Abstract
We have been working on functional genomics using C. elegans as a model organism. We first used cell-type specific markers and preexisting mutants to investigate how genotype-phenotype causal relationships are regulated. With the aid of transgenic methods, we analyzed various biological processes in C. elegans. We have developed efficient methods to isolate gene knockout strains. Thousands of strains isolated this way are used by many researchers and have revealed many biological mechanisms. We have also developed methods to examine the functions of genes in a comprehensive manner by integrating transgenes into chromosomes, designing conditional knockouts, and creating balancers for lethal mutations. A combination of these biological resources and techniques will be useful to understand the functions of genes in C. elegans, which has many genes that are orthologous to those of higher organisms including humans.
Keywords: Caenorhabditis elegans, functional genomics, transgenic, RNAi, knockout
1. Introduction
Medical and life science research studies involve many disciplinary approaches, including genetics. Strategies using genetics are characterized by tight causal relationships between genotypes and phenotypes, although these are often indirect. Many of the earlier findings of genetic studies were descriptive and did not reveal cellular and molecular mechanisms. Genetic studies of human diseases are also based on the causal relationships between some genetic variations and symptoms or disease susceptibility. Importantly, the GWAS (genome-wide association study) and other new technologies such as high-throughput genome sequencing are now able to handle an enormous number of genes and, thus, are rapidly changing not only medical researches, but also clinical practice (see ref. 1–3 and cancer moonshot. Bethesda, MD: National Cancer Institute (https://www.cancer.gov/research/key-initiatives/moonshot-cancer-initiative)). However, in spite of the huge volume of information on the relationships between genetic variations and diseases, an increased understanding of individual gene function and interactions among genes would help us to understand their influence on the outcome.
Genetics is also useful for investigating cellular and molecular mechanisms and genetic pathways. To fully understand the cellular and molecular mechanisms, it is useful to know the functions of genes and their products in relation to phenotypes found in experimental organisms and genetic epistatic analyses. To facilitate the acquisition of such experimental evidence, we have been using the nematode Caenorhabditis elegans (C. elegans), a small model organism. C. elegans has a large number of genes that are homologous to human genes and therefore, studies of gene function in C. elegans can help researchers understand the functions of human genes and their products. Moreover, C. elegans was the first model animal to have its whole genome sequenced.4) The availability of whole-genome sequences for many organisms, including both experimental animals and humans, has facilitated research to elucidate the functions of disease-related genes. Here, I describe how C. elegans can be used as a tool for gene function analyses, mainly focusing on gene knockout, RNA interference (double-stranded RNA-mediated gene silencing), and transgenics, with which many C. elegans researchers are working. We have been isolating deletion mutants of C. elegans and have distributed them to many laboratories worldwide through the National BioResource Project for the nematode (ref. 5, http://shigen.nig.ac.jp/c.elegans/index.jsp). By now, the cumulative number of publications using our mutants has exceeded 2,000, and it is difficult to summarize the entire contents of those papers by citing all the important publications. Therefore, here I will review the development of our functional genomics approaches using C. elegans and explain how C. elegans can be used for functional genomics studies by reviewing some publications that are relevant to our own experiences.
2. C. elegans as a model organism for genetic analyses
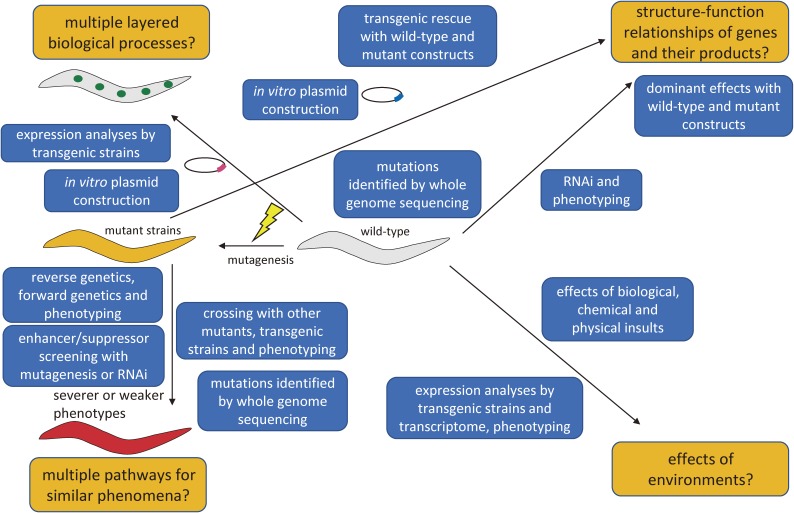
To describe how genetic analyses can be used to solve biological questions using C. elegans, a schematic diagram is shown (Fig. 1). Explanations of technical terms are described in the legend of Fig. 1 and historical examples are described in the following text.
Figure 1.
A schematic diagram showing questions to be addressed by functional genomics of C. elegans. Examples of important questions to be addressed by genetical analyses are indicated by orange boxes, whereas methods to solve the questions are indicated by blue boxes. Representative questions are originated by multiple layered biological processes, multiple pathways for similar phenomena, structure-function relationships of genes and their products, and effects of environments. Multiple layered biological processes mean that organisms have multiple layered regulations of causal relationships with molecular-, cellular-, tissue- and whole body-levels. Also, organisms have spatio-temporal regulations. Complicated mechanisms are unraveled, for example, by examining transgenic analyses mentioned in the text. Multiple pathways for similar phenomena mean that organisms have often multiple independent causes for seemingly similar phenotypes and these are categorized and unraveled by crossing with other strains (as epistatic analyses) and isolation of enhancers and suppressors to identify genetic interactions. Effects of environments are important subjects because gene regulations are essential for adaptation to the environment an organism is exposed. Explanation of blue boxes are followings. The phrase “expression analyses by transgenic strains” indicates that transgenic strains can be generated by injecting plasmids into the germline of the animals. The phrase “in vitro plasmid construction” means that researchers can generate varieties of plasmids with current molecular biological methods. The phrase “reverse genetics, forward genetics and phenotyping” means that researchers can isolate new mutants only by sequences and only by the inspection of phenotypes of interest as “reverse genetics” and “forward genetics”, respectively. The phrase “enhancer/suppressor screening with mutagenesis or RNAi” means that researchers screen for new mutants (or RNAi clones) which aggravate (enhancer) or attenuate (suppressor) the phenotypes of parent mutants. The phrase “crossing with other mutants, transgenic strains and phenotyping” means that researchers make double mutants etc. to see which phenotype is dominant, to dissect whether two causal genetic variations are interactive with each other. The phrase “mutations identified by whole genome sequencing” means that researchers can identify any types of mutations by analyzing the whole C. elegans genome using the next-generation sequencers and bioinformatics. The phrase “transgenic rescue with wild-type and mutant construct” means that researchers can microinject various plasmids to see whether the sequences can rescue the mutant phenotypes to understand whether some genetic variants retain wild-type function or not. The phrase “dominant effects with wild-type and mutant constructs” means that researchers can know by overexpression or ectopic expression of wild-type or mutant proteins in the wild-type animal background whether they have dominant phenotypes or not. The phrase “effects of biological, chemical and physical insults” means that researchers can expose worms to some biological (for example, bacterial infection), chemical (for example, toxic substances) and physical (for example, hot temperature) and see the effects by phenotypes. The phrase “expression analyses by transgenic strains and transcriptome, phenotyping” means that researchers can use transgenic reporters to see the potential changes of the expression patterns under some environmental changes in addition to the information about when and where the gene products may play roles. Also, researchers can use the worms to analyze RNA expression changes with microarray or RNA sequencing with a next-generation sequencer to find the expression changes under some environmental changes in addition to phenotypes.
Half a century ago, Dr. Sydney Brenner proposed the use of the nematode C. elegans as a model organism to understand development and behavior because of its simple structure and short life cycle, compared with those of the various other experimental model organisms. Researchers working on C. elegans have been trying to elucidate its biological processes as a whole. The best-known examples are cell-lineage description, morphological reconstitution of electron micrographs, and genome sequencing.4,6–8)
C. elegans is a small animal, only 1 mm long at the adult stage, and mainly proliferates as a hermaphrodite. The animals can be easily cultured on agar plates and fed with Escherichia coli. They can be stably stored for years in deep freezers or liquid nitrogen. The organism has only approximately 1,000 somatic cells, including 302 neurons and 7,000 synaptic connections that form neural networks.6,8) C. elegans worms grow from fertilized eggs to adulthood in only 3 days at 20 ℃. C. elegans has two sexes: hermaphrodite and male. Hermaphrodites have 5 pairs of autosomes and one pair of sex chromosomes (X), while males have only one sex chromosome and the same number of autosomes. Hermaphrodites have both oocytes and sperm, with which they self-fertilize to give rise to progeny, making it easy for researchers to obtain homozygous animals by culturing a single hermaphrodite. Males are found at a rate of approximately 1 in 500 self-fertilized hermaphrodites because of the loss of one X chromosome during the meiosis. The genome of C. elegans, approximately 100 Mb in size, is much smaller than the human genome of 3 Gb. Thanks to the organism’s small body and detailed basic descriptions with many experimental tools, researchers can use C. elegans as proliferative “test tubes”.
Brenner published a commemorative paper in 1973 in which he described hundreds of mutations mapped on 6 chromosomes (linkage groups) of C. elegans.9) These mutations served as genetic landmarks in the following analyses by researchers. After having obtained the genetic map, researchers tried to isolate many mutants with abnormalities, including cell lineage, apoptosis, behavior, and morphogenesis etc.10) Early efforts focused on the forward genetics, which involved isolating mutants with interesting phenotypes and then positionally cloning the causative genes. In the 1980s, it took researchers a long time to identify a gene that causes a given phenotype. Researchers added new genes to the genetic map, which eventually made it easier to identify other genes and sequence the genome.
In 1992, the C. elegans Sequencing Consortium reported its progress on chromosome III, and in 1998, the consortium published a paper that described the whole C. elegans genome sequence and the various characteristics of the genome structure.4,11) The genome analysis revealed that C. elegans has approximately 20,000 protein-coding genes and that they are homologous to protein-coding genes found in other organisms, including humans. In addition, many more non-coding RNA genes were found. During the genome sequencing process and after the release of the whole genome, researchers often used this information to achieve new insights into the biological processes. Interestingly, the C. elegans Sequencing Consortium joined the Human Genome Project, which expedited our present understanding of the genetic system and human diseases.12) After a number of bioinformatics analyses, a paper was published in 2011 that described an OrthoList of the C. elegans genome: 7,663 protein-coding genes (approximately 38% of C. elegans protein-coding genes) are orthologous to human genes, which are likely to be derived from common ancestors and have similar functions. Worms have many more homologous genes that constitute gene families related to human genes, which share sequence similarities but may not have the same functions.13) Thus, in spite of its simple structure, C. elegans has many molecular similarities to other higher eukaryotes and is useful for examining cellular and molecular functions in a multicellular system. With the wealth of information on genome structure, researchers began to focus more effort on reverse genetics (see below).
Another important characteristic of the C. elegans research is the sharing of strains and databases. C. elegans researchers have been sharing published strains through the Caenorhabditis Genetics Center (CGC) for decades (http://cbs.umn.edu/cgc/home). Researchers shared established data by constructing ACEDB (A C. elegans DataBase).14) Earlier on, researchers shared the database by copying the data through the community. In 2000, the successor database was established and became available through a website as WormBase (http://wormbase.org). It contains almost all published C. elegans research, so that anyone can search the accumulated data for information of interest. Through this standardization, researchers can avoid unnecessary trials and easily reproduce previous experiments to proceed to further analyses. By using C. elegans as a model organism, researchers can examine genotype-phenotype relationships in a reproducible manner by referring to prior knowledge, sharing strains among laboratories and examining a large number of clonal animals.
3. Analyses of nerve cell development as a beginning
I began my C. elegans research work in 1989 in the Chalfie laboratory at Columbia University. During my stay in the laboratory, we analyzed how gene expression and cell specification are regulated, focusing on touch receptor neurons. Touch receptor neurons, which are composed of 6 cells (two ALMs, AVM, PVM and two PLMs), had been analyzed by forward genetics, and important developmental and functional genes had been described.15–17) As an early phase of the project, we tried to develop a whole mount in situ hybridization method that allowed detection of gene expression at a single-cell resolution and was applicable to many genetic backgrounds without crossing strains to transgenic reporter strains such as lacZ fusion-expressing transgenic animals. We successfully detected the specific expression of two genes, ben-1 (benzimidazole resistant) and mec-7 (mechanosensory abnormality), both of which are β-tubulin, a major microtubule protein that forms a dimer with α-tubulin. Our efforts to understand gene functions via genotype-phenotype causal relationships began with an experiment on developmental neurobiology, which was published in 1993.18) In the course of the experiments, we used a combination of touch receptor neuron-specific markers including mec-7 and approximately 50 preexisting mutants that had shown some developmental defects in previous works. Those mutants, which were found as developmental determinants for cells other than touch receptor neurons, showed changes in touch receptor neuron development at a high frequency, which implied that many developmental and behavioral processes are regulated by a complicated combination of gene functions. The work also indicated that only a small portion of the markers, such as mec-7 and mec-4 genes, which are expressed in a touch receptor neuron-specific manner, and developmental mutants showed new biological insights into the causal relationships of development and behavior in C. elegans. The analysis revealed that additional genes are required to regulate the correct expression of touch receptor neuron-specific genes. The egl-44 (egg-laying defective) and egl-46 mutants had extra mec-7-positive cells in the head, which were transformed from FLP head sensory neurons. The lin-4 (abnormal cell lineage) mutants had ectopic mec-7-expressing cells in the mid-body, which were transformed from PVD mid-body sensory neurons. Loss-of-function mutations in lin-14 resulted in the loss of PVM neurons. ced-3 (cell death abnormality) and ced-4 mutants had extra mec-7-expressing cells that were survivors from apoptotic cells in the PLM cell lineage. sem-4 (sex muscle abnormal) mutants had ectopic mec-7-expressing cells in the tail, presumably transformed from PHC neurons near PLMs. Although the additional mutants found in this work did not show apparent touch-insensitive phenotypes, they were needed for correct differentiation of touch receptor neurons. Thus, given that many gene markers and many mutations are used in combination, it is unsurprising that enormous amounts of information would be obtained.
At that time, only classical gene expression markers were available. However, the introduction of the green fluorescent protein (GFP), which were originally obtained from a species of jellyfish (Aequorea victoria), as a neuronal visualization reporter for C. elegans mec-7 was enthusiastically appreciated by researchers in many fields of life science.19,20) Since the introduction of GFP as a biological reporter, many more fluorescent proteins with various fluorescent characteristics, including excitation/emission color spectra, have been developed,21) allowing, for example, double labeling of cells and/or molecules of interest as the experiment requires.22)
Although the labeling of cells/molecules had been much improved by that time, the collection of mutants defective for genes of interest was lagging far behind because of the difficult and laborious experimental procedures that the process required.23,24) However, the introduction of a method by Mello et al. to generate transgenic strains of C. elegans expedited the analysis by fluorescent protein reporters and functional analysis by ectopic expression of genes of interest.25) Because C. elegans bodies are transparent, they can be conveniently examined for fluorescence under the microscope, and thus they are useful for transgenic analyses.
4. Transgenic strains for functional genomics
Because transgenic strains have been readily available after year 1991, many researchers have worked with transgenic strains. We adapted the original methods developed by Mello et al. for our studies of functional analysis.
A. Integration of transgenes into chromosomes.
The original methods to isolate transgenic strains yield animals harboring extrachromosomal transgenes, usually called extrachromosomal arrays. Extrachromosomal arrays are often lost during development and between generations, and they cause mosaicism, which means loss of transgenes in subsets of somatic cells, and loss of transgenes in the progeny. The characteristics of extrachromosomal arrays themselves sometimes have an experimental advantage, e.g., for mosaic analyses, which utilize the mosaicism of transgenes as the random and differential presence or absence of proteins of interest, to determine whether phenotypes are caused by a cell-autonomous gene function. However, in many cases involving extrachromosomal transgenic strains, mosaic expression may result in, for example, partial rescue of the mutants and make experimental results unclear. To solve this problem, integration of extrachromosomal arrays into the chromosomes is useful.
We used UV irradiation to integrate extrachromosomal transgenes into a chromosome.26) After irradiation of transgenic strains with a UV crosslinker, which is used to fix DNAs and RNAs to nylon membranes for Southern and northern blotting, worms were allowed to produce F1 (daughter generation) animals. F1 animals were cultured in individual plates, and integrated strains were examined to determine whether the transgenic markers were segregated in the F2 (granddaughter) generation and later transmitted to all the progeny by inspection of the expression of fluorescent protein or other markers under a dissecting microscope. We were able to isolate strains that had stably integrated transgenes from up to 5% of parent extrachromosomal strains in this way.
B. Analyses of the effects of ectopic expression of human disease-causal genes.
After we obtained stable transgenic strains by integrating extrachromosomal transgenes, we applied the technique to a number of analyses. Two examples are shown here. We collaborated with Dr. T. Iwatsubo on the ectopic expression of α-synuclein, a causal gene of familial Parkinson’s disease and a major protein of Lewy bodies of sporadic Parkinson’s disease.27)
α-Synuclein is one of the best-known genes related to Parkinson’s disease and is enriched in synapses and nuclei, as suggested by the name. There is no homologous gene for α-synuclein in the C. elegans genome. We overexpressed α-synuclein using promoters that are active in C. elegans dopaminergic neurons.28) The α-synuclein accumulated in the cell bodies and neurites of dopamine neurons. We did not find neuronal loss in these transgenic strains. It is known that dopamine is important for C. elegans behavior in response to the presence or absence of food.29) α-Synuclein transgenic animals showed behavioral abnormalities similar to those of dopamine neuron-deficient mutants and were rescued by the administration of dopamine in the culture. Thus, the overexpression of α-synuclein in C. elegans dopamine neurons caused functional abnormalities within a shorter period than human disease progress.
Additional transgenic strains were used to screen for modulators of α-synuclein toxicity.30) Transgenic strains expressing α-synuclein in all neurons under the unc-51 promoter showed a weak Unc (uncoordinated) phenotype. We used 1,673 RNAi (RNA interference) clones to find the modifiers of the phenotype by looking for severe growth or motor defects. We found 10 genes contributing to the phenotype. Among these were four genes related to the endocytic pathway. The RNAi phenotypes were confirmed by crossing the transgenic animals with mutants. Furthermore, the accumulation of phosphorylated α-synuclein was observed in neuronal cell bodies in an AP-2 (a clathrin adaptor protein) subunit knockdown, mimicking synucleinopathy (α-synuclein-mediated neurodegenerative disorder).
C. Screening for splicing factors.
Another application of chromosomally integrated transgenic strains was to search for alternative splicing factors. It is important for multicellular organisms to have alternative splicing mechanisms to generate many more varieties of functional proteins than the number of genes in the genome by using distinct sets of exons from a gene. To address this issue, we collaborated with Dr. H. Kuroyanagi and Dr. M. Hagiwara to screen for C. elegans alternative splicing mutants. We chose the gene egl-15, which has mutually exclusive exons by alternative splicing and tissue-specific expression.31) We constructed reporters indicating which exon was expressed by alternative splicing, enabling us to detect changes in the splicing pattern by color. We injected plasmids and obtained extrachromosomal transgenic animals harboring egl-15A fused to RFP (red fluorescent protein) and egl-15B fused to GFP. EGL-15B protein was mainly expressed in the hypodermis and neurons in the wild-type background, whereas EGL-15A protein was mainly expressed in muscle cells. We then integrated the transgene into a chromosome. The transgenic animals showed predominantly “red” fluorescence because of a larger volume of muscle cells. Then, we mutagenized the transgenic animals and searched for “green” worms by using a fluorescence-activated worm sorter. We obtained green mutant worms, and among them were mutants that had lesions in a gene, asd-1 (alternative splicing defective), a homolog of the human FOX family splicing factors. We found that asd-1 was partially redundant with fox-1 (a FOX ortholog) and alternative splicing patterns were gradually changed by the dosage of these genes.
5. Isolation of gene knockout strains as a tool for reverse genetics
Once mutant phenotypes have been described, researchers can perform transgenic rescue experiments to examine protein structure-function relationships. With the aid of transgenic techniques, the in vivo functions of wild-type and mutant sequences can be compared in C. elegans by examining the rescuing activity of the mutant phenotypes. Transgenes with a domain swapped with other genes, including human homologs, may also reveal the structure-function relationships of the molecules of interest. Thus, using mutants, we can infer more definitive causal relationships rather than mere correlation.
C. elegans was the first animal model to have its whole genome sequenced. During the process, researchers were aware of the usefulness of the sequence information because sequencing revealed that many homologous genes, with which higher vertebrates share the same ancestral genes, are present in the C. elegans genome. Many laboratories tried to develop methods to isolate mutants that could be used for reverse genetics (see Fig. 1 legend).23,32) However, in the early stages of those trials, many people recognized that it was very laborious to isolate mutants. To overcome this problem, we tried to develop efficient mutagenesis and screening methods, which eventually allowed us to obtain thousands of mutants for gene function analyses (so-called “tm mutants” named after the allele names of the mutants isolated in our laboratory). We preferred to use deletion mutants because deletions are easy to detect by PCR (polymerase chain reaction) and agarose gel electrophoresis and are more likely to disrupt gene functions than point mutations.
A. ben-1 as both a humble and assertive gene.
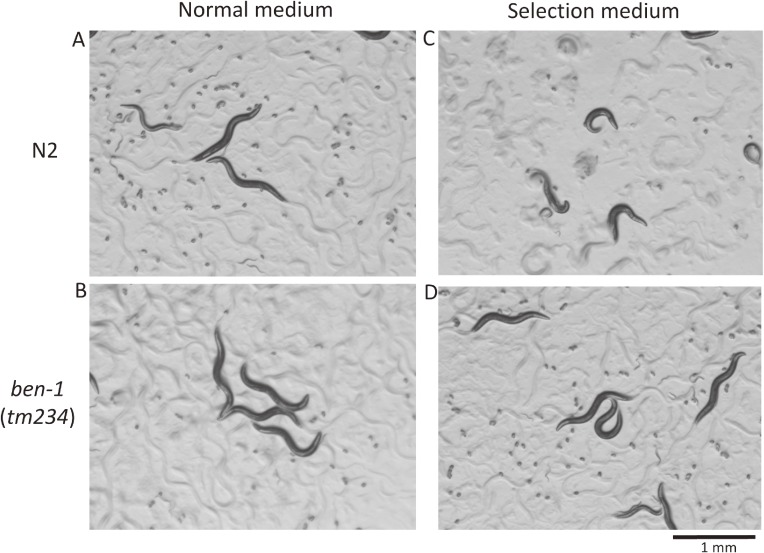
C. elegans β-tubulins are highly homologous with each other except in the C-terminal regions.33,34) While working at Columbia University, I learned of the interesting phenotypes of ben-1 mutants published by the Chalfie laboratory;33) wild-type animals are sensitive to an anti-tubulin drug benzimidazole and become sick, Dpy (dumpy) and Unc (uncoordinated movement), and eventually die. By contrast, ben-1 mutants are completely resistant to the chemical and paradoxically healthy on the media containing the drug. ben-1 mutants do not show any visible phenotype on their own, presumably because the ben-1 gene is compensated by the redundant function of other β-tubulin(s), and the mutants are completely healthy on the plates with or without the chemical (Fig. 2); the wild-type BEN-1 β-tubulin protein has dominant toxic effects on worm viability in the presence of the chemical. Moreover, the ben-1 gene is small in size, spanning only approximately 3 kb of the genome, and the only one gene that produces the Ben (benzimidazole-resistant) phenotype. We exploited the characteristics to isolate many mutations that showed resistance by random mutagenesis.35) C. elegans researchers often mutagenize worms with EMS (ethylmethanesulfonate) to isolate new mutations.10) However, EMS mutagenesis usually induces point mutations rather than deletions. Because genetic analyses usually require crossing between strains, it is more laborious to use mutants with a point mutation to examine genotypes. Thus, we chose a mutagenesis method with ultraviolet-activated trimethylpsoralen, which introduces crosslinks between neighboring nucleotides and results in deletions during DNA repair.36) We mutagenized worms under various conditions, cultured worms on selection media, and isolated many alleles. Then, we mapped the genome lesions in and around the ben-1 gene by PCR and direct sequencing. In this way, we determined how to mutagenize the worms to isolate appropriate deletion mutations that were readily detectable with PCR and agarose gel electrophoresis. The ease of isolating ben-1 mutants also helped our research later (see below).
Figure 2.
Paradoxical phenotypes observed for wild-type and ben-1 mutant animals. Animals raised on normal medium (A, B) or on selection medium containing benzimidazole (C, D) are shown. Only wild-type animals on selection medium show strong phenotypes (C). This phenotypic characteristic is very useful for negative selection of worms with the presence or absence of the wild-type ben-1 gene.
B. Sensitive detection of deletion mutants by PCR.
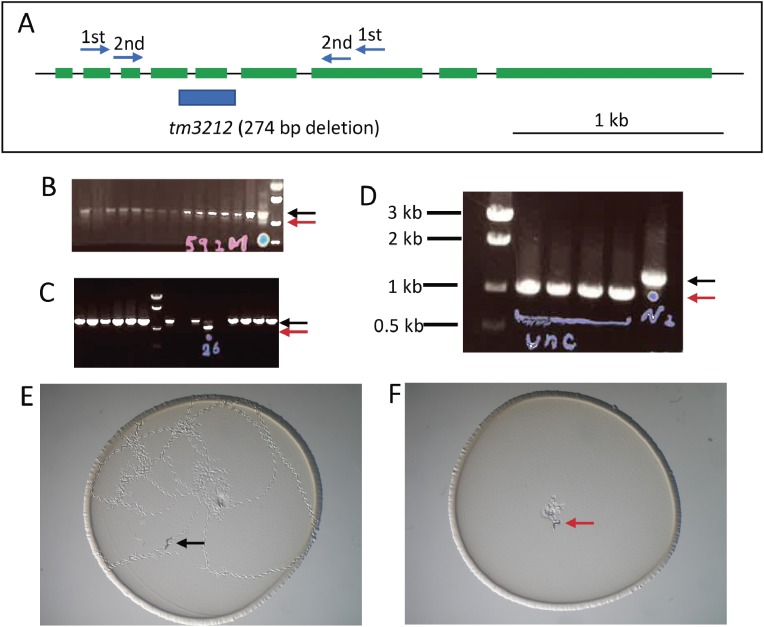
It was also necessary to detect deletion mutations within a large amount of wild-type DNA because random mutations occur at a low frequency in the gene of interest. PCR-amplified deletion mutant DNA fragments are shorter than those of wild-type animals. We developed a very sensitive PCR protocol called “stringent PCR”.35) We could preferentially amplify a small quantity of shorter (deletion mutant) targets over abundant longer (wild-type) targets, even if the shorter DNA is diluted 1:10,000 with wild-type DNA. When we detected mutant bands, we thawed frozen mutants and confirmed the deletion in growing progeny by sib selection (Fig. 3). After isolating the mutants of interest in this manner, researchers can easily handle the mutants, because the genotype can be identified using only PCR and agarose gel electrophoresis even from a single animal, without sequencing every time.
Figure 3.
An example of mutant isolation and phenotyping. (A) A set of nested primers for a gene (unc-18) was designed to span exons. The deletion site of an isolated mutant allele (blue bar) is shown under the gene structure (green: exons). (B) An example of PCR screening and agarose gel electrophoresis. Wild-type bands are shown by a black arrow, while a faint mutant band of a shorter size is seen (a red arrow in the lane with a blue dot). Frozen stocks for candidates were thawed and cultured in individual culture dishes and then again examined by PCR after laying eggs (C). Both the wild-type band (black arrow) and the mutant band (red arrow) are observed in a lane, indicated that the animal was heterozygous. Hatched worms from the culture above were again singled to individual dishes and analyzed again (D) and all the animals showing the Unc phenotype have only mutant bands (red arrow) in contrast to control wild-type (N2) band (black arrow). Phenotype observation of wild-type (E) and unc-18 mutant (F) animals placed at the center of the dishes and left for an hour. Only wild-type animals moved around on the bacterial lawn.
6. Simple knockout (KO) strains often serve as good biological materials for gene function analyses in addition to RNAi
In many cases, scientists in the C. elegans research community have used deletion mutants as simple loss-of-function alleles. As had already been shown in mouse,37) simple loss-of-function mutations in C. elegans often resulted in some phenotypes that were similar to those found by RNAi.
A. Control of the engulfment of apoptotic cells by surrounding phagocytic cells.
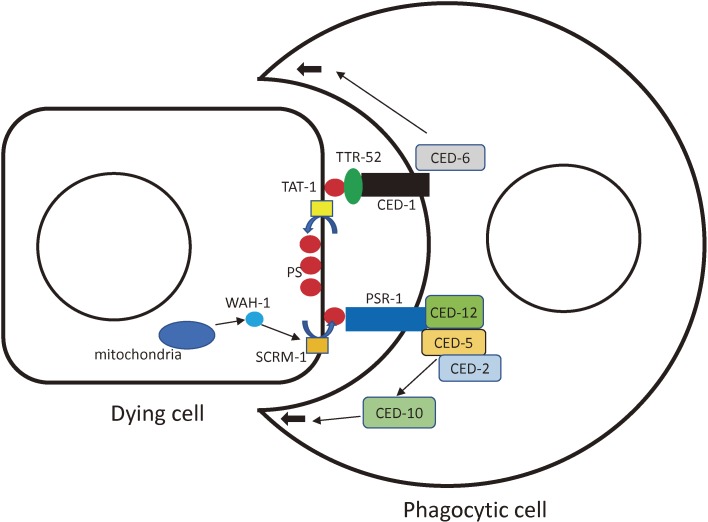
We conducted many interesting studies by generations and phenotyping of gene knockout strains. The signaling mechanisms underlying the signals from dying cells to phagocytes were examples of such analyses. We collaborated with Dr. D. Xue and Dr. X. Wang on the subject (Fig. 4). It is well known that apoptotic cells are removed by engulfment by neighboring phagocytic cells in C. elegans.38) Extensive genetic analyses on this mechanism have been conducted. We isolated many mutants of genes that may be involved in this process. In C. elegans, as in other organisms including humans, phosphatidylserine (PS) is the key molecule of apoptotic cell removal. PS, which is located in the inner leaflet of the plasma membrane of healthy cells, is exposed to the cell surface of apoptotic cells, serving as the “eat-me” signal. The membrane receptor, ced-1 on the phagocyte membrane is responsible for finding dying cells and presumably detecting the eat-me signal. When the ced-1 gene is lost, many more cell corpses, which are dying but not yet removed, are visible than in wild-type animals because of the delayed removal of dead cells.39) However, even in the ced-1 mutant background, cell corpses are removed eventually, although the process takes longer. A reasonable mechanism is that another pathway may transmit the presence of PS to phagocytes. Wang et al. showed that another membrane receptor, PSR-1, plays a role in detecting PS.40) When the psr-1 gene is knocked out by a deletion mutation (tm469), the presence of cell corpses in worms is prolonged (55% longer than those of wild-type animals). The rescue activity of the transgenic psr-1 gene is dependent on the expression of phagocytes because expression of PSR-1 by the ced-1 promoter, which is expressed in phagocytes but not in the dying cells, is necessary. The psr-1 mutation in conjunction with other mutations of ced-1, ced-6 and ced-7 enhanced the corpse-prolonged phenotype, but in conjunction with mutations of ced-2, ced-5, ced-10 or ced-12 it did not. Since the PSR-1 protein interacts with CED-5 and CED-12, which are cytoplasmic signaling molecules, PSR-1 is thought to be a PS receptor for apoptotic cell engulfment.
Figure 4.
A schematic drawing of the interaction between an apoptotic dying cell and an engulfing phagocytic cell. Illustrated are how phosphatidylserine (PS) is localized to inner and outer leaflets of the plasma membrane and how the PS signal is transmitted to the phagocytic cell through PSR-1 and CED-1 pathways. The molecules are summarized from the papers described in the text and also adapted from Wang et al. and a review by Conradt et al.76,77) Thin black arrows indicate the directions of signal transduction. Thick black arrows show the cellular extension of the phagocyte to engulf the dying cell. Blue arrows indicate movement of PS between inner and outer leaflets of plasma membrane.
Mechanisms of externalization of PS to the outside of the membrane were explored. It is believed that localization of phospholipids is dependent on phospholipid scramblase.41) Wang et al. showed that a mitochondrial factor WAH-1 promotes this externalization through a scramblase, as demonstrated by a deletion mutation of scrm-1 (SCRaMblase, tm698).42) The mechanisms of asymmetric localization of PS on the dying cells were also explored.43) We isolated deletion mutants for aminophospholipid translocases that may promote the inward movement of aminophospholipids such as PS. There are six homologs of the human phospholipid translocases (tat-1 to tat-6 (transbilayer amphipath transporters)). When worm germ cells treated with RNAi against tat-1 were labeled with annexin V, which binds to PS, faint staining was observed at the cell surface. RNAi against other tat genes did not produce the same phenotype. We used a tat-1 deletion mutant (tm1034) and a tat-3 deletion mutant (tm1275) to reproduce the experiment. Staining these mutants with annexin V resulted in a strong signal in the tat-1 mutant animals but never in the tat-3 mutant animals, confirming the RNAi experiments. Thus, the tat-1 gene underlies the restriction of PS to the inner leaflet of the plasma membrane in C. elegans. PS was able to induce removal of non-apoptotic cells. When six touch receptor neurons were labeled by GFP and examined, cells were more susceptible to loss in the tat-1 mutant background, suggesting that phagocytes sometimes attack cells that are not undergoing apoptosis if PS is on the membrane surface.
B. Longevity regulated by the FoxO pathway.
C. elegans is a popular organism for longevity analyses because of its short life cycle: 3 days to grow to adulthood and a total lifespan of 2 weeks. Direct screening for long-lived mutants is difficult because isolated mutant animals are too old to lay eggs and screening itself takes a long time. Furthermore, many unhealthy mutants show short-lifespans and it can be difficult to distinguish the different mechanisms affecting longevity. Thus, after finding the basic important pathways for longevity, it is better to screen for enhancers or suppressors of some preexisting longevity mutants with another phenotype, such as Daf (dauer formation-defective). Dr. P. Hu and colleagues tried to isolate such genes systematically; for example, eak (enhancers of akt-1), which may be among the daf-16 (a homolog of FoxO) pathway genes and is involved in longevity in parallel to akt-1 (akt kinase family). After identifying an interesting gene, eak-7, it is convenient and reliable to have another allele of the eak-7 mutant to test whether the phenotypes obtained by the forward genetics are reproducible by null mutations.44) An eak-7 deletion mutant (tm3188) enhanced the dauer arrest phenotype. This phenotype was suppressed by the daf-16 mutation, suggesting that the EAK-7 protein inhibits the DAF-16 protein. eak-7 mutants live longer than wild-type animals, as expected from the daf-16 inhibition.
C. Mechanistic insights into RNA interference.
RNAi became a very popular experimental technique among the C. elegans research community after an exciting paper from the Fire and Mello laboratories was published in 1998.45) Today, researchers working in many different research fields use the RNAi method to assess the effects of downregulation. We collaborated with Dr. C. Mello to explore the mechanisms of RNAi using our mutants, because RNAi knockdown of genes needed for RNAi is often difficult to interpret. We isolated a number of mutants of genes for DCR-1 (Dicer related) interactors.46) Among the interactors, PIR-1 (phosphatase interacting with RNA/RNP) and DRH-3 (Dicer related helicase) were essential proteins for RNAi. These proteins were shown to play roles in processing double-stranded RNAs.
We also obtained deletion mutants for all 27 Argonaute (AGO) genes and examined the roles of each clade in RNAi and related gene-silencing mechanisms. Single- and multiple-AGO mutants showed several distinct phenotypes in relation to the corresponding genetic pathways: chromosome segregation, fertility, and RNAi.47) Several AGO proteins interacted with amplified siRNAs and microRNAs, suggesting that these AGO proteins are limiting factors of both microRNA and exogenous and endogenous RNAi pathways.
D. Some examples concerning vesicular trafficking machinery.
We have been working on vesicular trafficking. The GON-1 protein is an ortholog of human ADAMTS9 and is necessary for C. elegans gonadal cells to proliferate and migrate. The gon-1 gene encodes a secretory matrix metalloprotease that digests the extracellular matrix and helps gonadal migration.48) We found that gon-1 (tm3146) mutants show not only a sterile phenotype but also the accumulation of abnormal membrane structures in the cytoplasm. We found that gon-1 is necessary for proper vesicular trafficking of many proteins, including secretory proteins, membrane proteins, and GPI-anchored proteins.49) We examined whether ADAMTS9 has a similar role in human cells by knocking down the gene in HEK293 cells. This was the case, and vesicular transport from the endoplasmic reticulum to the Golgi apparatus was severely impaired. Interestingly, the GON domain located in the C-terminus, but not the metalloprotease domain, plays a major role in vesicular trafficking. We also found that transfection of the C. elegans GON domain cDNA into HEK293 cells rescued the trafficking defects by knockdown of the ADAMTS9 gene. Thus, the GON domain is important for the secretory pathway in both C. elegans and humans.
E. Stem cell maintenance mechanisms.
To study disease mechanisms, we chose a gene bcl-7 (mammalian BCL (B cell lymphoma) gene homolog), which is homologous to human BCL7 gene family members (BCL7A, BCL7B and BCL7C). BCL7B is one of 28 genes that are deleted in Williams-Beuren syndrome, but little is known about its role, although clinical reports describe a high incidence of blood malignancies.50) We isolated a deletion mutant bcl-7 (tm5268) and analyzed the phenotypes. We found that bcl-7 is necessary for the maintenance of the stem cell characteristics of seam cells, which are stem cell-like cells in the C. elegans postembryonic lineage.51) Furthermore, the bcl-7 mutants are sterile. Interestingly, bcl-7 mutants have enlarged nuclei. We used a human cancer cell line, Kato III, and found that knocking down of human BCL7B results in a similar enlargement phenotype with the appearance of malignant morphological changes to the nuclei. Because C. elegans has only one bcl-7, it was a convenient model in which we explore the basic function of the BCL-7 protein to gain insights into the human BCL7 family.
F. Some discrepancies between phenotypes found in deletion mutants and RNAi.
We sometimes encounter discrepancies between phenotypes observed by RNAi and gene knockout strains. Theoretically, it is conceivable that some RNAi phenotypes are different from those generated by gene knockouts. For example, the redundant function of two homologous genes may indicate that removing only one gene in the genome does not result in any phenotype, but double mutants show some phenotypes. However, RNAi downregulates transcripts from both genes, resulting in visible phenotypes. In different contexts, some genes show very weak or no phenotypes by RNAi but strong phenotypes by gene knockout; such phenomena are described for many genes expressed in nerve cells, especially when RNAi is administered by feeding. To solve this problem, researchers sometimes use transgenic RNAi with neural promoters, which allows dsRNA to be expressed in neurons and to act cell-autonomously,52) or overexpress the sid-1 (systemic RNA interference defective) gene with a neuronal promoter,53) which helps the uptake of dsRNA by neuronal cells. In these situations, gene knockdown animals often show apparent phenotypes. It is also expected that maternal-effect genes may show distinct phenotypes between RNAi and gene KO. Homozygous mutants from heterozygous parents (by self-progeny) show weak phenotypes during the early embryonic stages in the gene KO strains and, thus, can be maintained as heterozygotes, whereas sometimes they show severe lethal phenotypes by RNAi. Because feeding parental animals with RNAi clones causes the degradation of maternal mRNAs in the embryo, this is the expected difference between RNAi and gene KO strains.
Another difference between RNAi and the mutants is that the phenotypes observed by RNAi are sometimes variable among experiments, from weak phenotypes to severe lethality (many examples are described in WormBase), depending on the researchers (differences of delivery methods of dsRNA) and the RNAi clones (the length and regions of inserts).
Other mechanisms may be envisaged. Gene KO may not represent all the possible phenotypes in which a gene is involved; when an essential gene is involved in two or more developmental stages, phenotypes that should appear in later stages are more difficult to observe. In other cases, some loss-of-function mutations may be compensated by homologous genes. The possibilities that certain phenomena may use more than two pathways should also be considered (typically, daf mutations; see below). To examine such genetic pathways, more sophisticated and intensive approaches are needed, reminiscent of the complicated situations of multifactorial diseases in humans.
We ourselves experienced some discrepancies in phenotypes between RNAi and gene knockout strains. For example, the vps-45 gene did not have a visible phenotype with RNAi clones but showed a temperature-sensitive lethal phenotype with gene knockout strains.54) Also, vps-33.1 and vps-33.2 knockouts showed phenotypes different from those previously reported by RNAi clones.55,56) The reasons why KO or RNAi of some genes induces different phenotypes are often difficult to explain.
7. Methods to help analyses of gene functions in C. elegans
As exemplified above, sometimes we should use multiple and more sophisticated techniques to reveal gene functions. We have tried several useful techniques, as shown below.
A. Single/low-copy integration of extrachromosomal transgenes.
The gene expression of C. elegans transgenic strains is silenced in germ cells. This occurs because multicopy transgenes are recognized as repetitive sequences and inactivated. Transgenic animals generated by microinjection of DNA solution into the germline syncytium usually contains multicopy repetitive extrachromosomal DNAs that are silenced in the germline but expressed in the somatic cells.57)
As mentioned above, multicopy transgenes are silenced in the germline. To solve this problem, a single-copy transgene can be integrated into the chromosome via Mos1 transposon excision.58) We have developed an easy method for integrating single- or low-copy transgenes.59) We used a ben-1 and vps-45 double mutant strain, which shows a benzimidazole-resistance and temperature-sensitive lethality phenotype as mentioned above. We added rescue DNA plasmids with the ben-1 and vps-45 genes when we generated transgenic strains. The extrachromosomal multicopy transgenic animals became wild-type for both the ben-1 and vps-45 mutations, and were sensitive to benzimidazole but viable at 20 ℃. If these transgenic strains are UV-irradiated and cultured on selection media, a small fraction of animals have single/low-copy transgenes in some chromosomes. Many of the transgenic animals retain the wild-type ben-1 gene, showing sensitivity to benzimidazole. In addition, only animals that maintain the vps-45 rescue transgene can survive. This can occur only when short transgenic DNAs that are devoid of the ben-1 fragment but retain the vps-45 fragment are integrated into a chromosome. Thus, by culturing transgenic animals on selection media at 20 ℃, researchers can easily obtain single/low-copy transgenic strains by simply waiting for animals to grow on the media.
Although this selection yields random integration again, we have now successfully integrated single/low-copy transgenes into defined loci using the genome editing method.60) We used the CRISPR/Cas9 method to cut a chromosomal integration site and a transgene sequence. In this way, we could integrate single/low-copy transgenes into the loci of interest, making it easier to generate multiple variations of mutations and transgenes to cross with other mutants or transgenic strains. Single-copy integration is also useful to avoid overexpression of toxic transgene products.61)
B. A conditional KO system.
In C. elegans, as mentioned above, lethal mutants are often difficult to use for behavior analyses. However, conditional knockout strains are routinely used in mice.62) We suspect that the use of conditional knockout strains is unpopular among C. elegans researchers because most C. elegans transgenic strains are extrachromosomal and multicopy. Multicopy transgenes are not suitable for conditional knockout; even if transgenes are floxed (loxP sequence from bacteriophage P1 is inserted and can be recombined between two loxP sites by Cre recombinase from bacteriophage P1), only some of the copies are removed by recombinases, resulting in the failure of knockout. After succeeding in single-copy integration, we next tried to perform conditional knockout experiments.61) Single-copy integrated floxed DNA fragments could be excised cleanly by crossing with Cre recombinase-expressing-transgenic strains driven by spatiotemporally specific promoters, enabling a conditional knockout. Although it was easy to remove the transgene in a spatiotemporally specific manner, we found that conditional experiments in C. elegans must be performed carefully. C. elegans grows very fast: it takes only 3 days for a fertilized egg to develop into an adult animal, and therefore, some proteins may not be removed in some cell types and tissues, while the same proteins are removed almost completely in other cell types. Accordingly, some phenotypes may appear incomplete. Applications should be carefully chosen with this limitation in mind.
C. A CALI system for stable proteins.
Instead of removing the genome sequence, we found that some proteins may be better analyzed by chromophore-assisted light inactivation (CALI). In C. elegans, mutants of many genes can be rescued by the wild-type transgene. We often find that GFP-fusion genes can also rescue the phenotypes, albeit partially. In such cases, we can replace GFP with KillerRed and rescue mutant phenotypes. KillerRed, a fluorescent protein-related protein, absorbs green light and generates superoxide: when overexpressed it can kill the expressing cells.63) In our hands, it was difficult to kill cells with KillerRed. However, inactivating fusion protein was easy and reproducible. Because the absorbance maximum is in the wavelength range of green light and the excitation light itself rarely affects cell function, protein function can be safely and easily switched on and off. For example, we expressed a KillerRed fusion protein in mutant animals with severe phenotypes; gon-1 mutants and bcl-7 mutants.49,51) We rescued the gon-1 mutant with transgenic expression of a KillerRed-GON protein. Using this strain, we investigate how the secretion process is regulated by the GON-1 protein. Another gene, bcl-7, is a homolog of human BCL7, and bcl-7 deletion mutants show sterile phenotypes because of abnormal somatic and germ cells. We rescued bcl-7 mutants completely with transgenes of a KillerRed-BCL-7 fusion and could inactivated the BCL-7 by illuminating the strain with a green light. Thus, we were able to examine when the protein is required for proper gonadal cell development. Because proteins appeared completely inactivated using this method, the slow turnover rate of the preexisting proteins of targeted genes is no longer a limitation.
D. Creation of new balancers that enable more reliable description of mutant phenotypes.
We isolated many deletion mutants and found a certain portion of mutants showing lethal or sterile phenotypes (NBRP for the nematode: http://shigen.nig.ac.jp/c.elegans/index.jsp). Because C. elegans animals grow and proliferate quickly as hermaphrodites, such mutants need to be balanced: otherwise, mutant animals are diluted by the segregated wild-type animals, which grow faster than mutants. To maintain lethal or sterile mutations, researchers often use balancer chromosomes, which are associated with some phenotypes and are used as heterozygote with mutant chromosome so that healthy heterozygous animals grow as a large population.64) Unfortunately, most C. elegans balancer chromosomes are translocations generated by γ-ray irradiation, which give rise to aneuploidy. Theoretically about half of the self-progeny die from the balancer itself. This is very inconvenient for analyzing, for example, embryonic lethal strains because the developmental stages at which the mutant phenotypes of interest and the developmental arrest by the balancer aneuploidy occur are very close together. There are a few crossover balancers created by γ-ray irradiation, which have complex rearrangements within the same chromosome and, thus, do not show any aneuploidy.64) We used the CRISPR/Cas9 technique to create crossover balancers.65) We induced chromosomal inversions twice in two overlapped regions in the same chromosome, which are suitable both for stable maintenance of covered mutant alleles and for phenotype analyses. We have now generated such crossover balancers to cover the whole C. elegans genome and labeled the balancers with fluorescent protein reporters (Dejima et al., manuscript in preparation). We expect that with these balancers, mutations with lethal phenotypes will be analyzed more readily and exactly.
8. Discussion
A. Homology analyses are useful, but we sometimes need more.
Genome sequence information is essential for medical and life science studies. We routinely search for homologies among genes and their products, and the findings are very useful to understand gene functions. In light of our limited experiences in functional genomics research, sometimes we need to know more. To reveal the disease susceptibility genes, an enormous number of studies are being performed, with sophisticated algorithms. One reason that makes it very difficult to examine the human genome is the high variation of genetic backgrounds among individuals. Even if a gene is related to some diseases, odds scores by statistical analyses are often low. Model organisms shed light on such enigmas. We found that an amino acid change in Argonaute proteins removes their catalytic function and changes the role of the subfamily into RNA receptors from RNase.47) DCR-1 was found to digest DNAs instead of dsRNA after it was cut by a caspase, CED-3.66) There are many cases in which alternative splicing changes not only gene expression but also protein function: for example, the ubiquitously expressed protein DAF-16 (a homolog of mammalian FOXO) produces different phenotypes when different alternatively spliced exons are removed.67) The function of GON-1, a homolog of human ADAMTS9 metalloprotease, made us recognize that an important domain (GON domain) in the C-terminus, which is not related to protease but plays a critical role for cells to normally secrete proteins, has been ignored because the metalloprotease domain is so obvious.49)
An rSNP (regulatory single nucleotide polymorphism, which has a variation in a gene regulatory sequence but not a protein coding sequence) of the human gene ADAMTS9 is regarded as a susceptibility gene for type 2 diabetes, but a knockout mouse strain was reported as fetal lethal.68) gon-1 null mutants show a number of severe phenotypes, including sterility, but we also found that certain genes, if downregulated, suppress even such severe phenotypes (Yoshina et al., in preparation). Medical research usually targets diseases with some symptoms detected after birth. In an extreme idea, some of the strains we are using may have such mutations in the vicinity of the same linked chromosome from the start and may not simply be “wild-type” strains. In the same way, humans have many combinations of variations, and this makes problems very difficult to solve. Interestingly, a small fraction of adult population has severe and high-penetrant Mendelian childhood disease mutations with no clinical manifestation, suggesting that they may harbor protective genetic variants.69)
B. Integrated genetics and future direction.
We have been using a model organism to understand gene functions (Fig. 1). What fundamental insights can we expect to obtain in the future to solve questions in the medical and life sciences? For example, it is intriguing to consider whether it is possible to simulate all the behavior of C. elegans as an initial move toward understanding the human mind as a connectome, perhaps as originally proposed by Brenner.9,70) We have the wiring diagram of C. elegans, showing how synaptic connections are formed among neurons.8) The C. elegans research community is analyzing the C. elegans body to reveal everything about the worm and to try to use the information to understand more general mechanisms of life, including development, behavior, cell biology, and even human diseases. However, we sometimes find unexpected phenomena beyond the inference from known information.
The reasons why biological analyses are so complicated exist in the organisms themselves. Biological phenomena are based on multilayered structures: molecules, cells, tissues and whole bodies in various time courses, which make biological analyses complicated. In addition, a similar phenomenon may be caused by signal transduction between distinct molecules, distinct tissues, and even by distinct environments. For example, with classical mutant analyses, we know that the phenomenon of dauer formation consists of at least four distinct pathways.71) This shows us that a phenomenon, or seemingly undistinguishable multiple phenomena, may be caused by completely independent pathways and should be examined carefully. We also found that a Wnt molecule may cause apparent opposite effects for bcl-7 mutant phenotypes, which showed a haploinsufficient phenotype.51) This is reminiscent of the fact that Williams-Beuren syndrome is caused by haploinsufficiency of chromosome 7q11.23.72) It is a common idea that gene expression is regulated in a temporally-specific and tissue/cell-type-specific manner, so a single line of evidence may not give a complete picture of the physiological and pathological states of the body.
We have been using transgenic expression, RNAi, and gene knockout strains in addition to mutants isolated by forward genetics. Moreover, we have shown that it is now possible to obtain drastically gene-modified strains, such as artificial structural changes of the chromosomes.65) To elucidate all the gene functions in a model organism will require integrated genetics, which is a combination of forward and reverse genetics, a systematic and large set of reduction-of-function studies using RNAi clones73) or gene knockout strains, which are or will be available for C. elegans in the near future as they are in yeast.74) Moreover, we should combine gain-of-function transgenic studies with reduction-of-function studies. We also need to combine enhancer screening and suppressor screening to reveal possible gene networks or genetic pathways. Recently, forward genetics with next-generation sequencing has enabled researchers to identify C. elegans mutations quickly and easily.75)
To understand the genetic pathways involved in all the layers of the different biological processes in C. elegans, we need to examine genotype-phenotype relationships for every possible phenomenon, for every possible developmental stage and every possible environment. In addition, although the overall structures of C. elegans and humans are very different, the genetic pathways are partly similar and information relating to those genetic pathways is valuable for understanding the nature of health and diseases in humans.
Acknowledgements
The author is grateful to Dr. M. Chalfie who introduced him the principle of genetics and the members of Mitani laboratory to accomplish the genetic system described in this review. The author is grateful to many collaborators who encouraged him to join various interesting studies and Dr. S. Miyazaki and Dr. Y. Kawakami for providing him with the research environment at the present address. This work was mainly supported by Ministry of Education, Culture, Sports, Science and Technology-Japan (MEXT), Japan Agency for Medical Research and Development (AMED), Japan Science and Technology Agency (JST), and Japan Society for the Promotion of Science (JSPS).
Abbreviations
- ACEDB
A C.elegans DataBase
- akt
akt kinase family
- asd-1
alternative splicing defective
- bcl
mammalian BCL (B cell lymphoma) gene homolog
- ben
benzimidazole resistant
- CALI
chromophore-assisted light inactivation
- ced
cell death abnormality
- C. elegans
Caenorhabditis elegans
- CGC
Caenorhabditis Genetics Center
- Daf
dauer formation-defective
- DCR
Dicer related
- Dpy
dumpy
- DRH
Dicer related helicase
- eak
enhancers of akt-1
- egl
egg-laying defective
- EMS
ethylmethanesulfonate
- fox
Feminizing gene on X
- GFP
green fluorescent protein
- GWAS
genome-wide association study
- KO
knockout
- lin
abnormal cell lineage
- mec
mechanosensory abnomarity
- PCR
polymerase chain reaction
- PIR
phosphatase interacting with RNA/RNP
- PS
phosphatidylserine
- RFP
red fluorescent protein
- RNAi
RNA interference
- rSNP
regulatory single nucleotide polymorphism
- scrm
SCRaMblase (phospholipid scramblase)
- sem
sex muscle abnormal
- sid
systemic RNA interference defective
- tat
transbilayer amphipath transporters
- Unc
uncoordinated movement
Profile
Shohei Mitani was born in Tottori prefecture in 1958. He graduated from the University of Tokyo School of Medicine in 1984, and obtained his Ph.D. in 1988 from the University of Tokyo. He started genetics study on Caenorhabidtis elegans at the Columbia University in the City of New York with a postdoctoral fellowship research abroad awarded by Japan Society for the Promotion of Science in 1989 and has been continuing the research. He was appointed to the professor and chair at the Department of Physiology in Tokyo Women’s Medical University School of Medicine in 2007. He was additionally appointed to the director of Tokyo Women’s Medical University Institute for Integrated Medical Sciences in 2010.
References
- 1).Hirschhorn J.N., Daly M.J. (2005) Genome-wide association studies for common diseases and complex traits. Nat. Rev. Genet. 6, 95–108. [DOI] [PubMed] [Google Scholar]
- 2).Pleasance E.D., Cheetham R.K., Stephens P.J., McBride D.J., Humphray S.J., Greenman C.D., Varela I., Lin M.L., Ordóñez G.R., Bignell G.R., Ye K., Alipaz J., Bauer M.J., Beare D., Butler A., Carter R.J., Chen L., Cox A.J., Edkins S., Kokko-Gonzales P.I., Gormley N.A., Grocock R.J., Haudenschild C.D., Hims M.M., James T., Jia M., Kingsbury Z., Leroy C., Marshall J., Menzies A., Mudie L.J., Ning Z., Royce T., Schulz-Trieglaff O.B., Spiridou A., Stebbings L.A., Szajkowski L., Teague J., Williamson D., Chin L., Ross M.T., Campbell P.J., Bentley D.R., Futreal P.A., Stratton M.R. (2010) A comprehensive catalogue of somatic mutations from a human cancer genome. Nature 463, 191–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3).Ng S.B., Buckingham K.J., Lee C., Bigham A.W., Tabor H.K., Dent K.M., Huff C.D., Shannon P.T., Jabs E.W., Nickerson D.A., Shendure J., Bamshad M.J. (2010) Exome sequencing identifies the cause of a mendelian disorder. Nat. Genet. 42, 30–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4).The C. elegans Sequencing Consortium (1998) Genome sequence of the nematode C. elegans: a platform for investigating biology. Science 282, 2012–2018. [DOI] [PubMed] [Google Scholar]
- 5).Mitani S. (2009) Nematode, an experimental animal in the national BioResource project. Exp. Anim. 58, 351–356. [DOI] [PubMed] [Google Scholar]
- 6).Sulston J.E., Horvitz H.R. (1977) Post-embryonic cell lineages of the nematode, Caenorhabditis elegans. Dev. Biol. 56, 110–156. [DOI] [PubMed] [Google Scholar]
- 7).Sulston J.E., Schierenberg E., White J.G., Thomson J.N. (1983) The embryonic cell lineage of the nematode Caenorhabditis elegans. Dev. Biol. 100, 64–119. [DOI] [PubMed] [Google Scholar]
- 8).White J.G., Southgate E., Thomson J.N., Brenner S. (1986) The structure of the nervous system of Caenorhabditis elegans. Philos. Trans. R. Soc. Lond. B Biol. Sci. 314, 1–340. [DOI] [PubMed] [Google Scholar]
- 9).Brenner S. (1974) The genetics of Caenorhabditis elegans. Genetics 77, 71–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10).Wood, W.B. ed. (1988) The Nematode Caenorhabditis elegans. Cold Spring Harbor Laboratory, NY. [Google Scholar]
- 11).Sulston J., Du Z., Thomas K., Wilson R., Hillier L., Staden R., Halloran N., Green P., Thierry-Mieg J., Qiu L., Dear S., Coulson A., Craxton M., Durbin R., Berks M., Metzstein M., Hawkins T., Ainscough R., Waterston R. (1992) The C. elegans genome sequencing project: a beginning. Nature 356, 37–41. [DOI] [PubMed] [Google Scholar]
- 12).International Human Genome Sequencing Consortium (2004) Finishing the euchromatic sequence of the human genome. Nature 431, 931–945. [DOI] [PubMed] [Google Scholar]
- 13).Shaye D.D., Greenwald I. (2011) OrthoList: a compendium of C. elegans genes with human orthologs. PLoS One 6, e20085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14).Eckman F.H., Durbin R. (1995) ACeDB and Macace. Methods Cell Biol. 48, 583–605. [PubMed] [Google Scholar]
- 15).Chalfie M., Sulston J. (1981) Developmental genetics of the mechanosensory neurons of Caenorhabditis elegans. Dev. Biol. 82, 358–370. [DOI] [PubMed] [Google Scholar]
- 16).Chalfie M., Sulston J.E., White J.G., Southgate E., Thomson J.N., Brenner S. (1985) The neural circuit for touch sensitivity in Caenorhabditis elegans. J. Neurosci. 5, 956–964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17).Chalfie M., Au M. (1989) Genetic control of differentiation of the C. elegans touch receptor neurons. Science 243, 1027–1033. [DOI] [PubMed] [Google Scholar]
- 18).Mitani S., Du H., Hall D.H., Driscoll M., Chalfie M. (1993) Combinatorial control of touch receptor neuron expression in Caenorhabditis elegans. Development 119, 773–783. [DOI] [PubMed] [Google Scholar]
- 19).Shimomura O., Johnson F.H., Saiga Y. (1962) Extraction, purification and properties of aequorin, a bioluminescent protein from the luminous hydromedusan, Aequorea. J. Cell. Comp. Physiol. 59, 223–239. [DOI] [PubMed] [Google Scholar]
- 20).Chalfie M., Tu Y., Euskirchen G., Ward W.W., Prasher D.C. (1994) Green fluorescent protein as a marker for gene expression. Science 263, 802–805. [DOI] [PubMed] [Google Scholar]
- 21).Giepmans B.N., Adams S.R., Ellisman M.H., Tsien R.Y. (2006) The fluorescent toolbox for assessing protein location and function. Science 312, 217–224. [DOI] [PubMed] [Google Scholar]
- 22).Gengyo-Ando K., Yoshina S., Inoue H., Mitani S. (2006) An efficient transgenic system by TA cloning vectors and RNAi for C. elegans. Biochem. Biophys. Res. Commun. 349, 1345–1350. [DOI] [PubMed] [Google Scholar]
- 23).Zwaal R.R., Broeks A., van Meurs J., Groenen J.T., Plasterk R.H. (1993) Target-selected gene inactivation in Caenorhabditis elegans by using a frozen transposon insertion mutant bank. Proc. Natl. Acad. Sci. U.S.A. 90, 7431–7435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24).Rushforth A.M., Saari B., Anderson P. (1993) Site-selected insertion of the transposon Tc1 into a Caenorhabditis elegans myosin light chain gene. Mol. Cell. Biol. 13, 902–910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25).Mello C.C., Kramer J.M., Stinchcomb D., Ambros V. (1991) Efficient gene transfer in C. elegans: extrachromosomal maintenance and integration of transforming sequences. EMBO J. 10, 3959–3970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26).Mitani S. (1995) Genetic regulation of mec-3 gene expression implicated in the specification of the mechanosensory neuron cell types in Caenorhabditis elegans. Dev. Growth Differ. 37, 551–557. [DOI] [PubMed] [Google Scholar]
- 27).Polymeropoulos M.H., Lavedan C., Leroy E., Ide S.E., Dehejia A., Dutra A., Pike B., Root H., Rubenstein J., Boyer R., Stenroos E.S., Chandrasekharappa S., Athanassiadou A., Papapetropoulos T., Johnson W.G., Lazzarini A.M., Duvoisin R.C., Di Iorio G., Golbe L.I., Nussbaum R.L. (1997) Mutation in the alpha-synuclein gene identified in families with Parkinson’s disease. Science 276, 2045–2047. [DOI] [PubMed] [Google Scholar]
- 28).Kuwahara T., Koyama A., Gengyo-Ando K., Masuda M., Kowa H., Tsunoda M., Mitani S., Iwatsubo T. (2006) Familial Parkinson mutant alpha-synuclein causes dopamine neuron dysfunction in transgenic Caenorhabditis elegans. J. Biol. Chem. 281, 334–340. [DOI] [PubMed] [Google Scholar]
- 29).Sawin E.R., Ranganathan R., Horvitz H.R. (2000) C. elegans locomotory rate is modulated by the environment through a dopaminergic pathway and by experience through a serotonergic pathway. Neuron 26, 619–631. [DOI] [PubMed] [Google Scholar]
- 30).Kuwahara T., Koyama A., Koyama S., Yoshina S., Ren C.H., Kato T., Mitani S., Iwatsubo T. (2008) A systematic RNAi screen reveals involvement of endocytic pathway in neuronal dysfunction in alpha-synuclein transgenic C. elegans. Hum. Mol. Genet. 17, 2997–3009. [DOI] [PubMed] [Google Scholar]
- 31).Kuroyanagi H., Kobayashi T., Mitani S., Hagiwara M. (2006) Transgenic alternative-splicing reporters reveal tissue-specific expression profiles and regulation mechanisms in vivo. Nat. Methods 3, 909–915. [DOI] [PubMed] [Google Scholar]
- 32).Plasterk R.H. (1995) Reverse genetics: from gene sequence to mutant worm. Methods Cell Biol. 48, 59–80. [DOI] [PubMed] [Google Scholar]
- 33).Driscoll M., Dean E., Reilly E., Bergholz E., Chalfie M. (1989) Genetic and molecular analysis of a Caenorhabditis elegans β-tubulin that conveys benzimidazole sensitivity. J. Cell Biol. 109, 2993–3003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34).Savage C., Hamelin M., Culotti J.G., Coulson A., Albertson D.G., Chalfie M. (1989) mec-7 is a beta-tubulin gene required for the production of 15-protofilament microtubules in Caenorhabditis elegans. Genes Dev. 3, 870–881. [DOI] [PubMed] [Google Scholar]
- 35).Gengyo-Ando K., Mitani S. (2000) Characterization of mutations induced by ethylmethanesulfonate, UV and trimethylpsoralen in the nematode Caenorhabditis elegans. Biochem. Biophys. Res. Commun. 269, 64–69. [DOI] [PubMed] [Google Scholar]
- 36).Yandell M.D., Edgar L.G., Wood W.B. (1994) Trimethylpsoralen induces small deletion mutations in Caenorhabditis elegans. Proc. Natl. Acad. Sci. U.S.A. 91, 1381–1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37).Thomas K.R., Capecchi M.R. (1987) Site-directed mutagenesis by gene targeting in mouse embryo-derived stem cells. Cell 51, 503–512. [DOI] [PubMed] [Google Scholar]
- 38).Ellis H.M., Horvitz H.R. (1986) Genetic control of programmed cell death in the nematode C. elegans. Cell 44, 817–829. [DOI] [PubMed] [Google Scholar]
- 39).Hedgecock E.M., Sulston J.E., Thomson J.N. (1983) Mutations affecting programmed cell deaths in the nematode Caenorhabditis elegans. Science 220, 1277–1279. [DOI] [PubMed] [Google Scholar]
- 40).Wang X., Wu Y.C., Fadok V.A., Lee M.C., Gengyo-Ando K., Cheng L.C., Ledwich D., Hsu P.K., Chen J.Y., Chou B.K., Henson P., Mitani S., Xue D. (2003) Cell corpse engulfment mediated by C. elegans phosphatidylserine receptor through CED-5 and CED-12. Science 302, 1563–1566. [DOI] [PubMed] [Google Scholar]
- 41).Zhou Q., Zhao J., Stout J.G., Luhm R.A., Wiedmer T., Sims P.J. (1997) Molecular cloning of human plasma membrane phospholipid scramblase. A protein mediating transbilayer movement of plasma membrane phospholipids. J. Biol. Chem. 272, 18240–18244. [DOI] [PubMed] [Google Scholar]
- 42).Wang X., Wang J., Gengyo-Ando K., Gu L., Sun C.L., Yang C., Shi Y., Kobayashi T., Shi Y., Mitani S., Xie X.S., Xue D. (2007) C. elegans mitochondrial factor WAH-1 promotes phosphatidylserine externalization in apoptotic cells through phospholipid scramblase SCRM-1. Nat. Cell Biol. 9, 541–549. [DOI] [PubMed] [Google Scholar]
- 43).Darland-Ransom M., Wang X., Sun C.L., Mapes J., Gengyo-Ando K., Mitani S., Xue D. (2008) Role of C. elegans TAT-1 protein in maintaining plasma membrane phosphatidylserine asymmetry. Science 320, 528–531. [DOI] [PubMed] [Google Scholar]
- 44).Alam H., Williams T.W., Dumas K.J., Guo C., Yoshina S., Mitani S., Hu P.J. (2010) EAK-7 controls development and life span by regulating nuclear DAF-16/FoxO activity. Cell Metab. 12, 30–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45).Fire A., Xu S., Montgomery M.K., Kostas S.A., Driever S.E., Mello C.C. (1998) Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 391, 806–811. [DOI] [PubMed] [Google Scholar]
- 46).Duchaine T.F., Wohlschlegel J.A., Kennedy S., Bei Y., Conte D., Jr., Pang K., Brownell D.R., Harding S., Mitani S., Ruvkun G., Yates J.R., 3rd, Mello C.C. (2006) Functional proteomics reveals the biochemical niche of C. elegans DCR-1 in multiple small-RNA-mediated pathways. Cell 124, 343–354. [DOI] [PubMed] [Google Scholar]
- 47).Yigit E., Batista P.J., Bei Y., Pang K.M., Chen C.C., Tolia N.H., Joshua-Tor L., Mitani S., Simard M.J., Mello C.C. (2006) Analysis of the C. elegans Argonaute family reveals that distinct Argonautes act sequentially during RNAi. Cell 127, 747–757. [DOI] [PubMed] [Google Scholar]
- 48).Blelloch R., Kimble J. (1999) Control of organ shape by a secreted metalloprotease in the nematode Caenorhabditis elegans. Nature 399, 586–590. [DOI] [PubMed] [Google Scholar]
- 49).Yoshina S., Sakaki K., Yonezumi-Hayashi A., Gengyo-Ando K., Inoue H., Iino Y., Mitani S. (2012) Identification of a novel ADAMTS9/GON-1 function for protein transport from the ER to the Golgi. Mol. Biol. Cell 23, 1728–1741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50).Amenta S., Moschovi M., Sofocleous C., Kostaridou S., Mavrou A., Fryssira H. (2004) Non-Hodgkin lymphoma in a child with Williams syndrome. Cancer Genet. Cytogenet. 154, 86–88. [DOI] [PubMed] [Google Scholar]
- 51).Uehara T., Kage-Nakadai E., Yoshina S., Imae R., Mitani S. (2015) The tumor suppressor BCL7B functions in the Wnt signaling pathway. PLoS Genet. 11, e1004921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52).Tavernarakis N., Wang S.L., Dorovkov M., Ryazanov A., Driscoll M. (2000) Heritable and inducible genetic interference by double-stranded RNA encoded by transgenes. Nat. Genet. 24, 180–183. [DOI] [PubMed] [Google Scholar]
- 53).Calixto A., Chelur D., Topalidou I., Chen X., Chalfie M. (2010) Enhanced neuronal RNAi in C. elegans using SID-1. Nat. Methods 7, 554–559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54).Gengyo-Ando K., Kuroyanagi H., Kobayashi T., Murate M., Fujimoto K., Okabe S., Mitani S. (2007) The SM protein VPS-45 is required for RAB-5-dependent endocytic transport in Caenorhabditis elegans. EMBO Rep. 8, 152–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55).Solinger J.A., Spang A. (2014) Loss of the Sec1/Munc18-family proteins VPS-33.2 and VPS-33.1 bypasses a block in endosome maturation in Caenorhabditis elegans. Mol. Biol. Cell 25, 3909–3925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56).Gengyo-Ando K., Kage-Nakadai E., Yoshina S., Otori M., Kagawa-Nagamura Y., Nakai J., Mitani S. (2016) Distinct roles of the two VPS33 proteins in the endolysosomal system in Caenorhabditis elegans. Traffic 17, 1197–1213. [DOI] [PubMed] [Google Scholar]
- 57).Mello C., Fire A. (1995) DNA transformation. Methods Cell Biol. 48, 451–482. [PubMed] [Google Scholar]
- 58).Frøkjaer-Jensen C., Davis M.W., Hopkins C.E., Newman B.J., Thummel J.M., Olesen S.P., Grunnet M., Jorgensen E.M. (2008) Single-copy insertion of transgenes in Caenorhabditis elegans. Nat. Genet. 40, 1375–1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59).Kage-Nakadai E., Kobuna H., Funatsu O., Otori M., Gengyo-Ando K., Yoshina S., Hori S., Mitani S. (2012) Single/low-copy integration of transgenes in Caenorhabditis elegans using an ultraviolet trimethylpsoralen method. BMC Biotechnol. 12, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60).Yoshina S., Suehiro Y., Kage-Nakadai E., Shohei Mitani S. (2016) Locus-specific integration of extrachromosomal transgenes in C. elegans with the CRISPR/Cas9 system. Biochem. Biophys. Rep. 5, 70–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61).Kage-Nakadai E., Imae R., Suehiro Y., Yoshina S., Hori S., Mitani S. (2014) A conditional knockout toolkit for Caenorhabditis elegans based on the Cre/loxP recombination. PLoS One 9, e114680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62).Skarnes W.C., Rosen B., West A.P., Koutsourakis M., Bushell W., Iyer V., Mujica A.O., Thomas M., Harrow J., Cox T., Jackson D., Severin J., Biggs P., Fu J., Nefedov M., de Jong P.J., Stewart A.F., Bradley A. (2011) A conditional knockout resource for the genome-wide study of mouse gene function. Nature 474, 337–342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63).Bulina M.E., Chudakov D.M., Britanova O.V., Yanushevich Y.G., Staroverov D.B., Chepurnykh T.V., Merzlyak E.M., Shkrob M.A., Lukyanov S., Lukyanov K.A. (2006) A genetically encoded photosensitizer. Nat. Biotechnol. 24, 95–99. [DOI] [PubMed] [Google Scholar]
- 64).Edgley, M.L., Baillie, D.L., Riddle, D.L. and Rose, A.M. (2006) Genetic balancers. WormBook, doi:10.1895/wormbook.1.89.1. [DOI] [PMC free article] [PubMed]
- 65).Iwata S., Yoshina S., Suehiro Y., Hori S., Mitani S. (2016) Engineering new balancer chromosomes in C. elegans via CRISPR/Cas9. Sci. Rep. 6, 33840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66).Nakagawa A., Shi Y., Kage-Nakadai E., Mitani S., Xue D. (2010) Caspase-dependent conversion of Dicer ribonuclease into a death-promoting deoxyribonuclease. Science 328, 327–334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67).Chen A.T., Guo C., Itani O.A., Budaitis B.G., Williams T.W., Hopkins C.E., McEachin R.C., Pande M., Grant A.R., Yoshina S., Mitani S., Hu P.J. (2015) Longevity genes revealed by integrative analysis of isoform-specific daf-16/FoxO mutants of Caenorhabditis elegans. Genetics 201, 613–629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68).Silver D.L., Hou L., Somerville R., Young M.E., Apte S.S., Pavan W.J. (2008) The secreted metalloprotease ADAMTS20 is required for melanoblast survival. PLoS Genet. 4, e1000003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69).Chen R., Shi L., Hakenberg J., Naughton B., Sklar P., Zhang J., Zhou H., Tian L., Prakash O., Lemire M., Sleiman P., Cheng W.Y., Chen W., Shah H., Shen Y., Fromer M., Omberg L., Deardorff M.A., Zackai E., Bobe J.R., Levin E., Hudson T.J., Groop L., Wang J., Hakonarson H., Wojcicki A., Diaz G.A., Edelmann L., Schadt E.E., Friend S.H. (2016) Analysis of 589,306 genomes identifies individuals resilient to severe Mendelian childhood diseases. Nat. Biotechnol. 34, 531–538. [DOI] [PubMed] [Google Scholar]
- 70).Seung, S. (2012) Connectome, How the brain’s wiring makes us who we are. Houghton Mifflin Harcourt Publishing, NY. [Google Scholar]
- 71).Hu, P.J. (2007) Dauer. WormBook, doi:10.1895/wormbook.1.144.1. [DOI] [PMC free article] [PubMed]
- 72).Pérez Jurado L.A., Peoples R., Kaplan P., Hamel B.C., Francke U. (1996) Molecular definition of the chromosome 7 deletion in Williams syndrome and parent-of-origin effects on growth. Am. J. Hum. Genet. 59 (4), 781–792. [PMC free article] [PubMed] [Google Scholar]
- 73).Kamath R.S., Fraser A.G., Dong Y., Poulin G., Durbin R., Gotta M., Kanapink A., Le Bot N., Moreno S., Sohrmann M., Welchman D.P., Zipperlen P., Ahringer J. (2003) Systematic functional analysis of the Caenorhabditis elegans genome using RNAi. Nature 421, 231–237. [DOI] [PubMed] [Google Scholar]
- 74).Giaever G., Chu A.M., Ni L., Connelly C., Riles L., Véronneau S., Dow S., Lucau-Danila A., Anderson K., André B., Arkin A.P., Astromoff A., El-Bakkoury M., Bangham R., Benito R., Brachat S., Campanaro S., Curtiss M., Davis K., Deutschbauer A., Entian K.D., Flaherty P., Foury F., Garfinkel D.J., Gerstein M., Gotte D., Güldener U., Hegemann J.H., Hempel S., Herman Z., Jaramillo D.F., Kelly D.E., Kelly S.L., Kötter P., LaBonte D., Lamb D.C., Lan N., Liang H., Liao H., Liu L., Luo C., Lussier M., Mao R., Menard P., Ooi S.L., Revuelta J.L., Roberts C.J., Rose M., Ross-Macdonald P., Scherens B., Schimmack G., Shafer B., Shoemaker D.D., Sookhai-Mahadeo S., Storms R.K., Strathern J.N., Valle G., Voet M., Volckaert G., Wang C.Y., Ward T.R., Wilhelmy J., Winzeler E.A., Yang Y., Yen G., Youngman E., Yu K., Bussey H., Boeke J.D., Snyder M., Philippsen P., Davis R.W., Johnston M. (2002) Functional profiling of the Saccharomyces cerevisiae genome. Nature 418, 387–391. [DOI] [PubMed] [Google Scholar]
- 75).Doitsidou M., Jarriault S., Poole R.J. (2016) Next-generation sequencing-based approaches for mutation mapping and identification in Caenorhabditis elegans. Genetics 204, 451–474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76).Wang X., Li W., Zhao D., Liu B., Shi Y., Chen B., Yang H., Guo P., Geng X., Shang Z., Peden E., Kage-Nakadai E., Mitani S., Xue D. (2010) Caenorhabditis elegans transthyretin-like protein TTR-52 mediates recognition of apoptotic cells by the CED-1 phagocyte receptor. Nat. Cell Biol. 12, 655–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77).Conradt B., Wu Y.C., Xue D. (2016) Programmed cell death during Caenorhabditis elegans Development. Genetics 203, 1533–1562. [DOI] [PMC free article] [PubMed] [Google Scholar]