Summary
A successful bioeconomy depends on the manifestation of biorefineries that entirely convert renewable resources to valuable products and energies. Here, the poorly exploited hemicellulose fraction (HF) from beech wood organosolv processing was applied for isobutanol production with Corynebacterium glutamicum. To enable growth of C. glutamicum on HF, we integrated genes required for d‐xylose and l‐arabinose metabolization into two of 16 systematically identified and novel chromosomal integration loci. Under aerobic conditions, this engineered strain CArXy reached growth rates up to 0.34 ± 0.02 h−1 on HF. Based on CArXy, we developed the isobutanol producer strain CIsArXy, which additionally (over)expresses genes of the native l‐valine biosynthetic and the heterologous Ehrlich pathway. CIsArXy produced 7.2 ± 0.2 mM (0.53 ± 0.02 g L−1) isobutanol on HF at a carbon molar yield of 0.31 ± 0.02 C‐mol isobutanol per C‐mol substrate (d‐xylose + l‐arabinose) in an anaerobic zero‐growth production process.
Introduction
The future shortage of fossil oil and energy resources raises the demand for a sustainable bioeconomy which mitigates greenhouse gas emissions, relies on alternative energies and exploits renewable material streams and value chains. Biorefineries play a key role in processing lignocellulosic materials (reviewed in Cherubini, 2010; Valdivia et al., 2016; Rabaçal et al., 2017) but require a high efficiency in holistically converting the input biomasses in an economic manner to marketable products and energies. Because of the complexity and variability of the lignocellulosic feed, side streams evoke from conversion technologies such as the organosolv processing or fast pyrolysis, which are tedious to exploit and therefore limit the overall efficiency of the applied biorefinery approach. With respect to their abundance, hemicelluloses, which constitute between 15% and 35% of lignocellulosic biomass (Sauer et al., 2014), have initiated much consideration for biotechnological applications (Álvarez et al., 2016). However, they are still commonly wasted (Gírio et al., 2010) due to their complexity, limited accessibility for microorganisms and potential to form toxic components (e.g. weak acids and furan derivatives).
During the organosolv processing, a mixture of lignocellulose, organic solvent (e.g. ethanol), water and catalysts (e.g. sulfuric acid) is heated to 180–210°C, which fractionizes fibres (cellulosic material) and a black liquor (containing lignin and hemicelluloses; reviewed in Brosse et al., 2017; Zhao et al., 2017). After recovery of the organic solvent by distillation, the black liquor is diluted with water to yield precipitated lignin and the remainder liquid HF (Zhao et al., 2009). Cellulosic fibres can be enzymatically saccharified and used for fermentation purposes (Zhao et al., 2017) and high purity lignin fractions for example for functionalized materials, fuels, biodegradable polymers or adhesives (Brosse et al., 2011; Liu et al., 2015). Typically, the HF comprises weak acids, sugars (e.g. d‐xylose, l‐arabinose, d‐glucose, d‐mannose, d‐galactose), furan derivatives, phenolic residues and other extractives, and was proposed to be used for fermentation and production of chemicals (e.g. xylitol, furfural) (Zhao et al., 2009). Still, due to its complexity, the HF remains difficult to access. The need for technologies that utilize the HF without further laborious treatments lies therefore at hand. Microorganisms generally possess a versatile metabolism allowing in principle the conversion of such complex substrate mixtures to value‐added products through fermentation processes.
In this study, we applied the industrial workhorse Corynebacterium glutamicum, which has a long tradition in biotechnological production of amino acids but is also exploited for the biosynthesis of organic acids, alcohols and specialty chemicals (Liebl, 2005; Becker and Wittmann, 2015). This Gram‐positive, facultatively anaerobic bacterium (Nishimura et al., 2007) is robust and accepted as suitable candidate for future biorefinery applications (Jojima et al., 2013). Previously, C. glutamicum has been engineered to produce isobutanol, a next‐generation biofuel and precursor for chemical synthesis of rubber and specialty chemicals, from glucose (Smith et al., 2010; Blombach et al., 2011; Yamamoto et al., 2013). Alternative carbon source utilization has been implemented in tailored strains (Leßmeier et al., 2015) and harnessed for production of e.g. l‐lysine from pretreated hemicellulosic materials (Gopinath et al., 2011). However, hemicelluloses such as the organosolv‐derived HF have not been assayed for isobutanol production so far. Although tools for genetic engineering, omics and systems level analysis of this industrial workhorse are available (Kirchner and Tauch, 2003; Eggeling and Bott, 2005; Wendisch et al., 2006; Burkovski, 2015; Cho et al., 2017; Lee and Wendisch, 2017), there is still a need for suitable chromosomal sites to integrate genetic information, such as synthetic operons, to expand the metabolism for enhanced substrate consumption or production purposes. This issue was the moving cause to systematically identify suitable gene integration loci in this study. We inserted synthetic operons for d‐xylose and l‐arabinose metabolization into two of these sites to enable aerobic growth and anaerobic isobutanol production on HF with engineered C. glutamicum strains.
Results and discussion
Identification of Corynebacterium glutamicum landing pads (CgLPs)
Metabolic engineering aims at enhancing the substrate or product spectrum of microorganisms, which is a crucial prerequisite to fully exploit their biotechnological potential. This essentially requires the integration of additional genetic information into the host chromosome to circumvent the inherent disadvantages of plasmid‐based gene expression. So far, no general strategy to identify suitable spots for insertion was formulated. To propose such gene integration loci (designated as C. glutamicum landing pads, CgLPs), we harnessed the knowledge about transcription units (Pfeifer‐Sancar et al., 2013), non‐essential gene clusters (Unthan et al., 2014) and prophage regions (Kalinowski, 2005). First, the three prophage regions of C. glutamicum [CGP1 (cg1507‐cg1524), CGP2 (cg1746‐cg1752) and CGP3 (cg1890‐cg2071)] were excluded from the search for relevant integration sites (Kalinowski, 2005). Although they were shown to be non‐relevant for ordinary growth under laboratory conditions, the overall function is to date not clarified in depth and a genetic stability is not guaranteed (Baumgart et al., 2013). Second, we contemplated non‐essential chromosome sections in the published list of deletable regions (Unthan et al., 2014). These provide ideal arrays for the integration of genes and exclude lethal effects that arise from disruption of essential genetic structures. Third, the non‐essential regions were analysed for suitability regarding knowledge about transcription start sites, operon structures and Rho‐independent termination sites (Pfeifer‐Sancar et al., 2013). In total, 16 landing pads were identified throughout the chromosome as suitable spots for integration of additional genetic information (cf. Table 1, Fig. S1). All CgLPs locate after a Rho‐independent terminator of the upstream gene and are succeeded by a downstream gene stop or start codon at > 50 bps spacing (Fig. 1, Table 1). The distance between the CgLP and the upstream gene terminator was chosen between 10 and 40 bps depending on the size of the intergenic region. Integration of synthetic gene constructs should in general provide a strong termination site to minimize downstream effects. Two of the identified integration loci, CgLP4 and CgLP12, were exemplarily used in this study for integration of synthetic operons for d‐xylose and l‐arabinose metabolization respectively (cf. Fig. 1, Table 1).
Table 1.
Compilation of identified C. glutamicum landing pads (CgLPs) for chromosomal integration of additional genetic information
| C. glutamicum Landing Pad | Base Positiona | Adjacenceb | Spacerc | Upstream gened | Downstream gene/operone | Experimental verification |
|---|---|---|---|---|---|---|
| CgLP1 | 97220 | ◁ ⌇ ⋂ ◄ | 20 | cg0121 | cg0120 | – |
| CgLP2 | 287966 | ► ⋂ ⌇ ◁ | 20 | cg0327 | cg0328 | – |
| CgLP3f | 558101 | ► ⋂ ⌇ ◁ | 40 | cg0634 (rplO)g | cg0635 | – |
| CgLP4 | 836158 | ► ⋂ ⌇ ▷ | 10 | cg0901 | cg0902 | xylAB |
| CgLP5 | 837445 | ► ⋂ ⌇ ▷ | 20 | cg0903 | cg0904 | – |
| CgLP6f | 857008 | ► ⋂ ⌇ ▷ | 20 | cg0928g | rrnB | – |
| CgLP7 | 1205320 | ◁ ⌇ ⋂ ◄ | 20 | cg1302 | cg1301 (cydA) | – |
| CgLP8 | 1427460 | ► ⋂ ⌇ ▷ | 40 | cg1538 (coaE)g | cg1540 | – |
| CgLP9 | 2741407 | ► ⋂ ⌇ ◁ | 40 | cg2880 | cg2883 | – |
| CgLP10f | 2971748 | ◁ ⌇ ⋂ ◄ | 40 | cg3112 (cysZ)g | cg3111 | – |
| CgLP11 | 3077633 | ► ⋂ ⌇ ▷ | 10 | cg3212 | cg3213 | yesh |
| CgLP12 | 3094266 | ► ⋂ ⌇ ▷ | 20 | cg3227 (lldD) | cg3228 | araBAD |
| CgLP13 | 3191992 | ► ⋂ ⌇ ▷ | 10 | cg3344 | cg3345 | yesh |
| CgLP14f | 3213531 | ▷ ⌇ ⋂ ◄ | 10 | cg3365 (rmpC) | cg3364 (trpA)g | – |
| CgLP15 | 3229705 | ◁ ⌇ ⋂ ◄ | 10 | cg3385 (rhcD2) | cg3384 | – |
| CgLP16 | 3248838 | ◁ ⌇ ⋂ ◄ | 40 | cg3397 | cg3396 | – |
a. Referring to the C. glutamicum ATCC 13032 complete genome NCBI reference sequence: NC_006958.1.
b. ⌇ = CgLP; ⋂ = Terminator loop; ◄, ► = upstream gene; ◁, ▷ = downstream gene; arrowheads indicate direction of adjacent genes.
c. Spacer between the predicted end of terminator site (Pfeifer‐Sancar et al., 2013) and the CgLP position.
d. Delivers the terminator site.
e. In succession of the CgLP.
f. Directly adjacent to the non‐essential gene cluster [outside location CgLP3 (80 bps), CgLP6 (39 bps), CgLP10 (342 bps), CgLP14 (123 bps)].
g. Gene outside (up‐ or downstream) the non‐essential gene cluster (Unthan et al., 2014); downstream gene is located inside the non‐essential gene cluster.
h. Were used in our laboratories and are evidentially feasible (data not shown).
Figure 1.
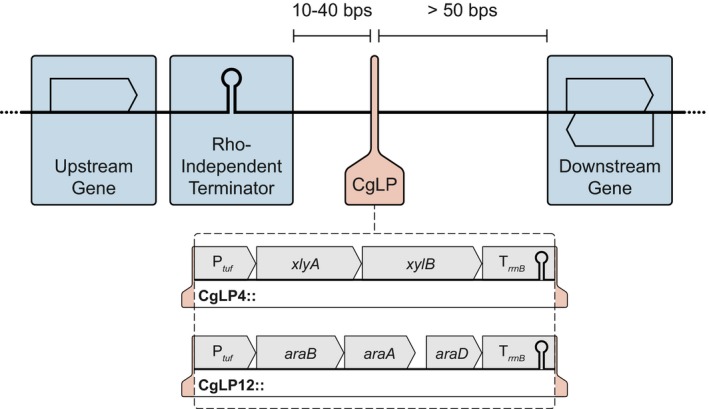
Schematic chromosomal location of C. glutamicum landing pads (CgLP) for chromosomal integration of genetic information. The synthetic operons Ptuf‐xylAB‐TrrnB and Ptuf‐araBAD‐TrrnB for d‐xylose and l‐arabinose metabolization were inserted exemplarily into CgLP4 and CgLP12 respectively. Ptuf: promoter of the C. glutamicum elongation factor EF‐TU (cg0587); TrrnB: terminator of the E. coli rrnB operon; xylAB: genes encoding XylA (xylose isomerase) of Xanthomonas campestris and XylB (xylulokinase) of C. glutamicum; araBAD: encoding AraB (l‐ribulokinase), AraA (l‐arabinose isomerase) and AraD (l‐ribulose‐5‐phosphate 4‐epimerase) of E. coli MG1655. Arrows indicate gene direction.
d ‐Xylose and l ‐arabinose metabolization in CArXy
To enable growth of C. glutamicum on d‐xylose and l‐arabinose as abundant components of the organosolv‐derived hemicellulose fraction, we integrated the synthetic operons Ptuf ‐xylAB‐TrrnB and Ptuf‐araBAD‐TrrnB into CgLP4 and CgLP12 respectively, yielding the strain CArXy (C. glutamicum Δpqo ΔilvE ΔldhA Δmdh CgLP4::(Ptuf‐xylAB‐TrrnB) CgLP12::(Ptuf‐araBAD‐TrrnB); cf. Fig. 1). Cloning, isolation and purification of plasmids, PCR fragments or genomic DNA, and procedures for strain construction are given in the Appendix S1, where a detailed list of the applied bacterial strains, plasmids and oligonucleotides (cf. Table S1) is also provided. In brief, the integration of both synthetic operons into the chromosome harnessed a previously published method (Schäfer et al., 1994) for plasmid‐based (pK19mobsacB) gene disruption and allelic exchange by homologous recombination. We designed homologous flanking regions of > 500 bps to specifically locate the additional genetic information to designated CgLPs. The two synthetic operons express the xylAB genes encoding XylA (xylose isomerase) of Xanthomonas campestris and XylB (xylulokinase) of C. glutamicum and araBAD encoding AraB (l‐ribulokinase), AraA (l‐arabinose isomerase) and AraD (l‐ribulose‐5‐phosphate 4‐epimerase) of E. coli MG1655 under control of the constitutive promoter of the C. glutamicum elongation factor EF‐TU (cg0587, Ptuf) and are terminated by the E. coli rrnB operon terminator (TrrnB) respectively, following already published operon architectures (Schneider et al., 2011; Meiswinkel et al., 2013).
First, we characterized growth of C. glutamicum CArXy in shaking flask cultivations for single and combined metabolization of d‐glucose, d‐xylose and l‐arabinose. CArXy reached a growth rate (μ) of 0.39 ± 0.03 h−1, a biomass/substrate yield (YX/S) of 0.52 ± 0.02 g CDW per g d‐glucose and showed a biomass‐specific uptake rate (qS) of 4.18 ± 0.16 mmol d‐glucose per g CDW per h (cf. Fig. 2A). All growth parameters were identical to previously described values (Buchholz et al., 2014) for the wild type of C. glutamicum and indicate that integration of both synthetic operons does not negatively interfere with the strain's vitality under standard cultivation conditions. Furthermore, C. glutamicum CArXy grew on d‐xylose and l‐arabinose with rates of 0.18 ± 0.02 h−1 and 0.16 ± 0.01 h−1, respectively (cf. Fig. 2B, C). Previous studies using plasmid‐based expression of araBAD (Schneider et al., 2011) or xylAB (Meiswinkel et al., 2013) yielded maximal rates of 0.31 h−1 or 0.20 h−1 respectively. In our experiments, a full consumption of the pentoses was not achieved at the end of cultivation (78 ± 7% of d‐xylose and 14 ± 4% of l‐arabinose metabolized). Poor l‐arabinose uptake can be explained by a high Monod constant (Schneider et al., 2011) and could be overcome by additional expression of the transporter araE, which was shown to also improve d‐xylose consumption (Sasaki et al., 2009). Combined supplementation of d‐glucose, d‐xylose and l‐arabinose showed a clear preference for the consumption of the hexose compared to the pentoses (cf. Fig. 2D), a fact that has been described previously for C. glutamicum (e.g. Kawaguchi et al., 2008; Radek et al., 2014). In contrast to the isomerase pathway, the Weimberg pathway enables a more carbon efficient utilization of d‐xylose and allows a parallel consumption of d‐xylose and d‐glucose in C. glutamicum (Radek et al., 2014, 2016). However, the maximal net generated biomass (4.7 ± 0.4 g CDW L−1) was doubled with respect to sole d‐glucose (2.2 ± 0.1 g CDW L−1), and the higher cell density allowed a full consumption of d‐xylose and 80% of l‐arabinose within the given cultivation time (cf. Fig. 2D).
Figure 2.
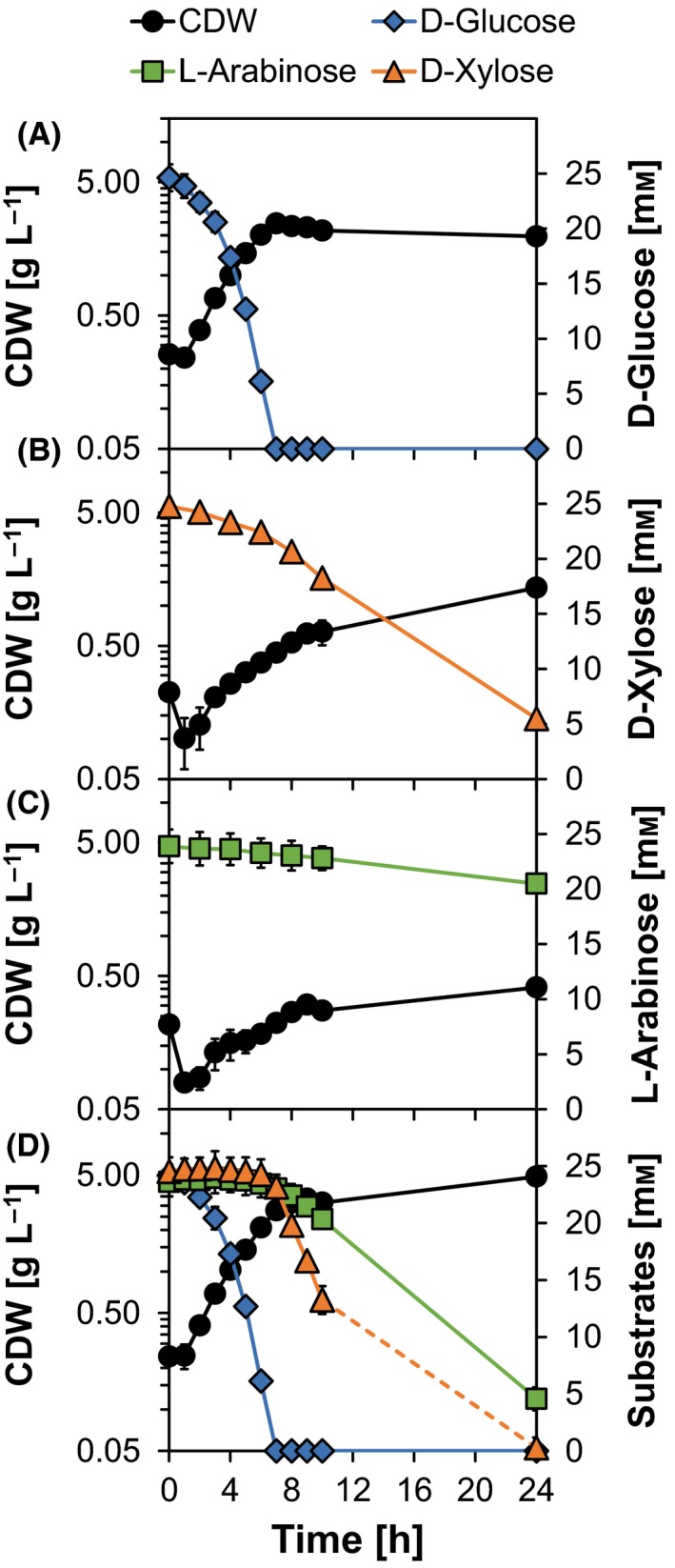
Shaking flask cultivations of the strain CArXy (C. glutamicum Δpqo ΔilvE ΔldhA Δmdh CgLP4::(Ptuf ‐xylAB‐TrrnB) CgLP12::(Ptuf ‐araBAD‐TrrnB)) in a modified CGXII minimal medium based on the literature (Eikmanns et al., 1991; Keilhauer et al., 1993) with either combined or single supplementation of 25 mM d‐glucose, d‐xylose and l‐arabinose. Bacterial growth (cell dry weight, CDW) and substrate consumption are depicted over time. Cultivations were performed in 50 ml medium in 500 ml baffled shaking flasks on a rotary shaker at 120 rpm and 30 °C. Detailed information concerning strain construction, medium, seed train and cultivation conditions is given in the Appendix S1. Error bars represent the standard deviation (SD) of three independent experiments.
In summary, the strain CArXy functionally expresses the synthetic operons in the identified CgLPs enabling d‐xylose and l‐arabinose metabolization without negatively influencing the cell's general viability under given conditions.
Aerobic growth on the hemicellulose fraction
The aqueous hemicellulose fraction (HF) was derived from a beech wood ethanol/water organosolv processing after lignin precipitation (without enzymatic hydrolysis and further purification procedures) as a black liquor with high viscosity (Ludwig et al., 2014). A description of the short pretreatment procedure extracting water‐soluble compounds is given in the Appendix S1. To investigate aerobic growth of C. glutamicum CArXy (cf. Table S1) on the HF, shaking flask cultivations were performed (cf. Fig. 3A, B, Fig. S2). In contrast to previous studies, in which engineered C. glutamicum was shown to proliferate on aci d‐pretreated lignocelluloses such as rice straw and wheat bran in minimal medium (Gopinath et al., 2011), growth in the presence of organosolv‐derived HF was only manifested upon additional supplementation of 5 g of yeast extract (YE) L−1 (data not shown) and was therefore included in all following experiments. In minimal medium with 5 g YE L−1 and 9.7 g HF L−1, 19.3 g HF L−1 or 38.7 g HF L−1 combined with 5 g YE L−1, CArXy showed growth rates of 0.14 ± 0.03 h−1, 0.34 ± 0.02 h−1, 0.33 ± 0.01 h−1 and 0.17 ± 0.02 h−1 and maximal net generated biomasses of 0.29 ± 0.06, 1.02 ± 0.15, 1.50 ± 0.11 and 2.19 ± 0.41 g CDW L−1 respectively. A consecutive consumption of acetate and the pentoses d‐xylose and l‐arabinose was found, and the depletion of acetate coincided with an arrest of the exponential growth phase (cf. Fig. 3A, B, Fig. S2).
Figure 3.
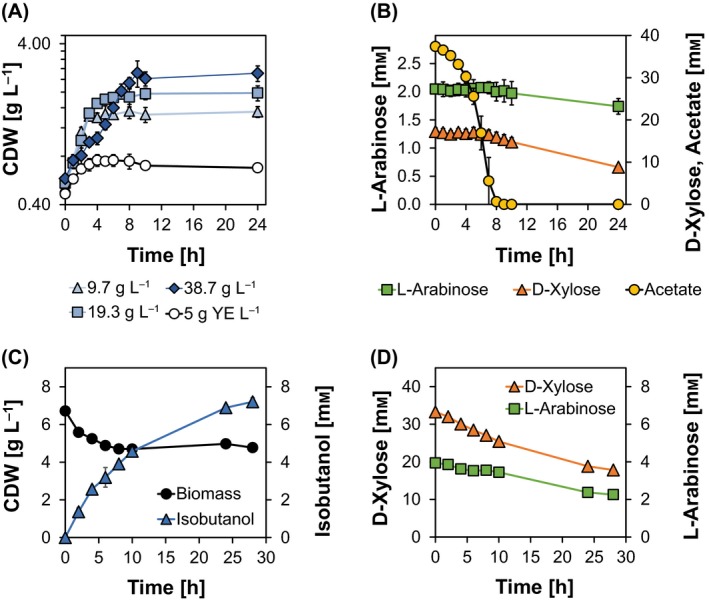
Aerobic cultivation (A, B) of the strain CArXy (C. glutamicum Δpqo ΔilvE ΔldhA Δmdh CgLP4::(Ptuf‐xylAB‐TrrnB) CgLP12::(Ptuf‐araBAD‐TrrnB)) and anaerobic isobutanol production (C, D) with CIsArXy (CArXy harbouring pJC4ilvBNCD‐pntAB and pBB1kivd‐adhA) using the hemicellulose fraction (HF). A. CArXy was cultivated in CGXII minimal medium supplemented with 5 g YE L−1 as reference (open circles) and variable concentrations of hemicellulose fraction (HF) [9.7 g HF L−1 (triangles), 19.3 g HF L−1 (squares) and 38.7 g HF L−1 (diamonds)] + 5 g YE L−1. B. Consumption of acetate (circles), d‐xylose (triangles) and l‐arabinose (squares) is depicted for the respective experiment using 38.7 g HF L−1. C. Zero‐growth isobutanol production was realized with the strain CIsArXy using 77.3 g HF L−1 + 5 g YE L−1 in sealed 100 ml flasks containing 50 mL CGXII medium. D. Metabolization of d‐xylose and l‐arabinose during the incubation is shown. Error bars represent SD of three independent experiments. Detailed information concerning strain construction, medium, seed train and cultivation conditions is given in the Appendix S1.
Although substrate consumption is still improvable, we show the capability of C. glutamicum to grow efficiently on HF which in general opens the opportunity to exploit this biorefinery side stream for microbial production of chemicals and fuels.
Two‐stage isobutanol production
To prove our concept, we aimed to utilize HF for the production of isobutanol under anaerobic conditions. Therefore, we transformed CArXy with the plasmids pJC4ilvBNCD‐pntAB and pBB1kivd‐adhA, which enabled isobutanol production in C. glutamicum (cf. Table S1, Blombach et al., 2011). Then, the resulting strain CIsArXy was applied in a zero‐growth production processes (Lange et al., 2016), where an aerobic stage was implemented to generate biomass that is used in a subsequent anaerobic, growth‐arrested phase to produce isobutanol at high cell densities (cf. Fig. 3C, D). Under anaerobic conditions, we observed a simultaneous metabolization of d‐xylose and l‐arabinose (cf. Fig. 3D, acetate was not consumed cf. Fig. S3), which directly served as substrate for isobutanol production (cf. Fig. 3C). No significant production of lactate or succinate (< 0.4 mM) was found. About 15.5 ± 0.6 mM (46 ± 1%) and 1.7 ± 0.0 mM (43 ± 1%) of d‐xylose and l‐arabinose were metabolized respectively, and CIsArXy produced 7.2 ± 0.2 mM of isobutanol within 28 h of cultivation. With respect to the analysed pentoses, a carbon molar product/substrate yield (YP/S) of 0.31 ± 0.02 C‐mol isobutanol per C‐mol substrate (d‐xylose + l‐arabinose) was achieved, which is already in the range of d‐glucose‐based processes with engineered C. glutamicum strains (0.15–0.52 C‐mol C‐mol−1; Blombach et al., 2011; Smith et al., 2010; Yamamoto et al., 2013). Isobutanol production based on the pentoses d‐xylose and l‐arabinose has so far not been demonstrated and therefore represents a promising example for the valorization of HF within a novel value chain. As a future perspective, a dual‐phase process (Lange et al., 2016) is apparent, where an aerobic growth based on acetate within the HF would be directly followed by an anaerobic isobutanol production phase based on the remaining pentoses.
Conclusions
In the presented study, we systematically identified 16 landing pads, which represent prominent loci for chromosomal integration of additional genetic information in C. glutamicum. As a proof of concept, we integrated synthetic operons into two CgLPs that enabled growth on d‐xylose and l‐arabinose as well as on a so far unexploited hemicellulose fraction derived from beech wood organosolv processing. For the first time, we showed isobutanol production with engineered C. glutamicum based on pentoses within this fraction. The work demonstrates the suitability to microbially convert complex side streams to valuable products, enabling a holistic exploitation of renewable resources in biorefinery approaches. Moreover, the proposed chromosomal integration loci can be prospectively used as basis for metabolic engineering in future studies.
Conflict of interest
None declared.
Supporting information
Appendix S1. Material and Methods.
Fig. S1. Novel proposed C. glutamicum landing pads (red, CgLPs) located in the genome of C. glutamicum ATCC 13032 (NCBI reference sequence NC_006958.1).
Fig. S2. Aerobic cultivation of the strain CArXy (C. glutamicum Δpqo ΔilvE ΔldhA Δmdh CgLP4::(Ptuf ‐xylAB‐TrrnB) CgLP12::(Ptuf ‐araBAD‐TrrnB)) in CGXII minimal medium supplemented with 5 g yeast extract (YE) L−1 as reference (open circles) and variable concentrations of hemicellulose fraction (HF, circles) [9.7 g HF L−1 (A), 19.3 g HF L−1 (B) and 38.7 g HF L−1 (C)] + 5 g YE L−1.
Fig. S3. Course of acetate concentration during the anaerobic isobutanol production with the strain CIsArXy (CArXy harboring pJC4ilvBNCD‐pntAB and pBB1kivd‐adhA) using the HF (cf. Fig. 3). Error bars represent SD of three independent experiments.
Table S1. List of bacterial strains, plasmids and oligonucleotides.
Acknowledgements
We thank Mira Lenfers‐Lücker (Institute of Biochemical Engineering, University of Stuttgart, Germany) for assistance during HPLC analysis and Dr.‐Ing. Susanne Zibek (Fraunhofer Institute for Interfacial Engineering and Biotechnology, Stuttgart, Germany) for providing of the hemicellulose fraction.
Microbial Biotechnology (2018) 11(1), 257–263
Funding information
This work was supported by a grant from the Ministry of Science, Research and the Arts of Baden‐Württemberg (Az: 33‐7533‐10‐5/84/1).
References
- Álvarez, C. , Reyes‐Sosa, F.M. , and Díez, B. (2016) Enzymatic hydrolysis of biomass from wood. Microb Biotechnol 9: 149–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumgart, M. , Unthan, S. , Rückert, C. , Sivalingam, J. , Grünberger, A. , Kalinowski, J. , et al (2013) Construction of a prophage‐free variant of Corynebacterium glutamicum ATCC 13032 for use as a platform strain for basic research and industrial biotechnology. Appl Environ Microbiol 79: 6006–6015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker, J. and Wittmann, C. (2015) Advanced biotechnology: metabolically engineered cells for the bio‐based production of chemicals and fuels, materials, and health‐care products. Angew Chem Int Ed 54, 3328–3350. [DOI] [PubMed] [Google Scholar]
- Blombach, B. , Riester, T. , Wieschalka, S. , Ziert, C. , Youn, J.‐W. , Wendisch, V.F. , and Eikmanns, B.J. (2011) Corynebacterium glutamicum tailored for efficient isobutanol production. Appl Environ Microbiol 77: 3300–3310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brosse, N. , Mohamad Ibrahim, M.N. , and Abdul Rahim, A. (2011) Biomass to bioethanol: initiatives of the future for lignin. ISRN Mater Sci 2011: 1–10. [Google Scholar]
- Brosse, N. , Hussin, M.H. and Rahim, A.A. (2017) Organosolv processes. Adv Biochem Eng Biotechnol https://doi.org/10.1007/10_2016_61. [DOI] [PubMed] [Google Scholar]
- Buchholz, J. , Graf, M. , Freund, A. , Busche, T. , Kalinowski, J. , Blombach, B. , and Takors, R. (2014) CO2/HCO3 − perturbations of simulated large scale gradients in a scale‐down device cause fast transcriptional responses in Corynebacterium glutamicum . Appl Microbiol Biotechnol 98: 8563–8572. [DOI] [PubMed] [Google Scholar]
- Burkovski, A. (2015) Corynebacterium glutamicum: From Systems Biology to Biotechnological Applications, 1st edn Norfolk, UK: Caister Academic Press. [Google Scholar]
- Cherubini, F. (2010) The biorefinery concept: using biomass instead of oil for producing energy and chemicals. Energy Convers Manag 51: 1412–1421. [Google Scholar]
- Cho, J.S. , Choi, K.R. , Prabowo, C.P.S. , Shin, J.H. , Yang, D. , Jang, J. , and Lee, S.Y. (2017) CRISPR/Cas9‐coupled recombineering for metabolic engineering of Corynebacterium glutamicum . Metab Eng 42: 157–167. [DOI] [PubMed] [Google Scholar]
- Eggeling, L. , and Bott, M. (2005) Handbook of Corynebacterium glutamicum, 1st edn Boca Raton, FL: CRC Press. [Google Scholar]
- Eikmanns, B.J. , Metzger, M. , Reinscheid, D. , Kircher, M. , and Sahm, H. (1991) Amplification of three threonine biosynthesis genes in Corynebacterium glutamicum and its influence on carbon flux in different strains. Appl Microbiol Biotechnol 34: 617–622. [DOI] [PubMed] [Google Scholar]
- Gírio, F.M. , Fonseca, C. , Carvalheiro, F. , Duarte, L.C. , Marques, S. , and Bogel‐Łukasik, R. (2010) Hemicelluloses for fuel ethanol: a review. Bioresour Technol 101: 4775–4800. [DOI] [PubMed] [Google Scholar]
- Gopinath, V. , Meiswinkel, T.M. , Wendisch, V.F. , and Nampoothiri, K.M. (2011) Amino acid production from rice straw and wheat bran hydrolysates by recombinant pentose‐utilizing Corynebacterium glutamicum . Appl Microbiol Biotechnol 92: 985–996. [DOI] [PubMed] [Google Scholar]
- Jojima, T. , Inui, M. , and Yukawa, H. (2013) Biorefinery applications of Corynebacterium glutamicum In Corynebacterium glutamicum: Biology and Biotechnology. Yukawa H., and Inui M. (eds). Berlin Heidelberg: Springer, pp. 149–172. [Google Scholar]
- Kalinowski, J. (2005) The genomes of amino acid‐producing Corynebacteria In Handbook of Corynebacterium glutamicum. Eggeling L., and Bott M. (eds). Boca Raton, FL: CRC Press, pp. 37–56. [Google Scholar]
- Kawaguchi, H. , Sasaki, M. , Vertès, A.A. , Inui, M. , and Yukawa, H. (2008) Engineering of an L‐arabinose metabolic pathway in Corynebacterium glutamicum . Appl Microbiol Biotechnol 77: 1053–1062. [DOI] [PubMed] [Google Scholar]
- Keilhauer, C. , Eggeling, L. , and Sahm, H. (1993) Isoleucine synthesis in Corynebacterium glutamicum: molecular analysis of the ilvB‐ilvN‐ilvC operon. J Bacteriol 175: 5595–5603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirchner, O. , and Tauch, A. (2003) Tools for genetic engineering in the amino acid‐producing bacterium Corynebacterium glutamicum . J Biotechnol 104: 287–299. [DOI] [PubMed] [Google Scholar]
- Lange, J. , Takors, R. , and Blombach, B. (2016) Zero‐growth bioprocesses – a challenge for microbial production strains and bioprocess engineering. Eng Life Sci 17: 1–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, J.‐H. and Wendisch, V.F. (2017) Production of amino acids – genetic and metabolic engineering approaches. Bioresour Technol 245: 1575–1587. [DOI] [PubMed] [Google Scholar]
- Leßmeier, L. , Zahoor, A. , Lindner, S.N. , and Wendisch, V.F. (2015) Metabolic engineering of Corynebacterium glutamicum for alternative carbon source utilization In Corynebacterium glutamicum: From Systems Biology to Biotechnological Applications. Burkovski A. (ed). Norfolk, UK: Caister Academic Press, pp. 57–70. [Google Scholar]
- Liebl, W. (2005) Corynebacterium taxonomy In Handbook of Corynebacterium glutamicum. Eggeling L., and Bott M. (eds). Boca Raton, FL: CRC Press, pp. 9–34. [Google Scholar]
- Liu, W.‐J. , Jiang, H. , and Yu, H.‐Q. (2015) Thermochemical conversion of lignin to functional materials: a review and future directions. Green Chem 17: 4888–4907. [Google Scholar]
- Ludwig, D. , Michael, B. , Hirth, T. , Rupp, S. , and Zibek, S. (2014) High solids enzymatic hydrolysis of pretreated lignocellulosic materials with a powerful stirrer concept. Appl Biochem Biotechnol 172: 1699–1713. [DOI] [PubMed] [Google Scholar]
- Meiswinkel, T.M. , Gopinath, V. , Lindner, S.N. , Nampoothiri, K.M. , and Wendisch, V.F. (2013) Accelerated pentose utilization by Corynebacterium glutamicum for accelerated production of lysine, glutamate, ornithine and putrescine. Microb Biotechnol 6: 131–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura, T. , Vertès, A.A. , Shinoda, Y. , Inui, M. , and Yukawa, H. (2007) Anaerobic growth of Corynebacterium glutamicum using nitrate as a terminal electron acceptor. Appl Microbiol Biotechnol 75: 889–897. [DOI] [PubMed] [Google Scholar]
- Pfeifer‐Sancar, K. , Mentz, A. , Rückert, C. , and Kalinowski, J. (2013) Comprehensive analysis of the Corynebacterium glutamicum transcriptome using an improved RNAseq technique. BMC Genom 14: 888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabaçal, M. , Ferreira, A.F. , Silva, C.A.M. , and Costa, M. (2017) Biorefineries – Targeting Energy, High Value Products and Waste Valorisation, 1st edn Cham: Springer International Publishing. [Google Scholar]
- Radek, A. , Krumbach, K. , Gätgens, J. , Wendisch, V. , Wiechert, W. , Bott, M. , et al (2014) Engineering of Corynebacterium glutamicum for minimized carbon loss during utilization of D‐xylose containing substrates. J Biotechnol 192: 156–160. [DOI] [PubMed] [Google Scholar]
- Radek, A. , Müller, M.‐F. , Gätgens, J. , Eggeling, L. , Krumbach, K. , Marienhagen, J. , and Noack, S. (2016) Formation of xylitol and xylitol‐5‐phosphate and its impact on growth of D‐xylose‐utilizing Corynebacterium glutamicum strains. J Biotechnol 231: 160–166. [DOI] [PubMed] [Google Scholar]
- Sasaki, M. , Jojima, T. , Kawaguchi, H. , Inui, M. , and Yukawa, H. (2009) Engineering of pentose transport in Corynebacterium glutamicum to improve simultaneous utilization of mixed sugars. Appl Microbiol Biotechnol 85: 105–115. [DOI] [PubMed] [Google Scholar]
- Sauer, M. , Steiger, M. , and Mattanovich, D. (2014) Biorefineries‐concepts for sustainability In Bioprocessing of Renewable Resources to Commodity Bioproducts. Bisaria V.S., and Kondo A. (eds). Hoboken, NJ: John Wiley & Sons Inc, pp. 3–27. [Google Scholar]
- Schäfer, A. , Tauch, A. , Jäger, W. , Kalinowski, J. , Thierbach, G. , and Pühler, A. (1994) Small mobilizable multi‐purpose cloning vectors derived from the Escherichia coli plasmids pK18 and pK19: selection of defined deletions in the chromosome of Corynebacterium glutamicum . Gene 145: 69–73. [DOI] [PubMed] [Google Scholar]
- Schneider, J. , Niermann, K. , and Wendisch, V.F. (2011) Production of the amino acids L‐glutamate, L‐lysine, L‐ornithine and L‐arginine from arabinose by recombinant Corynebacterium glutamicum . J Biotechnol 154: 191–198. [DOI] [PubMed] [Google Scholar]
- Smith, K.M. , Cho, K.‐M. , and Liao, J.C. (2010) Engineering Corynebacterium glutamicum for isobutanol production. Appl Microbiol Biotechnol 87: 1045–1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unthan, S. , Baumgart, M. , Radek, A. , Herbst, M. , Siebert, D. , Brühl, N. , et al (2014) Chassis organism from Corynebacterium glutamicum – a top‐down approach to identify and delete irrelevant gene clusters. Biotechnol J 10: 290–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valdivia, M. , Galan, J.L. , Laffarga, J. , and Ramos, J.‐L. (2016) Biofuels 2020: biorefineries based on lignocellulosic materials. Microb Biotechnol 9: 585–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wendisch, V.F. , Bott, M. , Kalinowski, J. , Oldiges, M. , and Wiechert, W. (2006) Emerging Corynebacterium glutamicum systems biology. J Biotechnol 124: 74–92. [DOI] [PubMed] [Google Scholar]
- Yamamoto, S. , Suda, M. , Niimi, S. , Inui, M. , and Yukawa, H. (2013) Strain optimization for efficient isobutanol production using Corynebacterium glutamicum under oxygen deprivation. Biotechnol Bioeng 110: 2938–2948. [DOI] [PubMed] [Google Scholar]
- Zhao, X. , Cheng, K. , and Liu, D. (2009) Organosolv pretreatment of lignocellulosic biomass for enzymatic hydrolysis. Appl Microbiol Biotechnol 82: 815–827. [DOI] [PubMed] [Google Scholar]
- Zhao, X. , Li, S. , Wu, R. , and Liu, D. (2017) Organosolv fractionating pre‐treatment of lignocellulosic biomass for efficient enzymatic saccharification: chemistry, kinetics, and substrate structures. Biofuels Bioprod Biorefining 11: 567–590. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix S1. Material and Methods.
Fig. S1. Novel proposed C. glutamicum landing pads (red, CgLPs) located in the genome of C. glutamicum ATCC 13032 (NCBI reference sequence NC_006958.1).
Fig. S2. Aerobic cultivation of the strain CArXy (C. glutamicum Δpqo ΔilvE ΔldhA Δmdh CgLP4::(Ptuf ‐xylAB‐TrrnB) CgLP12::(Ptuf ‐araBAD‐TrrnB)) in CGXII minimal medium supplemented with 5 g yeast extract (YE) L−1 as reference (open circles) and variable concentrations of hemicellulose fraction (HF, circles) [9.7 g HF L−1 (A), 19.3 g HF L−1 (B) and 38.7 g HF L−1 (C)] + 5 g YE L−1.
Fig. S3. Course of acetate concentration during the anaerobic isobutanol production with the strain CIsArXy (CArXy harboring pJC4ilvBNCD‐pntAB and pBB1kivd‐adhA) using the HF (cf. Fig. 3). Error bars represent SD of three independent experiments.
Table S1. List of bacterial strains, plasmids and oligonucleotides.


