Abstract
The association of kava product use with liver-related risks has prompted regulatory action in many countries. We studied the changes in gene expression of drug metabolizing enzymes in the livers of Fischer 344 male rats administered kava extract by gavage for 14 weeks. Analysis of 22,226 genes revealed that there were 14, 41, 110, 386, and 916 genes significantly changed in the 0.125, 0.25, 0.5, 1.0, and 2.0 g/kg treatment groups, respectively. There were 16 drug metabolizing genes altered in all three high-dose treatment groups, among which seven genes belong to cytochrome P450 isozymes. While gene expression of Cyp1a1, 1a2, 2c6, 3a1, and 3a3 increased; Cyp 2c23 and 2c40 decreased, all in a dose-dependent manner. Real-time PCR analyses of several genes verified these results. Our results indicate that kava extract can significantly modulate drug metabolizing enzymes, particularly the CYP isozymes, which could cause herb-drug interactions and may potentially lead to hepatotoxicity.
Keywords: Drug metabolizing enzyme, Drug metabolizing gene, Gene expression, Kava extract, Microarray, TaqMan assay
1. Introduction
Since the US Congress passed the Dietary Supplement Health and Education Act (DSHEA) for the US Food and Drug Administration (FDA) to regulate dietary supplements in 1994, herbal products represent the fastest growing segment of the VMH (Vitamin, Mineral supplements, and Herbal products) industry (Chan and Fu, 2007; Chan et al., 2007; FDA, 1994). Currently, there are a variety of herbal dietary supplements sold in the United States, including the most widely-used products, echinacea, St John’s wort, golden seal, ginseng (Panax ginseng, American ginseng, and Siberian ginseng), kava, Ginkgo biloba (ginkgo), Aloe vera, and mild thistle extract.
Although it is perceived that “natural” products are safe, evidence from clinic and scientific data suggests that the use of herbal plants is not without risk. It has been reported that a majority of herbal dietary supplements cause adverse health effects (Chan and Fu, 2007; Chan et al., 2007; Fu, 2007; Fu et al., 2007, 2008; Gurley et al., 2005, 2007; Hu et al., 2005; Singh, 2005). To date, safety issues concerning potential side-effects and toxic contamination of herbal products have not been adequately addressed and toxicological data on the identification of genotoxic and tumorigenic ingredients in many raw herbs are also lacking. Thus, assessment of the efficacy and safety of herbal plants and herbal dietary supplements is important for human health protection (FDA, 2001, 2004a,b; Fong, 2002; Fu et al., 2002).
Both herbal products, including herbal dietary supplements, and many therapeutic drugs require drug metabolizing enzymes for metabolism in order to exert their therapeutic effects. Herbal dietary supplements can also modulate drug metabolizing enzymes, particularly the cytochrome P450 isozymes (CYPs). As a consequence, concomitant administration of dietary supplements and therapeutic drugs very likely raise the potential for herb–drug interactions, which may lead to serious clinical consequences (Bressler, 2005; Gurley et al., 2005, 2007; Hu et al., 2005; Mathews et al., 2002; Schulze et al., 2003; Singh, 2005). For instance, several reports have indicated that G. biloba inhibites CYP activity, and when taken in combination with prescription and conventional medications may produce cytochrome P450-mediated herb–drug interactions (Bressler, 2005; Gurley et al., 2005, 2007; Hu et al., 2005; Matthias et al., 2007; Singh, 2005; Williamson, 2005; Wold et al., 2005; Zou et al., 2002).
Kava kava, prepared from the rhizome of the kava tropical shrub plant, Piper methysticum Forst. F., is a traditional beverage used for many centuries in the South Pacific (Dentali, 1997; Schulze et al., 2003; Singh, 1992, 2005; Smith et al., 1984). During recent years, kava has been used in Europe for treatment of anxiety and nervous disorders such as stress and restlessness, and in the United States as a natural alternative to anti-anxiety drugs and sleeping pills (Dentali, 1997). However, the potential hepatotoxicity of kava in humans has recently been reported (Brauer et al., 2001; Campo et al., 2002; Clough et al., 2003; De Smet, 2002; Ernst, 2006; Humberston et al., 2003; Parkman, 2002; Russmann et al., 2001, 2003; Saß et al., 2001). The association of kava product use with liver-related risks prompted regulatory agencies in many countries, including Germany, Switzerland, France, Canada, and the United Kingdom, to act. The actions range from warning consumers to removing kava-containing products from the marketplace. On March 25, 2002, the Center for Food Safety and Applied Nutrition (CFSAN) of the US Food and Drug Administration (FDA) issued a Consumer Advisory entitled “Kava-containing dietary supplements may be associated with severe liver injury”(CFSAN, 2002).
Kava-containing products remain popular in the United States and continue to be sold in health food stores and ethnic markets regardless of the fact that it was banned in several Western countries following reports of alleged hepatotoxicity. Kava extract was nominated for a chronic tumorigenicity bioassay conducted by the National Toxicology Program (NTP). The NTP conducted 14-week rat studies to characterize the toxicology of kava exposure in Fischer 344 rats. Groups of 10 male and 10 female rats were administered kava extract by gavage at 0, 0.125, 0.25, 0.5, 1.0, or 2.0 g/kg/day. Using these animal liver tissues, Clayton et al. (2007) reported the expression of hepatic cytochrome P450 by immunohistochemical analysis. It was determined that there was decreased expression of Cyp 2D1 (human CYP2D6 homolog) in 2.0 g/kg females and increased expression of Cyp 1A2, 2B1, and 3A1 in the 1.0 and 2.0 g/kg groups of both sexes. It was proposed that kava-induced hepatic functional changes in the rat might be relevant to human clinical cases of hepatotoxicity following exposure. Because only a limited number of antibodies were used for the immunohistochemical studies (Clayton et al., 2007), it is not known how many kava-induced drug metabolizing genes were significantly altered.
We have been interested in studying the toxicity and tumorigenicity induced by a variety of dietary supplements and Chinese herbal plants. Our study includes the identification of toxic contaminants and tumorigenic components, determination of the mechanisms leading to hepatotoxicity and tumorigenicity in experimental animals, and a DNA microarray study of gene expression profiles (Chan and Fu, 2007; Chan et al., 2006, 2007; Chou and Fu, 2006; Fu, 2007; Fu et al., 2002, 2007, 2008; Guo et al., 2006, 2007; Mei et al., 2006, 2007). In this paper, we examined the gene expression changes of drug metabolizing enzymes in the same livers of F344 male rats used for the 14-week study (oral treatment with kava extract). We found that compared to the control group, the expression of a large number of drug metabolizing genes is significantly altered in a dose-dependent manner after treatment with kava extract, and the changes in gene expression were further validated by real-time PCR analysis.
2. Material and methods
2.1. Kava extract preparation
Kava extract was formulated in the corn oil at 0, 25, 50, 100, 200, and 400 mg/ml and stored in sealed glass containers. Homogeneity of formulations was determined as indicated prior to the start of administration of dosages.
2.2. Rat liver tissue collection and RNA isolation
Six treatment groups of F344 rats, 10 male rats per group, were administered kava extract in corn oil by gavage at 0 (vehicle control), 0.125, 0.25, 0.5, 1.0, or 2.0 g/kg/day, 5 days per week for 14 weeks. At 14 weeks, animals were sacrificed and livers were collected. Animal handling and husbandry were conducted in accordance with guidelines of National Institutes of Health (NIH).
Total RNA from liver tissue was isolated using Trizol reagent (Invitrogen, Carlsbad, CA) and then further purified using an RNeasy system (Qiagen, Valencia, CA). The yield of the extracted RNA was determined spectrophotometrically by measuring the optical density at 260 nm. The purity and quality of extracted RNA were evaluated using the RNA 6000 LabChip and Agilent 2100 Bioanalyzer (Agilent Technologies, Palo Alto, CA). High quality RNA with RNA integrity numbers (RINs) greater than 8.5 was used for microarray experiments and TaqMan gene expression assays.
2.3. Microarray analysis
To identify signatures in gene expression profiles associated with effects of chemical treatments, we used microarray techniques to examine the gene expression patterns in rat livers exposed to various doses of kava extract. Gene expression profiling was performed using the Illumina Sentrix Rat Ref-12 Expression BeadChip platform that contains 22,226 probes (Illumina Inc., San Diego, CA).
2.3.1. Sample labeling and quality control of labeled aRNA
For each sample, 200 ng total RNA was labeled using a MessageAmp II-biotin enhanced kit (Applied Biosystems, Foster City, CA) according to the manufacturer’s instructions. Briefly, double stranded cDNA was synthesized using T7-oligo (dT) primers and followed by an in vitro transcription (IVT) reaction to amplify aRNA while biotin was incorporated into the synthesized aRNA probe. The aRNA probe was then purified and quantified using a NanoDrop spectrophotometer (Thermo Fisher Scientific, Waltham, MA). The size distributions of aRNA were made by running 200 ng of each of sample on the Bioanalyzer (Agilent) using the Eukaryotic mRNA Assay with smear analysis.
2.3.2. Sample hybridization
Biotinylated cRNA probe was hybridized to the Rat Raf-12 Sentrix Arrays (Illumina). Labeled aRNA (1.5 µg) was used for hybridization for each array. The hybridization, washing and scanning were performed according to the manufacturer’s instructions with the addition of a 10-min wash in High-temp wash buffer (Illumina) at 55 °C for 10 min in a Scigene Hybex Microarray Incubation System (Sci-Gene Corportaion, Sunnyvale, CA) with a water bath insert following the overnight hybridization.
2.3.3. Scanning and data outputs
The chips were scanned using a BeadScan 2.3.0.10 (Illumina) at a multiplier setting of “2.” The microarray images were registered and extracted automatically during the scan according to the manufacturer’s default settings.
2.3.4. Normalization
Raw microarray intensity data were background subtracted and normalized using the cubic spline normalization method.
2.3.5. Microarray data analysis
Gene expression data from the Illumina Rat Ref-12 were input into ArrayTrack, a software system developed by the FDA’s National Center for Toxicological Research for the management, analysis, visualization and interpretation of microarray data (http://www.fda.gov/nctr/science/centers/toxicoinformatics/ArrayTrack/). Clustering analysis was conducted within ArrayTrack. Additional calculations were performed within JMP 6 (SAS Institute, Cary, NC).
2.3.6. Detection of differentially expressed genes
A list of differentially expressed genes was identified using a two group t-test. The criteria were P-value <0.05 and a mean difference greater than 1.5-fold. No filtering related to gene detection was applied.
2.4. TaqMan gene expression assays
The gene expression of the following five drug metabolizing genes were confirmed by TaqMan assays (Applied Biosystems, Foster City, CA). The TaqMan probes are listed: Cyp1a1 (Rn00487218_m1); Cyp1a2 (Rn00561082_m1); Cyp2d1 (Rn01775090_mH); Cp2e1 (Rn00580624_m1); Cyp3a1 (Rn01640761_gH). The following two genes were used for endogenous controls: Polr2a (Rn01496541_m1) and Actb (Rn00667869_m1).
2.4.1. First strand cDNA synthesis
cDNA was prepared using a High-Capacity cDNA Archive Kit (Applied Biosystems). Briefly, total RNA (2 µg) was reverse-transcribed in a final volume of 20 µl with random primers at 25 °C for 10 min followed by 37 °C for 120 min according to the manufacturer’s instructions.
2.4.2. TaqMan assays
Each assay was run in triplicate for each RNA sample. Total cDNA (25 ng) in a 25 µl final volume was used for each assay. Assays were run with Universal Master Mix (2X) without AmpErase UNG on an Applied Biosystems 7000 Real-Time PCR System using universal cycling conditions (10 min at 95 °C; 15 s at 95 °C, 1 min 60 °C, 40 cycles).
2.4.3. Data normalization and analysis
Two endogenous control genes, β-actin (Actb) and RNA polymerase II A (Polr2a), were used for normalization. Each replicate cycle threshold (CT) was normalized to the average CT of the two endogenous controls on a per sample basis. The comparative CT method was used to calculate relative quantification of gene expression (Livak and Schmittgen, 2001). The following formula was used to calculate the relative amount of the transcripts in the kava-treated sample (treat) and the vehicle-treated sample (control), and both were normalized to the endogenous controls. ΔΔCT = Δ CT (treat) − Δ CT (control).
ΔCT is the difference in CT between the target gene and endogenous controls by subtracting the average CT of controls. The fold-change for each treated sample relative to the control sample = 2−ΔΔct.
2.4.4. Sensitivity detection and identification of differentially expressed gene
TaqMan assay’s quantification was decided by the CT number. The CT number for each reaction was determined by setting the same threshold value across all reactions. A gene was considered not detectable when CT > 32. A list of differentially expressed genes was identified using a two-tailed t-test. The criteria were P-value less than 0.05 and a mean difference equal to or greater than 2-fold. The statistical calculation was based on ΔCT values.
3. Results
3.1. DNA microarray data quality
In this study, the gene expression profiles of the liver of male rats treated with kava for 14 weeks were determined by whole genome wide gene expression microarray analysis (Illumina Sentrix Rat Ref-12 Expression BeadChip). There were one control group and five treatment groups in this study, each group containing four biological replicates, with a total of 24 microarrays performed. To assess the overall quality and reproducibility of the DNA microarray data, Pearson’s correlation coefficient of pair-wise log2 intensity correlation was calculated. Pearson’s correlation matrix of 24 arrays was calculated based on all data points (22,226 probes), no filtering applied. As shown in Table 1, the median rank of correlation was 0.989 across all 24 arrays with the range of 0.976–0.993, indicating the data from the microarrays were highly reproducible.
Table 1.
Assessment of microarrav data quality.
| L_13M | L_14M | L_15M | L_17M | L_22M | L_25M | L_27M | L_28M | L_2M | L_32M | L_34M | L_37M | L_39M | L_3M | L_41M | L_47M | L_48M | L_50M | L_51M | L_54M | L_56M | L_59M | L_5M | L_8M | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| L_13M_0.125 | 1.000 | 0.990 | 0.989 | 0.990 | 0.990 | 0.990 | 0.990 | 0.989 | 0.990 | 0.988 | 0.987 | 0.988 | 0.987 | 0.989 | 0.985 | 0.984 | 0.984 | 0.985 | 0.977 | 0.979 | 0.980 | 0.981 | 0.989 | 0.987 |
| L_14M_0.125 | 1.000 | 0.989 | 0.991 | 0.991 | 0.991 | 0.991 | 0.990 | 0.990 | 0.989 | 0.989 | 0.989 | 0.988 | 0.989 | 0.987 | 0.986 | 0.986 | 0.987 | 0.980 | 0.982 | 0.982 | 0.984 | 0.989 | 0.987 | |
| L_15M_0.125 | 1.000 | 0.990 | 0.989 | 0.990 | 0.990 | 0.990 | 0.989 | 0.987 | 0.987 | 0.989 | 0.988 | 0.990 | 0.985 | 0.984 | 0.985 | 0.986 | 0.978 | 0.980 | 0.980 | 0.982 | 0.989 | 0.987 | ||
| L_17M_0.125 | 1.000 | 0.991 | 0.991 | 0.991 | 0.991 | 0.991 | 0.989 | 0.989 | 0.989 | 0.988 | 0.990 | 0.986 | 0.985 | 0.986 | 0.987 | 0.980 | 0.982 | 0.981 | 0.983 | 0.990 | 0.989 | |||
| L_22M_0.25 | 1.000 | 0.993 | 0.992 | 0.992 | 0.991 | 0.989 | 0.990 | 0.989 | 0.989 | 0.989 | 0.987 | 0.987 | 0.986 | 0.987 | 0.981 | 0.983 | 0.983 | 0.985 | 0.989 | 0.988 | ||||
| L_25M_0.25 | 1.000 | 0.992 | 0.992 | 0.991 | 0.990 | 0.991 | 0.991 | 0.990 | 0.990 | 0.988 | 0.988 | 0.987 | 0.987 | 0.981 | 0.982 | 0.983 | 0.985 | 0.989 | 0.988 | |||||
| L_27M_0.25 | 1.000 | 0.991 | 0.991 | 0.989 | 0.990 | 0.989 | 0.989 | 0.989 | 0.987 | 0.987 | 0.986 | 0.988 | 0.981 | 0.983 | 0.983 | 0.985 | 0.989 | 0.988 | ||||||
| L_28M_0.25 | 1.000 | 0.991 | 0.989 | 0.990 | 0.990 | 0.989 | 0.990 | 0.986 | 0.986 | 0.986 | 0.987 | 0.980 | 0.982 | 0.982 | 0.984 | 0.989 | 0.988 | |||||||
| L_2M_0 | 1.000 | 0.988 | 0.988 | 0.988 | 0.987 | 0.990 | 0.985 | 0.984 | 0.985 | 0.986 | 0.978 | 0.980 | 0.980 | 0.982 | 0.990 | 0.988 | ||||||||
| L_32M_0.5 | 1.000 | 0.993 | 0.992 | 0.992 | 0.987 | 0.991 | 0.991 | 0.991 | 0.991 | 0.986 | 0.987 | 0.987 | 0.988 | 0.987 | 0.985 | |||||||||
| L_34M_0.5 | 1.000 | 0.992 | 0.992 | 0.987 | 0.991 | 0.992 | 0.990 | 0.991 | 0.986 | 0.986 | 0.986 | 0.988 | 0.987 | 0.986 | ||||||||||
| L_37M_0.5 | 1.000 | 0.993 | 0.988 | 0.992 | 0.990 | 0.991 | 0.991 | 0.985 | 0.987 | 0.987 | 0.988 | 0.987 | 0.986 | |||||||||||
| L_39M_0.5 | 1.000 | 0.986 | 0.991 | 0.991 | 0.991 | 0.990 | 0.985 | 0.986 | 0.986 | 0.987 | 0.987 | 0.985 | ||||||||||||
| L_3M_0 | 1.000 | 0.984 | 0.983 | 0.983 | 0.985 | 0.976 | 0.978 | 0.978 | 0.980 | 0.988 | 0.987 | |||||||||||||
| L_41M_1 | 1.000 | 0.992 | 0.993 | 0.992 | 0.989 | 0.990 | 0.990 | 0.991 | 0.984 | 0.983 | ||||||||||||||
| L_47M_1 | 1.000 | 0.992 | 0.991 | 0.990 | 0.990 | 0.991 | 0.992 | 0.983 | 0.982 | |||||||||||||||
| L_48M_1 | 1.000 | 0.991 | 0.990 | 0.991 | 0.990 | 0.990 | 0.984 | 0.983 | ||||||||||||||||
| L_50M_1 | 1.000 | 0.988 | 0.990 | 0.989 | 0.990 | 0.984 | 0.983 | |||||||||||||||||
| L_51M_2 | 1.000 | 0.993 | 0.993 | 0.992 | 0.977 | 0.976 | ||||||||||||||||||
| L_54M_2 | 1.000 | 0.992 | 0.993 | 0.979 | 0.978 | |||||||||||||||||||
| L_56M_2 | 1.000 | 0.993 | 0.979 | 0.978 | ||||||||||||||||||||
| L_59M_2 | 1.000 | 0.981 | 0.980 | |||||||||||||||||||||
| L_5M_0 | 1.000 | 0.987 | ||||||||||||||||||||||
| L_8M_0 | 1.000 |
The microarray data reproducibility was assessed by the Pearson’s correlation coefficient of pair-wise log2 intensity correlation. The median rank of correlation is 0.989 across all 24 arrays with the range of 0.976–0.993. Pearson’s correlation matrix of 24 arrays was calculated based on all data points (22,226 probes).
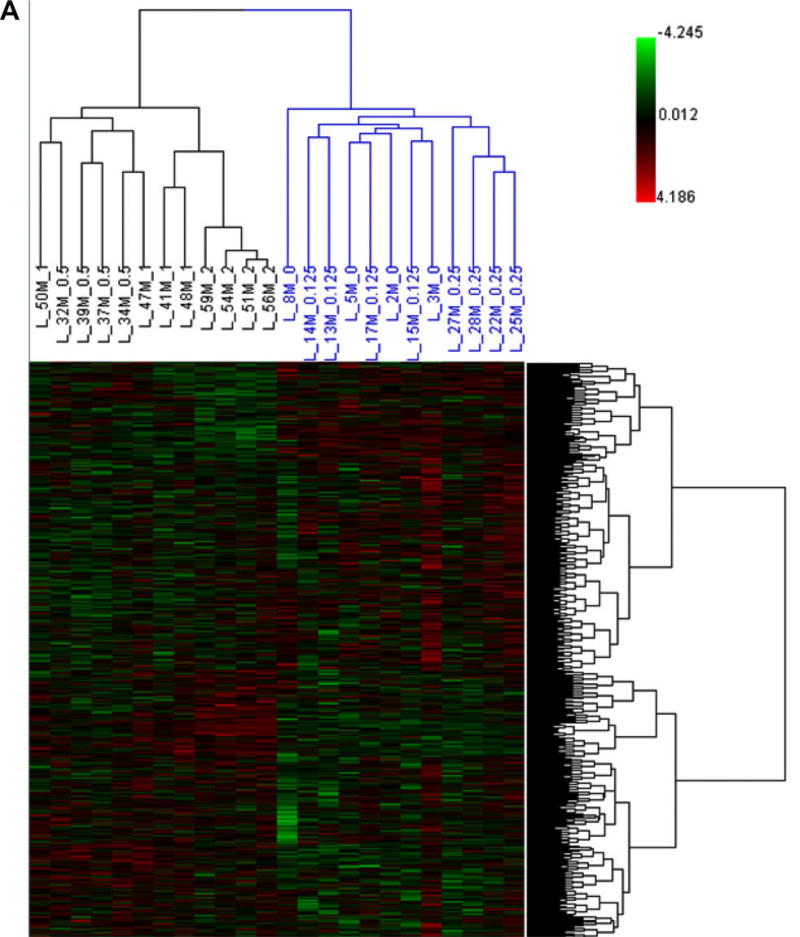
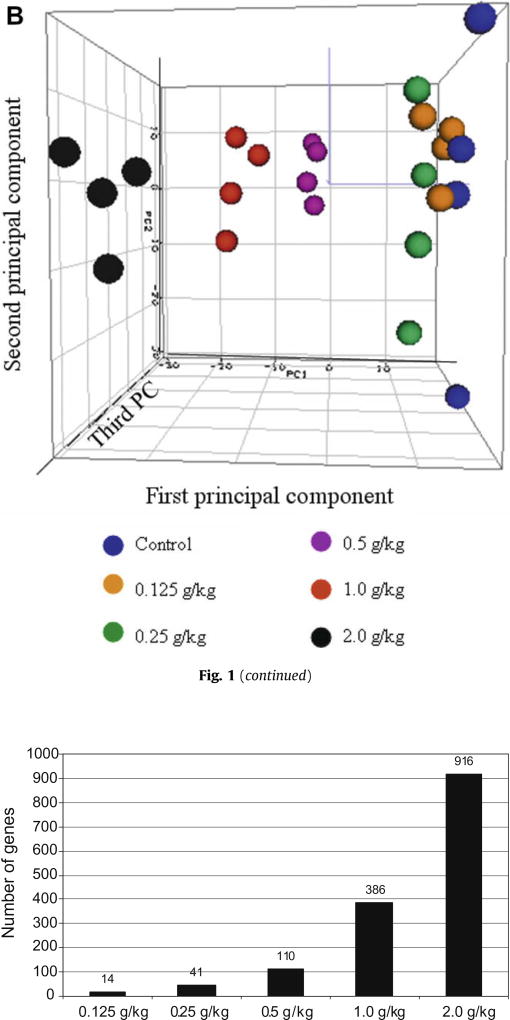
Hierarchical Cluster Analysis (HCA) (Fig. 1A) and Principal Component Analysis (PCA) (Fig. 1B) were employed to determine the gene expression profiles for control and the five kava-treated groups. HCA showed there were two large clusters and samples were clustered according to the treatment groups, in terms of high-dose groups (0.5, 1.0 and 2.0 g/kg kava treatment, in black) versus low-dose groups (0, 0.125 and 0.25 g/kg kava treatment, in blue). PCA revealed that the three high-dose groups (0.5, 1.0 and 2.0 g/kg kava treatment) were well separated, while the two low-dose groups, 0.125 g/kg and 0.25 g/kg, were less separated from the control group. The results indicated clear treatment effects were detectable in the high-dose treatment groups. It should be noted that clustering analysis is an unbiased data analysis procedure, because it does not consider of the assignment of doses of kava extract. More over, the entire data set, the data from a total of 22,226 probes, without a specific cut off, was applied for the clustering analysis.
Fig. 1.
(A) Hierarchical Cluster Analysis (HCA) of expression profiles for control and kava-treated groups. The log2 intensity of the entire gene set was scaled by Z-score transformation, and then these values were hierarchically clustered using 1 − r distance metric and average linkage. Each column represents the results from an individual animal. 0 (control), 0.125, 0.25, 0.5,1 and 2 represent 0.0 g/kg, 0.125 g/kg, 0.25 g/kg, 0.5 g/kg, 1.0 g/kg and 2.0 g/kg kava treatment, respectively. (B) Principal Component Analysis (PCA) of expression profiles for control and kava-treated groups. The intensity of the entire gene set was used; no specific cut off was applied for the analysis.
3.2. Analysis of differentially expressed genes
3.2.1. Total differentially expressed genes
A differentially expressed gene was identified based on the criteria of a fold-change greater than 1.5 (up or down) and a P-value less than 0.05 in comparison to the control group. Based on these two criteria, out of 22,226 probes there were 14, 41, 110, 386, and 916 genes, respectively, in the 0.125, 0.25, 0.5, 1.0, and 2.0 g/ kg treatment groups that satisfied the requirements (Table 2 and Fig. 2). These results demonstrate a dose-response relationship for the number of genes affected. The data shown in Table 2 indicate that there are nearly equal numbers of up- and down-regulated genes in each of the kava treatment groups, with the exception of the lowest dose treatment group where most of genes were down-regulated.
Table 2.
Numbers of genes altered in liver treated with various doses of kava extract.
| Treatment (g/kg) |
Total # of altered genes |
# of up-regulated genes |
# of down-regulated genes |
|---|---|---|---|
| 0.125 | 14 | 3 | 11 |
| 0.25 | 41 | 21 | 20 |
| 0.5 | 110 | 58 | 52 |
| 1.0 | 386 | 179 | 207 |
| 2.0 | 916 | 422 | 494 |
A gene was identified as significantly changed if the fold-change was greater than 1.5 and the P-value was less than 0.05 in comparison to control group. Each treatment group consisted of four biological replicates.
Fig. 2.
Number of genes altered in rat livers treated with various doses of kava extract for 14-weeks. There is an apparent difference in the number of genes that are significantly up- or down-regulated by kava treatments. A gene was identified as significantly changed if the fold-change (in comparison to control group) was greater than 1.5 and the P-value was less than 0.05. Each treatment group consisted of four biological replicates.
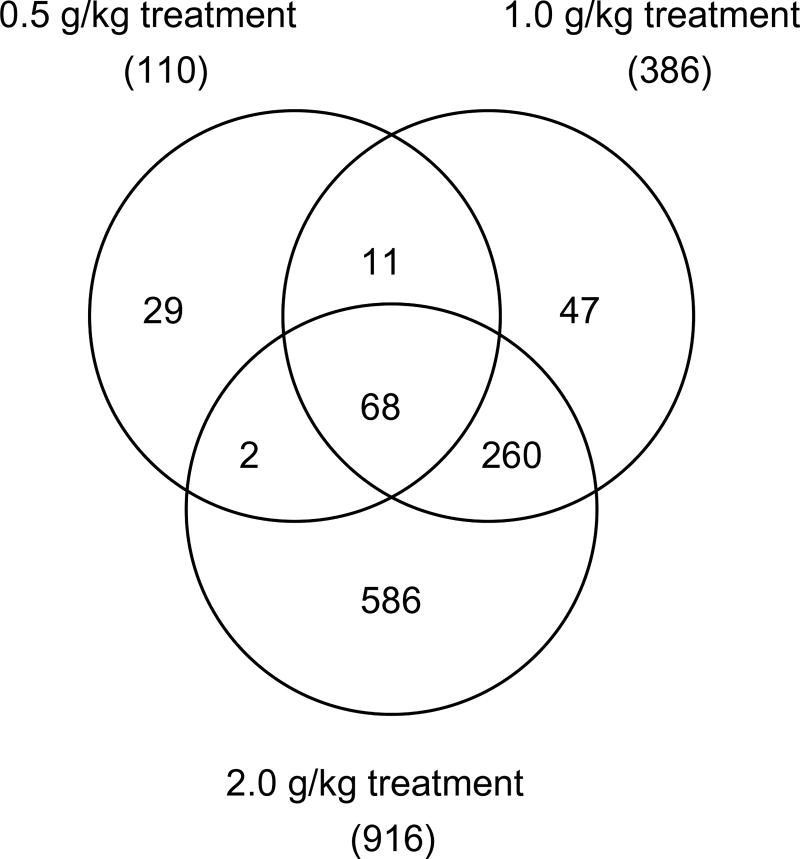
As the PCA result demonstrated in Fig. 1B, a clear separation was observed between the control group/low-dose groups and the three high-dose kava treatment groups (0.5, 1.0 and 2.0 g /kg). Since there were only a limited number of genes altered in the two low-dose groups (14 for 0.125 g/kg and 41 for 0.25 g/kg, Fig. 2), we focused our analysis on the three high-dose groups in comparison with the control group. As demonstrated in the Venn diagram (Fig. 3), there were 68 genes commonly regulated by all three higher dose kava treatments; while there were 328 genes regulated by both the highest dose (2.0 g/kg) and second highest dose groups (1.0 g/kg). Very interestingly, the majority of differentially expressed genes identified in the lower dose group were also found in the higher dose groups. There were 74% (81 out of 110) of the genes in the 0.5 g/kg treatment group that overlapped those in either the 1.0 g/kg or 2.0 g/kg treatment groups. There were 85% (328 out of 386) of the genes in the 1.0 g/kg treatment group that overlapped those in the 2.0 g/kg treatment groups. Without exception, all commonly regulated genes were changed in the same direction (up-regulated or down-regulated).
Fig. 3.
Numbers of differentially expressed genes regulated by 0.5, 1.0 and 2.0 g/kg kava treatments. A gene was identified as differentially expressed if the fold-change was greater than 1.5 (up- or down-regulated) and the P-value was less than 0.05 in comparison to the control group.
3.2.2. Total differentially expressed genes of drug metabolizing enzymes
Since metabolic activation of chemicals is very important for liver toxicity, we investigated the gene expression changes of drug metabolizing enzymes for 2.0 g/kg kava treatment in detail. Table 3 shows 72 drug metabolizing genes whose expression was significantly changed by 2.0 g/kg kava treatment. As tabulated in Table 3, among the 72 drug metabolizing enzyme associated genes, 19 genes were associated with Phase I metabolizing enzymes; 21 genes with Phase II metabolizing enzymes; and 32 genes with transporters (Phase III). Sixteen out of the 19 expressed Phase I metabolism associated genes belong to the CYPs. The other three non-CYP Phase I metabolism genes were aldehyde dehydrogenase family 1, member A1 (Aldh1a1), flavin containing monooxygenase 1 (Fmo1), and granzyme (Gzma).
Table 3.
Genes involved in 3 Phases of drug metabolism altered by 2.0 g/kg kava treatment in liver.
| Gene symbol | Gene description | Locus Link ID | Fold change | P-value |
|---|---|---|---|---|
| Phase I metabolism | ||||
| Aldh1a1 | aldehyde dehydrogenase family 1, member A1 | 24,188 | 2.8 | <0.001 |
| Cyp17a1 | cytochrome P450, family 17, subfamily a, polypeptide 1 | 25,146 | −2.2a | 0.005 |
| Cyp1a1 | cytochrome P450, family 1, subfamily a, polypeptide 1 | 24,296 | 214.0 | <0.001 |
| Cyp1a2 | cytochrome P450, family 1, subfamily a, polypeptide 2 | 24,297 | 6.4 | <0.001 |
| Cyp2c23 | cytochrome P450, family 2, subfamily c, polypeptide 23 | 83,790 | −1.7 | <0.001 |
| Cyp2c37 | cytochrome P450, 2c37 | 29,296 | 2.1 | 0.001 |
| Cyp2c40 | cytochrome P450, family 2, subfamily c, polypeptide 40 | 25,011 | −5.3 | <0.001 |
| Cyp2c55_predicted | cytochrome P450, family 2, subfamily c, polypeptide 55 (predicted) | 292,330 | 4.0 | <0.001 |
| Cyp2c6 | cytochrome P450, subfamily IIC6 | 246,070 | 4.0 | <0.001 |
| Cyp2t1 | cytochrome P450 monooxygenase CYP2T1 | 171,380 | −1.7 | <0.001 |
| Cyp3a1 | cytochrome P450, family 3, subfamily a, polypeptide 1 | 286,929 | 2.4 | <0.001 |
| Cyp3a13 | cytochrome P450, family 3, subfamily a, polypeptide 13 | 171,352 | −3.7 | <0.001 |
| Cyp3a3 | cytochrome P450, subfamily 3A, polypeptide 3 | 25,642 | 5.7 | <0.001 |
| Cyp4a12 | cytochrome P450, 4al2 | 266,674 | 1.5 | 0.005 |
| Cyp4f6 | cytochrome P450 4F6 | 266,689 | −1.7 | <0.001 |
| Cyp7b1 | cytochrome P450, family 7, subfamily b, polypeptide 1 | 25,429 | −2.2 | 0.001 |
| Cyp8b1 | cytochrome P450, family 8, subfamily b, polypeptide 1 | 81,924 | −1.6 | 0.036 |
| Fmo1 | flavin containing monooxygenase 1 | 25,256 | −3.8 | <0.001 |
| Gzma | granzyme A | 266,708 | −1.6 | 0.004 |
| Phase II metabolism | ||||
| Acsl4 | acyl-CoA synthetase long-chain family member 4 | 113,976 | 1.7 | <0.001 |
| Ces2 | carboxylesterase 2 (intestine, liver) | 171,118 | 8.5 | <0.001 |
| Ces6 | carboxylesterase 6 | 246,252 | 2.6 | <0.001 |
| Ephx1 | epoxide hydrolase 1, microsomal | 25,315 | 3.6 | <0.001 |
| Gnmt | glycine N-methyltransferase | 25,134 | −2.1 | <0.001 |
| Gsta2 | glutathione-S-transferase, alpha type2 | 24,422 | 2.9 | <0.001 |
| Gsta4_predicted | glutathione S-transferase, alpha 4 | 300,850 | 2.0 | <0.001 |
| Gstm1 | glutathione S-transferase, mu 1 | 24,423 | 1.8 | <0.001 |
| Gstm2 | glutathione S-transferase, mu 2 | 24,424 | 1.6 | 0.005 |
| Gstm3 | glutathione S-transferase, mu type 3 | 81,869 | −1.6 | 0.018 |
| Gstp1 | glutathione-S-transferase, pi 1 | 24,426 | 1.6 | 0.001 |
| Nqo1 | NAD(P)H dehydrogenase, quinone 1 | 24,314 | 11.4 | <0.001 |
| Sulf2_predicted | sulfatase 2 | 311,642 | −1.6 | 0.002 |
| Sult1b1 | sulfotransferase family 1B, member 1 | 64,305 | 1.8 | <0.001 |
| Sult1c1 | sulfotransferase family, cytosolic, 1C, member 1 | 65,185 | −2.0 | <0.001 |
| Ugt1a1 | UDP glycosyltransferase 1 family, polypeptide A1 | 24,861 | 1.7 | 0.005 |
| Ugt1a6 | UDP glycosyltransferase 1 family, polypeptide A6 | 113,992 | 10.0 | <0.001 |
| Ugt1a7 | UDP glycosyltransferase 1 family, polypeptide A7 | 154,516 | 3.5 | <0.001 |
| Ugt2b10_predicted | UDP glycosyltransferase 2 family, polypeptide BIO (predicted) | 305,264 | 2.1 | <0.001 |
| Ugt2b4 | UDP glycosyltransferase 2 family, polypeptide B4 | 83,808 | 1.5 | <0.001 |
| Ugt2b4 | UDP glycosyltransferase 2 family, polypeptide B4 | 83,808 | 1.8 | 0.001 |
| Phase III metabolism | ||||
| Abca8_predicted | ATP-binding cassette, subfamily A (ABC1), member 8a (predicted) | 303,638 | −1.8 | <0.001 |
| Abcb6 | ATP-binding cassette, subfamily B (MDR/TAP), member 6 | 140,669 | 1.9 | <0.001 |
| Abcb9 | ATP-binding cassette, subfamily B (MDR/TAP), member 9 | 63,886 | −2.0 | 0.047 |
| Abcc2 | ATP-binding cassette, subfamily C (CFTR/MRP), member 2 | 25,303 | 1.7 | <0.001 |
| Abcc3 | ATP-binding cassette, subfamily C (CFTR/MRP), member 3 | 140,668 | 15.9 | <0.001 |
| Abcc6 | ATP-binding cassette, subfamily C (CFTR/MRP), member 6 | 81,642 | −1.8 | <0.001 |
| Abcc8 | ATP-binding cassette, subfamily C (CFTR/MRP), member 8 | 25559 | −1.7 | 0.006 |
| Abcc9 | ATP-binding cassette, subfamily C (CFTR/MRP), member 9 | 25,560 | −1.8 | 0.027 |
| Slc11a1 | solute carrier family 11 (proton-coupled divalent metal ion transporters), member 1 | 316,519 | −1.7 | 0.001 |
| Slc13a3 | solute carrier family 13 (sodium-dependent dicarboxylate transporter), member 3 | 64,846 | −1.5 | 0.003 |
| Slc16a1 | solute carrier family 16 (monocarboxylic acid transporters), member 1 | 25,027 | 1.5 | 0.001 |
| Slc16a10 | solute carrier family 16 (monocarboxylic acid transporters), member 10 | 170566 | −1.9 | 0.002 |
| Slc16a11_predicted | solute carrier family 16 (monocarboxylic acid transporters), member 11 (predicted) | 287,450 | −1.6 | 0.001 |
| Slc16a6 | solute carrier family 16 (monocarboxylic acid transporters), member 6 | 303,772 | 1.5 | 0.002 |
| Slc17a1 | solute carrier family 17 (sodium phosphate), member 1 | 171,080 | 1.8 | 0.001 |
| Slc17a3 | solute carrier family 17 (sodium phosphate), member 3 | 266,730 | 1.9 | <0.001 |
| Slc22a7 | solute carrier family 22 (organic anion transporter), member 7 | 89,776 | −1.6 | <0.001 |
| Slc22a8 | solute carrier family 22 (organic anion transporter), member 8 | 83,500 | −1.6 | 0.002 |
| Slc25a17_predicted | solute carrier family 25 (mitochondrial carrier, peroxisomal membrane protein), member 17 (predicted) | 300,083 | 1.5 | 0.003 |
| Slc27a5 | solute carrier family 27 (fatty acid transporter), member 5 | 79,111 | −1.9 | <0.001 |
| Slc28a2 | solute carrier family 28 (sodium-coupled nucleoside transporter), member 2 | 60,423 | −1.5 | 0.007 |
| Slc2a2 | solute carrier family 2 (facilitated glucose transporter), member 2 | 25,351 | −1.6 | 0.002 |
| Slc34a2 | solute carrier family 34 (sodium phosphate), member 2 | 84,395 | −4.8 | <0.001 |
| Slc38a3 | solute carrier family 38, member 3 | 252,919 | −1.8 | <0.001 |
| Slc38a4 | solute carrier family 38, member 4 | 170,573 | −1.7 | <0.001 |
| Slc39a4_predicted | solute carrier family 39 (zinc transporter), member 4 (predicted) | 300,051 | 1.8 | 0.004 |
| Slc3a2 | solute carrier family 3 (activators of dibasic and neutral amino acid transport), member 2 | 50,567 | 2.9 | <0.001 |
| Slc4a2 | solute carrier family 4, member 2 | 24,780 | 1.6 | 0.006 |
| Slc6a9 | solute carrier family 6 (neurotransmitter transporter, glycine), member 9 | 116,509 | 1.5 | 0.001 |
| Slc7a5 | solute carrier family 7 (cationic amino acid transporter, y+ system), member 5 | 50,719 | 1.7 | 0.002 |
| Slco2b1 | solute carrier organic anion transporter family, member 2b1 | 140,860 | −1.6 | 0.002 |
| Tap1 | transporter 1, ATP-binding cassette, subfamily B (MDR/TAP) | 24,811 | −1.7 | <0.001 |
The symbol of minus (−) indicates down-regulation.
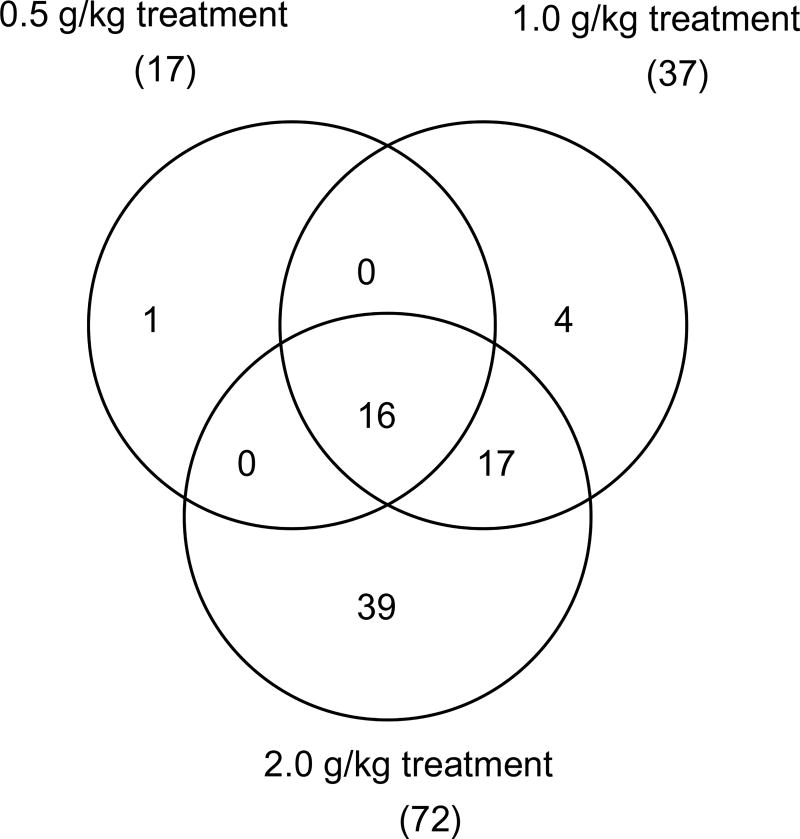
There were 17, 37, and 72 drug metabolizing enzymes that were significantly expressed by the 0.5, 1.0 and 2.0 g/kg kava treatments, respectively, with 16 regulated by all the three treatment groups and 33 in both the highest and second highest treatment groups (Fig. 4). The names of the 16 genes commonly regulated by the three high-dose kava treatments are listed in Table 4.
Fig. 4.
Numbers of drug metabolizing genes regulated by 0.5, 1.0 and 2.0 g/kg kava treatments. A gene was identified as differentially expressed if the fold-change was greater than 1.5 (up- or down-regulated) and the P-value was less than 0.05 in comparison to the control group.
Table 4.
Induction or inhibition of 16 common drug metabolizing genes by three kava treatments (0.5, 1.0 and 2.0 g/kg).
| Gene name | Fold change
|
|||
|---|---|---|---|---|
| Phase | 0.5 (g/kg) | 1.0 (g/kg) | 2.0 (g/kg) | |
| Aldh1a1 | I | 2.1 | 2.5 | 2.8 |
| Cyp1a | I | 7.8 | 35.2 | 214.0 |
| Cyp1a2 | I | 3.7 | 4.2 | 6.4 |
| Cyp2c6 | I | 2.1 | 3.0 | 4.0 |
| Cyp2c23 | I | −1.5a | −1.6 | −1.7 |
| Cyp2c40 | I | −2.5 | −4.3 | −5.3 |
| Cyp3a1 | I | 1.7 | 2.1 | 2.4 |
| Cyp3a3 | I | 1.7 | 2.8 | 5.7 |
| Ces2 | II | 3.1 | 5.5 | 8.5 |
| Ces6 | II | 1.6 | 2.3 | 2.6 |
| Ephx1 | II | 1.8 | 2.7 | 3.6 |
| Gsta2 | II | 1.6 | 2.3 | 2.9 |
| Gstm1 | II | 1.5 | 1.9 | 1.8 |
| Nqo1 | II | 3.7 | 7.5 | 11.4 |
| Ugt1a6 | II | 2.8 | 4.8 | 10.0 |
| Abcc3 | III | 5.7 | 12.3 | 15.9 |
The symbol of minus (−) indicates down-regulation.
(P < 0.01 for every identified gene).
3.2.3. Gene expression of the cytochrome P450 drug metabolizing enzymes
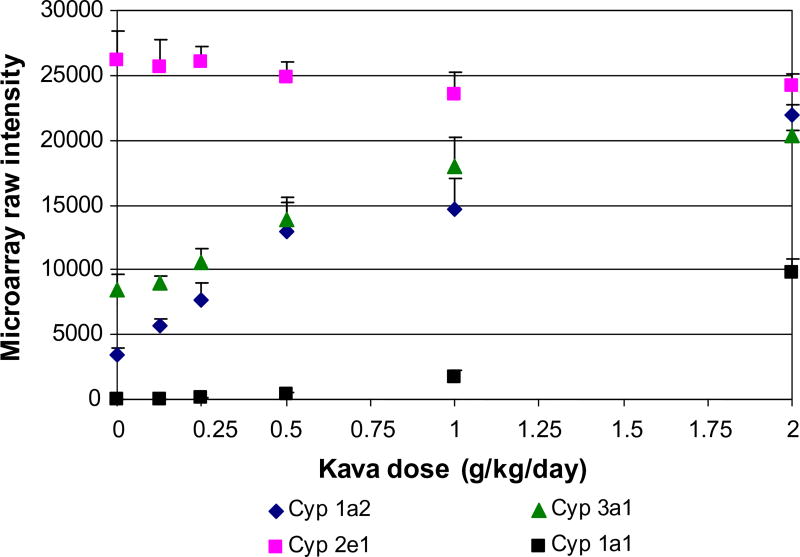
As shown in Table 4, among the 16 genes coded for drug metabolizing enzymes, seven belong to CYPs. As shown in Table 4, while gene expression of Cyp1a1, Cyp1a2, Cyp2c6, Cyp3a1, and Cyp3a3 increased; Cyp 2c23 and Cyp2c40 decreased, all in a dose-dependent manner. The expression changes of a few of these genes (Cyp1a1, 1a2, 3a1 and 2e1) are shown in Fig. 5. Cyp 2e1 did not show a significant change.
Fig. 5.
Gene expression of Cyp1a1, 1a2, 2e1 and 3a1 in the rat liver treated with various doses of kava extract. Intensity of four microarrays for each treatment group and control group were averaged. Data points represent mean + SD (n = 4).
3.3. Real-time PCR validation
TaqMan assays were used to verify the results of the gene expression changes measured using microarrays. Since the most important Phase I metabolizing enzymes are the CYPs, five of these isozymes, including Cyp1a1, 1a2, 2d1, 2e1 and 3a1 were selected for the TaqMan verification. As the results show in Table 5, based on triplicate measurements for each RNA sample, the genes of Cyp1a1, 1a2, and 3a1 are over expressed and the changes are dose-dependent. While there are slight decreases in gene expression of Cyp2d1 and Cyp2e1, these changes were not significant (P-values >0.05).
Table 5.
Gene expression induction of CYPs by TaqMan assay.
| Treatment(g/kg) | Cyp1a1
|
Cyp1a2
|
Cyp2d1
|
Cyp2e1
|
Cyp3a1
|
|||||
|---|---|---|---|---|---|---|---|---|---|---|
| Fold change | P-value | Fold change | P-value | Fold change | P-value | Fold change | P-value | Fold Change | P-value | |
| 0.125 | 4.35 | 0.0003 | 1.65 | 0.0089 | 0.74 | 0.0400 | 1.09 | 0.7213 | 1.01 | 0.9336 |
| 0.25 | 16.29 | 0.0015 | 2.21 | 0.0263 | 0.79 | 0.4228 | 0.98 | 0.5613 | 1.32 | 0.0726 |
| 0.5 | 89.50 | 0.0003 | 3.74 | 0.0021 | 0.68 | 0.2294 | 0.69 | 0.0585 | 1.96 | 0.0145 |
| 1.0 | 452.60 | 0.0003 | 4.60 | 0.4235 | 0.64 | 0.6829 | 0.55 | 0.4280 | 3.34 | 0.0022 |
| 2.0 | 4658.54 | 0.0001 | 12.76 | 0.0046 | 0.73 | 0.2744 | 0.60 | 0.7495 | 5.78 | 0.0014 |
4. Discussion
In this study, we analyzed gene expression changes in the livers of F344 rats following oral treatment with kava extract for 14 weeks. The results of Pearson’s correlation coefficient of pair-wise log2 intensity correlation (Table 1), the Principal Component Analysis, and the Hierarchical Cluster Analysis (Fig. 1) indicate that high quality, reproducible DNA microarray data were obtained. A total of 22,226 probes were analyzed. Not to our surprise, there were 72 drug metabolizing genes that were significantly altered by 2.0 g/kg kava treatment, among which 19, 21, and 32 were Phase I, Phase II, and Phase III drug metabolizing genes, respectively (Table 3).
Sixteen out of the 19 expressed Phase I metabolism associated genes belong to the CYPs (Table 3). The other three non-CYP Phase I metabolism genes include aldehyde dehydrogenase family 1, member A1 (Aldh1a1), flavin containing monooxygenase 1 (Fmo1), and granzyme (Gzma). There were 17, 37, and 72 drug metabolizing enzymes that were significantly altered in the 0.5, 1.0 and 2.0 g/kg kava treatments, respectively, with 16 drug metabolizing enzymes regulated by all the three treatment groups and 33 drug metabolizing genes in both the highest and second highest treatment groups (Fig. 4).
The mechanisms by which kava induces hepatotoxicity are not well understood. It has been proposed that kava induces hepatotoxicity through modulation of drug metabolizing enzymes, in particular, CYP enzymes, thus affecting drug metabolism (Bressler, 2005). When taken concomitantly with therapeutic drugs, kava can lead to herb–drug interactions (Bressler, 2005; Clouatre, 2004; Hu et al., 2005; Singh, 2005). CYP isozymes, the most important metabolizing enzymes, catalyze the oxidative, reductive, and peroxidative metabolism of a variety of both endogenous and exogenous chemicals. Singh (2005) reported that inhibition of CYP isozymes by kavalactones, constituents of kava, can result in pharmacokinetic interactions. Our study, reporting for the first time the analyses of the full spectrum of the gene expression profiles of rats treated with kava, found that a large number of drug metabolizing enzymes have significantly regulated gene expression. It is expected that our findings are important information for studying herb–drug interactions. Our findings also should be highly useful in helping determine the relationship between kava hepatotoxicity and kava-induced gene expression changes of metabolizing enzymes.
In this microarray study, we determined that gene expression of Cyp1a1, 1a2, 2c6, 3a1, and 3a3 increased; but Cyp 2c23 and 2c40 decreased, all in a dose-dependent manner (Table 4). It is worthy to note that Clayton et al. (2007) used the same liver tissues to conduct immunohistochemical analyses of the protein expression of CYP enzymes in liver of the control and 1.0 and 2.0 g/kg treatment groups. They found increased expression of Cyp 1a2, 2b1, and 3a1 in the male rats from the 1.0 and 2.0 g/kg treatment groups, but the expression of Cyp 2e1 remained unchanged. Our finding on the expression of Cyp1a2, 2e1 and 3a1 are consistent with those reported by Clayton et al. (2007) (Table 6) and by Mathews et al. (2002, 2005). In addition, we also found significant expression changes for Cyp1a1, 2c6, 3a3, 2c23, and 2c40. It is known that the metabolism of most xenobiotics is catalyzed by the Cyp1, Cyp2, and Cyp3 isozymes (Martignoni et al., 2006). Thus, human intake of kava extract would increase expression of Cyp1a1, Cyp2c6, Cyp3a3, resulting in increased metabolic activation of toxic xenobiotics. Unfortunately, the Cyp2b1 gene was not present on the rat microarray used in this study, so no comparative data between microarray analysis and immunohistochemical analysis is available for the expression of this gene.
Table 6.
Comparison of the expression induction of Cyp1a2, Cyp2e1 and Cyp3a1 between microarray data and immunohistochemical analysis.
| Dose (g/kg) | Microarray (Fold change)
|
Immunohistochemical analysisa
|
||||
|---|---|---|---|---|---|---|
| Cyp1a2 | Cyp2e1 | Cyp3a1 | Cyp1a2 | Cyp2e1 | Cyp3a1 | |
| 0 | 1.00 | 1.00 | 1.00 | 0.0 ± 0.0 | 2.6 ± 0.2 | 1.0 ±0.0 |
| 0.125 | 1.67*** | 1.01 | 1.08 | |||
| 0.25 | 2.22*** | 1.00 | 1.27* | |||
| 0.5 | 3.71*** | 1.05 | 1.65*** | |||
| 1.0 | 4.25*** | 1.11 | 2.15*** | 0.8 ± 0.2* | 2.6 ± 0.2 | 2.0 ± 0.0** |
| 2.0 | 6.39*** | 1.07 | 2.45*** | 2.0 ± 0.0** | 2.8 ± 0.2 | 2.8 ±0.2** |
Adopted from Clayton et al. (2007).
Differs from controls at P < 0.05.
Differs from controls at P < 0.01.
Differs from controls at P < 0.001.
“The NTP 14-week rat study found that hepatocellular hypertrophy was observed in all dosed rats and the severity was dose-related (Clayton et al., 2007). Hepatocellular hypetrophy was often concurrently associated with homogeneous glycogen depleted amphophilic-staining cytoplasm. Hepatocellular hypertrophy was considered a consequence of cytochrome oxidase induction. Serum cholesterol levels were significantly increased in the 0.5 g/kg, 1.0 g/kg, and 2.0 g/kg groups (Clayton et al., 2007). In addition, clinical chemistry evaluations showed that gamma glutamyl transpeptidase activities were increased in the 2.0 g/kg rats (unpublished data). At study termination, increases were noted in mean serum total protein and albumin in the 0.5 g/kg and higher dose groups (unpublished data). The changes were considered related to hepatic alteration resulting from kava administration. Mean serum glucose of the 2.0 g/kg rats was decreased significantly; that in the 1.0 g/kg rats were marginally decreased. At the present it is not known which gene expression changes are responsible for these pathological effects. It is conceivable to conclude that part of these drug metabolizing enzyme gene expression changes results in: (i) these pathological effects and (ii) the expression of hepatic cytochrome P450 by immunohistochemical analysis by Clayton et al. (2007).
Nevertheless, in this 14-week study, no hepatotoxicity was observed histopathologically. To interpret this no hepatotoxic effects mechanistically, there are three possible explanations: (i) kava extract does not induce hepatotoxicity in F344 rats. As such, the observed metabolizing gene expression does not contribute to kava-induced hepatotoxicity; (ii) kava-induced hepatotoxic effect requires latent time that is longer than 14 weeks so that hepatptoxicity cannot be observed histopathologically at 14 weeks; and (iii) kava-induced hepatotoxicity is through an idiosyncratic mechanism. It is currently not known which mechanism or mechanisms are involved in this 14-week NTP study with F344 rats. Since the NTP is conducting a two-year chronic tumorigenicity bioassay, the outcome of this study would shed light on whether the first two mechanisms are involved. Concerning the idiosyncratic mechanism, Clouatre (2004) in his review stated that “The direct toxicity of kava extracts is quite small under any analysis, yet the potential for drug interactions and/or the potentiation of the toxicity of other compounds is large. Presently, kava toxicity appears to be idiosyncratic”. Based on the reported kava-induced hepatotoxicity in humans, as addresses in the CFSAN/FDA’s 2002 warning (CFSAN, 2002), and based on the findings we report in this present paper, it is possible that kava-induced hepatotoixcity is idiosyncratic.
It is important to note the dramatic induction of Cyp1a1 detected by both the microarray analysis and TaqMan assay. Even though there are a number of reports on the study of modulation of CYPs by kava extract in rodents and humans (Anke and Ramzan, 2004a,b; Gurley et al., 2005; Mathews et al., 2002, 2005; Raucy, 2003; Russmann et al., 2005; Zou et al., 2002), none of them have reported that the Cyp1a1 gene was induced. It is known that the gene expression of Cyp1a1 is different from that of Cyp1a2 and 3a1 in that Cyp1a1 is primarily expressed in extrahepatic tissues and there is a low amount in the liver (Martignoni et al., 2006). Indeed, our microarray analysis shows a very low level of Cyp1a1 in the control liver (Fig. 5), confirming that the basal gene expression of Cyp1a1 in the liver is low. The Cyp1a1 isozyme can metabolize a number of xenobiotics, including those with flat and planar structures, which include the highly toxic and tumorigenic polycyclic aromatic hydrocarbons (PAH) (Martignoni et al., 2006). This is evidenced by reports indicating that both the tumorigenic bez-no[a]pyrene and dibenzo[a,l]pyrene are metabolized primarily by Cyp1a1 (Conney, 2003; Shou et al., 1996). Thus, induction of the Cyp1a1 gene by kava extract would enhance metabolic activation of the hazardous PAHs and jeopardize human health. This is the first report demonstrating that kava can induce the gene expression of Cpy1a1.
Approaches for the mechanistic study of chemical toxicology and carcinogenesis are well established. However, most of these studies, if not all, are with a single pure chemical. Currently, it is still a challenge to determine the mechanisms of toxicity induced by a mixture of many chemical components, such as kava and other herbal plant extracts. Under this circumstance, DNA micro-array followed by real-time PCR should be a highly practical approach for revealing the whole spectrum of modifying gene expression by a chemical mixture (e.g., kava). After obtaining this information at the gene level, several conventional methodologies, including determining protein expression by immunoblotting and enzyme activity determination by quantitative metabolism can be followed. Without obtaining the whole spectrum of gene expression change, some important pathways for study will most likely be missed. The detection of kava-induced Cyp1a1 gene over-expression is an obvious example. Consequently, our DNA microarray study on liver drug metabolizing enzymes of kava-treated male rats represents a good example of this advantage, e.g., measuring the effect of kava on the whole spectrum of liver genes, including all the genes of drug metabolizing enzymes, at the same time.
Currently, there are a large number of herbal dietary supplements sold in the United States, and their consumption is drastically increasing. To protect consumer health, the quality and safety of herbal plants and herbal dietary supplements have to be assured. A number of dietary supplements and active ingredients have been nominated by the US FDA and the US National Institutes of Health for the US NTP to determine their toxicity and tumorigenicity. Kava is one of the herbal dietary supplements that is being studied by the NTP chronic tumorigenicity bioassays. Our mechanistic study on the gene expression changes of drug metabolizing enzymes provides useful information and directions for further study of kava-induced hepatotoxicity.
Acknowledgments
We thank Drs. Frederick A. Beland, Robert Heflich and Tucker Patterson for their critical review of this manuscript. We also thank reviewers whose valuable suggestions improved the quality of the manuscript.
Footnotes
DISCLAIMER: This article is not an official guidance or policy statement of National Toxicology Program (NTP) or US Food and Drug Administration (FDA). No official support or endorsement by the US FDA and NTP is intended or should be inferred.
Conflict of interest statement
The authors declare that there is no conflict of interest.
References
- Anke J, Ramzan I. Kava hepatotoxicity: are we any closer to the truth? Planta Med. 2004a;70(3):193–196. doi: 10.1055/s-2004-815533. [DOI] [PubMed] [Google Scholar]
- Anke J, Ramzan I. Pharmacokinetic and pharmacodynamic drug interactions with Kava (Piper methysticum Forst. f.) J. Ethnopharmacol. 2004b;93(2–3):153–160. doi: 10.1016/j.jep.2004.04.009. [DOI] [PubMed] [Google Scholar]
- Brauer R, Pfab R, Becker K, Berger H, Stangl M. Fulminantes leberversagen nach einnahme des pfalzlichen heimittels kava–kava. Z. Gastroenterol. 2001;39:491. [Google Scholar]
- Bressler R. Herb–drug interactions: interactions between kava and prescription medications. Geriatrics. 2005;60(9):24–25. [PubMed] [Google Scholar]
- Campo JV, McNabb J, Perel JM, Mazariegos GV, Hasegawa SL, Reyes J. Kava-induced fulminant hepatic failure. J. Am. Acad. Child Adolesc. Psychiatr. 2002;41(6):631–632. doi: 10.1097/00004583-200206000-00001. [DOI] [PubMed] [Google Scholar]
- CFSAN. Center for Food Safety and Applied Nutrition (CFSAN): Kava-containing dietary supplements may be associated with severe liver injury. US Department of Health and Human Services, Food and Drug Administration; Rockville, Maryland: 2002. [March 25, 2002]. < http://www.cfsan.fda.gov/~dms/ds-warn.html>. [Google Scholar]
- Chan PC, Fu PP. Toxicity of Panax ginseng-An herbal medicine and dietary supplements. J. Food Drug Anal. 2007;(15):416–427. [Google Scholar]
- Chan PC, Xia Q, Fu PP. Cinkgo biloba leave extract: biological, medicinal, and toxicological effects. J. Environ. Sci. Health C Environ. Carcinog. Ecotoxicol. Rev. 2007;25(3):211–244. doi: 10.1080/10590500701569414. [DOI] [PubMed] [Google Scholar]
- Chen T, Guo L, Zhang L, Shi L, Fang H, Sun Y, Fuscoe JC, Mei N. Gene expression profiles distinguish the carcinogenic effects of aristolochic acid in target (kidney) and non-target (liver) tissues in rats. BMC Bioinform. 2006;7(Suppl. 2):S20. doi: 10.1186/1471-2105-7-S2-S20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou MW, Fu PP. Formation of DHP-derived DNA adducts in vivo from dietary supplements and Chinese herbal plant extracts containing carcinogenic pyrrolizidine alkaloids. Toxicol. Ind. Health. 2006;22(8):321–327. doi: 10.1177/0748233706071765. [DOI] [PubMed] [Google Scholar]
- Clayton NP, Yoshizawa K, Kissling GE, Burka LT, Chan PC, Nyska A. Immunohistochemical analysis of expressions of hepatic cytochrome P450 in F344 rats following oral treatment with kava extract. Exp. Toxicol. Pathol. 2007;58(4):223–236. doi: 10.1016/j.etp.2006.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clouatre DL. Kava kava: examining new reports of toxicity. Toxicol. Lett. 2004;150(1):85–96. doi: 10.1016/j.toxlet.2003.07.005. [DOI] [PubMed] [Google Scholar]
- Clough AR, Bailie RS, Currie B. Liver function test abnormalities in users of aqueous kava extracts. J. Toxicol. Clin. Toxicol. 2003;41(6):821–829. doi: 10.1081/clt-120025347. [DOI] [PubMed] [Google Scholar]
- Conney AH. Enzyme induction and dietary chemicals as approaches to cancer chemoprevention: the Seventh DeWitt S. Goodman Lecture. Cancer Res. 2003;63(21):7005–7031. [PubMed] [Google Scholar]
- De Smet PA. Safety concerns about kava not unique. Lancet. 2002;(360):1336. doi: 10.1016/S0140-6736(02)11347-X. [DOI] [PubMed] [Google Scholar]
- Dentali SJ. Herb Safety Review. Kava. Piper methysticum Forster f. (Piperaceae) Herb Research Foundation; Boulder, Co: 1997. p. 29. [Google Scholar]
- Ernst E. Herbal remedies for anxiety – a systematic review of controlled clinical trials. Phytomedicine. 2006;13(3):205–208. doi: 10.1016/j.phymed.2004.11.006. [DOI] [PubMed] [Google Scholar]
- FDA. Dietary Supplement Health and Education Act of 1994, Public Law 103-417, 103rd Congress. 1994 < http://www.fda.gov/opacom/laws/dshea.html>.
- FDA. FDA Advises Dietary Supplement Manufacturers to Remove Comfrey Products From the Market July 6, 2001. 2001 < http://www.cfsan.fda.gov/~dms/supplmnt.html>.
- FDA. FDA Public Health News, FDA Announced Major Initiatives for Dietary Supplements. 2004a. [November 5, 2004]. [Google Scholar]
- FDA. Final Rule Declaring Dietary Supplements Containing Ephedrine Alkaloids Adulterated Because They Present an Unreasonable Risk Federal Register. 2004b. [February 11, 2004]. [PubMed] [Google Scholar]
- Fong HH. Integration of herbal medicine into modern medical practices: issues and prospects. Integr. Cancer Ther. 2002;1(3):287–293. doi: 10.1177/153473540200100313. (discussion 293) [DOI] [PubMed] [Google Scholar]
- Fu PP. Quality assurance and safety of herbal dietary supplements. J. Food Drug Anal. 2007;(15):333–334. [Google Scholar]
- Fu PP, Xia Q, Chou MW, Lin G. Detection, hepatotoxicity, and tumorigenicity of pyrrolizidine alkaloids in Chinese herbal plants and herbal dietary supplements. J. Food Drug Anal. 2007;(15):400–415. [Google Scholar]
- Fu PP, Xia Q, Guo L, Yu H, Chan PC. Toxicity of kava kava. Environ. Carcinog. Ecotocicol. Rev. 2008;(26):1–24. doi: 10.1080/10590500801907407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu PP, Yang YC, Xia Q, Chou MW, Cui YY, Lin G. Pyrrolizidine alkaloids - tumorigenic components in Chinese herbal medicines and dietary supplements. J. Food Drug Anal. 2002;(10):198–211. [Google Scholar]
- Guo L, Lobenhofer EK, Wang C, Shippy R, Harris SC, Zhang L, Mei N, Chen T, Herman D, Goodsaid FM, et al. Rat toxicogenomic study reveals analytical consistency across microarray platforms. Nat. Biotechnol. 2006;24(9):1162–1169. doi: 10.1038/nbt1238. [DOI] [PubMed] [Google Scholar]
- Guo L, Mei N, Dial S, Fuscoe J, Chen T. Comparison of gene expression profiles altered by comfrey and riddelliine in rat liver. BMC Bioinform. 2007;8(Suppl. 7):S22. doi: 10.1186/1471-2105-8-S7-S22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurley BJ, Gardner SF, Hubbard MA, Williams DK, Gentry WB, Cui Y, Ang CY. Clinical assessment of effects of botanical supplementation on cytochrome P450 phenotypes in the elderly: St John’s wort, garlic oil, Panax ginseng and Cinkgo biloba. Drugs Aging. 2005;22(6):525–539. doi: 10.2165/00002512-200522060-00006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurley BJ, Swain A, Barone GW, Williams DK, Breen P, Yates CR, Stuart LB, Hubbard MA, Tong Y, Cheboyina S. Effect of goldenseal (Hydrastis canadensis) and kava kava (Piper methysticum) supplementation on digoxin pharmacokinetics in humans. Drug Metab. Dispos. 2007;35(2):240–245. doi: 10.1124/dmd.106.012708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Z, Yang X, Ho PC, Chan SY, Heng PW, Chan E, Duan W, Koh HL, Zhou S. Herb-drug interactions: a literature review. Drugs. 2005;65(9):1239–1282. doi: 10.2165/00003495-200565090-00005. [DOI] [PubMed] [Google Scholar]
- Humberston CL, Akhtar J, Krenzelok EP. Acute hepatitis induced by kava kava. J. Toxicol. Clin. Toxicol. 2003;41(2):109–113. doi: 10.1081/clt-120019123. [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−ΔΔ C(T)) method. Methods. 2001;25(4):402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Martignoni M, Groothuis GM, De Kanter R. Species differences between mouse, rat, dog, monkey and human CYP-mediated drug metabolism, inhibition and induction. Exp. Opin. Drug Metab. Toxicol. 2006;2(6):875–894. doi: 10.1517/17425255.2.6.875. [DOI] [PubMed] [Google Scholar]
- Mathews JM, Etheridge AS, Black SR. Inhibition of human cytochrome P450 activities by kava extract and kavalactones. Drug Metab. Dispos. 2002;30(11):1153–1157. doi: 10.1124/dmd.30.11.1153. [DOI] [PubMed] [Google Scholar]
- Mathews JM, Etheridge AS, Valentine JL, Black SR, Coleman DP, Patel P, So J, Burka LT. Pharmacokinetics and disposition of the kavalactone kawain: interaction with kava extract and kavalactones in vivo and in vitro. Drug Metab. Dispos. 2005;33(10):1555–1563. doi: 10.1124/dmd.105.004317. [DOI] [PubMed] [Google Scholar]
- Matthias A, Blanchfield JT, Penman KG, Bone KM, Toth I, Lehmann RP. Permeability studies of Kavalactones using a Caco-2 cell monolayer model. J. Clin. Pharm. Ther. 2007;32(3):233–239. doi: 10.1111/j.1365-2710.2007.00810.x. [DOI] [PubMed] [Google Scholar]
- Mei N, Guo L, Liu R, Fuscoe JC, Chen T. Gene expression changes induced by the tumorigenic pyrrolizidine alkaloid riddelliine in liver of Big Blue rats. BMC Bioinform. 2007;8(Suppl. 7):S4. doi: 10.1186/1471-2105-8-S7-S4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mei N, Guo L, Zhang L, Shi L, Sun YA, Fung C, Moland CL, Dial SL, Fuscoe JC, Chen T. Analysis of gene expression changes in relation to toxicity and tumorigenesis in the livers of Big Blue transgenic rats fed comfrey (Symphytum officinale) BMC Bioinform. 2006;7(Suppl. 2):S16. doi: 10.1186/1471-2105-7-S2-S16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkman CA. Another FDA warning: Kava supplements. Case Manager. 2002;13(4):26–28. doi: 10.1067/mcm.2002.126437. [DOI] [PubMed] [Google Scholar]
- Raucy JL. Regulation of CYP3A4 expression in human hepatocytes by pharmaceuticals and natural products. Drug Metab. Dispos. 2003;31(5):533–539. doi: 10.1124/dmd.31.5.533. [DOI] [PubMed] [Google Scholar]
- Russmann S, Barguil Y, Cabalion P, Kritsanida M, Duhet D, Lauterburg BH. Hepatic injury due to traditional aqueous extracts of kava root in New Caledonia. Eur. J. Gastroenterol. Hepatol. 2003;15(9):1033–1036. doi: 10.1097/00042737-200309000-00015. [DOI] [PubMed] [Google Scholar]
- Russmann S, Lauterburg BH, Barguil Y, Choblet E, Cabalion P, Rentsch K, Wenk M. Traditional aqueous kava extracts inhibit cytochrome P450 1A2 in humans: Protective effect against environmental carcinogens? Clin. Pharmacol. Ther. 2005;77(5):453–454. doi: 10.1016/j.clpt.2005.01.021. [DOI] [PubMed] [Google Scholar]
- Russmann S, Lauterburg BH, Helbling A. Kava hepatotoxicity. Ann. Int. Med. 2001;135(1):68–69. doi: 10.7326/0003-4819-135-1-200107030-00036. [DOI] [PubMed] [Google Scholar]
- Saß M, Schnabel S, Kroger J, Liebe S, Schareck WD. Acute liver failure from kavakava – a rare indication for liver transplantation. Z. Gastroenterol. 2001;39:491. [Google Scholar]
- Schulze J, Raasch W, Siegers CP. Toxicity of kava pyrones, drug safety and precautions – a case study. Phytomedicine. 2003;10(Suppl. 4):68–73. doi: 10.1078/1433-187x-00300. [DOI] [PubMed] [Google Scholar]
- Shou M, Krausz KW, Gonzalez FJ, Gelboin HV. Metabolic activation of the potent carcinogen dibenzo[a,l]pyrene by human recombinant cytochromes P450, lung and liver microsomes. Carcinogenesis. 1996;17(11):2429–2433. doi: 10.1093/carcin/17.11.2429. [DOI] [PubMed] [Google Scholar]
- Singh YN. Kava: an overview. J. Ethnopharmacol. 1992;37(1):13–45. doi: 10.1016/0378-8741(92)90003-a. [DOI] [PubMed] [Google Scholar]
- Singh YN. Potential for interaction of kava and St. John’s wort with drugs. J. Ethnopharmacol. 2005;100(1–2):108–113. doi: 10.1016/j.jep.2005.05.014. [DOI] [PubMed] [Google Scholar]
- Smith RM, Thakrar H, Arowolo TA, Shafi AA. High-performance liquid chromatography of kava lactones from Piper methysticum. J. Chromatogr. 1984;283:303–308. [Google Scholar]
- Williamson EM. Interactions between herbal and conventional medicines. Exp. Opin. Drug Saf. 2005;4(2):355–378. doi: 10.1517/14740338.4.2.355. [DOI] [PubMed] [Google Scholar]
- Wold RS, Lopez ST, Yau CL, Butler LM, Pareo-Tubbeh SL, Waters DL, Garry PJ, Baumgartner RN. Increasing trends in elderly persons’ use of nonvitamin, nonmineral dietary supplements and concurrent use of medications. J. Am. Diet Assoc. 2005;105(1):54–63. doi: 10.1016/j.jada.2004.11.002. [DOI] [PubMed] [Google Scholar]
- Zou L, Harkey MR, Henderson GL. Effects of herbal components on cDNA-expressed cytochrome P450 enzyme catalytic activity. Life Sci. 2002;71(13):1579–1589. doi: 10.1016/s0024-3205(02)01913-6. [DOI] [PubMed] [Google Scholar]