Abstract
Unrepaired DNA lesions block replication and threaten genomic stability. Several specialized translesion polymerases, including Pol θ, contribute to replicative bypass of these lesions. The role of Pol θ in double strand break repair is well understood, but its contribution to translesion synthesis is much less so. We describe the action of Pol θ on templates containing thymidine glycol (Tg), a major cytotoxic, oxidative DNA lesion which blocks DNA replication. Unrepaired Tg lesions are bypassed in human cells by specialized translesion polymerases by one of two distinct pathways: high-fidelity bypass by the combined action of Pol κ and Pol ζ or weakly mutagenic bypass by Pol θ. Here we report that in vitro bypass of Tg by Pol θ results in frameshift mutations (deletions) in a sequence-dependent fashion. Steady-state kinetic analysis indicated that one- and two-nucleotide deletions are formed 9- and 6-fold more efficiently, respectively, than correct, full-length bypass products. Sequencing of in vitro bypass products revealed that bypass preference followed the order two-nucleotide deletion > correct bypass > one-nucleotide deletion on a template where all three outcomes were possible. These results suggest that bypass of Tg by Pol θ results in mutations opposite the lesion, as well as frameshift mutations.
TOC Graphic
Introduction
Reactive oxygen species are formed in cells exposed to ionizing radiation and as a byproduct of cellular respiration. The most damaging of these species is hydroxyl radical, which attacks DNA to form oxidative lesions.1 Approximately 300 Tg lesions are formed in each human cell every day, and Tg is a major thymidine oxidation product in cells exposed to ionizing radiation.2–4 Tg can exist as one of four diastereomers, but γ-radiolysis of duplex DNA primarily forms the two cis diastereomers (Scheme 1).5 Epimerization at the C6 position results in interconversion between cis- and trans-diastereomers, although the equilibrium favors the cis-diastereomers by 3-5 fold.6,7 Tg is cytotoxic but not highly mutagenic.8,9 Experiments in human cell extracts and in mouse embryonic fibroblasts revealed that Tg is primarily repaired by base excision repair, although nucleotide excision repair may also play a role in removing this lesion.10–13 Tg lesions which escape repair strongly block progression of the replication fork, a potentially lethal event that requires the action of specialized translesion synthesis polymerases, which replicate past Tg and other blocking DNA lesions.9,14 Among the translesion synthesis polymerases, those of the Y-family (Pol η, Pol κ, Pol ι, and Rev1) along with Pol ζ of the B-family are the best characterized. However, growing evidence suggests that the A-family polymerase Pol θ also contributes to translesion synthesis in human cells.15–21 Herein, we describe the formation of frameshift mutations (deletions) during in vitro bypass of Tg by Pol θ.
Scheme 1.
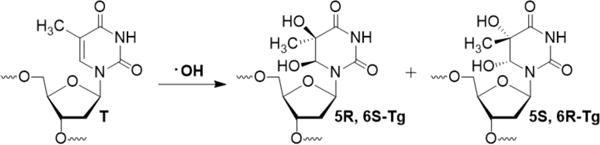
Formation of Tg by oxidation of T.
Bypass of Tg occurs by two distinct pathways in human cells.20 The primary pathway occurs with high fidelity and involves dA insertion opposite the lesion by Pol κ and subsequent extension by Pol ζ. A minor pathway requires only Pol θ, which conducts translesion synthesis and subsequent extension past the lesion, and is weakly mutagenic. The unique ability of Pol θ to insert opposite a lesion and conduct subsequent extension is not restricted to Tg. For instance, Pol θ conducts both steps during bypass of 3-deaza-3-methyl-2′-deoxyadenosine (3-deaza-3-methyl dA, a stable analogue of 3-methyl-2′-deoxyadenosine) in vivo and on templates containing abasic and oxidized abasic lesions in vitro.21–23 The contribution of Pol θ to translesion synthesis is potentially physiologically significant because Pol θ expression is upregulated in some cancers, and this upregulation correlates with poor prognosis and increased genomic instability.24 Additionally, Pol θ promotes resistance to ionizing radiation, as well as other DNA damaging agents such as methyl methanesulfonate, bleomycin, and H2O2.25,26 These observations have largely been attributed to the well-established role of Pol θ in double strand break repair.26–28 However, lesion bypass by Pol θ could conceivably augment these effects. Therefore, the ability of Pol θ to bypass DNA lesions and the mutational spectrum with which it does so may be relevant to cancer progression, tumor chemo- and radio-resistance, and even to the development of cancer by increasing mutational frequency during bypass of endogenous lesions (such as Tg).
We recently reported that in vitro bypass of abasic and oxidized abasic lesions by Pol θ gives significant numbers of one- and two-nucleotide deletions.23 These deletions were formed by sequence-dependent template misalignment during extension past the lesion. The cellular role for Pol θ in Tg bypass prompted us to explore whether template misalignment is also operative when this lesion is positioned within templates of appropriate sequence(s). Steady-state kinetic analysis revealed that template misalignment was more efficient than insertion of the correct nucleotide on two different primer-template complexes. Sequencing of Pol θ bypass products revealed a preference for two-nucleotide deletions over correct bypass and single-nucleotide deletions on a sequence where all three were possible. These results suggest that Pol θ bypass of Tg may give rise to mutations opposite the lesion site, as well as frameshift mutations generated by template misalignment.
Methods
General Methods
Oligonucleotides were synthesized on an Applied Biosystems Inc. 394 DNA synthesizer using reagents from Glen Research (Sterling, VA) and deprotected according to the manufacturer’s instructions. γ-32P-dATP was obtained from PerkinElmer. Protein purification was conducted using an AKTA FPLC and columns were from GE Healthcare. Sonication was done using a Branson SFX-150 sonifier. Rosetta 2 pLysS E. coli and C18 Sep Pak cartridges were from Millipore. The Quick Ligase Kit, Phusion polymerase, NEB Buffer 3.1, T4 polynucleotide kinase, Acc65I, EcoRI, and dNTPs were obtained from New England Biolabs. NP-40 substitute was from Sigma. Protease inhibitor cocktail (EDTA-free) was from Roche. SUMO Protease II was from LifeSensors. Dynabeads M-280 streptavidin beads were from ThermoFisher. Analysis of radio-labeled oligonucleotides was carried out using a Storm 860 Phosphorimager and ImageQuant 7.0 TL software. Pre-steady-state kinetics were carried out using a RQF-3 rapid quench instrument from Kintek. Colony sequencing was conducted by Genewiz (South Plainfield, NJ).
Tg-containing oligonucleotides were prepared using the TBDMS-protected 5R, 6S-Tg phosphoramidite (Glen Research) originally reported by Iwai.29 Removal of TBDMS protecting groups was conducted as reported previously using 300 μL of a solution containing triethylamine trihydrofluoride, triethylamine, and anhydrous N-methyl pyrrolidinone (in a 4:3:6 ratio by volume) at 65 °C for 1.5 hr.30,31 The biotinylated primer used for bypass sequencing experiments was synthesized previously.23 All oligonucleotides were purified by 20% denaturing polyacrylamide gel electrophoresis (PAGE) and desalted by C18 Sep Pak. Tg-containing oligonucleotides were characterized by MALDI-TOF MS. Primer-template complexes were prepared by mixing 32P-labeled primer with the appropriate template in a 1:1.5 ratio in phosphate buffered saline (10 mM sodium phosphate, 100 mM NaCl, pH 7.2), heating to 85 °C, and slowly cooling to 25 °C. Pol θ catalytic core (residues 1792-2590) was expressed and purified as previously described.32 See Supporting Information for a demonstration of purity. Unless otherwise specified, the active concentration of Pol θ (determined by active site titration) is written for all experiments.
Active site titration of Pol θ
The active fraction of Pol θ was determined by pre-steady state kinetic analysis as previously described.23 Briefly, Pol θ (5 nM protein concentration) was incubated with primer-template (12.5 nM) and dATP (500 μM) in reaction buffer for a fixed time and quenched with 80% formamide containing 100 mM EDTA. Samples were loaded on 20% denaturing PAGE, and analyzed by phosphorimaging. The fraction of product was plotted as a function of time and fit to the equation P=A(1-e−kt) + ksst where P is fraction of extended primer, A is the burst amplitude, k is the burst phase rate constant, kss is the steady-state rate constant, and t is the reaction time. This experiment was conducted 4 times and the active fraction was determined to be 43.2 ± 3.6%. See Supporting Information for a representative kinetic plot.
Steady-state kinetic analysis of Pol θ
Polymerase reactions were conducted with Pol θ (432 pM for 1-4 except for dGTP and dTTP insertion where 4.32 nM was used; 4.32 nM for 5-8, except for dGTP insertion on 6 which was 2 nM), primer-template complexes (50 nM), and various concentrations of the indicated dNTP at 25 °C in reaction buffer (10 mM Tris HCl pH 8, 25 mM KCl, 10 mM MgCl2, 1 mM BME). The concentration range of dNTP and the reaction time were selected such that reactions did not proceed past 20% completion (single-hit conditions, see Supporting Information for a list of concentrations and reaction times). In a typical experiment, a 2 × DNA-enzyme solution was prepared by mixing primer-template (1 μM, 10 μL), 10 × reaction buffer (20 μL), 10 × Pol θ (4.32 nM, 20 μL) in storage buffer (20 mM Tris HCl pH 7, 300 mM NaCl, 10% glycerol, 5 mM BME), and H2O (50 μL). The 2 × DNA-enzyme solution (3 μL) was mixed with the appropriate 2 × dNTP solution (3 μL) to initiate the reaction, which was quenched after a fixed time with 95% formamide loading buffer containing 25 mM EDTA (8 μL). An aliquot (4 μL) was loaded on a 20% denaturing PAGE, which was run at 55 W for approximately 3.5 h. The gel was analyzed by phosphorimaging, and the data were fit to the Michaelis-Menten equation. The kcat was determined by dividing Vmax by the active concentration of enzyme determined by active site titration.
Sequencing of Pol θ bypass products
Lesion bypass products were sequenced using a method reported by Sabouri with modifications previously reported.23,33 Briefly, primer-template complex (100 nM) was incubated with Pol θ (20 nM) and dNTPs (100 μM) at room temperature for 30 min. The biotinylated strand was isolated with Dynabeads M-280 streptavidin beads and amplified by PCR. The PCR product was purified by native PAGE, digested with Acc65I and EcoRI, and ligated into pBlueScript SK-plasmid, which was transformed into DH5 α cells. The cells were plated on LB medium with ampicillin, grown at 37 °C for 14 hr, and the plate was sent to Genewiz for sequencing of individual colonies.
Results
Primer-template design and steady-state kinetic analysis of translesion synthesis
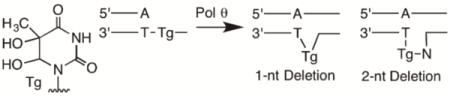
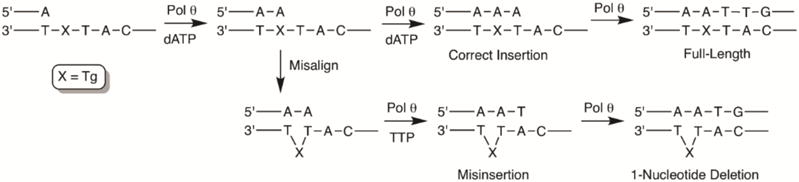
Oxidation of thymidine to Tg in duplex DNA primarily gives the two cis diastereomers (5R, 6S and 5S, 6R), which are formed in equal amounts.5 Because Pol θ bypasses these two diastereomers with equal efficiency in vivo,20 we prepared templates 1-4 containing the commercially available 5R, 6S diastereomer or T in the same position as a control (Chart 1). The template sequences were designed such that the template strand contained a T either one or two nucleotides downstream from Tg. Because dA is primarily inserted opposite Tg,20 template misalignment could be mediated by Watson-Crick base pairing between the dA inserted opposite the lesion and the template T present 1 or 2 nucleotides away. Extension from the misaligned primer could then result in one- or two-nucleotide deletions, depending upon the position of the downstream T (Scheme 2 and 3).
Chart 1.
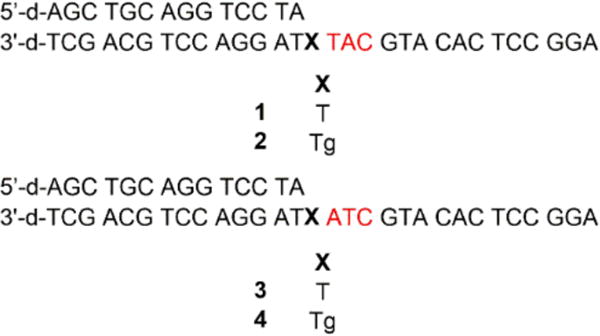
Primer-templates used for translesion synthesis
Scheme 2.
Formation of one-nucleotide deletions by template misalignment
Scheme 3.
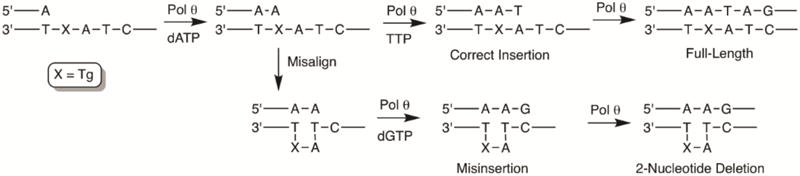
Formation of two-nucleotide deletions by template misalignment
We first confirmed the preference for dA insertion during translesion synthesis by conducting single nucleotide incorporation experiments on 1-4 (Figure 1). As expected, Pol θ preferentially inserted dA opposite both Tg and T, even when high concentrations (500 μM) of each dNTP were employed. In many cases, this concentration was above the corresponding Km, and the observations in Figure 1 are strongly dependent upon differences in kcat (Table 1). Weak misinsertion of dG was detectable only on templates containing T (1 and 3) but not Tg (2 and 4), while T insertion was even less efficient, and dC insertion was undetectable. These observations are consistent with larger kcat values for inserting dG opposite T (1) than opposite Tg (2). Despite this, Pol θ exhibits reduced fidelity during translesion synthesis, largely due to a 20-fold reduction in the efficiency of dA insertion opposite Tg (2) relative to T (1). This reduced fidelity is reflected in a larger insertion frequency (Fins, Table 1) for incorrect nucleotides (dG and T) opposite Tg than opposite T (Table 1). During incorporation opposite T (1), dA is preferred by 750-fold over dG and 7000-fold over T, while the preference for dA is reduced to 350-fold over dG and 625-fold over T during insertion opposite Tg (2). Similar relative effects on Pol θ fidelity were observed for steady-state kinetic analysis of dNTP incorporation opposite T or Tg in 3 and 4, respectively (Table S1). These results are inconsistent with a previous report showing that Pol θ inserts dA with equal efficiency opposite Tg and T and also exhibits increased fidelity when incorporating opposite Tg.20 We do note; however, that these results are consistent with the weak mutagenicity observed for cellular Tg bypass by Pol θ.20
Figure 1.
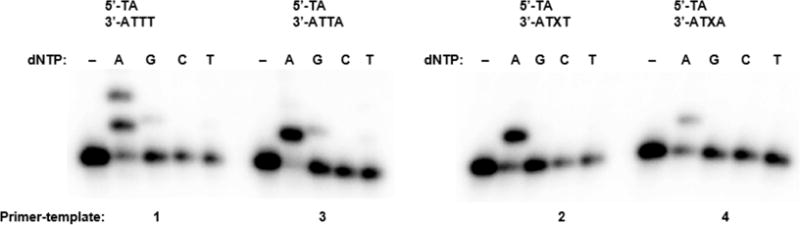
Single nucleotide insertion opposite Tg and T. Pol θ (432 pM) was incubated with 1-4 (50 nM) and the indicated dNTP (500 μM) for 5 min at 25 °C. The relevant portion of each primer-template complex is shown above. X = Tg.
Table 1.
Steady-state kinetic analysis of nucleotide insertion opposite T and Tg (1 and 2).a
| X | dNTP | kcat (× 10−2 s−1) | Km (μM) | kcat/Km (× 10−2 s−1/μM) | Fins b |
|---|---|---|---|---|---|
| T (1) | A | 108.5 ± 19.5 | 4.8 ± 0.6 | 22.7 | 1 |
| T (1) | T | 4.0 ± 0.2 | 1216 ± 54 | 0.003 | 0.00014 |
| T (1) | G | 9.8 ± 1.0 | 336 ± 3 | 0.03 | 0.0013 |
| Tg (2) | A | 114 ± 1 | 93 ± 13 | 1.2 | 1 |
| Tg (2) | T | 1.5 ± 0.1 | 751 ± 21 | 0.002 | 0.0016 |
| Tg (2) | G | 0.6 ± 0.1 | 432 ± 27 | 0.003 | 0.0028 |
Data are the average ± std. dev. of two independent experiments performed in triplicate. bFins = (kcat/Km)dNTP/(kcat/Km)dATP
Steady-state analysis of Tg bypass: template misalignment vs. error-free extension
Because dA is preferentially inserted opposite Tg, we prepared primer-templates 5-8 containing dA opposite either Tg or T (Chart 2). Notably, the template sequences for 6 and 8 differ only in the orientation of the two nucleotides downstream from Tg, with either 3′-TA flanking the lesion for 6 or 3′-AT for 8. If template misalignment occurred during extension past Tg as it does when Pol θ extends past abasic lesions,23 T would be misinserted during extension of 6 (Scheme 2) and dG would be misinserted during extension of 8 (Scheme 3). Consistent with this mechanism, for both 6 and 8, Pol θ inserted only the correct nucleotide and the nucleotide resulting from the aforementioned template misalignment (Figure 2). Misinsertion of other nucleotides was not detected, suggesting that the observed misinsertion events are dependent upon template misalignment (Scheme 2 and 3) and are not a result of low-fidelity synthesis by Pol θ.
Chart 2.

Primer-templates used for analyzing extension past Tg
Figure 2.
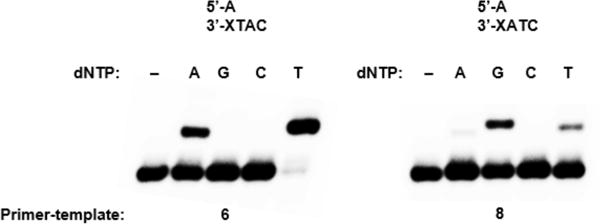
Single nucleotide insertion during extension past Tg. Pol θ (4.32 nM) was incubated with 6 or 8 (50 nM) and each individual dNTP (1 mM) for 5 min at 25 °C. The relevant sequence of each primer-template is shown.
Steady-state kinetic experiments on 6 (Tg) showed that insertion of T, presumably mediated by template-misalignment, was 9-fold more efficient than insertion of the correct nucleotide, dA (Table 2). However, both processes were considerably less efficient than correct bypass (dA incorporation) of control primer-template 5 (T). Correct bypass of Tg (dA incorporation) being 1800-fold less efficient and misalignment-mediated bypass (thymidine incorporation) being 200-fold less efficient. Similar results were obtained during steady-state kinetic experiments on 7 and 8 (Table 3). When Tg was present in the template (8), the product resulting from misalignment (dG incorporation) was again preferred over incorporation of the correct nucleotide (T), in this case by almost 6-fold (Table 3). The extension efficiency past Tg was also substantially reduced relative to 7, where Tg was replaced with T. Taken together, these data suggest that when Pol θ extends a primer past template Tg, template misalignment, followed by nucleotide misinsertion is more efficient than error-free extension when a thymidine is present one or two nucleotides downstream.
Table 2.
Steady-state kinetic analysis of extension past T and Tg (5 and 6).a
| X | dNTP | kcat (× 10−2 s−1) | Km (μM) | kcat/Km (× 10−2 s−1/μM) | Finsb |
|---|---|---|---|---|---|
| T (5) | A | 56.0 ± 3.3 | 3.7 ± 1.6 | 15.2 | ---- |
| Tg (6) | A | 1.8 ± 0.2 | 215 ± 9 | 0.008 | 1 |
| Tg (6) | T | 36.8 ± 1.0 | 503 ± 22 | 0.07 | 9.0 |
Data are the average ± std. dev. of two independent experiments performed in triplicate. bFins = (kcat/Km)dNTP/(kcat/Km)dATP
Table 3.
Steady-state kinetic analysis of extension past T and Tg (7 and 8).a
| X | dNTP | kcat (× 10−2 s−1) | Km (μM) | kcat/Km (× 10−2 s−1/μM) | Finsb |
|---|---|---|---|---|---|
| T (7) | T | 35.2 ± 7.5 | 1.0 ± 0.4 | 35.2 | ---- |
| Tg (8) | T | 1.1 ± 0.1 | 488 ± 24 | 0.002 | 1 |
| Tg (8) | G | 2.4 ± 0.03 | 177 ± 12 | 0.01 | 5.8 |
Data are the average ± std. dev. of two independent experiments performed in triplicate. bFins = (kcat/Km)dNTP/(kcat/Km)dATP
In principle, nucleotide insertion on a misaligned template could give rise to two outcomes: either the primer can realign and Pol θ can extend from the resulting mismatch, which would produce full-length product containing a promutagenic mismatch, or the misaligned primer can be extended, giving rise to deletions, as shown in Schemes 2 and 3. To determine which outcome is operative during Tg bypass, 1-4 were utilized in full-length extension experiments such that Pol θ conducted both translesion synthesis and subsequent extension (Figure 3). When incubated with controls 1 and 3, Pol θ rapidly extended the primer to form N+1 products, consistent with blunt-end addition of Pol θ.22 This was followed by slower addition of another nucleotide to form N+2 products. Extension of primers on Tg-containing templates 2 and 4; however, primarily gave rise to shorter products than the corresponding controls. The length of these products was consistent with the expected deletions for each sequence (one-nucleotide deletion for 2 and two-nucleotide deletion for 4). For 2, the apparent one-nucleotide deletion was nearly the exclusive product, whereas a mixture of products was formed by extension of 4, albeit with a preference for two-nucleotide deletion. These results suggest that Pol θ exhibits a preference for deletion formation during Tg bypass in appropriate sequences. However, interpretation of Figure 3 is complicated by blunt-end addition by Pol θ to the extension products, necessitating further support for deletion formation.33
Figure 3.
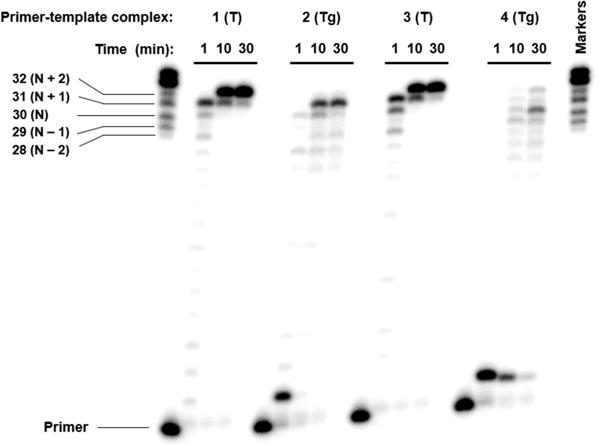
Translesion synthesis and extension past Tg. Pol θ (20 nM) was incubated with 1-4 (50 nM) and all four dNTPs (500 μM) at 25 °C for the indicated time.
Sequencing of Tg bypass products
To obtain direct support for misalignment-mediated deletion formation, we sequenced individual Tg bypass products using a previously reported method.33 A biotinylated primer was annealed to a template strand containing either T (9) or Tg (10). The template sequence was designed such that Tg was flanked by a downstream 5′-TTA, meaning that lesion bypass could be mediated by a Watson-Crick base pair with either the first or second T, resulting in either one-or two-nucleotide deletions. Pol θ was incubated with 9 or 10 in the presence of all four dNTPs. The biotinylated strand was isolated using streptavidin beads and amplified by PCR. The PCR product was subcloned into the pBlueScript plasmid and transformed into E. coli for sequencing of individual colonies. As expected, control 9 bearing T gave only full-length products (Table 4). No deletions were detected and dA was inserted opposite T with 100% frequency. In contrast, translesion synthesis of 10 was weakly mutagenic, with 3% (one event) insertion of T. This is consistent with the increased mutagenicity of Tg, although insertion of dA is still strongly preferred over T and dG as shown in steady-state kinetic experiments (Table 1). Extension beyond the lesion by Pol θ primarily resulted in 2-nucleotide deletions (72%) with a smaller number of full-length bypass events (22%). One nucleotide deletions were the minor product, accounting for fewer than 10% of bypass events. Notably, T insertion opposite Tg gave rise to full-length product, consistent with the proposed deletion mechanism. These results suggest that on sequences containing a downstream T within one or two nucleotides, Tg bypass by Pol θ is primarily mediated by a misaligned primer-template complex, which primarily gives rise to deletions.
Table 4.
Fidelity of Tg bypass by Pol θ
| Template nucleotide | #Colonies sequenced | dA opposite X | Full-length extension | One-nucleotide deletion | Two-nucleotide deletion |
|---|---|---|---|---|---|
| T (9) | 15 | 15 (100%) | 15 (100%) | 0 | 0 |
| Tg (10) | 36 | 35 (97%)a | 8 (22%) | 2 (6%) | 26 (72%) |
Misinsertion of T accounted for one full-length bypass event
Discussion
Tg is a major oxidative DNA lesion which is cytotoxic due to its strong replication-blocking properties.8,14 Replicative polymerases insert dA opposite the lesion in vitro but are impeded considerably in extending beyond the lesion.14,34 Crystal structures of the RB69 replicative polymerase with Tg show that this impediment arises from the nonplanar conformation of Tg, which causes the C5 methyl group to block incorporation of the next nucleotide.35,36 Replication of plasmid vectors in human cells indicated that the translesion synthesis polymerases Pol κ, Pol ζ, and Pol θ mediate the majority of Tg bypass, suggesting that although replicative polymerases insert a nucleotide opposite the lesion in vitro, they are somehow prevented from doing so in vivo.9,20 Bypass of Tg by translesion polymerases in eukaryotic cells is predominantly error-free, with only a small number of mutations resulting from insertion of an incorrect nucleotide opposite the lesion. Formation of frameshift deletions was not reported, although based on the observations presented above, the sequence employed in previous experiments could have given rise to frameshift deletions during Pol θ bypass of Tg.20 It is possible that frameshift mutations are suppressed in vivo or they are rapidly repaired. However, it is also possible that frameshift mutations are formed in cells when Pol θ bypasses Tg, but they were not detected in the elegant study which established the cellular role for Pol θ in Tg bypass.20
We report that when Tg is present in a template containing a T either one or two nucleotides downstream from the lesion, extension by Pol θ primarily forms deletions. Steady-state kinetic data indicate that template misalignment is 5- to 10-fold more efficient than correct bypass on the two local sequences studied. Sequencing of bypass products on a template that can yield one- and two-nucleotide deletions, as well as full-length products, showed that two-nucleotide deletions accounted for more than 70% of bypass events. On this sequence, correct bypass accounted for only 20% and one-nucleotide deletions accounted for less than 10% of bypass events. Although template misalignment is kinetically favored over full-length product formation, extension past Tg is markedly less efficient (200-fold for template-misalignment and 1,800-fold for correct bypass on 6 and 3,520-fold and 17,600-fold for 8) than synthesis on otherwise identical templates containing T. On the other hand, the efficiency of translesion synthesis (insertion opposite Tg) is reduced only modestly (15- to 20-fold). Structural data for Pol θ (and other mammalian polymerases) with Tg are lacking, so it is unclear whether the impediment which Pol θ experiences during the extension reaction arises from a steric clash with the C5 methyl group, as reported for the RB69 polymerase.35,36 In any case, the reduced efficiency with which Pol θ extends past Tg is not necessarily in conflict with the cellular role of Pol θ in Tg bypass, which is firmly established.20 Pol θ also exhibits a substantial reduction in efficiency for translesion synthesis (1000-fold) and extension (20-fold) during bypass of a stable analogue of 3-methyl-dA; however, Pol θ bypasses the lesion in cells.21
The differences between Tg bypass in the test tube and in cells raise interesting questions about the cellular regulation of this process. For example, Pol η and Pol ν bypass Tg lesions in vitro, with Pol η being particularly proficient.37,38 However, neither of these polymerases contributes to Tg bypass in cells.20 Evidently, the in vitro catalytic efficiency of Tg bypass is not the exclusive indicator of which polymerase(s) bypasses the lesion in cells. However, the ability of other polymerases to bypass Tg raises the question of why cells utilize Pol θ when it has a strong propensity to form frameshift mutations in vitro. Formation of even a small number of frameshift mutations during bypass of a lesion produced hundreds of times in each cell every day would be expected to contribute to genomic instability, especially in cells where Pol θ is upregulated. Frameshift mutations formed by Pol θ could possibly be repaired by mismatch repair, as occurs for frameshift mutations generated by replicative polymerases.39 If so, cells with defects in mismatch repair would likely produce greater numbers of frameshift mutations during Pol θ bypass of Tg. It is also possible that accessory proteins facilitate error-free bypass of Tg by Pol θ, although presently there is no evidence that either of these processes occur during Pol θ bypass of Tg. Therefore, the extent to which Pol θ induces in frameshifts mutations cells, or the specific factors which prevent it from doing so, may be highly relevant to the induction of genomic instability, both in healthy cells and in cancer cells, some of which upregulate Pol θ.24
Conclusions
Expression of Pol θ is upregulated in some cancers, and this upregulation correlates with poor prognosis.24 Growing evidence suggests that, in addition to its established role in double strand break repair, Pol θ is a translesion polymerase that bypasses multiple replication-blocking lesions in human cells.20,21,26 Therefore, the lesion bypass capability of Pol θ may be relevant not just to normal cell function, but also to tumor progression and resistance to treatment.40 Pol θ mediates a weakly mutagenic pathway of Tg bypass in human cells, resulting in approximately 5% nucleotide misinsertion opposite the lesion.20 Here we present evidence that, in addition to low frequency misinsertion events, Pol θ has a strong propensity to introduce frameshift mutations during Tg bypass on appropriate template sequences. We previously reported that a similar template misalignment mechanism gives rise to frameshift deletions during abasic lesion bypass by Pol θ.23 If unrepaired, such mutations would be expected to increase genomic instability, a contributor to cancer progression as well as radio- and chemo-resistance. Frameshift mutations may have gone undetected in the previous report on Pol θ bypass of Tg in human cells.20 It is therefore important to determine the frequency with which Pol θ induces frameshift deletions in vivo and whether additional proteins are involved in suppressing their formation or in repairing the mutations.
Supplementary Material
Chart 3.
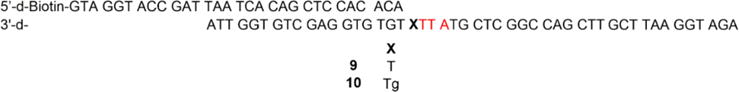
Primer-templates used for sequencing Tg bypass products
Acknowledgments
We thank Dr. Sylvie Doublié for providing the plasmid for Pol θ expression and April Averill for helpful discussions regarding Pol θ purification.
Funding Sources
We are grateful for financial support of this research from the National Institute of General Medical Science (NIH GM-063028).
Footnotes
Supporting Information
Experimental procedures, mass spectra of oligonucleotides containing thymidine glycol, representative active site titration of Pol θ. Supporting Information is available free of charge on the ACS Publications website.
The authors declare no competing financial interests.
References
- 1.Jovanovic SV, Simic MG. Mechanism of OH Radical Reactions with Thymine and Uracil Derivatives. J Am Chem Soc. 1986;108:5968–5972. doi: 10.1021/ja00279a050. [DOI] [PubMed] [Google Scholar]
- 2.Adelman R, Saul RL, Ames BN. Oxidative damage to DNA: relation to species metabolic rate and life span. Proc Natl Acad Sci U S A. 1988;85:2706–8. doi: 10.1073/pnas.85.8.2706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Breimer LH, Lindahl T. Thymine lesions produced by ionizing radiation in double-stranded DNA. Biochemistry. 1985;24:4018–4022. doi: 10.1021/bi00336a032. [DOI] [PubMed] [Google Scholar]
- 4.Cathcart R, Schwiers E, Saul RL, Ames BN. Thymine glycol and thymidine glycol in human and rat urine: a possible assay for oxidative DNA damage. Proc Natl Acad Sci U S A. 1984;81:5633–5637. doi: 10.1073/pnas.81.18.5633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Teebor G, Cummings A, Frenkel K, Shaw A, Voituriez L, Cadet J. Quantitative measurement of the diastereoisomers of cis thymidine glycol in gamma-irradiated DNA. Free Radic Res Commun. 1987;2:303–309. doi: 10.3109/10715768709065296. [DOI] [PubMed] [Google Scholar]
- 6.Lustig MJ, Cadet J, Boorstein RJ, Teebor GW. Synthesis of the diastereomers of thymidine glycol, determination of concentrations and rates of interconversion of their cis-trans epimers at equilibrium and demonstration of differential alkali lability within DNA. Nucleic Acids Res. 1992;20:4839–4845. doi: 10.1093/nar/20.18.4839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brown KL, Adams T, Jasti VP, Basu AK, Stone MP. Interconversion of the cis -5 R, 6 S -and trans -5 R, 6 R -thymine glycol lesions in duplex DNA. J Am Chem Soc. 2008;130:11701–11710. doi: 10.1021/ja8016544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Evans J, Maccabee M, Hatahet Z, Courcelle J, Bockrath R, Ide H, Wallace S. Thymine ring saturation and fragmentation products: lesion bypass, misinsertion and implications for mutagenesis. Mutat Res Toxicol. 1993;299:147–156. doi: 10.1016/0165-1218(93)90092-r. [DOI] [PubMed] [Google Scholar]
- 9.Yoon JH, Bhatia G, Prakash S, Prakash L. Error-free replicative bypass of thymine glycol by the combined action of DNA polymerases kappa and zeta in human cells. Proc Natl Acad Sci U S A. 2010;107:14116–14121. doi: 10.1073/pnas.1007795107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dianov GL, Thybo T, Dianova II, Lipinski LJ, Bohr VA. Single nucleotide patch base excision repair is the major pathway for removal of thymine glycol from DNA in human cell extracts. J Biol Chem. 2000;275:11809–11813. doi: 10.1074/jbc.275.16.11809. [DOI] [PubMed] [Google Scholar]
- 11.Klungland A, Höss M, Gunz D, Constantinou A, Clarkson SG, Doetsch PW, Bolton PH, Wood RD, Lindahl T. Base excision repair of oxidative DNA damage activated by XPG protein. Mol Cell. 1999;3:33–42. doi: 10.1016/s1097-2765(00)80172-0. [DOI] [PubMed] [Google Scholar]
- 12.Takao M, Kanno SI, Shiromoto T, Hasegawa R, Ide H, Ikeda S, Sarker AH, Seki S, Xing JZ, Le XC, Weinfeld M, Kobayashi K, Miyazaki J ichi, Muijtjens M, Hoeijmakers JHJ, Van der Horst G, Yasui A. Novel nuclear and mitochondrial glycosylases revealed by disruption of the mouse Nth1 gene encoding an endonuclease III homolog for repair of thymine glycols. EMBO J. 2002;21:3486–3493. doi: 10.1093/emboj/cdf350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Reardon JT, Bessho T, Kung HC, Bolton PH, Sancar A. In vitro repair of oxidative DNA damage by human nucleotide excision repair system: possible explanation for neurodegeneration in xeroderma pigmentosum patients. Proc Natl Acad Sci U S A. 1997;94:9463–9468. doi: 10.1073/pnas.94.17.9463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McNulty JM, Jerkovic B, Bolton PH, Basu AK. Replication inhibition and miscoding properties of DNA templates containing a site-specific cis-thymine glycol or urea residue. Chem Res Toxicol. 1998;11:666–673. doi: 10.1021/tx970225w. [DOI] [PubMed] [Google Scholar]
- 15.Haracska L, Unk I, Johnson RE, Johansson E, Burgers PM, Prakash S, Prakash L. Roles of yeast DNA polymerases delta and zeta and of Rev1 in the bypass of abasic sites. Genes Dev. 2001;15:945–54. doi: 10.1101/gad.882301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nair DT, Johnson RE, Prakash L, Prakash S, Aggarwal AK. Rev1 employs a novel mechanism of DNA synthesis using a protein template. Science. 2005;309:2219–2222. doi: 10.1126/science.1116336. [DOI] [PubMed] [Google Scholar]
- 17.Johnson RE, Washington MT, Haracska L, Prakash S, Prakash L. Eukaryotic polymerases iota and zeta act sequentially to bypass DNA lesions. Nature. 2000;406:1015–1019. doi: 10.1038/35023030. [DOI] [PubMed] [Google Scholar]
- 18.Shachar S, Ziv O, Avkin S, Adar S, Wittschieben J, Reissner T, Chaney S, Friedberg EC, Wang Z, Carell T, Geacintov N, Livneh Z. Two-polymerase mechanisms dictate error-free and error-prone translesion DNA synthesis in mammals. EMBO J. 2009;28:383–393. doi: 10.1038/emboj.2008.281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lone S, Townson SA, Uljon SN, Johnson RE, Brahma A, Nair DT, Prakash S, Prakash L, Aggarwal AK. Human DNA polymerase encircles DNA: implications for mismatch extension and lesion κbypass. Mol Cell. 2007;25:601–614. doi: 10.1016/j.molcel.2007.01.018. [DOI] [PubMed] [Google Scholar]
- 20.Yoon JH, Choudhury JR, Park J, Prakash S, Prakash L. A role for DNA polymerase θ in promoting replication through oxidative DNA lesion, thymine glycol, in human cells. J Biol Chem. 2014;289:13177–13185. doi: 10.1074/jbc.M114.556977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yoon JH, Roy Choudhury J, Park J, Prakash S, Prakash L. Translesion synthesis DNA polymerases promote error-free replication through the minor-groove DNA adduct 3-deaza-3-methyl adenine. J Biol Chem. 2017;292:18682–18688. doi: 10.1074/jbc.M117.808659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Seki M, Masutani C, Yang LW, Schuffert A, Iwai S, Bahar I, Wood RD. High-efficiency bypass of DNA damage by human DNA polymerase Q. EMBO J. 2004;23:4484–4494. doi: 10.1038/sj.emboj.7600424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Laverty DJ, Averill AM, Doublié S, Greenberg MM. The A-Rule and deletion formation during abasic and oxidized abasic site bypass by DNA polymerase θ. ACS Chem Biol. 2017;12:1584–1592. doi: 10.1021/acschembio.7b00211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lemée F, Bergoglio V, Fernandez-Vidal A, Machado-Silva A, Pillaire MJ, Bieth A, Gentil C, Baker L, Martin AL, Leduc C, Lam E, Magdeleine E, Filleron T, Oumouhou N, Kaina B, Seki M, Grimal F, Lacroix-Triki M, Thompson A, Roché H, Bourdon JC, Wood RD, Hoffmann JS, Cazaux C. DNA polymerase theta up-regulation is associated with poor survival in breast cancer, perturbs DNA replication, and promotes genetic instability. Proc Natl Acad Sci U S A. 2010;107:13390–5. doi: 10.1073/pnas.0910759107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yoshimura M, Kohzaki M, Nakamura J, Asagoshi K, Sonoda E, Hou E, Prasad R, Wilson SH, Tano K, Yasui A, Lan L, Seki M, Wood RD, Arakawa H, Buerstedde JM, Hochegger H, Okada T, Hiraoka M, Takeda S. Vertebrate POLQ and POLβ cooperate in base excision repair of oxidative DNA damage. Mol Cell. 2006;24:115–125. doi: 10.1016/j.molcel.2006.07.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yousefzadeh MJ, Wyatt DW, Takata KI, Mu Y, Hensley SC, Tomida J, Bylund GO, Doublié S, Johansson E, Ramsden DA, McBride KM, Wood RD. Mechanism of suppression of chromosomal instability by DNA polymerase POLQ. PLoS Genet. 2014;10 doi: 10.1371/journal.pgen.1004654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ceccaldi R, Liu JC, Amunugama R, Hajdu I, Primack B, Petalcorin MIR, O’Connor KW, Konstantinopoulos PA, Elledge SJ, Boulton SJ, Yusufzai T, D’Andrea AD. Homologous-recombination-deficient tumours are dependent on Polθ-mediated repair. Nature. 2015;518:258–262. doi: 10.1038/nature14184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chan SH, Yu AM, McVey M. Dual roles for DNA polymerase theta in alternative end-joining repair of double-strand breaks in Drosophila. PLoS Genet. 2010;6:1–16. doi: 10.1371/journal.pgen.1001005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Iwai S. Synthesis of thymine glycol containing oligonucleotides from a building block with the oxidized base. Angew Chemie - Int Ed. 2000;39:3874–3876. doi: 10.1002/1521-3773(20001103)39:21<3874::AID-ANIE3874>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- 30.Wincott F, Direnzo A, Shaffer C, Grimm S, Tracz D, Workman C, Sweedler D, Gonzalez C, Scaringe S, Usman N. Synthesis, deprotection, analysis and purification of RNA and ribosomes. Nucleic Acids Res. 1995;23:2677–2684. doi: 10.1093/nar/23.14.2677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Huang H, Imoto S, Greenberg MM. The mutagenicity of thymidine glycol in Escherichia coli is increased when it is part of a tandem lesion. Biochemistry. 2009;48:7833–41. doi: 10.1021/bi900927d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zahn KE, Averill AM, Aller P, Wood RD, Doublié S. Human DNA polymerase θ grasps the primer terminus to mediate DNA repair. Nat Struct Mol Biol. 2015;22:304–311. doi: 10.1038/nsmb.2993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sabouri N, Johansson E. Translesion synthesis of abasic sites by yeast DNA polymerase ε. J Biol Chem. 2009;284:31555–31563. doi: 10.1074/jbc.M109.043927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Clark JM, Beardsley GP. Functional effects of cis-thymine glycol lesions on DNA synthesis in vitro. Biochemistry. 1987;26:5398–5403. doi: 10.1021/bi00391a027. [DOI] [PubMed] [Google Scholar]
- 35.Aller P, Rould MA, Hogg M, Wallace SS, Doublié S. A structural rationale for stalling of a replicative DNA polymerase at the most common oxidative thymine lesion, thymine glycol. Proc Natl Acad Sci U S A. 2007;104:814–8. doi: 10.1073/pnas.0606648104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Aller P, Duclos S, Wallace SS, Doublié S. A crystallographic study of the role of sequence context in thymine glycol bypass by a replicative DNA polymerase serendipitously sheds light on the exonuclease complex. J Mol Biol. 2011;412:22–34. doi: 10.1016/j.jmb.2011.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kusumoto R, Masutani C, Iwai S, Hanaoka F. Translesion synthesis by human DNA polymerase η across thymine glycol. Biochemistry. 2002;41:6090–6099. doi: 10.1021/bi025549k. [DOI] [PubMed] [Google Scholar]
- 38.Takata K, Shimizu T, Iwai S, Wood RD. Human DNA polymerase N (POLN) is a low fidelity enzyme capable of error-free bypass of 5 S -thymine glycol. J Biol Chem. 2006;281:23445–23455. doi: 10.1074/jbc.M604317200. [DOI] [PubMed] [Google Scholar]
- 39.Iyer RR, Pluciennik A, Burdett V, Modrich PL. DNA mismatch repair: functions and mechanisms. Chem Rev. 2006;106:302–323. doi: 10.1021/cr0404794. [DOI] [PubMed] [Google Scholar]
- 40.Rouquette I, Lepage B, Oumouhou N, Walschaerts M, Leconte E, Schilling V, Gordien K, Brouchet L. DNA replication stress response involving PLK1, CDC6, POLQ, RAD51 and CLASPIN upregulation prognoses the outcome of early/mid-stage non-small cell lung cancer patients. Oncogenesis. 2012;1:1–10. doi: 10.1038/oncsis.2012.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


