Abstract
Epigenetic mechanisms control various functions throughout the body, from cell fate determination in development to immune responses and inflammation. Neuroinflammation is one of the prime contributors to the initiation and progression of neurodegeneration in a variety of diseases, including Alzheimer’s and Parkinson’s diseases. Because astrocytes are the largest population of glial cells, they represent an important regulator of CNS function, both in health and disease. Only recently have studies begun to identify the epigenetic mechanisms regulating astrocyte responses in neurodegenerative diseases. These epigenetic mechanisms, along with the epigenetic marks involved in astrocyte development, could elucidate novel pathways to potentially modulate astrocyte-mediated neuroinflammation and neurotoxicity. This review examines the known epigenetic mechanisms involved in regulation of astrocyte function, from development to neurodegeneration, and links these mechanisms to potential astrocyte-specific roles in neurodegenerative disease with a focus on potential opportunities for therapeutic intervention.
Keywords: astrocyte, epigenetic, neurodegeneration, neuroinflammation, histone, DNA methylation
Excessive neuroinflammation is one of the pathogenic hallmarks of neurodegenerative diseases and is thought to contribute both to the initiation and progression of neurodegeneration (Reviewed by [1]; and [2]). Therefore, understanding the regulatory mechanisms involved in components of the inflammatory response has been an intense area of study. Although the vast majority of research to date has focused on microglia as key regulators of neuroinflammation in neurodegeneration, existing and emerging data highlight the importance and contribution of astrocytes to the inflammation found in neurodegenerative diseases [3–6]. Astrocyte reactivity is a sensitive and robust response to many different types of pathology and is regulated by a wide array of processes, including epigenetic regulation. Growing evidence indicates that astrocytes are important in protection of neurons, while at the same time, they can also contribute to neuroinflammation and neurodegeneration when converted to a reactive state. However, many of the mechanisms surrounding astrocyte-induced neuroprotection or neurotoxicity are still not well-established. Therefore, greater understanding of these factors, including epigenetic alterations that regulate astrocyte functions, could better elucidate the precise role of astrocytes in neurodegenerative diseases and provide a pathway to target astrocytes for neuroprotection.
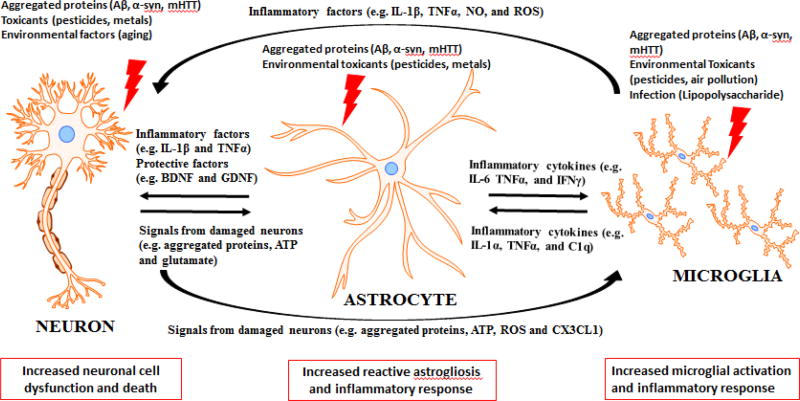
Astrocytes are the largest population of glial cells in the CNS and were traditionally viewed as only structural elements supporting brain structure (Hence their name, meaning “glue” in Greek. [7]). However, it is now obvious that astrocytes have many other important functions in the CNS. Proper astrocyte function is essential for normal development, including developmental synapse formation and pruning through both secreted factors and direct contact with the synapse [8, 9]. In adults, evidence indicates that hippocampal astrocytes have an important role in promoting neurogenesis from adult neural stem cells [10]. Similarly, astrocytes exhibit extensive processes that contact neural elements, especially synapses and nodes of Ranvier, and astrocyte end feet cover nearly all of the brain microvasculature. These contacts position astrocytes as the main conduit for neuron-glia interactions [11]. Astrocytes, far from being merely physical support cells, actively modulate synaptic glutamate levels, scavenge free radicals, produce neurotrophic factors, and regulate the blood brain barrier [7, 12, 13]. The importance of astrocytes for neuronal survival was confirmed by the study that found conditional ablation of astrocytes in adult mice led to significant neurodegeneration [14]. Additionally, multiple reports have demonstrated that reactive astrocytes may exacerbate inflammatory processes that can lead to neurodegeneration [3, 15]. Indeed, astrocytes exposed to an inflammatory stimulus also produce pro-inflammatory cytokines, such as interleukin-1β (IL-1β) and TNF-α, along with increased production of reactive oxygen species (ROS) [16]. This response is known as reactive astrogliosis, and is the mechanism by which astrocytes respond to injuries such as trauma, infection, misfolded protein accumulation, and excitotoxicity in neurodegenerative disease such as Parkinson’s disease (PD), Alzheimer’s disease (AD), Huntington’s disease (HD) and amyotrophic lateral sclerosis (ALS) (reviewed in [17]). The cell-to-cell communication of astrocytes, neurons, and microglia in neurodegenerative diseases is detailed in Figure 1, with the factors involved in spreading these damaging factors between cell types.
Figure 1. Factors involved in the cell-to-cell communication between neurons, astrocytes and microglia in neurodegenerative diseases.
Neurodegenerative diseases have several hallmarks in common, which include aggregated proteins and neuroinflammation. The three major cell types involved in neurodegenerative diseases are neurons, astrocytes and microglia. These cells communicate to each other and can influence the response to various factors. Neurons can be damaged by factors such as aggregated proteins (including amyloid β (Aβ), α-synuclein (α-syn), or mutant Huntingtin protein (mHTT)), toxicants, or environmental factors. These damaged neurons can signal to both microglia and astrocytes through aggregated proteins, ATP, reactive oxygen species (ROS), glutamate and Cx3cl1. Once microglia and astrocytes are exposed to damaging factors such as aggregated proteins or environmental toxicants they respond with increased inflammatory factors (such as inflammatory cytokines IL-1β, and TNFα while also producing reactive oxygen and nitrogen species) that can signal to the other two cells. Overall, the consequence of this cell-to-cell communication is that dysfunction in one of these cell types can lead to dysfunction in the other two as well. Astrocytes undergo reactive astrogliosis and microglia become activated, which along with the original damaging factor, can lead to increased neuronal cell dysfunction and death, and this culminates to produce the disease states found in neurodegenerative diseases.
Reactive astrogliosis is an evolutionary conserved response that is a constitutive, graded, and multi-staged process [18]. This reactive state is characterized by both molecular and morphological changes. These include increased expression of the glial fibrillary acidic protein (GFAP), vimentin, and nestin, accompanying a reduction in the number of primary branches and hypertrophy of the cell body and remaining processes [19]. Recently, Liddelow et al. demonstrated that pro-inflammatory microglia can induce conversion into reactive astrocytes, and that these reactive astrocytes are cytotoxic to neurons [20]. Complicating the issue however, Rappold and Tieu reported that astrocytes in vivo did not respond in a uniform manner, and instead represented a heterogeneous population [21], some of which are protective. Therefore, there is a need to elucidate the mechanisms surrounding astrocyte responses to different neurologic conditions and the various neurodegenerative diseases. A comparison of astrocyte gene expression in either a model of ischemia or an endotoxin inflammatory model demonstrated that at least 50% of the injury-altered gene expression was disease specific [22]. These points underscore the fact that astrocytes are a complex CNS cell-type that exhibits a differential, heterogeneous response depending on context. Thus, understanding the mechanisms leading to regulation and differential responses may provide insight into the pathogenic process of neurodegeneration and identify novel targets for intervention.
Emerging data demonstrate that epigenetic mechanisms regulate the innate and adaptive immune response, including macrophages, T lymphocytes and microglia. For instance, tri-methylation of histone 3 lysine 27 (H3K27) leads to an increased inflammatory phenotype of macrophages and microglia, also known as an M1 response, while the H3K27 histone demethylase jumonji-domain containing protein 3 (Jmjd3) is essential in promoting the anti-inflammatory M2 phenotype in microglia [23]. Treatment with lipopolysaccharide (LPS) as an inflammatory stimulus induced hypomethylation of sites in the TNFα promoter and increases production of this inflammatory factor [24]. Further, PD patients show hypomethylation of the tumor necrosis factor alpha (TNFα) promoter, a major inflammatory factor found in this disease [25]. Given recent research, it is likely that the different responses of astrocytes in neurodegenerative diseases arise, in part, from differential epigenetic regulation of various functions. However, the examination of the epigenetic regulation of astrocyte function has not been reviewed, including the relationship of these epigenetic mechanisms to neurodegenerative diseases. The focus of this review is to assess the current state of knowledge regarding epigenetic regulation of astrocytes and the potential role for this process in neurodegeneration.
Epigenetics in Neurodegenerative Disease
Although the concept of epigenetic regulation of gene expression is over 70 years old, the field of epigenetics has recently exploded, with 98% of epigenetic research published in the last 15 years [26]. Many CNS functions have significant epigenetic components, including neural stem cell fate determination, neural plasticity, and learning and memory (Reviewed by [27]; and [28]). With regard to neurodegenerative disease, dysregulation of DNA methylation, histone modifications, and miRNAs are present in multiple neurodegenerative diseases, including Alzheimer’s disease (AD), Parkinson’s disease (PD), amyotrophic lateral sclerosis (ALS), and stroke [29–32]. This is not surprising, as age is the major risk factor for several neurodegenerative diseases, including AD and PD, and increased age has been associated with alterations in DNA methylation [33]. AD patients exhibited higher methylation of repetitive DNA elements compared to controls, which could result in altered global DNA methylation levels [34]. In PD, epigenetic mechanisms have been suggested to play a substantial role, with decreased methylation of the α-synuclein (SNCA) gene [35]. This could be an important contributing factor leading to the increased accumulation of α-synuclein, which forms the basis of the Lewy bodies seen in PD patients [36]. Multiple studies have also investigated pharmacological intervention of epigenetic mechanisms in neurodegenerative diseases. For instance, Pan et al. (2016) reports that inhibiting DNA methyltransferases can be beneficial in Huntington’s disease [37]. Similarly, DNA methyltransferase inhibitors have been found to modulate the outcome in animal models of neurodegenerative diseases [38–41]. Histone deacylases are the most targeted epigenetic mechanism for neurodegenerative diseases, with multiple histone deacetylase inhibitors demonstrated to be neuroprotective in animal models of aging, AD, PD, Huntington’s disease (HD), Multiple sclerosis (MS) and ischemia [37, 42–77]. Other epigenetic therapies that have demonstrated the ability to alter the disease course in animal models of neurodegenerative diseases include Sirtuin 1 activators [78–82] and histone acetyltransferase (HAT) activators [83, 84]. MicroRNAs have also been implicated in multiple neurodegenerative diseases, including PD and AD (reviewed by [85]), with several miRNA therapies tested in animal models of neurodegeneration [86–93]. Table 1 outlines the current epigenetic therapeutic options that have been tested in animal models of neurodegenerative diseases and the effects of these drugs on disease course.
Table 1.
Epigenetic therapeutics tested in animal models of neurodegenerative diseases.
| Target | Therapeutic | Disease | Result | References |
|---|---|---|---|---|
| DNA methylation | S-adenosylmethionine (SAM) supplementation | AD |
|
38, 39 |
| ALS |
|
40 | ||
| DNA methyltansferase inhibitors | HD |
|
37 | |
| Ischemia |
|
41 | ||
| HDAC inhibition | Sodium Butyrate | AD |
|
48 |
| PD |
|
49, 50 | ||
| HD |
|
51 | ||
| Phenylbutyrate | AD |
|
52 | |
| PD |
|
53, 55, 56 | ||
| ALS |
|
57 | ||
| HD |
|
54 | ||
| Ischemia |
|
58 | ||
| Valproate/valproic acid | AD |
|
59 | |
| PD |
|
60 | ||
| ALS |
|
61 | ||
| HD |
|
62 | ||
| Ischemia |
|
63 | ||
| Trichostatin A (TSA) | AD |
|
64 | |
| PD |
|
65 | ||
| ALS |
|
66 | ||
| Ischemia |
|
67 | ||
| Suberoylanilide hydroxamic acid (SAHA) | AD |
|
68, 69 | |
| PD |
|
70 | ||
| Ischemia |
|
71 | ||
| HD |
|
72 | ||
| Sirtuin inhibitors | AD |
|
73 | |
| PD |
|
74, 75 | ||
| ALS |
|
75 | ||
| HD |
|
76, 77 | ||
| Ischemia |
|
75 | ||
| Sirtuin 1 activation | Resveratrol | AD |
|
78 |
| PD |
|
79 | ||
| ALS |
|
80 | ||
| HD |
|
81 | ||
| Ischemia |
|
82 | ||
| HAT activation | CBP/p300 activator | AD |
|
83 |
| SPV-106 | AD |
|
84 | |
| miRNA therapies | miRNA delivery | PD |
|
91 |
| HD |
|
92 | ||
| Ischemia |
|
93 | ||
| Antagomirs (Oligonucleotides that act as specific silencers of endogenous miRNAs) | AD |
|
86 | |
| ALS |
|
87, 88 | ||
| HD |
|
89 | ||
| Ischemia |
|
90 |
Multiple therapeutic interventions have been tested in animal models of neurodegenerative diseases with targets ranging from DNA methylation, histone deacetylase (HDAC) inhibition, histone deacetylase Sirt 1 activation, histone acetyltrasferase (HAT) activation, and microRNAs (miRNAs). Therapeutics directed to these targets have demonstrated the ability to attenuate disease phenotypes in Alzheimer's (AD), Parkinson's (PD), Huntington's (HD), amyotrophic lateral sclerosis (ALS) and cerebral ischemia.
Thus, epigenetic alterations may play a significant role in these complex neurodegenerative diseases through a variety of pathways. Understanding how epigenetic mechanisms contribute to the development or progression of these diseases could offer a novel approach to treatment. The following sections will discuss the known epigenetic mechanisms found in various astrocyte functions, from DNA methylation to the various histone modifications and finally the involvement of miRNAs. We also relate these findings to the overall role of epigenetics in the regulation of reactive astrocytes in neurodegenerative diseases.
Epigenetic Regulation of Astrocyte Development and Function
DNA Methylation
DNA methylation generally leads to repression of gene expression through the addition of methyl groups to the gene promoter which blocks binding of transcriptional enzymes [94]. A family of DNA methyltransferases (DNMTs) adds these methyl groups to gene promoters. DNMTs add methyl groups to CpG sites, or cytosine residues in DNA directly followed by a guanidine residue on the 3’ prime side of the same DNA strand. Regions with high numbers of CpG sites, at least 200bp region with higher than 60% observed-to-expected CpG ratio, are designated CpG islands and are mainly found in the gene promoter upstream of the transcription start site. Methylation of CpG islands in a gene promoter stably inhibits transcription of that gene. Therefore, DNA methylation is mainly viewed as a major source of genetic silencing.
Most of the studies focused on epigenetic regulation of astrocytes have primarily investigated the role of DNA methylation in astrocyte differentiation during development. These studies may not directly have a major impact on astrocytes in neurodegeneration, but this information can potentially serve as a roadmap to understand possible epigenetic mechanisms in astrocytes that could also appear in neurodegenerative diseases. In the developing brain, there is a tight regulation of fate determination for neural precursor cells (NPCs), and DNA methylation is one of the critical mediators that regulates their differentiation into astrocytes [95]. The promoter of the astrocyte specific gene GFAP is methylated at a specific CpG element that is known for binding of signaling transducer and activator of transcription 3 (STAT3) in development when astrocyte differentiation is not known to occur. However, in late-stage NPCs this particular cytosine is demethylated, allowing for leukemia inhibitory factor-induced activation of STAT3 to increase gene expression of GFAP, driving astrocyte differentiation [95]. Similarly, Hatada et al. found that many other astrocyte specific genes are demethylated in late-stage neural precursor cells that can facilitate expression of these genes when astrocyte differentiation is completed [96]. Further, inhibition of DNMTs induces astrocyte differentiation from neural progenitor cells [97]. Genetic knockout of DNA methyltransferase 1 (DNMT1) in postnatal calmodulin-kinase Iiα (CamKIIα) neuroblasts had no obvious effects [98]. However, conditional knockout of DNMT1 in nestin-positive NPCs induces GFAP and S100β gene expression along with activation of the JAK-STAT pathway, including STAT1 and STAT3 [99]. This indicates that DNMT1 methylates astrocyte-specific genes and regulates the JAK-STAT pathway in development. Similarly, conditional knockout of DNA methyltransferase 3a (DNMT3a) in nestin-positive cells results in neuromuscular defects and reduced postnatal re-methylation of the GFAP promoter in the cortex [100]. Thus, DNA methylation appears to be critically important in the differentiation of astrocytes from NPCs. Contrasting these findings however, Urayama et al. knocked out DNMT1, 3a and 3b in NPCs and found that hypomethylation of the GFAP promoter was not itself sufficient to induce expression, that differential chromatin accessibility was also involved in regulating the expression of GFAP, indicating that histone modifications (see next section) influences this process [101]. Further study is needed to reconcile these disparate findings.
The results of DNA methylation in astrocytes during development indicate that astrocytes express DNMTs and that astrocyte specific gene expression can be reduced via methylation. As these genes are differentially active in the adult brain, it is possible that similar mechanisms controlling gene expression in astrocytes exist in neurodegenerative diseases. For instance, the JAK/STAT pathway has been demonstrated to control reactive astrogliosis. Sriram et al. found that activation of the JAK2/STAT3 pathway in the MPTP mouse model of PD precedes, and likely drives, the upregulation of GFAP and reactive astrogliosis [102]. The activation of STAT3 following MPTP-induced neuronal damage was only found at the site of injury, preceded increased astrocyte reactivity, and STAT3 activation fell before the peak of reactive astrogliosis. The addition of the neuroprotective agent nomifensine significantly attenuated MPTP-induced expression of factors involved in the activation of STAT3 in astrocytes, including Tnf-α, Osm, Lif, and Ccl2 [103]. Further, conditionally knocking out STAT3 in astrocytes significantly reduced the amount of reactive astrocytes following MPTP [103]. These findings indicate that STAT3 is an important regulator of reactive astrogliosis, and the developmental data demonstrate that epigenetic mechanisms such as DNA methylation can influence the STAT3 pathway. Therefore, DNA methylation could potentially alter astrocyte reactivity and inflammatory status in neurodegenerative diseases through controlling the STAT3 pathway, along with other signaling pathways.
Although few studies have examined astrocyte-specific DNA methylation in CNS pathologies, it clearly plays at least some role in various neurodegenerative diseases. For instance, peroxisome proliferator-activated receptors (PPARs), which are prominently expressed in glial cells, have demonstrated neuroprotective properties in various in vitro and in vivo studies (Reviewed by [104]). PPARβ/δ-induced reduction of DNMTs in rat hippocampal astrocytes attenuates hypermethylation of CpG islands in the miR-181a gene, and the increased miR-181a can protect these cells from corticosterone-induced ER stress [105]. Another intriguing role concerns methylation-mediated vulnerability to neuroinflammation or degeneration. Peroxisome proliferator-activated receptor coactivator 1α (PGC-1α) is reduced in the dopaminergic neurons of PD patients compared to age-matched controls [106]. Palmitate, a common, pro-inflammatory fatty acid postulated to influence neurodegeneration, induces promoter methylation of PGC-1α in major cell types of the brain, including astrocytes [107]. Similarly, human AD patients have decreased global DNA methylation specifically in pyramidal neurons and astrocytes [108]. Mechanistically connecting these observations may provide novel insights into disease pathogenesis and open new avenues for targeted therapeutics.
Another factor that implicated in neurodegenerative diseases is the inward-rectifying potassium channel Kir4.1, with reduced levels of Kir4.1 in AD, ALS, and ischemia. Astrocytes have been demonstrated to protect neurons through the buffering of potassium, and astrocytes regulate potassium buffering in the cortex and hippocampus through Kir4.1 during development [109]. Some evidence indicates that expression of Kir4.1 in astrocytes is regulated by DNMT1-mediated DNA methylation [110]. Therefore, understanding the DNA methylation of Kir4.1 in disease could represent a new therapeutic option for several neurodegenerative diseases.
Astrocyte dysfunction has also been implicated in neuropsychiatric disorders such as major depression, with global methylation patterns altered in depressive patients that demonstrated changes in astrocyte specific genes [111]. Alcohol exposure alters neuronal plasticity in both the developing and mature CNS. Zhang et al. found that ethanol exposure led to altered DNA methylation levels in cultured primary rat astrocytes, with ethanol reducing DNMT3a protein level while not changing DNMT1 levels [112]. The same study found that reduced DNA methylation led to an increase in tissue plasminogen activator (tPA), which is known to modulate neuronal plasticity and is involved in complement regulation, a significant contributor to astrocyte reactivity in neurodegeneration [20, 113].
Overall, DNA methylation in astrocytes appears to be altered in numerous neurological disorders, which has broad implications for controlling both the neuroprotective and neurodegenerative qualities of astrocytes.
Histone modifications
Histones are nuclear proteins that are responsible for arranging DNA into euchromatin, a conformation that allows gene transcription, or into heterochromatin, a dense structure that blocks gene transcription [114]. DNA wraps around the histones to form a structure known as the nucleosome. There are five major classes of histones: H1/H5, H2A, H2B, H3, and H4. The core histones, which consist of H2A, H2B, H3 and H4, exist as dimers and form the complex that the DNA strand enwraps. These histone proteins contain tails that have residues capable of being acetylated, methylated, phosphorylated, ubiquitylated, and sumoylated. Depending on the site and type of modification, the histones can alter the DNA structure, into either euchromatin or hererochromatin, for a specific gene and change the expression of that gene. There are enzymes that add modifications, such as histone methyltransferases (HMTs), and enzymes that remove these modifications, including histone demethylases. A complex interplay between activating and repressing histone modifications is a major regulatory pathway for gene expression [115].
Histone Methylation
Methylation of histone tails by HMTs results in either increased or reduced gene expression depending on the specific amino acid methylated on the histone tails. For instance, H3K4 tri-methylation (H3K4me3) is associated with increased transcription, whereas H3K9me3 results in transcription repression. Histone methylation is not well studied in astrocytes during CNS dysfunction, but the existing information suggests an important role in astrocyte function. Astrocytes express many of the known enzymes that control the levels of histone methylation, including both HMTs and histone demethylases, and have different basal levels of tri-methylation of histone 3 lysine 27 (H3K27me3) [116]. Ezh2, for instance, is an HMT that is mainly responsible for tri-methylation of histone 3 lysine 27 (H3K27me3). During development Ezh2 is downregulated in neural stem cells during differentiation towards neurons and astrocytes, and forced expression of Ezh2 in recently differentiated astrocytes led to a partial de-differentiation back to neural stem cells [117, 118]. Similarly, mice with Ezh2 knockout specifically in the cortex demonstrated accelerated astrogliogenesis and astrocyte differentiation [119]. On the other hand, astrocytes also express histone demethylases, including the jumonji-domain containing proteins (Jmjds), with Jmjd3 and Jmjd5 being the most highly expressed [116]. Demethylases, such as jumonji-domain containing protein 2c (Jmjd2c), are also important in astrocyte differentiation in development, with Jmjd2c hypomorphic mutant mice had increased GFAP-positive cells in the developing brain [120]. Similarly, absence of the HMT ESET increased astrocyte formation in development and overexpression of ESET significantly reduced astrocyte differentiation [121].
Histone methylation is also important in astrocytes during CNS dysfunction, including ischemia, viral infection, and cancer. Chisholm et al. examined adult cortical astrocytes following an ischemic injury in rats contained higher H3K4me3 and less H3K9me3 compared to middle-aged astrocytes with the same treatment, indicating an overall lower level of gene transcription in aged astrocytes following ischemia [122]. This study went on to examine specific genes affected by ischemia in aged cortical astrocytes by the KEGG pathway analysis and found increased activation of the neuroprotective VEGF pathway. There was no observed difference in DNMT1 expression, but along with the differences in histone methylation, the authors found a decrease in histone deacetylase (HDAC) expression. One major caveat to this study was that the levels of these epigenetic mechanisms were not measured in normal aging in these rats, and some of the results that were found in this study could have been affected in the normal aging process.
Along with ischemia, the HIV virus can induce neurodegeneration in what is termed HIV-associated neurodegeneration (HAND). Narasipura et al. found that histone methylation is important for HIV latency in astrocytes, with H3K9me3 pushing HIV out of latency while inhibition of H3K9 methylation increasing pro-viral transcription [123]. Therefore, the expression of genes involved in reducing HIV pro-viral transcription is inhibited by tri-methylation of the H3K9 residue. Similarly, astroglial tumor progression is regulated to some extent by histone methylation, with higher H3K9me3 in control patients and higher H4K20me3 in grade 2 tumor brains [124]. The cytoplasmic levels of the HMT SUV39H1 increased in brain of astroglioma patients, whereas the nuclear levels of this HMT were inversely correlated with tumor grade and affected survival. Expression of the HMT Ezh2 positively correlates with astrocytic tumor grade, along with increased H3K27me3 and reduced H3K27 target gene expression [125]. The same study found that DNA methylation was also altered in astrocytic tumors. As it relates to astrocytes and inflammation, Li et al. found that the astrocyte protein S100β could increase nuclear factor kappa-light-chain-enhancer of activated B cells (NFκB) downstream inflammatory factors by increasing SET7/9-induced H3K4 methylation [126]. Taken together, histone methylation occurs in both developing and adult astrocytes and plays an important role in several CNS dysfunctions. However, there is still a dearth of knowledge on astrocyte specific histone methylation in the most common neurodegenerative diseases, including AD and PD. For instance, histone methylation is involved in the inflammatory response for several cell types, including macrophages and microglia, and inhibiting the histone methyltransferase Ezh2 in microglia leads to reduction in the inflammatory response to LPS [127]. Ezh2 is involved in reducing astrocyte differentiation but there has been no link to adult astrocytes. Therefore, it is plausible that a similar mechanism could be present in astrocytes. Because astrocytes contribute to the exaggerated inflammatory response in neurodegenerative diseases, histone methylation could play a role in promoting the production of inflammatory factors in astrocytes.
Histone Acetylation
Unlike methylation of histone tails that can result in either induction or repression of gene transcription, acetylation of histone tails is solely associated with active gene expression. As with histone methylation, astrocytes express the enzymes required for adding and removing acetyl groups to the histone tails, histone acetyltransferases (HATs) and histone deacetylases (HDACs). Astrocytes express various levels of the histone deacetylases including HDAC1–11 and the sirtuin family of HDACs [43, 128, 129]. The interplay between these two factors is important in the differentiation of astrocytes in development. Cheng et al. found that the HAT p300 is required for astrocyte differentiation in development by acetylating H3K9 and H3K14 [130]. Similarly, inhibition of HDACs by the addition of trichostatin A (TSA) prevents both neuronal and astrocyte differentiation [131]. Zhang et al. went on to demonstrate that control of astrocyte or oligodendrocyte differentiation is specifically through by HDAC3, and removal of HDAC3 induces robust astrocyte differentiation [132].
Stem cell differentiation research is another area in which regulation of astrocyte function by histone acetylation appears to play a key role. Mouse neuronal differentiation of embryonic and neural stem cells is increased with inhibition of HDACs through chemicals such as sodium butyrate and TSA [133, 134]. However, it becomes more complicated when studying human stem cells, with some indications that HDAC inhibition increases neuronal differentiation and other findings demonstrating that it increases astrocyte differentiation instead. Inhibition of HDACs by sodium butyrate and mocetinostat promoted 10 to 15-fold higher neural progenitor cells from induced pluripotent stem cells [135]. Majumder and co-workers reported that treatment of human neural stem cells with the DNA methyltransferase inhibitor 5-Aza-2-deoxycytidine along with the HDAC inhibitor TSA and the pro-astrocyte differentiation factor BMP2, there is increased astrocyte differentiation [97]. The most likely explanation for these mixed results is that multiple interacting factors, including histone acetylation, are involved in neuronal and astrocyte differentiation. Nonetheless, these findings generally indicate that increased histone acetylation is involved in astrocyte differentiation during rodent development and in cultured human stem cells, with increased histone acetylation or inhibition of HDACs leading to increased astrocyte differentiation.
Histone acetylation is currently the most researched epigenetic mechanism in CNS dysfunction and neurodegenerative diseases as it relates to astrocyte function. Similar to development, adult astrocytes rely on histone acetylation for various important functions, including filament organization, cell survival, and inflammatory response. Hsieh et al. found that ex vivo differentiated rat astrocytes have lower levels of global histone acetylation when compared to neural stem cells and neurons [133]. Interestingly, HDAC inhibition through sodium butyrate treatment led to a significant reduction in GFAP expression, while also reorganizing the entire astrocyte filament network with GFAP, vimentin and nestin redistributed together [136]. Specific knockdown of HDAC3 and HDAC6 by siRNA led to similar effects on reorganization of the astrocyte filament network and reduction of GFAP expression.
Reactive astrocytes in neurodegenerative diseases contribute to neuroinflammation through the production of inflammatory cytokines and reduced production of neurotrophic factors [3–6]. Conditioned media from LPS-activated microglia led to decreased total H3 and H4 acetylation in cultured astrocytes and increased astrocyte cell death from H2O2 treatment, while decreasing expression of the anti-oxidant factor Nrf2. This same study reported that the HDAC inhibitor valproic acid (VPA) attenuated the detrimental effects from LPS-treatment microglial conditioned media [137]. Another HDAC inhibitor suberoylanilide hydroxamic acid (SAHA) significantly reduced the IFNγ-induced inflammatory response in the human astrocytoma cell line U373-MG and primary human astrocytes by reducing activation of STAT3 and the chemokine I-TAC. The authors went on to determine that SAHA treatment led to protection of SHSY5Y neuroblastoma cell death from IFNγ-treated astrocyte conditioned media [138]. Although these studies did not directly identify epigenetic alterations, such as increased histone acetylation, reduced STAT3 activation has been linked to promoter acetylation of SOCS1 and SOCS3 in cancer cells, which could be the underlying mechanisms seen by Sadayuki et al [139]. Therefore, increased histone acetylation might affect reactive astrogliosis through reducing STAT3 signaling, similar to that seen with DNA methylation during development [95].
Additional evidence supporting a role for HDACs and HATs in the reduction of astrocyte inflammatory response, includes the observation that dimethyl fumarate (DMF) treatment reduces HDAC expression in astrocytes, and attenuates cytokine-induced HDAC gene expression and activity [140]. The same study went on to demonstrate that DMF treatment increased Nrf2 expression, possibly by increasing acetylation. Similarly, Soliman et al. found that sodium acetate, a product of Acetyl-CoA metabolism, treatment in primary mouse astrocytes resulted in H3K9 hyperacetylation and significantly reduced LPS-induced the pro-inflammatory cytokine TNFα while also increased IL-4 and IL-10 gene expression [141]. On the other hand, Beurel et al. found that specifically inhibiting HDAC6 with tubacin led to a reduction in LPS tolerance in cultured primary mouse astrocytes [142]. Endotoxin tolerance, including LPS, results from prior exposure to small amounts of the endotoxin that leads to a reduced effect in later exposures [143]. HDAC inhibition reduction in LPS tolerance opposes other research findings that demonstrated inhibition of HDACs or increasing acetylation resulted in reduced astrocyte inflammatory response [140, 144], and could be due to the fact that it is centric on LPS and mouse astrocytes do not have the signaling mechanisms for an LPS inflammatory response (TLR4 and MyD88) [145]. Alternatively, there may be differences in specific HDAC isoforms that contribute to these disparate findings.
Although most of the studies have focused on astrocyte-specific histone acetylation in vitro very few studies have investigated the importance of histone acetylation in astrocytes in vivo. Bailey et al. demonstrated that reduced histone H3 acetylation is found in rat astrocytes in vivo following traumatic brain injury (TBI) [146]. This reduction in astrocyte-specific histone acetylation could be one of the mechanisms that leads to the increased inflammatory response found in the CNS following TBI [147]. Additionally, HDAC inhibitors can increase the neurotrophic factors GDNF and BDNF gene expression, contributing to increased neuroprotection of dopaminergic neurons [148, 149]. Overall, increased histone acetylation appears to reduce astrocyte inflammatory response and lead to increased neuroprotection. Therefore, astrocyte-specific histone acetylation could be an important mediator in neurodegenerative diseases, and novel therapeutics that can alter histone acetylation in astrocytes could have beneficial effects, such as dimethyl fumarate.
Histone Phosphorylation, Ubiquitylation and Sumoylation
Other histone modifications include phosphorylation, ubiquitylation and sumoylation [150]. Very little work has been published regarding these modifications specifically in astrocytes, but the existing studies indicate that astrocyte-specific histone phosphorylation, ubiquitylation and sumoylation could be important factors to examine in neurodegenerative diseases. Histone phosphorylation is typically associated with DNA damage, with increased phosphorylation of the histone H2AX around sites of DNA damage. For example, oxidative stress that is generated by a neurological insult such as cerebral ischemia can generate higher levels of phosphorylated histone 2AX (H2AX) [151]. The same study went on to determine that pretreatment with an antioxidant reduced the levels of H2AX phosphorylation and the neuronal death. These results indicate that non-neurotoxic levels of insults can induce histone phosphorylation, and this histone modification could play a role in determining the vulnerability of neurons to insults such as oxidative stress. Increased phosphorylation of the H2A histone family member H2AX occurs in astrocytes of the hippocampus and cortex in AD patients compared to controls [152]. This indicates that astrocytes in the affected regions of AD patients may have higher astrocyte-specific DNA damage, and this could lead to reduced support for neuronal survival that basally comes from astrocytes. Similarly, Enomoto et al. found that cultured human astrocytes basally express low levels of histone phosphorylation, but histone phosphorylation of H2A subfamilies is involved in astrocyte apoptosis due to saline exposure [153]. Histone phosphorylation is apparently part of the complex interplay between other epigenetic marks, such as histone acetylation, histone methylation and DNA methylation. Increased histone phosphorylation has been found to maintain certain histone and DNA methylation marks early in a mouse zygote [154]. Similarly, evidence indicates that increased histone 3 Serine 10 (H3S10) phosphorylation during meiosis and mitosis is related to chromatin condensation and gene repression with increased methylation of H3K9 [155], but this modification can also be associated with gene activation in some instances. Phosphorylation of histone 3 serine 10 (H3S10), threonine 11(T11), and serine 28 (S28), has been linked to increased histone acetylation and subsequent activation of gene expression when examined [156–158]. As it relates to inflammation, phosphorylation of H3S28 has been demonstrated to displace polycomb repressive complexes (PRCs) and induce demethylation of H3K27 and activating transcription [156]. The reduction of H3K27 tri-methylation has been linked to reduced inflammation in macrophages and microglia [23], and therefore H3S28 phosphorylation could potentially reduce the inflammatory response in multiple cell types. Cytokine-induced histone phosphorylation by IkappaB kinase alpha (IKKα) subsequently increases histone acetylation in transcription factor nuclear factor kappa B (NFκB) responsive genes, which include pro-inflammatory cytokines [159]. Similarly, an animal model of ischemia found increased IKKα-induced histone phosphorylation in cerebral endothelial cells [160]. IKKα-induced histone phosphorylation has not been studied in astrocytes, however, these cells are known to produce higher levels of pro-inflammatory cytokines during neurodegenerative diseases and this mechanism could be partially controlled by IKKα-induced histone phosphorylation in NFκB responsive genes. Therefore, histone phosphorylation has been demonstrated to increase pro-inflammatory gene activation and astrocyte-specific histone phosphorylation is an emerging area for investigation in neurodegenerative disease due to the role of these cells in the inflammatory response.
Histone ubiquitylation is also found in response to DNA damage, however, ubiquitylation of H2A usually leads to gene silencing, whereas H2B ubiquitylation has been linked to gene activation. Histone ubiquitylation is important for regulating stem cell maintenance and differentiation, including neural stem cells that differentiate into astrocytes [161]. Unfortunately, very little is known about how histone ubiquitylation is involved in astrocytes as it relates to neurodegenerative diseases. Cultured epithelial cells exposed to the pro-inflammatory cytokine TNFα displayed an overall reduction in monoubiquitylation of H2B (H2Bub1), and this effect preceded the induction of NFκB target genes. Decreased H2Bub1 due to the reduction in the ubiquitylation enzyme RNF20 increased the inflammatory response from TNFα in mice by regulating NFκB target genes [162]. As expected, astrocytes express deubiquitylation enzymes and have been found to contain H2Bub1 [163]. Therefore, the inflammatory response of astrocytes in neurodegenerative diseases could partially rely on H2Bub1 and warrants further exploration of this novel mechanism.
Small ubiquitin-related modifier (SUMO) is a modification that is similar to ubiquitin, but does not have as large of a role in protein degradation. The majority of research into SUMOylation and transcription indicate that this modification can alter transcription factors while influencing neuronal differentiation and inflammation [164, 165]. Regulation of epigenetic marks by SUMOylation influences gene expression through two main functions: 1) SUMOylating enzymes adding SUMO groups to histone tails and competing with acetylation, and 2) addition of this modification to particular proteins, such as HDACs, that can control other epigenetic marks. Similar to ubiquitin, lysine residues on histone tails can be SUMOylated, and all four of the core histones can be SUMOylated [166, 167]. Along with direct gene repression, SUMOylation can inhibit chromatin-modifying enzymes that would normally open up the chromatin and allow gene transcription [168]. SUMOylation can also affect histone modifications through the degradation or recruitment of enzymes such as HDACs to act on the histone tails. The enzyme HDAC1 can be SUMOylated both in vitro and in vivo [169]. The same study found that SUMOylation is not required for HDAC1 activity, but it is involved in a complex interplay with acetylation and phosphorylation of HDAC1. Similarly, protein SUMOylation in macrophages can alter peroxisome proliferator-activated receptor gamma (PPARγ) binding to a complex with HDAC3 to repress inflammatory gene transcription [165]. Studies have also found that SUMOylation can repress gene transcription through opposing histone acetylation and ubiquitylation, and reduction of histone SUMOylation leads to higher levels of acetylation [167]. Although post-translational protein SUMOylation has been suggested to contribute to the pathogenesis of several neurodegenerative diseases, such as PD [170, 171] and AD [172], there limited studies on SUMOylation in any of these diseases. One of the major findings of SUMOylation in neurodegeneration was reported by Tao et al., who demonstrated that SUMOylation of HDAC1 was an endogenous protective mechanism against amyloid-β (Aβ) toxicity in a mouse model of AD [173]. The same study found that acute Aβ treatment in rats induced the expression of Protein inhibitor of activated STAT1 (PIAS1), which is a SUMO E3 ligase. However, this study did not go into the specific cell type responsible for this protection.
SUMO ligases are expressed in rat astrocytes, including SUMO-1 and PIAS1, and these factors are decreased with aging or exposure to the inflammatory stimulus lipopolysaccharide (LPS) [174]. The same study found that protein SUMOylation of the transcription factor CCAAT/enhancer-binding protein (C/EBP) regulates the pro-inflammatory NOS2 gene expression, with decreased sumoylation leading to increased NOS2 expression in cultured astrocytes. Hoppe et al. found that β-amyloid 42 (Aβ42) treated mouse astrocytes contained reduced levels of SUMO-1 conjugated proteins, and that over-expression of constitutively active SUMO-1 halted some of the morphological effects from Aβ42 [175]. Similarly, SUMOylation of liver X receptors (LXRs) in cultured astrocytes, by the SUMO ligases PIAS1 and HDAC4, reduced the interferon gamma (IFGγ)-induced activation of STAT1 and subsequent inflammatory response [176]. These results indicate that SUMOylation is involved in astrocyte gene expression and that SUMOylation can reduce inflammation, which points to a potential role for SUMOylation in neurodegeneration and suggests that it might be a possible therapeutic option to reduce CNS inflammation.
MicroRNAs
MicroRNAs (miRNAs) are small non-coding RNAs that do not form proteins, but can alter gene expression through multiple mechanisms [177]. Although not directly associated with gene expression, microRNAs generally prevent mRNA translation into a protein by binding to the 3’ untranslated end of mRNA, and they can directly lead to mRNA degradation [178]. Therefore, miRNAs represent another epigenetic regulation by altering gene translation into proteins. Over 5,000 different miRNAs have been detected and each contains a particular sequence that it will lead to the inhibition of specific targets, although one miRNA can regulate over one thousand mRNA targets [179]. Several studies have examined miRNAs in astrocytes and found that these molecules are involved in astrocyte differentiation, inflammatory response and various other mechanisms. As it relates to development, the miRNA-124 (miR-124) can directly downregulate Ezh2 expression and lead to increased neuronal differentiation over astrocyte differentiation [180]. Letzen et al. found multiple miRNAs that are potentially involved in astrocyte differentiation [181], including miR-16 that has been linked to GFAP, along with miR-184 and miR-118. These miRNAs potentially could prevent astrocyte differentiation and promote oligodendrocyte differentiation by inhibiting expression of key astrocyte genes. Interestingly, Vijayaraghava et al. found that there are regional and age-specific differences in the miRNA expression levels of laser-captured human astrocytes by analyzing 20 specific miRNAs related to inflammation and neurodegeneration or due to their proven involvement in astrocyte function [182]. This study went on to report that expression of certain miRNAs in astrocytes are specific to different regions of the human brain, including higher expression of pro-inflammatory miRNAs in white matter compared to grey matter. The authors also found 10 miRNAs that were more highly expressed in adult white matter astrocytes when compared to the fetal astrocytes, and that the majority of these miRNAs were involved in immune response or glutamate transport. One caveat of this study that the authors discuss is that the laser capture technique could potentially miss some astrocytes that do not highly express GFAP, and another is that this study is focused only on 20 known miRNAs and did not broadly examine miRNA expression in astrocytes. Together, these studies infer that miRNAs are important gene expression regulators in the differentiation of astrocytes and are differentially expressed in astrocytes of different brain regions.
As it relates to neurodegeneration, Ziu et al. demonstrated that several miRNAs were altered at different timepoints in cultured rat astrocytes following oxygen-glucose deprivation to simulate ischemic injury, including miR-21, 29b, 30b, 107, 137 and 210 [183]. Since some miRNAs are involved in inflammation, another study examined miRNA levels from rodents and primates following LPS and IFNγ treatment [184]. The authors found that miRNAs such as miR-146a, miR-146b, and miR-155 were increased in mouse cortical astrocytes following LPS and IFNγ treatment, whereas miR-351, 455 and 149 were decreased. These data indicate that there could be a miRNA component to reactive astrogliosis following an inflammatory stimulus. Similarly, the same study found that some miRNAs are specifically expressed in rodent (mouse and rat) cortical astrocytes following the inflammatory stimulus compared to non-human primates, and that these miRNAs focused on TNFα signaling pathways. Iyer et al. demonstrated that astrocytes treated with the inflammatory cytokine IL-1β had elevated levels of miR-146a, and downstream inflammatory factors were increased through miR-146a [185]. Because astrocytes secrete many factors, including exosomes, that can influence the health of neurons, it is interesting that the levels of miRNAs in astrocyte-derived exosomes can be altered in neurodegenerative diseases and that these miRNAs could potentially exert control on neuronal plasticity, and perhaps astrocyte activity [186]. In relation to PD, Song et al. found that overexpressing the stress protein HO-1 in astrocytes led to decreased miR-7 levels and increased α-synuclein in the substantia nigra and ventral tegmental area [187]. On the other hand, a recent study found that the miRNA content in astrocyte-derived exosomes was not altered in an ALS culture model, SOD1-G93A [188]. Taken together, these studies indicate that miRNAs play a complex role in control of astrocyte function in health and disease, but that miRNAs are certainly involved in astrocyte inflammatory response in CNS disorders [189]. Clearly, further study on the alteration of the miRNA network in specific neurodegenerative diseases and the functions that they control could help find novel therapeutic avenues to treat these diseases.
Conclusion
The extent of astrocyte involvement in the development and progression of neurodegenerative diseases is an active area of investigation, particularly following the development of new tools and understanding of reactive astrocyte pathways. Under basal conditions astrocytes support neurons through a variety of mechanisms, including neurotrophic support, glutamate uptake, and potassium buffering. However, reactive astrogliosis found in neurodegenerative diseases results in altered astrocyte functions to where they no longer provide support for neurons, and could potentially secrete harmful factors instead [20]. Astrocytes have garnered increased interest because they can contribute to the neuroinflammation found in neurodegenerative diseases, including AD and PD. However, the mechanisms that underlie the astrocyte-specific neuroinflammation and neurotoxicity found in these diseases are currently unknown. The field of epigenetics is still relatively new, particularly as it relates to the nervous system, and there are still significant gaps in knowledge regarding how epigenetic mechanisms can influence different cell types and responses in neurodegenerative diseases. Though much of what we know about this topic is on astrocyte development, it is clear that astrocytes in the adult brain express the various enzymes required for the different epigenetic mechanisms, including DNA methylation, histone modifications, and miRNAs, suggesting that these factors could be involved in the astrocyte response to inflammation and disease. Figure 2 summarizes the known roles of each epigenetic mark specifically in astrocytes.
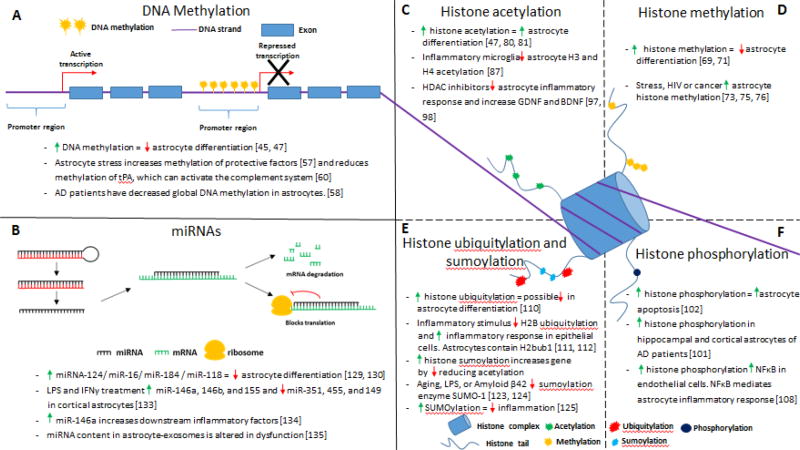
Figure 2. The different epigenetic mechanisms alter astrocyte development and functions, which could affect the astrocyte responses in neurodegenerative diseases.
(A) DNA methyltransferases add methyl groups to the promoter region of specific genes, which generally leads to repression of the gene expression. DNA methylation has been implicated in regulating astrocyte differentiation during development and is altered in astrocytes during disease states. (B) MicroRNAs control gene expression through several mechanisms. MicroRNAs begin as a double stranded oligonucleotide, called pri-miRNA, enzymes cleave off the post-transcriptional modification and the complementary sequence is degraded, leaving only the mature miRNA. Once the miRNA finds the target sequence it can either lead to degradation of the mRNA strand or it can inhibit translation of the mRNA sequence. Different miRNAs affect gene expression in astrocytes with several known miRNAs that affect astrocyte differentiation or function, along with altered levels of miRNAs in astrocytes during disease states. (C–F) Histone tails can be modified by various enzymes that add marks onto particular residues, and these marks can gene activity through the opening or closing of the DNA reading frame and allowing the transcription machinery to access the specific gene. (C) Acetylation of histone tails are generally associated with increased gene activation, and increased histone acetylation is involved in astrocyte differentiation and decreased histone acetylation in astrocytes is associated with an increased inflammatory response. (D) The effect of histone methylation depends on the residue that is methylated. Increased histone methylation has been demonstrated to decrease astrocyte differentiation, whereas increased histone methylation has been found increase in astrocytes during disease states. (E) Increased histone ubiquitylation has been linked to a possible role in reducing astrocyte differentiation, and decreased histone ubiquitylation is associated with an increased inflammatory response in epithelial cells. Similarly, small ubiquitin-like modifier (SUMOylation) is decreased in disease states, and increased SUMOylation has been found to decrease inflammation. (F) Histone phosphorylation is generally seen around areas of DNA damage, but also can affect gene expression. Evidence indicates that increased histone phosphorylation is associated with increased astrocyte apoptosis, and histone phosphorylation is increased in astrocytes of Alzheimer’s disease patients.
This review has focused on the roles of epigenetic mechanisms demonstrated to contribute to various functions of astrocytes, from astrocyte differentiation to inflammatory response. Many of these epigenetic changes in astrocytes have not been tied directly to the astrocyte response in neurodegenerative diseases, but focus more on regulation of specific astrocyte functions. These studies serve as a foundation that can be used to explore new avenues in the initiation, promotion and treatment of neurodegeneration. With the available knowledge regarding epigenetic mechanisms in astrocytes there is considerable evidence that these factors regulate astrocyte responses in neurodegenerative diseases and warrants further investigation.
Highlights.
Astrocytes are central mediators of homeostasis and neuroinflammation
Epigenetic mechanisms control astrocyte development and inflammatory state
Understanding of epigenetic regulation of astrocyte function in neurodegeneration may lead to new therapies
Acknowledgments
This work was supported in part by the following NIH Grants: R01ES021800, R01ES026057 and U01NS079249. Additional support was provided by the Michael J Fox Foundation and private support from the Glenn and Karen Leppo, the Richard Nicely and the Alan and Janice Woll Parkinson’s Disease Research Funds. The content of this article is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health or any other funding source.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Frank-Cannon TC, et al. Does neuroinflammation fan the flame in neurodegenerative diseases? Mol Neurodegener. 2009;4:47. doi: 10.1186/1750-1326-4-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Morales I, et al. Neuroinflammation in the pathogenesis of Alzheimer's disease. A rational framework for the search of novel therapeutic approaches. Front Cell Neurosci. 2014;8:112. doi: 10.3389/fncel.2014.00112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sofroniew MV. Multiple roles for astrocytes as effectors of cytokines and inflammatory mediators. Neuroscientist. 2014;20(2):160–72. doi: 10.1177/1073858413504466. [DOI] [PubMed] [Google Scholar]
- 4.Yasuda Y, et al. The effects of MPTP on the activation of microglia/astrocytes and cytokine/chemokine levels in different mice strains. J Neuroimmunol. 2008;204(1–2):43–51. doi: 10.1016/j.jneuroim.2008.08.003. [DOI] [PubMed] [Google Scholar]
- 5.Teismann P, et al. Pathogenic role of glial cells in Parkinson's disease. Mov Disord. 2003;18(2):121–9. doi: 10.1002/mds.10332. [DOI] [PubMed] [Google Scholar]
- 6.Forno LS, et al. Astrocytes and Parkinson's disease. Prog Brain Res. 1992;94:429–36. doi: 10.1016/s0079-6123(08)61770-7. [DOI] [PubMed] [Google Scholar]
- 7.Volterra A, Meldolesi J. Astrocytes, from brain glue to communication elements: the revolution continues. Nat Rev Neurosci. 2005;6(8):626–40. doi: 10.1038/nrn1722. [DOI] [PubMed] [Google Scholar]
- 8.Ullian EM, et al. Control of synapse number by glia. Science. 2001;291(5504):657–61. doi: 10.1126/science.291.5504.657. [DOI] [PubMed] [Google Scholar]
- 9.Hama H, et al. PKC signaling mediates global enhancement of excitatory synaptogenesis in neurons triggered by local contact with astrocytes. Neuron. 2004;41(3):405–15. doi: 10.1016/s0896-6273(04)00007-8. [DOI] [PubMed] [Google Scholar]
- 10.Song H, Stevens CF, Gage FH. Astroglia induce neurogenesis from adult neural stem cells. Nature. 2002;417(6884):39–44. doi: 10.1038/417039a. [DOI] [PubMed] [Google Scholar]
- 11.Halassa MM, et al. Synaptic islands defined by the territory of a single astrocyte. J Neurosci. 2007;27(24):6473–7. doi: 10.1523/JNEUROSCI.1419-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hamby ME, Sofroniew MV. Reactive astrocytes as therapeutic targets for CNS disorders. Neurotherapeutics. 2010;7(4):494–506. doi: 10.1016/j.nurt.2010.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Barreto GE, et al. Astrocytic-neuronal crosstalk: implications for neuroprotection from brain injury. Neurosci Res. 2011;71(2):107–13. doi: 10.1016/j.neures.2011.06.004. [DOI] [PubMed] [Google Scholar]
- 14.Cui W, et al. Inducible ablation of astrocytes shows that these cells are required for neuronal survival in the adult brain. Glia. 2001;34(4):272–82. doi: 10.1002/glia.1061. [DOI] [PubMed] [Google Scholar]
- 15.Niranjan R. The role of inflammatory and oxidative stress mechanisms in the pathogenesis of Parkinson's disease: focus on astrocytes. Mol Neurobiol. 2014;49(1):28–38. doi: 10.1007/s12035-013-8483-x. [DOI] [PubMed] [Google Scholar]
- 16.Hirsch EC, et al. The role of glial reaction and inflammation in Parkinson's disease. Ann N Y Acad Sci. 2003;991:214–28. doi: 10.1111/j.1749-6632.2003.tb07478.x. [DOI] [PubMed] [Google Scholar]
- 17.Maragakis NJ, Rothstein JD. Mechanisms of Disease: astrocytes in neurodegenerative disease. Nat Clin Pract Neurol. 2006;2(12):679–89. doi: 10.1038/ncpneuro0355. [DOI] [PubMed] [Google Scholar]
- 18.Sofroniew MV. Molecular dissection of reactive astrogliosis and glial scar formation. Trends Neurosci. 2009;32(12):638–47. doi: 10.1016/j.tins.2009.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sun D, Jakobs TC. Structural remodeling of astrocytes in the injured CNS. Neuroscientist. 2012;18(6):567–88. doi: 10.1177/1073858411423441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liddelow SA, et al. Neurotoxic reactive astrocytes are induced by activated microglia. Nature. 2017;541(7638):481–487. doi: 10.1038/nature21029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rappold PM, Tieu K. Astrocytes and therapeutics for Parkinson's disease. Neurotherapeutics. 2010;7(4):413–23. doi: 10.1016/j.nurt.2010.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zamanian JL, et al. Genomic analysis of reactive astrogliosis. J Neurosci. 2012;32(18):6391–410. doi: 10.1523/JNEUROSCI.6221-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tang Y, et al. Jmjd3 is essential for the epigenetic modulation of microglia phenotypes in the immune pathogenesis of Parkinson's disease. Cell Death Differ. 2014;21(3):369–80. doi: 10.1038/cdd.2013.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wilson AG. Epigenetic regulation of gene expression in the inflammatory response and relevance to common diseases. J Periodontol. 2008;79(8 Suppl):1514–9. doi: 10.1902/jop.2008.080172. [DOI] [PubMed] [Google Scholar]
- 25.Mogi M, et al. Interleukin (IL)-1 beta, IL-2, IL-4, IL-6 and transforming growth factor-alpha levels are elevated in ventricular cerebrospinal fluid in juvenile parkinsonism and Parkinson's disease. Neurosci Lett. 1996;211(1):13–6. doi: 10.1016/0304-3940(96)12706-3. [DOI] [PubMed] [Google Scholar]
- 26.Sweatt JD. The emerging field of neuroepigenetics. Neuron. 2013;80(3):624–32. doi: 10.1016/j.neuron.2013.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ravi B, Kannan M. Epigenetics in the nervous system: An overview of its essential role. Indian J Hum Genet. 2013;19(4):384–91. doi: 10.4103/0971-6866.124357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sultan FA, Day JJ. Epigenetic mechanisms in memory and synaptic function. Epigenomics. 2011;3(2):157–81. doi: 10.2217/epi.11.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hwang JY, Aromolaran KA, Zukin RS. Epigenetic mechanisms in stroke and epilepsy. Neuropsychopharmacology. 2013;38(1):167–82. doi: 10.1038/npp.2012.134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lardenoije R, et al. The epigenetics of aging and neurodegeneration. Prog Neurobiol. 2015;131:21–64. doi: 10.1016/j.pneurobio.2015.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rogers J, et al. The epigenetics of Alzheimer's disease--additional considerations. Neurobiol Aging. 2011;32(7):1196–7. doi: 10.1016/j.neurobiolaging.2011.02.025. [DOI] [PubMed] [Google Scholar]
- 32.Paez-Colasante X, et al. Amyotrophic lateral sclerosis: mechanisms and therapeutics in the epigenomic era. Nat Rev Neurol. 2015;11(5):266–79. doi: 10.1038/nrneurol.2015.57. [DOI] [PubMed] [Google Scholar]
- 33.Hernandez DG, et al. Distinct DNA methylation changes highly correlated with chronological age in the human brain. Hum Mol Genet. 2011;20(6):1164–72. doi: 10.1093/hmg/ddq561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bollati V, et al. DNA methylation in repetitive elements and Alzheimer disease. Brain Behav Immun. 2011;25(6):1078–83. doi: 10.1016/j.bbi.2011.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jowaed A, et al. Methylation regulates alpha-synuclein expression and is decreased in Parkinson's disease patients' brains. J Neurosci. 2010;30(18):6355–9. doi: 10.1523/JNEUROSCI.6119-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Matsumoto L, et al. CpG demethylation enhances alpha-synuclein expression and affects the pathogenesis of Parkinson's disease. PLoS One. 2010;5(11):e15522. doi: 10.1371/journal.pone.0015522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pan Y, et al. Inhibition of DNA Methyltransferases Blocks Mutant Huntingtin-Induced Neurotoxicity. Sci Rep. 2016;6:31022. doi: 10.1038/srep31022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tchantchou F, et al. S-adenosylmethionine mediates glutathione efficacy by increasing glutathione S-transferase activity: implications for S-adenosyl methionine as a neuroprotective dietary supplement. J Alzheimers Dis. 2008;14(3):323–8. doi: 10.3233/jad-2008-14306. [DOI] [PubMed] [Google Scholar]
- 39.Lee S, et al. Dietary supplementation with S-adenosyl methionine delayed amyloid-β and tau pathology in 3×Tg-AD mice. J Alzheimers Dis. 2012;28(2):423–31. doi: 10.3233/JAD-2011-111025. [DOI] [PubMed] [Google Scholar]
- 40.Suchy J, et al. Dietary supplementation with S-adenosyl methionine delays the onset of motor neuron pathology in a murine model of amyotrophic lateral sclerosis. Neuromolecular Med. 2010;12(1):86–97. doi: 10.1007/s12017-009-8089-7. [DOI] [PubMed] [Google Scholar]
- 41.Endres M, et al. DNA methyltransferase contributes to delayed ischemic brain injury. J Neurosci. 2000;20(9):3175–81. doi: 10.1523/JNEUROSCI.20-09-03175.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Baltan S, et al. Histone deacetylase inhibitors preserve white matter structure and function during ischemia by conserving ATP and reducing excitotoxicity. J Neurosci. 2011;31(11):3990–9. doi: 10.1523/JNEUROSCI.5379-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Baltan S. Histone deacetylase inhibitors preserve function in aging axons. J Neurochem. 2012;123(Suppl 2):108–15. doi: 10.1111/j.1471-4159.2012.07949.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chen SH, et al. Suberoylanilide hydroxamic acid, a histone deacetylase inhibitor, protects dopaminergic neurons from neurotoxin-induced damage. Br J Pharmacol. 2012;165(2):494–505. doi: 10.1111/j.1476-5381.2011.01575.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gray SG, Dangond F. Rationale for the use of histone deacetylase inhibitors as a dual therapeutic modality in multiple sclerosis. Epigenetics. 2006;1(2):67–75. doi: 10.4161/epi.1.2.2678. [DOI] [PubMed] [Google Scholar]
- 46.Stout KA, et al. Selective Enhancement of Dopamine Release in the Ventral Pallidum of Methamphetamine-Sensitized Mice. ACS Chem Neurosci. 2016;7(10):1364–1373. doi: 10.1021/acschemneuro.6b00131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nuutinen T, et al. Valproic acid stimulates clusterin expression in human astrocytes: Implications for Alzheimer's disease. Neurosci Lett. 2010;475(2):64–8. doi: 10.1016/j.neulet.2010.03.041. [DOI] [PubMed] [Google Scholar]
- 48.Fischer A, et al. Recovery of learning and memory is associated with chromatin remodelling. Nature. 2007;447(7141):178–82. doi: 10.1038/nature05772. [DOI] [PubMed] [Google Scholar]
- 49.Zhou W, et al. Phenylbutyrate up-regulates the DJ-1 protein and protects neurons in cell culture and in animal models of Parkinson disease. J Biol Chem. 2011;286(17):14941–51. doi: 10.1074/jbc.M110.211029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rane P, et al. The histone deacetylase inhibitor, sodium butyrate, alleviates cognitive deficits in pre-motor stage PD. Neuropharmacology. 2012;62(7):2409–12. doi: 10.1016/j.neuropharm.2012.01.026. [DOI] [PubMed] [Google Scholar]
- 51.Ferrante RJ, et al. Histone deacetylase inhibition by sodium butyrate chemotherapy ameliorates the neurodegenerative phenotype in Huntington's disease mice. J Neurosci. 2003;23(28):9418–27. doi: 10.1523/JNEUROSCI.23-28-09418.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ricobaraza A, et al. Phenylbutyrate ameliorates cognitive deficit and reduces tau pathology in an Alzheimer's disease mouse model. Neuropsychopharmacology. 2009;34(7):1721–32. doi: 10.1038/npp.2008.229. [DOI] [PubMed] [Google Scholar]
- 53.Gardian G, et al. Neuroprotective effects of phenylbutyrate against MPTP neurotoxicity. Neuromolecular Med. 2004;5(3):235–41. doi: 10.1385/NMM:5:3:235. [DOI] [PubMed] [Google Scholar]
- 54.Gardian G, et al. Neuroprotective effects of phenylbutyrate in the N171-82Q transgenic mouse model of Huntington's disease. J Biol Chem. 2005;280(1):556–63. doi: 10.1074/jbc.M410210200. [DOI] [PubMed] [Google Scholar]
- 55.Roy A, et al. Sodium phenylbutyrate controls neuroinflammatory and antioxidant activities and protects dopaminergic neurons in mouse models of Parkinson's disease. PLoS One. 2012;7(6):e38113. doi: 10.1371/journal.pone.0038113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Inden M, et al. Neurodegeneration of mouse nigrostriatal dopaminergic system induced by repeated oral administration of rotenone is prevented by 4-phenylbutyrate, a chemical chaperone. J Neurochem. 2007;101(6):1491–1504. doi: 10.1111/j.1471-4159.2006.04440.x. [DOI] [PubMed] [Google Scholar]
- 57.Ryu H, et al. Sodium phenylbutyrate prolongs survival and regulates expression of antiapoptotic genes in transgenic amyotrophic lateral sclerosis mice. J Neurochem. 2005;93(5):1087–98. doi: 10.1111/j.1471-4159.2005.03077.x. [DOI] [PubMed] [Google Scholar]
- 58.Qi X, et al. Sodium 4-phenylbutyrate protects against cerebral ischemic injury. Mol Pharmacol. 2004;66(4):899–908. doi: 10.1124/mol.104.001339. [DOI] [PubMed] [Google Scholar]
- 59.Kilgore M, et al. Inhibitors of class 1 histone deacetylases reverse contextual memory deficits in a mouse model of Alzheimer's disease. Neuropsychopharmacology. 2010;35(4):870–80. doi: 10.1038/npp.2009.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Monti B, et al. Valproic acid is neuroprotective in the rotenone rat model of Parkinson's disease: involvement of alpha-synuclein. Neurotox Res. 2010;17(2):130–41. doi: 10.1007/s12640-009-9090-5. [DOI] [PubMed] [Google Scholar]
- 61.Sugai F, et al. Benefit of valproic acid in suppressing disease progression of ALS model mice. Eur J Neurosci. 2004;20(11):3179–83. doi: 10.1111/j.1460-9568.2004.03765.x. [DOI] [PubMed] [Google Scholar]
- 62.Zádori D, et al. Valproate ameliorates the survival and the motor performance in a transgenic mouse model of Huntington's disease. Pharmacol Biochem Behav. 2009;94(1):148–53. doi: 10.1016/j.pbb.2009.08.001. [DOI] [PubMed] [Google Scholar]
- 63.Ren M, et al. Valproic acid reduces brain damage induced by transient focal cerebral ischemia in rats: potential roles of histone deacetylase inhibition and heat shock protein induction. J Neurochem. 2004;89(6):1358–67. doi: 10.1111/j.1471-4159.2004.02406.x. [DOI] [PubMed] [Google Scholar]
- 64.Francis YI, et al. Dysregulation of histone acetylation in the APP/PS1 mouse model of Alzheimer's disease. J Alzheimers Dis. 2009;18(1):131–9. doi: 10.3233/JAD-2009-1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Suo H, et al. NRSF is an essential mediator for the neuroprotection of trichostatin A in the MPTP mouse model of Parkinson's disease. Neuropharmacology. 2015;99:67–78. doi: 10.1016/j.neuropharm.2015.07.015. [DOI] [PubMed] [Google Scholar]
- 66.Yoo YE, Ko CP. Treatment with trichostatin A initiated after disease onset delays disease progression and increases survival in a mouse model of amyotrophic lateral sclerosis. Exp Neurol. 2011;231(1):147–59. doi: 10.1016/j.expneurol.2011.06.003. [DOI] [PubMed] [Google Scholar]
- 67.Kim HJ, et al. Histone deacetylase inhibitors exhibit anti-inflammatory and neuroprotective effects in a rat permanent ischemic model of stroke: multiple mechanisms of action. J Pharmacol Exp Ther. 2007;321(3):892–901. doi: 10.1124/jpet.107.120188. [DOI] [PubMed] [Google Scholar]
- 68.Peleg S, et al. Altered histone acetylation is associated with age-dependent memory impairment in mice. Science. 2010;328(5979):753–6. doi: 10.1126/science.1186088. [DOI] [PubMed] [Google Scholar]
- 69.Jia M, et al. Suberoylanilide hydroxamic acid, a histone deacetylase inhibitor, attenuates postoperative cognitive dysfunction in aging mice. Front Mol Neurosci. 2015;8:52. doi: 10.3389/fnmol.2015.00052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kontopoulos E, Parvin JD, Feany MB. Alpha-synuclein acts in the nucleus to inhibit histone acetylation and promote neurotoxicity. Hum Mol Genet. 2006;15(20):3012–23. doi: 10.1093/hmg/ddl243. [DOI] [PubMed] [Google Scholar]
- 71.Faraco G, et al. Pharmacological inhibition of histone deacetylases by suberoylanilide hydroxamic acid specifically alters gene expression and reduces ischemic injury in the mouse brain. Mol Pharmacol. 2006;70(6):1876–84. doi: 10.1124/mol.106.027912. [DOI] [PubMed] [Google Scholar]
- 72.Hockly E, et al. Suberoylanilide hydroxamic acid, a histone deacetylase inhibitor, ameliorates motor deficits in a mouse model of Huntington's disease. Proc Natl Acad Sci U S A. 2003;100(4):2041–6. doi: 10.1073/pnas.0437870100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Spires-Jones TL, et al. Inhibition of Sirtuin 2 with Sulfobenzoic Acid Derivative AK1 is Non-Toxic and Potentially Neuroprotective in a Mouse Model of Frontotemporal Dementia. Front Pharmacol. 2012;3:42. doi: 10.3389/fphar.2012.00042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Outeiro TF, et al. Sirtuin 2 inhibitors rescue alpha-synuclein-mediated toxicity in models of Parkinson's disease. Science. 2007;317(5837):516–9. doi: 10.1126/science.1143780. [DOI] [PubMed] [Google Scholar]
- 75.Chen X, et al. The sirtuin-2 inhibitor AK7 is neuroprotective in models of Parkinson's disease but not amyotrophic lateral sclerosis and cerebral ischemia. PLoS One. 2015;10(1):e0116919. doi: 10.1371/journal.pone.0116919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Chopra V, et al. The sirtuin 2 inhibitor AK-7 is neuroprotective in Huntington's disease mouse models. Cell Rep. 2012;2(6):1492–7. doi: 10.1016/j.celrep.2012.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Chin PC, et al. The c-Raf inhibitor GW5074 provides neuroprotection in vitro and in an animal model of neurodegeneration through a MEK-ERK and Akt-independent mechanism. J Neurochem. 2004;90(3):595–608. doi: 10.1111/j.1471-4159.2004.02530.x. [DOI] [PubMed] [Google Scholar]
- 78.Kelsey NA, Wilkins HM, Linseman DA. Nutraceutical antioxidants as novel neuroprotective agents. Molecules. 2010;15(11):7792–814. doi: 10.3390/molecules15117792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Guo YJ, et al. Resveratrol alleviates MPTP-induced motor impairments and pathological changes by autophagic degradation of α-synuclein via SIRT1-deacetylated LC3. Mol Nutr Food Res. 2016;60(10):2161–2175. doi: 10.1002/mnfr.201600111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Mancuso R, et al. Resveratrol improves motoneuron function and extends survival in SOD1(G93A) ALS mice. Neurotherapeutics. 2014;11(2):419–32. doi: 10.1007/s13311-013-0253-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kumar P, et al. Effect of resveratrol on 3-nitropropionic acid-induced biochemical and behavioural changes: possible neuroprotective mechanisms. Behav Pharmacol. 2006;17(5–6):485–92. doi: 10.1097/00008877-200609000-00014. [DOI] [PubMed] [Google Scholar]
- 82.Meng Z, et al. Resveratrol relieves ischemia-induced oxidative stress in the hippocampus by activating SIRT1. Exp Ther Med. 2015;10(2):525–530. doi: 10.3892/etm.2015.2555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Chatterjee S, et al. A novel activator of CBP/p300 acetyltransferases promotes neurogenesis and extends memory duration in adult mice. J Neurosci. 2013;33(26):10698–712. doi: 10.1523/JNEUROSCI.5772-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Wei W, et al. p300/CBP-associated factor selectively regulates the extinction of conditioned fear. J Neurosci. 2012;32(35):11930–41. doi: 10.1523/JNEUROSCI.0178-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Maciotta S, Meregalli M, Torrente Y. The involvement of microRNAs in neurodegenerative diseases. Front Cell Neurosci. 2013;7:265. doi: 10.3389/fncel.2013.00265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Lee ST, et al. miR-206 regulates brain-derived neurotrophic factor in Alzheimer disease model. Ann Neurol. 2012;72(2):269–77. doi: 10.1002/ana.23588. [DOI] [PubMed] [Google Scholar]
- 87.Koval ED, et al. Method for widespread microRNA-155 inhibition prolongs survival in ALS-model mice. Hum Mol Genet. 2013;22(20):4127–35. doi: 10.1093/hmg/ddt261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Stoica L, et al. Adeno-associated virus-delivered artificial microRNA extends survival and delays paralysis in an amyotrophic lateral sclerosis mouse model. Ann Neurol. 2016;79(4):687–700. doi: 10.1002/ana.24618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Boudreau RL, et al. Nonallele-specific silencing of mutant and wild-type huntingtin demonstrates therapeutic efficacy in Huntington's disease mice. Mol Ther. 2009;17(6):1053–63. doi: 10.1038/mt.2009.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Selvamani A, et al. An antagomir to microRNA Let7f promotes neuroprotection in an ischemic stroke model. PLoS One. 2012;7(2):e32662. doi: 10.1371/journal.pone.0032662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Saraiva C, Ferreira L, Bernardino L. Traceable microRNA-124 loaded nanoparticles as a new promising therapeutic tool for Parkinson's disease. Neurogenesis (Austin) 2016;3(1):e1256855. doi: 10.1080/23262133.2016.1256855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Lee ST, et al. Exosome-Based Delivery of miR-124 in a Huntington's Disease Model. J Mov Disord. 2017;10(1):45–52. doi: 10.14802/jmd.16054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Zeng LL, et al. Lentivirus-Mediated Overexpression of MicroRNA-210 Improves Long-Term Outcomes after Focal Cerebral Ischemia in Mice. CNS Neurosci Ther. 2016;22(12):961–969. doi: 10.1111/cns.12589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Siegfried Z, Simon I. DNA methylation and gene expression. Wiley Interdiscip Rev Syst Biol Med. 2010;2(3):362–71. doi: 10.1002/wsbm.64. [DOI] [PubMed] [Google Scholar]
- 95.Takizawa T, et al. DNA methylation is a critical cell-intrinsic determinant of astrocyte differentiation in the fetal brain. Dev Cell. 2001;1(6):749–58. doi: 10.1016/s1534-5807(01)00101-0. [DOI] [PubMed] [Google Scholar]
- 96.Forman HJ, Torres M. Reactive oxygen species and cell signaling: respiratory burst in macrophage signaling. Am J Respir Crit Care Med. 2002;166(12 Pt 2):S4–8. doi: 10.1164/rccm.2206007. [DOI] [PubMed] [Google Scholar]
- 97.Quinn PO. Treating adolescent girls and women with ADHD: gender-specific issues. J Clin Psychol. 2005;61(5):579–87. doi: 10.1002/jclp.20121. [DOI] [PubMed] [Google Scholar]
- 98.Fan G, et al. DNA hypomethylation perturbs the function and survival of CNS neurons in postnatal animals. J Neurosci. 2001;21(3):788–97. doi: 10.1523/JNEUROSCI.21-03-00788.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Fan G, et al. DNA methylation controls the timing of astrogliogenesis through regulation of JAK-STAT signaling. Development. 2005;132(15):3345–56. doi: 10.1242/dev.01912. [DOI] [PubMed] [Google Scholar]
- 100.Nguyen NH, et al. Propionate increases neuronal histone acetylation, but is metabolized oxidatively by glia. Relevance for propionic acidemia. J Neurochem. 2007;101(3):806–14. doi: 10.1111/j.1471-4159.2006.04397.x. [DOI] [PubMed] [Google Scholar]
- 101.Faraj BA, et al. Metabolism and disposition of methylphenidate-14C: studies in man and animals. J Pharmacol Exp Ther. 1974;191(3):535–47. [PubMed] [Google Scholar]
- 102.Sriram K, et al. Induction of gp130-related cytokines and activation of JAK2/STAT3 pathway in astrocytes precedes up-regulation of glial fibrillary acidic protein in the 1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine model of neurodegeneration: key signaling pathway for astrogliosis in vivo? J Biol Chem. 2004;279(19):19936–47. doi: 10.1074/jbc.M309304200. [DOI] [PubMed] [Google Scholar]
- 103.O'Callaghan JP, et al. Early activation of STAT3 regulates reactive astrogliosis induced by diverse forms of neurotoxicity. PLoS One. 2014;9(7):e102003. doi: 10.1371/journal.pone.0102003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Bordet R, et al. PPARs: a new target for neuroprotection. J Neurol Neurosurg Psychiatry. 2006;77(3):285–7. doi: 10.1136/jnnp.2005.077495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Ji J, et al. PPARβ/δ activation protects against corticosterone-induced ER stress in astrocytes by inhibiting the CpG hypermethylation of microRNA-181a. Neuropharmacology. 2017;113(Pt A):396–406. doi: 10.1016/j.neuropharm.2016.10.022. [DOI] [PubMed] [Google Scholar]
- 106.Zheng B, et al. PGC-1α, a potential therapeutic target for early intervention in Parkinson's disease. Sci Transl Med. 2010;2(52):52ra73. doi: 10.1126/scitranslmed.3001059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Su X, et al. PGC-1α Promoter Methylation in Parkinson's Disease. PLoS One. 2015;10(8):e0134087. doi: 10.1371/journal.pone.0134087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Phipps AJ, et al. Neurofilament-labeled pyramidal neurons and astrocytes are deficient in DNA methylation marks in Alzheimer's disease. Neurobiol Aging. 2016;45:30–42. doi: 10.1016/j.neurobiolaging.2016.05.003. [DOI] [PubMed] [Google Scholar]
- 109.Moroni RF, et al. Developmental expression of Kir4.1 in astrocytes and oligodendrocytes of rat somatosensory cortex and hippocampus. Int J Dev Neurosci. 2015;47(Pt B):198–205. doi: 10.1016/j.ijdevneu.2015.09.004. [DOI] [PubMed] [Google Scholar]
- 110.Nwaobi SE, Olsen ML. Correlating Gene-specific DNA Methylation Changes with Expression and Transcriptional Activity of Astrocytic KCNJ10 (Kir4.1) J Vis Exp. 2015;(103) doi: 10.3791/52406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Nagy C, et al. Astrocytic abnormalities and global DNA methylation patterns in depression and suicide. Mol Psychiatry. 2015;20(3):320–8. doi: 10.1038/mp.2014.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Zhang X, et al. Regulation of DNA methylation by ethanol induces tissue plasminogen activator expression in astrocytes. J Neurochem. 2014;128(3):344–9. doi: 10.1111/jnc.12465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Zhao XJ, et al. Tissue plasminogen activator mediates deleterious complement cascade activation in stroke. PLoS One. 2017;12(7):e0180822. doi: 10.1371/journal.pone.0180822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Venkatesh S, Workman JL. Histone exchange, chromatin structure and the regulation of transcription. Nat Rev Mol Cell Biol. 2015;16(3):178–89. doi: 10.1038/nrm3941. [DOI] [PubMed] [Google Scholar]
- 115.Berger SL. The complex language of chromatin regulation during transcription. Nature. 2007;447(7143):407–12. doi: 10.1038/nature05915. [DOI] [PubMed] [Google Scholar]
- 116.Smith SM, Kimyon RS, Watters JJ. Cell-type-specific Jumonji histone demethylase gene expression in the healthy rat CNS: detection by a novel flow cytometry method. ASN Neuro. 2014;6(3):193–207. doi: 10.1042/AN20130050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Sher F, et al. Differentiation of neural stem cells into oligodendrocytes: involvement of the polycomb group protein Ezh2. Stem Cells. 2008;26(11):2875–83. doi: 10.1634/stemcells.2008-0121. [DOI] [PubMed] [Google Scholar]
- 118.Sher F, Boddeke E, Copray S. Ezh2 expression in astrocytes induces their dedifferentiation toward neural stem cells. Cell Reprogram. 2011;13(1):1–6. doi: 10.1089/cell.2010.0052. [DOI] [PubMed] [Google Scholar]
- 119.Pereira JD, et al. Ezh2, the histone methyltransferase of PRC2, regulates the balance between self-renewal and differentiation in the cerebral cortex. Proc Natl Acad Sci U S A. 2010;107(36):15957–62. doi: 10.1073/pnas.1002530107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Sudo G, et al. Increase in GFAP-positive astrocytes in histone demethylase GASC1/KDM4C/JMJD2C hypomorphic mutant mice. Genes Cells. 2016;21(3):218–25. doi: 10.1111/gtc.12331. [DOI] [PubMed] [Google Scholar]
- 121.Patrick KS, et al. Influence of ethanol and gender on methylphenidate pharmacokinetics and pharmacodynamics. Clin Pharmacol Ther. 2007;81(3):346–53. doi: 10.1038/sj.clpt.6100082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Kumar A, et al. Chromatin remodeling is a key mechanism underlying cocaine-induced plasticity in striatum. Neuron. 2005;48(2):303–14. doi: 10.1016/j.neuron.2005.09.023. [DOI] [PubMed] [Google Scholar]
- 123.Liu J, et al. Microglial Hv1 proton channel promotes cuprizone-induced demyelination through oxidative damage. J Neurochem. 2015;135(2):347–56. doi: 10.1111/jnc.13242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Sepsa A, et al. Emerging role of linker histone variant H1× as a biomarker with prognostic value in astrocytic gliomas. A multivariate analysis including trimethylation of H3K9 and H4K20. PLoS One. 2015;10(1):e0115101. doi: 10.1371/journal.pone.0115101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Sharma V, et al. Genome-wide ChIP-seq analysis of EZH2-mediated H3K27me3 target gene profile highlights differences between low- and high-grade astrocytic tumors. Carcinogenesis. 2017;38(2):152–161. doi: 10.1093/carcin/bgw126. [DOI] [PubMed] [Google Scholar]
- 126.Li Y, et al. Role of the histone H3 lysine 4 methyltransferase, SET7/9, in the regulation of NF-kappaB-dependent inflammatory genes. Relevance to diabetes and inflammation. J Biol Chem. 2008;283(39):26771–81. doi: 10.1074/jbc.M802800200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Arifuzzaman S, et al. Selective inhibition of EZH2 by a small molecule inhibitor regulates microglial gene expression essential for inflammation. Biochem Pharmacol. 2017;137:61–80. doi: 10.1016/j.bcp.2017.04.016. [DOI] [PubMed] [Google Scholar]
- 128.Gräff J, Tsai LH. Histone acetylation: molecular mnemonics on the chromatin. Nat Rev Neurosci. 2013;14(2):97–111. doi: 10.1038/nrn3427. [DOI] [PubMed] [Google Scholar]
- 129.Sidorova-Darmos E, et al. Differential expression of sirtuin family members in the developing, adult, and aged rat brain. Front Aging Neurosci. 2014;6:333. doi: 10.3389/fnagi.2014.00333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Cheng PY, et al. Interplay between SIN3A and STAT3 mediates chromatin conformational changes and GFAP expression during cellular differentiation. PLoS One. 2011;6(7):e22018. doi: 10.1371/journal.pone.0022018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Marin-Husstege M, et al. Histone deacetylase activity is necessary for oligodendrocyte lineage progression. J Neurosci. 2002;22(23):10333–45. doi: 10.1523/JNEUROSCI.22-23-10333.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Zhang L, et al. Hdac3 Interaction with p300 Histone Acetyltransferase Regulates the Oligodendrocyte and Astrocyte Lineage Fate Switch. Dev Cell. 2016;36(3):316–30. doi: 10.1016/j.devcel.2016.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Hsieh J, et al. Histone deacetylase inhibition-mediated neuronal differentiation of multipotent adult neural progenitor cells. Proc Natl Acad Sci U S A. 2004;101(47):16659–64. doi: 10.1073/pnas.0407643101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Humphrey GW, et al. Complementary roles for histone deacetylases 1, 2, and 3 in differentiation of pluripotent stem cells. Differentiation. 2008;76(4):348–56. doi: 10.1111/j.1432-0436.2007.00232.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Yang J, et al. Suppression of histone deacetylation promotes the differentiation of human pluripotent stem cells towards neural progenitor cells. BMC Biol. 2014;12:95. doi: 10.1186/s12915-014-0095-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Seidl SE, Potashkin JA. The promise of neuroprotective agents in Parkinson's disease. Front Neurol. 2011;2:68. doi: 10.3389/fneur.2011.00068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Correa F, et al. Activated microglia decrease histone acetylation and Nrf2-inducible anti-oxidant defence in astrocytes: restoring effects of inhibitors of HDACs, p38 MAPK and GSK3β. Neurobiol Dis. 2011;44(1):142–51. doi: 10.1016/j.nbd.2011.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Hashioka S, Klegeris A, McGeer PL. The histone deacetylase inhibitor suberoylanilide hydroxamic acid attenuates human astrocyte neurotoxicity induced by interferon-γ. J Neuroinflammation. 2012;9:113. doi: 10.1186/1742-2094-9-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Xiong H, et al. Trichostatin A, a histone deacetylase inhibitor, suppresses JAK2/STAT3 signaling via inducing the promoter-associated histone acetylation of SOCS1 and SOCS3 in human colorectal cancer cells. Mol Carcinog. 2012;51(2):174–84. doi: 10.1002/mc.20777. [DOI] [PubMed] [Google Scholar]
- 140.Kalinin S, et al. Dimethyl fumarate regulates histone deacetylase expression in astrocytes. J Neuroimmunol. 2013;263(1–2):13–9. doi: 10.1016/j.jneuroim.2013.07.007. [DOI] [PubMed] [Google Scholar]
- 141.Soliman ML, Combs CK, Rosenberger TA. Modulation of inflammatory cytokines and mitogen-activated protein kinases by acetate in primary astrocytes. J Neuroimmune Pharmacol. 2013;8(1):287–300. doi: 10.1007/s11481-012-9426-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Beurel E. HDAC6 regulates LPS-tolerance in astrocytes. PLoS One. 2011;6(10):e25804. doi: 10.1371/journal.pone.0025804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Biswas SK, Lopez-Collazo E. Endotoxin tolerance: new mechanisms, molecules and clinical significance. Trends Immunol. 2009;30(10):475–87. doi: 10.1016/j.it.2009.07.009. [DOI] [PubMed] [Google Scholar]
- 144.Suh HS, et al. Histone deacetylase inhibitors suppress the expression of inflammatory and innate immune response genes in human microglia and astrocytes. J Neuroimmune Pharmacol. 2010;5(4):521–32. doi: 10.1007/s11481-010-9192-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Cahoy JD, et al. A transcriptome database for astrocytes, neurons, and oligodendrocytes: a new resource for understanding brain development and function. J Neurosci. 2008;28(1):264–78. doi: 10.1523/JNEUROSCI.4178-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Rojo AI, et al. Nrf2 regulates microglial dynamics and neuroinflammation in experimental Parkinson's disease. Glia. 2010;58(5):588–98. doi: 10.1002/glia.20947. [DOI] [PubMed] [Google Scholar]
- 147.Chiu CC, et al. Neuroinflammation in animal models of traumatic brain injury. J Neurosci Methods. 2016;272:38–49. doi: 10.1016/j.jneumeth.2016.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Wu X, et al. Histone deacetylase inhibitors up-regulate astrocyte GDNF and BDNF gene transcription and protect dopaminergic neurons. Int J Neuropsychopharmacol. 2008;11(8):1123–34. doi: 10.1017/S1461145708009024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Chen PS, et al. Valproate protects dopaminergic neurons in midbrain neuron/glia cultures by stimulating the release of neurotrophic factors from astrocytes. Mol Psychiatry. 2006;11(12):1116–25. doi: 10.1038/sj.mp.4001893. [DOI] [PubMed] [Google Scholar]
- 150.Sadakierska-Chudy A, Filip M. A comprehensive view of the epigenetic landscape. Part II: Histone post-translational modification, nucleosome level, and chromatin regulation by ncRNAs. Neurotox Res. 2015;27(2):172–97. doi: 10.1007/s12640-014-9508-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Crowe SL, et al. Rapid phosphorylation of histone H2A.X following ionotropic glutamate receptor activation. Eur J Neurosci. 2006;23(9):2351–61. doi: 10.1111/j.1460-9568.2006.04768.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Myung NH, et al. Evidence of DNA damage in Alzheimer disease: phosphorylation of histone H2AX in astrocytes. Age (Dordr) 2008;30(4):209–15. doi: 10.1007/s11357-008-9050-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Enomoto R, et al. Involvement of histone phosphorylation in apoptosis of human astrocytes after exposure to saline solution. Neurochem Int. 2004;44(6):459–67. doi: 10.1016/s0197-0186(03)00175-x. [DOI] [PubMed] [Google Scholar]
- 154.Lan J, et al. Histone and DNA methylation control by H3 serine 10/threonine 11 phosphorylation in the mouse zygote. Epigenetics Chromatin. 2017;10:5. doi: 10.1186/s13072-017-0112-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Minc E, et al. Localization and phosphorylation of HP1 proteins during the cell cycle in mammalian cells. Chromosoma. 1999;108(4):220–34. doi: 10.1007/s004120050372. [DOI] [PubMed] [Google Scholar]
- 156.Lau PN, Cheung P. Histone code pathway involving H3 S28 phosphorylation and K27 acetylation activates transcription and antagonizes polycomb silencing. Proc Natl Acad Sci U S A. 2011;108(7):2801–6. doi: 10.1073/pnas.1012798108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Lo WS, et al. Phosphorylation of serine 10 in histone H3 is functionally linked in vitro and in vivo to Gcn5-mediated acetylation at lysine 14. Mol Cell. 2000;5(6):917–26. doi: 10.1016/s1097-2765(00)80257-9. [DOI] [PubMed] [Google Scholar]
- 158.Zhong S, et al. Phosphorylation at serine 28 and acetylation at lysine 9 of histone H3 induced by trichostatin A. Oncogene. 2003;22(34):5291–7. doi: 10.1038/sj.onc.1206507. [DOI] [PubMed] [Google Scholar]
- 159.Yamamoto Y, et al. Histone H3 phosphorylation by IKK-alpha is critical for cytokine-induced gene expression. Nature. 2003;423(6940):655–9. doi: 10.1038/nature01576. [DOI] [PubMed] [Google Scholar]
- 160.Song YS, et al. Oxidative stress increases phosphorylation of IkappaB kinase-alpha by enhancing NF-kappaB-inducing kinase after transient focal cerebral ischemia. J Cereb Blood Flow Metab. 2010;30(7):1265–74. doi: 10.1038/jcbfm.2010.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Molofsky AV, et al. Bmi-1 dependence distinguishes neural stem cell self-renewal from progenitor proliferation. Nature. 2003;425(6961):962–7. doi: 10.1038/nature02060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Tarcic O, et al. RNF20 Links Histone H2B Ubiquitylation with Inflammation and Inflammation-Associated Cancer. Cell Rep. 2016;14(6):1462–76. doi: 10.1016/j.celrep.2016.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Atanassov BS, et al. ATXN7L3 and ENY2 Coordinate Activity of Multiple H2B Deubiquitinases Important for Cellular Proliferation and Tumor Growth. Mol Cell. 2016;62(4):558–71. doi: 10.1016/j.molcel.2016.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Ceballos-Chávez M, et al. Control of neuronal differentiation by sumoylation of BRAF35, a subunit of the LSD1-CoREST histone demethylase complex. Proc Natl Acad Sci U S A. 2012;109(21):8085–90. doi: 10.1073/pnas.1121522109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Pascual G, et al. A SUMOylation-dependent pathway mediates transrepression of inflammatory response genes by PPAR-gamma. Nature. 2005;437(7059):759–63. doi: 10.1038/nature03988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Shiio Y, Eisenman RN. Histone sumoylation is associated with transcriptional repression. Proc Natl Acad Sci U S A. 2003;100(23):13225–30. doi: 10.1073/pnas.1735528100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Nathan D, et al. Histone sumoylation is a negative regulator in Saccharomyces cerevisiae and shows dynamic interplay with positive-acting histone modifications. Genes Dev. 2006;20(8):966–76. doi: 10.1101/gad.1404206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Dhall A, et al. Sumoylated human histone H4 prevents chromatin compaction by inhibiting long-range internucleosomal interactions. J Biol Chem. 2014;289(49):33827–37. doi: 10.1074/jbc.M114.591644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Colombo R, et al. The adenovirus protein Gam1 interferes with sumoylation of histone deacetylase 1. EMBO Rep. 2002;3(11):1062–8. doi: 10.1093/embo-reports/kvf213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Eckermann K. SUMO and Parkinson's disease. Neuromolecular Med. 2013;15(4):737–59. doi: 10.1007/s12017-013-8259-5. [DOI] [PubMed] [Google Scholar]
- 171.Guerra de Souza AC, Prediger RD, Cimarosti H. SUMO-regulated mitochondrial function in Parkinson's disease. J Neurochem. 2016;137(5):673–86. doi: 10.1111/jnc.13599. [DOI] [PubMed] [Google Scholar]
- 172.Lee L, et al. SUMO and Alzheimer's disease. Neuromolecular Med. 2013;15(4):720–36. doi: 10.1007/s12017-013-8257-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Tao CC, et al. Epigenetic regulation of HDAC1 SUMOylation as an endogenous neuroprotection against Aβ toxicity in a mouse model of Alzheimer's disease. Cell Death Differ. 2017;24(4):597–614. doi: 10.1038/cdd.2016.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Akar CA, Feinstein DL. Modulation of inducible nitric oxide synthase expression by sumoylation. J Neuroinflammation. 2009;6:12. doi: 10.1186/1742-2094-6-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Hoppe JB, Salbego CG, Cimarosti H. SUMOylation: Novel Neuroprotective Approach for Alzheimer's Disease? Aging Dis. 2015;6(5):322–30. doi: 10.14336/AD.2014.1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Lee JH, et al. Differential SUMOylation of LXRalpha and LXRbeta mediates transrepression of STAT1 inflammatory signaling in IFN-gamma-stimulated brain astrocytes. Mol Cell. 2009;35(6):806–17. doi: 10.1016/j.molcel.2009.07.021. [DOI] [PubMed] [Google Scholar]
- 177.Mohr AM, Mott JL. Overview of microRNA biology. Semin Liver Dis. 2015;35(1):3–11. doi: 10.1055/s-0034-1397344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Valencia-Sanchez MA, et al. Control of translation and mRNA degradation by miRNAs and siRNAs. Genes Dev. 2006;20(5):515–24. doi: 10.1101/gad.1399806. [DOI] [PubMed] [Google Scholar]
- 179.Londin E, et al. Analysis of 13 cell types reveals evidence for the expression of numerous novel primate- and tissue-specific microRNAs. Proc Natl Acad Sci U S A. 2015;112(10):E1106–15. doi: 10.1073/pnas.1420955112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Hamill CE, et al. Exacerbation of dopaminergic terminal damage in a mouse model of Parkinson's disease by the G-protein-coupled receptor protease-activated receptor 1. Mol Pharmacol. 2007;72(3):653–64. doi: 10.1124/mol.107.038158. [DOI] [PubMed] [Google Scholar]
- 181.Letzen BS, et al. MicroRNA expression profiling of oligodendrocyte differentiation from human embryonic stem cells. PLoS One. 2010;5(5):e10480. doi: 10.1371/journal.pone.0010480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Rao VT, et al. MicroRNA Expression Patterns in Human Astrocytes in Relation to Anatomical Location and Age. J Neuropathol Exp Neurol. 2016;75(2):156–66. doi: 10.1093/jnen/nlv016. [DOI] [PubMed] [Google Scholar]
- 183.Ziu M, et al. Temporal differences in microRNA expression patterns in astrocytes and neurons after ischemic injury. PLoS One. 2011;6(2):e14724. doi: 10.1371/journal.pone.0014724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Mor E, et al. Species-specific microRNA roles elucidated following astrocyte activation. Nucleic Acids Res. 2011;39(9):3710–23. doi: 10.1093/nar/gkq1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Iyer A, et al. MicroRNA-146a: a key regulator of astrocyte-mediated inflammatory response. PLoS One. 2012;7(9):e44789. doi: 10.1371/journal.pone.0044789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Lafourcade C, et al. MiRNAs in Astrocyte-Derived Exosomes as Possible Mediators of Neuronal Plasticity. J Exp Neurosci. 2016;10(Suppl 1):1–9. doi: 10.4137/JEN.S39916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Song W, et al. Schizophrenia-like features in transgenic mice overexpressing human HO-1 in the astrocytic compartment. J Neurosci. 2012;32(32):10841–53. doi: 10.1523/JNEUROSCI.6469-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Jovičić A, Gitler AD. Distinct repertoires of microRNAs present in mouse astrocytes compared to astrocyte-secreted exosomes. PLoS One. 2017;12(2):e0171418. doi: 10.1371/journal.pone.0171418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Hoogland IC, et al. Systemic inflammation and microglial activation: systematic review of animal experiments. J Neuroinflammation. 2015;12:114. doi: 10.1186/s12974-015-0332-6. [DOI] [PMC free article] [PubMed] [Google Scholar]