Abstract
Background
Molecular mechanisms underlying psychological sequelae of exposure to stressful experiences, such as Post-Traumatic Stress Disorder (PTSD) and depression, are not well understood.
Methods
Using convergent evidence from animal and human transcriptomic and genomic studies, we aimed to identify genetic mechanisms underlying depression and anxiety after traumatic experiences.
Results
From a transcriptome-wide analysis in mice, we found the Ppm1f gene to be differentially expressed in the amygdala and medial prefrontal cortex (mPFC) a week after immobilization stress. Next, we found that PPM1F mRNA levels in human blood were down-regulated in cases with symptoms of comorbid PTSD and depression (PTSD&Dep), and consistently in cases with anxiety symptoms in a separate human dataset. Furthermore, we showed that a genetic variant of PPM1F, rs17759843, was associated with PTSD&Dep and with PPM1F expression in both human brain and blood. Given prior reported mechanistic links between PPM1F and CAMK2, we examined blood mRNA level of CAMK2G in human and found it to be lower in PTSD&Dep. We also found that Ppm1f protein levels and its colocalization with Camk2G were altered in the amygdala and mPFC of male mice. Additionally, we found that a systemic dose of corticosterone blocked the depressive-like phenotype elicited by stress in female mice. Lastly, corticosterone rescued the anxiety-like phenotype and mRNA levels of Ppm1f in the amygdala and mPFC in male mice and in the mPFC of female mice.
Discussion
Taken together, our data suggest a mechanistic pathway involving PPM1F and CAMK2G in stress and trauma-related manifestation of anxiety and depression across species.
Keywords: Stress, Anxiety, Depression, PPM1F, Camk2g, PTSD
Introduction
Exposure to stressful or traumatic life experiences increases risks of developing depressive disorders, anxiety disorders, or post-traumatic stress disorder (PTSD) (1). Anxiety and depressive disorders rank at the top of disabling mental illnesses causing tremendous economic costs to society (2). However, effectiveness of pharmacological treatments for anxiety and depressive disorders still remains rather limited with small effect sizes (3, 4). To facilitate efforts in developing novel treatments for these disorders, better insights into molecular mechanisms underlying anxiety and depression are needed.
With the goal of elucidating molecular mechanisms underlying anxiety and depression after traumatic experiences, we used convergent evidence from both animal and human studies. We note that among susceptible individuals, PTSD and depression are often comorbid (PTSD&Dep) (5). The shared symptoms and relatively high prevalence of PTSD and depression comorbidity suggest that examining their comorbidity in the aftermath of trauma may be a more powerful approach than examining either outcome alone. In addition, animal stress models leading to similar PTSD-like or depression-like behaviors can facilitate our efforts in uncovering their neurobiological mechanisms (6, 7).
Methods
Animals
All experiments were performed on adult wild-type C57BL/6J mice obtained from Jackson Labs. Male and female mice were group-housed in a temperature-controlled vivarium, with ad libitum access to food and water. See detailed methods for ethics protocols. Female mice were used only when explicitly stated, otherwise only male mice were used.
Immobilization Stress
Immobilization Stress (IMO) procedures, as previously described (8–10), were conducted in a room separate from housing and behavioral paradigms (11). All mice were habituated to the test chambers before the IMO stress and received exposure to handling and the training context completely separate and distinct from the IMO context (9). We chose the 6–8 days post-IMO time point because we wanted to compare these results with our previous findings studying gene regulation at this same time point (8,9,11,12).
Microarray hybridization and analysis
Mice were sacrificed under basal conditions (Home Cage Control Group) at 6 or 8 days after a 2 hour immobilization (IMO Stress Group). The reason for having two amygdala microarrays was to replicate the findings on day 6 with those on day 8 and get more robust results. Amygdala and medial prefrontal cortex (mPFC) tissue from both hemispheres was extracted by 1mm micropunch. Total RNA was extracted from the tissue and purified with the RNeasy Mini Kit catalog #74106 (Qiagen) following the manufacturer’s instructions. Illumina Mouse WG-6 v2 Expression BeadChip microarray was assayed for 45,281 transcripts. These microarray are available at GEO #GSE100086. Plots of principal variance component analysis and volcano plots are in Supplemental Figure S1.
Bioinformatics of the microarray analysis
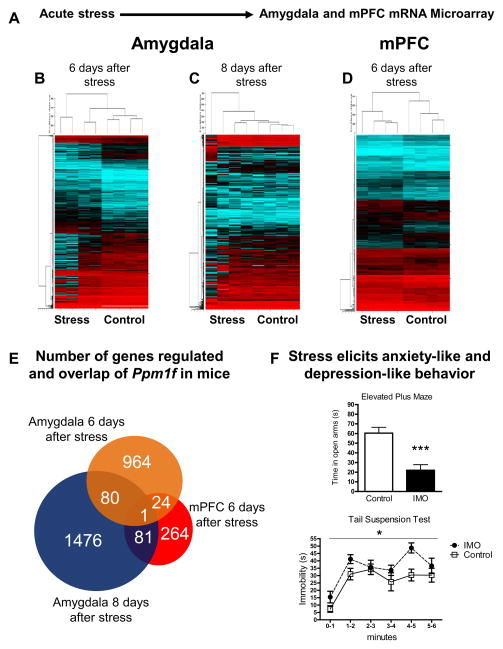
The top genes regulated after stress in the Amygdala and mPFC were further analyzed with DAVID Bioinformatics 6.8 to understand their functions. Note that Ppm1f was the only top gene present in all the microarrays (Figure 1A–E).
Figure 1. Ppm1f brain regulation after stress, anxiety-like and depression-like behaviors.
(A) Schematic of the experimental protocol (N=4 per group). (B) Amygdala microarray 6 days after acute immobilization stress (IMO). (C) Amygdala microarray 8 days after IMO stress. (D) mPFC microarray 6 days after IMO stress. (E) Gene regulation of the microarrays show overlap in the Ppm1f gene. (F) Top, 6 days after stress mice spend less time in the open arms in the elevated plus maze indicating enhanced anxiety-like behavior (p=0.0004, n=8 per group). Below, 6 days after stress mice present enhanced levels of immobility in the tail suspension test indicating enhanced depressive-like behavior (p=0.0361, n=8 per group).
Reverse Transcription and PCR Quantification
Protocol was followed as previously described (8).
RT-PCR arrays
RNA was isolated and reverse transcribed as outlined in the section above. We used a custom plate format 24 × 4 catalog #CAPM10412 (Applied Biosystems, Carlsbad, CA).
Elevated plus maze
The elevated plus maze was performed as previously described (12).
Tail suspension test
Mice were suspended with duct tape ~2 cm at the end of the tail. The session lasted for 6 minutes. Behavior was recorded by a video camera and a researcher blind to the experimental groups analyzed the immobility time.
Mouse fear conditioning
Mice were fear conditioned in standard rodent modular test chambers (ENV-008-VP; Med Associates Inc, St. Albans, VT) with an inside area of 30.5cm (L) × 24.1cm (W) × 21.0cm (H). Mice received 5 trials of a conditioned stimulus (CS; 30 second tone, 6 kHz, 70 dB) co-terminating with a footshock (500ms, 0.6mA) unconditioned stimulus (US).
Gene expression profiles in human GTP participants
Participants were recruited from Atlanta inner-city residents by the Grady Trauma Project (GTP) (13–15). This study was approved by the Institutional Review Board of Emory University School of Medicine and Grady Memorial Hospital.
PTSD symptoms were assessed with the modified PTSD Symptom Scale (PSS) (16). Depressive symptoms were measured with the Beck Depression Inventory (BDI)(17). Cases with current symptoms of comorbid PTSD and depression (PTSD&Dep) were defined as having PSS≥14 and BDI≥14. Controls were defined as not having either PTSD or depressive symptoms, as reflected by a PSS ≤7 and BDI ≤7, despite being exposed to trauma. See Supplementary Methods for rationale behind these cutoff scores. Of note, these participants were not part of a rigorously assessed clinical sample.
RNA was extracted from blood collected in the morning. All samples had RNA Integrity Number (RIN) ≥6. Raw probe intensities were generated on Illumina HumanHT-12 v3 or v4 BeadChip arrays. We performed normalization of probe intensities using the Supervised Normalization of Microarray algorithm (18), removing the effects of batch and RIN. Association between mRNA level of a gene of interest and PTSD&Dep status was examined using multiple linear regression adjusting for gender, age, and population substructure.
Center for Health Discovery and Well Being (CHDWB) replication sample and gene expression profiles
The CHDWB project evaluated effectiveness of a health- and prevention-focused approach (19). Anxiety symptoms were assessed with the GAD-7 scale (20), a well-validated and efficient tool for screening anxiety symptoms. A score of 5–9 on the GAD-7 indicates mild anxiety, 10–14 moderate, and 15–21 severe anxiety symptoms (20). We categorized participants with GAD-7 scores of ≥ 5 as having anxiety symptoms and those with GAD-7 scores ≤1 as controls.
RNA was extracted from whole blood and all samples had RIN >7. Probe intensities were generated on Illumina HT12 v3 or v4 beadchip arrays. We performed normalization using the Supervised Normalization of Microarrays (18). Human PPM1F microarrays from CHWDB are available at GEO #GSE61672.
PPM1F genotypes and principal components among GTP participants
DNA was extracted from saliva or blood, and genotyping was conducted using Illumina’s HumanOmni1-Quad. Standard quality control of the genotyping was performed using PLINK (21), removing individuals with >2% missing data and removing one in each pair of related individuals with an identity by descent proportion >0.12 (indicating cousins or a closer relation). We used principal component analysis to infer axes of ancestry and remove outlier subjects following methods previously described (22). Association between PPM1F single nucleotide polymorphisms (SNPs) and PTSD&Dep was examined using PLINK, adjusting for gender and 10 PCs. Multiple testing was addressed with permutation (23).
eQTL analysis
Association between rs17759843 and blood mRNA level of PPM1F was provided by the Blood eQTL browser (genenetwork.nl/bloodeqtlbrowser) (24). Quality control for this dataset was described in detail by Westra and colleagues (24).
Postmortem eQTL Analysis
Association between rs17759843 and brain mRNA level of PPM1F was provided by Brain Cloud, which has gene expression profiles from the dorsolateral prefrontal cortex of 269 subjects without neuropathological and neuropsychiatric diagnosis (25).
Results
Behavior and gene expression profiles in Amygdala and mPFC of stressed mice
We performed transcriptome-wide differential expression analysis of genes in the Amygdala and mPFC in mice 6 and 8 days after immobilization (IMO) stress versus controls. We examined gene expression profiles in the Amygdala at 6 days and 8 days after IMO stress to determine if the long-term gene expression changes found at 6 days were also sustained at 8 days. Figures 1A–D show the heatmaps revealing differences in the gene expression in stressed versus control mice groups. Of the probes that were differentially expressed at unadjusted p<0.05 in the Amygdala, we used the following criteria to select probes for follow-up: a) probe differentially expressed at unadjusted p-value <0.05 at both 6 days and 8 days after stress; b) probe with >1.5 fold change between cases and controls; c) probe is expressed at moderate to high levels in the Amygdala (through search in the Allen Brain Atlas). Supplemental Table S1 lists the 24 probes that met these criteria. Of note, without adjusting for multiple testing, there is an increased risk of false positives. Therefore, we performed replication of these probes in different mouse cohorts to determine if they were differentially expressed between cases and controls using RT-PCR. Of these 24 probes, 21 had available primers for the RT-PCR replication (Supplemental Table S2). Supplemental Table S3 lists the results of the PCR replication in the Amygdala. We found five probes whose altered mRNA levels were replicated with PCR; Ppm1f was one of them. Along with our replication studies, we showed that these gene expression changes, which occurred 6 to 8 days after IMO, may need an incubation period because no significant changes in gene expression were found 1 day after exposure to stress (Supplemental Table S3). Interestingly, although Ppm1f is significantly regulated 6–8 days post-stress, it is not regulated in the Amygdala 30 minutes or 2 hours after fear conditioning (FC) nor in the mPFC 2 hours after FC (Supplemental Figure S2). Thus, Ppm1f appears to have a dynamic expression after stress in the brain. Moreover, only five of these genes were significantly regulated after stress - Hbp1, Ssbp4, Fbxl5, Ppm1f and Fhl1 – at three different time points: 6, 7 and 8 days after IMO (Supplemental Table S3). Thus, we examined the expression of these five genes in human. Among them, only PPM1F was differentially expressed between cases with symptoms of PTSD&Dep versus controls (Supplemental Table S4; more detailed description to follow).
We followed similar criteria to select probes with unadjusted p-value <0.05 from the mPFC transcriptome-wide differential analysis for further replication in a mouse microarray (more details in Supplemental Methods). See Supplemental Table S5 for the list of genes that survived the filtering criteria and were followed up with RT-PCR replication. The number of top-regulated genes within these microarrays and their overlap are represented in Figure 1E. Ppm1f was the only gene highly regulated in all the three mouse microarray conditions.
Behaviorally, two groups of mice were exposed to IMO stress and 6 days later they were tested in the elevated plus maze (EPM, anxiety-like behavior) or the tail suspension test (TST, depressive-like behavior). The EPM results showed that stress reduced the time mice spent in the open arms (Figure 1F top, t=4.597 df=14, p=0.0004). Moreover, the TST analysis revealed that stress increased the time mice spent in immobility (Figure 1F down, f=2.280, df=7, p=0.0361).
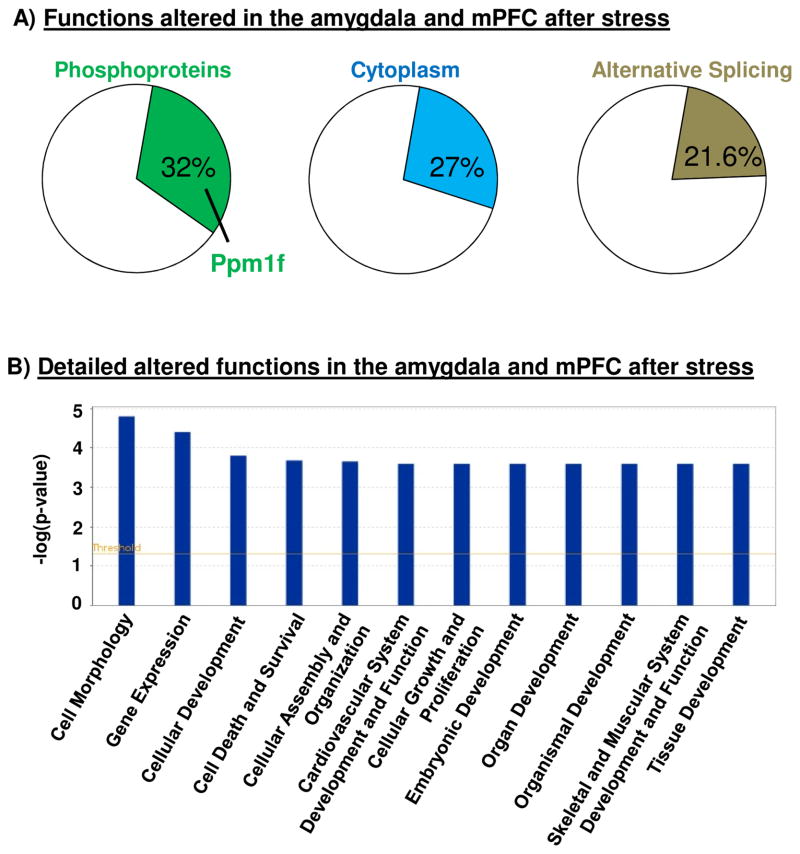
Brain functions altered after acute stress, network analysis and predicted interactions with drugs
We performed bioinformatics analyses of the shared genes that had altered expression in the microarray. The list of the genes studied with DAVID are in Supplemental Tables S1 and S5 and Supplemental Figure S3. We found that the top three functions altered in both the Amygdala and mPFC were phosphoproteins (including Ppm1f), alternative splicing, and splice variants (Figure 2A). A more detailed functional analysis with IPA showed the biological pathways that were most different between stress and control animals included cell morphology, gene expression, and cellular development (Figure 2B). Supplemental Figure S4 shows that PPM1F has direct relationships with the CaMK2 family and indirect relationships with brain-derived neurotrophic factor (BDNF), cAMP response element-binding (CREB), extracellular signal-regulated kinase 1/2 (Erk 1/2) and (Mitogen-activated protein kinase kinase) Map2K 1/2.
Figure 2. Bioinformatics analysis of the altered functions in the Amygdala and mPFC after exposure to stress in mice.
(A) Analysis with DAVID 6.8 revealed general altered functions 6 to 8 days after acute stress in both Amygdala and mPFC. Phosphoproteins (including Ppm1f), Cytoplasm and Alternative Splicing were the more affected functions in these analysis. (B) IPA analysis showed more detailed functions altered after a single exposure to 2 hours stress immobilization.
Examine expression of the above five genes in comorbid PTSD & Depression
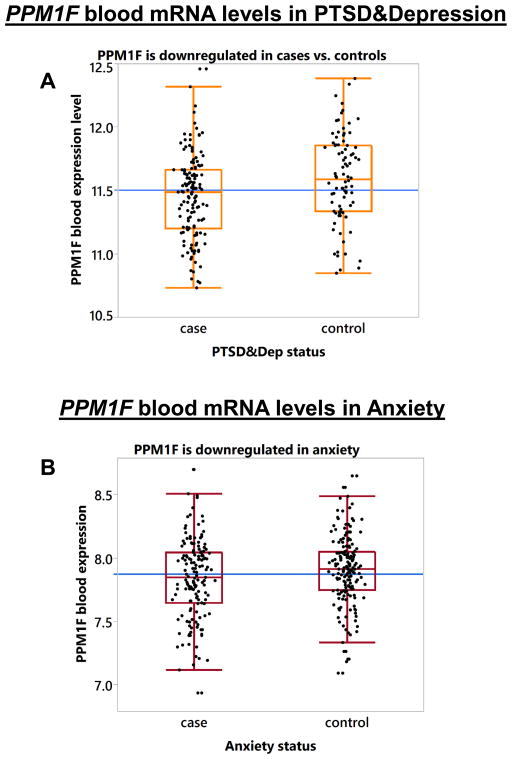
We next examined expression of the five genes identified to be significantly differentially regulated in mouse Amygdala after exposure to stress in 230 GTP human participants who were exposed to at least one traumatic experience. Their sociodemographic characteristics are presented in Supplemental Table S6. Of these participants, 142 were cases with PTSD&Dep symptoms and 88 were controls with no PTSD and no depressive symptoms. Among these five genes, PPM1F (ILMN_2059535) was significantly associated with PTSD&Dep after adjusting for gender, age, and population substructure (β=0.135; p=0.005; Bonferroni adjusted p=0.025; N=230). Cases with PTSD&Dep symptoms had lower PPM1F expression than controls (Figure 3A). Notably, PPM1F is highly expressed in the brain in both mice and humans (Supplemental Figure S5).
Figure 3. PPM1F blood mRNA levels in PTSD&Depression and in Anxiety.
(A) Box plot showing that PPM1F mRNA level is lower in cases with symptoms of PTSD&Depression versus controls after adjusting for gender, age, and population substructure (p=0.005, n=230). The black dots represent the PPM1F expression level of each sample. The blue horizontal line represents the mean of PPM1F level in the overall sample; (B) PPM1F mRNA level is significantly down-regulated in anxiety vs. controls after adjusting for gender, age, and ethnicity (p=0.044, n=316).
PPM1F in anxiety in a replication sample
Given that PTSD patients present with anxiety symptoms, we examined blood mRNA of PPM1F (ILMN_2059535) in anxiety in the CHWDB cohort, which has 151 cases with anxiety symptoms and 165 controls (N=316). Their characteristics are presented in Supplemental Table S7. We found that PPM1F was significantly down-regulated in patients with anxious symptomatology versus controls after adjusting for gender and ethnicity (p=0.044, N=316, Figure 3B, Supplemental Table S7), consistent with the association found in the GTP sample.
Association between PPM1F SNPs and PTSD&Dep
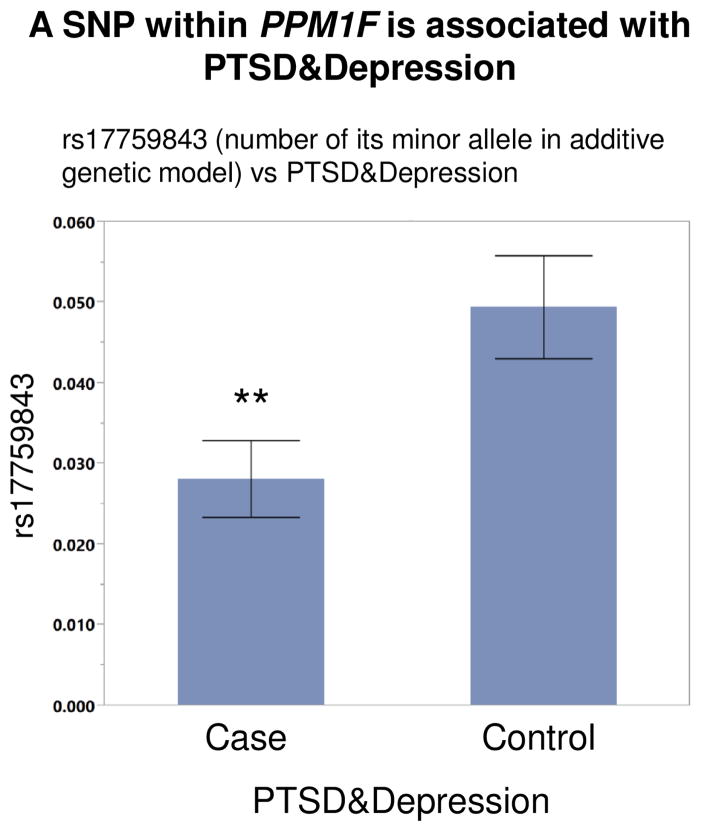
Having identified PPM1F mRNA expression level to be significantly associated with PTSD, depression and anxiety, we next investigated whether certain PPM1F SNPs influence this association. To this end, we examined the association between PPM1F SNPs and PTSD&Dep, and whether the identified significant SNPs influenced PPM1F expression level in blood and brain. In the GTP human dataset, there was a total of 14 tagging SNPs for PPM1F. Among these SNPs, rs17759843, located in the 3′UTR of PPM1F, was significantly associated with PTSD&Dep after adjusting for sex and 10 genetic principal components (OR=0.53; p=0.0046; permuted p=0.0463; N=2361; Figure 4). The minor allele of rs17759843 was associated with lower odds for comorbid PTSD&Dep (Figure 4). rs17759843 has a MAF of 0.018 and p-value for departure from HWE of 0.6 in the unaffected population and 1 in affected population.
Figure 4. A SNP within the PPM1F gene is associated with PTSD&Depression.
The minor allele of rs17759843, a PPM1F 3′UTR SNP, is significantly associated with lower odds for PTSD & Depression after adjusting for gender and population substructure (OR=0.53; p=0.005; permuted p=0.046; n=2361). Each error bar is constructed using 1 standard error from the mean.
PPM1F rs17759843 influences expression of PPM1F in human blood
Since we found rs17759843 significantly associated with PTSD&Dep, we examined whether this SNP is associated with human blood PPM1F mRNA level in 358 GTP participants. There was no significant association between this SNP and PPM1F mRNA level after adjusting for sex, age, and population substructure (p=0.635, N=358). However, when we looked at the Blood eQTL browser (24) of 5300 human samples, there was a highly significant association between rs17759843 and PPM1F (p=6.1 × 10−25, FDR=0.00; effect size Z-score = 10.31). Specifically, the minor allele of rs17759843 was associated with higher expression of PPM1F. This directionality is consistent with our genetic and gene expression findings in the GTP sample, in which the minor allele of rs17759843 was significantly associated with lower probability of having PTSD&Dep, and that controls had higher expression of PPM1F relative to that of PTSD&Dep cases. With a larger sample size and thus more power, we conclude that rs17759843 is indeed a blood cis-eQTL for PPM1F mRNA expression in humans.
rs17759843 influences expression of PPM1F in human brain
Since Ppm1f mRNA level was upregulated in the amygdala and downregulated in the mPFC of stressed mice, which is consistent with another published study (26), we examined whether rs17759843 is associated with brain mRNA level of PPM1F in the publicly available BrainCloud dataset, which has human gene expression profiles of the prefrontal cortex. There was a total of three probes for PPM1F in this dataset. Of these, two probes had significant association with PPM1F: Probe 8896: p=0.0032, Bonferroni-adjusted p=0.0097; and Probe 16009: p=0.0004, Bonferroni-adjusted p=0.0012. In sum, rs17759843 is significantly associated with PPM1F mRNA level in human prefrontal cortex.
CAMK2
Since PPM1F regulates calcium/calmodulin-dependent protein kinase 2 (CaMK2) in neuronal cells (27, 28) and fibroblasts (29), we examined blood mRNA level of CaMK2 in PTSD&Dep. After QC, the human dataset had three probes for CaMK2 (one for CaMK2D and two for CaMK2G; none for CaMK2A or CaMK2B). Of these three probes, the CaMK2G probe had significantly lower expression in cases with PTSD&Dep symptoms versus controls after adjusting for gender, age, and population substructure (β=0.072; p=0.0158; Bonferroni corrected p = 0.0474, N=230; Supplemental Figure S6A–B).
We analyzed Camk2g in the mouse brain because it was the CaMK2 isoform that was most significantly regulated in our human dataset. After stress Camk2g mRNA was marginally upregulated in the mouse amygdala 6 days after IMO (t=2.213, df=8, p=0.058, Supplemental Figure S6C). However, Camk2g expression was not regulated in mouse amygdala 2 hours after FC in mice with or without a previous exposure to stress (Supplemental Figure S6D).
Immunohistochemistry of Ppm1f and Camk2g
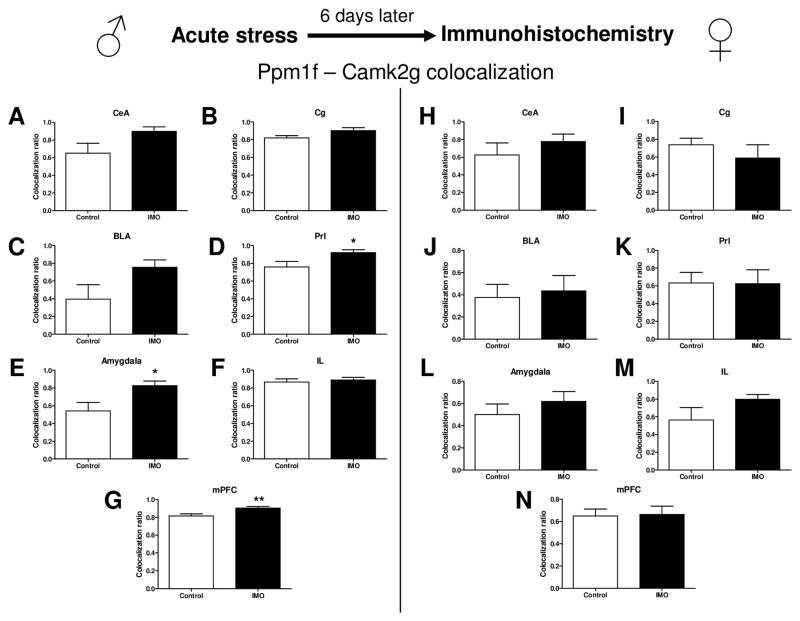
Ppm1f protein levels were upregulated in the Central amygdala (CeA), Basolateral amygdala (BLA), the summatory of CeA+BLA (Total Amygdala) and the mPFC in male mice 6 days after stress IMO (Supplemental Figure S7). Moreover, Ppm1f levels in the BLA were also upregulated in female mice 6 days after IMO (Supplemental Figure S8). Camk2g levels were upregulated in the CeA, BLA, Total Amygdala and mPFC in male mice 6 days after IMO (Supplemental Figure S9). However, there were no changes in Camk2g levels 6 days after IMO in female mice (Supplemental Figure S10). Figure 5 shows that IMO enhances the colocalization of Ppm1f and Camk2g in the Prelimbic cortex (PrL) (t= −2.308, df=12, p=0.046), Total Amygdala (t= −2.598, df=24, p=0.017) and the mPFC in male mice (t= −2.814, df=41, p=0.008) (Figure 5). See Figure 6 for representative images of these immunohistochemistry studies.
Figure 5. Stress immobilization and Ppm1f/Camk2g levels in the Amygdala and mPFC in both male and female mice: an immunohistochemistry study.
Quantification of Ppm1f and Camk2g levels reveals that both are more densely colocalized after traumatic stress in male mice in the Amygdala and mPFC. (A,H) CeA = Central amygdala, (B,I) Cg = Cingulate cortex, (C,J) BLA = Basolateral amygdala, (D,K) Prl= Prelimbic cortex, (E,L) (Total) Amygdala = CeA + BLA, (F,M) IL= Infralimbic cortex, (G,N) mPFC = Medial prefrontal cortex = Cg+Prl+IL. *p≤0.05, **p≤0.01. N=2 per group.
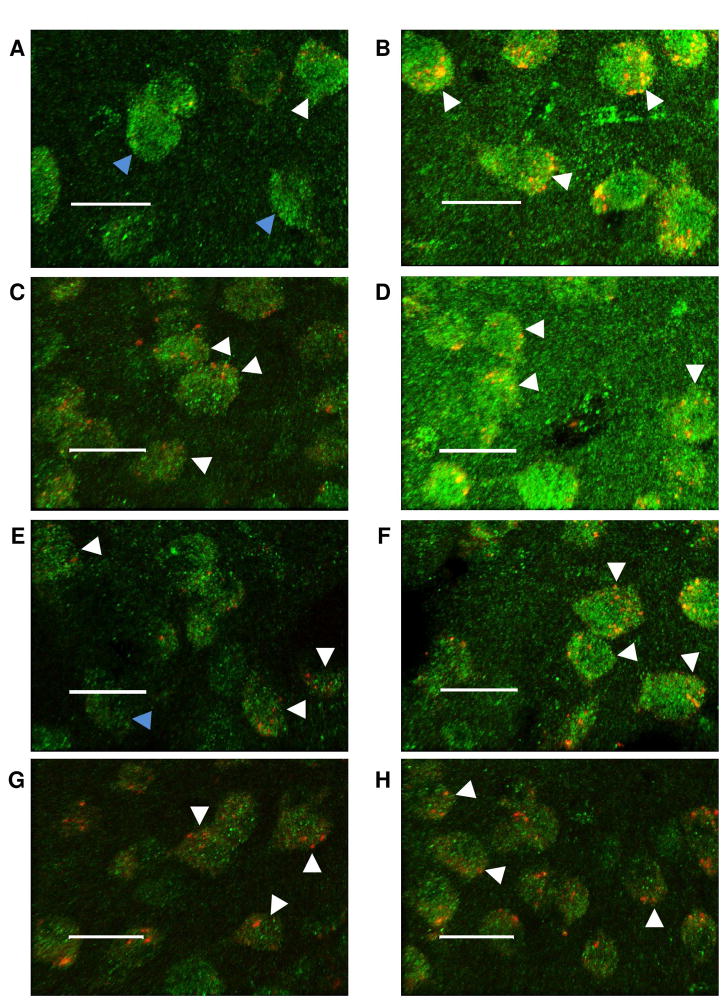
Figure 6. Representative images of the immunohistochemistry study evaluating the colocalization of Ppm1f (green) and Camk2g (red) in the Amygdala and mPFC.
Scale bars = 20μm. (A) Male Basolateral amygdala, group control. (B) Male Basolateral amygdala, group IMO. (C) Male Prelimbic cortex (PrL), group control. (D) Male PrL, group IMO. (E) Female Basolateral amygdala, group control. (F) Female Basolateral amygdala, group IMO. (G) Female Prelimbic cortex (PrL), group control. (H) Female PrL, group IMO. Blue arrow indicates no colocalization. White arrow indicates colocalization.
Corticosterone and Ppm1f
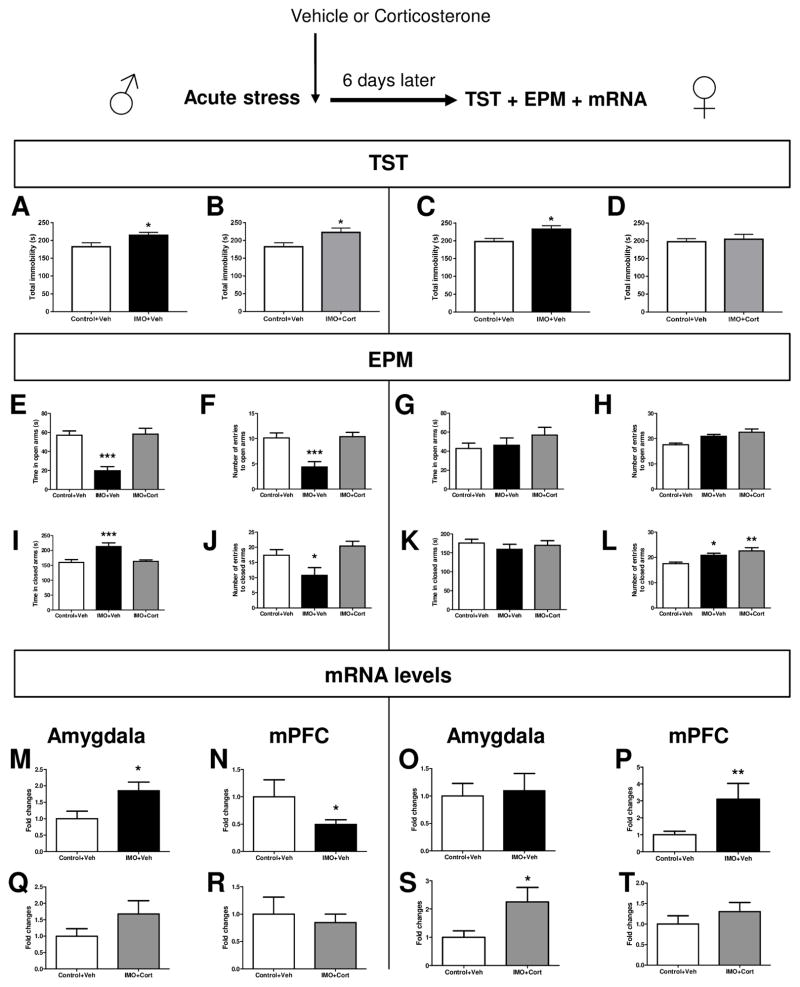
Vehicle or corticosterone was given 1 hour after IMO in male and female mice. 6 days later TST, EPM and mRNA levels were evaluated. In males, there was a significant increase in total immobility as measured by TST in IMO+Veh (t=−2.328, df=14, p=0.037, Figure 7A) and IMO+CORT (t=−2.45, df=14, p=0.029, Figure 7B) groups compared to Control+Veh group. In females, there was a significant increase in total immobility as measured by a TST in IMO+Veh (t=−2.394, df=8, p=0.044, Figure 7C] compared to Control+Veh group. Analysis in males of the EPM showed significant differences in the time in open arms [F(2,21)=10.358; p=0.001, post-hoc IMO-Veh p=0.001 vs the other groups], entries to open arms [F(2,21)=11.902; p=0.000, post-hoc p=0.0001 vs the other groups], time in closed arms [F(2,21)=9.691; p=0.001, post-hoc p=0.001 vs the other groups] and entries to closed arms [F(2,21)=5.762; p=0.010, post-hoc p=0.033 vs the other groups]. The EPM results in females showed a significant effect for treatment on the total number of entries to closed arms [F(2,21)=6.879; p=0.005]. Post-hoc comparisons revealed an increased number of entries to closed arms in IMO+Veh (p=0.023) and IMO+Cort group (p=0.002) compared to Control+Veh group. Analysis of the mRNA levels in males showed that the IMO+Veh group presented enhanced levels of Ppm1f compared to Control group (t=2.383, df=13, p=0.0331, Figure 7M) in the Amygdala and decreased levels in the mPFC (t=2.321, df=13, p=0.0371, Figure 7N). The mRNA levels in females were upregulated in mPFC in the group IMO+Veh vs Control+Veh (t= 3,248 df=13 p=0.0064, Figure 7P) and also in the group IMO+Cort vs Control+Veh in the Amygdala (t=2.579, df=14, p=0.0218, Figure 7S).
Figure 7. Ppm1f and corticosterone in anxiety-like behavior and depressive-like behavior in both male and female mice.
N=5–8 per group. Systemic corticosterone given 1 hour after stress did not rescue the depressive-like phenotype elicited by IMO in male mice (A, B). However, Corticosterone rescued the depressive-like behavior induced by IMO in female mice (C, D) *p≤0.05). EPM results showed that corticosterone rescued the anxiety-like behavior in male mice (E,F, I, J, *p≤0.05, **p≤0.01, ***p≤0.001 vs the other groups). In contrast, IMO did not induce an anxiety-like behavior in female mice (G, H, K). The number of entries in closed arms was increased in the IMO+Veh vs Control+Veh group (L, *p≤0.05, ***p≤0.001 vs Control+Veh). Analysis of the mRNA levels in males showed that the IMO+Veh group presented enhanced levels of ppm1f compared to Control group (p=0.0331, M) in the Amygdala and decreased levels in the mPFC (p=0.0371, N). Corticosterone rescued the ppm1f gene changes induced by IMO in the Amygdala (Q) and mPFC (R). The mRNA levels in females were upregulated in mPFC in the group IMO+Veh vs Control+Veh (P) and also in the group IMO+Cort vs Control+Veh in the Amygdala (S). Corticosterone rescued the ppm1f gene changes in the mPFC (T).
Discussion
Here, we found that Ppm1f mRNA levels were significantly altered in the amygdala and mPFC after stress exposure in a mouse model. Furthermore, we demonstrate that the human blood mRNA level of PPM1F was significantly down-regulated in PTSD&Dep. Moreover, we show that a genetic variant in the 3′UTR of PPM1F, rs17759843, was significantly associated with PTSD&Dep and with mRNA levels of PPM1F in human blood as well as brain. Additionally, we found that one of the substrates of PPM1F, CAMK2G, had significantly down-regulated blood mRNA levels in PTSD&Dep, which is consistent with CAMK2 being a substrate of PPM1F. Consistently, Ppm1f protein levels and its colocalization with Camk2G were altered in the Amygdala and mPFC of male mice. Taken together, our findings suggest a novel and critical role of PPM1F in association with stress, anxiety, and depressive symptoms following trauma exposure.
PPM1F is a protein phosphatase and a member of the PP2C family of Ser/Thr protein phosphatases, which are negative regulators of stress response pathways. Calcium/calmodulin-dependent protein kinase II gamma is one of the substrates of this phosphatase (27–29). Interestingly, Ppm1f expression in mice was not altered at early time points after an aversive learning paradigm nor 1 day after IMO stress. A potential interpretation is that these changes in basal expression of Ppm1f require about a week to develop in mice. Also, it is possible that Ppm1f changes may not be related to fear learning, per se, or are not detected at the timing of the collection of the brain tissue in these experiments. In fact, our data suggest that it may be a marker of the chronic effects of prior trauma exposure.
Ppm1f specifically dephosphorylates Camk2 in rat neurons in vitro and in cells (30). This results in an inactive form of CaMK2. CAMK2 is critical in serotonergic regulation of prefrontal cortex neuronal activity, which plays a major role in depression, suicide, anxiety, and schizophrenia (31–33), and which may explain the neuropsychiatric behavioral patterns seen in CaMK2 knockout mice (34). CaMK levels in the brain have been previously associated with anxiety (35), depression (36), and suicide victims (37). Our present data suggests that PPM1F could be an upstream regulator – activated by traumatic stress – of CaMK, promoting the development of anxiety and depression-like symptoms. However, the consequences of regulation of PPM1F by stress could have broader implications than described here because of ubiquitous expression of PPM1F in the body and brain. For example, PTSD and depression have been associated with higher rates of coronary heart disease (38–40) and knowledge of the underlying mechanisms for this association is still limited. Since CaMK2 plays an important role in the development and progression of cardiovascular disease (41–43), it is possible that the PPM1F/CaMK2 pathway may play a role in coronary diseases. Further study of PPM1F and CAMK2 may shed light on the mechanisms linking PTSD and depression to elevated risk for heart disease.
Using a different traumatic model than ours, previous studies have shown that exposing rats to a predator results in Amygdala upregulation but mPFC downregulation of phosphorylated Camk2 (pCamk2) one week later (44). Consistently, we found significant changes in mRNA levels of CaMK2G in blood in human with PTSD&Dep symptoms and in the Amygdala of mice exposed to stress. Concordantly, we found that stress also regulated Camk2g protein in the mouse brain. Moreover, Ppm1f protein level in the Amygdala is regulated by stress in male mice suggesting that IMO alters Ppm1f levels. Regarding directionality, the Ppm1f mRNA level and Ppm1f protein level are in the same direction in the Amygdala but in opposite direction in the mPFC. Interestingly, Ppm1f/Camk2g colocalization is regulated by stress in the mouse brain. Thus, it is possible that PPM1F levels may be regulating CaMK2G after stress, promoting the development of depression and anxiety behaviors.
We also examined whether post-stress manipulation, i.e. systemic administration of corticosterone, may decrease the stress-related behavioral and molecular phenotypes (44–46). Systemic corticosterone did not rescue the depressive-like phenotype elicited by IMO in male mice. However, corticosterone rescued the depressive-like behavior induced by IMO in female mice. EPM results showed that corticosterone rescued the anxiety-like behavior in male mice. In contrast, IMO did not induce an anxiety-like behavior in female mice. Analysis of the mRNA levels showed that corticosterone treatment shortly after stress rescued the genetic regulation of Ppm1f in the Amygdala and mPFC in males and in the mPFC in females. Thus, there appears to be relevant sex differences in Ppm1f regulation in the brain, acute exposure to traumatic stress and its treatment with corticosterone.
Our study should be interpreted in light of its limitations. First, the data obtained with IMO should be interpreted with caution. While most rodents develop anxiety and depression-like behavior after IMO, only a small percentage of the human population develops PTSD and/or depression. Second, rs17759843 is associated with blood expression of PPM1F in a sample of mostly European descent. rs17759843 has an MAF of 0.11 in Caucasians and 0.01 in African Americans per dbGAP HapMap populations. Whether this SNP is an eQTL in the African Americans remain to be determined, as we were likely underpowered to detect this relationship in our GTP dataset. Third, the Blood eQTL Browser provides statistics on bivariate association between rs17759843 and blood PPM1F expression. Hence, rs17759843 may be associated with PPM1F due to its linkage disequlibrium with other stronger eQTL SNPs for PPM1F. Fourth, we did not have access to human brain amygdala to examine association between rs17759843 and PPM1F expression. Fifth, we analyzed cases with significant current symptoms of PTSD&Dep but we did not have rigorous clinical assessment to determine if these cases had the diagnosis of PTSD or Major Depressive Disorder. Additionally, our cutoff score of 14 for the PSS and BDI, though based on published psychometric studies, may be lenient and thus could increase the risk of false positive. Sixth, the CHDWB replication sample uses the GAD-7, which is a good screening tool but could provide a false positive detection if not followed by an accurate clinical evaluation. Seventh, we found blood mRNA PPM1F level significantly down-regulated in cases with PTSD&Dep symptoms in the GTP sample and replicated this association in cases with anxiety symptoms in an independent CHDWB cohort. Our replication is only a quasi-replication since anxiety symptoms can be PTSD symptoms but they can also be symptoms of generalized anxiety disorder or social anxiety disorder. Lastly, only a subset of GTP participants with genetic data had gene expression data. However, the two datasets are comparable as reflected by the distribution of their sex, BDI, and PSS scores (Supplemental Table S8).
In conclusion, mRNA and protein levels of phosphatase Ppm1f and Camk2g are regulated in the brain after exposure to traumatic stress in male and female mouse models and are associated with chronic anxiety-related (PTSD) and depression symptoms in traumatized humans. Because of the widespread expression of PPM1F in the body and brain, these changes in expression of PPM1F by stress may possibly have other pathophysiological implications that should be examined in future studies. Insights into the role of CAMK2 and PPM1F following trauma exposure may contribute to discovering novel approaches to treat or prevent the disabling conditions of PTSD and depression.
Supplementary Material
Figure S1: Principal variance component analysis and volcano plots for mouse amygdala and mPFC microarrays at 6, 7, or 8 days post IMO stress. A) Amygdala expression 6 days after IMO; red circles denote stressed mice and blue circles control mice. B) Amygdala expression 8 days after IMO; red circles denote stressed mice, blue circles control mice. C) mPFC expression 6 days after IMO; red circles denote stressed mice, blue circles control mice. D) Volcano plot for amygdala microarray 6 days after IMO. E) Volcano plot for amygdala microarray 8 days after IMO. F) Volcano plot for mPFC microarray 6 days after IMO.
Figure S2. Ppm1f levels are not altered by fear conditioning. Mice were exposed to cued-fear conditioning (FC) and sacrificed at the time points shown below for amygdala (A and B) and medial prefrontal cortext (mPFC, C) microarray studies. The control group was home cage which was not exposed to FC. Results showed that a mild stress such as FC did not alter Ppm1f mRNA levels in the amygdala or mPFC. N=4 per group.
Figure S3. DAVID analysis. Functional analysis of the top genes regulated 6 and 8 days after stress in both the amygdala and mPFC. Results show that the Phosphoprotein, Cytoplasm and Alternative splicing are the three top categories involved in acute stress in mice.
Figure S4. Network analysis of the top gene candidates regulated after stress in amygdala and the mPFC. PPM1F has direct relationships with the CaMKII family (blue arrow) and indirect relationship with BDNF, Creb, ERK1/2 and MAP2K1/2.
Figure S5. PPM1F expression in the brain. These images modified from the Allen Brain Atlas show that PPM1F mRNA expression in both mice and humans at basal levels is ubiquitous including the amygdala and the medial prefrontal cortex (mPFC). Legend of the images: Green = low expression; Yellow = moderate expression; Red = high expression. White arrow = amygdala; blue arrow = mPFC.
Figure S6. Camk2g mRNA levels are altered after stress in mouse amygdala and its blood mRNA levels are associated with PTSD&Depression. A) Top associations between CAMK2 blood mRNA level and PTSD&Dep, adjusting for gender, age, and population substructure (n=230); B) CAMK2G has significantly decreased expression level in blood in PTSD&Dep cases versus controls after adjusting for gender, age, and population substructure. C) There was an increase, marginally significant, in amygdala CamK2G mRNA basal levels 6 days after stress exposure t=2,213 df=8, p=0.0578, n=8 control group, n=3 stress group. D) Fear conditioning does not change amygdala CamK2g mRNA levels two hours later, home cage n=8, fear conditioning, n=7, immobilization-fear conditioning n=4.
Figure S7. Ppm1f is upregulated in the central amygdala (CeA) (A), Basolateral amygdala (BLA) and in the amygdala (CeA+BLA) 6 days after stress immobilization (IMO) in male mice. Moreover, IMO also enhanced Ppm1f levels in the medial prefrontal cortex (mPFC: Cg+Prl+IL). P*p≤0.05, **p≤0.01, ***p≤0.001. A.U.= Arbitrary Units. N=7–8 per group.
Figure S8. Ppm1f is upregulated in the Basolateral amygdala (BLA) 6 days after stress immobilization (IMO) in female mice. P*p≤0.05. A.U.= Arbitrary Units. N=7–8 per group.
Figure S9. Camk2g is upregulated in the central amygdala (CeA) (A), Basolateral amygdala (BLA) and in the amygdala (CeA+BLA) 6 days after stress immobilization (IMO) in male mice. Moreover, IMO also enhanced Ppm1f levels in the medial prefrontal cortex (mPFC: Cg+Prl+IL). P*p≤0.05, **p≤0.01, ***p≤0.001. A.U.= Arbitrary Units. N=7–8 per group.
Figure S10. Camk2g levels are similar in control vs stress groups 6 days after stress immobilization (IMO) in female mice. A.U.= Arbitrary Units. N=7–8 per group.
Supplemental Table 1. Gene regulation in the amygdala microarrays. List of the genes that survived these criteria: a) Probe present at least one time in both lists (6 and 8 days after stress), b) >1.5 fold change compared to control, c) significant difference at p<.05 (nominal p-value), d) clearly present at moderate to high levels in amygdala (search in the Allen Brain Atlas).
Supplemental Table 2. RT-PCR arrays plates. This list shows the top hits of the microarray replicated by RT-PCR arrays including the controls used for these experiments (mGDC, RTC, PPC).
Supplemental Table 3. Amygdala microarray replications. From the initial gene list (Supplementary Figure 1) probes highlighted in red here are those replicated in two independent cohorts of mice - including ppm1f. Genes highlighted in yellow are genes replicated only in one cohort.
Supplementary Table 4. Associations between human blood mRNA levels of the five stress-regulated genes in mouse amygdala in PTSD&Dep, covarying for gender, age, and population substructure.
Supplemental Table 5. Gene regulation of the mPFC microarray. List of the genes that survived these criteria: a) >1.30 fold change (There are only 6 genes which survive a 1.5 cut-off criteria), b) significant at nominal p<.05, c) clearly present at moderate to high levels in amygdala (based on Allen Brain Atlas).
Supplemental Table 6. Sociodemographic characteristics of the 230 African American GTP participants.
Supplemental Table 7. Sociodemographic characteristics of the CHDWB cohort.
Supplemental Table 8: Comparing the GTP genetic dataset to GTP gene expression subset
Acknowledgments
We would like to thank Dr. Greg Gibson (Georgia Tech, USA) for his comments on this paper and data on Figure 3B. We are thankful to Dr. Antonio Armario and Dr. Roser Nadal for support on this work. We would also like to thank the staff and participants of the Grady Trauma Project for their contributions to this important work.
FINANCIAL DISCLOSURES
KJR received support from the Howard Hughes Medical Institute, RA and KJR were supported by R21MH101492-01, KJR by R01MH071537 and R01MH096764, and RA by a NARSAD Young Investigator Grant (22434), Ramon y Cajal RYC-2014-15784 MINECO and SAF2016-76565-R MINECO and FEDER. APW was supported by the Department of Veterans Affairs Career Development Award IK2CX000601. TJ has support from NARSAD and R01MH100122. The contents do not represent the views of the Department of Veterans Affairs or the United States Government. The UAB Animal Facility received funding from 2015 FEDER7S-20IU16-001945.
All authors report no biomedical financial interests or potential conflicts of interest.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.McEwen BS, Bowles NP, Gray JD, Hill MN, Hunter RG, Karatsoreos IN, et al. Mechanisms of stress in the brain. Nat Neurosci. 2015;18:1353–1363. doi: 10.1038/nn.4086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wittchen HU, Jacobi F, Rehm J, Gustavsson A, Svensson M, Jonsson B, et al. The size and burden of mental disorders and other disorders of the brain in Europe 2010. Eur Neuropsychopharmacol. 2011;21:655–679. doi: 10.1016/j.euroneuro.2011.07.018. [DOI] [PubMed] [Google Scholar]
- 3.Tham A, Jonsson U, Andersson G, Soderlund A, Allard P, Bertilsson G. Efficacy and tolerability of antidepressants in people aged 65 years or older with major depressive disorder - A systematic review and a meta-analysis. J Affect Disord. 2016;205:1–12. doi: 10.1016/j.jad.2016.06.013. [DOI] [PubMed] [Google Scholar]
- 4.Henssler J, Kurschus M, Franklin J, Bschor T, Baethge C. Long-Term Acute-Phase Treatment With Antidepressants, 8 Weeks and Beyond: A Systematic Review and Meta-Analysis of Randomized, Placebo-Controlled Trials. J Clin Psychiatry. 2017 doi: 10.4088/JCP.15r10545. [DOI] [PubMed] [Google Scholar]
- 5.Caramanica K, Brackbill RM, Liao T, Stellman SD. Comorbidity of 9/11-Related PTSD and Depression in the World Trade Center Health Registry 10–11 Years Postdisaster. J Trauma Stress. 2014;27:680–688. doi: 10.1002/jts.21972. [DOI] [PubMed] [Google Scholar]
- 6.Matar MA, Zohar J, Cohen H. Translationally relevant modeling of PTSD in rodents. Cell Tissue Res. 2013;354:127–139. doi: 10.1007/s00441-013-1687-6. [DOI] [PubMed] [Google Scholar]
- 7.Krishnan V, Nestler EJ. Linking molecules to mood: new insight into the biology of depression. Am J Psychiatry. 2010;167:1305–1320. doi: 10.1176/appi.ajp.2009.10030434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Andero R, Dias BG, Ressler KJ. A role for Tac2, NkB, and Nk3 receptor in normal and dysregulated fear memory consolidation. Neuron. 2014;83:444–454. doi: 10.1016/j.neuron.2014.05.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sawamura T, Klengel T, Armario A, Jovanovic T, Norrholm SD, Ressler KJ, et al. Dexamethasone Treatment Leads to Enhanced Fear Extinction and Dynamic Fkbp5 Regulation in Amygdala. Neuropsychopharmacology. 2016;41:832–846. doi: 10.1038/npp.2015.210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Marquez C, Belda X, Armario A. Post-stress recovery of pituitary-adrenal hormones and glucose, but not the response during exposure to the stressor, is a marker of stress intensity in highly stressful situations. Brain Res. 2002;926:181–185. doi: 10.1016/s0006-8993(01)03112-2. [DOI] [PubMed] [Google Scholar]
- 11.Andero R, Heldt SA, Ye K, Liu X, Armario A, Ressler KJ. Effect of 7,8-dihydroxyflavone, a small-molecule TrkB agonist, on emotional learning. Am J Psychiatry. 2011;168:163–172. doi: 10.1176/appi.ajp.2010.10030326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Andero R, Brothers SP, Jovanovic T, Chen YT, Salah-Uddin H, Cameron M, et al. Amygdala-dependent fear is regulated by Oprl1 in mice and humans with PTSD. Science translational medicine. 2013;5:188ra173. doi: 10.1126/scitranslmed.3005656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kilaru V, Iyer SV, Almli LM, Stevens JS, Lori A, Jovanovic T, et al. Genome-wide gene-based analysis suggests an association between Neuroligin 1 (NLGN1) and post-traumatic stress disorder. Translational psychiatry. 2016;6:e820. doi: 10.1038/tp.2016.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lowe SR, Quinn JW, Richards CA, Pothen J, Rundle A, Galea S, et al. Childhood trauma and neighborhood-level crime interact in predicting adult posttraumatic stress and major depression symptoms. Child Abuse Negl. 2016;51:212–222. doi: 10.1016/j.chiabu.2015.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gillespie CF, Bradley RG, Mercer K, Smith AK, Conneely K, Gapen M, et al. Trauma exposure and stress-related disorders in inner city primary care patients. General Hospital Psychiatry. 2009;31:505–514. doi: 10.1016/j.genhosppsych.2009.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Foa EB, Riggs DS, Dancu CV, Rothbaum BO. Reliability and validity of a brief instrument for assessing post-traumatic stress disorder. Journal of Traumatic Stress. 1993;6:459–473. [Google Scholar]
- 17.Beck A, Ward C, Mendelson M, Mock J, Erbaugh J. An inventory for measuring depression. Archives of General Psychiatry. 1961;4:561–571. doi: 10.1001/archpsyc.1961.01710120031004. [DOI] [PubMed] [Google Scholar]
- 18.Mecham BH, Nelson PS, Storey JD. Supervised normalization of microarrays. Bioinformatics. 2010;26:1308–1315. doi: 10.1093/bioinformatics/btq118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tabassum R, Cunningham L, Stephens EH, Sturdivant K, Martin GS, Brigham KL, et al. A Longitudinal Study of Health Improvement in the Atlanta CHDWB Wellness Cohort. Journal of personalized medicine. 2014;4:489–507. doi: 10.3390/jpm4040489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Spitzer RL, Kroenke K, Williams JBW, Lowe B. A Brief Measure for Assessing Generalized Anxiety Disorder: The GAD-7. Arch Intern Med. 2006;166:1092–1097. doi: 10.1001/archinte.166.10.1092. [DOI] [PubMed] [Google Scholar]
- 21.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira M, Bender D, et al. PLINK: a toolset for whole-genome association and population-based linkage analysis. American Journal of Human Genetics. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38:904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 23.Conneely KN, Boehnke M. So Many Correlated Tests, So Little Time! Rapid Adjustment of P Values for Multiple Correlated Tests. The American Journal of Human Genetics. 2007;81:1158–1168. doi: 10.1086/522036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Westra HJ, Peters MJ, Esko T, Yaghootkar H, Schurmann C, Kettunen J, et al. Systematic identification of trans eQTLs as putative drivers of known disease associations. Nat Genet. 2013;45:1238–1243. doi: 10.1038/ng.2756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Colantuoni C, Lipska BK, Ye T, Hyde TM, Tao R, Leek JT, et al. Temporal dynamics and genetic control of transcription in the human prefrontal cortex. Nature. 2011;478:519–523. doi: 10.1038/nature10524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zoladz PR, Park CR, Halonen JD, Salim S, Alzoubi KH, Srivareerat M, et al. Differential expression of molecular markers of synaptic plasticity in the hippocampus, prefrontal cortex, and amygdala in response to spatial learning, predator exposure, and stress-induced amnesia. Hippocampus. 2012;22:577–589. doi: 10.1002/hipo.20922. [DOI] [PubMed] [Google Scholar]
- 27.Strack S, Barban MA, Wadzinski BE, Colbran RJ. Differential inactivation of postsynaptic density-associated and soluble Ca2+/calmodulin-dependent protein kinase II by protein phosphatases 1 and 2A. Journal of neurochemistry. 1997;68:2119–2128. doi: 10.1046/j.1471-4159.1997.68052119.x. [DOI] [PubMed] [Google Scholar]
- 28.Yoshimura Y, Sogawa Y, Yamauchi T. Protein phosphatase 1 is involved in the dissociation of Ca2+/calmodulin-dependent protein kinase II from postsynaptic densities. FEBS letters. 1999;446:239–242. doi: 10.1016/s0014-5793(99)00226-4. [DOI] [PubMed] [Google Scholar]
- 29.Harvey BP, Banga SS, Ozer HL. Regulation of the multifunctional Ca2+/calmodulindependent protein kinase II by the PP2C phosphatase PPM1F in fibroblasts. The Journal of biological chemistry. 2004;279:24889–24898. doi: 10.1074/jbc.M400656200. [DOI] [PubMed] [Google Scholar]
- 30.Ishida A, Kameshita I, Fujisawa H. A novel protein phosphatase that dephosphorylates and regulates Ca2+/calmodulin-dependent protein kinase II. J Biol Chem. 1998;273:1904–1910. doi: 10.1074/jbc.273.4.1904. [DOI] [PubMed] [Google Scholar]
- 31.Dubovsky SL, Thomas M. Serotonergic mechanisms and current and future psychiatric practice. J Clin Psychiatry. 1995;56(Suppl 2):38–48. [PubMed] [Google Scholar]
- 32.Simpson MD, Lubman DI, Slater P, Deakin JF. Autoradiography with [3H]8-OH-DPAT reveals increases in 5-HT(1A) receptors in ventral prefrontal cortex in schizophrenia. Biol Psychiatry. 1996;39:919–928. doi: 10.1016/0006-3223(95)00026-7. [DOI] [PubMed] [Google Scholar]
- 33.Stockmeier CA. Neurobiology of serotonin in depression and suicide. Ann N Y Acad Sci. 1997;836:220–232. doi: 10.1111/j.1749-6632.1997.tb52362.x. [DOI] [PubMed] [Google Scholar]
- 34.Cai X, Gu Z, Zhong P, Ren Y, Yan Z. Serotonin 5-HT1A receptors regulate AMPA receptor channels through inhibiting Ca2+/calmodulin-dependent kinase II in prefrontal cortical pyramidal neurons. The Journal of biological chemistry. 2002;277:36553–36562. doi: 10.1074/jbc.M203752200. [DOI] [PubMed] [Google Scholar]
- 35.Jackson KJ, Damaj MI. L-type calcium channels and calcium/calmodulin-dependent kinase II differentially mediate behaviors associated with nicotine withdrawal in mice. J Pharmacol Exp Ther. 2009;330:152–161. doi: 10.1124/jpet.109.151530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li K, Zhou T, Liao L, Yang Z, Wong C, Henn F, et al. betaCaMKII in lateral habenula mediates core symptoms of depression. Science. 2013;341:1016–1020. doi: 10.1126/science.1240729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Choi K, Le T, Xing G, Johnson LR, Ursano RJ. Analysis of kinase gene expression in the frontal cortex of suicide victims: implications of fear and stress. Front Behav Neurosci. 2011;5:46. doi: 10.3389/fnbeh.2011.00046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kubzansky LD, Koenen KC, Jones C, Eaton WW. A prospective study of posttraumatic stress disorder symptoms and coronary heart disease in women. Health Psychol. 2009;28:125–130. doi: 10.1037/0278-6133.28.1.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kubzansky LD, Koenen KC, Spiro A, 3rd, Vokonas PS, Sparrow D. Prospective study of posttraumatic stress disorder symptoms and coronary heart disease in the Normative Aging Study. Arch Gen Psychiatry. 2007;64:109–116. doi: 10.1001/archpsyc.64.1.109. [DOI] [PubMed] [Google Scholar]
- 40.Whang W, Kubzansky LD, Kawachi I, Rexrode KM, Kroenke CH, Glynn RJ, et al. Depression and risk of sudden cardiac death and coronary heart disease in women: results from the Nurses’ Health Study. J Am Coll Cardiol. 2009;53:950–958. doi: 10.1016/j.jacc.2008.10.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pellicena P, Schulman H. CaMKII inhibitors: from research tools to therapeutic agents. Front Pharmacol. 2014;5:21. [Google Scholar]
- 42.Mollova MY, Katus HA, Backs J. Regulation of CaMKII signaling in cardiovascular disease. Front Pharmacol. 2015;6:178. doi: 10.3389/fphar.2015.00178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mattiazzi A, Bassani RA, Escobar AL, Palomeque J, Valverde CA, Vila Petroff M, et al. Chasing cardiac physiology and pathology down the CaMKII cascade. Am J Physiol Heart Circ Physiol. 2015;308:H1177–1191. doi: 10.1152/ajpheart.00007.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cohen H, Kozlovsky N, Matar MA, Zohar J, Kaplan Z. Distinctive hippocampal and amygdalar cytoarchitectural changes underlie specific patterns of behavioral disruption following stress exposure in an animal model of PTSD. European neuropsychopharmacology : the journal of the European College of Neuropsychopharmacology. 2014;24:1925–1944. doi: 10.1016/j.euroneuro.2014.09.009. [DOI] [PubMed] [Google Scholar]
- 45.Rao RP, Anilkumar S, McEwen BS, Chattarji S. Glucocorticoids protect against the delayed behavioral and cellular effects of acute stress on the amygdala. Biol Psychiatry. 2012;72:466–475. doi: 10.1016/j.biopsych.2012.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zohar J, Yahalom H, Kozlovsky N, Cwikel-Hamzany S, Matar MA, Kaplan Z, et al. High dose hydrocortisone immediately after trauma may alter the trajectory of PTSD: interplay between clinical and animal studies. European neuropsychopharmacology : the journal of the European College of Neuropsychopharmacology. 2011;21:796–809. doi: 10.1016/j.euroneuro.2011.06.001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1: Principal variance component analysis and volcano plots for mouse amygdala and mPFC microarrays at 6, 7, or 8 days post IMO stress. A) Amygdala expression 6 days after IMO; red circles denote stressed mice and blue circles control mice. B) Amygdala expression 8 days after IMO; red circles denote stressed mice, blue circles control mice. C) mPFC expression 6 days after IMO; red circles denote stressed mice, blue circles control mice. D) Volcano plot for amygdala microarray 6 days after IMO. E) Volcano plot for amygdala microarray 8 days after IMO. F) Volcano plot for mPFC microarray 6 days after IMO.
Figure S2. Ppm1f levels are not altered by fear conditioning. Mice were exposed to cued-fear conditioning (FC) and sacrificed at the time points shown below for amygdala (A and B) and medial prefrontal cortext (mPFC, C) microarray studies. The control group was home cage which was not exposed to FC. Results showed that a mild stress such as FC did not alter Ppm1f mRNA levels in the amygdala or mPFC. N=4 per group.
Figure S3. DAVID analysis. Functional analysis of the top genes regulated 6 and 8 days after stress in both the amygdala and mPFC. Results show that the Phosphoprotein, Cytoplasm and Alternative splicing are the three top categories involved in acute stress in mice.
Figure S4. Network analysis of the top gene candidates regulated after stress in amygdala and the mPFC. PPM1F has direct relationships with the CaMKII family (blue arrow) and indirect relationship with BDNF, Creb, ERK1/2 and MAP2K1/2.
Figure S5. PPM1F expression in the brain. These images modified from the Allen Brain Atlas show that PPM1F mRNA expression in both mice and humans at basal levels is ubiquitous including the amygdala and the medial prefrontal cortex (mPFC). Legend of the images: Green = low expression; Yellow = moderate expression; Red = high expression. White arrow = amygdala; blue arrow = mPFC.
Figure S6. Camk2g mRNA levels are altered after stress in mouse amygdala and its blood mRNA levels are associated with PTSD&Depression. A) Top associations between CAMK2 blood mRNA level and PTSD&Dep, adjusting for gender, age, and population substructure (n=230); B) CAMK2G has significantly decreased expression level in blood in PTSD&Dep cases versus controls after adjusting for gender, age, and population substructure. C) There was an increase, marginally significant, in amygdala CamK2G mRNA basal levels 6 days after stress exposure t=2,213 df=8, p=0.0578, n=8 control group, n=3 stress group. D) Fear conditioning does not change amygdala CamK2g mRNA levels two hours later, home cage n=8, fear conditioning, n=7, immobilization-fear conditioning n=4.
Figure S7. Ppm1f is upregulated in the central amygdala (CeA) (A), Basolateral amygdala (BLA) and in the amygdala (CeA+BLA) 6 days after stress immobilization (IMO) in male mice. Moreover, IMO also enhanced Ppm1f levels in the medial prefrontal cortex (mPFC: Cg+Prl+IL). P*p≤0.05, **p≤0.01, ***p≤0.001. A.U.= Arbitrary Units. N=7–8 per group.
Figure S8. Ppm1f is upregulated in the Basolateral amygdala (BLA) 6 days after stress immobilization (IMO) in female mice. P*p≤0.05. A.U.= Arbitrary Units. N=7–8 per group.
Figure S9. Camk2g is upregulated in the central amygdala (CeA) (A), Basolateral amygdala (BLA) and in the amygdala (CeA+BLA) 6 days after stress immobilization (IMO) in male mice. Moreover, IMO also enhanced Ppm1f levels in the medial prefrontal cortex (mPFC: Cg+Prl+IL). P*p≤0.05, **p≤0.01, ***p≤0.001. A.U.= Arbitrary Units. N=7–8 per group.
Figure S10. Camk2g levels are similar in control vs stress groups 6 days after stress immobilization (IMO) in female mice. A.U.= Arbitrary Units. N=7–8 per group.
Supplemental Table 1. Gene regulation in the amygdala microarrays. List of the genes that survived these criteria: a) Probe present at least one time in both lists (6 and 8 days after stress), b) >1.5 fold change compared to control, c) significant difference at p<.05 (nominal p-value), d) clearly present at moderate to high levels in amygdala (search in the Allen Brain Atlas).
Supplemental Table 2. RT-PCR arrays plates. This list shows the top hits of the microarray replicated by RT-PCR arrays including the controls used for these experiments (mGDC, RTC, PPC).
Supplemental Table 3. Amygdala microarray replications. From the initial gene list (Supplementary Figure 1) probes highlighted in red here are those replicated in two independent cohorts of mice - including ppm1f. Genes highlighted in yellow are genes replicated only in one cohort.
Supplementary Table 4. Associations between human blood mRNA levels of the five stress-regulated genes in mouse amygdala in PTSD&Dep, covarying for gender, age, and population substructure.
Supplemental Table 5. Gene regulation of the mPFC microarray. List of the genes that survived these criteria: a) >1.30 fold change (There are only 6 genes which survive a 1.5 cut-off criteria), b) significant at nominal p<.05, c) clearly present at moderate to high levels in amygdala (based on Allen Brain Atlas).
Supplemental Table 6. Sociodemographic characteristics of the 230 African American GTP participants.
Supplemental Table 7. Sociodemographic characteristics of the CHDWB cohort.
Supplemental Table 8: Comparing the GTP genetic dataset to GTP gene expression subset