Abstract
Human pegivirus (HPgV; originally called GB virus C/hepatitis G virus) is an RNA virus within the genus Pegivirus of the family Flaviviridae that commonly causes persistent infection. Worldwide, ~750 million people are actively infected (viraemic) and an estimated 0.75–1.5 billion people have evidence of prior HPgV infection. No causal association between HPgV and disease has been identified; however, several studies described a beneficial relationship between persistent HPgV infection and survival in individuals infected with human immunodeficiency virus. The beneficial effect appeared to be related to a reduction in host immune activation. HPgV replicates well in vivo (mean plasma viral loads typically >1×107 genome copies ml−1); however, the virus grows poorly in vitro and systems to study this virus are limited. Consequently, mechanisms of viral persistence and host immune modulation remain poorly characterized, and the primary permissive cell type(s) has not yet been identified. HPgV RNA is found in liver, spleen, bone marrow and PBMCs, including T-and B-lymphocytes, NK-cells, and monocytes, although the mechanism of cell-to-cell transmission is unclear. HPgV RNA is also present in serum microvesicles with properties of exosomes. These microvesicles are able to transmit viral RNA to PBMCs in vitro, resulting in productive infection. This review summarizes existing data on HPgV cellular tropism and the effect of HPgV on immune activation in various PBMCs, and discusses how this may influence viral persistence. We conclude that an increased understanding of HPgV replication and immune modulation may provide insights into persistent RNA viral infection of humans.
Introduction
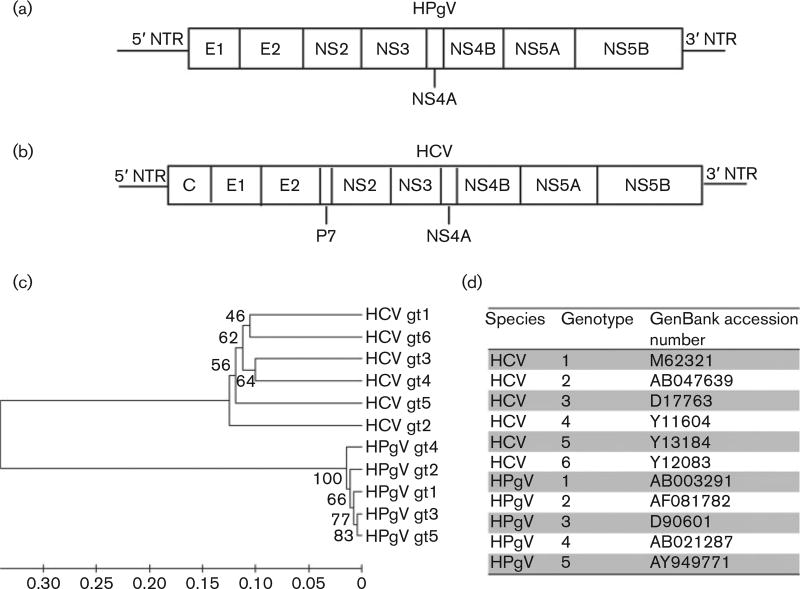
Human pegivirus (HPgV) is an RNA virus within the Pegivirus A species and family Flaviviridae (Adams et al., 2013; Stapleton et al., 2012a). The virus was originally called hepatitis G virus (HGV)/GB virus type C (GBV-C). The letters ‘GB’ represented the initials of a surgeon with acute hepatitis whose sera caused hepatitis in marmosets (Deinhardt et al., 1967). Subsequent studies found that this virus did not cause hepatitis and there was no direct evidence that the surgeon (‘G. B.’) was infected with HPgV. Thus, the virus (HGV/GBV-C), along with several related primate and bat viruses (GBV-A, GBV-Cczp, GBV-D), was assigned to a new genus (Pegivirus) and renamed as HPgV (Adams et al., 2013; Stapleton et al., 2012a). HPgV has a 9.4 kb positive-sense ssRNA genome that is organized similarly to hepatitis C virus (HCV) (Fig. 1). HPgV and HCV are unusual amongst RNA viruses in that they commonly cause persistent infection in immune-competent humans.
Fig. 1.
HPgV and HCV genome organization and phylogenetic relationships. (a, b) Whilst HPgV (a) is within the genus Pegivirus and HCV (b) is within the genus Hepacivirus of the family Flaviviridae, both viruses share many features, including a ssRNA positive-sense genome with 59 and 39 NTRs. Both viruses are capable of causing persistent human infection. (c, d) HPgV and HCV phylogenetic trees (unweighted pair group method with arithmetic mean) (c) were reconstructed in mega6 using alignments of isolates with GenBank numbers shown in (d). The original tree without bootstrapping is shown. gt, Genotype.
Although HPgV is not as efficient at establishing persistent infection as HCV, an estimated 25% of infections persist and the other 75% clear viraemia within 2 years of infection (Gutierrez et al., 1997; Tanaka et al., 1998a). The prevalence of HPgV viraemia in cross-sectional serum surveys of healthy blood donors in developed countries is between 1 and 5 %, whilst up to 20% of blood donors in developing countries have active infection (Mohr & Stapleton, 2009). The rate of HPgV viraemia is also higher amongst individuals with blood-borne or sexually transmitted infections. Based on cross-sectional prevalence data, there are ~750 million people with HPgV viraemia worldwide (Stapleton et al., 2014). Anti-HPgV antibodies are not usually detected during active infection, but antibody against the envelope glycoprotein E2 appears following viral clearance (Gutierrez et al., 1997; Tacke et al., 1997; Tanaka et al., 1998a; Thomas et al., 1997). These E2 antibodies appear to provide partial protection against reinfection (Elkayam et al., 1999; Tillmann et al., 1998). Thus, antibody to E2 represents a marker of prior infection, although antibody levels may decrease and become undetectable over time (Gutierrez et al., 1997; Tacke et al., 1997; Tanaka et al., 1998a). As 12–20% of blood donors from developed countries have HPgV E2 antibodies (Gutierrez et al., 1997; Tacke et al., 1997) and a higher percentage of healthy blood donors in developing countries have E2 antibodies (reviewed by Mohr & Stapleton, 2009), the data suggest that ~1.5–2.5 billion people are currently infected or have evidence of prior HPgV infection, making this virus a major contributor to the human virome. Thus, HPgV shares infection rates with many viruses, including papillomaviruses, herpesviruses and torque teno viruses (TTVs), frequently found in healthy individuals (Okamoto, 2009).
HPgV is transmitted by exposure to infected blood, sexual exposure or by maternal–foetal transmission (Bhanich Supapol et al., 2009; Hino et al., 1998; Kleinman, 2001; Lin et al., 1998; Ohto et al., 2000; Stapleton, 2003). Although HPgV has not been conclusively shown to cause any human disease (Bhattarai & Stapleton, 2012; Mohr & Stapleton, 2009), several studies observed an association between HPgV viraemia and an increased risk of non-Hodgkin’s lymphoma (Chang et al., 2014; Civardi et al., 1998; De Renzo et al., 2002; Ellenrieder et al., 1998; Giannoulis et al., 2004; Keresztes et al., 2003; Krajden et al., 2010; Michaelis et al., 2003; Nakamura et al., 1997). In contrast, HPgV is associated with a beneficial effect in human immunodeficiency virus (HIV) infection, and most studies and a meta-analysis found prolonged survival in HIV-infected individuals co-infected with HPgV compared with those without HPgV viraemia (Heringlake et al., 1998; Nunnari et al., 2003; Tillmann et al., 2001; Toyoda et al., 1998; Williams et al., 2004; Xiang et al., 2001; Zhang et al., 2006). A recent study of incident, transfusion-related HPgV infection in HIV-infected individuals confirmed a survival benefit in those subjects who acquire HPgV through blood transfusion (Vahidnia et al., 2012). As a result of this beneficial interaction in HIV co-infected subjects, numerous studies designed to understand how HPgV confers the ascribed survival benefit have been reported.
As with HCV, HPgV does not grow well in vitro, yet replicates extremely well in vivo with mean plasma viral loads typically > 1×107 genome copies ml−1. Nevertheless, due to the poor growth of the virus in vitro, study of this virus is difficult, resulting in several unanswered questions about HPgV persistence and how the virus interacts with the host.
Many acute viral infections induce sterilizing T- and B-cell immune responses that clear the infection and result in immunological memory leaving the host resistant to reinfection. Following acute infection by many DNA viruses, such as the herpesviruses, actively replicating virus is mostly cleared by the host immune response; however, residual foci of quiescent (latent) infection remain, providing a source for occasional reactivation (Welsh & Waggoner, 2013). Whilst HIV and other retroviruses have a DNA intermediate that integrates into host cell chromosomal DNA enabling persistence, it remains unclear how strictly cytoplasmic RNA viruses such as HCV and HPgV evade immune clearance. Several viral mechanisms have been proposed as contributors to immune escape for HCV, including the presence of two hypervariable regions on the envelope (E2) protein (one in a location shown to be involved in viral entry), mutation of immunodominant T-cell epitopes (Keck et al., 2009, 2014), interference of several HCV proteins with intracellular signalling pathways resulting in reduced innate and adaptive immune responses, and chronic stimulation of T-cells leading to T-cell exhaustion (reviewed in Burke & Cox, 2010; Lemon, 2010). In contrast, HPgV envelope proteins do not have any hypervariable regions; the virus displays limited sequence variability within persistently infected hosts in protein regions predicted to be T-cell epitopes, and is associated with reduced T-cell activation and exhaustion (Mohr & Stapleton, 2009; Stapleton et al., 2012b). Complicating studies of HPgV persistence, the receptor(s) for HPgV have not been identified and the primary permissive cell(s) are not fully characterized. Nevertheless, HPgV RNA is detected in several types of PBMCs (Chivero et al., 2014), and the relationship between HPgV infection and immune function, and how this relates to viral persistence in chronic infection in vivo, remain to be clarified. Understanding the life cycle and tropism of this relatively non-pathogenic virus may provide insights into virus-mediated interference in co-infected individuals and potentially identify a component of the microbiome that influences human immune responses. This review summarizes our current understanding of HPgV tropism and the contribution of HPgV infection of human immune cells (T-and B-lymphocytes, NK-cells, and monocytes) to host immune modulation.
HPgV tropism
HPgV was initially thought to be hepatotropic based on its identification in individuals with non-A, non-B, non-C hepatitis; however, subsequent epidemiological studies failed to find a conclusive association with either acute or chronic hepatitis (Fan et al., 1999; Pessoa et al., 1998; Tucker et al., 2000). Although several early studies found HPgV RNA in liver biopsies, virus was subsequently detected in spleen, bone marrow, cerebrospinal fluid and PBMCs, as noted above (Chivero et al., 2014; George et al., 2006; Kisiel et al., 2013; Mellor et al., 1998; Pessoa et al., 1998; Radkowski et al., 2000; Tucker et al., 2000). Studies attempting to find evidence of HPgV replication in liver tissue were either negative or inconclusive (Berg et al., 1999; Fan et al., 1999; Laskus et al., 1997). Specifically, RNA levels of the hepatotropic HCV were consistently higher in the liver of HCV/HPgV co-infected individuals relative to plasma, whilst the inverse was true for HPgV RNA. Furthermore, the median liver/serum ratio of HPgV RNA was <1.0, consistent with serum contamination of liver tissue (Pessoa et al., 1998), and several additional studies did not find evidence of HPgV replication in the liver (Berg et al., 1999; Fan et al., 1999; Laras et al., 1999; Pessoa et al., 1998). HPgV negative-strand RNA, indicative of viral RNA replication, was not detected in liver samples, but was detected in bone marrow and spleen in autopsy or biopsy samples (Laskus et al., 1997; Radkowski et al., 2000; Tucker et al., 2000). Furthermore, HPgV serum RNA levels did not decrease significantly following liver transplantation in an HCV/HPgV co-infected individual, whilst HCV RNA levels transiently decreased (Berg et al., 1999). Table 1 summarizes selected studies showing HPgV replication as measured by the negative-strand RNA replication intermediate.
Table 1.
Selected studies examining replication of HPgV
| Reference | Cell/tissue type | Positive-strand RNA | Negative-strand RNA |
|---|---|---|---|
| Mellor et al. (1998) | PBMCs | 11/20 | 0/20 |
| Mellor et al. (1998) | Hepatocytes | 8/13 | 0/13 |
| Laskus et al. (1997) | Liver | 6/10 | 0/10 |
| Laras et al. (1999) | Hepatocytes | 6/12 | 0/12 |
| Tucker et al. (2000) | Spleen | 3/3 | 3/3 |
| Tucker et al. (2000) | Bone marrow | 2/2 | 2/2 |
| Radkowski et al. (2000) | Bone marrow | 9/48 | 4/5 |
Life cycle
Based on similarities in genome organization and amino acid homology with HCV, it is likely that multiple attachment and entry receptors are involved in HPgV entry. HCV utilizes many receptors in vitro, including the low-density lipoprotein receptor (LDLr), scavenger receptor class B type I, CD81, claudin-1, occludin, DC-SIGN (dendritic cell-specific intercellular adhesion molecule-3-grabbing non-integrin), signal transducer Harvey rat sarcoma viral oncogene homologue, Niemann–Pick C1-like 1 and transferrin receptor 1 (Evans et al., 2007; Lozach et al. 2003; Martin & Uprichard, 2013; Moradpour et al., 2007; Ploss et al., 2009; Sainz et al., 2012; Zeisel et al., 2013; Zona et al., 2014). Although little is known about HPgV entry, early studies suggested that the LDLr is involved in HPgV and HCV entry (Agnello et al., 1999; Wünschmann et al., 2000). Consistent with this, HPgV RNA in plasma is associated with lipids (Xiang et al., 1998), including lipid-associated microvesicles that exhibit properties of exosomes (Bhattarai et al., 2013). Given the universal distribution of LDLr on human cells, lipid-associated virus and/or serum microvesicle uptake may involve the LDLr (Wünschmann et al., 2006). As there are no efficient in vitro replication systems, detailed studies on the HPgV life cycle have not been performed. Nevertheless, based on the related HCV, it is predicted that there is post-entry acidification of the endosomal vesicle leading to a conformational change in the virion with subsequent fusion of the viral and cellular membranes.
A potential alternative mechanism of HPgV entry may involve serum HPgV RNA-containing vesicles that have properties of exosomes (Chivero et al., 2014). Exosomes are microvesicles of endocytic origin that are emerging as novel particles employed by cells and viruses to transmit viral and cellular RNAs and proteins to cells (Meckes & Raab-Traub, 2011). As HPgV and other members of the genus Pegivirus do not appear to encode a nucleocapsid protein at the N terminus of the polyprotein (Xiang et al., 1998; Stapleton et al., 2011), cellular-derived microvesicles (which carry ~50% of the HPgV RNA found in plasma; Bhattarai et al., 2013) may be involved in virus release and/ or cellular infection. Supporting this hypothesis, HPgV RNA-containing microvesicles positive for an exosomal marker (cellular CD63) were able to deliver viral RNA to uninfected PBMCs and viral RNA replicated within these cells ex vivo (Chivero et al., 2014). Other viruses such as hepatitis A virus and HCV have also been shown to utilize exosomes for viral transmission and intercellular communication (Bukong et al., 2014; Dreux et al., 2012; Feng et al., 2013; Ramakrishnaiah et al., 2013).
HPgV infects diverse haematopoietic cell types in vitro
The primary site(s) of replication and cellular receptors for HPgV have not been identified, despite the passage of nearly 20 years since discovery. As noted above, early studies proposed that the virus was hepatotropic; however, several lines of evidence suggest that the virus is lymphotropic (Fogeda et al., 1999; George et al., 2006; Xiang et al., 2000, 2001). Specifically, PBMCs from HPgV-infected individuals maintained in culture ex vivo release virus into culture media and virus production can persist for at least 35 days in culture (Fogeda et al., 1999; George et al., 2003; Rydze et al., 2012). Furthermore, serum-derived virus from infected individuals is capable of infecting PBMCs in vitro (Chivero et al., 2014; Xiang et al., 2000).
In cell culture, HPgV replication is at best intermittent and produces low titres of intracellular RNA or virus released into cell culture media. The best-characterized in vitro methods described to date for HPgV replication utilize primary PBMCs (Fogeda et al., 1999; George et al., 2003, 2006; Rydze et al., 2012; Xiang et al., 2000). Unfortunately, production of HPgV by donor lymphocytes demonstrates considerable donor and isolate variability (George et al., 2003), and quantification of HPgV in various peripheral blood cell types detects only 1–10 copies per 100 cells on average. Thus, a very small proportion of PBMCs appear to support replication (Chivero et al., 2014). Long-term passage of HPgV in PBMCs for more than four or five passages has not been described even though the virus is non-cytopathic.
In a recent study, HPgV RNA was detected in T- and B-lymphocytes, NK-cells, and monocytes (Chivero et al., 2014). HPgV RNA was present in all 14 study subjects’ PBMCs, and in highly purified populations of T- and B-lymphocytes (>98% purity), consistent with previous studies (George et al., 2006; Ruiz et al., 2010). Amongst CD4+ and CD8+ T-cells, viral RNA was present in naive, central memory and effector memory subpopulations, with the highest concentrations reported in the naive (CD45RA+) T-cells compared with central memory or effector memory T-cells (Chivero et al., 2014). This finding is consistent with selective HPgV infection of naive T-cells that subsequently differentiate into effector or memory cells, or alternatively the infection of T-cells regardless of differentiation state. If infection is selective for naive T-cells, it would suggest that there are differences in expression of HPgV receptor(s) or that restriction factor(s) are expressed following differentiation. Alternatively, the data could be interpreted to suggest that HPgV infects haematopoietic precursor cells that maintain infection during differentiation into T- and B-cells. If the latter hypothesis is correct, HPgV infection may also reduce the frequency of naive cell differentiation into effector cells by decreasing activation and proliferation.
Although HPgV replicates efficiently in humans, with mean serum viral loads typically >1×107 genome equivalents ml−1, replication is inefficient in vitro and the production of virus by lymphocytes maintained in culture ex vivo is reduced following T-cell activation (George et al., 2003; Rydze et al., 2012). Consistent with an interaction between HPgV and cell activation, several clinical studies observed a reduction in T-cell activation and proliferation markers in HIV-infected subjects with HPgV co-infection compared with HIV-mono-infected subjects (Bhattarai et al., 2012a; Maidana-Giret et al., 2009; Rydze et al., 2012; Stapleton et al., 2012b). Further evidence consistent with HPgV reducing inflammation comes from a recent report which found that HPgV infection was associated with reduced mortality in individuals infected with ebolavirus (Lauck et al., 2015). Finally, higher concentrations of HPgV RNA are found in naive T-cells or non-proliferating cells (Chivero et al., 2014). Taken together, the data suggest that activation reduces HPgV replication, and that HPgV infection interferes with activation and proliferation of lymphocytes. During in vitro replication, this would reduce the proportion of cells harbouring virus within a cell culture over time, potentially contributing to the difficulty in detecting HPgV replication in cell culture systems.
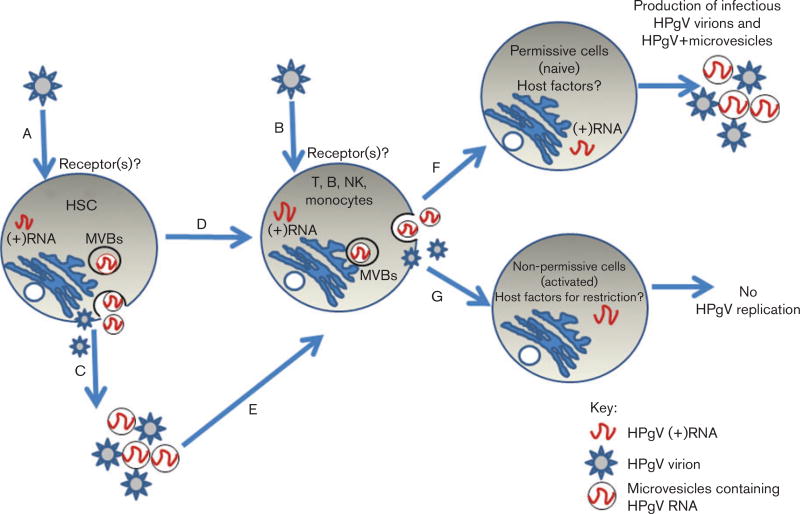
In addition to T- and B-lymphocytes, HPgV RNA is found in highly purified monocytes and NK-cell preparations (Chivero et al., 2014). As HPgV RNA is present in both lymphoid and myeloid cells, either wide tropism occurs or perhaps the primary target cell is a progenitor haematopoietic stem cell (HSC) that subsequently differentiates, carrying the HPgV genome. To examine this, we infected peripheral blood CD34+ cells, but this did not result in improved replication compared with PBMC cultures (E. T. Chivero, N. Bhattarai, E. L. Mohr & J. T. Stapleton, unpublished observation). However, this does not exclude the possibility of an HSC target cell, as the properties of peripherally derived stem cells differ considerably from bone-marrow-derived HSCs. Table 2 summarizes HPgV RNA localization studies of different cell types. Potential mechanisms by which HPgV transmits its RNA to cells are summarized in Fig. 2.
Table 2.
Selected studies examining HPgV RNA in different cell types
| Reference | PBMCs | Bone marrow | CD4+ T-cells | CD8+ T-cells | B-cells | Monocytes | NK-cells |
|---|---|---|---|---|---|---|---|
| Chivero et al. (2014) | 14/14 | nt | 13/13 | 13/13 | 7/10 | 4/5 | 4/5 |
| Kisiel et al.. (2013) | 11/23 | 18/23 | nt | nt | nt | nt | nt |
| Ruiz et al. (2010) | 1/1 | nt | 1/1 | 1/1 | 1/1 | nt | nt |
| George et al. (2006) | 9/9 | nt | 9/9 | 9/9 | 9/9 | nt | nt |
| Radkowski et al. (2000) | nt | 9/48 | nt | nt | nt | nt | nt |
| Fogeda et al. (1999) | 4/4 | nt | nt | nt | nt | nt | nt |
HPgV viral RNA was detected by reverse transcription-PCR.
nt, Not tested.
Fig. 2.
Model for HPgV tropism. HPgV may infect HSCs in spleen and/or bone marrow due to expression of a specific receptor that is not present in differentiated cells (A) or lymphocytes expressing yet unidentified common receptors (B). If HSCs are infected, HPgV genomes are passaged during differentiation (D) with the production of low-level virus. Alternatively, infected HSCs produce exosomes that deliver RNA to multiple cell types (C, E) that produce low-level virus. Host factors either promote (in resting cells F) or restrict (in activated cells; G) viral replication within each specific cell type. MVBs, multivesicular bodies.
HPgV negative-strand RNA (a marker of replication) has not been reproducibly found in PBMCs, presumably due to the very low viral RNA copy number in PBMCs (Chivero et al., 2014; Mellor et al., 1998). However, several lines of evidence strongly support HPgV replication in PBMCs. Specifically, PBMCs removed from infected individuals produce viral RNA over many weeks despite washing and/or treatment with trypsin (George et al., 2003; Rydze et al., 2012). Secondly, uninfected PBMCs inoculated with viral particles for 1 h and then treated with trypsin to remove virus adherent to the outside of cells demonstrate a subsequent increase in HPgV RNA over time, and release more viral RNA into culture supernatants than used in the initial inocula (Xiang et al., 2000; Chivero et al., 2014). The fact that HPgV negative-strand RNA is preferentially found in bone marrow and spleen rather than PBMCs (Radkowski et al., 2000; Tucker et al., 2000), and the finding of HPgV RNA in multiple lineages of peripheral blood white cells (references in Table 1), reinforce the possibility that a HSC precursor may serve as the primary target of HPgV infection, and that the virus persists and can be produced in cells during and following maturation into B- and T-lymphocytes, NK-cells and monocytes, as suggested in Fig. 2. If this model is true and HPgV replicates in stem cells without cytopathic effect, HPgV may be a useful platform for developing viral vectors for gene delivery similar to current adenoviral vectors. Nevertheless, until there is an animal model or efficient in vitro replication system, it will be difficult to further elucidate the cellular receptor(s) and target cell(s) for HPgV replication or test the use of HPgV as a gene therapy vector.
Host immune responses in chronic HPgV infection
T-cell activation
HIV infection results in chronic activation of T-cells, thus promoting activation-induced CD4+ T-cell death with subsequent immune dysfunction and progression to AIDS. Consequently, T-cells in HIV-infected individuals have increased expression of activation markers and lower CD4+ T-cell counts (Hunt et al., 2003). In contrast, HPgV infection is associated with significantly lower expression of surface markers of activation on T-cells (CD38, CD69 and CD25) in acutely HIV-infected individuals (Maidana-Giret et al., 2009) and chronically HIV-infected individuals compared with those without HPgV, independent of HIV treatment status (Bhattarai et al., 2012a; Stapleton et al., 2012b). HPgV replication ex vivo was reduced in activated PBMCs compared with cells without activation (Rydze et al., 2012), suggesting an interaction between HPgV and T-cell proliferation. Consistent with this hypothesis, HPgV viraemia was associated with reduced CD4+ T-cell expansion in HIV-infected individuals receiving recombinant IL-2 therapy (Stapleton et al., 2009). The CD4+ T-cell count did not increase significantly in subjects with HPgV infection, whilst it increased by >1000 cells mm−3 in those without HPgV. In addition, the proportion of naive CD8+ and CD4+ T-cells is increased relative to memory and effector cells in individuals with persistent HPgV infection compared with HPgV-negative controls (Stapleton et al., 2012b). As IL-2 is required for proliferation and differentiation of T-cells, and HPgV inhibits T-cell activation and IL-2 production, we hypothesize that the CD8+ T-cell cytotoxic functions that control viral infections are reduced in those with HPgV infection, contributing to HPgV persistence and high serum viral loads (typically >107 copies ml−1) (Tillmann et al., 2001). This is consistent with a beneficial effect in HIV infection, as HIV mediates disease by inducing chronic immune activation leading to lethal immune pathology (Welsh & Waggoner, 2013). This may be mediated in part by the finding that HPgV inhibits T-cell receptor signalling (Bhattarai et al., 2012b, 2013); however, the effects of HPgV on negative signalling molecules such as programmed death-1, 2B4 (CD244) or cytotoxic T-lymphocyte antigen-4 have not been studied.
HPgV is also associated with an increase in the number of double-negative (DN) T-cells (CD4−CD8−CD3+) (Bhattarai et al., 2012a). DN T-cells are associated with reduced immune activation and high levels of immune-suppressive cytokines, including TGF-β and IL-10, in HIV infection (Petitjean et al., 2012). In simian immunodeficiency virus (SIV) infection of non-human primates, DN T-cells are capable of performing CD4+ T-cell-like helper functions whilst remaining refractory to SIV infection and are associated with a lack of clinical disease progression (Milush et al., 2011; Sundaravaradan et al., 2012, 2013). Thus, the increased numbers of DN T-cells in HPgV infection may play important roles in reducing immune activation and maintenance of immune homeostasis, contributing to improved survival in HIV co-infection.
Although little is known regarding HPgV persistence and T-cell escape mechanisms, data related to HCV persistence may be relevant. Key factors in HCV persistence include the emergence of viral neutralization and T-cell escape mutations, and reduced virus-specific CD4+ T-cell function (reviewed by Rehermann, 2013). CD4+ T-cell help is critical in HCV persistence, as depletion of CD4+ T-cells from chimpanzees that had recovered from HCV infection abrogated the protective CD8+ T-cell-mediated immune response upon HCV rechallenge (Grakoui et al., 2003). In contrast to HCV, HPgV does not have a hypervariable region in the envelope proteins and neutralizing antibodies directed against viral structural proteins are not detected in chronically infected individuals. Thus, neutralizing antibody escape does not appear to be a strategy utilized by HPgV (Thomas et al., 1998). The critical roles of T-cell escape mutations and impaired CD4+ T-cell help during HPgV infection, and how these features mediate viral persistence, have not been studied.
B-cell function and antibody production
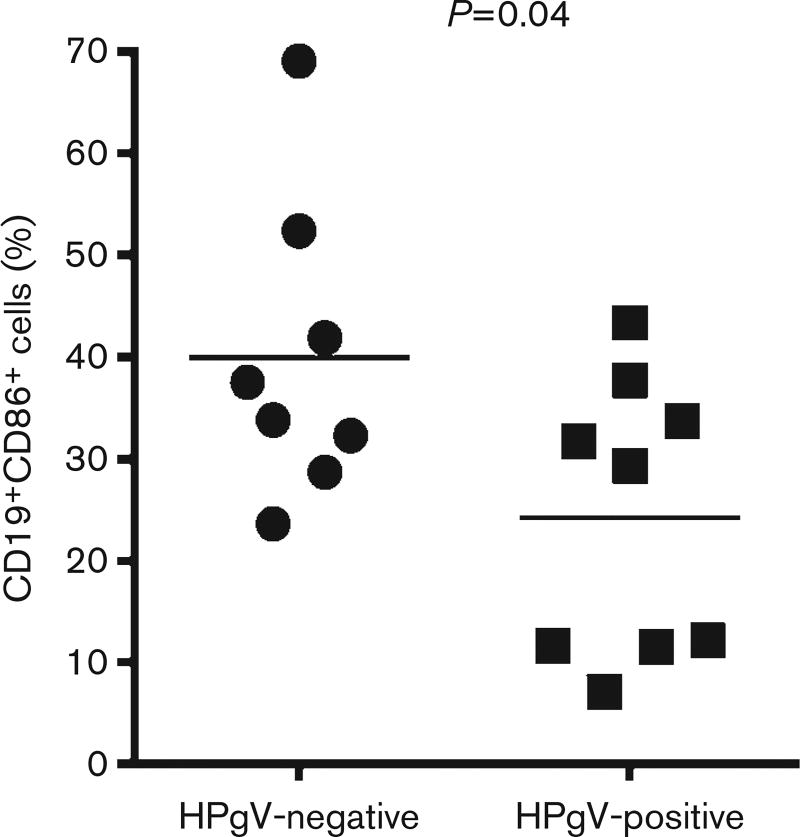
As noted above, antibodies against HPgV structural proteins are generally not detected during viraemia and most individuals develop conformation-dependent antibodies to E2 at or shortly following viral clearance (Gutierrez et al., 1997; Tacke et al., 1997; Tanaka et al., 1998a). However, anti-E2 antibodies are occasionally found during active viraemia and some of these individuals may be in the act of clearing viraemia (Schwarze-Zander et al., 2006; Tan et al., 1999; Williams et al., 2004). Some recent reports describe the detection of anti-HPgV peptide antibodies (Fernández et al., 2013; Gómara et al., 2011); however, no clear relationship between peptide-reactive antibodies in actively HPgV-infected or convalescent subjects has been reported. As anti-E2 antibody is usually inversely related to HPgV viraemia (Gutierrez et al., 1997; Tacke et al., 1997; Tanaka et al., 1998a), it is possible that HPgV suppresses the production of antibodies, contributing to persistent infection. There are some data suggesting that HPgV may reduce B-cell activation in addition to T-cell activation. Specifically, subjects with HPgV and HIV co-infection had reduced activation marker (CD86) expression on B-cells when compared with HIV-infected subjects without HPgV co-infection (Stapleton et al., 2013). To examine this further, PBMCs were isolated from HPgV-positive (n=9) and HPgV-negative control subjects (n=8), stimulated with soluble CD40 ligand (sCD40L) and activation was measured on B-cells (CD3−CD19+) by measuring surface CD86 expression. CD40–CD40L interaction is required for germinal centre formation, isotype switching, immunoglobulin production and induction of memory B-cells (Durie et al., 1994; Foy et al., 1994). We found a significant reduction in activation as measured by CD86 surface expression on B-cells following sCD40L stimulation (P=0.04; Fig. 3). Further research is required to understand specific B-cell pathways suppressed by HPgV.
Fig. 3.
HPgV inhibits sCD40L–mediated B-cell activation (CD86 expression). PBMCs isolated from HPgV-positive subjects (n=9) or HPgV-negative subjects (n = 8) were stimulated ex vivo with sCD40L (5 ng ml−1) overnight. Expression of the B-cell activation marker CD86 was quantified on CD19+ CD3− B-cells by flow cytometry. HPgV viraemic subjects had a significantly lower percentage of cells expressing the activation marker CD86 (P=0.04) compared with HPgV-negative subjects.
HPgV and NK-cells
Viral infection of NK-cells is relatively uncommon, although a few examples have been described. NK-cells are permissive for vaccinia infection in vitro (Kirwan et al., 2006; Rajagopalan & Long, 2000; Sánchez-Puig et al., 2004) and TTV genomes are present in NK-cells ex vivo (Maggi et al., 2001; Okamoto, 2009; Takahashi et al., 2002; Zhong et al., 2002). We found HPgV RNA in highly purified NK-cells obtained from four of five subjects (mean 42 genome equivalents per 104 cells) (Chivero et al., 2014). Viral RNA was shown to be taken up by NK-cells obtained from donors not infected with HPgV, and viral RNA increased and was released into culture supernatants by PBMCs in these studies (Chivero et al., 2014). As noted earlier, HPgV RNA is present in serum extracellular microvesicles and these may be involved in infection of various PBMCs (Bhattarai et al., 2013).
One clinical study suggested that HPgV infection may modulate NK-cell activation in addition to its effect on T-cells (Stapleton et al., 2013). This study found lower levels of the activation marker CD69 were detected on peripheral CD56bright NK-cells in HPgV/HIV co-infected individuals compared with HIV mono-infected individuals (Stapleton et al., 2013). CD56bright NK-cells are major cytokine producers (IFN-γ and TNF-α); thus the reduction suggests that HPgV may impair NK-cell function. To date, no studies examining the relationship between HPgV infection and NK function have been reported. As NK-cells act as rheostats modulating antiviral T-cells, interference with NK-cell function may also contribute to viral persistence (Welsh & Waggoner, 2013). In humans infected with the closely related HCV, NK-cell production of IFN-γ and TNF-α is suppressed (Ahlenstiel et al., 2010; Oliviero et al., 2009; Peppa et al., 2010), whilst cytotoxicity and degranulation is increased (Ahlenstiel et al., 2010; De Maria et al., 2007). Further study to examine the effect of HPgV infection on NK-cell function is needed, and these studies may identify critical and novel host immunomodulatory mechanisms pertinent to viral persistence.
HPgV: an ancient and successful human virus
Although HPgV was discovered only ~20 years ago (Leary et al., 1996; Simons et al., 1995), several lines of evidence suggest that it is an ancient virus that is well-adapted to growth in the human host. Genetically divergent isolates of HPgV have been isolated from different parts of the world with distribution extending to highly isolated populations, such as indigenous tribes in Papua New Guinea and Central and South America (Simmonds & Smith, 1999). The geographical distribution of HPgV genotypes reflects that of ancient human migrations, suggesting HPgV co-migration and viral diversification within the human hosts (Pavesi, 2001; Sharp & Simmonds, 2011). Two examples of this are the finding of isolates in Southeast Asia (genotype 3) that are most closely related to those of African origin, consistent with a major route of ancient human migrations from Africa to southeastern parts of the Asian continent (Pavesi, 2001), and the finding of genotype 3 viruses in indigenous (native) populations of South Americans, with a mixture of the Western European and North American genotype 2 and genotype 3 viruses in mixed-race South American populations (Loureiro et al., 2002; Tanaka et al., 1998b).
The lack of a causal relationship between HPgV infection and disease raises the possibility that HPgV is a symbiont or commensal of humans. If so, HPgV may provide some benefit to humans. There are considerable data suggesting a beneficial effect of HPgV infection on survival in HIV-positive subjects (Heringlake et al., 1998; Nunnari et al., 2003; Tillmann et al., 2001; Toyoda et al., 1998; Vahidnia et al., 2012; Williams et al., 2004; Xiang et al., 2001; Zhang et al., 2006). It is unlikely that the relationship between HPgV infection and immune activation was selected for by HIV, as HIV was only introduced into humans during the past century (Zhu et al., 1998). Mechanisms purported to contribute to slower HIV disease progression include the findings of reduced surface expression of the chemokine receptors CCR5 and CXCR4, entry co-receptors for HIV (Nattermann et al., 2003; Schwarze-Zander et al., 2007; Xiang et al., 2001), and high plasma levels of the ligands for these receptors, i.e. macrophage inflammatory protein (MIP)-1a, MIP-1b, RANTES (CCR5) and stromal-derived factor-1 (CXCR4) (Xiang et al., 2004), in chronic HPgV viraemic subjects compared with those without HPgV infection. Additionally, the relationship between HPgV infection and reduced immune activation (Bhattarai et al., 2012a; Maidana-Giret et al., 2009) suggests that this virus has anti-inflammatory effects that may be generally beneficial to humans. In contrast, this subtle reduction in immune function may contribute to the observed association between HPgV and non-Hodgkin’s lymphoma by reducing immune surveillance mechanisms (Chang et al., 2014; Civardi et al., 1998; De Renzo et al., 2002; Ellenrieder et al., 1998; Giannoulis et al., 2004; Keresztes et al., 2003; Krajden et al., 2010; Michaelis et al., 2003; Nakamura et al., 1997).
The association between HPgV infection and survival outcomes in HIV-infected individuals suggests that HPgV has a symbiotic relationship; however, due to the lack of an animal model, this has been difficult to assess directly. The application of next-generation sequencing has identified pegiviruses closely related to HPgV in Old World primates and numerous other species, raising the possibility that an animal infection model may be feasible (Kapoor et al., 2013; Sibley et al., 2014). An animal model would allow direct testing of the hypothesis that chronic infection is beneficial.
Conclusions and future directions
The mechanisms by which HPgV infects diverse blood cell types and persists in humans remain enigmatic, and require further clarification. First, although HPgV RNA is found in many cell types, negative-strand RNA is only found consistently in bone marrow and spleen (Radkowski et al., 2000; Tucker et al., 2000). Replication at these sites suggests the possibility that the primary permissive cell type is a HSC. Second, the mechanisms of HPgV persistence and clearance are not understood. Whilst HPgV effects on B-cells have not been studied in detail, failure to mount an antibody response to HPgV non-structural proteins and delayed development of E2 antibodies suggests impaired B-cell function. Furthermore, CD8+ T-cells play a critical role in the clearance of many viral infections. As HPgV infection inhibits CD8+ T-cell activation, further characterization may provide insight into persistence. In addition, host genetic factors that may contribute to viral persistence are also unknown and warrant further studies. Third, the effects of HPgV on immune cells may be shared amongst other viruses and characterization of these may provide novel insights into mechanisms used by other viruses to evade host immune responses. Finally, as HPgV is a non-cytopathic virus that persistently infects humans without illness, and appears to replicate in spleen and bone marrow tissues, HPgV may provide novel approaches to gene therapy.
Acknowledgments
We thank Drs Jinhua Xiang, James McLinden and Nirjal Bhattarai for helpful discussions related to this review. The work was supported in part by grants from the Department of Veterans Affairs, Veterans Health Administration, Office of Research and Development (Merit Review BX000207 and CX000821) to J. T. S. E. T. C. was a recipient of an International Fulbright Science and Technology Award.
Footnotes
Conflict of interest: J. T. S. holds patents related to HPgV (GB virus C).
References
- Adams MJ, King AMQ, Carstens EB. Ratification vote on taxonomic proposals to the International Committee on Taxonomy of Viruses (2013) Arch Virol. 2013;158:2023–2030. doi: 10.1007/s00705-013-1688-5. [DOI] [PubMed] [Google Scholar]
- Agnello V, Abel G, Elfahal M, Knight GB, Zhang QX. Hepatitis C virus and other flaviviridae viruses enter cells via low density lipoprotein receptor. Proc Natl Acad Sci U S A. 1999;96:12766–12771. doi: 10.1073/pnas.96.22.12766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahlenstiel G, Titerence RH, Koh C, Edlich B, Feld JJ, Rotman Y, Ghany MG, Hoofnagle JH, Liang TJ, et al. Natural killer cells are polarized toward cytotoxicity in chronic hepatitis C in an interferon-alfa-dependent manner. Gastroenterology. 2010;138:325–335. doi: 10.1053/j.gastro.2009.08.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg T, Müller AR, Platz KP, Höhne M, Bechstein WO, Hopf U, Wiedenmann B, Neuhaus P, Schreier E. Dynamics of GB virus C viremia early after orthotopic liver transplantation indicates extrahepatic tissues as the predominant site of GB virus C replication. Hepatology. 1999;29:245–249. doi: 10.1002/hep.510290121. [DOI] [PubMed] [Google Scholar]
- Bhanich Supapol W, Remis RS, Raboud J, Millson M, Tappero J, Kaul R, Kulkarni P, McConnell MS, Mock PA, et al. Mother-to-child transmission of GB virus C in a cohort of women coinfected with GB virus C and HIV in Bangkok, Thailand. J Infect Dis. 2009;200:227–235. doi: 10.1086/599793. [DOI] [PubMed] [Google Scholar]
- Bhattarai N, Stapleton JT. GB virus C: the good boy virus? Trends Microbiol. 2012;20:124–130. doi: 10.1016/j.tim.2012.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattarai N, Rydze RT, Chivero ET, Stapleton JT. GB virus C viremia is associated with higher levels of double-negative T cells and lower T-cell activation in HIV-infected individuals receiving antiretroviral therapy. J Infect Dis. 2012a;206:1469–1472. doi: 10.1093/infdis/jis515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattarai N, McLinden JH, Xiang JH, Kaufman TM, Stapleton JT. GB virus C envelope protein E2 inhibits TCR-induced IL-2 production and alters IL-2-signaling pathways. J Immunol. 2012b;189:2211–2216. doi: 10.4049/jimmunol.1201324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattarai N, McLinden JH, Xiang JH, Landay AL, Chivero ET, Stapleton JT. GB virus C particles inhibit T cell activation via envelope E2 protein-mediated inhibition of TCR signaling. J Immunol. 2013;190:6351–6359. doi: 10.4049/jimmunol.1300589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bukong TN, Momen-Heravi F, Kodys K, Bala S, Szabo G. Exosomes from hepatitis C infected patients transmit HCV infection and contain replication competent viral RNA in complex with Ago2-miR122-HSP90. PLoS Pathog. 2014;10:e1004424. doi: 10.1371/journal.ppat.1004424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burke KP, Cox AL. Hepatitis C virus evasion of adaptive immune responses: a model for viral persistence. Immunol Res. 2010;47:216–227. doi: 10.1007/s12026-009-8152-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang CM, Stapleton JT, Klinzman D, McLinden JH, Purdue MP, Katki HA, Engels EA. GBV-C infection and risk of NHL among U.S. adults. Cancer Res. 2014;74:5553–5560. doi: 10.1158/0008-5472.CAN-14-0209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chivero ET, Bhattarai N, Rydze RT, Winters MA, Holodniy M, Stapleton JT. Human pegivirus RNA is found in multiple blood mononuclear cells in vivo and serum-derived viral RNA-containing particles are infectious in vitro. J Gen Virol. 2014;95:1307–1319. doi: 10.1099/vir.0.063016-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Civardi G, Tanzi E, Ferrari B, Vallisa D, Zanetti A, Cavanna L. High prevalence of anti-HGV/E2 antibodies in HCV-positive patients with non Hodgkin’s lymphoma. Haematologica. 1998;83:957–958. [PubMed] [Google Scholar]
- De Maria A, Fogli M, Mazza S, Basso M, Picciotto A, Costa P, Congia S, Mingari MC, Moretta L. Increased natural cytotoxicity receptor expression and relevant IL-10 production in NK cells from chronically infected viremic HCV patients. Eur J Immunol. 2007;37:445–455. doi: 10.1002/eji.200635989. [DOI] [PubMed] [Google Scholar]
- De Renzo A, Persico E, de Marino F, di Giacomo Russo G, Notaro R, di Grazia C, Picardi M, Santoro L, Torella R, et al. High prevalence of hepatitis G virus infection in Hodgkin’s disease and B-cell lymphoproliferative disorders: absence of correlation with hepatitis C virus infection. Haematologica. 2002;87:714–718. [PubMed] [Google Scholar]
- Deinhardt F, Holmes AW, Capps RB, Popper H. Studies on the transmission of human viral hepatitis to marmoset monkeys. I. Transmission of disease, serial passages, and description of liver lesions. J Exp Med. 1967;125:673–688. doi: 10.1084/jem.125.4.673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreux M, Garaigorta U, Boyd B, Décembre E, Chung J, Whitten-Bauer C, Wieland S, Chisari FV. Short-range exosomal transfer of viral RNA from infected cells to plasmacytoid dendritic cells triggers innate immunity. Cell Host Microbe. 2012;12:558–570. doi: 10.1016/j.chom.2012.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durie FH, Foy TM, Masters SR, Laman JD, Noelle RJ. The role of CD40 in the regulation of humoral and cell-mediated immunity. Immunol Today. 1994;15:406–411. doi: 10.1016/0167-5699(94)90269-0. [DOI] [PubMed] [Google Scholar]
- Elkayam O, Hassoba HM, Ferrell LD, Garcia-Kennedy R, Gish RG, Wright TL, Laffler T, Traylor D, Hunt G, Rosenthal P. GB virus C (GBV-C/HGV) and E2 antibodies in children preliver and postliver transplant. Pediatr Res. 1999;45:795–798. doi: 10.1203/00006450-199906000-00002. [DOI] [PubMed] [Google Scholar]
- Ellenrieder V, Weidenbach H, Frickhofen N, Michel D, Prümmer O, Klatt S, Bernas O, Mertens T, Adler G, Beckh K. HCV and HGV in B-cell non-Hodgkin’s lymphoma. J Hepatol. 1998;28:34–39. doi: 10.1016/s0168-8278(98)80199-2. [DOI] [PubMed] [Google Scholar]
- Evans MJ, von Hahn T, Tscherne DM, Syder AJ, Panis M, Wölk B, Hatziioannou T, McKeating JA, Bieniasz PD, Rice CM. Claudin-1 is a hepatitis C virus co-receptor required for a late step in entry. Nature. 2007;446:801–805. doi: 10.1038/nature05654. [DOI] [PubMed] [Google Scholar]
- Fan X, Xu Y, Solomon H, Ramrakhiani S, Neuschwander-Tetri BA, Di Bisceglie AM. Is hepatitis G/GB virus-C virus hepatotropic? Detection of hepatitis G/GB virus-C viral RNA in liver and serum. J Med Virol. 1999;58:160–164. [PubMed] [Google Scholar]
- Feng Z, Hensley L, McKnight KL, Hu F, Madden V, Ping L, Jeong SH, Walker C, Lanford RE, Lemon SM. A pathogenic picornavirus acquires an envelope by hijacking cellular membranes. Nature. 2013;496:367–371. doi: 10.1038/nature12029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernández L, Bleda MJ, Gómara MJ, Haro I. Design and application of GB virus C (GBV-C) peptide microarrays for diagnosis of GBV-C/HIV-1 co-infection. Anal Bioanal Chem. 2013;405:3973–3982. doi: 10.1007/s00216-012-6585-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fogeda M, Navas S, Martín J, Casqueiro M, Rodríguez E, Arocena C, Carreño V. in vitro infection of human peripheral blood mononuclear cells by GB virus C/hepatitis G virus. J Virol. 1999;73:4052–4061. doi: 10.1128/jvi.73.5.4052-4061.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foy TM, Durie FH, Noelle RJ. The expansive role of CD40 and its ligand, gp39, in immunity. Semin Immunol. 1994;6:259–266. doi: 10.1006/smim.1994.1034. [DOI] [PubMed] [Google Scholar]
- George SL, Xiang JH, Stapleton JT. Clinical isolates of GB virus type C vary in their ability to persist and replicate in peripheral blood mononuclear cell cultures. Virology. 2003;316:191–201. doi: 10.1016/s0042-6822(03)00585-3. [DOI] [PubMed] [Google Scholar]
- George SL, Varmaz D, Stapleton JT. GB virus C replicates in primary T and B lymphocytes. J Infect Dis. 2006;193:451–454. doi: 10.1086/499435. [DOI] [PubMed] [Google Scholar]
- Giannoulis E, Economopoulos T, Mandraveli K, Giannoulis K, Nikolaides C, Zervou E, Papageorgiou E, Zoulas D, Tourkantonis A, et al. The prevalence of hepatitis C and hepatitis G virus infection in patients with B cell non-Hodgkin lymphomas in Greece: a Hellenic Cooperative Oncology Group Study. Acta Haematol. 2004;112:189–193. doi: 10.1159/000081270. [DOI] [PubMed] [Google Scholar]
- Gómara MJ, Fernández L, Pérez T, Tenckhoff S, Casanovas A, Tillmann HL, Haro I. Diagnostic value of anti-GBV-C antibodies in HIV-infected patients. Chem Biol Drug Des. 2011;78:277–282. doi: 10.1111/j.1747-0285.2011.01143.x. [DOI] [PubMed] [Google Scholar]
- Grakoui A, Shoukry NH, Woollard DJ, Han JH, Hanson HL, Ghrayeb J, Murthy KK, Rice CM, Walker CM. HCV persistence and immune evasion in the absence of memory T cell help. Science. 2003;302:659–662. doi: 10.1126/science.1088774. [DOI] [PubMed] [Google Scholar]
- Gutierrez RA, Dawson GJ, Knigge MF, Melvin SL, Heynen CA, Kyrk CR, Young CE, Carrick RJ, Schlauder GG, et al. Seroprevalence of GB virus C and persistence of RNA and antibody. J Med Virol. 1997;53:167–173. doi: 10.1002/(sici)1096-9071(199710)53:2<167::aid-jmv10>3.0.co;2-g. [DOI] [PubMed] [Google Scholar]
- Heringlake S, Ockenga J, Tillmann HL, Trautwein C, Meissner D, Stoll M, Hunt J, Jou C, Solomon N, et al. GB virus C/hepatitis G virus infection: a favorable prognostic factor in human immunodeficiency virus-infected patients? J Infect Dis. 1998;177:1723–1726. doi: 10.1086/517431. [DOI] [PubMed] [Google Scholar]
- Hino K, Moriya T, Ohno N, Takahashi K, Hoshino H, Ishiyama N, Katayama K, Yoshizawa H, Mishiro S. Mother-to-infant transmission occurs more frequently with GB virus C than hepatitis C virus. Arch Virol. 1998;143:65–72. doi: 10.1007/s007050050268. [DOI] [PubMed] [Google Scholar]
- Hunt PW, Martin JN, Sinclair E, Bredt B, Hagos E, Lampiris H, Deeks SG. T cell activation is associated with lower CD4+ T cell gains in human immunodeficiency virus-infected patients with sustained viral suppression during antiretroviral therapy. J Infect Dis. 2003;187:1534–1543. doi: 10.1086/374786. [DOI] [PubMed] [Google Scholar]
- Kapoor A, Simmonds P, Scheel TK, Hjelle B, Cullen JM, Burbelo PD, Chauhan LV, Duraisamy R, Sanchez Leon M, et al. Identification of rodent homologs of hepatitis C virus and pegiviruses. MBio. 2013;4:e00216–13. doi: 10.1128/mBio.00216-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keck ZY, Li SH, Xia J, von Hahn T, Balfe P, McKeating JA, Witteveldt J, Patel AH, Alter H, et al. Mutations in hepatitis C virus E2 located outside the CD81 binding sites lead to escape from broadly neutralizing antibodies but compromise virus infectivity. J Virol. 2009;83:6149–6160. doi: 10.1128/JVI.00248-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keck ZY, Angus AG, Wang W, Lau P, Wang Y, Gatherer D, Patel AH, Foung SK. Non-random escape pathways from a broadly neutralizing human monoclonal antibody map to a highly conserved region on the hepatitis C virus E2 glycoprotein encompassing amino acids 412–423. PLoS Pathog. 2014;10:e1004297. doi: 10.1371/journal.ppat.1004297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keresztes K, Takács M, Horányi M, Miltényi Z, Illés A. HCV and HGV infection in Hodgkin’s disease. Pathol Oncol Res. 2003;9:222–225. doi: 10.1007/BF02893381. [DOI] [PubMed] [Google Scholar]
- Kirwan S, Merriam D, Barsby N, McKinnon A, Burshtyn DN. Vaccinia virus modulation of natural killer cell function by direct infection. Virology. 2006;347:75–87. doi: 10.1016/j.virol.2005.11.037. [DOI] [PubMed] [Google Scholar]
- Kisiel E, Cortez KC, Pawełczyk A, Ośko IB, Kubisa N, Laskus T, Radkowski M. Hepatitis G virus/GBV-C in serum, peripheral blood mononuclear cells and bone marrow in patients with hematological malignancies. Infect Genet Evol. 2013;19:195–199. doi: 10.1016/j.meegid.2013.07.010. [DOI] [PubMed] [Google Scholar]
- Kleinman S. Hepatitis G virus biology, epidemiology, and clinical manifestations: implications for blood safety. Transfus Med Rev. 2001;15:201–212. doi: 10.1053/tmrv.2001.24589. [DOI] [PubMed] [Google Scholar]
- Krajden M, Yu A, Braybrook H, Lai AS, Mak A, Chow R, Cook D, Tellier R, Petric M, et al. GBV-C/hepatitis G virus infection and non-Hodgkin lymphoma: a case control study. Int J Cancer. 2010;126:2885–2892. doi: 10.1002/ijc.25035. [DOI] [PubMed] [Google Scholar]
- Laras A, Zacharakis G, Hadziyannis SJ. Absence of the negative strand of GBV-C/HGV RNA from the liver. J Hepatol. 1999;30:383–388. doi: 10.1016/s0168-8278(99)80094-4. [DOI] [PubMed] [Google Scholar]
- Laskus T, Radkowski M, Wang LF, Vargas H, Rakela J. Lack of evidence for hepatitis G virus replication in the livers of patients coinfected with hepatitis C and G viruses. J Virol. 1997;71:7804–7806. doi: 10.1128/jvi.71.10.7804-7806.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauck M, Bailey AL, Anderson KG, Goldberg TL, Sabeti PC, O’Connor DH. GB virus C coinfections in West African Ebola patients. J Virol. 2015;89:2425–2429. doi: 10.1128/JVI.02752-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leary TP, Muerhoff S, Simons JN, Pilot-Matias TJ, Erker JC, Chalmers ML, Schlauder GG, Dawson GJ, Desai SM, Mushahwar IK. Sequence and genomic organization of GBV-C: a novel member of the flaviviridae associated with human non-A-E hepatitis. J Med Virol. 1996;48:60–67. doi: 10.1002/(SICI)1096-9071(199601)48:1<60::AID-JMV10>3.0.CO;2-A. [DOI] [PubMed] [Google Scholar]
- Lemon SM. Induction and evasion of innate antiviral responses by hepatitis C virus. J Biol Chem. 2010;285:22741–22747. doi: 10.1074/jbc.R109.099556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin HH, Kao JH, Yeh KY, Liu DP, Chang MH, Chen PJ, Chen DS. Mother-to-infant transmission of GB virus C/ hepatitis G virus: the role of high-titered maternal viremia and mode of delivery. J Infect Dis. 1998;177:1202–1206. doi: 10.1086/515264. [DOI] [PubMed] [Google Scholar]
- Loureiro CL, Alonso R, Pacheco BA, Uzcátegui MG, Villegas L, León G, De Saéz A, Liprandi F, López JL, Pujol FH. High prevalence of GB virus C/hepatitis G virus genotype 3 among autochthonous Venezuelan populations. J Med Virol. 2002;68:357–362. doi: 10.1002/jmv.10211. [DOI] [PubMed] [Google Scholar]
- Lozach PY, Lortat-Jacob H, de Lacroix de Lavalette A, Staropoli I, Foung S, Amara A, Houles C, Fieschi F, Schwartz O, et al. DC-SIGN and L-SIGN are high affinity binding receptors for hepatitis C virus glycoprotein E2. J Biol Chem. 2003;278:20358–20366. doi: 10.1074/jbc.M301284200. [DOI] [PubMed] [Google Scholar]
- Maggi F, Fornai C, Zaccaro L, Morrica A, Vatteroni ML, Isola P, Marchi S, Ricchiuti A, Pistello M, Bendinelli M. TT virus (TTV) loads associated with different peripheral blood cell types and evidence for TTV replication in activated mononuclear cells. J Med Virol. 2001;64:190–194. doi: 10.1002/jmv.1035. [DOI] [PubMed] [Google Scholar]
- Maidana-Giret MT, Silva TM, Sauer MM, Tomiyama H, Levi JE, Bassichetto KC, Nishiya A, Diaz RS, Sabino EC, et al. GB virus type C infection modulates T-cell activation independently of HIV-1 viral load. AIDS. 2009;23:2277–2287. doi: 10.1097/QAD.0b013e32832d7a11. [DOI] [PubMed] [Google Scholar]
- Martin DN, Uprichard SL. Identification of transferrin receptor 1 as a hepatitis C virus entry factor. Proc Natl Acad Sci U S A. 2013;110:10777–10782. doi: 10.1073/pnas.1301764110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meckes DG, Jr, Raab-Traub N. Microvesicles and viral infection. J Virol. 2011;85:12844–12854. doi: 10.1128/JVI.05853-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellor J, Haydon G, Blair C, Livingstone W, Simmonds P. Low level or absent in vivo replication of hepatitis C virus and hepatitis G virus/GB virus C in peripheral blood mononuclear cells. J Gen Virol. 1998;79:705–714. doi: 10.1099/0022-1317-79-4-705. [DOI] [PubMed] [Google Scholar]
- Michaelis S, Kazakov DV, Schmid M, Dummer R, Burg G, Kempf W. Hepatitis C and G viruses in B-cell lymphomas of the skin. J Cutan Pathol. 2003;30:369–372. doi: 10.1034/j.1600-0560.2003.00083.x. [DOI] [PubMed] [Google Scholar]
- Milush JM, Mir KD, Sundaravaradan V, Gordon SN, Engram J, Cano CA, Reeves JD, Anton E, O’Neill E, et al. Lack of clinical AIDS in SIV-infected sooty mangabeys with significant CD4+ T cell loss is associated with double-negative T cells. J Clin Invest. 2011;121:1102–1110. doi: 10.1172/JCI44876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohr EL, Stapleton JT. GB virus type C interactions with HIV: the role of envelope glycoproteins. J Viral Hepat. 2009;16:757–768. doi: 10.1111/j.1365-2893.2009.01194.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moradpour D, Penin F, Rice CM. Replication of hepatitis C virus. Nat Rev Microbiol. 2007;5:453–463. doi: 10.1038/nrmicro1645. [DOI] [PubMed] [Google Scholar]
- Nakamura S, Takagi T, Matsuda T. Hepatitis G virus RNA in patients with B-cell non-Hodgkin’s lymphoma. Br J Haematol. 1997;98:1051–1052. [PubMed] [Google Scholar]
- Nattermann J, Nischalke HD, Kupfer B, Rockstroh J, Hess L, Sauerbruch T, Spengler U. Regulation of CC chemokine receptor 5 in hepatitis G virus infection. AIDS. 2003;17:1457–1462. doi: 10.1097/00002030-200307040-00006. [DOI] [PubMed] [Google Scholar]
- Nunnari G, Nigro L, Palermo F, Attanasio M, Berger A, Doerr HW, Pomerantz RJ, Cacopardo B. Slower progression of HIV-1 infection in persons with GB virus C co-infection correlates with an intact T-helper 1 cytokine profile. Ann Intern Med. 2003;139:26–30. doi: 10.7326/0003-4819-139-1-200307010-00009. [DOI] [PubMed] [Google Scholar]
- Ohto H, Ujiie N, Sato A, Okamoto H, Mayumi M & Vertical Transmission of Hepatitis Viruses Collaborative Study Group. Mother-to-infant transmission of GB virus type C/HGV. Transfusion. 2000;40:725–730. doi: 10.1046/j.1537-2995.2000.40060725.x. [DOI] [PubMed] [Google Scholar]
- Okamoto H. History of discoveries and pathogenicity of TT viruses. Curr Top Microbiol Immunol. 2009;331:1–20. doi: 10.1007/978-3-540-70972-5_1. [DOI] [PubMed] [Google Scholar]
- Oliviero B, Varchetta S, Paudice E, Michelone G, Zaramella M, Mavilio D, De Filippi F, Bruno S, Mondelli MU. Natural killer cell functional dichotomy in chronic hepatitis B and chronic hepatitis C virus infections. Gastroenterology. 2009;137:1151–1160. doi: 10.1053/j.gastro.2009.05.047. [DOI] [PubMed] [Google Scholar]
- Pavesi A. Origin and evolution of GBV-C/hepatitis G virus and relationships with ancient human migrations. J Mol Evol. 2001;53:104–113. doi: 10.1007/s002390010198. [DOI] [PubMed] [Google Scholar]
- Peppa D, Micco L, Javaid A, Kennedy PTF, Schurich A, Dunn C, Pallant C, Ellis G, Khanna P, et al. Blockade of immunosuppressive cytokines restores NK cell antiviral function in chronic hepatitis B virus infection. PLoS Pathog. 2010;6:e1001227. doi: 10.1371/journal.ppat.1001227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pessoa MG, Terrault NA, Detmer J, Kolberg J, Collins M, Hassoba HM, Wright TL. Quantitation of hepatitis G and C viruses in the liver: evidence that hepatitis G virus is not hepatotropic. Hepatology. 1998;27:877–880. doi: 10.1002/hep.510270335. [DOI] [PubMed] [Google Scholar]
- Petitjean G, Chevalier MF, Tibaoui F, Didier C, Manea ME, Liovat AS, Campa P, Müller-Trutwin M, Girard PM, et al. Level of double negative T cells, which produce TGF-β and IL-10, predicts CD8 T-cell activation in primary HIV-1 infection. AIDS. 2012;26:139–148. doi: 10.1097/QAD.0b013e32834e1484. [DOI] [PubMed] [Google Scholar]
- Ploss A, Evans MJ, Gaysinskaya VA, Panis M, You H, de Jong YP, Rice CM. Human occludin is a hepatitis C virus entry factor required for infection of mouse cells. Nature. 2009;457:882–886. doi: 10.1038/nature07684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radkowski M, Kubicka J, Kisiel E, Cianciara J, Nowicki M, Rakela J, Laskus T. Detection of active hepatitis C virus and hepatitis G virus/GB virus C replication in bone marrow in human subjects. Blood. 2000;95:3986–3989. [PubMed] [Google Scholar]
- Rajagopalan S, Long EO. Use of vaccinia virus for functional gene transfer in natural killer cells. Methods Mol Biol. 2000;121:265–272. doi: 10.1385/1-59259-044-6:265. [DOI] [PubMed] [Google Scholar]
- Ramakrishnaiah V, Thumann C, Fofana I, Habersetzer F, Pan QW, de Ruiter PE, Willemsen R, Demmers JAA, Stalin Raj V, et al. Exosome-mediated transmission of hepatitis C virus between human hepatoma Huh7.5 cells. Proc Natl Acad Sci U S A. 2013;110:13109–13113. doi: 10.1073/pnas.1221899110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehermann B. Pathogenesis of chronic viral hepatitis: differential roles of T cells and NK cells. Nat Med. 2013;19:859–868. doi: 10.1038/nm.3251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz V, Giordano M, Rivero CW, Minassian ML, Cuestas ML, Trinks J, Mathet VL, Oubiña JR. GB virus C quasispecies detected in plasma and lymphocyte subsets in a natural human infection. J Gen Virol. 2010;91:1687–1692. doi: 10.1099/vir.0.019877-0. [DOI] [PubMed] [Google Scholar]
- Rydze RT, Bhattarai N, Stapleton JT. GB virus C infection is associated with a reduced rate of reactivation of latent HIV and protection against activation-induced T-cell death. Antivir Ther. 2012;17:1271–1279. doi: 10.3851/IMP2309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sainz B, Jr, Barretto N, Martin DN, Hiraga N, Imamura M, Hussain S, Marsh KA, Yu X, Chayama K, et al. Identification of the Niemann-Pick C1-like 1 cholesterol absorption receptor as a new hepatitis C virus entry factor. Nat Med. 2012;18:281–285. doi: 10.1038/nm.2581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sánchez-Puig JM, Sánchez L, Roy G, Blasco R. Susceptibility of different leukocyte cell types to Vaccinia virus infection. Virol J. 2004;1:10. doi: 10.1186/1743-422X-1-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarze-Zander C, Blackard JT, Zheng H, Addo MM, Lin W, Robbins GK, Sherman KE, Zdunek D, Hess G, et al. GB virus C (GBV-C) infection in hepatitis C virus (HCV)/ HIV-coinfected patients receiving HCV treatment: importance of the GBV-C genotype. J Infect Dis. 2006;194:410–419. doi: 10.1086/505713. [DOI] [PubMed] [Google Scholar]
- Schwarze-Zander C, Neibecker M, Othman S, Welzel C, Schulz M, Voigt E, Vogel M, Wasmuth C, Luechters G, et al. GBV-C co-infection in HIV patients is associated with low CCR5 and CXCR4 surface expression on CD4 cells. Hepatology. 2007;46(Suppl. S1):891A. [Google Scholar]
- Sharp PM, Simmonds P. Evaluating the evidence for virus/host co-evolution. Curr Opin Virol. 2011;1:436–441. doi: 10.1016/j.coviro.2011.10.018. [DOI] [PubMed] [Google Scholar]
- Sibley SD, Lauck M, Bailey AL, Hyeroba D, Tumukunde A, Weny G, Chapman CA, O’Connor DH, Goldberg TL, Friedrich TC. Discovery and characterization of distinct simian pegiviruses in three wild African Old World monkey species. PLoS One. 2014;9:e98569. doi: 10.1371/journal.pone.0098569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmonds P, Smith DB. Structural constraints on RNA virus evolution. J Virol. 1999;73:5787–5794. doi: 10.1128/jvi.73.7.5787-5794.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simons JN, Leary TP, Dawson GJ, Pilot-Matias TJ, Muerhoff AS, Schlauder GG, Desai SM, Mushahwar IK. Isolation of novel virus-like sequences associated with human hepatitis. Nat Med. 1995;1:564–569. doi: 10.1038/nm0695-564. [DOI] [PubMed] [Google Scholar]
- Stapleton JT. GB virus type C/hepatitis G virus. Semin Liver Dis. 2003;23:137–148. doi: 10.1055/s-2003-39943. [DOI] [PubMed] [Google Scholar]
- Stapleton JT, Chaloner K, Zhang JY, Klinzman D, Souza IE, Xiang JH, Landay A, Fahey J, Pollard R, Mitsuyasu R. GBV-C viremia is associated with reduced CD4 expansion in HIV-infected people receiving HAART and interleukin-2 therapy. AIDS. 2009;23:605–610. doi: 10.1097/QAD.0b013e32831f1b00. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stapleton JT, Foung S, Muerhoff AS, Bukh J, Simmonds P. The GB viruses: a review and proposed classification of GBV-A, GBV-C (HGV), and GBV-D in genus Pegivirus within the family Flaviviridae. J Gen Virol. 2011;92:233–246. doi: 10.1099/vir.0.027490-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stapleton JT, Bukh J, Muerhoff AS, Foung S, Simmonds P. Assignment of human, simian and bat pegiviruses (previously described as GBV-A, GBV-C, and GBV-D) as members of a new genus (Pegivirus) within the Flaviviridae. 2012a http://www.ictvonline.org/proposals/2012.011a-dV.A.v2.Pegivirus.pdf.
- Stapleton JT, Chaloner K, Martenson JA, Zhang JY, Klinzman D, Xiang JH, Sauter W, Desai SN, Landay A. GB virus C infection is associated with altered lymphocyte subset distribution and reduced T cell activation and proliferation in HIV-infected individuals. PLoS One. 2012b;7:e50563. doi: 10.1371/journal.pone.0050563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stapleton JT, Martinson JA, Klinzman D, Xiang J, Desai SN, Landay A. GB virus C infection and B-cell, natural killer cell, and monocyte activation markers in HIV-infected individuals. AIDS. 2013;27:1829–1832. doi: 10.1097/QAD.0b013e328363089f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stapleton JT, Xiang J, McLinden JH, Bhattarai N, Chivero ET, Klinzman D, Kaufman TM, Chang Q. A novel T cell evasion mechanism in persistent RNA virus infection. Trans Am Clin Climatol Assoc. 2014;125:14–24. [PMC free article] [PubMed] [Google Scholar]
- Sundaravaradan V, Mir KD, Sodora DL. Double-negative T cells during HIV/SIV infections: potential pinch hitters in the T-cell lineup. Curr Opin HIV AIDS. 2012;7:164–171. doi: 10.1097/COH.0b013e3283504a66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundaravaradan V, Saleem R, Micci L, Gasper MA, Ortiz AM, Else J, Silvestri G, Paiardini M, Aitchison JD, Sodora DL. Multifunctional double-negative T cells in sooty mangabeys mediate T-helper functions irrespective of SIV infection. PLoS Pathog. 2013;9:e1003441. doi: 10.1371/journal.ppat.1003441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tacke M, Schmolke S, Schlueter V, Sauleda S, Esteban JI, Tanaka E, Kiyosawa K, Alter HJ, Schmitt U, et al. Humoral immune response to the E2 protein of hepatitis G virus is associated with long-term recovery from infection and reveals a high frequency of hepatitis G virus exposure among healthy blood donors. Hepatology. 1997;26:1626–1633. doi: 10.1002/hep.510260635. [DOI] [PubMed] [Google Scholar]
- Takahashi M, Asabe S, Gotanda Y, Kishimoto J, Tsuda F, Okamoto H. TT virus is distributed in various leukocyte subpopulations at distinct levels, with the highest viral load in granulocytes. Biochem Biophys Res Commun. 2002;290:242–248. doi: 10.1006/bbrc.2001.6183. [DOI] [PubMed] [Google Scholar]
- Tan D, Matsumoto A, Conry-Cantilena C, Melpolder JC, Shih JWK, Leuther M, Hess G, Gibble JW, Ness PM, Alter HJ. Analysis of hepatitis G virus (HGV) RNA, antibody to HGV envelope protein, and risk factors for blood donors coinfected with HGV and hepatitis C virus. J Infect Dis. 1999;179:1055–1061. doi: 10.1086/314722. [DOI] [PubMed] [Google Scholar]
- Tanaka E, Kiyosawa K, Shimoda K, Hino K, Tacke M, Schmolke S, Engel AM, Hess G. Evolution of hepatitis G virus infection and antibody response to envelope protein in patients with transfusion-associated non-A, non-B hepatitis. J Viral Hepat. 1998a;5:153–159. doi: 10.1046/j.1365-2893.1998.00095.x. [DOI] [PubMed] [Google Scholar]
- Tanaka Y, Mizokami M, Orito E, Ohba K, Nakano T, Kato T, Kondo Y, Ding X, Ueda R, et al. GB virus C/ hepatitis G virus infection among Colombian native Indians. Am J Trop Med Hyg. 1998b;59:462–467. doi: 10.4269/ajtmh.1998.59.462. [DOI] [PubMed] [Google Scholar]
- Thomas DL, Nakatsuji Y, Shih JW, Alter HJ, Nelson KE, Astemborski JA, Lyles CM, Vlahov D. Persistence and clinical significance of hepatitis G virus infections in injecting drug users. J Infect Dis. 1997;176:586–592. doi: 10.1086/514078. [DOI] [PubMed] [Google Scholar]
- Thomas DL, Vlahov D, Alter HJ, Hunt JC, Marshall R, Astemborski J, Nelson KE. Association of antibody to GB virus C (hepatitis G virus) with viral clearance and protection from reinfection. J Infect Dis. 1998;177:539–542. doi: 10.1086/514245. [DOI] [PubMed] [Google Scholar]
- Tillmann HL, Heringlake S, Trautwein C, Meissner D, Nashan B, Schlitt HJ, Kratochvil J, Hunt J, Qiu X, et al. Antibodies against the GB virus C envelope 2 protein before liver transplantation protect against GB virus C de novo infection. Hepatology. 1998;28:379–384. doi: 10.1002/hep.510280213. [DOI] [PubMed] [Google Scholar]
- Tillmann HL, Heiken H, Knapik-Botor A, Heringlake S, Ockenga J, Wilber JC, Goergen B, Detmer J, McMorrow M, et al. Infection with GB virus C and reduced mortality among HIV-infected patients. N Engl J Med. 2001;345:715–724. doi: 10.1056/NEJMoa010398. [DOI] [PubMed] [Google Scholar]
- Toyoda H, Fukuda Y, Hayakawa T, Takamatsu J, Saito H. Effect of GB virus C/hepatitis G virus coinfection on the course of HIV infection in hemophilia patients in Japan. J Acquir Immune Defic Syndr Hum Retrovirol. 1998;17:209–213. doi: 10.1097/00042560-199803010-00004. [DOI] [PubMed] [Google Scholar]
- Tucker TJ, Smuts HEM, Eedes C, Knobel GD, Eickhaus P, Robson SC, Kirsch RE. Evidence that the GBV-C/ hepatitis G virus is primarily a lymphotropic virus. J Med Virol. 2000;61:52–58. [PubMed] [Google Scholar]
- Vahidnia F, Petersen M, Stapleton JT, Rutherford GW, Busch M, Custer B. Acquisition of GB virus type C and lower mortality in patients with advanced HIV disease. Clin Infect Dis. 2012;55:1012–1019. doi: 10.1093/cid/cis589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welsh RM, Waggoner SN. NK cells controlling virus-specific T cells: rheostats for acute vs. persistent infections. Virology. 2013;435:37–45. doi: 10.1016/j.virol.2012.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams CF, Klinzman D, Yamashita TE, Xiang J, Polgreen PM, Rinaldo C, Liu C, Phair J, Margolick JB, et al. Persistent GB virus C infection and survival in HIV-infected men. N Engl J Med. 2004;350:981–990. doi: 10.1056/NEJMoa030107. [DOI] [PubMed] [Google Scholar]
- Wünschmann S, Medh JD, Klinzmann D, Schmidt WN, Stapleton JT. Characterization of hepatitis C virus (HCV) and HCV E2 interactions with CD81 and the low-density lipoprotein receptor. J Virol. 2000;74:10055–10062. doi: 10.1128/jvi.74.21.10055-10062.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wünschmann S, Muller HM, Stipp CS, Hemler ME, Stapleton JT. in vitro interaction between hepatitis C virus (HCV) envelope glycoprotein E2 and serum lipoproteins (LPs) results in enhanced cellular binding of both HCV E2 and LPs. J Infect Dis. 2006;194:1058–1067. doi: 10.1086/507647. [DOI] [PubMed] [Google Scholar]
- Xiang JH, Klinzman D, McLinden J, Schmidt WN, LaBrecque DR, Gish R, Stapleton JT. Characterization of hepatitis G virus (GB-C virus) particles: evidence for a nucleocapsid and expression of sequences upstream of the E1 protein. J Virol. 1998;72:2738–2744. doi: 10.1128/jvi.72.4.2738-2744.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang J, Wünschmann S, Schmidt W, Shao J, Stapleton JT. Full-length GB virus C (hepatitis G virus) RNA transcripts are infectious in primary CD4-positive T cells. J Virol. 2000;74:9125–9133. doi: 10.1128/jvi.74.19.9125-9133.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang JH, Wünschmann S, Diekema DJ, Klinzman D, Patrick KD, George SL, Stapleton JT. Effect of coinfection with GB virus C on survival among patients with HIV infection. N Engl J Med. 2001;345:707–714. doi: 10.1056/NEJMoa003364. [DOI] [PubMed] [Google Scholar]
- Xiang JH, George SL, Wünschmann S, Chang Q, Klinzman D, Stapleton JT. Inhibition of HIV-1 replication by GB virus C infection through increases in RANTES, MIP-1a, MIP-1b, and SDF-1. Lancet. 2004;363:2040–2046. doi: 10.1016/S0140-6736(04)16453-2. [DOI] [PubMed] [Google Scholar]
- Zeisel MB, Felmlee DJ, Baumert TF. Hepatitis C virus entry. Curr Top Microbiol Immunol. 2013;369:87–112. doi: 10.1007/978-3-642-27340-7_4. [DOI] [PubMed] [Google Scholar]
- Zhang W, Chaloner K, Tillmann HL, Williams CF, Stapleton JT. Effect of early and late GB virus C viraemia on survival of HIV-infected individuals: a meta-analysis. HIV Med. 2006;7:173–180. doi: 10.1111/j.1468-1293.2006.00366.x. [DOI] [PubMed] [Google Scholar]
- Zhong S, Yeo W, Tang M, Liu CL, Lin XR, Ho WM, Hui P, Johnson PJ. Frequent detection of the replicative form of TT virus DNA in peripheral blood mononuclear cells and bone marrow cells in cancer patients. J Med Virol. 2002;66:428–434. doi: 10.1002/jmv.2163. [DOI] [PubMed] [Google Scholar]
- Zhu T, Korber BT, Nahmias AJ, Hooper E, Sharp PM, Ho DD. An African HIV-1 sequence from 1959 and implications for the origin of the epidemic. Nature. 1998;391:594–597. doi: 10.1038/35400. [DOI] [PubMed] [Google Scholar]
- Zona L, Tawar RG, Zeisel MB, Xiao F, Schuster C, Lupberger J, Baumert TF. CD81-receptor associations - impact for hepatitis C virus entry and antiviral therapies. Viruses. 2014;6:875–892. doi: 10.3390/v6020875. [DOI] [PMC free article] [PubMed] [Google Scholar]