SUMMARY
Massively parallel single-cell RNA sequencing can precisely resolve cellular diversity in a high-throughput manner at low cost, but unbiased isolation of intact single cells from complex tissues, such as adult mammalian brains, is challenging. Here, we integrate sucrose-gradient assisted purification of nuclei with droplet microfluidics to develop a highly scalable single-nucleus RNA-Seq approach (sNucDrop-Seq), which is free of enzymatic dissociation and nuclei sorting. By profiling ~18,000 nuclei isolated from cortical tissues of adult mice, we demonstrate that sNucDrop-Seq not only accurately reveals neuronal and non-neuronal subtype composition with high sensitivity, but also enables in-depth analysis of transient transcriptional states driven by neuronal activity, at single-cell resolution, in vivo.
INTRODUCTION
A fundamental challenge in deciphering cell-type composition and cells’ functional states in complex mammalian tissues manifests in the extraordinary diversity of cell morphology, size and local microenvironment. While existing single-cell RNA-Seq approaches have proved to be powerful tools for interrogating cell types, dynamic states, and functional processes in vivo (Tanay and Regev, 2017), these methods require the preparation of intact, single-cell suspensions from freshly isolated tissues, which is only practical for easily-dissociated embryonic and young postnatal tissues. This requirement poses an even greater challenge for cells with complex morphology, such as mature neurons. Enzymatic treatment not only favors recovery of easily dissociated cell types, but also introduces aberrant transcriptional changes during the whole-cell dissociation process (Lacar et al., 2016; Wu et al., 2017). In addition, skeletal and cardiac muscle cells are frequently multinucleated and are large in size. For instance, each adult mouse skeletal muscle cell contains hundreds of nuclei and is ~5,000 μm in length and 10–50 μm in width (White et al., 2010). Thus, existing high-throughput single-cell capture and library preparation methods, including isolation of cells by fluorescence activated cell sorting (FACS) into multi-well plates, sub-nanoliter wells, or droplet microfluidic encapsulation, are not optimized to accommodate these unusually large cells. Isolating individual nuclei for transcriptome analysis is a promising strategy, as single-nucleus RNA-Seq methods avoid strong biases against cells of complex morphology and large size (Habib et al., 2016; Lacar et al., 2016; Lake et al., 2016; Zeng et al., 2016) and can be potentially standardized to accommodate the study of various tissues. However, current single-nucleus RNA-Seq methods primarily rely on fluorescence-activated nuclei sorting (FANS) (Habib et al., 2016; Lake et al., 2016) or Fluidigm C1 microfludics platform (Zeng et al., 2016) to capture nuclei, and thus cannot easily be scaled up to generate a comprehensive atlas of cell types in a given tissue, much less a whole organism.
DESIGN
An ideal solution to increase the throughput of single-nucleus RNA-Seq is to integrate nucleus purification with massively parallel single-cell RNA-Seq methods such as Drop-Seq (Macosko et al., 2015), InDrop (Klein et al., 2015), or commercial platforms such as 10× Genomics (Zheng et al., 2017). However, single-nucleus RNA-Seq is currently not supported on these droplet microfluidics platforms. Inefficient lysis of nuclear membranes and/or cellular debris contamination might contribute to this failure. Historically, nuclei of high purity can be isolated from solid tissues or from cell lines with fragile nuclei by centrifugation through a dense sucrose cushion to protect nucleus integrity and strip away cytoplasmic contaminants. The sucrose gradient ultracentrifugation approach has been adapted to isolate neuronal nuclei for profiling histone modifications, nuclear RNA, and DNA methylation at genome-scale (Johnson et al., 2017; Lister et al., 2013; Mo et al., 2015). Here, we develop “sucrose gradient-assisted single-nucleus Drop-Seq” (sNucDrop-Seq), a method that enables highly scalable profiling of nuclear transcriptomes at single cell resolution by integrating sucrose gradient ultracentrifugation-based nucleus purification with droplet microfluidics.
RESULTS
Validation of sNucDrop-Seq
To test whether this nucleus purification method supports single-nucleus RNA-Seq analysis, we isolated nuclei from cultured cells, as well as freshly isolated or frozen adult mouse brain tissues through dounce homogenization followed by sucrose gradient ultracentrifugation (Figure 1A and Figure S1A). After quality assessment and counting of nuclei, we performed emulsion droplet barcoding of the nuclei and library preparation. We found that the Drop-Seq platform yielded high quality cDNA libraries from both whole cells and nuclei (Figure S1B).
Figure 1. sNucDrop-Seq: a massively parallel single-nucleus RNA-Seq method.
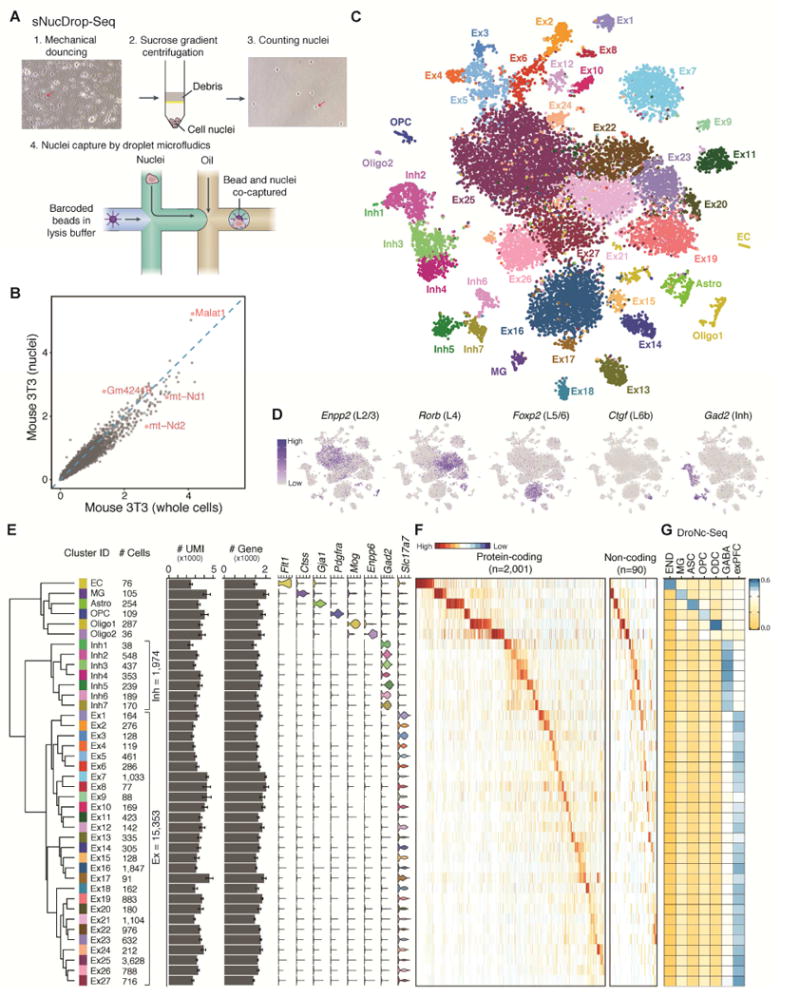
A) Overview of sNucDrop-Seq. Red arrows indicate representative nuclei before or after sucrose gradient centrifugation.
(B) Scatter plot comparing the average expression levels detected in NIH3T3 nuclei (y-axis, by sNucDrop-Seq) and cells (x-axis, by Drop-Seq). Red dots mark representative genes preferentially enriched in either nuclei or whole cells.
(C) Visualization by tSNE plot of clustering of 18,194 single-nucleus expression profiles from adult mouse cortices (n=17 mice). Ex, excitatory neurons; Inh, inhibitory neurons; Astro, astrocytes; OPC, oligodendrocyte precursor cells; Oligo, oligodendrocytes; MG, microglia; EC, endothelial cells.
(D) Marker gene expression, shown by re-coloring nuclei according to the expression level and projecting onto the tSNE plot in Figure 1A.
(E) Dendrogram showing relatedness of cell clusters, followed by (from left to right) cluster identification (ID), cell number per major cell type, UMIs per cluster (mean ± s.e.m.), number of genes detected per cluster (mean ± s.e.m.).
(F) Heatmap showing expression of cell-type specific protein-coding and long non-coding RNA markers in clusters defined in Figure 1E.
(G) Cell-type-specific expression signatures (identified by sNucDrop-Seq) agree with previously published work. Pairwise correlations of the average expression for the genes in each cell-type signature defined by sNucDrop-Seq and cell-types defined by DroNc-Seq in the mouse prefrontal cortex (Habib et al., 2017). DroNc-Seq clusters: exPFC, excitatory neurons; GABA, inhibitory neurons; ASC, astrocytes; OPC, oligodendrocyte precursor cells; ODC, oligodendrocytes; MG, microglia; END, endothelial cells.
We next validated the specificity of sNucDrop-Seq with species-mixing experiments, using nuclei isolated from cultured mouse and human cells. This analysis indicates that the rate of co-encapsulation of multiple nuclei per droplet (~2.6%) is comparable to standard Drop-Seq (Figure S1C). To assess the sensitivity of sNucDrop-Seq, we performed shallow sequencing of cultured mouse 3T3 cells at either single-cell (with Drop-Seq: detecting on average 3,325 genes with ~25,000 reads per cell for 1,160 cells with >800 genes detected) or single-nucleus (with sNucDrop-Seq: detecting on average 2,665 genes with ~23,000 reads per nucleus for 1,984 nuclei with >800 genes detected) resolution (Figure S1D). With standard Drop-Seq microfluidics devices and flow parameters, the capture rate of sNucDrop-Seq (1.9%, 1,829/95,000 barcoded beads) is comparable to that of Drop-Seq (1.5%, 1,160/77,000 barcoded beads). Comparative analysis reveals that mitochondria-derived RNAs (e.g. mt-Nd1, mt-Nd2) and nucleus-enriched long-noncoding RNAs (e.g. Malat1) were enriched in reads derived from Drop-Seq and sNucDrop-Seq, respectively (Figure 1B). Thus, integrating sucrose gradient centrifugation-based nuclear purification with the Drop-Seq microfluidic platform and workflow may support massively parallel single-nucleus RNA-Seq.
Application of sNucDrop-Seq to adult mouse cortex
To demonstrate the utility of sNucDrop-Seq in studying complex adult tissues, we analyzed nuclei isolated from adult mouse cerebral cortex (Table S1). The average expression profiles of single nuclei from two biologically independent replicates were well correlated (r=0.993; Figure S1E–F). Out of reads uniquely mapped to the genome (78% of all reads), 76% of reads were aligned to the expected strand of genic regions (25% exonic and 51% intronic), and the remaining 24% to intergenic regions or to the opposite strand of annotated genic regions. The relatively high proportion of intronic reads is similar to a previous single-nucleus RNA-Seq study of human cortex (49%) (Lake et al., 2016), reflecting the enrichment of nascent, preprocessed transcripts in nuclei. Because most exonic (91%) and intronic (86%) reads were mapped to the expected strand of annotated transcripts, we retained both exonic and intronic reads for downstream analyses. After initial quality filtering (>800 genes detected per nucleus), we retained 20,858 nuclei (15,471 uniquely mapped reads per nucleus), detecting, on average, 3,464 transcripts (unique molecular identifiers [UMIs]), and 1,662 genes per nucleus. After correcting for batch effects, we identified highly variable genes, and determined significant principal components (PC). We then performed graph-based clustering and visualized distinct groups of nuclei using non-linear dimensionality reduction with spectral t-distributed stochastic neighbor embedding (tSNE). After removing non-cortical cells (~7%, mostly striatal inhibitory neurons) and data points contributing toward potential noise (see Methods), our analysis segregated 18,194 cortical nuclei into 40 distinct clusters (Figure 1C). Each cluster contains nuclei from multiple animals, indicating the transcriptional identities of these cell-type-specific clusters are reproducible across biological replicates (Figure S2A).
On the basis of known markers for major cortical cell types, we identified 27 excitatory neuronal clusters (Ex 1-27: Slc17a7+), 7 inhibitory neuronal clusters (Inh 1–7: Gad2+), and six non-neuronal clusters (astrocytes [Astro: Gja1+], oligodendrocyte precursor cells [OPC: Pdgfra+], oligodendrocytes [Oligo1: Mog+; Oligo2: Enpp6+], microglia [MG: Ctss+], and endothelial cells [EC: Flt1+]) (Figure 1C–F). Consistent with previous studies (Lake et al., 2016; Madisen et al., 2015; Zeisel et al., 2015), known layer-specific marker genes (Layer (L) 2/3: Enpp2; L4: Rorb; L6a: Foxp2; L6b, Ctgf) can be readily detected in specific excitatory neuronal clusters (Figure 1D), revealing the anatomic locations of these glutamatergic excitatory neuronal subtypes. We also uncovered all major subclasses of cortical inhibitory neurons named for the neurochemical markers they express: somatostatin-expressing (Sst+: cluster Inh1-2) cells, parvalbumin-expressing (Pvalb+: cluster Inh3-4) cells, vasoactive intestinal peptide-expressing (Vip+: cluster Inh6) cells, and cells that express 5-hydroxytryptamine receptor 3A (Htr3a) but lack Vip expression (Tnfaip8l3+/Sema3c+/Vip−: cluster Inh5 and Inh7) (Figure 1C–E and S2B). This unbiased sampling strategy captured enough cells to resolve heterogeneity among non-neuronal cell types present in relatively low abundance in the adult cortex (Figure 1E and S2B), including two oligodendrocyte subtypes (Oligo1: Mog+/Enpp6−; Oligo2: Mog−/Enpp6+) recently identified through single-cell deep sequencing of full-length mRNAs (Tasic et al., 2016) (Figure S3C). Finally, the cell types and their signatures from sNucDrop-Seq were comparable to those obtained with DroNc-Seq (a recently published approach similar to sNucDrop-Seq) of mouse prefrontal cortex (Habib et al., 2017) (Figure 1G).
In addition to subtype-specific protein-coding marker genes (Figure 1F), we have identified a list of long non-coding RNAs that are specifically expressed in distinct cell clusters (Figure 1F and S2B). For instance, 1700016P03Rik is specifically detected in cluster Ex17 and Ex24, and this acts, mainly, as a primary, non-coding transcript encoding two microRNAs (Mir212 and Mir132), which are regulated by neuronal activity (Aten et al., 2016; Nudelman et al., 2010), raising the possibility that Ex17 and 24 clusters are specifically associated with activity-induced transcriptional states. Cell-type-specific non-coding RNA marker genes have also been identified for inhibitory neuronal subtypes (e.g. Dlx1as for Inh7) and non-neuronal cells (e.g. 4933406I18Rik for MG). The identification of both protein-coding and non-coding transcripts as cell-type-specific markers highlights the potential of sNucDrop-Seq in exploring the emerging role of non-coding RNAs at single-cell resolution, in vivo.
We next explored whether aggregation of our single-nucleus RNA-Seq data into subtype-specific transcriptome profiles, as a proof of concept, might enable analysis of differential mRNA processing in a cell-type-specific manner. Using a probabilistic model that quantitates the expression level of alternatively spliced genes (MISO) (Katz et al., 2010), we identified 263 differential exon processing events through pairwise comparison of cell-type-specific transcriptome profiles. In agreement with a previous single-cell RNA-Seq study (Tasic et al., 2016), Syntaxin binding protein 1 (Stxbp1) mRNA exhibited differential processing of a specific exon amongst excitatory and inhibitory neurons (Figure S2C). In particular, additional heterogeneity in the usage of this exon was detected among major inhibitory neuronal subtypes (Sst+ versus Pvalb+ in Figure S2C). Our cell-type-specific analysis also revealed differentially processed exon usage between abundant cortical cell-types such as excitatory neurons and relatively rare non-neuronal cells (e.g. Macf1 in Figure S2C). Thus, despite the shallow sequencing depth and 3′ bias, sNucDrop-Seq allows a population of highly heterogeneous cells to be analyzed together to discover cell-type-specific signatures of mRNA processing.
sNucDrop-Seq reveals composition of cortical inhibitory neurons
GABAergic interneurons are highly diverse in terms of morphology, connectivity and physiological properties (Kepecs and Fishell, 2014), but the relative composition of these neurons, particularly those of low abundance in the cortex, is not well established. To accurately measure the subtype composition and validate the specificity of cortical inhibitory neuronal subtypes identified by sNucDrop-Seq, we performed sub-clustering on cortical inhibitory neuronal nuclei (Inh1-7 in Figure 2A) together with non-cortical nuclei isolated from dorsal striatum (>95% striatal cells are GABAergnic neurons), identifying 17 sub-clusters (Figure 2A). Based on expression patterns of known marker genes, we first segregated these sub-clusters into cortical interneuron (cluster A-I in Figure 4A: Gad1+/Gad2+/Meis2−) and non-cortical (clusters in grey: Meis2+) clusters. Consistent with their striatal origin, many non-cortical cells express Ppp1r1b (also known as DARPP-32) (Figure 2A–B), a marker gene indicative of medium spiny neurons (MSNs, D1-type) in the striatum. Because sNucDrop-Seq samples nuclei in proportion to cells’ abundance in their native environment, this approach enables direct measurement of subtype composition. This analysis identified Pvalb-expressing subtypes (cluster D, E, and F: 40.2%) and Sst-expressing subtypes (cluster G, H, and I: 31.4%) as two major groups of cortical interneurons (Figure 2C–D), in complete accordance with previous observations derived from in situ hybridization (ISH)- or immunostaining-based analysis of mouse neocortex (Rudy et al., 2011). Beyond major interneuron subtypes, we identified 10.8% of cortical interneurons as an Ndnf-expressing subtype (cluster A), 8.3% as a Vip-expressing subtype (cluster B), and 9.3% as a synuclein gamma (Sncg)-expressing subtype (cluster C) (Figure 2C–D). On the basis of combinatorial expression of known marker genes, interneuron subtypes identified by sNucDrop-Seq parallel those identified in previous studies of mouse or human cortex (Lake et al., 2016; Tasic et al., 2016), revealing inhibitory neuronal heterogeneity in both cortical layer distribution and the developmental origin from subcortical regions of the medial or caudal ganglionic eminences (MGE or CGE) (Figure 2E–F and S3C). Therefore, sNucDrop-Seq resolves cellular heterogeneity and quantifies cell-type composition at the transcriptomic level, with high sensitivity.
Figure 2. sNucDrop-Seq reveals inhibitory neuronal subtypes and composition.
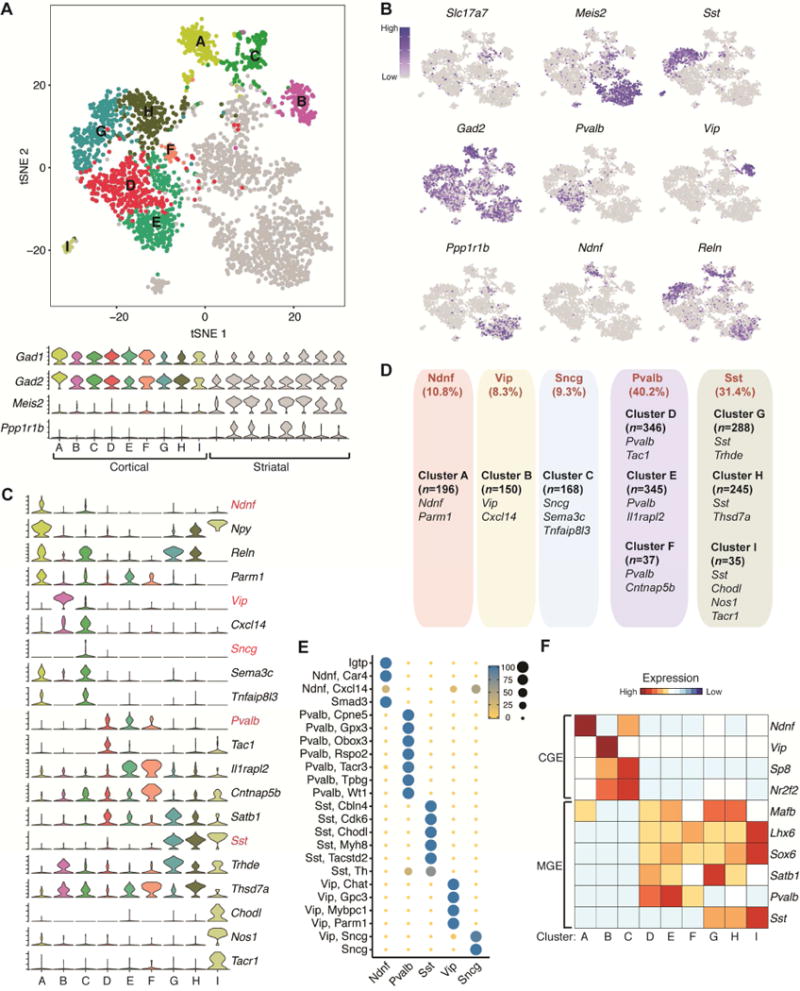
(A) Spectral tSNE plot of 1,810 cortical GABAergic neurons and 1,462 non-cortical nuclei. The cortical inhibitory neurons are colored (A-H) while non-cortical nuclei are marked in grey. Violin plots for two GABAergic neuronal markers (Gad1 and Gad2) and two striatal markers (Meis2 and Ppp1r1b) are shown in the lower panel.
(B) Marker gene expression shown by re-coloring and projecting onto the tSNE plot in Figure 2A.
(C) Violin plots showing select marker gene expression for inhibitory neuronal subtypes. Five mutually exclusive subtype-specific marker genes are highlighted in red.
(D) Summary of inhibitory neuronal subtypes identified by sNucDrop-Seq. GABAergic subtypes are grouped according to five major classes. Also shown are number of nuclei per subtype and representative marker genes associated with each.
(E) Congruence of five major GABAergic neuronal classes defined by sNucDrop-Seq in Figure 2D with subpopulations defined from single-cell RNA-Seq profiles in the mouse visual cortex (Tasic et al., 2016).
(F) Heatmap showing select marker genes that distinguish cortical inhibitory neurons originated from either CGE or MGE.
Figure 4. sNucDrop-Seq reveals cell-type specific transcriptional responses to PTZ-induced seizure.
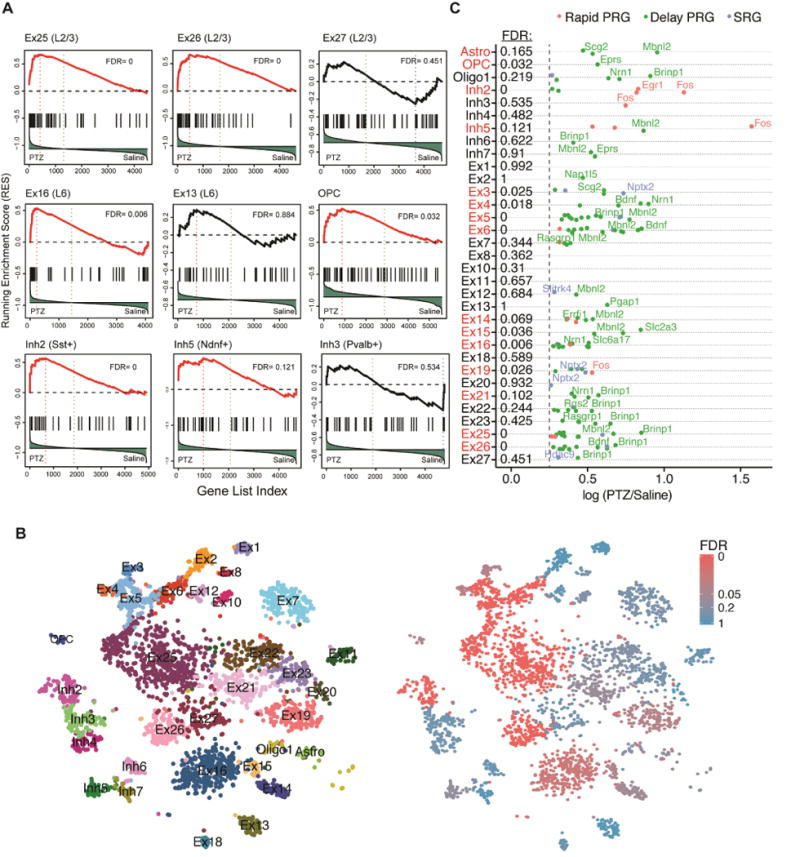
(A) GSEA of the ARG set in different cell clusters by the comparison between PTZ- and saline-treated neurons was performed as in Figure 3B. For each cell type, genes were ranked based on the differential expression between PTZ (left) and saline (right). The FDR for GSEA of ARGs is listed and clusters with FDR < 0.2 are highlighted in red.
(B) Spectral tSNE plot of 3,491 nuclei from PTZ-treated mouse cortex. The nuclei were colored per annotated cell types (left panel) or the FDR of GSEA analysis (right panel).
(C) Cell-type specific induction of ARGs in response to PTZ treatment. Rapid PRGs, delayed PRGs and SRGs are highlighted in red, green, and blue, respectively. X-axis indicates the natural log fold change of gene expression between PTZ and saline. The dash line denotes the cut-off of gene expression fold change (log 0.25). The FDR for GSEA of ARGs (as in Figure 4A) is listed and clusters with FDR < 0.2 are highlighted in red.
sNucDrop-Seq reveals layer-specific composition and activity-dependent transcriptional state of cortical excitatory neurons
For glutamatergic neuronal clusters, we associated each excitatory neuronal subtype with a combination of known markers indicative of their superficial-to-deep layer distribution (Figure 3A), revealing layer-specific composition of excitatory neuronal subtypes (Figure S3A–B). Thus, sNucDrop-Seq analysis captures transcriptomic distinctions between closely related subtypes in each cortical layer, which is in high concordance with subtypes previously identified in human (Lake et al., 2016) and mouse (Habib et al., 2017; Tasic et al., 2016; Zeisel et al., 2015) cortices (Figure S3C–G).
Figure 3. Excitatory neuronal subtypes resolve heterogeneity in cortical layer distribution and transcriptional states driven by neuronal activity.
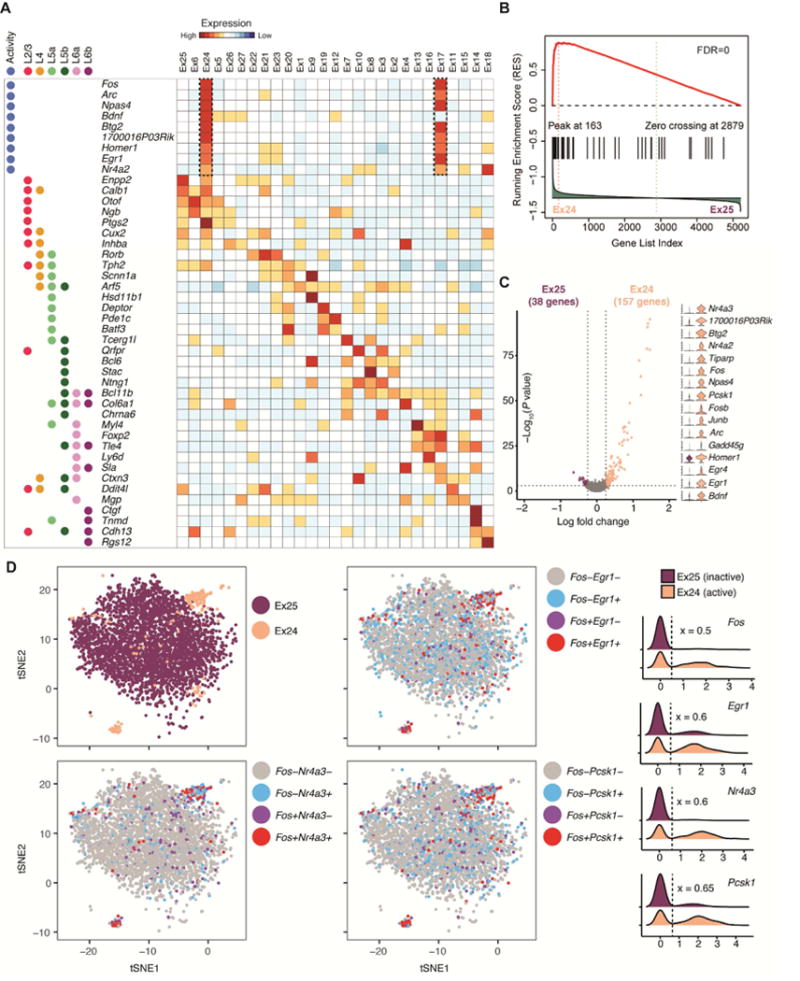
(A) Heatmap for layer-specific markers and neuronal activity-regulated genes showing cortical layer identity (L2/3/, L4, L5a/b, L6a/b), excitatory subtypes, and ARG expression.
(B) GSEA of ARGs between closely related neuronal sub-clusters. Genes were ranked based on the differential expression between active (left: Ex24) and inactive (right: Ex25). The plot of running enrichment score (RES) is shown in red (top panel). Vertical bars (in black) in the middle indicate a gene within the ARG gene set. The correlation of gene expression with sub-clusters is shown in the bottom histogram (in green).
(C) Differential expression between activated and inactivated excitatory neurons within upper-(Ex24 versus Ex25) layer sub-clusters. Shown on the right are violin plots of select marker gene expression.
(D) Co-expression analysis of neuronal activity-dependent genes in excitatory neuronal subtypes. Spectral tSNE plots (left) and density plots (right) illustrating expression of select ARGs in upper layer excitatory neuronal subtypes. The expression level threshold (x) for ARGs was determined post-hoc (the dashed line) and marked by the dash line in density. The nuclei in tSNE plots are re-colored by ARG expression.
In response to neuronal activity, excitatory neurons induce expression of hundreds of activity-regulated genes (ARGs), many of which regulate synaptic function and allow neuronal circuits to respond dynamically to experience (Flavell and Greenberg, 2008). Because induced mRNAs may remain in the cytoplasm for hours to days (Schwanhausser et al., 2011), it is challenging to capture the dynamic and transient neuronal activation process using whole-cell RNA-Seq. Direct comparison between single-cell and single-nucleus transcriptomic profiling of activated neurons demonstrated that single-nucleus RNA-Seq analysis not only avoids the aberrant activation of ARGs by whole-cell dissociation, but also reveals dynamics of transcriptional response to neuronal activity-inducing experience (Lacar et al., 2016). However, the reported low-throughput single-nucleus RNA-Seq analysis requires pre-enrichment of activated neuronal nuclei by sorting and the Fluidigm C1 platform, which is not easily scaled up to accommodate more samples.
On the basis of ARG expression, we explored whether sNucDrop-Seq analysis can resolve heterogeneity in activity-induced transcriptional states amongst closely related subtypes. We found that while nuclei in the Ex24 cluster (n=212 nuclei) express nearly identical layer-specific marker genes as E25 (n=3,628), Ex24 is specifically associated with high-level expression of ARGs (Figure 3A and S4A), including well-defined immediately early genes (IEGs) such as Fos, Arc, and Egr1, as well as other activity-regulated transcription factors (e.g. Npas4), genes encoding proteins that function at synapses (e.g. Homer1), and non-coding RNAs (e.g. 1700016P03Rik that encodes Mir132). A similar relationship was detected between Ex17 (n=91) and Ex16 (n=1,847). Interestingly, despite variations in number of nuclei and sequencing depth among samples (Table S1), the small percentage of putatively activated neurons (Ex17: ~0.5%; Ex24: ~1.2% of all nuclei) was found in nearly all animals (Figure S4B), raising the possibility that neuronal activity-induced transcriptional states can be reproducibly captured by sNucDrop-Seq. We next performed gene set enrichment analysis (GSEA) of a recently curated list of ARGs induced by acute or prolonged neuronal activity (Tyssowski et al., 2017). This analysis indicated that both Ex17 (false-discovery rate [FDR]=0) and Ex24 (FDR=0) are significantly enriched for this set of ARGs (Figure 3B). We next determined the genes specifically enriched in activated neurons in Ex17 (n=129 genes, as compared to other Ex16) or Ex24 (n= 157 genes, as compared to Ex25) neurons (Figure 3C). KEGG pathway analysis suggested that expression signatures of these two excitatory neuronal clusters are enriched for genes involved in the MAPK signaling pathway (adjusted P=5.74×10−3 for E×17 and 8.14×10−3 for Ex24), consistent with previous reports (Lacar et al., 2016; Tyssowski et al., 2017). We also observed heterogeneous expression patterns of ARGs (rapid IEGs: Fos and Egr1; delayed IEGs: Nr4a3 and Pcsk1) among nuclei in Ex24 (Fos+: 53% in Ex24 versus 5.7% in Ex25; P=1.57E-71, Fisher’s exact test) and Ex17 (Fos+: 55% in Ex17 versus 6.8% in Ex17; P=3.99E-31) excitatory neuronal clusters (Figure 3D and Table S2), in agreement with a continuum of transcriptional states of ARGs revealed by a recent analysis of Fos-positive neuronal nuclei isolated from adult mice exposed to a novel environment (Lacar et al., 2016). Together, these results indicate that sNucDrop-Seq can potentially identify activity-dependent transcriptional states at single-nucleus resolution in vivo.
Mapping cell-type-specific transcriptional response to the Pentylenetetralzol (PTZ)-induced seizure
To further examine cell-type-specific responses to neuronal activity by sNucDrop-Seq, we elicited large-scale neuronal activation with PTZ, a GABA(A) receptor antagonist that induces seizures coupled with ARG expression in cortex (Morgan et al., 1987; Yount et al., 1994). Mice were treated with PTZ or received an injection with saline as a control. One hour after injection, nuclei were immediately isolated for sNucDrop-Seq analysis (saline: 4,005 nuclei; PTZ: 3,491 nuclei). After filtering out the clusters that did not contain sufficient data points (7 clusters; see Methods), we determined that 15 out of 33 clusters are significantly enriched for the ARG gene set by GSEA (FDR<0.2) (Figure 4A–B). In addition to 11 excitatory neuronal clusters, two inhibitory neuronal clusters (Inh2 and Inh5) and two non-neuronal clusters (Astro and OPC) were significantly associated with expression of activity-regulated genes in response to PTZ treatment. Our analysis suggests that Sst-expressing inhibitory neurons (Inh2) are much more likely to express ARGs than Pvalb-expressing neurons (Inh3) in response to PTZ-induced seizure, which is in agreement with a recent cell-type-specific analysis of inhibitory neuronal response to PTZ using a synthetic IEG reporter system (Sorensen et al., 2016). Interestingly, relatively low-abundant interneuron subtypes, such as Ndnf-expressing cells (Inh5) and oligodendrocyte precursor cells, also exhibited significant transcriptional response to PTZ treatment (Figure 4A), suggesting conserved signaling pathways underlying neuronal activation in these cell-types.
The activity-dependent gene expression program is likely structured temporally into two major waves of gene activation. The first wave comprises primary response genes (PRGs, also called IEGs). The second wave comprises secondary response genes (SRGs), which require de novo translation for their induction and are likely to be regulated by PRG proteins (Tyssowski et al., 2017). Further analysis of top ranked PTZ-induced ARGs (separated into three groups: rapid PRG, delay PRG, and SRG) in each cluster indicates that rapid PRGs such as Fos and Egr1 are more likely to be detected in inhibitory neuronal clusters (Inh2 and Inh5), whereas delay PRGs (e.g. Bdnf, Mbnl2) and SRG (e.g. Nptx2) are more frequently detected in excitatory neuronal clusters (Figure 4C). This observation suggests that after one-hour of, rapid PRG expression is probably already attenuated in excitatory neurons, and inhibitory neurons are likely activated by the PTZ-induced seizure in a temporally distinct manner. We also found that clusters significantly associated with ARG gene set induction generally induced a stronger transcriptional response than others (that is, greater number of genes and amplitude of changes) (Figure S4C). Overall, these data should provide a rich resource for identification of genes whose dynamic expression in specific cell-types may be important for that cell-type’s functional response to neuronal activity.
DISCUSSION
In conclusion, sNucDrop-Seq is a robust approach for massively parallel analysis of nuclear RNA, at single-cell resolution. Because intact nuclear isolation can potentially be accomplished by mechanical douncing and sucrose gradient ultracentrifugation in almost any primary tissue, including frozen, archived human tissues, sNucDrop-Seq and similar approaches (Habib et al., 2017) pave the way to systematically identify cell-types, reveal subtype composition, and dissect dynamic functional states such as activity-dependent transcription in complex mammalian tissues.
Our study, with its focus on profiling cortical cells in their native environment and in proportion to their abundance, complements a recent single-cell RNA-Seq study of adult mouse visual cortex that deeply surveyed FACS-isolated single cells from a comprehensive collection of transgenic mouse lines (Tasic et al., 2016). We found that the subtypes of both neurons and non-neuronal cells identified in the two studies are highly consistent (Figure S3C). Overall, our unbiased sampling approach may provide a more complete and accurate description of the cell-type landscapes, whereas deep sequencing of full-length mRNAs from select cells may identify a more complete set of molecular markers for each subtype.
In addition to neuronal subtype classification, precise localization and quantification of the activity of all neurons across specific brain regions would provide insights into how neural information is processed in physiological and pathological states. In mammals, a snapshot of the activity of ensembles of neurons in the mammalian brain can be obtained by whole-brain tissue clearing coupled with immunolabeling of IEGs (Renier et al., 2016), which requires advanced instrumentation and has low temporal resolution (that is, IEG transcripts can outlast the end of activity by over 4 hours). Single-nucleus RNA-Seq approaches such as sNucDrop-Seq relies on isolation of nuclei, thereby enriching nascent transcripts and enabling the detection and quantification of neural activity (using expression of ARGs as proxies) at much higher temporal resolution. In addition, our results show that transcriptomic differences between closely related subtypes (e.g. between Ex24 and Ex25) may be largely driven by neuronal activity-dependent transcriptional programs, rather than differences in developmental origin or cell-type. This observation suggests that environmental stimuli may contribute to discontinuous transcriptomic differences, and future studies are needed to investigate how continuous variables, such as neuronal activity, can be best implemented as additional classifiers for cell-types and functional states.
LIMITATIONS
As with other single-cell or single-nucleus RNA-Seq approaches, sNucDrop-Seq relies on dissociation of individual cells/nuclei in dissected tissues, thereby discarding cells’ original anatomical location information. Thus, combining sequencing-based single-cell genomics approach with imaging-based multiplex fluorescence in situ hybridization (FISH) methods (Chen et al., 2016; Chen et al., 2015; Shah et al., 2016) that allows detection of a set of preselected genes in tissues, is a promising approach to acquire essential information about the precise anatomical location, to reconstruct a more complete cell-type atlas in complex mammalian tissues.
STAR METHODS
Detailed methods are provided in the online version of this paper and include the following:
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Chemicals, Peptides, and Recombinant Proteins | ||
| Dulbecco’s Modified Eagle’s Medium | Life Technologies | Cat#11965084 |
| Fetal Bovine Serum | Life Technologies | Cat#26140079 |
| L-glutamine | Life Technologies | Cat#25030081 |
| 0.05% Trypsin | Life Technologies | Cat#25300054 |
| Matrigel matrix | Corning | Cat#354230 |
| DMEM/F12 | Life Technologies | Cat#11320033 |
| TeSR-E8 Medium | Stem Cell Technologies | Cat#05940 |
| DPBS, no calcium, no magnesium | Invitrogen | Cat#14190136 |
| Sucrose | Sigma-Aldrich | Cat#S0389-1KG |
| 1M Tris-HCl, pH 8.0 | Invitrogen | Cat#15568-025 |
| MgAc2 | Sigma-Aldrich | Cat#M5661-50G |
| cOmplete™, EDTA-free Protease Inhibitor Cocktail | Roche | Cat#11873580001 |
| CaCl2 | Sigma-Aldrich | Cat#C1016-500G |
| Triton X-100 | Sigma-Aldrich | Cat#T8787-100mL |
| 0.5M EDTA, pH 8.0 | Invitrogen | Cat#15575-020 |
| NxGen RNase Inhibitor | Lucigen | Cat#30281-2 |
| Bovine Serum Albumin | Sigma-Aldrich | Cat#A8806-5G |
| Ficoll PM-400 | GE Healthcare/Fisher Scientific | Cat#45-001-745 |
| Sarkosyl | Sigma-Aldrich | Cat#L7414-50mL |
| DTT | Fermentas | Cat#R0862 |
| QX200 Droplet Generation Oil for EvaGreen | Bio-Rad | Cat#186-4006 |
| Perfluoro-1-octanol | Sigma-Aldrich | Cat#370533-25G |
| dNTPs | Clontech | Cat#639125 |
| Critical Commercial Assays | ||
| Maxima H Minus Reverse Transcriptase | ThermoFisher | Cat#EP0753 |
| KAPA HiFi hotstart readymix | KAPA Biosystems | Cat#KK2602 |
| Deposited Data | ||
| Raw and analyzed data | This paper | GEO: GSE106678 |
| Experimental Models: Cell Lines | ||
| NIH3T3 | ATCC | Cat#CRL-1658 |
| H7 (female, human embryonic stem cells) | WiCell | Cat#WA07 |
| Experimental Models: Organisms/Strains | ||
| Mouse: C57BL/6 | From Dr. Joe Zhou | N/A |
| Oligonucleotides | ||
| Template Switch Oligo: TSO: AAGCAGTGGTATCAACGCAGAGTGAATrGrGrG | Macosko et al., 2015 | N/A |
| TSO-PCR primer: AAGCAGTGGTATCAACGCAGAGT | Macosko et al., 2015 | N/A |
| Illumina Nextera XT i7 primers: AATGATACGGCGACCACCGAGATCTACACGCCTGTCCGCGGAAGCAGTGGTATCAACGCAGAGT*A*C | Macosko et al., 2015 | N/A |
| Cuatom Read 1 Primer: GCCTGTCCGCGGAAGCAGTGGTATCAACGCAGAGTAC | Macosko et al., 2015 | N/A |
| Software and Algorithms | ||
| Drop-seq_tools (v1.12) | Macosko et al., 2015 | http://mccarrolllab.com/dropseq/ |
| STAR v2.5.2a | Dobin et al., 2013 | https://github.com/alexdobin/STAR |
| Seurat v1.4 | Satija et al., 2015 | http://satijalab.org/seurat/ |
| Seurat v2.0 | Butler and Satija, 2017 | http://satijalab.org/seurat/ |
| GSEA | Subramanian et al., 2005 | http://software.broadinstitute.org/gsea/index.jsp |
| DBSCAN | Ester et al., 1996 | https://cran.r-project.org/web/packages/dbscan/index.html |
| MISO | Katz et al., 2010 | https://miso.readthedocs.io/en/fastmiso/ |
| Random Forest | Liaw and Wiener, 2002 | https://www.stat.berkeley.edu/~breiman/RandomForests/ |
| ggplot2 | Wickham, 2016 | https://cran.r-project.org/web/packages/ggplot2/ |
| Other | ||
| Detailed Bench Protocol | This paper | Methods S1 |
| Tube, Thinwall, Polypropylene, 38.5 mL, 25 × 89 mm (qty. 50) | Beckman Coulter | 326823 |
| Glass 15mL Dounce Tissue Grinder Set with Two Glass Pestles, Grinding Chamber O.D. × L: 22 × 94mm (Case of 2) | Wheaton | 357544 |
| SW 28 Ti Rotor, Swinging Bucket, Aluminum, 6 × 38.5 mL, 28,000 rpm, 141,000 × g | Beckman Coulter | 342207 |
| Barcoded Beads | ChemGenes | MACOSKO-2011-10 |
| Aquapel-coated PDMS Microfluidic Device | uFluidix | Custom (described) |
| Syringe Pumps | KD Scientific | 78-8100 |
| 40μm Sterile Cell Strainer | Fisher Scientific | 22-363-547 |
| Medical Grade Polyethylene Micro Tubing | Scientific Commodities | BB31695-PE/2 |
| SPRISelect Beads | Beckman Coulter | B23318 |
| 75-cycle High Output v2 Kit | Illumina | FC-404-2005 |
| 10-micron carboxylated polystyrene beads | Bangs Labs | #PC06N-11355 |
CONTACT FOR REAGENT AND RESOURCE SHARING
Requests should be addressed to and will be fulfilled by Lead Contact Hao Wu (haowu2@pennmedicine.upenn.edu).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Cell Lines and Culture Conditions
Mouse NIH3T3 cells were purchased from ATCC and were grown in Dulbecco’s Modified Eagle’s Medium (DMEM) (Life Technologies) supplemented with 10% fetal bovine serum (FBS) (Life Technologies) and 2 mM L-glutamine (Life Technologies) at 37°C in 5% CO2. The culture was passaged every 2–3 days using 0.05% Trypsin (Life Technologies). Female, human embryonic stem cells (H7) have been procured from WiCell (Madison, WI) and maintained at 37°C in 5% CO2 on growth factor-reduced Matrigel matrix (Corning) coated six-well tissue culture plates. The six-well plates were coated with diluted (1:30) Matrigel matrix in DMEM/F12 (Life Technologies). H7 cells (between passage 40 and 70) were maintained in TeSR-E8 medium (Stem Cell Technologies) and passaged every 5–6 days as small aggregates using an enzymatic digestion-free method (0.5 mM ETDA in DPBS without CaCl2 and MgCl2 (Sigma-Aldrich)).
Animals
Experiments were conducted in accordance with the ethical guidelines of the National Institutes of Health and with the approval of the Institutional Animal Care and Use Committee of the University of Pennsylvania. Mice were group-housed in cages of three to five in a 12-h light/dark cycle with food and water provided ad libitum. All mice used for experiments were naive to behavioral assays and other procedures. For pentylenetetrazole (PTZ)-induced seizures, 10-week old, wild type, male F1 mice from a C56BL/6J to FVB/NJ breeding were used. All others were 6-week-old, wild type, male mice in a pure C57BL/6 background. For PTZ treatment, mice were injected intraperitoneally with PTZ at 50 mg/kg body weight or received an injection with saline as a control. One hour after injection, the mice were sacrificed.
METHOD DETAILS
Isolation and purification of nuclei
Mouse brains (postnatal 6–10 weeks) were rapidly resected on ice. Cortices were freshly processed or flash frozen in liquid nitrogen for 2 minutes and subsequently kept at −80°C before nuclear isolation. Nuclei were isolated and purified as previously described with some modifications (Johnson et al., 2017). Briefly, 14 mL of sucrose cushion (1.8 M sucrose (Sigma-Aldrich, RNase & DNase free, ultra pure grade), 10 mM Tris-HCl pH 8.0 (Invitrogen), 3 mM MgAc2 (Sigma-Aldrich), protease inhibitor cocktail (Sigma-Aldrich)) was added to the bottom of centrifuge tubes (Beckman Coulter). Using a glass homogenizer (Wheaton), a freshly isolated or frozen mouse cortex sample was subjected to dounce homogenization (21 times with loose pestle followed by 7 times with tight pestle) in 12 mL of homogenization buffer (0.32M sucrose, 5 mM CaCl2 (Sigma-Aldrich), 3mM MgAc2, 10 mM Tris-HCl pH 8.0, 0.1% Triton X-100 (Sigma-Aldrich), 0.1 mM EDTA (Invitrogen), protease inhibitor cocktail). For in vitro cultured cells, cell pellets (~5 million cells) were resuspended in homogenization buffer and dounced 20 times with a loose pestle. Homogenates (~12 mL) were layered onto the sucrose cushion in the centrifuge tubes, and 10 mL of homogenization buffer was added atop of the homogenates. The tubes were then centrifuged in a Beckman Coulter L7-65 Ultracentrifuge at 25,000 rpm at 4°C for 2 hours using a Beckman Coulter SW28 swinging bucket rotor (Beckman Coulter). The supernatant was carefully removed via aspiration. 1 mL of chilled DPBS with protease and RNase inhibitor (Lucigen) was added to resuspend the nuclear pellet, and nuclei were subsequently transferred to a 1.5-mL tube. Nuclei were pelleted at 5,000 rpm for 10 min at 4°C, and then resuspended in 0.01% BSA (Sigma-Aldrich) in DPBS. After resuspension, nuclei were filtered through a 40-μm cell strainer (Fisher Scientific), visually inspected for morphology and quality assurance, and counted using a Fuchs-Rosenthal counting chamber before droplet microfluidic encapsulation. For mouse cortices, we obtain 3.45 ± 2.00 ×106 nuclei, per round of isolation (based on 11 measurements). The nuclear isolation efficiency for in vitro cultured cells is ~84% (number of nuclei/number of input cells × 100).
Single-nucleus RNA-Seq library preparation and sequencing
The nuclear suspension was diluted to a concentration of 100 nuclei/μL in DPBS containing 0.01% BSA. Approximately 1.25 mL of this single-nucleus suspension was loaded for each sNucDrop-Seq run. The single-nucleus suspension was then co-encapsulated with barcoded beads (ChemGenes) using an Aquapel-coated PDMS microfluidic device (uFluidix) connected to syringe pumps (KD Scientific) via polyethylene tubing with an inner diameter of 0.38 mm (Scientific Commodities). Barcoded beads were resuspended in lysis buffer (200 mM Tris-HCl pH8.0, 20 mM EDTA, 6% Ficoll PM-400 (GE Healthcare/Fisher Scientific), 0.2% Sarkosyl (Sigma-Aldrich), and 50 mM DTT (Fermentas; freshly made on the day of run) at a concentration of 120 beads/μL. The flow rates for cells and beads were set to 4,000 μL/hour, while QX200 droplet generation oil (Bio-rad) was run at 15,000 μL/hour. A typical run lasts ~20 min. Droplet breakage with Perfluoro-1-octanol (Sigma-Aldrich), reverse transcription and exonuclease I treatment were performed, as previously described, with minor modifications (Macosko et al., 2015). Specifically, up to 120,000 beads, 200 μL of reverse transcription (RT) mix (1× Maxima RT buffer (ThermoFisher), 4% Ficoll PM-400, 1 mM dNTPs (Clontech), 1 U/μL RNase inhibitor, 2.5 μM Template Switch Oligo (TSO: AAGCAGTGGTATCAACGCAGAGTGAATrGrGrG) (Macosko et al., 2015), and 10 U/ μL Maxima H Minus Reverse Transcriptase (ThermoFisher)) were added. The RT reaction was incubated at room temperature for 30 minutes, followed by incubation at 42°C for 150 minutes. To determine an optimal number of PCR cycles for amplification of cDNA, an aliquot of 6,000 beads (corresponding to ~100 nuclei) was amplified by PCR in a volume of 50 μL (25 μL of 2x KAPA HiFi hotstart readymix (KAPA biosystems), 0.4 μL of 100 μM TSO-PCR primer (AAGCAGTGGTATCAACGCAGAGT (Macosko et al., 2015), 24.6 μL of nuclease-free water) with the following thermal cycling parameter (95°C for 3 min; 4 cycles of 98°C for 20 sec, 65°C for 45 sec, 72°C for 3 min; 9 cycles of 98°C for 20 sec, 67°C for 45 sec, 72°C for 3 min; 72°C for 5 min, hold at 4°C). After two rounds of purification with 0.6x SPRISelect beads (Beckman Coulter), amplified cDNA was eluted with 10 μL of water. 10% of amplified cDNA was used to perform real-time PCR analysis (1 μL of purified cDNA, 0.2 μL of 25 μM TSO-PCR primer, 5 μL of 2x KAPA FAST qPCR readymix, and 3.8 μL of water) to determine the additional number of PCR cycles needed for optimal cDNA amplification (Applied Biosystems QuantStudio 7 Flex). We then prepared PCR reactions per total number of barcoded beads collected for each sNucDrop-Seq run, adding 6,000 beads per PCR tube, and ran the aforementioned program to enrich the cDNA for 4 + 10 to 12 cycles. We then tagmented cDNA using the Nextera XT DNA sample preparation kit (Illumina, cat# FC-131-1096), starting with 550 pg of cDNA pooled in equal amounts, from all PCR reactions for a given run. Following cDNA tagmentation, we further amplified the library with 12 enrichment cycles using the Illumina Nextera XT i7 primers along with the P5-TSO hybrid primer (AATGATACGGCGACCACCGAGATCTACACGCCTGTCCGCGGAAGCAGTGGTATCAACGCAGAGT*A*C) (Macosko et al., 2015). After quality control analysis using a Bioanalyzer (Agilent), libraries were sequenced on an Illumina NextSeq 500 instrument using the 75-cycle High Output v2 Kit (Illumina). We loaded the library at 2.0 pM and provided Custom Read1 Primer (GCCTGTCCGCGGAAGCAGTGGTATCAACGCAGAGTAC) at 0.3 μM in position 7 of the reagent cartridge. The sequencing configuration was 20 bp (Read1), 8 bp (Index1), and 50 or 60 bp (Read2). In total, 17 mouse cortex samples were analyzed with sNucDrop-Seq in four sequencing runs.
Single-cell RNA-Seq library preparation and sequencing
Drop-Seq was performed as previously described, with default settings (Macosko et al., 2015). The cell suspension was diluted to 100 cells/μL with DPBS containing 0.01% BSA and 1 mL cell suspension was loaded for each Drop-seq run. After cell capture, reverse transcription, exonuclease treatment, cDNA amplification and tagmentation, libraries were diluted to 1.9 pmol, and equal amounts of distinctively indexed libraries were mixed and subjected to paired-end sequencing on Illumina NextSeq 500 sequencer (read1: 20 bp; read2: 60 bp). 10× Genomics single-cell 3′ libraries were constructed, as previously described (Zheng et al., 2017), with recommended settings using Chromium single cell 3′ v2 reagent kits by the Center for Applied Genomics of The Children’s Hospital of Philadelphia (CHOP). It is worth noting that the 10x Genomics single-cell 3′ solution workflow supports cDNA amplification only from whole cells, possibly due to inefficient lysis of nuclei purified via sucrose gradient ultracentrifugation. 10× genomics recently released a specific nucleus isolation and purification protocol (https://support.10xgenomics.com/single-cell-gene-expression/index/doc/) that may support single-nucleus RNA-Seq analysis on the 10× genomics platform.
Microfluidics device for sNucDrop-Seq
To examine that the channel depth of the microfluidics device (custom-made by μFluidix) used for sNucDrop-Seq, droplet volume was measured using 10 μm carboxylated polystyrene fluorescent beads (Bangs Labs), as previously described (described by Macosko et al., 2015). The beads were first suspended at a concentration of 1,000 beads/μL in lysis buffer (200 mM Tris-HCl pH8.0, 20 mM EDTA, 6% Ficoll PM-400, 0.2% Sarkosyl, and 50 mM DTT). Drop-Seq was performed as previously described, with default settings (Macosko et al., 2015), except the syringe pump intended for cells was loaded with DPBS. The total number of beads encapsulated in several hundred droplets was determined, and droplet volume was calculated, as follows:
We calculated the droplet volume (1.2 nL), and estimated the radius of droplets (1/2 of channel depth) as 65μm. This confirms that the channel depth of microfluidics devices used for sNucDrop-Seq experiments is approximately ~130μm.
It is worth noting that while both sNucDrop-Seq (this study) and DroNc-Seq (Habib et al., 2017) have adapted the Drop-Seq pipeline for single-nucleus RNA-Seq analysis of mammalian brains, there are several technical differences between the two. First, different nuclei purification methods were employed (sNucDrop-Seq: sucrose-gradient ultracentrifugation; DroNc-Seq: a commercially available kit from Sigma). Second, while sNucDrop-Seq utilizes standard Drop-Seq microfluidics devices (channel depth: 125 um), DroNc-Seq requires a custom-made, modified device (channel depth: 75 um). Because of the reduced channel depth, the DroNc-Seq specific device is more likely to clog and requires pre-filtering of barcoded beads to enrich beads of smaller sizes (causing 50% loss of this expensive reagent). Third, re-analysis of raw DroNc-Seq data using our own pipeline (utilizing both exonic and intronic reads) suggests that sNucDrop-Seq requires less reads per nucleus (~50% reads) to achieve similar cell-type specific gene expression signatures and clustering assignment as compared to DroNc-Seq (the performance is benchmarked by pairwise comparison between single-nucleus analysis and the deeply sequenced single-cell analysis study (Tasic et al., 2016). Thus, while sNucDrop-Seq requires special instrument such as ultracentrifuge, sNucDrop-Seq is more cost-effective than DroNc-Seq.
QUANTIFICATION AND STATISTICAL ANALYSIS
Preprocessing of sNucDrop-Seq data
Paired-end sequencing reads of Single-nucleus RNA-seq were processed using publicly available the Drop-Seq Tools v1.12 software (Macosko et al., 2015) with some modifications. Briefly, each mRNA read (read2) was tagged with the cell barcode (bases 1 to 12 of read 1) and unique molecular identifier (UMI, bases 13 to 20 of read 1), trimmed of sequencing adaptors and poly-A sequences, and aligned using STAR v 2.5.2a to the mouse (mm10, Gencode release vM13) or a concatenation of the mouse and human (for the species-mixing experiment) reference genome assembly. Because a substantial proportion (~50%) of reads derived from nuclear transcriptomes of mouse cortices were mapped to the intronic regions, the intronic reads were retained for downstream analysis. A custom Perl script was implemented in the Drop-Seq Tools pipeline to retrieve both exonic and intronic reads mapped to predicted strands of annotated genes. Uniquely mapped reads were grouped by cell barcodes. Cell barcodes were corrected for possible bead synthesis errors, using the DetectBeadSynthesisErrors program from the Drop-Seq Tools v1.12 software. To generate digital expression matrix, a list of UMIs in each gene (as rows), within each cell (as columns), was assembled, and UMIs within ED = 1 were merged together. The total number of unique UMI sequences was counted, and this number was reported as the number of transcripts of that gene for a given cell.
Comparison of Drop-Seq (whole cells) and sNucDrop-Seq (nuclei)
Only mouse NIH3T3 cells or nuclei that expressed >800 genes were retained for analysis. For each gene, the average normalized expression level was calculated as log (normalized UMI counts + 1).
Clustering and marker gene identification
Raw digital expression matrices were combined and loaded into the R package Seurat. For normalization, UMI counts for all nuclei were scaled by library size (total UMI counts), multiplied by 10,000 and transformed to log space. Only genes found to be expressing in >10 cells were retained. Nuclei with a relatively high percentage of UMIs mapped to mitochondrial genes (>=0.1) were discarded. Moreover, nuclei with fewer than 800 or more than 6,000 detected genes were omitted, resulting in 20,858 nuclei that pass filter.
Before clustering, batch effects from multiple animals were regressed out using the function RegressOut in R package Seurat (Macosko et al., 2015). The highly variable genes were identified using the function MeanVarPlot with the parameters: x.low.cutoff = 0.0125, x.high.cutoff = 3 and y.cutoff = 0.8, resulting in an output of 1,932 highly variable genes. The expression level of highly variable genes in the nuclei was scaled and centered along each gene, and was conducted to principal component analysis. We then used two methods to assess the number of PCs to be utilized in downstream analysis: 1) The cumulative standard deviations of each PC were plotted using the function PCElbowPlot in Seurat to identify the ‘knee’ point at a PC number after which successive PCs explain diminishing degrees of variance, and 2) the significance for each gene’s association with each PC was accessed by the function JackStraw in Seurat. Based on these two methods, we selected first 30 PCs for two-dimensional t-distributed stochastic neighbor embedding (tSNE), implemented by the Seurat software with the default parameters. Based on the tSNE map, twenty-one clusters were identified using the function FindCluster in Seurat with the resolution parameter set to 2. Clusters that co-express both non-neuronal and neuronal markers, representing cell doublets, were removed. Based on expression of well-established marker genes, we assigned 19,241 nuclei to cerebral cortex (93.1% of our data) and 1,415 nuclei to non-cortical neurons (6.9%). To perform clustering analysis on 19,241 cortical cells, we identified 1,968 highly variable genes and selected first 40 PCs. The nuclei were classified into 44 clusters with the resolution parameter set to 4. To identify clusters with highly similar identity, we performed pairwise comparison to identify differentially expressed genes using the function FinderMarkers in Seurat, with likelihood-ratio test. We merged the clusters exhibiting less than 10 genes with an average expression difference greater than 2-fold between clusters. Based on two-dimensional coordinates of nuclei in the tSNE plot, a density-based clustering method implemented in the DBSCAN R package was used to identify outliers, filtering out 882 nuclei (4.5% of 19,241 nuclei) with the reachability distance parameter (eps) setting to 0.9 and minimal number of nuclei within that eps radius setting to 10. Finally, we excluded clusters containing expression markers for more than one canonical cell type. As a result of these steps, we were able to assign 18,194 (94.6% of cortical nuclei) nuclei into 40 clusters.
To identify the marker genes, differential expression analysis was performed by the function FindAllMarkers in Seurat with likelihood-ratio test. Differentially expressed genes that were expressed at least in 10% cells within the cluster and with a fold change more than 0.5 (log scale) were considered to be marker genes. In total, 2,001 protein-coding genes and 90 long non-coding RNAs were identified for 40 clusters. For the marker genes, average gene expression for each cluster was determined, and Euclidean distances between all pairs was calculated. This dataset was used as input for complete linkage hierarchical clustering and dendrogram assembly. To generate a heatmap of marker genes across clusters, the average expression level of marker genes within each cluster were calculated. For each cluster, the average expression was centered and scaled by each gene. Next, the hclust function in the R was used to generate the cluster dendrogram with the “ward.D” method.
Sub-clustering of cortical and non-cortical inhibitory neurons
Cortical inhibitory neuronal nuclei from cluster Inh 1–7 were first combined with non-cortical nuclei. We then identified 2,139 highly variable genes in a total of 3,425 nuclei and performed PCA analysis. The significance of PCs was examined by the Jackstraw function in Seurat. First 23 significant PCs with P-value < 1E-8 were selected for clustering analysis and 19 sub-clusters were identified with the resolution setting to 2. After merging clusters, identifying/removing outliers (DBSCAN), and filtering cell doublets, we identified 9 cortical inhibitory neuronal subgroups (1,810 nuclei), and 8 sub-clusters for non-cortical nuclei (1,462 nuclei).
Identification of differentially expressed genes
Differential gene expression analysis between PTZ treatment and saline groups or between active and inactive neurons was performed using the function FindMarkers in Seurat, using a likelihood ratio test. For the PTZ and saline comparison, genes with a fold-change of more than 0.25 (log scale) and a P-value less than 0.01 were considered to be differentially expressed, while genes showing 0.25-fold change (log-scale) with P-value < 1E-3 were defined as differentially expressed between active and inactive neurons. For the PTZ and saline comparison within each cluster, only the clusters containing at least 20 nuclei of both two conditions, PTZ and saline, were tested.
Gene Set Enrichment Analysis of neuronal activity-regulated genes (ARGs)
The list of 172 ARGs was previously defined (Tyssowski et al., 2017). These ARGs were classified into 3 groups: rapid primary response genes (rapid PRG), delayed primary response genes (delayed PRG) and secondary response genes (SRG) based on the expression pattern across the time points in response to the KCl stimulation of cultured primary neurons (Tyssowski et al., 2017). For each cluster, only genes that were expressed in at least of 10% nuclei were considered and the expression matrix of those genes in each cluster was loaded into the GSEA analysis (www.broadinstitue.org/gsea/index.jsp) (Subramanian et al., 2005). Clusters with an FDR < 0.25 were considered significant.
Alternative splicing analysis
To identify the cell-type specific alternative splicing events, we classify the 40 clusters into 8 cell types: excitatory neuronal layer 2/3 (Cluster Ex5, Ex6, Ex24, Ex25, Ex26, and Ex27), excitatory neuronal layer 4 (cluster Ex21, Ex22, and Ex23), excitatory neuronal layer 5/6 (Cluster Ex1, Ex2, Ex3, Ex4, Ex7, Ex8, Ex9, Ex10, Ex12, Ex19 and Ex20), excitatory neuronal layer 6 (Cluster Ex11, Ex13, Ex14, Ex15, Ex16, Ex17 and Ex18), inhibitory neuronal Sst+ (Cluster Inh1 and Inh2), inhibitory neuronal Pvalb+ (Cluster Inh3 and Inh4), other inhibitory neurons (Cluster Inh5, Inh6 and Inh7), and non-neuronal cell types (Cluster EC, MG, Astro, OPC, Oligo1 and Oligo2). The bam files of the nuclei from the same cell type were merged and sorted by Samtools. Pairwise comparison of differential splicing events between cell types was performed using MISO (Katz et al., 2010). Splicing events with a bayes factor (>10), ∆PSI > 0.2; at least 1 read supports the inclusion or exclusion isoform, and at least 10 reads supports either of these events.
Comparison of sNucDrop-Seq data to other single-cell/nucleus RNA-Seq data sets
To assess the validity of sNucDrop-seq results, we compare our data with recently published single-nucleus RNA-Seq data from mouse prefrontal cortex (Habib et al., 2017) or single-cell RNA-Seq data derived from select mouse visual cortex (Tasic et al., 2016). To compare cell-type specific expression signatures defined by sNucDrop-Seq and those of other data sets, we computed the pairwise Pearson correlation coefficients between each pair of cell types in other data sets and our sNucDrop-Seq data set for a common set of genes. To generate a common marker gene list for the pairwise comparison, we identified 1,986 (95.0%) and 1,919 (91.8%) common marker genes for the DroNuc-Seq data set (Figure 1G) and the single-cell data set (Figure S3C), respectively. Average natural log transformed scaled UMI counts or TPM counts were used to generate the DroNc-Seq (Habib et al., 2017) and single-cell RNA-Seq (Tasic et al., 2016) gene expression matrix, respectively. R function cor.test was used to calculate the pairwise Pearson correlation coefficients.
To further examine whether GABAergic neuronal subclusters identified in this study agree with a previous single-cell RNA-Seq study of well-characterized transgenic mouse lines (Tasic et al., 2016), we adopted a random forest classifier-based method (Shekhar et al., 2016) (Figure 2E). First, we used five major GABAergic neuron sub-types defined by sNucDrop-Seq to train a random forest classifier on our sNucDrop-seq dataset. We first sampled 60% of nuclei from each cell type to build the training set of 1,204 nuclei, and the remaining 40% nuclei (770) were used as test set for evaluating the performance of the trained classifier. We then identified the 1,182 most highly variable genes using the Seurat function MeanVarPlot in with the following parameters: x.low.cutoff = 0.0125, x.high.cutoff = 4 and y.cutoff = 0.8. The digital expression matrix comprising 1,182 genes across 1,204 nuclei was used to build the classifier. We trained a random forest classifier using the R package randomForest with the ntree parameter set to 1,000. By applying this classifier to assign the nuclei in the test set, 96.9% of the nuclei were correctly mapped to their classes. Then the classifier was used to map the cells in single cell dataset from transgenic Cre lines (Tasic et al., 2016).
By utilizing a recently published computational strategy for integrated analysis of multiple data sets (implemented in Seurat v2.0), we further performed comparative analysis of the DroNc-Seq data set (Habib et al., 2017) and this study (Figures S3E–G). First, we downloaded the raw DroNc-seq data (mouse prefrontal cortex) and generated the digital expression matrix using our pipeline that utilizes both exonic and intronic reads. After filtering low quality nuclei, 5,441 nuclei from DroNc-seq and all 18,194 nuclei from our dataset with > 800 genes expressed were imported into Seurat. Then, the union of the top 1,000 genes with the highest dispersion from both datasets was subject to a canonical correlation analysis (CCA) to identify common sources of variation between the two datasets. The first 15 dimensions of the CCA was chosen to align the two datasets. Nuclei whose expression level cannot be explained by those 15 CCA were removed. The remaining nuclei, that passed filter, were subject to clustering with the resolution parameter set to 1.2. We assigned 23,213 (98.2% of input data) comprised 5,337 DroNuc-seq nuclei and 17,876 scNucDrop-seq nuclei into 18 clusters.
Identification of nuclei expressing ARGs
To evaluate the fraction and heterogeneity of ARG-expressing nuclei in cortical excitatory neurons (Figure 3D), we examined the pattern of 4 selected ARGs (rapid PRGs: Fos and Egr1; delayed PRGs: Nr4a3 and Pcsk1). Based on the distribution of gene expression levels between active (Ex24) and inactive (Ex25) neurons, we determined the baseline expression level of these ARGs, post-hoc. The nuclei whose expression level was higher than the baseline ARG expression level were classified as ARG-expressing nuclei. We then examined whether the proportions of ARG-expressing nuclei were higher in the active neurons compared to the inactive neurons using Fisher’s exact test (Table S2).
To compare the proportion of IEG-expressing nuclei across all the cell types (Figure S4A), mean and standard deviation (SD) of selected ARGs were computed. Nuclei whose expression level of ARGs was higher than mean+2SD are defined as ARG-expressing nuclei. Then the percentage of ARG-expressing nuclei was reported for each cluster, defined in Figure 1C.
DATA AND SOFTWARE AVAILABILITY
The next-generation sequencing data reported in this study have been deposited to the Gene Expression Omnibus (GEO). The accession number for this data is GSE106678. R markdown scripts for data analysis are available from the corresponding author, upon request.
ADDITIONAL RESOURCES
Detailed Protocol
A detailed bench protocol is supplied in an attached file, entitled Methods S1.
Supplementary Material
Acknowledgments
We thank X. Kang for technical support. H.W. is supported by National Human Genome Research Institute (NHGRI) grant R00HG007982, National Heart Lung and Blood Institute (NHLBI) grant DP2HL142044, and the Penn Epigenetics Institute Pilot Grant. Z.Z is supported by National Institute of Mental Health (NIMH) grant R56MH111719. E.F. is supported by the T32 Training Program in Cell and Molecular Biology (T32-GM007229). D.Y.K. is supported by the T32 Training Program in Neurodevelopmental disabilities (T32-NS007413). S.T. is supported by the NRAS Individual Predoctoral Fellowship (F32-NS100433).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
AUTHOR CONTRIBUTIONS
H.W. and Z.Z. conceived the study. H.W., P.H., E.F., D.K., and S.T. performed experiments. H.W. and P.H. carried out data analysis. H.W. wrote the manuscript with input from other authors.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
References
- Aten S, Hansen KF, Hoyt KR, Obrietan K. The miR-132/212 locus: a complex regulator of neuronal plasticity, gene expression and cognition. RNA Dis. 2016;3 [PMC free article] [PubMed] [Google Scholar]
- Chen F, Wassie AT, Cote AJ, Sinha A, Alon S, Asano S, Daugharthy ER, Chang JB, Marblestone A, Church GM, et al. Nanoscale imaging of RNA with expansion microscopy. Nature methods. 2016;13:679–684. doi: 10.1038/nmeth.3899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen KH, Boettiger AN, Moffitt JR, Wang S, Zhuang X. RNA imaging. Spatially resolved, highly multiplexed RNA profiling in single cells. Science. 2015;348:aaa6090. doi: 10.1126/science.aaa6090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flavell SW, Greenberg ME. Signaling mechanisms linking neuronal activity to gene expression and plasticity of the nervous system. Annu Rev Neurosci. 2008;31:563–590. doi: 10.1146/annurev.neuro.31.060407.125631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habib N, Avraham-Davidi I, Basu A, Burks T, Shekhar K, Hofree M, Choudhury SR, Aguet F, Gelfand E, Ardlie K, et al. Massively parallel single-nucleus RNA-seq with DroNc-seq. Nature methods. 2017;14:955–958. doi: 10.1038/nmeth.4407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habib N, Li Y, Heidenreich M, Swiech L, Avraham-Davidi I, Trombetta JJ, Hession C, Zhang F, Regev A. Div-Seq: Single-nucleus RNA-Seq reveals dynamics of rare adult newborn neurons. Science. 2016;353:925–928. doi: 10.1126/science.aad7038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson BS, Zhao YT, Fasolino M, Lamonica JM, Kim YJ, Georgakilas G, Wood KH, Bu D, Cui Y, Goffin D, et al. Biotin tagging of MeCP2 in mice reveals contextual insights into the Rett syndrome transcriptome. Nature medicine. 2017;23:1203–1214. doi: 10.1038/nm.4406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katz Y, Wang ET, Airoldi EM, Burge CB. Analysis and design of RNA sequencing experiments for identifying isoform regulation. Nature methods. 2010;7:1009–1015. doi: 10.1038/nmeth.1528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kepecs A, Fishell G. Interneuron cell types are fit to function. Nature. 2014;505:318–326. doi: 10.1038/nature12983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein AM, Mazutis L, Akartuna I, Tallapragada N, Veres A, Li V, Peshkin L, Weitz DA, Kirschner MW. Droplet barcoding for single-cell transcriptomics applied to embryonic stem cells. Cell. 2015;161:1187–1201. doi: 10.1016/j.cell.2015.04.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacar B, Linker SB, Jaeger BN, Krishnaswami S, Barron J, Kelder M, Parylak S, Paquola A, Venepally P, Novotny M, et al. Nuclear RNA-seq of single neurons reveals molecular signatures of activation. Nat Commun. 2016;7:11022. doi: 10.1038/ncomms11022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lake BB, Ai R, Kaeser GE, Salathia NS, Yung YC, Liu R, Wildberg A, Gao D, Fung HL, Chen S, et al. Neuronal subtypes and diversity revealed by single-nucleus RNA sequencing of the human brain. Science. 2016;352:1586–1590. doi: 10.1126/science.aaf1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lister R, Mukamel EA, Nery JR, Urich M, Puddifoot CA, Johnson ND, Lucero J, Huang Y, Dwork AJ, Schultz MD, et al. Global epigenomic reconfiguration during Mammalian brain development. Science. 2013;341:1237905. doi: 10.1126/science.1237905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macosko EZ, Basu A, Satija R, Nemesh J, Shekhar K, Goldman M, Tirosh I, Bialas AR, Kamitaki N, Martersteck EM, et al. Highly Parallel Genome-wide Expression Profiling of Individual Cells Using Nanoliter Droplets. Cell. 2015;161:1202–1214. doi: 10.1016/j.cell.2015.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madisen L, Garner AR, Shimaoka D, Chuong AS, Klapoetke NC, Li L, van der Bourg A, Niino Y, Egolf L, Monetti C, et al. Transgenic mice for intersectional targeting of neural sensors and effectors with high specificity and performance. Neuron. 2015;85:942–958. doi: 10.1016/j.neuron.2015.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mo A, Mukamel EA, Davis FP, Luo C, Henry GL, Picard S, Urich MA, Nery JR, Sejnowski TJ, Lister R, et al. Epigenomic Signatures of Neuronal Diversity in the Mammalian Brain. Neuron. 2015;86:1369–1384. doi: 10.1016/j.neuron.2015.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan JI, Cohen DR, Hempstead JL, Curran T. Mapping patterns of c-fos expression in the central nervous system after seizure. Science. 1987;237:192–197. doi: 10.1126/science.3037702. [DOI] [PubMed] [Google Scholar]
- Nudelman AS, DiRocco DP, Lambert TJ, Garelick MG, Le J, Nathanson NM, Storm DR. Neuronal activity rapidly induces transcription of the CREB-regulated microRNA-132, in vivo. Hippocampus. 2010;20:492–498. doi: 10.1002/hipo.20646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renier N, Adams EL, Kirst C, Wu Z, Azevedo R, Kohl J, Autry AE, Kadiri L, Umadevi Venkataraju K, Zhou Y, et al. Mapping of Brain Activity by Automated Volume Analysis of Immediate Early Genes. Cell. 2016;165:1789–1802. doi: 10.1016/j.cell.2016.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudy B, Fishell G, Lee S, Hjerling-Leffler J. Three groups of interneurons account for nearly 100% of neocortical GABAergic neurons. Dev Neurobiol. 2011;71:45–61. doi: 10.1002/dneu.20853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwanhausser B, Busse D, Li N, Dittmar G, Schuchhardt J, Wolf J, Chen W, Selbach M. Global quantification of mammalian gene expression control. Nature. 2011;473:337–342. doi: 10.1038/nature10098. [DOI] [PubMed] [Google Scholar]
- Shah S, Lubeck E, Zhou W, Cai L. In Situ Transcription Profiling of Single Cells Reveals Spatial Organization of Cells in the Mouse Hippocampus. Neuron. 2016;92:342–357. doi: 10.1016/j.neuron.2016.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shekhar K, Lapan SW, Whitney IE, Tran NM, Macosko EZ, Kowalczyk M, Adiconis X, Levin JZ, Nemesh J, Goldman M, et al. Comprehensive Classification of Retinal Bipolar Neurons by Single-Cell Transcriptomics. Cell. 2016;166:1308–1323 e1330. doi: 10.1016/j.cell.2016.07.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorensen AT, Cooper YA, Baratta MV, Weng FJ, Zhang Y, Ramamoorthi K, Fropf R, LaVerriere E, Xue J, Young A, et al. A robust activity marking system for exploring active neuronal ensembles. Elife. 2016;5 doi: 10.7554/eLife.13918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanay A, Regev A. Scaling single-cell genomics from phenomenology to mechanism. Nature. 2017;541:331–338. doi: 10.1038/nature21350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tasic B, Menon V, Nguyen TN, Kim TK, Jarsky T, Yao Z, Levi B, Gray LT, Sorensen SA, Dolbeare T, et al. Adult mouse cortical cell taxonomy revealed by single cell transcriptomics. Nat Neurosci. 2016;19:335–346. doi: 10.1038/nn.4216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyssowski KM, Saha RN, DeStefino NR, Cho J, Jones RD, Chang SM, Romeo P, Wurzelmann MK, Ward JM, Dudek SM, Gray JM. Distinct neuronal activity patterns induce different gene expression programs. In bioRxiv. 2017 doi: 10.1016/j.neuron.2018.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White RB, Bierinx AS, Gnocchi VF, Zammit PS. Dynamics of muscle fibre growth during postnatal mouse development. BMC Dev Biol. 2010;10:21. doi: 10.1186/1471-213X-10-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu YE, Pan L, Zuo Y, Li X, Hong W. Detecting Activated Cell Populations Using Single-Cell RNA-Seq. Neuron. 2017;96:313–329 e316. doi: 10.1016/j.neuron.2017.09.026. [DOI] [PubMed] [Google Scholar]
- Yount GL, Ponsalle P, White JD. Pentylenetetrazole-induced seizures stimulate transcription of early and late response genes. Brain Res Mol Brain Res. 1994;21:219–224. doi: 10.1016/0169-328x(94)90252-6. [DOI] [PubMed] [Google Scholar]
- Zeisel A, Munoz-Manchado AB, Codeluppi S, Lonnerberg P, La Manno G, Jureus A, Marques S, Munguba H, He L, Betsholtz C, et al. Brain structure. Cell types in the mouse cortex and hippocampus revealed by single-cell RNA-seq. Science. 2015;347:1138–1142. doi: 10.1126/science.aaa1934. [DOI] [PubMed] [Google Scholar]
- Zeng W, Jiang S, Kong X, El-Ali N, Ball AR, Jr, Ma CI, Hashimoto N, Yokomori K, Mortazavi A. Single-nucleus RNA-seq of differentiating human myoblasts reveals the extent of fate heterogeneity. Nucleic Acids Res. 2016;44:e158. doi: 10.1093/nar/gkw739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng GX, Terry JM, Belgrader P, Ryvkin P, Bent ZW, Wilson R, Ziraldo SB, Wheeler TD, McDermott GP, Zhu J, et al. Massively parallel digital transcriptional profiling of single cells. Nat Commun. 2017;8:14049. doi: 10.1038/ncomms14049. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


