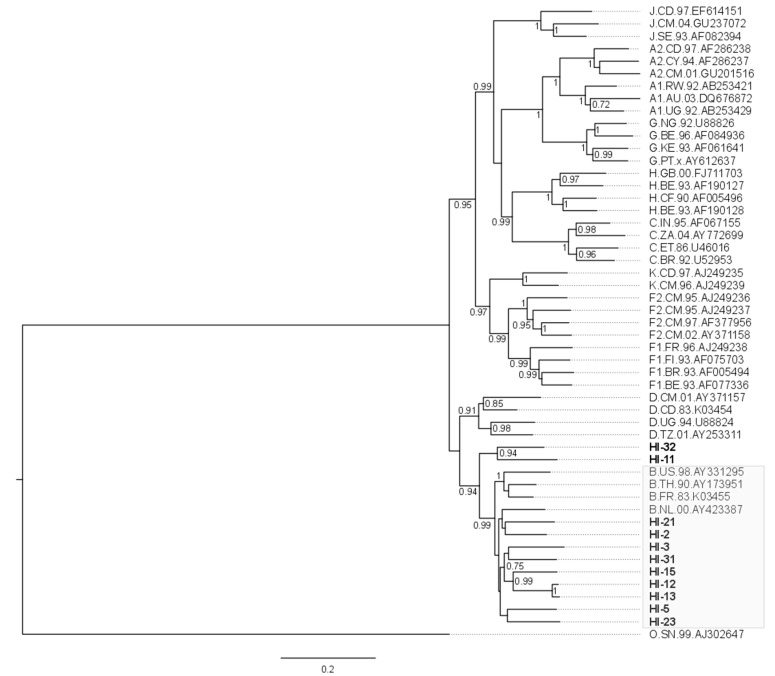
Figure 1.
Phylogenetic maximum likehood analysis of HIV near full-length genomes obtained in this study. The analysis was conducted with 1000 bootstrap iterations and included eleven HIV-1 proviral sequences from Hospital Federal de Ipanema, Rio de Janeiro (represented in bold) and references of HIV-1 subtypes (represented by the subtype, country, year and GenBank accession number). Only bootstrap values greater than 0.7 are shown. The gray box highlights the sequences classified as subtype B, among which nine of the eleven sequences were placed. The two remaining sequences represented unique recombinant forms comprising subtype B and other subtypes.