Abstract
Cholangiocarcinoma (CCA) is the most common primary biliary malignancy with poor prognosis. Less understanding of its etiology and pathogenesis makes the diagnosis and therapy difficult. Recently, accumulating evidences have demonstrated that deregulated expression of non-coding RNAs (ncRNAs) is closely associated with the etiopathogenesis of CCA. NcRNAs which lack open reading frame are a heterogeneous class of transcribed RNA molecules, including microRNAs, long non-coding RNAs and circular RNAs. Several studies have shown ncRNAs dysregulation is a common central event occurring in CCA and has the potential of being therapy targets. Moreover, ncRNAs can be easily detected in cancer tissues and biofluids, representing valuable tools for diagnosis. In this review, we illustrate the role of ncRNA in the CCA and discuss their potential clinical value.
Keywords: Cholangiocarcinoma, Non-noncoding RNAs, Circular RNAs, Biomarkers, Therapeutic targets.
Introduction
As the first most common primary biliary malignancy and the second most common primary hepatic malignancy, CCA accounts for 3% of gastrointestinal tumors 1. Cholangiocarcinoma can be separated into three distinct tumor types by anatomical locations: intrahepatic cholangiocarcinoma (iCCA), perihilar cholangiocarcinoma (pCCA) and distal cholangiocarcinoma (dCCA). Despite common association with the biliary tract, each type of CCA is differ in clinical presentation 2. So indistinguishable and aggressive the CCA is, patients suffered from it have a bad clinical outcome with median survival time less than 24 months 3. Despite recent improvement in medicine, the surgical extirpation or liver transplantation is still the only therapy which can cure the patients at early stages. As no specific biomarker or special clinical manifestation, CCA is typically characterized by advanced diagnosis. Molecular targeting agents do nothing in the CCA management, which largely result from the poor illustration of CCA molecular mechanism 4. For solving these problems, elucidating the molecular pathogenesis of CCA may be crucial.
NcRNAs are a heterogeneous class of RNA molecules, which account for 98% human RNAs 5. Despite lacking the Open reading frame, ncRNAs have risen to prominence in gene expression. Accumulating evidences suggest that ncRNAs involved in numerous biological processes in cancers. NcRNAs can largely divided into miRNAs (microRNAs), lncRNAs (long non-coding RNAs) and circRNAs (circular RNAs). The majority studies about CCA are mainly on miRNAs, which length are less than 200nt 10-43. They can directly regulate gene expression through “ceRNA” manner at post transcriptional level 6. Interacted with miRNAs, some lncRNAs like H19 or HULC can influence CCA biological processes 68-70. Although no circRNA has been proved to be associated with CCA pathogenesis so far. Bioinformatics had shown that was possible. This review article provides the understanding of ncRNAs which were specifically associated with CCA and discusses the potential utility in clinic.
The short NcRNAs-MicroRNAs deregulation in CCA
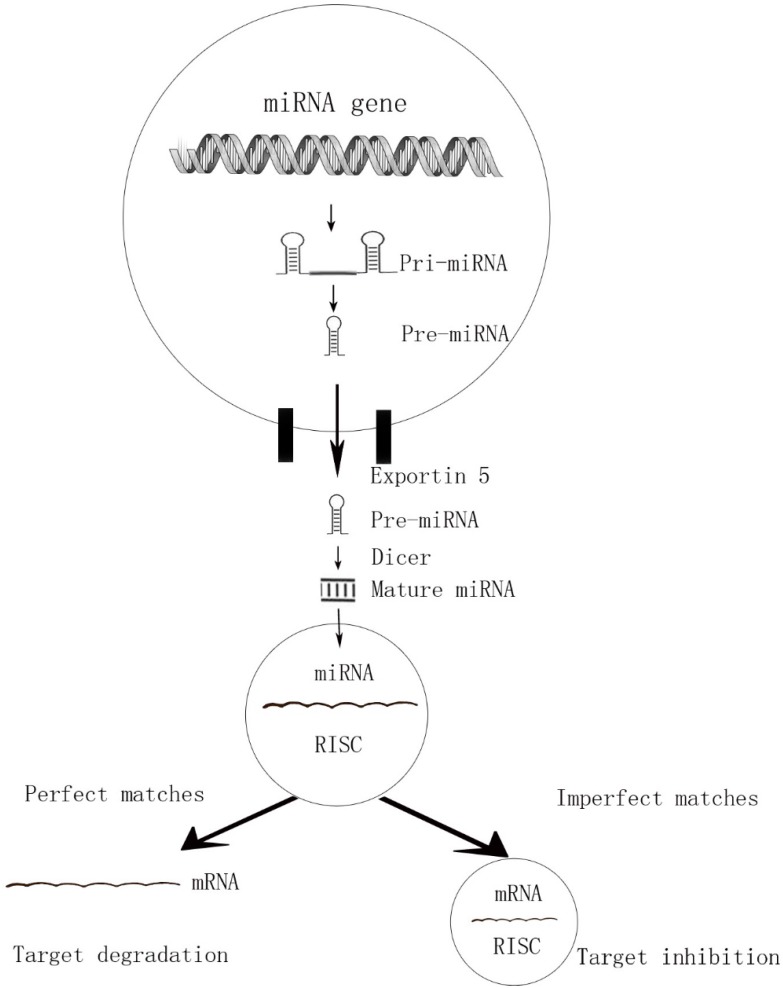
MicroRNAs are endogenous, short, highly conserved non-coding single-stranded RNAs of about 18-25nt in length. By binding to 3' untranslated region (3' -UTR) of targeted mRNAs, the miRNAs are incorporated into miRNA-associated RNA-induced silencing complex (miRISC) and play acritical role in gene expression at posttranscriptional level 7. This interaction causing either degradation (perfect matches) or repression (imperfect target duplexes) of the translation 8, 9. Due to the imperfect matches, a single miRNA can target hundreds of potential messenger RNAs (mRNA) targets and a single mRNA can converge on more than one miRNAs (Figure 1). That is why one particular miRNA can regulate many genes and influence multiple cell progresses.
Figure 1.
The perfect matches and imperfect matches of miRNAs
Cell cycle, proliferation and apoptosis
The first article in this area is by Meng et al. Using q-PCR, they found that miR-21, miR-141, and miR-200b were highly overexpressed in malignant cells compared with normal cholangiocytes 10. Downregulated miR-141 reduced the CCA cell proliferation, which means it could be a potential therapy target. Mir-29b was found to be downregulated in malignant KMCH CCA cell lines 11. By suppressing antiapoptotic member Mcl-1, mir-29b significantly sensitized the cells to TNF (tumor necrosis factor) and induced cells apoptosis. Mcl-1 is an anti-apoptotic protein of Bcl-2 family. Its overexpressed was commonly observed in cancers including cholangiocarcinoma 12-13. The following study of the same group showed the c-Myc, Hedgehog and NF-kappaB may be responsible for miR-29 dysregulation 14.
Another study by Selaru et al identified miR-494 level was low in CCA tissues and up-regulated expression of miR-494 decreased cell proliferation. Furthermore, miR-494 could reduce CDK6, CDK4, CCND1, CCNE2 and HDAC1 levels, resulting in G1/S arrest 15. After that, miR-494 was shown also can induce G2/M cell cycle arrest as well 16. MiR-494 modulated six G2 /M transitional genes, including Polo-like Kinase 1 (PLK1), pituitary tumor-transforming gene 1 (PTT G1), Cyclin B1 (CCNB1), cell-division cycle 2 (CDC2), cell-division cycle 20 (CDC20) and topoisomerase II α (TOP 2A). Those two researches confirmed that miR-494 plays a global regulatory role in cell proliferation and the miR-494 based therapies may have an extraordinary effect in CCA treatment. Wang et al compared the miRNA levels in CCA samples with normal bile duct and identified miR-138 was downregulated 17. The dual luciferase reporter assay showed that RhoC was a direct target of miR-138. Treatment with miR-138 mimics significant decreased Rhoc expression and resulted in decreased proliferation and migration in CCA cells. Farnesoid X receptor (FXR) was an important nuclear receptor in cell proliferation and migration. And it had been reported that FXR could act as a tumor suppressor in many tumors 18-19. Zhong et al demonstrated FXR expression was lower in tumor tissues than normal tissues, while miR-421 was up-regulated. Moreover, they confirmed that miR-421 can suppress FXR expression both in mRNA and protein levels and result in promoting proliferation in CCA cell finally 20. Encoded by the MEN1, Menin is a tumor-suppressor protein. Ehrlich et al found that Menin's expression was decreased in CCA, negative regulated by miR-24. Moreover, a negative feedback loop was formed by these two molecules to fulfill their antitumor functions 21.
As mentioned above, there is a strong relationship between miRNAs and CCA molecular pathogenesis. But most studies focused on a single time point of CCA. Using real-time miRNA activity monitoring and activity technology, Chen et al examined dynamic activity of six miRNAs. The date showed that those six miRNAs' function was temporal and the expression levels varied over time 22. Although some limits in this research, it takes us a new angle in CCA pathogenesis study.
Migration and Invasion
Chusorn et al. found miR-21 to be markedly overexpressed in CCA tissues compared with normal tissues 23. Knockout of miR-21 decreased the metastatic potential of CCA cells through the regulation of PDCD4 and TIMP3. TIMP3 is a member of Matrix metalloproteinases, which play a crucial role in cancer invasion and metastasis 24. The sensitivity and specificity of the miR-21 in diagnosis were evaluated as 95% and 100% respectively, which means it could be a potential biomarker. Moreover, another study showed that RECK was also a target of miR-21 25. Ars2 is a protein that was essential for microRNA biogenesis. Ars2 and miR-21 were both found overexpressed in CCAs. Knockdown Ars2 blocker decreased miR-21 levels, result in tumor formation suppression. But, overexpressed Ars2 could not increase miR-21 level and get tumor formation promoted 26. This study indicated that there must be some unknown complexity multifactorial processes in CCA molecular pathogenesis.
The phenomenon that epithelial cells converse into mesenchymal-like cells calls EMT (epithelial-to-mesenchymal transition). EMT is pivotal in cancer invasion and metastasis. It has been reported that many miRNAs may prompt cell migration and invasion by assisting EMT progress. In 2012, Oishi et al examined the miR-200c in 23 ICC samples and cell lines. MiR-200c was downregulated in tumor tissues. Activated MiR-200c targeted NCAM1 and reduced ICC cells EMT progress 27. Liu et al. demonstrated that miR-21 contributed to EMT activation by suppressing E-cadherin, N-cadherin and Vimentin 28. In agreement with Liu's result, another team found that miR-21 conjunction with KLF4 could also contribute to EMT activation by Akt/ERK1/2 pathway29 . Down-regulation of miR-204 has been observed in ICC by Qiu et al 30. Its overexpression suppressed ICC cell EMT process, as well as the migration and invasion. The direct target was Slug. Employing the TCGA database, Canu et al also found that miR-204 was essential in CCA pathogenesis31 . Then, they validated it in CC cell lines. A group of 25 patients with extrahepatic cholangiocarcinoma (EHCC) exhibited higher level of miR-221 than mean value 32. Overexpressed miR-221 prompted cell migration and invasion. Moreover, the miR-221/β-catenin formed a positive feedback loop which get PTEN reinforced, resulting in the maintenance of EMT.
Inflammation
Inflammation is a causative factor in the evolution of CCA. A variety of cytokines, growth factors, tyrosine kinases are believed to contribute to cholangiocarcinogenesis under conditions of inflammation. Among them, interleukin-6 (IL-6) is an essential one. It can promote cell survival and provide mitogenic signals in autocrine or paracrine way. Meng et al demonstrated that IL-6 was overexpressed in CCA 33. And so was the let-7a. let-7a enforced IL-6 activity by promoting Stat-3 phosphorylation. The target was neurofibromatosis 2 (NF2) gene, which encode the tumor suppressor gene merlin. This is the first study explored the role of miRNA in chronic inflammation and malignancy. After that, the same group identified that IL-6 could contribute to tumor growth by targeting miR-37034. Using allelic loss technology, An et al confirmed this result 35. The let-7c/miR-99a/miR-125b cluster was found downregulated in CCA. Elevated level of let-7c/miR-99a/miR-125b inhibited CCA cell migration and invasion in vivo and vitro. The cluster reduced the IL-6/STAT3 inflammatory pathway by suppressing IL-6, IL-6R and IGF1R 36. COX-2 which plays a pivotal role in prostacyclin formation was another essential inflammation factor. Zhang et al reported that miR-101 was decreased in CCA specimens. Overexpression of it suppressed tumor growth in xenograft model. Their dates suggested that miR-101 reduced VEGF and COX-2 levels, resulting in decreasing vascular density. Lack of blood supply killed tumor cells 37. Lu et al found that miR-21 promote CCA cell growth and invasion in vitro 38. Furthermore, they identify that miR-21 directly targeted 15-PGDH (15-Hydroxyprostaglandin dehydrogenase), which was vital for COX-2 based PGE 2 (prostaglandin E2) conversion. Moreover, the PGE2 could further stimulate miR-21 expression, which formed a positive feedback loop. Thus, miR-21/COX2/PGE2 signaling blocking may provide a promising therapeutic strategy for the treatment of CCA.
The molecular clock regulates cells behavioral and physiological processes over the 24 hours. Many studies have shown that cell's transcriptome and proteome perform a circadian manner 39-40. The proteins BMAL1 and CLOCK and NPAS2 are found to be responsible for circadian regulation. The disorder of cellular circadian rhythm may contribute to cancer carcinogenesis 41. In this regard, Meng et al predicted that miRNA-141 which was overexpressed in iCCA targets CLOCK gene directly by bioinformatics technology 42. Han et al found miR-34a was expressed rhythmically in CCA cells 43. Suppression of miR-34a reduce CCA cell proliferation, migration and invasion by increasing Per1(Period) level. The Per1 proteins inhibited the transcription of CLOCK and BMAL1, having a negative effect on autonomous oscillation 44.
Chemoresistance
Remarkable multidrug resistance of CCA is a multifactorial process which resulted from numerous genetic and epigenetic processes on survival/apoptosis balance. Overexpressed miRNA-21 and miRNA-200b were contribute to the gemcitabine chemoresistance of CCA. Those two miRNAs appeared to be mediated by activation of PI3k/PTEN signaling 10. Using microRNA array, Okamoto et al identified that miR-29b, miR-205, miR-221 was downregulated in CCA. Enforcement of three miRNAs restored gemcitabine sensitivity in CCA cell lines. Using bioinformatics, they predicted that miR-29b and miR-221 targets PIK3R1, while miR-205 suppresses MMP-2 44. Meng et al found that miRNA-let7a could increase gemcitabine toxicity by targeting NF2 in vivo and vitro 33. Peng et al reported that miR-200b/c was downregulated in CCA tissues. Elevated miR-200b/c level got CCA cell sensitivity to 5-fluorouracil. The target was SUZ12/ROCK245. Li et al demonstrated that the miR-199a-3p played an important role in CCA cell lines cisplatin sensitivity. Overexpressed miR-199a-3p could increase apoptosis of CCA cells under cisplatin condition and vice versa. Further study identified that mTOR was the direct target 46. Asukai et al found that miR130a-3p was upregulated in CCA cell lines 47. Reduced miR130a-3p Expression restored Gemcitabine Resistance in cell lines. PPARG was the direct target (Table 1).
Table 1.
MiRNAs dysregulation with targets in CCA
| Expression | miRNA | Disease | Target | Function | Reference |
|---|---|---|---|---|---|
| Up | miR-21 | CCA, ICC | 1* | 2* | 10 23-26 28-29 38 48 51-53 |
| miR-221 | EHCC | PTEN | invasion, migration, EMT | 41 | |
| miR-25 | CCA | DR4 | 54 | ||
| miR-26a | CCA | 3* | proliferation, EMT | 55 67 68 | |
| miR-31 | ICC | RASA1 | proliferation, apoptosis | 49 | |
| miR-106a | CCA | migration | 56 | ||
| miR-141 | ICC | CLOCK | proliferation, physiological rhythm | 10 | |
| miR-200b | CCA | PI3k, PTEN | multidrug resistance | 10 | |
| miR-210 | CCA | Mnt | proliferation | 57 | |
| miR-199a-3p | CCA | mTOR | multidrug resistance | 46 | |
| miR-421 | CCA | FXR | proliferation, migration | 20 | |
| miR-130a-3p | CCA | PPARG | gemcitabine resistance | 47 | |
| Let-7a | CCA | NF2 | cell survival, mitogenic, multidrug resistance | 33 | |
| Down | miR-205 | CCA | MMP-2 | multidrug resistance | 44 |
| miR-221 | CCA | PIK3R1 | multidrug resistance | 44 | |
| miR-29b | CCA | 4* | apoptosis, multidrug resistance | 11 14 44 51 | |
| miR-34a | CCA | Per1, Smad4 | proliferation, invasion, migration, cell cycle | 43 58 | |
| miRNA-101 | CCA | VEGF, COX-2 | vascular density, growth | 37 | |
| miR-124 | ICC | SMYD3 | invasion, migration | 59 | |
| miR-138 | CCA | RhoC | proliferation, migration, cell cycle | 17 | |
| miR-144 | CCA | LIS1 | proliferation, invasion, migration | 60 | |
| miR-148a | CCA | DNMT-1 | proliferation | 61 | |
| miR-200b/c | CCA | SUZ12, ROCK2 | invasion, migration, multidrug resistance | 45 | |
| miR-204 | ICC | Slug, Bcl-2 | EMT, invasion, migration, apoptosis | 30 62 | |
| miR-214 | ICC | Twist | EMT, migration | 63 | |
| miR-320 | ICC | Mcl-1/Bcl-2 | apoptosis | 64 | |
| miR-370 | CCA | MAP3K8 | proliferation | 34 35 | |
| miR-373 | HCCA | MBD2 | epigenetic | 65 | |
| miR-376c | ICC | GRB2 | proliferation, migration | 66 | |
| miR-494 | CCA | 5* | proliferation, cell cycle | 15 16 | |
| miR-410 | CCA | XIAP | proliferation | 50 | |
| miR-141 | CCA | proliferation | 10 | ||
| miR-200c | ICC | NCAM1 | EMT | 27 | |
| miR-24 | CCA | MEN1 | 21 | ||
| Let-7a/miR-99a/miR-125b | CCA | IL-6, IL-6R, IGF1R | invasion, migration | 36 |
1* PI3k, PDCD4, TIMP3, RECK, TPM1, 15-PGDH, PTPN14, PTEN and KLF4
2* proliferation, invasion, migration, apoptosis, EMT and multidrug resistance
3* GSK-3b, TGFβ1 and KRT19
4* Mcl-1, MMP-2c-Myc, Hedgehog, NF-kappaB and PIK3R1
5* CDK6, CDK4, CCND1, CCNE2 and HDAC1
The long NcRNAs-Long non-coding RNAs deregulation in CCA
Long non-coding RNAs (lncRNAs) are a subgroup of non-coding RNA with size larger than 200 nucleotides. Transcribed by RNA polymerase II, lncRNAs are 5' capped and 3' adenylated 69. According to their association with mRNA, lncRNAs can be divided into five categories: (1) sense (2) antisense (3) intronic (4) intergenic (5) bidirectional 70. LncRNAs can regulate gene expression via many ways, including epigenetic regulation, transcriptional activation and posttranscriptional modification of mRNA 71. Moreover, recent reports have demonstrated that lncRNAs could sponge miRNAs, result in their targeting proteins alteration post-transcriptionally 72. On account of those mechanisms, the potential regulatory scenario of lncRNAs is enormous. Thus, lncRNAs have been valued by scholars and garnered intense research in recent years. By now, although the functions of some lncRNAs have been clearly illustrated, most of them remain unknown.
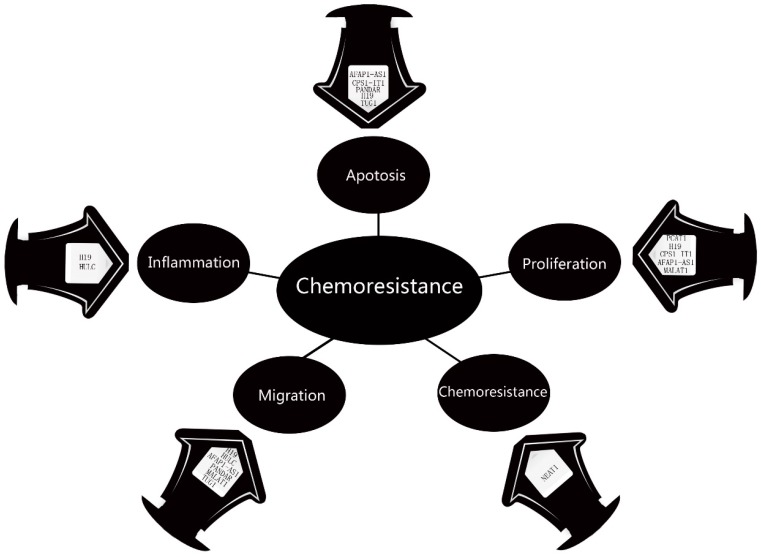
Wang et al in 2014 examined the lncRNAs profile in 77 ICC tissues. Microarray studies found that 2773 lncRNAs were upregulated in ICC tissues, while 2392 lncRNAs were downregulated. Using bioinformatics, they built 4 lncRNA -mRNA pairs in ICC: RNA43085 and SULF1, RNA47504 and KDM8, RNA58630 and PCSK6, and RNA40057 and CYP2D6. Moreover, the CYP2D6 and PCSK6 were negatively correlated with overall survival rate, while CYP2D6 and RNA40057 were positively 73. In 2015, Ma et al focused on abnormally genes and lncRNAs in ICC and found that CPS1 (Carbamoyl‑phosphate synthase 1) and its lncRNA CPS1‑IT1 (CPS1 intronic transcript 1) were upregulated in ICC. Knockdown of CPS1 and CPS1‑IT1 resulted in diminished proliferation and promoted apoptosis of ICC cells. Moreover, their levels were positively correlation with poor liver function and lymphatic invasion, which resulted in bad clinic outcome 74. Thus, CPS1 and CPS1‑IT1 may have the potential to be biomarker for ICC. In investigating the role of inflammation and oxidative stress in the development of CCA, Wang et al demonstrated that the H19 and HULC levels were elevated under the hypoxic or inflammatory condition. Inhibition of those two lncRNAs in CCA cells suppressed their migration and invasion potential, whereas increased expression had the opposite effect. By ponging let-7a/let-7b and miR-372/miR-373, H19 and HULC activate the IL-6, CXCR4 pathway may be responsible for this procedure 75. Mutations of BRCA-1 associated protein-1 (BAP1) was frequent in ICC. Parasramka et al found high level of BAP1 could make CCA cell resistance to gemcitabine. The direct target was lncRNA NEAT-176. NEAT-1 interruption significantly restored cell response to gemcitabine, which means it could be a potential sensitizer. Actin filament associated protein 1 antisense RNA1 (AFAP1-AS1) was another important lncRNA in CCA pathogenesis. Both Shi et al and Lu et al found it was up-regulated in CCA cell. AFAP1-AS1 knockdown inhibited G0/G1 cell cycle arrest and S-G2/M transition, by increasing AFAP1 and interrupting cell stress filament formation 77. Lu et al demonstrated that AFAP1-AS1 could inhibit cell proliferation by regulating C-myc and Cycling D178. Moreover, it also could downregulate C-myc and Cycling D1, resulted in cell migration blocking. Thus, AFAP1-AS1 may have the potential to be therapeutic target for ICC. The fact that lncRNA MALAT1 prompt CCA cell proliferation and migration have been confirmed by Tan et al 79 and Wang et al80. The pathways were MALAT1/miR-204/CXCR4 and PI3K/Akt pathway. Our team found lncRNA PANDAR 81, CCAT182, TUG183, PCAT184 and H1985 were upregulated in CCA cells. These five lncRNAs were all correlated with poor prognosis in CCA, such as more frequent lymph node invasion, advanced TNM stage and more postoperative recurrence. Moreover, Obstruction of PANDAR or TUG1 induced cell apoptosis and erased EMT progress. Silencing of H19 inhibited cell proliferation and promoted cell apoptosis. PCAT1 induces cell proliferation and apoptosis by miR-122/Wnt/β-catenin-signaling pathway. In conclusion, current study suggests that these lncRNAs might serve as prognostic therapy targets for CCA patients (Figure 2).
Figure 2.
The Dysregulation of lncRNAs in CCA
The Novel NcRNAs-Circular RNAs deregulation in CCA
Circular RNAs (circRNAs) represent a novel class of ncRNA molecules They are a kind of single-stranded RNA ring without 5' capping and 3' polyadenylation. Seemingly the product of mistaken RNA splicing, circRNAs have not be recognized until 1979 86. Due to the covalently closed loop, circRNAs are more stable, and confer high resistance to RNase. CircRNAs are specific expression in tissues and much higher abundance than their linear transcripts in some genes 77. Several studies indicated that circRNAs can regulate gene expression by competing with endogenous RNA (ceRNA) at multiple levels 88-89. For instance, CDR1as (cerebellum degeneration-related antigen 1 antisense) is abundance in human and mice brain. It harbors 74 conventional miR-7 binding sites 90. By sponging miR-7, CDR1as plays profound roles in diseases, including colorectal cancer 91, hepatocellular carcinoma 92 and diabetes 93. Although there is still no research reported the relationship between circRNAs and CCA. Using bioinformatics, Kulcheski et al predicted that ciR-SRY/miR-138/RhoC pathway may be essential for CCA pathology 94. So, it is reasonable to speculate that circRNAs can operate as a chief regulator of CCA.
Conclusions
CCA carcinogenesis is result from disorders of numerous oncogenes, tumor-suppressor genes and the related molecules. Revealing the molecular mechanisms underlying it may contribute to searching the effective therapy targets and biomarkers. Thus, the ncRNAs become a research hotspot gradually. In this review, we overviewed ncRNA biogenesis and their specific regulatory mechanisms in CCA. The ncRNA therapeutic may affect many aspects of the tumors by targeting hundreds of transcripts accurately and effectivity. The liver first pass effect may also be benefit for this therapy 95. Many ncRNAs can be steady in extracellular vesicles. Thus, extracellular vesicles may be reconstructed to deliver the tumor-targeting drug 96. There has been a initial attempt in rat model. And the result was quite inspired 97. Moreover, the detection of ncRNAs will provide an accurate technology for early detection and diagnosis of CCAs. As biliary drainage widely used in patients with obstructive jaundice, it is easy to get bile from the CCA penitents. Li et al reported that the technology of detecting bile will be more sensitive than blood 98. However, the exploration on ncRNAs is still in its infancy. More investigations are required to solve many problems if we want to translate ncRNAs into clinical use.
Acknowledgments
This study was funded by National Natural Science Foundation of China (Grant No. 81602088), Health and Family Planning Commission Research Project of Heilongjiang Province (Grant No. 2016-049), Heilongjiang Postdoctoral Science Foundation (Grant No. LBH-Z16096) and Innovative Science Foundation of Harbin Medical University (Grant No. 2016LCZX09).
References
- 1.Rizvi S, Gores GJ. Pathogenesis, diagnosis, and management of cholangiocarcinoma. Gastroenterology. 2013;145(6):1215–1229. doi: 10.1053/j.gastro.2013.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sandhu DS, Shire AM, Roberts LR. Epigenetic DNA hypermethylation in cholangiocarcinoma: potential roles in pathogenesis, diagnosis and identification of treatment targets. Liver International. 2008;28(1):12–27. doi: 10.1111/j.1478-3231.2007.01624.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Farley DR, Weaver AL, Nagorney DM. “Natural history” of unresected. cholangiocarcinoma: patient outcome after noncurative intervention. Mayo Clinic Proceedings. 1995;70(5):425–429. doi: 10.4065/70.5.425. [DOI] [PubMed] [Google Scholar]
- 4.Chen JS, Hsu C, Chiang NJ. et al. A KRAS mutation status-stratified randomized phase II trial of gemcitabine and oxaliplatin alone or in combination with cetuximab in advanced biliary tract cancer. Annals of Oncology. 2015;26(5):943–949. doi: 10.1093/annonc/mdv035. [DOI] [PubMed] [Google Scholar]
- 5.Mattick JS, Makunin IV. Non-coding RNA. Human Mol Genet. 2006;15(1):R17–R29. doi: 10.1093/hmg/ddl046. [DOI] [PubMed] [Google Scholar]
- 6.Salmena L, Poliseno L, Tay Y. et al. A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language? Cell. 2011;146(3):353–358. doi: 10.1016/j.cell.2011.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Krol J, Loedige I, Filipowicz W. The widespread regulation of microRNA biogenesis, function and decay. Nat Rev Genet. 2010;11(9):597–610. doi: 10.1038/nrg2843. [DOI] [PubMed] [Google Scholar]
- 8.Rhoades MW, Reinhart BJ, Lim LP. et al. Prediction of plant MicroRNA targets. Cell. 2002;110(4):513–520. doi: 10.1016/s0092-8674(02)00863-2. [DOI] [PubMed] [Google Scholar]
- 9.Finnegan EJ, Matzke MA. The small RNA world. Journal of cell science. 2003;116(23):4689–4693. doi: 10.1242/jcs.00838. [DOI] [PubMed] [Google Scholar]
- 10.Meng F, Henson R, Lang M. et al. Involvement of human micro-RNA in growth and response to chemotherapy in human cholangiocarcinoma cell lines. Gastroenterology. 2006;130(7):2113–2129. doi: 10.1053/j.gastro.2006.02.057. [DOI] [PubMed] [Google Scholar]
- 11.Mott JL, Kobayashi S, Bronk SF. et al. Mir-29 regulates Mcl-1 protein expression and apoptosis. Oncogene. 2007;26(42):6133–6140. doi: 10.1038/sj.onc.1210436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kaufmann SH, Karp JE, Svingen PA. et al. Elevated expression of the apoptotic regulator Mcl-1 at the time of leukemic relapse. Blood. 1998;91(3):991–1000. [PubMed] [Google Scholar]
- 13.Kobayashi S, Werneburg NW, Bronk SF. et al. Interleukin-6 contributes to Mcl-1 upregulation and TRAIL resistance via an Akt-signaling pathway in cholangiocarcinoma cells. Gastroenterology. 2005;128(7):2054–2065. doi: 10.1053/j.gastro.2005.03.010. [DOI] [PubMed] [Google Scholar]
- 14.Mott JL, Kurita S, Cazanave SC. et al. Transcriptional suppression of mir-29b-1/mir-29a promoter by c-Myc, hedgehog, and NF-kappaB. International journal of biochemistry & cell biology. 2010;110(5):1155–1164. doi: 10.1002/jcb.22630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Olaru AV, Ghiaur G, Yamanaka S. et al. MicroRNA down-regulated in human cholangiocarcinoma control cell cycle through multiple targets involved in the G1/S checkpoint. Hepatology. 2011;54(6):2089–2098. doi: 10.1002/hep.24591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yamanaka S, Campbell NR, An F. et al. Coordinated effects of microRNA-494 induce G 2 /M arrest in human cholangiocarcinoma. Cell Cycle. 2012;11(14):2729–2738. doi: 10.4161/cc.21105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang Q, Tang H, Yin S. et al. Downregulation of microRNA-138 enhances the proliferation, migration and invasion of cholangiocarcinoma cells through the upregulation of RhoC/p-ERK/MMP-2/MMP-9. Oncology Reports. 2013;29(5):2046–2052. doi: 10.3892/or.2013.2304. [DOI] [PubMed] [Google Scholar]
- 18.Yang F, Huang X, Yi T. et al. Spontaneous development of liver tumors in the absence of the bile acid receptor farnesoid X receptor. Cancer Research. 2007;67(3):863–867. doi: 10.1158/0008-5472.CAN-06-1078. [DOI] [PubMed] [Google Scholar]
- 19.Swales KE, Korbonits M, Carpenter R. et al. The farnesoid X receptor is expressed in breast cancer and regulates apoptosis and aromatase expression. Cancer Research. 2006;66(20):10120–10126. doi: 10.1158/0008-5472.CAN-06-2399. [DOI] [PubMed] [Google Scholar]
- 20.Zhong XY, Yu JH, Zhang WG. et al. MicroRNA-421 functions as an oncogenic miRNA in biliary tract cancer through down-regulating farnesoid X receptor expression. Gene. 2012;493(1):44–51. doi: 10.1016/j.gene.2011.11.028. [DOI] [PubMed] [Google Scholar]
- 21.Ehrlich L, Hall C, Venter J. et al. MiR-24 Inhibition Increases Menin Expression and Decreases Cholangiocarcinoma Proliferation. American Journal of Pathology. 2017;187(3):570–580. doi: 10.1016/j.ajpath.2016.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen X, Chen J, Liu X. et al. The real-time dynamic monitoring of microRNA function in cholangiocarcinoma. Plos One. 2014;9(6):e99431. doi: 10.1371/journal.pone.0099431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Selaru FM, Olaru AV, Kan T. MicroRNA-21 is overexpressed in human cholangiocarcinoma and regulates programmed cell death 4 and tissue inhibitor of metalloproteinase 3. Hepatology. 2009;49(5):1595–1601. doi: 10.1002/hep.22838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jiang Y, Goldberg ID, Shi YE. Complex roles of tissue inhibitors of metalloproteinases in cancer. Oncogene. 2002;21(14):2245–2252. doi: 10.1038/sj.onc.1205291. [DOI] [PubMed] [Google Scholar]
- 25.Namwat N, Chusorn P, Loilome W. et al. Expression profiles of oncomir miR-21 and tumorsuppressor let-7a in the progression of opisthorchiasis-associated cholangiocarcinoma. Asian Pacific Journal of Cancer Prevention. 2011;13:65–69. [PubMed] [Google Scholar]
- 26.He Q, Cai L, Shuai L. et al. Ars2 is overexpressed in human cholangiocarcinomas and its depletion increases PTEN and PDCD4 by decreasing microRNA-21. Molecular Carcinogenesis. 2013;2(4):286–296. doi: 10.1002/mc.21859. [DOI] [PubMed] [Google Scholar]
- 27.Oishi N, Kumar MR, Roessler S. et al. Transcriptomic profiling reveals hepatic stem-like genesignatures and interplay of miR-200c and epithelial-mesenchymal transition in intrahepatic cholangiocarcinoma. Hepatology. 2012;56(5):1792–1803. doi: 10.1002/hep.25890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu Z, Jin ZY, Liu CH. et al. MicroRNA-21 regulates biological behavior by inducing EMT in human cholangiocarcinoma. International Journal of Experimental Pathology. 2015;8(5):4684–4694. [PMC free article] [PubMed] [Google Scholar]
- 29.Liu CH, Huang Q, Jin ZY. et al. MiR-21 and KLF4 jointly augment epithelial-mesenchymal transition via the Akt/ERK1/2 pathway. International Journal of Oncology. 2017;50(4):1109–1115. doi: 10.3892/ijo.2017.3876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Qiu YH, Wei YP, Shen NJ. et al. MiR-204 inhibits epithelial to mesenchymal transition by targeting slug in intrahepatic cholangiocarcinoma cells. Journal of Cellular Biochemistry. 2013;32(5):1331–1341. doi: 10.1159/000354531. [DOI] [PubMed] [Google Scholar]
- 31.Canu V, Sacconi A, Lorenzon L. et al. MiR-204 down-regulation elicited perturbation of a gene target signature common to human cholangiocarcinoma and gastric cancer. Oncotarget. 2017;8(18):29540–29557. doi: 10.18632/oncotarget.15290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li J, Yao L, Li G. et al. MiR-221 Promotes Epithelial-Mesenchymal Transition through Targeting PTEN and Forms a Positive Feedback Loop with β-catenin/c-Jun Signaling Pathway in Extra-Hepatic Cholangiocarcinoma. Plos One. 2015;10(10):e0141168. doi: 10.1371/journal.pone.0141168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Meng F, Henson R, Wehbe-Janek H. et al. The microRNA let-7a modulates interleukin-6-dependent STAT-3 survival signaling in malignant human cholangiocytes. Journal of Biological Chemistry. 2007;282(11):8256–8264. doi: 10.1074/jbc.M607712200. [DOI] [PubMed] [Google Scholar]
- 34.Meng F, Wehbe-Janek H, Henson R. et al. Epigenetic regulation of microRNA-370 by interleukin-6 in malignant human cholangiocytes. Oncogene. 2008;27(3):378–386. doi: 10.1038/sj.onc.1210648. [DOI] [PubMed] [Google Scholar]
- 35.An F, Yamanaka S, Allen S. et al. Silencing of miR-370 in human cholangiocarcinoma by allelic loss and interleukin-6 induced maternal to paternal epigenotype switch. PLoS One. 2012;7(10):e45606. doi: 10.1371/journal.pone.0045606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lin KY, Ye H, Han BW. et al. Genome-wide screen identified let-7c/miR-99a/miR-125b regulating tumor progression and stem-like properties in cholangiocarcinoma. Oncogene. 2016;35(26):3376–3386. doi: 10.1038/onc.2015.396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang J, Han C, Zhu H. et al. MiR-101 inhibits cholangiocarcinoma angiogenesis through targeting vascular endothelial growth factor (VEGF) American Journal of Pathology. 2013;182(5):1629–1639. doi: 10.1016/j.ajpath.2013.01.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lu L, Byrnes K, Han C. et al. MiR-21 targets 15-PGDH and promotes cholangiocarcinoma growth. Molecular Cancer Research. 2014;12(6):890–900. doi: 10.1158/1541-7786.MCR-13-0419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vollmers C, Gill S, DiTacchio L. et al. Time of feeding and the intrinsic circadian clock drive rhythms in hepatic gene expression. Proceedings of The National Academy of Sciences of The United States of America. 2009;106(50):21453–21458. doi: 10.1073/pnas.0909591106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schmutz I, Albrecht U, Ripperger JA. The role of clock genes and rhythmicity in the liver. Molecular and Cellular Endocrinology. 2012;349(1):38–44. doi: 10.1016/j.mce.2011.05.007. [DOI] [PubMed] [Google Scholar]
- 41.Khapre RV, Samsa WE, Kondratov RV. Circadian regulation of cell cycle: Molecular connections between aging and the circadian clock. Annals of Internal Medicine. 2010;42(6):404–415. doi: 10.3109/07853890.2010.499134. [DOI] [PubMed] [Google Scholar]
- 42.Meng F, Henson R, Lang M. et al. Involvement of human micro-RNA in growth and response to chemotherapy in human cholangiocarcinoma cell lines. Gastroenterology. 2006;130(7):2113–2129. doi: 10.1053/j.gastro.2006.02.057. [DOI] [PubMed] [Google Scholar]
- 43.Han Y, Meng F, Venter J. et al. MiR-34a-dependent overexpression of Per1 decreases cholangiocarcinoma growth. Journal of Hepatology. 2016;64(6):1295–1304. doi: 10.1016/j.jhep.2016.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cermakian N, Sassone-Corsi P. Multilevel regulation of the circadian clock. Nature Reviews Molecular Cell Biology. 2000;1(1):59–67. doi: 10.1038/35036078. [DOI] [PubMed] [Google Scholar]
- 45.Peng F, Jiang J, Yu Y. et al. Direct targeting of SUZ12/ROCK2 by miR-200b/c inhibits cholangiocarcinoma tumourigenesis and metastasis. British Journal of Cancer. 2013;109(12):3092–3104. doi: 10.1038/bjc.2013.655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li Q, Xia X, Ji J. et al. MiR-199a-3p enhances cisplatin sensitivity of cholangiocarcinoma cells by inhibiting mTOR signaling pathway and expression of MDR1. Oncotarget. 2017 doi: 10.18632/oncotarget.16834. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Asukai K, Kawamoto K, Eguchi H. et al. Micro-RNA-130a-3p Regulates Gemcitabine Resistance via PPARG in Cholangiocarcinoma. Annals of Surgical Oncology. 2017;24(8):2344–2352. doi: 10.1245/s10434-017-5871-x. [DOI] [PubMed] [Google Scholar]
- 48.Ma Y, Ouyang J, Wei J, Involvement of Host Non-Coding RNAs in the Pathogenesis of the Influenza Virus. International Journal of Moecular Sciences; 2016. p. 18. (1) pii: E39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hu C, Huang F, Deng G. et al. miR-31 promotes oncogenesis in intrahepatic cholangiocarcinoma cells via the direct suppression of RASA1. Experimental and Therapeutic Medicine. 2013;6(5):1265–1270. doi: 10.3892/etm.2013.1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Meng F, Wehbe-Janek H, Henson R. et al. Epigenetic regulation of microRNA-370 by interleukin-6 in malignant human cholangiocytes. Oncogene. 2008;27(3):378–386. doi: 10.1038/sj.onc.1210648. [DOI] [PubMed] [Google Scholar]
- 51.Okamoto K, Miyoshi K, Murawaki Y. miR-29b, miR-205 and miR-221 enhance chemosensitivity to gemcitabine in HuH28 human cholangiocarcinoma cells. Plos One. 2013;8(10):e77623. doi: 10.1371/journal.pone.0077623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chusorn P, Namwat N, Loilome W. et al. Overexpression of microRNA-21 regulating PDCD4 during tumorigenesis of liver fluke-associated cholangiocarcinoma contributes to tumor growth and metastasis. Tumor Biology. 2013;34(3):1579–1588. doi: 10.1007/s13277-013-0688-0. [DOI] [PubMed] [Google Scholar]
- 53.Wang LJ, He CC, Sui X. et al. MiR-21 promotes intrahepatic cholangiocarcinoma proliferation and growth in vitro and in vivo by targeting PTPN14 and PTEN. Oncotarget. 2015;6(8):5932–5946. doi: 10.18632/oncotarget.3465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Razumilava N, Bronk SF, Smoot RL. et al. MiR-25 targets TNF-related apoptosis inducing ligand (TRAIL) death receptor-4 and promotes apoptosis resistance in cholangiocarcinoma. Hepatology. 2012;55(2):465–475. doi: 10.1002/hep.24698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhang J, Han C, Wu T. MicroRNA-26a promotes cholangiocarcinoma growth by activating β-catenin. Gastroenterology. 2012;143(1):246–256. doi: 10.1053/j.gastro.2012.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cheng Q, Feng F, Zhu L. et al. Circulating miR-106a is a Novel Prognostic and Lymph Node Metastasis Indicator for Cholangiocarcinoma. Scientific Reports. 2015;5:16103. doi: 10.1038/srep16103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yang H, Li TW, Peng J. et al. A mouse model of cholestasis-associated cholangiocarcinoma and transcription factors involved in progression. Gastroenterology. 2011;141(1):378–388. doi: 10.1053/j.gastro.2011.03.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Qiao P, Li G, Bi W. et al. MicroRNA-34a inhibits epithelial mesenchymal transition in human cholangiocarcinoma by targeting Smad4 through transforming growth factor-beta/Smad pathway. BMC Cancer. 2015;15:469. doi: 10.1186/s12885-015-1359-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zeng B, Li Z, Chen R. et al. Epigenetic regulation of miR-124 by hepatitis C virus core protein promotes migration and invasion of intrahepatic cholangiocarcinoma cells by targeting SMYD3. Febs Letters. 2012;586(19):3271–3278. doi: 10.1016/j.febslet.2012.06.049. [DOI] [PubMed] [Google Scholar]
- 60.Yang R, Chen Y, Tang C. et al. MicroRNA-144 suppresses cholangiocarcinoma cell proliferation and invasion through targeting platelet activating factor acetylhydrolase isoform 1b[J] BMC Cancer. 2014;14:917. doi: 10.1186/1471-2407-14-917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Braconi C, Huang N, Patel T. MicroRNA-dependent regulation of DNA methyltransferase-1 and tumor suppressor gene expression by interleukin-6 in human malignant cholangiocytes. Hepatology. 2010;51(3):881–890. doi: 10.1002/hep.23381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chen L, Yan HX, Yang W. et al. The role of microRNA expression pattern in human intrahepatic cholangiocarcinoma. Journal of Hepatology. 2009;50(2):358–369. doi: 10.1016/j.jhep.2008.09.015. [DOI] [PubMed] [Google Scholar]
- 63.Li B, Han Q, Zhu Y. et al. Down-regulation of miR-214 contributes to intrahepatic cholangiocarcinoma metastasis by targeting Twist. Febs Journal. 2012;279(13):2393–2398. doi: 10.1111/j.1742-4658.2012.08618.x. [DOI] [PubMed] [Google Scholar]
- 64.Chen L, Yan HX, Yang W. et al. The role of microRNA expression pattern in human intrahepatic cholangiocarcinoma. Journal of Hepatology. 2009;50(2):358–369. doi: 10.1016/j.jhep.2008.09.015. [DOI] [PubMed] [Google Scholar]
- 65.Chen Y, Luo J, Tian R. et al. MiR-373 negatively regulates methyl-CpG-binding domain protein 2 (MBD2) in hilar cholangiocarcinoma. Digestive Diseases and Sciences. 2011;56(6):1693–1701. doi: 10.1007/s10620-010-1481-1. [DOI] [PubMed] [Google Scholar]
- 66.Iwaki J, Kikuchi K, Mizuguchi Y. et al. MiR-376c down-regulation accelerates EGF-dependent migration by targeting GRB2 in the HuCCT1 human intrahepatic cholangiocarcinoma cell line. Plos One. 2013;8(7):e69496. doi: 10.1371/journal.pone.0069496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zheng Z, Guan M, Jia Y, The coordinated roles of miR-26a and miR-30c in regulating TGFβ1-induced epithelial-to-mesenchymal transition in diabetic nephropathy. Scientific Reports; 2016. p. 1122. 6: 37492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wang P, Lv L. MiR-26a induced the suppression of tumor growth of cholangiocarcinoma via KRT19 approach. Oncotarget. 2016;7(49):81367–81376. doi: 10.18632/oncotarget.13229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.J. L. Rinn, H. Y. Chang. Genome regulation by long noncoding RNAs. Annual review of biochemistry. 2012;81(1):145–166. doi: 10.1146/annurev-biochem-051410-092902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.R. L. Siegel, K. D. Miller, et al. Cancer Statistics. CA-A Cancer Journal for Clinicians. 2017;67(1):7–30. doi: 10.3322/caac.21387. [DOI] [PubMed] [Google Scholar]
- 71.Jiajia Chen, Zhijun Miao, Boxin Xue. et al. J. Long Non-coding RNAs in Urologic Malignancies: Functional Roles and Clinical Translation. Cancer. 2016;7(13):1842–1855. doi: 10.7150/jca.15876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Salmena L, Poliseno L, Tay Y. et al. A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language? Cell. 2011;146(3):353–358. doi: 10.1016/j.cell.2011.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wang J, Xie H, Ling Q. et al. Coding-noncoding gene expression in intrahepatic cholangiocarcinoma. Translational Research. 2016;168:107–121. doi: 10.1016/j.trsl.2015.07.007. [DOI] [PubMed] [Google Scholar]
- 74.Ma SL, Li AJ, Hu ZY. et al. Co-expression of thecarbamoyl-phosphatesynthase 1 gene and its long non-coding RNA correlates with poor prognosis of patients with intrahepatic cholangiocarcinoma. Molecular Medicine Reports. 2015;12(6):7915–7926. doi: 10.3892/mmr.2015.4435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wang WT, Ye H, Wei PP. et al. LncRNAs H19 and HULC activated by oxidative stress, promote cell migration and invasion in cholangiocarcinoma through a ceRNA manner. Journal of Hematology& Oncology. 2016;9(1):117. doi: 10.1186/s13045-016-0348-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Tan X, Huang Z. Long Non-Coding RNA MALAT1 Interacts With miR-204 to Modulate Human Hilar Cholangiocarcinoma Proliferation, Migration, and Invasion by Targeting CXCR4. Journal of cellular biochemistry. 2017;16(1):22. doi: 10.1002/jcb.25862. [DOI] [PubMed] [Google Scholar]
- 77.Shi X, Zhang H, Wang M. et al. LncRNA AFAP1-AS1 promotes growth and metastasis of cholangiocarcinoma cells. Oncotarget. 2017 doi: 10.18632/oncotarget.16880. DOI:10.18632/oncotarget.16880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Lu X, Zhou C, Li R. et al. Long Noncoding RNA AFAP1-AS1 Promoted Tumor Growth and Invasion in Cholangiocarcinoma. Cellular physiology and biochemistry: international journal of experimental cellular physiology, biochemistry, and pharmacology. 2017;42(1):222–230. doi: 10.1159/000477319. [DOI] [PubMed] [Google Scholar]
- 79.Tan X, Huang Z. Long Non-Coding RNA MALAT1 Interacts With miR-204 to Modulate Human Hilar Cholangiocarcinoma Proliferation, Migration, and Invasion by Targeting CXCR4. Journal of Cellular Biochemistry. 2017 doi: 10.1002/jcb.25862. DOI:10.1002/jcb.25862. [DOI] [PubMed] [Google Scholar]
- 80.Wang C, Mao ZP, Wang L. et al. Long non-coding RNA MALAT1 promotes cholangiocarcinoma cell proliferation and invasion by activating PI3K/Akt pathway. Neoplasma. 2017 doi: 10.4149/neo_2017_510. DOI:10.4149/neo_2017_510. [DOI] [PubMed] [Google Scholar]
- 81.Xu Y, Jiang X. Upregulated long noncoding RNA PANDAR predicts an unfavorable prognosis and promotes tumorigenesis in cholangiocarcinoma. Onco Targets and Therapy. 2017;10:2873–2883. doi: 10.2147/OTT.S137044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Jiang XM, Li ZL, Li JL. et al. LncRNA CCAT1 as the unfavorable prognostic biomarker for cholangiocarcinoma[J] European Review for Medical and Pharmacological Sciences. 2017;21:1242–1247. [PubMed] [Google Scholar]
- 83.Xu Y, Leng K, Li Z, The prognostic potential and carcinogenesis of long non-coding RNA TUG1 in human cholangiocarcinoma.Oncotarget. 2017. [DOI] [PMC free article] [PubMed] [Retracted]
- 84.Zhang F, Wan M, Xu Y. et al. Long noncoding RNA PCAT1 regulates extrahepatic cholangiocarcinoma progression via the Wnt/β-catenin-signaling pathway. Biomedicine & Pharmacotherapy. 2017;94:55–62. doi: 10.1016/j.biopha.2017.07.025. [DOI] [PubMed] [Google Scholar]
- 85.Xu Y, Wang Z, Jiang X. Overexpression of long noncoding RNA H19 indicates a poor prognosis for cholangiocarcinoma and promotes cell migration and invasion by affecting epithelial-mesenchymal transition. Biomedicine & Pharmacotherapy. 2017;92:17–23. doi: 10.1016/j.biopha.2017.05.061. [DOI] [PubMed] [Google Scholar]
- 86.Hsu MT, Coca-Prados M. Electron microscopic evidence for the circular form of RNA in the cytoplasm of eukaryotic cells. Nature. 1979;280(5720):339–340. doi: 10.1038/280339a0. [DOI] [PubMed] [Google Scholar]
- 87.Jeck WR, Sorrentino JA, Wang K. et al. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA. 2013;19(2):141–157. doi: 10.1261/rna.035667.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Hansen TB, Jensen TI, Clausen BH. et al. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495(4771):384–388. doi: 10.1038/nature11993. [DOI] [PubMed] [Google Scholar]
- 89.Memczak S, Jens M, Elefsinioti A. et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495(4771):333–338. doi: 10.1038/nature11928. [DOI] [PubMed] [Google Scholar]
- 90.Guo JU, Agarwal V, Guo H. et al. Expanded identification and characterization of mammalian circular RNAs. Genome Biology. 2014;15(7):409. doi: 10.1186/s13059-014-0409-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Weng W, Wei Q, Toden S. et al. Circular RNA ciRS-7 - A promising prognostic biomarker and a potential therapeutic target in colorectal cancer. Clinical Cancer Research. 2017 doi: 10.1158/1078-0432.CCR-16-2541. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Xu L, Zhang M, Zheng X. et al. The circular RNA ciRS-7 (Cdr1as) acts as a risk factor of hepatic microvascular invasion in hepatocellular carcinoma. Journal of Cancer Research and Clinical Oncology. 2017;143(1):17–27. doi: 10.1007/s00432-016-2256-7. [DOI] [PubMed] [Google Scholar]
- 93.Latreille M, Hausser J, Stützer I. et al. MicroRNA-7a regulates pancreatic beta cell function. Journal of clinical investigation. 2014;124(6):2722–2735. doi: 10.1172/JCI73066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Kulcheski FR, Christoff AP, Margis R. Circular RNAs are miRNA sponges and can be used as a new class of biomarker. Journal of Biotechnology. 2016;238:42–51. doi: 10.1016/j.jbiotec.2016.09.011. [DOI] [PubMed] [Google Scholar]
- 95.Gradilone SA, O'Hara SP, Masyuk TV. et al. MicroRNAs and benign biliary tract diseases. Seminars in Liver Disease. 2015;35(1):26–35. doi: 10.1055/s-0034-1397346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Gradilone SA. Extracellular vesicles as therapeutic carriers of microRNAs for cholangiocarcinoma. Hepatology. 2017;65(2):404–406. doi: 10.1002/hep.28925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Li L, Piontek K, Ishida M. et al. Extracellular vesicles carry microRNA-195 to intrahepatic cholangiocarcinoma and improve survival in a rat model. Hepatology (Baltimore, Md.) 2017;65(2):501–514. doi: 10.1002/hep.28735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Li L, Masica D, Ishida M. et al. Human bile contains microRNA-laden extracellular vesicles that can be used for cholangiocarcinoma diagnosis. Hepatology. 2014;60(3):896–907. doi: 10.1002/hep.27050. [DOI] [PMC free article] [PubMed] [Google Scholar]