Summary
Two bacterial consortia were enriched from uncontaminated soil by virtue of their ability to grow on 1,4‐dioxane (dioxane) as a sole carbon and energy source. Their specific dioxane degradation rates at 30°C, pH = 7 (i.e. 5.7 to 7.1 g‐dioxane per g‐protein per day) were comparable to those of two dioxane‐metabolizing archetypes: Pseudonocardia dioxanivorans CB1190 and Mycobacterium dioxanotrophicus PH‐06. Based on 16S rRNA sequencing, Mycobacterium was the dominant genus. Acetylene inhibition tests suggest that dioxane degradation was mediated by monooxygenases. However, qPCR analyses targeting the tetrahydrofuran/dioxane monooxygenase gene (thmA/dxmA) (which is, to date, the only sequenced dioxane monooxygenase gene) were negative, indicating that other (as yet unknown) catabolic gene(s) were responsible. DNA sequence analyses also showed threefold to sevenfold enrichment of group 5 and group 6 soluble di‐iron monooxygenase (SDIMO) genes relative to the original soil samples. Whereas biodegradation of trace levels of dioxane is a common challenge at contaminated sites, both consortia degraded dioxane at low initial concentrations (300 μg l−1) below detectable levels (5 μg l−1) in bioaugmented microcosms prepared with impacted groundwater. Overall, this work shows that dioxane‐degrading bacteria (and the associated natural attenuation potential) exist even in some uncontaminated soils, and may be enriched to broaden bioaugmentation options for sites experiencing insufficient dioxane catabolic capacity.
Introduction
1,4‐Dioxane (dioxane) is a cyclic ether that is widely used as a stabilizer for chlorinated solvents, especially for 1,1,1‐trichloroethane (1,1,1‐TCA). Dioxane can be present as a solvent in paints, as well as in cosmetics and detergents (Zenker et al., 2003; Mohr et al., 2010), and it was classified as a probable human carcinogen (Class B2) by the United States Environmental Protection Agency (U.S. EPA) (U.S.EPA, 2014). As a result, some states have limited dioxane concentrations in drinking water to ≤ 0.35 μg l−1 based on 1 × 10−6 cancer risk assessments (U.S.EPA, 2014). Removal of dioxane from contaminated sites is usually challenging because of the compound's physical and chemical characteristics, including extreme water miscibility, limited volatility and low tendency to be retarded by sorption (Zenker et al., 2003).
Different remediation strategies have been proposed to remediate dioxane‐impacted sites, including advanced oxidation processes (Ikehata et al., 2016), photocatalytic degradation (Barndõk et al., 2016), phytoremediation (Aitchison et al., 2000), bioaugmentation (Isaka et al., 2016) and biostimulation (Lippincott et al., 2015; Li et al., 2017). Among these approaches, advanced chemical oxidation may offer effective source zone treatment, but with relatively high energy consumption and operational costs. Conversely, biodegradation approaches (when applicable) are more economical and environmentally friendly, and can also lead to dioxane mineralization without the accumulation of toxic by‐products.
Although dioxane is relatively recalcitrant, a number of strains capable of degrading dioxane have been isolated (Table 1). Most of these strains degrade dioxane cometabolically [i.e. dioxane degradation is induced by another substrate, and it does not serve as carbon or energy source to support bacterial growth (Alvarez and Illman, 2006)]. Nearly all of these strains were isolated from wastewater treatment plants or dioxane‐impacted sites. However, the potential for encountering dioxane degraders in uncontaminated environments, which is important to assess the distribution and potential ubiquity of dioxane degraders and the associated natural attenuation potential, has not been reported in the literature.
Table 1.
Bacteria capable of metabolizing or cometabolizing dioxane
| Bacterial strain | Dioxane metabolisma | Cosubstrate | Inoculum source | References |
|---|---|---|---|---|
| Rhodococcus ruber 219 | + | − | WWTP effluent | (Bernhardt and Diekmann, 1991) |
| Pseudonocardia dioxanivorans CB1190 | + | − | Industrial sludge from a dioxane contaminated site | (Parales et al., 1994) |
| Pseudonocardia benzenivorans B5 | + | − | Soil contaminated by various chlorinated, aromatic compounds | (Kämpfer and Kroppenstedt, 2004) |
| Mycobacterium sp. PH‐06 | + | − | River sediment contaminated by dioxane | (Kim et al., 2009) |
| Afipia sp. D1 | + | − | Soil samples near a dioxane producing factory | (Sei et al., 2013b) |
| Mycobacterium sp. D6 | + | − | Same as above | (Sei et al., 2013b) |
| Mycobacterium sp. D11 | + | − | Same as above | (Sei et al., 2013b) |
| Pseudonocardia sp. D17 | + | − | Same as above | (Sei et al., 2013b) |
| Acinetobacter baumannii DD1 | + | − | WWTP sludge | (Huang et al., 2014) |
| Rhodanbacter AYS5 | + | − | Industrial sludge | (Pugazhendi et al., 2015) |
| Xanthobacter flavus DT8 | + | − | Activated sludge of pharmaceutical plants | (Chen et al., 2016) |
| Pseudonocardia tetrahydrofuranoxydans sp. K1 | − | Tetrahydrofuran | WWTP sludge | (Kohlweyer et al., 2000) |
| Pseudonocardia sp. ENV478 | − | Tetrahydrofuran | Industrial wastewater treatment system | (Vainberg et al., 2006) |
| Rhodococcus ruber T1 | − | Tetrahydrofuran | Landfill soil | (Sei et al., 2013a) |
| Rhodococcus ruber T5 | − | Tetrahydrofuran | WWTP Sludge | (Sei et al., 2013a) |
| Flavobacterium sp. | − | Tetrahydrofuran | A contaminated groundwater plume | (Sun et al., 2011) |
| Mycobacterium sp. JOB5 | − | Propane | Soil samples with hydrocarbons | (Ooyama and Foster, 1965) |
| Mycobacterium sp. ENV421 | − | Propane | Turf soil enriched with propane | (Masuda, 2009) |
| Rhodococcus ruber ENV 425 | − | Propane | Turf soil samples enriched with propane | (Steffan et al., 1997) |
| Pseudomonas mendocina KR1 | − | Toluene | Algal‐bacterial mat from Colorado River in Austin | (Whited and Gibson, 1991) |
| Rhodococcus RR1 | − | Toluene | Gasoline contaminated aquifer | (Stringfellow and Alvarez‐Cohen, 1999) |
| Ralstonia pickettii PKO1 | − | Toluene | Soil microcosms amended with BTEX | (Kukor and Olsen, 1990) |
| Burkholderia cepacia G4 | − | Toluene | Water and soil samples | (Nelson et al., 1986) |
| Methylosinus trichosporium OB3b | − | Methane | Mud, river and stream water, soil samples | (Whittenbury et al., 1970) |
WWTP, wastewater treatment plant; BTEX, benzene, toluene, ethylbenzene and xylenes.
a. ’+’ and ‘−’ indicate this condition is applicable and not applicable respectively.
The critical role of monooxygenases in dioxane degradation has been emphasized by several studies (Mahendra and Alvarez‐Cohen, 2006; Kim et al., 2009), and one gene cluster that is responsible for dioxane degradation by Pseudonocardia dioxanivorans CB1190 has been identified and sequenced (i.e. thmADBC) (Sales et al., 2011). This gene cluster codes for dioxane monooxygenase, which is a soluble di‐iron monooxygenase (SDIMO) that initiates dioxane degradation (Grostern et al., 2012). SDIMOs are multicomponent bacterial enzymes that also catalyse the oxidation of various priority pollutants such as chlorinated solvents, aromatic hydrocarbons, alkanes and alkenes (Notomista et al., 2003). Therefore, investigating the presence and diversity of SDIMO genes in various environments is important to advance understanding of intrinsic bioremediation potential.
In this study, we describe the enrichment of two consortia from garden soils with no known previous exposure to dioxane. We compare their dioxane degradation capabilities with those of two archetype degraders [Pseudonocardia dioxanivorans CB1190 (Parales et al., 1994) and Mycobacterium dioxanotrophicus PH‐06 (Kim et al., 2009; He et al., 2017)]. Both high (500 mg l−1) and low (300 μg l−1) initial dioxane concentrations are considered to assess the performance of these consortia over a broad range of scenarios at impacted sites. We also examine the microbial communities before and after enrichment with dioxane through phylogenetic gene sequencing analyses, and investigate the suspected enzymes that initiate the degradation process using acetylene inactivation tests (Prior and Dalton, 1985), qPCR assays and SDIMO sequence analyses (Coleman et al., 2006).
Results and discussion
Dioxane degradation rates for both consortia were similar to those of two archetype degraders
Using surface soil samples (5 g) with no known history of dioxane exposure as bacterial seeds, two consortia were enriched over 3 months following 12 weekly cycles of 10‐fold serial dilution and growth in 100 ml of ammonium mineral salts (AMS) medium (Parales et al., 1994; Li et al., 2010) amended with 100 mg l−1 dioxane as sole carbon source. These two consortia were capable of degrading 500 mg l−1 dioxane (in AMS medium) as sole source of carbon and energy to non‐detect levels (< 5 μg l−1) within 1 week (Fig. 1), with a doubling time of approximately 24 h at 30°C and pH = 7. After normalizing rates to the biomass concentrations, the specific degradation rates were 7.12 ± 0.18 g‐dioxane per g‐protein per day for Consortium A and 5.68 ± 0.07 g‐dioxane per g‐protein per day for Consortium B. These rates are comparable to those of two archetype dioxane degraders, Pseudonocardia dioxanivorans CB1190 (6.21 ± 0.68 g‐dioxane per g‐protein per day) and Mycobacterium dioxanotrophicus PH‐06 (7.35 ± 0.20 g‐dioxane per g‐protein per day) (Fig. 1). For both consortia, dioxane removal was coupled to bacterial growth (cell yield coefficients were 0.11 g‐protein per g‐dioxane for Consortium A and 0.09 g‐protein per g‐dioxane for Consortium B) (Fig. 1), indicating metabolic (rather than cometabolic) degradation of dioxane as sole carbon and energy source.
Figure 1.
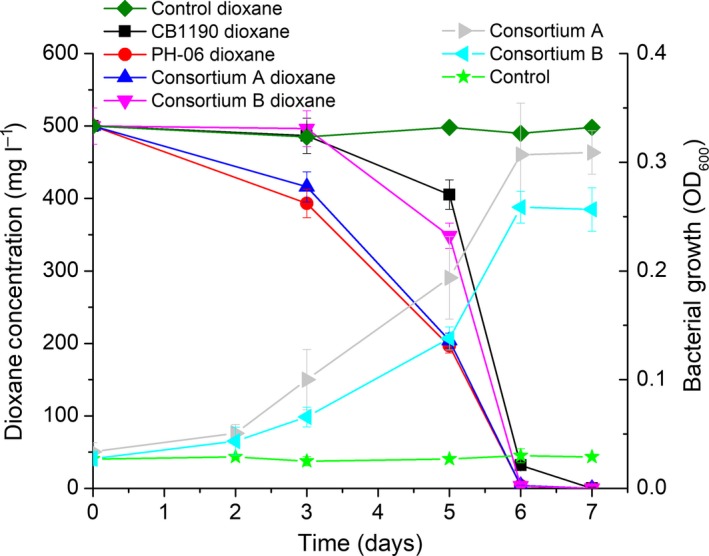
Dioxane degradation by enriched consortia and two archetypes (left) and concurrent growth of consortia (right). Experiments were conducted using AMS medium with an initial dioxane concentration of 500 mg l−1. The initial biomass densities (measured as protein content) were 25 mg l−1 for CB1190, 32 mg l−1 for PH‐06, 19 mg l−1 for Consortium A and 36 mg l−1 for Consortium B. The controls contained autoclaved bacteria.
Dioxane‐impacted groundwater from a site in Seattle, WA, that was not exhibiting intrinsic biodegradation capabilities was used to assess the potential of these consortia for bioaugmentation. Addition of both consortia (48 mg l−1 for Consortium A and 18 mg l−1 for Consortium B) resulted in dioxane removal below detection levels (< 5 μg l−1) within 3 days, even when the initial dioxane concentration was low (i.e. 300 μg l−1) (Fig. 2). This is an encouraging result because such low dioxane concentrations may represent a bioremediation challenge at impacted sites, as they might be insufficient to induce and/or sustain an active degrading indigenous population (Adamson et al., 2014; Rodriguez, 2016). The specific dioxane biodegradation rates of these two consortia (0.062 ± 0.003 g‐dioxane per g‐protein per day for Consortium A and 0.056 ± 0.003 g‐dioxane per g‐protein per day for Consortium B) are comparable with those of two archetype degraders (0.052 ± 0.002 g‐dioxane per g‐protein per day for CB1190 and 0.10 ± 0.005 g‐dioxane per g‐protein per day for PH‐06) when the initial dioxane concentration was similarly low.
Figure 2.
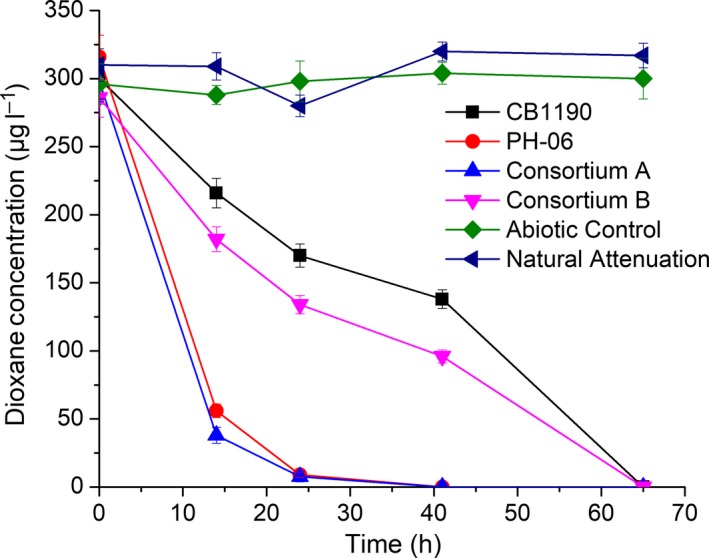
Dioxane degradation in various bioaugmented microcosms. Experiments were conducted using groundwater (initial dioxane concentration was 300 µg l−1) from a dioxane‐impacted site in Seattle, Washington, USA, T = 15°C. The added bacterial biomass measured as protein content was 20 mg l−1 for CB1190, 32 mg l−1 for PH‐06, 48 mg l−1 for Consortium A and 18 mg l−1 for Consortium B.
A key challenge to consider in bioaugmentation is the subsurface distribution of injected bacteria, which is hindered by filtration through the aquifer material – especially when the cultures aggregate. This is an important limitation for bioaugmentation with the archetype dioxane degraders CB1190 and PH‐06, which form clumps in suspension (Kim et al., 2009; Grostern et al., 2012). Interestingly, these two consortia do not clump as much as CB1190 and PH‐06 (Fig. S1), which could make them superior candidates for in situ bioaugmentation (i.e. easier perfusion through porous media) at sites where the indigenous dioxane biodegradation capacity is insufficient. Furthermore, this unprecedented enrichment of two consortia from uncontaminated soil based on their capability to grow on dioxane as sole carbon source indicates that pristine environments may harbour dioxane degraders and exhibit associated natural attenuation capacity.
Dioxane is a synthetic organic (rather than naturally occurring) compound to which the original soil biota had no known previous exposure. Thus, the observed metabolic capacity may seem counterintuitive. Nevertheless, all reported dioxane‐degrading bacteria can also degrade other cyclic ethers such as tetrahydrofuran (THF) and some naturally occurring compounds. For example, cyclic ethers containing 5‐ and 6‐membered rings are widespread in nature and are used in the biosynthesis of certain antibiotics (Rutkowski and Brzezinski, 2013; Martín et al., 2014), and the capacity to degrade these antibiotics may serve a protective role (Topp et al., 2013). Additionally, lactones, which can be intermediates of both dioxane and THF biodegradation (Bernhardt and Diekmann, 1991; Sales et al., 2013) (Fig. S2), are also commonly encountered in nature as chemical signals for bacterial communication (i.e. quorum sensing) (Keller and Surette, 2006). Many bacteria produce lactonases to degrade these signals and interrupt quorum sensing (Fig. S2), which enhances their ability to compete against the bacteria that produce them (Dong et al., 2001; Wang et al., 2004). This raises the possibility that biodegradation of simple cyclic ethers such as THF and dioxane (which are lactone precursors) may be a fortuitous result of backward evolution of catabolic pathways (Caetano‐Anollés et al., 2009). Specifically, we cannot rule out the possibility that if an organism is capable of growing on lactones that became depleted, this could impose selective pressure to transform other compounds (e.g. dioxane) into a lactone for enhanced survival.
Microbial characterization in both dioxane‐degrading consortia
Phylogenetic analysis by 16S rRNA sequences revealed a decrease in microbial diversity after enrichment on dioxane. For Consortium A, the number of genera present decreased from 288 in the original soil samples to 47 genera after enrichment. The corresponding decrease in Consortium B was from 340 to 50. The dominant genus in the original soils samples was Acidobacterium (33% in Consortium A and 30% in Consortium B) (Table 2). After enrichment, the dominant genus in both consortia is Mycobacterium (56% in Consortium A and 49% in Consortium B) (Fig. 3). Enrichment significantly changed the microbial community structure. For example, the top three genera in the original soil samples disappeared, while the top 3 genera in enriched consortia were minor populations in the original soil samples (Table 2). The observed population shift with a clear predominance of Mycobacterium spp., suggests that some species from this genus were involved in dioxane biodegradation.
Table 2.
Three most abundant genera before and after enrichment
| Consortium A | Consortium B | ||||
|---|---|---|---|---|---|
| Genus name | Percentage (%) | Genus name | Percentage (%) | ||
| Before | After | Before | After | ||
| Acidobacterium | 31.9 a | 0.00 | Acidobacterium | 29.81 | 0.00 |
| Pirellula | 3.6 | 0.00 | Chloroflexus | 6.67 | 0.00 |
| Holophaga | 3.6 | 0.00 | Dongia | 2.56 | 0.00 |
| Mycobacterium | 0.1 | 56.0 | Mycobacterium | 0.2 | 48.8 |
| Aminobacter | 0.04 | 10.5 | Chitinophaga | 0.01 | 15.5 |
| Ralstonia | 0.08 | 5.9 | Aminobacter | 0.03 | 9.2 |
Figure in bold represents significant change before and after enrichment.
Figure 3.
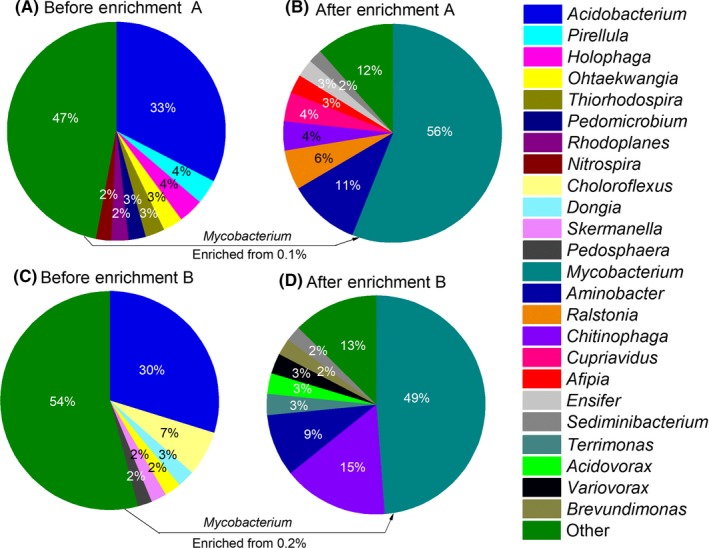
16S rRNA sequencing analyses of the two original soil samples and enriched consortia. The sum of all bacteria genera representing < 2% of total community is indicated as ‘Other’. These sequence data were submitted to the GenBank database under accession number SRP103870.
Various Mycobacterium strains capable of degrading dioxane metabolically or cometabolically have been previously isolated from environmental samples with known dioxane exposure history (Table 1). The predominance of Mycobacterium spp, in these consortia supports the notion that this is an important and commonly encountered genus of dioxane degraders. Furthermore, we show that Mycobacterium may be more prevalent in some environments than the more widely studied dioxane‐degrading actinomycetes belonging to Pseudonocardia and Rhodococcus genera.
Among other genera present that make at least 2% of these consortia (Fig. 3) (i.e. Aminobacter (10%), Ralstonia (6%), Chitinophaga (4%), Cupriavidus (4%), Afipia (3%), Ensifer (3%) and Sediminibacterium (2%) in Consortium A; Chitinophaga (16%), Aminobacter (9%), Terrimonas (3%), Acidovorax (3%), Variovorax(3%), Brevundimonas (2%) and Sediminibacterium (2%) in Consortium B), only Afipia encompasses a strain (i.e. Afipia sp. D1) that was reported to degrade dioxane as sole carbon and energy source (Sei et al., 2013b). Aminobacter and Sediminibacterium were the only genera that were enriched in both consortia, with Aminobacter reaching much higher abundance. It is still uncertain if and how the other genera identified in these consortia contribute to the biodegradation of dioxane and/or its potential by‐products. Our unsuccessful efforts to isolate dioxane‐degrading strains using the conventional enrichment and dilution to extinction approach (Xing et al., 2010) suggest that the dominant Mycobacteria and other coexisting bacterial species might constitute complex syntrophic communities.
Acetylene inhibition test indicates the involvement of monooxygenases
Acetylene, a known monooxygenase inhibitor (Prior and Dalton, 1985), was used to determine whether monooxygenases could play a role in dioxane degradation by the two consortia. When exposed to 8% (v/v) of acetylene in headspace, dioxane removal significantly decreased (by > 90% over 7 days) compared to the untreated controls (Fig. S3). These results suggest that monooxygenases played a crucial role on dioxane degradation, likely catalysing the initial biotransformation steps. This corroborates previous reports about the importance of monooxygenases in initiating the aerobic degradation of dioxane (Mahendra and Alvarez‐Cohen, 2006; Li et al., 2013b). Therefore, information on the responsible monooxygenase genes can advance understanding and assessment/ of dioxane biodegradation and natural attenuation potential at contaminated sites.
The thmA/dxmA biomarker was not detected in these two consortia
The acetylene inhibition test indicates the involvement of monooxygenases, although it is still unclear which specific monooxygenases initiated dioxane catabolism. Up to date, despite the successful isolation of numerous dioxane‐degrading bacteria in previous work (Table 1), little is known about the genetic basis for dioxane biodegradation. In fact, only one enzyme responsible for initiating dioxane metabolism has been discovered. This tetrahydrofuran monooxygenase/dioxane monooxygenase is encoded by the gene cluster thmADBC, which is located on plasmid pPSED02 in Pseudonocardia dioxanivorans CB1190 (Grostern et al., 2012; Sales et al., 2013). A primer/probe set for the thmA/dxmA biomarker designed to target the nucleotide sequence coding for the hydroxylase alpha subunit (i.e. the enzyme's active site) was shown to be an excellent indicator of dioxane degradation capacity (Li et al., 2013a), and no false positives have been observed. However, this thmA/dxmA biomarker was not detected in either of our two consortia using qPCR analysis (limit of detection = 12 copies per ng genomic DNA; 16S rRNA from the consortia and thmA/dxmA from CB1190 were used as positive controls, Fig. S4). Therefore, other monooxygenase genes were likely responsible for dioxane degradation in these consortia.
The role of soluble di‐iron monooxygenase (SDIMO) on dioxane biodegradation
A nested PCR analysis using two degenerate primers to target the hydroxylase alpha subunit of SDIMOs (Coleman et al., 2006) was performed to gain insights into which enzymes were responsible for dioxane degradation by these consortia. SDIMOs were present in both consortia as indicated by agarose gel electrophoresis (Fig. S5).
Soluble di‐iron monooxygenases are divided into six groups, based on their subunit organization and composition, substrate specificity and sequence similarity (Coleman et al., 2006), and the only sequenced dioxane monooxygenase gene cluster to date (i.e. thmADBC) belongs to group 5 (Grostern et al., 2012). To gain insight about the specific SDIMOs harboured by these two consortia, the purified products obtained from the second PCR run were sequenced and compared to genes databases (NCBI) encompassing all of the currently reported SDIMOs. The diversity of SDIMO genes in both consortia decreased compared with the original soil samples. The dominant SDIMO genes in Consortium A corresponded to a group 6 SDIMO (98.81%); while in Consortium B, SDIMO genes from both groups 5 (47.27%) and 6 (51.88%) were observed. These represent threefold to sevenfold increases, respectively, relative to the original soil samples (Fig. 4). In both consortia, the relative abundance of thmA/dxmA gene was negligible (0.03%), which is consistent with the negative amplification of these genes by qPCR (Fig. S4).
Figure 4.
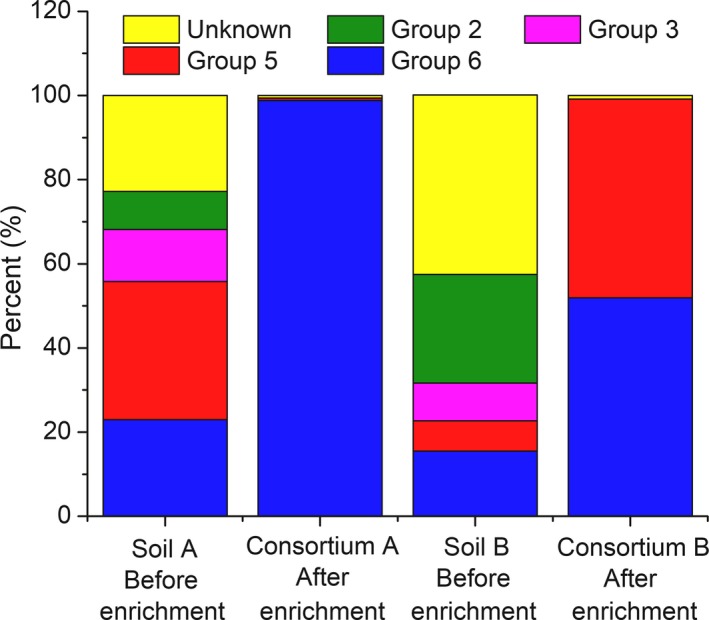
SDIMO gene sequencing of nested PCR products from the original soil samples and enriched consortia. ‘Group 2′, ‘Group 3′, ‘Group 5′ and ‘Group 6′ indicate a high similarity (> 93%) with groups 2, 3, 5 and 6 soluble di‐iron monooxygenase hydroxylase (SDIMOs) alpha subunit genes respectively. ‘Unknown’ indicates no significant similarities with any SDIMO gene sequences available on NCBI. The percentages bars represent the fractions of different SDIMO gene groups in the two consortia. These sequence data were submitted to the GenBank database under accession number SRP103872.
The prevalence of such SDIMOs is consistent with the observed enrichment of Mycobacterium spp., as previous studies have reported that groups 5 and 6 SDIMO genes are harboured by Mycobacterium spp. These include group 5 SDIMO genes in Mycobacterium semegmatis MC2 155 and Mycobacterium goodii strain 12523 (Furuya et al., 2011), and group 6 SDIMO genes in Mycobacterium chubuense NBB4 (Coleman et al., 2006, 2011), Mycobacterium sp. TY‐6 (Kotani et al., 2006) and Mycobacterium sp. ENV421 (Masuda et al., 2012). This underscores the need for further research on SDIMO genes and enzymes involved in dioxane biodegradation to develop novel biomarkers that can be useful for forensic analysis and performance assessment of bioremediation and natural attenuation at dioxane‐impacted sites.
Overall, these data suggest that dioxane catabolic capacity may be found even in some uncontaminated soils and that such bacteria can be enriched from such soils to broaden bioaugmentation applications to bioremediate sites experiencing negligible dioxane degradation potential.
Experimental procedures
Enrichment of dioxane‐degrading bacteria consortia
Two dioxane‐degrading bacteria consortia (designated as Consortium A and Consortium B) were enriched from garden soil that had no known previous exposure to dioxane. Soil samples were obtained from the Rice University campus (Houston, TX, USA) at a depth of 5 cm from the soil surface. Two samples (corresponding to the two consortia) were collected at two different spots (10 m away) at different seasons (in September 2015 and March 2016). A dark soil sample collected from an area near bushes and trees was used for the enrichment of Consortium A, while a lighter yellowish soil sample from a grassy area was used to enrich Consortium B. No dioxane was detected in these soils (detection limit < 5 μg l−1).
Soil samples (5 g of soil in 10 ml of deionized water) were vortexed vigorously on a Vortex‐Genie® 2 (MO BIO Laboratories, Inc. Carlsbad, CA, USA, at the maximum speed for 10 min, and the aqueous phase was used as the initial inoculum of the enrichment. A series of 10‐fold dilutions (Xing et al., 2010) were performed for each sample each week, using ammonium mineral salts (AMS) medium (Parales et al., 1994; Li et al., 2010) amended with 100 mg l−1 of dioxane as the sole source of carbon. Amphotericin B solution (2.5 mg l−1) (Sigma‐Aldrich, St Louis, MO, USA) was added to the medium to avoid the growth of fungus, which could confound assessment of dioxane biodegradation by bacteria. All samples were kept at 30°C while shaking on a Digital Linear Shaker (Scilogex, Rocky Hill, CT, USA) at 150 rpm.
After 12 cycles of enrichment for approximately 3 months, rapid dioxane biodegradation was sustained at relatively constant rates by both consortia. However, isolation of pure strains that metabolize dioxane was unsuccessful using agar plates prepared with 1.5% molecular grade agar and AMS amended with 100 mg l−1 of dioxane. The failure to isolate dioxane degraders was partially due to the fact that some bacteria in the enriched consortia grew on the plates using agar, rather than dioxane, as carbon source. Thus, the enriched consortia were used for further studies.
Dioxane degradation rates and bioaugmentation studies
Dioxane degradation capabilities by these consortia were benchmarked against those of two archetype degraders: Pseudonocardia dioxanivorans CB1190 (Parales et al., 1994) and Mycobacterium dioxanotrophicus PH‐06 (Kim et al., 2009; He et al., 2017). Both reference strains (CB1190 or PH‐06) and the two consortia were grown in AMS medium with dioxane as the only carbon source, and were harvested in exponential stage by centrifuging and washing with AMS medium for three times. Batch biodegradation assays were conducted in 250 ml bottles amended with 100 ml of AMS medium with an initial dioxane concentration of 500 mg l−1, while shaking similarly on a Digital Linear Shaker (Scilogex) at 150 rpm. The initial biomass concentrations (measured as protein content) were 25 mg l−1 for CB1190, 32 mg l−1 for PH‐06, 19 mg l−1 for Consortium A and 30 mg l−1 for Consortium B.
Microcosms prepared with dioxane‐impacted groundwater from a site in Seattle, WA, USA, that was not exhibiting intrinsic biodegradation capabilities. A low initial concentration of dioxane (300 μg l−1) was used to assess the potential of these consortia for bioaugmentation of contaminated sites where dioxane might be present at insufficient concentrations to induce and/or sustain an active degrading indigenous community. The concentrations of cocontaminants in the groundwater, such as chlorinated solvents and BTEX, were very low (< 6 μg l−1 for organic contaminants and < 15 mg l−1 for heavy metals) or non‐detectable (Table S1). The initial bacterial biomass concentrations in this study were 20 mg l−1 for CB1190, 35 mg l−1 for PH‐06, 48 mg l−1 for Consortium A and 18 mg l−1 for Consortium B. This study was conducted at 15°C to mimic the actual average groundwater temperature.
Dioxane and biomass analytical procedures
To measure the dioxane concentrations in the original soil samples, 5 g of soil was mixed with 10 ml of deionized water and vortexed vigorously on a Vortex‐Genie® 2 (MO BIO Laboratories, Inc.) at the maximum speed for 10 min. The extracted dioxane concentration in the aqueous phase was measured as detailed below, and the total dioxane mass contained in the 10 ml of extract was divided by the initial 5 g of soil to calculate the soil concentration.
Liquid samples were filtered through 0.22 μm syringe filters, and dioxane was extracted by the liquid/liquid frozen micro‐extraction method using dichloromethane as the solvent (Li et al., 2011). Dioxane concentrations were measured using an Agilent 7820A gas chromatograph equipped with a 5977E mass spectrum detector.
Total biomass was quantified as the total protein concentration using the Pierce™ BCA Protein Assay Kit (Thermo Fisher Scientific, Rockford, IL, USA). Serial dilutions of the bovine serum albumin (BSA) standard were prepared for calibration.
Bacterial growth on dioxane was assessed per increase in optical density at 600 nm (OD600). Even though both consortia clumped to a much lower extent than the two archetype dioxane degraders (CB1190 and PH‐06), the bacterial suspensions were not as uniform as that of other non‐aggregating bacteria. Thus, before the measurement of OD600, the bacterial cultures were put into a 1.5 ml centrifuge tube and sonicated for 10 min in a 5510 Branson Ultrasonic Cleaner (Sigma‐Aldrich) at 40 kHZ. The OD600 was then measured with a Ultrospec™ 2100 pro UV/Visible Spectrophotometer (GE Healthcare, Little Chalfont, UK).
Microbial community analyses
16S rRNA gene sequencing was used to determine the microbial community composition of the original soil samples and the enriched two consortia. DNA from the original soil samples was extracted using PowerSoil® DNA Isolation Kit (MO BIO Laboratories, Inc.). DNA from bacteria was extracted from bacteria biomass harvested in exponential growth phase, when half or more of the added dioxane (100 mg l−1) was consumed. The extraction kit was the UltraClean® Microbial DNA Isolation Kit (MO BIO, Carlsbad, CA, USA). The V4 region of the 16S rRNA gene was amplified by PCR using the forward 515F and reverse 806R primers (Caporaso et al., 2011). Sequencing was performed at MR DNA (www.mrdnalab.com, Shallowater, TX, USA) by Illumina MiSeq paired‐end sequencing (approximately 2 × 300 bp as the read length). Sequence data were processed using MR DNA analysis pipeline. Operational taxonomic units (OTUs) were defined by clustering at 3% divergence (97% similarity) (Konstantinidis and Tiedje, 2005). Final OTUs were taxonomically classified using BLASTn against the RDPII (http://rdp.cme.msu.edu) and NCBI (www.ncbi.nlm.nih.gov ) databases.
Real‐time quantitative PCR (qPCR)
qPCR analysis of the bacterial DNA was conducted to determine the presence and abundance of 16S rRNA and thmA/dxmA genes (Li et al., 2013a). 16S rRNA was used as a positive control to show amplification of genes unrelated to degradation of dioxane. qPCR mixture contained 10 ng of genomic DNA, 300 nM of forward and reverse primers, 150 nM of fluorogenic probe, 10 μl of TaqMan universal master mix II (Applied Biosystems, Foster City, CA, USA) and DNA‐free water, to a total volume of 20 μl. qPCR was performed in a CFX 96™ Real‐Time System (Bio‐Rad, Hercules, CA, USA) with the following temperature set up: 50°C for 2 min, 95°C for 10 min and 40 cycles of 95°C for 15 s and 60°C for 10 min. Serial dilutions (10−4 to 10 ng of DNA μl−1) of the extracted genomic DNA of CB1190 were used to prepare the calibration curves and determine the amplification efficiency for the tested genes.
Analysis of soluble di‐iron monooxygenase (SDIMO) genes
Previously designed degenerate primers, NVC57, NVC58, NVC65 and NVC66, to target conserved regions in the soluble di‐iron monooxygenases (SDIMO) alpha subunit gene (Coleman et al., 2006) were used to examine the presence and diversity of SDIMO genes in the original soil samples and the two consortia. A nested PCR strategy was used to increase the PCR product yield. In the first run, the PCR mixture contained 1 μl of NVC65 and NVC58 primer mixture (10 μM), 20 ng of the extracted genomic DNA, 12.5 μL of KAPA HiFi HotStart ReadyMix (2X) (KAPA Biosystems, Wilmington, MA, USA) and nuclease‐free water to yield a total volume of 25 μl. PCR was performed in a Bio‐Rad T100™ Thermal Cycler (Bio‐Rad) with the following temperature profile: initial denaturation (94°C, 5 min), then 29 amplification cycles (94°C for 30 s, 55°C for 30 s, 72°C for 1 min per kb) and a final extension (72°C for 5 min) (Coleman et al., 2006). The length of the PCR products in the first run was checked by 1% agarose gel, and DNA bands of the correct size (1100 bp) were excised and purified. Twenty nanograms of the purified PCR product was used as the DNA template in the second run, with the second set of primers (NVC57 and NVC66). The purified product (420 bp) from the second PCR was sent to MR DNA (www.mrdnalab.com, Shallowater, TX, USA) for Illumina MiSeq paired‐end sequencing (approximately 2 × 300 bp as the read length). Sequence data were processed using MR DNA analysis pipeline. Operational taxonomic units (OTUs) were defined by clustering at 3% divergence (97% similarity) (Konstantinidis and Tiedje, 2005). A database including all of the currently reported SDIMO genes on NCBI was created and used to taxonomically classify the final OTUs.
Statistical analysis
All treatments were conducted in triplicate. Statistical significance of differences between experimental treatments was assessed using two‐tailed unpaired Student's t‐test at the 95% confidence level.
Conflict of Interest
None declared.
Acknowledgements
This work was funded by SERDP (Grant # ER‐2301).
Microbial Biotechnology (2018) 11(1), 189–198
Funding Information
SERDP (Grant # ER‐2301).
References
- Adamson, D.T. , Mahendra, S. , Walker, K.L. Jr , Rauch, S.R. , Sengupta, S. , and Newell, C.J. (2014) A multisite survey to identify the scale of the 1, 4‐dioxane problem at contaminated groundwater sites. Environ Sci Technol Lett 1: 254–258. [Google Scholar]
- Aitchison, E.W. , Kelley, S.L. , Alvarez, P.J. , and Schnoor, J.L. (2000) Phytoremediation of 1, 4‐dioxane by hybrid poplar trees. Water Environ Res 72: 313–321. [Google Scholar]
- Alvarez, P. and Illman, W. (2006) Bioremediation and Natural Attenuation. Hoboken, NJ: Wiley‐Interscience. [Google Scholar]
- Barndõk, H. , Hermosilla, D. , Han, C. , Dionysiou, D.D. , Negro, C. , and Blanco, Á. (2016) Degradation of 1, 4‐dioxane from industrial wastewater by solar photocatalysis using immobilized NF‐TiO2 composite with monodisperse TiO2 nanoparticles. Appl Catal B 180: 44–52. [Google Scholar]
- Bernhardt, D. , and Diekmann, H. (1991) Degradation of dioxane, tetrahydrofuran and other cyclic ethers by an environmental Rhodococcus strain. Appl Microbiol Biotechnol 36: 120–123. [DOI] [PubMed] [Google Scholar]
- Caetano‐Anollés, G. , Yafremava, L.S. , Gee, H. , Caetano‐Anollés, D. , Kim, H.S. , and Mittenthal, J.E. (2009) The origin and evolution of modern metabolism. Int J Biochem Cell Biol 41: 285–297. [DOI] [PubMed] [Google Scholar]
- Caporaso, J.G. , Lauber, C.L. , Walters, W.A. , Berg‐Lyons, D. , Lozupone, C.A. , Turnbaugh, P.J. , et al (2011) Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc Natl Acad Sci 108: 4516–4522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, D.‐Z. , Jin, X.‐J. , Chen, J. , Ye, J.‐X. , Jiang, N.‐X. , and Chen, J.‐M. (2016) Intermediates and substrate interaction of 1, 4‐dioxane degradation by the effective metabolizer Xanthobacter flavus DT8. Int Biodeter Biodegradation 106: 133–140. [Google Scholar]
- Coleman, N.V. , Bui, N.B. , and Holmes, A.J. (2006) Soluble di‐iron monooxygenase gene diversity in soils, sediments and ethene enrichments. Environ Microbiol 8: 1228–1239. [DOI] [PubMed] [Google Scholar]
- Coleman, N.V. , Yau, S. , Wilson, N.L. , Nolan, L.M. , Migocki, M.D. , Ly, M.a. , et al (2011) Untangling the multiple monooxygenases of Mycobacterium chubuense strain NBB4, a versatile hydrocarbon degrader. Environ Microbiol Rep 3, 297–307. [DOI] [PubMed] [Google Scholar]
- Dong, Y.‐H. , Wang, L.‐H. , Xu, J.‐L. , Zhang, H.‐B. , Zhang, X.‐F. , and Zhang, L.‐H. (2001) Quenching quorum‐sensing‐dependent bacterial infection by an N‐acyl homoserine lactonase. Nature 411: 813–817. [DOI] [PubMed] [Google Scholar]
- Furuya, T. , Hirose, S. , Osanai, H. , Semba, H. , and Kino, K. (2011) Identification of the monooxygenase gene clusters responsible for the regioselective oxidation of phenol to hydroquinone in mycobacteria. Appl Environ Microbiol 77: 1214–1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grostern, A. , Sales, C.M. , Zhuang, W.‐Q. , Erbilgin, O. , and Alvarez‐Cohen, L. (2012) Glyoxylate metabolism is a key feature of the metabolic degradation of 1,4‐dioxane by Pseudonocardia dioxanivorans strain CB1190. Appl Environ Microbiol 78: 3298–3308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He, Y. , Wei, K. , Si, K. , Mathieu, J. , Li, M. and Alvarez, P.J. (2017) Whole‐genome sequence of the 1,4‐dioxane‐degrading bacterium Mycobacterium dioxanotrophicus PH‐06. Genome Announc 5: e00625‐17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, H. , Shen, D. , Li, N. , Shan, D. , Shentu, J. , and Zhou, Y. (2014) Biodegradation of 1, 4‐dioxane by a novel strain and its biodegradation pathway. Water Air Soil Pollut 225: 1–11. [Google Scholar]
- Ikehata, K. , Wang‐Staley, L. , Qu, X. and Li, Y. (2016) Treatment of groundwater contaminated with 1,4‐dioxane, tetrahydrofuran, and chlorinated volatile organic compounds using advanced oxidation processes. Ozone: Sci Eng 38, 413–424. [Google Scholar]
- Isaka, K. , Udagawa, M. , Sei, K. , and Ike, M. (2016) Pilot test of biological removal of 1, 4‐dioxane from a chemical factory wastewater by gel carrier entrapping Afipia sp. strain D1. J Hazard Mater 304: 251–258. [DOI] [PubMed] [Google Scholar]
- Kämpfer, P. , and Kroppenstedt, R.M. (2004) Pseudonocardia benzenivorans sp. nov. Int J Syst Evol Microbiol 54: 749–751. [DOI] [PubMed] [Google Scholar]
- Keller, L. , and Surette, M.G. (2006) Communication in bacteria: an ecological and evolutionary perspective. Nat Rev Microbiol 4: 249–258. [DOI] [PubMed] [Google Scholar]
- Kim, Y.‐M. , Jeon, J.‐R. , Murugesan, K. , Kim, E.‐J. , and Chang, Y.‐S. (2009) Biodegradation of 1, 4‐dioxane and transformation of related cyclic compounds by a newly isolated Mycobacterium sp. PH‐06. Biodegradation 20: 511–519. [DOI] [PubMed] [Google Scholar]
- Kohlweyer, U. , Thiemer, B. , Schräder, T. , and Andreesen, J.R. (2000) Tetrahydrofuran degradation by a newly isolated culture of Pseudonocardia sp. strain K1. FEMS Microbiol Lett 186: 301–306. [DOI] [PubMed] [Google Scholar]
- Konstantinidis, K.T. , and Tiedje, J.M. (2005) Genomic insights that advance the species definition for prokaryotes. Proc Natl Acad Sci 102: 2567–2572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotani, T. , Kawashima, Y. , Yurimoto, H. , Kato, N. , and Sakai, Y. (2006) Gene structure and regulation of alkane monooxygenases in propane‐utilizing Mycobacterium sp. TY‐6 and Pseudonocardia sp. TY‐7. J Biosci Bioeng 102: 184–192. [DOI] [PubMed] [Google Scholar]
- Kukor, J.J. , and Olsen, R.H. (1990) Molecular cloning, characterization, and regulation of a Pseudomonas pickettii PKO1 gene encoding phenol hydroxylase and expression of the gene in Pseudomonas aeruginosa PAO1c. J Bacteriol 172: 4624–4630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, M. , Fiorenza, S. , Chatham, J.R. , Mahendra, S. , and Alvarez, P.J. (2010) 1, 4‐Dioxane biodegradation at low temperatures in Arctic groundwater samples. Water Res 44: 2894–2900. [DOI] [PubMed] [Google Scholar]
- Li, M. , Conlon, P. , Fiorenza, S. , Vitale, R.J. , and Alvarez, P.J. (2011) Rapid analysis of 1,4‐dioxane in groundwater by frozen micro‐extraction with gas chromatography/mass spectrometry. Groundwater Monit Remediat 31: 70–76. [Google Scholar]
- Li, M. , Mathieu, J. , Liu, Y. , Van Orden, E.T. , Yang, Y. , Fiorenza, S. , and Alvarez, P.J. (2013a) The abundance of tetrahydrofuran/dioxane monooxygenase genes (thmA/dxmA) and 1, 4‐dioxane degradation activity are significantly correlated at various impacted aquifers. Environ Sci Technol Lett 1: 122–127. [Google Scholar]
- Li, M. , Mathieu, J. , Yang, Y. , Fiorenza, S. , Deng, Y. , He, Z. , et al (2013b) Widespread distribution of soluble di‐iron monooxygenase (SDIMO) genes in arctic groundwater impacted by 1, 4‐dioxane. Environ Sci Technol 47: 9950–9958. [DOI] [PubMed] [Google Scholar]
- Li, M. , Liu, Y. , He, Y. , Mathieu, J. , Hatton, J. , DiGuiseppi, W. , and Alvarez, P.J. (2017) Hindrance of 1, 4‐dioxane biodegradation in microcosms biostimulated with inducing or non‐inducing auxiliary substrates. Water Res 112: 217–225. [DOI] [PubMed] [Google Scholar]
- Lippincott, D. , Streger, S.H. , Schaefer, C.E. , Hinkle, J. , Stormo, J. , and Steffan, R.J. (2015) Bioaugmentation and propane biosparging for in situ biodegradation of 1, 4‐dioxane. Groundwater Monit Remediat 35: 81–92. [Google Scholar]
- Mahendra, S. , and Alvarez‐Cohen, L. (2006) Kinetics of 1, 4‐dioxane biodegradation by monooxygenase‐expressing bacteria. Environ Sci Technol 40: 5435–5442. [DOI] [PubMed] [Google Scholar]
- Martín, T. , Padrón, J.I. , and Martín, V.S. (2014) Strategies for the synthesis of cyclic ethers of marine natural products. Synlett 25: 12–32. [Google Scholar]
- Masuda, H. (2009) Identification and characterization of monooxygenase enzymes involved in 1, 4‐dioxane degradation in Pseudonocardia sp. strain ENV478, Mycobacterium sp. strain ENV421, and Nocardia sp. strain ENV425: Rutgers The State University of New Jersey‐New Brunswick and University of Medicine and Dentistry of New Jersey.
- Masuda, H. , McClay, K. , Steffan, R. , and Zylstra, G. (2012) Characterization of three propane‐inducible oxygenases in Mycobacterium sp. strain ENV421. Lett Appl Microbiol 55: 175–181. [DOI] [PubMed] [Google Scholar]
- Mohr, T.K. , Stickney, J.A. and DiGuiseppi, W.H. (2010) Environmental Investigation and Remediation: 1, 4‐dioxane and Other Solvent Stabilizers. Boca Raton, Florida, USA: CRC Press. [Google Scholar]
- Nelson, M.J. , Montgomery, S. , O'neill, E. and Pritchard, P. (1986) Aerobic metabolism of trichloroethylene by a bacterial isolate. Appl Environ Microbiol 52, 383–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Notomista, E. , Lahm, A. , Di Donato, A. , and Tramontano, A. (2003) Evolution of bacterial and archaeal multicomponent monooxygenases. J Mol Evol 56: 435–445. [DOI] [PubMed] [Google Scholar]
- Ooyama, J. , and Foster, J. (1965) Bacterial oxidation of cycloparaffinic hydrocarbons. Antonie Van Leeuwenhoek 31: 45–65. [DOI] [PubMed] [Google Scholar]
- Parales, R.E. , Adamus, J.E. , White, N. , and May, H.D. (1994) Degradation of 1,4‐dioxane by an actinomycete in pure culture. Appl Environ Microbiol 60: 4527–4530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prior, S. , and Dalton, H. (1985) Acetylene as a suicide substrate and active site probe for methane monooxygenase from Methylococcus capsulatus (Bath). FEMS Microbiol Lett 29: 105–109. [Google Scholar]
- Pugazhendi, A. , Banu, J.R. , Dhavamani, J. and Yeom, I.T. (2015) Biodegradation of 1, 4‐dioxane by Rhodanobacter AYS5 and the role of additional substrates. Ann Microbiol 65: 2201–2208. [Google Scholar]
- Rodriguez, F.J.B. (2016) Evaluation of 1, 4‐dioxane Biodegradation Under Aerobic and Anaerobic Conditions. South Carolina, USA: Clemson University. [Google Scholar]
- Rutkowski, J. and Brzezinski, B. (2013) Structures and properties of naturally occurring polyether antibiotics. Biomed Res Int 2013, (Epub ahead of print) https://dx.doi.org/10.1155/2013/162513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sales, C.M. , Mahendra, S. , Grostern, A. , Parales, R.E. , Goodwin, L.A. , Woyke, T. , et al (2011) Genome sequence of the 1, 4‐dioxane‐degrading Pseudonocardia dioxanivorans strain CB1190. J Bacteriol 193: 4549–4550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sales, C.M. , Grostern, A. , Parales, J.V. , Parales, R.E. , and Alvarez‐Cohen, L. (2013) Oxidation of the cyclic ethers 1, 4‐dioxane and tetrahydrofuran by a monooxygenase in two Pseudonocardia species. Appl Environ Microbiol 79: 7702–7708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sei, K. , Oyama, M. , Kakinoki, T. , Inoue, D. , and Ike, M. (2013a) Isolation and characterization of tetrahydrofuran‐degrading bacteria for 1, 4‐dioxane‐containing wastewater treatment by co‐metabolic degradation. J Water Environ Technol 11: 11–19. [Google Scholar]
- Sei, K. , Miyagaki, K. , Kakinoki, T. , Fukugasako, K. , Inoue, D. , and Ike, M. (2013b) Isolation and characterization of bacterial strains that have high ability to degrade 1, 4‐dioxane as a sole carbon and energy source. Biodegradation 24: 665–674. [DOI] [PubMed] [Google Scholar]
- Steffan, R.J. , McClay, K. , Vainberg, S. , Condee, C.W. , and Zhang, D. (1997) Biodegradation of the gasoline oxygenates methyl tert‐butyl ether, ethyl tert‐butyl ether, and tert‐amyl methyl ether by propane‐oxidizing bacteria. Appl Environ Microbiol 63: 4216–4222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stringfellow, W.T. , and Alvarez‐Cohen, L. (1999) Evaluating the relationship between the sorption of PAHs to bacterial biomass and biodegradation. Water Res 33: 2535–2544. [Google Scholar]
- Sun, B. , Ko, K. , and Ramsay, J.A. (2011) Biodegradation of 1, 4‐dioxane by a Flavobacterium . Biodegradation 22: 651–659. [DOI] [PubMed] [Google Scholar]
- Topp, E. , Chapman, R. , Devers‐Lamrani, M. , Hartmann, A. , Marti, R. , Martin‐Laurent, F. , et al (2013) Accelerated biodegradation of veterinary antibiotics in agricultural soil following long‐term exposure, and isolation of a sulfamethazine‐degrading sp. J Environ Qual 42: 173–178. [DOI] [PubMed] [Google Scholar]
- U.S.EPA (2014) Technical Fact Sheet – 1,4‐dioxane, EPA 505‐F‐14‐011. Seattle, WA, USA: U.S.EPA; https://www.epa.gov/sites/production/files/2014-03/documents/ffrro_factsheet_contaminant_14-dioxane_january2014_final.pdf [Google Scholar]
- Vainberg, S. , McClay, K. , Masuda, H. , Root, D. , Condee, C. , Zylstra, G.J. , and Steffan, R.J. (2006) Biodegradation of ether pollutants by Pseudonocardia sp. strain ENV478. Appl Environ Microbiol 72: 5218–5224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, L.‐H. , Weng, L.‐X. , Dong, Y.‐H. , and Zhang, L.‐H. (2004) Specificity and enzyme kinetics of the quorum‐quenching N‐acyl homoserine lactone lactonase (AHL‐lactonase). J Biol Chem 279: 13645–13651. [DOI] [PubMed] [Google Scholar]
- Whited, G.M. , and Gibson, D. (1991) Separation and partial characterization of the enzymes of the toluene‐4‐monooxygenase catabolic pathway in Pseudomonas mendocina KR1. J Bacteriol 173: 3017–3020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whittenbury, R. , Phillips, K. , and Wilkinson, J. (1970) Enrichment, isolation and some properties of methane‐utilizing bacteria. Microbiology 61: 205–218. [DOI] [PubMed] [Google Scholar]
- Xing, D. , Cheng, S. , Logan, B.E. , and Regan, J.M. (2010) Isolation of the exoelectrogenic denitrifying bacterium Comamonas denitrificans based on dilution to extinction. Appl Microbiol Biotechnol 85: 1575–1587. [DOI] [PubMed] [Google Scholar]
- Zenker, M.J. , Borden, R.C. , and Barlaz, M.A. (2003) Occurrence and treatment of 1, 4‐dioxane in aqueous environments. Environ Eng Sci 20: 423–432. [Google Scholar]


