ABSTRACT
Mycoplasma gallisepticum is the most virulent and economically important Mycoplasma species for poultry worldwide. Currently, M. gallisepticum strain differentiation based on sequence analysis of 5 loci remains insufficient for accurate outbreak investigation. Recently, whole-genome sequences (WGS) of many human and animal pathogens have been successfully used for microbial outbreak investigations. However, the massive sequence data and the diverse properties of different genes within bacterial genomes results in a lack of standard reproducible methods for comparisons among M. gallisepticum whole genomes. Here, we proposed the development of a core genome multilocus sequence typing (cgMLST) scheme for M. gallisepticum strains and field isolates. For development of this scheme, a diverse collection of 37 M. gallisepticum genomes was used to identify cgMLST targets. A total of 425 M. gallisepticum conserved genes (49.85% of M. gallisepticum genome) were selected as core genome targets. A total of 81 M. gallisepticum genomes from 5 countries on 4 continents were typed using M. gallisepticum cgMLST. Analyses of phylogenetic trees generated by cgMLST displayed a high degree of agreement with geographical and temporal information. Moreover, the high discriminatory power of cgMLST allowed differentiation between M. gallisepticum strains of the same outbreak. M. gallisepticum cgMLST represents a standardized, accurate, highly discriminatory, and reproducible method for differentiation among M. gallisepticum isolates. cgMLST provides stable and expandable nomenclature, allowing for comparison and sharing of typing results among laboratories worldwide. cgMLST offers an opportunity to harness the tremendous power of next-generation sequencing technology in applied avian mycoplasma epidemiology at both local and global levels.
KEYWORDS: strain typing, cgMLST, genomes, Mycoplasma gallisepticum, whole-genome sequence
INTRODUCTION
Mycoplasma gallisepticum is the most virulent avian Mycoplasma species, affecting chickens and turkeys worldwide (1). Recently, M. gallisepticum has expanded its host range to include house finches (Haemorhous mexicanus), causing major population losses in North America at early years following host shift (2, 3). M. gallisepticum-affected poultry usually suffer from air sacculitis with or without complications. M. gallisepticum infections are among the costliest diseases in commercial poultry production due to carcass condemnation and downgrading in processing plants and reduced meat and egg production efficiency (1). As a result, maintenance of M. gallisepticum-free breeder flocks is the method of choice to control M. gallisepticum. This strategy requires regular surveillance and monitoring and effective outbreak investigation tools in order to quickly identify sources of infection and allow containment before further spread (1, 4, 5). In addition, the use of live commercial M. gallisepticum vaccines as an alternative strategy for M. gallisepticum control in areas with high density of poultry operations complicates early diagnosis and outbreak investigation efforts. Consequently, the need for more efficient epidemiological investigation tools has intensified, especially to differentiate among vaccine strains and field strains (6–9).
Several sequence-based methods were developed and widely used to replace the traditional DNA fingerprinting techniques and minimize the need for M. gallisepticum isolation, which is often a challenging task due to the fastidious nature of mycoplasmas (10, 11). The first gene-targeted sequence typing (GTS) scheme was developed in 2005. It used the sequence information of 3 characterized variable surface proteins (mgc2, pvpA, and gapA) and one predicted surface protein (MGA_0319) to differentiate between 67 different M. gallisepticum strains and isolates. The total number of nucleotides used for sequence analysis from these 4 targets was 1,886 nucleotides (0.9% of the whole M. gallisepticum genome) (12). In 2007, a single-locus typing scheme was developed based on the variable intergenic spacer region (IGSR) (660 nucleotides) between 23S rRNA and 16S rRNA (13). In addition to improved reproducibility, the most valuable advantage of these sequence typing techniques over DNA fingerprinting techniques is that M. gallisepticum isolation is not required (10). Later, IGSR was used in combination with GTS to improve the discriminatory power (unpublished observation). This combination allowed differentiation among strains from different outbreaks. However, their discriminatory power was insufficient for differentiation between related outbreak strains. This limitation was clearly evident in vaccine-related outbreaks and the M. gallisepticum outbreak in house finches between 1994 and 2011 (3, 14). Additionally, the accuracy and degree of long-term evolutionary relatedness is difficult to infer between different sequence types due to the variable nature of the GTS loci (15–17).
In contrast, whole-genome sequences (WGS) have a higher level of discriminatory power than conventional molecular typing methods, such as multilocus sequence typing (MLST), pulsed-field gel electrophoresis (PFGE), and random amplified polymorphic DNA (RAPD) (18). The emergence of next-generation sequencing and its continuously decreasing costs paved the way for implementing the use of WGS of many human and animal pathogens in routine microbial diagnosis and epidemiological investigation of clinical outbreaks (19). In addition, WGS proved to be useful in comparative and evolutionary genetic studies (20, 21).
Recently, a core genome multilocus sequence typing approach (cgMLST) was proposed as a standard reproducible method for WGS-based strain differentiation and epidemiological investigation. It has been used to study the epidemiology of several microbial pathogens, including Listeria monocytogenes (22, 23), Neisseria meningitides (24), Mycobacterium tuberculosis (25), methicillin-resistant Staphylococcus aureus (26), Francisella tularensis (27), Escherichia coli (28), and Enterococcus faecium (29). cgMLST provides an efficient, accurate, and reproducible method for differentiation among strains and field isolates of the same species with stable and expandable nomenclature. This allows for comparing isolates from different outbreaks and sharing the typing results between different laboratories worldwide through web-based databases (29). Moreover, it could provide sufficient discriminatory power for outbreak investigations and a more reliable evaluation for the degree of relatedness between isolates. Here, we are describing the development and evaluation of a cgMLST scheme for typing M. gallisepticum strains and field isolates. This is the first application of the cgMLST typing approach on an important poultry pathogen. This newly developed assay could improve applied avian mycoplasma epidemiology and serve as an example for other important animal pathogens.
MATERIALS AND METHODS
Study set.
A total of 81 M. gallisepticum WGS were used in this study. This collection is temporally diverse, spanning a range between 1950 and 2014. It is also geographically diverse, originating from 5 countries (United States, United Kingdom, Jordan, Australia, and Israel) on 4 continents (North America, Europe, Asia, and Australia). Among this collection, 18 WGS were house finch M. gallisepticum isolates, one was from American goldfinch (Spinus tristis), and 62 were from poultry (turkey and chicken).
This M. gallisepticum WGS collection included all WGS data of M. gallisepticum available at GenBank (21 genomes) as of July 2017 (https://www.ncbi.nlm.nih.gov/genome/?term=NC_017502). This GenBank collection included 7 draft genomes related to a TS-11 vaccine outbreak in Georgia during 2007 and 2008. It also included 8 complete genomes from house finches, encompassing the first isolate of M. gallisepticum from house finches in 1994 to 2008 and from eastern and western U.S. populations. Additionally, we used 10 more M. gallisepticum WGS from house finches and one American goldfinch originating from a relatively limited geographical area (Alabama) and a limited period of time during August of 2011, which could represent a natural outbreak in a wild bird population. A large collection of poultry M. gallisepticum isolates were sequenced during the current study, including several reference and vaccine isolates, six isolates from Israel collected between 1999 and 2001, eight isolates from the United Kingdom, 12 isolates from North Carolina, including 9 from the same outbreak between1999 and 2001 that were previously investigated using RAPD (30), and several isolates from 15 different U.S. states.
WGS and assembly.
A total of 60 M. gallisepticum isolates were sequenced and assembled. Forty-five M. gallisepticum isolates were sequenced at the molecular and cellular imaging center (MCIC) of the Ohio State University, Ohio Agricultural Research and Development Center (OARDC) in Wooster, Ohio. They were obtained from different sources as mentioned above and detailed in Data Set S1 in the supplemental material. Most of the strains and isolates were received frozen in modified Frey's broth medium (31). Frozen cultures were thawed at room temperature, and 200 μl was transferred to 3 to 5 ml of the same broth and incubated at 37°C for one to several days. A few of the received isolates were lyophilized. For lyophilized isolates, 1 ml of modified Frey's broth was used for reconstitution and then transferred to 3 to 5 ml of broth and incubated at 37°C for one to several days. Two vaccine strains, M. gallisepticum 6/85 (Mycovac-L, Intervet, Inc. Omaha, NE, USA) and M. gallisepticum TS-11 (Merial Select, Inc., Gainesville, GA, USA), were used in this study. Vaccine was thawed according to the manufacturer's recommendations, transferred to 3 to 5 ml of modified Frey's broth, and incubated until color change. A total volume of 3 to 5 ml of modified Frey's broth color-changed culture was centrifuged at 415 × g for 30 min. The pellets were suspended in 200 μl of phosphate-buffered saline (PBS) for genomic DNA extraction using the QIAamp DNA minikit (Qiagen, Valencia, CA) by following the manufacturer's instructions. Species identity of these isolates was confirmed using real-time PCR (32). DNA extracts from isolates were quantified using a Qubit fluorometric analysis double-stranded DNA HS (high sensitivity) scheme kit (Invitrogen). Paired-end libraries for next-generation sequencing were prepared using the Illumina Nextera XT DNA library preparation kit by following the manufacturer's protocols (Illumina, Inc., San Diego, CA). Sequencing was performed on an Illumina MISeq (Illumina, Inc., San Diego, CA). DNA extraction and sequencing methods for the remaining 15 M. gallisepticum isolates were described by Lu and Delaney et al. (3, 14). SPAdes Genome Assembler (33) implemented within PATRIC from the pathosystems resource integration center (34) was used for assembly of 56 WGS reads, and the Velvet de novo assembler was used for assembling only 4 WGS reads. The PATRIC-generated assemblies for all samples were evaluated using QUAST (http://quast.bioinf.spbau.ru/) (35).
cgMLST development using SeqSphere+.
The first step in cgMLST scheme development is to define the core genome that will serve as the typing target. We selected SeqSphere+ for our cgMLST scheme development, as it provides a robust, detailed, and highly customizable approach for core genome target definition that helps in development of stable cgMLST schemes, which can be used for investigation of multiple outbreaks and serve as a standard typing approach for M. gallisepticum. SeqSphere+ version 4 (36) is commercial software that adopted the concept of conventional MLST and developed a genome-wide allele-based gene-by-gene typing platform that can be used by laboratory personnel who are not bioinfomaticians. The detailed steps for cgMLST development using SeqSphere+ were described in several studies (24, 26, 28, 29, 35).
Briefly, the cgMLST scheme was developed using SeqSphere+ version 4 (Ridom GmbH, Münster, Germany; http://www.ridom.de/seqsphere/index.shtml). A total of 37 M. gallisepticum whole-genome sequences were used for cgMLST scheme development and listed as query and reference genomes in Data Set S1 under the genome use column. These 37 M. gallisepticum samples were selected to represent the entire diversity of the M. gallisepticum population based on ad hoc cgMLST analysis of 81 different M. gallisepticum WGSs used in this study. This ad hoc cgMLST uses 370 M. gallisepticum core genes identified after filtration of the reference genome and blasting the 81 genomes against the reference genome to select shared targets. The minimum spanning tree generated based on that ad hoc cgMLST was used for typing the 81 M. gallisepticum genomes and identifying the degree of relatedness among these genomes. One or more representative genomes from each cluster or individual genome with 100 or more different alleles from the closest neighbor were selected to be included as a query genome for stable cgMLST scheme development. This collection included 32 poultry M. gallisepticum genomes and five house finch M. gallisepticum genomes, nine complete genomes available at GenBank, and 28 draft genomes sequenced in this study. The well-characterized reference strain MG01/GA/R low (accession number GCF_000092585.1) was used as a reference for developing the cgMLST scheme. FASTA files of genome assemblies were loaded into SeqSphere+. Only contigs/scaffolds of the draft genomes of ≥200 bp were included in the analysis. cgMLST target gene set identification involves two main steps. In the first step, MG01/GA/R low reference genome filtration was used to exclude unfit gene targets for MLST typing. This step included the following filters: a homologous gene filter to exclude all genes with high DNA similarity within a genome (with >90% identity and >100-bp overlap); a start codon filter to exclude all genes that are devoid of the translation start codon at the beginning of the gene; a minimum-length filter to exclude all genes with length of <50 bp; a stop codon filter to exclude all genes that are devoid of stop codons, have multiple stop codons, or have a stop codon that is not located at the end of the gene; and a gene overlap filter that excludes the shorter of two genes overlapping by >4 (22, 26, 28). In the second step, we query genomes using pairwise comparison to select shared fit targets between the reference genome and 36 M. gallisepticum query genomes (core genome targets) using BLAST v2.2.12 (37). All of the reference genome-filtered genes that were found in all query genomes with a sequence identity of ≥90% and 100% overlap and passed the (default) SeqSphere+ parameter stop codon percentage filter (to exclude all genes with internal stop codons in >20% of the query genomes) formed the targets of the final cgMLST scheme. The MLST+ target definer (version 1.0) function of SeqSphere+, with default parameters, was used to perform all of these genome-wide gene-by-gene comparisons. The final selected genes were examined for the allele numbers and their nucleotide diversity percent.
cgMLST scheme evaluation.
Forty-four nonquery genomes were used for scheme validation and identification of the percentage of good cgMLST targets in a diverse set of samples. They are listed as evaluation genomes under the genome use column in Data Set S1.
Comparison between cgMLST and core genome-SNP typing method for M. gallisepticum WGS.
A total of 81 WGS were typed using the newly developed cgMLST, including 1 reference genome, 36 query genomes, and 44 nonquery genomes. To evaluate the resolution of the M. gallisepticum cgMLST, SNP analysis was performed by mapping the core genome genes in 81 M. gallisepticum isolates to identify single-nucleotide variants from reference strain MG01/GA/R low core genome genes and predict the phylogeny of these samples based on core genome SNPs. This analysis was performed using the tool find nucleotide variants in SeqSphere+ with default filters (filter out insertions/deletions [InDels] and filter out neighbor SNP window within 10 bp) to minimize possible effects of recombination-induced SNPs. For each sample, a concatenated FASTA sequence containing the 425 cgMLST target gene sequences that were conserved in all 81 samples, in the order and orientation of the cgMLST target gene sequences of the reference strain MG01/GA/R low, were mapped to the reference sequence, and a table of all variants, their positions, and the variant nucleotides in each sample was generated. A neighbor-joining (NJ) phylogenetic tree was generated based on the core genome SNP after exclusion of target genes that are not found in all 81 samples (101 genes).
Typing using the 4-GTS analysis.
To evaluate the currently used GTS scheme (mgc2, pvpA, gapA, and MGA_0319) compared to the newly developed cgMLST scheme, we extracted the 4 gene targets that are currently used for typing of M. gallisepticum clinical samples. In order to perform this analysis, the 4 genes had to be present in all 81 genomes, and SNP-based analysis was performed with recombination filters turned off due to high variability within these targets. A phylogenetic tree was generated using the neighbor-joining tree with the option missing values are own category. This analysis was performed using SeqSphere+.
Accession number(s).
The raw nucleotide sequence reads generated in this study were submitted to the Short Read Archive (SRA) database of National Center for Biotechnology Information (NCBI) under the BioProject accession number PRJNA401291.
RESULTS
Whole-genome sequencing and assembly.
A total of 45 M. gallisepticum isolates were successfully sequenced in this study using the Illumina MiSeq platform. An additional 15 M. gallisepticum isolates were sequenced in a previous study (14). Sixty WGS assemblies used in this study were evaluated using QUAST (35). Raw reads were quality filtered and assembled de novo, generating assemblies with a mean size of 0.954534 Mbp (including aligned and nonaligned bases), a mean number of 74.9 contigs, and average calculated coverage of 45.98× (minimum, 20.5×; maximum, 67.68×). On average, 88.9% of the genome assembly for all samples was aligned to the reference genome.
Development of M. gallisepticum cgMLST scheme.
Following the default setting of SeqSphere+ software for definition of core genome targets, the number of core genome targets was 425 cgMLST (475,581 bases), representing 49.85% of the genome sequence. The number of accessory targets was 242 (286,803 bases), representing 30.06%. One hundred four targets were discarded due to filters applied for the reference genome (start codon filter [3 targets], stop codon filter [21 targets], and homologous gene filter [80 targets]). A complete list of the core genome genes can be found in Data Set S2 in the supplemental material.
Evaluation of the M. gallisepticum cgMLST scheme.
To evaluate the selected cgMLST target genes, 44 samples were loaded into SeqSphere+ and typed according to the 425 cgMLST targets. For all samples, more than 95.3% (405 good targets) were found, with a mean of 97.67% ± 1.04% standard deviation (SD). In M. gallisepticum cgMLST, the frameshift quality filter was the main cause for target failure, resulting in exclusion of 85.6% of the total failed targets from the analysis. Most mycoplasmas are known for frequent mutations resulting in frameshift and phase variation for certain groups of proteins as an adaptive mechanism for host immune evasion (30, 38–43).
Twenty-nine targets seemed to have higher frequency of frameshift than other genes. The mean nucleotide variability for this group was 7% and the mean number of alleles for this group was 23, indicating that these frameshift mutations became fixed in the population. Therefore, we turned off the frameshift quality filter for the 29 targets with frequent failure. Following this modification, for all samples, more than 97.4% (414 genes) of the 425 cgMLST target genes were found in all samples (good targets) with a mean of 99.5% ± 0.58% SD. The average number of alleles for 425 targets was 20 alleles, and the average nucleotide variability within a representative subset of genes was 6.3%.
CT threshold identification for M. gallisepticum cgMLST scheme.
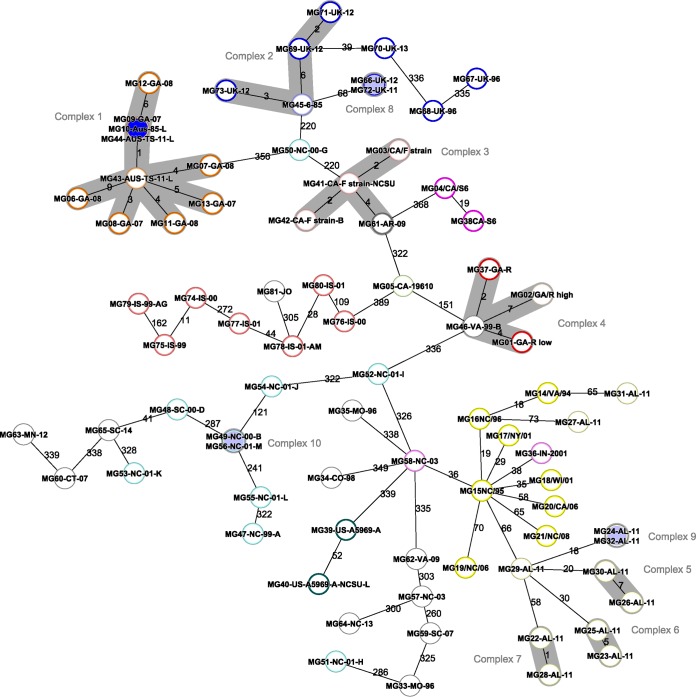
The cluster typing (CT) threshold is the maximum number of allele differences that can be found between highly clonal outbreak samples. It is used to improve the ability of the cgMLST scheme to differentiate between epidemiologically related and nonrelated samples. For M. gallisepticum, the maximum number of allele differences observed within the well-defined TS-11 vaccine-like outbreak samples was 10 alleles. Therefore, the CT threshold for the M. gallisepticum cgMLST scheme became 10 allele differences. Based on this threshold, 10 different clusters with more than one sample (Fig. 1) were identified in the 81 typed samples.
FIG 1.
Minimum spanning tree analysis based on the allelic profiles of 425 single-copy core genome targets from 81 clinical and reference M. gallisepticum samples using SeqSphere+ software. Ten different clonal clusters with fewer than 10 different alleles are marked with gray color. The core genome allelic profiles were determined by loading de novo assemblies into the M. gallisepticum cgMLST scheme developed in this study. The distance matrix underlying the network was built from all pairwise allelic profile comparisons of 425 cgMLST targets using the pairwise ignoring-missing-values option in SeqSphere+ software. Isolates that are discussed in the text are color coded according to the key shown in Fig. 2. The numbers on the connecting lines illustrate the numbers of target genes with differing alleles.
Comparison between cgMLST and SNP-based typing method.
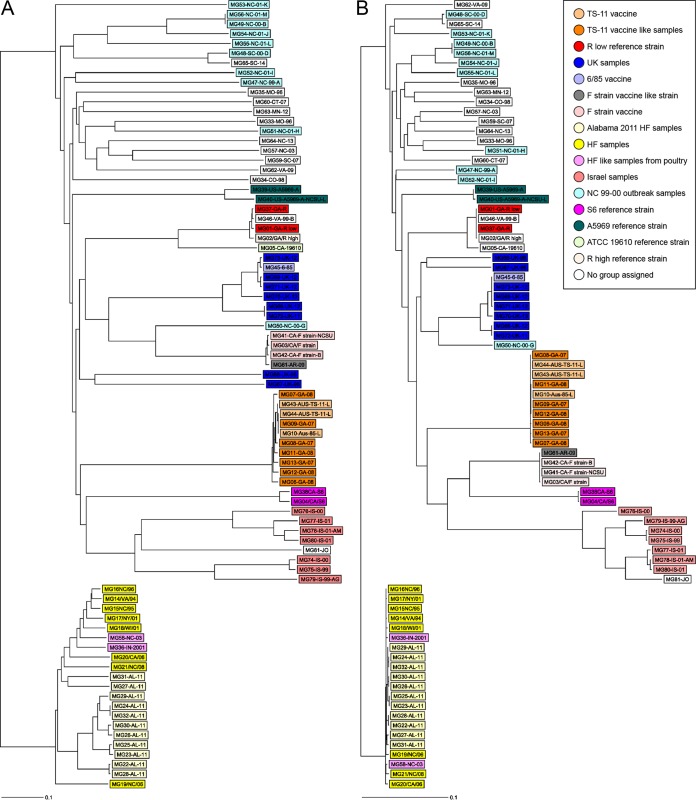
To evaluate the resolution of the M. gallisepticum cgMLST, we compared the degree of genetic relatedness of 81 samples based on allelic profiles using the newly developed cgMLST and based on the single-nucleotide polymorphism (SNP) level of the same core genome genes identified in this study and used for cgMLST. Only 324 cgMLST target genes that were present in all 81 samples were used. A total of 5,741 filtered SNPs out of 18,823 nonfiltered SNPs were identified from the alignment of these 324 genes. A neighbor-joining (NJ) tree based on 5,741 filtered SNP was generated and compared to the NJ tree built using the cgMLST profiles of the 81 samples (Fig. 2A and B). This revealed the high degree of congruence between the topologies of the two trees with relatively close resolution.
FIG 2.
Side-by-side core genome allele-based and core genome SNP-based rooted neighbor-joining tree for 81 clinical and reference M. gallisepticum samples using SeqSphere+ software. Note the high degree of congruence among the two trees. (A) The neighbor-joining (NJ) phylogenetic tree based on the allelic profiles of 425 single-copy core genome targets from 81 clinical and reference M. gallisepticum samples using SeqSphere+ software. Core genome allelic profiles were determined by loading de novo assemblies into the M. gallisepticum cgMLST scheme developed in this study. The distance matrix was built from all pairwise allelic profile comparisons of 425 cgMLST targets, using the pairwise ignoring-missing-values option in SeqSphere+ software. Using this option, genes with at least one missing value are not completely removed from the comparison but are ignored only during a pairwise comparison in case of a missing value. (B) Core genome SNP-based tree was built based on 5,741 filtered SNP out of 18,823 total SNP. SNP were identified after alignment of the concatenated 324 cgMLST target gene sequences after exclusion of targets with missing values present in all 81 samples, using the tool find single-nucleotide variants in Ridom SeqSphere+ software with the default setting for filtering out InDels and a neighboring SNP filtering window of 10 bases. Groups of samples are color coded according to the key.
Typing using the 4-GTS analysis.
Querying the 4 gene targets (mgc2, pvpA, gapA, and MGA_0319) in the 81 genomes resulted in retrieving gapA and MGA_0319 in 100% of the genomes, mgc2 in 75% of the genomes, and pvpA in only 24% of the genomes. This could be due to the high variability of the latter two genes and the presence of multiple nucleotide InDels. Moreover, pvpA may have a 37-nucleotide internal repeat, which results in size variation of that gene (12). Therefore, we decided to exclude mgc2 and pvpA from our analysis and rely on only 2 genes (gapA and MGA_0319). A total of 65 SNP where discovered within these two genes and were used for distance calculation and building a phylogenetic tree for the 81 samples (see Fig. S1). The 65-SNP tree has a degree of congruence with the cgMLST and core genome SNP tree, where it successfully grouped related samples together as in cgMLST. However, it lacked sufficient discriminatory power to differentiate between these related samples, in contrast to cgMLST and core genome SNP analysis.
DISCUSSION
We have developed the cgMLST scheme as a standardized typing approach for M. gallisepticum WGS. The core genome was identified to be 425 genes based on analysis of 37 different M. gallisepticum genomes, which is close to the previous estimation of an M. gallisepticum core genome of 409 genes and core proteome of 481 genes (41). We tested the applicability of the newly developed cgMLST for studying the epidemiology of M. gallisepticum using 81 M. gallisepticum samples. Figure 2A shows the closeness of house finch and American goldfinch samples (heavy and light yellow), representing a monophyletic clade as previously reported (3, 42). Furthermore, cgMLST allowed differentiation between the Alabama house finch outbreak samples, where two samples (MG 27/Al-11 and MG31/Al-11) appeared to have a greater number of different alleles than the rest of the samples within the group (Fig. 1), suggesting a different source of infection for these two samples than the prevailing samples within this region at that time of the year. Interestingly, two samples (MG36/IN/2001 and MG58/NC-03) isolated from commercial poultry operations (turkey farms) were closely related to the house finch group. This suggests that house finch M. gallisepticum could have infected poultry, which has been also reported previously (42). This also confirms that wild birds could act as a reservoir and a source of M. gallisepticum infection, which emphasizes the need for stricter biosecurity measures to prevent wild birds from being on farm premises.
Figure 1 shows that the TS-11-like samples (MG06/GA-08, MG07/GA-08, MG08/GA-07, MG09/GA-07, MG11/GA/08, MG12/GA/08, and MG13/GA/07) are closely related to the TS-11 vaccine samples (MG10/AUS/85, MG43/AUS/TS-11, and MG44/AUS/TS-11), forming one clonal complex that is different from all other samples by at least 358 alleles. This is consistent with the geographical origin of TS-11 vaccine, which is from Australia, a different origin than that of all other samples in this study except the U.S. TS-11-like samples (38). This also suggests that the vaccine was the shared origin of this outbreak in the United States. Most UK samples also are closely related, and some of these samples (MG69/UK/12, MG71/UK/12, and MG73/UK/12) appeared to be closely related to the 6/85 vaccine strain, indicating a possible common origin for these samples. All samples from Israel, despite being diverse, were close together, and the sample from Jordan was located within this group. All laboratory and reference strains and vaccine samples from different sources (MG37/GA/R, MG41/CA/F, and MG42/CA/F) used in this study were close to their corresponding reference sequences from GenBank (MG01/GA/R low and MG03/CA/F), with only a few allele differences, usually fewer than 10 alleles.
During an M. gallisepticum outbreak in North Carolina between 1999 and 2001, M. gallisepticum isolates were analyzed by random amplified polymorphic DNA (RAPD), and at least eight different RAPD types were identified. M. gallisepticum RAPD type B accounted for more than 90% of the samples and caused significant clinical disease in chickens and turkeys (30). Nine samples (MG47/NC/99-A, MG49/NC/00-B, MG50/NC/00-G, MG51/NC/01-H, MG52/NC/01-I, MG53/NC/01-K, MG54/NC/01-J, MG55/NC/01-L, and MG56/NC/01-M) of this North Carolina outbreak were different using RAPD and were different using cgMLST, except for 2 samples (MG49/NC/00-B andMG56/NC/01-M) that were identical on cgMLST, one from a chicken breeder flock and one from a backyard flock, indicating the inaccuracy of RAPD typing. Also, the difference of cgMLST type for these samples indicates that multiple and different scenarios and sources of infection were involved in the transmission of M. gallisepticum within that outbreak.
In order to maximize the portability of the M. gallisepticum cgMLST and allow national and international comparison of results with stable and expandable nomenclature, we have defined cluster type (CT) thresholds for M. gallisepticum cgMLST to differentiate between clonal and nonclonal isolates.
According to the M. gallisepticum cgMLST CT threshold (10 allele difference), 10 different clusters with more than one sample (Fig. 1) were identified in the 81 typed samples, indicating that members of these clusters that share the same CT are considered indistinguishable. Beyond the identified threshold, the degree of relatedness between isolates decreases with the increasing number of different alleles accordingly.
The cgMLST typing results were compared to core genome SNP typing results, and a high degree of agreement was detected between the two methods. Both of them had high reliability and discriminatory power and matched the related epidemiological information for all samples. The same closely related groups of samples (house finch samples, including Alabama samples, TS-11 vaccine samples, TS-11 vaccine-like samples, UK samples, Israel samples, 6/85 vaccine-like samples, F vaccine samples, and F vaccine-like samples) were observed in both trees with similar degrees of relatedness.
In addition, we used the newly developed cgMLST scheme to evaluate the currently used GTS scheme using the same set of samples. The lack of discriminatory power of the GTS scheme to differentiate between related samples compared to cgMLST and cg-SNP analysis was apparent. Moreover, some samples were clustered within different clusters than those of the cgMLST and core genome SNP analysis. For example, three poultry samples (MG54-NC-01-J, MG55-NC-01-L, and MG47-NC-99-A) were typed as house finch samples (see Fig. S1 in the supplemental material), while according to cgMLST and core genome SNP analysis they are related to poultry samples (Fig. 2A and B). It is important to acknowledge that the actual discriminatory power of GTS may be higher than that shown in our analysis, as our analysis was based on only two genes, because the other two were accidentally missed from the WGS of some samples and we could not include them in the analysis. However, the results of this GTS scheme should be carefully interpreted, and its use for typing of isolates should be replaced by cgMLST whenever possible. This also suggests that further research is needed to find a more reliable method for typing clinical samples directly.
Currently, the cgMLST scheme could be applied only for typing M. gallisepticum isolates, as whole-genome sequence generation is still dependent on the availability of isolates. Therefore, an alternative sequencing approach that can be applied directly to clinical samples without isolation warrants further investigation to overcome difficulties in M. gallisepticum isolation.
The known fast evolutionary rate of mycoplasma among prokaryotes (3) coupled with the genetic information from large numbers of core genes may be instrumental for studying the epidemiological as well as evolutionary changes within the mycoplasma genome.
In this study, core genome SNP typing and cgMLST performed equally in typing our samples and their results were similar, evidencing the validity of cgMLST to be used for M. gallisepticum epidemiology. However, cgMLST has several advantages over core genome SNP typing in addition to the general drawbacks of the SNP typing approach (17). cgMLST has a stable typing approach because it allows typing of samples even when a few genes are missing, as missing genes will result in missing only a few alleles with minimal effect on the results. On the other hand, in the SNP-based typing approach, missing a few genes will result in missing higher numbers of SNPs within those genes, which could drastically affect the result if included in the analysis. If excluded, we lose the ability to replicate the analysis. In addition, the allele typing approach provides more buffers for the effects of misleading horizontal signals through considering the recombination change as a single evolutionary event that will lead to only one allele difference, in contrast to the SNP-based typing, which counts each SNP as a single event that incorrectly leads to greater distance differences between typed strains (26, 29, 39). Another important buffer is the large number of defined core genome targets, which increases the reliability of the typing results. Yet another buffer is the cluster typing threshold definition that is equivalent to the sequence type of traditional MLST. This CT threshold allows buffering any misleading differences during typing of closely related strains that may result from either sequencing or assembly errors, recombination, or microevolutionary events. Therefore, this approach might be biologically more relevant than approaches that consider only point mutations.
Handling and analysis of next-generation data were always challenging for clinicians and laboratory personnel; however, these issues are not as challenging with cgMLST. The numeric allele typing data of cgMLST data are easier to handle and less computationally demanding than the SNP data. This fact increases the availability of public databases with a user-friendly interface and expandable nomenclature, like those of cgMLST.org and BIGSdb. These databases would allow studying M. gallisepticum epidemiology on local, global, and long-term scales following the leading example of conventional MLST at the PubMLST database (40). Moreover, it opens the door for sharing other typing and clinical metadata besides identification, for example, information about virulence and antimicrobial susceptibility of the typed strains in addition to field data. cgMLST may represent a standard scheme for identification of M. gallisepticum isolates and offers an opportunity to harness the tremendous power of next-generation sequencing in applied avian mycoplasma epidemiology at a global level.
Supplementary Material
ACKNOWLEDGMENTS
We thank all of the researchers listed here, who provided us with Mycoplasma gallisepticum strains and field isolates used in this study: Janet Bradbury of The University of Liverpool, UK, Inna lysnyansky of the Kimron Veterinary Institute, Israel, and Naola Ferguson-Noel of the Poultry Diagnostic and Research Center at the University of Georgia. We also thank Dag Harmsen, Department for Periodontology, University of Münster, Münster, Germany, for his critical review and comments on the manuscript. In addition, we thank Tea Meulia and Maria Elena Hernandez Gonzalez at the Molecular and Cellular Imaging Center of The Ohio State University for their technical support and help at the sequencing core facility.
This work was financially supported by The Ohio State University, OH, USA, Alexandria University, Alexandria, Egypt, and The Ministry of Higher Education, Egypt.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JCM.01145-17.
REFERENCES
- 1.Raviv Z, Ley DH. 2013. Mycoplasma gallisepticum. Infection diseases of poultry, 13th ed John Wiley & Sons, New York, NY. [Google Scholar]
- 2.Ley DH, Berkhoff JE, McLaren JM. 1996. Mycoplasma gallisepticum isolated from house finches (Carpodacus mexicanus) with conjunctivitis. Avian Dis 40:480–483. doi: 10.2307/1592250. [DOI] [PubMed] [Google Scholar]
- 3.Delaney NF, Balenger S, Bonneaud C, Marx CJ, Hill GE, Ferguson-Noel N, Tsai P, Rodrigo A, Edwards SV. 2012. Ultrafast evolution and loss of CRISPRs following a host shift in a novel wildlife pathogen, Mycoplasma gallisepticum. PLoS Genet 8:e1002511. doi: 10.1371/journal.pgen.1002511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kleven S. 1997. Changing expectations in the control of Mycoplasma gallisepticum. Acta Vet Hung 45:299–305. [PubMed] [Google Scholar]
- 5.Kleven SH. 2008. Control of avian Mycoplasma infections in commercial poultry. Avian Dis 52:367–374. doi: 10.1637/8323-041808-Review.1. [DOI] [PubMed] [Google Scholar]
- 6.Abdelwhab EM, Abdelmagid MA, El-Sheibeny LM, El-Nagar HA, Arafa A, Selim A, Nasef SA, Aly MM, Hafez HM. 2011. Detection and molecular characterization of Mycoplasma gallisepticum field infection in TS-11-vaccinated broiler breeders. J Appl Poult Res 20:390–396. doi: 10.3382/japr.2009-00130. [DOI] [Google Scholar]
- 7.El Gazzar M, Laibinis VA, Ferguson-Noel N. 2011. Characterization of a ts-11–like Mycoplasma gallisepticum isolate from commercial broiler chickens. Avian Dis 55:569–574. doi: 10.1637/9689-021711-Reg.1. [DOI] [PubMed] [Google Scholar]
- 8.Kempf I. 1997. DNA amplification methods for diagnosis and epidemiological investigations of avian mycoplasmosis. Acta Vet Hung 45:373–386. [PubMed] [Google Scholar]
- 9.Whithear KG. 1996. Control of avian mycoplasmoses by vaccination. Rev Sci Tech Int Off Epizoot 15:1527–1553. doi: 10.20506/rst.15.4.985. [DOI] [PubMed] [Google Scholar]
- 10.Ghanem M, El-Gazzar M. 2016. Development of multilocus sequence typing (MLST) assay for Mycoplasma iowae. Vet Microbiol 195:2–8. doi: 10.1016/j.vetmic.2016.08.013. [DOI] [PubMed] [Google Scholar]
- 11.Power EGM. 1996. RAPD typing in microbiology–a technical review. J Hosp Infect 34:247–265. doi: 10.1016/S0195-6701(96)90106-1. [DOI] [PubMed] [Google Scholar]
- 12.Ferguson NM, Hepp D, Sun S, Ikuta N, Levisohn S, Kleven SH, García M. 2005. Use of molecular diversity of Mycoplasma gallisepticum by gene-targeted sequencing (GTS) and random amplified polymorphic DNA (RAPD) analysis for epidemiological studies. Microbiol Read Engl 151:1883–1893. doi: 10.1099/mic.0.27642-0. [DOI] [PubMed] [Google Scholar]
- 13.Raviv Z, Callison S, Ferguson-Noel N, Laibinis V, Wooten R, Kleven SH. 2007. The Mycoplasma gallisepticum 16S-23S rRNA intergenic spacer region sequence as a novel tool for epizootiological studies. Avian Dis 51:555–560. doi: 10.1637/0005-2086(2007)51[555:TMGSRI]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- 14.Lu A. 2013. The recent evolution of Mycoplasma gallisepticum in house finches. BA thesis Harvard University, Cambridge, MA. [Google Scholar]
- 15.El-Gazzar MM, Wetzel AN, Raviv Z. 2012. The genotyping potential of the Mycoplasma synoviae vlhA gene. Avian Dis 56:711–719. doi: 10.1637/10200-041212-Reg.1. [DOI] [PubMed] [Google Scholar]
- 16.Maiden MC, Bygraves JA, Feil E, Morelli G, Russell JE, Urwin R, Zhang Q, Zhou J, Zurth K, Caugant DA, Feavers IM, Achtman M, Spratt BG. 1998. Multilocus sequence typing: a portable approach to the identification of clones within populations of pathogenic microorganisms. Proc Natl Acad Sci U S A 95:3140–3145. doi: 10.1073/pnas.95.6.3140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sheppard SK, Jolley KA, Maiden MCJ. 2012. A gene-by-gene approach to bacterial population genomics: whole genome MLST of Campylobacter. Genes 3:261–277. doi: 10.3390/genes3020261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kwong JC, Mercoulia K, Tomita T, Easton M, Li HY, Bulach DM, Stinear TP, Seemann T, Howden BP. 2016. Prospective whole-genome sequencing enhances national surveillance of Listeria monocytogenes. J Clin Microbiol 54:333–342. doi: 10.1128/JCM.02344-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pérez-Losada M, Cabezas P, Castro-Nallar E, Crandall KA. 2013. Pathogen typing in the genomics era: MLST and the future of molecular epidemiology. Infect Genet Evol 16:38–53. doi: 10.1016/j.meegid.2013.01.009. [DOI] [PubMed] [Google Scholar]
- 20.Medini D, Serruto D, Parkhill J, Relman DA, Donati C, Moxon R, Falkow S, Rappuoli R. 2008. Microbiology in the post-genomic era. Nat Rev Microbiol 6:419–430. [DOI] [PubMed] [Google Scholar]
- 21.Ruan Z, Feng Y. 2016. BacWGSTdb, a database for genotyping and source tracking bacterial pathogens. Nucleic Acids Res 44:D682–D687. doi: 10.1093/nar/gkv1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schmid D, Allerberger F, Huhulescu S, Pietzka A, Amar C, Kleta S, Prager R, Preußel K, Aichinger E, Mellmann A. 2014. Whole genome sequencing as a tool to investigate a cluster of seven cases of listeriosis in Austria and Germany, 2011–2013. Clin Microbiol Infect 20:431–436. doi: 10.1111/1469-0691.12638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen Y, Gonzalez-Escalona N, Hammack TS, Allard MW, Strain EA, Brown EW. 2016. Core genome multilocus sequence typing for identification of globally distributed clonal groups and differentiation of outbreak strains of Listeria monocytogenes. Appl Environ Microbiol 82:6258–6272. doi: 10.1128/AEM.01532-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bratcher HB, Corton C, Jolley KA, Parkhill J, Maiden MC. 2014. A gene-by-gene population genomics platform: de novo assembly, annotation and genealogical analysis of 108 representative Neisseria meningitidis genomes. BMC Genomics 15:1138. doi: 10.1186/1471-2164-15-1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kohl TA, Diel R, Harmsen D, Rothgänger J, Walter KM, Merker M, Weniger T, Niemann S. 2014. Whole-genome-based Mycobacterium tuberculosis surveillance: a standardized, portable, and expandable approach. J Clin Microbiol 52:2479–2486. doi: 10.1128/JCM.00567-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Leopold SR, Goering RV, Witten A, Harmsen D, Mellmann A. 2014. Bacterial whole-genome sequencing revisited: portable, scalable, and standardized analysis for typing and detection of virulence and antibiotic resistance genes. J Clin Microbiol 52:2365–2370. doi: 10.1128/JCM.00262-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Antwerpen MH, Prior K, Mellmann A, Höppner S, Splettstoesser WD, Harmsen D. 2015. Rapid high resolution genotyping of Francisella tularensis by whole genome sequence comparison of annotated genes (“MLST+”). PLoS One 10:e0123298. doi: 10.1371/journal.pone.0123298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mellmann A, Harmsen D, Cummings CA, Zentz EB, Leopold SR, Rico A, Prior K, Szczepanowski R, Ji Y, Zhang W, McLaughlin SF, Henkhaus JK, Leopold B, Bielaszewska M, Prager R, Brzoska PM, Moore RL, Guenther S, Rothberg JM, Karch H. 2011. Prospective genomic characterization of the German enterohemorrhagic Escherichia coli O104:H4 outbreak by rapid next generation sequencing technology. PLoS One 6:e22751. doi: 10.1371/journal.pone.0022751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Been M de, Pinholt M, Top J, Bletz S, Mellmann A, Schaik W van, Brouwer E, Rogers M, Kraat Y, Bonten M, Corander J, Westh H, Harmsen D, Willems RJL. 2015. Core genome multilocus sequence typing scheme for high-resolution typing of Enterococcus faecium. J Clin Microbiol 53:3788–3797. doi: 10.1128/JCM.01946-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sanei B, Barnes HJ, Vaillancourt JP, Ley DH. 2007. Experimental infection of chickens and turkeys with Mycoplasma gallisepticum reference strain S6 and North Carolina field isolate RAPD type B. Avian Dis 51:106–111. doi: 10.1637/0005-2086(2007)051[0106:EIOCAT]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- 31.Kleven SH. 2008. Mycoplasmosis, p 59–64. In A laboratory manual for the isolation, identification, and characterization of avian pathogens, 5th ed American Association of Avian Pathologists, Jacksonville, FL. [Google Scholar]
- 32.Raviv Z, Kleven SH. 2009. The development of diagnostic real-time TaqMan PCRs for the four pathogenic avian mycoplasmas. Avian Dis 53:103–107. doi: 10.1637/8469-091508-Reg.1. [DOI] [PubMed] [Google Scholar]
- 33.Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, Pyshkin AV, Sirotkin AV, Vyahhi N, Tesler G, Alekseyev MA, Pevzner PA. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wattam AR, Abraham D, Dalay O, Disz TL, Driscoll T, Gabbard JL, Gillespie JJ, Gough R, Hix D, Kenyon R, Machi D, Mao C, Nordberg EK, Olson R, Overbeek R, Pusch GD, Shukla M, Schulman J, Stevens RL, Sullivan DE, Vonstein V, Warren A, Will R, Wilson MJC, Yoo HS, Zhang C, Zhang Y, Sobral BW. 2013. PATRIC, the bacterial bioinformatics database and analysis resource. Nucleic Acids Res 42:D581–D591. doi: 10.1093/nar/gkt1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gurevich A, Saveliev V, Vyahhi N, Tesler G. 2013. QUAST: quality assessment tool for genome assemblies. Bioinformatics 29:1072–1075. doi: 10.1093/bioinformatics/btt086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jünemann S, Sedlazeck FJ, Prior K, Albersmeier A, John U, Kalinowski J, Mellmann A, Goesmann A, von Haeseler A, Stoye J, Harmsen D. 2013. Updating benchtop sequencing performance comparison. Nat Biotechnol 31:294–296. doi: 10.1038/nbt.2522. [DOI] [PubMed] [Google Scholar]
- 37.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J Mol Biol 215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 38.Ricketts C, Pickler L, Maurer J, Ayyampalayam S, García M, Ferguson-Noel NM. 2017. Identification of strain-specific sequences that distinguish a Mycoplasma gallisepticum vaccine strain from field isolates. J Clin Microbiol 55:244–252. doi: 10.1128/JCM.00833-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Maiden MCJ, van Rensburg MJJ, Bray JE, Earle SG, Ford SA, Jolley KA, McCarthy ND. 2013. MLST revisited: the gene-by-gene approach to bacterial genomics. Nat Rev Microbiol 11:728–736. doi: 10.1038/nrmicro3093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jolley KA, Maiden MC. 2010. BIGSdb: Scalable analysis of bacterial genome variation at the population level. BMC Bioinformatics 11:595. doi: 10.1186/1471-2105-11-595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fisunov GY, Alexeev DG, Bazaleev NA, Ladygina VG, Galyamina MA, Kondratov IG, Zhukova NA, Serebryakova MV, Demina IA, Govorun VM. 2011. Core proteome of the minimal cell: comparative proteomics of three mollicute species. PLoS One 6:e21964. doi: 10.1371/journal.pone.0021964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hochachka WM, Dhondt AA, Dobson A, Hawley DM, Ley DH, Lovette IJ. 2013. Multiple host transfers, but only one successful lineage in a continent-spanning emergent pathogen. Proc R Soc Lond B Biol Sci 280:20131068. doi: 10.1098/rspb.2013.1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Theiss P, Wise KS. 1997. Localized frameshift mutation generates selective, high-frequency phase variation of a surface lipoprotein encoded by a mycoplasma ABC transporter operon. J Bacteriol 179:4013–4022. doi: 10.1128/jb.179.12.4013-4022.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.