-231 cells.
Figure 3.
MTH-3 induces G2/M phase arrest of MDA-MB-231 cells. (A) Cells were exposed to 10 μM MTH-3 for 24 h. The cell cycle distribution was detected using flow cytometric analysis and cell cycle distribution was quantified. (B) Cells were exposed to 10 μM MTH-3 and then incubated for 0, 4, 8, 16 and 24 h. The protein levels of cyclin B1, CDK1 and β-actin were determined by western blotting. C, control; M, MTH-3 exposure. (C) CDK1 activity was examined as described in Materials and methods. Data are presented as the mean ± SD of three independent experiments. The different letters (a–b) show statistically significant differences (P<0.05) in each group by the Duncan's test.
MTH-3 elicits cell apoptosis of MDA-MB-231 cells
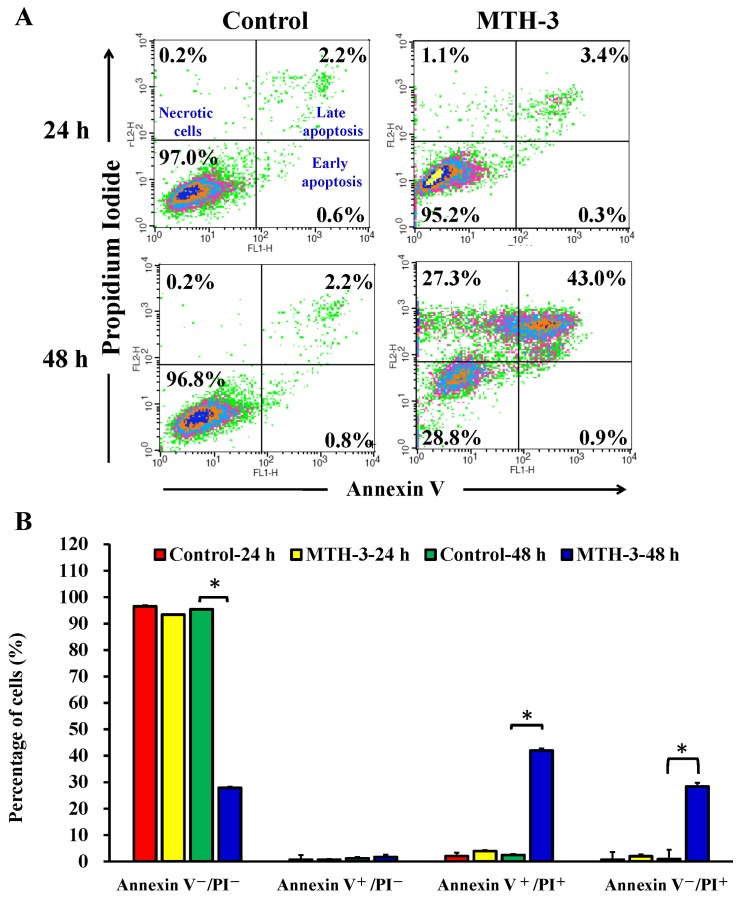
To further explore whether the inhibition of cell viability results from the induction of apoptosis in MDA-MB-231 cells, MTH-3-treated cells were detected with Annexin V/PI double staining (Fig. 4). Treatment with 10 μM MTH-3 for 48 h significantly increased the population of Annexin V-positive cells (Fig. 4), indicating that MTH-3 induced apoptosis in MDA-MB-231 cells. However, the necrotic cells (Annexin V+/PI+) increased rapidly after 48 h of 10 μM MTH-3 exposure.
Figure 4.
MTH-3 induces apoptosis of MDA-MB-231 cells. Cells were incubated with 10 μM MTH-3 for 24 and 48 h. Cells were collected and stained with Annexin V/propidium iodide (PI) before analysis with flow cytometry. The Annexin V-positive cells were counted, and data are presented as the mean ± SD of three independent experiments. *P<0.05 indicates statistically significant differences by the Duncan's test.
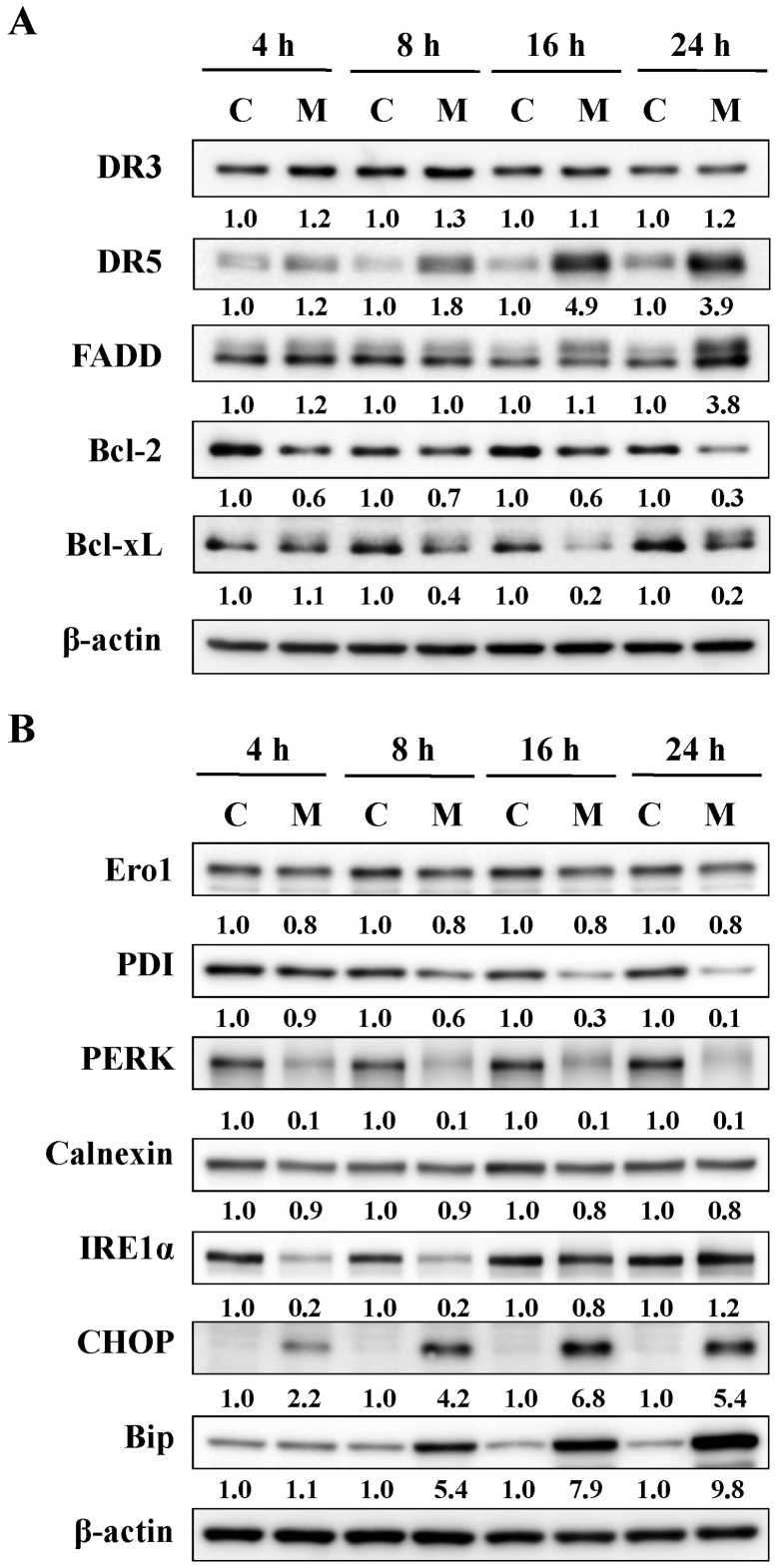
MTH-3 activates death receptor, mitochondrial and ER stress-mediated apoptotic pathways in MDA-MB-231 cells
The effects of MTH-3 on apoptosis-related proteins in MDA-MB-231 cells were investigated. Our results demonstrated that MTH-3 upregulated the levels of DR5 and FADD, and it downregulated the levels of Bcl-2 and Bcl-xL (Fig. 5A). Furthermore, our findings also revealed that MTH-3 markedly increased the levels of CHOP and Bip, as well as decreased the levels of Ero1, PDI, PERK, calnexin and IRE1α (Fig. 5B). These results suggest that MTH-3 induced apoptosis through death receptor (extrinsic pathway) and mitochondria (intrinsic pathway)-dependent pathways and possibly by modulating ER stress mechanism in MDA-MB-231 cells.
Figure 5.
MTH-3 activates death receptor-mediated, mitochondrial and ER stress-regulated apoptosis pathways in MDA-MB-231 cells. Cells were exposed to 10 μM MTH-3 for 0, 4, 8, 16 and 24 h, and cell lysates were collected for western blot analysis. (A) Death receptor-mediated (DR3, DR5 and FADD) and mitochondrial (Bcl-2 and Bcl-xL) apoptosis pathways, and (B) ER stress (Ero1, PDI, PERK, calnexin, IRE1α, CHOP and Bip) were performed. β-actin served as an internal control. C, control; M, MTH-3 exposure.
MTH-3 stimulates autophagy in MDA-MB-231 cells
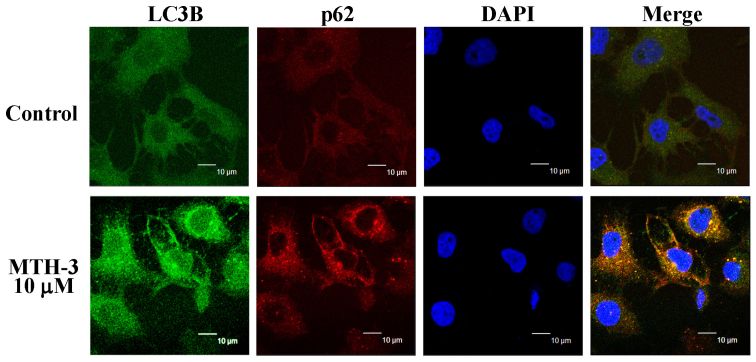
To confirm if autophagy is involved in the inhibition of MDA-MB-231 cell viability, cells with or without MTH-3 exposure were detected with LC3B and p62 double immunostaining. MTH-3 at 10 μM increased the LC3B (FITC; green color) and p62 (PE; red color) protein expression (Fig. 6), indicating that MTH-3 induced autophagy through increasing LC3B/p62 signaling in MDA-MB-231 cells.
Figure 6.
MTH-3 induces LC3B and p62 expression of MDA-MB-231 cells. Cells were treated with 10 μM MTH-3 for 24 h. Cells were collected and stained with LC3B-FITC antibody (green color) and p62-PE antibody (red color) and analyzed with confocal microscope. DAPI dye (blue color) is for nuclear acid (nuclear) staining. Scale bar, 10 μm.
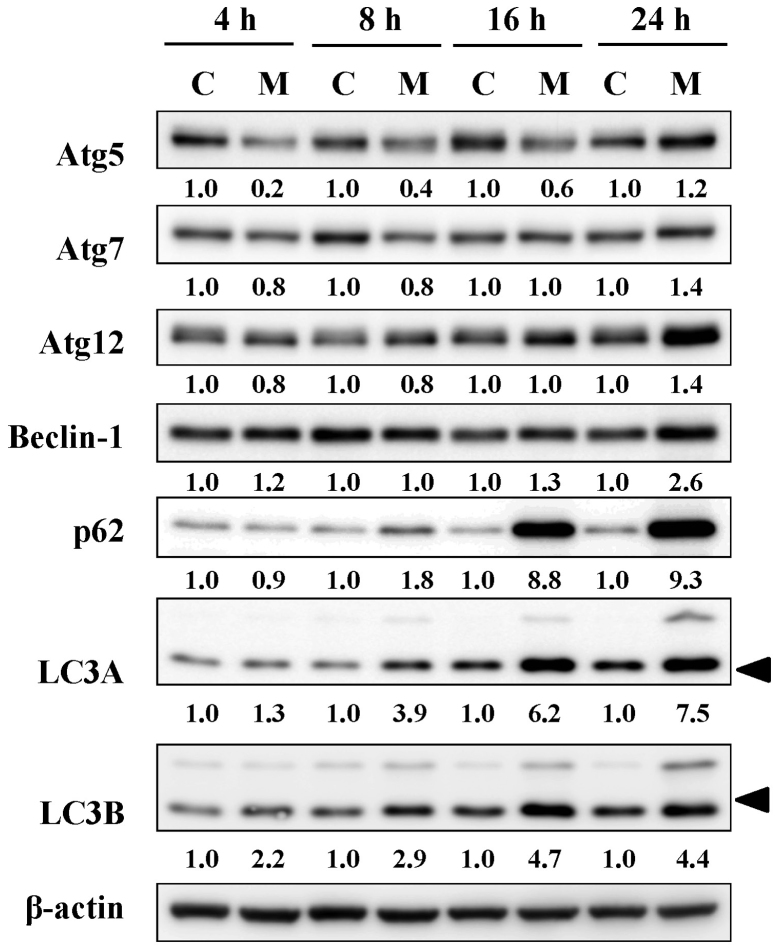
MTH-3 alters the levels of autophagy-associated proteins in MDA-MB-231 cells
Based on the results of autophagy, its related signals were further employed by immunoblotting analysis. MTH-3 treatment induced the levels of Atg5, Atg7, Atg12, Beclin-1, p62 and LC3B in a time-dependent manner (Fig. 7). These data demonstrated that MTH-3 induced autophagy by activating Atg family proteins in MDA-MB-231 cells.
Figure 7.
MTH-3 alters the protein levels of autophagy-related proteins in MDA-MB-231 cells. Cells were incubated with 10 μM MTH-3 for 4, 8, 16 and 24 h, and cell lysates were collected for western blot analysis to probe autophagic signals (Atg5, Atg7, Atg12, Beclin-1, p62, LC3A and LC3B). β-actin was an internal control. C, control; M, MTH-3 exposure.
MTH-3 modulates cell death-related gene expression in MDA-MB-231 cells by cDNA microarray analysis
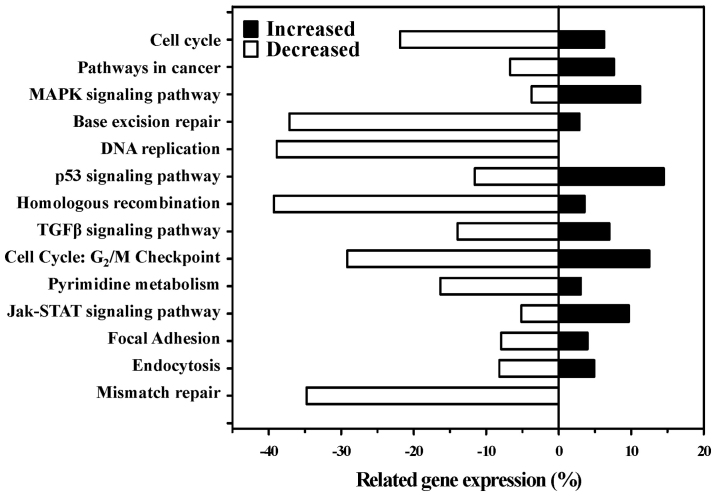
After treatment with 10 μM MTH-3 for 24 h, cells were collected, and cDNA microarray analysis was performed. The analysis showed that 97 genes (69 genes, upregulated; 28 genes, down-regulated) were expressed at least by 2.5-fold compared with the untreated control (Table I). The top alteration in gene expression scored by the number of pathway networks from GeneGo analysis program (Fig. 8). These genes may also be involved in cell death and cytotoxic responses in MTH-3-treated MDA-MB-231 cells.
Table I.
The >2.5-fold changes in mRNA levels in MDA-MB-231 cells following a 24-h treatment with 10 μM MTH-3 as identified using DNA microarray.
| ID | log2 (ratio) | Gene_symbol | Description |
|---|---|---|---|
| PH_hs_0049600 | 6.643856 | HSPA6 | Heat shock 70 kDa protein 6 (HSP70B') |
| PH_hs_0006387 | 6.274261 | ZFAND2A | zinc finger, AN1-type domain 2A |
| PH_hs_0004421 | 5.381376 | PPP1R15A | Protein phosphatase 1, regulatory subunit 15A |
| PH_hs_0000305 | 4.941673 | MMP10 | Matrix metallopeptidase 10 (stromelysin 2) |
| PH_hs_0046245 | 4.763129 | RN7SK | RNA, 7SK small nuclear |
| PH_hs_0000076 | 4.587356 | IL12A | Interleukin 12A |
| PH_hs_0027902 | 4.286664 | ABL2 | v-abl Abelson murine leukemia viral oncogene homolog 2 |
| PH_hs_0010276 | 4.189167 | DUSP1 | Dual specificity phosphatase 1 |
| PH_hs_0031719 | 4.146525 | CCL26 | Chemokine (C-C motif) ligand 26 |
| PH_hs_0000156 | 4.093858 | DUSP2 | Dual specificity phosphatase 2 |
| PH_hs_0011943 | 4.063702 | HMOX1 | Heme oxygenase (decycling) 1 |
| PH_hs_0045501 | 4.039442 | EID3 | EP300 interacting inhibitor of differentiation 3 |
| PH_hs_0004561 | 3.997336 | GEM | GTP binding protein overexpressed in skeletal muscle |
| PH_hs_0042334 | 3.931415 | MT4 | Metallothionein 4 |
| PH_hs_0048553 | 3.866096 | MYCT1 | myc target 1 |
| PH_hs_0000684 | 3.853854 | DNAJB9 | DnaJ (Hsp40) homolog, subfamily B, member 9 |
| PH_hs_0035404 | 3.763571 | SAT1 | Spermidine/spermine N1-acetyltransferase 1 |
| PH_hs_0000057 | 3.698185 | ATF3 | Activating transcription factor 3 |
| PH_hs_0025319 | 3.562429 | C3orf52 | Chromosome 3 open reading frame 52 |
| PH_hs_0033101 | 3.555868 | DDIT3 | DNA-damage-inducible transcript 3 (CHOP) |
| PH_hs_0002700 | 3.513438 | OSGIN1 | Oxidative stress induced growth inhibitor 1 |
| PH_hs_0037472 | 3.480422 | MALAT1 | Metastasis associated lung adenocarcinoma transcript 1 |
| PH_hs_0035765 | 3.427173 | GDF15 | Growth differentiation factor 15 |
| PH_hs_0002492 | 3.366024 | SAT1 | Spermidine/spermine N1-acetyltransferase 1 |
| PH_hs_0062199 | 3.356707 | AKR1C1|LOC101060798 | Aldo-keto reductase family 1, member C1|aldo-keto reductase family 1 member C2-like |
| PH_hs_0000852 | 3.324182 | SESN2 | Sestrin 2 |
| PH_hs_0023008 | 3.242113 | FRS2 | Fibroblast growth factor receptor substrate 2 |
| PH_hs_0004751 | 3.219326 | MMP1 | Matrix metallopeptidase 1 (interstitial collagenase) |
| PH_hs_0031143 | 3.213328 | VIMP | VCP-interacting membrane protein |
| PH_hs_0025525 | 3.198476 | CLU | Clusterin |
| PH_hs_0024315 | 3.075314 | DNAJB4 | DnaJ (Hsp40) homolog, subfamily B, member 4 |
| PH_hs_0035614 | 3.062771 | RC3H1 | Ring finger and CCCH-type domains 1 |
| PH_hs_0027152 | 3.037995 | RMND5A | Required for meiotic nuclear division 5 homolog A (S. cerevisiae) |
| PH_hs_0021974 | 3.010862 | DNAJC3 | DnaJ (Hsp40) homolog, subfamily C, member 3 |
| PH_hs_0061784 | 2.967357 | CDKN1A | Cyclin-dependent kinase inhibitor 1A (p21, Cip1) |
| PH_hs_0035466 | 2.962064 | AKR1C3|AKR1C1 | Aldo-keto reductase family 1, member C3|aldo-keto reductase family 1, member C1 |
| PH_hs_0027162 | 2.960759 | SLC3A2 | Solute carrier family 3 (activators of dibasic and neutral amino acid transport), member 2 |
| PH_hs_0022919 | 2.960552 | CLCF1 | Cardiotrophin-like cytokine factor 1 |
| PH_hs_0000255 | 2.916655 | SRGN | Serglycin |
| PH_hs_0024155 | 2.904033 | CDKN1A | Cyclin-dependent kinase inhibitor 1A (p21, Cip1) |
| PH_hs_0043719 | 2.894684 | HMGCS1 | 3-hydroxy-3-methylglutaryl-CoA synthase 1 (soluble) |
| PH_hs_0045838 | 2.838192 | SLC6A6 | Solute carrier family 6 (neurotransmitter transporter, taurine), member 6 |
| PH_hs_0014155 | 2.836392 | HSPA1B | Heat shock 70 kDa protein 1B |
| PH_hs_0044272 | 2.829317 | CLK1 | CDC-like kinase 1 |
| PH_hs_0048881 | 2.809371 | FKBP4 | FK506 binding protein 4, 59 kDa |
| PH_hs_0020147 | 2.803912 | CLK1 | CDC-like kinase 1 |
| PH_hs_0028987 | 2.768552 | TCF21 | Transcription factor 21 |
| PH_hs_0042409 | 2.76703 | DNAJB1 | DnaJ (Hsp40) homolog, subfamily B, member 1 |
| PH_hs_0001262 | 2.748306 | SENP5 | SUMO1/sentrin specific peptidase 5 |
| PH_hs_0060828 | 2.734692 | TRIB3 | Tribbles homolog 3 (Drosophila) |
| PH_hs_0023556 | 2.733421 | C21orf91 | Chromosome 21 open reading frame 91 |
| PH_hs_0061012 | 2.731293 | ZBTB21 | Zinc finger and BTB domain containing 21 |
| PH_hs_0029660 | 2.695316 | AKR1C1 | Aldo-keto reductase family 1, member C1|aldo-keto reductase family 1 |
| PH_hs_0037242 | 2.683231 | MALAT1 | Metastasis associated lung adenocarcinoma transcript 1 (non-protein coding) |
| PH_hs_0002812 | 2.667718 | C18orf25 | Chromosome 18 open reading frame 25 |
| PH_hs_0027209 | 2.665362 | GADD45B | Growth arrest and DNA-damage-inducible, β |
| PH_hs_0002971 | 2.664712 | ZNF77 | Zinc finger protein 77 |
| PH_hs_0003180 | 2.646292 | SMIM13 | Small integral membrane protein 13 |
| PH_hs_0000694 | 2.625719 | RND3 | Rho family GTPase 3 |
| PH_hs_0023711 | 2.599232 | HSPA5 | Heat shock 70 kDa protein 5 |
| PH_hs_0023894 | 2.583817 | TRIB3 | Tribbles homolog 3 (Drosophila) |
| PH_hs_0060053 | 2.574976 | ZNF121 | Zinc finger protein 121 |
| PH_hs_0014119 | 2.571605 | BRF2 | BRF2, subunit of RNA polymerase III transcription initiation factor, BRF1-like |
| PH_hs_0033027 | 2.547837 | SIK1 | Salt-inducible kinase 1 |
| PH_hs_0024236 | 2.547678 | ATP2A2 | ATPase, Ca++ transporting, cardiac muscle, slow twitch 2 |
| PH_hs_0042225 | 2.541029 | DUSP5 | Dual specificity phosphatase 5 |
| PH_hs_0044921 | 2.534876 | HSPA1A | Heat shock 70 kDa protein 1A |
| PH_hs_0000566 | 2.528881 | SLC25A25 | Solute carrier family 25, member 25 |
| PH_hs_0030976 | 2.516291 | NFKBIB | Nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, β |
| PH_hs_0014995 | −3.653241 | METTL7A | Methyltransferase like 7A |
| PH_hs_0023845 | −3.269308 | BBS2 | Bardet-Biedl syndrome 2 |
| PH_hs_0009437 | −3.05235 | TOP2A | Topoisomerase (DNA) II α 170 kDa |
| PH_hs_0047352 | −3.043277 | MARCKS | Myristoylated alanine-rich protein kinase C substrate |
| PH_hs_0047965 | −2.959225 | PHLDA1 | Pleckstrin homology-like domain, family A, member 1 |
| PH_hs_0040619 | −2.891495 | MXD3 | MAX dimerization protein 3 |
| PH_hs_0012629 | −2.890238 | H1F0 | H1 histone family, member 0 |
| PH_hs_0004988 | −2.878231 | LMNB1 | Lamin B1 |
| PH_hs_0035609 | −2.788184 | ETV1 | Ets variant 1 |
| PH_hs_0049449 | −2.729758 | GPR39 | G protein-coupled receptor 39 |
| PH_hs_0027843 | −2.724437 | FAM20C | FAmily with sequence similarity 20, member C |
| PH_hs_0027863 | −2.718276 | LRRC45 | Leucine rich repeat containing 45 |
| PH_hs_0007383 | −2.717289 | F2R | Coagulation factor II (thrombin) receptor |
| PH_hs_0036878 | −2.71449 | PIF1 | PIF1 5′-to-3′ DNA helicase homolog (S. cerevisiae) |
| PH_hs_0047697 | −2.688182 | ARF6 | ADP-ribosylation factor 6 |
| PH_hs_0048993 | −2.677322 | NRP1 | Neuropilin 1 |
| PH_hs_0031540 | −2.66121 | GNG2 | Guanine nucleotide binding protein (G protein), gamma 2 |
| PH_hs_0010634 | −2.659899 | TXNIP|LOC101060503 | Thioredoxin interacting protein|thioredoxin-interacting protein-like |
| PH_hs_0028935 | −2.621805 | CCDC85B | Coiled-coil domain containing 85B |
| PH_hs_0000866 | −2.612763 | OMA1 | OMA1 zinc metallopeptidase homolog (S. cerevisiae) |
| PH_hs_0030800 | −2.552826 | FANCF | Fanconi anemia, complementation group F |
| PH_hs_0025966 | −2.55207 | CTDSP1 | CTD small phosphatase 1 |
| PH_hs_0023862 | −2.551096 | CBY1 | Chibby homolog 1 (Drosophila) |
| PH_hs_0047571 | −2.546813 | PDP1 | Pyruvate dehyrogenase phosphatase catalytic subunit 1 |
| PH_hs_0028200 | −2.537288 | CENPI | Centromere protein I |
| PH_hs_0003147 | −2.533627 | PDGFC | Platelet derived growth factor C |
| PH_hs_0035337 | −2.514458 | OMA1 | OMA1 zinc metallopeptidase homolog (S. cerevisiae) |
| PH_hs_0038982 | −2.502536 | LOC100134259 | Uncharacterized LOC100134259 |
Figure 8.
The possible canonical pathways from MDA-MB-231 cells after exposure to MTH-3 by cDNA microarray. Cells were treated with 10 μM MTH-3 for 24 h, and then were harvested before total RNA was extracted for cDNA microarray assay. The changes in gene expression scored by the number of pathways from GeneGo analysis.
Discussion
Previous studies have demonstrated the anticancer potential of curcumin in regulating cell cycle, autophagy, apoptosis and survival, proliferation, angiogenesis, invasion and metastasis (19–23). Guan et al (43) demonstrated that curcumin reduced Akt kinase in MDA-MB-231 cells accompanied by a decrease in cell proliferation and migration as well as an increase in autophagic activity; moreover, AMPK-mediated activation of autophagy contributes to anticancer effects through Akt degradation. In the present study, we also checked the growth inhibition effect of curcumin on MDA-MB-231 cells. Our data indicated that the half maximal inhibitory concentration (IC50) value of curcumin on MDA-MB-231 cells is 38.77±3.35 μM. Strikingly, the IC50 value of MTH-3 on MDA-MB-231 cells is 5.37±1.22 μM (data not shown). Our results demonstrated that the MTH-3 had highly cytotoxic effects on MDA-MB-231 cells. Moreover, we also found that MTH-3 was non-cytotoxic on non-tumorigenic epithelial mammary MCF10A cells and human skin fibroblast Detroit 551 cells, respectively (data not shown). These are only preliminary data and further study is needed to validate the findings.
There are no reports regarding that the effects of MTH-3 on cell cycle arrest, autophagy and apoptosis and associated gene expression in human breast cancer cells. This study is first to demonstrate that MTH-3 induced cytotoxic effect on induction of G2/M phase arrest, autophagy and apoptosis in human breast adenocarcinoma MDA-MB-231 cells. The data demonstrated that MTH-3 induced growth inhibitory effects through G2/M phase arrest, apoptosis and autophagy in MDA-MB-231 cells. Our results showed that MTH-3 induced G2/M phase arrest through regulating cyclin B1 and CDK1 signaling. G2/M phase progression has been reported to regulate CDK1 and CDK2 kinases that are activated primarily in association with cyclins A and B (44). Furthermore, MTH-3 inhibited the CDK1 activity and the protein expression of CDK1 in MDA-MB-231 cells. However, neither effect is positively correlated because CDK1 activity might be involved in kinase activation rather than CDK1/cdc2 protein level (32,33). Previous studies also demonstrated that curcumin inhibited cell proliferation through induction of G0/G1 phase arrest of cancer cells (45,46), but our finding indicated that MTH-3 induced G2/M phase arrest upon different types of cancer cell lines. However, the results are in agreement with previous studies to show that curcumin inhibited cell proliferation by inducing G2/M phase arrest in human glioblastoma U87 cells (47) and in Bcl-2 overex-pressed MCF-7 cells (48). Further research is required to verify the mechanism of MTH-3 action in different breast cancer cell lines (such as MCF-7 and MDA-MB-453 cells).
It is well documented that apoptosis plays an important role in the maintenance of tissue homeostasis for the elimination of excessive cells (49,50). Induction of apoptosis of cancer cells by anticancer drugs such as etoposide, cisplatin and paclitaxel have been used for treatment of cancer in target cells (51). Apoptosis-associated signaling pathways include extrinsic (death receptor), intrinsic (mitochondria-dependent) and ER stress (unfolded protein response) signals (52,53). Our results demonstrated that MTH-3 promoted the protein levels of DR5, and FADD and downregulated the levels of Bcl-2 and Bcl-xL in MDA-MB-231 cells. MTH-3 also promoted the protein levels of CHOP and Bip, and it reduced the levels of Ero1, PDI, PERK, calnexin and IRE1α in MDA-MB-231 cells. Our novel findings suggest that both extrinsic and intrinsic pathways, and ER stress signals were involved in MTH-3-treated cells in vitro. This agrees with a previous study reporting that the major targets of apoptotic initiation are mediated by dysfunction of cellular organelles (mitochondria, ER, lysosomes and golgi apparatus) (54).
Autophagy is another major clearance route for intracellular protein (55). Recently, curcumin can induce autophagy in cancer cells (56,57). Our results showed that MTH-3 significantly increased protein expression of autophagy markers LC3B, Atg complex (Atg5, Atg7 and Atg12) and Beclin-1, as well as GFP-LC3 puncta formation, suggesting that LC3B was recruited to the autophagosomal membrane during autophagosome formation. Our data strongly suggest that MTH-3 activated autophagy in MDA-MB-231 cells.
From gene expression profiles by DNA microarray, we found that cellular and molecular responses to MTH-3 treatment are multi-faceted and mediated by various regulatory pathways in MDA-MB-231 cells. MTH-3 regulated the expression of important genes in cell cycle, pathways in cancer, MAPK signaling, base excision repair, DNA replication, p53 signaling, homologous recombination, TGF-β signaling, G2/M checkpoint, pyrimidine metabolism, Jak-STAT signaling, focal adhesion, endocytosis and mismatch repair pathways. The gene regulation may be responsible for inhibiting the proliferation of MDA-MB-23 cells. Cyclins associate with cyclin-dependent protein kinases (CDKs) and CDK inhibitor (CKI) can control the procedure of cell cycle to arrest the cell cycle and inhibit the cell growth of cancer cells (44,58). Our results from gene expression profiles indicated that MTH-3 changed the expression of cyclin and cyclin-dependent kinase inhibitor gene CDKN1A, suggesting a change in cyclin, cyclin-dependent kinase inhibitors which could finally lead to cell cycle G2/M phase arrest.
Heme oxygenase-1 (HO-1) has been implicated in cellular defense against oxidative stress and has anti-inflammation function (59,60). A recent study has demonstrated that curcumin inhibits appoptosin-induced apoptosis by upregulating HO-1 expression in SH-SY5Y cells (61). Curcumin-induced HO-1 expression also prevents H2O2-induced cell death in wild-type and HO-2 knockout adipose-derived mesenchymal stem cells (62). In this study of the gene expression profiles, MTH-3 upregulated the expression of heme oxygenase 1 (HMOX1) gene, suggesting that MTH-3 might have anti-inflammation and cell protection function.
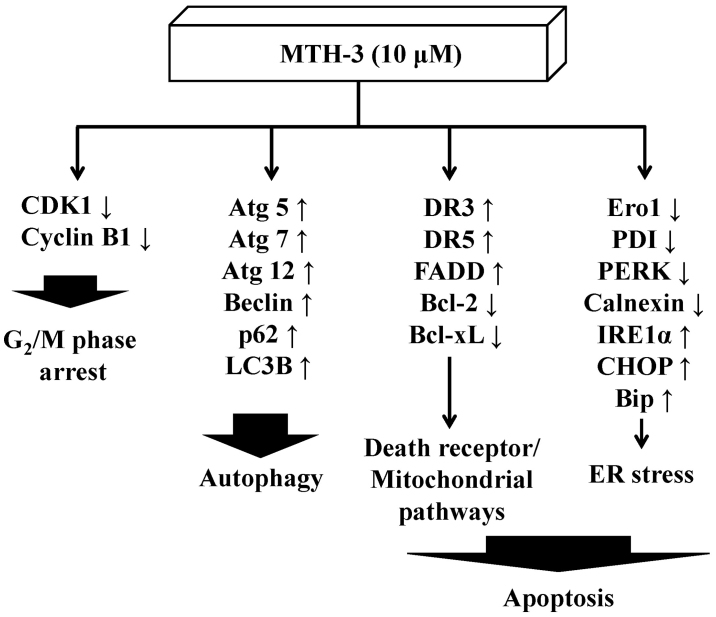
In conclusion, the molecular signaling pathways are summarized in Fig. 9. This study is the first report to provide an approach regarding the bis(hydroxymethyl) alkanoate curcuminoid derivative, MTH-3 tends to inhibit human breast adenocarcinoma MDA-MB-231 cells. Based on the presented novel findings, the efficacy of MTH-3 might be sufficient to further investigate the potential of breast cancer treatment.
Figure 9.
The proposed model shows that MTH-3 induces G2/M phase arrest, autophagy and apoptotic cell death (death receptor/mitochondrial pathways and ER stress) in human breast adenocarcinoma MDA-MB-231 cells.
Acknowledgments
The present study was supported by research grants from the National Science Council of the Republic of China awarded to S.-C.K. and by China Medical University under the Aim for Top University Plan of the Ministry of Education, Taiwan (CHM106-2).
References
- 1.Aseyev O, Ribeiro JM, Cardoso F. Review on the clinical use of eribulin mesylate for the treatment of breast cancer. Expert Opin Pharmacother. 2016;17:589–600. doi: 10.1517/14656566.2016.1146683. [DOI] [PubMed] [Google Scholar]
- 2.Cornejo-Moreno BA, Uribe-Escamilla D, Salamanca-Gómez F. Breast cancer genes: Looking for BRACA's lost brother. Isr Med Assoc J. 2014;16:787–792. [PubMed] [Google Scholar]
- 3.Koo T, Kim IA. Brain metastasis in human epidermal growth factor receptor 2-positive breast cancer: From biology to treatment. Radiat Oncol J. 2016;34:1–9. doi: 10.3857/roj.2016.34.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mouh FZ, Mzibri ME, Slaoui M, Amrani M. Recent progress in triple negative breast cancer research. Asian Pac J Cancer Prev. 2016;17:1595–1608. doi: 10.7314/APJCP.2016.17.4.1595. [DOI] [PubMed] [Google Scholar]
- 5.Hurvitz S, Mead M. Triple-negative breast cancer: Advancements in characterization and treatment approach. Curr Opin Obstet Gynecol. 2016;28:59–69. doi: 10.1097/GCO.0000000000000239. [DOI] [PubMed] [Google Scholar]
- 6.Zeichner SB, Terawaki H, Gogineni K. A review of systemic treatment in metastatic triple-negative breast cancer. Breast Cancer (Auckl) 2016;10:25–36. doi: 10.4137/BCBCR.S32783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang Y, Cao S, Chen Y. Molecular treatment of different breast cancers. Anticancer Agents Med Chem. 2015;15:701–720. doi: 10.2174/1871520615666150129211901. [DOI] [PubMed] [Google Scholar]
- 8.Tomao F, Papa A, Zaccarelli E, Rossi L, Caruso D, Minozzi M, Vici P, Frati L, Tomao S. Triple-negative breast cancer: New perspectives for targeted therapies. Onco Targets Ther. 2015;8:177–193. doi: 10.2147/OTT.S67673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gilani RA, Phadke S, Bao LW, Lachacz EJ, Dziubinski ML, Brandvold KR, Steffey ME, Kwarcinski FE, Graveel CR, Kidwell KM, et al. UM-164: A potent c-Src/38 kinase inhibitor with in vivo activity against triple-negative breast cancer. Clin Cancer Res. 2016;22:5087–5096. doi: 10.1158/1078-0432.CCR-15-2158. [DOI] [PubMed] [Google Scholar]
- 10.Zheng B, Yang L, Wen C, Huang X, Xu C, Lee KH, Xu J. Curcumin analog L3 alleviates diabetic atherosclerosis by multiple effects. Eur J Pharmacol. 2016;775:22–34. doi: 10.1016/j.ejphar.2016.02.016. [DOI] [PubMed] [Google Scholar]
- 11.Ferreira N, Saraiva MJ, Almeida MR. Natural polyphenols as modulators of TTR amyloidogenesis: in vitro and in vivo evidences towards therapy. Amyloid. 2012;19(Suppl 1):39–42. doi: 10.3109/13506129.2012.668502. [DOI] [PubMed] [Google Scholar]
- 12.Vilekar P, King C, Lagisetty P, Awasthi V, Awasthi S. Antibacterial activity of synthetic curcumin derivatives: 3,5-bis(benzylidene)-4-piperidone (EF24) and EF24-dimer linked via diethylenetriaminepentacetic acid (EF2DTPA) Appl Biochem Biotechnol. 2014;172:3363–3373. doi: 10.1007/s12010-014-0741-5. [DOI] [PubMed] [Google Scholar]
- 13.Bhutani MK, Bishnoi M, Kulkarni SK. Anti-depressant like effect of curcumin and its combination with piperine in unpredictable chronic stress-induced behavioral, biochemical and neurochemical changes. Pharmacol Biochem Behav. 2009;92:39–43. doi: 10.1016/j.pbb.2008.10.007. [DOI] [PubMed] [Google Scholar]
- 14.Cianciulli A, Calvello R, Porro C, Trotta T, Salvatore R, Panaro MA. PI3k/Akt signalling pathway plays a crucial role in the anti-inflammatory effects of curcumin in LPS-activated microglia. Int Immunopharmacol. 2016;36:282–290. doi: 10.1016/j.intimp.2016.05.007. [DOI] [PubMed] [Google Scholar]
- 15.Choudhury AK, Raja S, Mahapatra S, Nagabhushanam K, Majeed M. Synthesis and evaluation of the anti-oxidant capacity of curcumin glucuronides, the major curcumin metabolites. Antioxidants. 2015;4:750–767. doi: 10.3390/antiox4040750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yang H, Xu W, Zhou Z, Liu J, Li X, Chen L, Weng J, Yu Z. Curcumin attenuates urinary excretion of albumin in type II diabetic patients with enhancing nuclear factor erythroid-derived 2-like 2 (Nrf2) system and repressing inflammatory signaling efficacies. Exp Clin Endocrinol Diabetes. 2015;123:360–367. doi: 10.1055/s-0035-1545345. [DOI] [PubMed] [Google Scholar]
- 17.Lee D, Kim IY, Saha S, Choi KS. Paraptosis in the anti-cancer arsenal of natural products. Pharmacol Ther. 2016;162:120–133. doi: 10.1016/j.pharmthera.2016.01.003. [DOI] [PubMed] [Google Scholar]
- 18.Borges GA, Rêgo DF, Assad DX, Coletta RD, De Canto Luca G, Guerra EN. In vivo and in vitro effects of curcumin on head and neck carcinoma: A systematic review. J Oral Pathol Med. 2017;46:3–20. doi: 10.1111/jop.12455. [DOI] [PubMed] [Google Scholar]
- 19.Sordillo PP, Helson L. Curcumin and cancer stem cells: Curcumin has asymmetrical effects on cancer and normal stem cells. Anticancer Res. 2015;35:599–614. [PubMed] [Google Scholar]
- 20.Iqbal B, Ghildiyal A, Sahabjada, Singh S, Arshad M, Mahdi AA, Tiwari S. Antiproliferative and apoptotic effect of curcumin and TRAIL (TNF related apoptosis inducing ligand) in chronic myeloid leukaemic cells. J Clin Diagn Res. 2016;10:XC01–XC05. doi: 10.7860/JCDR/2016/18507.7579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang L, Cheng X, Gao Y, Bao J, Guan H, Lu R, Yu H, Xu Q, Sun Y. Induction of ROS-independent DNA damage by curcumin leads to G2/M cell cycle arrest and apoptosis in human papillary thyroid carcinoma BCPAP cells. Food Funct. 2016;7:315–325. doi: 10.1039/C5FO00681C. [DOI] [PubMed] [Google Scholar]
- 22.Huang YT, Lin YW, Chiu HM, Chiang BH. Curcumin induces apoptosis of colorectal cancer stem cells by coupling with CD44 marker. J Agric Food Chem. 2016;64:2247–2253. doi: 10.1021/acs.jafc.5b05649. [DOI] [PubMed] [Google Scholar]
- 23.Kantara C, O'Connell M, Sarkar S, Moya S, Ullrich R, Singh P. Curcumin promotes autophagic survival of a subset of colon cancer stem cells, which are ablated by DCLK1-siRNA. Cancer Res. 2014;74:2487–2498. doi: 10.1158/0008-5472.CAN-13-3536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ji JL, Huang XF, Zhu HL. Curcumin and its formulations: Potential anti-cancer agents. Anticancer Agents Med Chem. 2012;12:210–218. doi: 10.2174/187152012800228733. [DOI] [PubMed] [Google Scholar]
- 25.Shehzad A, Wahid F, Lee YS. Curcumin in cancer chemo-prevention: Molecular targets, pharmacokinetics, bioavailability, and clinical trials. Arch Pharm (Weinheim) 2010;343:489–499. doi: 10.1002/ardp.200900319. [DOI] [PubMed] [Google Scholar]
- 26.Chang PY, Peng SF, Lee CY, Lu CC, Tsai SC, Shieh TM, Wu TS, Tu MG, Chen MY, Yang JS. Curcumin-loaded nanoparticles induce apoptotic cell death through regulation of the function of MDR1 and reactive oxygen species in cisplatin-resistant CAR human oral cancer cells. Int J Oncol. 2013;43:1141–1150. doi: 10.3892/ijo.2013.2050. [DOI] [PubMed] [Google Scholar]
- 27.Douglass BJ, Clouatre DL. Beyond yellow curry: Assessing commercial curcumin absorption technologies. J Am Coll Nutr. 2015;34:347–358. doi: 10.1080/07315724.2014.950392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hsieh MT, Chang LC, Hung HY, Lin HY, Shih MH, Tsai CH, Kuo SC, Lee KH. New bis(hydroxymethyl) alkanoate curcuminoid derivatives exhibit activity against triple-negative breast cancer in vitro and in vivo. Eur J Med Chem. 2017;131:141–151. doi: 10.1016/j.ejmech.2017.03.006. [DOI] [PubMed] [Google Scholar]
- 29.Peng SF, Lee CY, Hour MJ, Tsai SC, Kuo DH, Chen FA, Shieh PC, Yang JS. Curcumin-loaded nanoparticles enhance apoptotic cell death of U2OS human osteosarcoma cells through the Akt-Bad signaling pathway. Int J Oncol. 2014;44:238–246. doi: 10.3892/ijo.2013.2175. [DOI] [PubMed] [Google Scholar]
- 30.Liao CL, Lai KC, Huang AC, Yang JS, Lin JJ, Wu SH, Gibson Wood W, Lin JG, Chung JG. Gallic acid inhibits migration and invasion in human osteosarcoma U-2 OS cells through suppressing the matrix metalloproteinase-2/-9, protein kinase B (PKB) and PKC signaling pathways. Food Chem Toxicol. 2012;50:1734–1740. doi: 10.1016/j.fct.2012.02.033. [DOI] [PubMed] [Google Scholar]
- 31.Tsai SC, Lu CC, Lee CY, Lin YC, Chung JG, Kuo SC, Amagaya S, Chen FN, Chen MY, Chan SF, et al. AKT serine/threonine protein kinase modulates bufalin-triggered intrinsic pathway of apoptosis in CAL 27 human oral cancer cells. Int J Oncol. 2012;41:1683–1692. doi: 10.3892/ijo.2012.1605. [DOI] [PubMed] [Google Scholar]
- 32.Liu CY, Yang JS, Huang SM, Chiang JH, Chen MH, Huang LJ, Ha HY, Fushiya S, Kuo SC. Smh-3 induces G2/M arrest and apoptosis through calcium-mediated endoplasmic reticulum stress and mitochondrial signaling in human hepatocellular carcinoma Hep3B cells. Oncol Rep. 2013;29:751–762. doi: 10.3892/or.2012.2166. [DOI] [PubMed] [Google Scholar]
- 33.Yang JS, Hour MJ, Huang WW, Lin KL, Kuo SC, Chung JG. MJ-29 inhibits tubulin polymerization, induces mitotic arrest, and triggers apoptosis via cyclin-dependent kinase 1-mediated Bcl-2 phosphorylation in human leukemia U937 cells. J Pharmacol Exp Ther. 2010;334:477–488. doi: 10.1124/jpet.109.165415. [DOI] [PubMed] [Google Scholar]
- 34.Yang JS, Lin CA, Lu CC, Wen YF, Tsai FJ, Tsai SC. Carboxamide analog ITR-284 evokes apoptosis and inhibits migration ability in human lung adenocarcinoma A549 cells. Oncol Rep. 2017;37:1786–1792. doi: 10.3892/or.2017.5374. [DOI] [PubMed] [Google Scholar]
- 35.Ho CC, Huang AC, Yu CS, Lien JC, Wu SH, Huang YP, Huang HY, Kuo JH, Liao WY, Yang JS, et al. Ellagic acid induces apoptosis in TSGH8301 human bladder cancer cells through the endoplasmic reticulum stress- and mitochondria-dependent signaling pathways. Environ Toxicol. 2014;29:1262–1274. doi: 10.1002/tox.21857. [DOI] [PubMed] [Google Scholar]
- 36.Yuan CH, Horng CT, Lee CF, Chiang NN, Tsai FJ, Lu CC, Chiang JH, Hsu YM, Yang JS, Chen FA. Epigallocatechin gallate sensitizes cisplatin-resistant oral cancer CAR cell apoptosis and autophagy through stimulating AKT/STAT3 pathway and suppressing multidrug resistance 1 signaling. Environ Toxicol. 2017;32:845–855. doi: 10.1002/tox.22284. [DOI] [PubMed] [Google Scholar]
- 37.Chiang JH, Yang JS, Lu CC, Hour MJ, Chang SJ, Lee TH, Chung JG. Newly synthesized quinazolinone HMJ-38 suppresses angiogenetic responses and triggers human umbilical vein endo-thelial cell apoptosis through p53-modulated Fas/death receptor signaling. Toxicol Appl Pharmacol. 2013;269:150–162. doi: 10.1016/j.taap.2013.03.007. [DOI] [PubMed] [Google Scholar]
- 38.Huang WW, Chiu YJ, Fan MJ, Lu HF, Yeh HF, Li KH, Chen PY, Chung JG, Yang JS. Kaempferol induced apoptosis via endoplasmic reticulum stress and mitochondria-dependent pathway in human osteosarcoma U-2 OS cells. Mol Nutr Food Res. 2010;54:1585–1595. doi: 10.1002/mnfr.201000005. [DOI] [PubMed] [Google Scholar]
- 39.Lu CC, Yang JS, Chiang JH, Hour MJ, Lin KL, Lee TH, Chung JG. Cell death caused by quinazolinone HMJ-38 challenge in oral carcinoma CAL 27 cells: Dissections of endoplasmic reticulum stress, mitochondrial dysfunction and tumor xenografts. Biochim Biophys Acta. 2014;1840:2310–2320. doi: 10.1016/j.bbagen.2014.02.022. [DOI] [PubMed] [Google Scholar]
- 40.King YA, Chiu YJ, Chen HP, Kuo DH, Lu CC, Yang JS. Endoplasmic reticulum stress contributes to arsenic trioxide-induced intrinsic apoptosis in human umbilical and bone marrow mesenchymal stem cells. Environ Toxicol. 2016;31:314–328. doi: 10.1002/tox.22046. [DOI] [PubMed] [Google Scholar]
- 41.Kuo YJ, Yang JS, Lu CC, Chiang SY, Lin JG, Chung JG. Ethanol extract of Hedyotis diffusa willd upregulates G0/G1 phase arrest and induces apoptosis in human leukemia cells by modulating caspase cascade signaling and altering associated genes expression was assayed by cDNA microarray. Environ Toxicol. 2015;30:1162–1177. doi: 10.1002/tox.21989. [DOI] [PubMed] [Google Scholar]
- 42.Wu RS, Liu KC, Tang NY, Chung HK, Ip SW, Yang JS, Chung JG. cDNA microarray analysis of the gene expression of murine leukemia RAW 264.7 cells after exposure to propofol. Environ Toxicol. 2013;28:471–478. doi: 10.1002/tox.20742. [DOI] [PubMed] [Google Scholar]
- 43.Guan F, Ding Y, Zhang Y, Zhou Y, Li M, Wang C. Curcumin suppresses proliferation and migration of MDA-MB-231 breast cancer cells through autophagy-dependent Akt degradation. PLoS One. 2016;11:e0146553. doi: 10.1371/journal.pone.0146553. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 44.Dorée M, Hunt T. From Cdc2 to Cdk1: When did the cell cycle kinase join its cyclin partner? J Cell Sci. 2002;115:2461–2464. doi: 10.1242/jcs.115.12.2461. [DOI] [PubMed] [Google Scholar]
- 45.Chen ZQ, Jie X, Mo ZN. Curcumin inhibits growth, induces G1 arrest and apoptosis on human prostatic stromal cells by regulating Bcl-2/Bax. Zhongguo Zhong Yao Za Zhi. 2008;33:2022–2025. In Chinese. [PubMed] [Google Scholar]
- 46.Srivastava RK, Chen Q, Siddiqui I, Sarva K, Shankar S. Linkage of curcumin-induced cell cycle arrest and apoptosis by cyclin-dependent kinase inhibitor p21(/WAF1/CIP1) Cell Cycle. 2007;6:2953–2961. doi: 10.4161/cc.6.23.4951. [DOI] [PubMed] [Google Scholar]
- 47.Cheng C, Jiao JT, Qian Y, Guo XY, Huang J, Dai MC, Zhang L, Ding XP, Zong D, Shao JF. Curcumin induces G2/M arrest and triggers apoptosis via FoxO1 signaling in U87 human glioma cells. Mol Med Rep. 2016;13:3763–3770. doi: 10.3892/mmr.2016.5037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Berrak O, Akkoc Y, Arisan ED, Coker-Gurkan A, Obakan-Yerlikaya P, Palavan-Unsal N. The inhibition of PI3K and NFkappaB promoted curcumin-induced cell cycle arrest at G2/M via altering polyamine metabolism in Bcl-2 overexpressing MCF-7 breast cancer cells. Biomed Pharmacother. 2016;77:150–160. doi: 10.1016/j.biopha.2015.12.007. [DOI] [PubMed] [Google Scholar]
- 49.Negroni A, Cucchiara S, Stronati L. Apoptosis, necrosis, and necroptosis in the gut and intestinal homeostasis. Mediators Inflamm. 2015;2015:250762. doi: 10.1155/2015/250762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yang Y, Jiang G, Zhang P, Fan J. Programmed cell death and its role in inflammation. Mil Med Res. 2015;2:12. doi: 10.1186/s40779-015-0039-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Friesen C, Fulda S, Debatin KM. Cytotoxic drugs and the CD95 pathway. Leukemia. 1999;13:1854–1858. doi: 10.1038/sj.leu.2401333. [DOI] [PubMed] [Google Scholar]
- 52.Zhang YS, Shen Q, Li J. Traditional Chinese medicine targeting apoptotic mechanisms for esophageal cancer therapy. Acta Pharmacol Sin. 2016;37:295–302. doi: 10.1038/aps.2015.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tameire F, Verginadis II, Koumenis C. Cell intrinsic and extrinsic activators of the unfolded protein response in cancer: Mechanisms and targets for therapy. Semin Cancer Biol. 2015;33:3–15. doi: 10.1016/j.semcancer.2015.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ferri KF, Kroemer G. Organelle-specific initiation of cell death pathways. Nat Cell Biol. 2001;3:E255–E263. doi: 10.1038/ncb1101-e255. [DOI] [PubMed] [Google Scholar]
- 55.Dong Z, Liang S, Hu J, Jin W, Zhan Q, Zhao K. Autophagy as a target for hematological malignancy therapy. Blood Rev. 2016;30:369–380. doi: 10.1016/j.blre.2016.04.005. [DOI] [PubMed] [Google Scholar]
- 56.Gupta SC, Kismali G, Aggarwal BB. Curcumin, a component of turmeric: From farm to pharmacy. Biofactors. 2013;39:2–13. doi: 10.1002/biof.1079. [DOI] [PubMed] [Google Scholar]
- 57.Ye MX, Li Y, Yin H, Zhang J. Curcumin: Updated molecular mechanisms and intervention targets in human lung cancer. Int J Mol Sci. 2012;13:3959–3978. doi: 10.3390/ijms13033959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sánchez-Martínez C, Gelbert LM, Lallena MJ, de Dios A. Cyclin dependent kinase (CDK) inhibitors as anticancer drugs. Bioorg Med Chem Lett. 2015;25:3420–3435. doi: 10.1016/j.bmcl.2015.05.100. [DOI] [PubMed] [Google Scholar]
- 59.Kozakowska M, Szade K, Dulak J, Józkowicz A. Role of heme oxygenase-1 in postnatal differentiation of stem cells: a possible cross-talk with microRNAs. Antioxid Redox Signal. 2014;20:1827–1850. doi: 10.1089/ars.2013.5341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Murakami A. Dose-dependent functionality and toxicity of green tea polyphenols in experimental rodents. Arch Biochem Biophys. 2014;557:3–10. doi: 10.1016/j.abb.2014.04.018. [DOI] [PubMed] [Google Scholar]
- 61.Zheng KM, Zhang J, Zhang CL, Zhang YW, Chen XC. Curcumin inhibits appoptosin-induced apoptosis via upregulating heme oxygenase-1 expression in SH-SY5Y cells. Acta Pharmacol Sin. 2015;36:544–552. doi: 10.1038/aps.2014.166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Cremers NA, Lundvig DM, van Dalen SC, Schelbergen RF, van Lent PL, Szarek WA, Regan RF, Carels CE, Wagener FA. Curcumin-induced heme oxygenase-1 expression prevents H2O2 induced cell death in wild type and heme oxygenase-2 knockout adipose-derived mesenchymal stem cells. Int J Mol Sci. 2014;15:17974–17999. doi: 10.3390/ijms151017974. [DOI] [PMC free article] [PubMed] [Google Scholar]