ABSTRACT
Microscopic diagnosis of malaria using Giemsa-stained blood smears is the standard of care in resource-limited settings. These smears represent a potential source of DNA for PCR testing to confirm Plasmodium infections or for epidemiological studies of archived samples. Therefore, we assessed the use of DNA extracts from stained blood smears for the detection of Plasmodium species using real-time PCR. We extracted DNA from archived blood smears and corresponding red blood cell pellets collected from asymptomatic children in southwestern Uganda in 2010. We then performed real-time PCR followed by high-resolution melting (HRM) to identify Plasmodium species, and we compared our results to those of microscopy. We analyzed a total of 367 blood smears and corresponding red blood cell pellets, including 185 smears (50.4%) that were positive by microscopy. Compared to microscopy, PCR-HRM analysis of smear DNA had a sensitivity of 93.0% (95% confidence interval [CI], 88.2 to 96.2%) and a specificity of 96.7% (95% CI, 93.0 to 98.8%), and PCR-HRM analysis of pellet DNA had a sensitivity of 100.0% (95% CI, 98.0 to 100.0%) and a specificity of 94.0% (95% CI, 89.4 to 96.9%). Identification of positive PCR-HRM results to the species level revealed Plasmodium falciparum (92.0%), Plasmodium ovale (5.6%), and Plasmodium malariae (2.4%). PCR-HRM analysis of DNA extracts from Giemsa-stained thick blood smears or corresponding blood pellets had high sensitivity and specificity for malaria diagnosis, compared to microscopy. Therefore, blood smears can provide an adequate source of DNA for confirmation of Plasmodium species infections and can be used for retrospective genetic studies.
KEYWORDS: diagnostics, high-resolution melting, malaria, PCR
INTRODUCTION
Malaria remains a major global cause of childhood deaths, particularly in sub-Saharan Africa (1). In Uganda, malaria is a leading cause of morbidity and deaths, accounting for 25 to 40% of all outpatient visits to health facilities in all age groups, and is responsible for nearly one-half of inpatient pediatric deaths (2). The standard laboratory method for the diagnosis of malaria is microscopic examination of blood smears; however, microscopic examination is insensitive and nonspecific, especially when levels of parasitemia are low or mixed infections are present (3, 4). PCR is an alternative method to diagnose malaria and is increasingly being used for species confirmation and epidemiological studies in reference laboratory centers (5). Compared to microscopy, PCR is more sensitive, particularly in cases with low levels of parasitemia, and is becoming increasingly available (6). PCR followed by high-resolution melting (HRM) has been employed previously for malaria species identification; each species produces a diagnostic amplicon-specific melting profile (7, 8). HRM is useful in resource-limited settings due to its simple and fast workflow and high accuracy.
Effective PCR testing for Plasmodium species identification depends in part on the quality of the DNA obtained from blood samples. Whole blood and red blood cell pellets are reliable sources of high-quality DNA, but such samples require careful transportation, handling, and storage (9, 10). Alternatively, blood spots on filter paper and Giemsa-stained blood smears have been used as sources of DNA in several molecular and epidemiological studies (11–13). An advantage of blood smears is that they can provide both microscopic and molecular diagnoses. Additionally, long-term storage of these smears allows for retrospective studies (14). For example, archived blood smears could be used to determine changes in infecting malaria species and the progression of drug resistance over time. Therefore, our objective was to determine whether field-based PCR testing in a setting in which the disease is endemic can be accurately performed with a variety of sample sources. To do so, we used real-time PCR testing combined with HRM analysis to compare Plasmodium species detection from blood smears and corresponding red blood cell pellets collected in southwestern Uganda.
MATERIALS AND METHODS
Study area.
The study was conducted at the Epicentre Mbarara Research Centre, a research arm of Médecins sans Frontières, using Giemsa-stained thick blood smears from a previous cross-sectional malaria prevalence survey carried out between January and March 2010 (15). Samples were obtained from asymptomatic children less than 5 years of age, with no history of fever in the previous 72 h, from the districts of Ibanda, Isingiro, Kiruhura, and Mbarara, all of which are located in southwestern Uganda. Blood smears were stored at room temperature (25°C to 28°C) in plastic or wooden slide boxes, with two sachets of desiccant in each slide box. Ethical approval to conduct this study was obtained from the institutional review committees of the Mbarara University of Science and Technology and the University of Virginia.
Tested samples.
We selected a convenience sample of Giemsa-stained thick blood smears and corresponding red blood cell pellets from a total of over 2,000 participant samples collected in the original study. We first selected all positive smears that had an available corresponding red blood cell pellet (n = 185), and we then selected a similar number of pairs of available negative smears and red blood cell pellets (n = 182). The smears had been read previously by two independent experienced blood smear microscopists, using a final magnification of ×1,000. A third, more experienced microscopist served as the tie-breaker in cases with discrepancies in interpretation between the first two microscopists.
DNA extractions.
We used sterile surgical blades to scrape material from the thick smears, which contained approximately 10 to 20 μl of whole blood, onto aluminum foil sheets (6 by 6 cm) before transfer of the contents into 1.5-ml Eppendorf tubes (5). We then followed the Qiagen protocol for DNA extraction from dry blood spots on filter paper, according to the manufacturer's instructions (Qiagen, Hilden, Germany). We thawed the corresponding red blood cell pellets, which had been stored at −80°C, at room temperature for 30 min, after which 100 μl of the pellet material was transferred into clean 1.5-ml Eppendorf tubes. We extracted and purified DNA by using the QIAamp DNA minikit for DNA preparations from whole blood, according to the manufacturer's instructions (Qiagen). We eluted DNA in 100 μl and stored it at −20oC until use.
HRM plasmid and parasite DNA controls.
We obtained the Plasmodium species plasmid controls from the American Type Culture Collection (ATCC) (Manassas, VA, USA). The plasmids included Plasmodium falciparum (MRA-177-Pf small subunit [SSU] rRNA nest 1 PCR plasmid clone 8; lot 5946054), Plasmodium malariae (MRA-179-Pm SSU rRNA nest 1 PCR plasmid clone 34; lot 61909614), Plasmodium ovale (MRA-180-Po SSU rRNA nest 1 PCR plasmid clone 54; lot 59467055), and Plasmodium vivax (MRA-178-Pv SSU rRNA nest 1 PCR plasmid clone 16; lot 58067149). We obtained the P. falciparum laboratory clone (strain Dd2, MRA-331) from BEI Resources (Manassas, VA, USA).
PCR cycling and HRM.
We used a 2× HRM master mix (Rotor-Gene Probe PCR kit; Qiagen Benelux) and primers targeting the Plasmodium DNA, i.e., PL1473 F18 (5′-TAA CGA ACG AGA TCT TAA-3′) and PL1679 R18 (5′-GTT CCT CTA AGA AGC TTT-3′) (8). These primers hybridize to regions of the 18S rRNA genes that are conserved across human Plasmodium species and surround variable regions, which allows the differentiation of species during HRM analysis. We prepared reagents on a cooled sample rack, and each 25-μl PCR mixture included 12.5 μl of Rotor Gene Probe PCR master mix, 0.7 μM (final concentrations) of the forward and reverse primers (PL1473 F18 and PL1679 R18, respectively), 3 μl of template DNA, and 6 μl of RNase-free water in the final reaction mixture. We performed PCR cycling using the following conditions: 95°C for 5 min, followed by 40 cycles of 95°C for 10 s, 57°C for 30 s, and 72°C for 10 s. We performed HRM analysis of the resulting PCR product using a ramp from 65°C to 95°C, increasing 0.1°C in each step. Thermocycling, fluorescent detection, and HRM steps were performed in a Rotor-Gene Q real-time PCR instrument, using a 72-well rotor (Qiagen).
Plasmodium species determination.
We used Rotor-Gene Q software (version 2.3.1, build 49; Qiagen) for primary analysis. The software was used first to normalize the raw fluorescence values relative to signals resulting from positive and negative controls, which provided signal levels between 0% and 100%. For HRM analysis, the software plotted the negative of the change in fluorescence versus temperature (−dF/T). Within this −dF/T plot, each Plasmodium species produced a characteristic thermal profile and a peak allowing species differentiation. We set a manual fluorescence threshold based on control samples; only fluorescence data above this threshold were considered. The species-specific peaks were grouped into bins, which were used to assign genotypes for automatic assignment of tested samples.
Sequence alignment.
The 18S rRNA genes (chromosomes 1 [PF3D7_0112300], 5 [PF3D7_0531600], 7 [PF3D7_0725600], and 13 [PF3D7_1371000]) were obtained from the P. falciparum 3D7 genome in PlasmoDB (16). The P. malariae PCR amplicon sequence was obtained by Sanger sequencing of the plasmid. Sequences were compared using global alignment with free end gaps, with Geneious software (version 9.1.5; Biomatters Ltd.). Approximate melting temperatures were determined with Geneious 9.1.5. The P. malariae UG01 plasmid sequence and 18S rRNA gene sequences from PlasmoDB (chromosomes 3 [PmUG01_03031900] and 10 [PmUG01_03031900]) were 99% identical, with no change in melting temperature. Therefore, only the plasmid sequence was used for alignment with P. falciparum genes.
Statistical analysis.
We used Stata software (version 13.0; Stata, College Station, TX, USA) to construct two-by-two tables to determine the sensitivity and specificity of PCR results from blood smears and cell pellets. Microscopy was used as the gold standard. We used the Mann-Whitney U test for comparison of parasite concentrations between positive and negative PCR-HRM results. We also evaluated the ability of parasite concentrations to discriminate positive PCR-HRM results using the area under the receiver operating characteristic curve. We considered two-sided P values of <0.05 to be significant.
RESULTS
Validation of control results.
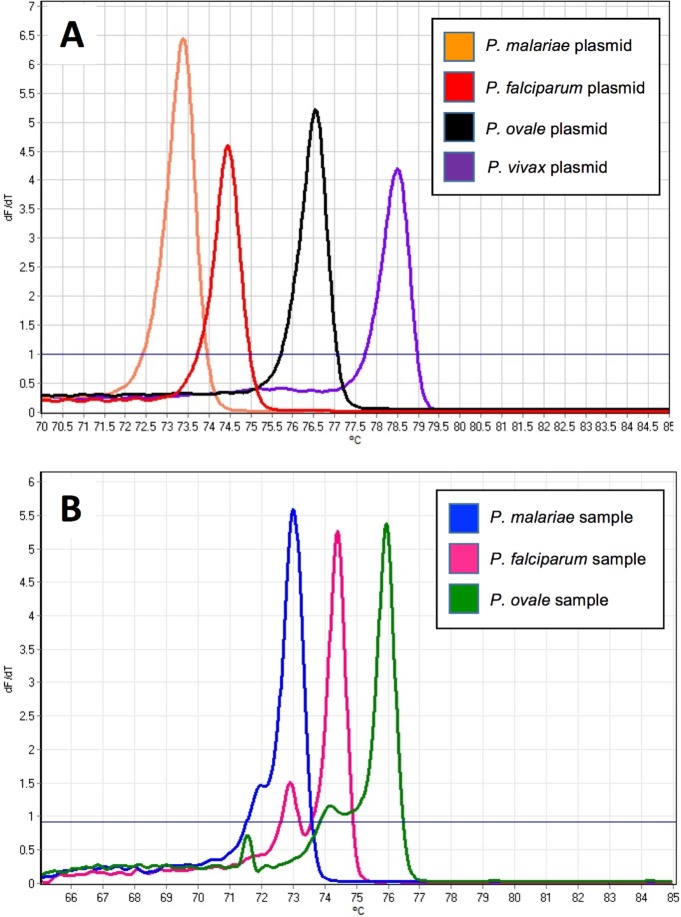
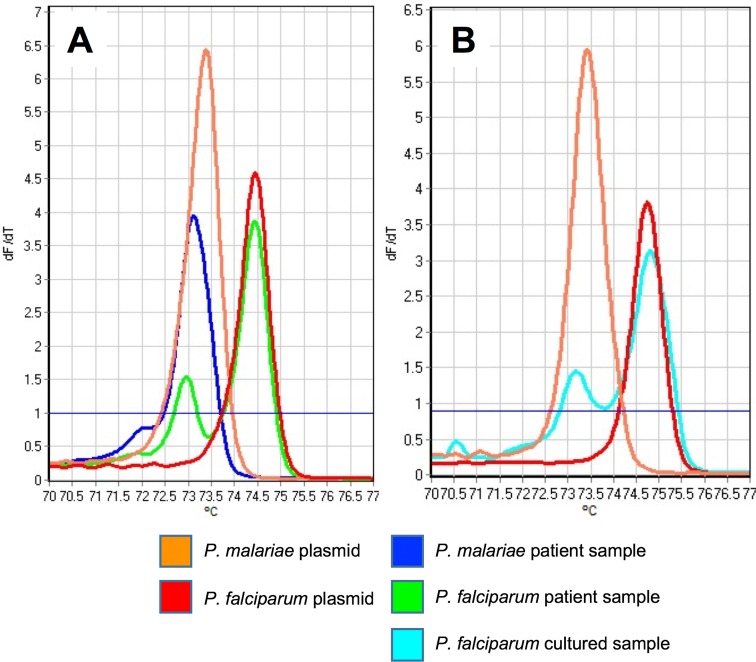
HRM curves for the positive-control plasmids from the four Plasmodium species provided the basis for initial interpretations of the patient sample results (Fig. 1A and B). The melting temperature ranges for HRM using these standards were 74.1 to 74.9°C for P. falciparum, 73.1 to 73.9°C for P. malariae, 76.5 to 79.0°C for P. ovale, and 78.6 to 81.0°C for P. vivax. P. falciparum melting profiles from patient samples did not match the control curves generated from the plasmids; rather, they had two peaks instead of one (Fig. 1B and 2A). Initially, we characterized this result as indicating coinfection with P. malariae and P. falciparum, with the first, smaller peak corresponding to P. malariae and the second, larger peak corresponding to P. falciparum. However, further experiments performed under the same PCR conditions but using DNA purified from a pure culture of a P. falciparum laboratory clone demonstrated the two peaks we observed in patient samples (Fig. 2B). Accordingly, we required detection of both peaks for a diagnosis of P. falciparum infection in patient samples.
FIG 1.
Comparison of melting profiles for controls and patient samples. (A) Melt peaks of control plasmids used to determine malaria species. (B) Melting curves generated from nucleic acids derived from blood pellets. The data shown were generated by taking the negative of the first derivative of the raw melting data (y axis) derived from each control plasmid and plotting it against temperature (x axis). The fluorescence threshold (solid blue line) was used as a noise filter.
FIG 2.
Comparison of melting profiles for controls, patient samples, and cultured parasites. (A) Melting curves for P. malariae and P. falciparum control plasmids and patient samples. Note the double peak shown by the P. falciparum sample. (B) Melting curves for the same P. malariae and P. falciparum control plasmids, compared to the melting curve generated using a laboratory-cultured sample of P. falciparum. The double peak displayed by P. falciparum samples is therefore diagnostic of the species, rather than indicating double infections in patient samples.
To determine whether multiple alleles with different melting temperatures could account for this finding, we aligned sequences from the four 18S rRNA alleles present in the P. falciparum genome. First, this comparison revealed two closely related gene copies on P. falciparum chromosomes 5 and 7 and another two related copies on chromosomes 1 and 13 (Fig. 3). Second, the predicted melting temperatures of these alleles confirmed that only two melt peaks would be observed (∼2°C away from each other). Furthermore, alignment of the P. malariae 18S rRNA PCR product with the P. falciparum alleles (Fig. 3) confirmed that, although there were significant differences present in the sequence, the predicted melting temperature corresponded to that of the lower P. falciparum peak (Fig. 1B, 2, and 3).
FIG 3.
Alignment of 18S rRNA sequences for P. falciparum (Pf) and P. malariae (Pm). The PCR products from the four 18S RNA alleles predicted from the P. falciparum genome were aligned with the 18S RNA product from P. malariae obtained from control plasmid sequences, using Geneious 9.1.5 (24). The total product sizes are between 214 and 225 bp. The level of identity between P. falciparum chromosomes 1 and 13 is 97.8% (estimated melting temperature is 71.9°C), that between P. falciparum chromosomes 5 and 7 is 100% (estimated melting temperature is 73.4°C), that between P. falciparum chromosome 1 and 13 and P. malariae is 75.4%, and that between P. falciparum chromosomes 5 and 7 and P. malariae is 72.3%. The estimated P. malariae melting temperature is 71.8°C. Discordant bases are highlighted.
Species identification.
Using our PCR-HRM approach, we analyzed a total of 367 blood smears and corresponding red blood cell pellets. Among the 185 microscopy-positive samples, all were identified by PCR-HRM using DNA from the pellets but 13 (7.0%) of 185 samples were found to be negative by PCR-HRM using DNA from the smears (Table 1). Among the 182 microscopy-negative samples, 6 (3.3%) of 182 smears and 11 (6.0%) of 182 cell pellets were found to be positive by PCR-HRM. As expected, the 6 newly positive samples from smears were also detected as positive in cell pellets. PCR-HRM assessment of smear-derived DNA had a sensitivity of 93.0% (95% confidence interval [CI], 88.2 to 96.2%) and a specificity of 96.7% (95% CI, 93.0 to 98.8%). PCR-HRM assessment of pellet-derived DNA had a sensitivity of 100% (95% CI, 98.0 to 100%) and a specificity of 94.0% (95% CI, 89.4 to 96.9%). From the Plasmodium-positive PCR results, we identified P. falciparum (92.0%), P. ovale (5.6%), and P. malariae (2.4%) but not P. vivax.
TABLE 1.
Microscopy versus smear and pellet PCR-HRM results for the diagnosis of malaria species in 367 patient samples from southwestern Uganda
| Sample type and PCR result | No. with microscopy result of: |
Sensitivity (95% CI) (%) | Specificity (95% CI) (%) | |
|---|---|---|---|---|
| Positive | Negative | |||
| Any malaria species | ||||
| Smear | ||||
| Positive | 172 | 6 | 93.0 (88.2–96.2) | 96.7 (93.0–98.8) |
| Negative | 13 | 176 | ||
| Pellet | ||||
| Positive | 185 | 11 | 100.0 (98.0–100.0) | 94.0 (89.4–96.9) |
| Negative | 0 | 171 | ||
| Plasmodium falciparum | ||||
| Smear | ||||
| Positive | 161 | 4 | 88.5 (82.9–92.7) | 97.8 (94.6–99.4) |
| Negative | 21 | 181 | ||
| Pellet | ||||
| Positive | 165 | 0 | 91.71 (86.6–95.2) | 100.0 (98.0–100.0) |
| Negative | 15 | 187 | ||
| Plasmodium malariae | ||||
| Smear | ||||
| Positive | 1 | 4 | 50.0 (1.2–98.7) | 98.9 (97.2–99.7) |
| Negative | 1 | 361 | ||
| Pellet | ||||
| Positive | 1 | 4 | 50.0 (1.2–98.7) | 98.9 (97.2–99.7) |
| Negative | 1 | 361 | ||
| Plasmodium ovale | ||||
| Smear | ||||
| Positive | 4 | 4 | 100.0 (39.8–100.0) | 98.9 (97.2–99.7) |
| Negative | 0 | 359 | ||
| Pellet | ||||
| Positive | 4 | 7 | 100.0 (39.8–100.0) | 98.1 (96.1–99.2) |
| Negative | 0 | 356 | ||
Association between levels of parasitemia and PCR-HRM results.
The overall median parasite concentration for the cohort was 32 parasites/μl (interquartile range [IQR], 0 to 7,470 parasites/μl). Samples that were smear microscopy positive and smear PCR-HRM positive (n = 161) had a median parasite concentration of 9,142 parasites/μl (IQR, 2,205 to 27,377 parasites/μl), whereas samples that were smear microscopy positive but smear PCR-HRM negative (n = 13) had a median parasite concentration of 1,064 parasites/μl (IQR, 441 to 4,282 parasites/μl) (P < 0.001). Samples that were smear PCR-HRM negative but pellet PCR-HRM positive (n = 18) had a median parasite concentration of 679 parasites/μl (IQR, 0 to 2,632 parasites/μl), whereas samples that were smear PCR-HRM positive and pellet PCR-HRM positive (n = 178) had a median parasite concentration of 7,602 parasites/μl (IQR, 1,297 to 23,505 parasites/μl) (P < 0.001). The area under the receiver operating characteristic curve for parasite concentrations was 0.961 (95% CI, 0.939 to 0.983) to predict a positive PCR-HRM result from smears and 0.972 (95% CI, 0.953 to 0.991) to predict a positive PCR-HRM result from pellets.
DISCUSSION
To our knowledge, this is the first study to directly compare PCR results with DNA obtained from smears versus cell pellets for malaria diagnosis. Both DNA sources performed well, with sensitivity and specificity values of >90% in comparison with microscopy. The smears and pellets were initially collected under field conditions, without a plan for using them as a source for DNA extraction, and were stored at room temperature. Therefore, our findings suggest that blood smears are an acceptable source of DNA for large-scale retrospective studies, especially when highly sensitive methods such as PCR-HRM analysis are used.
Similar studies that used DNA from thick blood smears achieved sensitivities of approximately 65% using nested PCR (17, 18). In comparison, we detected a higher diagnostic sensitivity for smear DNA using PCR-HRM (93.0%), which was similar to the 100% sensitivity noted using PCR-HRM to detect malaria in clinical blood samples from Malaysia (7). Although this difference could be due to PCR methodologies, our results more closely resembled those achieved by studies that used the same protocol for DNA purification. We used a column-based method, which improved extraction and purification, compared with other extraction protocols, and yielded sensitivities ranging from 95% to 100% (5, 19). This difference highlights the importance of the DNA extraction method, which can significantly affect PCR performance.
There was only a small difference in diagnostic sensitivity using DNA from smears versus pellets (93% versus 100%). While microscopy is considered the gold standard for Plasmodium detection, our PCR methodology using DNA from either sample type identified additional Plasmodium infections even in a number of microscopy-negative samples (20). This demonstrates the greater diagnostic sensitivity of PCR. In our study, no amplification was observed for 7% of DNA extracts from positive smears, whereas amplification was observed for all DNA extracts from pellets. This may be due to a difference in the levels of parasitemia and a greater quantity of DNA obtained from the pellets, compared to blood smears. Some DNA might have been lost during smear preparation and processing, as well as during the 5 years of storage.
The samples utilized in this study came from a field survey of asymptomatic participants, and our results compare favorably to prior PCR studies using blood slides. Our detection rate was associated with parasite concentration and was slightly lower than the 99.1% sensitivity for real-time PCR analysis of DNA extracts from thick blood smears collected and stored in Europe (5). This difference could be due to the 5 years of sample storage prior to our analyses, which might have had negative effects on the quantity and quality of the resulting DNA. Variations in methods for fixation and storage of smears, including dehemoglobinization, also might have contributed to DNA degradation over time. For example, dust accumulation and high ambient temperatures might have negatively affected the quality of the DNA. Furthermore, thick smears are not normally fixed prior to staining, but this step might be critical for DNA preservation for future use. Such details could be more rigorously evaluated in future studies.
In our study, P. falciparum accounted for 92% of the infections identified by PCR, with the remainder including P. malariae and P. ovale but not P. vivax (Table 1). In a 2010 cohort study of symptomatic cases in Kampala, 94% of malaria cases were similarly caused by P. falciparum (including mixed infections), 4.6% by P. malariae, 0.8% by P. ovale, and 0.5% by P. vivax (21). In contrast, similar work from northern Uganda in 2011 showed that, in samples collected from asymptomatic participants, P. falciparum parasite prevalence was 55.2% by PCR, compared to only 37.5% by microscopy (22). In a more recent study of asymptomatic infections in children, conducted in 2014 in the same area in southwestern Uganda as our study, there were fewer P. falciparum cases than in our study of samples from 2010 (54% versus 92%) and an increased prevalence of non-P. falciparum cases (23). This change in malaria epidemiology over time may reflect climate fluctuations that influence species prevalence, changes in diagnostic methods that more readily reveal non-P. falciparum species, or a true shift in species prevalence (23).
While the P. falciparum plasmid control in our experiment showed a single peak, melting curves from patient samples revealed two peaks. This initially raised concerns regarding interpretation of the data, as it appeared that there were many coinfections with P. falciparum and P. malariae. However, further experiments using DNA from a P. falciparum laboratory clone cultured in vitro revealed the same two peaks. Our assay targeted the 18S rRNA gene and P. falciparum has four copies of that gene, including two related copies on chromosomes 5 and 7 and another two related copies on chromosomes 11 and 13 (Fig. 3). Close examination of the regions of these genes that were targeted by our PCR primers revealed that the two sets of similar genes had distinct melting temperatures (∼2°C difference), which corresponded to the melting temperature difference between our observed peaks (Fig. 2).
The level of identity between P. malariae and P. falciparum amplicons was ∼77%, which was higher than the identity levels for the other species (∼71% and ∼62% for P. ovale and P. vivax, respectively) (24). However, we do not expect that this would have been enough identity to cause a heteroduplex between P. malariae and P. falciparum PCR products to form. Therefore, we concluded that the two peaks were generated from multiple copies of the 18S rRNA gene present in the P. falciparum genome, rather than coinfection with P. malariae. Accordingly, both peaks were required to confirm P. falciparum in our study. Interestingly, this two-peak phenomenon was not noted in the PCR-HRM study of malaria in Malaysia (7); perhaps the Malaysian parasites did not contain the extra 18S rRNA copies or the parasite levels present in the Malaysian samples were too low to detect the smaller peak using these methods. Although we had no dual infections, we expect that dual infections with P. falciparum and P. malariae would have either two peaks (a large combined P. falciparum and P. malariae peak followed by a second P. falciparum peak) or three distinct peaks (one peak for P. malariae and two peaks for P. falciparum).
There were several limitations in our study, including the inability to quantify the loss of DNA from blood smears during scraping, different storage conditions, and the presence of possible inhibitors of DNA extraction and/or PCR due to the chemical properties of Giemsa staining. The lack of amplification or false-negative results seen for some of the blood smear samples might have occurred as a result of DNA loss during smear processing, from preparation through staining to microscopic examination. Furthermore, differences in parasite concentrations and sample volumes (approximately 10 to 20 μl of blood was used for blood smear preparation, compared to 100 μl for cell pellets) likely decreased DNA amounts in blood smear samples, compared to pellets.
Despite these limitations, the novelty and importance of this work are 4-fold. (i) PCR-HRM can serve as a tool to investigate archived samples collected in settings in which disease is endemic, with implications for public health. For example, archived samples could be used as a source of DNA for further epidemiological studies of species type and antimalarial drug resistance. (ii) As was the case here, the use of PCR-HRM to analyze DNA extracted from clinical blood smears can build research capacity for investigators in settings in which disease is endemic who have access to archived smears, which can serve as a reservoir of parasite DNA. (iii) The clinical use of molecular diagnostic techniques in most clinical settings in which disease is endemic is not yet routine but is increasingly available. Where resources are available, the use of PCR-HRM can reduce the subjectivity of microscopy and improve diagnostic sensitivity and accuracy, compared to microscopy. (iv) Using traditional PCR methods to detect species is more difficult, because the PCR product would need to be sequenced (which is less available in resource-limited settings) or size polymorphisms in the product would need to be detected using gel electrophoresis. The latter approach is more subjective, much less sensitive, and more resource and time intensive (each sample would need to be run on a single lane of a gel). Quantitative PCR using probes is more expensive than HRM and also requires specialized instrumentation, which often is more problematic than the Rotor-Gene instrument used for PCR-HRM analysis. Taken together, these advantages of PCR-HRM suggest that it is a viable option in many clinical and research laboratories in areas in which disease is endemic.
In conclusion, PCR-HRM using DNA extracts from blood smears had >90% sensitivity and specificity for malaria diagnoses, compared to microscopy. Therefore, Giemsa-stained thick blood smears provide an acceptable source of DNA for confirmation of Plasmodium species infections and can be used for antimalarial resistance screening of archived samples from prior studies. Potential future uses for this method include the use of archived smears for widespread screening of antimalarial resistance.
ACKNOWLEDGMENTS
We thank the Epicentre Mbarara Research Centre laboratory staff for support with DNA extraction. We also thank Eric Houpt from the Division of Infectious Diseases and International Health at the University of Virginia for access to laboratory equipment for PCR-HRM analysis.
C.C.M. received a University of Virginia Henry Rose Carter Foundation Award to support this work. K.C.C., M.N., and L.T. all received University of Virginia Center for Global Health scholarships to participate in this study. K.K. received support from the Uganda Research Student Support Fund at the Epicentre Mbarara Research Centre.
REFERENCES
- 1.White NJ, Pukrittayakamee S, Hien TT, Faiz MA, Mokuolu OA, Dondorp AM. 2014. Malaria. Lancet 383:723–735. doi: 10.1016/S0140-6736(13)60024-0. [DOI] [PubMed] [Google Scholar]
- 2.Kassam R, Collins JB, Liow E, Rasool N. 2015. Narrative review of current context of malaria and management strategies in Uganda (part I). Acta Trop 152:252–268. doi: 10.1016/j.actatropica.2015.07.028. [DOI] [PubMed] [Google Scholar]
- 3.Amexo M, Tolhurst R, Barnish G, Bates I. 2004. Malaria misdiagnosis: effects on the poor and vulnerable. Lancet 364:1896–1898. doi: 10.1016/S0140-6736(04)17446-1. [DOI] [PubMed] [Google Scholar]
- 4.Ohrt C, Purnomo, Sutamihardja MA, Tang D, Kain KC. 2002. Impact of microscopy error on estimates of protective efficacy in malaria-prevention trials. J Infect Dis 186:540–546. doi: 10.1086/341938. [DOI] [PubMed] [Google Scholar]
- 5.Cnops L, Van Esbroeck M, Bottieau E, Jacobs J. 2010. Giemsa-stained thick blood films as a source of DNA for Plasmodium species-specific real-time PCR. Malar J 9:370. doi: 10.1186/1475-2875-9-370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kamya MR, Gasasira AF, Yeka A, Bakyaita N, Nsobya SL, Francis D, Rosenthal PJ, Dorsey G, Havlir D. 2006. Effect of HIV-1 infection on antimalarial treatment outcomes in Uganda: a population-based study. J Infect Dis 193:9–15. doi: 10.1086/498577. [DOI] [PubMed] [Google Scholar]
- 7.Chua KH, Lim SC, Ng CC, Lee PC, Lim YA, Lau TP, Chai HC. 2015. Development of high resolution melting analysis for the diagnosis of human malaria. Sci Rep 5:15671. doi: 10.1038/srep15671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mangold KA, Manson RU, Koay ES, Stephens L, Regner M, Thomson RB Jr, Peterson LR, Kaul KL. 2005. Real-time PCR for detection and identification of Plasmodium spp. J Clin Microbiol 43:2435–2440. doi: 10.1128/JCM.43.5.2435-2440.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pieroni P, Mills CD, Ohrt C, Harrington MA, Kain KC. 1998. Comparison of the ParaSight-F test and the ICT Malaria Pf test with the polymerase chain reaction for the diagnosis of Plasmodium falciparum malaria in travellers. Trans R Soc Trop Med Hyg 92:166–169. doi: 10.1016/S0035-9203(98)90730-1. [DOI] [PubMed] [Google Scholar]
- 10.Zhong KJ, Kain KC. 1999. Evaluation of a colorimetric PCR-based assay to diagnose Plasmodium falciparum malaria in travelers. J Clin Microbiol 37:339–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Edoh D, Steiger S, Genton B, Beck HP. 1997. PCR amplification of DNA from malaria parasites on fixed and stained thick and thin blood films. Trans R Soc Trop Med Hyg 91:361–363. doi: 10.1016/S0035-9203(97)90109-7. [DOI] [PubMed] [Google Scholar]
- 12.Long GW, Fries L, Watt GH, Hoffman SL. 1995. Polymerase chain reaction amplification from Plasmodium falciparum on dried blood spots. Am J Trop Med Hyg 52:344–346. doi: 10.4269/ajtmh.1995.52.344. [DOI] [PubMed] [Google Scholar]
- 13.Singh B, Cox-Singh J, Miller AO, Abdullah MS, Snounou G, Rahman HA. 1996. Detection of malaria in Malaysia by nested polymerase chain reaction amplification of dried blood spots on filter papers. Trans R Soc Trop Med Hyg 90:519–521. doi: 10.1016/S0035-9203(96)90302-8. [DOI] [PubMed] [Google Scholar]
- 14.Alger J, Acosta MC, Lozano C, Velasquez C, Labrada LA. 1996. Stained smears as a source of DNA. Mem Inst Oswaldo Cruz 91:589–591. doi: 10.1590/S0074-02761996000500009. [DOI] [PubMed] [Google Scholar]
- 15.De Beaudrap P, Nabasumba C, Grandesso F, Turyakira E, Schramm B, Boum Y, Etard JF. 2011. Heterogeneous decrease in malaria prevalence in children over a six-year period in south-western Uganda. Malar J 10:132. doi: 10.1186/1475-2875-10-132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Plasmodium Genome Database Collaborative. 2001. PlasmoDB: an integrative database of the Plasmodium falciparum genome: tools for accessing and analyzing finished and unfinished sequence data. Nucleic Acids Res 29:66–69. doi: 10.1093/nar/29.1.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Di Santi SM, Kirchgatter K, Brunialti KC, Oliveira AM, Ferreira SR, Boulos M. 2004. PCR-based diagnosis to evaluate the performance of malaria reference centers. Rev Inst Med Trop Sao Paulo 46:183–187. doi: 10.1590/S0036-46652004000400002. [DOI] [PubMed] [Google Scholar]
- 18.Scopel KK, Fontes CJ, Nunes AC, Horta MF, Braga EM. 2004. Low sensitivity of nested PCR using Plasmodium DNA extracted from stained thick blood smears: an epidemiological retrospective study among subjects with low parasitaemia in an endemic area of the Brazilian Amazon region. Malar J 3:8. doi: 10.1186/1475-2875-3-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vince A, Poljak M, Seme K. 1998. DNA extraction from archival Giemsa-stained bone-marrow slides: comparison of six rapid methods. Br J Haematol 101:349–351. doi: 10.1046/j.1365-2141.1998.00702.x. [DOI] [PubMed] [Google Scholar]
- 20.Wang B, Han SS, Cho C, Han JH, Cheng Y, Lee SK, Galappaththy GN, Thimasarn K, Soe MT, Oo HW, Kyaw MP, Han ET. 2014. Comparison of microscopy, nested-PCR, and real-time-PCR assays using high-throughput screening of pooled samples for diagnosis of malaria in asymptomatic carriers from areas of endemicity in Myanmar. J Clin Microbiol 52:1838–1845. doi: 10.1128/JCM.03615-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Clark TD, Njama-Meya D, Nzarubara B, Maiteki-Sebuguzi C, Greenhouse B, Staedke SG, Kamya MR, Dorsey G, Rosenthal PJ. 2010. Incidence of malaria and efficacy of combination antimalarial therapies over 4 years in an urban cohort of Ugandan children. PLoS One 5:e11759. doi: 10.1371/journal.pone.0011759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Proietti C, Pettinato DD, Kanoi BN, Ntege E, Crisanti A, Riley EM, Egwang TG, Drakeley C, Bousema T. 2011. Continuing intense malaria transmission in northern Uganda. Am J Trop Med Hyg 84:830–837. doi: 10.4269/ajtmh.2011.10-0498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Roh ME, Oyet C, Orikiriza P, Wade M, Kiwanuka GN, Mwanga-Amumpaire J, Parikh S, Boum Y. 2016. Asymptomatic Plasmodium infections in children in low malaria transmission setting, southwestern Uganda. Emerg Infect Dis 22:1494–1498. doi: 10.3201/eid2208.160619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gardner MJ, Hall N, Fung E, White O, Berriman M, Hyman RW, Carlton JM, Pain A, Nelson KE, Bowman S, Paulsen IT, James K, Eisen JA, Rutherford K, Salzberg SL, Craig A, Kyes S, Chan MS, Nene V, Shallom SJ, Suh B, Peterson J, Angiuoli S, Pertea M, Allen J, Selengut J, Haft D, Mather MW, Vaidya AB, Martin DM, Fairlamb AH, Fraunholz MJ, Roos DS, Ralph SA, McFadden GI, Cummings LM, Subramanian GM, Mungall C, Venter JC, Carucci DJ, Hoffman SL, Newbold C, Davis RW, Fraser CM, Barrell B. 2002. Genome sequence of the human malaria parasite Plasmodium falciparum. Nature 419:498–511. doi: 10.1038/nature01097. [DOI] [PMC free article] [PubMed] [Google Scholar]