ABSTRACT
Fungal bloodstream infections are a significant problem in the United States, with an attributable mortality rate of up to 40%. An early diagnosis to direct appropriate therapy has been shown to be critical to reduce mortality rates. Conventional phenotypic methods for fungal detection take several days, which is often too late to impact outcomes. Herein, we describe a cost-effective multiplex assay platform for the rapid detection and differentiation of major clinically relevant Candida species directly from blood culture. This approach utilizes a novel biotin-labeled polymer-mediated signal amplification process combined with targeting rRNA to exploit phylogenetic differences for sensitive and unambiguous species identification; this assay detects seven pathogenic Candida species (C. albicans, C. glabrata, C. parapsilosis, C. tropicalis, C. krusei, C. lusitaniae, and C. guilliermondii) simultaneously with very high specificity to the species level in less than 80 min with the limits of detection at 1 × 103 to 10 × 103 CFU/ml or as few as 50 CFU per assay. The performance of the described assay was verified with 67 clinical samples (including mixed multiple-species infections as well), with an overall 100% agreement with matrix-assisted laser desorption ionization (MALDI) mass spectrometry-based reference results. By providing a species identity rapidly, the clinician is aided with information that may direct appropriate therapy sooner and more accurately than current approaches, including PCR-based tests.
KEYWORDS: AMPED, blood culture, Candida, Candida detection, molecular diagnostics, signal amplification
INTRODUCTION
Candidemia is one of the major nosocomial bloodstream infections in the United States and worldwide. The main causative agent for this infection are the fungal Candida species, and the top seven species (C. albicans, C. glabrata, C. parapsilosis, C. tropicalis, C. krusei, C. lusitaniae, and C. guilliermondii) account for more than 95% of the candidemia cases globally (1–4). This infection is opportunistic and may be life threatening for immunocompromised and critically ill patients, such as those with cancers, ongoing chemotherapy, and broad-spectrum antibiotic treatment, as well as those undergoing organ transplant or other major surgeries, particularly, those with the use of central venous and arterial catheters (5–8). In addition to the most common species C. albicans, it has also been reported that C. parapsilosis and C. tropicalis are more frequently associated with neonatal and pediatric patients with low birth weights, parental malnutrition, and hematological malignancies (9–11).
It is well established that the rapid initiation of appropriate therapy is critical for the treatment of fungal bloodstream infections (3, 4, 12). It has also been shown that individual Candida species exhibit unique antimicrobial susceptibility profiles, and so species information is an attractive approach to more rapidly determine the appropriate treatment for patients suspected of having a Candida bloodstream infection. Over the last decades, the prevalence of different species in specific regions and antifungal susceptibility have been closely studied and monitored by the ARTEMIS global antifungal surveillance program team to provide general guidance for local clinicians (3, 4). The focus has been on in vitro susceptibility testing for two major antifungal drugs, namely, fluconazole and voriconazole, as the differential cost is significant for these two drugs (13). More recently, the Infectious Disease Society of America (IDSA) has updated the practice guidelines for the treatment of Candida bloodstream infection (14). Although echinocandins have been recommended as the frontline therapy, the follow-up treatment is more dependent on the specific species and the associated antifungal susceptibility. Therefore, a low-cost and rapid detection of Candida at the species level is valuable for the efficacious treatment of candidemia.
The current gold standards for Candida species isolation and identification are still conventional culture-based methods, which take up to several days to get results. In addition, the conventional biochemical, immunological, and serological assays have often been shown to have poor sensitivity and specificity (15, 16, 17). Recently, mass spectrometry (such as matrix-assisted laser desorption ionization–time of flight mass spectrometry [MALDI-TOF MS]) testing has improved the accuracy on colonies isolated from positive blood cultures and has shown some promise, but there are some inconsistencies in pretreatment procedures and systems (18, 19, 20, 21). PCR and other nucleic acid amplification methods have been developed for detecting Candida species directly from a blood draw (22), and although they have significantly shortened the time to an answer, they are still controversial with some limitations. These methods still cannot provide accurate species-specific information (23, 24, 25). Some other molecular-based assays are either not rapid enough, associated with poor specificity, too costly for reagents and instrumentation, or the process cannot be easily automated (23, 24, 25). However, in general, nucleic acid-based molecular methods could provide more accurate and useful species-specific information if the detection probes are properly designed compared with that from morphology-based culture methods.
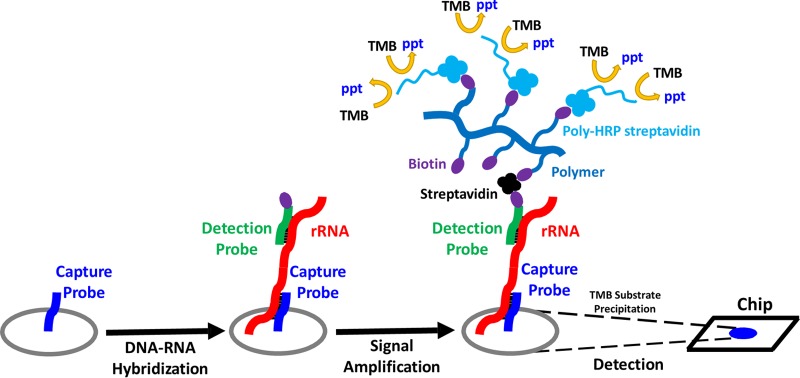
Here, we report the development of a rapid and cost-efficient molecular assay based on combining a novel signal amplification process (26) with our chip-array technology (27, 28) to target rRNA for detecting and differentiating seven clinically relevant Candida species simultaneously with high specificity. The amplification process is mediated by a high-molecular-weight polysaccharide polymer conjugated with multiple biotin molecules. Using a multivalent bridge, streptavidin, this polymer is conjugated to a biotin-labeled DNA detection probe hybridized to rRNA sequences in Candida to allow for amplification of the single probe biotin into ∼80 biotin signaling events (Fig. 1). More importantly, the overall high sensitivity of this process eliminates the need for target amplification approaches such as PCR, reducing the test expense and complexity. Furthermore, this process is easily automated on our Portrait device (originally developed for an isothermal amplification process) (29) to provide a rapid and accurate approach for clinicians to identify Candida infection with valuable species information.
FIG 1.
Diagram showing the principle of the AMPED method. TMB, 3,3′,5,5′-tetramethylbenzidine; ppt, precipitation.
MATERIALS AND METHODS
Chemicals, reagents, and blood culture medium.
All chemicals and reagents were purchased from Sigma-Aldrich (St. Louis, MO, USA) or Fisher Scientific (Pittsburgh, PA, USA) unless otherwise indicated below. 3,3′,5,5′-Tetramethylbenzidine (TMB)-enhanced horseradish peroxidase (HRP) membrane substrate was obtained from SurModics (Eden Prairie, MN, USA). Unspun human whole blood (EDTA, sodium) was purchased from Biological Specialty Corporation (Colmar, PA, USA). BD Bactec instrumented blood culture system and blood culture bottles were from BD Diagnostics (Sparks, MD, USA).
AMP polymer preparation.
AMP polymer (originally termed PED polymer) (26) was prepared in-house using the protocol described below. One milligram per milliliter of amino dextran (500 kDa; Molecular Probes) was reacted with 500 μM sulfo-N-hydroxysuccinamide (NHS)-biotin (Pierce) in 0.1 M borate buffer (pH 8.5) at room temperature for 3 h with constant mixing. The biotinylated dextran was purified and desalted using a PD-10 column (Fisher GE Healthcare) equilibrated in 0.1 M borate buffer (pH 8.5) according to the manufacturer's instructions. The polymer solution was then diluted to 5 ml in 0.1 M borate buffer (pH 8.5) and reacted with 100 μM sulfo-NHS-acetate at room temperature for 3 h with constant mixing. The reaction mixture was passed over another PD-10 column equilibrated with 1× phosphate-buffered saline ([PBS] pH 7.0). The biotinylated and acetylated AMP polymer was eluted with 2.5 ml of 1× PBS (pH 7.0) and stored at 4°C until further use.
AMPED Candida identification assay probe design.
DNA probes were designed against the 28S rRNA of the target Candida species. The 28S rRNA sequences of relevant species obtained from GenBank were aligned and analyzed using the CLC sequence viewer (CLC Bio, Aarhus, Denmark). A desired candidate region with maximal mismatches between different target species was chosen for capture probe design to distinguish different target Candida species from other closely related species. Detection probes were then designed against the immediate upstream region of the corresponding capture probes. BLAST analyses were performed for all capture probe sequences to determine any potential cross-reactivity. Capture probes were designed using MeltCalc (30, 31), which uses nearest neighbor calculations to optimize the discrimination of all the mismatches. Criteria were set for a melting temperature (Tm) of 65 to 70°C under our assay condition of 825 mM monovalent cation. Higher Tm probes (up to 74°C) were also required for some capture probes, depending on the secondary structure of that region. Each probe was screened to maximize the sensitivity and specificity against other closely related species. All the capture and detection probes used in this study are listed in Table 1. These oligonucleotide probes were ordered from Integrated DNA Technologies (Coralville, IA, USA).
TABLE 1.
List of detection and capture probe sets
| Target species | Probe |
Accession no. | Location (nt)a | ||
|---|---|---|---|---|---|
| Type | Name | Sequence | |||
| C. albicans | Capture | Calb853-CP7 | /5Ilink12//iSp18/CGCAGCGGCCGCTCCAGAGAGAGCAGCATGC | MF767829.1 | 835–865 |
| Paired detection | Calb-DP12 | AAAATACCAAGTCTGATCTCAAGCCCTTCCCTTT/3BioEG/ | MF767829.1 | 800–834 | |
| C. glabrata | Capture | Cgla284rCP3 | /5Ilink12//iSp18/ACTCTTCGAGCACCCTTTACAA | AB499020.1 | 185–206 |
| Paired detection | Cand234DP1 | mAmCmGmGmGmATTCTCACCCTC/3BioEG/ | AB499020.1 | 158–173 | |
| C. tropicalis | Capture | Ctro264rCP2 | /5Ilink12//iSp18/TTACATAGGCCTGGATCAT | KY928442.1 | 198–216 |
| Paired detection | Cand234DP1 | mAmCmGmGmGmATTCTCACCCTC/3BioEG/ | KY928442.1 | 173–189 | |
| C. parapsilosis | Capture | Cpar1260-CP5 | /5Ilink12//iSp18/CGCTAGTCCACTCCTAAAGGAGGTCCTAC | GQ254874.1 | 457–485 |
| Paired detection | Cpar1260-DP1 | CTACGTTCACTTTCATTACGCGTACGGGTTTTACA/3BioTEG/ | GQ254874.1 | 422–456 | |
| C. krusei | Capture | Ckru262rCP1 | /5Ilink12//iSp18/CACTGCTTCCGCCGGCATCCC | LC015645.1 | 173–193 |
| Paired detection | Cand234DP1 | mAmCmGmGmGmATTCTCACCCTC/3BioEG/ | LC015645.1 | 156–172 | |
| C. guilliermondii | Capture | Cgui2600-CP2 | /5Ilink12//iSp18/ACCGCCGGTTCTGCTGGGTATGGTAAAG | NG_042640.1 | 1896–1923 |
| Paired detection | Cgui2600-DP1 | GCCCAAGACACCCGATCCTTAGAGCCAATCCTTA/3BioEG/ | NG_042640.1 | 1862–1895 | |
| C. lusitaniae | Capture | Clus3900-CP1 | /5Ilink12//iSp18/ACCGCCGCCAAACGCCGCCTTG | JQ689030.1 | 3030–3051 |
| Paired detection | Clus3900-DP1 | TATGGTCCACATCGTATTTGTATCCAACTG/3BioTEG/ | JQ689030.1 | 3000–3029 | |
| S. pombe/SPC | Capture | Spombe2600-CP1 | /5Ilink12//iSp18/AGTCCAGCAACCGTTCAGGTTCCAAG | Z19136.1 | 2019–2044 |
| Paired detection | Spombe2600-DP1 | GCCCAACGTACCCAACCCTTAGAGCCAATCCTTA/3BioEG/ | Z19136.1 | 1985–2018 | |
nt, nucleotide.
Strains and blood culture process.
All the strains used in this study were purchased or were gifts from the American Type Culture Collection (Manassas, VA, USA), Microbiologics, Inc. (St. Cloud, MN, USA), or ARUP Laboratories (Salt Lake City, UT, USA). All samples were subcultured on yeast extract-peptone-dextrose (YEPD) agar plates. To prepare spiked blood cultures, 5 to 7 ml of human blood was injected into BD Bactec bottles. The bottles were then seeded with 3 to 5 isolated colonies of the strain of interest from subcultures suspended in 100 μl of 1× PBS and placed in a BD Bactec 9240 instrumented blood culture system until microbial activity was detected. Blood cultures were serially diluted and plated out to determine the culture titers and then aliquoted and stored at −80°C before use.
Chip production.
Crystalline silicon wafers were coated with the polymer amine-functional T-structure polydimethylsiloxane ([TSPS] United Chemical Technologies, Bristol, PA, USA) and cured at 150°C for 24 h. The TSPS-coated wafers were further prepared by soaking in a solution of poly-(Lys-Phe) (50 mg/liter) in 1× PBS (pH 6) containing NaCl (2 mol/liter) overnight at room temperature. The poly-(Lys-Phe)-coated wafers were then washed and incubated with 10 μM succimidyl-4-formyl benzoate ([SFB] Sigma) for 2 h at room temperature, and then washed thoroughly with water, dried with a stream of nitrogen, and stored at room temperature before use. Capture probes contain a reactive hydrazide group on the 5′ end designed to interact with and attach to the aldehyde-functionalized surface of the silicon wafers with a 12-carbon atom spacer to separate the surface from the capture probe sequence, and they were spotted (75 nl) on the SFB-coated silicon wafers in spotting buffer (0.1 M phosphate buffer [pH 7.8], 10% glycerol) using a nanoliter dispenser (BioDot, Irvine, CA, USA). The probe concentration spotted was 300 nM for all the species, except for 100 nM for the C. krusei probe and 50 nM for the C. lusitaniae probe. To orient the chips for subsequent processing, a fiducial marker (carboxylated polystyrene microspheres) was also printed. After incubating for 2 h, the wafers were washed with 0.1% sodium dodecyl sulfate (SDS), dried, and scribed into 6.5-mm2 chips using DTX Scribe and Break (Dynatek International, Santa Rosa, CA, USA).
Assay process.
For manual assays, chips were attached to the bottoms of the wells of a 96-well plate using double-sided tape, were covered with a microplate sealer, and were preheated on the heater block in a 60°C oven for 5 to 10 min. Sixty microliters of fungal lysis buffer (30% N-lauroyl sarcosine with 20 nM each detection probe) was mixed well with 60 μl of blood culture sample in PCR tubes and lysed at 95°C for 30 min. Then, 100 μl from each of the lysed samples was transferred into the wells of the preheated plate with chips, which was sealed and incubated on the heated block for 20 min at 60°C. After this incubation, the polymer-enhanced detection was performed at room temperature. Briefly, chips were washed 3 times with wash buffer B (0.1× SSC, 0.05%Tween 20), and 100 μl of streptavidin (10 μg/ml in 1× hybridization buffer) was added to each well and incubated for 4 min. Chips were further washed 3 times with buffer B, and 100 μl of AMP-polymer (1:1,000 dilution in 1× hybridization buffer) was added and incubated for 4 min. Again, the chip solution was removed and 100 μl of poly-HRP streptavidin (1:1,000 dilution in Guardian buffer) was added and incubated for 4 min. The chips were washed as before, and 100 μl of BioFX-enhanced small particle TMB was added and incubated for 10 min. Finally, chips were washed 2 times with distilled water and dried 2 times with ethanol. An image was taken using a charge-coupled-device (CCD) camera for each chip.
Clinical specimen testing.
A total of 67 frozen specimens from blood culture-positive bottles seeded directly with whole blood of patients suspected with candidemia were collected over 6 months in the Midwest region of the United States. These specimens were characterized in a reference clinical laboratory using MALDI-TOF MS and were identified at the species level. There were a total of 60 Candida specimens, representing the 5 most prevalent species, with 22 of C. albicans, 25 of C. glabrata, 7 of C. parapsilosis, 3 of C. tropicalis, and 3 of C. krusei. There were no C. lusitaniae, C. guilliermondii, or other nontarget Candida species detected in the collection. Two other specimens in this collection were mixed-species infections with C. albicans plus C. dubliniensis or C. glabrata. Furthermore, there were also five specimens identified as bacterial infections rather than fungal infections in this collection. These clinical specimens were tested using the AMPED method according to the assay process described.
RESULTS
Novel signal amplification approach targeting rRNA.
Previously, we described the polymer-based enzymatic detection (AMPED) approach and its application for the detection of single-copy genomic DNA targets in Staphylococcus species, in which the signal is amplified to provide a 10- to 100-fold improvement in the assay limit of detection, allowing for direct-from-blood culture detection (26). Here, we have applied the AMPED approach to target multicopy rRNA for the detection of fungal Candida species in positive blood culture bottles from patients suspected with candidemia as an attempt to further improve the assay limit of detection. Because phylogenetic differences in Candida species are significant, unambiguous detection to the species level is possible (32). The AMPED process is briefly depicted in Fig. 1.
AMPED Candida identification assay analytical performance.
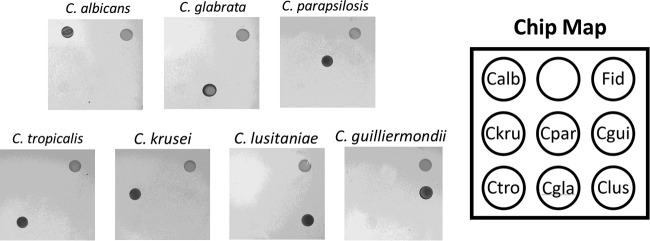
To validate the sensitivity and specificity of the final probe set for the identification of Candida species, negative blood culture bottles were spiked with cultured cells, incubated until they became positive as indicated by an alarm on the Bactec blood culture instrument, and then tested in the described assay (see Materials and Methods). For each of the species present in the array, an unambiguous signal was observed with no cross-reaction with any of the other target species (Fig. 2).
FIG 2.
Reactivity panel of AMPED Candida identification assay target species. (Left) Chip images for each target species. (Right) Chip map. Calb, C. albicans; Ckru, C. krusei; Cpar, C. parapsilosis; Cgui, C. guilliermondii; Ctro, C. tropicalis; Cgla, C. glabrata; Clus, C. lusitaniae; Fid, fiducial (colored carboxylated polystyrene microspheres spotted for easy orientation identification).
Additionally, we determined the reactivity of this AMPED Candida identification assay with other closely related Candida species. All other Candida species tested showed no cross-reactivity, except C. orthopsilosis and C. metapsilosis, which reacted with the C. parapsilosis probe (Table 2, top). However, these results may be expected, as both are members of C. parapsilosis complex strains and they have identical sequence in the probe region. Also, there was no cross-reactivity observed for other non-Candida fungal and bacterial species tested (Table 2, middle and bottom), including the budding yeast Saccharomyces cerevisiae and the fission yeast Schizosaccharomyces pombe, which will be used as a sample preparation control strain in our automated assay system.
TABLE 2.
List of strains in exclusivity test panel
| Species | Source and strain | Input (CFU/ml) | Reactivity |
|---|---|---|---|
| Non-target Candida | |||
| C. auris | CDC 0391-3090 | ∼1.0E+7 | None |
| C. catenulata | ATCC 10565 | ∼1.0E+6 | None |
| C. dubliniensis | ATCC MYA-646 | ∼1.0E+6 | None |
| C. duobushaemulonii | CDC 0391 | ∼1.0E+7 | None |
| C. haemulonii | ATCC 22991 | 4.60E+6 | None |
| C. kefyr | Microbiologic 2512 | 2.75E+5 | None |
| C. metapsilosis | ATCC 14054 | 8.56E+6 | C. parapsilosis probe |
| C. norvegensis | ATCC 96301 | 1.61E+6 | None |
| C. orthopsilosis | ATCC 20503 | 1.35E+7 | C. parapsilosis probe |
| C. rugosa | ATCC 20263 | 3.30E+6 | None |
| C. utilis | ATCC 9905 | 1.81E+7 | None |
| C. viswanathii | ATCC 28269 | 1.87E+7 | None |
| Non-Candida fungi | |||
| Aspergillus flavus | ATCC 9643 | ∼1.0E+6 | None |
| Aspergillus fumigatus | ATCC 204305 | ∼1.0E+6 | None |
| Aspergillus niger | ATCC 6275 | ∼1.0E+6 | None |
| Aspergillus terreus | ATCC 1012 | ∼1.0E+6 | None |
| Fusarium proliferatum | ATCC 201904 | ∼1.0E+6 | None |
| Rhizopus oryzae | ATCC 56536 | ∼1.0E+6 | None |
| Saccharomyces cerevisiae | ATCC MYA-796 | ∼1.0E+6 | None |
| Schizosaccharomyces pombe | ATCC 10667 | ∼1.0E+6 | None |
| Bacteria | |||
| Acinetobacter baumannii | ATCC 19606 | 6.15E+7 | None |
| Bacillus subtilis | ATCC 23857 | ∼1.0E+6 | None |
| Bacteroides fragilis | ATCC 23745 | 5.00E+6 | None |
| Enterococcus faecalis | ATCC 29212 | 7.80E+7 | None |
| Enterobacter cloacae | ATCC 13047 | 1.86E+9 | None |
| Escherichia coli | ATCC 43888 | ∼1.0E+6 | None |
| Klebsiella pneumoniae | ATCC 13883 | 8.95E+7 | None |
| Klebsiella oxytoca | ATCC 49134 | 1.71E+9 | None |
| Morganella morganii | ATCC 25829 | 4.30E+7 | None |
| Serratia marcescens | ATCC 13880 | 5.00E+6 | None |
| Staphylococcus aureus | ATCC 700699 | 3.70E+7 | None |
| Staphylococcus epidermidis | ATCC 700576 | 4.70E+7 | None |
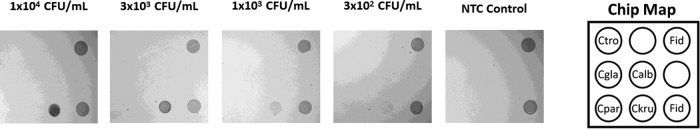
The limit of detection (LOD) was determined for this AMPED Candida identification assay by titrating known quantities of CFU into negative blood cultures for each target. A visual LOD ranging from 1.0 × 103 to 5.0 × 104 CFU/ml was determined for all the target species (Table 3, column 4). Chip images for the C. krusei LOD determination are shown in Fig. 3 below. Actually, the LOD of C. krusei is lower than 1.0 × 103 CFU/ml, but the signal is faint below these levels (Fig. 3). These limits of detection are far below the measured blood culture titers of alarmed Candida positive bottles with C. albicans alarm positive at the lowest level (2.7 × 105 CFU)/ml and with all other species alarm positive at 3.1 × 106 CFU/ml or higher (Table 3). The titers measured here confirm the titers previously reported from the Bactec blood culture instrument (33).
TABLE 3.
Limit of detection of each target Candida species
| Species | Strain | Titer at alarm (CFU/ml) | Limit of detection (CFU/ml) |
|---|---|---|---|
| C. albicans | ARUP1 | 2.7 × 105 | 1.0 × 103 |
| C. glabrata | ATCC 2001 | 4.2 × 106 | 5.0 × 104 |
| C. tropicalis | ATCC 750 | 3.0 × 107 | 5.0 × 103 |
| C. parapsilosis | ATCC 22019 | 9.4 × 106 | 1.0 × 104 |
| C. krusei | ATCC 24210 | 1.1 × 107 | 1.0 × 103 |
| C. guilliermondii | ARUP2 | 4.8 × 106 | 1.0 × 104 |
| C. lusitaniae | ATCC 60247 | 3.1 × 106 | 1.0 × 104 |
FIG 3.
Limit detection of C. krusei. (Left) Chip images showing the detection of C. krusei with different cell inputs. (Right) Chip map. Calb, C. albicans; Ckru, C. krusei; Cpar, C. parapsilosis; Ctro, C. tropicalis; Cgla, C. glabrata; Fid, fiducial (colored carboxylated polystyrene microspheres spotted for easy orientation identification).
Clinical specimen verification.
We verified 67 clinical specimens from frozen blood culture-positive bottles from patients suspected with candidemia on our AMPED Candida identification assay platform. We correctly detected 62 of the 67 total specimens as positive (including 2 mixed infections) and the other 5 as negative (non-Candida infections) in perfect (100%) concordance with the reference method (MALDI-TOF MS) (Table 4).
TABLE 4.
Comparison of clinical specimen and reference identifications by MALDI-TOF MS and the AMPED Candida assay
| Species | No. positively identified |
|
|---|---|---|
| MALDI-TOF MS | AMPED assay | |
| Reference Candida species (MALDI-TOF MS) | ||
| C. albicans | 22 | 22 |
| C. glabrata | 25 | 25 |
| C. parapsilosis | 7 | 7 |
| C. tropicalis | 3 | 3 |
| C. krusei | 3 | 3 |
| C. lusitaniae | 0 | 0 |
| C. guilliermondii | 0 | 0 |
| Mixed-infection specimens | ||
| C. albicans and C. dubliniensisa | 1 | 1 |
| C. albicans and C. glabratab | 1 | 1 |
| Non-Candida infection specimens | ||
| Histoplasma capsulatum | 1 | 0 |
| Malassezia pachydermatis | 2 | 0 |
| Rhodotorula species | 1 | 0 |
| Gram-positive rods resembling Bacillusc | 1 | 0 |
C. dubliniensis was not detected in this study as it is not a target species.
Both C. albicans and C. glabrata were detected.
No yeast growth.
DISCUSSION
We have herein described a novel multiplex Candida identification test that combines a sensitive modified silicon chip with an array of nucleic acid probes and a signal amplification method, AMPED, for the detection of multicopy rRNA sequences, allowing for the direct detection of Candida species from positive blood cultures. An advantage of nucleic acid detection approaches that do not require target amplification, such as by PCR, is that the level of multiplexing is only limited by the probe density on the chip. Primer interactions that harm the sensitivity and limit the number of primer sets that may be used in PCR-based multiplex designs are also eliminated. The AMPED approach uses inexpensive, stable, and easy-to-produce biotinylated polymers. This polymer is highly water soluble, chemically simple, and uncharged, limiting nonspecific interactions. Additionally, we have demonstrated that the approach is efficient, with the signal amplification proportional to the number of biotin molecules present on the polymer (26).
The AMPED Candida identification test displayed excellent analytical specificity for species-level detection of seven important Candida species. No cross talk was observed between the target species or with other key pathogenic Candida species, with the exception of C. orthopsilosis and C. metapsilosis which cross-react with the C. parapsilosis probes; however, these species belong to the same complex of strains. The limits of detection of 103 to 104 CFU/ml for this assay are 30- to 5,000-fold below those reported at alarm positivity for Candida species, including C. albicans, which has been observed elsewhere and measured here to alarm positive at ∼3 × 104 CFU/ml, or 30- to 100-fold lower than the other Candida species (Table 3) (33). These limits of detection permit excellent clinical sensitivity in detecting alarm-positive blood cultures, as evidenced herein with 100% sensitivity compared with MALDI reference data in a retrospective study of 62 Candida-positive blood cultures. To achieve these limits of detection using the described AMPED method, it is critical to target multicopy genes such as rRNA. A previous work focused on the detection of single-copy genomic sequences specific to Staphylococcus using the AMPED approach found lower limits of detection (LLODs) of 2 × 106 to 6 × 106 copies/ml using the same chip formulation (26). This suggests that rRNA is contributing to an up to 6,000-fold improvement in detection sensitivity. Variability in the LODs across the probe set is likely related to differences in secondary structures for the targeted regions of the Candida species, as it has been observed that sensitivity can vary by up to 200-fold in targeting structured RNA (34). We applied an approach used in a previous work aimed at targeting the GC-rich, and therefore highly structured, Mycobacterium tuberculosis genome in which the probe sets were longer, with higher Tms, than those for targeting less-structured nucleic acid targets under the same assay conditions (35). Combined with a coaxial stack of the capture and detect probes, the secondary structures could be more effectively unfolded or invaded to enable highly sensitive detection (34).
The AMPED identification assay was designed to provide species-specific information to aid in making appropriate treatment decisions as early as possible from positive blood cultures. The rapid initiation of appropriate therapy has significant positive outcomes in patients with fungal bloodstream infections. The determination of drug susceptibility profiles using phenotypic methods is time consuming; however, the determination of the Candida species identity has been shown to be a good indicator of what treatments may be efficacious. On the basis of the local epidemiology information from national or regional surveys, some basic guidelines for the treatment of candidemia have been provided to local clinicians for selecting empirical antifungal therapy with species-level information by some government agencies (14, 36, 37). For example, C. tropicalis, like C. albicans, is susceptible to most antifungal agents available on the market but with a relatively high-level sensitivity to fluconazole; thus, the most cost-effective drug, fluconazole, is the best choice (38, 39). C. glabrata and C. krusei display increased resistance to fluconazole; therefore, it has been recommended that treatment should be with echinocandins (14, 36, 37). C. parapsilosis has been the key species for catheter-related bloodstream infections. The removal of catheters and the use of echinocandins as the frontline therapy are recommended for C. parapsilosis infection patients. C. lusitaniae and C. guilliermondii are infrequently observed but display resistance to amphotericin B or fluconazole, respectively, making echinocandins the treatment of choice. More recently, outbreaks of Candida auris across different continents have been observed with potentially even broader drug resistance (40, 41). We are currently adding the ability to detect Candida auris in the AMPED format to improve the diagnostic potential of this test.
Currently, there are several other systems on the market for fungal and Candida identification with different advantages and limitations. AdvanDx offers a peptide nucleic acid fluorescence in situ hybridization (PNA FISH)-based test for the identification of a few key Candida species from positive blood culture bottles (42). This is a rapid method, but the process is not automated and it provides limited species-specific information. BioFire's BCID test is a large blood culture identification panel (including both fungal and bacterial species) with good sensitivity and specificity (43). However, the system is too costly for small clinical hospitals. T2 Biosystems' Candida panel enables a more rapid detection of candidemia directly from whole blood samples but with limited target species numbers and species-specific information (22, 44). Although MALDI-TOF MS has some improved accuracy, it cannot detect mixed infections with the current libraries available on the market, and also, the sample preparation procedure is inconsistent (18, 19, 20, 21). Despite some limitations (such as the need for a blood culture step), the AMPED method does have its own advantages. In addition to the low reagent cost, it provides more accurate species-specific information and also detects more species, including mixed-species infections, with the possibility to further expand the multiplex in a straightforward manner.
ACKNOWLEDGMENTS
We thank Brian Hicke, Georges Frech, Chris Pasko, Wendy Smith, and Amber Raigne for the early stage development of this assay.
REFERENCES
- 1.Pfaller MA, Castanheira M. 2016. Nosocomial candidiasis: antifungal stewardship and the importance of rapid diagnosis. Med Mycol 54:1–22. doi: 10.1093/mmy/myv076. [DOI] [PubMed] [Google Scholar]
- 2.Papon N, Courdavault V, Clastre M, Bennett RJ. 2013. Emerging and emerged pathogenic Candida species: beyond the Candida albicans paradigm. PLoS Pathog 9:e1003550. doi: 10.1371/journal.ppat.1003550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pfaller MA, Diekema DJ, Gibbs DL, Newell VA, Meis JF, Gould IM, Fu W, Colombo AL, Rodriguez-Noriega E, Global Antifungal Surveillance Study. 2007. Results from the ARTEMIS DISK Global Antifungal Surveillance study, 1997 to 2005: an 8.5-year analysis of susceptibilities of Candida species and other yeast species to fluconazole and voriconazole determined by CLSI standardized disk diffusion testing. J Clin Microbiol 45:1735–1745. doi: 10.1128/JCM.00409-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pfaller MA, Diekema DJ, Gibbs DL, Newell VA, Ellis D, Tullio V, Rodloff A, Fu W, Ling TA. Global Antifungal Surveillance Group. 2010. Results from the ARTEMIS DISK Global Antifungal Surveillance Study, 1997 to 2007: a 10.5-year analysis of susceptibilities of Candida species to fluconazole and voriconazole as determined by CLSI standardized disk diffusion. J Clin Microbiol 48:1366–1377. doi: 10.1128/JCM.02117-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blumberg HM, Jarvis WR, Soucie JM, Edwards JE, Patterson JE, Pfaller MA, Rangel Frausto MS, Rinaldi MG, Saiman L, Wiblin RT, Wenzel RP. National Epidemiology of Mycoses Survey (NEMIS) Study Group. 2001. Risk factors for Candida bloodstream infections in surgical intensive care unit patients: the NEMIS prospective multicenter study. The National Epidemiology of Mycosis Survey. Clin Infect Dis 33:177–186. doi: 10.1086/321811. [DOI] [PubMed] [Google Scholar]
- 6.Mermel LA, Allon M, Bouza E, Craven DE, Flynn P, O'Grady NP, Raad II, Rijnders BJ, Sherertz RJ, Warren DK. 2009. Clinical practice guidelines for the diagnosis and management of intravascular catheter-related infection: update by the Infectious Diseases Society of America. Clin Infect Dis 49:1–45. doi: 10.1086/599376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brown GD, Denning DW, Gow NA, Levitz SM, Netea MG, White TC. 2012. Hidden killers: human fungal infections. Sci Transl Med 4:165rv113. doi: 10.1126/scitranslmed.3004404. [DOI] [PubMed] [Google Scholar]
- 8.Pfaller MA, Diekema DJ. 2010. Epidemiology of invasive mycoses in North America. Crit Rev Microbiol 36:1–53. doi: 10.3109/10408410903241444. [DOI] [PubMed] [Google Scholar]
- 9.Taira CL, Okay TS, Delgado AF, Ceccon ME, de Almeida MT, Del Negro GM. 2014. A multiplex nested PCR for the detection and identification of Candida species in blood samples of critically ill paediatric patients. BMC Infect Dis 14:406–412. doi: 10.1186/1471-2334-14-406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chai ALY, Denning DW, Warn P. 2010. Candida tropicalis in human disease. Crit Rev Microbiol 36:282–298. doi: 10.3109/1040841X.2010.489506. [DOI] [PubMed] [Google Scholar]
- 11.Nucci M, Queiroz-Telles F, Alvarado-Matute T, Tiraboschi IN, Cortes J, Zurita J, Guzman Blanco M, Santolaya ME, Thompson L, Sifuentes-Osornio J, Echevarria JI, Colombo AL, Latin American Invasive Mycosis Network. 2013. Epidemiology of candidemia in Latin America: a laboratory-based survey. PLoS One 8:e59373. doi: 10.1371/journal.pone.0059373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schelenz S. 2008. Management of candidiasis in the intensive care unit. J Antimicrob Chemother 61:i31–i34. doi: 10.1093/jac/dkm430. [DOI] [PubMed] [Google Scholar]
- 13.Alexander BD, Ashley ED, Reller LB, Reed SD. 2006. Cost savings with implementation of PNA FISH testing for identification of Candida albicans in blood cultures. Diagn Microbiol Infect Dis 54:277–282. doi: 10.1016/j.diagmicrobio.2005.10.011. [DOI] [PubMed] [Google Scholar]
- 14.Pappas PG, Kauffman CA, Andes DR, Clancy CJ, Marr KA, Ostrosky-Zeichner L, Reboli AC, Schuster MG, Vazquez JA, Walsh TJ, Zaoutis TE, Sobel JD. 2016. Clinical practice guideline for the management of candidiasis: 2016 update by the Infectious Diseases Society of America. Clin Infect Dis 62:e1–e50. doi: 10.1093/cid/civ1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bacconi A, Richmond GS, Baroldi MA, Laffler TG, Blyn LB, Carolan HE, Frinder MR, Toleno DM, Metzgar D, Gutierrez JR, Massire C, Rounds M, Kennel NJ, Rothman RE, Peterson S, Carroll KC, Wakefield T, Ecker JDJ, Sampath R. 2014. Improved sensitivity for molecular detection of bacterial and Candida infections in blood. J Clin Microbiol 52:3164–3174. doi: 10.1128/JCM.00801-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fenollar F, Raoult D. 2007. Molecular diagnosis of bloodstream infections caused by non-cultivable bacteria. Int J Antimicrob Agents 30(Suppl 1):7–15. doi: 10.1016/j.ijantimicag.2007.06.024. [DOI] [PubMed] [Google Scholar]
- 17.Lamy B, Roy P, Carret G, Flandrois JP, Delignette-Muller ML. 2002. What is the relevance of obtaining multiple blood samples for culture? A comprehensive model to optimize the strategy for diagnosing bacteremia. Clin Infect Dis 35:842–850. doi: 10.1086/342383. [DOI] [PubMed] [Google Scholar]
- 18.Cassagne C, Cella AL, Suchon P, Normand AC, Ranque S, Piarroux R. 2013. Evaluation of four pretreatment procedures for MALDI-TOF MS yeast identification in the routine clinical laboratory. Med Mycol 51:371–377. doi: 10.3109/13693786.2012.720720. [DOI] [PubMed] [Google Scholar]
- 19.Turhan O, Ozhak-baysan B, Zaragoza O, Er H, Saritas ZE, Ongut G, Ogunc D, Colak D, Cuenca-Estrella M. 2017. Evaluation of MALDI-TOF-MS for the identification of yeast isolates causing bloodstream infection. Clin Lab 63:699–703. doi: 10.7754/Clin.Lab.2016.161101. [DOI] [PubMed] [Google Scholar]
- 20.Haas Grenouillet M, F, Loubersac S, Ariza B, Pepin-Puget L, Alvarez-Moreno CA, Valderrama-Beltran SL, Lavergne RA, Le Pape P, Morio F. 2016. Identification of cryptic Candida species by MALDI-TOF mass spectrometry, not all MALDI-TOF systems are the same: focus on the C. parapsilosis species complex. Diagn Microbiol Infect Dis 86:385–386. doi: 10.1016/j.diagmicrobio.2016.08.028. [DOI] [PubMed] [Google Scholar]
- 21.Bader O. 2017. Fungal species identification by MALDI-TOF mass spectrometry. Methods Mol Biol 1508:323–337. doi: 10.1007/978-1-4939-6515-1_19. [DOI] [PubMed] [Google Scholar]
- 22.Neely LA, Audeh M, Phung NA, Min M, Suchocki A, Plourde D, Blanco M, Demas V, Skewis LR, Anagnostou T, Coleman JJ, Wellman P, Mylonakis E, Lowery TJ. 2013. T2 magnetic resonance enables nanoparticle-mediated rapid detection of candidemia in whole blood. Sci Transl Med 5:182ra54. doi: 10.1126/scitranslmed.3005377. [DOI] [PubMed] [Google Scholar]
- 23.Arvanitis M, Anagnostou T, Fuchs BB, Caliendo AM, Mylonakis E. 2014. Molecular and nonmolecular diagnostic methods for invasive fungal infections. Clin Microbiol Rev 27:490–526. doi: 10.1128/CMR.00091-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wellinghausen N, Siegel D, Winter J, Gebert S. 2009. Rapid diagnosis of candidemia by real-time PCR detection of Candida DNA in blood samples. J Med Microbiol 58:1106–1111. doi: 10.1099/jmm.0.007906-0. [DOI] [PubMed] [Google Scholar]
- 25.White PL, Shetty A, Barnes RA. 2003. Detection of seven Candida species using the Light-Cycler system. J Med Microbiol 52:229–238. doi: 10.1099/jmm.0.05049-0. [DOI] [PubMed] [Google Scholar]
- 26.Klonoski J, Mondesire R, Rea L, Ward DC, Jenison RD. 2010. Enhanced detection of Staphylococcal genomes in positive blood cultures using a polymeric enzyme complex. Anal Biochem 396:284–289. doi: 10.1016/j.ab.2009.09.035. [DOI] [PubMed] [Google Scholar]
- 27.Jenison RD, Yang S, Haeberli A, Polisky B. 2001. Interference-based detection of nucleic acid targets on optically coated silicon. Nat Biotechnol 19:62–65. doi: 10.1038/83530. [DOI] [PubMed] [Google Scholar]
- 28.Jenison RD, La H, Haeberli A, Ostroff R, Polisky B. 2001. Silicon-based biosensors for repaid detection of protein or Nucleic acid targets. Clin Chem 47:1894–1900. [PubMed] [Google Scholar]
- 29.Rea L, Hicke B, Lindsey W, McMahon M, Owen C, Jenison RD. 2012. Point-of-care molecular diagnostic testing. IVD Technol 18:17–24. [Google Scholar]
- 30.Schutz E, von Ahsen N. 1999. Spreadsheet software for the thermodynamic melting point prediction of oligonucleotide hybridization with and without mismatches. Biotechniques 27:1218–1224. [DOI] [PubMed] [Google Scholar]
- 31.von Ahsen N, Oellerich VW, Armstrong VW, Schutz E. 1999. Application of thermodynamic nearest neighbor mode to estimate nucleic acid stability and optimize probe design: prediction of melting points of different mutations of apolipoprotein B 3500 and factor V Leiden with a hybridization probe genotyping assay in the LightCycler. Clin Chem 45:2094–2101. [PubMed] [Google Scholar]
- 32.Merseguel KB, Nishikaku AS, Rodgigues AM, Padovan AC, e Ferreira RC, de Azevedo Melo AS, da Silva Briones MR, Colombo AL. 2015. Genetic diversity of medically important and emerging Candida species causing invasive infection. BMC Infect Dis 15:57–67. doi: 10.1186/s12879-015-0793-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.BioFire Diagnostics, LLC. 510(k) summary. FDA no. K143171. http://www.accessdata.fda.gov/cdrh_docs/pdf14/k143171.pdf.
- 34.Buvoli A, Buvoli M, Leinwand LA. 2000. Enhanced detection of tRNA isoacceptors by combinatorial oligonucleotide hybridization. RNA 6:912–918. doi: 10.1017/S1355838200000339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ao W, Aldous S, Woodruff E, Hicke B, Rea L, Kreiswirth B, Jenison R. 2012. Rapid detection of rpoB gene mutations conferring rifampin resistance in Mycobacterium tuberculosis. J Clin Microbiol 50:2433–2440. doi: 10.1128/JCM.00208-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Arendrup MC, Boekhout T, Akova M, Meis JF, Cornely OA, Lortholary O, European Society of Clinical Microbiology and Infectious Diseases Fungal Infection Study Group, European Confederation of Medical Mycology. 2014. ESCMID and ECMM joint clinical guidelines for the diagnosis and management of rare invasive yeast infections. Clin Microbiol Infect 20(Suppl 3):76–98. doi: 10.1111/1469-0691.12360. [DOI] [PubMed] [Google Scholar]
- 37.Ullmann AJ, Akova M, Herbrecht R, Viscoli C, Arendrup MC, Arikan-Akdagli S, Bassetti M, Bille J, Calandra T, Castagnola E, Cornely OA, Donnelly JP, Garbino J, Groll AH, Hope WW, Jensen HE, Kullberg BJ, Lass-Flörl C, Lortholary O, Meersseman W, Petrikko G, Richardson MD, Roilides E, Verweij PE, Cuenca-Estrella M, ESCMID Fungal Infection Study Group. 2012. ESCMID guideline for the diagnosis and management of Candida diseases 2012: adults with haematological malignancies and after haematopoietic stem cell transplantation (HCT). Clin Microbiol Infect 18:53–67. doi: 10.1111/1469-0691.12041. [DOI] [PubMed] [Google Scholar]
- 38.Castanheira M, Messer SA, Jones RN, Farrell DJ, Pfaller MA. 2014. Activity of echinocandins and triazoles against a contemporary (2012) worldwide collection of yeast and moulds collected from invasive infections. Int J Antimicrob Agents 44:320–326. doi: 10.1016/j.ijantimicag.2014.06.007. [DOI] [PubMed] [Google Scholar]
- 39.Pfaller MA, Messer SA, Rhomberg PR, Castanheira M. 2017. Activity of a long-acting echinocandin (CD101) and seven comparator antifungal agents tested against a global collection of contemporary invasive fungal isolates in the SENTRY 2014 antifungal surveillance program. Antimicrob Agents Chemother 61:e02045-16. doi: 10.1128/AAC.02045-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lockhart SR, Etienne KA, Vallabhaneni S, Farooqi J, Chowdhary A, Govender NP, Colombo AL, Calvo B, Cuomo CA, Desjardins CA, Berkow EL, Castanheira M, Magobo RE, Jabeen K, Asghar RJ, Meis JF, Jackson B, Chiller T, Litvintseva AP. 2017. Simultaneous emergence of multidrug-resistant Candida auris on 3 continents confirmed by whole-genome sequencing and epidemiological analyses. Clin Infect Dis 64:134–140. doi: 10.1093/cid/ciw691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chowdhary A, Sharma C, Meis JF. 2017. Candida auris: a rapidly emerging cause of hospital-acquired multidrug-resistant fungal infections globally. PLoS Pathog 13:e1006290. doi: 10.1371/journal.ppat.1006290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stone NR, Gorton RL, Barker K, Ramnarain P, Kibbler CC. 2013. Evaluation of PNA-FISH yeast traffic light for rapid identification of yeast directly from positive blood cultures and assessment of clinical impact. J Clin Microbiol 51:1301–1302. doi: 10.1128/JCM.00028-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Salimnia H, Fairfax MR, Lephart PR, Schreckenberger P, DesJarlais SM, Johnson JK, Robinson G, Carroll KC, Greer A, Morgan M, Chan R, Loeffelholz M, Valencia-Shelton F, Jenkins S, Schuetz AN, Daly JA, Barney T, Hemmert A, Kanack KJ. 2016. Evaluation of the FilmArray blood culture identification panel: results of a multicenter controlled trial. J Clin Microbiol 54:687–698. doi: 10.1128/JCM.01679-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hamula CL, Hughes K, Fisher BT, Zaoutis TE, Singh IR, Velegraki A. 2016. T2Candida provides rapid and accurate species identification in pediatric cases of candidemia. Am J Clin Pathol 145:858–861. doi: 10.1093/ajcp/aqw063. [DOI] [PubMed] [Google Scholar]