ABSTRACT
Real-time quantitative PCR (qPCR) assay of sputum or nasopharyngeal specimens has shown promising results in the detection of pneumococcal community-acquired pneumonia (PncCAP). We applied qPCR for the autolysin gene (lytA) and compared sputum and nasopharyngeal swab (NPS) pneumococcal loads in elderly patients with community-acquired pneumonia (CAP), and specifically in patients with PncCAP, to those in patient groups with other respiratory diseases. We studied patients aged ≥65 years with radiologically confirmed CAP, clinical CAP not retrospectively radiologically confirmed, other acute respiratory infections, or stable chronic lung disease. Pneumococcal etiology of CAP was ascertained by using a combination of multiple diagnostic methods. We analyzed sputum and NPS specimens by lytA qPCR with 104 pneumococcal genome equivalents (GE)/ml as a cutoff for positivity. Among PncCAP patients, lytA qPCR detected pneumococci in 94% of the sputum samples and in large quantities (mean, 6.82 ± 1.02 log10 GE/ml) but less frequently in NPS (44%) and in smaller quantities (5.55 ± 0.92 log10 GE/ml). In all other patient groups, ≤10% of the sputum samples and <5% of the NPS samples were lytA qPCR positive; but when they were positive, the sputum pneumococcal loads were similar to those in the PncCAP patients, suggesting a pneumococcal etiology in these patients. This was supported by other pneumococcal assay results. Overall, sputum lytA qPCR positivity was more common in PncCAP patients than in the other patient groups, but the quantitative results were mainly similar. NPS lytA qPCR was less sensitive than sputum lytA qPCR in detecting PncCAP.
KEYWORDS: community-acquired pneumonia, nasopharynx, PCR, sputum, Streptococcus pneumoniae
INTRODUCTION
Streptococcus pneumoniae is the most frequently detected bacterial etiologic agent in community-acquired pneumonia (CAP) (1), and the incidence of pneumococcal pneumonia in the elderly increases with age (2, 3). When conventional identification methods such as blood and sputum cultures are used, pneumococcal pneumonia remains underdiagnosed (4). Newer diagnostic methods such as pneumococcal urine antigen detection and PCR on respiratory specimens have consequently been applied to increase the sensitivity of case detection (5–8). A real-time quantitative PCR (qPCR) assay with the autolysin gene (lytA) as the target and applied to sputum (8, 9) or nasopharyngeal specimens (8–10) has shown promising results in the diagnosis of pneumococcal CAP (PncCAP). Here, we studied the genomic pneumococcal loads in sputum samples and nasopharyngeal swab (NPS) specimens from elderly patients with CAP, and specifically in patients with PncCAP (J. Jokinen et al., submitted for publication) (11), but also in patients with acute respiratory tract infections other than CAP and those with chronic lung disease (CLD), and compared the results between the groups.
(Data from this study were presented at the 10th International Symposium on Pneumococci and Pneumococcal Diseases, Glasgow, Scotland, 26 to 30 June 2016.)
MATERIALS AND METHODS
Study participants.
We studied noninstitutionalized patients ≥65 years old participating in a 2-year Finnish CAP epidemiological (FinCAP Epi) study in 2005 to 2007 (11). CAP was defined as any new opacity in a chest X-ray compatible with pneumonia in a symptomatic patient and confirmed retrospectively radiologically by at least two of three reviewers (two radiologists from Tampere University Hospital and one international reviewer). Clinical CAP cases that were not retrospectively radiologically confirmed were designated rejected CAP cases. Patients with an acute respiratory infection (ARI) and no radiological signs of pneumonia and no diagnosis of pneumonia in the 30 days preceding and the 30 days following enrollment were enrolled as one comparison group, and patients with a CLD (International Classification of Diseases 10th revision discharge diagnosis of chronic bronchitis, lung emphysema, or chronic obstructive pulmonary disease) were enrolled as another comparison group. The CLD patients underwent spirometry and were in stable condition without concurrent exacerbation. Informed consent was obtained from all subjects or their next of kin/guardians. The study protocol was approved by the ethical review board of the Pirkanmaa Hospital District, the Tampere City Health Center, and the institutional review board at the National Institute for Health and Welfare (THL).
Microbiological assessment.
The collection and processing of study samples, including blood specimens for aerobic and anaerobic blood cultures and serology, sputum, NPS, and urine, as well as the assays carried out to detect pneumococci, have been described in detail previously (11). The sputum samples were spontaneously produced or induced with a NaCl nebulizer. Sputum quality was assessed by microscopy after Gram staining and was rated high (high-quality [HQ] sputum) if the ratio of leukocytes to epithelial cells was >1. This criterion for HQ sputum differs from that described by Murray and Washington (12) but has been used previously (13). However, all sputum samples were processed and assayed as planned, irrespective of their quality. Part of each sputum sample was stored for viral analyses, and the rest was treated with Sputolysin (dithiothreitol). Part of the processed sputum was stored at −70°C for DNA extraction and real-time qPCR, and another part was plated undiluted and diluted 1/100 on sheep blood agar, chocolate agar, and blood agar plates with gentamicin. Alpha-hemolytic colonies suspected to be S. pneumoniae were identified primarily by using optochin sensitivity and, if needed, by using bile solubility and the quellung reaction with pneumococcal omniserum. NPS samples were collected with a calcium alginate swab in STGG (skim milk, tryptone, glucose, and glycerol) medium and stored at −70°C. DNA was extracted from sputum and NPS samples with the MagNA Pure LC DNA isolation kit III (bacteria, fungi) and a MagNA Pure LC instrument (Roche Diagnostics, Germany). Real-time lytA qPCR was carried out with the LightCycler 2.0 instrument (Roche Diagnostics) and a previously described method (14) in which a 173-bp fragment of the lytA gene is amplified and detected and a melting curve program is used to check the specificity of probe binding. The sample volume in each reaction mixture was 5 μl, and each qPCR run included a standard containing 500 pneumococcal genome equivalents (GE). No internal process control was used. The data were analyzed with LightCycler software version LCS4 4.0.5.415 (Roche Diagnostics). An external standard curve created by amplifying four replicates of six serial dilutions of purified pneumococcal DNA (5 × 105, 5 × 104, 5 × 103, 5 × 102, 5 × 101, and 5 GE) was used for quantification. Ten thousand GE/ml of sample (50 GE/qPCR) was used as a cutoff for positivity for both specimen types, as this was the lowest concentration that could be detected in at least 95% of the assays when dilution series of four independently extracted negative patient samples with a known amount of added pneumococcal DNA were analyzed in six replicates as part of the validation of the lytA qPCR method carried out separately for both specimen types.
During the first year of this study, clinical CAP patients were assayed for certain viruses, including the influenza A and B viruses (sputum PCR and serology), respiratory syncytial virus (RSV; sputum PCR and serology), and parainfluenza virus type 1 (PIV1), PIV2, and PIV3 (sputum PCR) (11).
Criteria for PncCAP.
We used our previously published definition of PncCAP (11), which was constructed by utilizing latent class analysis (LCA) to determine true disease status on the basis of imperfect tests and is presented in detail elsewhere (Jokinen et al., submitted). PncCAP was defined as (i) encapsulated pneumococci cultured from blood, (ii) encapsulated pneumococci cultured from HQ sputum, or (iii) at least two of the following: (i) a ≥2-fold increase in serum antipneumococcal surface adhesin A (PsaA) and/or anti-choline-binding protein A (CbpA) antibodies, (ii) a positive pneumococcal urine antigen test, or (iii) detection of pneumococci in sputum of any quality or NPS by culture (encapsulated) or lytA qPCR (11).
Data analysis.
Pearson's chi-square test or Fisher's exact test, where appropriate, was used for the comparison of categorical variables. The qPCR results were log10 transformed and compared between and within the patient groups. The Student t test was used for comparisons of means, and the correlation of pneumococcal genomic loads in sputum and NPS samples was calculated by using Pearson's correlation coefficient. A P value of <0.05 was considered statistically significant. Statistical analyses were performed with SPSS version 22 (SPSS Inc., Chicago, IL).
The CURB-65 score, a measure of severity of pneumonia, was calculated as described by Lim et al. (15).
RESULTS
When 104 GE/ml was used as the cutoff for positivity, pneumococci were detected by lytA qPCR in sputum samples from 44/47 (94%) PncCAP cases, 10/103 (10%) CLD cases, and <5% of the cases in the other groups. NPS specimens were positive for lytA in 23/52 (44%) PncCAP cases and in <5% of those in all other groups (Table 1). There were 0 PncCAP, 7 non-PncCAP, 2 rejected CAP, 1 ARI, and 1 CLD cases with a lytA qPCR result below the cutoff but greater than zero in sputum and 12, 7, 3, 2, and 11 in NPS, respectively. The log10-transformed lytA qPCR results of the sputum and NPS samples with a result greater than zero among the patient groups are shown in Fig. 1A and B, respectively. The samples with a quantitative result below the cutoff were considered negative and were not included in the following analyses.
TABLE 1.
Percentage of positive casese and pneumococcal loads in the different patient groups assayed by using lytA qPCR
| Patient group | Sputum |
NPS |
||||||
|---|---|---|---|---|---|---|---|---|
| n | No. (%) lytA qPCR positive | Mean log10 GE/ml ± SDa | P valueb | n | No. (%) lytA qPCR positive | Mean log10 GE/ml ± SDa | P valueb | |
| Confirmed CAP | 224 | 50 (22) | 6.74 ± 1.09 | 306 | 25 (8) | 5.44 ± 0.97 | ||
| PncCAPc | 47 | 44 (94) | 6.82 ± 1.02 | 52 | 23 (44) | 5.55 ± 0.92 | ||
| Non-PncCAP | 177 | 6 (3)d | 6.16 ± 1.45 | 0.33 | 254 | 2 (1)d | 4.08 ± 0.00 | 0.04 |
| Rejected CAP | 106 | 3 (3)d | 7.70 ± 0.41 | 0.15 | 157 | 6 (4)d | 6.08 ± 1.19 | 0.25 |
| ARI | 65 | 2 (3)d | 6.94 ± 0.38 | 0.87 | 80 | 1 (1)d | 4.99 | |
| CLD | 103 | 10 (10)d | 5.80 ± 1.70 | 0.09 | 121 | 1 (1)d | 6.11 | |
Only lytA qPCR positive cases included.
Quantitative results compared to PncCAP.
PncCAP was defined as (i) encapsulated pneumococci cultured from blood, (ii) encapsulated pneumococci cultured from HQ sputum (a leukocyte/epithelial cell ratio of >1), or (iii) at least two of the following: (i) a ≥2-fold increase in serum anti-PsaA and/or anti-CbpA antibodies, (ii) a positive pneumococcal urine antigen test, (iii) detection of pneumococci from sputum of any quality or NPS by culture (encapsulated) or lytA qPCR.
P < 0.001 compared to PncCAP.
The cutoff for positivity was 104 GE/ml.
FIG 1.
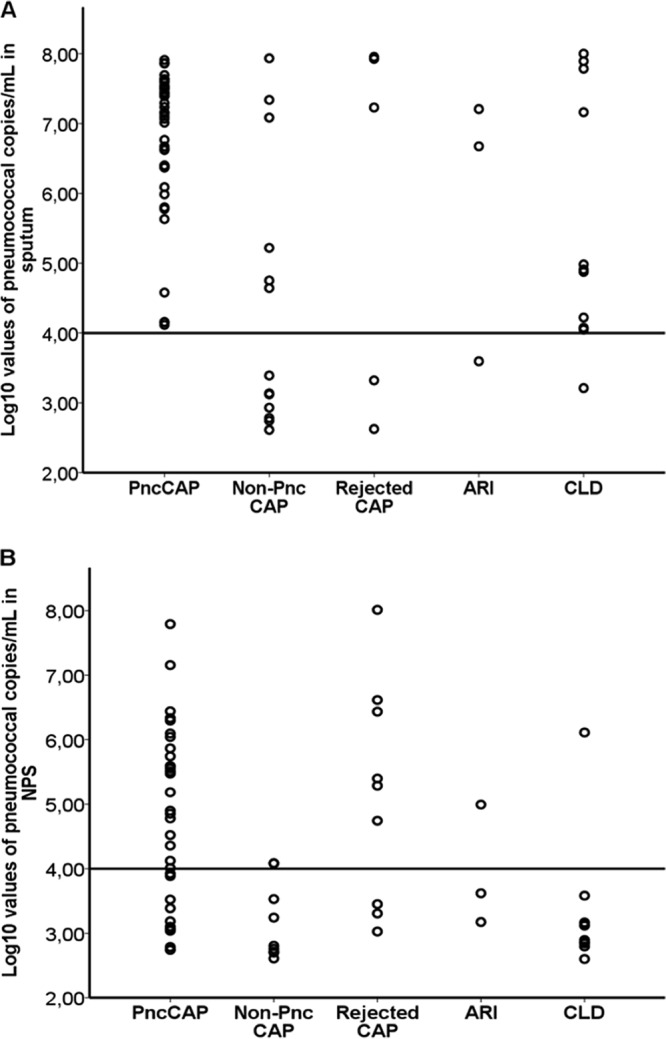
Log10-transformed results of lytA qPCR assays of sputum (A) and NPS (B) samples from patients with PncCAP, non-PncCAP, clinical CAP that was not radiologically confirmed (rejected CAP), ARI, and CLD. All results greater than zero are included. The horizontal line shows the cutoff for positivity (104 GE/ml) that was used in this study.
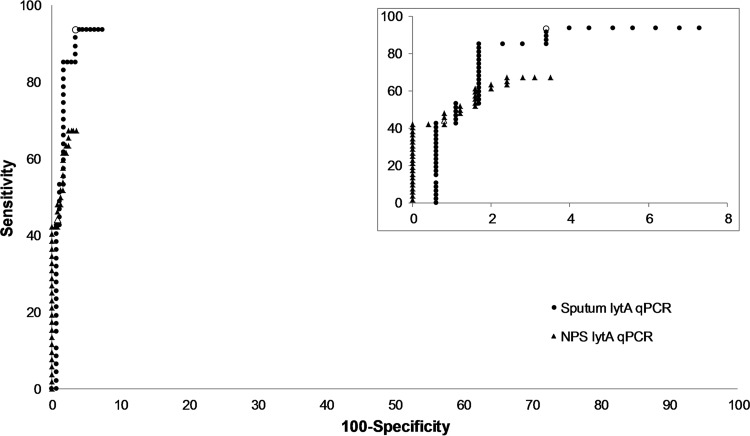
The diagnostic performance of lytA qPCR for the detection of PncCAP among CAP patients is presented in Fig. 2. At the predefined cutoff of 104 GE/ml of sample, the sensitivity and specificity of the lytA qPCR test were 94 and 97% for sputum samples and 44 and 99% for NPS samples, respectively. The criteria for PncCAP (Jokinen et al., submitted) (11) included lytA qPCR from sputum or NPS but only in combination with a ≥2-fold increase in paired serum anti-PsaA and/or anti-CbpA antibodies or a positive pneumococcal urine antigen test. There were four cases in which the PncCAP diagnosis depended on a positive sputum lytA qPCR result, and in three of these, pneumococcal serology or the urine antigen test was missing. When lytA qPCR positivity was left out of the criteria for PncCAP, the sensitivity and specificity of the lytA qPCR test with sputum were similar (93 and 94%, respectively). There were no cases in which the diagnosis depended on a positive NPS lytA qPCR result.
FIG 2.
Diagnostic performance (sensitivity versus 100 − specificity) of lytA qPCR applied to sputum and NPS samples for detection of PncCAP among patients with CAP and the sensitivity and specificity of lytA qPCR at the cutoff for positivity (104 GE/ml) for sputum (○), and NPS (Δ). The inset contains the same data but with a smaller x-axis scale.
While only ≤10% of the non-PncCAP, rejected CAP, ARI, or CLD cases were positive for pneumococcus by lytA qPCR, when positive their mean pneumococcal load (± standard deviation) in sputum did not differ statistically significantly from that of the PncCAP cases (Table 1). This was true also when only HQ sputum samples were included in the analysis (data not shown). The five patients with ARI or rejected CAP that had a positive lytA qPCR result and a high pneumococcal density in sputum (Table 1) had encapsulated pneumococci cultured from their sputum samples, which all were of high quality. Two of these patients also had a positive urine antigen test result. This suggests a pneumococcal etiology in these five patients, as they fulfilled the etiological case definition but not the radiological CAP definition.
In the subgroup of patients with positive lytA qPCR results (≥104 GE/ml) with both sputum and NPS, the lytA qPCR results showed no correlation between sputum and NPS pneumococcal genomic loads (n = 23, Pearson's correlation r = 0.19, and P = 0.39 for all cases; n = 19, r = 0.14, and P = 0.56 for CAP cases; n = 18, r = 0.17, and P = 0.51 for PncCAP cases) (Fig. 3).
FIG 3.
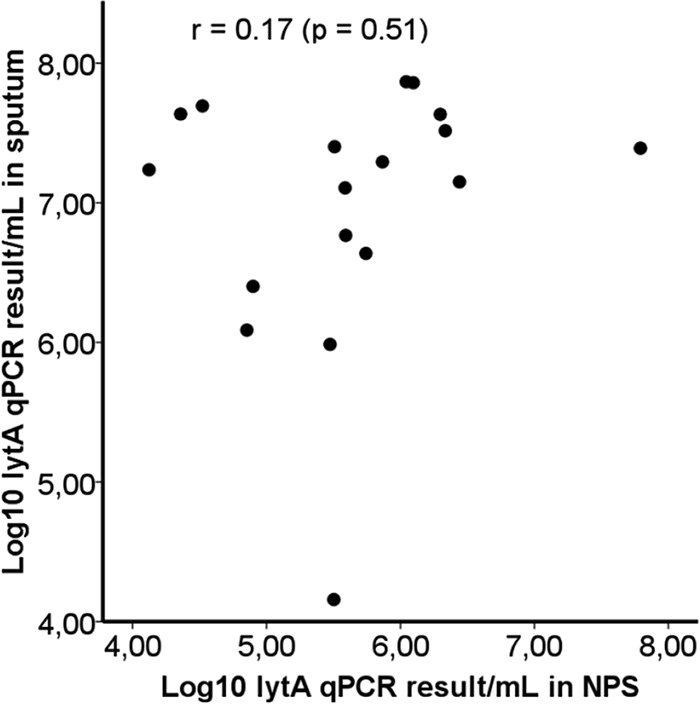
Correlation of sputum lytA qPCR results with NPS lytA qPCR results among patients with PncCAP and a positive lytA qPCR result with both sputum and NPS samples (n = 18).
In the patients with a positive sputum lytA qPCR result, the mean pneumococcal genomic copy numbers were higher in HQ sputum than in low-quality (LQ) sputum among all cases (n = 53, 6.78 ± 1.15 versus n = 11, 5.91 ± 1.35 log10 GE/ml, P = 0.03); however, among CAP cases only (Table 2) or among PncCAP cases only (n = 38, 6.88 ± 0.96 versus n = 5, 6.15 ± 1.37 log10 GE/ml, P = 0.13), the difference was not statistically significant.
TABLE 2.
Factors and their association with sputum pneumococcal loads among CAP cases assayed by lytA qPCR
| Factor | Yes |
No |
P valueb | P valuec | ||||
|---|---|---|---|---|---|---|---|---|
| n | No. (%) lytA qPCR positive | Mean log10 GE/ml ± SDa | n | No. (%) lytA qPCR positive | Mean log10 GE/ml ± SDa | |||
| Male | 121 | 31 (26) | 6.50 ± 1.25 | 103 | 19 (18) | 7.13 ± 0.59 | 0.20 | 0.02 |
| Current smoker | 27 | 8 (30) | 6.69 ± 1.33 | 185 | 38 (21) | 6.70 ± 1.08 | 0.28 | 0.98 |
| HQ sputumd | 169 | 40 (24) | 6.86 ± 0.98 | 53 | 9 (17) | 6.13 ± 1.40 | 0.31 | 0.17 |
| Encapsulated pneumococci cultured from sputum | 40 | 37 (93) | 7.22 ± 0.57 | 184 | 13 (7) | 5.38 ± 1.07 | <0.001 | <0.001 |
| Influenza A virus-positive sputum PCR and/or serology | 7 | 2 (29) | 6.24 ± 0.62 | 96 | 21 (22) | 6.80 ± 1.09 | 0.65 | 0.49 |
| Influenza B virus-positive sputum PCR and/or serology | 0 | NA | NA | 101 | 23 (23) | 6.75 ± 1.06 | NA | NA |
| RSV-positive sputum PCR and/or serology | 14 | 4 (29) | 7.59 ± 0.26 | 88 | 19 (22) | 6.58 ± 1.09 | 0.51 | 0.002 |
| PIV1, -2, or -3-positive sputum PCR | 7 | 4 (57) | 7.27 ± 0.64 | 104 | 21 (20) | 6.69 ± 1.07 | 0.04 | 0.31 |
| Any viruse | 28 | 10 (36) | 7.19 ± 0.69 | 76 | 13 (17) | 6.42 ± 1.20 | 0.04 | 0.07 |
| Antibiotic use at visit only | 41 | 13 (32) | 5.75 ± 1.21 | 138f | 34 (25) | 7.06 ± 0.82 | 0.37 | 0.002 |
| Antibiotic use within 2 wk before visit | 39 | 3 (8) | 7.47 ± 0.07 | 138f | 34 (25) | 7.06 ± 0.82 | 0.03 | 0.40 |
| Hospitalization | 184 | 38 (21) | 6.72 ± 1.15 | 40 | 12 (30) | 6.81 ± 0.90 | 0.20 | 0.81 |
| CURB-65 score of 3, 4, or 5g | 50 | 12 (24) | 6.92 ± 0.78 | 167 | 37 (22) | 6.66 ± 1.18 | 0.78 | 0.47 |
Only qPCR-positive cases included.
Proportions of lytA qPCR-positive samples compared with the Pearson chi-square test or Fisher exact test. Statistically significant values are in boldface.
Genomic loads compared with the Student t test. Statistically significant values are in boldface.
More than one leukocyte per epithelial cell.
Positive sputum PCR for PIV1, -2, or -3 or positive sputum PCR or serology for influenza A virus, influenza B virus, or RSV.
No use of antibiotics at visit or within 2 weeks before visit.
CURB-65: confusion, urea concentration of >7 mmol/liter, respiratory rate of ≥30/min, low blood pressure (systolic <90 mm Hg or diastolic ≤60 mm Hg), age of ≥65 years (15).
Pneumococci were cultured from 50 (22%) CAP cases' sputum and 37 (12%) CAP cases' NPS samples, and the cultured pneumococci were encapsulated in 40 (80%) and 32 (86%) of the cases, respectively; the lytA qPCR result was positive in 37 (93%) and 22 (69%) of these, respectively, and negative in all cases with unencapsulated pneumococci cultured from the same sample type. Among the CAP cases with a negative pneumococcal culture, the lytA qPCR was positive in 13 (7%) and 3 (1%) cases with sputum and NPS samples, respectively. In the patients with a positive sputum lytA qPCR result, the mean pneumococcal genomic load in sputum was greater among the cases with pneumococci (encapsulated) cultured from sputum than among those with no pneumococci cultured from sputum (Table 2). The same was true of NPS samples (5.59 ± 0.93 versus 4.33 ± 0.49 log10 GE/ml, P = 0.03).
The lytA qPCR positivity and quantitative results of those with a positive lytA qPCR result were also explored for sputum in some selected subgroups of CAP patients (Table 2). The CAP patients with any respiratory viral coinfection (influenza A or B virus, RSV, PIV1, PIV2, or PIV3) were more often lytA qPCR positive and had a slightly higher pneumococcal genomic load than those with no viruses detected, although the difference in the quantitative result was not statistically significant. However, among the patients with RSV, the mean pneumococcal load was significantly greater (Table 2). The pneumococcal genomic load of the CAP patients with a CURB-65 score of 3 to 5 did not differ from that of the patients with a CURB-65 score 1 or 2, and the pneumococcal genomic loads in hospitalized and nonhospitalized patients were similar (Table 2). The CAP patients who had received antibiotics at their acute-phase visit before sputum sampling had a lower mean sputum pneumococcal genomic load than those who were not exposed to antibiotics at the visit or within 2 weeks before the visit (Table 2). The analyses presented in Table 2 were also conducted by including only the CAP cases with no antimicrobial exposure in the 2 weeks before the visit or at the visit (n = 138; lytA qPCR positive n = 34). The results were similar to those in Table 2, except that there was no difference between the pneumococcal loads of females and males when only patients with no antimicrobial use were included (data not shown).
DISCUSSION
In the present study of elderly patients, pneumococci were detected by lytA qPCR in the majority of the PncCAP patients' sputum samples and less frequently and in smaller quantities in NPS. In all other patient groups, the prevalence of pneumococci by lytA qPCR was low. However, when the test was positive, a high pneumococcal density was observed in the sputum of ARI and rejected CAP patients, suggesting a pneumococcal etiology in these patients.
The diagnostic performance of the lytA qPCR test was studied by comparing it to the criteria used for PncCAP in the FinCAP Epi study (11). The criteria were derived by LCA with a focus on an optimal case definition for vaccine trial purposes (Jokinen et al., submitted), and they included lytA qPCR positivity with sputum or NPS in combination with a positive urine antigen test result or a ≥2-fold increase in paired serum anti-PsaA and/or anti-CbpA antibodies. Therefore, comparison of lytA qPCR test performance against these criteria may not be optimal for the determination of lytA qPCR sensitivity. On the basis of the LCA model, the sensitivity of lytA qPCR with sputum was 90% (Jokinen et al., submitted), which is likely to be closer to the real sensitivity of the test. Also, the criterion for HQ sputum used in our definition of PncCAP (11) and in the present study differs from that described by Murray and Washington (12). We have previously evaluated the significance of sputum quality in sputum culture for the diagnosis of PncCAP by using more stringent criteria for HQ sputum (a leukocyte/epithelial cell ratio of >5 and ≤2.5 epithelial cells/field) and found that cultures positive for encapsulated pneumococci from HQ and LQ sputum samples showed a similar concordance with other pneumococcal diagnostic tests if the other test was positive (16). If the other pneumococcal diagnostic test was negative, encapsulated pneumococci were isolated from LQ sputum samples less often than from HQ sputum samples (16). As our previous results did not support the concept that encapsulated pneumococci cultured from an LQ sputum sample would more probably be a false-positive indicator of pneumococcal etiology in CAP than the same finding in an HQ sample (16), we applied a less stringent criterion for HQ sputum in the PncCAP definition (Jokinen et al., submitted) (11) in the present study.
In a recent study, Strålin et al. (8) used a lytA qPCR assay and reported a prevalence of positive results with sputum samples (94% with a cutoff of 105 copies/ml and 97% with a cutoff of 104 copies/ml) from adult PncCAP patients (median age, 71 years) similar to that in the present study. They also reported a similar mean sputum pneumococcal load (6.71 ± 1.01 log10 copies/ml) in their qPCR-positive cases. In nasopharyngeal aspirates (NPA), they detected positive results among PncCAP patients by using their lytA qPCR assay more often (62% with a cutoff of 104 copies/ml) than we did in NPS specimens, but the mean nasopharyngeal pneumococcal load (5.35 ± 1.69 log10 copies/ml) among the qPCR-positive cases was similar to that reported here (8). NPA and NPS sampling methods for detecting pneumococcal pharyngeal carriage by culture have been compared in children by Rapola et al. (17), and they found no significant difference in the rate of pneumococcus isolation between NPA and NPS by culture. Among the patients with non-PncCAP and a positive lytA qPCR result, we detected a greater mean pneumococcal load in sputum than Strålin et al. did (8). In agreement with their results, we found no correlation between the pneumococcal genomic copy numbers in sputum and nasopharyngeal specimens. However, Albrich et al. (9) previously noted a good correlation between the genomic pneumococcal loads in sputum and NPS from HIV-infected adults with CAP. The studies of Strålin et al. (8) and Albrich et al. (9) did not include patients with respiratory tract infections other than CAP.
The mean pneumococcal load detected by qPCR was greater in patients with a positive sputum culture (encapsulated pneumococci) than in patients with no pneumococci cultured. This is in accordance with previous studies where greater pneumococcal genomic loads have been observed in culture-positive sputum specimens (5, 9, 18). Gadsby et al. (5), however, noted that their culture-negative group was more frequently exposed to antibiotics, which was also associated with lower bacterial loads. Werno et al. (18) did not find any significant effect of antibiotic use on genomic pneumococcal loads, even though they detected pneumococci less often in the sputum of patients exposed to antibiotics prior to admission. We have previously studied the effect of antibiotic use on pneumococcal diagnostic tests with the same study population as here (19) and found that antibiotic use within 2 weeks before the acute-phase visit, and specifically when still ongoing at enrollment, but not antibiotic use at the visit, was associated with lower sputum lytA qPCR positivity. In the present study, patients who had received antibiotics at the acute-phase visit, but not those who had gotten antibiotics within 2 weeks before the visit, had a significantly lower mean pneumococcal genomic load in their sputum than those with no antibiotic use.
In all CAP cases in which unencapsulated pneumococci were cultured, the lytA qPCR result with the same sample type was negative. This implies that the unencapsulated cultured isolates were, in fact, not true pneumococci and they were disregarded in the further analyses.
It is well known that respiratory virus infections play a role in respiratory bacterial infections (20). Respiratory virus coinfection has also been associated with greater nasopharyngeal pneumococcal loads in children with CAP (21) and in ARI patients with a high HIV prevalence (22). Alpkvist et al. (23), however, found no association between viral coinfection and greater nasopharyngeal pneumococcal loads in adult patients. In the present study, virus infection was detected by sputum PCR and/or serology and CAP patients with a respiratory viral coinfection had a slightly, although not statistically significantly, greater sputum pneumococcal genomic load than CAP patients with no viruses detected. Among the CAP patients with RSV, the pneumococcal load was, however, significantly greater. Interestingly, in mice, RSV infection has been shown to decrease the clearance of pneumococci from the lungs and increase pulmonary inflammation (24).
A high nasopharyngeal pneumococcal density has been found to be associated with greater disease severity (23, 25). However, when the sputum pneumococcal load and disease severity were compared by Werno et al. (18), no association was found except for those patients who were previous and current smokers; they were more often in a higher pneumonia severity index risk class when the pneumococcal load in sputum was >103 CFU/ml. We analyzed the pneumococcal loads in the sputum samples of CAP patients, their CURB-65 scores, and their hospitalization status for associations and found none.
The present study confirms the results of the study of Strålin et al. (8), which identified the lytA qPCR assay of sputum as a useful method for the diagnosis of PncCAP. In contrast to the study of Albrich et al. (9), the lytA qPCR assay of NPS did not perform as well as the lytA qPCR assay of sputum samples in the present study of elderly persons. The quantitative result obtained with NPS may be less reliable because of variations in the quantity of the actual sample and in the recovery of pneumococci from STGG. The lytA qPCR assay is a more rapid method than culture. We did not try to identify optimal cutoff values for the diagnosis of PncCAP by using lytA qPCR but used a predefined cutoff that was the lowest concentration that could be consistently detected. The strengths of the present study are that it was a prospective follow-up study with systematic data collection and that it included different patient groups. Thus, we had qPCR results available also for patient groups with respiratory tract infections other than CAP or a CLD. However, a limitation of this study was the low number of patients, especially in the PncCAP group. In addition, only a few patients in the present study had pneumococcal bacteremia. A potential limitation of the lytA qPCR assay is that lytA has been found in some nonpneumococcal streptococcal isolates of the Streptococcus mitis group (26, 27). Thus, the lytA qPCR assay potentially overestimates the pneumococcus as the etiological agent of CAP. In the present study, 3% of the non-PncCAP cases' sputum samples and 1% of their NPS samples were positive by lytA qPCR assay but the etiological agent in these was not determined.
In conclusion, pneumococci were detected by lytA qPCR in the majority of the PncCAP patients' sputum samples when 104 GE/ml was used as the cutoff for positivity. In all other patient groups, the prevalence of pneumococci by lytA qPCR was low. Sputum was superior to NPS as a sample type in the detection of PncCAP by lytA qPCR, and lytA qPCR analysis of sputum continues to be a promising diagnostic tool in the detection of pneumococcal etiology in CAP. A trend toward greater pneumococcal loads in sputum samples from CAP patients with a viral coinfection, particularly RSV, was seen.
ACKNOWLEDGMENTS
We sincerely thank all of the participating elderly subjects for consenting to the study during an acute illness.
This study was conducted in collaboration with GlaxoSmithKline Biologicals SA, which supported this study, and the GSK coauthors of this paper participated in study design, interpretation of results, and manuscript writing.
A.S., A.A.P., and J.J. are employees of the National Institute for Health and Welfare, which has received research funding from the GSK group of companies. W.P.H. was an employee and T.P. and V.V. are employees of the GSK group of companies. T.P. and V.V. own shares in the GSK group of companies.
REFERENCES
- 1.Musher DM, Roig IL, Cazares G, Stager CE, Logan N, Safar H. 2013. Can an etiologic agent be identified in adults who are hospitalized for community-acquired pneumonia: results of a one-year study. J Infect 67:11–18. doi: 10.1016/j.jinf.2013.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jokinen C, Heiskanen L, Juvonen H, Kallinen S, Kleemola M, Koskela M, Leinonen M, Ronnberg PR, Saikku P, Sten M, Tarkiainen A, Tukiainen H, Pyorala K, Makela PH. 2001. Microbial etiology of community-acquired pneumonia in the adult population of 4 municipalities in eastern Finland. Clin Infect Dis 32:1141–1154. doi: 10.1086/319746. [DOI] [PubMed] [Google Scholar]
- 3.Gutierrez F, Masia M, Mirete C, Soldan B, Rodriguez JC, Padilla S, Hernandez I, Royo G, Martin-Hidalgo A. 2006. The influence of age and gender on the population-based incidence of community-acquired pneumonia caused by different microbial pathogens. J Infect 53:166–174. doi: 10.1016/j.jinf.2005.11.006. [DOI] [PubMed] [Google Scholar]
- 4.Ruiz-González A, Falguera M, Nogues A, Rubio-Caballero M. 1999. Is Streptococcus pneumoniae the leading cause of pneumonia of unknown etiology? A microbiologic study of lung aspirates in consecutive patients with community-acquired pneumonia. Am J Med 106:385–390. [DOI] [PubMed] [Google Scholar]
- 5.Gadsby NJ, Russell CD, McHugh MP, Mark H, Conway Morris A, Laurenson IF, Hill AT, Templeton KE. 2016. Comprehensive molecular testing for respiratory pathogens in community-acquired pneumonia. Clin Infect Dis 62:817–823. doi: 10.1093/cid/civ1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Smith MD, Sheppard CL, Hogan A, Harrison TG, Dance DA, Derrington P, George RC, South West Pneumococcus Study Group. 2009. Diagnosis of Streptococcus pneumoniae infections in adults with bacteremia and community-acquired pneumonia: clinical comparison of pneumococcal PCR and urinary antigen detection. J Clin Microbiol 47:1046–1049. doi: 10.1128/JCM.01480-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Said MA, Johnson HL, Nonyane BA, Deloria-Knoll M, O'Brien KL, AGEDD Adult Pneumococcal Burden Study Team, Andreo F, Beovic B, Blanco S, Boersma WG, Boulware DR, Butler JC, Carratala J, Chang FY, Charles PG, Diaz AA, Dominguez J, Ehara N, Endeman H, Falco V, Falguera M, Fukushima K, Garcia-Vidal C, Genne D, Guchev IA, Gutiérrez F, Hernes SS, Hoepelman AI, Hohenthal U, Johansson N, Kolek V, Kozlov RS, Lauderdale TL, Marekovic I, Masia M, Matta MA, Miro O, Murdoch DR, Nuermberger E, Paolini R, Perello R, Snijders D, Plecko V, Sorde R, Strålin K, van der Eerden MM, Vila-Corcoles A, Watt JP. 2013. Estimating the burden of pneumococcal pneumonia among adults: a systematic review and meta-analysis of diagnostic techniques. PLoS One 8:e60273. doi: 10.1371/journal.pone.0060273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Strålin K, Herrmann B, Abdeldaim G, Olcen P, Holmberg H, Molling P. 2014. Comparison of sputum and nasopharyngeal aspirate samples and of the PCR gene targets lytA and Spn9802 for quantitative PCR for rapid detection of pneumococcal pneumonia. J Clin Microbiol 52:83–89. doi: 10.1128/JCM.01742-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Albrich WC, Madhi SA, Adrian PV, Telles JN, Paranhos-Baccala G, Klugman KP. 2014. Genomic load from sputum samples and nasopharyngeal swabs for diagnosis of pneumococcal pneumonia in HIV-infected adults. J Clin Microbiol 52:4224–4229. doi: 10.1128/JCM.01553-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Albrich WC, Madhi SA, Adrian PV, van Niekerk N, Mareletsi T, Cutland C, Wong M, Khoosal M, Karstaedt A, Zhao P, Deatly A, Sidhu M, Jansen KU, Klugman KP. 2012. Use of a rapid test of pneumococcal colonization density to diagnose pneumococcal pneumonia. Clin Infect Dis 54:601–609. doi: 10.1093/cid/cir859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Palmu AA, Saukkoriipi A, Snellman M, Jokinen J, Torkko P, Ziegler T, Kaijalainen T, Hausdorff WP, Verlant V, Kilpi TM. 2014. Incidence and etiology of community-acquired pneumonia in the elderly in a prospective population-based study. Scand J Infect Dis 46:250–259. doi: 10.3109/00365548.2013.876509. [DOI] [PubMed] [Google Scholar]
- 12.Murray PR, Washington JA. 1975. Microscopic and bacteriologic analysis of expectorated sputum. Mayo Clin Proc 50:339–344. [PubMed] [Google Scholar]
- 13.Yang S, Lin S, Khalil A, Gaydos C, Nuemberger E, Juan G, Hardick J, Bartlett JG, Auwaerter PG, Rothman RE. 2005. Quantitative PCR assay using sputum samples for rapid diagnosis of pneumococcal pneumonia in adult emergency department patients. J Clin Microbiol 43:3221–3226. doi: 10.1128/JCM.43.7.3221-3226.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sheppard CL, Harrison TG, Morris R, Hogan A, George RC. 2004. Autolysin-targeted LightCycler assay including internal process control for detection of Streptococcus pneumoniae DNA in clinical samples. J Med Microbiol 53:189–195. doi: 10.1099/jmm.0.05460-0. [DOI] [PubMed] [Google Scholar]
- 15.Lim WS, van der Eerden MM, Laing R, Boersma WG, Karalus N, Town GI, Lewis SA, Macfarlane JT. 2003. Defining community acquired pneumonia severity on presentation to hospital: an international derivation and validation study. Thorax 58:377–382. doi: 10.1136/thorax.58.5.377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kilpi TMK, Erkkilä L, Palmu AA, Snellman M, Jokinen J, Leinonen M. 2007. Diagnostic value of culture of high-quality (HQ) versus low-quality (LQ) sputum for diagnosis of pneumococcal (Pnc) community acquired pneumonia (CAP) in elderly, abstr D-894. Abstr 47th Intersci Conf Antimicrob Agents Chemother. [Google Scholar]
- 17.Rapola S, Salo E, Kiiski P, Leinonen M, Takala AK. 1997. Comparison of four different sampling methods for detecting pharyngeal carriage of Streptococcus pneumoniae and Haemophilus influenzae in children. J Clin Microbiol 35:1077–1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Werno AM, Anderson TP, Murdoch DR. 2012. Association between pneumococcal load and disease severity in adults with pneumonia. J Med Microbiol 61:1129–1135. doi: 10.1099/jmm.0.044107-0. [DOI] [PubMed] [Google Scholar]
- 19.Saukkoriipi A, Palmu AA, Jokinen J, Verlant V, Hausdorff WP, Kilpi TM. 2015. Effect of antimicrobial use on pneumococcal diagnostic tests in elderly patients with community-acquired pneumonia. Eur J Clin Microbiol Infect Dis 34:697–704. doi: 10.1007/s10096-014-2278-5. [DOI] [PubMed] [Google Scholar]
- 20.Hament JM, Kimpen JL, Fleer A, Wolfs TF. 1999. Respiratory viral infection predisposing for bacterial disease: a concise review. FEMS Immunol Med Microbiol 26:189–195. doi: 10.1111/j.1574-695X.1999.tb01389.x. [DOI] [PubMed] [Google Scholar]
- 21.Vu HT, Yoshida LM, Suzuki M, Nguyen HA, Nguyen CD, Nguyen AT, Oishi K, Yamamoto T, Watanabe K, Vu TD. 2011. Association between nasopharyngeal load of Streptococcus pneumoniae, viral coinfection, and radiologically confirmed pneumonia in Vietnamese children. Pediatr Infect Dis J 30:11–18. doi: 10.1097/INF.0b013e3181f111a2. [DOI] [PubMed] [Google Scholar]
- 22.Wolter N, Tempia S, Cohen C, Madhi SA, Venter M, Moyes J, Walaza S, Malope-Kgokong B, Groome M, du Plessis M, Magomani V, Pretorius M, Hellferscee O, Dawood H, Kahn K, Variava E, Klugman KP, von Gottberg A. 2014. High nasopharyngeal pneumococcal density, increased by viral coinfection, is associated with invasive pneumococcal pneumonia. J Infect Dis 210:1649–1657. doi: 10.1093/infdis/jiu326. [DOI] [PubMed] [Google Scholar]
- 23.Alpkvist H, Athlin S, Naucler P, Herrmann B, Abdeldaim G, Slotved HC, Hedlund J, Strålin K. 2015. Clinical and microbiological factors associated with high nasopharyngeal pneumococcal density in patients with pneumococcal pneumonia. PLoS One 10:e0140112. doi: 10.1371/journal.pone.0140112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stark JM, Stark MA, Colasurdo GN, LeVine AM. 2006. Decreased bacterial clearance from the lungs of mice following primary respiratory syncytial virus infection. J Med Virol 78:829–838. doi: 10.1002/jmv.20631. [DOI] [PubMed] [Google Scholar]
- 25.Albrich WC, Madhi SA, Adrian PV, van Niekerk N, Telles JN, Ebrahim N, Messaoudi M, Paranhos-Baccala G, Giersdorf S, Vernet G, Mueller B, Klugman KP. 2014. Pneumococcal colonisation density: a new marker for disease severity in HIV-infected adults with pneumonia. BMJ Open 4:e005953. doi: 10.1136/bmjopen-2014-005953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Simões AS, Tavares DA, Rolo D, Ardanuy C, Goossens H, Henriques-Normark B, Linares J, de Lencastre H, Sá-Leão R. 2016. lytA-based identification methods can misidentify Streptococcus pneumoniae. Diagn Microbiol Infect Dis 85:141–148. doi: 10.1016/j.diagmicrobio.2016.03.018. [DOI] [PubMed] [Google Scholar]
- 27.Whatmore AM, Efstratiou A, Pickerill AP, Broughton K, Woodard G, Sturgeon D, George R, Dowson CG. 2000. Genetic relationships between clinical isolates of Streptococcus pneumoniae, Streptococcus oralis, and Streptococcus mitis: characterization of “atypical” pneumococci and organisms allied to S. mitis harboring S pneumoniae virulence factor-encoding genes. Infect Immun 68:1374–1382. [DOI] [PMC free article] [PubMed] [Google Scholar]