Abstract
Human genome variation may cause differences in traits and disease risks. Disease-causal/susceptible genes and variants for both common and rare diseases can be detected by comprehensive whole-genome analyses, such as whole-genome sequencing (WGS), using next-generation sequencing (NGS) technology and genome-wide association studies (GWAS). Here, in addition to the application of an NGS as a whole-genome analysis method, we summarize approaches for the identification of functional disease-causal/susceptible variants from abundant genetic variants in the human genome and methods for evaluating their functional effects in human diseases, using an NGS and in silico and in vitro functional analyses. We also discuss the clinical applications of the functional disease causal/susceptible variants to personalized medicine.
Keywords: next-generation sequencing (NGS) technology, whole-genome sequencing (WGS), whole-exome sequencing (WES), genome-wide association study (GWAS), functional disease-causal/susceptible variants
Introduction
The concordance rate of human genome DNA sequences among individuals is reported to be 99.5%.1) The differences in human genomes are attributed to genetic variations such as single-nucleotide polymorphisms (SNPs), short insertions/deletions (in/dels), short tandem repeats (STR), copy number variations (CNV), etc. Any of these variations may cause differences in traits and disease risk.
At the end of 20th century, the primary comprehensive investigative method for disease causal genes or variants was linkage analysis using genetic markers such as microsatellites or restriction fragment length polymorphisms. Disease causal genes or variants for some rare diseases that affect a small percentage of the population, e.g., Fukuyama muscular dystrophy, could be identified using this method.2) From early in the first decade of the 2000s, the primary comprehensive method of investigation of disease susceptibility genes, especially for common diseases whose prevalence is relatively high, has been genome-wide association studies (GWASs). GWASs involve statistical-genetics methods that identify disease-susceptibility loci by performing case–control association studies that simultaneously analyze 0.5–5 million SNPs (“tag SNPs”) in samples from affected patients and controls using a commercially available DNA array. As of the present day, tens of thousands of genetic variants that are associated with susceptibility to common human diseases and traits have been identified by GWAS.3) Up to November 2016, 2,610 studies and 29,382 unique SNP-trait associations have been registered in the National Human Genome Research Institute Catalog of published GWAS (http://www.ebi.ac.uk/gwas/). However, these variants often explain only relatively small proportions of the heritability of the associated diseases, which has led to the concept of “missing heritability”.4)
From around 2005, novel and large-scale DNA sequencing methods, termed next-generation sequencing (NGS) technologies, were developed by the combination of advanced laboratory methodologies and bioinformatics tools. The use of NGS is currently providing breakthroughs in many biomedical research areas including human genetics. In particular, NGS methods has become indispensable for the identification of disease-causal/susceptible variants from abundant genetic variants in the human genome.
In this review, we discuss the application of NGS, approaches for the identification and evaluation of functional disease-causal/susceptible variants, and the clinical applications of these variants to personalized medicine.
1. Applications of next-generation sequencing to whole-genome analyses
In 2001, the first draft sequences that provided an overall view of the human genome, which were obtained using an automated Sanger sequencing method, were reported by the International Human Genome Sequencing Consortium (IHGSC) and Celera Genomics.5,6) Subsequently, the next challenge was the sequencing of individual, personal genomes. From around 2010, NGS had developed to the point where it allows the investigation of whole genomes or transcriptomes in a relatively short period of time.7,8) To date, the sequencing of individual human genomes is enhancing the understanding of how genetic differences affect human traits and diseases. In this chapter, the applications of NGS are discussed, especially those in human whole-genome analyses.
Whole genome sequencing and whole exome sequencing.
Using NGS, whole genome sequencing (WGS), which determines the entire genomic DNA sequence of an individual at a single time, and whole exome sequencing (WES), which is a method for sequencing of all of the exons of expressed human genes, have made strong contributions to clarifying the complexity of human genome variation.
From 2008, WGS using massive parallel sequencing technologies has provided extensive information regarding genetic variants.9–11) A Japanese research group has also reported the first WGS analysis of Japanese individuals. Their analysis identified 3,132,608 single nucleotide variations (SNVs), 5,319 deletions smaller than 10 kb, CNVs, and rearrangements with high accuracy.12) As of the end of 2016, WGS data have been acquired using NGS from tens of thousands of individuals, including both healthy individuals and patients around the world, and have been subsequently analyzed.
Compared with WGS, WES is currently the more popular platform for the discovery of rare disease causal genes or variants, because WES often detects variants that lead to amino acid changes in proteins and can efficiently identify such causal genes or variants without the complexity and high cost associated with WGS. The first WES analyses reported the successful identification of disease causal variants for specific rare diseases.13–15) Subsequently, since 2014, 228 novel rare disease causal genes have been discovered using WES analyses,16) and the number of novel rare disease causal genes identified by WES is now increasing exponentially.
Large scale human genome sequencing projects.
Several large-scale sequencing projects using NGS are ongoing. Two such global projects are the 1000 Genomes Project and the Exome Sequencing Project, which aim to create the largest public catalogue of human variations and genotype data in the world.17,18) WGS data from these projects are currently contributing to a number of studies in the field of human genetics. Additionally, several population-specific projects are also ongoing. In Iceland, genetic analyses supported by deCODE Genetics have been performed for Icelanders, and analyzed data of WGS (median depth of 20×) has been reported recently.19) In the UK, Genomics England (http://www.genomicsengland.co.uk), which is owned by the UK Department of Health, plans to perform WGS of 100,000 participant samples over a four-year period. In Asia, the GenomeAsia 100K project (http://genomeasia100k.com) also plans to sequence 100,000 individuals from 12 South Asian countries and at least 7 North and East Asian countries. Additionally, in Japan, the Tohoku Medical Megabank Organization (ToMMo) has performed the WGS for 1,070 healthy Japanese individuals (32.4× on average), and constructed a Japanese population reference panel (1KJPN).20) Some of these projects are linking genome sequencing with personalized medicine, cohort studies, and bio-banking.
2. Screening of whole-genome data for candidate functional disease-causal/susceptible variants using large scale bioinformatics analysis
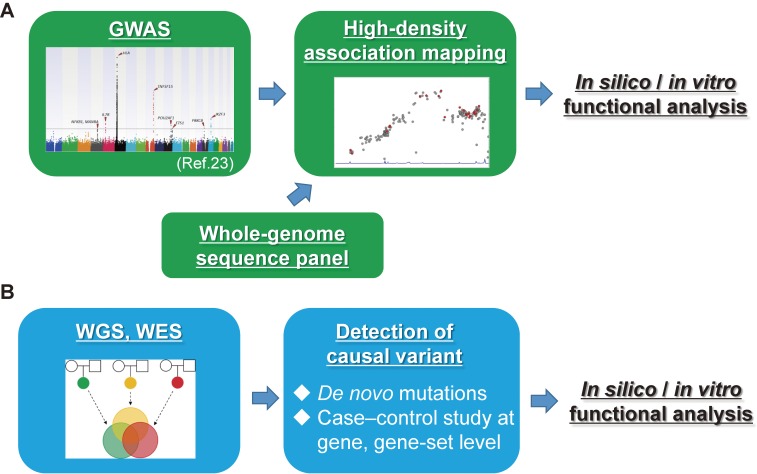
As mentioned above, GWAS and NGS allow genome-wide detection of functional disease-causal/susceptible variants for both common and rare diseases. In this chapter, the flow of screening methods for the detection of functional disease-causal/susceptible variants of human genome sequences is shown (Figure 1).
Figure 1.
Flow of screening of the whole-genome for candidate functional disease-causal/susceptible variants. (A) Flow of screening for common disease-susceptible variants. Susceptibility gene loci are detected using GWAS. High-density association mapping is then performed by genotype imputation analysis and is followed by a case–control association study using the data of the GWAS and a whole-genome sequencing panel that is acquired from the general population using NGS. In silico/in vitro functional analyses are needed for the identification of disease-susceptible variants. (B) Flow of screening for rare disease-causal rare variants. All variants are detected by WES and WGS, and candidate disease-causal variants are then identified using large-scale bioinformatics analysis. In silico/in vitro functional analyses are needed for functional evaluation of disease-causal variants.
Disease susceptible variants for common diseases.
GWASs focus on identifying significant associations among about 1 million genotyped common “tag SNPs”, whose allele frequencies are relatively high, with susceptibility to human traits or diseases. The association signals of the “tag SNPs” are reflected by a few functional disease susceptible variants that show linkage disequilibrium (LD) with the tag SNPs (sometimes the tag SNP itself is also identified as a functional disease susceptible variant), but which have not been directly genotyped. Therefore, disease susceptibility gene loci can be identified by GWASs, but functional disease susceptible variants for common diseases cannot.
Recently, in order to identify unknown genetic factors for disease susceptibility, missing genotyping data of SNPs that are not represented in commercial genome-wide SNP typing platforms have been predicted by referring to WGS data from thousands of healthy individuals. “SNP imputation analysis” using software such as IMPUTE2 can predict missing genotyping data of SNPs that are not represented in commercial genome-wide SNP typing platforms by using a reference panel of known haplotypes in a population such as the panel of the 1000 Genomes Project or that of 1KJPN.17,20,21) Subsequently, variants with P-values less than those of the “tag SNPs” located in the same LD blocks, can be detected by case–control association studies for all of the variants in the disease susceptible gene regions. Such “high-density association mapping” has been able to explain some of the remaining “missing heritability” for human traits and disorders such as that for human height and body mass index.22) This evidence indicates that the roles of functional variants in not only rare but also common diseases need to be investigated, and that NGS can strongly contribute to such studies.
To date, hundreds of studies that have involved SNP imputation analysis and subsequent high-density association mapping have been reported. Such analyses have also been done in Japan focusing on protein kinase C beta (PRKCB), which is a susceptibility gene for primary biliary cholangitis (PBC) detected by a GWASs in the Japanese population, using the 1KJPN panel constructed by ToMMo as a reference.20,23)
This type of screening, however, cannot be applied to all human genes. The HLA genes encode cell-surface signal transduction molecules that present specific antigen peptides to T cell antigen receptors and Natural Killer cell receptors. HLA loci are strongly associated with susceptibility to various autoimmune or immune-related diseases.24) The HLA locus is highly polymorphic (consisting of tens of thousands of HLA alleles) because of natural selection caused by a wide range of pathogens.25) This complexity interferes with the accurate detection of disease susceptibility genes by GWASs, e.g., SNPs in the HLA-DRB locus could not be installed on the general GWAS chip because the gene copy number polymorphism in HLA-DRB causes deviation from the Hardy-Weinberg equilibrium (HWE) in the case–control association study for each SNP. In order to correctly detect disease susceptibility genes in HLA loci, it is necessary to perform an HLA genotyping method such as Luminex HLA-SSO. Recently, several tools that can predict HLA alleles from existing GWAS data have been established. Among these methods, a Japanese group successfully and accurately imputed HLA alleles from several GWAS datasets using HIBAG.26) Additionally, NGS-based 4–8 digit allelic level HLA typing methods, such as NXTypeTM workflow (One Lambda, Inc.), have been developed.27)
Disease-causal variants for rare diseases.
For the detection of functional disease-causal rare variants for rare diseases, two types of approaches based on WGS and WES are used.
The first approach is that of trio-based studies, in which the genomic DNA of the patient and both unaffected parents is sequenced. Using this approach, the following can be detected: i) “de novo mutations,” i.e., new germline mutations that are not inherited from either parent; ii) “newly recessive disease-causing mutations,” which are mutations that are inherited from both parents, each of whom carries one copy of the disease-causing allele; and iii) “compound heterozygous mutations,” which are mutations that are inherited by receiving one disease-causing allele that is benign when present in each of the heterozygous unaffected parents. The number of trios that need to be investigated depends on the morbidity rate of the disease. For example, WES of seven trios detected disease causal variants in ATPase Na+/K+ Transporting Subunit Alpha 3 (ATP1A3) for alternating hemiplegia of childhood (AHC), which is a rare, severe neurodevelopmental syndrome characterized by recurrent hemiplegic episodes and distinct neurological manifestations. Some of these ATP1A3 mutations were found to cause at least 74% of AHC by subsequent sequence analysis of ATP1A3 in 98 other patients.28) On the other hand, the Epi4K and EuroEPINOMICS-RES Consortiums performed the largest de novo mutation screening study in epilepsy using a total of 356 probands with epileptic encephalopathies. These studies detected novel genes with a significant excess of de novo mutations in dynamin 1 (DNM1), gamma-aminobutyric acid type A receptor beta3 subunit (GABRB3), and other genes in epilepsy patients.29,30)
The second approach is case–control association studies, which looks for enrichment of the variant, gene, or gene-set level. One such method is a burden test whereby the total number of disease-causing alleles in patients for a particular gene, pathway, or data sets, is compared with the number of the same alleles in controls. This method can be performed using software such as the sequence kernel association test (SKAT).31–35) For example, significant associations with hemoglobin level and hematocrit were detected for three rare missense variants in pyruvate kinase, liver and RBC (PKLR) using SKAT.36)
Somatic mutations.
There exist not only inherited variants or new mutations in gametes, but also mutations that arise post-zygotically, e.g., postzygotic mutations are found in some cells in nearly all tissues, and somatic mutations are found in specific tissue types. Some studies have reported the contributions of such mutations to human diseases. Using a combination of PCR with a peptide nucleic acid that strongly hybridizes to the target region and inhibits target amplification by PCR, and the ultra-sensitive droplet digital PCR method, a guanine nucleotide-binding protein, Q polypeptide (GNAQ) mutation was detected in patients with Sturge–Weber syndrome (SWS).37) In addition to CNVs, germline variants, and epigenetic factors, accumulation of diverse types of postzygotic and somatic mutations have also been reported to lead to the development and evolution of cancer. Such postzygotic and somatic mutations can be detected by sequencing a sufficiently large number of tumor-normal tissue pairs.38,39) To date, several consortia, such as The Cancer Genome Atlas (TCGA) and The International Cancer Genome Consortium (ICGC), have analyzed cancer genomes.40,41) Additionally, WGS of about 300 liver cancers from Japanese individuals has been performed in Japan.42,43)
3. Identification of functional disease-causal/susceptible variants by in silico and in vitro functional analyses
Even if appropriate screening of functional disease-causal/susceptible variants is performed using whole-genome analysis, this does not mean that the “finish line” has been reached. There are two reasons why completion of whole-genome analysis is not “the goal”. One reason is the inaccuracy of variant calling by current NGS platforms. Although improvements in both the “wet” side (reagents, methods, etc.) and the “dry” side (algorisms, software, etc.) of whole genome analysis have resulted in a dramatic reduction in the false calling rate, this analysis is not yet perfect. The second reason is the existence of LD. Due to the LD between GWAS tag-SNPs and other SNPs, several (sometimes tens of) SNPs show similar levels of associations with the most significantly associated GWAS tag-SNP after SNP imputation analysis, especially in the case of common variants. Therefore, to identify the functional disease-causal/susceptible variants, evaluation of each SNP by in silico- and in vitro-functional analyses should be done.
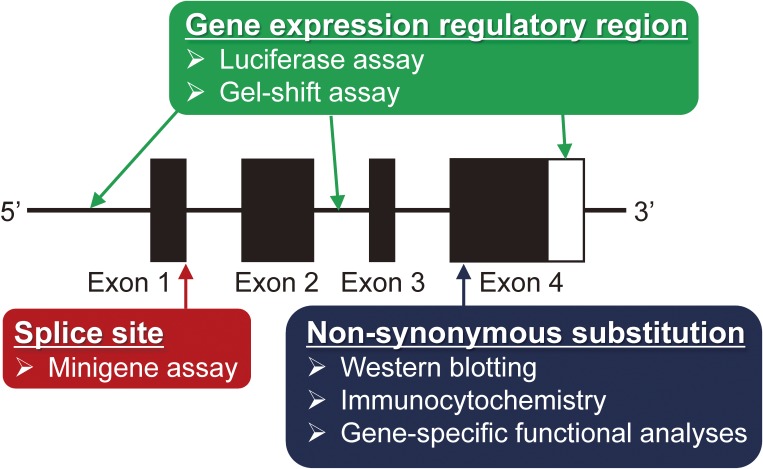
In this chapter, in silico analyses using several valuable computer tools for narrowing down the final candidate functional disease-causal/susceptible variants (Table 1), and in vitro functional analyses for identification of functional disease-causal/susceptible variants (Figure 2) are described.
Table 1.
Available computer tools for in silico functional analysis
| Location of the variant | Notes | Tools | URL | Refs |
|---|---|---|---|---|
| Exon (Non-synonymous variants) | Prediction of the protein structure damage by the mis-sense variants | Polyphen2 | http://genetics.bwh.harvard.edu/pph2/ | 46 |
| MuPIT | http://mupit.icm.jhu.edu/MuPIT_Interactive/ | 47 | ||
| Splice site | Evaluate the functional change by splicing | Human Splice Finder | http://www.umd.be/HSF/ | 48 |
| Gene expression regulatory region | Summarize the regulatory elements for gene expression | HaploReg v4.1 | http://archive.broadinstitute.org/mammals/haploreg/haploreg.php | 55 |
| Regulome DB | http://www.regulomedb.org/ | 56 | ||
| Comprehensive information | UCSC genome browser | http://genome.ucsc.edu/index.html | 57 | |
| ZENBU | http://fantom.gsc.riken.jp/zenbu/ | 58 | ||
| Prediction of TF binding | TRANSFAC | http://www.gene-regulation.com/pub/databases.html | 59 | |
| 3′UTR | Prediction of miRNA binding to 3′UTRs | PolymiRTS | http://compbio.uthsc.edu/miRSNP/ | 61 |
| RegRNA | http://regrna2.mbc.nctu.edu.tw/ | 62 | ||
| Coding, splicing isoform | Prediction of mRNA folding | RNAsnp | http://rth.dk/resources/rnasnp/ | 64 |
| mfold | http://unafold.rna.albany.edu/ | 65 | ||
| All | Database of e-QTLs | GTEx portal | http://gtexportal.org/home/ | 70, 71 |
Figure 2.
The required in vitro functional analysis depends on the genomic location of each variant. i) Western blotting, immunocytochemistry, and gene-specific functional analyses are need for analysis of candidate disease-causal/susceptible variants that are located in exons. ii) Mini-gene assays are need for analysis of candidate disease-causal/susceptible variants that are located in splice sites. Additional functional analyses similar to those required for variants located in exons are also needed if the abnormal splicing influences the amino acid sequence. iii) Luciferase assays and gel-shift assays are needed for analysis of candidate disease-causal/susceptible variants that are located in a gene expression regulatory region such as a gene expression promoter, enhancer, insulator, or a 3′-untranslated region (UTR).
Non-synonymous variants.
Non-synonymous variants (including nonsense and missense variants) are translated into a change in protein structure or function based on the amino acid change involved. Most of the variants that have been identified by WGS or WES for rare diseases have been shown to be non-synonymous variants in thousands of studies. For example, using WES, six homozygous and two compound-heterozygous non-synonymous mutations in activating transcription factor 6A (ATF6A), which is related to the unfolded protein response (UPR) and cellular endoplasmic reticulum (ER) homeostasis, were identified in ten families with achromatopsia.44) In addition, in Japan, using WES and follow-up studies, 11 MIRAGE (myelodysplasia, infection, restriction of growth, adrenal hypoplasia, genital phenotypes, and enteropathy) syndrome patients were recently found to have non-synonymous mutations in Sterile Alpha Motif Domain Containing 9 (SAMD9).45)
In order to evaluate the functional change in gene products encoded by final candidate functional disease-causal/susceptible variants that are missense variants, several computer tools, e.g., Polyphen2 and MuPIT, can be used for the prediction of the protein structural damage caused by these missense variants.46,47) Such structural damage of gene products often induces loss of gene product stability, abnormal gene product cellular localization, or abnormal gene product function. The function of the gene product of each candidate functional disease-causal/susceptible missense variant can be evaluated by in vitro analyses. The stability and cellular localization of the gene product can be analyzed by western blotting, which detects specific protein expression in cells (if the expression level is not sufficient for blotting, it is better to use immunoprecipitation), and immunocytochemistry, which detects the intracellular localization of specific proteins in cells. If the variants do not cause changes in protein stability and localization in these in vitro functional analyses, more detailed functional analyses that are specific for the functions of the gene products are needed.
As described above, WES of AHC patients detected de novo disease causal variants in ATP1A3. These variants showed higher damaging scores in predictions using Polyphen2. Although these ATP1A3 variants did not influence the stability of the ATP1A3 protein, in vitro functional analysis indicated consistent reductions in the ATPase activities of the ATP1A3 gene products encoded by these ATP1A3 variants.28)
Variants in splicing regulatory motifs.
Not only non-synonymous variants but also variants that are located in splicing regulatory motifs (essential splice sites, brunch sites, edge of the exons, exonic/intronic splicing enhancers, and exonic/intronic splicing silencers) also strongly impact on the functions of gene products. This is because exon skipping, which is caused by the abnormal regulation of splicing, may induce frame shifts or deletions of amino acid sequences.
If the final candidate functional disease-causal/susceptible variants are located in splicing regulatory motifs, several computer tools (e.g., Human Splice Finder) can be used to predict the resulting functional changes.48) For in vitro functional evaluation of the exon skipping caused by the lower splicing efficiency of these variants, a minigene assay can be used. In this assay, the RNA resulting from a minigene (a gene fragment that contains some of the exons and introns) that has been transfected into cells is detected using RT-PCR. If a difference in the formation of splicing isoforms can be observed among alleles of candidate disease-causal/susceptible variants in a minigene assay, then such a difference may induce a drastic change in the amino acid sequence of the encoded protein. In this case, it is also necessary to perform the in vitro functional analyses described above for non-synonymous variants (e.g., western blotting, immunocytochemistry, etc.).
For example, a variant that is located at the edge of the 9th exon in transcription factor 4 (TCF4) was detected by WES of an undiagnosed patient (who had features of a wide mouth, high cheekbones, deep-set eyes, limited speech, and severe intellectual disabilities, but who did not have the characteristic hyperventilation and epilepsy) and their unaffected parents. By analysis using a minigene assay and subsequent western blotting, this variant was found to cause an extension of exon 9 and to induce a frame shift, and the gene product encoded by the disease causal allele was degraded by the proteasome.49)
Additionally, although CD72, which is an inhibitory receptor for B cell antigen receptor signaling pathways, was not detected as a susceptibility gene by GWASs, it showed significant association with susceptibility to systemic lupus erythematosus in a Japanese study, and it had a 13 bp variable number tandem repeat (VNTR) in the 8th intron. This variant was significantly associated with the ratio of the quantity of the full-length isoform (flCD72) and the exon 8 skipping isoform (CD72Δex8) by a minigene assay.50) Additional in vitro functional analysis revealed that the CD72Δex8 protein product is localized in the ER and induces the UPR.51)
Variants in gene expression regulatory regions.
To date, thousands of GWAS for common traits or diseases have been reported, in which most of the tag SNPs installed on the GWAS chip fell outside of coding sequences, and often the genotypes of the SNPs were significantly associated with the expression levels of genes that were located near the SNPs.52) Therefore, for the identification of functional common-disease-causal/susceptible variants in disease susceptibility gene loci, the most appropriate approach is to focus on the variants that are located in gene expression regulatory elements (promoters, enhancers, and insulators). Differences in transcription factor (TF) binding to the SNP site between major and minor alleles should be investigated because they may influence gene expression efficiencies. Additionally, regulation of gene expression by binding of TFs is active in the nucleosome-depleted chromatin region, called the “open chromatin region,” where TF binding can be assessed. These regions are characterized by DNase hyper-sensitivity sites and by epigenetic histone markers such as the H3K4Me1, H3K27Ac, and H3K4Me3 marks.53,54) Therefore, as well as TF binding, the location of candidate disease causal/susceptible variants in these markers should be checked.
Several computer tools for prediction include information regarding the location of SNPs in gene expression regulatory regions (e.g., HaploReg v4.1, Regulome DB, UCSC genome browser, and ZENBU).55–58) For the prediction of differences in specific TF binding to the variant, TRANSFAC professional can be used.59) In vitro functional evaluation of SNPs that are located in gene expression regulatory regions can be done using a luciferase gene reporter assay, which assesses transcriptional activity in cells that are transfected with a genetic construct containing the luciferase gene together with the specific promoters or enhancers, and an electrophoretic mobility shift assay (also referred to as a “gel-shift assay”), which is an affinity electrophoresis technique for assessment of TF–DNA interaction. In order to identify the specific TF involved, a super-shift assay, which assesses TF-specific antibody–TF–DNA interaction, is appropriate.
Rs4979462, which is located in the nuclear factor 1 (NF-1)-related gene expression enhancer of tumor necrosis factor superfamily member 15 (TNFSF15), which is a PBC susceptibility gene in the Japanese population, was identified as a functional disease susceptible variant using in silico and in vitro functional analyses.23,60)
Variants in the 3′-UTR.
In addition to gene expression regulatory elements, the 3′ untranslated region (3′ UTR), which is the part of the mRNA that immediately follows the translation termination codon, also contributes to the regulation of gene expression by binding to microRNAs (miRNAs). miRNAs are small non-coding RNA molecules that resemble small interfering RNAs (siRNAs) of RNA interference (RNAi).
There are several computer tools for the prediction of the binding of miRNA to the sequence containing each SNP (e.g., PolymiRTS and RegRNA).61,62) For the in vitro functional evaluation of SNPs that are located in 3′-UTRs, a luciferase gene reporter assay using a genetic construct containing the luciferase gene together with the 3′-UTR of the specific gene can be used to assess the inhibition of gene expression by the miRNA for each allele.
For example, although it was not detected by GWASs, a SNP in the 3′-UTR of NLR family, pyrin domain containing 3 (NLRP3), which controls the activation of inflammatory caspase-1 by forming NLRP3-inflammasomes, was found to be significantly associated with susceptibility to Japanese food-induced anaphylaxis. A difference in NLRP3 mRNA stability due to this SNP was detected in a luciferase assay.63)
Variants that influence mRNA folding.
The folding of RNA into a secondary structure is important for its function. Variants that are located in the coding region can influence the stability of RNA folding. Through a combination of thermodynamics, sequence comparison, and experiments, method for secondary structure prediction have improved. Among several prediction tools for secondary structure, RNAsnp web server can predict the effect of variants on mRNA folding.64) Splicing isoforms that are influenced by variants also can be compared using mfold.65)
Expression-quantitative trait loci.
A quantitative trait locus (QTL) is a genetic locus that correlates with variation in the degree of a phenotype. In particular, a genomic locus that is important for variation in gene expression is called an expression QTL (eQTL). Based on the physical distance between the eQTL and the affected gene, an eQTL can be classified as a cis-eQTL (a distance of 250 kb to 1 Mb in natural populations and of 1–5 Mb in segregating populations) or a trans-eQTL (further away from each other or located on different chromosomes).66,67) Recently, cis- and trans-eQTLs have been systematically identified by the development of statistical frameworks for eQTL analysis and by increasing sample sizes.68,69)
The data from these analyses have already been supplied as computer tools from some projects. The Genotype-Tissue Expression (GTEx) Project provides a resource for human gene expression and regulation and its relationship to genetic variation, in which data were acquired from 53 tissues and 544 donors.70,71) For evaluation of the function of SNPs that are located in gene expression regulatory regions, eQTL analysis for each SNP provides indispensable evidence as a kind of “in vivo data in humans”.
4. Applications of the variants to personalized medicine
To date, thousands of large-scale genetic studies aimed at complete understanding of the genetic background to human diseases have been done around the world. However, the application of the variants identified by such large-scale genetic studies to personalized medicine are still under consideration. In this chapter, their future application as well as existing examples will be described.
Systemic understanding of genes or variants in human disease.
Due to expansion of the findings obtained using GWAS data, a number of reports of meta-analyses have been published recently. Meta-analysis aggregates information, thereby leading to a higher statistical power. By using this powerful approach, a number of disease susceptibility genes whose p-values had not reached a statistically significant level in individual studies could be detected. Of these meta-analysis reports, one world-wide collaboration team reported the largest meta-analysis of rheumatoid arthritis (RA) susceptibility, in which a total of >100,000 subjects of European and Asian ancestry (29,880 RA cases and 73,758 controls) was analyzed, and which successfully connected biological RA susceptible genes to drug targets.72)
Additionally, in order to explore the systemic contribution of genes or variants to each disease, pathway analyses or gene-set analyses using omics data have been performed, especially in WGS/WES for rare diseases and in large-scale GWASs for common diseases. However, these analyses depend on existing incomplete data (i.e., many interactions between genes in databases are sometimes based on a specific cell type or disease). It is also possible that data from literature that includes non-reproducible experiments might be contained in these databases. For these reasons, care should be taken when such kinds of systemic analyses are used.
As well as studies at the gene level, natural human genetic variants should be included in the above-described powerful approaches for their accuracy and statistical power, because human genetic variants have been reported to have dynamic effects on gene expression depending on the cell-type, tissue-type, developmental stage or environmental condition.73,74)
Prediction of disease onset or adverse effects by genetic variants.
Based on accumulated evidence regarding variants that have been associated with susceptibility to specific diseases and their functions, some of these variants are already being used as “predictors”, especially in the field of pharmacogenetics.
The first examples of the use of such a predictor is in Stevens–Johnson syndrome (SJS) and toxic epidermal necrolysis (TEN), which are severe adverse drug reactions characterized by necrosis of the epidermis. Their incidence rates are 2–3 per million per year, and the average mortality rate is 25–35%.75) The drug carbamazepine is known to be the main causative agent of SJS/TEN. HLA-B*15:02 was initially reported to be associated with susceptibility to carbamazepine-induced SJS/TEN in Han Chinese in Taiwan.76) The carrier frequency of HLA-B*15:02 in these cases was 100%, and at the present time this allele is used as a biomarker before medication in Taiwan. In Japan, both HLA-A*31:01 and HLA-B*15:11 have been reported as risk factors for carbamazepine-induced SJS/TEN,77,78) and these alleles may be used as predictors in the future in Japanese as well as in Taiwanese populations.
A second example of the use of such a predictor is in the treatment response to hepatitis C virus (HCV) infection. Prior to the recent appearance of more effective drugs, combination therapy with pegylated interferon (PEG-IFN) and ribavirin (RBV) was used for the treatment of chronic HCV infection.79) However, in spite of effective treatment with PEG-IFN and RBV combination therapy, 20–50% of these patients did not achieve a sustained virological response (SVR).80) In 2009, three independent research groups performed GWASs for chronic HCV treatment responses and the clearance of virus after acute infection, and SNPs located near the interleukin 28B (IL28B) gene, which encodes IFN-λ3, showed significant associations.81–83) The addition of genetic testing using IL-28B genotypes into the clinical management of chronic HCV infection has already been used at the bedside.84) It has been possible to detect by GWASs not only genes that are significantly associated with the response to PEG-IFN and RBV combination therapy but also genes that are associated with adverse effects such as RBV-induced hemolytic anemia. Inosine triphosphatase (ITPA), which encodes a protein that hydrolyzes inosine triphosphate (ITP), was identified by GWAS as a susceptibility gene for RBV-induced hemolytic anemia.85,86) A functional study showed that the ITP that accumulated in the erythrocytes of individuals with anemia-protective ITPA genotypes conferred protection against RBV-induced ATP reduction by substituting for erythrocyte GTP, which was depleted by RBV.87) In addition to prediction of the treatment response based on IL28B, ITPA genotyping is also useful for the prediction of the adverse effect of PEG-IFN and RBV combination therapy for HCV infection.
The third example of the use of such predictors is that of Tamoxifen, which is a prodrug that is mainly metabolized via cytochrome P450 (CYP) enzymes. Tamoxifen is the most commonly used drug for the treatment of estrogen receptor-positive breast cancer, and it works as a selective estrogen receptor modulator. However, there are individual differences in tamoxifen effects and adverse drug reactions.88) The enzymatic activity of hepatic CYP2D6, which accounts for 2–3% of the total liver CYPs, varies among individuals due to CYP2D6 polymorphisms that include over 105 variants whose functions have been partly evaluated in in vitro and in vivo functional studies.89–93) Up to now, CYP2D6 genotyping has not been recommended for prediction because of controversial results in clinical trials.94,95) However, further studies of the association of CYP variants with individual differences in response to Tamoxifen will be of clinical value in the future.
Genetic variants as the targets of therapies or drugs.
Functional disease-causal/susceptible variants themselves can also be used as targets of therapies or drugs. Duchenne’s muscular dystrophy is an X-linked recessive muscle disorder with severe progressive muscle wasting, leading to early death, which affects 1 in 3,500 newborn boys.96) Mutation in the dystrophin gene (DMD), which is the cause of Duchenne’s muscular dystrophy, leads to disruption of the open reading frame, dystrophin deficiency at the myofiber membrane, and continued fiber degeneration.97–101) As a therapeutic approach for patients with Duchenne’s muscular dystrophy, antisense oligonucleotides that induce specific exon skipping (around the 45th to the 55th exon where the causal variants are located) during pre-mRNA splicing, and subsequent reading-frame correction and production of transcripts, have been suggested.102) Among such antisense oligonucleotides for these exons, a 2′-O-methyl phosphorothioate oligoribonucleotide (PRO051) that induces exon 51 skipping was administered to patients with Duchenne’s muscular dystrophy. This oligonucleotide showed dose-dependent molecular efficacy in these patients.103,104)
In the future, it may be possible to correct disease causal/susceptible variants using brand-new biological techniques. CRISPR–Cas9 is a technology for editing genomic DNA that is in widespread use in experimental and applied systems.105,106) To date, there have been some reports regarding the correction of risk alleles of disease causal/susceptible variants in cells by using CRISPR–Cas9-mediated genome editing in vitro (e.g., Fanconi anemia).107) Additionally, the muscular dystrophy-causal genotype (exon 23 frame shifts) in the dystrophin gene could be corrected by CRISPR–Cas9-mediated genome editing and was able to improve muscle function in mdx mouse models.108–110) Improvements in such genome editing technologies will lead to implementation of therapies whose targets are the risk alleles of disease-causal/susceptible variants.
Personal genomic information in hospitals.
Due to the evolution of technologies and reductions in cost, personal genomics using NGS is becoming more common in healthcare. However, there are several issues that remain to be overcome. The first problem is the low sensitivity of most genetic tests because of heterogeneity in etiologies and families, even in monogenic diseases. Therefore, most monogenic disease tests require sequencing approaches. Because of the higher costs, WGS and WES services are poorly covered by health insurance. The second problem is the interpretation of large quantities of variants detected by sequencing in the hospital. Data-sharing and the establishment of standard classification can help overcome this issue, as well as the problem of time-effective point-of-care. The last problem is the ethical debate with regard to the returning of results. It is easy to imagine the existence of secondary genetic findings in personal genomic tests in the future.
In conclusion, for application of these findings to personalized medicine, because human genome variations that are related to human diseases are attributed to genetic variants, there should be a focus not only on genes but also on each variant and their functional role. Before the “NGS-era”, it had been difficult to accurately identify disease causal/susceptible variants from whole genome sequences. In other words, we are now passing through the gate of human genome variation-related personalized medicine. By performing systemic research regarding disease causal/susceptible variants including their functions, we will come to understand the molecular pathogenic mechanisms of diseases, establish novel drugs based on the information obtained about the variants, develop prediction kits for disease onset and adverse effects using genotyping of the variants, and solve the problems related to missing heritability.
Acknowledgements
We sincerely thank our past and present colleagues who were involved with these studies, especially Prof. Minoru Nakamura (Nagasaki Medical Center), Prof. Masao Nagasaki (Tohoku University), Prof. Naoyuki Tsuchiya (Tsukuba University), Prof. David B. Goldstein (Columbia University), Dr. Mayumi Ueta (Kyoto Prefectural Medical University), Dr. Mayumi Tamari (RIKEN), and all of the members of the Department of Human Genetics, Graduate School of Medicine, the University of Tokyo. Our research was supported by Grants-in-Aid for Scientific Research from the Japan Society for the Promotion of Science to Y. H. (#15K19314), Grants-in-Aid for Scientific Research from the Japan Agency for Medical Research and Development to K. T. (H26-kanenjitsu-kanen-ippan-004 and Platform Program for Promotion of Genome Medicine, 16km0405205h0101), and grants from the Takeda Science Foundation, SENSHIN Medical Research Foundation, Uehara Memorial Foundation, and Japan Research Foundation for Clinical Pharmacology to Y.H.
Biographies
Profile
Yuki Hitomi was born in Kumamoto in 1980. He graduated from the University of Tokyo in 2003, and received his Ph.D. from the University of Tokyo in 2008. He has worked as a postdoctoral scholar at the Center for Human Genome Variation, Duke University, under the supervision of Prof. David B. Goldstein. Since 2013, he has been an Assistant Professor at the University of Tokyo. His research interests include the functional impacts of disease causal/susceptible variants that are hidden within the abundant genetic variants in the human genome. His current research theme is a post-GWAS study for complex diseases, in particular for diseases such as primary biliary cholangitis and Stevens–Johnson syndrome.
Katsushi Tokunaga graduated from the Faculty of Science, the University of Tokyo in 1977. He started his research projects at the Graduate School of Science, the University of Tokyo and obtained his Ph.D. Since 1983, he has been an Assistant Professor in the Faculty of Science and University of Tokyo Hospital, and Head of Research Section, Japanese Red Cross Central Blood Center. He has been Professor and Chairman of the Department of Human Genetics, Graduate School of Medicine, the University of Tokyo since 1995. His research interests include genome-wide searches for susceptibility genes to various human complex diseases including HLA-disease associations. He is a member of the Science Council of Japan, President of the Japanese Society for Histocompatibility and Immunogenetics, and Director of the Japan Society of Human Genetics. He is also the Editor-in-Chief of ‘Human Genome Variation’ and an Advisory Editor for other journals, including ‘HLA’.
References
- 1).Levy S., Sutton G., Ng P.C., Feuk L., Halpern A.L., Walenz B.P., Axelrod N., Huang J., Kirkness E.F., Denisov G., Lin Y., MacDonald J.R., Pang A.W., Shago M., Stockwell T.B., Tsiamouri A., Bafna V., Bansal V., Kravitz S.A., Busam D.A., Beeson K.Y., McIntosh T.C., Remington K.A., Abril J.F., Gill J., Borman J., Rogers Y.H., Frazier M.E., Scherer S.W., Strausberg R.L., Venter J.C. (2007) The diploid genome sequence of an individual human. PLoS Biol. 5, e254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2).Kobayashi K., Nakahori Y., Miyake M., Matsumura K., Kondo-Iida E., Nomura Y., Segawa M., Yoshioka M., Saito K., Osawa M., Hamano K., Sakakihara Y., Nonaka I., Nakagome Y., Kanazawa I., Nakamura Y., Tokunaga K., Toda T. (1998) An ancient retrotransposal insertion causes Fukuyama-type congenital muscular dystrophy. Nature 394, 388–392. [DOI] [PubMed] [Google Scholar]
- 3).Welter D., MacArthur J., Morales J., Burdett T., Hall P., Junkins H., Klemm A., Flicek P., Manolio T., Hindorff L., Parkinson H. (2014) The NHGRI GWAS Catalog, a curated resource of SNP-trait associations. Nucleic Acids Res. 42, D1001–D1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4).Manolio T.A., Collins F.S., Cox N.J., Goldstein D.B., Hindorff L.A., Hunter D.J., McCarthy M.I., Ramos E.M., Cardon L.R., Chakravarti A., Cho J.H., Guttmacher A.E., Kong A., Kruglyak L., Mardis E., Rotimi C.N., Slatkin M., Valle D., Whittemore A.S., Boehnke M., Clark A.G., Eichler E.E., Gibson G., Haines J.L., Mackay T.F., McCarroll S.A., Visscher P.M. (2009) Finding the missing heritability of complex diseases. Nature 461, 747–753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5).Lander E.S., Linton L.M., Birren B., Nusbaum C., Zody M.C., Baldwin J., Devon K., Dewar K., Doyle M., FitzHugh W., Funke R., Gage D., Harris K., Heaford A., Howland J., Kann L., Lehoczky J., LeVine R., McEwan P., McKernan K., Meldrim J., Mesirov J.P., Miranda C., Morris W., Naylor J., Raymond C., Rosetti M., Santos R., Sheridan A., Sougnez C., Stange-Thomann Y., Stojanovic N., Subramanian A., Wyman D., Rogers J., Sulston J., Ainscough R., Beck S., Bentley D., Burton J., Clee C., Carter N., Coulson A., Deadman R., Deloukas P., Dunham A., Dunham I., Durbin R., French L., Grafham D., Gregory S., Hubbard T., Humphray S., Hunt A., Jones M., Lloyd C., McMurray A., Matthews L., Mercer S., Milne S., Mullikin J.C., Mungall A., Plumb R., Ross M., Shownkeen R., Sims S., Waterston R.H., Wilson R.K., Hillier L.W., McPherson J.D., Marra M.A., Mardis E.R., Fulton L.A., Chinwalla A.T., Pepin K.H., Gish W.R., Chissoe S.L., Wendl M.C., Delehaunty K.D., Miner T.L., Delehaunty A., Kramer J.B., Cook L.L., Fulton R.S., Johnson D.L., Minx P.J., Clifton S.W., Hawkins T., Branscomb E., Predki P., Richardson P., Wenning S., Slezak T., Doggett N., Cheng J.F., Olsen A., Lucas S., Elkin C., Uberbacher E., Frazier M., Gibbs R.A., Muzny D.M., Scherer S.E., Bouck J.B., Sodergren E.J., Worley K.C., Rives C.M., Gorrell J.H., Metzker M.L., Naylor S.L., Kucherlapati R.S., Nelson D.L., Weinstock G.M., Sakaki Y., Fujiyama A., Hattori M., Yada T., Toyoda A., Itoh T., Kawagoe C., Watanabe H., Totoki Y., Taylor T., Weissenbach J., Heilig R., Saurin W., Artiguenave F., Brottier P., Bruls T., Pelletier E., Robert C., Wincker P., Smith D.R., Doucette-Stamm L., Rubenfield M., Weinstock K., Lee H.M., Dubois J., Rosenthal A., Platzer M., Nyakatura G., Taudien S., Rump A., Yang H., Yu J., Wang J., Huang G., Gu J., Hood L., Rowen L., Madan A., Qin S., Davis R.W., Federspiel N.A., Abola A.P., Proctor M.J., Myers R.M., Schmutz J., Dickson M., Grimwood J., Cox D.R., Olson M.V., Kaul R., Raymond C., Shimizu N., Kawasaki K., Minoshima S., Evans G.A., Athanasiou M., Schultz R., Roe B.A., Chen F., Pan H., Ramser J., Lehrach H., Reinhardt R., McCombie W.R., de la Bastide M., Dedhia N., Blöcker H., Hornischer K., Nordsiek G., Agarwala R., Aravind L., Bailey J.A., Bateman A., Batzoglou S., Birney E., Bork P., Brown D.G., Burge C.B., Cerutti L., Chen H.C., Church D., Clamp M., Copley R.R., Doerks T., Eddy S.R., Eichler E.E., Furey T.S., Galagan J., Gilbert J.G., Harmon C., Hayashizaki Y., Haussler D., Hermjakob H., Hokamp K., Jang W., Johnson L.S., Jones T.A., Kasif S., Kaspryzk A., Kennedy S., Kent W.J., Kitts P., Koonin E.V., Korf I., Kulp D., Lancet D., Lowe T.M., McLysaght A., Mikkelsen T., Moran J.V., Mulder N., Pollara V.J., Ponting C.P., Schuler G., Schultz J., Slater G., Smit A.F., Stupka E., Szustakowki J., Thierry-Mieg D., Thierry-Mieg J., Wagner L., Wallis J., Wheeler R., Williams A., Wolf Y.I., Wolfe K.H., Yang S.P., Yeh R.F., Collins F., Guyer M.S., Peterson J., Felsenfeld A., Wetterstrand K.A., Patrinos A., Morgan M.J., de Jong P., Catanese J.J., Osoegawa K., Shizuya H., Choi S., Chen Y.J., Szustakowki J., International Human Genome Sequencing Consortium (2001) Initial sequencing and analysis of the human genome. Nature 409, 860–921. [DOI] [PubMed] [Google Scholar]
- 6).Venter J.C., Adams M.D., Myers E.W., Li P.W., Mural R.J., Sutton G.G., Smith H.O., Yandell M., Evans C.A., Holt R.A., Gocayne J.D., Amanatides P., Ballew R.M., Huson D.H., Wortman J.R., Zhang Q., Kodira C.D., Zheng X.H., Chen L., Skupski M., Subramanian G., Thomas P.D., Zhang J., Gabor Miklos G.L., Nelson C., Broder S., Clark A.G., Nadeau J., McKusick V.A., Zinder N., Levine A.J., Roberts R.J., Simon M., Slayman C., Hunkapiller M., Bolanos R., Delcher A., Dew I., Fasulo D., Flanigan M., Florea L., Halpern A., Hannenhalli S., Kravitz S., Levy S., Mobarry C., Reinert K., Remington K., Abu-Threideh J., Beasley E., Biddick K., Bonazzi V., Brandon R., Cargill M., Chandramouliswaran I., Charlab R., Chaturvedi K., Deng Z., Di Francesco V., Dunn P., Eilbeck K., Evangelista C., Gabrielian A.E., Gan W., Ge W., Gong F., Gu Z., Guan P., Heiman T.J., Higgins M.E., Ji R.R., Ke Z., Ketchum K.A., Lai Z., Lei Y., Li Z., Li J., Liang Y., Lin X., Lu F., Merkulov G.V., Milshina N., Moore H.M., Naik A.K., Narayan V.A., Neelam B., Nusskern D., Rusch D.B., Salzberg S., Shao W., Shue B., Sun J., Wang Z., Wang A., Wang X., Wang J., Wei M., Wides R., Xiao C., Yan C., Yao A., Ye J., Zhan M., Zhang W., Zhang H., Zhao Q., Zheng L., Zhong F., Zhong W., Zhu S., Zhao S., Gilbert D., Baumhueter S., Spier G., Carter C., Cravchik A., Woodage T., Ali F., An H., Awe A., Baldwin D., Baden H., Barnstead M., Barrow I., Beeson K., Busam D., Carver A., Center A., Cheng M.L., Curry L., Danaher S., Davenport L., Desilets R., Dietz S., Dodson K., Doup L., Ferriera S., Garg N., Gluecksmann A., Hart B., Haynes J., Haynes C., Heiner C., Hladun S., Hostin D., Houck J., Howland T., Ibegwam C., Johnson J., Kalush F., Kline L., Koduru S., Love A., Mann F., May D., McCawley S., McIntosh T., McMullen I., Moy M., Moy L., Murphy B., Nelson K., Pfannkoch C., Pratts E., Puri V., Qureshi H., Reardon M., Rodriguez R., Rogers Y.H., Romblad D., Ruhfel B., Scott R., Sitter C., Smallwood M., Stewart E., Strong R., Suh E., Thomas R., Tint N.N., Tse S., Vech C., Wang G., Wetter J., Williams S., Williams M., Windsor S., Winn-Deen E., Wolfe K., Zaveri J., Zaveri K., Abril J.F., Guigó R., Campbell M.J., Sjolander K.V., Karlak B., Kejariwal A., Mi H., Lazareva B., Hatton T., Narechania A., Diemer K., Muruganujan A., Guo N., Sato S., Bafna V., Istrail S., Lippert R., Schwartz R., Walenz B., Yooseph S., Allen D., Basu A., Baxendale J., Blick L., Caminha M., Carnes-Stine J., Caulk P., Chiang Y.H., Coyne M., Dahlke C., Mays A., Dombroski M., Donnelly M., Ely D., Esparham S., Fosler C., Gire H., Glanowski S., Glasser K., Glodek A., Gorokhov M., Graham K., Gropman B., Harris M., Heil J., Henderson S., Hoover J., Jennings D., Jordan C., Jordan J., Kasha J., Kagan L., Kraft C., Levitsky A., Lewis M., Liu X., Lopez J., Ma D., Majoros W., McDaniel J., Murphy S., Newman M., Nguyen T., Nguyen N., Nodell M., Pan S., Peck J., Peterson M., Rowe W., Sanders R., Scott J., Simpson M., Smith T., Sprague A., Stockwell T., Turner R., Venter E., Wang M., Wen M., Wu D., Wu M., Xia A., Zandieh A., Zhu X. (2001) The sequence of the human genome. Science 291, 1304–1351. [DOI] [PubMed] [Google Scholar]
- 7).Morozova O., Marra M.A. (2008) Applications of next-generation sequencing technologies in functional genomics. Genomics 92, 255–264. [DOI] [PubMed] [Google Scholar]
- 8).Pareek C.S., Smoczynski R., Tretyn A. (2011) Sequencing technologies and genome sequencing. J. Appl. Genet. 52, 413–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9).Tucker T., Marra M., Friedman J.M. (2009) Massively parallel sequencing: the next big thing in genetic medicine. Am. J. Hum. Genet. 85, 142–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10).Bentley D.R., Balasubramanian S., Swerdlow H.P., Smith G.P., Milton J., Brown C.G., Hall K.P., Evers D.J., Barnes C.L., Bignell H.R., Boutell J.M., Bryant J., Carter R.J., Keira Cheetham R., Cox A.J., Ellis D.J., Flatbush M.R., Gormley N.A., Humphray S.J., Irving L.J., Karbelashvili M.S., Kirk S.M., Li H., Liu X., Maisinger K.S., Murray L.J., Obradovic B., Ost T., Parkinson M.L., Pratt M.R., Rasolonjatovo I.M., Reed M.T., Rigatti R., Rodighiero C., Ross M.T., Sabot A., Sankar S.V., Scally A., Schroth G.P., Smith M.E., Smith V.P., Spiridou A., Torrance P.E., Tzonev S.S., Vermaas E.H., Walter K., Wu X., Zhang L., Alam M.D., Anastasi C., Aniebo I.C., Bailey D.M., Bancarz I.R., Banerjee S., Barbour S.G., Baybayan P.A., Benoit V.A., Benson K.F., Bevis C., Black P.J., Boodhun A., Brennan J.S., Bridgham J.A., Brown R.C., Brown A.A., Buermann D.H., Bundu A.A., Burrows J.C., Carter N.P., Castillo N., Chiara E., Catenazzi M., Chang S., Neil Cooley R., Crake N.R., Dada O.O., Diakoumakos K.D., Dominguez-Fernandez B., Earnshaw D.J., Egbujor U.C., Elmore D.W., Etchin S.S., Ewan M.R., Fedurco M., Fraser L.J., Fuentes Fajardo K.V., Scott Furey W., George D., Gietzen K.J., Goddard C.P., Golda G.S., Granieri P.A., Green D.E., Gustafson D.L., Hansen N.F., Harnish K., Haudenschild C.D., Heyer N.I., Hims M.M., Ho J.T., Horgan A.M., Hoschler K., Hurwitz S., Ivanov D.V., Johnson M.Q., James T., Huw Jones T.A., Kang G.D., Kerelska T.H., Kersey A.D., Khrebtukova I., Kindwall A.P., Kingsbury Z., Kokko-Gonzales P.I., Kumar A., Laurent M.A., Lawley C.T., Lee S.E., Lee X., Liao A.K., Loch J.A., Lok M., Luo S., Mammen R.M., Martin J.W., McCauley P.G., McNitt P., Mehta P., Moon K.W., Mullens J.W., Newington T., Ning Z., Ling Ng B., Novo S.M., O’Neill M.J., Osborne M.A., Osnowski A., Ostadan O., Paraschos L.L., Pickering L., Pike A.C., Pike A.C., Chris Pinkard D., Pliskin D.P., Podhasky J., Quijano V.J., Raczy C., Rae V.H., Rawlings S.R., Chiva Rodriguez A., Roe P.M., Rogers J., Rogert Bacigalupo M.C., Romanov N., Romieu A., Roth R.K., Rourke N.J., Ruediger S.T., Rusman E., Sanches-Kuiper R.M., Schenker M.R., Seoane J.M., Shaw R.J., Shiver M.K., Short S.W., Sizto N.L., Sluis J.P., Smith M.A., Ernest Sohna Sohna J., Spence E.J., Stevens K., Sutton N., Szajkowski L., Tregidgo C.L., Turcatti G., Vandevondele S., Verhovsky Y., Virk S.M., Wakelin S., Walcott G.C., Wang J., Worsley G.J., Yan J., Yau L., Zuerlein M., Rogers J., Mullikin J.C., Hurles M.E., McCooke N.J., West J.S., Oaks F.L., Lundberg P.L., Klenerman D., Durbin R., Smith A.J. (2008) Accurate whole human genome sequencing using reversible terminator chemistry. Nature 456, 53–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11).Wheeler D.A., Srinivasan M., Egholm M., Shen Y., Chen L., McGuire A., He W., Chen Y.J., Makhijani V., Roth G.T., Gomes X., Tartaro K., Niazi F., Turcotte C.L., Irzyk G.P., Lupski J.R., Chinault C., Song X.Z., Liu Y., Yuan Y., Nazareth L., Qin X., Muzny D.M., Margulies M., Weinstock G.M., Gibbs R.A., Rothberg J.M. (2008) The complete genome of an individual by massively parallel DNA sequencing. Nature 452, 872–876. [DOI] [PubMed] [Google Scholar]
- 12).Fujimoto A., Nakagawa H., Hosono N., Nakano K., Abe T., Boroevich K.A., Nagasaki M., Yamaguchi R., Shibuya T., Kubo M., Miyano S., Nakamura Y., Tsunoda T. (2010) Whole-genome sequencing and comprehensive variant analysis of a Japanese individual using massively parallel sequencing. Nat. Genet. 42, 931–936. [DOI] [PubMed] [Google Scholar]
- 13).Ng S.B., Turner E.H., Robertson P.D., Flygare S.D., Bigham A.W., Lee C., Shaffer T., Wong M., Bhattacharjee A., Eichler E.E., Bamshad M., Nickerson D.A., Shendure J. (2009) Targeted capture and massively parallel sequencing of 12 human exomes. Nature 461, 272–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14).Ng S.B., Buckingham K.J., Lee C., Bigham A.W., Tabor H.K., Dent K.M., Huff C.D., Shannon P.T., Jabs E.W., Nickerson D.A., Shendure J., Bamshad M.J. (2010) Exome sequencing identifies the cause of a Mendelian disorder. Nat. Genet. 42, 30–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15).Hoischen A., van Bon B.W., Gilissen C., Arts P., van Lier B., Steehouwer M., de Vries P., de Reuver R., Wieskamp N., Mortier G., Devriendt K., Amorim M.Z., Revencu N., Kidd A., Barbosa M., Turner A., Smith J., Oley C., Henderson A., Hayes I.M., Thompson E.M., Brunner H.G., de Vries B.B., Veltman J.A. (2010) De novo mutations of SETBP1 cause Schinzel-Giedion syndrome. Nat. Genet. 42, 483–485. [DOI] [PubMed] [Google Scholar]
- 16).Boycott K.M., Dyment D.A., Sawyer S.L., Vanstone M.R., Beaulieu C.L. (2014) Identification of genes for childhood heritable diseases. Annu. Rev. Med. 65, 19–31. [DOI] [PubMed] [Google Scholar]
- 17).1000 Genomes Project Consortium (2012) An integrated map of genetic variation from 1,092 human genomes. Nature 491, 56–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18).Fu W., O’Connor T.D., Jun G., Kang H.M., Abecasis G., Leal S.M., Gabriel S., Rieder M.J., Altshuler D., Shendure J., Nickerson D.A., Bamshad M.J., NHLBI Exome Sequencing Project. Akey J.M. (2013) Analysis of 6,515 exomes reveals the recent origin of most human protein-coding variants. Nature 493, 216–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19).Gudbjartsson D.F., Helgason H., Gudjonsson S.A., Zink F., Oddson A., Gylfason A., Besenbacher S., Magnusson G., Halldorsson B.V., Hjartarson E., Sigurdsson G.T., Stacey S.N., Frigge M.L., Holm H., Saemundsdottir J., Helgadottir H.T., Johannsdottir H., Sigfusson G., Thorgeirsson G., Sverrisson J.T., Gretarsdottir S., Walters G.B., Rafnar T., Thjodleifsson B., Bjornsson E.S., Olafsson S., Thorarinsdottir H., Steingrimsdottir T., Gudmundsdottir T.S., Theodors A., Jonasson J.G., Sigurdsson A., Bjornsdottir G., Jonsson J.J., Thorarensen O., Ludvigsson P., Gudbjartsson H., Eyjolfsson G.I., Sigurdardottir O., Olafsson I., Arnar D.O., Magnusson O.T., Kong A., Masson G., Thorsteinsdottir U., Helgason A., Sulem P., Stefansson K. (2015) Large-scale wholegenome sequencing of the Icelandic population. Nat. Genet. 47, 435–444. [DOI] [PubMed] [Google Scholar]
- 20).Nagasaki M., Yasuda J., Katsuoka F., Nariai N., Kojima K., Kawai Y., Yamaguchi-Kabata Y., Yokozawa J., Danjoh I., Saito S., Sato Y., Mimori T., Tsuda K., Saito R., Pan X., Nishikawa S., Ito S., Kuroki Y., Tanabe O., Fuse N., Kuriyama S., Kiyomoto H., Hozawa A., Minegishi N., Douglas Engel J., Kinoshita K., Kure S., Yaegashi N., ToMMo Japanese Reference Panel Project. Yamamoto M. (2015) Rare variant discovery by deep whole-genome sequencing of 1,070 Japanese individuals. Nat. Commun. 6, 8018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21).Howie B.N., Donnelly P., Marchini J. (2009) A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet. 5, 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22).Yang J., Bakshi A., Zhu Z., Hemani G., Vinkhuyzen A.A., Lee S.H., Robinson M.R., Perry J.R., Nolte I.M., van Vliet-Ostaptchouk J.V., Snieder H., LifeLines Cohort Study. Esko T., Milani L., Mägi R., Metspalu A., Hamsten A., Magnusson P.K., Pedersen N.L., Ingelsson E., Soranzo N., Keller M.C., Wray N.R., Goddard M.E., Visscher P.M. (2015) Genetic variance estimation with imputed variants finds negligible missing heritability for human height and body mass index. Nat. Genet. 47, 1114–1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23).Kawashima M., Hitomi Y., Aiba Y., Nishida N., Kojima K., Kawai Y., Nakamura H., Tanaka A., Zeniya M., Hashimoto E., Ohira H., Yamamoto K., Abe M., Nakao K., Yamagiwa S., Kaneko S., Honda M., Umemura T., Ichida T., Seike M., Sakisaka S., Harada M., Yokosuka O., Ueno Y., Senju M., Kanda T., Shibata H., Himoto T., Murata K., Miyake Y., Ebinuma H., Taniai M., Joshita S., Nikami T., Ota H., Kouno H., Kouno H., Nakamura M., Kohjima M., Komatsu T., Komeda T., Ohara Y., Muro T., Yamashita T., Yoshizawa K., Kamitsukasa Y., Nakamura Y., Shimada M., Hirashima N., Sugi K., Ario K., Takesaki E., Naganuma A., Mano H., Yamashita H., Matsushita K., Yamauchi K., Makita F., Nishimura H., Furuta K., Takahashi N., Masaki N., Tanaka T., Tamura S., Mori A., Yagi S., Shirabe K., Komori A., Migita K., Ito M., Nagaoka S., Abiru S., Yatsuhashi H., Yasunami M., Shimoda S., Harada K., Egawa H., Maehara Y., Uemoto S., Kokubo N., Takikawa H., Ishibashi H., Chayama K., Mizokami M., Nagasaki M., Tokunaga K., Nakamura M. (2017) Genome-wide association studies identify PRKCB as genetic susceptibility locus for primary biliary cirrhosis in the Japanese population. Hum. Mol. Genet. 26, 650–659. [DOI] [PubMed] [Google Scholar]
- 24).Tsuchiya N., Ohashi J. (2015) Human immune system diversity and its implications in diseases. J. Hum. Genet. 60, 655–656. [DOI] [PubMed] [Google Scholar]
- 25).Barreiro L.B., Quintana-Murci L. (2010) From evolutionary genetics to human immunology: how selection shapes host defence genes. Nat. Rev. Genet. 11, 17–30. [DOI] [PubMed] [Google Scholar]
- 26).Khor S.S., Yang W., Kawashima M., Kamitsuji S., Zheng X., Nishida N., Sawai H., Toyoda H., Miyagawa T., Honda M., Kamatani N., Tokunaga K. (2016) High-accuracy imputation for HLA class I and II genes based on high-resolution SNP data of population-specific references. Pharmacological J. 15, 530–537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27).Barone J.C., Saito K., Beutner K., Campo M., Dong W., Goswami C.P., Johnson E.S., Wang Z.X., Hsu S. (2015) HLA-genotyping of clinical specimens using Ion Torrent-based NGS. Hum. Immunol. 76, 903–909. [DOI] [PubMed] [Google Scholar]
- 28).Heinzen E.L., Swoboda K.J., Hitomi Y., Gurrieri F., Nicole S., de Vries B., Tiziano F.D., Fontaine B., Walley N.M., Heavin S., Panagiotakaki E., European Alternating Hemiplegia of Childhood (AHC) Genetics Consortium. Biobanca e Registro Clinico per l’Emiplegia Alternante (I.B.AHC) Consortium. European Network for Research on Alternating Hemiplegia (ENRAH) for Small and Medium-sized Enterpriese (SMEs) Consortium. Fiori S., Abiusi E., Di Pietro L., Sweney M.T., Newcomb T.M., Viollet L., Huff C., Jorde L.B., Reyna S.P., Murphy K.J., Shianna K.V., Gumbs C.E., Little L., Silver K., Ptáček L.J., Haan J., Ferrari M.D., Bye A.M., Herkes G.K., Whitelaw C.M., Webb D., Lynch B.J., Uldall P., King M.D., Scheffer I.E., Neri G., Arzimanoglou A., van den Maagdenberg A.M., Sisodiya S.M., Mikati M.A., Goldstein D.B. (2012) De novo mutations in ATP1A3 cause alternating hemiplegia of childhood. Nat. Genet. 44, 1030–1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29).Epi4K Consortium. Epilepsy Phenome/Genome Project. Allen A.S., Berkovic S.F., Cossette P., Delanty N., Dlugos D., Eichler E.E., Epstein M.P., Glauser T., Goldstein D.B., Han Y., Heinzen E.L., Hitomi Y., Howell K.B., Johnson M.R., Kuzniecky R., Lowenstein D.H., Lu Y.F., Madou M.R., Marson A.G., Mefford H.C., Esmaeeli Nieh S., O’Brien T.J., Ottman R., Petrovski S., Poduri A., Ruzzo E.K., Scheffer I.E., Sherr E.H., Yuskaitis C.J., Abou-Khalil B., Alldredge B.K., Bautista J.F., Berkovic S.F., Boro A., Cascino G.D., Consalvo D., Crumrine P., Devinsky O., Dlugos D., Epstein M.P., Fiol M., Fountain N.B., French J., Friedman D., Geller E.B., Glauser T., Glynn S., Haut S.R., Hayward J., Helmers S.L., Joshi S., Kanner A., Kirsch H.E., Knowlton R.C., Kossoff E.H., Kuperman R., Kuzniecky R., Lowenstein D.H., McGuire S.M., Motika P.V., Novotny E.J., Ottman R., Paolicchi J.M., Parent J.M., Park K., Poduri A., Scheffer I.E., Shellhaas R.A., Sherr E.H., Shih J.J., Singh R., Sirven J., Smith M.C., Sullivan J., Lin Thio L., Venkat A., Vining E.P., Von Allmen G.K., Weisenberg J.L., Widdess-Walsh P., Winawer M.R. (2013) De novo mutations in epileptic encephalopathies. Nature 501, 217–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30).EuroEPINOMICS-RES Consortium. Epilepsy Phenome/Genome Project. Epi4k Consortium (2014) De novo mutations in synaptic transmission genes including DNM1 cause epileptic encephalopathies. Am. J. Hum. Genet. 95, 360–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31).Ionita-Laza I., Lee S., Makarov V., Buxbaum J., Lin X. (2013) Sequence kernel association tests for the combined effect of rare and common variants. Am. J. Hum. Genet. 92, 841–853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32).Lee S., Emond M.J., Bamshad M.J., Barnes K.C., Rieder M.J., Nickerson D.A., NHLBI GO Exome Sequencing Project-ESP Lung Project Team. Christiani D.C., Wurfel M.M., Lin X. (2012) Optimal unified approach for rare variant association testing with application to small sample case-control whole-exome sequencing studies. Am. J. Hum. Genet. 91, 224–237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33).Lee S., Wu M.C., Lin X. (2012) Optimal tests for rare variant effects in sequencing association studies. Biostatistics 13, 762–775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34).Wu M.C., Lee S., Cai T., Li Y., Boehnke M., Lin X. (2011) Rare variant association testing for sequencing data using the sequence kernel association test (SKAT). Am. J. Hum. Genet. 89, 82–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35).Wu M.C., Kraft P., Epstein M.P., Taylor D.M., Chanock S.J., Hunter D.J., Lin X. (2010) Powerful SNP set analysis for case-control genomewide association studies. Am. J. Hum. Genet. 86, 929–942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36).Chami N., Chen M.H., Slater A.J., Eicher J.D., Evangelou E., Tajuddin S.M., Love-Gregory L., Kacprowski T., Schick U.M., Nomura A., Giri A., Lessard S., Brody J.A., Schurmann C., Pankratz N., Yanek L.R., Manichaikul A., Pazoki R., Mihailov E., Hill W.D., Raffield L.M., Burt A., Bartz T.M., Becker D.M., Becker L.C., Boerwinkle E., Bork-Jensen J., Bottinger E.P., O’Donoghue M.L., Crosslin D.R., de Denus S., Dubé M.P., Elliott P., Engström G., Evans M.K., Floyd J.S., Fornage M., Gao H., Greinacher A., Gudnason V., Hansen T., Harris T.B., Hayward C., Hernesniemi J., Highland H.M., Hirschhorn J.N., Hofman A., Irvin M.R., Kähönen M., Lange E., Launer L.J., Lehtimäki T., Li J., Liewald D.C., Linneberg A., Liu Y., Lu Y., Lyytikäinen L.P., Mägi R., Mathias R.A., Melander O., Metspalu A., Mononen N., Nalls M.A., Nickerson D.A., Nikus K., O’Donnell C.J., Orho-Melander M., Pedersen O., Petersmann A., Polfus L., Psaty B.M., Raitakari O.T., Raitoharju E., Richard M., Rice K.M., Rivadeneira F., Rotter J.I., Schmidt F., Smith A.V., Starr J.M., Taylor K.D., Teumer A., Thuesen B.H., Torstenson E.S., Tracy R.P., Tzoulaki I., Zakai N.A., Vacchi-Suzzi C., van Duijn C.M., van Rooij F.J., Cushman M., Deary I.J., Velez Edwards D.R., Vergnaud A.C., Wallentin L., Waterworth D.M., White H.D., Wilson J.G., Zonderman A.B., Kathiresan S., Grarup N., Esko T., Loos R.J., Lange L.A., Faraday N., Abumrad N.A., Edwards T.L., Ganesh S.K., Auer P.L., Johnson A.D., Reiner A.P., Lettre G. (2016) Exome genotyping identifies pleiotropic variants associated with red blood cell traits. Am. J. Hum. Genet. 99, 8–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37).Uchiyama Y., Nakashima M., Watanabe S., Miyajima M., Taguri M., Miyatake S., Miyake N., Saitsu H., Mishima H., Kinoshita A., Arai H., Yoshiura K., Matsumoto N. (2016) Ultra-sensitive droplet digital PCR for detecting a low-prevalence somatic GNAQ mutation in Sturge-Weber syndrome. Sci. Rep. 6, 22985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38).Stratton M., Campbell P.J., Futreal A. (2009) The cancer genome. Nature 458, 719–724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39).Garraway L.A., Lander E.S. (2013) Lessons from the cancer genome. Cell 153, 17–37. [DOI] [PubMed] [Google Scholar]
- 40).Cancer Genome Atlas Research Network (2008) Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature 455, 1061–1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41).International Cancer Genome Consortium. Hudson T.J., Anderson W., Artez A., Barker A.D., Bell C., Bernabé R.R., Bhan M.K., Calvo F., Eerola I., Gerhard D.S., Guttmacher A., Guyer M., Hemsley F.M., Jennings J.L., Kerr D., Klatt P., Kolar P., Kusada J., Lane D.P., Laplace F., Youyong L., Nettekoven G., Ozenberger B., Peterson J., Rao T.S., Remacle J., Schafer A.J., Shibata T., Stratton M.R., Vockley J.G., Watanabe K., Yang H., Yuen M.M., Knoppers B.M., Bobrow M., Cambon-Thomsen A., Dressler L.G., Dyke S.O., Joly Y., Kato K., Kennedy K.L., Nicolás P., Parker M.J., Rial-Sebbag E., Romeo-Casabona C.M., Shaw K.M., Wallace S., Wiesner G.L., Zeps N., Lichter P., Biankin A.V., Chabannon C., Chin L., Clément B., de Alava E., Degos F., Ferguson M.L., Geary P., Hayes D.N., Hudson T.J., Johns A.L., Kasprzyk A., Nakagawa H., Penny R., Piris M.A., Sarin R., Scarpa A., Shibata T., van de Vijver M., Futreal P.A., Aburatani H., Bayés M., Botwell D.D., Campbell P.J., Estivill X., Gerhard D.S., Grimmond S.M., Gut I., Hirst M., López-Otín C., Majumder P., Marra M., McPherson J.D., Nakagawa H., Ning Z., Puente X.S., Ruan Y., Shibata T., Stratton M.R., Stunnenberg H.G., Swerdlow H., Velculescu V.E., Wilson R.K., Xue H.H., Yang L., Spellman P.T., Bader G.D., Boutros P.C., Campbell P.J., Flicek P., Getz G., Guigó R., Guo G., Haussler D., Heath S., Hubbard T.J., Jiang T., Jones S.M., Li Q., López-Bigas N., Luo R., Muthuswamy L., Ouellette B.F., Pearson J.V., Puente X.S., Quesada V., Raphael B.J., Sander C., Shibata T., Speed T.P., Stein L.D., Stuart J.M., Teague J.W., Totoki Y., Tsunoda T., Valencia A., Wheeler D.A., Wu H., Zhao S., Zhou G., Stein L.D., Guigó R., Hubbard T.J., Joly Y., Jones S.M., Kasprzyk A., Lathrop M., López-Bigas N., Ouellette B.F., Spellman P.T., Teague J.W., Thomas G., Valencia A., Yoshida T., Kennedy K.L., Axton M., Dyke S.O., Futreal P.A., Gerhard D.S., Gunter C., Guyer M., Hudson T.J., McPherson J.D., Miller L.J., Ozenberger B., Shaw K.M., Kasprzyk A., Stein L.D., Zhang J., Haider S.A., Wang J., Yung C.K., Cros A., Liang Y., Gnaneshan S., Guberman J., Hsu J., Bobrow M., Chalmers D.R., Hasel K.W., Joly Y., Kaan T.S., Kennedy K.L., Knoppers B.M., Lowrance W.W., Masui T., Nicolás P., Rial-Sebbag E., Rodriguez L.L., Vergely C., Yoshida T., Grimmond S.M., Biankin A.V., Bowtell D.D., Cloonan N., DeFazio A., Eshleman J.R., Etemadmoghadam D., Gardiner B.B., Kench J.G., Scarpa A., Sutherland R.L., Tempero M.A., Waddell N.J., Wilson P.J., McPherson J.D., Gallinger S., Tsao M.S., Shaw P.A., Petersen G.M., Mukhopadhyay D., Chin L., DePinho R.A., Thayer S., Muthuswamy L., Shazand K., Beck T., Sam M., Timms L., Ballin V., Lu Y., Ji J., Zhang X., Chen F., Hu X., Zhou G., Yang Q., Tian G., Zhang L., Xing X., Li X., Zhu Z., Yu Y., Yu J., Yang H., Lathrop M., Tost J., Brennan P., Holcatova I., Zaridze D., Brazma A., Egevard L., Prokhortchouk E., Banks R.E., Uhlén M., Cambon-Thomsen A., Viksna J., Ponten F., Skryabin K., Stratton M.R., Futreal P.A., Birney E., Borg A., Børresen-Dale A.L., Caldas C., Foekens J.A., Martin S., Reis-Filho J.S., Richardson A.L., Sotiriou C., Stunnenberg H.G., Thoms G., van de Vijver M., van’t Veer L., Calvo F., Birnbaum D., Blanche H., Boucher P., Boyault S., Chabannon C., Gut I., Masson-Jacquemier J.D., Lathrop M., Pauporté I., Pivot X., Vincent-Salomon A., Tabone E., Theillet C., Thomas G., Tost J., Treilleux I., Calvo F., Bioulac-Sage P., Clément B., Decaens T., Degos F., Franco D., Gut I., Gut M., Heath S., Lathrop M., Samuel D., Thomas G., Zucman-Rossi J., Lichter P., Eils R., Brors B., Korbel J.O., Korshunov A., Landgraf P., Lehrach H., Pfister S., Radlwimmer B., Reifenberger G., Taylor M.D., von Kalle C., Majumder P.P., Sarin R., Rao T.S., Bhan M.K., Scarpa A., Pederzoli P., Lawlor R.A., Delledonne M., Bardelli A., Biankin A.V., Grimmond S.M., Gress T., Klimstra D., Zamboni G., Shibata T., Nakamura Y., Nakagawa H., Kusada J., Tsunoda T., Miyano S., Aburatani H., Kato K., Fujimoto A., Yoshida T., Campo E., López-Otín C., Estivill X., Guigó R., de Sanjosé S., Piris M.A., Montserrat E., González-Díaz M., Puente X.S., Jares P., Valencia A., Himmelbauer H., Quesada V., Bea S., Stratton M.R., Futreal P.A., Campbell P.J., Vincent-Salomon A., Richardson A.L., Reis-Filho J.S., van de Vijver M., Thomas G., Masson-Jacquemier J.D., Aparicio S., Borg A., Børresen-Dale A.L., Caldas C., Foekens J.A., Stunnenberg H.G., van’t Veer L., Easton D.F., Spellman P.T., Martin S., Barker A.D., Chin L., Collins F.S., Compton C.C., Ferguson M.L., Gerhard D.S., Getz G., Gunter C., Guttmacher A., Guyer M., Hayes D.N., Lander E.S., Ozenberger B., Penny R., Peterson J., Sander C., Shaw K.M., Speed T.P., Spellman P.T., Vockley J.G., Wheeler D.A., Wilson R.K., Hudson T.J., Chin L., Knoppers B.M., Lander E.S., Lichter P., Stein L.D., Stratton M.R., Anderson W., Barker A.D., Bell C., Bobrow M., Burke W., Collins F.S., Compton C.C., DePinho R.A., Easton D.F., Futreal P.A., Gerhard D.S., Green A.R., Guyer M., Hamilton S.R., Hubbard T.J., Kallioniemi O.P., Kennedy K.L., Ley T.J., Liu E.T., Lu Y., Majumder P., Marra M., Ozenberger B., Peterson J., Schafer A.J., Spellman P.T., Stunnenberg H.G., Wainwright B.J., Wilson R.K., Yang H. (2010) International network of cancer genome projects. Nature 464, 993–998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42).Fujimoto A., Totoki Y., Abe T., Boroevich K.A., Hosoda F., Nguyen H.H., Aoki M., Hosono N., Kubo M., Miya F., Arai Y., Takahashi H., Shirakihara T., Nagasaki M., Shibuya T., Nakano K., Watanabe-Makino K., Tanaka H., Nakamura H., Kusuda J., Ojima H., Shimada K., Okusaka T., Ueno M., Shigekawa Y., Kawakami Y., Arihiro K., Ohdan H., Gotoh K., Ishikawa O., Ariizumi S., Yamamoto M., Yamada T., Chayama K., Kosuge T., Yamaue H., Kamatani N., Miyano S., Nakagama H., Nakamura Y., Tsunoda T., Shibata T., Nakagawa H. (2012) Whole-genome sequencing of liver cancers identifies etiological influences on mutation patterns and recurrent mutations in chromatin regulators. Nat. Genet. 44, 760–764. [DOI] [PubMed] [Google Scholar]
- 43).Fujimoto A., Furuta M., Totoki Y., Tsunoda T., Kato M., Shiraishi Y., Tanaka H., Taniguchi H., Kawakami Y., Ueno M., Gotoh K., Ariizumi S., Wardell C.P., Hayami S., Nakamura T., Aikata H., Arihiro K., Boroevich K.A., Abe T., Nakano K., Maejima K., Sasaki-Oku A., Ohsawa A., Shibuya T., Nakamura H., Hama N., Hosoda F., Arai Y., Ohashi S., Urushidate T., Nagae G., Yamamoto S., Ueda H., Tatsuno K., Ojima H., Hiraoka N., Okusaka T., Kubo M., Marubashi S., Yamada T., Hirano S., Yamamoto M., Ohdan H., Shimada K., Ishikawa O., Yamaue H., Chayama K., Miyano S., Aburatani H., Shibata T., Nakagawa H. (2016) Whole-genome mutational landscape and characterization of noncoding and structural mutations in liver cancer. Nat. Genet. 48, 500–509. [DOI] [PubMed] [Google Scholar]
- 44).Kohl S., Zobor D., Chiang W.C., Weisschuh N., Staller J., Gonzalez Menendez I., Chang S., Beck S.C., Garcia Garrido M., Sothilingam V., Seeliger M.W., Stanzial F., Benedicenti F., Inzana F., Héon E., Vincent A., Beis J., Strom T.M., Rudolph G., Roosing S., Hollander A.I., Cremers F.P., Lopez I., Ren H., Moore A.T., Webster A.R., Michaelides M., Koenekoop R.K., Zrenner E., Kaufman R.J., Tsang S.H., Wissinger B., Lin J.H. (2015) Mutations in the unfolded protein response regulator ATF6 cause the cone dysfunction disorder achromatopsia. Nat. Genet. 47, 757–765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45).Narumi S., Amano N., Ishii T., Katsumata N., Muroya K., Adachi M., Toyoshima K., Tanaka Y., Fukuzawa R., Miyako K., Kinjo S., Ohga S., Ihara K., Inoue H., Kinjo T., Hara T., Kohno M., Yamada S., Urano H., Kitagawa Y., Tsugawa K., Higa A., Miyawaki M., Okutani T., Kizaki Z., Hamada H., Kihara M., Shiga K., Yamaguchi T., Kenmochi M., Kitajima H., Fukami M., Shimizu A., Kudoh J., Shibata S., Okano H., Miyake N., Matsumoto N., Hasegawa T. (2016) SAMD9 mutations cause a novel multisystem disorder, MIRAGE syndrome, and are associated with loss of chromosome 7. Nat. Genet. 48, 792–797. [DOI] [PubMed] [Google Scholar]
- 46).Adzhubei I.A., Schmidt S., Peshkin L., Ramensky V.E., Gerasimova A., Bork P., Kondrashov A.S., Sunyaev S.R. (2010) A method and server for predicting damaging missense mutations. Nat. Methods 7, 248–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47).Niknafs N., Kim D., Kim R., Diekhans M., Ryan M., Stenson P.D., Cooper D.N., Karchin R. (2013) MuPIT interactive: webserver for mapping variant positions to annotated, interactive 3D structures. Hum. Genet. 132, 1235–1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48).Desmet F.O., Hamroun D., Lalande M., Collod-Beroud G., Claustres M., Beroud C. (2009) Human Splicing Finder: an online bioinformatics tool to predict splicing signals. Nucleic Acids Res. 37, e67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49).Need A.C., Shashi V., Hitomi Y., Schoch K., Shianna K.V., McDonald M.T., Meisler M.H., Goldstein D.B. (2012) Clinical application of exome sequencing in undiagnosed genetic conditions. J. Med. Genet. 49, 353–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50).Hitomi Y., Tsuchiya N., Kawasaki A., Ohashi J., Suzuki T., Kyogoku C., Fukazawa T., Bejrachandra S., Siriboonrit U., Chandanayingyong D., Suthipinittharm P., Tsao B.P., Hashimoto H., Honda Z., Tokunaga K. (2004) CD72 polymorphisms associated with alternative splicing modify susceptibility to human systemic lupus erythematosus through epistatic interaction with FCGR2B. Hum. Mol. Genet. 13, 2907–2917. [DOI] [PubMed] [Google Scholar]
- 51).Hitomi Y., Adachi T., Tsuchiya N., Honda Z., Tokunaga K., Tsubata T. (2012) Human CD72 splicing isoform responsible for resistance to systemic lupus erythematosus regulates serum immunoglobulin level and is localized in endoplasmic reticulum. BMC Immunol. 13, 72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52).Frazer K.A., Murray S.S., Schork N.J., Topol E.J. (2009) Human genetic variation and its contribution to complex traits. Nat. Rev. Genet. 10, 241–251. [DOI] [PubMed] [Google Scholar]
- 53).Kimura H. (2013) Histone modifications for human epigenome analysis. J. Hum. Genet. 58, 439–445. [DOI] [PubMed] [Google Scholar]
- 54).Maurano M.T., Humbert R., Rynes E., Thurman R.E., Haugen E., Wang H., Reynolds A.P., Sandstrom R., Qu H., Brody J., Shafer A., Neri F., Lee K., Kutyavin T., Stehling-Sun S., Johnson A.K., Canfield T.K., Giste E., Diegel M., Bates D., Hansen R.S., Neph S., Sabo P.J., Heimfeld S., Raubitschek A., Ziegler S., Cotsapas C., Sotoodehnia N., Glass I., Sunyaev S.R., Kaul R., Stamatoyannopoulos J.A. (2012) Systematic localization of common disease-associated variation in regulatory DNA. Science 337, 1190–1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55).Ward L.D., Kellis M. (2012) HaploReg: a resource for exploring chromatin states, conservation, and regulatory motif alterations within sets of genetically linked variants. Nucleic Acids Res. 40, D930–D934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56).Boyle A.P., Hong E.L., Hariharan M., Cheng Y., Schaub M.A., Kasowski M., Karczewski K.J., Park J., Hitz B.C., Weng S., Cherry J.M., Snyder M. (2012) Annotation of functional variation in personal genomes using RegulomeDB. Genome Res. 22, 1790–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57).Kent W.J., Sugnet C.W., Furey T.S., Roskin K.M., Pringle T.H., Zahler A.M., Haussler D. (2002) The human genome browser at UCSC. Genome Res. 12, 996–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58).Severin J., Lizio M., Harshbarger J., Kawaji H., Daub C.O., Hayashizaki Y., FANTOM Consortium. Bertin N., Forrest A.R. (2014) Interactive visualization and analysis of large-scale sequencing datasets using ZENBU. Nat. Biotechnol. 32, 217–219. [DOI] [PubMed] [Google Scholar]
- 59).Wingender E., Dietze P., Karas H., Knüppel R. (1996) TRANSFAC: a database on transcription factors and their DNA binding sites. Nucleic Acids Res. 24, 238–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60).Hitomi Y., Kawashima M., Aiba Y., Nishida N., Matsuhashi M., Okazaki H., Nakamura M., Tokunaga K. (2015) Human primary biliary cirrhosis-susceptible allele of rs4979462 enhances TNFSF15 expression by binding NF-1. Hum. Genet. 134, 737–747. [DOI] [PubMed] [Google Scholar]
- 61).Bhattacharya A., Ziebarth J.D., Cui Y. (2014) PolymiRTS Database 3.0: linking polymorphisms in microRNAs and their target sites with human diseases and biological pathways. Nucleic Acids Res. 42, D86–D91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62).Chang T.H., Huang H.Y., Hsu J.B., Weng S.L., Horng J.T., Huang H.D. (2013) An enhanced computational platform for investigating the roles of regulatory RNA and for identifying functional RNA motifs. BMC Bioinformatics 14 (Suppl 2), S4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63).Hitomi Y., Ebisawa M., Tomikawa M., Imai T., Komata T., Hirota T., Harada M., Sakashita M., Suzuki Y., Shimojo N., Kohno Y., Fujita K., Miyatake A., Doi S., Enomoto T., Taniguchi M., Higashi N., Nakamura Y., Tamari M. (2009) Associations of functional NLRP3 polymorphisms with susceptibility to food-induced anaphylaxis and aspirin-induced asthma. J. Allergy Clin. Immunol. 124, 779–785. [DOI] [PubMed] [Google Scholar]
- 64).Sabarinathan R., Tafer H., Seemann S.E., Hofacker I.L., Stadler P.F., Gorodkin J. (2013) The RNAsnp web server: predicting SNP effects on local RNA secondary structure. Nucleic Acids Res. 41, W475–W479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65).Zuker M. (2003) Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 31, 3406–3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66).de Koning D.J., Haley C.S. (2005) Genetical genomics in humans and model organisms. Trends Genet. 21, 377–381. [DOI] [PubMed] [Google Scholar]
- 67).Ge B., Pokholok D.K., Kwan T., Grundberg E., Morcos L., Verlaan D.J., Le J., Koka V., Lam K.C., Gagné V., Dias J., Hoberman R., Montpetit A., Joly M.M., Harvey E.J., Sinnett D., Beaulieu P., Hamon R., Graziani A., Dewar K., Harmsen E., Majewski J., Göring H.H., Naumova A.K., Blanchette M., Gunderson K.L., Pastinen T. (2009) Global patterns of cis variation in human cells revealed by high-density allelic expression analysis. Nat. Genet. 41, 1216–1222. [DOI] [PubMed] [Google Scholar]
- 68).He X., Fuller C.K., Song Y., Meng Q., Zhang B., Yang X., Li H. (2013) Sherlock: detecting gene-disease associations by matching patterns of expression QTL and GWAS. Am. J. Hum. Genet. 92, 667–680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69).Westra H.J., Peters M.J., Esko T., Yaghootkar H., Schurmann C., Kettunen J., Christiansen M.W., Fairfax B.P., Schramm K., Powell J.E., Zhernakova A., Zhernakova D.V., Veldink J.H., Van den Berg L.H., Karjalainen J., Withoff S., Uitterlinden A.G., Hofman A., Rivadeneira F., ’t Hoen P.A., Reinmaa E., Fischer K., Nelis M., Milani L., Melzer D., Ferrucci L., Singleton A.B., Hernandez D.G., Nalls M.A., Homuth G., Nauck M., Radke D., Völker U., Perola M., Salomaa V., Brody J., Suchy-Dicey A., Gharib S.A., Enquobahrie D.A., Lumley T., Montgomery G.W., Makino S., Prokisch H., Herder C., Roden M., Grallert H., Meitinger T., Strauch K., Li Y., Jansen R.C., Visscher P.M., Knight J.C., Psaty B.M., Ripatti S., Teumer A., Frayling T.M., Metspalu A., van Meurs J.B., Franke L. (2013) Systematic identification of trans eQTLs as putative drivers of known disease associations. Nat. Genet. 45, 1238–1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70).GTEx Consortium (2013) The Genotype-Tissue Expression (GTEx) project. Nat. Genet. 45, 580–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71).GTEx Consortium (2015) Human genomics. The Genotype-Tissue Expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science 348, 648–660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72).Okada Y., Wu D., Trynka G., Raj T., Terao C., Ikari K., Kochi Y., Ohmura K., Suzuki A., Yoshida S., Graham R.R., Manoharan A., Ortmann W., Bhangale T., Denny J.C., Carroll R.J., Eyler A.E., Greenberg J.D., Kremer J.M., Pappas D.A., Jiang L., Yin J., Ye L., Su D.F., Yang J., Xie G., Keystone E., Westra H.J., Esko T., Metspalu A., Zhou X., Gupta N., Mirel D., Stahl E.A., Diogo D., Cui J., Liao K., Guo M.H., Myouzen K., Kawaguchi T., Coenen M.J., van Riel P.L., van de Laar M.A., Guchelaar H.J., Huizinga T.W., Dieudé P., Mariette X., Bridges S.L., Jr., Zhernakova A., Toes R.E., Tak P.P., Miceli-Richard C., Bang S.Y., Lee H.S., Martin J., Gonzalez-Gay M.A., Rodriguez-Rodriguez L., Rantapää-Dahlqvist S., Arlestig L., Choi H.K., Kamatani Y., Galan P., Lathrop M., RACI consortium. GARNET consortium. Eyre S., Bowes J., Barton A., de Vries N., Moreland L.W, Criswell L.A., Karlson E.W., Taniguchi A., Yamada R., Kubo M., Liu J.S., Bae S.C., Worthington J., Padyukov L., Klareskog L., Gregersen P.K., Raychaudhuri S., Stranger B.E., De Jager P.L., Franke L., Visscher P.M., Brown M.A., Yamanaka H., Mimori T., Takahashi A., Xu H., Behrens T.W., Siminovitch K.A., Momohara S., Matsuda F., Yamamoto K., Plenge R.M. (2014) Genetics of rheumatoid arthritis contributes to biology and drug discovery. Nature 506, 376–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73).Ackermann M., Sikora-Wohlfeld W., Beyer A. (2013) Impact of natural genetic variation on gene expression dynamics. PLoS Genet. 9, e1003514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74).Francesconi M., Lehner B. (2014) The effects of genetic variation on gene expression dynamics during development. Nature 505, 208–211. [DOI] [PubMed] [Google Scholar]
- 75).Kaniwa N., Saito Y. (2013) Pharmacogenomics of severe cutaneous adverse reactions and drug-induced liver injury. J. Hum. Genet. 58, 317–326. [DOI] [PubMed] [Google Scholar]
- 76).Chung W.H., Hung S.I., Hong H.S., Hsih M.S., Yang L.C., Ho H.C., Wu J.Y., Chen Y.T. (2004) A marker for Stevens-Johnson syndrome. Nature 428, 486. [DOI] [PubMed] [Google Scholar]
- 77).Ozeki T., Mushiroda T., Yowang A., Takahashi A., Kubo M., Shirakata Y., Ikezawa Z., Iijima M., Shiohara T., Hashimoto K., Kamatani N., Nakamura Y. (2011) Genome-wide association study identifies HLA-A*3101 allele as a genetic risk factor for carbamazepine-induced cutaneous adverse drug reactions in Japanese population. Hum. Mol. Genet. 20, 1034–1041. [DOI] [PubMed] [Google Scholar]
- 78).Kaniwa N., Saito Y., Aihara M., Matsunaga K., Tohkin M., Kurose K., Furuya H., Takahashi Y., Muramatsu M., Kinoshita S., Abe M., Ikeda H., Kashiwagi M., Song Y., Ueta M., Sotozono C., Ikezawa Z., Hasegawa R., JSAR research group (2010) HLA-B*1511 is a risk factor for carbamazepine-induced Stevens-Johnson syndrome and toxic epidermal necrolysis in Japanese patients. Epilepsia 51, 2461–2465. [DOI] [PubMed] [Google Scholar]
- 79).Wang L.S., D’Souza L.S., Jacobson I.M. (2016) Hepatitis C-A clinical review. J. Med. Virol. 88, 1844–1855. [DOI] [PubMed] [Google Scholar]
- 80).Ghany M.G., Strader D.B., Thomas D.L., Seeff L.B. (2009) AASLD practice guidelines—diagnosis, management, and treatment of hepatitis C: an update. Hepatology 49, 1335–1374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81).Ge D., Fellay J., Thompson A.J., Simon J.S., Shianna K.V., Urban T.J., Heinzen E.L., Qiu P., Bertelsen A.H., Muir A.J., Sulkowski M., McHutchison J.G., Goldstein D.B. (2009) Genetic variation in IL28B predicts hepatitis C treatment-induced viral clearance. Nature 461, 399–401. [DOI] [PubMed] [Google Scholar]
- 82).Tanaka Y., Nishida N., Sugiyama M., Kurosaki M., Matsuura K., Sakamoto N., Nakagawa M., Korenaga M., Hino K., Hige S., Ito Y., Mita E., Tanaka E., Mochida S., Murawaki Y., Honda M., Sakai A., Hiasa Y., Nishiguchi S., Koike A., Sakaida I., Imamura M., Ito K., Yano K., Masaki N., Sugauchi F., Izumi N., Tokunaga K., Mizokami M. (2009) Genome-wide association of IL28B with response to pegylated interferon-alpha and ribavirin therapy for chronic hepatitis C. Nat. Genet. 41, 1105–1109. [DOI] [PubMed] [Google Scholar]
- 83).Suppiah V., Moldovan M., Ahlenstiel G., Berg T., Weltman M., Abate M.L., Bassendine M., Spengler U., Dore G.J., Powell E., Riordan S., Sheridan D., Smedile A., Fragomeli V., Müller T., Bahlo M., Stewart G.J., Booth D.R., George J. (2009) IL28B is associated with response to chronic hepatitis C interferon-alpha and ribavirin therapy. Nat. Genet. 41, 1100–1104. [DOI] [PubMed] [Google Scholar]
- 84).Puri C.P. (2011) Interleukin 28B polymorphisms and hepatitis C—translating the association into clinical decision making. J. Clin. Exp. Hepatol. 1, 27–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85).Fellay J., Thompson A.J., Ge D., Gumbs C.E., Urban T.J., Shianna K.V., Little L.D., Qiu P., Bertelsen A.H., Watson M., Warner A., Muir A.J., Brass C., Albrecht J., Sulkowski M., McHutchison J.G., Goldstein D.B. (2010) ITPA gene variants protect against anaemia in patients treated for chronic hepatitis C. Nature 464, 405–408. [DOI] [PubMed] [Google Scholar]
- 86).Tanaka Y., Kurosaki M., Nishida N., Sugiyama M., Matsuura K., Sakamoto N., Enomoto N., Yatsuhashi H., Nishiguchi S., Hino K., Hige S., Itoh Y., Tanaka E., Mochida S., Honda M., Hiasa Y., Koike A., Sugauchi F., Kaneko S., Izumi N., Tokunaga K., Mizokami M. (2011) Genome-wide association study identified ITPA/DDRGK1 variants reflecting thrombocytopenia in pegylated interferon and ribavirin therapy for chronic hepatitis C. Hum. Mol. Genet. 20, 3507–3516. [DOI] [PubMed] [Google Scholar]
- 87).Hitomi Y., Cirulli E.T., Fellay J., McHutchison J.G., Thompson A.J., Gumbs C.E., Shianna K.V., Urban T.J., Goldstein D.B. (2011) Inosine triphosphate protects against ribavirin-induced adenosine triphosphate loss by adenylosuccinate synthase function. Gastroenterology 140, 1314–1321. [DOI] [PubMed] [Google Scholar]
- 88).Early Breast Cancer Trialists’ Collaborative Group (EBCTCG) Davies C., Godwin J., Gray R., Clarke M., Cutter D., Darby S., McGale P., Pan H.C., Taylor C., Wang Y.C., Dowsett M., Ingle J., Peto R. (2011) Relevance of breast cancer hormone receptors and otherfactors to the efficacy of adjuvant tamoxifen: patient-level meta-analysis ofrandomised trials. Lancet 378, 771–784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89).Zanger U.M., Fischer J., Raimundo S., Stüven T., Evert B.O., Schwab M., Eichelbaum M. (2001) Comprehensive analysis of thegenetic factors determining expression and function of hepatic CYP2D6. Pharmacogenetics 11, 573–585. [DOI] [PubMed] [Google Scholar]
- 90).Sim S.C., Ingelman-Sundberg M. (2013) Update on allele nomenclature for humancytochromes P450 and the human cytochrome P450 allele (CYP-allele) nomenclature database. Methods Mol. Biol. 987, 251–259. [DOI] [PubMed] [Google Scholar]
- 91).Sachse C., Brockmöller J., Bauer S., Roots I. (1997) Cytochrome P450 2D6 variants in a Caucasian population: allele frequencies and phenotypic consequences. Am. J. Hum. Genet. 60, 284–295. [PMC free article] [PubMed] [Google Scholar]
- 92).Griese E.U., Zanger U.M., Brudermanns U., Gaedigk A., Mikus G., Mörike K., Stüven T., Eichelbaum M. (1998) Assessment of the predictive power of genotypes for the in-vivo catalytic function of CYP2D6 in a German population. Pharmacogenetics 8, 15–26. [DOI] [PubMed] [Google Scholar]
- 93).Raimundo S., Toscano C., Klein K., Fischer J., Griese E.U., Eichelbaum M., Schwab M., Zanger U.M. (2004) A novel intronic mutation, 2988G > A, with high predictivity for impaired function of cytochrome P450 2D6 in white subjects. Clin. Pharmacol. Ther. 76, 128–138. [DOI] [PubMed] [Google Scholar]
- 94).Regan M.M., Leyland-Jones B., Bouzyk M., Pagani O., Tang W., Kammler R., Dell’orto P., Biasi M.O., Thürlimann B., Lyng M.B., Ditzel H.J., Neven P., Debled M., Maibach R., Price K.N., Gelber R.D., Coates A.S., Goldhirsch A., Rae J.M., Viale G., Breast International Group (BIG) 1-98 Collaborative Group (2012) CYP2D6 genotype and tamoxifen response in postmenopausal women with endocrine-responsive breast cancer: the Breast International Group 1–98 trial. J. Natl. Cancer Inst. 104, 441–451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95).Hertz D.L., McLeod H.L., Irvin W.J. (2012) Tamoxifen and CYP2D6: a contradiction of data. Oncologist 17, 620–630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96).Emery A.E.H. (1991) Population frequencies of inherited neuromuscular diseases — a world survey. Neuromuscul. Disord. 1, 19–29. [DOI] [PubMed] [Google Scholar]
- 97).Aartsma-Rus A., Van Deutekom J.C., Fokkema I.F., Van Ommen G.J., Den Dunnen J.T. (2006) Entries in the Leiden Duchenne muscular dystrophy mutation database: an overview of mutation types and paradoxical cases that confirm the reading-frame rule. Muscle Nerve 34, 135–144. [DOI] [PubMed] [Google Scholar]
- 98).Muntoni F., Torelli S., Ferlini A. (2003) Dystrophin and mutations: one gene, several proteins, multiple phenotypes. Lancet Neurol. 2, 731–740. [DOI] [PubMed] [Google Scholar]
- 99).Hoffman E.P., Fischbeck K.H., Brown R.H., Johnson M., Medori R., Loike J.D., Harris J.B., Waterston R., Brooke M., Specht L., Kupsky W., Chamberlain J., Caskey T.C., Shapiro F., Kunkel L.M. (1988) Characterization of dystrophin in muscle-biopsy specimens from patients with Duchenne’s or Becker’s muscular dystrophy. N. Engl. J. Med. 318, 1363–1368. [DOI] [PubMed] [Google Scholar]
- 100).Koenig M., Beggs A.H., Moyer M., Scherpf S., Heindrich K., Bettecken T., Meng G., Müller C.R., Lindlöf M., Kaariainen H., de la Chapellet A., Kiuru A., Savontaus M.L., Gilgenkrantz H., Récan D., Chelly J., Kaplan J.C., Covone A.E., Archidiacono N., Romeo G., Liechti-Gailati S., Schneider V., Braga S., Moser H., Darras B.T., Murphy P., Francke U., Chen J.D., Morgan G., Denton M., Greenberg C.R., Wrogemann K., Blonden L.A., van Paassen M.B., van Ommen G.J., Kunkel L.M. (1989) The molecular basis for Duchenne versus Becker muscular dystrophy: correlation of severity with type of deletion. Am. J. Hum. Genet. 45, 498–506. [PMC free article] [PubMed] [Google Scholar]
- 101).Monaco A.P., Bertelson C.J., Liechti-Gallati S., Moser H., Kunkel L.M. (1988) An explanation for the phenotypic differences between patients bearing partial deletions of the DMD locus. Genomics 2, 90–95. [DOI] [PubMed] [Google Scholar]
- 102).Aartsma-Rus A., van Vliet L., Hirschi M., Janson A.A., Heemskerk H., de Winter C.L., de Kimpe S., van Deutekom J.C., Hoen P.A., van Ommen G.J. (2009) Guidelines for antisense oligonucleotide design and insight into splicemodulating mechanisms. Mol. Ther. 17, 548–553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103).van Deutekom J.C., Janson A.A., Ginjaar I.B., Frankhuizen W.S., Aartsma-Rus A., Bremmer-Bout M., den Dunnen J.T., Koop K., van der Kooi A.J., Goemans N.M., de Kimpe S.J., Ekhart P.F., Venneker E.H., Platenburg G.J., Verschuuren J.J., van Ommen G.J. (2007) Local dystrophin restoration with antisense oligonucleotide PRO051. N. Engl. J. Med. 357, 2677–2686. [DOI] [PubMed] [Google Scholar]
- 104).Goemans N.M., Tulinius M., van den Akker J.T., Burm B.E., Ekhart P.F., Heuvelmans N., Holling T., Janson A.A., Platenburg G.J., Sipkens J.A., Sitsen J.M., Aartsma-Rus A., van Ommen G.J., Buyse G., Darin N., Verschuuren J.J., Campion G.V., de Kimpe S.J., van Deutekom J.C. (2011) Systemic administration of PRO051 in Duchenne’s muscular dystrophy. N. Engl. J. Med. 364, 1513–1522. [DOI] [PubMed] [Google Scholar]
- 105).Pennisi E. (2013) The CRISPR craze. Science 341, 833–836. [DOI] [PubMed] [Google Scholar]
- 106).Ledford H. (2015) CRISPR, the disruptor. Nature 522, 20–24. [DOI] [PubMed] [Google Scholar]
- 107).Osborn M., Lonetree C.L., Webber B.R., Patel D., Dunmire S., McElroy A.N., DeFeo A.P., MacMillan M.L., Wagner J., Balzar B.R., Tolar J. (2016) CRISPR/Cas9 targeted gene editing and cellular engineering in Fanconi anemia. Stem Cells Dev. 25, 1591–1603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108).Long C., Amoasii L., Mireault A.A., McAnally J.R., Li H., Sanchez-Ortiz E., Bhattacharyya S., Shelton J.M., Bassel-Duby R., Olson E.N. (2016) Postnatal genome editing partially restores dystrophin expression in a mouse model of muscular dystrophy. Science 351, 400–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109).Nelson C.E., Hakim C.H., Ousterout D.G., Thakore P.I., Moreb E.A., Castellanos Rivera R.M., Madhavan S., Pan X., Ran F.A., Yan W.X., Asokan A., Zhang F., Duan D., Gersbach C.A. (2016) In vivo genome editing improves muscle function in a mouse model of Duchenne muscular dystrophy. Science 351, 403–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110).Tabebordbar M., Zhu K., Cheng J.K., Chew W.L., Widrick J.J., Yan W.X., Maesner C., Wu E.Y., Xiao R., Ran F.A., Cong L., Zhang F., Vandenberghe L.H., Church G.M., Wagers A.J. (2016) In vivo gene editing in dystrophic mouse muscle and muscle stem cells. Science 351, 407–411. [DOI] [PMC free article] [PubMed] [Google Scholar]