Abstract
Avermectins are commercially important anthelmintic antibiotics produced by Streptomyces avermitilis. The homologous TetR-family transcriptional regulators AvaR1 and AvaR2 in this species were identified previously as receptors of avenolide, a novel butenolide-type autoregulator signal required for triggering avermectin biosynthesis. AvaR2 was found to be an important pleiotropic regulator in repression of avermectin and avenolide production and cell growth, whereas the regulatory role of AvaR1 remains unclear. Investigation of AvaR1 function in the present study showed that it had no effect on cell growth or morphological differentiation, but inhibited avenolide and avermectin production mainly through direct repression of aco (the key enzyme gene for avenolide biosynthesis) and aveR (the cluster-situated activator gene). AvaR1 also directly repressed its own gene (avaR1) and two adjacent homologous genes (avaR2 and avaR3). Binding sites of AvaR1 on these five target promoter regions completely overlapped those of AvaR2, leading to the same consensus binding motif. However, AvaR1 and AvaR2 had both common and exclusive target genes, indicating that they cross-regulate diverse physiological processes. Ten novel identified AvaR1 targets are involved in primary metabolism, stress responses, ribosomal protein synthesis, and cyclic nucleotide degration, reflecting a pleiotropic role of AvaR1. Competitive EMSAs and GST pull-down assays showed that AvaR1 and AvaR2 competed for the same binding regions, and could form a heterodimer and homodimers, suggesting that AvaR1 and AvaR2 compete and cooperate to regulate their common target genes. These findings provide a more comprehensive picture of the cellular responses mediated by AvaR1 and AvaR2 regulatory networks in S. avermitilis.
Keywords: Streptomyces avermitilis, avermectins, AvaR1, AvaR2, avenolide
Introduction
Streptomycetes have the useful capability of producing a variety of antibiotics that have broad applications in medicine and agriculture. Initiation of antibiotic biosynthesis is often accompanied by morphological development and triggered by low-molecular-weight signaling molecules (termed autoregulators) at nanomolar concentrations (Bibb, 2005). Effects of autoregulator signals are transmitted by their cognate receptor proteins, which are usually members of the TetR-family transcriptional regulators. Interaction of an autoregulator with its receptor alters the receptor's DNA-binding activity, leading to derepression of target genes involved in antibiotic biosynthesis and/or morphological differentiation (Folcher et al., 2001; Willey and Gaskell, 2011).
Twenty-four autoregulators have been described to date in 12 Streptomyces species, and have been classified into five groups. The three major groups are the γ-butyrolactones (GBLs), furans, and γ-butenolides. Each of the other two groups has a single member: N-methylphenylalanyl-dehydrobutyrine diketopiperazine and PI factor [2,3-diamino-2,3-bis (hydroxymethyl)-1,4-butanediol], respectively (Niu et al., 2016). The largest group is the GBLs, whose 14 members share a 2,3-disubstituted-GBL skeleton and differ in regard to stereochemistry, chain length, and branching of C2 fatty acid side chains (Choi et al., 2003; Takano, 2006). The first-described and best-studied GBL is A-factor from Streptomyces griseus. It binds to receptor ArpA to initiate streptomycin production and sporulation (Horinouchi and Beppu, 2007). Many Streptomyces genomes contain multiple genes encoding ArpA-like GBL receptor homologs, e.g., ScbR, ScbR2, CprA, and CprB in model species S. coelicolor; BarA and BarB in S. virginiae; JadR2 and JadR3 in S. venezuelae (Niu et al., 2016). The identified GBL/receptor pairs are SCBs/ScbR (controlling SCB and coelimycin biosynthesis) (Takano et al., 2001, 2005; Gomez-Escribano et al., 2012), VBs/BarA (controlling virginiamycin biosynthesis) (Nakano et al., 1998), and SVB1/JadR3 (controlling jadomycin biosynthesis) (Zou et al., 2014). ScbR2 and JadR2 are designated “pseudo” GBL receptors because they do not recognize GBLs; rather, they bind and respond to antibiotics as ligands to coordinate antibiotic biosynthesis (Xu et al., 2010; Wang W. et al., 2014). CprA stimulates both antibiotic production and sporulation in S. coelicolor, whereas CprB depresses antibiotic production (Onaka et al., 1998). BarB controls an early process of virginiamycin biosynthesis (Matsuno et al., 2004). The signals recognized by CprA, CprB, and BarB remain to be clarified.
The important industrial species Streptomyces avermitilis produces avermectins, which are efficient, broad-spectrum anthelmintic agents (Burg et al., 1979; Egerton et al., 1979). AveR, the only ave cluster-situated regulator (CSR), is essential for activation of ave structural genes (Guo et al., 2010). A novel γ-butenolide-type autoregulator, termed avenolide, was found to function as a signal eliciting avermectin biosynthesis at concentration 4 nM, whereas GBLs had no such effect (Kitani et al., 2011). Two butenolides (SRB1, SRB2) from S. rochei were subsequently identified as autoregulators that trigger lankacidin and lankamycin production (Arakawa et al., 2012). The pathway for avenolide biosynthesis remains to be fully elucidated, but has been shown to require aco (sav_3706) and cyp17 (sav_3704), which respectively encode an acyl-CoA oxidase and a cytochrome P450 hydroxylase (Kitani et al., 2011). aco/cyp17 homologs have been found in other species, including S. fradiae, S. ghanaensis, and S. griseoauranticus. Thus, butenolide-type autoregulators appear to be widely distributed among Streptomyces species, and it is of interest to elucidate their signaling cascades.
Streptomyces avermitilis contains an avaR (S. avermitilis autoregulator receptors) locus that includes avaR1 (sav_3705), avaR2 (sav_3702), and avaR3 (sav_3703). AvaR3 has an extra 75-amino acid stretch that is not present in typical GBL receptors, and promotes avermectin production through a yet-unknown regulatory mechanism (Miyamoto et al., 2011). AvaR2 is a homolog of pseudo GBL receptors ScbR2 and JadR2. Recent studies in our lab have clarified the roles of AvaR2. It is an important pleiotropic regulator of cell growth, secondary metabolism, primary metabolism, ribosomal protein synthesis, and stress responses. It inhibits avermectin and avenolide production mainly through direct repression of aveR and aco. Unlike ScbR2 and JadR2, AvaR2 binds, and responds to not only endogenous avenolide autoregulator, but also exogenous antibiotic signals that modulate its DNA-binding activity (Zhu et al., 2016). AvaR1 is a close homolog of genuine GBL receptors ArpA and ScbR. Studies by Nihira's group showed that AvaR1 acts as an avenolide receptor and binds specifically to the aco promoter (Kitani et al., 2011). Disruption of avaR1 in the wild-type (WT) strain KA320 increased avenolide production, but had no effect on avermectin production (Sultan et al., 2016). On the other hand, Tang's group reported that avaR1 deletion increased avermectin production in an avermectin high-producing strain, but had no effect on aco expression, and that AvaR1 did not bind to the promoter region of aveR (Wang J. et al., 2014). These contrasting findings for AvaR1, and our observation that AvaR2 also fuctions as an avenolide receptor, illustrate the need to further elucidate the roles of AvaR1 and its relationship with AvaR2 in S. avermitilis.
Results of in vitro and in vivo experiments described here clearly indicate that AvaR1 directly represses expression of aveR and aco, and thereby inhibits production of avermectin and avenolide. AvaR1 also plays a pleiotropic role in primary metabolism, ribosomal protein synthesis, stress responses, and other physiological processes. AvaR1 and AvaR2 have both common and differing target genes, and presumably cross-regulate diverse physiological processes. When AvaR1 and AvaR2 co-exist, they compete and cooperate on the same binding site in both homodimer and heterodimer form. On the basis of our findings, we propose a model in which AvaR1 and AvaR2 coordinate avermectin and avenolide biosynthesis in response to avenolide signal.
Materials and methods
Plasmids, strains, and growth conditions
Plasmids and strains used or constructed in the present study are listed in Table 1. S. avermitilis WT strain ATCC31267 was used as original host for gene manipulations. Culture conditions of S. avermitilis strains for avermectin production, sporulation, phenotype observation, mycelial growth, and protoplast regeneration were as described previously (Liu et al., 2015).
Table 1.
Strains and plasmids used in this study.
| Strain or plasmid | Description | Source or reference |
|---|---|---|
| S. avermitilis | ||
| ATCC31267 | Wild-type (WT) strain | Laboratory stock |
| ΔavaR1 | avaR1 deletion mutant | This study |
| CavaR1 | avaR1 complemented strain | This study |
| OavaR1 | avaR1 overexpression strain | This study |
| ΔavaR1/avaR1-3FLAG | avaR1 complemented strain with AvaR1-3FLAG | This study |
| WT/pKC1139 | WT strain containing control vector pKC1139 | This study |
| WT/pSET152 | WT strain containing control vector pSET152 | This study |
| ΔavaR2 | avaR2 deletion mutant | Zhu et al., 2016 |
| ΔavaR1R2 | avaR1 avaR2 double deletion mutant | This study |
| E. coli | ||
| JM109 | Routine cloning host and host for lux reporter system | Laboratory stock |
| ET12567 | Non-methylating strain | MacNeil and Klapko, 1987 |
| BL21 (DE3) | Protein overexpression host | Novagen |
| Plasmids | ||
| pKC1139 | Multiple-copy, temperature-sensitive E. coli-Streptomyces shuttle vector | Bierman et al., 1992 |
| pSET152 | Integrative E. coli-Streptomyces shuttle vector | Bierman et al., 1992 |
| pET-28a (+) | His6-tagged protein expression vector | Novagen |
| pGEX-4T-1 | GST-tagged protein expression vector | GE Healthcare |
| pJL117 | Vector carrying ermE*p (Streptomyces strong constitutive promoter) | Li et al., 2010 |
| pΔavaR1 | avaR1 deletion vector based on pKC1139 | This study |
| pKC1139-ermp-avaR1 | avaR1 overexpression vector based on pKC1139 | This study |
| pSET152-avaR1 | avaR1 complemented vector based on pSET152 | This study |
| pET28-avaR1 | His6-AvaR1 expression vector based on pET-28a (+) | Zhu et al., 2016 |
| pIJ10500 | Vector carrying 3×flag fragment | Pullan et al., 2011 |
| pSET152-avaR1-3FLAG | avaR1 complemented vector with avaR1-3×flag on pSET152 | This study |
| pCS26-Pac | Vector carrying promoterless lux reporter | Tahlan et al., 2007 |
| pOaveRlux | pCS26-Pac carrying aveR promoter-controlled lux reporter | Zhu et al., 2016 |
| pACYC184 | Protein expression vector in reporter system | Tahlan et al., 2007 |
| pAvaR1 | AvaR1 expression vector in reporter system | This study |
| pGEX-avaR2 | GST-AvaR2 expression vector based on pGEX-4T-1 | This study |
| pET28-avaR2 | His6-AvaR2 expression vector based on pET-28a (+) | Zhu et al., 2016 |
Escherichia coli JM109 was used for DNA cloning. E. coli ET12567 (MacNeil and Klapko, 1987) was used to generate non-methylated plasmids for transformation into S. avermitilis. Antibiotics were added as described previously (Zhao et al., 2007).
Construction of S. avermitilis mutants
To construct an avaR1 gene deletion mutant, two fragments flanking avaR1 were amplified by PCR using WT genomic DNA as template. A 379-bp 5′ flanking region (positions −336 to +43 relative to the avaR1 start codon) was amplified with primers ZJY107 and ZJY108, and a 368-bp 3′ flanking region (positions +645 to +1012) was amplified with primers ZJY109 and ZJY110. The two fragments were assembled by fusion PCR with primers ZJY107 and ZJY110 and cloned into pKC1139 to generate avaR1-deletion vector pΔavaR1, which was introduced into WT protoplasts. The desired avaR1-deleted mutant, termed ΔavaR1, was selected as described previously (Zhao et al., 2007), and confirmed by PCR with primer pairs ZJY115/ZJY116 (flanking the exchange regions) and ZJY117/ZJY118 (located within the deletion region) (Figure S1), followed by DNA sequencing.
For complementation of ΔavaR1, a 921-bp DNA fragment carrying the avaR1 open reading frame (ORF) and its native promoter was obtained by PCR using primers ZJY111 and ZJY112. The PCR product was ligated into pSET152 to generate avaR1-complemented vector pSET152-avaR1, which was then introduced into ΔavaR1 to obtain complemented strain CavaR1. For overexpression of avaR1, a 708-bp fragment containing the avaR1 ORF and a 195-bp fragment containing promoter ermE*p from pJL117 were ligated simultaneously into pKC1139 to produce AvaR1 overexpression vector pKC1139-ermp-avaR1, which was introduced into WT strain to construct avaR1 overexpression strain OavaR1.
To express 3×flag-tagged AvaR1 in S. avermitilis, the avaR1 gene carrying its own promoter was amplified with primers ZJY121 and ZJY122, and 3×flag fragment was amplified from pIJ10500 with primers ZJY45 and ZJY46. The resulting 993-bp avaR1 gene and the 87-bp FLAG fragment were ligated simultaneously into pSET152 to generate pSET152-avaR1-3FLAG, which was transformed into ΔavaR1 to obtain recombinant strain ΔavaR1/avaR1-3FLAG for expression of C-terminally 3×flag-tagged AvaR1. All primers used are listed in Table S1.
To construct an avaR1 avaR2 double deletion mutant, vector pΔavaR1 was transformed into protoplasts of avaR2 deletion mutant ΔavaR2 (Zhu et al., 2016). The desired mutant, ΔavaR1R2, was isolated by selection of the ΔavaR1 mutant.
Analysis of avermectin production and cell growth
Fermentation of S. avermitilis strains, and HPLC conditions for quantitative analysis of avermectin yield, were as described previously (Chen et al., 2007). For cell growth determination, mycelia from 50 mL cell cultures grown in soluble fermentation medium FM-II (Guo et al., 2010) were collected by centrifugation, dried at 80°C to constant weight, and weighed.
Quantitative real-time RT-PCR (qRT-PCR) analysis
Mycelia of S. avermitilis WT, ΔavaR1, and ΔavaR2 grown in FM-I fermentation medium (Chen et al., 2007) were harvested at various times for RNA isolation. Extraction of total RNAs, synthesis of cDNAs, and qRT-PCR analysis of transcription levels of various genes using the primers listed in Table S1 were performed as described previously (Luo et al., 2014). Housekeeping gene 16S rRNA from WT was used as internal control. Gene expression was determined in triplicate.
Western blotting
Total protein of ΔavaR1/avaR1-3FLAG was prepared from cultures grown in FM-I for various durations. Western blotting was performed as described previously (Guo et al., 2010). Mouse ANTI-FLAG mAb (M2; Sigma, USA) was used at ratio 1:5,000.
Overexpression and purification of GST-AvaR2
To prepare GST-AvaR2 protein, the 657-bp avaR2 coding region was amplified using primers ZJY135 and ZJY136. The PCR product was digested with BamHI/EcoRI and cloned into expression vector pGEX-4T-1 to generate pGEX-avaR2, which was confirmed by DNA sequencing, and then transformed into E. coli BL21 (DE3) for overexpression of GST-tagged recombinant protein. Cells were induced with IPTG, and those containing GST-AvaR2 protein were collected, resuspended in lysis buffer (Luo et al., 2014), disrupted by sonication on ice, and centrifuged. Soluble GST-AvaR2 present in supernatant was purified with glutathione-sepharose beads (CWBIO, China) according to the manufacturer's protocol, and stored at −80°C.
Electrophoretic mobility shift assays (EMSAs)
DNA probes carrying promoter regions of tested genes were amplified by PCR with their respective primers (Table S1). The 3′ ends of PCR products were labeled with digoxigenin (DIG) using terminal transferase, and incubated individually with various quantities of His6-AvaR1 and/or GST-AvaR2 in a binding reaction. Overexpression and purification of His6-AvaR1, and EMSA conditions, were as described previously (Zhu et al., 2016). To confirm specificity of AvaR1-probe interaction, a ~200-fold excess of nonspecific DNA or each unlabeled specific probe was added to the binding mixture before incubation.
Chromatin immunoprecipitation-quantitative PCR (ChIP-qPCR)
Mycelia of ΔavaR1/avaR1-3FLAG cultured in FM-II for various times were harvested, and processed as described previously (Zhu et al., 2016).
Bioluminescence detection in E. coli
The plasmid pOaveRlux containing aveRp controlled lux reporter genes was constructed previously in our lab (Zhu et al., 2016). For expression of AvaR1, the avaR1 gene containing its ribosome-binding site (RBS) sequence and ORF was amplified with primers ZJY127 and ZY128. After cleavage with BamHI, the 736-bp avaR1 gene fragment was ligated into EcoRV/BamHI-digested pACYC184 (Tahlan et al., 2007) to give pAvaR1. Control vector pACYC184 and AvaR1 expression vector pAvaR1 were separately transformed into E. coli strain bearing pOaveRlux. Bioluminescence of E. coli reporter cultures was detected as described previously (Zhu et al., 2016).
DNase I footprinting
A fluorescence labeling procedure (Zianni et al., 2006) was used for these assays with some modifications (Zhu et al., 2016). In brief, 5′ FAM fluorescence-labeled probes corresponding to upstream regions of AvaR1 target genes were synthesized by PCR using primers listed in Table S1, and then incubated with various concentrations of His6-AvaR1. Followed by DNase I digestion, DNA samples were extracted and subjected to capillary electrophoresis by loading into a 3730XL DNA Genetic Analyzer. Electropherograms were analyzed using GeneMarker software program v2.2 (Applied Biosystems).
GST pull-down assay
pGEX-avaR2 (for expressing GST-AvaR2) was introduced into E. coli BL21 (DE3)/pET28-avaR2 (for expressing His6-AvaR2) and BL21 (DE3)/pET28-avaR1 (for expressing His6-AvaR1) (Zhu et al., 2016), respectively. Bacteria containing pGEX-4T-1 and pET28-avaR2, or pGEX-4T-1 and pET28-avaR1, were used as negative controls. Following IPTG induction, cells containing both GST and His6-tagged proteins were disrupted in lysis buffer (Luo et al., 2014) by sonication on ice, and then centrifuged. The lysate containing total protein was incubated with glutathione-sepharose beads overnight at 4°C, and washed three times with PBS buffer (0.113 M NaH2PO4, 0.387 M Na2HPO4, 1.5 M NaCl). The beads were boiled with SDS sample buffer, and the eluted bound proteins were subjected to SDS-PAGE and Western blotting with anti-GST or anti-His antibody (Tiangen, China).
Results
AvaR1 is a negative regulator of avermectin production
To clarify the function of AvaR1 in S. avermitilis, we constructed avaR1 deletion mutant ΔavaR1 by homologous recombination (Figure S1). HPLC analysis of fermentation products showed that avermectin yield of ΔavaR1 grown in FM-I for 10 days was ~2 fold higher than that of WT (Figure 1A). When an intact avaR1 gene in integrative vector pSET152 was introduced into ΔavaR1, avermectin yield in complemented strain CavaR1 was restored to WT level. Overexpression of avaR1 by introduction of plasmid pKC1139-ermp-avaR1 into WT (strain OavaR1) resulted in 30% reduction of avermectin yield. Vector control strains WT/pSET152 and WT/pKC1139 produced closely amount of avermectins to that of WT (Figure 1A). These findings indicate that AvaR1 has an inhibitory effect on avermectin production.
Figure 1.
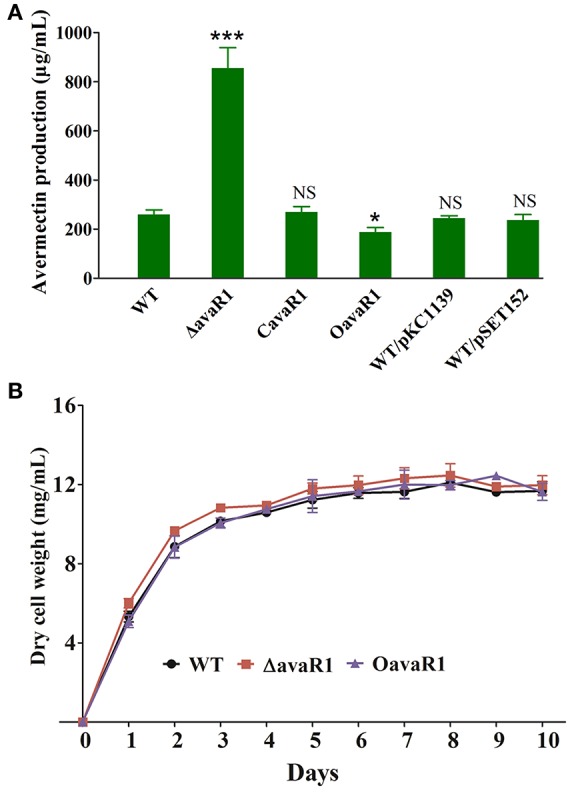
Effects of deletion and overexpression of avaR1 on avermectin production and cell growth in S. avermitilis. (A) Comparison of avermectin production in WT, avaR1 deletion mutant (ΔavaR1), complemented strain (CavaR1), and overexpression strain (OavaR1) cultured in FM-I for 10 days. WT/pKC1139 and WT/pSET152: vector control strains. Error bars: standard deviation (SD) from three replicate experiments. NS, not significant; *, P < 0.05; ***, P < 0.001 for comparison with WT (Student's t-test). (B) Growth curves of WT, ΔavaR1, and CavaR1 in FM-II. Error bars: SD from three replicates.
To assess the effect of AvaR1 on cell growth, we measured biomasses of WT, ΔavaR1, and OavaR1 cultured in FM-II. Deletion and overexpression of avaR1 had no effect on biomass (Figure 1B), indicating that altered avermectin yield in ΔavaR1 and OavaR1 did not result from changes in growth. ΔavaR1 and OavaR1 grew normally on YMS, MM and RM14 plates (Figure S2), indicating that AvaR1 is not involved in regulation of morphological differentiation.
AvaR1 represses expression of ave, aco, cyp17, and avaR genes
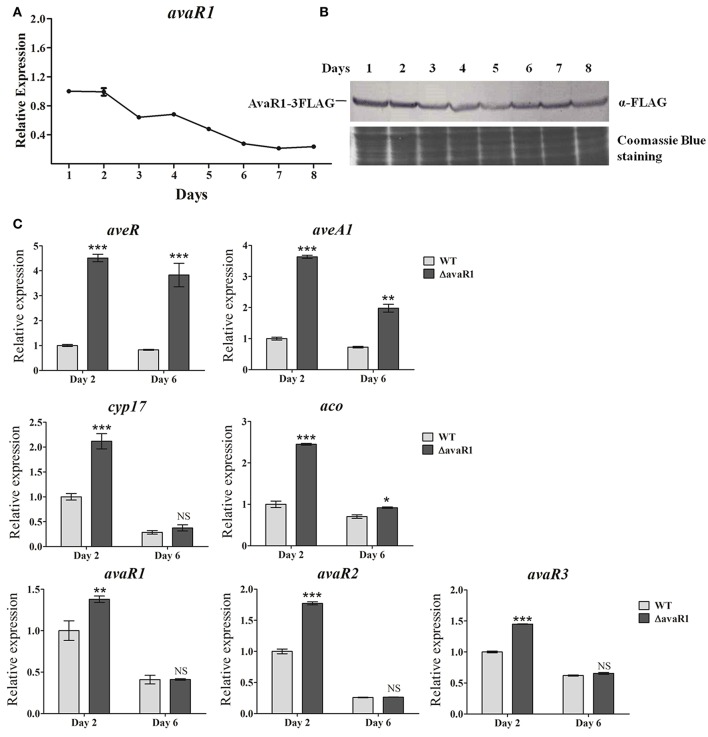
To clarify the relationship between AvaR1 and avermectin production, we examined avaR1 expression profiles of WT cultured in FM-I. avaR1 transcript was monitored by qRT-PCR throughout the avermectin fermentation process (Figure 2A). Transcript level was highest on day 1, remained high on day 2, then declined gradually, and was low from day 6 onward. We next examined AvaR1 expression profile by Western blotting. To prevent cross immunoreactions among homologous AvaR1, AvaR2, and AvaR3, a 3×flag sequence was fused to the 3′ end of avaR1 in integrative vector pSET152 and transformed into ΔavaR1. Expression of fusion protein AvaR1-3FLAG in ΔavaR1 restored avermectin yield (Figure S3), indicating that AvaR1-3FLAG complemented AvaR1 function, and that AvaR1 expression profile could be monitored with ANTI-FLAG mAb in recombinant strain ΔavaR1/avaR1-3FLAG. AvaR1 protein was detectable throughout the fermentation process, and its level was maximal on day 1 (Figure 2B), consistent with its transcriptional profile. These findings suggest that AvaR1 functions throughout the entire fermentation process, and plays its regulatory role mainly in the early stage.
Figure 2.
Expression analysis of avaR1 and related genes. (A) Transcriptional profile of avaR1 during avermectin production process in WT grown in FM-I. Relative value of avaR1 on day 1 was assigned as 1. Error bars: SD from three replicates. (B) Western blotting analysis of AvaR1 protein expression profile during fermentation process. AvaR1 temporal expression in strain ΔavaR1/avaR1-3FLAG grown in FM-I was analyzed using ANTI-FLAG mAb. 100 μg total protein was added in each lane. Loading control: Coomassie Blue staining of total protein. (C) qRT-PCR analysis of aveR, aveA1, cyp17, aco, and three avaR genes in WT and ΔavaR1 grown in FM-I. Value for each gene was expressed relative to that of WT on day 2, which was assigned as 1. avaR1, 125-bp transcript amplified from the remainder avaR1 ORF in ΔavaR1 with primers ZJY129 and ZJY130. Error bars: SD from three replicates. NS, not significant; *, P < 0.05; **, P < 0.01; ***, P < 0.001 (Student's t-test).
To test the possibility that AvaR1 regulates avermectin production through ave genes, we performed qRT-PCR analysis using RNAs isolated from WT and ΔavaR1 cultured in FM-I at 2 days (early exponential phase) and 6 days (stationary phase). Transcription levels of CSR gene aveR and structural gene aveA1 were strongly upregulated in ΔavaR1 on both days, particularly on day 2 (Figure 2C), consistent with avermectin overproduction in ΔavaR1, indicating that AvaR1 represses expression of ave genes.
AvaR1, AvaR2, and AvaR3 are members of the TetR-family regulators, which are typically autoregulated (Yu et al., 2010). AvaR2, AvaR3, Cyp17, and Aco have been shown to affect avermectin production in various ways (Kitani et al., 2011; Miyamoto et al., 2011; Zhu et al., 2016). We determined transcription levels of cyp17, aco, avaR1, avaR2, and avaR3 by qRT-PCR. All five genes were upregulated in ΔavaR1 on day 2 (Figure 2C). On day 6, aco transcription was slightly increased in ΔavaR1, whereas there was no notable change in expression of cyp17 or the three avaR genes. These findings indicate that AvaR1 is autorepressed and functions as a repressor of cyp17, aco, avaR2, and avaR3, mainly in the early stage of fermentation. In view of previous findings that Aco and Cyp17 are required for avenolide biosynthesis (Kitani et al., 2011), and that AvaR3 promotes avermectin production (Miyamoto et al., 2011), it is possible that elevated expression of aco, cyp17, and avaR3 also contributed to enhanced avermectin production in ΔavaR1.
AvaR1 binds specifically to upstream regions of aveR, aco, and three avaR genes
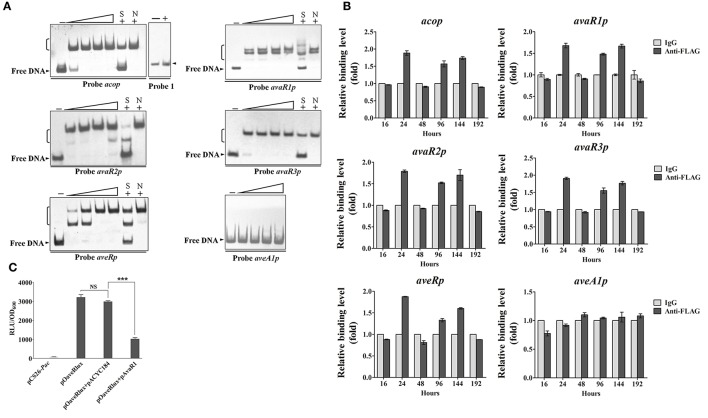
AvaR1 was reported to bind to the promoter regions of its own gene and of aco (Kitani et al., 2011; Sultan et al., 2016). To confirm these findings, and to determine whether the regulatory effect of AvaR1 on aveR, aveA1, cyp17, avaR2, and avaR3 is direct, we performed EMSAs using soluble His6-AvaR1 protein (Zhu et al., 2016) and probable promoter regions of these genes. Because cyp17 is co-transcribed with avaR1, we used the probes aveRp, aveA1p, acop, avaR1p, avaR2p, and avaR3p, constructed in our previous study (Zhu et al., 2016), for EMSAs. His6-AvaR1 did not bind to negative control probe 1, but formed complexes with probes acop and avaR1p (Figure 3A), as reported previously. His6-AvaR1 also bound specifically to probes avaR2p and avaR3p. No shifted band was observed for probe aveA1p. In contrast to the findings of Wang J. et al. (2014), probe aveRp generated clearly retarded signals (Figure 3A). Binding specificity was confirmed by competitive assays with a ~200-fold excess of unlabeled specific probe (lane S) and nonspecific probe 1 (lane N). These findings indicate that AvaR1 directly regulates expression of aco, avaR1, avaR2, avaR3, and aveR via binding to their promoter regions. The increased expression of aveA1 in ΔavaR1 was presumably an indirect effect of elevated aveR expression. cyp17 and avaR1 are co-transcribed; therefore, cyp17 is also a target gene of AvaR1, and upregulation of cyp17 in ΔavaR1 resulted from AvaR1 autorepression.
Figure 3.
Interaction of AvaR1 with target promoters in vitro and in vivo. (A) In vitro EMSAs of His6-AvaR1 with probes acop, avaR1p, avaR2p, avaR3p, aveRp, and aveA1p described previously (Zhu et al., 2016). Each reaction mixture contained 0.3 nM labeled probe. Specific (lanes S) and nonspecific (lanes N) competition assays were performed using ~200-fold excess of unlabeled competitor DNAs. Lanes 2–5 contained 10, 20, 50, and 100 nM His6-AvaR1, respectively. Lanes –: EMSAs without His6-AvaR1. 100 nM His6-AvaR1 was used for competition assays and negative control probe 1 (sig25 promoter region) (Lanes +). Arrowheads: free probes. Brackets: AvaR1-DNA complexes. (B) In vivo ChIP-qPCR assays. ANTI-FLAG mAb against AvaR1-3FLAG was used to immunoprecipitate AvaR1-3FLAG-DNA complexes from 16-, 24-, 48-, 96-, 144-, and 192-h cultures treated with formaldehyde. IgG-coprecipitated complexes were used as negative control. Enrichment level of target DNA in control at each time point was assigned as 1. The y axis represents relative fold binding of target DNA compared with control. Error bars: SD from three replicate experiments. (C) Bioluminescence levels of E. coli reporter cultures containing various plasmid combinations. pCS26-Pac and pACYC184 were used as vector controls. Values were expressed as relative light units (RLU). Error bars: SD from three replicates. NS, not significant; ***, P < 0.001 (Student's t-test).
In a search for corresponding evidence in vivo, we performed ChIP-qPCR experiments using samples from ΔavaR1/avaR1-3FLAG grown in FM-II for various durations, and mouse ANTI-FLAG mAb against AvaR1-3FLAG. No aveA1p enrichment was detected (Figure 3B), confirming that aveA1 is indirectly regulated by AvaR1. In comparison with control aveA1p, AvaR1 bound to acop, avaR1p, avaR2p, avaR3p, and aveRp at 24, 96, and 144 h, with strongest binding to each of these five target promoters at 24 h (Figure 3B), consistent with the maximal expression of AvaR1 on day 1. These findings confirm dynamic binding of AvaR1 to these target promoters in vivo. Target DNA enrichment levels of AvaR1 were <2-fold relative to that of negative control IgG, suggesting that DNA-binding affinity of AvaR1 is low in vivo, and that AvaR1 is easily released from target promoters after interaction with avenolide.
To clarify the apparent discrepancy between our observed interaction of AvaR1 with aveR promoter region and the findings of Wang J. et al. (2014), we examined the regulatory relationship of AvaR1 with aveR using an E. coli fluorescence-reporter system (Tahlan et al., 2007). In this system, pOaveRlux was constructed previously based on pCS26-Pac for expressing aveRp-controlled lux reporter (Zhu et al., 2016), and pAvaR1 was constructed in this study based on pACYC184 for expressing AvaR1. pOaveRlux produced a high level of bioluminescence in E. coli, whereas promoterless vector pCS26-Pac produced only a background level (Figure 3C). Expression vector pAvaR1 clearly reduced bioluminescence of the transformant bearing pOaveRlux, whereas control vector pACYC184 had no such effect (Figure 3C). These findings are consistent with those from EMSA and ChIP-qPCR experiments, and demonstrate that AvaR1 directly represses aveR transcription.
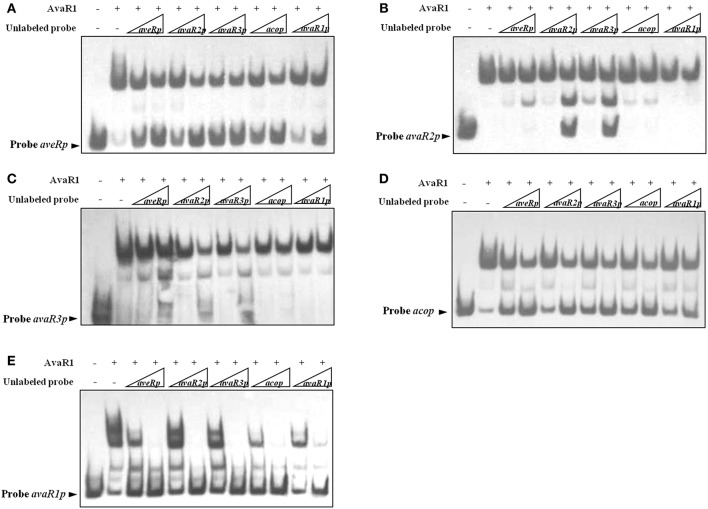
Affinity of AvaR1 binding to its target promoters
To compare affinity of AvaR1 for its five target promoters (acop, avaR1p, avaR2p, avaR3p, aveRp), we performed competitive EMSAs using labeled probes and excess unlabeled probes (specific competitor). In experiments using 50-fold excess of unlabeled probes, the dissociation of AvaR1 from labeled aveRp caused by unlabeled probe avaR1p was less than that of the other four unlabeled probes (Figure 4A). Further analysis using labeled probes avaR2p, avaR3p, and avaR1p (Figures 4B,C,E) confirmed that affinity of AvaR1 to avaR1p was the lowest among the five DNA targets. In experiments using 250-fold excesses, unlabeled probes avaR3p and avaR2p dissociated most AvaR1 from labeled avaR2p, whereas the other three unlabeled probes had little or no such effect (Figure 4B), indicating a higher relative affinity of AvaR1 for avaR3p and avaR2p. The competitive ability of 50-fold excess of unlabeled probe avaR3p was slightly higher than that of the same excess of avaR2p (Figure 4B). These findings indicate that AvaR1 had the strongest affinity for avaR3p. Comparison of intensities of AvaR1-avaR3p complex corresponding to unlabeled probes aveRp and acop (Figure 4C) indicated AvaR1 affinity in the order aveRp > acop. There were no striking differences of intensities of AvaR1-acop complex among the five unlabeled probes (Figure 4D). The above findings, taken together, indicate that binding affinity of AvaR1 to its target promoters is in the order avaR3p > avaR2p > aveRp > acop > avaR1p; however, the differences for avaR3p vs. avaR2p, and acop vs. avaR1p, are slight.
Figure 4.
Relative affinities of AvaR1 for various target promoters. (A) EMSA of His6-AvaR1 with labeled probe aveRp and unlabeled probes (aveRp, avaR2p, avaR3p, acop, avaR1p). (B) EMSA of His6-AvaR1 with labeled probe avaR2p and unlabeled probes. (C) EMSA of His6-AvaR1 with labeled probe avaR3p and unlabeled probes. (D) EMSA of His6-AvaR1 with labeled probe acop and unlabeled probes. (E) EMSA of His6-AvaR1 with labeled probe avaR1p and unlabeled probes. For competition assays, labeled probe (0.3 nM) and unlabeled competitor probe (50- and 250-fold) were added with His6-AvaR1 (50 nM). Arrowheads: free labeled probes.
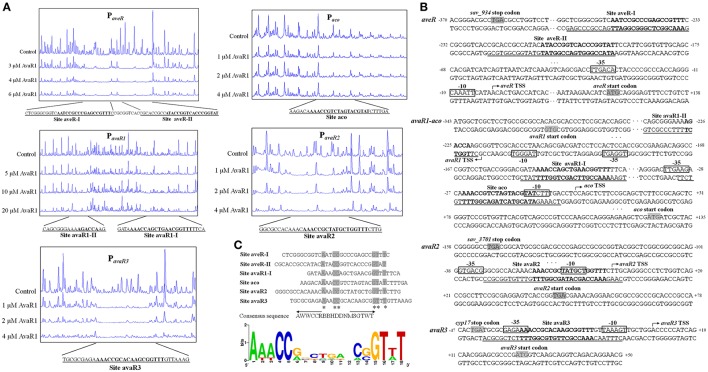
Identification of precise AvaR1 binding sites
The transcriptional start sites (TSSs) of aveR, aco, and three avaR genes were determined previously (Zhuo et al., 2010; Miyamoto et al., 2011; Sultan et al., 2016; Zhu et al., 2016). We performed DNase I footprinting assays to identify precise AvaR1 binding sites and elucidate the mechanism whereby AvaR1 regulates its five targets. AvaR1 was found to protect two sites (aveR-I, aveR-II), seperated by 9 nt, on the aveR promoter region (Figure 5A). These sites are far upstream of the aveR TSS. Site aveR-I corresponds to positions −262 to −233 nt, and site aveR-II −223 to −193 nt, relative to the the aveR TSS (Figure 5B). The mechanism whereby AvaR1 represses aveR is unkown. Interactions among AvaR1 and the two binding sites may prevent RNA polymerase acess to the aveR promoter, or recruit other repressors. The protected site of AvaR1 on the aco promoter region (site aco) overlaps the putative −10 and −35 promoter regions, and extends from −31 to −4 nt relative to the aco TSS (Figures 5A,B), suggesting that AvaR1 directly represses aco by impeding RNA polymerase binding to the aco promoter. avaR1 promoter region was protected at two sites: site avaR1-I (positions −93 to −68 nt relative to the avaR1 TSS) and site avaR1-II (+19 to +2 nt) (Figures 5A,B). AvaR1 protected a region from −32 to +3 nt relative to the avaR2 TSS on the avaR2 promoter region (site avaR2), and from −41 to −8 nt relative to the avaR3 TSS on the avaR3 promoter region (site avaR3) (Figures 5A,B). Site avaR1-I is located upstream of the potential −35 region of the avaR1 promoter, and sites avaR1-II, avaR2, and avaR3 are close to or overlap the potential −10 region of their respective promoters, suggesting that AvaR1 represses three avaR genes through mechanisms analogous to those for aveR and aco repression.
Figure 5.
Identification of AvaR1 binding sites. (A) DNase I footprinting assay of AvaR1 on target promoter regions. Protection fluorograms were acquired with increasing amounts of His6-AvaR1. Top fluorograms: control reactions with 10 μM BSA. (B) Nucleotide sequences of target promoter regions and AvaR1 binding sites. Numbers: distance (nt) from respective TSS. Shaded areas: translational start codons. Bent arrows: TSSs. Boxes: potential −10 and −35 regions. Solid lines: AvaR1 binding sites. Boldface: ARE-like sequences. (C) Analysis of consensus AvaR1 binding sequence using the WebLogo program (http://weblogo.berkeley.edu). Asterisks: consensus bases. Arrows: inverted repeats. Height of each letter is proportional to appearance frequency of corresponding base.
ARE (autoregulatory element) sequences are found in genuine and pseudo GBL receptor binding sites in many Streptomyces species (Wang et al., 2011; Willey and Gaskell, 2011). AvaR1 is a receptor of γ-butenolide-type avenolide, not of GBLs; however, the protected sites of AvaR1 on the five promoter regions all contain ARE-like sequences. Site avaR1-II contains a half-length ARE, and the other sites contain a full-length ARE. Interestingly, the binding sites of AvaR1 completely overlap those of AvaR2 on the five target promoter regions (Figure S4; Zhu et al., 2016), with the same protection sequences on aveRp, acop, and avaR1p, and three or four nucleotides more than the AvaR2 binding sequences on avaR3p and avaR2p. WebLogo program analysis of AvaR1 binding sites including full-length AREs produced an 18-bp consensus ARE-like palindromic sequence (5′-AWWCCRBBHDDNMSGTWT-3′) (Figure 5C), identical to that of the AvaR2 consensus binding motif (Figure S4; Zhu et al., 2016).
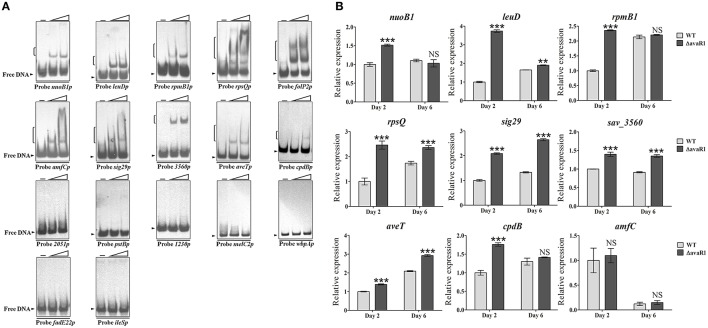
AvaR1 and AvaR2 have both common and exclusive target genes
AvaR1 and AvaR2 have an identical consensus binding motif, and are therefore expected to have an identical predicted regulon based on use of the same web-based application. Because the consensus binding motif of AvaR1 and AvaR2 was deduced only from five common target promoter regions, we could not rule out the possibility that AvaR1 and AvaR2 have different targets. To test the possibility that AvaR1 binds to other AvaR2 targets in addition to the above five common targets, 11 identified AvaR2 target genes (nuoB1, leuD, rpmB1, rpsQ, folP2, amfC, sig29, sav_3560, sav_2051, pstB, and sav_1230) (Zhu et al., 2016) were subjected to EMSAs with purified His6-AvaR1 protein. AvaR1 bound to the promoter regions of nuoB1, leuD, rpmB1, rpsQ, folP2, amfC, sig29, and sav_3560, but not to those of sav_2051, pstB, or sav_1230 (Figure 6A), indicating that AvaR1 and AvaR2 have both common and exclusive targets. To follow up on this finding, melC2, wbpA, fadE22, ileS, aveT (sav_3619), and cpdB listed in putative AvaR2 regulon (Zhu et al., 2016) were subjected to EMSAs with His6-AvaR1. Among these genes, melC2, wbpA, fadE22, and ileS were found not to be AvaR2 targets (Zhu et al., 2016). aveTp and cpdBp had the ability to interact with AvaR1, but the other four promoter probes did not (Figure 6A). aveT encodes a TetR-family transcriptional activator for avermectin production and morphological differentiation (Liu et al., 2015), and cpdB encodes a putative 2′,3′-cyclic-nucleotide 2′-phosphodiesterase. We performed EMSAs to test the possibility that AvaR2 binds to these two novel AvaR1 targets, and observed that it bound to aveTp, but not to cpdBp (Figure S5A).
Figure 6.
Confirmation of new AvaR1 target genes. (A) EMSAs of His6-AvaR1 protein with 17 putative binding promoter regions. Each lane contained 0.3 nM labeled probe. Lanes –, EMSAs without His6-AvaR1. Lanes 2 to 3 contained 50 and 200 nM His6-AvaR1, respectively. (B) qRT-PCR analysis of newly identified AvaR1 target genes in WT and ΔavaR1 strains. WT value of each gene on day 2 was assigned as 1. Error bars: SD from three replicates. NS, not significant; **, P < 0.01; ***, P < 0.001 (Student's t-test).
The regulatory roles of AvaR1 in expression of the 10 newly identified target genes were evaluated by qRT-PCR analysis. AvaR1 repressed transcription of nuoB1, leuD, rpmB1, rpsQ, sig29, sav_3560, aveT, and cpdB; i.e., transcription levels of these targets were higher in ΔavaR1 than in WT on day 2, or on days 2 and 6 (Figure 6B). amfC encodes an aerial mycelium-associated protein (Yonekawa et al., 1999). amfC transcript levels were very similar in WT and ΔavaR1, consistent with the absence of morphological differences between these two strains. folP2 expression was not detected in either WT or ΔavaR1 on day 2 or 6 under our fermentation conditions. The effect of AvaR2 on aveT expression was also examined by qRT-PCR. AvaR2 activated transcription of aveT (Figure S5B), in contrast to the repressing effect of AvaR1 on aveT (Figure 6B).
Taken together, the identified common targets of AvaR1 and AvaR2 are aveR, aco, avaR1, avaR2, avaR3, nuoB1, leuD, folP2, rpmB1, rpsQ, amfC, sig29, sav_3560, and aveT. cpdB is a target of AvaR1, but not of AvaR2. sav_2051, pstB and sav_1230 are targets of AvaR2, but not of AvaR1. The above targets are involved in secondary metabolism, primary metabolism, morphological differentiation, ribosomal protein synthesis, stress responses, and other processes (Liu et al., 2015; Zhu et al., 2016), indicating that AvaR1 and AvaR2 cross-regulate a wide range of physiological processes.
Avermectin production level was much higher in single deletion mutant ΔavaR1 than in ΔavaR2, and levels in double deletion mutants ΔavaR1R2-1,-2, and -3 were intermediate (Figure S6), consistent with the finding that AvaR1 and AvaR2 have different target genes, which may affect avermectin production in different ways.
AvaR1 and AvaR2 compete and cooperate on the same binding site
AvaR1 and AvaR2 are TetR-family transcriptional regulators, which generally function as homodimers to bind to palindromic sequences (Yu et al., 2010). Because the two proteins are homologs, it is conceivable that they could form a heterodimer. Because they have common target genes, they may also compete or cooperate for DNA binding. The apparent molecular weights of His6-AvaR1 and His6-AvaR2 were similar (~35 kDa). To evaluate possible competition or cooperation of AvaR1 and AvaR2 on the same binding site, we expressed and purified GST-tagged AvaR2 protein from E. coli to separate the complexes formed by His6-AvaR1 or GST-AvaR2 using the same DNA probe in EMSAs. His6-AvaR1 and GST-AvaR2 were added both separately and together with probe aveRp2, which contains only one palindromic sequence: site aveR-II. When applied separately, both proteins retarded aveRp2 (Figure 7A). In the presence of 0.1 μM His6-AvaR1, an increase of GST-AvaR2 concentration resulted in reduction of AvaR1-aveRp2 complex and formation of a new complex, located between AvaR1-aveRp2 and AvaR2-aveRp2 (Figure 7A, left), which was most likely formed by AvaR1/AvaR2 heterodimer with probe aveRp2. When GST-AvaR2 was added at a concentration of 0.6 μM, the complex AvaR2-aveRp2 became predominant, and the above-mentioned new complex and AvaR1-aveRp2 disappeared (Figure 7A, left). In the presence of 0.1 μM GST-AvaR2, an increase of His6-AvaR1 concentration resulted in disappearance of AvaR2-aveRp2 complex and appearance of AvaR1-aveRp2 and a new band, most likely AvaR1/AvaR2-aveRp2 (Figure 7A, right). As His6-AvaR1 concentration increased, intensity of the new band declined and that of AvaR1-aveR2p became stronger. When His6-AvaR1 was added at a concentration of 0.2 μM, the new band disappeared and AvaR1-aveRp2 complex became predominant (Figure 7A, right). These findings suggest that AvaR1 and AvaR2 not only compete for the same DNA site, but also cooperate as a heterodimer for DNA binding.
Figure 7.
Relationships between AvaR1 and AvaR2. (A) Competitive EMSAs of probe aveRp2 with His6-AvaR1 and GST-AvaR2 proteins. 0.3 nM labeled probe aveRp2 was incubated with the indicated concentrations of His6-AvaR1 and GST-AvaR2. (B) GST pull-down assays of AvaR1 and AvaR2 from E. coli whole cell lysate. His6- and GST-tagged proteins were co-expressed in E. coli, recovered by sonication and centrifugation, and subjected to GST pull-down and Western blotting analysis with anti-GST and anti-His antibodies, respectively. Lanes 1: cell lysate before induction by IPTG. Lanes 2: cell lysate after induction. Lanes 3: GST pull-down.
To assess possible interaction between AvaR1 and AvaR2 under physiological conditions, we co-expressed His6-AvaR2 or His6-AvaR1 with GST or GST-AvaR2 in E. coli, and performed in vivo GST pull-down experiments. His6-AvaR2 was pulled down by GST-AvaR2, but not by GST (Figure 7B, left), indicating that AvaR2 interacts with itself in vivo, most likely to form a homodimer. His6-AvaR1 was also pulled down by GST-AvaR2, but not by GST (Figure 7B, right), supporting the conclusion from experiments shown in Figure 7A that AvaR1 interacts directly with AvaR2 to form a heterodimer.
Discussion
Autoregulator/receptor regulatory systems are widespread among Streptomyces species, and play key roles in eliciting secondary metabolites or/and morphological differentiation (Bibb, 2005; Niu et al., 2016). Unlike typical GBL signaling molecules applied by many other Streptomyces species, S. avermitilis uses avenolide, a γ-butenolide autoregulator, to trigger avermectin production (Kitani et al., 2011). AvaR1 was shown to be an avenolide receptor (Kitani et al., 2011); however, its role in regulation of avermectin biosynthesis remains unclear. In this study, we elucidated the molecular mechanism underlying this role, and demonstrated that AvaR1 is a direct repressor of avenolide and avermectin biosynthesis as well as other physiological processes including primary metabolism, ribosomal protein synthesis, and stress responses.
AvaR1 strongly inhibits avermectin production in WT strain ATCC31267. Results of our qRT-PCR, EMSA, and ChIP-qPCR analyses revealed that this inhibitory effect is due mainly to direct repression of the cluster-situated activator gene aveR. Our findings are in contrast to reports by other groups that AvaR1 indirectly controls aveR expression (Wang J. et al., 2014), and that AvaR1 has no effect on avermectin production in WT strain KA320 (Sultan et al., 2016). Our DNase I footprinting assays showed that AvaR1 binds to two sites on the far upstream region of aveR: one extending from −262 to −233 nt, the other from −223 to −193 nt, relative to the aveR TSS. In the EMSA studies by Wang J. et al. (2014), the aveR promoter probe used extended from positions −109 to +113 nt relative to the aveR TSS, and did not contain two AvaR1-binding sites. This explains their finding that AvaR1 did not bind to the aveR promoter region. The differences between our findings and those of Nihira's group regarding effect of AvaR1 on avermectin production may be due to differences in experimental strains and growth conditions used. Nihira's group used KA320 as WT strain (Sultan et al., 2016), and described it as being isogenic to S. avermitilis MA-4680–also known as ATCC31267, the WT strain used in our study. However, in a previous report, they described KA320 as being phenotypically unstable and requiring frequent reisolation (Kitani et al., 2009). In contrast, strain ATCC31267 we used was stable throughout our experiments, and AseI restriction patterns of our ATCC31267 chromosome were identical to the published data (Guo et al., 2010). Properties of CSR gene aveR also differed between the WT strains used by our group and Nihara's. In our 2010 study (Guo et al., 2010), overexpression of aveR promoted avermectin production in ATCC31267, in accordance with the general rule that overexpression of a cluster-situated activator gene leads to increased production of the corresponding antibiotic. In contrast, the 2009 study by Nihara's group found that addition of aveR abolished avermectin production in KA320 and its derivative K139 (Kitani et al., 2009). It therefore seems likely that differences in genetic background exist between KA320 and our ATCC31267, which account for the different findings. Genome resequencing of the two strains can clarify such genetic differences.
The aco gene encodes a key enzyme for avenolide production (Kitani et al., 2011). In the present study, AvaR1 directly repressed aco expression, consistent with previous findings by Nihira's group (Kitani et al., 2011; Sultan et al., 2016). Wang et al. (2015) confirmed the role of AvaR1 as a direct repressor of aco using a genetic biosensor. AvaR1 also inhibits transcription of cyp17, which is involved in avenolide production. Thus, AvaR1 is an important regulator in control of avenolide level. Our 2016 study showed that AvaR2 is also a key repressor of avenolide production (Zhu et al., 2016). The increase of aco transcription level was >130-fold in avaR2 deletion mutant ΔavaR2 (Zhu et al., 2016), but <2-fold in avaR1 deletion mutant ΔavaR1, indicating that AvaR2 plays a role dominant over that of AvaR1 in control of avenolide production.
AvaR1 and AvaR2 are homologs, both act as receptors of avenolide signal (Kitani et al., 2011; Zhu et al., 2016), and bind to the same sequence on aveR and aco promoter regions. We therefore investigated possible interaction or cooperation between these two proteins to regulate avermectin and avenolide production. Competitive EMSAs and GST pull-down assays revealed coexistence of AvaR1/AvaR2 heterodimer with AvaR1 and AvaR2 homodimers. The EMSA results also showed that the heterodimer-DNA complex reduced or disappeared when concentration of one protein was much higher than that of another protein, suggesting that interconversion among AvaR1/AvaR2 heterodimer and the AvaR1 and AvaR2 homodimers is a highly dynamic process. Analogously, Li et al. (2015, 2017) demonstrated that ScbR and ScbR2 share some common binding sites, and can form a heterodimer in addition to their respective homodimers. These examples of heterodimer formation between homologous TetR-family regulators indicate that regulatory mechanisms in this family are complex, and that the heterodimers may play regulatory roles of which the corresponding homodimers are not capable. For example, ScbR/ScbR2 heterodimer has an exclusive target sco5158, which is not a target of ScbR or ScbR2 homodimer (Li et al., 2017). We observed that AvaR1/AvaR2 heterodimer and the AvaR1 and AvaR2 homodimers all bind to the same DNA probe aveRp2; however, further investigation is needed to test the possibility that AvaR1/AvaR2 heterodimer has exclusive targets as well.
Competitive EMSAs indicated that AvaR1 and AvaR2 homodimers compete for the same binding region. In vivo ChIP-qPCR experiments revealed that maximal DNA enrichment level of AvaR2, in comparison with control IgG, was ~10.5-fold for aveRp, ~23.3-fold for acop, ~8.4-fold for avaR1p, ~4.1-fold for avaR2p, and ~4.6-fold for avaR3p (Zhu et al., 2016). Corresponding values for AvaR1 were <2-fold for all five target genes, indicating that in vivo DNA-binding affinity of AvaR2 is stronger than that of AvaR1, and that the role of AvaR2 in regulating these common targets is dominant over that of AvaR1. AvaR2 showed stronger binding affinity for aveRp than for avaR3p, avaR2p, acop, or avaR1p. Its binding to aveRp was strongest at 24 h, and that to the other four target promoters was strongest at 48 h, with subsequent gradual decrease in binding strength during the fermentation process (Zhu et al., 2016). AvaR1 showed little difference in affinity among these five promoters; it bound to them at 24, 96, and 144 h, with somewhat higher binding strength at 24 h. The dynamic binding of AvaR1 and AvaR2 to the five targets was consistent with their expression profiles, i.e., maximal expression occurred at 24 h for AvaR1 and at 48 h for AvaR2; AvaR2 level decreased sharply after 96 h and was barely detectable from 120 h onward, whereas AvaR1 level was stable from 72 h onward.
Our findings on AvaR1 and AvaR2 expression and their regulatory relationship, taken together, indicate that they have a sequential cooperative mechanism for control of avenolide and avermectin production in response to avenolide signal (Figure 8). During the early growth phase of S. avermitilis, aveRp is repressed by AvaR1 and (preferentially) AvaR2 to strongly inhibit avermectin biosynthesis (24 h); AvaR1 (24 h) and AvaR2 (48 h) sequentially repress aco expression to strictly control avenolide level and avenolide synthesis remains at basal level during this period. When avenolide level exceeds a threshold value, its receptors AvaR1 and AvaR2 are sequentially released from acop, leading to sharply increased avenolide concentration which becomes sufficient to trigger avermectin biosynthesis by releasing AvaR1 and AvaR2 from aveRp. This concept is supported by our observation that avermectin production is usually detectable by HPLC after 48 h. After avenolide has performed its role in triggering avermectin production, it is presumably degraded by a yet-unknown mechanism. During the late growth stage (≥144 h), acop and aveRp are bound again mainly by AvaR1 rather than AvaR2, to avoid overproduction of avermectins and ensure appropriate avermectin concentration in cells. The repression of acop and aveRp by AvaR1 during late stage is advantageous in that the relatively low repression strength of AvaR1 ensures continuous, but not excessive, synthesis of avermectins.
Figure 8.

Working model for the regulatory roles of AvaR1 and AvaR2 in control of avenolide and avermectin production. Solid-line bars: direct repression. Solid-line arrow: direct activation. Dashed-line arrows: production of avermectin or avenolide.
Although AvaR1 and AvaR2 have common targets, they each also have exclusive targets, and their regulatory mechanisms on common target aveT are different. Thus, the regulatory functions of these two proteins differ somewhat. For example, cell growth is promoted by avaR2 deletion (Zhu et al., 2016), but unaffected by avaR1 deletion. These findings are consistent with general economic principles of metabolic regulation, i.e., a given microorganism does not contain two identical regulatory factors. The AvaR1 targets are involved in diverse cellular processes, indicating the pleiotropic effects of AvaR1 on S. avermitilis physiology. Elevated expression of AvaR1 target genes leuD, nuoB1, rpmB1, and rpsQ in ΔavaR1 may promote avermectin production by providing more precursors, increasing availability of energy, or enhancing protein synthesis, analogously to their effects in ΔavaR2 (Zhu et al., 2016). AveT acts as an activator of avermectin production. Therefore, elevated expression of aveT in ΔavaR1 may enhance avermectin production. The cpdB product converts 2′,3′-cyclic nucleotide to 3′-nucleotide, and increased cpdB expression may provide more precursors for ATP synthesis and thereby promote avermectin biosynthesis. Our findings that AvaR1 regulates sig29 (whose homolog sigI in S. coelicolor is involved in osmotic stress) (Homerova et al., 2012) and sav_3560 (one component of a two-component system) suggest that AvaR1 responds to extracellular stimuli as part of adaptation to complex natural environments. More extensive studies of regulatory roles of AvaR1 and AvaR2 in the future will provide a comprehensive picture of the cellular responses triggered by avenolide and other signals, and mediated by AvaR1 and AvaR2, in S. avermitilis.
Author contributions
YW and JZ wrote the manuscript and designed the research. JZ performed experiments. ZC and JL contributed study materials.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors are grateful to Dr. S. Anderson for English editing of the manuscript.
Footnotes
Funding. This study was supported by the National Key Research and Development Program of China (grant no. 2017YFD0201207).
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2017.02577/full#supplementary-material
References
- Arakawa K., Tsuda N., Taniguchi A., Kinashi H. (2012). The butenolide signaling molecules SRB1 and SRB2 induce lankacidin and lankamycin production in Streptomyces rochei. Chembiochem 13, 1447–1457. 10.1002/cbic.201200149 [DOI] [PubMed] [Google Scholar]
- Bibb M. J. (2005). Regulation of secondary metabolism in streptomycetes. Curr. Opin. Microbiol. 8, 208–215. 10.1016/j.mib.2005.02.016 [DOI] [PubMed] [Google Scholar]
- Bierman M., Logan R., O'Brien K., Seno E. T., Rao R. N., Schoner B. E. (1992). Plasmid cloning vectors for the conjugal transfer of DNA from Escherichia coli to Streptomyces spp. Gene 116, 43–49. 10.1016/0378-1119(92)90627-2 [DOI] [PubMed] [Google Scholar]
- Burg R. W., Miller B. M., Baker E. E., Birnbaum J., Currie S. A., Hartman R., et al. (1979). Avermectins, new family of potent anthelmintic agents: producing organism and fermentation. Antimicrob. Agents Chemother. 15, 361–367. 10.1128/AAC.15.3.361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z., Wen J., Song Y., Wen Y., Li J. (2007). Enhancement and selective production of avermectin B by recombinants of Streptomyces avermitilis via intraspecific protoplast fusion. Chin. Sci. Bull. 52, 616–622. 10.1007/s11434-007-0119-y [DOI] [Google Scholar]
- Choi S. U., Lee C. K., Hwang Y. I., Kinosita H., Nihira T. (2003). γ-butyrolactone autoregulators and receptor proteins in non-Streptomyces actinomycetes producing commercially important secondary metabolites. Arch. Microbiol. 180, 303–307. 10.1007/s00203-003-0591-y [DOI] [PubMed] [Google Scholar]
- Egerton J. R., Ostlind D. A., Blair L. S., Eary C. H., Suhayda D., Cifelli S., et al. (1979). Avermectins, new family of potent anthelmintic agents: efficacy of the B1a component. Antimicrob. Agents Chemother. 15, 372–378. 10.1128/AAC.15.3.372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folcher M., Gaillard H., Nguyen L. T., Nguyen K. T., Lacroix P., Bamas-Jacques N., et al. (2001). Pleiotropic functions of a Streptomyces pristinaespiralis autoregulator receptor in development, antibiotic biosynthesis, and expression of a superoxide dismutase. J. Biol. Chem. 276, 44297–44306. 10.1074/jbc.M101109200 [DOI] [PubMed] [Google Scholar]
- Gomez-Escribano J. P., Song L., Fox D. J., Yeo V., Bibb M. J., Challis G. L. (2012). Structure and biosynthesis of the unusual polyketide alkaloid coelimycin P1, a metabolic product of the cpk gene cluster of Streptomyces coelicolor M145. Chem. Sci. 3, 2716–2720. 10.1039/c2sc20410j [DOI] [Google Scholar]
- Guo J., Zhao J., Li L., Chen Z., Wen Y., Li J. (2010). The pathway-specific regulator AveR from Streptomyces avermitilis positively regulates avermectin production while it negatively affects oligomycin biosynthesis. Mol. Genet. Genomics 283, 123–133. 10.1007/s00438-009-0502-2 [DOI] [PubMed] [Google Scholar]
- Homerova D., Sevcikova B., Rezuchova B., Kormanec J. (2012). Regulation of an alternative sigma factor σI by a partner switching mechanism with an anti-sigma factor PrsI and an anti-anti-sigma factor ArsI in Streptomyces coelicolor A3(2). Gene 492, 71–80. 10.1016/j.gene.2011.11.011 [DOI] [PubMed] [Google Scholar]
- Horinouchi S., Beppu T. (2007). Hormonal control by A-factor of morphological development and secondary metabolism in Streptomyces. Proc. Jpn. Acad. Ser. B Phys. Biol. Sci. 83, 277–295. 10.2183/pjab.83.277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitani S., Ikeda H., Sakamoto T., Noguchi S., Nihira T. (2009). Characterization of a regulatory gene, aveR, for the biosynthesis of avermectin in Streptomyces avermitilis. Appl. Microbiol. Biotechnol. 82, 1089–1096. 10.1007/s00253-008-1850-2 [DOI] [PubMed] [Google Scholar]
- Kitani S., Miyamoto K. T., Takamatsu S., Herawati E., Iguchi H., Nishitomi K., et al. (2011). Avenolide, a Streptomyces hormone controlling antibiotic production in Streptomyces avermitilis. Proc. Natl. Acad. Sci. U.S.A. 108, 16410–16415. 10.1073/pnas.1113908108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L., Guo J., Wen Y., Chen Z., Song Y., Li J. (2010). Overexpression of ribosome recycling factor causes increased production of avermectin in Streptomyces avermitilis strains. J. Ind. Microbiol. Biotechnol. 37, 673–679. 10.1007/s10295-010-0710-0 [DOI] [PubMed] [Google Scholar]
- Li X., Wang J., Li S., Ji J., Wang W., Yang K. (2015). ScbR- and ScbR2-mediated signal transduction networks coordinate complex physiological responses in Streptomyces coelicolor. Sci. Rep. 5:14831. 10.1038/srep14831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Wang J., Shi M., Wang W., Corre C., Yang K. (2017). Evidence for the formation of ScbR/ScbR2 heterodimers and identification of one of the regulatory targets in Streptomyces coelicolor. Appl. Microbiol. Biotechnol. 101, 5333–5340. 10.1007/s00253-017-8275-8 [DOI] [PubMed] [Google Scholar]
- Liu W., Zhang Q., Guo J., Chen Z., Li J., Wen Y. (2015). Increasing avermectin production in Streptomyces avermitilis by manipulating the expression of a novel TetR-family regulator and its target gene product. Appl. Environ. Microbiol. 81, 5157–5173. 10.1128/AEM.00868-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo S., Sun D., Zhu J., Chen Z., Wen Y., Li J. (2014). An extracytoplasmic function sigma factor, σ25, differentially regulates avermectin and oligomycin biosynthesis in Streptomyces avermitilis. Appl. Microbiol. Biotechnol. 98, 7097–7112. 10.1007/s00253-014-5759-7 [DOI] [PubMed] [Google Scholar]
- MacNeil D. J., Klapko L. M. (1987). Transformation of Streptomyces avermitilis by plasmid DNA. J. Ind. Microbiol. 2, 209–218. 10.1007/BF01569542 [DOI] [Google Scholar]
- Matsuno K., Yamada Y., Lee C. K., Nihira T. (2004). Identification by gene deletion analysis of barB as a negative regulator controlling an early process of virginiamycin biosynthesis in Streptomyces virginiae. Arch. Microbiol. 181, 52–59. 10.1007/s00203-003-0625-5 [DOI] [PubMed] [Google Scholar]
- Miyamoto K. T., Kitani S., Komatsu M., Ikeda H., Nihira T. (2011). The autoregulator receptor homologue AvaR3 plays a regulatory role in antibiotic production, mycelial aggregation and colony development of Streptomyces avermitilis. Microbiology 157, 2266–2275. 10.1099/mic.0.048371-0 [DOI] [PubMed] [Google Scholar]
- Nakano H., Takehara E., Nihira T., Yamada Y. (1998). Gene replacement analysis of the Streptomyces virginiae barA gene encoding the butyrolactone autoregulator receptor reveals that BarA acts as a repressor in virginiamycin biosynthesis. J. Bacteriol. 180, 3317–3322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu G., Chater K. F., Tian Y., Zhang J., Tan H. (2016). Specialised metabolites regulating antibiotic biosynthesis in Streptomyces spp. FEMS Microbiol. Rev. 40, 554–573. 10.1093/femsre/fuw012 [DOI] [PubMed] [Google Scholar]
- Onaka H., Nakagawa T., Horinouchi S. (1998). Involvement of two A-factor receptor homologues in Streptomyces coelicolor A3(2) in the regulation of secondary metabolism and morphogenesis. Mol. Microbiol. 28, 743–753. 10.1046/j.1365-2958.1998.00832.x [DOI] [PubMed] [Google Scholar]
- Pullan S. T., Chandra G., Bibb M. J., Merrick M. (2011). Genome-wide analysis of the role of GlnR in Streptomyces venezuelae provides new insights into global nitrogen regulation in actinomycetes. BMC Genomics 12:175. 10.1186/1471-2164-12-175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sultan S. P., Kitani S., Miyamoto K. T., Iguchi H., Atago T., Ikeda H., et al. (2016). Characterization of AvaR1, a butenolide-autoregulator receptor for biosynthesis of a Streptomyces hormone in Streptomyces avermitilis. Appl. Microbiol. Biotechnol. 100, 9581–9591. 10.1007/s00253-016-7781-4 [DOI] [PubMed] [Google Scholar]
- Tahlan K., Ahn S. K., Sing A., Bodnaruk T. D., Willems A. R., Davidson A. R., et al. (2007). Initiation of actinorhodin export in Streptomyces coelicolor. Mol. Microbiol. 63, 951–961. 10.1111/j.1365-2958.2006.05559.x [DOI] [PubMed] [Google Scholar]
- Takano E. (2006). γ-Butyrolactones: Streptomyces signalling molecules regulating antibiotic production and differentiation. Curr. Opin. Microbio. 9, 287–294. 10.1016/j.mib.2006.04.003 [DOI] [PubMed] [Google Scholar]
- Takano E., Chakraburtty R., Nihira T., Yamada Y., Bibb M. J. (2001). A complex role for the γ-butyrolactone SCB1 in regulating antibiotic production in Streptomyces coelicolor A3(2). Mol. Microbiol. 41, 1015–1028. 10.1046/j.1365-2958.2001.02562.x [DOI] [PubMed] [Google Scholar]
- Takano E., Kinoshita H., Mersinias V., Bucca G., Hotchkiss G., Nihira T., et al. (2005). A bacterial hormone (the SCB1) directly controls the expression of a pathway-specific regulatory gene in the cryptic type I polyketide biosynthetic gene cluster of Streptomyces coelicolor. Mol. Microbiol. 56, 465–479. 10.1111/j.1365-2958.2005.04543.x [DOI] [PubMed] [Google Scholar]
- Wang J., Wang W., Wang L., Zhang G., Fan K., Tan H., et al. (2011). A novel role of ‘pseudo’ γ-butyrolactone receptors in controlling gamma-butyrolactone biosynthesis in Streptomyces. Mol. Microbiol. 82, 236–250. 10.1111/j.1365-2958.2011.07811.x [DOI] [PubMed] [Google Scholar]
- Wang J., Zhang F., Pu J., Zhao J., Zhao Q., Tang G. (2014). Characterization of AvaR1, an autoregulator receptor that negatively controls avermectins production in a high avermectin-producing strain. Biotechnol. Lett. 36, 813–819. 10.1007/s10529-013-1416-y [DOI] [PubMed] [Google Scholar]
- Wang W., Ji J., Li X., Wang J., Li S., Pan G., et al. (2014). Angucyclines as signals modulate the behaviors of Streptomyces coelicolor. Proc. Natl. Acad. Sci. U.S.A. 111, 5688–5693. 10.1073/pnas.1324253111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W., Li X., Li Y., Li S., Fan K., Yang K. (2015). A genetic biosensor for identification of transcriptional repressors of target promoters. Sci. Rep. 5:15887. 10.1038/srep15887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willey J. M., Gaskell A. A. (2011). Morphogenetic signaling molecules of the streptomycetes. Chem. Rev. 111, 174–187. 10.1021/cr1000404 [DOI] [PubMed] [Google Scholar]
- Xu G., Wang J., Wang L., Tian X., Yang H., Fan K., et al. (2010). “Pseudo” γ-butyrolactone receptors respond to antibiotic signals to coordinate antibiotic biosynthesis. J. Biol. Chem. 285, 27440–27448. 10.1074/jbc.M110.143081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yonekawa T., Ohnishi Y., Horinouchi S. (1999). Involvement of amfC in physiological and morphological development in Streptomyces coelicolor A3(2). Microbiology 145, 2273–2280. 10.1099/00221287-145-9-2273 [DOI] [PubMed] [Google Scholar]
- Yu Z., Reichheld S. E., Savchenko A., Parkinson J., Davidson A. R. (2010). A comprehensive analysis of structural and sequence conservation in the TetR family transcriptional regulators. J. Mol. Biol. 400, 847–864. 10.1016/j.jmb.2010.05.062 [DOI] [PubMed] [Google Scholar]
- Zhao J., Wen Y., Chen Z., Song Y., Li J. (2007). An adpA homologue in Streptomyces avermitilis is involved in regulation of morphogenesis and melanogenesis. Chin. Sci. Bull. 52, 623–630. 10.1007/s11434-007-0105-4 [DOI] [Google Scholar]
- Zhu J., Sun D., Liu W., Chen Z., Li J., Wen Y. (2016). AvaR2, a pseudo γ-butyrolactone receptor homologue from Streptomyces avermitilis, is a pleiotropic repressor of avermectin and avenolide biosynthesis and cell growth. Mol. Microbiol. 102, 562–578. 10.1111/mmi.13479 [DOI] [PubMed] [Google Scholar]
- Zhuo Y., Zhang W., Chen D., Gao H., Tao J., Liu M., et al. (2010). Reverse biological engineering of hrdB to enhance the production of avermectins in an industrial strain of Streptomyces avermitilis. Proc. Natl. Acad. Sci. U.S.A. 107, 11250–11254. 10.1073/pnas.1006085107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zianni M., Tessanne K., Merighi M., Laguna R., Tabita F. R. (2006). Identification of the DNA bases of a DNase I footprint by the use of dye primer sequencing on an automated capillary DNA analysis instrument. J. Biomol. Tech. 17, 103–113. [PMC free article] [PubMed] [Google Scholar]
- Zou Z., Du D., Zhang Y., Zhang J., Niu G., Tan H. (2014). A γ-butyrolactone-sensing activator/repressor, JadR3, controls a regulatory mini-network for jadomycin biosynthesis. Mol. Microbiol. 94, 490–505. 10.1111/mmi.12752 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.