Abstract
Antibiotic resistance is compromising our ability to treat bacterial infections. Clinical microbiology laboratories guide appropriate treatment through antimicrobial susceptibility testing (AST) of patient bacterial isolates. However, increasingly, pathogens are developing resistance to a broad range of antimicrobials, requiring AST of alternative agents for which no commercially available testing methods are available. Therefore, there exists a significant AST testing gap in which current methodologies cannot adequately address the need for rapid results in the face of unpredictable susceptibility profiles. To address this gap, we developed a multi-component, microscopy-based AST platform (MAST) capable of AST determinations after only a 2 hour incubation. MAST consists of a solid-phase microwell growth surface in 384-well plate format; inkjet printing-based application of both antimicrobials and bacteria at any desired concentrations; automated microscopic imaging of bacterial replication; and a deep learning approach for automated image classification and determination of antimicrobial minimal inhibitory concentrations. In evaluating a susceptible strain set, 95.8% were within ±1 and 99.4% were within ±2, two-fold dilutions, respectively, of reference broth microdilution MIC values. 98.3% of results were in categorical agreement. We conclude that MAST offers promise for rapid, accurate, and flexible AST to help address the antimicrobial testing gap.
Keywords: Antimicrobials, Inkjet printing, Susceptibility testing, Machine-learning
Introduction
The rapid emergence of clinical antimicrobial resistance presents a significant challenge for treatment of bacterial infection. To guide appropriate therapy and preserve efficacy of existing antimicrobials, hospital-based clinical microbiology laboratories perform antimicrobial susceptibility testing (AST) of bacterial isolates from patients to determine which drugs will be active against an infecting organism. The gold standard methods used for AST require manual preparation of an antimicrobial doubling dilution series in agar or broth that is tested for antimicrobial effect and minimal inhibitory concentration (MIC) determination. MIC values are defined as the lowest concentration of antimicrobial that results in visible growth inhibition of an organism after 16–20 hours of incubation.1 MIC results are interpreted based on consensus guidelines to classify the pathogen as susceptible (S), intermediate (I), or resistant (R) to the tested antimicrobial.2
However, technical complexity of manual dilution testing, even for a few isolates or drugs, precludes use of these reference methods in hospital-based clinical laboratories.3 Therefore, alternative testing methods are typically used including various types of pre-formulated antimicrobial dilution panels such as Sensititre (ThermoFisher, Waltham, MA); methods that extrapolate MICs based on growth kinetics in the presence of a limited number of different antimicrobial concentrations, such as Vitek 2 (bioMérieux, Durham, NC); and diffusion based methods that can predict MIC-based susceptibility categories for a subset of organism-drug combinations.3 However, these tests are limited in that they incorporate only predetermined panels of relatively common antimicrobials; do not include recently approved agents developed for multidrug-resistant pathogens;4 require a long (>8–20 hour) incubation; and for MIC surrogate methods, may not always correlate with reference MIC results, a problem particularly noted for multidrug-resistant organisms.5–7
Importantly, delay in institution of active, appropriate therapy, ultimately guided by AST, is associated with significantly increased morbidity and mortality especially in patients with bloodstream and other serious infections.8 With emergence of multidrug-resistant pathogens, empiric therapy (treatment without antimicrobial susceptibility data) is now increasingly unreliable. Therefore, there exists a significant AST gap in which current methodologies cannot adequately address the need for rapid results in the face of unpredictable susceptibility profiles.4
To address this gap, we have combined use of inkjet-based digital dispensing technology, automated microscopy, and deep learning to develop a prototype microscopy-based antimicrobial susceptibility testing (MAST) platform. Importantly, the platform permits performance of AST for any antimicrobial at will after a 2-hour incubation using commercially-available equipment and consumables. Here we describe development and validation of the MAST prototype.
Materials and Methods
Bacterial strains and antimicrobials
Escherichia coli ATCC 25922, Enterobacter cloacae ATCC 13047, Klebsiella pneumoniae ATCC 13883, Pseudomonas aeruginosa ATCC 27853, and Acinetobacter baumannii 17978 were obtained from the American Type Culture Collection (Manassas, VA). Strains were stored at −80°C in tryptic soy broth (BD Diagnostics, Franklin Lakes, NJ) containing 50% glycerol (Sigma-Aldrich, St. Louis, MO).
Meropenem was from Ark Pharm (Arlington Heights, IL), cefepime was from Chem Impex (Wood Dale, IL), gentamicin was from Alfa Aesar (Haverhill, MA), and ciprofloxacin was from US Biological (Salem, MA). Antibiotic stock solutions were prepared in water for manual dilution-based testing or in water containing 0.3% polysorbate-20 as required for liquid handling by the HP D300 digital dispenser (HP Inc., Palo Alto, CA).9 We previously determined through extensive analysis that polysorbate-20 at the concentrations used in assay wells has no effect on MIC determinations for all antibiotics examined.5, 10
Broth microdilution (BMD) susceptibility testing
BMD was performed according to Clinical Laboratory and Standards Institute (CLSI) guidelines.1 Specifically, serial two-fold dilutions of antimicrobials were manually prepared in cation-adjusted Mueller-Hinton broth (CAMHB, BD Diagnostics) in sterile, polystyrene 96-well plates (Evergreen Scientific, Los Angeles, CA) in a 50 μL volume. Bacteria were grown overnight at 35±2°C in ambient air on tryptic soy agar containing 5% sheep’s blood. Several colonies were suspended in sterile 0.9% saline (Sigma-Aldrich) to a density corresponding to 0.5 McFarland as measured using a DensiCHEK Plus Colorimeter (bioMérieux, Durham, NC). This suspension was diluted 1:150 in sterile CAMHB, and 50 μL was added to antibiotic-containing wells resulting in an inoculum density of approximately 5 × 105 colony forming units (CFU) ml−1 in a 100 μL assay volume. Inoculated plates were incubated at 35±2°C in ambient air for 16–20 hours. Minimal inhibitory concentration (MIC) was defined as the lowest antimicrobial concentration resulting in complete inhibition of growth as determined visually.1 BMD experiments were performed in triplicate for each antibiotic/organism combination with the modal result recorded as the reference MIC.5
Preparation of solid microwell surfaces in assay plates
In preliminary experiments, we evaluated several agents for solidifying CAMHB including BD Bacto agar (1.5% w/v, BD Diagnostics), gellan gum (1% w/v, trade name GELRITE, Research Products International, Mt. Prospect, IL), polyacrylamide (15% v/v, ThermoFisher Scientific, Waltham, MA) and poloxamer 407 (15% w/v, trade name Pluronic F-127, Sigma-Aldrich). We identified 15% poloxamer 407 as the ideal solidifying agent (described in the “Preparation of solid microwell surfaces in microtiter plates” section of results), and all future experiments were therefore conducted with 15% poloxamer 407 in CAMHB (hereafter called CAMHB-P). Prior to use, CAMHB-P was centrifuged at 4,000 × g at 4°C for 10 minutes to remove small particles of media/poloxamer 407 which may interfere with microscopy. The cleared solution was kept on ice, and 10 μL was added to wells of clear polystyrene 384-well plates (Greiner Bio-One, Monroe, NC) using a manual pipette. Immediately after preparation, plates were centrifuged at 3,500 × g at 4°C for 5 minutes to ensure complete coverage of wells with CAMHB-P and stored at −80°C until use.
Digital dispensing of bacterial suspensions
Prior to dispensing, organisms were grown for 16–20 hours at 35±2°C in ambient air on tryptic soy agar containing 5% sheep’s blood. Several colonies were suspended in 0.9% NaCl containing 0.3% polysorbate-20 (NaCl-P20) and adjusted to the desired density using a handheld colorimeter. Suspensions were directly loaded into D4+ or T8+ cassettes (HP Inc.), and dispensed into wells of 384-well plates at volumes indicated using the HP D300 Digital Dispenser (HP Inc.).
Quantification of bacterial dispensing precision
Precision of bacterial inkjet dispensing was quantified by two complementary methods: colony forming unit (CFU) determination and microscopic cell counting. For CFU quantification, 50, 150, 250, and 350 nL of a 0.5±0.05 McFarland suspension of bacteria in NaCl-P20 were dispensed into wells of a 384-well plate containing 50 μL of CAMHB. The inoculated media was diluted 1:1000 in sterile CAMHB, and 100 μL was plated on Mueller-Hinton plates (Remel, Lenexa, KS). Plates were incubated for 16–20 hours at 37°C in ambient air, and colonies were counted to determine CFU/ml according to CLSI guidelines for inoculum density determination.1 Each experiment was performed in triplicate with three wells sampled per experiment. Standard curves comparing dispensed volume to bacterial density (CFU/ml) were generated using a custom Python (version 3.5.0) script, Matplotlib (version 2.0.0)11 and NumPy (version 1.11.3).12
For quantification by direct imaging, a 0.5 McFarland suspension in NaCl-P20 was diluted 1:5, and 200 nL was dispensed into the center of each of 240 wells of a 384-well plate containing CAMHB-P. The outer two rows and columns of wells were not used to avoid edge effects resulting from evaporative loss. Plates were kept at room temperature prior to imaging at the Harvard Center for Biological Imaging (HCBI, Harvard University, Cambridge, MA) using a Zeiss Cell Observer microscope (Zeiss, Oberkochen, Germany) operating in brightfield mode with a 40X air objective (0.6 NA, 2.9mm working distance) and an automated mechanical stage. The x and y position of each well was determined automatically using a pre-loaded plate map. Z-position for optimal focus was adjusted manually. A single field corresponding to the center of each well was imaged as a z-series of 40 μm with a step size of 2 μm. Image stacks were projected using the extended depth of focus module within the Zeiss Zen Blue software. Individual bacteria in resulting images were manually segmented and counted using the particle analysis function in ImageJ.13 Twelve wells were imaged in each of three independent experiments. A bar graph was generated using a custom Python script (version 3.5.0) and the Matplotlib library.11
Microscopy-based AST (MAST)
Antimicrobials were applied by digital dispensing using the HP D300, as previously described,10 into 384-well plates (Greiner Bio-One) equilibrated to 35±2°C containing 10 μL of solid CAMHB-P. Final antimicrobial concentrations in solidified wells ranged from 0.004–1 μg/ml for ciprofloxacin and meropenem, 0.016–4 μg/ml for cefepime, and 0.03–8 μg/ml for gentamicin. Two-hundred nL of bacteria in NaCl-P20 suspended at a density of 0.1 McFarland was then delivered to the center of each well by digital dispensing of bacterial suspensions. Immediately after inoculation, plates were incubated at 35±2°C for 2 hours, and held at room temperature for 2 hours during transport and imaging.
Plates were imaged with a Zeiss Cell Observer inverted microscope with settings described in the “quantification of bacterial dispensing” section. Following manual selection of the z-position for the first well in the series, the microscope stage independently advanced to subsequent wells based on a plate map with the z-position updated automatically using the autofocus feature of the microscope control software (Zeiss Zen Blue software version 2.1 and 2.3). For each well, a z-series of 40 μm (step size = 2 μm) was automatically collected and projected to form a single image using the extended depth of focus module in the Zeiss Zen Blue software (version 2.1 and 2.3). Custom Python scripts were used to export individual images as .jpeg files with the appropriate organism name, antibiotic concentration, and well location encoded in the filename.
Image classification by deep learning
Six replicate dilution series for each antibiotic/organism combination were imaged in three independent MAST experiments yielding 3,202 training images. Each image was manually classified as “growth” or “inhibition” based on known morphological characteristics of growing or inhibited cells.14–16 Each annotated image had dimensions 2048x2048 pixels (px), and was cropped into 64 non-overlapping 256x256 px images, which were further augmented by random blur and rotation to generate 512 augmented images per annotated image, for a total of 1,639,424 training images. From this data, 80% was used for training a convolutional neural network (ConvNet), and 20% was used for validation. During training, each 256x256 px image was additionally augmented ‘on the fly’ by random crop to generate a 220x220 px input image for the network. ConvNet architecture followed the VGG-style17 with small (3x3 pixel) stacked convolutional kernels with stride and pad equal to 1 pixel. The network consisted of 8 convolutional layers, 3 fully connected layers, and 4 max pool layers. The number of feature maps was increased by a factor of two after every spatial pooling layer, resulting in an overall bi-pyramidal architecture. All convolutional layers were batch normalized18, with learned scale and shift, followed by a rectified linear unit (ReLU) non-linearity. The first two fully connected layers were regularized by drop-out, with a drop-out probability of 0.5. The final activation was a 2-way softmax, corresponding to the categories “growth” and “inhibition”.
Training was done using mini-batch stochastic gradient descent (batch size 32), based on backpropagation with momentum. The loss function was cross-entropy, with additional regularization in the form of L2 weight decay. All networks were trained from random initializations using the Xavier initialization scheme19. The initial learning rate was set to 0.0003, and decayed according to an inverse schedule. Momentum was 0.9, and weight decay was 0.0001.
For the purposes of evaluating whole images, all image crops for a given image were evaluated to calculate three parameters: mean image inhibition probability; median image inhibition probability, and proportion of crops with inhibition probability >0.5.
Optimization of MIC calls
To optimize the ability to call MICs based on ConvNet output, we collected a separate dataset, independent of the training and validation datasets, which we refer to as the optimization dataset. We used the MAST assay to collect images from all combinations of antibiotics and organisms listed in “Bacterial strains and antimicrobials” on three separate days (180 MIC assays x 10 antibiotic concentrations = 1800 images). Images in the optimization dataset were evaluated using our trained ConvNet and all output parameters were recorded (mean image inhibition probability, median inhibition probability, and proportion of crops with inhibition probability >0.5).
For each output parameter, a custom Python script was used to model results from each dilution series as a sigmoidal curve of the form:
For each antibiotic/parameter combination, we iteratively set thresholds (ranging between 0 and 0.99) that represented the point on the sigmoidal curve above which all results would be called “inhibited”. We then calculated MIC accuracy at each threshold and identified the optimal parameter and threshold combination that resulted in the highest MIC accuracy. If the highest accuracy was achieved at multiple potential threshold values for a given parameter, the median of those values was designated as the optimal threshold cutoff value.
Proof-of-principle application of MAST assay for end-to-end MIC determination
A verification dataset, independent of the training, validation, and optimization datasets, was collected using the MAST assay on three separate days using all combinations of antibiotics and organisms listed in “Bacterial strains and antimicrobials.” Each of these 1800 images was evaluated using our ConvNet-based pipeline, and MIC’s for the 180 dilution series were calculated using the optimal parameter and thresholds determined in the “Optimization of MIC calls” experiments described above. A single dilution series (n = 10 images) was omitted from further analyses based on an autofocus failure which compromised curve fitting and MIC determination.
Log2 differences between MAST and BMD MICs were used to evaluate precision and accuracy of MAST. Taking into account the inherent variability in BMD, log2 differences of ±1 were considered to be in essential agreement, and thus equivalent as defined by CLSI guidelines20. Each MAST result was also assigned a categorical interpretation of susceptible, intermediate, or resistant based on CLSI criteria2 and compared to the categorical result from BMD to determine categorical agreement.
Results
The rate-limiting step in traditional AST readout is the threshold for bulk microbial growth detection, either by optical density determination (as in the Vitek2) or human visualization of bacterial growth (in reference AST methods). Therefore, the fastest phenotypic AST readout presumably should be approached by microscopic visualization of the effects of antimicrobials on the replication of individually resolved bacterial cells. The idea of MAST was born on this premise.
The technical requirements for MAST included the need to visualize microorganisms on a solidified microwell surface; the ability to apply doubling dilution series of any antimicrobials desired; the ability to deliver organisms consistently at desired concentrations; and the acquisition and automated classification of images as “growth” or “inhibition” with subsequent MIC calls. Investigation of each MAST component is discussed in turn along with preliminary validation of the end-to-end platform concept.
Preparation of solid microwell surfaces in microtiter plates
For efficient automated microscopic observation, bacteria must be located in a reproducible 3D location within microwells. In traditional liquid media used for AST testing, cells are distributed randomly throughout the volume of fluid and are highly mobile based on Brownian motion or intrinsic motility, and thus unsuitable for single cell imaging. Therefore, we considered use of a solid growth surface containing standard nutrient medium (CAMHB) for image acquisition. Requirements for the solidifying agent included: formation of surfaces in microwells with consistent height; absence of inhibitory effects on bacterial growth; and amenability to pipetting.
We first evaluated standard microbiological agar. However, the solution was difficult to pipette consistently due to rapid and practically irreversible solidification at <40°C. Next, we investigated gellan gum, an anionic polysaccharide alternative to agar;2 polyacrylamide; and poloxamer 407, a hydrophilic, nonionic copolymer, as alternative solidifying agents. Gellan gum supported bacterial growth, but preparation of consistent microwell surfaces was not possible due to phase transition characteristics similar to traditional agar. Polyacrylamide surfaces were exceptionally easy to prepare, but proved inhibitory to bacteria.
By contrast, a solution of 15% poloxamer 407 was identified as an ideal solidifying agent. Aqueous solutions of poloxamer 407 are liquid at 4°C allowing for facile pipetting of solutions kept on ice. Solidification occurs at ~20°C and is thermally reversible, allowing for preparation of plates at room temperature (with poloxamer 407 solution on ice) followed by centrifugation at 4°C to ensure substrate is evenly distributed on the bottom of wells. Prepared plates can be frozen indefinitely with no effect on the integrity of growth surfaces. Further, CAMHB solidified with poloxamer 407 was found to support growth of all common Gram-negative bacterial pathogens tested (data not shown).
Digital dispensing of bacteria
Use of solid surfaces contained within single wells of 384-well plates allows for multiple organism/antibiotic combinations to be tested in a single experiment. However, use of small wells presented a technical challenge in that: (1) organisms must be quantitatively delivered without disturbing the integrity of the growth surface; (2) volumes must be small (nL size) to avoid flooding of the surface that would lead to uneven distribution of organisms and associated requirement for drying; and (3) delivery must be spatially precise to allow efficient positioning of an automated microscope over fields containing organisms. Manual pipetting or pin transfer cannot satisfy these requirements. Therefore, we investigated bacterial cell dispensing using the HP D300 as an alternative method.
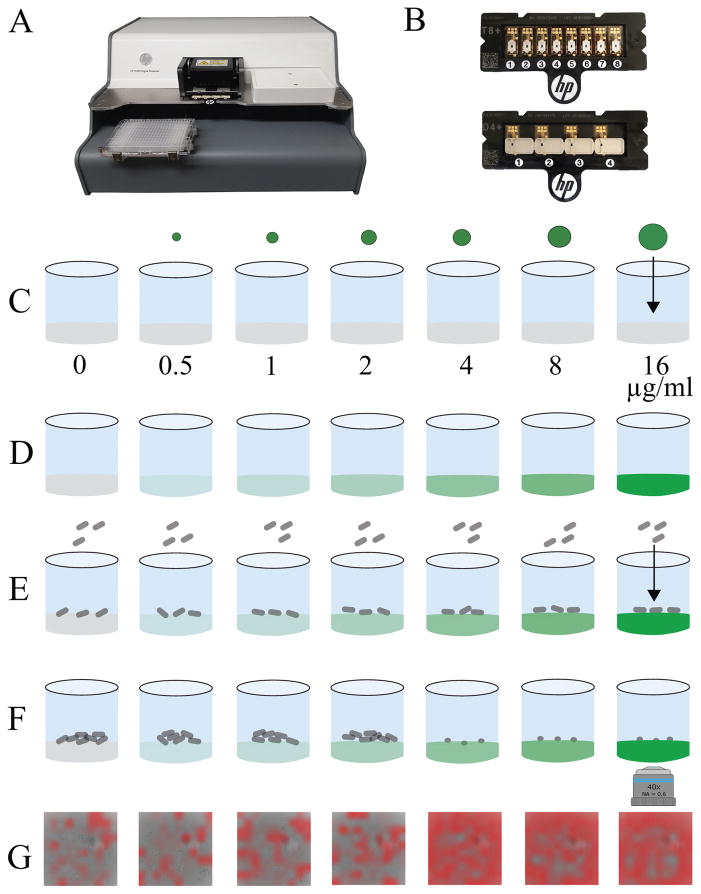
Bacterial cell dispensing is a novel application of inkjet printing technology. Of note, the HP D300 inkjet printer (Fig. 1A) used in our studies was designed to dispense droplet volumes ranging from 11 picoliters to 10 microliters per manufacturer’s specification9 from a single stock solution loaded into a reagent cassette (Fig. 1B). We previously used this technology to prepare doubling dilution series of antimicrobials in 384-well plates in liquid media,10 and in MAST used the same technique to apply antimicrobial dilutions to solid surfaces (Fig. 1C–D). We furthermore hypothesized that we could also use this technology to deliver any desired quantity of microorganism into microwells from a single bacterial suspension by choosing the appropriate droplet dispense size (Fig. 1E).
Figure 1. The microscopy-based antimicrobial susceptibility testing (MAST) assay.
The HP D300 digital dispenser (A) and disposable small volume T8+ (B, top) or large volume D4+ (B, bottom) cassettes are used for antibiotic and cell dispensing. (C) Solid surfaces are prepared in single wells of a 384-well plate using CAMHB solidified with poloxamer 407 (CAMHB-P), and varying size droplets of antimicrobial are digitally dispensed into each well creating (D) a doubling dilution series. (E) Bacteria are dispensed on top of the antibiotic-containing well surfaces and incubated at 35±2°C for 2 hours. (F) Images of cells are collected by automated microscopy and (G) classified as “growth” or “inhibition” using deep learning. Areas defined by the ConvNet as showing inhibition are highlighted with red overlay. The minimal inhibitory concentration (MIC) of antimicrobial is defined as the lowest concentration resulting in bacterial growth inhibition (F, third well from right, corresponding to G, third image from right).
Therefore, here we evaluated the ability of the HP D300 to dispense bacteria using two complementary techniques:
First, we evaluated the ability of the HP D300 to quantitatively deliver bacteria to microwells by measuring the number of cells dispensed in a given volume through CFU determination. We performed this evaluation with 5 organisms representative of the most common Gram-negative pathogens (E. coli, K. pneumoniae, E. cloacae, P. aeruginosa, and A. baumannii) to discern whether unique species-specific physical properties might impact ability of cells to travel through the cassette channel, thus affecting dispense accuracy.
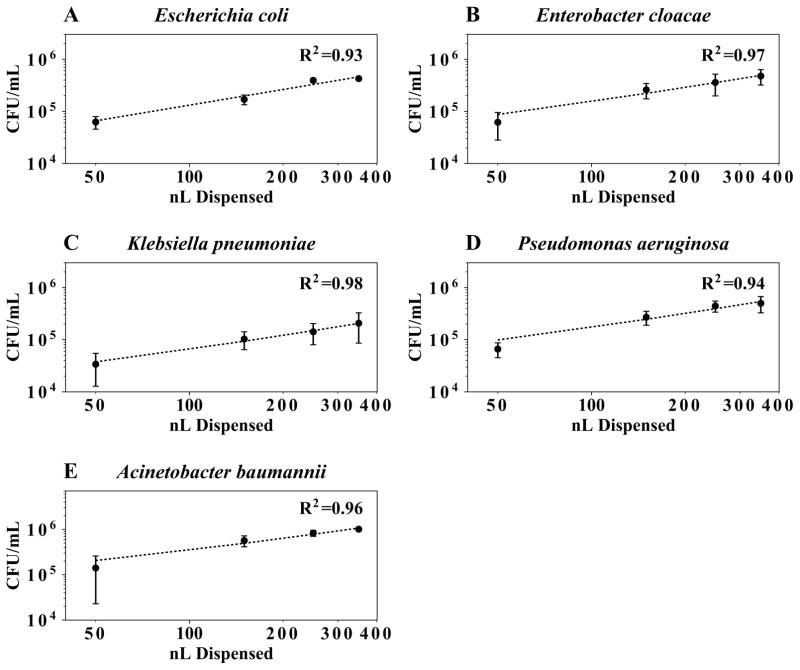
In linear dynamic range studies, we dispensed between 50 and 350 nL of a bacterial suspension and quantified the number of cells delivered to a well. We constructed a standard curve and found that dispense volume was predictive of CFU (average R2 = 0.96), indicating precise and reproducible cell dispensing (Fig. 2). Experiments were repeated on three separate days, and the day-to-day coefficient of variation was, on average, 34%. For each dispense volume, precision was evaluated for three wells spanning a series of 96-wells. No significant difference in bacterial dispense numbers was detected across wells (ANOVA, p > 0.05).
Figure 2. Digital dispensing of bacteria suspensions.
Standardized suspensions of bacteria were dispensed into liquid media using the HP D300. The number of bacteria dispensed was quantified by plate count (A–E). Each point is the mean of three independent experiments with error bars representing one standard deviation from the mean. There was a linear relationship between dispense volume and CFU dispensed for all organisms with an average R2 = 0.96 (dotted lines).
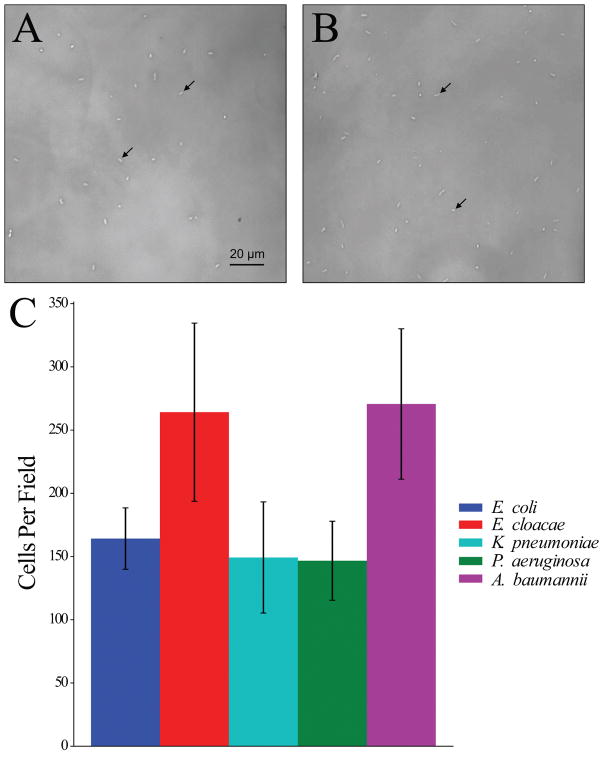
Second, after considering quantitative precision of dispensing, we evaluated spatial precision in a manner relevant to MAST. We dispensed bacteria into the center of wells and counted the number of digitally dispensed bacteria in a single central field using the Cell Observer microscope. This methodology allowed us to simultaneously evaluate spatial precision, quantitative precision, and reproducibility of digital dispensing. We found that 100% of wells imaged contained bacteria in the central field and that cells were well separated and evenly dispersed (See Fig. 3A–B for representative images). The average number of bacteria per field in the single selected central field across wells ranged, on average, from 150 to 260 (1.5 to 2.5 cells per 1000 μm2). The day-to-day coefficient of variation was 23% across all species tested (Figure 3C). There was no significant difference in bacterial dispense numbers across all wells (ANOVA, p > 0.05).
Figure 3. Representative cell densities immediately after digital dispensing.
Standardized suspensions of (A) E. coli ATCC 25922 (average of 164 cells/field or 1.6 cells/1000 μm2) or (B) E. cloacae ATCC 13047 (average of 264 cells/field or 2.58 cells/1000 μm2) were dispensed onto solid microwell surfaces using the HP D300 digital dispensing system and visualized using a Zeiss Cell Observer microscope. Arrows indicate individual cells. (C) Number of cells visible in the central field of 12 representative wells was quantified on 3 separate days. Error bars indicate one standard deviation from the mean.
During these studies, we found that bacteria could be clearly resolved in images using a long working distance 40X objective mounted on an inverted microscope, with brightfield optics, and standard polystyrene 384-well plates. Notably, use of optical quality 384-well plates with thin plastic film bottoms did not visibly improve image resolution of bacteria growing on CAMHB-P surfaces. Further, phase-contrast and differential interference contrast (DIC) imaging of wells surprisingly led to inferior bacterial image resolution, perhaps because of optical properties and the thickness of plastic well bottom and poloxamer separating the objective and bacterial growth surface (total working distance approximately 2.9 mm)
Microscopy-based antimicrobial susceptibility testing (MAST)
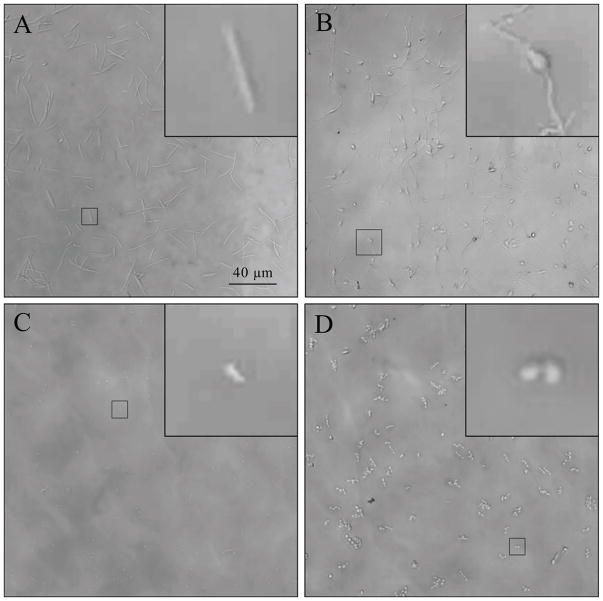
We then combined bacterial and antibiotic dispensing with automated microscopy to perform microscale, "agar dilution" AST assays inside wells of a 384-well plate. Here, bacterial growth was monitored by automated microscopy (Fig. 1F) and images were classified as “growth” or “inhibition” using a machine-learning algorithm to determine the minimal inhibitory concentration. For proof-of-concept studies, we chose to use four different antibiotics with clinical utility against Gram-negative bacteria. Each of these drugs was also specifically chosen based on unique and well-defined alterations in cell morphology at their MIC, all of which were observed using the MAST assay.14–16 Exposure to ciprofloxacin, a fluoroquinolone class, DNA gyrase inhibitor, resulted in elongated cells that failed to divide (Figure 4A). Exposure to cefepime, a cephalosporin β-lactam-based cell wall synthesis inhibitor, resulted in filamented bacteria with a pronounced central bulge (Figure 4B). Gentamicin, an aminoglycoside class protein synthesis inhibitor, blocked cell growth, but did not appreciably alter morphology (Figure 4C). Exposure to meropenem, a carbapenem β-lactam-based cell wall synthesis inhibitor, resulted in rounded spheroplasts (Figure 4D). We presumed that this range of cell morphologies would appropriately challenge automated image classification methods discussed in subsequent sections.
Figure 4. Representative morphologies of inhibited E. coli ATCC 25922.
Antibiotics and E. coli ATCC25922 were dispensed into microwells and automatically imaged with a Zeiss Cell Observer microscope after <4 hour incubation. Panels represent the central field of a microwell containing (A) ciprofloxacin, (B) cefepime, (C) gentamicin, (D) meropenem at the MIC. Insets A–C show close-up views of an individual cell. Inset D shows a close-up of two cells.
Image classification by deep learning
To call MICs, at least one image field must be analyzed per microwell in a doubling dilution series. When testing multiple drugs against multiple organisms with a typical dilution series for each drug including 10 doubling dilutions, manual inspection and classification of the resultant large image sets would be time-prohibitive and inconsistent with future clinical translation. Therefore, we considered use machine-learning for automated classification of MAST images.
Specifically, we examined the ability of a deep convolutional neural network (ConvNet) to classify images and for accurate determination of MIC values following only a two hour incubation period. Importantly, this accelerated time window contrasts with the 16–20 hour incubation period required for reference MIC determinations and similar extended incubation periods required for commercial AST methods. ConvNet programming structure is inspired by the organization of neurons in the visual cortex, and these models have recently achieved human-level performance on large-scale image classification tasks.21, 22 This approach takes a raw image as input and learns to extract relevant features for the particular task at hand. In other words, it does not rely on pre-conceived human biases regarding what features are likely important for optimal discrimination, but rather simultaneously learns which features to extract and how to combine them to achieve accurate classification, based on optimizing its output relative to a labeled training set.
Our network was trained and validated on a set of 3,202 full images (1024x1024 px) collected in three independent experiments in an attempt to capture the totality of biological and technical variability in the MAST assay and produce a model that will generalize to a diverse set of conditions. After training, the network reached a peak classification accuracy of 90% on the held out validation set at the per-image-crop (220x220px) level. After mapping image crops back to whole images, regions representing growth or inhibition were manually inspected and found to correspond to expected growth/no growth morphologies (data not shown).
Optimization of MIC calls
To determine MICs, each image, which corresponds to a specific antimicrobial dilution, must be classified as growth or inhibition. Our image classification algorithm first evaluated 64 non-overlapping crops from each image and determined mean inhibition probability, median inhibition probability, and number of crops with inhibition probability above 0.5. However, it was unknown which of these parameters and what threshold cutoff for each parameter would result in the most accurate MIC calls. Furthermore, we noted that absolute values of these parameters varied by approximately 20% on a day-to-day basis, suggesting the need for an adaptive algorithm that would be robust to these differences. Specifically, effects of biological and technical variability were mitigated by modeling the results for each parameter (median inhibition probability, mean inhibition probability, and number of crops with inhibition probability > 0.5) across each dilution series as a sigmoidal curve.
We collected an optimization dataset for the purpose of determining the best way to evaluate the ConvNet output. We then iteratively evaluated all possible discrimination thresholds normalized to each sigmoid curve for each antibiotic/organism/parameter for correct MIC prediction. Although the ConvNet was trained on images classified based on morphological characteristics of cells, here we evaluated and optimized performance against reference BMD MIC values determined for each dilution series.
Upon optimization of the discriminatory threshold for each parameter for all antibiotic/organism combinations, we identified the proportion of image crops with inhibition probability >0.5 as the optimal parameter for MIC determination. Importantly, we confirmed our prediction that a sigmoid curve would robustly model our data (average R2 = 0.93).
Proof-of-principle, end-to-end testing of MAST Assay
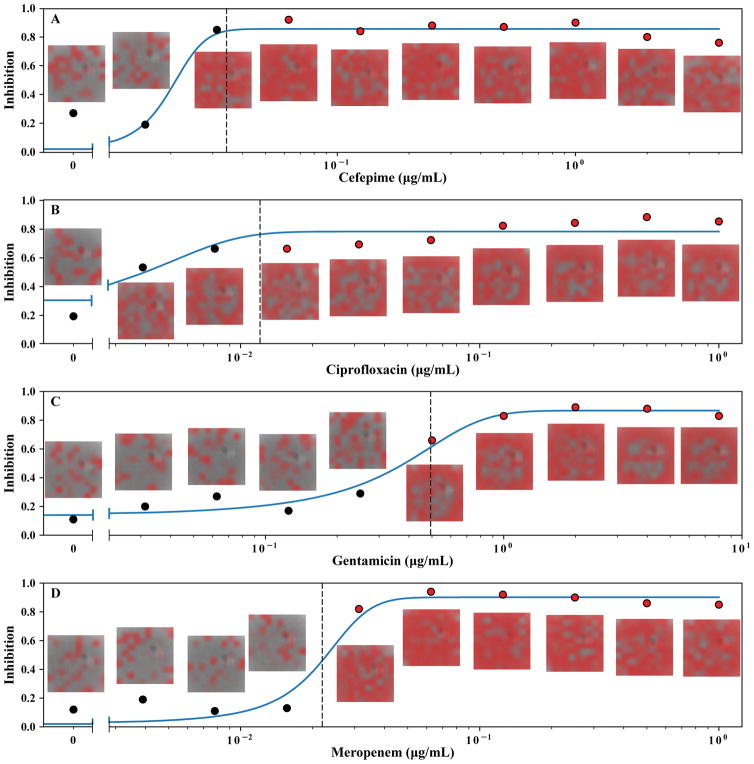
To evaluate the “end-to-end” MAST assay, we collected a large verification dataset which was independent of the training, validation, and optimization datasets. We then compared performance of our optimized MAST assay (the test method), which included only a two-hour growth incubation, to BMD (the reference method), which used a standard 16–20 hour growth incubation. Sigmoidal curve fitting was equally robust in this phase across all antimicrobial/organism combinations (average R2 = 0.91, Fig. 5).
Figure 5. Graphical representation of MAST MIC assay output for E. coli ATCC 25922.
Each point in (A–D) represents ConvNet output (fraction of image crops with inhibition probability >0.5) from the adjacent image at the indicated antibiotic concentration. Red overlay in images indicates areas where the ConvNet algorithm detected bacterial inhibition. Solid blue line represents a sigmoid fit to the ConvNet data. Dashed line represents the threshold delineating growth (black points) and inhibition (red points).
Notably, MAST MIC essential agreement (±1 two-fold dilution) was 95.8% compared to BMD. 99.4% of MIC results were within ±2 two-fold dilutions of modal reference BMD results (Table 1). MIC determinations for each antibiotic considered separately were similarly accurate and ranged from 93.3–100%. Notably, a 100% MIC accuracy was found for cefepime despite this antibiotic presenting a substantial challenge to the classification algorithm due to filamentation of cells near the MIC (Fig. 4B). Furthermore, categorical agreement (interpretation of organisms as susceptible, intermediate, or resistant) for the susceptible strain set between MAST and BMD was 98.3% (Table 1), indicating a very low level of false resistance (major errors).
Table 1.
Evaluation of MAST Accuracy.
| Antimicrobial | Number of measurements with Log2 difference from reference MICa | Essential Agreement % (CI) | Categorical Agreement % (CI) | |||||
|---|---|---|---|---|---|---|---|---|
|
| ||||||||
| −2 | −1 | 0 | 1 | 2 | 3 | |||
| Ciprofloxacin | 0 | 7 | 19 | 16 | 1 | 1 | 95.5 (84.5–98.7) | 100 (90.6–100) |
| Cefepime | 0 | 8 | 19 | 6 | 0 | 0 | 100 (89.3–100) | 93.3 (84.3–99.4) |
| Gentamicin | 3 | 9 | 16 | 17 | 0 | 0 | 93.3 (81.4–97.8) | 100 (92–100) |
| Meropenem | 1 | 7 | 14 | 21 | 1 | 0 | 95.5 (84.5–98.7) | 100 (92.1–100) |
|
| ||||||||
| Total n (%)b | 4 (2.4) | 31 (18.7) | 68 (41) | 60 (36.1) | 2 (1.2) | 1 (0.6) | 95.8 (91.5–98) | 98.3 (95.2–99.4) |
Reference MIC derived from broth microdilution
CI = 95% confidence interval
Discussion
Extensive antimicrobial resistance amongst Gram-negative pathogens reduces reliability of empiric antimicrobial therapy, underscoring the critical importance of timely AST in the clinical microbiology laboratory to guide therapy. Importantly, delay in institution of active therapy is correlated with increased mortality during serious infection.8 However, laboratory tests in current use require 8–24 hours for results.
Rapid AST systems are a potential solution to address this issue and may be approached broadly through two pathways: phenotypic testing or genotypic testing. Genotypic assays call resistance based on the presence of specific resistance elements. However, such assays are limited to evaluating known resistance determinants and thus are insensitive to novel mechanisms. Further, they are typically unable to determine exact MICs and thereby direct therapy based on known pharmacodynamic relationships. Therefore genotypic methods lack sensitivity and specificity.
In contrast, rapid phenotypic assays have the capability of determining exact MICs. Furthermore, well-established and validated pharmacodynamic relationships between drug exposure in vivo and the MIC are predictive of response to therapy.23 Accordingly, CLSI guidelines recognize phenotypic testing as the current methodology for determining susceptibility.2 For Gram-negative organisms in particular, guidelines have moved away from detecting or inferring the presence of specific resistance elements to guide therapy.24
Here, we developed and performed preliminary validation of a rapid phenotypic AST method based on observation of individual cells. Microscopic AST methods present logistical and technical challenges, one of which is the need to immobilize cells for observation. In most existing rapid AST instruments, this is accomplished by suspension of cells in a matrix or restriction of cells to a microfluidic channel.25 These procedures are complex and have no predicate in current clinical microbiology practice.
In contrast, the MAST assay immobilizes bacteria by dispensing cells on a solid microwell surfaces analogous to 1000-fold miniaturized agar plates, and in essence, replicates the existing agar dilution reference method.1 Using inkjet technology, inoculation of an entire assay plate (up to 240 wells) can be accomplished quickly (<1 minute) in an operator-independent manner with a single pipetting step. The steps required for MAST assay performance (including preparation of bacterial suspensions) are similar to those for currently used AST systems and could be performed by a technologist without additional training.
Clinical automated AST systems typically include only a fixed and limited set of antimicrobials; furthermore, only a limited number of antimicrobial dilutions are tested per drug.3 In contrast, MAST allows preparation of plates dynamically using any number of antimicrobials with dilution series of any size in a high-density 384-well format. Similar to bacterial dispensing, an antimicrobial doubling dilution series can be prepared very quickly (<10 seconds) with a single pipetting step to load antimicrobial stock solution into a dispensing cassette as we described previously.10 Flexibility to prepare any doubling dilution series at will allows for testing antimicrobials at concentrations relevant to multiple species that may have different breakpoints on a single microplate.2 Concentrations can easily be tailored to include changes in breakpoints that are recommended by agencies such as CLSI to accommodate evolving understanding of the relationships between MIC and clinical outcome.4 Ability to test antimicrobials at low concentrations ensures that standard quality control organism MIC ranges are on-scale so that assay performance can be appropriately monitored, something that is often not possible with current clinical systems which only include dilution series bracketing clinical breakpoints.2, 5
The large amount of image data generated by MAST led to the need for automated interpretation capabilities. Therefore, we developed a machine learning-based image classification algorithm. In this first stage conception of MAST, a ConvNet was trained based on human classification of a large set of training images collected after 2 hours of incubation. Importantly, images were collected from five organisms exposed to varying concentrations of four antibiotics over three separate days, such that the training set captured the biological and technical variability that might be seen in a clinical setting.
Ultimately, our image analysis pipeline contained three levels of image interpretation: (1) ConvNet image classification which returned per-image-crop probability of inhibition, a feature that in itself predicted growth status in image crops with > 90% accuracy. However, this output did not make a direct prediction regarding whole images. (2) Results from all image crops for each image were therefore pooled to provide image statistics (mean inhibition probability, median inhibition probability, and proportion of image crops with inhibition > 0.5). (3) Data from all images in a dilution series were then modeled as a sigmoid curve and classified based on thresholds that optimized the accuracy of MIC calls.
This optimized algorithm was then applied to a new set of images and accuracy compared to the broth microdilution MIC. Importantly, accuracy met essential agreement metrics established by the FDA26 and standards in the field for evaluating new antimicrobial susceptibility testing methodology in comparison to the BMD reference methodology.27 Specifically, the essential agreement was 95.8% (±1 dilution from the reference MIC), exceeding the recommended 95% essential agreement threshold.
In this preliminary analysis, a susceptible strain set was tested. Even so, it is notable that we found only 3 interpretive errors. All of these interpretive errors were found with cefepime and P. aeruginosa ATCC 27853, which has a reference MIC only one doubling dilution below the categorical breakpoint2. Moreover, the method appeared to perform equally well testing Escherichia coli, Enterobacter cloacae, Klebsiella pneumoniae, P. aeruginosa and A. baumannii, constituting the overwhelming majority of Gram-negative pathogens causing human infection. These organisms are also often associated with problematic drug resistance and therefore of high relevance for accelerated AST testing.28 Importantly, although our image analysis pipeline is not optimized for processing speed, a single 10-step doubling dilution series can still be evaluated in as little as 70 seconds through use of a dedicated graphics processing unit (GPU), allowing rapid image classification. Automated MIC calls based on sigmoidal modeling of ConvNet output are essentially instantaneous.
In a first generation MAST platform, we were able to achieve a high degree of accuracy after only two hours incubation. Nevertheless, classification performance could potentially be improved even further through several measures: (1) further training targeted to individual species or antimicrobials. Specifically, we suspect that the several different morphologies associated with both species and antimicrobial effects may have marginally compromised accuracy. In the future, species-specific models could be trained as many clinical laboratories now have systems such as MALDI-TOF which allow speciation of organisms within minutes. (2) Shifts up or down one doubling dilution of MAST modal MIC’s compared with BMD may relate to inherent differences observed in other contexts between solid and broth assays (i.e., reference agar dilution versus broth microdilution tests).1 (3) Use of a two hour incubation was a high bar, and it is likely that even without further modification MAST could achieve higher essential agreement if incubation periods were marginally longer. (4) Lastly, our human classification of the training data set was labor intensive and may not have been consistently accurate over the large numbers of images examined. Therefore, the underlying ConvNet accuracy could likely be improved through training based on modal reference MIC for each isolate-antimicrobial combination rather than visual classification of each individual image. This would reduce time and potential error inherent to human subjective interpretation and allow for use of much larger training datasets
Notably, typical rapid AST systems require custom-made consumables.25 In contrast, the MAST assay relies entirely on off-the-shelf consumables including standard polystyrene 384-well plates, poloxamer 407, and antibiotic powder. When fully utilized, a MAST assay plate can accommodate testing of 4 pathogens against 6 antibiotics each, resulting in a consumable cost of <$3.50 per antibiotic/organism combination (inclusive of the D300 cassette, microtiter plate, poloxamer 407, and antibiotic) based on list price. Furthermore, all required instrumentation is commercially available. Importantly, MAST utilizes only a subset of features of the HP D300 and Zeiss Cell Observer microscope. Indeed for the latter, the only features used were standard light microscopy optics, a long working distance 40X lens, and a mechanical stage. Therefore, instrumentation could conceivably be simplified to further reduce costs during development of a next generation platform contained within a single instrument.
Here, we demonstrate proof of principle for a flexible AST platform that allows highly accurate MIC determination after only a 2-hour incubation. Importantly, the system allows determination of AST for any concentration of any antimicrobial at will in an operator-independent manner. This flexibility extends to incorporation of new antimicrobials that may not be available in commercial panels for years yet will likely be required to treat clinically emerging multidrug-resistant pathogens. Further, MAST is a true MIC methodology that allows all relevant concentrations of antimicrobial to be tested without the need for MIC extrapolations that may not work reliably for resistant pathogens.29 Taken together, proof-of-principle was obtained for the MAST platform that will address a critical unmet need for flexible, extremely rapid AST diagnostics.
Acknowledgments
This work was conducted with support from Harvard Catalyst | The Harvard Clinical and Translational Science Center (National Center for Research Resources and the National Center for Advancing Translational Sciences, National Institutes of Health Award UL1 TR001102) and financial contributions from Harvard University and its affiliated academic healthcare centers. Thea Brennan-Krohn was supported by the Eunice Kennedy Shriver National Institute of Child Health and Human Development of the National Institutes of Health pediatric infectious diseases research training grant, T32HD055148. The content is solely the responsibility of the authors and does not necessarily represent the official views of Harvard Catalyst, Harvard University and its affiliated academic healthcare centers, or the National Institutes of Health.
We thank the Harvard Center for Biological Imaging for infrastructure (Cell Observer Microscope) and technical support. J.E.K. was provided with an HP D300 digital dispenser and associated consumables for use in his research laboratory, and received an honoraria and travel support from Tecan, Inc. (Morrisville, NC). The Kirby laboratory also received HP D300 consumables from HP Inc. Tecan and HP had no role in study design, data collection/interpretation, manuscript preparation, or decision to publish.
References
- 1.Clinical and Laboratory Standards Institute. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically; approved standard - tenth edition. CLSI document M07-A10. Clinical and Laboratory Standards Institute; Wayne, PA: 2015. [Google Scholar]
- 2.Clinical and Laboratory Standards Institute. Performance standards for antimicrobial susceptibility testing; twenty-seventh informational supplement. Clinical and Laboratory Standards Institute; Wayne, PA: 2017. CLSI document M100–S27. [Google Scholar]
- 3.Jorgensen JH, Ferraro MJ. Antimicrobial susceptibility testing: a review of general principles and contemporary practices. Clin Infect Dis. 2009;49(11):1749–55. doi: 10.1086/647952. [DOI] [PubMed] [Google Scholar]
- 4.Humphries RM, Hindler JA. Emerging Resistance, New Antimicrobial Agents … but No Tests! The Challenge of Antimicrobial Susceptibility Testing in the Current US Regulatory Landscape. Clin Infect Dis. 2016;63(1):83–8. doi: 10.1093/cid/ciw201. [DOI] [PubMed] [Google Scholar]
- 5.Smith KP, Brennan-Krohn T, Weir S, et al. Improved Accuracy of Cefepime Susceptibility Testing for Extended-Spectrum-Beta-Lactamase-Producing Enterobacteriaceae with an On-Demand Digital Dispensing Method. J Clin Microbiol. 2017;55(2):470–478. doi: 10.1128/JCM.02128-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Flynt LK, Veve MP, Samuel LP, et al. Comparison of Etest to Broth Microdilution for Testing of Susceptibility of Pseudomonas aeruginosa to Ceftolozane-Tazobactam. J Clin Microbiol. 2017;55(1):334–335. doi: 10.1128/JCM.01920-16. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 7.Brennan-Krohn T, Smith KP, Kirby JE. The Poisoned Well: Enhancing the predictive value of antimicrobial susceptibility testing in the era of multidrug-resistance. J Clin Microbiol. 2017 May 3; doi: 10.1128/JCM.00511-17. pii: JCM.00511–17. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schwaber MJ, Carmeli Y. Mortality and delay in effective therapy associated with extended-spectrum beta-lactamase production in Enterobacteriaceae bacteraemia: a systematic review and meta-analysis. J Antimicrob Chemother. 2007;60(5):913–20. doi: 10.1093/jac/dkm318. [DOI] [PubMed] [Google Scholar]
- 9.Tecan Inc. [Accessed April 26, 2017];Tecan D300e Digital Dispenser - Specification. http://ww3.tecan.com/mandant/files/doc/526/BR_Tecan_D300e_Specifications_399178_V1-0.pdf.
- 10.Smith KP, Kirby JE. Verification of an Automated, Digital Dispensing Platform for At-Will Broth Microdilution-Based Antimicrobial Susceptibility Testing. J Clin Microbiol. 2016;54(9):2288–93. doi: 10.1128/JCM.00932-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hunter JD. Matplotlib: A 2D graphics environment. Computing in Science and Engineering. 2007;9(3):90–95. [Google Scholar]
- 12.van der Walt S, Colbert SC, Varoquaux G. The NumPy Array: A Structure for Efficient Numerical Computation. Computing in Science and Engineering. 2011;13(2):22–30. [Google Scholar]
- 13.Schindelin J, Rueden CT, Hiner MC, et al. The ImageJ ecosystem: An open platform for biomedical image analysis. Mol Reprod Dev. 2015;82(7–8):518–29. doi: 10.1002/mrd.22489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pucci MJ, Boice-Sowek J, Kessler RE, et al. Comparison of cefepime, cefpirome, and cefaclidine binding affinities for penicillin-binding proteins in Escherichia coli K-12 and Pseudomonas aeruginosa SC8329. Antimicrob Agents Chemother. 1991;35(11):2312–7. doi: 10.1128/aac.35.11.2312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mason DJ, Power EG, Talsania H, et al. Antibacterial action of ciprofloxacin. Antimicrob Agents Chemother. 1995;39(12):2752–8. doi: 10.1128/aac.39.12.2752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Horii T, Kobayashi M, Sato K, et al. An in-vitro study of carbapenem-induced morphological changes and endotoxin release in clinical isolates of gram-negative bacilli. J Antimicrob Chemother. 1998;41(4):435–42. doi: 10.1093/jac/41.4.435. [DOI] [PubMed] [Google Scholar]
- 17.Simonyan K, Zisserman A. Very deep convolutional networks for large-scale image recognition. Conference paper at ICLR 2015. 2015 https://arxiv.org/pdf/1409.1556v6.pdf.
- 18.Ioffe S, Szegedy C. Batch Normalization: Accelerating Deep Network Training by Reducing Internal Covariate Shift. Proceedings of the 32nd International Conference on Machine Learning, PMLR. 2015;37:448–456. [Google Scholar]
- 19.Glorot X, Bengio Y. Understanding the difficulty of training deep feedforward neural networks. Proceedings of the 13th International Conference on Artificial Intelligence and Statistics(AISTATS) 2010;9:249–256. [Google Scholar]
- 20.Clinical and Laboratory Standards Institute. Verification of Commercial Microbial Identification and Antimicrobial Susceptibility Testing Systems. CLSI document M52. Clinical and Laboratory Standards Institute; Wayne, PA: 2015. [Google Scholar]
- 21.He K, Zhang X, Ren S, Sun J. Delving Deep into Rectifiers: Surpassing Human-Level Performance on ImageNet Classification. Proceedings of the International Conference on Computer Vision; 2015; pp. 1026–1034. [Google Scholar]
- 22.LeCun Y, Bengio Y, Hinton G. Deep learning. Nature. 2015;521(7553):436–44. doi: 10.1038/nature14539. [DOI] [PubMed] [Google Scholar]
- 23.Onufrak NJ, Forrest A, Gonzalez D. Pharmacokinetic and Pharmacodynamic Principles of Anti-infective Dosing. Clin Ther. 2016;38(9):1930–47. doi: 10.1016/j.clinthera.2016.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Clinical and Laboratory Standards Institute. Performance standards for antimicrobial susceptibility testing; nineteenth informational supplement. CLSI document M100-S19. Clinical and Laboratory Standards Institute; Wayne, PA: 2009. [Google Scholar]
- 25.Pulido MR, Garcia-Quintanilla M, Martin-Pena R, et al. Progress on the development of rapid methods for antimicrobial susceptibility testing. J Antimicrob Chemother. 2013;68(12):2710–7. doi: 10.1093/jac/dkt253. [DOI] [PubMed] [Google Scholar]
- 26.Food and Drug Administration. Guidance for Industry and FDA. Class II Special Controls Guidance Document: Antimicrobial Susceptibility Test (AST) Systems. Food and Drug Administration; Rockville, MD: 2009. [Google Scholar]
- 27.Clark RB, Lewinski MA, Loeffelholz MJ, et al. Cumulative Techniques and Procedures in Clinical Microbiology 31A: Verification and Validation of Procedures in the Clinical Microbiology Laboratory. American Society for Microbiology; Washington, DC: 2009. [Google Scholar]
- 28.Boucher HW, Talbot GH, Bradley JS, et al. Bad bugs, no drugs: no ESKAPE! An update from the Infectious Diseases Society of America. Clin Infect Dis. 2009;48(1):1–12. doi: 10.1086/595011. [DOI] [PubMed] [Google Scholar]
- 29.Smith KP, Kirby JE. How inkjet printing technology can defeat multidrug-resistant pathogens. Future Microbiol. 2016;11:1375–1377. doi: 10.2217/fmb-2016-0163. [DOI] [PubMed] [Google Scholar]