Abstract
The potential role of osteoblasts in bone and bone marrow (BM) metastases in neuroblastoma (NBL) remains unclear. In this study, we examined the effect of NBL cells on the osteoblastic differentiation of bone marrow-derived mesenchymal stromal cells (BMMSC). We show that the presence of NBL cells enhanced the osteoblastic differentiation of BMMSC driven by bone morphogenetic protein (BMP)-4, in the absence of any effect on NBL cell proliferation. Expression profiles of BMMSC driven towards osteoblastic differentiation revealed an increase in vascular endothelial growth factor A (Vegfa) expression in the presence of NBL cells. We demonstrated that NBL cells increased BMMSC-derived VEGFA mRNA and protein and that this was enhanced by BMP-4. However, in similar conditions, neither the addition of an mVEGFA blocking antibody nor exogenous recombinant (r) mVEGFA affected osteoblastic differentiation. In contrast, siRNA-mediated knock-down of VEGFA in BMMSC prevented osteoblastic differentiation in BMP-4-treated co-cultures, an effect that was not reversed in the presence of rmVEGFA. An analysis of murine bones injected with hNBL cells revealed an increase of mVEGFA producing cells near tumor cells concomitantly with an increase in Vegfa and Runx2 mRNA. This coincided with an increase in osteoclasts, in Rankl/Opg mRNA ratio and with the formation of osteolytic lesions. Thus NBL cells promote osteoblastogenesis in the BM by increasing VEGFA expression in BMMSC. Our study provides a new insight into the role of VEGFA in NBL metastases by pointing to the role of stroma-derived intracrine VEGFA in osteoblastogenesis.
Keywords: Metastasis, Osteoblastogenesis, Vascular endothelial cell growth factor, Neuroblastoma, Mesenchymal cells
Introduction
Bone and bone marrow (BM) are frequent sites of metastasis in pediatric and adult cancers, leading to significant morbidity and mortality. It is estimated that over 350,000 people die with bone metastases each year in the United States (1). Additionally, bone and BM metastases nearly always confer a poorer prognosis and present unique challenges in treatment. It is now clear that interactions between tumor cells and the bone and the BM microenvironment play a central role in promoting the homing of tumor cells to the BM and their ability to form bone metastasis (2–4).
In the bone microenvironment, tumor cells typically accelerate the normal process of bone remodeling, a controlled balance of bone resorption by osteoclasts coupled with bone formation by osteoblasts. One important aspect of bone remodeling is the stimulation of osteoblast differentiation and bone formation by growth factors released from the bone matrix during bone resorption. When tumor cells populate the bone, they accelerate bone turnover and hijack growth factors released from the bone matrix, now at increased rates, to support their replication and survival. Among the best examples of tumor-mediated acceleration of bone turnover is the secretion of parathyroid hormone-related peptide (PTHrP) and the receptor activator of nuclear factor-kB ligand (RANKL) by breast cancer cells (5). Accelerated osteolysis then leads to a “vicious cycle” in which tumor cells fuel bone turnover and are fed by the associated growth factor release.
The cancer-induced vicious cycle is therefore tumor-mediated acceleration of normal bone turnover in which tumor cells disrupt the controlled balance and usurp physiological mechanisms. For example, RANKL, a quintessential factor required for osteoclast differentiation and activation, continues to be present on the surface of osteoblasts and stromal cells, except now in larger numbers, upregulated by tumor cells, and feeding the accelerated bone resorption (1). Osteoblasts therefore play a role in promoting accelerated bone turnover in sites of tumor metastases, whether the ultimate phenotype is osteolytic or osteoblastic. Osteoblasts also play a critical role by forming the osteoblastic niche that is essential for the homing and initial establishment of metastatic tumor cells in the BM microenvironment (6).
In the BM, osteoblasts originate from mesenchymal stromal cells (MSC), a population of pluripotent progenitor cells able to form bone, muscle, fat, and cartilage. Three pathways are known to control the differentiation of BM-derived mesenchymal stromal cells (BMMSC) into osteoblasts: 1) the transformation growth factor-beta (TGF-β)/bone morphogenetic protein (BMP) pathway, 2) the Wnt signaling pathway (7), and 3) the Notch pathway (8). BMP proteins are members of the TGF-β superfamily of proteins that have been implicated in proliferation, differentiation, apoptosis, and migration of BMMSC (9). Among them, BMP-2 and -4 are osteo-inductive cytokines that play an essential early role in the formation of bone and cartilage under the control of Runt related transcription factor 2 (RUNX2), a master transcription factor driving osteoblastogenesis (10).
Neuroblastoma (NBL) is the most common extra-cranial solid tumor of childhood (11, 12) and frequently metastasizes to the bone and BM. The incidence of BM metastasis in patients with stage IV disease is approximately 70% while metastasis to the bone occurs in 56% of children. In contrast, in patients with stage IVs disease (<1 year old), bone marrow metastasis is common (35%), whereas bone metastasis is never seen, thus suggesting the presence of 2 separate processes (13). Metastatic NBL in the bone is primarily an osteolytic process, in which there is an increase in osteoclastogenesis leading to net bone loss. Two pathways which promote osteoclast activation in NBL have been reported (4): 1) RANKL produced by osteoblasts and by NBL cells themselves (13, 14), and 2) interleukin-6 (IL-6) released by BMMSC when in the presence of tumor cells, as reported by our laboratory (15).
Beyond the production of RANKL, little is known of the contribution of osteoblasts to NBL bone metastasis. In this study, we asked whether NBL cells inhibit or promote osteoblastogenesis by examining their interaction with MSC within the context of their differentiation into osteoblasts via RUNX2 and BMP. We provide evidence that as metastatic osteolytic lesions are established, NBL cells promote rather than inhibit osteoblastogenesis.
Materials and Methods
Reagents
Recombinant mouse (m)BMP-4 was from R&D Systems® (Minneapolis, MN). Recombinant mVEGFA protein was from Life Technologies (Grand Island, NY). A neutralizing antibody against mVEGFA (R&D Systems®) was used for blocking experiments at a concentration of 0.15 μg/mL.
Cell culture
The human (h)NBL cell lines CHLA-255 and SK-N-BE(2) were originally provided by Dr. C.P. Reynolds (Texas Tech University). CHLA-255 cells were grown in IMDM containing 20% (vivo) FBS with no additional supplementation. SK-N-BE(2) cells were grown in RPMI containing 10% FBS and L-glutamine. hNBL cell lines were confirmed by genotype analysis using the AmpFISTR Identifier Kit PCR reagents (Life Technologies) and Gene Mapper ID v3.2. The mNBL cell line THMYCN was provided by W.A. Weiss (The University of California San Francisco). The murine MC3T3E1, ST2 and ST2/Rx2Dox cell lines were grown in α-MEM and RPMI, respectively, and supplemented with 10% (v:v) FBS. Primary mBMMSC were isolated from the long bones of 6–8 week-old Balb/C mice and characterized as previously described (16). These cells remained primarily undifferentiated when passaged in culture with a low (<5%) level of spontaneous differentiation into osteoblasts or adipocytes. The cells were passaged within four weeks of initial plating and all experiments were conducted using mBMMSC within the first 20 passages. Cell were grown in IMDM containing 15% (v:v) FBS and 15% (v:v) heat treated horse serum supplemented with hydrocortisone (10−6 mol/L), 2-mercaptoethanol (10−4 mol/L) and 1% (v:v) penicillin-streptomycin.
Cell co-culture assays
Co-culture experiments were performed using the Transwell™ system with a 0.4μm pore insert for a non-contact, non-migration system. In all co-culture experiments, hNBL cells were plated in the inserts at a density of 20,000 cells/cm2, while MSC were plated in the wells at approximately 70% confluence. Cultures were treated with recombinant BMP-4 (final concentration of 100ng/mL) 24 hours before the addition of NBL cells. BMMSC were then analyzed after 4 days of co-culture. In the co-culture experiments used for the gene expression analysis, the previously described ST2/Rx2Dox cell system was used (17). For other experiments, we used a BMP-4 driven system with primary mBMMSC or MC3T3E1 cells.
Cell viability assay
CellTiter-Glo Luminescent cell viability assay (Promega #G7570) was used to evaluate the effect of BMP-4 on NBL cells viability.
Alkaline phosphatase (AP) assays
AP enzymatic activity was assessed qualitatively using the Millipore™ Alkaline Phosphatase Detection Kit (Billerica, MA). Images were quantified by pixel count using the MetaMorph® Microscopy Automation & Image Analysis Software, or by OD reading at 550 nm after dissolution of the AP stain in DMSO. AP enzymatic activity was assessed quantitatively using the Millipore™Quantitative AP ES Characterization Kit.
RNA gene expression analysis
Total RNA was isolated using the QiagenRNeasy® MiniKit (Hilden, Germany) and 1μg was reverse-transcribed using the Invitrogen Superscript III Kit. Gene expression profiling was performed in biological triplicates and technical triplicates using the Illumina MouseRef-8 v2.0 BeadChip™ platform, which contains probes for over 19,100 unique genes derived from the National Center for Biotechnology Information Reference Sequence (NCBI RefSeq) Database. The data were corrected for background using Genome Studio and normalized by shifting to the 75th percentile and transformed to median of control samples. Probe level data was summarized to gene level. To find differentially expressed genes, a t-test (p<0.05) was used and genes were further ranked by fold change. Differentially expressed genes were used to identify the enriched pathways using Ingenuity Pathway Analysis. The data has been deposited in the Gene Expression Omnibus (GEO) of the National Center for Biotechnology Information under the Series accession number (GSE 8541). RNA was isolated from formalin-fixed, paraffin-embedded tissue sections using the RNeasy FFPE kit from Qiagen following manufacturer instructions.
Quantitative RT-PCR
cDNA was created using the Invitrogen Superscript III Kit. Murine primer probes for Gapdh, β-actin, Vegfa, Alpl (AP), Opg, Rankl and Runx2 were from Life Technologies and q-PCR was performed on an ABI Prism Thermal Cycler.
ELISA assays
mVEGFA protein levels in both cell lysates and co-culture supernatants were assessed using a Duo-Set Immunoassay (R&D Systems) or an ELISA Kit (Life Technologies). mVEGFA levels in cell lysates were normalized to the total amount of proteins in the sample.
siRNA-mediated downregulation of VEGFA
Primary mBMMSC were transfected with siRNA directed against mVegfa with Lipofectamine RNAiMax transfection reagent (Life Technologies). siRNA sequences were purchased from Life Technologies (s233656 and s233657) and the BLOCK-It™AlexaFluor Red Fluorescent Control sequence (Life Technologies) was used as both the transfection control and the scramble control per manufacturer’s instructions. siRNA experiments were performed with each sequence individually and pooled. Cells were plated in 12-well plates without antibiotics for at least one day and grown to approximately 50–70% confluence. OPTIMEM reduced serum medium was used and the total transfection time was 18 hours. Co-culture experiments were then performed as described above.
Intrafemoral injections
Eight week-old Nu/Nu mice received intrafemoral injections following a protocol approved by the Institution Animal Care Utilization Committee at the Saban Research Institute of Children’s Hospital Los Angeles and previously described by us (18). Mice were monitored weekly by X-ray (Faxitron) to detect osteolytic lesions and were sacrificed at 5 weeks for histological analysis.
Histology and immunohistochemistry
Hind limbs were dissected and fixed in 4% (v:v) paraformaldehyde overnight at 4°C and decalcified for four weeks at 4°C in a solution containing 5% (w:v) EDTA and 10% (v:v) formalin. The decalcified samples were dehydrated and embedded in paraffin. Serial 5 μm-thick sections were processed for hematoxylin-eosin staining or for immunohistochemistry and tartrate resistant acid phosphatase (TRAP) staining. Tyrosine hydroxylase (TH) and mVEGFA protein expressions were detected after proteinase K (20 μg/ml) antigen retrieval using a rat anti-hTH (Abcam, Cambridge, MA) and a goat anti-mVEGFA antibody (R&D Systems) at 1:750 and 1:50 dilutions, respectively, followed by incubations with biotinylated secondary antibodies at 1:250 dilution (Vector Laboratories, Burlingame, CA) and visualized with an avidin-biotin peroxidase complex Vectastain ABC and ImPACT™DAB peroxidase (Vector Laboratories). TRAP staining was performed using the Acid Phosphatase Leukocyte kit from Sigma-Aldrich (St. Louis, MO). The sections were counterstained with methyl green. Images were acquired with a Zeiss Axiovert 200M microscope equipped with a Hamamatsu ORCA ER digital camera. Quantification of the amount of VEGFA-expressing cells and TRAP-positive cells was performed under 10 and 20× objectives and expressed as the total number of cells per section.
Statistical analysis
Statistical analysis of in vitro studies was performed using the GraphPad Prism® Software Package. For in vivo experiments, VEGFA and TRAP cell counts were examined at the 5 week time point and means were calculated across sections and mice. All values are expressed as mean ± standard deviation (SD). Differences between means were evaluated by ANOVA analysis and the Neuman-Keuls Multiple Comparison Analysis.
Results
NBL cells enhance BMP-4-induced osteoblastic differentiation of BMMSC
To first explore whether NBL cells influenced osteoblast development, we co-cultured hNBL cells in the presence of mBMMSC and examined their ability to induce the differentiation of mBMMSC into osteoblasts over a four-day period. Using AP staining to measure osteoblastogenesis, the results revealed a modest, 1.2 fold increase in the presence of either CHLA-255 or SK-N-BE(2) cells (Fig. 1a, left). However, when a similar experiment was done in the presence of BMP-4, which alone increased AP stain by only 1.1 fold, CHLA-255 and SKNBE(2) cells stimulated AP activity in the co-cultured BMMSC by as much as 1.7 and 1.6 fold, suggesting a cooperative activity between BMP-4 and a NBL-borne factor(s) in promoting osteoblastogenesis (Fig. 1a, right). These observations were confirmed by separate experiments (Fig. 1b) that measured the AP enzymatic activity and demonstrated an absence of increase in AP enzymatic activity in co-cultures in the absence of BMP-4 but as shown in previous experiments, a cooperative effect of NBL cells in the presence of BMP-4. While BMP-4 alone increased AP activity by 2.7 fold, it enhanced AP activity by 4.6 and 5.3 fold in the presence of CHLA-255 and SK-N-BE(2) cells, respectively. Additional evidence supporting a cooperative effect between NBL cells and BMP-4 on osteoblastogenesis was obtained by quantitative gene expression analysis of Alpl by qRT-PCR (Fig. 1c), which revealed an absence of effect of NBL cells on Alpl expression in the absence of BMP-4 but a significant increase in Alpl expression in the presence of BMP-4 and NBL cells. We found that BMP-4 had no effect on the survival of NBL cells (Figure 1d), suggesting that its activity was not related to a stimulation of NBL cell proliferation or inhibition of apoptosis. To further test the effect of co-culture with NBL cells on BMMSC, we examined the expression of 2 genes regulators of osteogenesis, Runx2 and Osteocalcin (Ocn) in BMP-4 treated BMMSC cultured in the presence and absence of CHLA-255 or SK-N-BE(2) cells (Fig. 1e–f). The presence of CHLA-255 or SK-N-BE(2) NBL cells increased the expression of Runx2 by 1.6 and 2.3 fold, respectively, and Ocn by 5 and 4 fold, respectively which is consistent with the increase in AP activity observed previously. From these data, we conclude that although NBL cells are unable to induce osteoblastogenesis in BMMSC alone, they cooperatively enhance BMP-4 induced osteoblastogenesis.
Figure 1. NBL cells enhance BMP-4-induced osteoblastic differentiation of primary mBMMSC.
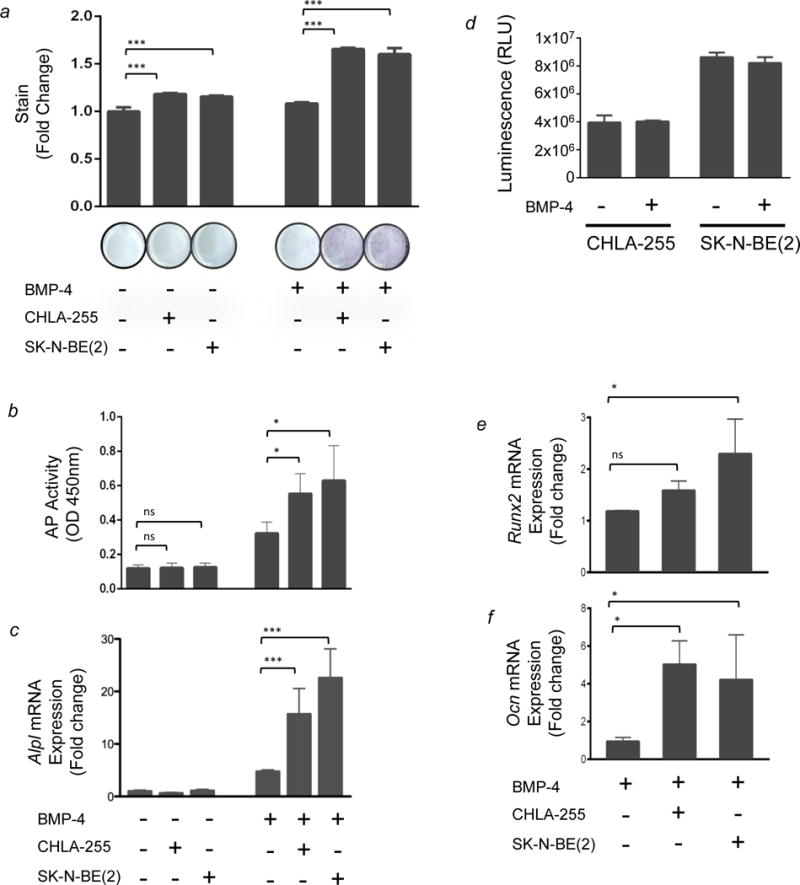
Primary mBMMSC were cultured in the presence or absence of NBL cells in insert wells (0.4 μm pore size) that permit the diffusion of soluble factors but prevent contact with mBMMSC. Cultures were treated with 100 ng/mL rBMP-4 as indicated 24 hours before the addition of NBL cells. BMMSC were then subjected to AP histochemical staining after 4 days. (a) Top: AP staining was quantified and normalized for the average value measured in cultures of mBMMSC cultured without NBL cells and in the absence of BMP-4. Columns 1–3 show that in the absence of BMP-4 but with CHLA-255 or SK-N-BE (2), there was a 1.2 fold increase in AP stain (p< 0.001). Columns 4–6 show that in the presence of rBMP-4 and CHLA-255 or SK-N-BE(2), there was a 1.7 and 1.6 fold increase, respectively (p <0.001). The data represent the mean fold increase of triplicate samples from one experiment representative of four experiments with similar results. Bottom: representative images of wells stained for AP. (b) Cells were cultured as described above and AP activity was measured by an enzymatic assay as described in the Materials and Methods. BMP-4 alone increased AP activity by 2.7 fold. However, it enhanced AP activity by 4.6 and 5.3 fold in the presence of CHLA-255 and SK-N-BE(2) cells, respectively, compared to mBMMSC cultured alone (p<0.05). The data represent the AP enzymatic activity as measured by OD 450 nm from triplicate samples. (c) RT-qPCR analysis of Alpl mRNA expression: ΔCT values were normalized for the average value measured in cultures of mBMMSC cultured alone and in the absence of BMP-4 (column 1). Columns 5 and 6 show a 16 fold increase with CHLA-255 + BMP-4 and a 23 fold increase with SKNBE(2) + BMP-4, p<0.001. (d) NBL tumor cells grown in the absence or presence of BMP-4. Cell cultures were treated with 100ng/mL of BMP-4 and cell viability was assessed after 4 days by Cell Titer Glo Assay®. The y-axis depicts mean (±SD) relative luminescence units (RLU). (e and f) RT-qPCR analysis of Runx2 and Ocn expression in co-culture experiments performed in the presence of BMP-4 in the conditions shown at the bottom of the lower panel. ΔCT values were normalized for the average value measured in cultures of mBMMSC cultured alone and in the absence of BMP-4. Columns 2 and 3 show a 1.6 and 2.3 fold increase in Runx2 expression with the addition of CHLA-255 and SK-N-BE(2), respectively. Similarly, there is a 5 and 4 fold increase in Ocn expression with the addition of CHLA-255 and SK-N-BE(2), respectively. The data are from triplicate samples in one experiment. In all panels, ns = not significant; * = p<0.05; *** = p<0.001.
Co-culture of BMMSC with NBL tumor cells leads to differential gene expression in BMMSC driven towards osteoblastogenesis
To shed light on the mechanism by which NBL cells enhance osteoblastogenesis, we profiled mRNA expression in mBMMSC induced to osteoblastic differentiation in the presence versus absence of NBL cells. To facilitate robust, reproducible results, we employed the ST2/Rx2dox cell line, in which doxycycline-induced Runx2 expression promotes differentiation of BM-derived pluripotent MSC into osteoblasts (17). We first validated the doxycycline-induced phenotype of these cell by showing that treatment of these cells with doxycycline in the absence of NBL cells resulted in the upregulation of several genes specifically expressed by osteoblasts during Runx2-induced differentiation, such as Alpl, Ocn1 and 2, and Sp7 (Osterix) (p<0.05, Supplemental Fig. S1). In contrast, genes involved in muscle differentiation were down-regulated. We then performed a comparative analysis of the gene expression of Runx2-induced ST2/Rx2Dox cells cultured in the absence or presence of each of three NBL cell lines – CHLA-255, SKNBE(2), or THMYCN. Of approximately 19,000 genes represented on the microarray chip, a subset of 493 genes were found to be differentially regulated (p<0.05) in doxycycline-treated ST2/Rx2Dox cells cultured in the presence versus the absence of NBL cells. To examine a global pattern of differential expression among these genes, a hierarchical clustering using Euclidean distance and average linkage revealed a clear separation between mBMMSC cultured in the absence of any NBL cells as compared to those cultured in the presence of CHLA-255, SKNBE(2) or THMYCN (Figure 2a). The results are in agreement with our previous studies which suggest a cooperative effect between NBL cells and BMP-4. To understand the biological implication of such differential expression, we carried out detailed pathway analysis in Ingenuity Pathway Analysis (IPA). IPA generates networks based on functional interactions reported in the literature and then associates these networks into biological pathways based on statistical significance and assigns a p value to each pathway. Our global canonical pathway analysis suggested the role of tissue factors as the most significant pathway enriched in the set of genes which was differentially expressed in the presence of NBL cells (p=2.1 × 103, Fig. 2b). Interestingly, of the eight genes in this group, VEGFA has been widely reported to be associated with angiogenesis and progression of cancer and has been a therapy target, including in neuroblastoma (19–21). Furthermore, because VEGFA has been recently reported to modulate osteoblastogenesis (22, 23), we further investigated the detailed role of VEGFA in the NBL-BMMSC interaction.
Figure 2. Co-culture of ST2/Rx2Dox murine mesenchymal cells with NBL tumor cells leads to differential gene expression in mesenchymal cells driven towards osteoblastogenesis.
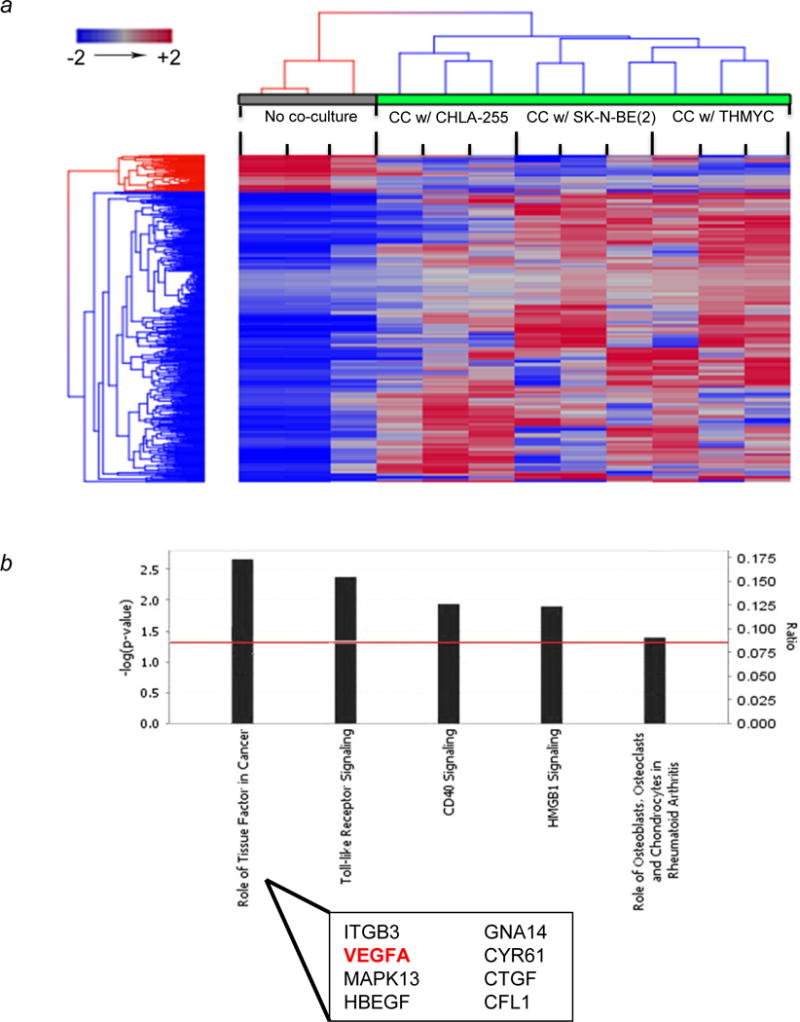
(a) Hierarchical clustering of differentially expressed genes (p<0.05) from ST2/Rx2Dox cells show a distinct pattern between co-culture (green bar) and control (gray bar). ST2/Rx2Dox cells were co-cultured with each of the three NBL cell lines as indicated in Materials and Methods. Euclidean distance and average linkage were used for clustering. Each single column represents technical triplicates. Each group of three columns represents biological triplicates for the same condition. (b) Top canonical pathways involved by differentially expressed genes in co-culture (p<0.05). The significance of canonical pathways was determined by Benjamin-Hocheberg corrected p value (indicated by red line as threshold). The ratio represents the number of differentially expressed genes involved in each canonical pathway. ITGB3, Integrin beta-3 glycoprotein IIIa; MAPK13, Mitogen-activated protein kinase 13; HBEGF, Heparin binding EGF-life growth factor; GNA14, Guanine nucleotide binding protein-14; CYR61, Cysteine-rich angiogenic inducer 61; CTGF, Connective Tissue Growth Factor; CFL1, Cofilin 1.
NBL cells induce an increase in mVEGFA expression in BMMSC
The increased expression of Vegfa mRNA in stromal cells co-cultured in the presence versus absence of NBL cells which was detected by gene array was then verified in primary mBMMSC cultured in the presence of hNBL cells. As in Figure 1, experiments were performed both in the presence and absence of BMP-4 (Fig. 3a). An RT-qPCR analysis revealed a ~2-fold increase in Vegfa mRNA expression in mBMMSC when co-cultured with hNBL cells compared to mBMMSC cultured alone. By ELISA, we observed an increase in mVEGFA protein expression in BMMSC co-cultured in the presence of NBL cells that was enhanced by BMP-4 (Figs. 3b and c). The increase in mVEGFA protein was noted in both the co-culture medium and the cell lysate. Thus, mVEGFA expression was increased most when BMMSC were cultured in the presence of BMP-4 and NBL cells, an effect which parallels the differentiation effect we observed in which co-culture with NBL cells enhanced BMP-4 induced osteoblastogenesis in mBMMSC.
Figure 3. NBL cells induce an increase in mVEGFA expression in primary mBMMSC.
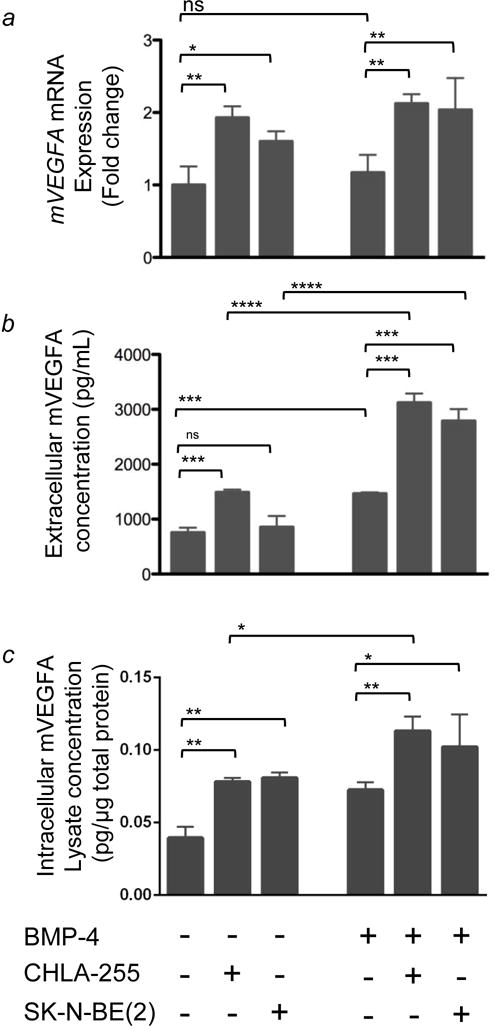
BMMSC were co-cultured in the presence or absence of human NBL cells as described in Figure 1. (a) mVegfa mRNA expression in BMMSC was quantified by qRT-PCR. Columns 5 and 6 show a 2 fold increase in mVEGFA in BMMSC cultured in the presence CHLA-255+BMP-4 and SKNBE(2)+BMP-4, respectively, compared to BMMSC cultured in the absence of NBL cells. The data represent the mean fold change (±SD) of gene expression from triplicate samples (n=3) and represent one of two experiments showing similar results. (b) Extracellular mVEGFA protein levels in the medium of BMMSC cultured in conditions indicated above. The data represent the mean protein level (±SD) of triplicate samples and is representative of similar results from 3 experiments. (c) Intracellular mVEGFA protein levels in BMMSC lysate expressed as pg/μg of total protein. Data is from triplicate samples from one experiment. In all panels, * = p<0.05, ** = p<0.01, *** = p<0.001, **** = p<0.0001
Role of intracellular VEGFA in osteoblastogenesis
We then tested whether exogenous recombinant mVEGFA added to MSC cultured alone could substitute for NBL cells in enhancing osteoblastogenesis. For these experiments we used MC3T3E1 cells that are predetermined to osteoblastogenesis, easier to transfect and responded better to BMP-4. This experiment (Fig. 4a) indicated that the addition of exogenous rmVEGFA had no effect on osteoblastogenesis, whether the cells were cultured in the absence or presence of BMP-4, suggesting that extracellular VEGFA may not be important. Consistent with these results, the addition of a functional blocking antibody against mVEGFA did not inhibit osteoblastogenesis when added to BMMSC co-cultured with CHLA-255 NBL cells in the presence of BMP-4 (Fig. 4b). Together, these data suggested that extracellular VEGFA does not mediate NBL-induced osteoblast differentiation from co-cultured BMMSC. Instead it raised the possibility that intracellular VEGFA plays a role, as recently suggested (23). To investigate this possibility, we knocked down (KD) mVegfa expression in mBMMSC by siRNA and examined the ability of NBL cells to enhance AP expression in the presence of BMP-4. We first showed that transfection of BMMSC with siRNA duplexes targeting Vegfa reduced mVEGFA protein expression when compared with a scramble sequence control. In particular, the siRNA virtually blocked the NBL-induction of VEGFA in the co-culture setting (Fig. 4c). VEGFA KD mBMMSC and scramble controls were then tested for AP expression in the absence or presence of NBL cells. In these experiments, mVEGFA knockdown significantly suppressed NBL-enhanced osteoblastogenesis, as shown by a ~2 fold decrease in AP activity (Fig. 4d). We then tested whether exogenous recombinant VEGFA could rescue the inhibition of osteoblastogenesis caused by Vegfa siRNA described above. In these experiments, the addition of recombinant VEGFA to cells transfected with Vegfa siRNA did not rescue the VEGFA KD phenotype in any of the experimental conditions (Fig.4e). Finally, we examined the potential link between VEGFA and RUNX2, a downstream transcription factor regulated by VEGFA (23). In BMMSC co-cultured with CHLA-255 and treated with BMP-4, Vegfa siRNA transfection led to a statistically significant decrease in Vegfa mRNA expression compared to transfection with the scramble sequence and a 20% decrease in Runx2 mRNA expression that was not statistically significant but not inconsistent with Runx2 being a downstream target of Vegfa as reported by others (23) (Fig. 4f).
Figure 4. Role of intracellular VEGFA in NBL-enhanced osteoblastogenesis.
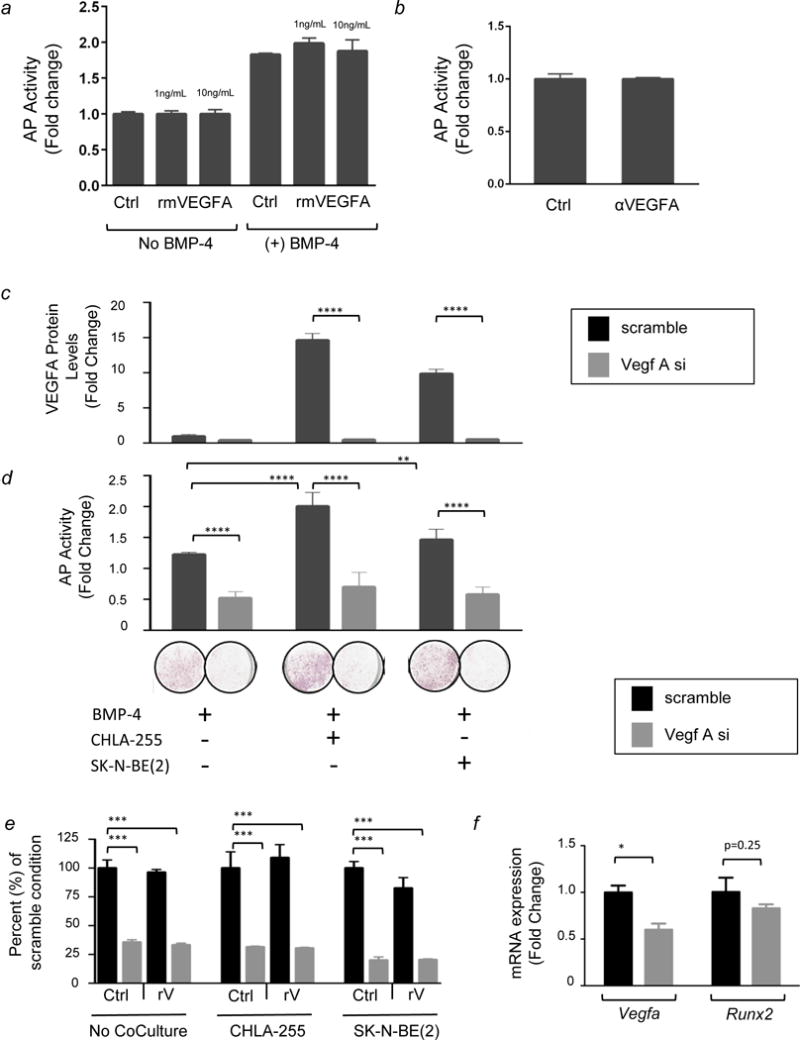
(a) AP activity in MC3T3E1 cells treated with rmVEGFA in the presence or absence of BMP-4 as indicated. The data represent the mean fold increase (±SD) when compared to untreated MC3T3E1 cells (column 1) of triplicate samples and are representative of three experiments showing similar results. (b) AP activity in MC3T3E1 cells treated with BMP-4 and co-cultured with NBL cells (CHLA-255) in the absence or presence of a functional blocking anti-mVEGFA antibody. The data represent the mean fold change (±SD) in triplicate samples from one experiment and are representative of three experiments showing similar results. (c) Extracellular mVEGFA protein expression in MC3T3E1 cells transfected with siRNA and cultured in the conditions indicated above. The data represent the mean fold change (±SD) compared to MC3T3E1 cells transfected with the scramble sequence and cultured alone in the presence of BMP-4. Samples were tested in triplicate and experiments were repeated twice. The data shown are mean values from one representative experiment. (d) Top: AP activity in MC3T3E1 cells in conditions described above. The data represent the mean fold change (±SD) compared to MC3T3E1 cells cultured alone in absence of BMP-4. Samples were tested in triplicate and experiments were repeated twice. Bottom: Representative photograph of wells stained for AP protein. The data are representative of one experiment repeated three times with triplicate samples. (e) AP expression in MC3T3E1 cells in the same conditions shown above. Data are expressed as percent of the AP activity in cells transfected with the scramble siRNA in each culture condition, respectively. (f) mRNA expression of Vegfa and Runx2 in MC3T3E1 cells treated with BMP-4, co-cultured with CHLA-255, and transfected with the scramble sequence or with Vegfa siRNA. Data represent duplicate samples from one experiment. In all panels: ** = p<0.01, **** = p<0.0001
Increased expression of mVEGFA in stromal cells in the bone of mice injected with NBL cells coincides with osteoclast activation and osteolysis
To test the relevance of observations made in the co-culture systems in vivo, we modeled NBL bone metastasis by injecting CHLA-255 NBL cells into the femur of nu/nu mice as previously described (18). Histological analysis of the femur confirmed the presence of NBL cells, which stain positive for TH (Fig. 5). Serial sections were then stained with anti-mVEGFA antibody, and we noted the presence of mVEGFA positive cells in the proximity of TH positive tumor cells (Fig. 5) that were not seen in control femurs without tumor (see mock control in Fig. 6a). Quantification of the number of VEGFA expressing cells revealed a significant increase in the number of VEGFA expressing cells around week 5 after tumor injection (Fig. 6a–c). Histological and X-rays assessment of bone resorption demonstrated a parallel increase in the number of TRAP-stained (Fig. 6c–e) as well as extensive osteolysis in the femurs injected with NBL cells as compared to saline control (Fig. 6 g–h). Consistent with this increase in osteoclast activity, we documented a change in the Rankl: Opg mRNA ratio between 0.89 ± 0.33 in mock injected femurs and 1.82 ± 0.41 in NBL cells injected femurs (Fig. 6i). We also documented a 2 fold increase in Vegfa mRNA expression in tumor cell injected versus saline (mock) injected sections, associated with a similar increase in Runx2 mRNA (Fig 6j). These in vivo experiments suggest that NBL cells induce VEGFA expression in host bone marrow stromal cells. In turn, these cells may undergo osteoblastic differentiation, consequentially pre-osteoclasts in the bone microenvironment.
Figure 5. Expression of mVEGFA in bones injected with human CHLA-255 NBL cells.
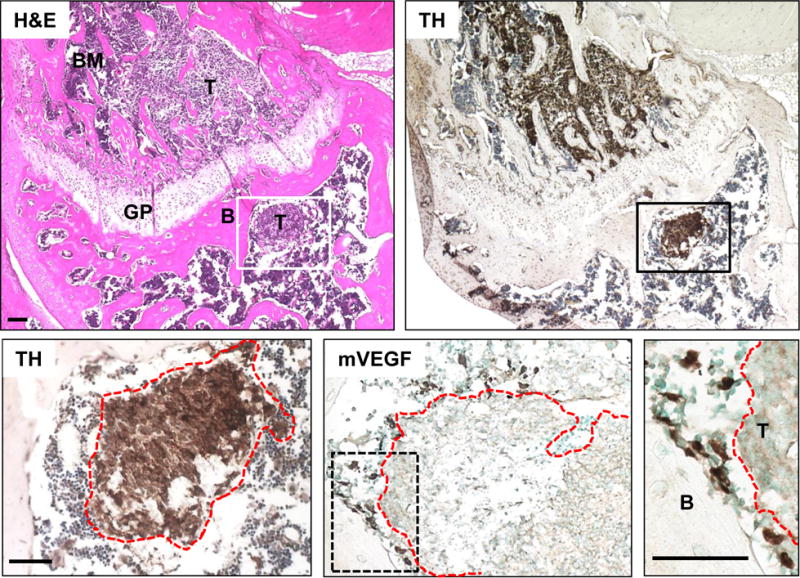
H&E stain of a section through a femur three weeks post injection with CHLA-255 cells in the bone marrow cavity (upper left). NBL cells were detected with an anti-tyrosine hydroxylase (TH) antibody (upper right). Detail of a tumor nodule outlined in red (lower left). Immunohistochemistry on an adjacent section with an anti-mouse VEGFA antibody, positive cells from the host tissue are surrounding the tumor (lower middle). Detail showing VEGFA-positive cells in close proximity of the bone, and surrounding the tumor border (lower right).
B, bone; T, tumor; GP, growth plate. Scale bar=50μm.
Figure 6. Expression of mVEGFA in bones injected with human CHLA-255 NBL cells coincides with osteoclast activation leading to osteolysis.
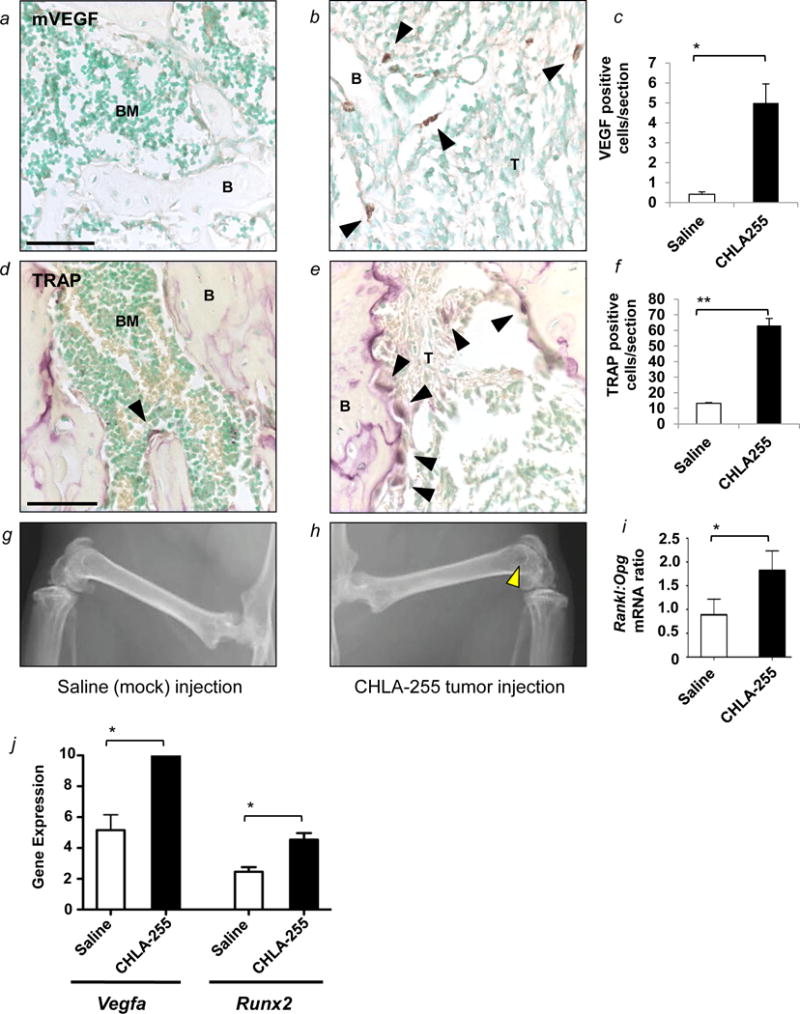
(a and b) Representative images of the distribution of mVEGFA-positive cells present in the BM cavity of mice injected with saline (mock) or CHLA-155 cells. Black arrowheads point to isolated positive cells. (c) Mean number (±SD) of mVEGFA positive cells per section from 3 sections per mouse from 2 mice. (d and e) Representative images of the distribution of TRAP-positive cells in bone sections stained for TRAP. Black arrowheads point to mature osteoclasts. (f) Mean number (±SD) of TRAP-positive cells per tissue section from 2–6 sections per mouse from 2 mice. (g and h) Representative radiographs from the femurs of mice from each group (each side). The yellow arrowhead points to osteolysis in the distal femur. (i) Mean (±SD) Rankl:Opg ratio in mRNA extracted from tissue sections in the 2 groups. (j) RT-qPCR analysis of Vegfa and Runx2 mRNA expression (ΔCT values) in mock (saline) and tumor (CHLA255) injected femurs. In panels: B = bone; T = tumor cells; V = vessel. Scale bars=50μm. * = p<0.05, ** = p<0.01
Discussion
In this study, we examined the influence of NBL cells on BMMSC during osteoblastogenesis and found that NBL cells act cooperatively with BMP-4 in enhancing osteoblastogenesis. Global gene expression analysis of BMMSC induced to an osteoblastic fate identified distinct gene expression patterns in BMMSC when cultured in the presence versus absence of NBL cells, and biological analysis pointed to Vegfa as a likely player in this process. Our data then demonstrated that NBL cells induce an increase in stromal Vegfa which then leads to an increase in both intracellular and extracellular VEGFA protein. This induction of Vegfa expression was necessary for NBL cells to enhance osteoblastogenesis because suppression of Vegfa expression prevented an increase in osteoblastogenesis in BMP-4-treated BMMSC cultured in the presence of NBL cells. We also demonstrated that it is intracellular, and not extracellular, VEGFA which is necessary for tumor enhancement of BMP-4 driven osteoblastogenesis. In vivo experiments in mice then revealed an increase in mVEGFA expressing host cells that surrounded NBL tumor cells. This coincided with an increase in osteoclastogenesis and the development of X-rays documented osteolytic lesions. These results bring to light a novel element in the interaction between NBL cells and BMMSC within the BM microenvironment beyond the stimulation of IL-6 production by BMMSC which we previously reported (15, 24). In this interaction, NBL cells stimulate the production of intracellular VEGFA in BMMSC thereby enhancing their differentiation into osteoblasts.
The role of osteoblastogenesis in cancer remains incompletely understood as both osteoblast promoting and osteoblast inhibiting activities of tumor cells have been reported (18, 25, 26). Here we demonstrate that NBL cells, although unable to induce osteoblastogenesis on their own, enhanced the process in the presence of an osteoblast-inducing factor such as BMP-4. The enhancement of osteoblastogenesis in bone metastasis has been described in prostate cancer, in which osteoblasts are activated following the homing of tumor cells to the bone, resulting in excessive and irregular bone deposition (27). The source of BMP-4 in the bone was not examined in this study. However, it is endogenously produced by stromal cells and present in the bone matrix where it is released upon matrix degradation by osteoclasts (1, 28, 29). Therefore the concomitant increase in VEGFA positive cells and osteoclasts in the bone of mice injected with NBL cells is consistent with a possible contribution of BMP-4 released from the bone matrix to osteoblastogenesis in vivo. The stimulation of osteoblasts by prostate cancer cells is mediated by activation of the Wnt signaling pathway and the production of BMP-2 and TGF-β by tumor cells (30). The role of Wnt in prostate cancer metastases has been well described as early inhibition of the Wnt pathway leads to osteolysis and establishment of the tumor which is then followed by decreased levels of Wnt inhibitors (dickkopfs and soluble frizzled related receptors) and an osteoblastic response (27). In contrast, in osteolytic metastases seen in breast cancer and multiple myeloma, tumor cells activate osteoclasts and can produce inhibitors of osteoblastogenesis (5, 31). Indeed, unilaterally targeting osteoclast activation with drugs such as bisphosphonates does not completely prevent the progression of bone lesions (32, 33) and in both multiple myeloma and breast cancer, it has been shown that tumor cells also produce factors that inhibit osteoblasts such as dickkopf 1 (DKK1) (25, 34). Additionally, breast cancer cells suppress osteoblast differentiation (35) and induce osteoblast apoptosis via the production of TGF-β (36). Our data show an enhancement of osteoblast differentiation by NBL cells which involves an up regulation of intracellular VEGFA in MSC, a novel role of host-derived VEGFA in metastatic cancer.
Tumor-derived extracellular VEGFA and its role in angiogenesis and NBL cell survival has been the main focus of attention in the past (21), but the role of host, stromal-derived VEGFA is less described. Our data show that not only tumor cells but also stromal cells can be an important source of VEGFA in the tumor-host microenvironment. This has been previously shown in chronic lymphocytic leukemia (CLL), where CLL cells stimulate the production of VEGFA by BMMSC which in turn provides VEGFR2 expressing CLL cells a survival advantage and resistance to apoptosis (37). In breast cancer, epithelial and stromal cultures derived from human breast carcinomas showed increased expression of stromal-produced VEGF compared to normal breast epithelial and stromal cultures (38). Whether stromal-derived VEGFA secreted in the extracellular milieu by BMMSC has a similar protective effect on NBL cells is presently unknown but investigated in our laboratory. Here we describe a novel, intracellular role for VEGFA in the tumor-host microenvironment, in which it is intracellular, not extracellular, VEGFA that affects the bone balance in osteolytic metastasis, potentially creating a favorable osteoblastic niche which ultimately leads to osteolytic metastasis.
It has been recently reported that VEGFA is expressed in osteoblast precursor cells and has osteo-inductive properties (22, 39). VEGFA knockout mice have distinct defects in skeletal development (40, 41), consistent with VEGFA playing a role in normal skeletal development and mesenchymal-derived VEGFA preferentially have been shown to drive osteoblastogenesis and suppress adipogenesis in vitro and in vivo, via an intracrine VEGFA-mediated signaling loop (23). This intracrine loop includes the stimulation of the Runx2 transcription factor and the inhibition of the PPARPγ transcription factor, which promotes adipocyte differentiation and drives osteoclastogenesis in vitro and in vivo. Consistently, we found a similar link between Vegfa and Runx2 expression in our experiments. Our data demonstrate that osteolytic cancer cells may also use such an intracrine loop to enhance osteoblastogenesis in the bone marrow microenvironment.
Finally, our in vivo experiments shed some light onto the effect of tumor on the relation between osteoblastogenesis and osteoclast activation in the BM microenvironment. Our data demonstrate that the increase in osteoclasts and radiographically detectable osteolysis coincides with the increase in VEGFA expressing host cells in close proximity to tumor cells. This suggests a potential relationship between the production of host-derived VEGFA and the stimulation of osteoclastogenesis. Although the exact nature of the VEGFA producing stromal cells remains to be determined, the data indicate that these cells may be critical for the early establishment of NBL cells in the BM microenvironment; however, their role may later become secondary, when the production of OAFs like IL-6 becomes dominant (24).
Presently it is unknown what factor(s) are released by NBL cells leading to the upregulation of VEGFA in BMMSC. We have previously shown that Galectin-3-Binding Protein (Gal3-BP), a self-adhesive glycoprotein that promotes cell adhesion to matrix proteins, is secreted by neuroblastoma cells and leads to an upregulation of Interleukin-6 (IL-6) by BMMSC (42, 43). It has been recently shown that the knockdown of the Gal3-BP in breast cancer cells leads to reduced VEGF expression which is restored by providing recombinant Gal3-BP (44). It is thus conceivable that Gal-3BP produced by NBL cells not only up regulates IL-6 but also VEGF in BMMSC. This possibility is presently explored in our laboratory.
Our study points to the potential use of VEGFA inhibitors as modulators not only of tumor-derived VEGFA but also of microenvironment-derived VEGFA expression. In many cases, inhibition of VEGFA receptors through blocking antibodies or inhibition of secreted VEGFA through neutralizing antibodies have not reversed the survival advantage provided by VEGFA producing tumor cells (9, 22, 23). Strategies which block VEGFA in an intracrine manner may be as important as strategies blocking extracellular VEGFA.
Supplementary Material
Novelty and impact.
The role of osteoblasts in bone and marrow metastases is not fully defined. Here we show that osteoblastogenesis is enhanced by tumor cells in a cooperative manner with bone morphogenetic protein 4, an osteo-inductive cytokine. Further, this effect requires the upregulation of intracellular, host-derived VEGFA by tumor cells and coincides with osteoclast activation in vivo. Our data suggest that strategies that target intracrine VEGFA may be as important as strategies that block extracellular VEGFA.
Acknowledgments
The authors thank Dr. G. Esteban Fernandez for his excellent technical support with image analysis and Ms. J. Rosenberg for typing and editing the manuscript. J. HaDuong contributed to the design of the study, performed the experiments described in Figures 1 through 3, contributed to the data described in Figures 4 and 6, analyzed and interpreted the data, and wrote the manuscript. L. Blavier contributed to the data described in Figures 1 and 3, performed the experiments described in Figure 4 and the in vivo work described in Figures 5 and 6 and critically reviewed and edited the manuscript. V. Punj contributed to the analysis of the experiment conducted in Figure 2. S. Baniwal and B. Frenkel contributed to the design of the experiment in Figure 2 as well as the reviewing and editing of the manuscript. J. Malvar and R. Sposto contributed to the statistical analysis of the data except for that in Figure 2. Y.A. DeClerck generated the concept, contributed to the design, supervised the performance of the experiments and the data analysis, contributed to the writing, and critically reviewed and edited the manuscript.
Funding: Josephine HaDuong was a recipient of the Ruth L. Kirschstein National Research Service Award (NRSA) (2-T32-CA 09659 to YAD). This work was in part supported by grant P01-CA81403 (Project 1 to YAD).
Abbreviations
- AP
alkaline phosphatase
- BM
bone marrow
- BMMSC
bone marrow-derived mesenchymal stromal cells
- BMP
bone morphogenetic protein
- h
human
- m
mouse
- MSC
mesenchymal stromal cells
- NBL
neuroblastoma
- OPG
Osteoprotegerin
- RANKL
receptor activator of nuclear factor kB ligand
- RUNX2
Runt related transcription factor 2
- TME
tumor microenvironment
- TRAP
tartrate resistant acid phosphatase
- VEGFA
vascular endothelial growth factor A
References
- 1.Roodman GD. Mechanisms of bone metastasis. The New England journal of medicine. 2004;350(16):1655–64. doi: 10.1056/NEJMra030831. [DOI] [PubMed] [Google Scholar]
- 2.Weilbaecher KN, Guise TA, McCauley LK. Cancer to bone: a fatal attraction. Nature reviews Cancer. 2011;11(6):411–25. doi: 10.1038/nrc3055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Guise TA, Mundy GR. Cancer and bone. Endocrine reviews. 1998;19(1):18–54. doi: 10.1210/edrv.19.1.0323. [DOI] [PubMed] [Google Scholar]
- 4.Sohara Y, Shimada H, DeClerck YA. Mechanisms of bone invasion and metastasis in human neuroblastoma. Cancer letters. 2005;228(1–2):203–9. doi: 10.1016/j.canlet.2005.01.059. [DOI] [PubMed] [Google Scholar]
- 5.Roodman GD. Biology of osteoclast activation in cancer. Journal of clinical oncology: official journal of the American Society of Clinical Oncology. 2001;19(15):3562–71. doi: 10.1200/JCO.2001.19.15.3562. [DOI] [PubMed] [Google Scholar]
- 6.Mendez-Ferrer S, Michurina TV, Ferraro F, Mazloom AR, Macarthur BD, Lira SA, Scadden DT, Ma’ayan A, Enikolopov GN, Frenette PS. Mesenchymal and haematopoietic stem cells form a unique bone marrow niche. Nature. 2010;466(7308):829–34. doi: 10.1038/nature09262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hall CL, Keller ET. The role of Wnts in bone metastases. Cancer metastasis reviews. 2006;25(4):551–8. doi: 10.1007/s10555-006-9022-2. [DOI] [PubMed] [Google Scholar]
- 8.Sethi N, Dai X, Winter CG, Kang Y. Tumor-derived JAGGED1 promotes osteolytic bone metastasis of breast cancer by engaging notch signaling in bone cells. Cancer cell. 2011;19(2):192–205. doi: 10.1016/j.ccr.2010.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gazit D, Ebner R, Kahn AJ, Derynck R. Modulation of expression and cell surface binding of members of the transforming growth factor-beta superfamily during retinoic acid-induced osteoblastic differentiation of multipotential mesenchymal cells. Molecular endocrinology. 1993;7(2):189–98. doi: 10.1210/mend.7.2.8385738. [DOI] [PubMed] [Google Scholar]
- 10.Phimphilai M, Zhao Z, Boules H, Roca H, Franceschi RT. BMP signaling is required for RUNX2-dependent induction of the osteoblast phenotype. Journal of bone and mineral research: the official journal of the American Society for Bone and Mineral Research. 2006;21(4):637–46. doi: 10.1359/jbmr.060109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Maris JM, Hogarty MD, Bagatell R, Cohn SL. Neuroblastoma. Lancet. 2007;369(9579):2106–20. doi: 10.1016/S0140-6736(07)60983-0. [DOI] [PubMed] [Google Scholar]
- 12.Cheung NK, Dyer MA. Neuroblastoma: developmental biology, cancer genomics and immunotherapy. Nature reviews Cancer. 2013;13(6):397–411. doi: 10.1038/nrc3526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Granchi D, Amato I, Battistelli L, Avnet S, Capaccioli S, Papucci L, Donnini M, Pellacani A, Brandi ML, Giunti A, Baldini N. In vitro blockade of receptor activator of nuclear factor-kappaB ligand prevents osteoclastogenesis induced by neuroblastoma cells. International journal of cancer Journal international du cancer. 2004;111(6):829–38. doi: 10.1002/ijc.20308. [DOI] [PubMed] [Google Scholar]
- 14.Michigami T, Ihara-Watanabe M, Yamazaki M, Ozono K. Receptor activator of nuclear factor kappaB ligand (RANKL) is a key molecule of osteoclast formation for bone metastasis in a newly developed model of human neuroblastoma. Cancer research. 2001;61(4):1637–44. [PubMed] [Google Scholar]
- 15.Sohara Y, Shimada H, Minkin C, Erdreich-Epstein A, Nolta JA, DeClerck YA. Bone marrow mesenchymal stem cells provide an alternate pathway of osteoclast activation and bone destruction by cancer cells. Cancer research. 2005;65(4):1129–35. doi: 10.1158/0008-5472.CAN-04-2853. [DOI] [PubMed] [Google Scholar]
- 16.Bergfeld SA, Blavier L, Declerck YA. Bone Marrow-Derived Mesenchymal Stromal Cells Promote Survival and Drug Resistance in Tumor Cells. Molecular cancer therapeutics. 2014 doi: 10.1158/1535-7163.MCT-13-0400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Baniwal SK, Shah PK, Shi Y, Haduong JH, Declerck YA, Gabet Y, Frenkel B. Runx2 promotes both osteoblastogenesis and novel osteoclastogenic signals in ST2 mesenchymal progenitor cells. Osteoporosis international: a journal established as result of cooperation between the European Foundation for Osteoporosis and the National Osteoporosis Foundation of the USA. 2012;23(4):1399–413. doi: 10.1007/s00198-011-1728-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sohara Y, Shimada H, Scadeng M, Pollack H, Yamada S, Ye W, Reynolds CP, DeClerck YA. Lytic bone lesions in human neuroblastoma xenograft involve osteoclast recruitment and are inhibited by bisphosphonate. Cancer research. 2003;63(12):3026–31. [PubMed] [Google Scholar]
- 19.Klement G, Baruchel S, Rak J, Man S, Clark K, Hicklin DJ, Bohlen P, Kerbel RS. Continuous low-dose therapy with vinblastine and VEGF receptor-2 antibody induces sustained tumor regression without overt toxicity. The Journal of clinical investigation. 2000;105(8):R15–24. doi: 10.1172/JCI8829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kerbel RS, Viloria-Petit A, Klement G, Rak J. ‘Accidental’ anti-angiogenic drugs. anti-oncogene directed signal transduction inhibitors and conventional chemotherapeutic agents as examples. European journal of cancer. 2000;36(10):1248–57. doi: 10.1016/s0959-8049(00)00092-7. [DOI] [PubMed] [Google Scholar]
- 21.Das B, Yeger H, Tsuchida R, Torkin R, Gee MF, Thorner PS, Shibuya M, Malkin D, Baruchel S. A hypoxia-driven vascular endothelial growth factor/Flt1 autocrine loop interacts with hypoxia-inducible factor-1alpha through mitogen-activated protein kinase/extracellular signal-regulated kinase 1/2 pathway in neuroblastoma. Cancer research. 2005;65(16):7267–75. doi: 10.1158/0008-5472.CAN-04-4575. [DOI] [PubMed] [Google Scholar]
- 22.Tang W, Yang F, Li Y, de Crombrugghe B, Jiao H, Xiao G, Zhang C. Transcriptional regulation of Vascular Endothelial Growth Factor (VEGF) by osteoblast-specific transcription factor Osterix (Osx) in osteoblasts. The Journal of biological chemistry. 2012;287(3):1671–8. doi: 10.1074/jbc.M111.288472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu Y, Berendsen AD, Jia S, Lotinun S, Baron R, Ferrara N, Olsen BR. Intracellular VEGF regulates the balance between osteoblast and adipocyte differentiation. The Journal of clinical investigation. 2012;122(9):3101–13. doi: 10.1172/JCI61209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ara T, Song L, Shimada H, Keshelava N, Russell HV, Metelitsa LS, Groshen SG, Seeger RC, DeClerck YA. Interleukin-6 in the bone marrow microenvironment promotes the growth and survival of neuroblastoma cells. Cancer research. 2009;69(1):329–37. doi: 10.1158/0008-5472.CAN-08-0613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tian E, Zhan F, Walker R, Rasmussen E, Ma Y, Barlogie B, Shaughnessy JD., Jr The role of the Wnt-signaling antagonist DKK1 in the development of osteolytic lesions in multiple myeloma. The New England journal of medicine. 2003;349(26):2483–94. doi: 10.1056/NEJMoa030847. [DOI] [PubMed] [Google Scholar]
- 26.Kakonen SM, Mundy GR. Mechanisms of osteolytic bone metastases in breast carcinoma. Cancer. 2003;97(3 Suppl):834–9. doi: 10.1002/cncr.11132. [DOI] [PubMed] [Google Scholar]
- 27.Hall CL, Bafico A, Dai J, Aaronson SA, Keller ET. Prostate cancer cells promote osteoblastic bone metastases through Wnts. Cancer research. 2005;65(17):7554–60. doi: 10.1158/0008-5472.CAN-05-1317. [DOI] [PubMed] [Google Scholar]
- 28.Cao X, Chen D. The BMP signaling and in vivo bone formation. Gene. 2005;357(1):1–8. doi: 10.1016/j.gene.2005.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nishimura R, Hata K, Matsubara T, Wakabayashi M, Yoneda T. Regulation of bone and cartilage development by network between BMP signalling and transcription factors. Journal of biochemistry. 2012;151(3):247–54. doi: 10.1093/jb/mvs004. [DOI] [PubMed] [Google Scholar]
- 30.Logothetis CJ, Lin SH. Osteoblasts in prostate cancer metastasis to bone. Nature reviews Cancer. 2005;5(1):21–8. doi: 10.1038/nrc1528. [DOI] [PubMed] [Google Scholar]
- 31.Kakonen SM, Selander KS, Chirgwin JM, Yin JJ, Burns S, Rankin WA, Grubbs BG, Dallas M, Cui Y, Guise TA. Transforming growth factor-beta stimulates parathyroid hormone-related protein and osteolytic metastases via Smad and mitogen-activated protein kinase signaling pathways. The Journal of biological chemistry. 2002;277(27):24571–8. doi: 10.1074/jbc.M202561200. [DOI] [PubMed] [Google Scholar]
- 32.Theriault RL, Lipton A, Hortobagyi GN, Leff R, Gluck S, Stewart JF, Costello S, Kennedy I, Simeone J, Seaman JJ, Knight RD, Mellars K, Heffernan M, Reitsma DJ. Pamidronate reduces skeletal morbidity in women with advanced breast cancer and lytic bone lesions: a randomized, placebo-controlled trial. Protocol 18 Aredia Breast Cancer Study Group. Journal of clinical oncology: official journal of the American Society of Clinical Oncology. 1999;17(3):846–54. doi: 10.1200/JCO.1999.17.3.846. [DOI] [PubMed] [Google Scholar]
- 33.Lluch A, Cueva J, Ruiz-Borrego M, Ponce J, Perez-Fidalgo JA. Zoledronic acid in the treatment of metastatic breast cancer. Anti-cancer drugs. 2014;25(1):1–7. doi: 10.1097/CAD.0000000000000020. [DOI] [PubMed] [Google Scholar]
- 34.Bu G, Lu W, Liu CC, Selander K, Yoneda T, Hall C, Keller ET, Li Y. Breast cancer-derived Dickkopf1 inhibits osteoblast differentiation and osteoprotegerin expression: implication for breast cancer osteolytic bone metastases. International journal of cancer Journal international du cancer. 2008;123(5):1034–42. doi: 10.1002/ijc.23625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mercer RR, Miyasaka C, Mastro AM. Metastatic breast cancer cells suppress osteoblast adhesion and differentiation. Clinical & experimental metastasis. 2004;21(5):427–35. doi: 10.1007/s10585-004-1867-6. [DOI] [PubMed] [Google Scholar]
- 36.Mastro AM, Gay CV, Welch DR, Donahue HJ, Jewell J, Mercer R, DiGirolamo D, Chislock EM, Guttridge K. Breast cancer cells induce osteoblast apoptosis: a possible contributor to bone degradation. Journal of cellular biochemistry. 2004;91(2):265–76. doi: 10.1002/jcb.10746. [DOI] [PubMed] [Google Scholar]
- 37.Fontanini G, Lucchi M, Vignati S, Mussi A, Ciardiello F, De Laurentiis M, De Placido S, Basolo F, Angeletti CA, Bevilacqua G. Angiogenesis as a prognostic indicator of survival in non-small-cell lung carcinoma: a prospective study. Journal of the National Cancer Institute. 1997;89(12):881–6. doi: 10.1093/jnci/89.12.881. [DOI] [PubMed] [Google Scholar]
- 38.Speirs V, Atkin SL. Production of VEGF and expression of the VEGF receptors Flt-1 and KDR in primary cultures of epithelial and stromal cells derived from breast tumours. British journal of cancer. 1999;80(5–6):898–903. doi: 10.1038/sj.bjc.6690438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mayr-Wohlfart U, Waltenberger J, Hausser H, Kessler S, Gunther KP, Dehio C, Puhl W, Brenner RE. Vascular endothelial growth factor stimulates chemotactic migration of primary human osteoblasts. Bone. 2002;30(3):472–7. doi: 10.1016/s8756-3282(01)00690-1. [DOI] [PubMed] [Google Scholar]
- 40.Zelzer E, McLean W, Ng YS, Fukai N, Reginato AM, Lovejoy S, D’Amore PA, Olsen BR. Skeletal defects in VEGF(120/120) mice reveal multiple roles for VEGF in skeletogenesis. Development. 2002;129(8):1893–904. doi: 10.1242/dev.129.8.1893. [DOI] [PubMed] [Google Scholar]
- 41.Zelzer E, Olsen BR. Multiple roles of vascular endothelial growth factor (VEGF) in skeletal development, growth, and repair. Current topics in developmental biology. 2005;65:169–87. doi: 10.1016/S0070-2153(04)65006-X. [DOI] [PubMed] [Google Scholar]
- 42.Fukaya Y, Shimada H, Wang LC, Zandi E, DeClerck YA. Identification of galectin-3-binding protein as a factor secreted by tumor cells that stimulates interleukin-6 expression in the bone marrow stroma. The Journal of biological chemistry. 2008;283(27):18573–81. doi: 10.1074/jbc.M803115200. [DOI] [PubMed] [Google Scholar]
- 43.Silverman AM, Nakata R, Shimada H, Sposto R, DeClerck YA. A galectin-3-dependent pathway upregulates interleukin-6 in the microenvironment of human neuroblastoma. Cancer research. 2012;72(9):2228–38. doi: 10.1158/0008-5472.CAN-11-2165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Piccolo E, Tinari N, Semeraro D, Traini S, Fichera I, Cumashi A, La Sorda R, Spinella F, Bagnato A, Lattanzio R, D’Egidio M, Di Risio A, Stampolidis P, Piantelli M, Natoli C, Ullrich A, Iacobelli S. LGALS3BP, lectin galactoside-binding soluble 3 binding protein, induces vascular endothelial growth factor in human breast cancer cells and promotes angiogenesis. Journal of molecular medicine. 2013;91(1):83–94. doi: 10.1007/s00109-012-0936-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


